Abstract
Background
The gut-lung axis is generally recognized, but there are few large studies of the gut microbiome and incident respiratory disease in adults.
Objective
We sought to investigate the association and predictive capacity of the gut microbiome for incident asthma and chronic obstructive pulmonary disease (COPD).
Methods
Shallow metagenomic sequencing was performed for stool samples from a prospective, population-based cohort (FINRISK02; N = 7115 adults) with linked national administrative health register–derived classifications for incident asthma and COPD up to 15 years after baseline. Generalized linear models and Cox regressions were used to assess associations of microbial taxa and diversity with disease occurrence. Predictive models were constructed using machine learning with extreme gradient boosting. Models considered taxa abundances individually and in combination with other risk factors, including sex, age, body mass index, and smoking status.
Results
A total of 695 and 392 statistically significant associations were found between baseline taxonomic groups and incident asthma and COPD, respectively. Gradient boosting decision trees of baseline gut microbiome abundance predicted incident asthma and COPD in the validation data sets with mean area under the curves of 0.608 and 0.780, respectively. Cox analysis showed that the baseline gut microbiome achieved higher predictive performance than individual conventional risk factors, with C-indices of 0.623 for asthma and 0.817 for COPD. The integration of the gut microbiome and conventional risk factors further improved prediction capacities.
Conclusions
The gut microbiome is a significant risk factor for incident asthma and incident COPD and is largely independent of conventional risk factors.
Key words: Gut, microbiome, metagenomics, asthma, COPD
Asthma and chronic obstructive pulmonary disease (COPD) represent the vast majority of chronic respiratory diseases worldwide, causing a considerable burden on health and economy.1,2 Both asthma and COPD are recognized as heterogeneous diseases with diverse phenotypes and various underlying mechanisms.3, 4, 5, 6 Currently, spirometry-confirmed airflow limitation is the most common reference standard for establishing diagnoses of asthma and COPD, yet a negative spirometry test result does not rule out the disease.7,8 Other criteria that complement evaluation include self-reported symptoms, medical history, physical examination, and other diagnoses such as infection, interstitial lung disease, and others.7,9 Despite rapidly changing assessments and treatments, both asthma and COPD remain largely underdiagnosed and thus undertreated, leading to lesser quality of life and poorer disease outcomes.3,9
With recent advances in high-throughput sequencing, improved characterization of the human respiratory and gastrointestinal microbiome has been followed by growing recognition of the link between human microbiota and chronic respiratory disease.10,11 The gut microbiome is by far the largest and most studied microbial community in the human body.11,12 Although the lung microbiome has become well characterized only recently, the link between the lung microbiome and respiratory diseases has been generally acknowledged.10,13, 14, 15 “Dysbiotic” changes in both airway and gut microbiome have been linked to respiratory diseases; however, the precise mechanism or causal pathway is, as yet, not well understood.16, 17, 18, 19 Emerging evidence suggests cross-talk between gut microbiome and the lungs, via changes to immune responses as well as an interaction of microbiota between the sites, in a hypothesized “gut-lung axis.”11,20
Existing studies on the association between gut microbiota and asthma have focused mainly on disease development during childhood,21, 22, 23 which is driven by evidence of the influence of early-life microbial exposures on immune function.24,25 Previous cross-sectional studies have reported compositional and functional differences of the gut microbiome between adult patients with asthma and healthy controls.26, 27, 28, 29 However, little is known about whether and to what extent the gut microbiome affects the prospective risk of developing incident asthma in adults. For COPD, there have been far fewer studies on the link between the gut microbiome and disease. Recently, the first analysis of gut microbiome in COPD by Bowerman et al30 reported that the fecal microbiome and metabolome differentiate patients with COPD and healthy controls, which suggests a possible avenue for further investigation using prospective population-scale data sets. Finally, it is only in recent years that methodological and technological advances have opened up the possibility of using large-scale microbial data to predict human respiratory disease,22,31 but the feasibility of such measures is yet to be evaluated for COPD.
Here, we report association analysis and predictive modeling of the gut microbiome and incident asthma and COPD using stool samples from more than 7000 participants of a prospective population-based cohort (FINRISK 2002) with electronic health records (EHRs) over approximately 15 years of follow-up.32 Specifically, we (1) describe the gut microbial composition from shallow shotgun metagenomic sequencing and assess the associations with incident asthma and COPD, (2) use machine learning approaches to quantify the predictive capacities of the gut microbiome at baseline for incident respiratory disease, and (3) construct integrated models of the gut microbiome and conventional risk factors and evaluate their predictive performance.
Methods
Study design and participants
The FINRISK 2002 study was a population-based nationwide survey carried out in Finland in 2002, consisting of random samples of the population aged 25 to 74 years drawn from the National Population Information System.32 The sampling was stratified by sex, region, and 10-year age group so that each stratum had 250 participants. The survey included self-administered questionnaires, health examinations conducted at the study sites by trained personnel, and collection of biological samples. The overall participation rate was 65.5% (n = 8798). The participants were followed up through linkage to national administrative electronic registers that proved highly reliable.33, 34, 35 Inclusion criteria have been described elsewhere.32 The present study excluded participants who did not have linked EHRs of clinical end points, who had prior diagnoses of the diseases for prediction, who were pregnant at baseline, and who had prescription of antibiotics use defined as ATC code of J01 up to 6 months before baseline. The incident cases of asthma and COPD were identified according to International Classification of Diseases, Tenth Revision diagnosis codes (Finnish modification) from linked EHRs, which were last followed up by December 31, 2016. COPD cases were defined using International Classification of Diseases codes J43|J44; asthma cases were defined using International Classification of Diseases codes J45|J46, or the Social Insurance Institution of Finland (Kela) reimbursement code 203 for asthma medication, or medicine purchases with Anatomical Therapeutic Chemical codes R03BA|R03BC|R03DC|R03AK. Covariates included baseline age, sex, body mass index (BMI), and smoking. Written informed consent was obtained from all participants. The Coordinating Ethics Committee of the Helsinki and Uusimaa Hospital District approved the FINRISK 2002 study protocols (reference no. 558/E3/2001). The study was conducted according to the World Medical Association’s Declaration of Helsinki on ethical principles.
Sample collection
During the baseline survey, stool samples were collected by willing participants at home using an ad hoc kit constructed in-house in the Finnish Institute for Health and Welfare (THL) with detailed instructions and a scoop method. The participants were advised to collect the sample preferably in the morning, but any time convenient to the participant was considered acceptable. The samples were mailed overnight between Monday and Thursday to the laboratory of the Finnish Institute for Health and Welfare for storing at −20°C. Special care was taken to avoid delayed transit at the post office over the weekend. The sample collection was done under winter conditions, with average temperatures well below 0°C in Finland from January through March 2002, and no special arrangements were made regarding the temperature during transportation. The possible short-term exposure of samples to room temperature after collection and the extended time before freezing might cause variations in detection and relative abundances of rare taxa,36,37 which is relatively minor given the average environmental temperatures well below 0°C and the focus on common taxa in this study. The frozen stool samples were transferred to University of California San Diego for sequencing in 2017.
DNA extraction, sequence processing, and taxonomic profiling
DNA extraction was carried out using the MagAttract PowerSoil DNA kit (Qiagen, Venlo, The Netherlands) according to the Earth Microbiome Project protocols.38 Libraries were prepared using a miniaturized version of the KAPA HyperPlus Illumina-compatible prep kit according to manufacturer’s protocol.39 The extracted DNA was normalized to 5 ng total input per sample with an Echo 550 acoustic liquid-handling robot (Labcyte Inc, San Jose, Calif). A 1/10 scale enzymatic fragmentation, end-repair, and adapter-ligation reactions were performed with a Mosquito HV liquid-handling robot (TTP Labtech, Melbourn, United Kingdom). Sequencing adapters were performed on the basis of the iTru protocol40 by ligating universal adapter stubs and adding sample-specific barcoded sequences in a subsequent PCR step. Amplified and barcoded libraries were quantified with the PicoGreen assay and pooled in approximately equimolar ratios. An Illumina HiSeq 4000 instrument was used to perform shallow shotgun metagenomics sequencing41 to a mean depth of approximately 106 reads/sample. The stool shotgun sequencing was successfully performed in 7231 individuals. The metagenomic sequences were processed using an automated Snakemake workflow pipeline (https://github.com/tanaes/snakemake_assemble).39,42 Removal of low-quality sequences and adapters was performed using Atropos.43 Host reads were removed with Bowtie244 by mapping against human genome assembly GRCh38. Samples with total reads lower than 400,000 were removed to preserve the quality of data while retaining most of the disease cases.
The raw shotgun metagenomes were mapped to an index database based on taxonomic nomenclature introduced and updated in the Genome Taxonomy Database (GTDB) release 8945 using default parameters in the k-mer–based metagenomic classification tool Centrifuge 1.0.4.46 In total, 151 phyla, 338 classes, 925 orders, 2,254 families, 7,906 genera, and 24,705 species were uniquely identified on the basis of GTDB taxonomy. The relative abundances of a bacterial taxon at phylum, class, order, family, genus, and species levels were computed as the proportion of reads assigned to the clade rooted at this taxon among total classified reads. The relative abundance of a taxon with no reads assigned was considered as zero in the metagenome. The present analyses focused on common and relatively abundant microbial taxa with relative abundances greater than 0.01% in more than 1% of samples. Three measures of microbial diversity were calculated: Shannon’s alpha diversity, Chao1 richness, and Pielou’s evenness (R packages vegan v2.5.5 and otuSummary v0.1.1). To overcome the sample comparison biases of compositional data, the centered log-ratio (CLR) transformation was performed on abundance data (R package compositions v1.4.2), of which zeros were substituted with 1/10 of nonzero minimum abundance.
Machine learning and statistical analysis
A machine learning framework was used to develop prediction models at different taxonomic levels separately. The samples were randomly partitioned into 2 subsets: (1) a training data set (70% of samples) for developing models and (2) a validation data set (30% of samples) for evaluating prediction performance. We resampled the data 10 times and performed the same training and validation procedure for each sampling partition. In each training data set, we first selected microbial indicators for predicting incident asthma and COPD; we analyzed the relationships between taxon-level abundance and incident disease using logistic regression adjusted for age and sex, Cox regression for time to disease onset adjusted for age and sex, and Spearman correlation. These approaches have been widely used to explore the relationship between microbiome features and disease-related traits in previous studies.30,31,47 Logistic regression naturally models binary outcomes; Cox regression takes into account the time until events occur; Spearman correlation is a nonparametric measure of the strength and direction of nonlinear correlation. To include any taxa with potential predictive signals, we considered taxa that were associated with incident diseases at a significance threshold of P less than .05 by any of the above approaches for further analyses. To avoid overfitting of feature selection, we did not use algorithms that take into account all the features simultaneously. The selected taxa together with diversity measurements were considered as microbial predictors for developing gradient boosting decision tree model, an ensemble method of sequential and additive training of trees. Each tree fits the residuals of the previous tree in sequence to minimize errors, which makes gradient boosting a highly efficient method. In addition, gradient boosting decision trees are robust to correlated features that naturally exist in microbiome abundance data and apply regularization to reduce overfitting. Gradient boosting decision tree models were implemented with XGBoost 0.82 through 5-fold cross-validation to determine optimal hyperparameters with Bayesian optimization (R package mlrMBO 1.1.2). XGBoost models were developed with objective “binary:logistic” and parameters considered include eta[0.001, 0.5], max_depth[3,10], min_child_weight[5,100], subsample[0.8,0.95], colsample_bytree[0.6,0.85], gamma[0,5], lambda[0.0001, 1], max_delta_step[0,8], early_stopping_rounds[10,20], and nrounds[100,1500]. The optimal setting was then trained on the whole training data to build the final model used in validation. For asthma and COPD, the predicted values from the optimal gradient boosting model of gut microbial features were used as the gut microbiome scores in the validation data set where the scores were used for further Cox analyses for each disease condition. We additionally performed ridge logistic regression to compare the prediction performance using the same samples for training and testing. The gradient-boosted trees-based models outperformed those based on ridge logistic regression. A similar trend of prediction performance across taxonomic levels was observed with both methods. The final performance across various models and partitions was assessed in the validation data sets.
Wilcoxon rank-sum test was performed to compare differences in patient characteristics and microbial diversity metrics between incident cases and noncases across all samples for each disease. Cox regression with adjustment of age and sex was used to assess the association between taxon-level CLR abundance and incident disease using all samples. Benjamini-Hochberg correction was used to control for multiple testing at each taxonomic rank, and false-discovery rate less than 0.05 was considered as statistical significance. We additionally applied Benjamini-Yekutieli correction across all taxonomic levels and reported corrected P values.
Cox models of conventional risk factors and in combination with the gut microbiome score were built using the time from baseline to the occurrence of the disease or end of follow-up in the validation data set (R package survival 2.44). Sensitivity analyses considered income and education level as risk factors and were performed in the validation data set using samples with complete data of risk factors. Association of risk factors was assessed separately and in combination using Cox models for incident asthma and COPD. Education was classified into 3 groups, low, middle, and high education, on the basis of years at school tertiles adjusted for birth cohort as reported in the questionnaire. Income level was represented as an ordinal variable of 1 to 9 according to household’s income before tax deduction with the following cutoffs: less than 50,000 FIM (<8,400€), 50,001 to 100,000 FIM (8,401-16,820€), 100,001 to 150,000 FIM (16,821-25,230€), 150,001 to 200,000 FIM (25,231-33,640€), 200,001 to 250,000 FIM (33,641-42,050€), 250,001-300,000 FIM (42,051-50,460€), 300,001-350,000 FIM (50,461-58,870€), 350,001-400,000 FIM (58,871-67,280€), and more than 400,000 FIM (>67,280€). Statistical analyses were carried out with R 3.6.1.
Data and code availability
The FINRISK data for the present study are available with a written application to the THL Biobank as instructed on the website of the Biobank (https://thl.fi/en/web/thl-biobank/for-researchers). A separate permission is needed from FINDATA (https://www.findata.fi/en/) for use of the EHR data. Custom code for analysis in this study is available at https://github.com/dpredprj/gut_respiratory_link.
Results
A total of 7115 FINRISK02 participants with baseline gut microbiome profiles and EHR linkage were available for the present study. A summary description of the cohort is given in the Methods section, and baseline characteristics are reported in Table I. After quality control and exclusion criteria were applied, 435 and 145 incident cases of asthma and COPD, respectively, occurred during a median follow-up of 14.8 years after gut microbiome sampling at baseline. Notably, more males than females developed COPD, and incident COPD cases displayed older baseline age than noncases (P < .001). The age of onset of incident COPD was older compared with that of incident asthma (P < .001). A higher BMI was observed in asthma cases versus noncases (P = .002), whereas there was no difference in BMI between COPD cases and noncases. For both COPD and asthma, a higher proportion of current smokers during the survey year were observed in disease cases than in noncases.
Table I.
Characteristics of study participants
| Characteristic | Asthma |
COPD |
||
|---|---|---|---|---|
| Incident cases (n = 435) | Noncases (n = 5244) | Incident cases (n = 145) | Noncases (n = 5932) | |
| Sex: female, n (%) | 252 (57.9) | 2740 (52.3) | 43 (29.7) | 3204 (54) |
| Baseline age (y) | 50.9 (40.5-60.5) | 50.5 (39.2-59.3) | 59.5 (53.6-66.5) | 50.5 (39.2-59.5) |
| Age at first event (y) | 57.6 (46.7-67.3) | — | 69.1 (61.3-73.6) | — |
| BMI (kg/m2) | 26.7 (24-30.7) | 26.3 (23.7-29.3) | 26.6 (23.5-29.6) | 26.4 (23.7-29.5) |
| Current smoker, n (%) | 151 (34.8) | 1192 (22.8) | 105 (72.9) | 1313 (22.2) |
| Ex-smoker, n (%) | 94 (21.6) | 1181 (22.5) | 32 (22.1) | 1321 (22.3) |
Continuous variables are presented as median (interquartile range).
Gut microbiome composition and taxon-level abundances
Individual gut microbiome compositions were characterized by shallow shotgun metagenomic sequencing of stool samples (see the Methods section). The present study focused on microbial taxa whose relative abundance exceeded 0.01% in at least 1% of samples; this yielded 46 phyla, 71 classes, 124 orders, 232 families, 617 genera, and 1224 species, as classified according to the Genome Taxonomy Database (GTDB) release 89.45 Most of the gut microbiota were dominated by the Firmicutes_A and Bacteroidota phyla (Fig 1, A), which mostly comprised members of classes Clostridia and Bacteroidia, respectively. At the genus level, Faecalibacterium and Agathobacter in phylum Firmicutes_A, as well as Bacteroides, Bacteroides_B, and Prevotella in phylum Bacteroidota, were most abundant in most samples (Fig 1, B).
Fig 1.
Gut microbiome composition and characteristics. A, Gut microbiome profiles at phylum level. B, Box plots of the 20 most abundant genera sorted by mean relative abundance. C, Shannon’s, Pielous’s, and Chao1 indices at genus level between cases and noncases. Median values are represented by horizontal lines. D, Principal-component analysis on CLR-transformed abundances at genus level. E, Genera associated with incident asthma or COPD surpassing a false-discovery rate threshold of 5% (PFDR < 0.05) with Benjamini-Hochberg correction. Only the top 10 most abundant genera for each of combination of positive or negative associations, with COPD or asthma.
Baseline alpha-diversity measures differed between incident asthma cases and noncases (P < .01), with lower values of Shannon’s, Chao1, and Pielou’s indices in individuals who went on to develop asthma (Fig 1, C). There was no statistical difference in alpha-diversity indices between COPD cases and noncases. Principal- component analysis of the CLR-transformed abundances showed no clear separation between incident cases and noncases (Fig 1, D), suggesting that the association of incident asthma and COPD with the gut microbiome was unlikely related to the whole microbial community and may be attributable to specific microbial taxa.
We assessed the association between baseline taxon-level microbial abundances and incident respiratory diseases using Cox regression, based on CLRs (see the Methods section). At 5% false-discovery rate, statistically significant associations of incident asthma were found in 5 phyla, 5 classes, 18 orders, 111 families, 257 genera, and 299 species (see Table E1 in this article’s Online Repository at www.jacionline.org); for incident COPD, we found associations with 5 phyla, 7 classes, 32 orders, 57 families, 133 genera, and 158 species (see Table E2 in this article’s Online Repository at www.jacionline.org). Of the asthma- and COPD-associated taxa, 76% and 68.6% showed positive associations with disease incidence, respectively. A number of highly abundant genera were associated with incident asthma, such as Bacteroides, Faecalibacterium, Agathobacter, Blautia_A, and Roseburia (Fig 1, E). Among the most abundant COPD-associated genera, increased abundance of Faecalicatena, Oscillibacter, Lawsonibacter, Flavonifractor, and Streptomyces and reduced abundances of Lachnospira, ER4, KLE1615, Eubacterium_F, and Coprococcus were associated with incident COPD.
Gut microbiome and gradient boosting decision trees to predict incident asthma and COPD
To investigate whether the baseline gut microbiome was predictive of incident asthma and COPD, we train and validate prediction models via the machine learning algorithm of gradient boosting decision trees. These models were trained with 5-fold cross-validation in 70% of the individuals and then the performances were validated in the remaining 30% (see the Methods section); all performance metrics given are based on the 30% validation set unless otherwise specified. Models were developed at different taxonomic levels separately and for a combination of all taxonomic levels (see Fig E1 in this article’s Online Repository at www.jacionline.org). To assess sampling variation, we resampled training and testing partitions at different taxonomic levels 10 times and report mean values of prediction performance.
The best performance was obtained at individual taxonomic levels, rather than their combination, for both asthma and COPD prediction. Generally better prediction performance was attained at lower taxonomic levels, particularly for COPD where the highest average area under the operating characteristic curve (AUC) was at species level (mean AUC, 0.780), followed by genus (mean AUC, 0.734) and family (mean AUC, 0.688) levels. For prediction of incident asthma, the best performance was obtained at family level (mean AUC, 0.608), with slight attenuation of AUC scores obtained at genus (mean AUC, 0.592) and species (mean AUC, 0.593) levels.
The gut microbiome had greater predictive value than individual conventional risk factors
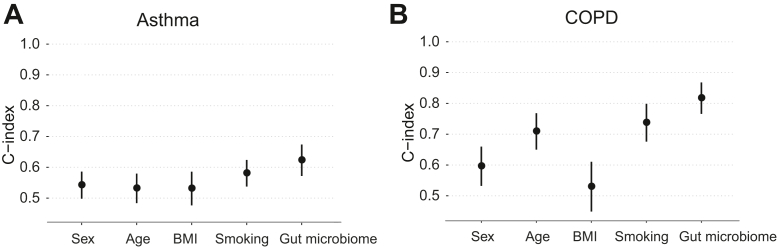
To compare the predictive value of conventional risk factors and the gut microbiome for incident asthma and COPD, we first conducted univariate analysis using Cox models. We used the optimal cross-validated gradient boosting model at family and species level for asthma and COPD, respectively, and refer to the resultant score as a “gut microbiome score” for each condition. We found that the gut microbiome score had a relatively high predictive capacity with C-indices of 0.623 for asthma and 0.817 for COPD, which were each greater than those of other risk factors (Fig 2). Smoking status at baseline was associated with increased risk of both asthma (hazard ratio [HR], 2.21; 95% CI, 1.53-3.20; P < .001) and COPD (HR, 8.16; 95% CI, 4.55-14.64; P < .001) compared with nonsmoking (Table II). Increased incidence of COPD was also associated with male sex (HR, 2.19; 95% CI, 1.25-3.82; P = .01) and older baseline age (HR, 1.07 per year; 95% CI, 1.04-1.10; P < .001). The gut microbiome score was associated with increased incidence of both asthma (HR, 1.44 per SD; 95% CI, 1.23-1.67; P < .001) and COPD (HR, 1.39 per SD; 95% CI, 1.30-1.49; P < .001). In sensitivity analysis that additionally accounted for income and education levels, the findings of higher predictive capacity in C-index of the gut microbiome score than other individual risk factors further held for both asthma and COPD (see Table E3 in this article’s Online Repository at www.jacionline.org), and higher income and high education level were associated with lower risk of COPD (HR, 0.63, 95% CI, 0.53-0.76, P < .001, and HR, 0.3, 95% CI, 0.14-0.67, P = .003, respectively).
Fig 2.
Predictive capacity of each risk factor separately for (A) incident asthma or (B) COPD. Univariate Cox models were used for each of sex, baseline age, BMI, smoking, and gut microbiome individually. Points and error bars represent the C-indices and 95% CIs.
Table II.
Association of risk factors separately and jointly for incident asthma and COPD
| Covariate | Asthma |
COPD |
||||||
|---|---|---|---|---|---|---|---|---|
| Univariable |
Multivariable |
Univariable |
Multivariable |
|||||
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | |
| Sex: male | 0.71 (0.49-1.03) | .07 | 0.67 (0.46-0.97) | .03 | 2.19 (1.25-3.82) | .01 | 1.35 (0.76-2.4) | .31 |
| Baseline age (y) | 0.99 (0.98-1.01) | .28 | 1.00 (0.98-1.01) | .75 | 1.07 (1.04-1.1) | <.001 | 1.1 (1.07-1.13) | <.001 |
| BMI (kg/m2) | 1.02 (0.99-1.06) | .22 | 1.03 (0.99-1.07) | .13 | 1.02 (0.97-1.08) | .48 | 0.99 (0.92-1.06) | .8 |
| Smoking (yes) | 2.21 (1.53-3.2) | <.001 | 2.06 (1.4-3.03) | <.001 | 8.16 (4.55-14.64) | <.001 | 11.07 (5.81-21.09) | <.001 |
| Gut microbiome | 1.44 (1.23-1.67) | <.001 | 1.34 (1.15-1.57) | <.001 | 1.39 (1.3-1.49) | <.001 | 1.18 (1.08-1.29) | <.001 |
Gut microbiome score is represented as microbiome-based predictions per SD. All analyses were performed in the validation set.
Integrated prediction models of the gut microbiome and conventional risk factors
When integrating risk factors and gut microbiome score, the Cox model for asthma showed that current smoking status and gut microbiome were associated with higher risk (HR, 2.06, 95% CI, 1.40-3.03, P < .001, and HR, 1.34 per SD, 95% CI, 1.15-1.57, P < .001, respectively), and male sex was associated with lower risk (HR, 0.67, 95% CI, 0.46-0.97, P = .03), whereas there were no associations for baseline age and BMI at a statistical significance level (Table II). For COPD, baseline age, current smoking status, and gut microbiome score were statistically significant predictors (HR, 1.1 per year, 95% CI, 1.07-1.13, P < .001; HR, 11.07, 95% CI, 5.81-21.09, P < .001; and HR, 1.18 per SD, 95% CI, 1.08-1.29, P < .001, respectively). Although consistent with the individual predictive power of the gut microbiome score, the multivariable Cox model showed that the risk associated with current smokers at baseline was significantly greater than that for other risk factors for COPD. Sensitivity analyses additionally adjusting the Cox model for income and education level confirmed similar estimates of effects of sex, age, BMI, smoking, and the gut microbiome score in the combined model (see Table E3 in this article’s Online Repository at www.jacionline.org), whereas no statistical significance was detected for income and education.
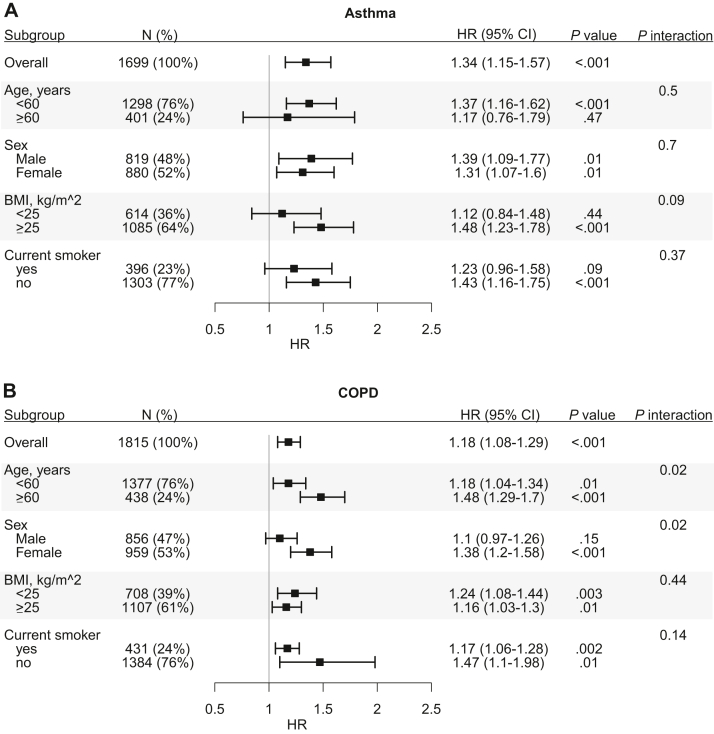
In subgroup analyses, the gut microbiome score association patterns were generally consistent with those above (Fig 3). For COPD, where current smoking status had a relatively large HR, the gut microbiome score was independently associated with incident COPD in both current smokers and nonsmokers. In individuals who indicated past smoking but who were not current smokers at survey (n = 414), we found that the gut microbiome score was not associated with incident COPD (HR, 1.22 per SD; 95% CI, 0.89-1.68; P = .22) but that, in individuals who reported never smoking (n = 970), there was an association with incident COPD (HR, 1.40 per SD; 95% CI, 1.02-1.91; P = .04). Finally, in COPD, we observed evidence for statistical interactions of the gut microbiome score with age and sex (Fig 3).
Fig 3.
Subgroup analyses for (A) incident asthma or (B) COPD. Cox models were applied to test for interactions between gut microbiome and patient characteristic subgroups. Points and error bars represent HRs per SD and 95% CIs of gut microbiome score across subgroups.
The integrated models showed improved predictive capacity for both incident asthma and COPD (Fig 4). For asthma, a reference model of age, sex, and BMI yielded C-index of 0.567; addition of smoking status and then gut microbiome score increased the C-index further to 0.626 and 0.656, respectively. For COPD, the reference model of age, sex, and BMI yielded C-index of 0.735; addition of smoking status and then gut microbiome score increased the C-index further to 0.855 and 0.862, respectively.
Fig 4.
Predictive capacity of integrated models for (A) incident asthma and (B) COPD. “Ref” is a reference model that jointly considers age, sex, and BMI. Points and error bars represent the C-indices and 95% CIs. Analysis of deviance based on the log partial likelihood, ∗∗P < .01; ∗∗∗P < .001.
Discussion
In this prospective study, we investigated the association and predictive capacity of the gut microbiome for future chronic respiratory diseases, asthma and COPD, in adults using shotgun metagenomics. We demonstrated that the gut microbiome is associated with incident asthma and COPD and evaluated the relative contributions of traditional risk factors and a gut microbiome score. We then constructed integrated risk models that maximized predictive performance. Taken together, our findings indicate that the gut microbiome is a potentially substantive biomarker with clinical validity for both asthma and COPD.
The gut and lung microbial communities, although residing in distal sites, are dominated by broadly similar bacterial phyla, including Firmicutes and Bacteroidetes, but differ in local compositions and total microbial biomass.11 Some of our findings are relevant to previous microbial studies of the respiratory tract. For example, Haemophilus and Streptococcus have been previously found to be positively associated with respiratory illnesses in the airways.18,48,49 In our gut microbiome samples, we also found positive associations between Streptococcus and incident asthma; however, we found that multiple Haemophilus spp. were negatively associated with incident COPD. An increased abundance of Pseudomonas spp. from the airway microbiome was previously reported in COPD exacerbations50,51 and impaired pulmonary function.52,53 Consistent with this, we found positive associations of the Pseudomonas, Pseudomonas_A, and Pseudomonas_E genera (all part of Pseudomonas according to the NCBI taxonomy) with incident asthma and COPD. These findings support the emerging evidence of possible functional links between the respiratory tract and the gastrointestinal tract; however, the underlying mechanisms by which microorganisms between the sites may interact remain unclear.54,55
Despite increasing recognition of the existence of gut-lung cross-talk, the role of the gut microbiota in respiratory disease has been primarily studied in children. Its relevance in adults has been unclear. Previous studies have demonstrated that the early-life gut microbial alteration and maturation patterns influence the risk of asthma development in childhood.22,23,56 In our data, we found that higher abundances of Escherichia,31 Enterococcus, Clostridium, Veillonella, and B fragilis were associated with increased incidence of asthma in adulthood, consistent with that observed for childhood asthma.22,57,58 In contrast to previous findings showing that the relative abundances of Faecalibacterium, Roseburia, and Flavonifractor were decreased in childhood asthma,22,57 we found positive associations with adult-onset asthma. We confirmed previous findings that increased abundances of Clostridium and Eggerthella lenta in the adult gut microbiome were associated with asthma.27 The relationship between the gut microbiome and COPD is even less understood. A recent study reported that Streptococcus sp000187445 was enriched in patients with COPD and was correlated with reduced lung function,30 which was also confirmed by a positive association with incident COPD in our study.
Regarding consideration of causality in observational studies, it is challenging to determine whether the composition of the gut microbiome is a cause or consequence of respiratory disease. In this respect, one strength of our study was the use of baseline gut microbiome and incident disease systematically identified through EHRs. The follow-up using EHRs was nearly complete in all samples (except for the small number of participants who moved abroad permanently). Using machine learning models, we found that the baseline gut microbiome had moderate predictive capacities in distinguishing incident cases from noncases for asthma and COPD, suggesting that there are detectable changes in the gut microbiome antecedent to the onset of symptomatic disease. This does not confirm causality or eliminate other possibilities. For example, disease-associated host changes and gut microbial alteration may influence each other and operate simultaneously.54 We also showed that the association between gut microbiome–based predictions and incident asthma or COPD was largely independent of age, sex, BMI, and smoking, all of which can influence susceptibility to respiratory diseases.59, 60, 61, 62 Moreover, interactions of gut microbiome by sex and age were found for COPD with relatively weak signals, suggesting different impact of gut microbiome on age and sex groups, consistent with findings in other settings.63, 64, 65
Importantly, our study affirms the large body of evidence that smoking is associated with respiratory illness, especially COPD. Despite many ways to characterize the smoking phenotype, we found that individuals who reported being current smokers were at high risk of future asthma and COPD. The association between smoking and gut microbiota is well established, and smoking cessation has been shown to have profound, putatively causal effects on the gut microbiome.66 Our results show that, particularly for COPD, the gut microbiome is both a substantial independent predictor of future disease and that its predictive power is partially explained by smoking behavior. As such, our findings are both consistent with previous studies and take us a step closer to delineating which and to what extent particular gut microbial taxa sit along the causal path from smoking behavior to future asthma and COPD. For the latter, larger prospective studies will be necessary but population-scale gut microbiome and e-health studies are underway. There are other traditional risk factors that could be investigated in future studies, such as family history, environmental pollutants, exposure to allergens or irritants, and other lifestyles. Family history of asthma and/or allergy has been linked to increased risk of childhood asthma in particular, and the impact of paternal asthma continues to young adulthood.67, 68, 69 Although previous studies have suggested that the impact of family history of asthma and allergy decreases with age and paternal allergic disease is not associated with late-onset asthma with a cutoff age of 12 years,70,71 the effect of family history on adult-onset asthma could be further explored in the future.
There are limitations of the present study. First, despite a relatively large sample size, our study was enrolled from a single European country (Finland), and the generalizability of the findings to other geographically and culturally distinct settings will require further investigation in external cohorts with baseline gut microbiome and long-term respiratory disease data. Furthermore, only 1 time point of the gut microbiome was sampled per individual, which did not allow for dynamic or temporal assessment of gut microbiome alterations along with incident disease onset. Changes in diet and environmental exposures (apart from smoking) can induce changes in gut microbiota and should be considered in future studies. Limitations also concern the disease phenotyping in the present study, where incident cases were identified by a combination of EHRs of diagnosis codes, medicine purchases, and insurance reimbursements (see the Methods section). This might be subject to possible misclassification of borderline cases due to incorrect diagnostic labeling, particularly overlabeling of asthma. For example, patients with COPD might be mislabeled as asthma due to smoking-related stigma of COPD and better medication reimbursements of asthma. Inaccurate diagnosis may also be attributed to the asthma-COPD overlap syndrome characterized by coexistence of clinical features of both asthma and COPD.72 Future studies should consider differential diagnostic characteristics of asthma, COPD, and potentially a third group of asthma-COPD overlap syndrome for improved disease phenotyping.73, 74, 75 Although the asthma and COPD phenotypes can be difficult to diagnose or indeed overlap in some individuals, our study takes a pragmatic approach and future clinical cohorts may be necessary to precisely quantify disease-specific effects. Finally, although formal lung function test results (FEV1, FVC) may further improve prediction, it was not feasible to perform whole-scale clinical examination of airflow obstruction at the population level. Regardless, our study demonstrates that future exploration of the influence of the gut microbiome in severity and progression of asthma and COPD is warranted, and may lead to further clinically significant findings.
Our study supports the role of gut microbiome in adult respiratory disease and as potential biomarkers that might aid in risk profiling of asthma and COPD. The underlying mechanisms and causal links by which gut microbiota influence the lung, and vice versa, remain to be established.
Key message.
-
•
The gut microbiome features are associated with incident COPD and adult-onset asthma.
-
•
The gut microbiome is potentially a predictive biomarker for primary prevention of asthma and COPD.
Footnotes
V.S. was supported by the Finnish Foundation for Cardiovascular Research and by Juho Vainio Foundation. M.I. was supported by the Munz Chair of Cardiovascular Prediction and Prevention. A.S.H. was supported by the Academy of Finland (grant no. 321356). L.L. was supported by the Academy of Finland (grant nos. 295741 and 328791). T.N. was supported by the Emil Aaltonen Foundation, the Finnish Foundation for Cardiovascular Research, the Sigrid Jusélius Foundation, and the Academy of Finland (grant no. 321351). This study was supported by the Victorian government’s Operational Infrastructure Support program and by core funding from the British Heart Foundation (grant nos. RG/13/13/30194 and RG/18/13/33946) and the NIHR Cambridge Biomedical Research Centre (grant no. BRC-1215-20014). The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care. This work was supported by Health Data Research UK, which is funded by the UK Medical Research Council, the Engineering and Physical Sciences Research Council, the Economic and Social Research Council, the Department of Health and Social Care (England), the Chief Scientist Office of the Scottish Government Health and Social Care Directorates, the Health and Social Care Research and Development Division (Welsh Government), the Public Health Agency (Northern Ireland), the British Heart Foundation, and Wellcome.
Disclosure of potential conflict of interest: V. Salomaa has received honoraria from Sanofi for consulting and also has ongoing research collaboration with Bayer Ltd (all outside this study). The rest of the authors declare that they have no relevant conflicts of interest.
Contributor Information
Yang Liu, Email: yang.liu2@baker.edu.au, yangl25@student.unimelb.edu.au.
Michael Inouye, Email: minouye@baker.edu.au, mi336@medschl.cam.ac.uk.
Supplementary data
References
- 1.Halpin D.M.G., Criner G.J., Papi A., Singh D., Anzueto A., Martinez F.J., et al. Global Initiative for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease. The 2020 GOLD Science Committee Report on COVID-19 and Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med. 2021;203:24–36. doi: 10.1164/rccm.202009-3533SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global Initiative for Asthma, Global strategy for asthma management and prevention, 2020. Available from: www.ginasthma.org
- 3.Papi A., Brightling C., Pedersen S.E., Reddel H.K. Asthma. Lancet. 2018;391:783–800. doi: 10.1016/S0140-6736(17)33311-1. [DOI] [PubMed] [Google Scholar]
- 4.Barnes P.J. Therapeutic approaches to asthma-chronic obstructive pulmonary disease overlap syndromes. J Allergy Clin Immunol. 2015;136:531–545. doi: 10.1016/j.jaci.2015.05.052. [DOI] [PubMed] [Google Scholar]
- 5.Mirza S., Benzo R. Chronic obstructive pulmonary disease phenotypes: implications for care. Mayo Clin Proc. 2017;92:1104–1112. doi: 10.1016/j.mayocp.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuruvilla M.E., Lee F.E., Lee G.B. Understanding asthma phenotypes, endotypes, and mechanisms of disease. Clin Rev Allergy Immunol. 2019;56:219–233. doi: 10.1007/s12016-018-8712-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCracken J.L., Veeranki S.P., Ameredes B.T., Calhoun W.J. Diagnosis and management of asthma in adults: a review. JAMA. 2017;318:279–290. doi: 10.1001/jama.2017.8372. [DOI] [PubMed] [Google Scholar]
- 8.Polverino F., Celli B. The challenge of controlling the COPD epidemic: unmet needs. Am J Med. 2018;131:1–6. doi: 10.1016/j.amjmed.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Riley C.M., Sciurba F.C. Diagnosis and outpatient management of chronic obstructive pulmonary disease: a review. JAMA. 2019;321:786–797. doi: 10.1001/jama.2019.0131. [DOI] [PubMed] [Google Scholar]
- 10.Budden K.F., Shukla S.D., Rehman S.F., Bowerman K.L., Keely S., Hugenholtz P., et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir Med. 2019;7:907–920. doi: 10.1016/S2213-2600(18)30510-1. [DOI] [PubMed] [Google Scholar]
- 11.Chotirmall S.H., Gellatly S.L., Budden K.F., Mac Aogain M., Shukla S.D., Wood D.L., et al. Microbiomes in respiratory health and disease: an Asia-Pacific perspective. Respirology. 2017;22:240–250. doi: 10.1111/resp.12971. [DOI] [PubMed] [Google Scholar]
- 12.Shreiner A.B., Kao J.Y., Young V.B. The gut microbiome in health and in disease. Curr Opin Gastroenterol. 2015;31:69–75. doi: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huffnagle G.B., Dickson R.P., Lukacs N.W. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. 2017;10:299–306. doi: 10.1038/mi.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yatera K., Noguchi S., Mukae H. The microbiome in the lower respiratory tract. Respir Investig. 2018;56:432–439. doi: 10.1016/j.resinv.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z., Bafadhel M., Haldar K., Spivak A., Mayhew D., Miller B.E., et al. Lung microbiome dynamics in COPD exacerbations. Eur Respir J. 2016;47:1082–1092. doi: 10.1183/13993003.01406-2015. [DOI] [PubMed] [Google Scholar]
- 16.Huang Y.J., Nariya S., Harris J.M., Lynch S.V., Choy D.F., Arron J.R., et al. The airway microbiome in patients with severe asthma: associations with disease features and severity. J Allergy Clin Immunol. 2015;136:874–884. doi: 10.1016/j.jaci.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y.J., Sethi S., Murphy T., Nariya S., Boushey H.A., Lynch S.V. Airway microbiome dynamics in exacerbations of chronic obstructive pulmonary disease. J Clin Microbiol. 2014;52:2813–2823. doi: 10.1128/JCM.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teo S.M., Mok D., Pham K., Kusel M., Serralha M., Troy N., et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teo S.M., Tang H.H.F., Mok D., Judd L.M., Watts S.C., Pham K., et al. Airway microbiota dynamics uncover a critical window for interplay of pathogenic bacteria and allergy in childhood respiratory disease. Cell Host Microbe. 2018;24:341–352.e5. doi: 10.1016/j.chom.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dang A.T., Marsland B.J. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 2019;12:843–850. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 21.Barcik W., Boutin R.C.T., Sokolowska M., Finlay B.B. The role of lung and gut microbiota in the pathology of asthma. Immunity. 2020;52:241–255. doi: 10.1016/j.immuni.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stokholm J., Blaser M.J., Thorsen J., Rasmussen M.A., Waage J., Vinding R.K., et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun. 2018;9:141. doi: 10.1038/s41467-017-02573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Depner M., Taft D.H., Kirjavainen P.V., Kalanetra K.M., Karvonen A.M., Peschel S., et al. Maturation of the gut microbiome during the first year of life contributes to the protective farm effect on childhood asthma. Nat Med. 2020;26:1766–1775. doi: 10.1038/s41591-020-1095-x. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y.J., Boushey H.A. The microbiome in asthma. J Allergy Clin Immunol. 2015;135:25–30. doi: 10.1016/j.jaci.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamburini S., Shen N., Wu H.C., Clemente J.C. The microbiome in early life: implications for health outcomes. Nat Med. 2016;22:713–722. doi: 10.1038/nm.4142. [DOI] [PubMed] [Google Scholar]
- 26.Barcik W., Pugin B., Westermann P., Perez N.R., Ferstl R., Wawrzyniak M., et al. Histamine-secreting microbes are increased in the gut of adult asthma patients. J Allergy Clin Immunol. 2016;138:1491–1494.e7. doi: 10.1016/j.jaci.2016.05.049. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q., Li F., Liang B., Liang Y., Chen S., Mo X., et al. A metagenome-wide association study of gut microbiota in asthma in UK adults. BMC Microbiol. 2018;18:114. doi: 10.1186/s12866-018-1257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Begley L., Madapoosi S., Opron K., Ndum O., Baptist A., Rysso K., et al. Gut microbiota relationships to lung function and adult asthma phenotype: a pilot study. BMJ Open Respir Res. 2018;5 doi: 10.1136/bmjresp-2018-000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hevia A., Milani C., Lopez P., Donado C.D., Cuervo A., Gonzalez S., et al. Allergic patients with long-term asthma display low levels of Bifidobacterium adolescentis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowerman K.L., Rehman S.F., Vaughan A., Lachner N., Budden K.F., Kim R.Y., et al. Disease-associated gut microbiome and metabolome changes in patients with chronic obstructive pulmonary disease. Nat Commun. 2020;11:5886. doi: 10.1038/s41467-020-19701-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salosensaari A., Laitinen V., Havulinna A.S., Meric G., Cheng S., Perola M., et al. Taxonomic signatures of cause-specific mortality risk in human gut microbiome. Nat Commun. 2021;12:2671. doi: 10.1038/s41467-021-22962-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borodulin K., Tolonen H., Jousilahti P., Jula A., Juolevi A., Koskinen S., et al. Cohort profile: the national FINRISK study. Int J Epidemiol. 2018;47:696–i. doi: 10.1093/ije/dyx239. [DOI] [PubMed] [Google Scholar]
- 33.Pajunen P., Koukkunen H., Ketonen M., Jerkkola T., Immonen-Räihä P., Kärjä-Koskenkari P., et al. The validity of the Finnish Hospital Discharge Register and Causes of Death Register data on coronary heart disease. Eur J Cardiovasc Prev Rehabil. 2005;12:132–137. doi: 10.1097/00149831-200504000-00007. [DOI] [PubMed] [Google Scholar]
- 34.Tolonen H., Salomaa V., Torppa J., Sivenius J., Immonen-Räihä P., Lehtonen A. The validation of the Finnish Hospital Discharge Register and Causes of Death Register data on stroke diagnoses. Eur J Cardiovasc Prev Rehabil. 2007;14:380–385. doi: 10.1097/01.hjr.0000239466.26132.f2. [DOI] [PubMed] [Google Scholar]
- 35.Sund R. Quality of the Finnish Hospital Discharge Register: a systematic review. Scand J Public Health. 2012;40:505–515. doi: 10.1177/1403494812456637. [DOI] [PubMed] [Google Scholar]
- 36.Guo Y., Li S.-H., Kuang Y.-S., He J.-R., Lu J.-H., Luo B.-J., et al. Effect of short-term room temperature storage on the microbial community in infant fecal samples. SciRep. 2016;6 doi: 10.1038/srep26648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flores R., Shi J., Yu G., Ma B., Ravel J., Goedert J.J., et al. Collection media and delayed freezing effects on microbial composition of human stool. Microbiome. 2015;3:33. doi: 10.1186/s40168-015-0092-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marotz L., Schwartz T., Thompson L., Humphrey G., Gogul G., Gaffney J., et al. 2018. Earth Microbiome Project (EMP) high throughput (HTP) DNA extraction protocol. Version 07112018. [Google Scholar]
- 39.Sanders J.G., Nurk S., Salido R.A., Minich J., Xu Z.Z., Zhu Q., et al. Optimizing sequencing protocols for leaderboard metagenomics by combining long and short reads. Genome Biol. 2019;20:226. doi: 10.1186/s13059-019-1834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glenn T.C., Nilsen R.A., Kieran T.J., Sanders J.G., Bayona-Vásquez N.J., Finger J.W., et al. Adapterama I: universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext) PeerJ. 2019;7 doi: 10.7717/peerj.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hillmann B., Al-Ghalith G.A., Shields-Cutler R.R., Zhu Q., Gohl D.M., Beckman K.B., et al. Evaluating the information content of shallow shotgun metagenomics. mSystems. 2018;3 doi: 10.1128/mSystems.00069-18. e00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Köster J., Rahmann S. Snakemake--a scalable bioinformatics workflow engine. Bioinformatics. 2012;28:2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- 43.Didion J.P., Martin M., Collins F.S. Atropos: specific, sensitive, and speedy trimming of sequencing reads. PeerJ. 2017;5 doi: 10.7717/peerj.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parks D.H., Chuvochina M., Waite D.W., Rinke C., Skarshewski A., Chaumeil P.A., et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 46.Kim D., Song L., Breitwieser F.P., Salzberg S.L. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26:1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson M.A., Verdi S., Maxan M.E., Shin C.M., Zierer J., Bowyer R.C.E., et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun. 2018;9:2655. doi: 10.1038/s41467-018-05184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hufnagl K., Pali-Schöll I., Roth-Walter F., Jensen-Jarolim E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin Immunopathol. 2020;42:75–93. doi: 10.1007/s00281-019-00775-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Dwyer D.N., Dickson R.P., Moore B.B. The lung microbiome, immunity, and the pathogenesis of chronic lung disease. J Immunol. 2016;196:4839–4847. doi: 10.4049/jimmunol.1600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Millares L., Ferrari R., Gallego M., Garcia-Nunez M., Perez-Brocal V., Espasa M., et al. Bronchial microbiome of severe COPD patients colonised by Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis. 2014;33:1101–1111. doi: 10.1007/s10096-013-2044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Vidal C., Almagro P., Romani V., Rodriguez-Carballeira M., Cuchi E., Canales L., et al. Pseudomonas aeruginosa in patients hospitalised for COPD exacerbation: a prospective study. Eur Respir J. 2009;34:1072–1078. doi: 10.1183/09031936.00003309. [DOI] [PubMed] [Google Scholar]
- 52.Garcia-Clemente M., de la Rosa D., Maiz L., Giron R., Blanco M., Olveira C., et al. Impact of Pseudomonas aeruginosa infection on patients with chronic inflammatory airway diseases. J Clin Med. 2020;9:3800. doi: 10.3390/jcm9123800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davies G., Wells A.U., Doffman S., Watanabe S., Wilson R. The effect of Pseudomonas aeruginosa on pulmonary function in patients with bronchiectasis. Eur Respir J. 2006;28:974–979. doi: 10.1183/09031936.06.00074605. [DOI] [PubMed] [Google Scholar]
- 54.Budden K.F., Gellatly S.L., Wood D.L., Cooper M.A., Morrison M., Hugenholtz P., et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 55.Zhang D., Li S., Wang N., Tan H.Y., Zhang Z., Feng Y. The cross-talk between gut microbiota and lungs in common lung diseases. Front Microbiol. 2020;11:301. doi: 10.3389/fmicb.2020.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arrieta M.C., Stiemsma L.T., Dimitriu P.A., Thorson L., Russell S., Yurist-Doutsch S., et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aab2271. 307ra152. [DOI] [PubMed] [Google Scholar]
- 57.Chiu C.Y., Cheng M.L., Chiang M.H., Kuo Y.L., Tsai M.H., Chiu C.C., et al. Gut microbial-derived butyrate is inversely associated with IgE responses to allergens in childhood asthma. Pediatr Allergy Immunol. 2019;30:689–697. doi: 10.1111/pai.13096. [DOI] [PubMed] [Google Scholar]
- 58.Vael C., Nelen V., Verhulst S.L., Goossens H., Desager K.N. Early intestinal Bacteroides fragilis colonisation and development of asthma. BMC Pulm Med. 2008;8:19. doi: 10.1186/1471-2466-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han M.K., Postma D., Mannino D.M., Giardino N.D., Buist S., Curtis J.L., et al. Gender and chronic obstructive pulmonary disease: why it matters. Am J Respir Crit Care Med. 2007;176:1179–1184. doi: 10.1164/rccm.200704-553CC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zein J.G., Erzurum S.C. Asthma is different in women. Curr Allergy Asthma Rep. 2015;15:28. doi: 10.1007/s11882-015-0528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zammit C., Liddicoat H., Moonsie I., Makker H. Obesity and respiratory diseases. Int J Gen Med. 2010;3:335–343. doi: 10.2147/IJGM.S11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sears M.R. Smoking, asthma, chronic airflow obstruction and COPD. Eur Respir J. 2015;45:586–588. doi: 10.1183/09031936.00231414. [DOI] [PubMed] [Google Scholar]
- 63.O’Toole P.W., Jeffery I.B. Gut microbiota and aging. Science. 2015;350:1214–1215. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- 64.Haro C., Rangel-Zúñiga O.A., Alcalá-Díaz J.F., Gómez-Delgado F., Pérez-Martínez P., Delgado-Lista J., et al. Intestinal microbiota is influenced by gender and body mass index. PLoS One. 2016;11 doi: 10.1371/journal.pone.0154090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fransen F., van Beek A.A., Borghuis T., Meijer B., Hugenholtz F., van der Gaast-de Jongh C., et al. The impact of gut microbiota on gender-specific differences in immunity. Front Immunol. 2017;8:754. doi: 10.3389/fimmu.2017.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biedermann L., Zeitz J., Mwinyi J., Sutter-Minder E., Rehman A., Ott S.J., et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guilbert T.W., Mauger D.T., Lemanske R.F., Jr. Childhood asthma-predictive phenotype. J Allergy Clin Immunol Pract. 2014;2:664–670. doi: 10.1016/j.jaip.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 68.Smit H.A., Pinart M., Antó J.M., Keil T., Bousquet J., Carlsen K.H., et al. Childhood asthma prediction models: a systematic review. Lancet Respir Med. 2015;3:973–984. doi: 10.1016/S2213-2600(15)00428-2. [DOI] [PubMed] [Google Scholar]
- 69.Paaso E.M., Jaakkola M.S., Lajunen T.K., Hugg T.T., Jaakkola J.J. The importance of family history in asthma during the first 27 years of life. Am J Respir Crit Care Med. 2013;188:624–626. doi: 10.1164/rccm.201212-2236LE. [DOI] [PubMed] [Google Scholar]
- 70.Paaso E.M., Jaakkola M.S., Rantala A.K., Hugg T.T., Jaakkola J.J. Allergic diseases and asthma in the family predict the persistence and onset-age of asthma: a prospective cohort study. Respir Res. 2014;15:152. doi: 10.1186/s12931-014-0152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu T., Valdez R., Yoon P.W., Crocker D., Moonesinghe R., Khoury M.J. The association between family history of asthma and the prevalence of asthma among US adults: National Health and Nutrition Examination Survey, 1999-2004. Genet Med. 2009;11:323–328. doi: 10.1097/GIM.0b013e31819d3015. [DOI] [PubMed] [Google Scholar]
- 72.Leung J.M., Sin D.D. Asthma-COPD overlap syndrome: pathogenesis, clinical features, and therapeutic targets. BMJ. 2017;358:j3772. doi: 10.1136/bmj.j3772. [DOI] [PubMed] [Google Scholar]
- 73.Maniscalco M., Paris D., Melck D.J., Molino A., Carone M., Ruggeri P., et al. Differential diagnosis between newly diagnosed asthma and COPD using exhaled breath condensate metabolomics: a pilot study. Eur Respir J. 2018;51 doi: 10.1183/13993003.01825-2017. [DOI] [PubMed] [Google Scholar]
- 74.Miravitlles M. Diagnosis of asthma-COPD overlap: the five commandments. Eur Respir J. 2017;49 doi: 10.1183/13993003.00506-2017. [DOI] [PubMed] [Google Scholar]
- 75.Bouwens J.D.M., Bischoff E.W.M.A., in ’t Veen J.C.C.M., Schermer T.R. Diagnostic differentiation between asthma and COPD in primary care using lung function testing. NPJ Prim Care Respir Med. 2022;32:32. doi: 10.1038/s41533-022-00298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The FINRISK data for the present study are available with a written application to the THL Biobank as instructed on the website of the Biobank (https://thl.fi/en/web/thl-biobank/for-researchers). A separate permission is needed from FINDATA (https://www.findata.fi/en/) for use of the EHR data. Custom code for analysis in this study is available at https://github.com/dpredprj/gut_respiratory_link.