Abstract
Recently, an increasing number of young never-smokers are diagnosed with lung cancer. The aim of this study is to investigate the genetic predisposition of lung cancer in these patients and discover candidate pathogenic variants for lung adenocarcinoma in young never-smokers. Peripheral blood was collected from 123 never-smoking east-Asian patients diagnosed with lung adenocarcinoma before the age of 40. Whole-exome sequencing (WES) was conducted on genomic DNA extracted from peripheral blood cells. As a result, 3,481 single nucleotide variants were identified. By bioinformatical tools and the published gene list associated with genetic predisposition of cancer, pathogenic variants were detected in ten germline genes: ATR, FANCD2, FANCE, GATA2, HFE, MSH2, PDGFRA, PMS2, SDHB, and WAS. Patients with pathogenic variants were more likely to occur in females (9/10, 90.0%) and have stage IV lung adenocarcinoma (4/10, 40%). Furthermore, germline mutations in 17 genes (ASB18, B3GALT5, CLEC4F, COL6A6, CYP4B1, C6orf132, EXO1, GATA4, HCK, KCP, NPHP4, PIGX, PPIL2, PPP1R3G, RRBP1, SALL4, and TTC28), which occurred in at least two patients, displayed potentially pathogenic effects. Gene ontology analysis further showed that these genes with germline mutations were mainly located in nucleoplasm and associated with DNA repair-related biological processes. The study provides spectrum of pathogenic variants and functional explanation for genetic predisposition of lung adenocarcinoma in young never-smokers, which sheds a light on prevention and early diagnosis of lung cancer.
Supplementary Information
The online version contains supplementary material available at 10.1007/s43657-022-00062-1.
Keywords: Lung adenocarcinoma, Germline mutation, Never-smoker, Young age, DNA repair
Introduction
Lung cancer is a major cause of cancer death worldwide (Bray et al. 2018). An estimated 25% of lung cancer patients are never-smokers (Parkin et al. 2005). Lung cancers from never-smokers are generally associated with young age at diagnosis and adenocarcinoma histology (Toh et al. 2006). Recently, there is an increasing trend of young never-smoking patients with lung cancer (Pelosof et al. 2017; Zhang et al. 2020). To reduce the morbidity, it is critical to elucidate the potential causes behind the phenomenon.
Most of lung cancer occurs accompanying the accumulation of genetic alterations, including somatic or germline mutations in oncogenes and tumor suppressor genes. Traditionally, tobacco smoking was considered to be the most significant contributor for lung cancer, resulting in somatic genetic mutations and ultimately lung cancer (Park et al. 2017). Therefore, smoking cessation campaign has substantially decreased the incidence of lung cancer in America (Jemal et al. 2018). For those nonsmoking individuals, germline alterations might play a significant role in the carcinogenesis of early-onset nonsmoking lung adenocarcinoma. Some previous studies have delineated some germline mutations, including BRCA1/2 mutation in breast and ovarian cancer, as well as germline defects of DNA mismatch repair in colorectal cancer (Thavaneswaran et al. 2019). Nevertheless, there is still a lack of evidence about the actual role of germline mutations in nonsmoking lung cancer.
To confirm the effect of genetic predisposition in these patients, we performed whole-exome sequencing (WES) to peripheral blood cells from 123 young (≤ 40 years old) never-smoking east-Asian patients with lung adenocarcinoma and analyzed their germline variants. The study provides candidate germline mutations for genetic predisposition of lung adenocarcinoma in young never-smokers.
Methods
Patients
East-Asian patients, who were hospitalized in Fudan University Shanghai Cancer Center from 2007 to 2016, were enrolled. Inclusion criteria were listed as follows: (1) age at diagnosis ≤ 40 years; (2) never-smokers, defined as patients who reported never-smoking any cigarettes in their lifetimes; (3) pathologically confirmed primary lung adenocarcinoma; (4) written informed consent was obtained. The diagnosis of lung adenocarcinoma was made by at least two pulmonary pathologists after reviewing morphology from hematoxylin–eosin-staining slides and immunohistochemical diagnostic makers. Any disagreements were solved by discussion. This study was conducted in line with the Helsinki Declaration. The Institutional Review Board of Fudan University Shanghai Cancer Center has approved this study (IRB#090977-1).
Whole-Exome Sequencing
WES was conducted on genomic DNA extracted from peripheral blood cells. Genomic DNA was fragmented and hybridized for enrichment. The pre-capture libraries were captured using the SureSelect capture library kit (Agilent Technologies, Santa Clara, CA, USA). Paired-end sequencing was performed using the Illumina HiSeq system (Illumina, San Diego, CA, USA) according to the manufacturer’s protocols for 2*150 paired-end sequencing.
Mutational Analyses
Whole-exome sequencing data after base calling were cleaned for adapters and mapped to human genome reference (GRCh38/hg38) with the Burrows-Wheeler Aligner. The standard GATK3 haplotype pipeline was performed to identify single nucleotide variations (SNVs) and indels. High-frequency coding alterations were prioritized by ranking each SNV or indel with its occurrence across samples. Gene-level alterations were collapsed by considering all SNVs and indels within all coding exons. Genetic variants were compared to the dbSNP database (Wheeler et al. 2007) to excluded undeleterious single nucleotide polymorphisms (SNPs). Global minor allele frequency was set to be 0.1% for common SNP mutations, which was the threshold in our study. Frequently mutated genes were identified by ranking these summarized occurrences.
SIFT (Sim et al. 2012), Polyphen-2 (Adzhubei et al. 2010), and CADD (Rentzsch et al. 2021) were used to determine the possible pathogenic germline variants in exon regions. Damaging variants of SIFT, and probably damaging or possibly damaging variants of Polyphen-2 were considered to be potentially pathogenic. In addition, identified variants were categorized into pathogenic and benign variants according to PHRED calculated by CADD, and 20 was the cutoff value for PHRED. For stop-gain variants, SIFT and Polyphen-2 were unable to predict their pathogenicity, leaving CADD as the only tool to determine. For other variants, potentially pathogenic mutations required confirmation by at least two out of three in silico tools (SIFT, Polyphen-2, and CADD).
A list of 152 known cancer susceptibility genes reported by Huang et al. (2018) in a recent pan-cancer study was used as candidate pathogenic genes for analysis. Variants were considered to be certainly pathogenic if they were part of the 152 candidate genes. Potentially pathogenic variants were defined as those occurring twice or more. All the recurrent variants were validated by Sanger sequencing of PCR-amplified products.
Statistical Analyses
Statistical analyses were conducted using SPSS software (version 25.0, IBM, Armonk, NY, America). All identified germline mutations in this study were listed in Supplementary Table 1, and clinical data were also included in Supplementary Table 2. The raw data of WES from 123 young nonsmoking patients with lung adenocarcinoma have been uploaded to the in the Genome Sequence Archive of the BIG Data Center at the Beijing Institute of Genomics, Chinese Academy of Science, China. They are accessible under HRA001459 (http://bigd.big.ac.cn/gsa-human/). All tests were two-tailed, and the statistical difference was set at p < 0.05. DAVID Bioinformatics Resources were used for gene ontology analysis (Huang et al. 2007).
Results
Patient Characteristics
A total of 123 east-Asian patients were enrolled in this study, and baselines were listed in Table 1. There were 87 (70.7%) females and 36 (29.3%) males, with a median age at diagnosis of 36 years old (range 24–40). Fifteen (12.2%) patients had stage 0 lung adenocarcinoma, 46 (37.4%) patients had stage I, 25 (20.3%) patients had stage III, and 37 (30.1%) patients had stage IV. Pathological results revealed adenocarcinoma in situ in 15 (12.2%) patients, minimally invasive adenocarcinoma in 24 (19.5%) patients, and invasive adenocarcinoma in 84 (68.3%) patients.
Table 1.
Characteristics of patients with young never-smoking lung adenocarcinoma who received whole-exome sequencing
| Variables | Patients (N = 123) |
|---|---|
| Age, years | |
| Median (IQR) | 36 (33, 39) |
| Mean ± SD | 35.0 ± 4.4 |
| Range | 24–40 |
| Sex | |
| Female | 87 (70.7) |
| Male | 36 (29.3) |
| Previous malignant history | |
| Yes | 12 (9.8) |
| No | 111 (90.2) |
| Malignant family history | |
| Yes | 29 (23.6) |
| No | 94 (76.4) |
| Multiple lesions | |
| Yes | 25 (20.3) |
| No | 98 (79.7) |
| TNM stage | |
| 0 | 15 (12.2) |
| I | 46 (37.4) |
| II | 0 (0) |
| III | 25 (20.3) |
| IV | 37 (30.1) |
| Adenocarcinoma subtype | |
| Adenocarcinoma in situ | 15 (12.2) |
| Minimally invasive a enocarcinoma | 24 (19.5) |
| Invasive adenocarcinoma | 84 (68.3) |
IQR interquartile range, SD standard deviation
Identification of Pathogenic Mutations
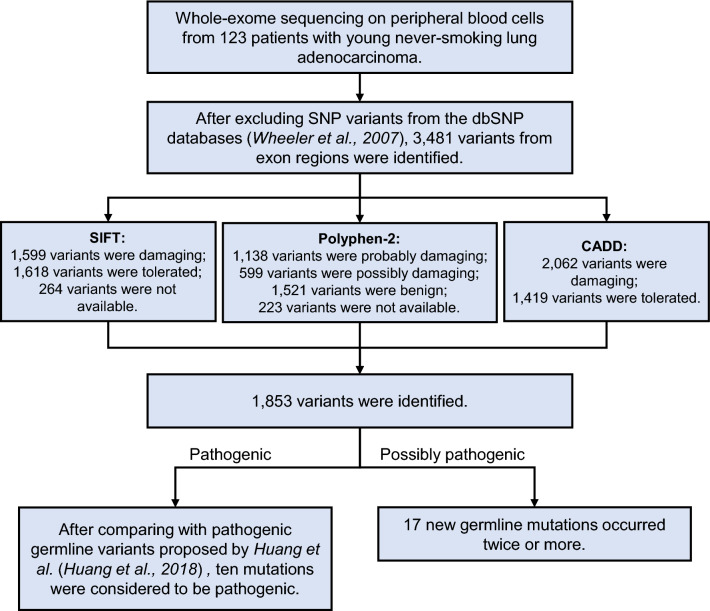
After analyzing data from whole-exome sequencing on peripheral blood cells from 123 enrolled patients and excluding undeleterious SNPs from dbSNP database (Wheeler et al. 2007), 3481 variants were identified. The workflow of study is shown in Fig. 1. SIFT, Polyphen-2, and CADD were used for further in silico prediction. Evaluations based on SIFT revealed that 1,599 variants were damaging, 1,618 variants were tolerated, and 264 variants were not available (Supplementary Table 1). According to the results from Polyphen-2, 1138 variants were probably damaging, 599 variants were possibly damaging, 1521 variants were benign, and 223 variants were not available (Supplementary Table 1). Analysis of CADD demonstrated 2062 damaging variants and 1419 tolerated variants (Supplementary Table 1). Taking together, 1853 variants might be associated with the genetic predisposition of lung adenocarcinoma in young never-smokers. After comparing them with the 152 genes that contributed to cancer susceptibility proposed by Huang et al. (2018) and validating mutations by Sanger sequencing, ten mutations (ATR-L1673V, FANCD2-E369Q, FANCD2-E369Q, GATA2-H169R, HFE-Q82X, MSH2-V817M, PDGFRA-E494V, PMS2-T231S, SDHB-P37L, and WAS-P315L) were pathogenic variants in young never-smokers with lung adenocarcinoma (Table 2).
Fig. 1.
The workflow of this study
Table 2.
Pathogenic variants belonging to the 152 known predisposition genes*
| Gene | Position | Transcript | Mutation | Amino acid change | Type | Patient case ID | Age | Sex | Family history | TNM stage |
|---|---|---|---|---|---|---|---|---|---|---|
| ATR | Chr3: 142,507,945 | NM_001184 | c.C5017G | p.L1673V | SNV | WGC094725 | 40 | F | Yes | IA1 |
| FANCD2 | Chr3: 10,043,835 | NM_001018115 | c.G1105C | p.E369Q | SNV | WGC094712 | 37 | F | No | IA2 |
| FANCE | Chr6: 35,457,943 | NM_021922 | c.C928T | p.P310S | SNV | WGC089849 | 31 | F | No | IA3 |
| GATA2 | Chr3: 128,486,092 | NM_001145662 | c.A506G | p.H169R | SNV | WGC094715 | 38 | F | No | 0 |
| HFE | Chr6: 26,091,008 | NM_000410 | c.C244T | p.Q82X | Stop-gain | WGC089936 | 39 | F | Yes | IA1 |
| MSH2 | Chr2: 47,478,510 | NM_000251 | c.G2449A | p.V817M | SNV | WGC089901 | 31 | F | No | IVB |
| PDGFRA | Chr4: 54,273,653 | NM_001347827 | c.A1481T | p.E494V | SNV | WGC089872 | 36 | M | No | IVB |
| PMS2 | Chr7: 5,989,934 | NM_001322008 | c.C692G | p.T231S | SNV | WGC094710 | 33 | F | No | 0 |
| SDHB | Chr1: 17,044,851 | NM_003000 | c.C110T | p.P37L | SNV | WGC089881 | 39 | F | No | IVB |
| WAS | ChrX: 48,688,672 | NM_000377 | c.C944T | p.P315L | SNV | WGC089895 | 39 | F | No | IVB |
*The genes were proposed by Huang et al. (2018)
SNV single nucleotide variation, F female, M male
The Characteristics of Patients with Pathogenic Mutations
Furthermore, we also investigated the characteristics of patients with pathogenic mutations (Table 2). In total, there were 10 patients (10/123, 8.1%) with identified pathogenic mutations. A majority of them (9/10, 90%) were females, and most of them (8/10, 80%) did not have a family malignant history. Nine patients had mutation types of SNV, and the remaining one patient harbored a stop-gain mutation. Stage IV lung adenocarcinoma was found in four out of ten patients.
Identification of Potentially Pathogenic Mutations and Functions of the Genes Harboring Them
In addition, we also investigated other potentially pathogenic germline mutations. We assumed that germline mutations with a high frequency were more likely to be pathogenic. Therefore, we identified 17 new potentially pathogenic mutations (ASB18-A159T, B3GALT5-R119X, CLEC4F-L257W, COL6A6-D657H, CYP4B1-L114P, C6orf132-P192S, EXO1-N99H, GATA4-P36A, HCK-P456H, KCP-A1345T, NPHP4-Y913H, PIGX-G4T, PPIL2-W346S, PPP1R3G-R353H, RRBP1-C1516T, SALL4-Y399C and RRBP1-L506F), which occurred twice (Table 3). Sixteen out of 17 variants were SNV.
Table 3.
Potentially pathogenic mutations which occurred in at least two patients
| Gene | Transcript | Mutation | Amino acid change | Type | Reported in ClinVar | Reported in COSMIC |
|---|---|---|---|---|---|---|
| ASB18 | NM_212556 | c.G475A | p.A159T | SNV | No | No |
| B3GALT5 | NM_001278650 | c.C343T | p.R115X | Stop-gain | No | No |
| CLEC4F | NM_001258027 | c.T770G | p.L257W | SNV | No | No |
| COL6A6 | NM_001102608 | c.G1969C | p.D657H | SNV | No | No |
| CYP4B1 | NM_001319161 | c.T341C | p.L114P | SNV | No | No |
| C6orf132 | NM_001164446 | c.C574T | p.P192S | SNV | No | No |
| EXO1 | NM_003686 | c.A295C | p.N99H | SNV | No | No |
| GATA4 | NM_001308093 | c.C106G | p.P36A | SNV | Likely benign | No |
| HCK | NM_001172129 | c.C1367A | p.P456H | SNV | No | No |
| KCP | NM_001135914 | c.G4033A | p.A1345T | SNV | No | No |
| NPHP4 | NM_001291594 | c.T2737C | p.Y913H | SNV | No | No |
| PIGX | NM_001166304 | c.G4T | p.A2S | SNV | No | No |
| PPIL2 | NM_001317996 | c.G1037C | p.W346S | SNV | No | No |
| PPP1R3G | NM_001145115 | c.G1058A | p.R353H | SNV | No | Yes |
| RRBP1 | NM_004587 | c.C1516T | p.L506F | SNV | No | No |
| SALL4 | NM_001318031 | c.A1196G | p.Y399C | SNV | No | No |
| TTC28 | NM_001145418 | c.C4517T | p.S1506L | SNV | No | Yes |
SNV single nucleotide variation
We further examined the functions of 27 genes with certainly and potentially pathogenic mutations. Gene ontology analysis showed that they mainly located in nucleoplasm and their functions were tightly associated with DNA repair or DNA damage (Table 4). Notably, KEGG pathways predominantly included Fanconi anemia and mismatch repair. The former was a rare genetic disease leading to defective response to DNA damage (Walden and Deans 2014).
Table 4.
Gene ontology analysis of 27 potential cancer predisposition genes
| Category | Term | FDR value |
|---|---|---|
| KEGG pathway | Fanconi anemia pathway | 7 × 10–3 |
| Biological process | Somatic hypermutation of immunoglobulin genes | 5.4 × 10–2 |
| Cellular component | Nucleoplasm | 1.8 × 10–2 |
| Biological process | DNA repair | 8 × 10–3 |
| Biological process | DNA damage | 9 × 10–3 |
| KEGG pathway | Mismatch repair | 2.3 × 10–2 |
FDR false discovery rate
Discussion
An increasing number of young never-smoking patients with lung cancer is observed in recent studies (Cufari et al. 2017; Pelosof et al. 2017; Zhang et al. 2019), which seems inconsistent with the previous definition of a high-risk population with lung cancer. However, few studies investigate the etiology of the increasing young never-smoking patients with lung cancer. This study aimed to investigate the genetic predisposition of lung adenocarcinoma in young never-smokers. In this study, we performed WES in 123 never-smoking young patients with lung adenocarcinoma. We identified ten pathogenic germline mutations (ATR, FANCD2, FANCE, GATA2, HFE, MSH2, PDGFRA, PMS2, SDHB, and WAS). Moreover, we only detected pathogenic mutations in 8.1% of young never-smoking patients with lung adenocarcinoma, implicating that genetic predisposition might not be a critical factor for carcinogenesis in these patients. Further etiological studies are warranted to reveal the mechanism behind the young never-smokers with lung adenocarcinoma.
In west countries, the percentage of never-smokers in lung cancer patients is about 10% to 20% (Sun et al. 2007), but it is as high as 50% to 63% in east-Asian population (Fu et al. 2019; Kim et al. 2019; Li et al. 2014). Lung adenocarcinoma in never-smokers is considered to be a different disease with distinct clinicopathological factors and molecular features (Sun et al. 2007). The genetic risk factors for lung cancer in never-smokers are poorly understood. Some genetic variations might contribute to the carcinogenesis of non-small cell lung cancer regardless of smoking history. In 2005, Bell et al. (2005b) reported a family with multiple cases of non-small cell lung cancer associated with the germline EGFR-T790M mutation. Subsequent studies identified relevant mutations in HER2, TP53, and BRCA2 (Parry et al. 2017; Yamamoto et al. 2014b). However, the germline mutations mentioned above were not detected in this study. The possible reason might be the different inclusion criteria of patients. Unlike the enrollment of patients with non-small cell lung cancer without limiting ages and smoking history in previous studies, our study considered young never-smoking patients with lung adenocarcinoma as targeted population. Therefore, our study demonstrated that young never-smoking lung adenocarcinoma was distinct from traditional lung cancer in genetic predisposition. More importantly, this study provides a unique gene list related to genetic predisposition of lung adenocarcinoma in young never-smokers, and it could guide prevention of lung cancer.
Not only tumor suppressor genes (TSGs) but also oncogenes consist of pathogenic gene mutations contributing to genetic predisposition of lung cancer. Typical TSGs for lung cancer include TP53 (Couto et al. 2017), BRCA1 (Cedrés et al. 2018), BRCA2 (Wang et al. 2014), and so on. As for oncogenes, EGFR T790M (Bell et al. 2005a; Gazdar et al. 2014; Helena et al. 2014; Oxnard et al. 2012; Thomas et al. 2013; Tibaldi et al. 2011) and HER2 G660D (Yamamoto et al. 2014a) are reported to be germline mutations in non-small cell lung cancer, although they act as oncogenes in lung cancer. In addition, Chen et al. also reported R331W missense mutation in YAP1, a typical oncogene, is a germline risk allele in lung cancer (Chen et al. 2015).
In this study, we identified 10 pathogenic mutations in 10 patients (10/123, 8.1%). In 2017, Couto et al. (2017) found only four patients (4/45, 8.9%) with germline TP53-R377H in 45 lung cancer patients. One study investigated germline mutations in 369 never-smokers with lung cancer and only detected two cases (2/369, 0.5%) harboring germline EGFR T790M (Girard et al. 2010). Moreover, our study indicated that there were 8.1% of young never-smoking lung adenocarcinoma with identifiable pathogenic mutations, and only 25% of patients with pathogenic mutations had a history of family malignancy. Given the rarity of pathogenic germline mutations in lung cancer, germline sequencing should not be routinely advocated at this time, regardless of the family malignant history. The gene ontology analysis of the genes with these pathogenic germline mutations showed that the patients carrying these mutations would have a damaged DNA repair function and excessive mutations in immune-related genes. It partly explains why these patients developed cancer predispositions and had early onset of lung adenocarcinoma without a harmful habit.
However, there are some limitations to this study. First, given the fact that it is a single-institutional study, selection bias is inevitable. Moreover, our results are based on the Chinese population, and future studies focusing on other races are needed to confirm our conclusions. Second, this study lacks experimental validation of potentially pathogenic variants, because germline mutations are difficult to validate by cell and animal experiments. The main purpose of this study was to give a gene list related to genetic predisposition of lung adenocarcinoma in young never-smokers. Third, we only performed WES to identify possible germline mutations, instead of whole-genome sequencing. It is reported that gene mutations might be underestimated by WES (Rusch et al. 2018). Future researchers need take this into consideration while using the data.
Conclusion
Pathogenic mutations (ATR, FANCD2, FANCE, GATA2, HFE, MSH2, PDGFRA, PMS2, SDHB, and WAS) were identified in 8.1% of young never-smoking patients with lung adenocarcinoma. Germline mutations in 17 genes (ASB18, B3GALT5, CLEC4F, COL6A6, CYP4B1, C6orf132, EXO1, GATA4, HCK, KCP, NPHP4, PIGX, PPIL2, PPP1R3G, RRBP1, SALL4, and TTC28), which occurred in at least two patients, displayed potentially pathogenic role in young never-smokers with lung adenocarcinoma. The study provided evidences for the predisposition of lung adenocarcinoma in young never-smokers, which could be used to guide prevention and early diagnosis of lung cancer.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81772466), Shanghai Rising-Star Program (21QC1400600), Ministry of Science and Technology of the People’s Republic of China (2017YFA0505500; 2016YFA0501800), and Science and Technology Commission of Shanghai Municipality (19XD1401300).
Authors’ contributions
Conceptualization: FF, XT, ZJ, YS, LS, and YZ. Methodology: FF, XT, ZJ, LS, YS, and YZ. Formal analysis and investigation: FF, XT, ZJ, ZG, YZ, YL. Writing—original draft: FF, XT, ZJ, HH, YS, and YZ. Writing—review and editing: FF, XT, ZJ, ZG, YZ, YL, HH, LS, YS, and YZ. Funding acquisition: YS and YZ. Resources: FF, XT, ZJ, ZG, YZ, YL, Hong Hu, LS, YS, and YZ. Supervision: LS, YS, and YZ.
Data Availability
All the identified germline mutations in this study were listed in Supplementary Table 1, and clinical data were also included in Supplementary Table 2. The raw data of WES from 123 young nonsmoking patients with lung adenocarcinoma have been uploaded to the in the Genome Sequence Archive of the BIG Data Center at the Beijing Institute of Genomics, Chinese Academy of Science, China. They are accessible under HRA001459 (http://bigd.big.ac.cn/gsa-human/).
Code Availability
The codes are available from the corresponding authors upon reasonable request.
Declarations
Conflicts of Interest
None.
Ethics Approval
The Institutional Review Board of Fudan University Shanghai Cancer Center has approved this study (IRB#090977-1).
Consent to Participate
Informed consent was obtained from the patients included in this study.
Consent to Publish
Not applicable.
Footnotes
Fangqiu Fu, Xiaoting Tao, and Zhonglin Jiang contributed equally to the work.
Contributor Information
Libing Shen, Email: shenlibing@ihug.org.cn.
Yihua Sun, Email: sun_yihua76@hotmail.com.
Yang Zhang, Email: fduzhangyang1987@hotmail.com.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DW, Gore I, Okimoto RA, Godin-Heymann N, Sordella R, Mulloy R, Sharma SV, Brannigan BW, Mohapatra G, Settleman J. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37(12):1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- Bell DW, Gore I, Okimoto RA, Godin-Heymann N, Sordella R, Mulloy R, Sharma SV, Brannigan BW, Mohapatra G, Settleman J, Haber DA. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37(12):1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Cedrés S, Felip E, Cruz C, Martinez de Castro A, Pardo N, Navarro A, Martinez-Marti A, Remon J, Zeron-Medina J, Balmana J. Activity of HSP90 inhibiton in a metastatic lung cancer patient with a germline BRCA1 mutation. JNCI J Natl Cancer Inst. 2018;110(8):914–917. doi: 10.1093/jnci/djy012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-Y, Yu S-L, Ho B-C, Su K-Y, Hsu Y-C, Chang C-S, Li Y-C, Yang S-Y, Hsu P-Y, Ho H, Chang Y-H, Chen C-Y, Yang H-I, Hsu C-P, Yang T-Y, Chen K-C, Hsu K-H, Tseng J-S, Hsia J-Y, Chuang C-Y, Yuan S, Lee M-H, Liu C-H, Wu G-I, Hsiung CA, Chen Y-M, Wang C-L, Huang M-S, Yu C-J, Chen K-Y, Tsai Y-H, Su W-C, Chen H-W, Chen JJW, Chen C-J, Chang G-C, Yang P-C, Li K-C. R331W missense mutation of oncogene YAP1 is a germline risk allele for lung adenocarcinoma with medical actionability. J Clin Oncol. 2015;33(20):2303–2310. doi: 10.1200/JCO.2014.59.3590. [DOI] [PubMed] [Google Scholar]
- Couto PP, Bastos-Rodrigues L, Schayek H, Melo FM, Lisboa RGC, Miranda DM, Vilhena A, Bale AE, Friedman E, De Marco L. Spectrum of germline mutations in smokers and non-smokers in Brazilian non-small-cell lung cancer (NSCLC) patients. Carcinogenesis. 2017;38(11):1112–1118. doi: 10.1093/carcin/bgx089. [DOI] [PubMed] [Google Scholar]
- Cufari ME, Proli C, De Sousa P, Raubenheimer H, Al Sahaf M, Chavan H, Shedden L, Niwaz Z, Leung M, Nicholson AG, Anikin V, Beddow E, McGonigle N, Dusmet ME, Jordan S, Ladas G, Lim E. Increasing frequency of non-smoking lung cancer: presentation of patients with early disease to a tertiary institution in the UK. Eur J Cancer. 2017;84:55–59. doi: 10.1016/j.ejca.2017.06.031. [DOI] [PubMed] [Google Scholar]
- Fu F, Zhang Y, Wen Z, Zheng D, Gao Z, Han H, Deng L, Wang S, Liu Q, Li Y, Shen L, Shen X, Zhao Y, Zhao Z, Ye T, Xiang J, Zhang Y, Sun Y, Hu H, Chen H. Distinct prognostic factors in patients with stage I non-small cell lung cancer with radiologic part-solid or solid lesions. J Thorac Oncol. 2019;14(12):2133–2142. doi: 10.1016/j.jtho.2019.08.002. [DOI] [PubMed] [Google Scholar]
- Gazdar A, Robinson L, Oliver D, Xing C, Travis WD, Soh J, Toyooka S, Watumull L, Xie Y, Kernstine K. Hereditary lung cancer syndrome targets never smokers with germline EGFR gene T790M mutations. J Thorac Oncol. 2014;9(4):456–463. doi: 10.1097/JTO.0000000000000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard N, Lou E, Azzoli CG, Reddy R, Robson M, Harlan M, Orlow I, Yatabe Y, Nafa K, Ladanyi M, Viale A, Kris MG, Riely G, Miller V, Klein RJ, Matsuo K, Pao W. Analysis of genetic variants in never-smokers with lung cancer facilitated by an internet-based blood collection protocol: a preliminary report. Clin Cancer Res. 2010;16(2):755–763. doi: 10.1158/1078-0432.CCR-09-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helena AY, Arcila ME, Fleischut MH, Stadler Z, Ladanyi M, Berger MF, Robson M, Riely GJ. Germline EGFR T790M mutation found in multiple members of a familial cohort. J Thorac Oncol. 2014;9(4):554–558. doi: 10.1097/JTO.0000000000000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KL, Mashl RJ, Wu Y, Ritter DI, Wang J, Oh C, Paczkowska M, Reynolds S, Wyczalkowski MA, Oak N, Scott AD, Krassowski M, Cherniack AD, Houlahan KE, Jayasinghe R, Wang LB, Zhou DC, Liu D, Cao S, Kim YW, Koire A, McMichael JF, Hucthagowder V, Kim TB, Hahn A, Wang C, McLellan MD, Al-Mulla F, Johnson KJ, Cancer Genome Atlas Research N. Lichtarge O, Boutros PC, Raphael B, Lazar AJ, Zhang W, Wendl MC, Govindan R, Jain S, Wheeler D, Kulkarni S, Dipersio JF, Reimand J, Meric-Bernstam F, Chen K, Shmulevich I, Plon SE, Chen F, Ding L. Pathogenic germline variants in 10,389 adult cancers. Cell. 2018;173(2):355–370.e314. doi: 10.1016/j.cell.2018.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Miller KD, Ma J, Siegel RL, Fedewa SA, Islami F, Devesa SS, Thun MJ. Higher lung cancer incidence in young women than young men in the United States. N Engl J Med. 2018;378(21):1999–2009. doi: 10.1056/NEJMoa1715907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Goo JM, Kim YT, Park CM. Validation of the eighth edition clinical T categorization system for clinical stage IA resected lung adenocarcinomas: prognostic implications of the ground-glass opacity component. J Thorac Oncol. 2019;15(4):580–588. doi: 10.1016/j.jtho.2019.12.110. [DOI] [PubMed] [Google Scholar]
- Li S, Li L, Zhu Y, Huang C, Qin Y, Liu H, Ren-Heidenreich L, Shi B, Ren H, Chu X, Kang J, Wang W, Xu J, Tang K, Yang H, Zheng Y, He J, Yu G, Liang N. Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: a comprehensive mutation profiling from 5125 Chinese cohorts. Br J Cancer. 2014;110(11):2812–2820. doi: 10.1038/bjc.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxnard GR, Miller VA, Robson ME, Azzoli CG, Pao W, Ladanyi M, Arcila ME. Screening for germline EGFR T790M mutations through lung cancer genotyping. J Thorac Oncol. 2012;7(6):1049–1052. doi: 10.1097/JTO.0b013e318250ed9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YR, Bae SH, Ji W, Seo EJ, Lee JC, Kim HR, Jang SJ, Choi CM. GAB2 amplification in squamous cell lung cancer of non-smokers. J Korean Med Sci. 2017;32(11):1784–1791. doi: 10.3346/jkms.2017.32.11.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Parry EM, Gable DL, Stanley SE, Khalil SE, Antonescu V, Florea L, Armanios M. Germline mutations in DNA repair genes in lung adenocarcinoma. J Thorac Oncol. 2017;12(11):1673–1678. doi: 10.1016/j.jtho.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosof L, Ahn C, Gao A, Horn L, Madrigales A, Cox J, McGavic D, Minna JD, Gazdar AF, Schiller J. Proportion of never-smoker non-small cell lung cancer patients at three diverse institutions. J Natl Cancer Inst. 2017 doi: 10.1093/jnci/djw295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch P, Schubach M, Shendure J, Kircher M. CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13(1):31. doi: 10.1186/s13073-021-00835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch M, Nakitandwe J, Shurtleff S, Newman S, Zhang Z, Edmonson MN, Parker M, Jiao Y, Ma X, Liu Y, Gu J, Walsh MF, Becksfort J, Thrasher A, Li Y, McMurry J, Hedlund E, Patel A, Easton J, Yergeau D, Vadodaria B, Tatevossian RG, Raimondi S, Hedges D, Chen X, Hagiwara K, McGee R, Robinson GW, Klco JM, Gruber TA, Ellison DW, Downing JR, Zhang J. Clinical cancer genomic profiling by three-platform sequencing of whole genome, whole exome and transcriptome. Nat Commun. 2018;9(1):3962. doi: 10.1038/s41467-018-06485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(Web Server issue):W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers—a different disease. Nat Rev Cancer. 2007;7(10):778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- Thavaneswaran S, Rath E, Tucker K, Joshua AM, Hess D, Pinese M, Ballinger ML, Thomas DM. Therapeutic implications of germline genetic findings in cancer. Nat Rev Clin Oncol. 2019;16(6):386–396. doi: 10.1038/s41571-019-0179-3. [DOI] [PubMed] [Google Scholar]
- Thomas A, Xi L, Carter CA, Rajan A, Khozin S, Szabo E, Dennis PA, Giaccone G, Raffeld M. Concurrent molecular alterations in tumors with germ line epidermal growth factor receptor T790M mutations. Clin Lung Cancer. 2013;14(4):452. doi: 10.1016/j.cllc.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibaldi C, Giovannetti E, Vasile E, Boldrini L, Gallegos-Ruiz MI, Bernardini I, Incensati R, Danesi R, Cappuzzo F, Peters GJ. Inherited germline T790M mutation and somatic epidermal growth factor receptor mutations in non-small cell lung cancer patients. J Thorac Oncol. 2011;6(2):395–396. doi: 10.1097/JTO.0b013e3182059a6f. [DOI] [PubMed] [Google Scholar]
- Toh CK, Gao F, Lim WT, Leong SS, Fong KW, Yap SP, Hsu AA, Eng P, Koong HN, Thirugnanam A, Tan EH. Never-smokers with lung cancer: epidemiologic evidence of a distinct disease entity. J Clin Oncol. 2006;24(15):2245–2251. doi: 10.1200/JCO.2005.04.8033. [DOI] [PubMed] [Google Scholar]
- Walden H, Deans AJ. The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu Rev Biophys. 2014;43:257–278. doi: 10.1146/annurev-biophys-051013-022737. [DOI] [PubMed] [Google Scholar]
- Wang Y, McKay JD, Rafnar T, Wang Z, Timofeeva MN, Broderick P, Zong X, Laplana M, Wei Y, Han Y. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet. 2014;46(7):736–741. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Geer LY, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Ostell J, Miller V, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35(Database issue):D5-12. doi: 10.1093/nar/gkaa892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Higasa K, Sakaguchi M, Shien K, Soh J, Ichimura K, Furukawa M, Hashida S, Tsukuda K, Takigawa N. Novel germline mutation in the transmembrane domain of HER2 in familial lung adenocarcinomas. J Natl Cancer Inst. 2014;106(1):djt338. doi: 10.1093/jnci/djt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Higasa K, Sakaguchi M, Shien K, Soh J, Ichimura K, Furukawa M, Hashida S, Tsukuda K, Takigawa N, Matsuo K, Kiura K, Miyoshi S, Matsuda F, Toyooka S. Novel germline mutation in the transmembrane domain of HER2 in familial lung adenocarcinomas. J Natl Cancer Inst. 2014;106(1):djt338. doi: 10.1093/jnci/djt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jheon S, Li H, Zhang H, Xie Y, Qian B, Lin K, Wang S, Fu C, Hu H. Results of low-dose computed tomography as a regular health examination among Chinese hospital employees. J Thorac Cardiovasc Surg. 2019;160(3):824–831. doi: 10.1016/j.jtcvs.2019.10.145. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jheon S, Li H, Zhang H, Xie Y, Qian B, Lin K, Wang S, Fu C, Hu H, Zheng Y, Li Y, Chen H. Results of low-dose computed tomography as a regular health examination among Chinese hospital employees. J Thorac Cardiovasc Surg. 2020;160(3):824–831. doi: 10.1016/j.jtcvs.2019.10.145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the identified germline mutations in this study were listed in Supplementary Table 1, and clinical data were also included in Supplementary Table 2. The raw data of WES from 123 young nonsmoking patients with lung adenocarcinoma have been uploaded to the in the Genome Sequence Archive of the BIG Data Center at the Beijing Institute of Genomics, Chinese Academy of Science, China. They are accessible under HRA001459 (http://bigd.big.ac.cn/gsa-human/).
The codes are available from the corresponding authors upon reasonable request.