Key Points
-
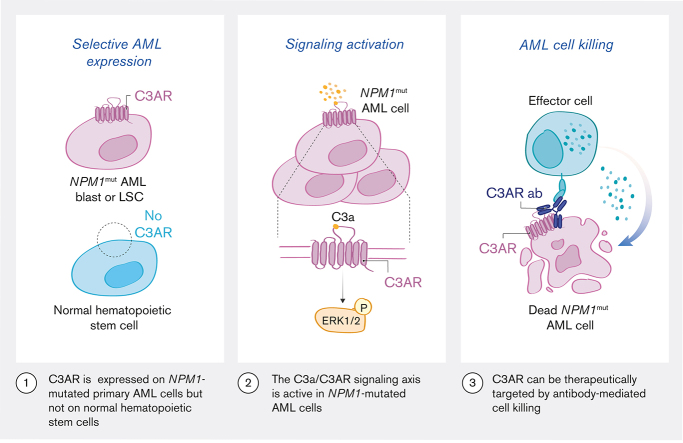
•
C3AR is selectively expressed on NPM1-mutated primary AML cells compared to both NPM1 wild type cases and normal hematopoietic stem cells.
-
•
The C3a/C3AR signaling axis is active in NPM1-mutated AML cells, and C3AR can be therapeutically targeted with antibodies.
Visual Abstract
Abstract
Mutated nucleophosmin 1 (NPM1) is the most common genetic alteration in acute myeloid leukemia (AML), found in ∼30% of cases. Although mutations in this gene are considered favorable according to current risk stratification guidelines, a large fraction of patients will experience relapse, demonstrating the urgent need for new treatment options. Therefore, we aimed to identify cell surface proteins specifically expressed on NPM1-mutated AML cells, allowing for potential targeting with antibody-based therapies. Herein, we report on an arrayed flow cytometry–based screen directed to 362 cell surface markers. In comparing the cell surface expression on NPM1-mutated AML cells with primitive (CD34+ CD38−) normal bone marrow cells, we identified the complement receptor C3AR as being specifically expressed in NPM1-mutated AML. By flow cytometry and single-cell RNA sequencing, we further show that normal hematopoietic stem and progenitor cells lack detectable C3AR gene and protein expression, making it particularly suitable as a target for antibody therapy. We also demonstrate that C3AR in combination with GPR56 distinguishes the leukemic stem cells (LSCs) in NPM1-mutated AML from the normal hematopoietic stem cells, defining the LSC population, as shown by transplantation into immunodeficient mice. Mechanistically, the stimulation of C3AR-expressing cells with C3a, the ligand of C3AR, leads to the activation of ERK1/2 and increased survival of AML cells, suggesting that this is an important signaling axis in this subtype of AML. Finally, we show that antibodies directed against C3AR efficiently elicit natural killer cell–mediated killing of primary AML cells ex vivo, highlighting C3AR as a candidate therapeutic target in NPM1-mutated AML.
Introduction
Acute myeloid leukemia (AML) is a highly heterogenous disease caused by acquired genetic alterations arising in a hematopoietic stem cell (HSC) or myeloid progenitor cell. This leads to aberrant self-renewal and faulty differentiation capacity, resulting in an expansion of immature malignant myeloid cells.1,2 The overall prognosis of AML is poor, especially with older patients, with survival rates of <10% for patients aged >70 years.2,3 Chemotherapy remains the standard treatment in AML,4 but it often fails to eradicate the leukemic stem cells (LSCs), which serve as a reservoir for disease relapse.5 Hence, novel therapies selectively targeting the LSC may provide new means to improve prognosis and achieve a cure. In this context, recombinant antibodies provide attractive therapies given their target antigen specificity and general minimal toxicity.6 An ideal target for an antibody-based targeted treatment in AML should be selectively expressed on the LSCs but not on normal HSCs,6,7 thereby allowing replenishment of normal hematopoiesis. In addition, restricted expression in nonhematopoietic tissues is critical for limiting potential side effects on other vital organs.
AML can be stratified into different genetic subgroups based on chromosomal aberrations and recurring mutations.8,9 The most common genetic subgroup of AML, which constitutes ∼30% of all AML cases, is defined by mutations in the nucleophosmin 1 (NPM1) gene, encoding a nucleocytoplasmic shuttling protein.2,8,9 NPM1-mutated AMLs are distinct from AMLs with other genetic alterations, as they are almost exclusively negative for the primitive marker CD34.5,10,11 Two well-known AML cell surface markers that are highly expressed in the majority of NPM1-mutated AML blasts are CD33 and CD123 (IL3RA),12, 13, 14 but they are not specific to this genetic subgroup and are also expressed on normal HSCs. Furthermore, very few cell surface markers that enrich for LSC activity in CD34-negative/NPM1-mutated AMLs have been described. Examples include the transmembrane tyrosine kinase c-Kit (CD117), especially in combination with the natural killer (NK) cell receptor 2B4 (CD244/NKR2B4),11,15 CD20016 and the adhesion G protein–coupled receptor G1 (GPR56).17, 18, 19 However, as therapeutic targets, these are limited by their expression on normal HSCs.11,16,18
To identify novel cell surface markers specifically upregulated on NPM1-mutated AML cells compared with primitive normal bone marrow (NBM) cells, we performed an arrayed cell surface marker screen using 362 phycoerythrin (PE)-conjugated monoclonal antibodies. With this approach, we identified the complement component 3a receptor 1 (C3AR) as being specifically upregulated on NPM1-mutated AML cells and show that C3AR-expressing AML cells respond to stimulation with their ligand, C3a. Importantly, we show that anti-C3AR antibodies and NK cells mediate specific cell killing of primary NPM1-mutated AML cells ex vivo, through antibody-dependent cellular cytotoxicity (ADCC). Combined, our results demonstrate that C3AR is a candidate therapeutic target in NPM1-mutated AML.
Materials and Methods
Patient samples
Bone marrow (BM) or peripheral blood (PB) samples from patients with AML were collected after written informed consent in accordance with the Declaration of Helsinki. Mononuclear cells (MNCs) were isolated using Lymphoprep (GE Healthcare Bio-Sciences AB) and samples were viably frozen. Clinical and genetic characteristics of the NPM1-mutated samples from patients were characterized by flow cytometry are summarized in supplemental Table 1. In addition, BM samples from healthy donors were prepared in the same way. The study was approved by a regional ethics committee.
Cell surface receptor screen
The LEGENDScreen Human PE Kit containing an arrayed library of 361 unique PE-labeled antibodies against human cell surface receptors was used as a base for the screen. The screen plates containing lyophilized antibodies were diluted in double-distilled water according to the manufacturer’s instructions, and an antibody cocktail of either anti-CD3 and anti-CD19 (for the AML samples) or anti-CD34 and anti-CD38 (for normal BM samples) was added to each well. An anti-IL1RAP antibody was also added to the screen, because we have shown previously that IL1RAP is a therapeutic marker on AML LSCs,20,21 bringing the total number of antibodies assessed to 362. BM or PB MNCs from NPM1-mutated AML samples (n = 4), or MNCs from NBM (n = 4) were thawed and resuspended in phosphate-buffered saline (PBS) with 2% fetal bovine serum (FBS) and added to the wells. Plates were incubated in the dark for 20 minutes at 4°C, washed twice, and resuspended in PBS with 2% FBS. Just before analysis on an LSR Fortessa with an HTS-sampler (BD Biosciences), the viability dye 7AAD (Biolegend) was added to each well. For details regarding data analysis, screen validation, and cell surface marker evaluation in NBM, see supplemental Methods. All antibodies used are listed in supplemental Table 2.
C3AR GPR56 transplantation experiments
To assess the engraftment potential of primary NPM1-mutated AML cells expressing C3AR+ GPR56+ compared with C3AR+ GPR56−, fluorescence-activated cell sorter-sorted cells were transplanted into irradiated NSGS immunodeficient mice. For a detailed description of xenograft experiments, see supplemental Methods.
C3a stimulations
The C3AR-expressing leukemia cell line OCI-AML3 and the C3AR-negative cell line NALM-6 were both cultured according to the vendor’s instructions (German Collection of Microorganisms and Cell Cultures GmbH) but were starved by resuspension in serum-free minimal essential medium–alpha (Merck) or serum-free RPMI 1640 medium (Thermo Fisher Scientific), respectively, the day before addition of C3a. Primary AML samples were resuspended in StemSpan serum-free expansion medium (Stem Cell Technologies, Canada) after thawing. Thereafter, OCI-AML3 cells, primary AML cells, and NALM-6 cells were cultured in 96-well plates at 37°C, 5% CO2, for 3 days in different concentrations of C3a (0, 10, 100, 1000, 5000, and 10 000 ng/mL) purified from human serum (Comp Tech or Merck). Viable cell number was evaluated using CountBright Absolute Counting Beads (Thermo Fisher Scientific) and the viability dye Draq7 (Biostatus) on a LSRFortessa (BD Biosceinces).
C3a phospho–flow cytometry
Primary AML cells were thawed and washed twice in RPMI 1640 medium (Thermo Fisher Scientific) without supplements and left to incubate for 15 minutes at 37°C, 5% CO2, before 1-minute or 5-minute stimulation with 100 ng/mL of human C3a (Comp Tech). Cells were fixed with 1.6% paraformaldehyde for 10 minutes at room temperature before permeabilization with 90% ethanol at −80°C. Subsequently, cells were thoroughly washed with PBS to remove any residual ethanol, resuspended in 2% FBS, and stained with anti-pERK1/2 antibody. Samples were analyzed on a LSRFortessa (BD Biosciences) instrument. All antibodies used are listed in supplemental Table 2.
ADCC assay
For the ADCC assays, KG-1 cells or primary AML samples from patients with AML were stained with PKH26 (Sigma-Aldrich) according to the manufacturer’s instruction, before resuspension in RPMI 1640 medium (Thermo Fisher Scientific) with 20% heat-inactivated FBS (Nordic Biolabs, Sweden). Cells were plated in a flat-bottom 96-well plate with different concentrations of anti-C3AR antibody or isotype control antibody. The plates were incubated for 30 minutes at 37°C, 5% CO2, before addition of freshly thawed NK cells at a 10:1 ratio to target cells. NK cells were purified from leukocyte concentrates using magnetic beads (Miltenyi Biotec). Subsequently, the plates were incubated overnight. CountBright Absolute Counting Beads (Thermo Fisher Scientific) and 4′,6-diamidino-2-phenylindole (Sigma-Aldrich) were added to the wells before analyses on a LSRFortessa (BD Biosciences). For details on the ADCC in vivo repopulation analysis, see supplemental Methods. All antibodies used are listed in supplemental Table 2
Bulk RNA sequencing and external data
RNA was extracted form PB or BM samples from patients with AML at diagnosis or relapse (n = 112) using the RNeasy mini kit (Qiagen) according to the manufacturer’s instructions (H. Lilljebjörn, C. Orsmark-Pietras, H. Thorsson, M. Rissler, and T. Fioretos, unpublished data, December 2022). Complementary DNA libraries were prepared using the Truseq RNA library prep kit v2 (Illumina) and sequenced by paired-end 2 × 150 bp sequencing on a NextSeq 500 (Illumina). Details on the gene expression analysis of in-house and external sequencing data are provided in the supplemental Methods.
Single-cell RNA sequencing (scRNA-seq)
In total, 20 BM or PB samples (12 NPM1-mutated AML diagnostic samples, 5 NBM MNC samples, and 3 CD34-enriched NBM samples) were subjected to scRNA-seq (H. Lilljebjörn, C. Orsmark-Pietras, H. Thorsson, M. Rissler, and T. Fioretos, unpublished data, December 2022). Cryopreserved cells were thawed and resuspended in Draq7-buffer (Biolegend) before sorting on an Aria Fusion (BD Biosciences). Samples were gated on viability; cell doublets and cell debris were excluded. The sorted cells were subjected to scRNA-seq by preparation of libraries using the Chromium Single Cell 3′ Reagent Kits v3 (10X Genomics).22 The libraries were sequenced on a NovaSeq 6000 System (Illumina). Details on the single-cell gene expression analysis are provided in the supplemental Methods.
Complement factor D (CFD) enzyme-linked immunosorbent assay
NPM1-mutated AML samples (n = 4) and NBM MNC samples (n = 4) were thawed and washed twice in Iscove modified Dulbecco medium, before resuspension in S7 media.23 One million cells per mL were plated and cell culture supernatants were collected for analysis after 24 hours. Samples were diluted 1:10 and the amount of CFD was evaluated using an enzyme-linked immunosorbent assay (Abcam), according to manufacturer’s instructions.
Results
Antibody-based screening identifies cell surface markers specifically upregulated in NPM1-mutated AML samples
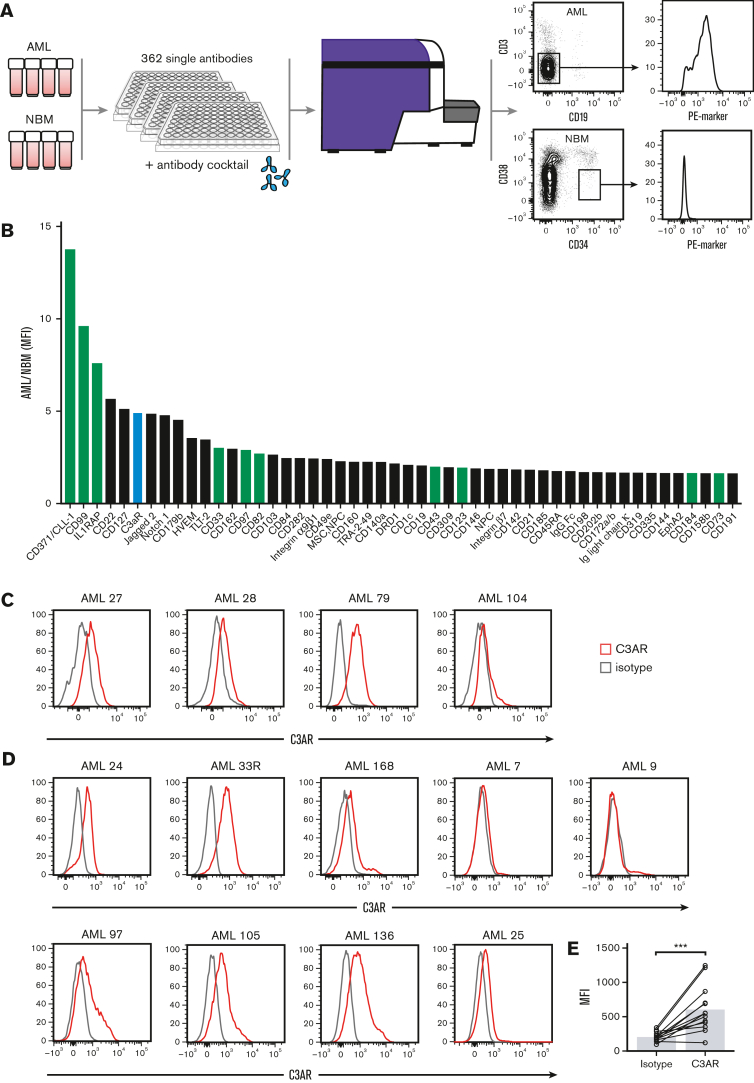
To identify cell surface molecules specifically upregulated in NPM1-mutated AML samples compared with healthy primitive BM cells, we performed a flow cytometry–based screen using antibodies directed to 362 cell surface proteins (Figure 1A). By comparing the CD3−CD19− cell fraction of 4 NPM1-mutated AML samples with the CD34+CD38low cell fraction of 4 NBM samples for each marker, a ranked list of cell surface markers with high expression in AML compared with NBM was obtained (Figure 1B). Several known AML cell surface markers were among the top 50 hits: CD371/CLL-1,24,25 CD99,26 IL1RAP,21,27 CD33,28 CD82,29 CD43,30 CD123,31 CD184 (CXCR4),32 and CD73,33 thus validating our approach to select for relevant markers. Of the novel markers, C3AR displayed the highest selective expression in the AML samples (Figure 1B). CD22 and CD127 scored above C3AR in the ranked list (Figure 1B), but these markers did not show consistent receptor expression when samples were reanalyzed individually (supplemental Figure 1). In contrast, validations confirmed that C3AR displayed a consistent expression on all 4 NPM1-mutated AML samples included in the screen (Figure 1C). Thus, C3AR was selected for further studies.
Figure 1.
Antibody-based screening identifies cell surface markers specifically upregulated on NPM1-mutated AML cells. (A) Schematic illustration showing the arrayed antibody screen performed on CD3− CD19− myeloid-enriched blasts from NPM1-mutated AML samples and CD34+ CD38low stem cell–enriched NBM samples. The antibody library contained 362 PE-marked antibodies. In total, 4 primary AML samples and 4 NBM samples were used in the screen. (B) Screening results showing the top-ranked cell surface markers in AML compared with NBM cells, calculated as the ratio of the median MFI of the CD3− CD19− AML cells over the median MFI of the NBM CD34+CD38low cells for each marker. The green bars illustrate known LSC markers, the lavender bars illustrate novel markers, and the red bar shows C3AR that was chosen for further investigation. (C) Expression of C3AR in the primary NPM1-mutated samples used in the screen. (D) Validation of C3AR expression in 9 additional NPM1-mutated AML samples. C3AR stain in red and isotype control in gray. (E) MFI of C3AR in all NPM1-mutated AML samples (n = 13), compared with isotype control. ∗∗∗P ≤ .001 in a Wilcoxon nonparametric paired test. MFI, median fluorescence intensity.
Next, we measured C3AR expression in 9 additional NPM1-mutated AML samples by flow cytometry (Figure 1D). Overall, most of the samples (11/13) showed expression of C3AR, with a significantly higher median fluorescence intensity compared with the isotype control (P < .001) (Figure 1E). These data suggest that C3AR is expressed in the vast majority of AML cases with NPM1 mutations.
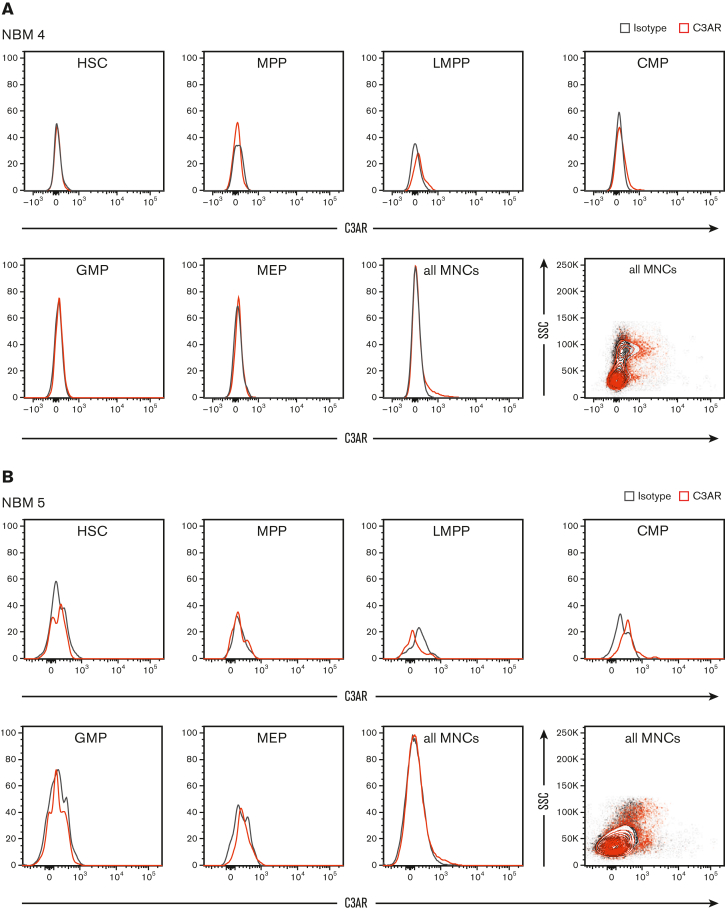
C3AR is upregulated in NPM1-mutated AML cells but absent on normal hematopoietic stem and progenitor cells (HSPC)
Given the consistent expression of C3AR in NPM1-mutated AML, we next investigated its specificity by analyzing the expression in NBM cells. Different fractions of NBM MNCs were analyzed by flow cytometry, demonstrating that C3AR is not expressed in any of the immature cell fractions (Figure 2; supplemental Figure 2). A weak expression of C3AR was noted in a subpopulation of MNCs. Because it is known that C3AR is expressed on monocytes and other myeloid innate immune cells,34,35 we investigated whether the C3AR-expressing MNCs in the NBM were indeed monocytes. Further flow cytometric analysis revealed that the bulk CD14pos monocyte population displayed a weak C3AR-expression. Out of the different monocyte subsets, CD14highCD16+ (intermediate monocytes) and CD14+CD16+ (nonclassical monocytes) had the highest expression, whereas the CD14highCD16− (classical monocytes) displayed weak expression (supplemental Figure 3).
Figure 2.
C3AR is not expressed on normal HSPCs. (A-B) flow cytometric analysis of C3AR expression in different fractions of NBM MNCs from 2 healthy donors. C3AR stain in red and isotype control in gray. The MNCs are shown both as a histograms and contour plots. CMP, common myeloid progenitor (Lin− CD34+ CD38+ CD123+ CD45RA−); GMP, granulocyte-macrophage progenitor (Lin− CD34+ CD38+ CD123+ CD45RA+); HSC, hematopoietic stem cell (Lin− CD34+ CD38− CD90+ CD45RA−); LMPP, lymphoid-primed multipotent progenitor (Lin− CD34+ CD38− CD45RA+); MEP, megakaryocyte-erythroid progenitor (Lin− CD34+ CD38+ CD123− CD45RA−); MPP, multipotent progenitor (Lin− CD34+ CD38− CD90− CD45RA−).
Collectively, our data suggest that therapeutic targeting of C3AR would selectively target the NPM1-mutated leukemic cells and a minority of the monocytes that express CD16 (<10%), thus sparing the vital HSPC populations.
RNA-seq reveals C3AR1 to be specifically upregulated in NPM1-mutated AMLs
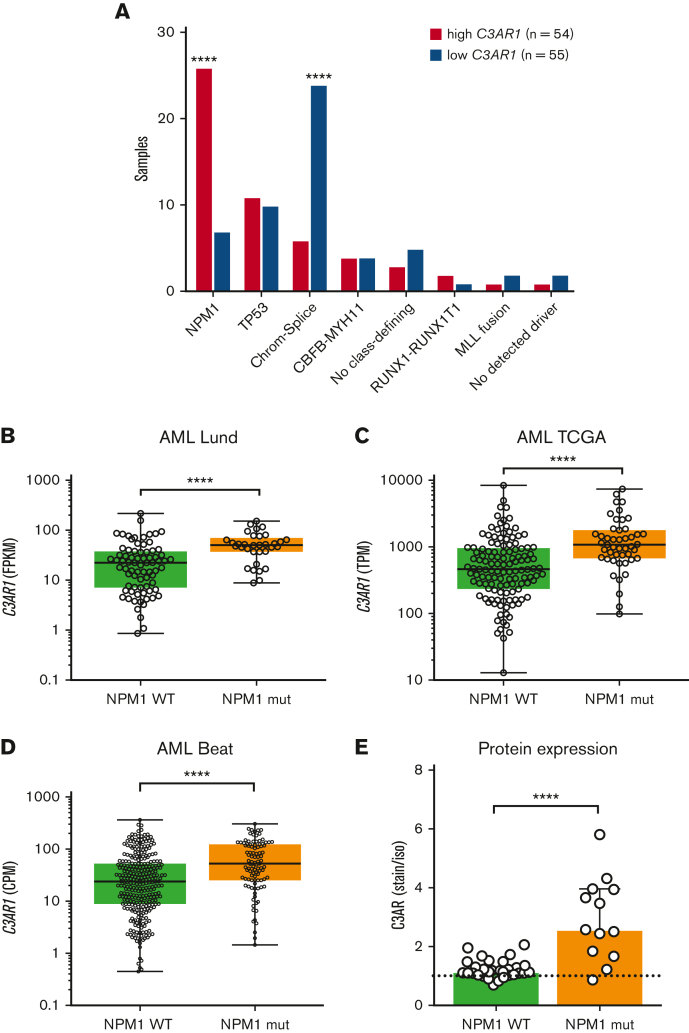
Next, we investigated whether the gene expression of C3AR (C3AR1) is specific to NPM1-mutated AMLs, or whether it is also expressed in AMLs characterized by other genetic alterations. We compared the C3AR1 expression in different genetic subgroups of AML using a cohort of AML samples analyzed by RNA-seq (referred to as the Lund cohort; H. Lilljebjörn, C. Orsmark-Pietras, H. Thorsson, M. Rissler, and T. Fioretos, unpublished data, December 2022). We divided the samples into 2 groups, C3AR1 high (n = 54) and C3AR1 low (n = 55) and determined the number of C3AR1-high and C3AR1-low cases in each genetic subgroup (Figure 3A). This analysis showed that C3AR1 high is most common in AML cases with mutated NPM1 (P ≤ .0001). The opposite was observed for AML samples classified into the chromatin-spliceosome subgroup, which had a higher proportion of C3AR1-low–expressing samples (P ≤ .0001).
Figure 3.
C3AR1 is specifically upregulated in NPM1-mutated AML cells. (A) Number of cases with high vs low C3AR1 mRNA expression in each genetic subgroup in a cohort of AML samples (n = 109) diagnosed and analyzed by RNA-seq in Lund, Sweden. AML subgroups with <3 patients were excluded from the analysis. High C3AR1 in magenta and low C3AR1 in purple. (B) FPKM values for C3AR1 in NPM1-mutated cases vs cases with NPM1 WT in the entire AML Lund cohort (n = 112). (C) TPM values for C3AR1 in NPM1-mutated cases vs cases with NPM1 WT, in the publicly available TCGA AML data set (n = 173). (D) CPM values of C3AR1 in NPM1-mutated cases vs cases with NPM1 WT, in the Beat AML data set (n = 400). C3AR1 expression in panels B-D are represented as box plots. (E) C3AR protein expression calculated as fold change of C3AR MFI over isotype control MFI in NPM1-mutated AML cases and NPM1 WT cases, analyzed by flow cytometry, in the CD3− CD19− myeloid-enriched compartment (n = 43). Bars show median with interquartile range. ∗∗∗∗P ≤ .0001 using a Fisher exact test in panel A. ∗∗∗∗P ≤ .0001 using a nonparametric Mann-Whitney U test in panels B-E. Chrom-splice, chromatin-spliceosome; CPM, counts per million mapped reads; FPKM, fragments per kilobase of exon model per million reads mapped; MFI, median fluorescence intensity; mRNA, messenger RNA; TCGA, The Cancer Genome Atlas; TPM, transcripts per million.
Furthermore, we compared the expression of C3AR1 in NPM1-mutated with all NPM1 wild-type (WT) cases in the Lund cohort and in 2 external data sets from The Cancer Genome Atlas36 and Beat AML37 (Figure 3B-D). This revealed a significantly higher expression of C3AR1 in NPM1-mutated compared with WT cases (P ≤ .0001 all cohorts).
Finally, to validate the correlation at the protein level, we assessed C3AR expression in NPM1 WT AML cases (n = 30) and compared it with the expression in NPM1-mutated cases (n = 13) (Figure 3E). This analysis demonstrates that the expression of C3AR is significantly higher in NPM1-mutated compared with WT cases (P < .0001). Looking at individual AML subgroups, none of the other genetic subtypes consistently express C3AR, either in the myeloid (CD3− CD19−) or stem cell–enriched (CD34+ CD38−) compartments (supplemental Figure 4). We thus conclude that C3AR expression correlates with mutations in NPM1.
scRNA-seq confirms expression of C3AR1 in NPM1-mutated AML cells and monocytes
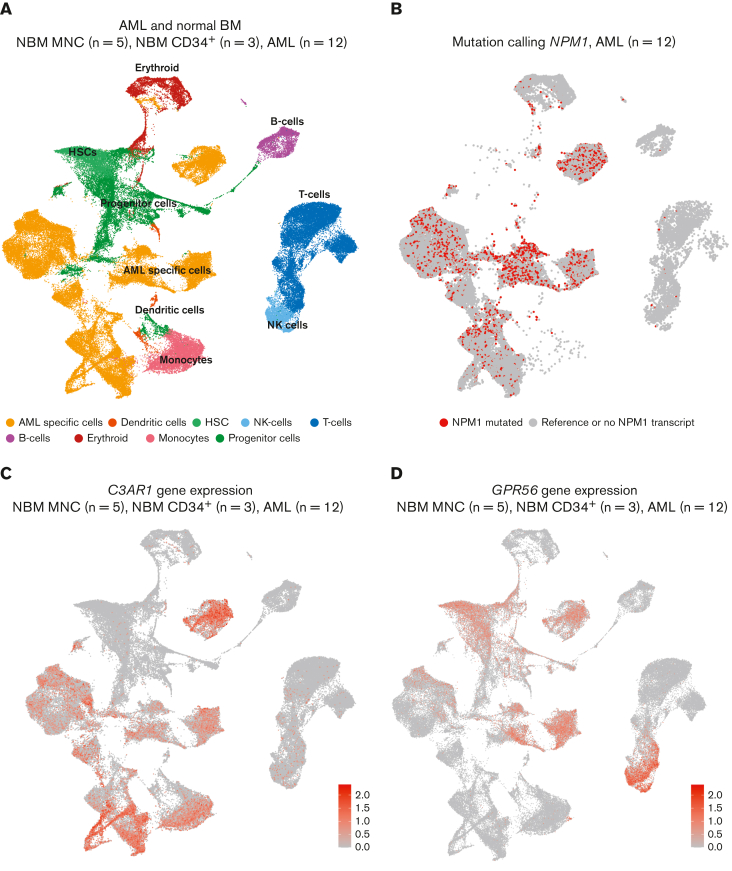
Because bulk RNA-seq does not fully recapitulate the heterogeneity of the samples sequenced, we performed scRNA-seq to investigate the expression of C3AR1 in AML and NBM. We sequenced NPM1-mutated AML samples (n = 12), along with NBM MNC samples (n = 5) and NBM MNC samples enriched for CD34 (n = 3). Gene expression signatures of all samples revealed distinct cell clusters of normal hematopoietic cells as well as major AML-specific clusters (Figure 4A). To determine which clusters contained the malignant cells, we performed mutation calling for all cells that had reads covering the NPM1-mutation site. This analysis showed that the cells with detected NPM1 mutations were mainly located within the AML-specific cell clusters (Figure 4B), confirming that these clusters constitute the actual leukemic cells whereas other cell clusters likely comprise residual normal hematopoiesis (predominantly NK cells, T cells, B cells, and erythroid cells). Assessing C3AR1 expression in all samples sequenced (Figure 4C), 2 main clusters expressing C3AR1 were observed, an AML-specific cluster and a monocyte cluster. No expression was seen in the HSC or progenitor populations (which almost exclusively contained cells from the NBM samples; Figure 4C; supplemental Figure 5). In concordance with this, the NBM samples analyzed by bulk RNA-seq in the Beat AML data set, showed much lower expression of C3AR1 in the CD34+ cells compared with the MNCs (supplemental Figure 6), in which the monocytes are found.
Figure 4.
scRNA-seq confirms C3AR1 expression in NPM1-mutated AML blasts and in normal monocytes. (A) UMAP showing NPM1-mutated AMLs (n = 12), NBM MNCs (n = 5), and NBM CD34+ (n = 3) samples combined. (B) UMAP showing mutation calling of NPM1 in the same AML cells and projection as in panel A. Red: cells with detected NPM1 mutations. Gray: cells with WT NPM1 or no detected NPM1 transcript. (C) UMAP showing the expression of C3AR1 in same cells and projection as in panel A. Red indicates high expression. (D) UMAP showing the expression of GPR56 in same cells and projection as in panel A. Red indicates high expression. UMAP, uniform manifold approximation and projection for dimension reduction.
These data are consistent with the protein expression data assessed by flow cytometry, showing C3AR expression on NPM1-mutated AML cells (Figure 1C-D) and monocytes (supplemental Figure 3) but not on immature NBM cells (Figure 2; supplemental Figure 2).
Coexpression of GPR56 and C3AR defines NPM1-mutated AML cells with leukemia-initiating capacity
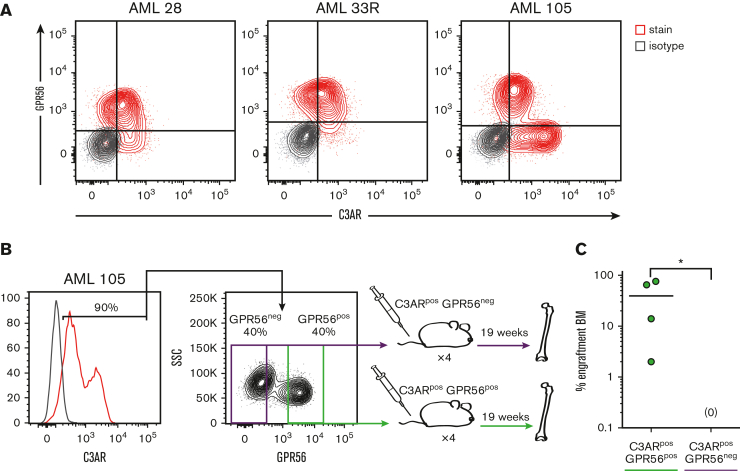
Because the cell surface protein GPR56 has been identified as an important LSC marker in NPM1-mutated AML,17, 18, 19 we investigated its coexpression with C3AR. Analysis of the scRNA-seq data set showed that GPR56 is expressed in certain AML cell populations but also in normal HSCs, NK cells, and some T cells (Figure 4D; supplemental Figure 7). Further flow cytometric analysis of 3 NPM1-mutated AML samples showed coexpression of C3AR and GPR56 in a portion of the AML cells (Figure 5A). GPR56 is known to enrich for LSC activity in AML cells18 but is also highly expressed on normal HSPCs. Considering our observation that C3AR is expressed on AML cells but not on normal immature cell populations, we speculated that GPR56+ C3AR+ could provide a combination of markers to define AML cells with increased LSC activity within the NPM1-mutated subtype. Therefore, we sorted and transplanted C3AR+ GPR56+ and C3AR+ GPR56− primary NPM1-mutated AML cells and assessed human engraftment in BM 19 weeks after transplantation (Figure 5B). Remarkably, all mice that underwent transplantion with C3AR+ GPR56+ cells showed human engraftment, whereas the C3AR+ GPR56− cells did not engraft in any of the mice (Figure 5C; supplemental Figure 8). In summary, these data show that the combination of C3AR and GPR56 can be used to enrich for LSCs in NPM1-mutated AML.
Figure 5.
Coexpression of GPR56 and C3AR marks NPM1-mutated AML cells with leukemia-initiating capacity. (A) Protein coexpression of C3AR and GPR56 in 3 NPM1-mutated AML samples. Marker stains in red and isotype in gray. (B) Schematic illustration showing the experimental setup for the C3AR/GPR56 transplant experiments; the myeloid-enriched (CD3−CD19−) cells expressing C3AR from a NPM1-mutated AML case were sorted based on GPR56 expression and transplanted into immunodeficient NSGS mice, which were sacrificed at 19 weeks. (C) Leukemic engraftment in the BM of mice that had received transplantation with C3AR+ GPR56+ and C3AR+ GPR56− cells, as the percentage of BM cells positive for human CD45 and CD33. Line at median; ∗P ≤ .05 using a nonparametric Mann-Whitney U test.
Stimulation with C3a leads to activation of ERK1/2 in primary NPM1-mutated AML cells and increases survival of OCI-AML3 cells
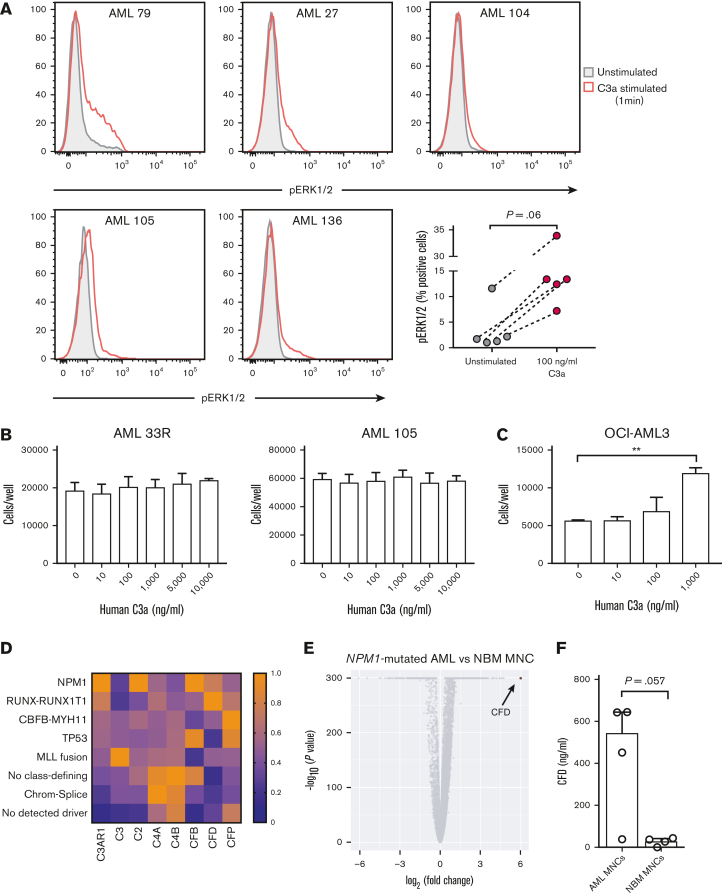
Next, we wanted to investigate whether C3AR-expressing AML cells respond to stimulation with the ligand C3a. Because it is known that binding of C3a to C3AR leads to activation of the MAP kinase pathway in immune cells,38,39 we investigated whether this was also true in primary AML cells. By phospho–flow cytometry, we found increased phosphorylation of ERK1/2 in C3AR-expressing AML cells following 1 minute of C3a stimulation (Figure 6A). After 5-minute stimulation, this phosphorylation was abolished in all but 1 sample in which the phosphorylation had decreased compared with the 1-minute time point (supplemental Figure 9A), indicating that C3AR-mediated activation by C3a is very rapid but transient.
Figure 6.
C3AR is biologically functional in NPM1-mutated AML. (A) Expression of pERK1/2 after 1-minute stimulation with 100 ng/mL of human C3a in 5 primary AML samples. Unstimulated shown in filled blue and stimulated in red. P = .06 using a Wilcoxon nonparametric paired test. (B) Number of primary NPM1-mutated AML cells and OCI-AML3 cells (C) after a 3-day culture with increasing concentrations of human C3a. Primary AML cells (100 000) and OCI-AML3 cells (20 000) were seeded per well in serum-free medium. The experiments were performed in triplicate. Bars in panels B-C show mean cell numbers with standard deviation. (D) Heat map showing the median FPKM value of C3-related complement genes in each AML subgroup, relative to the highest median value of that gene, in the Lund AML Cohort. AML subgroups with <3 patients were excluded from the analysis (n = 109). (E) Relative gene expression of bulk NPM1-mutated AML cells (n = 12 cases) compared with NBM MNCs (n = 5 cases), as assessed by scRNA-seq. (F) Amount of CFD in the cell culture supernatant 24 hour after seeding. One million cells per mL were seeded from 4 NPM1-mutated AML samples and 4 NBM MNC samples in S7 media. Bars show median with interquartile range. ∗∗P ≤ .01 using a Student t test in panel C. A nonparametric Mann-Whitney U test was used in panel F. chrom-splice, chromatin-spliceosome; FPKM, fragments per kilobase of exon model per million reads mapped.
To evaluate whether C3a-stimulation also leads to expansion of C3AR-expressing AML cells, we cultured 5 primary NPM1-mutated AML samples in increasing concentrations of human C3a (Figure 6B; supplemental Figure 9B). After 3 days in culture, no differences in cell numbers were seen, possibly owing to poor survival of primary AML cells in culture. To circumvent this problem, we repeated the experiment using serum-starved NPM1-mutated OCI-AML3 cells, which also express C3AR (supplemental Figure 9C). After 3 days in culture, the number of OCI-AML3 cells was more than twofold higher in the wells that had received 1000 ng/mL of C3a compared with control (Figure 6C). For the NPM1 WT cell line NALM-6, which does not express C3AR (supplemental Figure 9C), no increase in cell number after C3a stimulation was observed (supplemental Figure 9D).
Because C3AR-expressing AML cells were shown to respond to C3a stimulation by increased phosphorylation of ERK1/2, we also investigated whether the NPM1-mutated AML cells could produce C3a in an autocrine manner. First, we analyzed expression of C3, the precursor for C3a, in our scRNA-seq data (supplemental Figure 10). In this data set, the expression of C3 in normal hematopoietic cells and NPM1-mutated AML cells was low or absent. However, a lack of detectable gene expression using this method could be explained by limited sensitivity (a median of 2097 genes/cell were detected in each sample investigated). Therefore, we also analyzed the expression of C3 in the bulk RNA-seq data from the Lund cohort. This revealed expression of C3 in the vast majority of AML samples and in all analyzed genetic subtypes (Figure 6D; supplemental Figure 11). Although these data do not suggest high autocrine expression of C3 in AML, C3 is one of the most abundant plasma proteins in the blood40 and can thus be provided externally. Therefore, we next investigated whether AML cells can produce components of C3-convertases, the enzyme complexes that cleave C3 into its biologically active counterparts, C3a and C3b. To address this, we analyzed genes encoding the components of the classical pathway C3-convertase (C2 and C4A/C4B), as well as the alternative pathway C3-convertase (complement factor B [CFB] and CFD) (Figure 6D; supplemental Figure 11).
Notably, NPM1-mutated AMLs showed a relatively high expression of alternative pathway C3-convertase genes, CFB and CFD. In fact, CFD, encoding the protein CFD was the most upregulated gene in NPM1-mutated AML blasts compared with NBM NCs in our scRNA-seq data set (Figure 6E). Interestingly, CFD is known to be the rate-determining enzyme in the alternative complement pathway.41
To investigate whether the NPM1-mutated AML cells and NBM MNCs also produce the CFD protein, we measured the amount of CFD in the cell culture media after 24 hours (Figure 6F). Expression of CFD by NBM MNCs (n = 4) was detected in all but 1 sample (3/4), with a median CFD expression of 31.8 ng/mL. However, in the supernatants from the cultured AML cells (n = 4), the median CFD expression was ∼17 times higher (median = 547.4 ng/mL). One AML sample had notably lower expression than the other 3, possibly owing to poor survival of this sample in culture or because of biological differences between samples. Nevertheless, these data suggest that NPM1-mutated AMLs cells may have the ability to generate active C3a from the bloodstream by their own production of the alternative pathway C3-convertase.
In conclusion, our data show that C3AR-expressing AML cells respond to C3a-stimualtion with activation of the MAP kinase pathway and that these cells produce CFD, the rate-limiting enzyme of alternative pathway complement activation.
C3AR can be specifically targeted with an anti-C3AR antibody
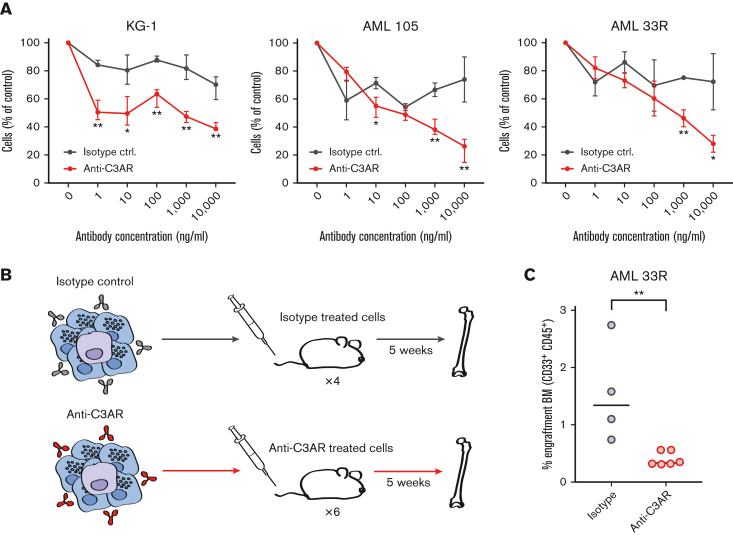
Finally, we evaluated the therapeutic potential of C3AR. The selective expression of this marker on NPM1-mutated AML cells but not on normal HSPCs or AMLs with NPM1 WT, suggests that C3AR can be utilized for antibody-based targeting in this subgroup of AML. Hence, we investigated, using an ADCC assay, whether C3AR-antibodies would be able to elicit cell killing by attracting human NK cells. For the C3AR-expressing AML cell line KG-1 (supplemental Figure 9C), and 5 primary NPM1-mutated AML samples, in 7 individual experiments, we observed selective killing of AML cells compared with treatment with an isotype control (Figure 7A; supplemental Figure 12A). Importantly, when residual primary AML cells from an ADCC assay were transplanted into immunodeficient mice (Figure 7B), the cells treated with C3AR-antibodies engrafted at lower levels than isotype control–treated cells (Figure 7C; supplemental Figure 12B), demonstrating that C3AR-antibodies are able to efficiently kill AML cells with leukemia-initiating capacity. Collectively, our results suggest that C3AR constitutes a novel therapeutic target in NPM1-mutated AML, representing, to the best of our knowledge, the first selectively expressed marker in this subtype that is targetable with antibodies while sparing normal HSPCs.
Figure 7.
C3AR can be specifically targeted with anti-C3AR antibodies. (A) ADCC assay on AML cell line KG-1 and 2 primary NPM1-mutated AML samples. The red line shows the percentage of AML cells after overnight incubation with a polyclonal anti-C3AR antibody and NK cells, compared with control without antibody. The black line shows the isotype-treated cells under the same experimental conditions. Error bars show mean with standard deviation. (B) Schematic illustration of the ADCC in vivo repopulation assay; residual cells from an ex vivo ADCC experiment on a primary AML sample were transplanted into immunodeficient NSG mice, which were sacrificed after 5 weeks. AML cells are depicted in purple and NK cells in blue. (C) Leukemic engraftment after ADCC, as the percentage of CD45+CD33+ human cells in the BM of mice that received transplantation with isotype-treated or anti-C3AR–treated AML cells, as in panel B. Antibody (10 μg/mL) was used. Line at median. ∗P ≤ .05 and ∗∗P ≤ .01 using a Student t test in panel A and a nonparametric Mann-Whitney U test in panel C.
Discussion
AML with mutated NPM1 is the most common molecular subgroup in AML, comprising ∼30% of all AML cases.8,9 When NPM1 mutations are observed without FLT3-ITD in AML with a normal karyotype, this is associated with a favorable prognosis according to current risk stratification guidelines.9 However, a recent study based on the Swedish AML Registry showed that the favorable prognosis of NPM1-mutated cases without FLT3-ITD is only seen in patients aged <60 years. For patients aged 60 to 75 years, the median overall survival is only 3.3 years. Moreover, if NPM1 mutations and FLT3-ITD are present, the prognosis is poor regardless of age (median overall survival <2 years).42 Hence, new treatment alternatives are urgently needed to improve survival rates for NPM1-mutated AML.
In this study, we searched for novel cell surface markers in NPM1-mutated AML that could form the basis for the development of an antibody-based treatment. In most subtypes of AML, the LSCs are enriched within the primitive (CD34+CD38−) population.43 However, NPM1-mutated AML typically lacks CD34-expression,44,45 making it impossible to use CD34 as an LSC maker. Furthermore, most studies of LSCs in AML have focused on AML samples with >10% CD34+ blasts,5 hence disregarding CD34-negative AML samples with NPM1 mutations. Therefore, we, instead, focused on identifying markers with expression within the entire myeloid compartment of NPM1-mutated AMLs but not in the primitive NBM cells. Using an arrayed antibody screen, we identified the complement protein C3a receptor C3AR as the highest and most consistently expressed selective marker in NPM1-mutated AMLs, with nondetectable expression in NBM HSPCs.
C3AR is a G protein–coupled receptor with high expression on myeloid innate immune cells, especially on granular cells and monocytes.35 It binds to the anaphylatoxin, C3a, an inflammatory molecule released following complement activation.46 In cancer, increased inflammation as a result of dysregulated complement has been suggested to play an important role in disease initiation and progression,39,46, 47, 48, 49, 50 and autocrine signaling involving tumor-derived complement factors has been identified in solid tumors.51 We show that genes encoding 2 of the complement proteins that make up the C3-convertase of the alternative pathway, CFB and CFD, are expressed in NPM1-mutated AML. In fact, the gene encoding the serine protease CFD, which is the rate-limiting enzyme of the alternative complement pathway, was the most upregulated gene in the NPM1-mutated AML cells analyzed by scRNA-seq, compared with NBM MNCs. This is remarkable as CFD usually has the lowest plasma concentration of all complement proteins and is predominantly produced by adipocytes.41 However, we show that NPM1-mutated AML cells can readily produce CFD. This suggests that the AML cells have the ability to generate active C3a from the abundant amounts of C3 in the blood40 by producing the alternative pathway C3-convertase, thus providing a previously unexplored mechanism of complement activation in this subtype of AML. As of now, complement involvement in leukemia has not been as extensively studied, as for many solid tumors51; herein, however, we provide evidence that the C3a/C3AR signaling axis is functional in primary NPM1-mutated AML cells, suggesting that cells would benefit from a proinflammatory environment with high levels of C3a.
We, and others, have shown that LSCs expressing, for example, IL1RAP20,21 or CD12352 on the cell surface can be specifically targeted with antibodies and killed by ADCC, with several clinical trials currently ongoing. Similarly, we show herein that C3AR-expressing NPM1-mutated AML cells can be specifically targeted ex vivo using C3AR antibodies and NK cells. Given that we observed a consistent expression of C3AR on the bulk leukemic cell population, an antibody targeting C3AR would be anticipated to kill AML cells, including the critical LSCs, while sparing normal HSPCs. Indeed, our results support the ability to target LSCs as demonstrated by reduced ability of C3AR-antibody–treated AML cells to initiate leukemia in immunodeficient mice.
In concordance with previous studies, we also found expression of C3AR on normal monocytes.34,35 However, cell surface expression of C3AR was mainly limited to CD16-expressing monocytes, suggesting that C3AR-antibodies would spare the classical (CD16neg) monocytes that constitute the majority of the monocytic population. Furthermore, as normal HSPCs do not express C3AR, the affected monocytes would be expected to rapidly replenish after the end of treatment.
Recently, the G protein–coupled receptor GPR56 was identified as a cell surface marker of LSCs on both CD34+ and CD34− cells.18 In particular, NPM1 FLT3-ITD DNMT3A triple-mutated AML shows an enrichment for LSCs in the CD34− GPR56+ compartment.19 However, although GPR56 may provide a marker to study LSCs, GPR56 is also expressed on normal CD34+ cells,18 making it less ideal as a therapeutic target. Herein, we show that C3AR+GPR56+ cells but not C3AR+GPR56− cells from a patient with primary NPM1-mutated AML engrafted in immunodeficient mice. Collectively, these data demonstrate that C3AR can discriminate AML cells from normal HSPCs, whereas GPR56 can be used to enrich for cells with LSCs activity within this population.
In conclusion, our results demonstrate that complement receptor C3AR is specifically upregulated on NPM1-mutated AML cells. Importantly, targeting C3AR by antibodies could provide new therapeutic opportunities in this AML subgroup, possibly through a dual mode of action, by inhibition of C3a signaling and by marking leukemic cells for effector-cell–mediated immune destruction.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Acknowledgments
The authors thank the Center for Translational Genomics at Lund University and Clinical Genomics Lund, SciLifeLab, for providing single-cell sequencing service. The authors also thank the investigators of The Cancer Genome Atlas (TCGA) study on AML36 and the Beat AML program37 (dbGaP study accession: phs001657.v1.p1) for sharing transcriptomic data used for validation of differential gene expression analysis (data were accessed as detailed in supplemental Methods).
This work was supported by the Swedish Childhood Cancer Fund, Sweden; the Swedish Cancer Society, Sweden; the Swedish Research Council, Sweden; Governmental Funding of Clinical Research within the National Health Service (ALF-grant), Sweden; the Knut and Alice Wallenberg Foundation, Sweden; the Cancera Foundation, Sweden; and the Mats Paulsson Foundation, Sweden.
Authorship
Contribution: S.v.P. and T.F. designed the study; S.v.P. made the figures; S.v.P., H.T., P.P.-M., N.P.-M., C.S., N.L., H.Å., and M.R. performed experiments; S.v.P., H.T., P.P.-M., N.P.-M., C.S., R.H., V. R., M.J., C.O.-P., H.L., and T.F. analyzed the data; G.J., V.L., H.L., and C.O.-P. collected patient material and clinical data; A.M.B. and B.K. provided intellectual input; S.v.P. and T.F. wrote the manuscript; and all other authors reviewed the manuscript.
Footnotes
The data sets generated during the current study fall under the GDPR regulations for sharing of personal data and will therefore be deposited at EGA-SE once this repository is complete. Until then, the data sets are available from the corresponding authors upon request through the following DOIs: https://doi.org/10.17044/scilifelab.21557163 (RNA-seq data set) and https://doi.org/10.17044/scilifelab.21557247 (scRNA-seq data set).
The full-text version of this article contains a data supplement.
Contributor Information
Sofia von Palffy, Email: sofia.von_palffy@med.lu.se.
Thoas Fioretos, Email: thoas.fioretos@med.lu.se.
Supplementary Material
References
- 1.Murati A, Brecqueville M, Devillier R, Mozziconacci MJ, Gelsi-Boyer V, Birnbaum D. Myeloid malignancies: mutations, models and management. BMC Cancer. 2012;12 doi: 10.1186/1471-2407-12-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 3.Shallis RM, Wang R, Davidoff A, Ma X, Zeidan AM. Epidemiology of acute myeloid leukemia: recent progress and enduring challenges. Blood Rev. 2019;36:70–87. doi: 10.1016/j.blre.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Khwaja A, Bjorkholm M, Gale RE, et al. Acute myeloid leukaemia. Nat Rev Dis Primers. 2016;2:16010. doi: 10.1038/nrdp.2016.10. [DOI] [PubMed] [Google Scholar]
- 5.Thomas D, Majeti R. Biology and relevance of human acute myeloid leukemia stem cells. Blood. 2017;129(12):1577–1585. doi: 10.1182/blood-2016-10-696054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majeti R. Monoclonal antibody therapy directed against human acute myeloid leukemia stem cells. Oncogene. 2011;30(9):1009–1019. doi: 10.1038/onc.2010.511. [DOI] [PubMed] [Google Scholar]
- 7.Gasiorowski RE, Clark GJ, Bradstock K, Hart DN. Antibody therapy for acute myeloid leukaemia. Br J Haematol. 2014;164(4):481–495. doi: 10.1111/bjh.12691. [DOI] [PubMed] [Google Scholar]
- 8.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 11.Quek L, Otto GW, Garnett C, et al. Genetically distinct leukemic stem cells in human CD34- acute myeloid leukemia are arrested at a hemopoietic precursor-like stage. J Exp Med. 2016;213(8):1513–1535. doi: 10.1084/jem.20151775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Propris MS, Raponi S, Diverio D, et al. High CD33 expression levels in acute myeloid leukemia cells carrying the nucleophosmin (NPM1) mutation. Haematologica. 2011;96(10):1548–1551. doi: 10.3324/haematol.2011.043786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu YR, Zhu HH, Ruan GR, et al. NPM1-mutated acute myeloid leukemia of monocytic or myeloid origin exhibit distinct immunophenotypes. Leuk Res. 2013;37(7):737–741. doi: 10.1016/j.leukres.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Ehninger A, Kramer M, Rollig C, et al. Distribution and levels of cell surface expression of CD33 and CD123 in acute myeloid leukemia. Blood Cancer J. 2014;4:e218. doi: 10.1038/bcj.2014.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnone M, Konantz M, Hanns P, et al. Acute myeloid leukemia stem cells: the challenges of phenotypic heterogeneity. Cancers (Basel) 2020;12(12):3742. doi: 10.3390/cancers12123742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho JM, Dobson SM, Voisin V, et al. CD200 expression marks leukemia stem cells in human AML. Blood Adv. 2020;4(21):5402–5413. doi: 10.1182/bloodadvances.2020001802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daria D, Kirsten N, Muranyi A, et al. GPR56 contributes to the development of acute myeloid leukemia in mice. Leukemia. 2016;30(8):1734–1741. doi: 10.1038/leu.2016.76. [DOI] [PubMed] [Google Scholar]
- 18.Pabst C, Bergeron A, Lavallee VP, et al. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood. 2016;127(16):2018–2027. doi: 10.1182/blood-2015-11-683649. [DOI] [PubMed] [Google Scholar]
- 19.Garg S, Reyes-Palomares A, He L, et al. Hepatic leukemia factor is a novel leukemic stem cell regulator in DNMT3A, NPM1, and FLT3-ITD triple-mutated AML. Blood. 2019;134(3):263–276. doi: 10.1182/blood.2018862383. [DOI] [PubMed] [Google Scholar]
- 20.Agerstam H, Karlsson C, Hansen N, et al. Antibodies targeting human IL1RAP (IL1R3) show therapeutic effects in xenograft models of acute myeloid leukemia. Proc Natl Acad Sci U S A. 2015;112(34):10786–10791. doi: 10.1073/pnas.1422749112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Askmyr M, Agerstam H, Hansen N, et al. Selective killing of candidate AML stem cells by antibody targeting of IL1RAP. Blood. 2013;121(18):3709–3713. doi: 10.1182/blood-2012-09-458935. [DOI] [PubMed] [Google Scholar]
- 22.Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pabst C, Krosl J, Fares I, et al. Identification of small molecules that support human leukemia stem cell activity ex vivo. Nat Methods. 2014;11(4):436–442. doi: 10.1038/nmeth.2847. [DOI] [PubMed] [Google Scholar]
- 24.van Rhenen A, van Dongen GA, Kelder A, et al. The novel AML stem cell associated antigen CLL-1 aids in discrimination between normal and leukemic stem cells. Blood. 2007;110(7):2659–2666. doi: 10.1182/blood-2007-03-083048. [DOI] [PubMed] [Google Scholar]
- 25.Bakker AB, van den Oudenrijn S, Bakker AQ, et al. C-type lectin-like molecule-1: a novel myeloid cell surface marker associated with acute myeloid leukemia. Cancer Res. 2004;64(22):8443–8450. doi: 10.1158/0008-5472.CAN-04-1659. [DOI] [PubMed] [Google Scholar]
- 26.Chung SS, Eng WS, Hu WH, et al. CD99 is a therapeutic target on disease stem cells in myeloid malignancies. Sci Transl Med. 2017;9(374) doi: 10.1126/scitranslmed.aaj2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barreyro L, Will B, Bartholdy B, et al. Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood. 2012;120(6):1290–1298. doi: 10.1182/blood-2012-01-404699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hauswirth AW, Florian S, Printz D, et al. Expression of the target receptor CD33 in CD34+/CD38-/CD123+ AML stem cells. Eur J Clin Invest. 2007;37(1):73–82. doi: 10.1111/j.1365-2362.2007.01746.x. [DOI] [PubMed] [Google Scholar]
- 29.Nishioka C, Ikezoe T, Furihata M, et al. CD34+/CD38 acute myelogenous leukemia cells aberrantly express CD82 which regulates adhesion and survival of leukemia stem cells. Int J Cancer. 2013;132(9):2006–2019. doi: 10.1002/ijc.27904. [DOI] [PubMed] [Google Scholar]
- 30.Gillissen MA, de Jong G, Kedde M, et al. Patient-derived antibody recognizes a unique CD43 epitope expressed on all AML and has antileukemia activity in mice. Blood Adv. 2017;1(19):1551–1564. doi: 10.1182/bloodadvances.2017008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jordan CT, Upchurch D, Szilvassy SJ, et al. The interleukin-3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells. Leukemia. 2000;14(10):1777–1784. doi: 10.1038/sj.leu.2401903. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Saavedra E, Tang R, et al. Targeting primary acute myeloid leukemia with a new CXCR4 antagonist IgG1 antibody (PF-06747143) Sci Rep. 2017;7(1):7305. doi: 10.1038/s41598-017-07848-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakobsen JS, Laursen LG, Schuster MB, et al. Mutant CEBPA directly drives the expression of the targetable tumor-promoting factor CD73 in AML. Sci Adv. 2019;5(7) doi: 10.1126/sciadv.aaw4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin U, Bock D, Arseniev L, et al. The human C3a receptor is expressed on neutrophils and monocytes, but not on B or T lymphocytes. J Exp Med. 1997;186(2):199–207. doi: 10.1084/jem.186.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Zhang H, He YW. The Complement receptors C3aR and C5aR are a new class of immune checkpoint receptor in cancer immunotherapy. Front Immunol. 2019;10:1574. doi: 10.3389/fimmu.2019.01574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research N. Ley TJ, Miller C, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562(7728):526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reis ES, Mastellos DC, Hajishengallis G, Lambris JD. New insights into the immune functions of complement. Nat Rev Immunol. 2019;19(8):503–516. doi: 10.1038/s41577-019-0168-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ajona D, Ortiz-Espinosa S, Pio R. Complement anaphylatoxins C3a and C5a: emerging roles in cancer progression and treatment. Semin Cell Dev Biol. 2019;85:153–163. doi: 10.1016/j.semcdb.2017.11.023. [DOI] [PubMed] [Google Scholar]
- 40.Ricklin D, Reis ES, Mastellos DC, Gros P, Lambris JD. Complement component C3-The “Swiss Army Knife” of innate immunity and host defense. Immunol Rev. 2016;274(1):33–58. doi: 10.1111/imr.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barratt J, Weitz I. Complement factor D as a strategic target for regulating the alternative complement pathway. Front Immunol. 2021;12:712572. doi: 10.3389/fimmu.2021.712572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Juliusson G, Jadersten M, Deneberg S, et al. The prognostic impact of FLT3-ITD and NPM1 mutation in adult AML is age-dependent in the population-based setting. Blood Adv. 2020;4(6):1094–1101. doi: 10.1182/bloodadvances.2019001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 44.Falini B, Brunetti L, Sportoletti P, Martelli MP. NPM1-mutated acute myeloid leukemia: from bench to bedside. Blood. 2020;136(15):1707–1721. doi: 10.1182/blood.2019004226. [DOI] [PubMed] [Google Scholar]
- 45.Alcalay M, Tiacci E, Bergomas R, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;106(3):899–902. doi: 10.1182/blood-2005-02-0560. [DOI] [PubMed] [Google Scholar]
- 46.Roumenina LT, Daugan MV, Petitprez F, Sautes-Fridman C, Fridman WH. Context-dependent roles of complement in cancer. Nat Rev Cancer. 2019;19(12):698–715. doi: 10.1038/s41568-019-0210-0. [DOI] [PubMed] [Google Scholar]
- 47.Reis ES, Mastellos DC, Ricklin D, Mantovani A, Lambris JD. Complement in cancer: untangling an intricate relationship. Nat Rev Immunol. 2018;18(1):5–18. doi: 10.1038/nri.2017.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merle NS, Church SE, Fremeaux-Bacchi V, Roumenina LT. Complement system part I - molecular mechanisms of activation and regulation. Front Immunol. 2015;6:262. doi: 10.3389/fimmu.2015.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Afshar-Kharghan V. The role of the complement system in cancer. J Clin Invest. 2017;127(3):780–789. doi: 10.1172/JCI90962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aykut B, Pushalkar S, Chen R, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574(7777):264–267. doi: 10.1038/s41586-019-1608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho MS, Vasquez HG, Rupaimoole R, et al. Autocrine effects of tumor-derived complement. Cell Rep. 2014;6(6):1085–1095. doi: 10.1016/j.celrep.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie LH, Biondo M, Busfield SJ, et al. CD123 target validation and preclinical evaluation of ADCC activity of anti-CD123 antibody CSL362 in combination with NKs from AML patients in remission. Blood Cancer J. 2017;7(6):e567. doi: 10.1038/bcj.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.