ABSTRACT
Bacterial flagella are involved in infection through their roles in host cell adhesion, cell invasion, auto-agglutination, colonization, the formation of biofilms, and the regulation and secretion of nonflagellar bacterial proteins that are involved in the virulence process. In this study, we constructed a fusion protein vaccine (FliCD) containing the Clostridioides difficile flagellar proteins FliC and FliD. The immunization of mice with FliCD induced potent IgG and IgA antibody responses against FliCD, protected mice against C. difficile infection (CDI), and decreased the C. difficile spore and toxin levels in the feces after infection. Additionally, the anti-FliCD serum inhibited the binding of C. difficile vegetative cells to HCT8 cells. These results suggest that FliCD may represent an effective vaccine candidate against CDI.
KEYWORDS: Clostridioides difficile infection (CDI), chimeric protein vaccine, FliCD, hyperimmune serum
INTRODUCTION
Clostridioides difficile (C. difficile) is an anaerobic, spore-forming, Gram-positive bacterium, and it is the leading cause of antibiotic-associated diarrhea and colitis (1, 2). C. difficile produces three protein toxins, including toxin A (TcdA), toxin B (TcdB), and binary toxin (CDT). The first two are the major virulence factors of C. difficile that cause C. difficile infection (CDI) symptoms (3, 4). CDI is spread by bacterial spores that are found in the feces, and infections occur in all areas of the world (5, 6). In the United States, there is an occurrence of 8.3 cases per 10,000 patient days, suggesting that CDI is associated with a large burden on the health care system (7). Currently, few antibiotics are available for the treatment of CDI, and none of them are fully effective (8). Antibiotic treatment is often followed by recurrent infection, which leads to the use of nontraditional therapies (9, 10).
The flagella of most pathogens increase the occurrence of interactions between the pathogen and the epithelial mucosal surface by facilitating bacteria to chemotaxis toward specific signals. Moreover, bacterial flagella are involved in infection through their roles in host cell adhesion, cell invasion, auto-agglutination, colonization, the formation of biofilms, and the regulation and secretion of nonflagellar bacterial proteins that are involved in the virulence process (11). C. difficile flagellin FliC is the major structural component of the flagellar filament, and the assembly of a flagellum requires other proteins, which are called hook-associated proteins (HAP1, HAP2, and HAP3). The fliD gene encodes the structural component HAP2 of the flagellar cap at the distal end of the filament (12–14). Both the FliC and FliD proteins are implicated in the attachment of C. difficile to the mucus layer of the intestine (15). Researchers also found that flagellated, motile C. difficile adheres more efficiently to the epithelium cell wall of axenic mice than do nonflagellated strains of the same serogroup (16). Interestingly, a different study showed that both fliC and fliD mutant strains adhered better than did the wild-type 630Δerm strain to human intestine-derived Caco-2 cells, and they were also more virulent in hamsters (17). These conflicting reports suggest a complex role for flagella in CDI.
Previously, FliC immunization provided partial protection against CDI in mice and hamsters (18). In this study, we constructed a fusion protein vaccine (FliCD) containing FliC and FliD and evaluated its immunogenicity and protection in a mouse model of CDI.
RESULTS
Homology of FliC and FliD in major toxinotypes and ribotypes of C. difficile strains.
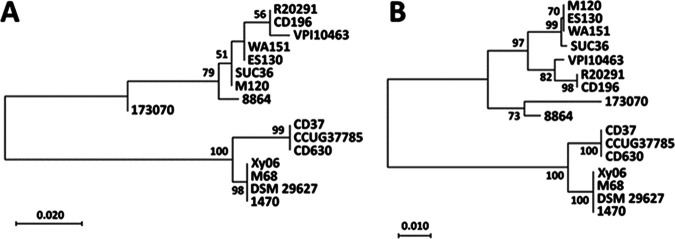
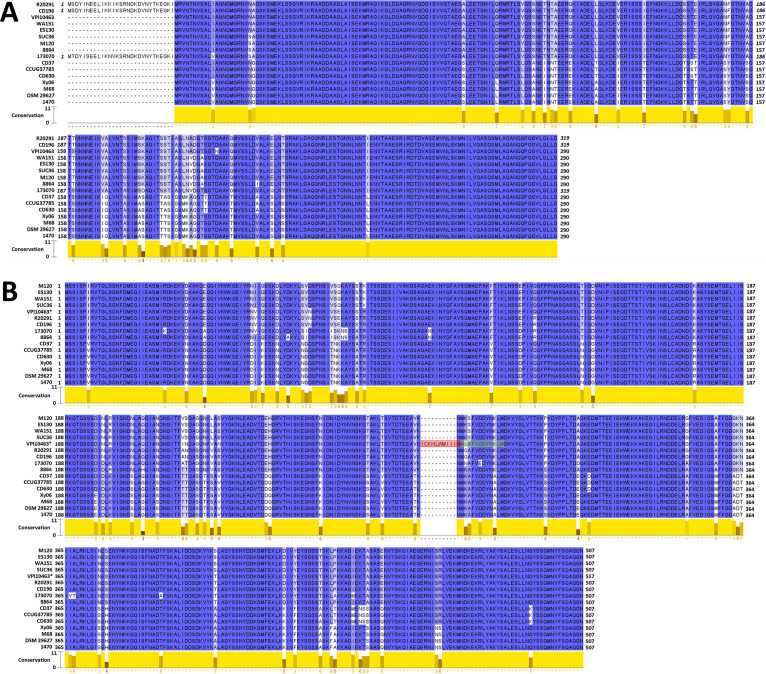
Bacterial flagellin is highly variable across species. An optional vaccine candidate should be conserved. We investigated the homology of FliC and FliD proteins in major toxionotypes, including A−B+CDT+, A+B+CDT−, A−B+CDT−, and A−B−CDT−, as well as major ribotypes, including the RT027, RT078, RT017, RT012, RT003, and RT009 C. difficile strains (Table 1). Maximum likelihood phylogenetic trees were generated using the FliC (Fig. 1A) and FliD (Fig. 1B) amino acid sequences. Both the RT027 FliC and FliD sequences cluster together, as do the RT017 FliC and FliD sequences. As shown more explicitly in the MUSCLE alignments of FliC (Fig. 2A) and FliD (Fig. 2B), there are no sequence variations within either ribotype group. This is in line with a previous study that observed no sequence differences between FliC and FliD sequences in RT027 and RT176 isolates (19). There does not appear to be a strong correlation between toxinotype and either FliC or FliD sequence relatedness among the strains that we selected.
TABLE 1.
C. difficile strains used for the homology analysis of FliC and FliD
| Toxinotype | Strain | Ribotype | Database | Accession no./barcode | Reference |
|---|---|---|---|---|---|
| A+B+CDT+ | R20291 | RT027 | GenBank | FN545816.1 | 30 |
| CD196 | RT027 | GenBank | FN538970.1 | 31 | |
| M120 | RT078 | GenBank | FN665653.1 | 32 | |
| A+B+CDT− | VPI 10463 | RT003 | Enterobase | CLO_AA6882AA | 33 |
| CD630 | RT012 | GenBank | AM180355.1 | 34 | |
| A−B+CDT− | M68 | RT017 | GenBank | NC_017175.1 | 35 |
| DSM 29627 | RT017 | GenBank | CP016102.1 | 36 | |
| Xy06 | RT017 | GenBank | NZ_JANFNF000000000.1 | GenBank | |
| 1470 | RT017 | GenBank | NZ_OEZL00000000.1 | 38 | |
| 8864 | RT59 | GenBank | NZ_OEZE00000000.1 | 39 | |
| SUC36 | RT078 | GenBank | NZ_OEZZ00000000.1 | 40 | |
| ES130 | SLO101 | GenBank | NZ_OEZV00000000.1 | 40 | |
| WA151 | SKO098 | GenBank | NZ_OEZY00000000.1 | 40 | |
| 173070 | RT015 | GenBank | NZ_OEZH00000000.1 | 40 | |
| A−B−CDT− | CD37 | RT09 | GenBank | NZ_AHJJ00000000.1 | 41 |
| CCUG37785 | ND | GenBank | NZ_JAGKRT000000000.1 | 42 |
FIG 1.
FliC and FliD phylogeny. The amino acid sequences of FliC (A) and FliD (B) were aligned with the MUSCLE algorithm in MegaX before the computation of a maximum likelihood tree with 500 bootstrap replicates (bootstrap values >50 displayed). Scale bars indicate 0.020 and 0.010 substitutions per site for FliC and FliD, respectively.
FIG 2.
FliC and FliD homology. Jalview software was used to visualize the MUSCLE alignments of the FliC (A) and FliD (B) sequences. Conservation scores between 0 (no agreement) and 11 (identical amino acids) were reported for each amino acid position, as determined through Jalview (see Materials and Methods).
To better illustrate the sequence diversity, FliC (Fig. 2A) and FliD (Fig. 2B) sequences were aligned using MUSCLE and visualized in Jalview software. Most sequence variations in FliC were observed between the amino acid positions 108 and 245 (using R20291 FliC as a reference), which is in concordance with the results of a previous study that reported the N terminus and C terminus of FliC to be more conserved than the center region (20). Three strains, namely, R20291, CD196, and 173070, encode FliC proteins with an additional 29 amino acid residues on their N termini. These residues were absent in all of the other strains that were examined. The R20291 and CD196 sequences are identical on account of both strains being ribotype RT027, but these sequences are also quite similar to the 173070 N terminus sequence (86% identical), despite strain 173070 being classified as a singleton in the phylogenetic tree (Fig. 1A).
In FliD, a flagellar cap protein (21), sequence variations are distributed more widely throughout the sequence (Fig. 2B). Flagellar caps assume a structural role by preventing the loss of flagellin monomers while also facilitating the excretion of various proteins (22, 23). The exposure of FliD to the external environment, such as to immune cells, may provide selective pressure for mutations throughout the protein that facilitate immune evasion. In strain VPI 10463, the fliD gene was unique among the strains examined, which accounts for the observation that the amino acid residues 286 to 295 of VPI 10463 FliD do not align with any of the other sequences that were examined. Upon viewing the annotated genome of VPI 10463, the fliD gene was found to be split roughly in half between two adjacent open-reading frames (ORFs). The end of the amino acid sequence that is encoded by the first ORF is highlighted in red in Fig. 2B, whereas the beginning of the second translated ORF is highlighted in green. These two ORFs are not on the same reading frame, but they do overlap at the highlighted regions. The fragmentation of the VPI 10463 fliD gene could potentially compromise the flagellin structure of this strain. Overall, FliC and FliD are rather conserved among the selected C. difficile strains, which is also in concordance with the results of previous studies (16, 20).
The immunization of mice with FliCD induces significant anti-FliCD responses in mice.
Gene sequences encoding full-length FliC and FliD from C. difficile R20291 were bridged with a linker (ggt ggc tct ggt), synthesized, and cloned into pHis1525 in Bacillus megaterium (24).
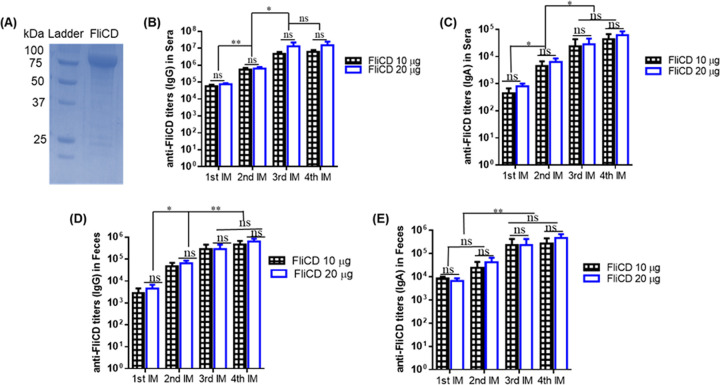
Recombinant FliCD with a 6×His tag (97 kDa) was purified via Ni-NTA affinity chromatography to a purity of >95% (Fig. 3A).
FIG 3.
(A) Expression and purification of protein FliCD. The gene sequence encoding FliCD was synthesized and cloned in Bacillus megatarium. FliCD was purified from bacterial lysate via Ni-affinity chromatography and analyzed via SDS-PAGE. (B–E) FliCD immunizations via the intraperitoneal (i.p.) route induce anti-FliCD antibody responses. Groups of C57BL/6 mice (n = 10) were immunized 4 times at 12-day intervals with 10 or 20 μg of FliCD with alum as an adjuvant. Sera and feces were collected, and anti-FliCD IgG/IgA titers were measured via standard ELISA. The data are presented as the mean ± SEM (*, P < 0.05; **, P < 0.01; ns, not significant).
The immunizations of mice with 10 or 20 μg FliCD with alum as an adjuvant via the intraperitoneal (i.p.) route induced high levels of IgG and IgA antibody responses against FliCD in sera (Fig. 3B and C) and in feces (Fig. 3D and E). However, no significant increases of anti-FliCD antibodies were observed between the third and fourth immunizations. Also, the titers were not significantly higher in the sera or feces of mice immunized with 20 μg FliCD, compared to 10 μg FliCD, after the third and fourth immunizations. To determine the natures of the anti-FliCD immune responses, we determined an anti-FliCD IgG isotype (Fig. 4). Immunizations with FliCD induced significantly high levels of IgG1, IgG2C, IgG2b, and IgG2a anti-FliCD isotypes, indicating balanced Th2 (IgG1) and Th1 (IgG2a, IgG2b, IgG2C) immune responses.
FIG 4.
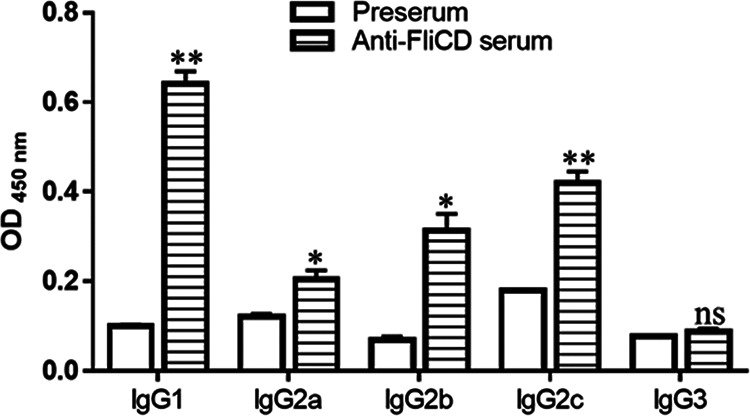
Anti-FliCD IgG isotypes of sera from mice immunized with FliCD. Mice were immunized with FliCD three times, and serum samples were collected. The anti-FliCD IgG isotypes of the pooled serum samples from the third immunization were measured via standard ELISA. The data are presented as the mean ± SEM (n = triplicate) (*, P < 0.05; **, P < 0.01; ns, not significant).
Immunization with FliCD protects mice against C. difficile infections and decreases the spore and toxin levels in their feces.
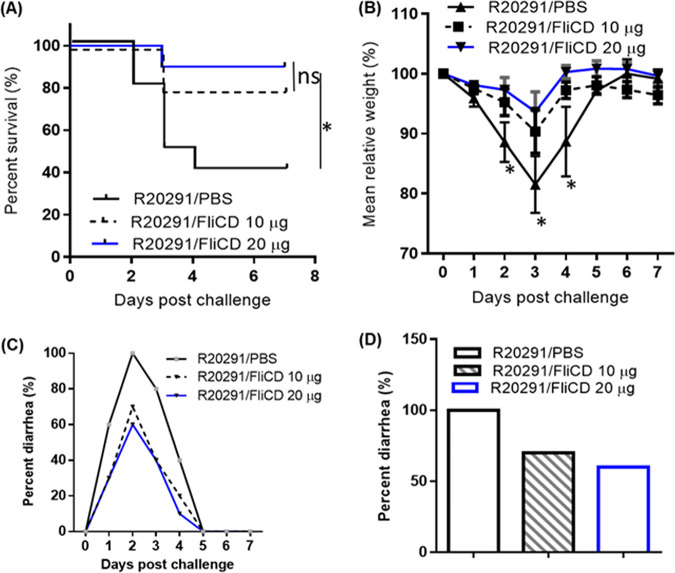
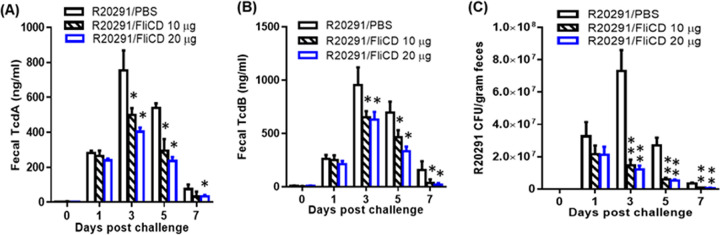
The protection efficacy of FliCD immunization was further evaluated in a mouse model of CDI. 3 groups of mice (n = 10) were immunized thrice via the i.p. route with 10 μg/20 μg of FliCD or PBS with alum at 12-day intervals. After the third immunization, the mice were challenged with 106 spores of C. difficile R20291, which is a hypervirulent strain of ribotype 027. The control (PBS-immunized) mice exhibited significant disease symptoms, including weight loss (Fig. 5B) and severe diarrhea (Fig. 3C and D). Approximately 60% of the mice succumbed by day 4 (Fig. 5A). In contrast, the FliCD-immunized mice developed much less severe disease symptoms, including less weight loss (Fig. 3B) and lower diarrhea rates (Fig. 5C and D) as well as a significantly higher survival rate (80% for the 10 μg FliCD-immunized mice and 90% for the 20 μg FliCD-immunized mice) (Fig. 5A). The FliCD-immunized mice excreted significantly smaller amounts of TcdA (Fig. 6A) and TcdB (Fig. 6B) in their feces, compared to the PBS group. The fecal samples of the FliCD-immunized mice contained significantly fewer R20291 spores, compared to the control group (Fig. 6C).
FIG 5.
Immunizations of mice with FliCD provide mice with significant protection against infection by C. difficile strain R20291. Mice were challenged with C. difficile R20291 spores (106/mouse) 14 days after the third immunization of the groups of mice (n = 10) with FliCD at 10 or 20 μg/mouse/immunization or with PBS in the presence of alum. (A) Kaplan-Meier survival plots (P = 0.02 between the R20291/PBS and R20291/FliCD 20 μg groups; P = 0.0628 between the R20291/PBS and R20291/FliCD 10 μg groups). (B) Mean relative weight of all surviving mice (up to the day of death). The data are presented as the mean ± SEM (*, P < 0.05). (C and D) Frequency of diarrhea.
FIG 6.
Immunizations of mice with FliCD decrease the C. difficile spores and toxins in feces after a challenge with C. difficile spores. The TcdA (A) or TcdB (B) levels in feces were determined via ELISA. (C) R20291 spore concentrations in feces. The data are presented as the mean ± SEM (n = 10) (*, P < 0.05; **, P < 0.01 versus PBS).
Anti-FliCD serum protects mice against C. difficile infection and decreases the spore and toxin levels in their feces.
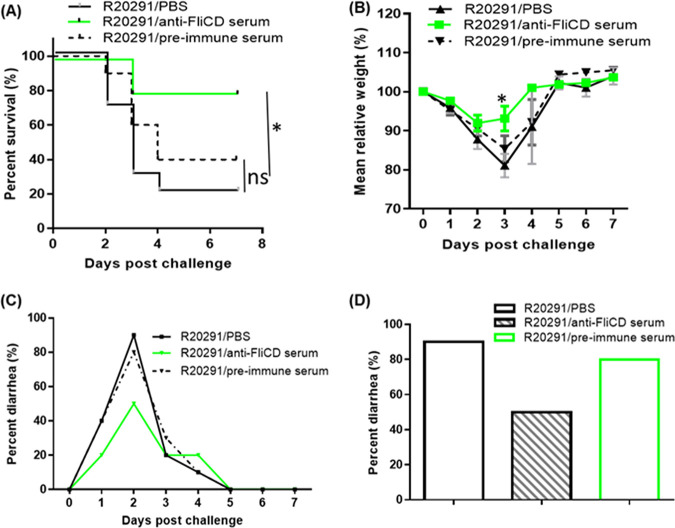
To elucidate how FliCD-immunized mice gain resistance to CDI, we tested whether the hyperimmune serum (anti-FliCD serum) provides protection against infection. Anti-FliCD serum (IgG titer of 107) was collected from mice that were immunized 4 times with 10 μg FliCD. 3 groups of mice (n = 10) were i.p. administered 400 μL of anti-FliCD serum, preimmune serum, or PBS 4 h before infection with C. difficile R20291 spores (106) in the mouse model of infection. The majority of the PBS and preimmune serum groups developed diarrhea (90% in the PBS group and 80% in the preimmune serum group) (Fig. 7C and D) and significant weight loss (Fig. 7B), with survival rates of 20% and 40% being observed in the PBS group and the preimmune serum group, respectively (Fig. 7A), whereas the mice that were administered 400 μL of hyperimmune serum developed much less severe disease symptoms, including less weight loss (Fig. 7B) and a lower diarrhea rate (50%) (Fig. 7C) as well as a significantly higher survival rate (80%) (Fig. 7A).
FIG 7.
Anti-FliCD hyperimmune serum provides mice with significant protection against infection by C. difficile strain R20291. Three groups of mice (n = 10) were i.p. administered 400 μL of anti-FliCD sera (IgG titer of 107), preimmune sera, or PBS 4 h before infection with C. difficile R20291 (107 spores) in the mouse model of infection. (A) Kaplan-Meier survival plots (P = 0.0079 between groups R20291/PBS and R20291/anti-FliCD; P = 0.078 between groups R20291/PBS and R20291/preimmune serum). (B) Mean relative weight of all surviving mice (up to the day of death). The data are presented as the mean ± SEM. (C and D) Frequency of diarrhea (*, P < 0.05).
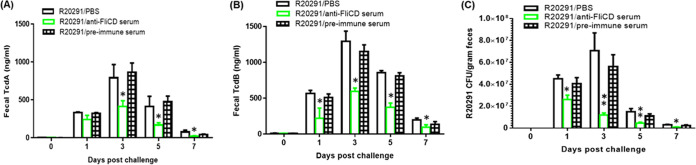
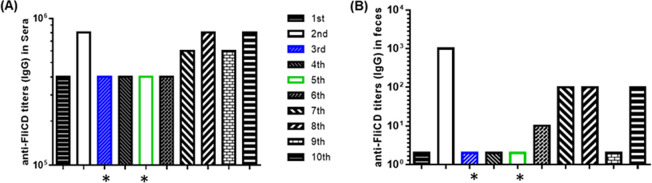
The mice that were administered the anti-FliCD serum excreted significantly smaller amounts of TcdA (Fig. 8A) and TcdB (Fig. 8B) in the feces that were collected on days 1 to 7 postinfection, compared to those of the PBS and the preimmune sera groups. The fecal samples that were collected from the mice that were administered the anti-FliCD serum contained significantly fewer R20291 spores, compared to the PBS and the preimmune serum groups (Fig. 8C). High levels of anti-FliCD antibodies were also detected in the sera and the feces that were collected from the mice that were administered the anti-FliCD serum (Fig. 9). The third and fifth mice in this group, whose weight loss values were 20%, were among those with the lowest anti-FliCD titers in the sera and feces, affirming the protection of the anti-FliCD antibodies against CDI in the mice.
FIG 8.
Anti-FliCD titers in sera and feces from mice administered with anti-FliCD hyperimmune serum and challenged with R20291 spores. (A) Sera and (B) feces were collected on day 4 postinfection, and the anti-FliCD IgG titers were measured via standard ELISA. The data are presented as the mean ± SEM (n = triplicate). (*, the sera and feces of the moribund mice were collected before the mice were euthanized).
FIG 9.
Anti-FliCD titers in the sera and feces from mice receiving anti-FliCD hyperimmune serum and being challenged with R20291 spores. (A) Sera and (B) feces were collected on day 4 postinfection, and the anti-FliCD IgG titers were measured via standard ELISA.The data are presented as the mean ± SEM (n = triplicate). (*, the sera and feces of the moribund mice were collected before the mice were euthanized).
Anti-FliCD serum inhibits the binding of C. difficile to HCT8 cells.
When the anti-FliCD serum was diluted 1 to 50 or 1 to 100 in the cell medium, the adherence rate of the C. difficile R20291 vegetative cells to the HCT8 cells significantly decreased, compared with that of HCT8 cells treated with preserum (5.0 ± 0.7% or 7.5 ± 0.9% versus 13.6 ± 0.7%). When the serum was diluted 1 to 500, the adherence rate decreased to 10.9 ± 0.6%, but this decrease is not statistically significant (Fig. 10).
FIG 10.
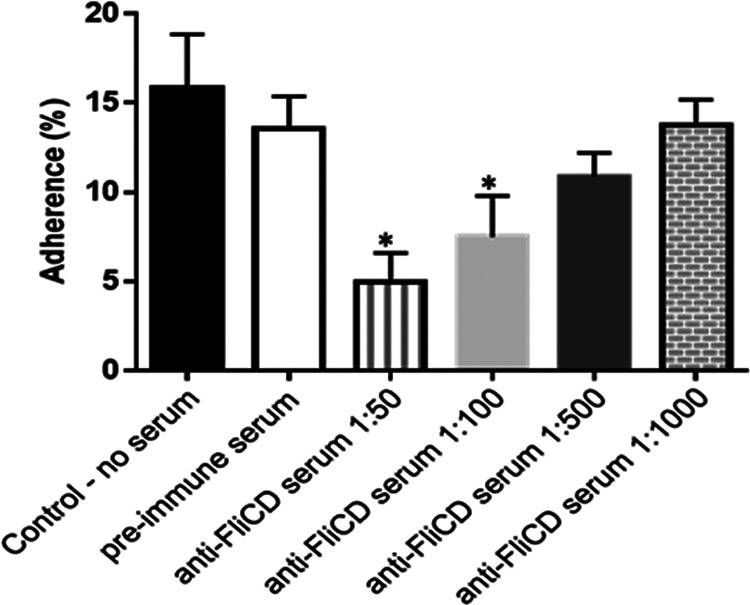
Anti-FliCD serum inhibits the adhesion of C. difficile to HCT8 cells. C. difficile R20291 vegetative cells (1.5 × 106) in 100 μL BHI were preincubated with anti-FliCD hyperimmune serum (IgG titer 107) at different serum dilutions (1/50, 1/100, 1/500 and 1/1,000) or with PBS/preimmune serum (1/50) for 30 min before being added to confluent HCT-8 cells (1 × 105/well) in a 24-well plate in an anaerobic chamber. The plate was incubated at 37°C for 100 min. After incubation, the cell-R20291 mixture was washed three times with PBS via centrifugation to collect unbound R20291 cells. The percentage of R20291 adhesion was calculated using the following formula: (initial CFU/mL − unbound CFU/mL)/ initial CFU/mL. The experiments were independently repeated thrice. The data are presented as the mean ± SEM. (*, P < 0.05, versus treatment with preimmune serum).
DISCUSSION
Both FliC and FliD may play an important role in the cell adherence, colonization, invasion and pathogenicity of C. difficile (15). In this study, we found that both FliCD immunizations and hyperimmune anti-FliCD serum protected mice against C. difficile infections in a mouse model (Fig. 5 and Fig. 7) and decreased the C. difficile spore and toxin levels observed in the feces of mice that were challenged with C. difficile spores. These results indicate that anti-FliCD antibodies from active or passive immunizations may either decrease C. difficile toxin production and sporulation on a per cell basis or reduce C. difficile colonization. Our further analyses support the later scenario. In vitro, we found that anti-FliCD serum inhibited the adherence of C. difficile R20291 vegetative cells to HCT8 cells (Fig. 10). This was also supported by our in vivo experiment. We found that the anti-FliCD titers in the sera and feces that were collected from the C. difficile-induced moribund mice were among the mice with lower titers (Fig. 9). However, further experiments are needed to determine whether FliCD antibodies can affect C. difficile toxin production and sporulation.
Interestingly, another study showed that both the fliC and fliD mutant strains of CD630Δerm lost flagella but adhered to human intestine-derived Caco-2 cells better than did the wild-type strain, and they were also more virulent in hamsters (17), which might be partially caused by the relatively increased toxin production in the mutant strains. Their data also suggest that neither FliC nor FliD is required for the cecal colonization of hamsters. More work is clearly needed to further understand the phenotypic differences between a complete loss of flagella by gene silence and the direct binding of flagella proteins by antibodies.
In addition, it has been reported that C. difficile FliC can activate TLR5 in vitro (25–27), indicating that it may stimulate the TLR5 pathway in vivo and thereby provide the mice with additional protection by directly or indirectly affecting C. difficile colonization and toxin production, as has been demonstrated by another study, which showed that the activation of TLR5 pathway by Salmonella enterica serovar Typhimurium flagellin is critical in protecting mice against CDI (28). Finally, it is reported that flagella production is phase variable in C. difficile (29). Antibodies generated against flagellar components presumably may not affect this flagella-off subpopulation.
In summary, we constructed a fusion protein vaccine, namely, FliCD, and showed its potent efficacy as a new vaccine candidate in the mouse model of CDI. Our data showed that not only did FliCD fusion protein represent an effective vaccine candidate but also anti-FliCD serum may represent an alternative therapy against CDI.
MATERIALS AND METHODS
Homology analysis of FliC and FliD.
C. difficile strains (Table 1) were chosen for analysis based on the results of previous studies (30–42). The genomes of each strain were accessed through GenBank (National Center for Biotechnology Information) or the Enterobase Clostridioides database (43). The amino acid sequences for FliC and FliD were mined from each genome before the performance of MUSCLE alignments in MegaX software (44) using the default parameters. Maximum likelihood phylogenetic trees were constructed in MegaX with 500 bootstrap replicates. The cluster patterns of the phylogenetic trees were used to order the sequences of FliC or FliD for a second MUSCLE alignment on the MPI Bioinformatics Toolkit server (45, 46), as this application produces an output file that is suitable for visualization using Jalview (47). Jalview calculates conservation scores for MUSCLE alignments according to a previously defined algorithm (48) that assesses both the amino acid identity as well as the physicochemical properties of the amino acids at a given position to produce a score between 0 (no similarities) and 11 (identical amino acids).
Animals.
The animal studies followed the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and were approved by the Institute Animal Care and Use Committee (IACUC) at the University of South Florida. Wild-type C57BL/6 mice were purchased from Charles River Laboratories.
Expression and purification of the recombinant fusion protein FliCD.
Gene sequences encoding FliC and FliD from C. difficile R20291 (49) were bridged with a linker (ggt ggc tct ggt) sequence, synthesized by Geneart (Germany), and cloned between the BsrGI and EagI restriction sites of pHis1525 (24). FliCD was expressed in B. megaterium and purified as described previously (50).
Preparation of C. difficile spores.
The sporulation of the C. difficile R20291 strain was induced in Clospore medium as described previously (51). Briefly, an overnight 20 mL C. difficile culture in Columbia Broth was inoculated into 500 mL of Clospore medium and incubated for 1 to 2 weeks at 37°C in an anaerobic incubator. The spore suspension was centrifuged at 10,000 × g for 20 min, and the pellet was washed 5 times with sterile water and suspended in 10 mL of sterile ddH2O. The spore suspension was layered onto the top of 10 mL of 50% (wt/vol) sucrose in water in a 15 mL tube. The gradient was centrifuged at 3,200 × g for 20 min, after which the spore pellet at the bottom was washed five times with ddH2O to remove the sucrose and was resuspended in sterile ddH2O. The spore preparations were >99% pure (52), and the spore concentration was determined via serial dilution on TCCA or BHI plates.
Immunization and mouse model of CDI.
Female C57BL/6 mice were housed under the same conditions at a seminatural light cycle of 14 h:10 h (light:dark) in a specific pathogen-free (SPF) environment. 3 groups of mice (n = 10) were immunized via the i.p. route with 10 μg or 20 μg FliCD or PBS with alum as an adjuvant 3 times at 12-day intervals. Sera were collected, and anti-FliCD IgG titers were determined via ELISA. 7 days after the final immunization, the mice were given drinking water containing a mixture of 5 antibiotics, including kanamycin (40 mg/kg), gentamicin (3.5 mg/kg), colistin (4.2 mg/kg), metronidazole (21.5 mg/kg), and vancomycin (4.5 mg/kg) for 4 days, and they then received autoclaved water for 2 days, and this was followed by the i.p injection of a dose of clindamycin (10 mg/kg) before a challenge with 106 C. difficile R20291 spores/mouse via oral gavage as described previously (53). After infection, the mice were monitored daily for a week for survival, weight changes, diarrhea, and other symptoms of the disease. Diarrhea was defined as wet tails and loose or watery feces. The death count included the number of mice that died after infection and the number of mice that were euthanized when their weight loss was >20%.
Evaluation of anti-FliCD serum in the protection of mice against CDI.
Mice (n = 10) were immunized 4 times at 12-day intervals via the i.p. route with 10 μg of FliCD in PBS with alum as an adjuvant. 14 days after the fourth immunization, serum was collected and defined as hyperimmune anti-FliCD serum (IgG titer of 107). The mouse model of C. difficile infection was established as described above, except that the mice were challenged with 107 C. difficile R20291 spores. 4 hours prior to infection with spores, 400 μL of hyperimmune serum, preimmune serum, or PBS were administered (i.p.) to each mouse in 3 groups, respectively.
ELISA for anti-FliCD IgG.
ELISA was performed as previously described (50). Briefly, Costar 96-well ELISA plates were coated with 100 μL/well of FliCD (0.5 μg/mL) at 4°C overnight. Following the washing of the unbound material, the plates were blocked with 300 μL of blocking buffer (PBS + 5% dry milk) at room temperature for 2 h. After washing, 100 μL of 10-fold diluted sera or fecal samples were added into each well of the plates and were incubated for 1.5 h at room temperature. Following washing with PBS, 100 μL of mouse IgG-HRP (1:3,000) were added to each well and incubated for 30 min to 1 h. After a washing step with PBS, substrate TMB was added to allow for color development at room temperature for 5 to 30 min. The reaction was stopped via the addition of H2SO4 to each well, and the OD values at 450 nm were recorded using a spectrophotometer. The anti-toxin or anti-FliCD IgG titer of a given sample (a serum or fecal sample from an immunized mouse) is defined as the dilution factor at which the OD450nm is greater than or equal to twice that of the serum or fecal samples that were collected from mice before immunization.
Quantification of C. difficile spores in mouse feces.
Fecal samples were collected on days 0, 1, 3, 5, and 7 postinfection. 50 mg of feces were dissolved in 500 μL of sterile water for 16 h at 4°C, and they were then treated with 500 μL of absolute ethanol (Sigma-Aldrich) for 1 h at room temperature to kill vegetative cells. The samples were vortexed, serially diluted, and plated onto selective medium supplemented with taurocholate (0.1% wt/vol), cefoxitin (8 μg/mL), and d-cycloserine (250 μg/mL). The plates were incubated anaerobically at 37°C for 48 h. The colonies were counted. The data are expressed as CFU/gram of feces.
Quantitation of C. difficile toxins in mouse feces.
After a challenge with C. difficile spores, feces were collected and dissolved in PBS (0.1 g/mL) containing a protease inhibitor cocktail. Supernatants were collected after centrifugation and were used for the determination of the TcdA/TcdB concentrations via ELISA. Briefly, 96-well Costar microplates were coated with 100 μL of the anti-TcdA antibody (1 μg/mL) and the anti-TcdB antibody (1 μg/mL) overnight in phosphate-buffered saline (PBS) at 4°C. The next day, each well was blocked with 300 μL of blocking buffer (PBS + 5% dry milk) at RT for 2 h. Next, standards and samples were added to each well (100 μL) in duplicate and incubated for 90 min at 25°C. After another set of washings, HRP-chicken anti-C. difficile TcdA/TcdB (1:5,000 dilution in PBS, Gallus Immunotech, Shirley, MA) was added to the wells for 30 min at RT. A final set of 3-washing preceded the addition of the TMB microwell peroxidase substrate for 20 min at RT in the dark. The reaction was stopped with 2 M H2SO4, and the absorbance was measured using a plate reader at 450 nm.
Adhesion inhibition assays.
The adherence of C. difficile R20291 vegetative cells to human gut epithelial cells was assessed as described previously (54). Briefly, HCT-8 cells were grown to 95% confluence (1 × 105/well) in a 24-well plate and were then moved into an anaerobic chamber. This was followed by infection with 1.5 × 106 log phase R20291 vegetative cells at a multiplicity of infection (MOI) of 15:1. The plate was incubated at 37°C for 100 min in an anaerobic chamber. R20291 vegetative cells in 100 μL of BHI medium were preincubated with hyperimmune serum (IgG titer of 107) at different serum dilutions (1/50, 1/100, 1/500 and 1/1,000) for 30 min before being added to the cells. After incubation, the cell-C. difficile mixture was washed three times with 1×PBS via centrifugation at 800 × g for 1 min to remove any unbound R20291. The supernatants were collected after the centrifugation of each wash step to enumerate any R20291 that did not adhere to the cells. The R20291 colonies in the supernatant were enumerated on prereduced BHI agar. As a control, the R20291 strain was incubated with either PBS or preimmune sera (1/50), and adhesion assays were performed in triplicate. The percentage of R20291 adhesion was estimated using the following formula: (initial CFU/mL – eluted CFU/mL) / initial CFU/mL.
Statistical analysis.
Survival curves were analyzed via Kaplan-Meier with a log-rank test of significance. The data for comparisons between two groups were analyzed using Student’s unpaired t tests for statistical significance. The data for comparisons between more than two groups were analyzed using a one-way analysis of variance (ANOVA) with a post hoc analysis via Bonferroni tests. The data are expressed as the mean ± the standard error of the mean (SEM). Differences were considered to be statistically significant if P < 0.05 (*). All statistical analyses were performed using the GraphPad Prism software package.
Data availability.
The accession numbers for sequences used in Table 1 are listed as follows: CLO_AA6882AA, NZ_OEZL00000000.1, NZ_OEZE00000000.1, NZ_OEZZ00000000.1, NZ_OEZV00000000.1, NZ_OEZY00000000.1, NZ_OEZH00000000.1, NZ_AHJJ00000000.1, NZ_JAGKRT000000000.1, NZ_JANFNF000000000.1.
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health (grants R21-AI113470, R01-AI132711, and R01-AI149852) and the Anthony Gagliardi Foundation. We thank Soumyadeep Chakraborty and Anastasia Tomatsidou on our team for their critical comments.
The manuscript was written through the contributions of all authors. X.S. conceptualized and designed the project as well as participated in the data analysis. S.W., X.J., J.H., K.Z., Z.D., H.M.L.W.P., and S.Z. performed experiments and data analysis. S.W. and X.S. wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Xingmin Sun, Email: sun5@usf.edu.
Nancy E. Freitag, University of Illinois at Chicago
REFERENCES
- 1.Curry SR. 2017. Clostridium difficile. Clin Lab Med 37:341–369. 10.1016/j.cll.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Moreno MA, Furtner F, Rivara FP. 2013. Clostridium difficile: a cause of diarrhea in children. JAMA Pediatr 167:592–592. 10.1001/jamapediatrics.2013.2551. [DOI] [PubMed] [Google Scholar]
- 3.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713. 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 4.Guh AY, Kutty PK. 2018. Clostridioides difficile infection. Ann Intern Med 169:ITC49–ITC64. 10.7326/AITC201810020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouwknegt M, van Dorp S, Kuijper E. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:2368–2368. 10.1056/NEJMc1505190. [DOI] [PubMed] [Google Scholar]
- 6.Lessa FC, Gould CV, McDonald LC. 2012. Current status of Clostridium difficile infection epidemiology. Clinical Infectious Diseases 55:S65–S70. 10.1093/cid/cis319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marra AR, Perencevich EN, Nelson RE, Samore M, Khader K, Chiang H-Y, Chorazy ML, Herwaldt LA, Diekema DJ, Kuxhausen MF, Blevins A, Ward MA, McDanel JS, Nair R, Balkenende E, Schweizer ML. 2020. Incidence and outcomes associated with Clostridium difficile infections: a systematic review and meta-analysis. JAMA Netw Open 3:e1917597. 10.1001/jamanetworkopen.2019.17597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drekonja DM, Butler M, MacDonald R, Bliss D, Filice GA, Rector TS, Wilt TJ. 2011. Comparative effectiveness of Clostridium difficile treatments a systematic review. Ann Intern Med 155:839–847. 10.7326/0003-4819-155-12-201112200-00007. [DOI] [PubMed] [Google Scholar]
- 9.Bagdasarian N, Rao K, Malani PN. 2015. Diagnosis and treatment of Clostridium difficile in adults: a systematic review. JAMA 313:398–408. 10.1001/jama.2014.17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao K, Malani PN. 2020. Diagnosis and treatment of Clostridioides (Clostridium) difficile infection in adults in 2020. JAMA 323:1403–1404. 10.1001/jama.2019.3849. [DOI] [PubMed] [Google Scholar]
- 11.Lillehoj EP, Kim BT, Kim KC. 2002. Identification of Pseudomonas aeruginosa flagellin as an adhesin for Muc1 mucin. Am J Physiol Lung Cell Mol Physiol 282:L751–L756. 10.1152/ajplung.00383.2001. [DOI] [PubMed] [Google Scholar]
- 12.Blair DF, Dutcher SK. 1992. Flagella in prokaryotes and lower eukaryotes. Curr Opin Genet Dev 2:756–767. 10.1016/s0959-437x(05)80136-4. [DOI] [PubMed] [Google Scholar]
- 13.Homma M, Kutsukake K, Iino T, Yamaguchi S. 1984. Hook-associated proteins essential for flagellar filament formation in Salmonella typhimurium. J Bacteriol 157:100–108. 10.1128/jb.157.1.100-108.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Postel S, Deredge D, Bonsor DA, Yu X, Diederichs K, Helmsing S, Vromen A, Friedler A, Hust M, Egelman EH, Beckett D, Wintrode PL, Sundberg EJ. 2016. Bacterial flagellar capping proteins adopt diverse oligomeric states. Elife 5. 10.7554/eLife.18857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tasteyre A, Barc MC, Collignon A, Boureau H, Karjalainen T. 2001. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect Immun 69:7937–7940. 10.1128/IAI.69.12.7937-7940.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tasteyre A, Karjalainen T, Avesani V, Delmée M, Collignon A, Bourlioux P, Barc MC. 2001. Molecular characterization of fliD gene encoding flagellar cap and its expression among Clostridium difficile isolates from different serogroups. J Clin Microbiol 39:1178–1183. 10.1128/JCM.39.3.1178-1183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dingle TC, Mulvey GL, Armstrong GD. 2011. Mutagenic analysis of the Clostridium difficile flagellar proteins, FliC and FliD, and their contribution to virulence in hamsters. Infect Immun 79:4061–4067. 10.1128/IAI.05305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghose C, Eugenis I, Sun X, Edwards AN, McBride SM, Pride DT, Kelly CP, Ho DD. 2016. Immunogenicity and protective efficacy of recombinant Clostridium difficile flagellar protein FliC. Emerging Microbes & Infections 5:1–10. 10.1038/emi.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karpiński P, Wultańska D, Piotrowski M, Brajerova M, Mikucka A, Pituch H, Krutova M. 2022. Motility and the genotype diversity of the flagellin genes fliC and fliD among Clostridioides difficile ribotypes. Anaerobe 73:102476. 10.1016/j.anaerobe.2021.102476. [DOI] [PubMed] [Google Scholar]
- 20.Tasteyre A, Karjalainen T, Avesani V, Delmée M, Collignon A, Bourlioux P, Barc MC. 2000. Phenotypic and genotypic diversity of the flagellin gene (fliC) among Clostridium difficile isolates from different serogroups. J Clin Microbiol 38:3179–3186. 10.1128/JCM.38.9.3179-3186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevenson E, Minton NP, Kuehne SA. 2015. The role of flagella in Clostridium difficile pathogenicity. Trends Microbiol 23:275–282. 10.1016/j.tim.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Yonekura K, Maki-Yonekura S, Namba K. 2001. Structure analysis of the flagellar cap–filament complex by electron cryomicroscopy and single-particle image analysis. J Struct Biol 133:246–253. 10.1006/jsbi.2000.4345. [DOI] [PubMed] [Google Scholar]
- 23.Kutsukake K. 1994. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol Gen Genet 243:605–612. 10.1007/BF00279569. [DOI] [PubMed] [Google Scholar]
- 24.Malten M, Hollmann R, Deckwer W-D, Jahn D. 2005. Production and secretion of recombinant Leuconostoc mesenteroides dextransucrase DsrS in Bacillus megaterium. Biotechnol Bioeng 89:206–218. 10.1002/bit.20341. [DOI] [PubMed] [Google Scholar]
- 25.Yoshino Y, Kitazawa T, Ikeda M, Tatsuno K, Yanagimoto S, Okugawa S, Yotsuyanagi H, Ota Y. 2013. Clostridium difficile flagellin stimulates toll-like receptor 5, and toxin B promotes flagellin-induced chemokine production via TLR5. Life Sci 92:211–217. 10.1016/j.lfs.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 26.Ghose C, Verhagen JM, Chen X, Yu J, Huang Y, Chenesseau O, Kelly CP, Ho DD. 2013. Toll-like receptor 5-dependent immunogenicity and protective efficacy of a recombinant fusion protein vaccine containing the nontoxic domains of Clostridium difficile toxins A and B and Salmonella enterica serovar typhimurium flagellin in a mouse model of Clostridium difficile disease. Infect Immun 81:2190–2196. 10.1128/IAI.01074-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batah J, Kobeissy H, Bui Pham PT, Denève-Larrazet C, Kuehne S, Collignon A, Janoir-Jouveshomme C, Marvaud J-C, Kansau I. 2017. Clostridium difficile flagella induce a pro-inflammatory response in intestinal epithelium of mice in cooperation with toxins. Sci Rep 7:3256. 10.1038/s41598-017-03621-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarchum I, Liu M, Lipuma L, Pamer EG. 2011. Toll-like receptor 5 stimulation protects mice from acute Clostridium difficile colitis. Infect Immun 79:1498–1503. 10.1128/IAI.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trzilova D, Warren MAH, Gadda NC, Williams CL, Tamayo R. 2022. Flagellum and toxin phase variation impacts intestinal colonization and disease development in a mouse model of Clostridioides difficile infection. Gut Microbes 14:2038854. 10.1080/19490976.2022.2038854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol 10:R102–15. 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groß U, Brzuszkiewicz E, Gunka K, Starke J, Riedel T, Bunk B, Spröer C, Wetzel D, Poehlein A, Chibani C, Bohne W, Overmann J, Zimmermann O, Daniel R, Liesegang H. 2018. Comparative genome and phenotypic analysis of three Clostridioides difficile strains isolated from a single patient provide insight into multiple infection of C. difficile. BMC Genomics 19:1–14. 10.1186/s12864-017-4368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HMB, Quail MA, Rance R, Brooks K, Churcher C, Harris D, Bentley SD, Burrows C, Clark L, Corton C, Murray V, Rose G, Thurston S, van Tonder A, Walker D, Wren BW, Dougan G, Parkhill J. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci USA 107:7527–7532. 10.1073/pnas.0914322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tasteyre A, Barc M-C, Karjalainen T, Dodson P, Hyde S, Bourlioux P, Borriello P. 2000. A Clostridium difficile gene encoding flagellin. Microbiology 146:957–966. 10.1099/00221287-146-4-957. [DOI] [PubMed] [Google Scholar]
- 34.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang H, Holden MTG, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38:779–786. 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 35.Cairns MD, Preston MD, Lawley TD, Clark TG, Stabler RA, Wren BW. 2015. Genomic epidemiology of a protracted hospital outbreak caused by a toxin A-negative Clostridium difficile sublineage PCR ribotype 017 strain in London, England. J Clin Microbiol 53:3141–3147. 10.1128/JCM.00648-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riedel T, Wetzel D, Hofmann JD, Plorin SPEO, Dannheim H, Berges M, Zimmermann O, Bunk B, Schober I, Spröer C, Liesegang H, Jahn D, Overmann J, Groß U, Neumann-Schaal M. 2017. High metabolic versatility of different toxigenic and non-toxigenic Clostridioides difficile isolates. Int J Med Microbiol 307:311–320. 10.1016/j.ijmm.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 37.Li CHJ, Zhu D, Meng X, Chakraborty S, Harmanus C, Wang S, Peng Z, Smits WK, Wu A, Sun X. 2022. Genomic and phenotypic characterization of a Clostridioides difficile strain of the epidemic ST37 type from China.
- 38.Depitre C, Delmee M, Avesani V, L'Haridon R, Roels A, Popoff M, Corthier G. 1993. Serogroup F strains of Clostridium difficile produce toxin B but not toxin A. J Med Microbiol 38:434–441. 10.1099/00222615-38-6-434. [DOI] [PubMed] [Google Scholar]
- 39.Soehn F, Wagenknecht-Wiesner A, Leukel P, Kohl M, Weidmann M, von Eichel-Streiber C, Braun V. 1998. Genetic rearrangements in the pathogenicity locus of Clostridium difficile strain 8864–implications for transcription, expression and enzymatic activity of toxins A and B. Mol Gen Genet 258:222–232. 10.1007/s004380050726. [DOI] [PubMed] [Google Scholar]
- 40.Janezic S, Dingle K, Alvin J, Accetto T, Didelot X, Crook DW, Lacy DB, Rupnik M. 2020. Comparative genomics of Clostridioides difficile toxinotypes identifies module-based toxin gene evolution. Microb Genom 6. 10.1099/mgen.0.000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brouwer MS, et al. 2012. Draft genome sequence of the nontoxigenic Clostridium difficile strain CD37. Am Soc Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S, Heuler J, Wickramage I, Sun X. 2022. Genomic and phenotypic characterization of the nontoxigenic Clostridioides difficile strain CCUG37785 and demonstration of its therapeutic potential for the prevention of C. difficile infection. Microbiol Spectr 10:e01788-21. 10.1128/spectrum.01788-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frentrup M, et al. 2020. A publicly accessible database for genome sequences supports tracing of transmission chains and epidemics. [DOI] [PMC free article] [PubMed]
- 44.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V. 2018. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430:2237–2243. 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 46.Gabler F, Nam S-Z, Till S, Mirdita M, Steinegger M, Söding J, Lupas AN, Alva V. 2020. Protein sequence analysis using the MPI Bioinformatics Toolkit. Curr Protoc Bioinformatics 72:e108. 10.1002/cpbi.108. [DOI] [PubMed] [Google Scholar]
- 47.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Livingstone CD, Barton GJ. 1993. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput Appl Biosci 9:745–756. 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- 49.Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP. 2012. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl Environ Microbiol 78:4683–4690. 10.1128/AEM.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y-K, Yan Y-X, Kim HB, Ju X, Zhao S, Zhang K, Tzipori S, Sun X. 2015. A chimeric protein comprising the glucosyltransferase and cysteine proteinase domains of toxin B and the receptor binding domain of toxin A induces protective immunity against Clostridium difficile infection in mice and hamsters. Hum Vaccin Immunother 11:2215–2222. 10.1080/21645515.2015.1052352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perez J, Springthorpe VS, Sattar SA. 2011. Clospore: a liquid medium for producing high titers of semi-purified spores of Clostridium difficile. J AOAC Int 94:618–626. 10.1093/jaoac/94.2.618. [DOI] [PubMed] [Google Scholar]
- 52.Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192:4983–4990. 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X, Katchar K, Goldsmith JD, Nanthakumar N, Cheknis A, Gerding DN, Kelly CP. 2008. A mouse model of Clostridium difficile-associated disease. Gastroenterology 135:1984–1992. 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 54.Joshi LT, Phillips DS, Williams CF, Alyousef A, Baillie L. 2012. Contribution of spores to the ability of Clostridium difficile to adhere to surfaces. Appl Environ Microbiol 78:7671–7679. 10.1128/AEM.01862-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The accession numbers for sequences used in Table 1 are listed as follows: CLO_AA6882AA, NZ_OEZL00000000.1, NZ_OEZE00000000.1, NZ_OEZZ00000000.1, NZ_OEZV00000000.1, NZ_OEZY00000000.1, NZ_OEZH00000000.1, NZ_AHJJ00000000.1, NZ_JAGKRT000000000.1, NZ_JANFNF000000000.1.