ABSTRACT
Antimicrobial resistance (AMR) of bacterial pathogens, including enterococci, is a global concern, and plasmids are crucial for spreading and maintaining AMR genes. Plasmids with linear topology were identified recently in clinical multidrug-resistant enterococci. The enterococcal linear-form plasmids, such as pELF1, confer resistance to clinically important antimicrobials, including vancomycin; however, little information exists about their epidemiological and physiological effects. In this study, we identified several lineages of enterococcal linear plasmids that are structurally conserved and occur globally. pELF1-like linear plasmids show plasticity in acquiring and maintaining AMR genes, often via transposition with the mobile genetic element IS1216E. This linear plasmid family has several characteristics enabling long-term persistence in the bacterial population, including high horizontal self-transmissibility, low-level transcription of plasmid-carried genes, and a moderate effect on the Enterococcus faecium genome alleviating fitness cost and promoting vertical inheritance. Combining all of these factors, the linear plasmid is an important factor in the spread and maintenance of AMR genes among enterococci.
KEYWORDS: Enterococcus faecium, conjugation, linear plasmid, multidrug resistance, vancomycin resistance
INTRODUCTION
Enterococcus faecium, one of “the Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species (ESKAPE) bugs,” is a major cause of nosocomial infection and frequently presents with multidrug resistance (1). Vancomycin resistance in enterococci is a serious clinical problem owing to the paucity of treatment options. Vancomycin-resistant E. faecium (VREfm) is the major species among vancomycin-resistant enterococci (VRE) (2). The propagation of VREfm involves the spread of both clones (i.e., clonal complex 17 [CC17]) and mobile genetic elements (MGEs), such as plasmids containing vancomycin-resistant genes (3, 4). Particularly for E. faecium, conjugative plasmids are the most important reservoirs and vehicles for vancomycin resistance genes (5, 6). A comprehensive understanding of enterococcal plasmids, which confer various ecological characteristics, is therefore of great clinical value.
In 2019, we reported an enterococcal linear-form plasmid (pELF) harboring vancomycin resistance gene clusters from a Japanese VRE isolate (7). In addition to the nosocomial transmission of VRE through a pELF1-like plasmid, pELF2, reports have confirmed the importance of the linear plasmid in the regional spread of VRE (8, 9). The host range is assumed to be broad, and the self-transmissibility of pELF to enterococci has been confirmed experimentally and clinically (7, 8). The efficient propagation of drug resistance genes complicates both the treatment and control of disease transmission; thus, to improve clinical outcomes, it is essential that we elucidate the dynamics of plasmid spread. However, the reports and epidemiological data of pELF1-like plasmids are scant, with no information on their biological effects on host enterococci. Here, we reported the detection of several pELF1-like plasmid-carrying strains from clinical isolates in Japan. Along with pELF1-like plasmid resources in a public database, we performed an in-depth molecular epidemiology analysis based on whole-genome sequencing (WGS). Furthermore, we analyzed the impact of pELF1-like plasmids on host enterococcus fitness at the transcriptional level. This study provides an integrated characterization of enterococcal linear plasmids at the epidemiologic, phenomic, and transcriptomic levels.
RESULTS
Prevalence of pELF1-like plasmids in clinical enterococcal isolates in Japan.
We analyzed a total of 1,769 strains of VRE (385 strains) and vancomycin-susceptible enterococci (VSE; 1,384 strains) (Table 1). VRE strains were isolated in Japan since the early 2000s, whereas VSE strains were isolated from October 2009 to May 2018 at a single core medical institute in Gunma Prefecture.
TABLE 1.
Enterococcal strains for detection of pELF1-like plasmidsa
| Enterococcus species | No. of VRE strains by van genotype |
No. of VSE strainsb | No. of isolates, % with pELF1-like plasmids | ||||
|---|---|---|---|---|---|---|---|
| vanA | vanB | vanC | vanD | vanM | |||
| E. faecium | 51 (8) | 240 (5) | 9 | 2 | 324 (3) | 626 (16), 2.6% | |
| E. faecalis | 11 | 53 | 4 | 958 | 1,026 | ||
| E. avium | 1 | 1 | 31 | 33 | |||
| E. gallinarum | 10 | 5 | 15 | ||||
| E. raffinosus | 36 | 36 | |||||
| E. casseliflavus | 3 | 25 | 28 | ||||
| E. durans/E. hirae | 5 | 5 | |||||
| Total no. of isolates | 63 (8) | 294 (5) | 13 | 13 | 2 | 1,384 (3) | 1,769 (16), 0.9% |
Parentheses indicate the number of strains carrying pELF1-like linear plasmids.
Strains that do not show vancomycin resistance as a phenotype.
The identification of the plasmids was performed via colony PCR using primers designed for paired-end detection of pELF1-like plasmids (see Table S1 in the supplemental material). Through PCR screening, pELF1-like plasmids were identified in 16 strains (0.9% of the total strains), of which all were E. faecium (2.6%) (Table 1); also, 3 of the 1,384 VSE strains (0.2%) and 13 of the 385 VRE strains (3.4%) harbored pELF1-like plasmids. Of the 13 VRE strains, 8 were of the VanA type and 5 were of the VanB type.
Characteristics of the pELF1-like plasmid-carrying strains.
We characterized the 18 pELF1-like plasmid-carrying strains, including AA708 (pELF1) and KUHS13 (pELF2) (Table 2), based on the available epidemiological information. VRE strains carrying the pELF1-like plasmid were detected nationwide (see Fig. S1A in the supplemental material). Among the 13 pELF1-like plasmids carrying VRE strains, 6 strains (JHP9, JHP10, JHP35, JHP36, JHP38, and JHP80) had been isolated in the early 2000s. The pELF1-like plasmid was also detected in the seven VRE strains isolated after 2010. Three pELF1-like plasmid-carrying VSE strains were detected in 2014 and 2015, and two of these strains (GK923 and GK961) were detected on the same patient on different days.
TABLE 2.
The Japanese clinical E. faecium strains carrying pELF1-like plasmids
| Strain | van genotype | Date of isolation | Prefecture of origin | Source | Size of chromosome (bp) | GC content (%) | MLST (atpA-ddl-gdh-purk-gyd-pstS-adk) | Location of van | Resistance genes in the genome | Replicon type of the circular plasmids which the host possesseda | Virulence genesb | Accession no. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| JHP9 | vanA | 2000 | Chiba | Sputum | 2,835,106 | 38.2 | 16 (1-2-1-1-1-1-1) | Circular plasmid (45,504 bp, Rep_trans) | ant(6)-Ia, aph(3′)-III, dfrG, erm(B), msr(C), tet(M), VanHAX | Inc18 (repUS7), RepA_N (rep17, repUS15), Rep_trams (rep14) | acm, efaAfm, espfm, hylEfm | SAMD00466802 |
| JHP10 | vanA | 2000 | Chiba | Stool | 2,839,030 | 38.2 | 16 (1-2-1-1-1-1-1) | Circular plasmid (45,488 bp, RepA_N, Rep_trans) | ant(6)-Ia, aph(3′)-III, dfrG, erm(B), msr(C), tet(M), VanHAX | Inc18 (repUS7), RepA_N (rep17, repUS15) | acm, efaAfm, espfm, hylEfm | SAMD00466803 |
| JHP35 | vanA | 2002 | Aichi | Unknown | 2,813,406 | 38.1 | 1902 (pstS_null strain; 3-1-1-1-1-0-1) | Circular plasmid (43,555 bp, Inc18, Rep_trans) | aac(6′)-aph(2″), erm(B), msr(C), tet(M), VanHAX | Inc18 (rep1, rep2), RepA_N (repUS15), Rep3 (rep11a), Rep_trans (rep14a, rep14b, repUS43) | acm, efaAfm | SAMD00466804 |
| JHP36 | vanA | 2002 | Aichi | Unknown | 2,905,854 | 38.4 | 1902 (pstS_null strain; 3-1-1-1-1-0-1) | Circular plasmid (42,609 bp, Inc18, Rep_trans) | erm(B), msr(C), tet(M), VanHAX | Inc18 (rep1, rep2), RepA_N (repUS15), Rep3 (rep11a rep29), Rep_trans (rep14a, rep14b) | acm, efaAfm | SAMD00466805 |
| JHP38 | vanA | 2002 | Aichi | Unknown | 2,907,603 | 38.1 | 1902 (pstS_null strain; 3-1-1-1-1-0-1) | Circular plasmid (43,559 bp, Inc18, Rep_trans) | erm(B), msr(C), tet(M), VanHAX | Inc18 (rep1, rep2), RepA_N (repUS15), Rep3 (rep11a), Rep_trans (rep14a, rep14b) | acm, efaAfm | SAMD00466806 |
| JHP80 | vanB | 2001–2002 | Akita | Unknown | 2,921,286 | 38.4 | 17 (1-1-1-1-1-1-1) | Chromosome | ant(6)-Ia, aph(3′)-III, erm(B), msr(C), VanHBX | Inc18 (rep2), RepA_N(rep17) | acm, efaAfm, espfm | SAMD00466807 |
| AA55 | vanA | 2010 | Okinawa | Unknown | 2,843,325 | 38.0 | 2060 (15-1-1-1-1-20-82) | Circular plasmid (50,850 bp, RepA_N, Rep3) | aac(6′)-aph(2″), aph(3′)-III, erm(B), msr(C), VanHAX | Inc18 (rep2), RepA_N (rep17, repUS15), Rep3 (rep11a) | acm, efaAfm, espfm | SAMD00466808 |
| AA94 | vanB | 2011 | Okayama | Unknown | 2,749,946 | 38.3 | 18 (7-1-1-1-5-1-1) | Circular plasmid (321,364 bp, RepA_N) | ant(6)-Ia, dfrG, erm(B), msr(C), tet(L), VanHBX | RepA_N (rep17, repUS15), Rep_trans (rep14a) | acm, efaAfm | SAMD00466809 |
| AA96 | vanB | 2011 | Okayama | Unknown | 2,759,577 | 38.5 | 78 (15-1-1-1-1-1-1) | Circular plasmid (319,076 bp, RepA_N) | ant(6)-Ia, aph(3′)-III, erm(B), msr(C), tet(L), VanHBX | Inc18 (rep2), RepA_N (repUS15), Rep3 (rep18b), Rep1 (repUS12) | acm, efaAfm | SAMD00466810 |
| AA242 | vanA | 2013 | Tokyo | Unknown | 2,921,512 | 38.5 | 203 (15-1-1-1-1-20-1) | Linear plasmid | erm(A), msr(C), spc, VanHAX | Inc18 (rep2), RepA_N (rep17), Rep3 (rep11a) | acm, efaAfm, espfm | SAMD00466811 |
| AA290 | vanA | 2013 | Chiba | Unknown | 2,797,787 | 38.6 | 80 (9-1-1-1-12-1-1) | Circular plasmid (33,993 bp, RepA_N) | ant(6)-Ia, aph(3′)-III, erm(B), msr(C), VanHAX | RepA_N (rep17), Rep3 (rep11a, rep18a) | acm, efaAfm, espfm | SAMD00466812 |
| AA708c | vanA, vanM | 2014 | Saitama | Stool | 2,786,019 | 38.0 | 78 (15-1-1-1-1-1-1) | Linear plasmid | aac(6′)-aph(2″), ant(6)-Ia, aph(3′)-III, erm(B), msr(C), VanHAX, vanHMX | RepA_N (rep17, repUS15), Rep3 (rep11a) | acm, efaAfm, espfm | SAMD00190298 |
| AA610 | vanB | 2017 | Not available | Unknown | 2,821,420 | 38.7 | 2061 (15-1-1-1-1-195-1) | Circular plasmid (276,002 bp, RepA_N) | aph(3′)-III, erm(B), msr(C), VanHBX | Inc18 (rep2), RepA_N (rep17), Rep3 (rep11a) | acm, efaAfm, espfm | SAMD00466813 |
| KUHS13c | vanA | 2017 | Osaka | Stool | 2,802,719 | 38.1 | 17 (1-1-1-1-1-1-1) | Linear plasmid | ant(6)-Ia, dfrG, erm(A), erm(B), msr(C), spc, tet(L), VanHAX | RepA_N (repUS15) | acm, efaAfm, espfm | SAMD00190298 |
| AA818 | vanB | 2019 | Saitama | Hospital environment (button) | 2,825,547 | 38.4 | 17 (1-1-1-1-1-1-1) | Chromosome | aph(3′)-III, dfrG, erm(B), msr(C), tet(L), VanHBX | Inc18 (rep2), RepA_N (repUS15), Rep3 (rep11a), Rep_trans (repUS43), Rep1 (repUS12) | acm, efaAfm, espfm, hylEfm | SAMD00466814 |
| GK923 | - (VSE) | 2014 | Gunma | Urine | 2,836,077 | 38.1 | 78 (15-1-1-1-1-1-1) | aph(3′)-III, erm(B), msr(C), tet(M) | Inc18 (rep2), RepA_N (rep17, repUS15), Rep3 (rep11a) | acm, efaAfm, espfm, hylEfm | SAMD00466815 | |
| GK941 | - (VSE) | 2015 | Gunma | Urine | 2,833,359 | 38.1 | 78 (15-1-1-1-1-1-1) | aph(3′)-III, erm(B), msr(C), tet(M) | Inc18 (rep2), RepA_N (rep17, repUS15), Rep3 (rep11a) | acm, efaAfm, espfm, hylEfm | SAMD00466816 | |
| GK961 | - (VSE) | 2015 | Gunma | Urine | 2,834,237 | 38.1 | 78 (15-1-1-1-1-1-1) | aph(3′)-III, erm(B), msr(C), tet(M) | Inc18 (rep2), RepA_N (rep17, repUS15) | acm, efaAfm, espfm, hylEfm | SAMD00466817 |
The replicon type was examined by default settings using the PlasmidFinder (2.0.1).
The virulence genes were identified using the Virulence Finder (2.0).
These strains have already been reported previously.
Using WGS with multilocus sequence typing (MLST), we found that all 16 strains belonged to clonal complex 17 (CC17), which is the major type of clinical isolates and an important factor of nosocomial infections (Table 2) (3, 10).
Except for strain JHP35, all VanA-type VREs were resistant to both vancomycin (≥512 mg/L) and teicoplanin (≥32 mg/L), whereas all VanB-type VREs were relatively less resistant to vancomycin (32 to 64 mg/L) and susceptible to teicoplanin, preserving the characteristics of each resistant type (see Table S2 in the supplemental material). All strains were resistant to ampicillin, which is consistent with the characteristics of CC17 (3).
According to the PCR results, the presence of the pELF1-like plasmid was confirmed in all 16 strains (Table 3). Each pELF1-like plasmid was named using the strain name (e.g., pELF_JHP9). We examined the replicon of the plasmids using PlasmidFinder (Table 2) (11) and found that all the strains contained one or more circular plasmids, suggesting that pELF1-like plasmids coexist with various circular plasmids belonging to the Inc18-, RepA_N-, Rep3-, and rolling-circle replicating (RCR)-type (Rep_trans and Rep1) plasmids (Table 2). Regarding virulence genes, all strains had efaAfm and acm on their chromosomes, but none were identified on the pELF1-like plasmids (Table 2) (12–14).
TABLE 3.
The pELF1-like plasmids found in the Japanese clinical isolates of E. faecium
| Plasmid name | van genotype | Date of isolation | Prefecture of origin | Plasmid topology | Size of plasmid (bp) | GC content (%) | No. of CDSs | Resistance genes on the plasmid | Accession no. |
|---|---|---|---|---|---|---|---|---|---|
| pELF_JHP9 | vanA | 2000 | Chiba | Linear | 82,303 | 33.6 | 101 | AP026568 | |
| pELF_JHP10 | vanA | 2000 | Chiba | Linear | 82,303 | 33.6 | 101 | AP026575 | |
| pELF_JHP35 | vanA | 2002 | Aichi | Linear | 110,099 | 34.5 | 141 | tet(M), erm(B) | AP026581 |
| pELF_JHP36 | vanA | 2002 | Aichi | Linear | 88,997 | 33.3 | 113 | AP026588 | |
| pELF_JHP38 | vanA | 2002 | Aichi | Linear | 88,998 | 33.3 | 113 | AP026595 | |
| pELF_JHP80 | vanB | 2001–2002 | Akita | Linear | 88,683 | 33.3 | 109 | AP026602 | |
| pELF_AA55 | vanA | 2010 | Okinawa | Linear | 87,193 | 33.2 | 108 | AP026606 | |
| pELF_AA94 | vanB | 2011 | Okayama | Linear | 88,777 | 33.3 | 110 | AP026614 | |
| pELF_AA96 | vanB | 2011 | Okayama | Linear | 87,216 | 33.2 | 109 | AP026619 | |
| pELF_AA242 | vanA | 2013 | Tokyo | Linear | 132,791 | 33.8 | 162 | vanA, ant(9)-Ia, erm(A) | AP026630 |
| pELF_AA290 | vanA | 2013 | Chiba | Linear | 89,119 | 33.3 | 109 | AP026634 | |
| pELF1 | vanA, vanM | 2014 | Saitama | Linear | 143,316 | 34.8 | 184 | vanA, vanM, ,ant(6)-Ia, aph(3′)-III, erm(B) | LC495616 |
| pELF_AA610 | vanB | 2017 | Not applicable | Linear | 89,199 | 33.5 | 103 | AP026643 | |
| pELF2 | vanA | 2017 | Osaka | Linear | 108,102 | 33.7 | 131 | vanA, ant(9)-Ia, erm(A) | AP022343 |
| pELF_AA818 | vanB | 2019 | Saitama | Linear | 86,780 | 33.2 | 111 | AP026652 | |
| pELF_GK923 | 2014 | Gunma | Linear | 76,985 | 34.6 | 97 | AP026657 | ||
| pELF_GK941 | 2015 | Gunma | Linear | 76,985 | 34.6 | 97 | AP026662 | ||
| pELF_GK961 | 2015 | Gunma | Linear | 76,985 | 34.6 | 97 | AP026668 |
Comparison of pELF1-like plasmid genetic structures based on WGS.
pELF1-like plasmids are unique in that their opposing ends have different structures (known as a hybrid-type linear plasmid) (7). We investigated the structure of the terminal ends of all pELF1-like plasmids using WGS and found that the nucleotide sequences at both ends were highly conserved and presumed to have different structures, consistent with pELF1 (see Fig. S2 in the supplemental material).
Few reports describe the diversity of the genetic structure of pELF1-like plasmids. Accordingly, we conducted a search of the NCBI database based on the whole nucleotide sequence of pELF2, which is actually widespread in clinical practice and was thought to be a better model for understanding the characteristics of linear plasmids (8, 9). We selected plasmids with linear topology from enterococci as candidates and identified 14 pELF1-like plasmids with conserved terminal ends from a variety of countries, including Denmark, Norway, China, the United States, Switzerland, India, and the Netherlands (Table 4; Fig. S1B). The hosts of these 14 pELF1-like plasmids were all E. faecium, and all but E7196 belonged to CC17.
TABLE 4.
The putative pELF1-like plasmids and their E. faecium hosts found in the public databasea
| Plasmid name | Date of isolation | Country of origin | Size of chromosome (bp) | GC content of chromosome (%) | MLST | Plasmid topology | Size of plasmid (bp)b | GC content of linear plasmid (%) | No. of CDSs | Resistance genes | Accession no. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| pV24-2 | 2013 | Denmark | 2,720,495 | 38.2 | 80 | Linear | 97,469 | 33.5 | 121 | CP036153 | |
| pVVEswe-S2 | 2014 | Norway | 2,851,771 | 38.0 | 203 | Linear | 89,798 | 33.4 | 107 | CP041272 | |
| pVVEswe-R2 | 2014 | Norway | 2,851,579 | 38.0 | 203 | Linear | 89,846 | 33.4 | 107 | CP041263 | |
| pSRR24 | 2017 | China | 2,796,178 | 38.0 | 78 | Linear | 123,020 | 34.6 | 141 | vanM | CP038997 |
| UAMSEF_20 plasmid unnamed 2 | 2018 | United States | 2,912,170 | 38.1 | 80 | Linear | 81,272 | 33.5 | 100 | CP035662 | |
| UAMSEF_09 plasmid unnamed 2 | 2018 | United States | 2,912,202 | 38.1 | 80 | Linear | 81,204 | 33.5 | 99 | CP035656 | |
| pIHVA-EV0426-12 | 2018 | Japan | 2,762,621 | 38.1 | 78 | Linear | 109,115 | 33.8 | 136 | vanA, ant(9)-Ia, erm(A) | LC566215 |
| pELF_USZ | 2018 | Switzerland | 2,896,894 | 38.0 | 203 | Linear | 103,363 | 34.4 | 127 | OU016038.1 | |
| VB3025 plasmid unnamed 2 | 2019 | India | 2,855,454 | 38.0 | 80 | Linear | 142,820 | 34.0 | 171 | vanA, ant(6)-Ia, aph(3′)-III, erm(A), erm(B), optrA, tet(S) | CP040238 |
| E6988 plasmid 3 | Netherlands | 2,725,409 | 38.1 | 17 | Linear | 89,294 | 33.3 | 107 | LR135245 | ||
| E7098 plasmid 3 | Netherlands | 2,679,986 | 38.1 | 78 | Linear | 153,828 | 34.5 | 184 | vanA, ant(6)-Ia, aph(3′)-III, erm(B), lnu(B), tet(L) | LR135256 | |
| E7196 plasmid 2 | Netherlands | 2,697,633 | 38.2 | 656 | Linear | 164,074 | 34.2 | 191 | vanA, erm(B) | LR135271 | |
| E7471 plasmid 2 | Netherlands | 2,882,360 | 38.0 | 18 | Linear | 81,276 | 33.6 | 101 | LR135332 | ||
| E8414 plasmid 3 | Netherlands | 3,004,008 | 37.9 | 117 | Linear | 88,760 | 33.2 | 117 | LR135490 |
Plasmids with presumed linear topology were retrieved from the NCBI database.
The nucleotide sequence sizes of the plasmids with both ends modified with reference to pELF1 are described.
pELF1-like plasmid size was relatively diverse, with the shortest length being 76 kb for pELF_GK923, GK941, and GK961 and the longest being 164 kb for E7196 (Table 3 and 4). The minimum number of coding sequences (CDSs) was 97 for pELF_GK923, GK941, and GK961, and the maximum number was 191 for E7196.
We also found that the GC content of the pELF1-like plasmid was ~4% lower than that of the host genome (mean GC content, 33.9% versus 38.1%) (Table 2 to 4). Furthermore, pELF1-like plasmids exhibited lower GC content than circular plasmids harbored by multiple Enterococcus spp., including E. faecium (Mann-Whitney test, P < 0.01) (see Fig. S3 and Table S3 in the supplemental material).
We performed a core plasmid gene analysis with 18 plasmids, including pELF1 and pELF2, along with 14 putative pELF1-like plasmids confirmed by a public database search (Fig. 1; see Table S4 in the supplemental material) (9, 15). At a BLASTP result of 80% (considered to cover most core genes), a rapid large-scale prokaryote pangenome analysis (Roary) estimated the core genome to consist of 57 CDSs, which was 56.4% of the CDSs in pELF_JHP9 (Table S4). According to the annotation data, most of the 57 core genes were hypothetical proteins (see Table S5 in the supplemental material). Among the core genes, we identified some encoding replication proteins and several toxin-antitoxin (TA) system-related genes.
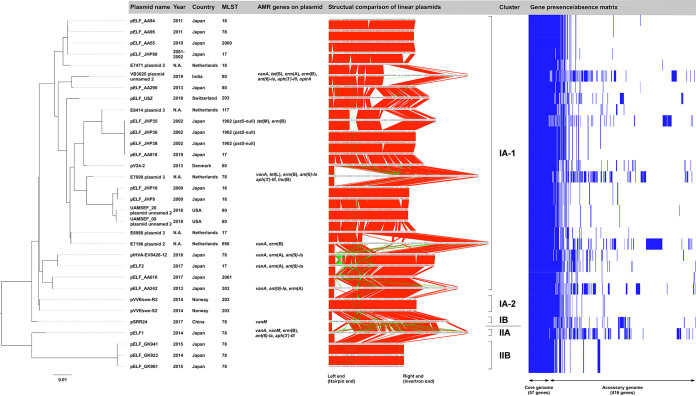
FIG 1.
Comprehensive phylogenetic analysis of pELF1-like plasmids. A core plasmid gene analysis was performed using Roary, and phylogenetic trees were generated in RAxML. Plasmid names, isolation years, countries, MLST, AMR gene possession status, and representative figures are shown along with the phylogenetic tree results. Synteny block lineages in the same orientation are shown in red, while those in the reverse orientation are shown in green. The gene presence/absence matrix was constructed using Phandango.
We performed a phylogenetic analysis based on the core gene sequences and identified several genetically distinct plasmid clusters (Fig. 1), which could be classified into two major groups, namely, cluster I and cluster II, and further classified into subclusters. Cluster I accounted for the majority of plasmids, among which cluster IA-1 comprised 25 of the 32 pELF1-like plasmids. Cluster II contained four pELF1-like plasmids, including pELF1, pELF_GK923, 941, and 961, which were harbored by three VSE strains. Cluster IA-1 accounted for pELF1-like plasmids detected worldwide, while cluster IA-2 accounted for the plasmids from Norway, cluster IB accounted for those from China, and cluster II accounted for those from Japan. pELF_AA94 and pELF_AA96 (harbored by strains detected in the same year in the same prefecture in Japan) belonged to the same cluster, and their synteny block lineage suggested a high degree of similarity (pairwise nucleotide identity, 98.2%); however, their host MLST types were different. This finding suggests the occurrence of plasmid transfer in clinical settings. Similarly, pELF_JHP36 and pELF_AA818 were highly similar plasmids (pairwise nucleotide identity, 97.5%), with different host MLST types. Their detection was 17 years apart, and they were detected in different regions of Japan. Likewise, the pELF_JHP9/pELF_JHP10 from Japan and UAMSEF_20 plasmid unnamed 2/UAMSEF_09 plasmid unnamed 2 from the United States belonged to the same cluster (pairwise nucleotide identity, 90.8%). These findings suggest that pELF1-like plasmids are structurally stable and occur worldwide.
We also found that the pELF1-like plasmids detected in Japan were clustered into different lineages (Fig. 1), although with highly conserved genetic structures. We could not confirm any major plasmid rearrangement, such as inversion, which would change the order of core genes (Fig. 1). Most of the synteny blocks displayed in reverse orientation were insertion sequences (ISs), with the only exception being the reverse orientation of the antimicrobial resistance (AMR) region containing the vanA gene cluster between pELF2 and pIHVA-EV0426-12 (Fig. 1). While the pELF1-like plasmid backbone was highly conserved, its diversity resulted from MGEs (Fig. 1 and 2).
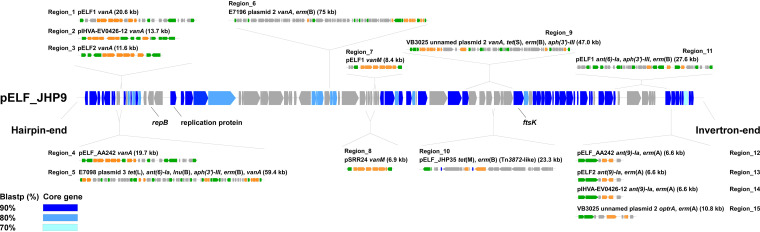
FIG 2.
Genetic structure of the core genes and AMR regions of pELF1-like plasmids. The locations of the core genes of the 32 pELF1-like plasmids and the insertion sites of the AMR regions of each pELF1-like plasmid were described in pELF_JHP9. The core genes obtained for each BLASTP cutoff value are shown in blue-gradient panels. For the AMR region, the plasmid name, drug resistance gene name, size (kb), and genetic structure schemas are noted along with the insertion position. The gray panel shows noncore genes. The green panels represent transposition/site-specific recombination-related genes, and the orange panels represent AMR genes.
Analysis of the AMR region generating the diversity of pELF1-like plasmids.
Next, we focused our analysis on the AMR region comprising accessory plasmid genes. A Resfinder search showed that 9 of 32 pELF1-like plasmids harbor AMR genes (Table 3 and 4). Since the order of CDSs in the pELF1-like plasmids was extremely well preserved, the insertion sites of AMR regions were assessed based on pELF_JHP9, one of the shortest plasmids belonging to cluster IA-1 (Fig. 1 and 2; see Fig. S4 and S5 in the supplemental material).
We identified 15 AMR regions in 32 pELF1-like plasmids. Nine of the AMR regions contained van gene clusters with IS1216E on at least one of the terminal ends, indicating that the IS1216E-translocatable unit is involved in AMR transmission onto pELF1-like plasmids (Region_1 to 9) (Fig. S4). Five of the AMR regions had IS1216E at both ends, with four in the same orientation and one in the opposite orientation (Region_1, 4 to 6, 9) (Fig. S4). Five of the AMR regions containing the vanA gene cluster were concentrated to the region near the left hairpin end, and the AMR region containing the vanM gene cluster was concentrated to the center of the pELF1-like plasmid (Region_7, 8; Fig. 2 and Fig. S4). Similar to the AMR region containing the van gene cluster, IS1216E was also present at the extremities of the AMR region containing ant(6)-Ia, aph(3′)-III, and erm(B) of pELF1 (Region_11) (Fig. 2 and Fig. S5). pELF2 and pIHVA-EV0426-12 possessed an AMR region similar to those containing ant(9)-Ia and erm(A) of pELF_AA242 (Region_12 to 14) (Fig. 2 and Fig. S5). VB3025 plasmid unnamed 2 possessed a similar AMR region but with optrA instead of ant(9)-Ia (Region 15) (Fig. 2 and Fig. S5). These four AMR regions were located near the core gene at the invertron end of the plasmid (Fig. 2). In these AMR regions, the presence of XerC1 and -C2 suggested their involvement in transitions (Fig. S5). pELF_JHP35 harbored Tn916, whose orf9 was disrupted by Tn917. AMR region closely resembled the Tn3872-like element of CGSP14 (Streptococcus pneumoniae, Region 10) (Fig. S5 and 6) (16).
Phenotypic analysis of fitness cost and stability of pELF1-like plasmids in enterococcal hosts.
Plasmids allow for the development of environmentally adaptive traits, such as drug resistance genes, but they also impose fitness costs (17). Fitness cost is an important factor of long-term stability in plasmids, especially under conditions where the genes encoding for drug resistance are redundant (18, 19). We analyzed the fitness cost and stability of pELF1-like plasmids using pELF2, which harbors several AMR genes, including vanA. This pELF1-like plasmid belongs to cluster IA-1 and is known to be spreading in some regions of Japan (Fig. 1) (8).
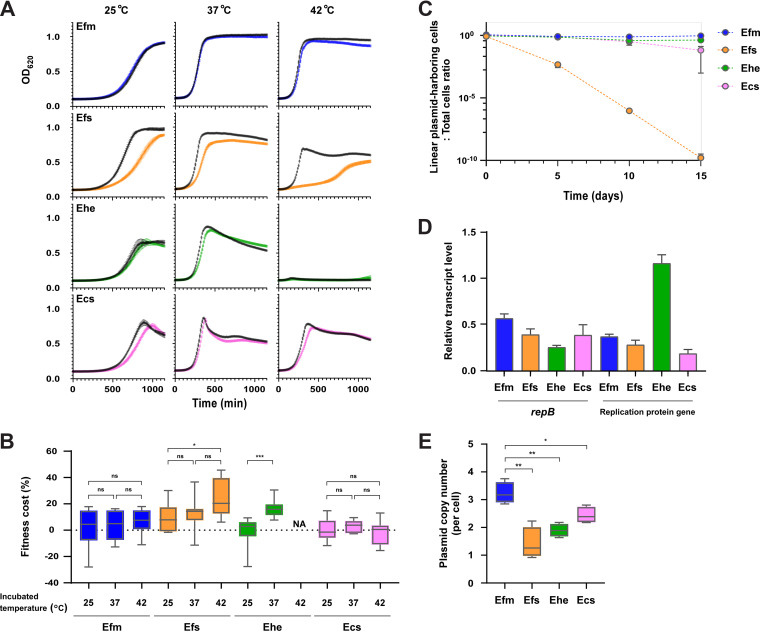
To investigate host-specific differences, we tested four strains under nonselective conditions at temperatures mimicking those of the human body (37°C), environmental wastewater (25°C), or chicken body (42°C) (Fig. 3A and B) (20–22). In E. faecium, there was no significant reduction in the growth curve related to pELF2 at any temperature. In contrast, a clear decrease in the growth curve of E. faecalis was observed at all temperatures. pELF2 imposed a fitness cost on all four enterococcus species; however, the fitness costs for E. faecium and Enterococcus casseliflavus were the lowest, with 4.1% and 3.2%, respectively, at 37°C (see Table S6 in the supplemental material). For these two species, the fitness cost did not increase at any of the temperatures (Fig. 3B). In contrast, we observed several times higher fitness costs for E. faecalis and Enterococcus hirae. To confirm whether this physiological effect was pELF2 specific, experiments with another pELF1-like plasmid (pELF_AA242) were performed, which confirmed similar growth effects and fitness costs (Fig. S7A and B in the supplemental material; Table S6). For E. faecalis—the most prevalent species among human enterococcus infections—we changed the host strain to OG1RF (accession number CP002621) and analyzed the growth effect of the pELF1-like plasmids (Fig. S7C and D). As with the FA2-2 (E. faecalis) host, we observed significantly lower growth rates and higher fitness costs, which implies that the fitness cost imposed by the pELF1-like plasmid involves host-specific factors. Accordingly, we assessed codon usage frequency; the codon adaptation index (CAI) between host genomes and MGE is hypothesized to correspond with the inhibition of gene translation and to negatively impact fitness costs (23). In E. faecium, there was no significant difference in CAI between genes on the chromosome and pELF2. The difference was greatest in E. faecalis (median CAI of E. faecalis chromosomal genes versus pELF2 genes, 0.5090 versus 0.4690; P < 0.0001 [Mann-Whitney test]) (see Fig. S8 in the supplemental material).
FIG 3.
Analysis of the stability and effect of the pELF1-like plasmid on the growth of enterococci. The effects of pELF1-like plasmid (pELF2) carriage on the growth curve (A) and plasmid-imposed fitness cost on four Enterococcus spp. (B) are shown. For the analysis of growth and fitness cost, nine biological replicates of the experiment were conducted. (a) Growth curves are shown as absorbance (OD620) ± SE. The black curve indicates pELF2 noncarrying strains, and the colored curve indicates pELF2-carrying strains. (b) Fitness cost is shown in a box-and-whisker diagram. The whiskers indicate the minimum to maximum values. *, P ≤ 0.05; ***, P ≤ 0.001; ns, not significant; NA, not applicable (Mann-Whitney test). (C) The stability of the pELF1-like plasmid was evaluated in a 15-day passaging experiment in a liquid medium without selection pressure. After 5, 10, and 15 days, the number of pELF1-like plasmid-harboring and nonharboring strains was measured, and the ratio (±SE) of the plasmid-harboring cells to total cells was calculated. Three independent biological experiments were conducted. (D) The transcript level of repB and replication protein genes were measured using reverse transcription qPCR (RT-qPCR). The bar charts show the relative transcript ratio compared with mRNA levels of each gyrB gene, along with ±SE. (E) Plasmid copy number was measured using qPCR and shown in a box-and-whisker diagram. *, P ≤ 0.05; **, P ≤ 0.01 (unpaired t test; P = 0.0019, P = 0.0010, and P = 0.0176, respectively). Blue symbols indicate E. faecium (Efm; BM4105RF), orange symbols indicate E. faecalis (Efs; FA2-2), green symbols indicate E. hirae (Ehe; ATCC 9790RF), and pink symbols indicate E. casseliflavus (Ecs; KT06RF).
We assessed the stability of pELF1-like plasmids in four Enterococcus spp. via long-term passage assay under nonselective conditions. During 15-day passages, pELF2 maintained the highest stability in E. faecium, followed by E. hirae and E. casseliflavus (Fig. 3C). However, in E. faecalis, the percentage of pELF1-like plasmid-carrying strains markedly decreased as passaging progressed, falling below the detection limit at 30 passages (15 days). To confirm whether fitness improved within a declining pELF2-carrying E. faecalis population, we investigated the genetic structure of pELF2 and fitness cost in E. faecalis after 5 days of passage (day 5-evolved strains). Pulsed-field gel electrophoresis (PFGE) showed a marked reduction in the size of pELF2 in some of the day 5-evolved strains (1.1 and 1.2 strains) (see Fig. S9A in the supplemental material), which also implies that pELF2 is unstable in E. faecalis. Somewhat surprisingly, given the eventual disappearance of pELF2 from the E. faecalis population, day 5-evolved strains had a 20 to 30% reduction in fitness cost compared with parental strains (Fig. S9B). Furthermore, the plasmid copy number of these day 5-evolved strains was higher than that of the parent strain (Fig. S9C).
Since the overexpression of replication-associated proteins is known to impair genome replication and lead to fitness costs, we examined the transcription levels of repB and the replication protein gene on pELF2 in four different hosts. We found that the transcription levels of repB and the replication protein gene were lower than those of gyrB on each chromosome (Fig. 3D) (24). Using quantitative PCR (qPCR), we tested whether the copy number of the pELF1-like plasmid varied between Enterococcus spp. (Fig. 3E; see Table S7 in the supplemental material). Consistent with the reverse transcriptase quantitative PCR (RT-qPCR) results, the plasmid copy number appeared to be tightly controlled in all four Enterococcus spp., as with other conjugative plasmids (25). However, we observed interspecific differences in the number of plasmid copies, with E. faecium possessing significantly more pELF1-like plasmid copies (3.2 copies/cell) than the other three species. The lowest number of plasmid copies was observed in E. faecalis, which has the highest pELF2 fitness cost.
The impact of pELF1-like plasmids on E. faecium.
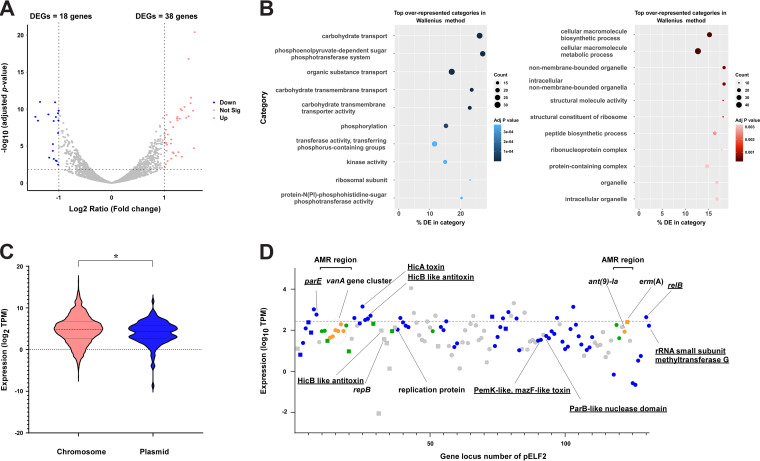
Plasmid carriage impacts bacterial physiology, e.g., metabolic regulation, but the effects of pELF1-like plasmids on gene transcription are unclear (26, 27). To analyze the interaction between the pELF1-like plasmid and the host chromosome, we compared the levels of chromosomal gene transcription between pELF2-carrying E. faecium and noncarrying E. faecium (28). Of the 2,515 genes, 56 genes (~2%) showed a >2-fold change in the transcript level, including 38 upregulated and 18 downregulated genes (adjusted P < 0.05) (Fig. 4A; see Table S8 in the supplemental material), indicating a modest effect of pELF1-like plasmid carriage (29). Among the upregulated genes, we identified glutamine-fructose 6-phosphate aminotransferase (glmS_1) and glutamine synthase (glnA). These genes are involved in the synthesis of glucosamine 6-phosphate, which participates in UDP-NAcGlc synthesis and metabolism of the cell wall structure. Of the downregulated genes, 50% of them were related to carbohydrate transport and metabolism (Table S8). A gene ontology (GO) analysis showed that carbohydrate transport and the phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS; especially mannose-specific PTS) were significantly downregulated in the clusters of orthologous group (Fig. 4B) (30). Similar to the transcriptome analysis of K. pneumoniae plasmid acquisition, pELF2 carriage influenced metabolism-related genes (27). To investigate its effect on carbohydrate availability, we conducted growth experiments under carbohydrate-limited conditions. Contrary to the transcriptome sequencing (RNA-seq) results, we did not observe any change in growth in the presence of glucose or mannose with or without the pELF1-like plasmid (see Fig. S10 in the supplemental material).
FIG 4.
Transcriptome analysis of the effect of pELF1-like plasmid carriage on the E. faecium host genome. We assessed the effect of pELF2 carriage on the E. faecium host (BM4105RF) via transcriptomic analysis. (A) Changes in the transcriptome of E. faecium possessing pELF2. Each dot indicates an open reading frame, with red circles indicating upregulation (log2 fold change, >1; adjusted P < 0.05) and blue circles indicating downregulation (log2 fold change, <–1; adjusted P < 0.05). (B) Gene enrichment analysis was performed with GOseq based on DESeq2 results. The blue and red bubble charts show the results of the downregulated and upregulated gene analysis. (C) Transcript abundances of genes on the chromosome (red) and the pELF1-like plasmid (blue, log2 of transcript per million [TPM]). The transcript abundances of the genes on the plasmid were calibrated with the copy number of the plasmid. *, P < 0.05 (Kolmogorov-Smirnov test). (D) The TPM values of CDS of pELF2 are plotted in the order of gene locus number. The circles indicate the plus strand and the squares indicate the minus strand. Dark blue indicates core genes; among noncore genes, IS and recombinase are shown in green, drug resistance genes are shown in orange, and other genes are shown in gray. The dashed line indicates the top 20% TPM values.
Transcriptome analysis of pELF1-like plasmid genes in E. faecium.
A particular plasmid has been reported to have lower transcription levels than host chromosomes and lower host costs (17, 25, 31). We assessed the transcriptional profile of genes on pELF2 and found that the transcript level of genes on the pELF1-like plasmid was lower than that on the chromosomes (P = 0.01, Kolmogorov-Smirnov test) (Fig. 4C).
pELF2 is a natural plasmid that has spread in some areas of Japan and possesses multiple drug resistance genes (8). We compared the transcript levels of the plasmid core genes, noncore genes, and genes on MGEs (including AMR genes), but we found no significant differences (see Fig. S11 in the supplemental material). The low transcription levels of the entire pELF2 gene set implicated the regulation of the transcription of relatively expensive core genes, such as transfer-related genes, while they also suggested that the entire gene set, including accessory genes, may not impose a high cost on E. faecium.
Of the genes on pELF2 in the top 20% of transcript per million (TPM) values, 65% were core genes, and four genes encoding the TA system were identified (Fig. 4D and Table S5). The TA system is considered to contribute to robust vertical propagation by postsegregational killing (PSK), which is particularly important in low-copy-number plasmids (32). According to TASer and BLAST searches, multiple putative type II TA system-related genes are present in pELF2, of which 5 were core genes (33). The type II TA system is the most abundant type in bacterial genomes, including MGEs (34). HicAB was classified as a type IIA TA system (harbored by many bacteria), and transcriptome analysis showed high transcription levels (35). Similarly, the transcript levels of the RelE/ParE family toxin and RelB/DinJ family antitoxin were comparably high in the top 20% of TPM values (36). From these results, we hypothesized that these relatively highly transcribed TA systems contributed to the stable inheritance of pELF1-like plasmids in E. faecium (37–39).
Comparative analysis of pELF1-like plasmid-carrying E. faecium host genomes.
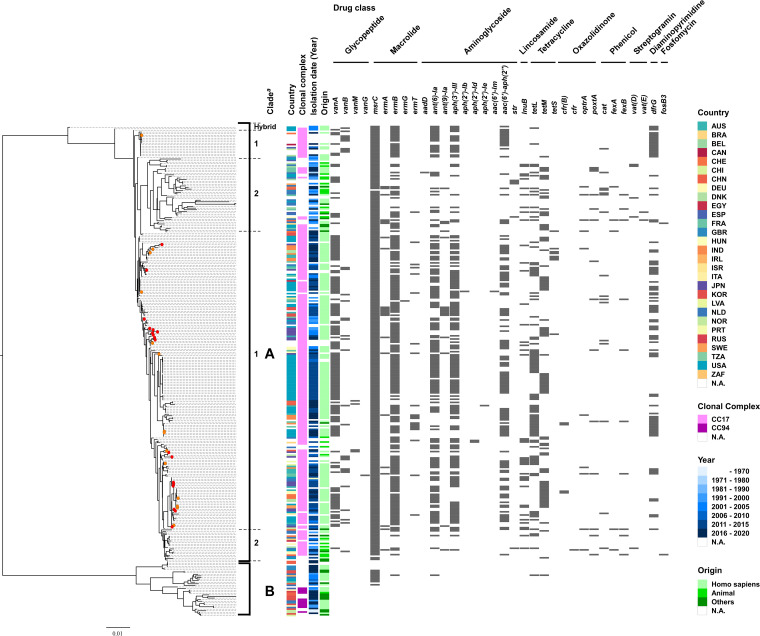
The above analyses of epidemiological data and physiological effects suggest that the plasmid is adapted to E. faecium. However, the stability of plasmids varies even within the same bacterial species with strain-specific traits (40). To determine the genetic background of the pELF1-like plasmid-carrying E. faecium, we conducted a phylogenetic analysis of the complete genome of 309 E. faecium strains, including 32 pELF1-like plasmid-carrying strains (Fig. 5 and Table S9). The phylogenetic tree was divided into two major groups, with the smaller group containing strains of clade B (41, 42); therefore, these two groups were considered to correspond to the previously reported clade classification. The pELF1-like plasmid-harboring strains were considered to belong to clade A (3, 42). Even within clade A, they were well dispersed and not concentrated to any particular lineage. These results indicate that the pELF1-like plasmid could be carried by various lineages of E. faecium belonging to the MDR clade.
FIG 5.
Genomic comparison of pELF1-like plasmid-harboring versus nonharboring E. faecium strains based on the core genome. Phylogenetic tree analysis based on core genome SNPs was performed on 309 strains of E. faecium, including 32 strains harboring pELF1-like plasmids. Core genes were identified using the Roary, with a cutoff value of BLASTP of 95%. The panel shows the country, clonal complex, year, and origin of each strain, as well as the resistance gene status. For the clonal complex, CC17 and CC94 were described based on the MLST profile in PubMLST (https://pubmlst.org/organisms/enterococcus-faecium). The red circles in the phylogenetic tree indicate pELF1-like plasmid-harboring strains of Japanese origin, and the orange circles indicate pELF1-like plasmid-harboring strains in the public database. aThe clade was determined based on the phylogenetic tree of Lebreton et al. (42).
DISCUSSION
MGEs, such as plasmids, play an important role in the efficient propagation of drug resistance genes in enterococci. Several reports on enterococcal pELF1-like plasmids have shown that they confer vancomycin resistance and promote carbohydrate availability; although they have a significant impact in clinical practice, their biological effects on the host enterococci are poorly understood (8, 9, 15). In this study, we conducted a comprehensive analysis of pELF1-like plasmids based on growth phenotype and transcriptomics coupled with molecular epidemiology.
We found that 3.4% of VREs and 0.2% of VSEs harbored pELF1-like plasmids, and all of them were E. faecium (Table 1). As early as the beginning of 2000 in Japan, pELF1-like plasmids were detected in vancomycin-resistant E. faecium (Table 2). In combination with the results of public database searches, phylogenetic analysis based on the core genome of the 32 pELF1-like plasmids showed that (i) multiple lineages with a high degree of structural similarity exist and (ii) cluster I pELF1-like plasmids occur globally (Fig. 1). The genetic structure and order of CDSs of the pELF1-like plasmids, detected over ~20 years, were highly conserved (Fig. 1). The pELF1-like plasmid also had the plasticity to acquire accessory genes (Fig. 2). The MGEs containing the AMR genes were transferred to several pELF1-like plasmids (Fig. 2). Of the 15 AMR regions on the pELF1-like plasmid, IS1216E was present at a comparatively high frequency (10 of 15 AMR regions) (Fig. 2 and Fig. S4 and S5). The IS6 family, to which IS1216 belongs, is involved in the propagation of AMR genes, both Gram positive and negative, and is reportedly responsible for many transitions between plasmids and chromosomes (39, 43, 44). According to the MobileElementFinder-based search, pELF1-like plasmids with AMR genes retained more MGEs, including ISs (AMR-holding plasmid versus nonholding plasmid, 7.25 versus 0.64 [mean number of MGEs per pELF]) (see Table S10 in the supplemental material) (45). These findings infer that IS-associated AMR regions are primarily responsible for the diversity of pELF1-like plasmids.
To confirm their adaptation to enterococci, especially E. faecium, we investigated the phenotypic effects of pELF1-like plasmids on Enterococcus sp. growth. The persistence of a conjugative plasmid in a bacterial population is believed to be determined by (i) the frequency of conjugative transmission, (ii) the fitness cost imposed by the plasmid, and (iii) the loss of the plasmid during cell segregation (18, 19). Therefore, both horizontal transmission and vertical inheritance are involved in the long-term plasmid stability. The pELF1-like plasmids exhibit a high transfer frequency in both solid and liquid mediums, which contributes to their persistence (7, 8). In addition, the fitness cost of the plasmid must be sufficiently low for stable vertical inheritance (17). As with the results of the plasmid stability experiments, we observed a lower impact on growth and fitness cost related to pELF1-like plasmids in E. faecium under all temperature conditions (Fig. 3A to C; Table S6). We observed similar trends in fitness cost for the other pELF1-like plasmid (pELF_AA242), suggesting that their effect on enterococci could be a general phenomenon (Fig. S7).
Various origins of fitness cost have been suggested, of which one is the disruption of genome replication resulting from the overexpression of replication-associated genes (17, 24). Plasmid copy number is also considered to increase fitness cost, but the pELF1-like plasmid had no noticeable effect on the transcription of replication-related proteins and its copy number was relatively low (Fig. 3D and E). This result was observed not only for E. faecium but also for E. faecalis, which showed a high fitness cost associated with pELF2. Since most of the fitness costs are considered to arise from the expression of genes on the plasmid, the effective control of transcription is crucial (17). Transcriptome analysis revealed that the transcription of entire genes on the pELF2 plasmid was suppressed to a lower level than that in the chromosomal genome, and the effect of plasmid carriage on host chromosomal genes was limited (Fig. 4). Together with the lack of significant differences in CAI, these results could explain the lower fitness cost in E. faecium (Fig. S8).
Meanwhile, pELF2 imposed a high fitness cost in E. faecalis. In the long-term passage assays, pELF2 derivatives were obtained easily, indicating its instability in E. faecalis (Fig. S9). They showed changes in plasmid size and copy number with decreasing fitness cost; however, the changes do not appear to be sufficient for the establishment of the plasmids in the E. faecalis population (Fig. 3C and Fig. S9).
Given the influence of pELF1-like plasmid genes on plasmid stabilization, we hypothesized that a large number of highly transcribed TA systems promote stable vertical inheritance (Fig. 4D and Table S5), although it may also be related to the expansion of the host range of plasmids, as reported previously (46). Consistent with previous reports of nosocomial transmission, we confirmed that the pELF1-like plasmid was maintained by several Enterococcus spp. (8). The ability to be retained by several Enterococcus spp. would likely promote plasmid diversity and contribute to the long-term survival of plasmids in a variety of settings, including clinical practice (47). Thus, the pELF1-like plasmid is a low-cost plasmid (especially for E. faecium), with high stability, high frequency of self-transmissibility, and the ability to carry AMR genes, which make it an ideal candidate for spreading drug resistance between Enterococcus spp.
The core genome analysis revealed that various lineages of E. faecium belonging to the MDR clade carry pELF1-like plasmids (clade A1) (Fig. 5). It is worth noting that, of the 32 pELF1-like plasmids in this analysis, all pELF1-like plasmids with vancomycin resistance genes were detected after 2010. More recently, Egan et al. (48) reported evidence of the regional spread of pELF1-like plasmids carrying the vanA gene cluster in Ireland. These findings indicate that pELF1-like plasmids have only recently become important vehicles for vancomycin resistance genes, at least in some regions (8, 9).
In this study, we combined molecular epidemiological, phenotypic, and transcriptomic analyses to provide an integrated characterization of the pELF1-like plasmid. Our characterization of the relationship between pELF1-like plasmids and E. faecium, the most abundant VRE, highlights the importance of further analysis. We acknowledge some limitations to our research. Our in silico and in vitro analyses neglected the context-specific effects of plasmids in vivo, although this limitation was unavoidable given that the adaptation of pELF1-like plasmids to enterococci in vivo remains unclear. In addition, many of the strains used were clade A medical strains, with clade B or enterococci of environmental origin being underrepresented. Future studies should also consider the spread of pELF1-like plasmids in animals and food from a One Health perspective.
MATERIALS AND METHODS
Enterococcus strains and drug susceptibility test.
The bacterial strains used in this study were stored at the Department of Bacteriology, Gunma University Graduate School of Medicine, Gunma, Japan (Table 1 and 2). All strains were detected at medical institutions in Japan. Unless otherwise noted, enterococci were cultured in Todd-Hewitt broth (THB; Difco Laboratories, Detroit, MI) at 37°C under a static condition. MICs were determined using the agar dilution method, and interpretation of MIC results was based on the Clinical and Laboratory Standards Institute guidelines (http://clsi.org/). NCBI was used to search for and retrieve genomes (49).
Conjugation assay.
Transconjugants were obtained via filter mating experiments as reported previously (7). Vancomycin (5 mg/L for E. faecium, E. faecalis, and E. hirae experiments; 12 mg/L for E. casseliflavus experiments) or erythromycin (16 mg/L) was used for transconjugant selection along with both rifampicin (50 mg/L) and fusidic acid (50 mg/L). KUHS13 (accession number SAMD00202474) and AA242 (accession number SAMD00466811) were used as donor strains, and FA2-2 (E. faecalis; accession number SAMN00809127), BM4105RF (E. faecium; accession number SAMN09464428), ATCC 9790RF (E. hirae), and KT06RF (E. casseliflavus) were used as recipient strains. In the second conjugation experiment, we used BM4105SS (E. faecium) as a recipient strain. Vancomycin (5 mg/L) was used for transconjugant selection along with both streptomycin (516 mg/L) and spectinomycin (256 mg/L). Before experimentation, we confirmed the presence of pELF1-like plasmids using PCR and PFGE.
Colony PCR.
We performed colony PCR to detect pELF1-like plasmids. Briefly, the bacterial colony was collected with the tip of a toothpick and used directly for PCR amplification. Quick Taq hot start (HS) dye mix was used as a PCR reagent (Toyobo, Tokyo, Japan). PCR primer sets, constructed for the predicted terminal ends of the plasmids, are listed in Table S1. The thermal cycling conditions were set to 94°C for 3 min and 40 cycles of 94°C for 30 s, 60°C for 30 s, and 68°C for 1 min 30 s. The presence of a pELF-like plasmid was verified by electrophoresis based on the size of the PCR product.
Growth kinetics and plasmid stability analysis.
Bacterial strains were grown overnight in 5 mL THB. After the bacterial culture was diluted to an absorbance (optical density at 620 nm [OD620]) of 0.2, 1 μL of the dilution was added to 299 μL of THB and incubated at 37°C for 24 h. The absorbance (OD595) was measured kinetically every 10 min in a Sunrise plate reader (Tecan, Männedorf, Switzerland) while incubating at 25°C, 37°C, or 42°C without shaking. Growth rates (h) and fitness costs were determined using the growth rate (GR) program (ver. 5.1) and the formula of Tedim et al. (50, 51). Briefly, the GR of the pELF1-like plasmid-carrying strain was normalized by the GR of the pELF1-like plasmid noncarrying strain (relative to 1; relative GR [RGR]). Plasmid fitness cost (%) was calculated as (1 – RGR) × 100. For both the growth curve and fitness cost analyses, we used nine biological replicates for each strain. Statistical analyses were performed using an unpaired t test for growth rate and Mann-Whitney test for fitness cost.
The stability of the pELF1-like plasmid was evaluated through a 15-day passaging experiment in a liquid medium without selection pressure. Every 12 h, 5 μL of the bacterial culture was transferred to 5 mL of fresh medium and incubated continuously at 37°C. After 5, 10, and 15 days, the number of pELF1-like plasmid-harboring and nonharboring strains was measured using vancomycin resistance as an indicator, and the ratio of pELF1-like plasmid-harboring cells to total cells was calculated. We conducted three independent biological experiments.
A carbon source-limited M1 medium was prepared as reported by Zhang et al. (52). The bacteria were cultured in a THB medium and diluted to an absorbance (OD620) of 0.2. After centrifugation (13 000 × g for 3 min), the supernatant was discarded and the bacterial pellet was washed twice with phosphate-buffered saline (PBS). The bacteria were inoculated with M1, M1 with 10 mM glucose, or M1 with 10 mM mannose and incubated at 37°C for 24 h. The absorbance (OD595) was measured kinetically using the Sunrise plate reader (Tecan).
Pulsed-field gel electrophoresis.
PFGE was performed as described previously (7). Briefly, a 1% agarose gel block embedded with enterococci was treated with lysozyme solution (10 mg/mL; Roche, Basel, Switzerland) at 37°C for 6 h, followed by treatment with proteinase K solution (60 mAnson-U/mL; Merck Millipore, Darmstadt, Germany) at 50°C for 48 h. After the treated plugs were washed, they were subjected to PFGE using a CHEF Mapper (Bio-Rad Laboratories, Richmond, CA) according to the manufacturer’s instructions (pulse from 5.3 to 66 s during 19.5 h at 6.0 V/cm and 4°C).
Plasmid copy number.
The copy number of pELF1-like plasmids was calculated using qPCR as described by San Milan et al. (53). Briefly, the copy number per chromosome was calculated as (1 + Ec)Ctc/(1 + Ep)Ctp × Sc/Sp, where Ec and Ep are the efficiencies of the chromosome and plasmid qPCR amplification (relative to 1), respectively; Ctc and Ctp are the threshold cycles of the chromosome and plasmid, respectively; and Sc and Sp are the amplicon size (bp) of the chromosome and plasmid, respectively. DNA was extracted from cells cultured in THB medium containing 5 mg/L vancomycin and used for analysis immediately after entering the stationary phase. Considering its proximity to oriC for each species, gyrB was used as the internal control, and the replication-protein gene was used as the target gene for the pELF1-like plasmid. We conducted four biological replicates of this experiment and used an unpaired t test for statistical analysis. The primers used are listed in Table S1.
Whole-genome sequencing analysis of pELF1-like plasmid-carrying isolates.
Total DNA was prepared using a Gentra Puregene yeast/bacterium kit according to the manufacturer’s protocol (Qiagen, Hilden, Germany). Short reads were obtained using a MiniSeq system (Illumina) with a high-output reagent kit (300 cycles). The library for sequencing (150-bp paired-end, insert size, 500 to 900 bp) was prepared using a Nextera XT DNA library prep kit (Illumina). The DNA library for the Nanopore MinION device was prepared using an SQK-RBK004 rapid barcoding kit according to the manufacturer’s protocol and then sequenced on a flow cell (R9.4.1; Oxford Nanopore Technologies, Oxford, UK). The long reads were base called using Guppy and were assembled de novo using Canu (ver. 2.1.1) and then polished by short reads using Pilon (ver. 1.20.1) (54). To obtain functional annotations, the assembled sequences were submitted to the RAST and DFAST annotation pipelines (55, 56).
For the core genome analysis, the genomes of all strains were annotated in Prokka (ver. 1.14.6) (57) for homogeneity. The core genes were identified and aligned in Roary (ver. 3.13.0) (58). From the results of the core genome analysis, the phylogenetic tree was constructed using RAxML (ver. 8.2.4) under the GTRGAMMA model and then visualized in the Figtree software (ver. 1.4.4, http://tree.bio.ed.ac.uk/software/figtree/) (59). We used Phandango and genome matcher (ver. 3.0.2) for visualization of the bacterial data set and genome comparison (60, 61). MLST, antibiotic-resistance genes, replicon-type, toxin-antitoxin system, virulence genes, and MGEs in whole-genome data were detected using Staramr (ver. 0.5.1; https://github.com/phac-nml/staramr), Resfinder (ver. 4.1 server), ABRicate (https://github.com/tseemann/abricate), PlasmidFinder (ver. 2.1 server), TASer (https://shiny.bioinformatics.unibe.ch/apps/taser/), Virulence finder (ver. 2.0), ISfinder, and MobileElementFinder (ver. 1.0.3) (10, 11, 14, 33, 45, 62–64). For the pELF comparison, pairwise nucleotide identity was calculated using MAFFT (ver. 7.450). CAI was calculated using the “cai” function of the EMBOSS package (65). The genes encoding the ribosomal proteins of each of the four Enterococcus spp. were used as a reference set (66).
Transcriptome analysis.
Bacteria used for RT-qPCR and RNA-seq were incubated overnight in brain heart infusion (BHI) medium at 37°C. The cultures were diluted 1,000-fold in fresh BHI medium and incubated until the late exponential phase. After treatment with RNA protect bacterial reagent (Qiagen), RNA extraction was performed from 1 mL of the bacterial solution using an RNeasy minikit (Qiagen) according to the manufacturer’s protocol. For RT-qPCR analysis, cDNA was synthesized using the PrimeScript RT master mix (TaKaRa Bio, Shiga, Japan), and real-time PCR was performed using the Luna universal qPCR master mix (New England BioLabs, Ipswich, MA) on an ABI 7500 fast RT-PCR system (Thermo Fisher, Waltham, MA). The primers used in this analysis are listed in Table S1. For the RNA-seq analysis, RNA samples were outsourced to Novogene (Beijing, China) for library preparation and sequencing (https://jp.novogene.com/). After the strand-specific libraries were prepared, they were sequenced using an Illumina NovaSeq 6000 sequencer.
RNA data analysis was conducted using the Galaxy platform (67). Reads were mapped to BM4105RF/pELF2 nucleotide sequences (accession numbers CP030110 and AP022343) using Bowtie2 (ver. 2.4.2) (68), and the numbers of aligned reads were counted with FeatureCounts (ver. 2.0.1) (69). DESeq2 (ver. 2.11.40.6) was used to determine the differentially expressed genes (DEGs) (28). In this analysis, significance was set at an adjusted P value of <0.05, and a log2 fold change of >1 and <–1 was considered upregulated and downregulated, respectively. For the GO analysis, we used GOseq (ver. 1.44.0) (30). Comparative analysis of plasmid and chromosomal gene transcription levels was performed using TPM values based on Kallisto Quant RNA-seq data, corrected for plasmid copy number (70). The statistical analysis of gene transcript abundances between chromosomes and pELF1-like plasmids was performed using the Kolmogorov-Smirnov test.
Data availability.
All assembly data are available from the NCBI database (JHP9, accession number SAMD00466802; JHP10, accession number SAMD00466803; JHP35, accession number SAMD00466804; JHP36, accession number SAMD00466805; JHP38, accession number SAMD00466806; JHP80, accession number SAMD00466807, AA55, accession number SAMD00466808; AA94, accession number SAMD00466809; AA96, accession number SAMD00466810; AA242, accession number SAMD00466811; AA290, accession number SAMD00466812; AA610 accession number SAMD00466813; AA818, accession number SAMD00466814; GK923, accession number SAMD00466815; GK941, accession number SAMD00466816; GK961, accession number SAMD00466817).
ACKNOWLEDGMENTS
We are grateful to Reika Kawabata-Iwakawa (Division of Integrated Oncology Research, Gunma University Initiative for Advanced Research [GIAR], Gunma University), Yohei Morishita, and Saori Fujimoto (Laboratory for Analytical Instruments, Education and Research Support Center, Gunma University Graduate School of Medicine) for their helpful discussions and assistance with the RNA integrity measurement.
This study was supported by grants from the Japanese Ministry of Health, Labor and Welfare (Research Program on ensuring Food Safety, 21KA1004); grants from the Japan Agency for Medical Research and Development (AMED) (JP22fk0108604 and JP22wm0225008 to H.T.; JP22gm1610003, JP22fk0108133, JP22fk0108139, JP22fk0108642, JP22wm0225004, JP22wm0225008, JP22wm0225022, JP22wm0325003, JP22wm0325022 and JP22wm0325037 to M.S.); grants from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (22K07067 to H.T.; 20K07509 and 21H03622 to M.S. Grant-in-Aid for Early-Career Scientists 22K16368 to Y. Hashimoto); and the grant from Gunma Foundation and Health Science (research grants Provided by Gunma Foundation and Health Science).
Y. Hashimoto, M.S., and H.T. conceived and designed the study. Y. Hashimoto, S.K., Y. Hirahara, T.N., J.K., H.H., and K.T. collected bacterial isolates and performed bacterial characterizations. Y. Hashimoto and M.S. performed the collection and analysis of WGS and transcriptome data. Y. Hashimoto conducted all other efforts. H.T. supervised the study. All authors read and approved the manuscript for publication.
There are no financial conflicts of interest to be disclosed.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Rice LB. 2008. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis 197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 2.Miller WR, Murray BE, Rice LB, Arias CA. 2020. Resistance in vancomycin-resistant enterococci. Infect Dis Clin North Am 34:751–771. doi: 10.1016/j.idc.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willems RJL, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, Grundmann H, Bonten MJM. 2005. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis 11:821–828. doi: 10.3201/1106.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freitas AR, Tedim AP, Francia MV, Jensen LB, Novais C, Peixe L, Sánchez-Valenzuela A, Sundsfjord A, Hegstad K, Werner G, Sadowy E, Hammerum AM, Garcia-Migura L, Willems RJ, Baquero F, Coque TM. 2016. Multilevel population genetic analysis of vanA and vanB Enterococcus faecium causing nosocomial outbreaks in 27 countries (1986-2012). J Antimicrob Chemother 71:3351–3366. doi: 10.1093/jac/dkw312. [DOI] [PubMed] [Google Scholar]
- 5.Johnsen PJ, Townsend JP, Bøhn T, Simonsen GS, Sundsfjord A, Nielsen KM. 2009. Factors affecting the reversal of antimicrobial-drug resistance. Lancet Infect Dis 9:357–364. doi: 10.1016/S1473-3099(09)70105-7. [DOI] [PubMed] [Google Scholar]
- 6.San Millan A. 2018. Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol 26:978–985. doi: 10.1016/j.tim.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto Y, Taniguchi M, Uesaka K, Nomura T, Hirakawa H, Tanimoto K, Tamai K, Ruan G, Zheng B, Tomita H. 2019. Novel multidrug-resistant enterococcal mobile linear plasmid pELF1 encoding vanA and vanM gene clusters from a Japanese vancomycin-resistant enterococci isolate. Front Microbiol 10:2568. doi: 10.3389/fmicb.2019.02568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashimoto Y, Kita I, Suzuki M, Hirakawa H, Ohtaki H, Tomita H. 2020. First report of the local spread of vancomycin-resistant enterococci ascribed to the interspecies transmission of a vanA gene cluster-carrying linear plasmid. mSphere 5:e00102-20. doi: 10.1128/mSphere.00102-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujiya Y, Harada T, Sugawara Y, Akeda Y, Yasuda M, Masumi A, Hayashi J, Tanimura N, Tsujimoto Y, Shibata W, Yamaguchi T, Kawahara R, Nishi I, Hamada S, Tomono K, Kakeya H. 2021. Transmission dynamics of a linear vanA-plasmid during a nosocomial multiclonal outbreak of vancomycin-resistant enterococci in a non-endemic area, Japan. Sci Rep 11:14780. doi: 10.1038/s41598-021-94213-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E, Van Embden JDA, Willems RJL. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J Clin Microbiol 40:1963–1971. doi: 10.1128/JCM.40.6.1963-1971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh KV, Coque TM, Weinstock GM, Murray BE. 1998. In vivo testing of an Enterococcus faecalis efaA mutant and use of efaA homologs for species identification. FEMS Immunol Med Microbiol 21:323–331. doi: 10.1111/j.1574-695X.1998.tb01180.x. [DOI] [PubMed] [Google Scholar]
- 13.Nallapareddy SR, Weinstock GM, Murray BE. 2003. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol Microbiol 47:1733–1747. doi: 10.1046/j.1365-2958.2003.03417.x. [DOI] [PubMed] [Google Scholar]
- 14.Tetzschner AMM, Johnson JR, Johnston BD, Lund O, Scheutz F, Dekker JP. 2020. In silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J Clin Microbiol 58:e01269-20. doi: 10.1128/JCM.01269-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boumasmoud M, Dengler Haunreiter V, Schweizer TA, Meyer L, Chakrakodi B, Schreiber PW, Seidl K, Kühnert D, Kouyos RD, Zinkernagel AS. 2022. Genomic surveillance of vancomycin-resistant Enterococcus faecium reveals spread of a linear plasmid conferring a nutrient utilization advantage. mBio 13:e03771-21. doi: 10.1128/mbio.03771-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding F, Tang P, Hsu M-H, Cui P, Hu S, Yu J, Chiu C-H. 2009. Genome evolution driven by host adaptations results in a more virulent and antimicrobial-resistant Streptococcus pneumoniae serotype 14. BMC Genomics 10:158. doi: 10.1186/1471-2164-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.San Millan A, MacLean RC. 2017. Fitness costs of plasmids: a limit to plasmid transmission. Microbiol Spectr 5:5.5.02. doi: 10.1128/microbiolspec.MTBP-0016-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart FM, Levin BR. 1977. The population biology of bacterial plasmids: a PRIORI conditions for the existence of conjugationally transmitted factors. Genetics 87:209–228. doi: 10.1093/genetics/87.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subbiah M, Top EM, Shah DH, Call DR. 2011. Selection pressure required for long-term persistence of blaCMY-2-positive IncA/C plasmids. Appl Environ Microbiol 77:4486–4493. doi: 10.1128/AEM.02788-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rozwandowicz M, Brouwer MSM, Mughini-Gras L, Wagenaar JA, Gonzalez-Zorn B, Mevius DJ, Hordijk J. 2019. Successful host adaptation of IncK2 plasmids. Front Microbiol 10:2384. doi: 10.3389/fmicb.2019.02384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guardabassi L, Dalsgaard A. 2004. Occurrence, structure, and mobility of Tn1546-like elements in environmental isolates of vancomycin-resistant enterococci. Appl Environ Microbiol 70:984–990. doi: 10.1128/AEM.70.2.984-990.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oravcova V, Mihalcin M, Zakova J, Pospisilova L, Masarikova M, Literak I. 2017. Vancomycin-resistant enterococci with vanA gene in treated municipal wastewater and their association with human hospital strains. Sci Total Environ 609:633–643. doi: 10.1016/j.scitotenv.2017.07.121. [DOI] [PubMed] [Google Scholar]
- 23.Carroll AC, Wong A. 2018. Plasmid persistence: costs, benefits, and the plasmid paradox. Can J Microbiol 64:293–304. doi: 10.1139/cjm-2017-0609. [DOI] [PubMed] [Google Scholar]
- 24.San Millan A, Toll-Riera M, Qi Q, MacLean RC. 2015. Interactions between horizontally acquired genes create a fitness cost in Pseudomonas aeruginosa. Nat Commun 6:6845. doi: 10.1038/ncomms7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.San Millan A, Toll-Riera M, Qi Q, Betts A, Hopkinson RJ, McCullagh J, MacLean RC. 2018. Integrative analysis of fitness and metabolic effects of plasmids in Pseudomonas aeruginosa PAO1. ISME J 12:3014–3024. doi: 10.1038/s41396-018-0224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lang KS, Johnson TJ. 2015. Transcriptome modulations due to A/C2 plasmid acquisition. Plasmid 80:83–89. doi: 10.1016/j.plasmid.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Buckner MMC, Saw HTH, Osagie RN, McNally A, Ricci V, Wand ME, Woodford N, Ivens A, Webber MA, Piddock LJV. 2018. Clinically relevant plasmid-host interactions indicate that transcriptional and not genomic modifications ameliorate fitness costs of Klebsiella pneumoniae carbapenemase-carrying plasmids. mBio 9:e02303-17. doi: 10.1128/mBio.02303-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vial L, Hommais F. 2020. Plasmid-chromosome cross-talks. Environ Microbiol 22:540–556. doi: 10.1111/1462-2920.14880. [DOI] [PubMed] [Google Scholar]
- 30.Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park C, Zhang J. 2012. High expression hampers horizontal gene transfer. Genome Biol Evol 4:523–532. doi: 10.1093/gbe/evs030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerdes K, Rasmussen PB, Molin S. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci USA 83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akarsu H, Bordes P, Mansour M, Bigot D-J, Genevaux P, Falquet L. 2019. TASmania: a bacterial toxin-antitoxin systems database. PLoS Comput Biol 15:e1006946. doi: 10.1371/journal.pcbi.1006946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraikin N, Goormaghtigh F, Melderen LV, Margolin W. 2020. Type II toxin-antitoxin systems: evolution and revolutions. J Bacteriol 202:e00763-19. doi: 10.1128/JB.00763-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makarova KS, Grishin NV, Koonin EV. 2006. The HicAB cassette, a putative novel, RNA-targeting toxin-antitoxin system in archaea and bacteria. Bioinformatics (Oxford, England) 22:2581–2584. doi: 10.1093/bioinformatics/btl418. [DOI] [PubMed] [Google Scholar]
- 36.Jurėnas D, Fraikin N, Goormaghtigh F, Van Melderen L. 2022. Biology and evolution of bacterial toxin-antitoxin systems. Nat Rev Microbiol 20:335–350. doi: 10.1038/s41579-021-00661-1. [DOI] [PubMed] [Google Scholar]
- 37.Moritz EM, Hergenrother PJ. 2007. Toxin-antitoxin systems are ubiquitous and plasmid-encoded in vancomycin-resistant enterococci. Proc Natl Acad Sci USA 104:311–316. doi: 10.1073/pnas.0601168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arredondo-Alonso S, Top J, McNally A, Puranen S, Pesonen M, Pensar J, Marttinen P, Braat JC, Rogers MRC, van Schaik W, Kaski S, Willems RJL, Corander J, Schürch AC. 2020. Plasmids shaped the recent emergence of the major nosocomial pathogen Enterococcus faecium. mBio 11:e03284-19. doi: 10.1128/mBio.03284-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Che Y, Yang Y, Xu X, Břinda K, Polz MF, Hanage WP, Zhang T. 2021. Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc Natl Acad Sci USA 118:e2008731118. doi: 10.1073/pnas.2008731118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Gelder L, Ponciano JM, Joyce P, Top EM. 2007. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology (Reading) 153:452–463. doi: 10.1099/mic.0.2006/001784-0. [DOI] [PubMed] [Google Scholar]
- 41.Palmer KL, Godfrey P, Griggs A, Kos VN, Zucker J, Desjardins C, Cerqueira G, Gevers D, Walker S, Wortman J, Feldgarden M, Haas B, Birren B, Gilmore MS. 2012. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 3:e00318-11. doi: 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lebreton F, van Schaik W, Manson McGuire A, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJL, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534-13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shan X, Li X-S, Wang N, Schwarz S, Zhang S-M, Li D, Du X-D. 2020. Studies on the role of IS1216E in the formation and dissemination of poxtA-carrying plasmids in an Enterococcus faecium clade A1 isolate. J Antimicrob Chemother 75:3126–3130. doi: 10.1093/jac/dkaa325. [DOI] [PubMed] [Google Scholar]
- 45.Johansson MHK, Bortolaia V, Tansirichaiya S, Aarestrup FM, Roberts AP, Petersen TN. 2021. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed Web tool: MobileElementFinder. J Antimicrob Chemother 76:101–109. doi: 10.1093/jac/dkaa390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loftie-Eaton W, Yano H, Burleigh S, Simmons RS, Hughes JM, Rogers LM, Hunter SS, Settles ML, Forney LJ, Ponciano JM, Top EM. 2016. Evolutionary paths that expand plasmid host-range: implications for spread of antibiotic resistance. Mol Biol Evol 33:885–897. doi: 10.1093/molbev/msv339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porse A, Schønning K, Munck C, Sommer MOA. 2016. Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol Biol Evol 33:2860–2873. doi: 10.1093/molbev/msw163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Egan SA, Kavanagh NL, Shore AC, Mollerup S, Samaniego Castruita JA, O'Connell B, McManus BA, Brennan GI, Pinholt M, Westh H, Coleman DC. 2022. Genomic analysis of 600 vancomycin-resistant Enterococcus faecium reveals a high prevalence of ST80 and spread of similar vanA regions via IS1216E and plasmid transfer in diverse genetic lineages in Ireland. J Antimicrob Chemother 77:320–330. doi: 10.1093/jac/dkab393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.NCBI Resource Coordinators. 2016. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 44:D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall BG, Acar H, Nandipati A, Barlow M. 2014. Growth rates made easy. Mol Biol Evol 31:232–238. doi: 10.1093/molbev/mst187. [DOI] [PubMed] [Google Scholar]
- 51.Tedim AP, Lanza VF, Rodríguez CM, Freitas AR, Novais C, Peixe L, Baquero F, Coque TM. 2021. Fitness cost of vancomycin-resistant Enterococcus faecium plasmids associated with hospital infection outbreaks. J Antimicrob Chemother 76:2757–2764. doi: 10.1093/jac/dkab249. [DOI] [PubMed] [Google Scholar]
- 52.Zhang X, Vrijenhoek JEP, Bonten MJM, Willems RJL, van Schaik W. 2011. A genetic element present on megaplasmids allows Enterococcus faecium to use raffinose as carbon source. Environ Microbiol 13:518–528. doi: 10.1111/j.1462-2920.2010.02355.x. [DOI] [PubMed] [Google Scholar]
- 53.San Millan A, Heilbron K, MacLean RC. 2014. Positive epistasis between co-infecting plasmids promotes plasmid survival in bacterial populations. ISME J 8:601–612. doi: 10.1038/ismej.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: Rapid Annotations using Subsystems Technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanizawa Y, Fujisawa T, Nakamura Y. 2018. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics (Oxford, England) 34:1037–1039. doi: 10.1093/bioinformatics/btx713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics (Oxford, England) 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 58.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, Harris SR. 2018. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34:292–293. doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M. 2008. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics 9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 66.Sharp PM, Li W-H. 1987. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 70.Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aac.01619-22-s0001.pdf, PDF file, 2.7 MB (2.7MB, pdf)
Data Availability Statement
All assembly data are available from the NCBI database (JHP9, accession number SAMD00466802; JHP10, accession number SAMD00466803; JHP35, accession number SAMD00466804; JHP36, accession number SAMD00466805; JHP38, accession number SAMD00466806; JHP80, accession number SAMD00466807, AA55, accession number SAMD00466808; AA94, accession number SAMD00466809; AA96, accession number SAMD00466810; AA242, accession number SAMD00466811; AA290, accession number SAMD00466812; AA610 accession number SAMD00466813; AA818, accession number SAMD00466814; GK923, accession number SAMD00466815; GK941, accession number SAMD00466816; GK961, accession number SAMD00466817).