ABSTRACT
The extracellular capsule is a virulence factor present in many facultative pathogens, but its role in antimicrobial resistance remains controversial. To shed light on this debate, we tested six antibiotics on four Klebsiella pneumoniae species complex strains. Noncapsulated strains exhibited increased tolerance to polymyxins, but not to other antibiotics, as measured using the MIC. Our results urge caution on the use of therapeutic agents that target the capsule and may result in selection for its inactivation.
KEYWORDS: multidrug resistance, antimicrobial compounds, capsule, Klebsiella, ESKAPE pathogen, colistin
INTRODUCTION
In the last 20 years, the rise in multidrug resistant (MDR) infections has become a global health crisis that threatens the ability to treat many bacterial, viral, and fungal infections (1). The antimicrobial-resistant ESKAPE bacterial pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) are recognized by the World Health Organization as capable of panresistance and are currently a key focus of public health research (2). Among the ESKAPE pathogens, Klebsiella pneumoniae causes a wide range of infections, mostly in immunocompromised individuals (3). Virulence factors in hypervirulent Klebsiella strains include adhesins, lipopolysaccharide, and iron-scavenging systems (4, 5), but the best known is the polysaccharide capsule, a feature common to all ESKAPE pathogens. The capsule protects the cell from the bactericidal action of host serum, impairs phagocytosis (6), and increases survival in the presence of biotic stresses, such as killing by the type VI secretion system (7). Some studies have asserted that the capsule protects against antimicrobial peptides (AMPs) (8, 9), small naturally occurring peptides with broad inhibitory effects against bacteria. Among AMPs, polymyxin B and E (also known as colistin) are last-resort compounds commonly used in the clinic. However, the literature presents conflicting results on the defensive role of the capsule against AMPs, and specifically, polymyxins (8–12). The exogenous addition of purified capsule significantly increases cell survival by acting as a decoy (9). On the other hand, strains with loss-of-function mutations in wcaJ (11, 13) or wzc (13) are more resistant to polymyxins.
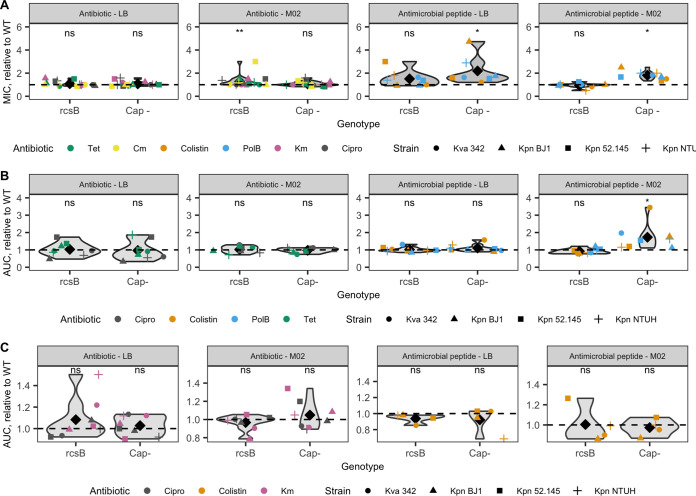
To test the impact of the capsule in antimicrobial resistance, we determined the MICs for four representative strains of the Klebsiella pneumoniae species complex (Table 1) and two types of isogenic mutants that produce a different quantity of capsule: the ΔrcsB mutant, which is capsulated but produces quantitatively less capsule than the wild type, and the ΔwcaJ or Δcps mutants, which are noncapsulated (see Fig. S1 in the supplemental material). These mutants were each compared to the respective wild type (WT). We assessed the MIC in the presence of last-resort polymyxins (polymyxin B and colistin) and four other conventional antibiotics with diverse mechanisms of action (Table S1) in a nutrient-rich (LB) and a nutrient-limited (M02) environment. Analyses of the 144 different combinations of antibiotic, strain, and environment revealed that noncapsulated strains displayed a significantly higher MIC than the WT in both media when treated with polymyxins but not the other antibiotics (Fig. 1A; Fig. S2). The difference between capsulated and noncapsulated variants was consistent across strains, but in the nutrient-limited medium, we observed that it was particularly high in the hypervirulent strain K. pneumoniae NTUH. Furthermore, in this strain, such a difference in MIC was also observed when it was exposed to kanamycin. Stepwise linear regression analyses confirmed that the MIC values were strongly affected by the type of antibiotic and the nutrient conditions, as expected, as well as by the capsule genotype (linear regression, R2 = 0.87; P < 0.01 for all factors; Table S3). Despite the expected differences across media in the MIC values (20, 21), the decrease in susceptibility of the noncapsulated strains was observed in both the nutrient-rich and nutrient-limited conditions, showing that it does not rely uniquely on the cellular metabolic state.
TABLE 1.
Strains used in this studya
| Strain | Species | Isolation source | ST | K serotype | O serotype | rmpA | Capsule mutants | Genome size (MB) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| NTUH-K2044 | K. pneumoniae | Liver abscess, Taiwan | ST23 | K1 | O1v2 | rmpA, rmpA | ΔrcsB, ΔwcaJ | 5.25 | 14 |
| BJ1 | K. pneumoniae | Liver abscess, France | ST380 | K2 | O1v1 | rmpA | ΔrcsB, ΔwcaJ | 5.26 | 15 |
| CIP 52.145 | K. pneumoniae | Indonesia | ST66 | K2 | O1 | ΔrcsB, Δcps | 5.44 | 16 | |
| 342 | Klebsiella variicola | Maize, USA | ST146 | K30 | O3/O3a | ΔrcsB, ΔwcaJ | 5.64 | 17 |
FIG 1.
The capsule impacts the MIC and growth when the cell is challenged with antibiotics. (A) MIC relative to the wild type. Each point represents the mean across all replicates for each genotype from each strain with each antimicrobial compound. Black diamonds depict the mean. (B) Area under the curve (AUC) relative to the wild type when grown at subinhibitory concentrations (0.5× MIC) of tetracycline, colistin, polymyxin B, and ciprofloxacin. Qualitatively similar results are obtained when treated with 0.25 and 0.2 MIC (see Fig. S4 in the supplemental material). (C) AUC relative to the wild type of bacterial survival when treated with 10× MIC of kanamycin, colistin, and ciprofloxacin. Qualitatively similar results were obtained for treatment with 50× MIC. Wilcoxon rank sum two-sided test (difference from 1). *, P < 0.05; **, P < 0.01; ns, not significant (P ≥ 0.05). Tet, tetracycline; Cm, chloramphenicol; PolB, polymyxin B; Km, kanamycin; Cipro, ciprofloxacin. LB indicates the nutrient-rich medium, whereas M02 corresponds to the nutrient-limited medium, with glucose as the sole carbon source. Strain Klebsiella variicola 342 was not considered for the ΔrcsB statistical calculations, since it did not impact the capsule production (see Fig. S1 in the supplemental material). The statistics were not affected when strain K. pneumoniae CIP 52.145 and its Δcps mutant were removed.
The decreased susceptibility of the noncapsulated bacteria to polymyxins may have been due to a smaller effect on the growth rate or a decreased killing rate. To test this hypotheses, we first assessed the area under the curve (AUC) of the growth curves in the presence of subinhibitory concentrations of each polymyxin and of two other antibiotics, the bacteriostatic tetracycline and the bactericidal ciprofloxacin. There was no significant difference between the capsulated and noncapsulated strains in the nutrient-rich medium when they were grown with the antimicrobials (0.5× MIC; Fig. 1B). However, in the nutrient-limited medium, the noncapsulated variants grew better in the presence of polymyxins (0.5× MIC; Fig. 1B), even in the presence of larger absolute amounts of antimicrobial peptides (Table S2), compared to the capsulated strains. This growth advantage is all the more remarkable as there is a fitness cost of not producing a capsule in M02 medium (22) (Fig. S3). Qualitatively similar results were obtained when strains were grown at 0.1× and 0.25× MIC (Fig. S4). A stepwise regression model indicated that the antibiotics, strain, and then nutrients influenced the AUC, while no effect due to genotype was observed (linear regression, R2 = 0.55; Table S3).
To test whether the decreased susceptibility of noncapsulated strains was due to a decreased death rate, we measured the bacterial survival when exposed to high concentrations of antibiotics (10× or 50× MIC). As expected, the bacteria died quickly (Fig. S5). No significant differences were observed between capsulated and noncapsulated cells, irrespective of the growth medium (Fig. 1C). We conclude that the presence of the capsule does not impact the death rate at high antimicrobial concentrations. Interestingly, regrowth, and thus tolerance, was readily observed when bacterial cells were exposed to high doses of colistin in LB, as previously reported (13) (Fig. S5A). In contrast, no regrowth was observed in M02 medium. Similarly, we found surviving subpopulations of the K2 strains (K. pneumoniae BJ1 and K. pneumoniae CIP 52.145) when cells were grown in kanamycin and ciprofloxacin, in agreement with previous experiments (23, 24) (Fig. S5B).
The decreased susceptibility of noncapsulated strains supports recent research suggesting that noncapsulated variants are more resistant to polymyxins (11) and that capsule inactivation is an efficient mechanism of resistance (13). Yet several groups have independently shown that the exogenously added capsule, or other exopolysaccharides, can bind AMPs, including polymyxins (9, 25). This was interpreted as evidence that the capsule provides resistance by sequestering the AMPs, thereby potentially preventing their entrance into the cell and reducing the polymyxin killing action. In light of our experiments, we speculate that when the capsule is added exogenously, this results in titration of the AMPs, and bacteria can withstand larger doses of the antimicrobial. When the capsule is in its natural biological role, i.e., attached to the outer membrane, this results in higher concentrations of antimicrobials near the cell. The increased concentration of antimicrobial peptides near the outer membrane is particularly helpful for antimicrobial peptides like colistin, whose mode of action is to bind to lipopolysaccharides and phospholipids in the outer cell membrane, leading to the disruption of the outer cell membrane and cell lysis (26, 27). The detrimental effect of the capsule was mostly observed in low-nutrient conditions, where the capsule is most expressed (Fig. S1). We hypothesize that the slower growth in the nutrient-limited medium was associated with slower renewal of the cell wall (and capsule), which could have resulted in higher concentrations of the antimicrobial peptides than in situations of faster growth. As a result, the bactericidal action of the peptides was increased.
To conclude, our observations may have important clinical implications. First, our results confirm previously observed data (19) showing that hypervirulent strains (i.e., K. pneumoniae NTUH and BJ1), despite their hypermucoviscosity, do not seem to exhibit increased tolerance to antibiotics, as measured using the MIC. Second, the use of therapies that may target the capsule, such as conjugate vaccines (28) or phage therapy, which oftentimes results in capsule inactivation (29, 30) and potentially in reduced virulence, could be a double-edged sword. We show that it may result in decreased intrinsic susceptibility to last-resort antimicrobials, while increasing the spread of plasmid-borne antibiotic resistance (31). Finally, these results challenge the view that capsules systematically protect against antimicrobials. Instead, we advise that the effects of capsules on resistance be systematically tested, since they may depend on the type of antimicrobial agent and on the species.
Data availability.
Data used in this study are available at the following link: https://doi.org/10.6084/m9.figshare.21400050.v1.
ACKNOWLEDGMENTS
We thank Jean-Marc Ghigo and Christophe Beloin for plasmid pKOBEG199 and the original KmFRT cassette, and we thank Didier Mazel for the gift of plasmid pMPIII, containing a flippase to excise kanamycin resistance, necessary for building the Δcps mutant in K. pneumoniae CIP 52.145. We are grateful to Manuel Ares Arroyo and Javier Pizarro Cerda for critical reading of the manuscript and to the Microbial Evolutionary Genomics lab for helpful discussions.
F.D. was funded by an ANR (Agence National de la Recherche) JCJC grant (ANR 18 CE12 0001 01 ENCAPSULATION) awarded to O.R. The laboratory is funded by a Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” grant (ANR-10-LABX-62-IBEID) and the FRM (EQU201903007835). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare that we do not have any competing interests in relation to the work described.
O.R. conceived, designed, and coordinated the study. F.D. and O.R. carried out the experimental lab work, performed data analysis, and wrote the first draft of the manuscript. E.P.C.R. critically revised the manuscript. All authors gave final approval for publication and agree to be held accountable for the work performed herein.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Antimicrobial Resistance Collaborators. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399:629–655. 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Oliveira DMP, Forde BM, Kidd TJ, Harris PNA, Schembri MA, Beatson SA, Paterson DL, Walker MJ. 2020. Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev 33:e00181-19. 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paczosa MK, Mecsas J. 2016. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661. 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu M-C, Lin T-L, Hsieh P-F, Yang H-C, Wang J-T. 2011. Isolation of genes involved in biofilm formation of a Klebsiella pneumoniae strain causing pyogenic liver abscess. PLoS One 6:e23500. 10.1371/journal.pone.0023500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortés G, Borrell N, de Astorza B, Gómez C, Sauleda J, Albertí S. 2002. Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect Immun 70:2583–2590. 10.1128/IAI.70.5.2583-2590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flaugnatti N, Isaac S, Lemos Rocha LF, Stutzmann S, Rendueles O, Stoudmann C, Vesel N, Garcia-Garcera M, Buffet A, Sana TG, Rocha EPC, Blokesch M. 2021. Human commensal gut proteobacteria withstand type VI secretion attacks through immunity protein-independent mechanisms. Nat Commun 12:5751. 10.1038/s41467-021-26041-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campos MA, Vargas MA, Regueiro V, Llompart CM, Albertí S, Bengoechea JA. 2004. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun 72:7107–7114. 10.1128/IAI.72.12.7107-7114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llobet E, Tomás JM, Bengoechea JA. 2008. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology (Reading) 154:3877–3886. 10.1099/mic.0.2008/022301-0. [DOI] [PubMed] [Google Scholar]
- 10.Held TK, Adamczik C, Trautmann M, Cross AS. 1995. Effects of MICs and sub-MICs of antibiotics on production of capsular polysaccharide of Klebsiella pneumoniae. Antimicrob Agents Chemother 39:1093–1096. 10.1128/AAC.39.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pal S, Verma J, Mallick S, Rastogi SK, Kumar A, Ghosh AS. 2019. Absence of the glycosyltransferase WcaJ in Klebsiella pneumoniae ATCC13883 affects biofilm formation, increases polymyxin resistance and reduces murine macrophage activation. Microbiology (Reading) 165:891–904. 10.1099/mic.0.000827. [DOI] [PubMed] [Google Scholar]
- 12.Fleeman RM, Macias LA, Brodbelt JS, Davies BW. 2020. Defining principles that influence antimicrobial peptide activity against capsulated Klebsiella pneumoniae. Proc Natl Acad Sci USA 117:27620–27626. 10.1073/pnas.2007036117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janssen AB, Doorduijn DJ, Mills G, Rogers MRC, Bonten MJM, Rooijakkers SHM, Willems RJL, Bengoechea JA, van Schaik W. 2020. Evolution of colistin resistance in the Klebsiella pneumoniae complex follows multiple evolutionary trajectories with variable effects on fitness and virulence characteristics. Antimicrob Agents Chemother 65:e01958-20. 10.1128/AAC.01958-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu K-M, Li L-H, Yan J-J, Tsao N, Liao T-L, Tsai H-C, Fung C-P, Chen H-J, Liu Y-M, Wang J-T, Fang C-T, Chang S-C, Shu H-Y, Liu T-T, Chen Y-T, Shiau Y-R, Lauderdale T-L, Su I-J, Kirby R, Tsai S-F. 2009. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol 191:4492–4501. 10.1128/JB.00315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blin C, Passet V, Touchon M, Rocha EPC, Brisse S. 2017. Metabolic diversity of the emerging pathogenic lineages of Klebsiella pneumoniae. Environ Microbiol 19:1881–1898. 10.1111/1462-2920.13689. [DOI] [PubMed] [Google Scholar]
- 16.Nassif X, Fournier JM, Arondel J, Sansonetti PJ. 1989. Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect Immun 57:546–552. 10.1128/iai.57.2.546-552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fouts DE, Tyler HL, DeBoy RT, Daugherty S, Ren Q, Badger JH, Durkin AS, Huot H, Shrivastava S, Kothari S, Dodson RJ, Mohamoud Y, Khouri H, Roesch LFW, Krogfelt KA, Struve C, Triplett EW, Methé BA. 2008. Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet 4:e1000141. 10.1371/journal.pgen.1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. 2021. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 12:4188. 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nucci A, Rocha EPC, Rendueles O. 2022. Adaptation to novel spatially-structured environments is driven by the capsule and alters virulence-associated traits. Nat Commun 13:4751. 10.1038/s41467-022-32504-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 21.Pontes MH, Groisman EA. 2020. A physiological basis for nonheritable antibiotic resistance. mBio 11:e00817-20. 10.1128/mBio.00817-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buffet A, Rocha EPC, Rendueles O. 2021. Nutrient conditions are primary drivers of bacterial capsule maintenance in Klebsiella. Proc Biol Sci 288:20202876. 10.1098/rspb.2020.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren H, He X, Zou X, Wang G, Li S, Wu Y. 2015. Gradual increase in antibiotic concentration affects persistence of Klebsiella pneumoniae. J Antimicrob Chemother 70:3267–3272. 10.1093/jac/dkv251. [DOI] [PubMed] [Google Scholar]
- 24.Abokhalil RN, Elkhatib WF, Aboulwafa MA, Hassouna NA. 2020. Persisters of Klebsiella pneumoniae and Proteus mirabilis: a common phenomenon and different behavior profiles. Curr Microbiol 77:1233–1244. 10.1007/s00284-020-01926-3. [DOI] [PubMed] [Google Scholar]
- 25.Sabnis A, Ledger EVK, Pader V, Edwards AM. 2018. Antibiotic interceptors: creating safe spaces for bacteria. PLoS Pathog 14:e1006924. 10.1371/journal.ppat.1006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen CH, Lu TK. 2020. Development and challenges of antimicrobial peptides for therapeutic applications. Antibiotics (Basel) 9:24. 10.3390/antibiotics9010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrade FF, Silva D, Rodrigues A, Pina-Vaz C. 2020. Colistin update on its mechanism of action and resistance, present and future challenges. Microorganisms 8:1716. 10.3390/microorganisms8111716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feldman MF, Mayer Bridwell AE, Scott NE, Vinogradov E, McKee SR, Chavez SM, Twentyman J, Stallings CL, Rosen DA, Harding CM. 2019. A promising bioconjugate vaccine against hypervirulent Klebsiella pneumoniae. Proc Natl Acad Sci USA 116:18655–18663. 10.1073/pnas.1907833116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hesse S, Rajaure M, Wall E, Johnson J, Bliskovsky V, Gottesman S, Adhya S. 2020. Phage resistance in multidrug-resistant Klebsiella pneumoniae ST258 evolves via diverse mutations that culminate in impaired adsorption. mBio 11:e02530-19. 10.1128/mBio.02530-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song L, Yang X, Huang J, Zhu X, Han G, Wan Y, Xu Y, Luan G, Jia X. 2021. Phage selective pressure reduces virulence of hypervirulent Klebsiella pneumoniae through mutation of the wzc gene. Front Microbiol 12:739319. 10.3389/fmicb.2021.739319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haudiquet M, Buffet A, Rendueles O, Rocha EPC. 2021. Interplay between the cell envelope and mobile genetic elements shapes gene flow in populations of the nosocomial pathogen Klebsiella pneumoniae. PLoS Biol 19:e3001276. 10.1371/journal.pbio.3001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aac.00127-23-s0001.pdf, PDF file, 0.6 MB (660KB, pdf)
Data Availability Statement
Data used in this study are available at the following link: https://doi.org/10.6084/m9.figshare.21400050.v1.