Summary
Cancer genomes harbor a broad spectrum of structural variants (SVs) driving tumorigenesis, a relevant subset of which escape discovery using short-read sequencing. We employed Oxford Nanopore Technologies (ONT) long-read sequencing in a paired diagnostic and post-therapy medulloblastoma to unravel the haplotype-resolved somatic genetic and epigenetic landscape. We assembled complex rearrangements, including a 1.55-Mbp chromothripsis event, and we uncover a complex SV pattern termed templated insertion (TI) thread, characterized by short (mostly <1 kb) insertions showing prevalent self-concatenation into highly amplified structures of up to 50 kbp in size. TI threads occur in 3% of cancers, with a prevalence up to 74% in liposarcoma, and frequent colocalization with chromothripsis. We also perform long-read-based methylome profiling and discover allele-specific methylation (ASM) effects, complex rearrangements exhibiting differential methylation, and differential promoter methylation in cancer-driver genes. Our study shows the advantage of long-read sequencing in the discovery and characterization of complex somatic rearrangements.
Keywords: long read sequencing, cancer genomics, templated insertions, complex rearrangements, epigenetic signatures, Nanopore methylation calling, chromothripsis
Graphical abstract

Highlights
-
•
Methods for the application of long-read sequencing in cancer genomics
-
•
Discovery of a complex genomic pattern termed templated insertion thread (TI thread)
-
•
TI threads are characterized by short insertions that are self- and cross-linked
-
•
Long-read-based methylome profiling reveals allele-specific methylation
Long-read sequencing in paired diagnostic and post-therapy medulloblastoma samples uncovers a complex DNA rearrangement pattern termed templated insertion thread (TI thread), characterized by short insertions showing prevalent self- and cross-chaining into amplified structures of up to 50 kbp in size. Pan-cancer screening using short reads discovers TI threads in multiple tumor types, with enrichment in dedifferentiated liposarcoma. Our study provides methods for long-read (epi)genome profiling and the discovery and characterization of complex rearrangements in cancer.
Introduction
Cancer genomic landscapes are shaped by a diversity of somatic rearrangement patterns, ranging from simple deletions, duplications, and reciprocal translocations to structural variants (SVs) formed via complex DNA rearrangements, including breakage-fusion-bridge cycles and chromothripsis events.1,2,3,4 SVs are the most common source of cancer-driver mutation, outnumbering point mutations for the generation of cancer drivers in the majority of common cancers.2 However, owing to technical difficulties with respect to their discovery and characterization,5 their structure and patterns remain underexplored compared with point mutations.2 This is particularly true for complex DNA rearrangements, the characterization of which remains an important challenge, with short-read (Illumina) sequencing data only partially resolving their sequence structures.3
Initial efforts to classify somatic SVs uncovered a variety of common somatic rearrangement patterns, which suggests that a wide variety of rearrangement processes are active in cancer. Using non-negative matrix factorization, Nik-Zainal et al.6 initially described six signatures of rearrangement in breast cancers sequenced using Illumina technology. More recent pan-cancer studies,3,7 again pursued using short-read data, combined simple SVs (e.g., deletion type, duplication type, and inversion-type) into discrete higher-level patterns based on breakpoint junction connectivity, resulting in over a dozen SV signatures. This included patterns of intermediate rearrangement complexity, such as templated insertion (TI) chains comprising up to 10 breakpoints. However, more complex rearrangement patterns have so far largely resisted systematic classification based on breakpoint junction connectivity. An important reason for this is difficulty in assembling short-read data into coherent structural segments to study patterns of somatic rearrangements. This problem is exacerbated by repetitive sequences in the genome, in which SV breakpoints are readily missed by Illumina whole-genome sequencing (WGS). This leaves open the possibility that important patterns of structural rearrangement have not yet been discovered and are elusive due to the predominant use of short-read sequencing in cancer genomics.2
Here we sought to evaluate the utility of long-read sequencing technology,8,9,10,11 in particular Oxford Nanopore technology (ONT), to reveal patterns of somatic structural variation. The technological choice was motivated by the fact that long-read sequencing of 1000 Genomes Project samples showed a greatly increased number of confidently discovered SVs in repetitive regions, improved sensitivity for SVs smaller than 1 kbp in size, and advantages for investigating complex SV patterns by facilitating haplotype-resolved genomic sequence assembly.12,13,14 ONT additionally shows great promise in cancer epigenomics, as, from the same long reads, both genetic and DNA methylome data can be obtained, the latter of which is quantified through measuring current changes within the nanopore,15 which should allow integrated characterization of genetic and epigenetic changes in tumors at single (long) molecule level. However, there is a current lack in suitable computational methods and hence a need in exploring and devising approaches leveraging long-read data in cancer genomes, with the complications of intra-tumor heterogeneity (ITH) in primary cancer samples, normal cell contamination, aneuploidy, and complex SVs, and variation in tumor methylation levels.
To address the current lack of long-read analytical methods to explore cancer genomes, we performed ONT sequencing of a childhood medulloblastoma and devised methods to enable characterizing SV and methylome patterns in these data. The tumor arose in a patient carrying a germline TP53 mutation (Li-Fraumeni syndrome; OMIM: 151623), previously associated with Sonic Hedgehog subgroup medulloblastoma (SHH-MB) and somatic chromothripsis.16,17 We reveal the fully assembled haplotype-resolved structure of a complex chromothripsis event.16,18 We further uncover a novel complex rearrangement pattern, termed TI thread, which copies and concatenates a substantial number of short subkilobase-sized TIs in forward and reverse orientation, resulting in massively amplified sequences ranging up to several tens of kilobases in size. While not initially discovered by Illumina WGS, we demonstrate that common features associated with TI threads allow their discovery in cancer genomes sequenced with short reads. A search for these patterns in 2,569 short-read cancer genomes from the Pan-Cancer Analysis of Whole Genomes (PCAWG) consortium2 reveals the footprints of TI threads in 3% of cancer genomes, with a particular abundance in liposarcoma (74%), glioblastoma (24%), osteosarcoma (22%), and melanoma (14%). TI threads can occasionally be found near overexpressed oncogenes, suggesting that cancer cells could exploit this somatic SV pattern to promote tumor evolution. Last, by integrating genomic and epigenomic readouts, we performed haplotype-resolved genome-wide analysis of CpG methylation. We associate a subset of the somatic DNA rearrangements, including TI threads, with functional consequences, and demonstrate the ability to explain aberrant gene-expression patterns, such as allele-specific expression and gene fusions, by integrating genomic and epigenetic long-read data.
Results
ONT-based integrated phasing and SV discovery in a medulloblastoma patient
We sequenced the primary medulloblastoma (sample ID: LFS_MB_P) to ∼30× ONT coverage, and generated ∼15× for a tumor specimen taken during relapse (LFS_MB_1R) and a paired blood control sample, respectively, with a median mapped read length of 5 kbp (Table S1). We developed workflows and algorithms to analyze both genetic and epigenetic alterations in these samples (STAR Methods). Making use of short-read data generated at 45–48× coverage for these samples17,19,20 (Table S2), we discovered single-nucleotide variants (SNVs) as well as short insertions and deletions (InDels), where ONT reads have limitations due to their relatively high error rate. As expected, germline variant calling confirmed a TP53 mutation (TP53:c.395A>G, p.Lys132Arg), consistent with Li-Fraumeni syndrome, coupled with somatic inactivation of the wild-type TP53 allele via somatic deletion in the tumor samples. To facilitate allele-specific analysis, we devised a haplotype-phasing approach that generates initial haplotype blocks from ONT reads using WhatsHap,21 which then are integrated with statistical haplotype-phasing data from the 1000 Genomes Project22 using ShapeIt.23 Haplotype switch errors are corrected by leveraging somatic copy-number alterations (SCNA) in the tumor that result in allelic shifts away from the normal 1:1 haplotype ratio (Figure S1). In regions of the genome without SCNAs, we estimate an N50 phased block length of 4.68 Mbp using this approach (STAR Methods). The estimated proportion of the somatic genome that is haplotype resolvable using our phased germline variant call set is 91.1% for the primary tumor and 89.9% for the relapse sample, respectively.
Haplotype-phased assembly of complex somatic rearrangements
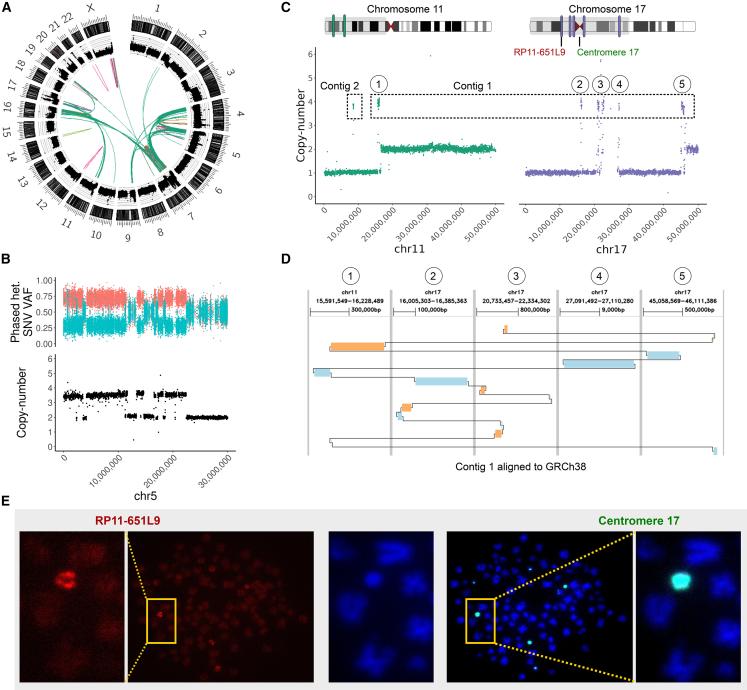
We integrated ONT-based somatic SV calling with Illumina-based SCNAs and variant detection to achieve haplotype-resolved reconstruction of the somatic SV landscape of this tumor (STAR Methods). In the primary tumor, we found 697 somatic SVs, including 106 deletion-type SVs, 107 duplication-type SVs, 189 inversion-type SVs, 295 inter-chromosomal rearrangements, and a copy-number profile with many sub-clonal changes, indicating heterogeneity within the tumor (Figures S2 and S3). Most of these rearrangements arose from two distinct chromothripsis events: one involving chromosomes 4, 5, 7, 9, 16, 19, and X, and the other chromosomes 11 and 17, respectively (Figures 1A, 1B, and S4). We next explored targeted phased assembly of the genomic outcomes of both chromothripsis events (STAR Methods). We constructed SV contigs for the chromothripsis event spanning chromosomes 4, 5, 7, 9, 16, 19, and X, and generated a phased assembly of fragments originating from chromosome 11 and 17 (denoted CS11-17, Figures 1C and 1D). The CS11-17 segment, present in both primary tumor and relapse, has a size of 1.55 Mbp; wild-type TP53 located on the 17p-arm region of the chromothriptic haplotype has been lost. We estimated an average copy number of 3–4 copies for CS11-17, consistent with fluorescence in situ hybridization (FISH) experiments (Table S3). FISH further reveals extensive ITH of CS11-17 copy numbers, which range from 1 to 7 (Tables S3–S6). We performed sequence-level characterization of CS11-17, and partially resolved peri-centromeric regions at its flanks (Figures 1C and 1D), which could provide the necessary sequence context for homology-based integration into the normal genome as observed previously for double minutes18 (Figure S5A). FISH analysis on metaphase spreads did not detect classical double-minute chromosomes (Figure S5B), but we identified structures that could represent marker chromosomes or ring chromosomes (Figure 1E). We also failed to identify reads supporting reintegration of this structure into a chromosomal context, possibly due to limitations in read depth and the ONT read length achieved for these primary patient samples, or problems in resolving low-variant allele frequency SVs in conjunction with ITH, especially in complex regions that exhibit repetitive segments larger than the ONT read length (Figure S6).24 We further validated this structure by using an orthogonal method for detecting circular DNA enrichment via purification and sequencing of extrachromosomal circular DNA (Circle-seq),25 and demonstrate that CS11-17 is potentially circular (Figure S7).
Figure 1.
Haplotype-phased assembly of an inter-chromosomal chromothripsis event
(A) A circos plot of the primary tumor showing, from outside to inside, the chromosome ideograms, read-depth, large (>10 Mbp) SVs, and inter-chromosomal rearrangements: orange, deletion-type; violet, duplication-type; light green, head-to-head inversion-type; pink, tail-to-tail inversion-type; dark green, inter-chromosomal.
(B) Chromosome 5 exhibits a pattern of oscillating copy-number states (lower panel) and alternating heterozygous allele frequencies (upper panel) common to chromothripsis.
(C) The CS11-17 assembly contains two contigs with amplified segments from chromosome 11 and chromosome 17. Segments from chromosome 11 are in green, segments from chromosome 17 in purple. The part of the chromosomes displayed (1–50 Mbp) is shown with a gray background in the chromosome ideograms as well as the locations of the amplified segments (green and purple segments).
(D) A chained alignment of contig 1 of the CS11-17 assembly against GRCh38. Forward matches are in blue, reverse matches in ochre. Matches are clustered within 1 Mbp, and distinct alignment regions are separated by a vertical gray line. Numbers 1–5 correspond to amplicons labeled as 1–5 in (C).
(E) FISH analysis identifies potential marker or ring chromosomes associated with the CS11-17 structure by means of localized signals of the red RP11-651L9 probe (chr17:16,169,409–16,359,715), shown in the left two panels, and the green centromere 17 probe, shown in the right two panels. The boxed structure (yellow) contains a putative ring or marker chromosome with enlarged views in the outer panels.
ONT sequencing reveals a novel complex rearrangement pattern denoted TI thread
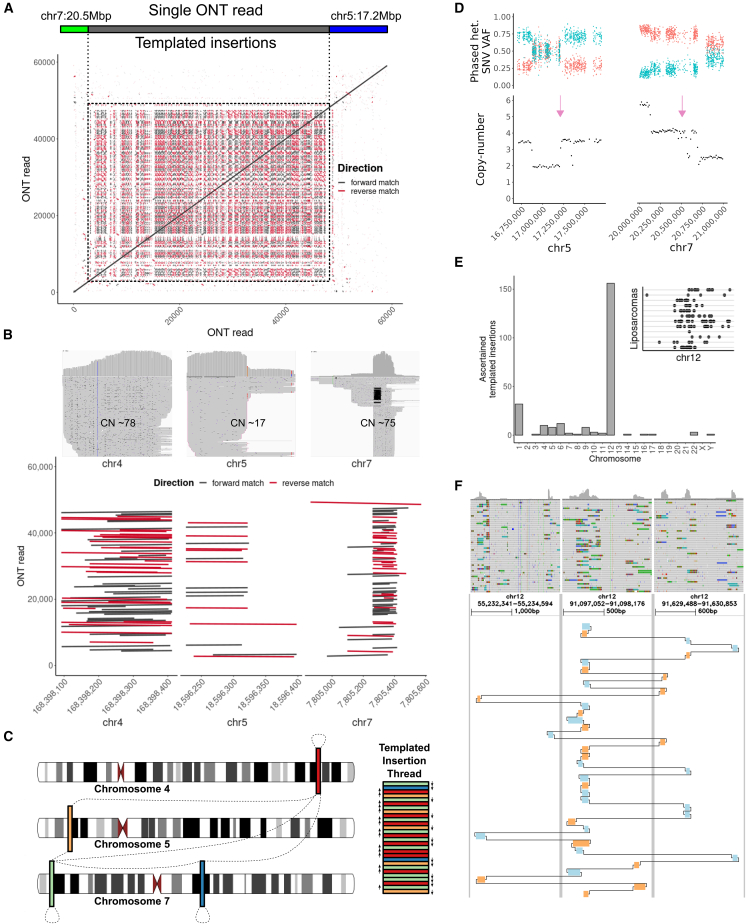
Notably, the somatic SVs seen in the primary tumor included a highly unusual pattern of inter-chromosomal DNA rearrangement not matching previously described somatic SV classes. This rearrangement pattern involves short DNA segments, mostly 100 bp to 1 kbp in size, that are concatenated by a structural rearrangement process in forward and reverse order, into a complex, highly amplified sequence comprising up to 50 kbp of DNA and dozens to hundreds of breakpoint junctions (Figure 2A). We found two such structures in the primary tumor with a length of the source sequence segments ranging from 144 to 3,637 bp, with all source segments with an estimated total copy number greater than 10 being between 225 and 403 bp in size. The total length of the resulting somatic amplicon structure is 50.3 kbp for the first structure (Figure 2B) and 39.9 kbp for the second structure (Figures S8 and S9). Both of these structures result in inter-chromosomal adjacencies, via concatenation of TIs stemming from distinct chromosomes. We obtained additional support for this rearrangement structure using raw long reads, targeted assembly, and de novo assembly approaches, including Flye26 and Shasta27 (Figure S10), using long-read sequencing in a matched patient-derived xenograft model (Figure S11), as well as indirectly via short reads using depth of coverage and split reads (Figures 2B and S12). Sequence analysis of these structures, and leveraging the full length of the ONT reads, suggests that these structures have likely emerged from TIs3 through a copy-and-paste process with no apparent regularity in the alignment of the concatenated source sequence segments (Figures 2C and S13). Based on the complexity and genomic appearance of the respective rearrangements, we term this novel pattern TI thread.
Figure 2.
TI threads
(A) Self-alignment of a single ONT read that spans the entire length of the TI thread, displaying an array of repetitive short sequence matches reflecting the copying and concatenation of few source sequence segments.
(B) Matched Illumina data show a characteristic coverage increase (upper panel) in Integrative Genomics Viewer (IGV).28 An alignment of the ONT read (y axis) against selected TI source sequences (x axis) shows how the ONT read aligns across these source sequences multiple times in seemingly random order.
(C) A scheme showing how TIs are copied and pasted in direct adjacency and random order into a growing TI thread. Arrows next to the TI thread indicate the segment orientation and dashed lines show discovered adjacencies among individual TIs.
(D) The colocalization of the beginning and the end of the TI thread (purple arrow) with chromothripsis segments on chromosome 5 (left) and chromosome 7 (right).
(E) Analysis of 2,569 cancer genomes reveals that liposarcomas often harbor TI threads, preferentially on chromosome 12 (main panel). The inset shows the distribution of TIs along chromosome 12 where each horizontal line is a distinct liposarcoma sample.
(F) A liposarcoma validation sample (P1) sequenced using long reads confirms the TI thread signature. Chained alignment matches to GRCh38 are shown for a single ONT read with forward matches in blue and reverse matches in ochre. Aligned segments show strong coverage increases in the matched Illumina short-read data (top panel; IGV28) with SV-supporting reads and soft-clips.
A comparison with previously described rearrangement patterns shows that the TI thread pattern shares features with the chains of TIs pattern previously described by Li et al. using PCAWG data,3 genomic shards described by Bignell et al. in bacterial artificial chromosomes,29 and the tandem short template jumps signature previously uncovered by Umbreit et al. in cell cultures,30 albeit with clear differences. While all these patterns concatenate TIs originating from distinct genomic locations, the most distinguishing feature of TI threads is the prevalent self-concatenation of TIs in a zigzag fashion, which result in short amplicons of remarkably high copy number (Figures 2B, 2C, S14, and S15); by comparison, the units comprising chains of TIs occur only once (no self-concatenation) in the previously described patterns.3,30 As an additional discriminating feature, chains of TIs as described by Li et al.3 comprise from 1 to 10 concatenated units, compared with >50 units included within a single TI thread in this medulloblastoma sample (see Figure S14).
We performed further analyses of the spanning ONT reads and found the TI threads to colocalize with chromothriptic rearrangements (Figure 2D). It is therefore possible that the rearrangement processes resulting in both event classes share some commonality, either with one event triggering the other, or with both chromothripsis and TI threads enabled by the same initiating DNA lesion. Analysis of the repeat units (source sequence segments) becoming self-concatenated in TI threads did not reveal any biases toward a specific sequence context; in the majority of cases, individual units originate from non-repetitive sequence (STAR Methods). A breakpoint junction analysis of TI threads shows a predominance of 0- to 5-bp microhomology length (Figure S16), indicative of alternative end-joining (alt-EJ) repair or microhomology-mediated end-joining (MMEJ). Notably, Circle-seq analysis of the respective sample (STAR Methods) did not reveal circular enrichment of the TI threads and thus provided no evidence for circular intermediates during TI thread formation. Interestingly, comparative alignment of ONT reads from the same sample revealed evidence for ITH with respect to the unit composition of TI threads, with clear differences in concatenated unit numbers becoming evident; this suggests that sites of TI thread events may be prone to undergo further somatic rearrangements generating further genetic heterogeneity (Figures S17–S19).
We notably did not identify TI threads in the relapse sample. A comparison of somatic mutations between primary and relapse showed that only 34% of all somatic SNVs are shared between both specimens (Figures S20 and S21). Among the relapse-specific acquired somatic SNVs is a 2-bp frameshift insertion in the tumor-suppressor gene SUFU (Figure S22). These data suggest that primary and relapse evolved from a distant common ancestor. The TI threads detected in the primary tumor, which may have played a driver role in the early stages of tumor development, do not appear to provide a selective advantage upon treatment. Alternatively, tumor cells with these rearrangements might have been eradicated during treatment.
Graph-based discovery of TI threads in Illumina WGS data
Most previously sequenced cancer genomes have been generated using short reads, which, compared with long reads, display poor sensitivity toward <1 kbp-sized rearrangements,13 the predominant rearrangement type within TI threads. Irrespective of this, we hypothesized that the distinguishing features of TI threads should be discoverable in short-read data once explicitly sought for, to allow further analysis of this novel SV pattern in large short-read based cancer genome cohorts. To address this hypothesis, we first closely examined the Illumina WGS reads from LFS_MB_P at the sites of TI threads. Indeed, we find specific short-read alignment patterns characteristic of self- and cross-linked sequence segments at the respective rearranged sites, with an exceptionally high copy number of source segments and paired-end as well as split-read support for rearrangement junctions (Figure S12). Encouraged by this observation, we devised the graph-based algorithm rayas, to enable the discovery and characterization of TI threads in short-read WGS data (STAR Methods). The algorithm combines read-depth and split-read patterns to identify rearrangement graphs, allowing the specification of 1:n relationships, whereby a single TI source sequence (i.e., a node in the graph) can contribute to different rearrangement adjacencies (i.e., edges in the graph; Figures S23). Application of rayas to the primary and relapse samples led to the re-discovery of both TI threads in the primary medulloblastoma, and confirmed the absence of these structures in the relapse.
Pan-cancer landscape of TI threads
The ability of TI threads to amplify short sequences suggests a potentially broader relevance in cancer, since amplified DNA sequences could potentially act as cancer drivers, such as by focally amplifying DNA regulatory sequences or altering the gene regulatory context to result in ectopic expression.2,31,32 To enable a wider characterization of this SV pattern, we used rayas to interrogate 2,569 cancer genomes from the PCAWG consortium.2 We found 169 TI threads in 76 (∼3%) cancer genomes, which suggests that this somatic rearrangement pattern arises in distinct cancers (Figure S24; Table S7). Across cancers, the distribution of this pattern is highly heterogeneous, with 74% of liposarcomas, 24% of glioblastoma, and 14% of melanomas exhibiting TI threads, versus 7% of leiomyosarcomas (Figure S24). We caution that, due to the lower sensitivity of short reads for detecting complex SVs involving short repeat units,13 future studies with larger cohorts of cancer samples sequenced with long reads will likely offer increased sensitivity for the detection of TI threads in cancer genomes.
On average, TI threads consist of four distinct source segments with a median unit size of 558 bp, and median number of concatenated units of 53.1, indicating that high copy number amplification is the norm rather than the exception for this SV pattern. We next analyzed the 76 cancer genomes bearing TI threads in further detail, to determine features that may potentially correlate with the occurrence of TI threads. Additionally, 65 out of these 76 cancer genomes (86%) were previously classified as having at least one chromothripsis event.2 The association of TI threads with chromothripsis is significant across 2,569 cancers, when adjusting for tumor histology, gender, and ancestry (p = 1.15 × 10−5, logistic regression). Interestingly we find a strong enrichment of TIs on chromosome 12 in liposarcoma samples, with a propensity toward the 12q15 chromosome band (Figure 2E). Liposarcomas often form supernumerary ring or giant marker chromosomes that include multiple copies of the target oncogenes (MDM2, CDK4, among others) on chromosome 12, a chromosome that frequently undergoes chromothripsis in this cancer type.19,33,34 Recent studies also identified chromosome 12 as a hotspot for seismic amplification and tyfonas in liposarcoma.7,35 These data suggest that TI threads could arise in association with supernumerary ring or giant marker chromosomes, possibly triggered by the same initiating lesions or through a common rearrangement process.
To confirm the co-occurrence of TI threads and giant marker chromosomes, we used rayas to interrogate 17 short-read-sequenced liposarcoma samples from the NCT/DKTK Master cohort.36 Rayas identified evidence for TI threads in six out of seven (86%) dedifferentiated liposarcoma patients from the NCT/DKTK master project cohort, which is consistent with the results generated in the PCAWG data, but 0% (N = 10) in myxoid liposarcomas that are driven by a chimeric fusion gene (FUS-DDIT3) instead of genomic rearrangements affecting chromosome 12q37 (Figure 2E). We generated low-coverage ONT data on two selected liposarcoma samples, a primary dedifferentiated liposarcoma (P1) and a skin metastasis of a liposarcoma (P2), allowing us to further characterize these complex SVs and achieve technical validation of the patterns identified with rayas. For both samples, lorax confirmed the TI thread pattern, thus verifying our ability to discover TI threads in short-read datasets using a graph-based approach (Figures 2F, S25, and S26). Notably, P2 revealed multiple independent occurrences of the TI thread structure in the tumor genome, leading to an even more increased overall copy number because multiple integration sites, evident by different adjacent genomic sequences, contributed to the overall copy number of TI source segments (Figure S27). Therefore, we conclude that TI threads may cause genetic instability at the locus of integration, leading to further copy-number rearrangements and multiple TI thread integrations.
Telomere analysis of derivative chromosomal segments
Critical telomere shortening is one mechanism implicated in triggering complex structural rearrangements such as chromothripsis events.38,39 Prompted by complex inter-chromosomal rearrangement seen in this medulloblastoma patient, we explored telomeric sequences associated with the resulting derivative chromosome structures, an analysis normally inaccessible to short reads. We devised a method to identify telomeric motifs, repeats of TTAGGG, TGAGGG, TCAGGG, TTGGGG, or their reverse complement, in error-prone ONT reads and applied this method to the long-read data of the primary tumor and the relapse sample (STAR Methods). Using this approach, we confidently detected five structural rearrangements involving telomeric sequences—three in the primary tumor and two in relapse—where a telomeric sequence of one chromosome is fused to a rearranged segment of another chromosome (Figure S28). For one of these telomeres, we identified a highly complex rearrangement pattern, involving the chromosome 5p-telomere and several short sequence segments from chromosome 4, 5, and 7 (Figure S28A), reminiscent of chains of TIs. For this event, telomere crisis may have initiated the complex SV pattern present throughout chromosome 4, 5 and 7, including chromothripsis and the above-mentioned TI threads. Telomere fusions can also stabilize altered chromosomes after catastrophic events such as chromothripsis,40 which would suggest an alternative sequence of events, with chromothripsis and TI threads causing unprotected break sites healed through telomere addition. Another complex SV event observed in the primary tumor likely fused chromosome 19 to the telomere of chromosome 16q, an event that could be resolved unambiguously only by using the CHM13 telomere-to-telomere (CHM T2T) assembly41 as a reference sequence (Figure S28). We further investigated whether eroded telomeres were preferentially fused with genomic loci active in transcription, as has been suggested previously,42 but our small number of telomere fusions do not provide sufficient evidence for conclusive findings. Telomeres can erode more rapidly in cells of Li-Fraumeni syndrome patients compared with healthy individuals, which is thought to lead to an increased frequency of telomeric fusions43 and possibly to have contributed to the complex SV patterns observed in this study.
Differential methylation from long-read data
ONT sequencing allows for direct assessment of the methylation likelihood of cytosine bases,15 providing the opportunity to characterize global DNA methylation levels in this medulloblastoma sample and to integrate DNA methylome and somatic rearrangement data. We quantified DNA methylation at base-level resolution using Nanopolish, which yields good correlation (pearson-R2 0.9111 in primary tumor, 0.8500 in relapse) with methylation rates obtained through the HumanMethylation450 array platform (Figure S29).
We attempted to identify patterns of variation in DNA methylation by comparing methylation rates between primary tumor and relapse sample using pycoMeth.44 We find that directly testing methylation rates of gene promoter regions (as defined in STAR Methods) yields poor power, with only 25 gene promoters called as differentially methylated (false discovery rate [FDR] ≤ 0.05, absolute methylation rate difference >0.5). We therefore applied two segmentation approaches, testing for differential methylation in segments defined using pycoMeth’s CGI finder and pycoMeth’s de novo methylome segmentation method Meth_Seg, respectively (STAR Methods). The between-sample segmentation identified 443,244 methylation-based segments as well as 357,702 CpG-dense regions. Differential methylation calling on the segmented methylation calls revealed 1,785 individual segments, or 23,576 CpG sites, called as differentially methylated (Figure 3A), with an average length of 690 bp per segment (FDR ≤ 0.05, absolute methylation rate difference >0.5; Figure S30). Of these CpG sites, 2,921 (12.39%) intersect with gene promoters, revealing 366 genes with differential promoter methylation. Furthermore, 784 genes are associated with differentially methylated regions (DMRs) within 5 kbp of transcription start site (TSS), six of which were previously annotated as medulloblastoma-driver genes45 representing a significant enrichment (Fisher’s exact test statistic, 10.3; p = 5.0 × 10−5). Furthermore, 601 (2.55%) CpG sites intersect with 47 enhancers active in the cerebellum. Among these, we detected hypermethylation in an enhancer and promoter region of the neuritin 1 gene (NRN1) (Figure 3B), previously identified as downregulated in treatment-resistant medulloblastoma46 and linked with tumor-growth-suppressive features in esophageal cancer.47 We also observed a 329-bp region in the promoter of PTCH1 (Figure 3C), a key driver in Sonic Hedgehog medulloblastoma,48 which is methylated in the relapsed tumor and heterozygously deleted in both samples. Overall, analysis of the ONT data provides a comprehensive picture of the tumor methylome, whereby a large number of effects escape discovery through commonly used array-based systems, with 76% of the between-sample DMRs inaccessible to the HumanMethylation450 BeadArray, and 66% inaccessible to the MethylationEPIC BeadArray (Figure S31).
Figure 3.
Functional analysis of primary tumor and relapse sample
(A) Number of CpGs in regions found to be differentially methylated in the sample comparison (primary tumor vs relapse) as well as ASM in the two samples. Colors represent an estimation of discoverability with short-read sequencing methods. CpGs in low-complexity regions (soft-masked in reference) are more difficult to map using only short reads. CpGs not phaseable with short reads are further than 150 bp from a phased heterozygous non C>T variants.
(B) Methylation of NRN1 promoter and enhancer in the primary tumor sample.
(C) Heterozygous deletion in promoter of PTCH1 (tumor-suppressor gene and driver in medulloblastoma) with differential methylation in the remaining haplotype.
(D) Predicted gene fusion pairs from Arriba validated using ONT long-read information, thresholded by confidence as reported by Arriba. Fusion pairs in the “supported by individual reads” category are supported by at least one genomic read with a chimeric alignment including both genes. Pairs in the “explainable using genomic breakpoints” category have a plausible explanation by following a graph of structural variations that connect the two genes. The category “high confidence read support” refers to pairs where both these criteria are met.
(E) Example of a gene fusion pair that can be explained using genomic breakpoints but with no individual genomic read that covers both genes. Two separate insertions of a total length of 42,797 bp appear to be involved in the fusion of LINC01091 and FKBP9 such that, even in ONT reads, there was no read extending across the entire gene fusion.
(F) PCDH17 (tumor-suppressor gene) promoter with ASM pattern in the primary tumor sample.
Resolving expression effects using ONT data
Leveraging Illumina RNA sequencing data generated for both primary tumor and relapse, we assessed whether differential methylation measured in gene promoters is associated with expression changes. Gene-expression analysis revealed 1,657 genes with differential expression between the two samples (absolute log fold change >2 [a-l2fc], STAR Methods; Table S8), including in six known medulloblastoma-driver genes.45 Of the total 366 promoter-linked DMRs (321 are expressed in both samples) and 41 overlap with differentially expressed genes; the overlap between differential expression and DMR effects is statistically significant (Fisher’s exact test statistic, 2.58; p = 3.5 × 10−7). While 74 of the 1,657 genes also show copy-number differences between tumor samples that correlate with the expression change (Spearman R, 0.31, p = 6.5 × 10−3), only two of those intersect with the promoter DMR genes. As previously described, promoter methylation has a mostly negative relation to expression49; 33 out of the 41 pairs (80.5%) are negatively correlated (Spearman R, −0.30; p = 5.3 × 10−2) between methylation and expression levels (Figure S32). When copy-number differences are considered, correlation is stronger (partial Spearman R, −0.33; p = 3.7 × 10−2). Discovered methylation effects also include alternative transcript promoter methylation, such as in TBX1, which is regulated by Sonic Hedgehog50 with two separate promoter-linked DMRs, one hypermethylated and one hypomethylated in primary tumor, but underexpressed (5.29 l2fc) in the primary tumor compared with the relapsed tumor (Figure S33).
We further sought to integrate the transcriptomic data with the long ONT reads to look for supporting data for gene fusion events (see Table S8), previously described to be prevalent in SHH-medulloblastoma.51 We inferred gene fusion events from transcriptomic reads using Arriba on the primary tumor, and identified 127 putative gene fusion pairs, of which 103 pairs are supported by genomic evidence, either directly through individual chimeric read alignments of ONT reads near the fusion breakpoints (53) or by tracing SVs called from long and short genomic reads (19) or both (31) (STAR Methods). Breaking down predictions by Arriba confidence shows increased traceability for higher confidence fusion calls (Figure 3D). Tracing SVs across a limited number of ONT reads allows us to explain long and complex gene fusions, with insertions in the magnitude of tens of kilobases (Figure 3E). Among these, we observe a translocation involving NCOR1 and AC087379.1, genes on the CS11-17 structure. NCOR1, a tumor-suppressor gene, has previously been reported in loss-of-function fusions in SHH medulloblastoma51; the NCOR1-AC087379.1 fusion detected here is out of frame and therefore would be predicted to disrupt NCOR1.
Allele-specific methylation and expression
ONT sequencing gives the unique opportunity to phase long methylation called reads, allowing high-resolution allele-specific methylation (ASM) analyses along the cancer genome. Using the same methylome segmentation and FDR cutoff as for DMR analysis (STAR Methods), we identified 1,068 differentially methylated segments between the haplotypes of the primary tumor sample, spanning a total of 28,803 CpGs, with an average segment length of 1,361 bp (Figure S30). Due to the lower sequencing depth in the relapse sample, the number of segments passing the significance threshold with ASM is lower, resulting in 146 differentially methylated segments (spanning 7,262 CpGs; Figure 3A). While the detection power in the relapse sample is reduced owing to lower read-depth, 370 of the 1,068 ASM segments (34.64%) found in the primary tumor show the same effect in the relapse sample with regard to sign and methylation rate difference (STAR Methods). To illustrate the benefit of using non-bisulfite-converted long reads for this analysis, we separated out CpGs close to heterozygous variants (≤150 bp away) versus CpGs further away from heterozygous variants (excluding C>T variants as those cannot be distinguished from methylation calls in bisulfite sequencing) observing that we can get 29,192 (390%) more CpGs confidently linked to ASM effects than theoretically possible in a whole-genome bisulfite sequencing analysis on a platform such as HiSeq 3000 (Figure 3A).
In the primary tumor sample, a total of 278 gene promoters and 26 enhancers intersected with segments with ASM, and 46 gene promoters and three enhancers in the relapse sample. Among these, we observe promoter methylation of PCDH17 (Figure 3F), a tumor-suppressor gene in which aberrant promoter methylation was previously observed in different tumors.52,53,54,55,56 We also detected longer segments, such as a 26,751-bp-long region found as part of a larger ∼250-kbp-long region on chromosome 15 spanning three protein-coding genes as well as a 53 non-coding genes, including the SNORD116 and SNORD115 clusters, which is partially methylated in one haplotype and fully methylated in the other. The full list of genes with sample-specific or allele-specific methylation can be found in Table S9. As we are unable to confirm a significant relationship between ASM and proximity to somatic variants, it is likely that a sizable fraction of ASM detected is associated with germline variation.
It is known that ASM plays an important role in the regulation of allele-specific expression (ASE)57 and the number of ASM loci is increased in cancer, caused by disease-associated regulatory single-nucleotide polymorphisms (SNPs).58 We therefore investigated whether ASM is associated with gene-expression levels, by performing ASE analysis. Using the phased variants from the blood sample, we were able to compute ASE rates using WASP59 (STAR Methods), focusing on the variants in the gene promoter region as defined for ASM. We observed a total of 896 genes with significant ASE effects (combined haplotype test, p < 0.05). After multiple testing correction, 220 genes remained significant (FDR < 0.05), of which a total of 71 genes were previously implicated in medulloblastoma, including the previously described ZIC1 driver gene,45 which is also a potential drug target.60 Of the 896 nominally significant genes, 312 (34.8%; Fisher’s exact test statistic, 2.6; p = 3.25 × 10−37) correspond with a copy-number increase in the matching major allele of >0.65. When subsetting the 896 ASE effects to genes with significant ASM, we found that 18 (2%) also contain strong (>0.5 absolute methylation rate difference) promoter ASM effects, and promoter methylation is also associated with reduced expression (Pearson R, −0.59; p = 5.2 × 10−3; Figure 3E). Among these, only four could also be explained by allelic copy number, and ASM/ASE correlation is stronger when copy-number effects are considered (partial correlation Pearson R, −0.60; p = 1.0 × 10−2). Again, we observed a significant overlap between ASE and ASM genes (Fisher’s exact test statistic, 2.9; p = 4.0 × 10−5, using all genes expressed in primary tumor as background).
Haplotype-resolved functional interpretation of complex rearrangements
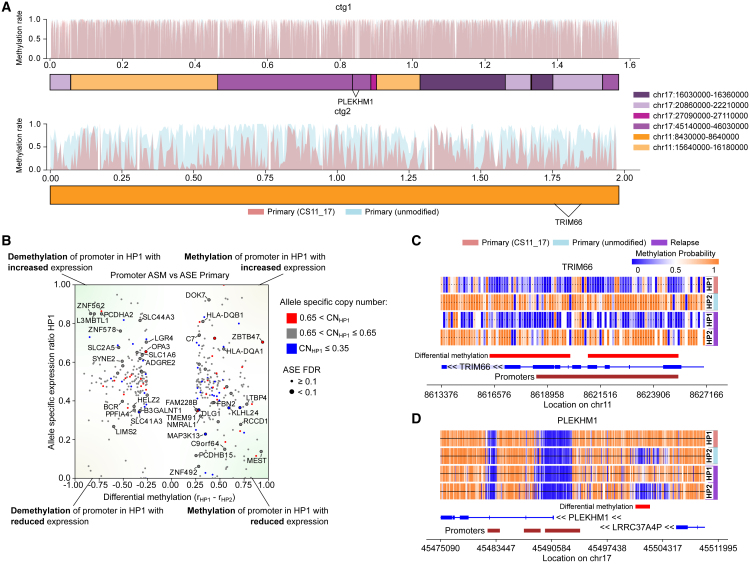
We notably observed ASM also in association with the chromothripsis event resulting in the complex CS11-17 structural segment. Since the CS11-17 rearrangement occurs in only one haplotype, we searched for ASM between the CS11-17 haplotype and the corresponding wild-type (non-rearranged) haplotype stretches. We found a global pattern of demethylation of the entire CS11-17 haplotype in contig 2 (Figure 4A) in both primary tumor and relapse, including demethylation of TRIM66 and STK33, while the wild-type haplotype in both primary and relapse, as well as both haplotypes in blood, retain normal methylation levels. On contig 1 of CS11-17, the promoter regions of SPATA32, USP22, and MAP3K14-AS1 are demethylated on the corresponding wild-type haplotype in the primary tumor, while being methylated on CS11-17 as well as on both of the unaffected haplotypes in the relapse (Figure 4B). No ASE is found for the genes on the demethylated contig 2 of CS11-17. USP22 on contig 1 of CS11-17 shows higher ASE in the demethylated allele, and MAP3K14-AS1 in the methylated allele, most likely driven by the higher copy number of the chromothriptic haplotype.
Figure 4.
Methylation of complex genomic rearrangements
(A) Methylation rates of chromothriptic contig CS11-17 in the primary tumor sample show global demethylation of contig 2, containing genes TRIM66 and STK33, to a methylation rate of 42% on the CS11-17 haplotype from 76% in the corresponding genomic ranges on the non-chromothriptic haplotype. While contig 1 displays some allele-specific differences, no significant global effects are detected.
(B) ASE correlates with promoter-linked ASM in primary tumor (Pearson R, −0.38, p value 6.6 × 10−3 for all ASM effects displayed and nominally significant ASE effects, Pearson R, −0.61, p = 5.1 × 10−3 when considering only ASM with >0.5 absolute methylation rate difference).
(C) Demethylation of CS11-17 haplotype of contig 2 effect shown on TRIM66 promoter.
(D) ASM of promoter of gene PLEKHM1 on contig 1.
Functional annotation of the TI threads and telomere SVs
We next performed similar functional annotation of the TI threads and the telomere insertions. The TI threads appear to retain their original methylation state with only a slight reduction in methylation rate measured (average methylation rate reduction structure 1, 0.16; structure 2, 0.09; Figure S34). Interestingly, the first TI thread (Figure 2B) lands in an intronic region of BASP1, which was previously implicated in metastatic medulloblastoma in a mouse model specifically by transposon insertion mutagenesis.61 While TI threads represent a different class of insertion, we notably do observe differences in splicing of BASP1 between the samples. Within the relapse sample, which does not harbor the TI thread, we find three splice junctions that are not used in the primary tumor: junction 1 (5:17,260,615–17,275,208), Fisher’s exact test p = 1.5 × 10−23; junction 2 (5:17,228,332–17,275,208), p = 2.0 × 10−22; junction 3 (5:17,263,478-17,275,208), p = 4.4 × 10−10. The junction used for the main BASP1 isoform (BASP-201) is more frequently used in the primary tumor as compared with the relapse (Table S10; Figure S35). To further explore the functional relevance of the observed TI threads, we also searched for potential gene dysregulation effects within the transcriptomic data available for liposarcoma samples in PCAWG.2 We identified one liposarcoma sample (donor ID: DO219945), which harbors a TI thread on chromosome 12 whose breakpoints intersect the coding sequence of proliferation-associated protein 2G4 (PA2G4), which can act as a contextual tumor suppressor,62 in association with reduced PA2G4 expression (Figure S36A). Another liposarcoma sample (donor ID: DO219967) shows strong overexpression of CCND3, a known sarcoma oncogene, and BYSL, a gene associated with tumor prognosis,63 and both genes have an estimated copy number of 49 with the TI thread in their immediate vicinity (Figure S36B). These examples suggest a possibly relevant role of TI threads in cancer, illustrating the need to routinely generate long reads to fully characterize complex somatic SVs with respect to cancer-related genes in tumor genomes.
Analyzing the telomere-associated SVs, we find that four SVs observed in the primary tumor and relapse samples (Figure S28) harbor a breakpoint junction in intronic regions of protein-coding genes, namely TLL1, THADA, and MYPOP in the primary tumor and LUZP2 in the relapse sample. The MYPOP and TLL1 SVs also show short TIs between the telomeric part and the above-mentioned genes, with TI source sequences originating from intronic regions of various other genes (Figure S28). We performed differential expression analysis between the primary tumor and relapse and found that TLL1 showed a slightly reduced expression in the primary tumor (−1.15 l2fc), whereas LUZP2 and MYPOP displayed a reduced expression in relapse (−1.16 l2fc and −1.08 l2fc, respectively). Additionally, MYPOP is found to be amplified in the haplotype where the telomere-associated SV is found (allele-specific copy-number ratio 0.7) with a matching ASE rate (0.75), while only 23.7% of the reads in the major allele contain the SV, suggesting subclonality. This amplification extends across most of chromosome 19q and is exclusive to the primary tumor (Figure S37).
Discussion
Interrogating cancer genomes using long reads
We describe the haplotype-resolved genetic and epigenetic profile of a diagnosis and post-therapy medulloblastoma using long reads and present new computational methods for targeted de novo assembly and complex SV characterization, as well as phasing, segmentation, and investigation of ONT methylome profiles. We used an integrated phasing approach that combines long reads with statistical phasing enabling the targeted assembly of a 1.55-Mbp chromothripsis event spanning 14 breakpoints. Furthermore, by leveraging the joint genetic and epigenetic readout of ONT data, we revealed haplotype-specific and chromothripsis-related methylation changes, analyses challenging to pursue with short reads due to the sparsity of germline heterozygous SNPs and limitations in read and phased block length. The combination of long-read genetic and phased methylation information from ONT reads can be used to detect aberrant expression patterns arising from allelic expression imbalance or gene fusion events at greater level of detail. In the future, deep coverage and highly accurate long-read data will be needed to achieve the complete de novo assembly of cancer genomes, especially in the context of ITH, contamination of normal cells, and large numbers of complex rearrangements.
TI threads
We describe a new complex DNA rearrangement pattern, termed TI thread, consisting predominantly of short segments (<1 kbp) that are copied and (self-)concatenated into amplified, highly repetitive somatic sequences of up to 50 kbp in size. Umbreit et al. did not detect self-concatenating insertions of high copy number in the cell cultures of their in vitro study, and their recently described tandem short template jump pattern30 therefore bears differences to the TI thread pattern described here. However, Umbreit et al.30 generated orthogonal validation data from a renal cell carcinoma, which included an example of a chained rearrangement with a zigzag pattern of TIs involving at least a few self-concatenations. These validation data, therefore, further support the TI thread pattern defined in our study, which also is further substantiated through the discovery and validation of TI threads in two liposarcoma samples as well as a patient-derived xenograft model. Future analysis of larger sample sets using long reads will be required to delineate the full extent and scope of concatenated insertions in cancers, which is likely to be currently underestimated since short TI source sequences often escape copy-number segmentation methods, leading to erroneous reconstructions of TI threads. Notably, tandem short template jumps,30 like TI threads, show an association with chromothripsis, which leaves the possibility of a continuum of concatenated insertion patterns arising in conjunction with complex DNA rearrangement processes. The observed multiple integrations of TI threads in a liposarcoma sample suggest that TI threads occasionally undergo genetic instability at the respective locus, which results in further rearrangements in tumor evolution altering the copy number of affected regions and possibly inducing further chaining events.
We demonstrate using a new graph-based method, rayas, that TI threads can be identified in short-read WGS data, which is important as it allows further study of this complex rearrangement pattern in existing large short-read cancer genomic cohorts, as has been done previously for other complex rearrangement types such as chromothripsis.64 We describe a remarkable enrichment of this pattern in several adult cancers, with the strongest prevalence in liposarcomas (74% of cancer samples affected) and a clear colocalization of these events with genomic regions undergoing giant marker chromosome formation and chromothripsis. We did not identify any additional medulloblastoma samples with TI threads in the PCAWG short-read dataset, which is perhaps explained by the relatively low portion of medulloblastoma samples contained in the PCAWG cohort exhibiting chromothripsis (∼12%; N = 145),65 which, in medulloblastoma, is tightly linked with germline TP53 mutations.16 One note of caution is that discovery of regions of high structural rearrangement complexity as seen in TI threads using short reads is obscured by somatic SV calling pipelines because multiple distinct SVs co-occur at the same SV breakpoint leading to algorithmic clustering and SV merging issues. This is contrary to long reads that have the capability to fully resolve the complex structure and composition of structural rearrangements in cancer genomes. While rayas can overcome this issue in part, it is likely that short-read WGS masks additional cases of TI threads, especially where they involve short (<1 kb) TI units or repeat-rich DNA, given the relatively poor sensitivity of Illumina reads for calling such SVs13 and limitations in short-read-based haplotype-reconstruction methods.
Telomere-associated SVs
The long-read data also enabled investigation of the association of complex SVs and telomeric repeats, an analysis that revealed the fusion of telomeres with chromosomes that underwent chromothripsis. Some of these events were captured in a single long ONT read connecting a telomere to various SV rearrangements, reminiscent of SV mutations stabilized by independent telomere fusions. The assignment of telomeric repeats to chromosomal haplotypes also highlighted the need for continuous reference improvements, as some of these events could only be unambiguously resolved using the new CHM13 telomere-to-telomere (T2T) assembly.41 A comparable analysis on short-read data failed to resolve the telomere-associated complex rearrangements; only three out of the five SV-to-telomere junctions showed confident telomeric repeat motifs in an unmapped mate or a soft-clipped read. This underscores the critical need for long-read sequencing to investigate telomere-associated structural rearrangements, a key mutational process associated with telomere crisis.38
ASM
ASM analysis uncovered a large number of haplotype-specific effects, many of which reside in regions with sparse germline variants or highly repetitive sequence context, showing the potential of methylation analysis from long reads. Methylation effects were further associated with complex SV patterns (CS11-17 contig 2 and TI threads). However, due to the high tissue specificity of DNA methylation, a systematic analysis of association between methylation change and somatic variants would require sequencing of a tissue-matched normal sample in order to exclude effects related to germline variation. Furthermore, while we find the coverage in our primary tumor sample (30×) sufficient for ASM analysis, the more limited coverage of the relapse sample (19×) affects the discoverability of ASM effects, which requires reads to be split between haplotypes for testing.
Conclusions
In summary, our study shows the benefits of using long reads in refining complex and repetitive rearrangement patterns such as TI threads and telomere-associated SVs, and of integrating these with ASM and expression changes. The computational methods developed in our study provide the foundation for a more broad application of long reads in cancer genomics to uncover new somatic mutation patterns and pave the way for deciphering the complex relationship of genetic and epigenetic changes in cancer biology.
Limitations of the study
Despite the unprecedented view into genetic and epigenetic patterns that ONT long reads enable, several future challenges remain. (1) Our strategy focused on targeted assemblies of high copy number regions due to the moderate long-read sequencing coverage (up to 30-fold). While long-read sequencing remains costly compared with Illumina sequencing, future gains in throughput will enable studies in larger sample panels with coverages suitable for uncovering SVs in the context of ITH. (2) Our assemblies failed to resolve peri-centromeric regions involved in the CS11-17 chromothripsis region exceeding the available read length. As ONT read lengths are determined by the sample preparation protocol, this suggests that “ultra-long” preparations may prove beneficial to characterize somatic SVs contained within repeat-rich regions, once available for routine application. (3) Further computational methods development will be needed to achieve the assembly of entire derivative chromosomes in cancer, including new algorithms for SV-aware haplotyping and multi-allelic assemblies. (4) A larger number of long-read datasets will be required to comprehensively characterize the TI thread landscape across tumor genomes and to characterize relationships with genetic instability as well as their potential functional effects. Our short-read analyzes of TI threads in the PCAWG cohort do not provide end-to-end reconstructions, and the complexity and frequency of TI threads in this short-read dataset is therefore likely to represent an underestimate. (5) From this study, primarily focused on a single patient, genes highlighted in the functional analysis are to be understood as anecdotal observations, prior to replication in larger cohorts.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| Qubit 1× dsDNA BR Assay kit | Thermo Fisher | Cat#Q33266 |
| FEMTO Pulse - Genomic DNA 165 kb Kit | Agilent | FP-1002-0275 |
| NEBNext FFPE Repair Mix | New England Biolabs | Cat # M6630 |
| NEBNext Ultra II End repair/dA-tailing Module | New England Biolabs | Cat # E7546 |
| NEBNext Quick Ligation Module | New England Biolabs | Cat # E6056 |
| SPRI Select beads | Beckman Coulter | B23319 |
| PromethION Ligation sequencing gDNA kit | Oxford Nanopore Technologies | SQK-LSK109 |
| PromethION R9.4.1 flow cells | Oxford Nanopore Technologies | FLO-PRO002 |
| MinION Ligation sequencing gDNA kit (patient-derived xenograft) | Oxford Nanopore Technologies | SQK-LSK110 |
| MinION R9.4.1 flow cells (patient-derived xenograft) | Oxford Nanopore Technologies | FLO-MIN106D |
| MinION Ligation sequencing gDNA kit (liposarcoma) | Oxford Nanopore Technologies | SQK-LSK109 |
| MinION R9.4.1 flow cells (liposarcoma) | Oxford Nanopore Technologies | FLO-MIN106D |
| Flow cell wash kit XL | Oxford Nanopore Technologies | EXP_WSH004-XL |
| BAC clone for FISH | RZPD | BAC clone RP11 651L9 |
| REPLI-g mini Kit | Qiagen | Cat # 150023 |
| Plasmid-Safe™ ATP-Dependent DNase | Epicentre | Cat # E3101K |
| NEBNext Ultra II FS DNA Library Prep kit | New England Biolabs | Cat # E7805S |
| Deposited data | ||
| Medulloblastoma raw data | This paper | EGA: EGAS00001006576 |
| Liposarcoma raw data | This paper | EGA: EGAS00001006629 |
| Medulloblastoma FISH imaging data & read alignment related to Figures 2 and S14. | This paper | https://www.ebi.ac.uk/biostudies/bioimages/studies/S-BIAD611 (https://doi.org/10.5281/zenodo.7658888) |
| Software and algorithms | ||
| Guppy | Oxford Nanopore Technologies | https://nanoporetech.com/ |
| Minimap2 | Li et al.66 | https://github.com/lh3/minimap2 |
| Bioconda | Grüning et al.67 | https://github.com/bioconda/ |
| Bwa | Li and Durbin68 | https://github.com/lh3/bwa |
| Delly | Rausch et al.69 | https://github.com/dellytools/delly |
| Sniffles | Sedlazeck et al.70 | https://github.com/fritzsedlazeck/Sniffles |
| WhatsHap | Patterson et al.21 | https://github.com/whatshap/whatshap |
| ShapeIt | Delaneau et al.23 | https://github.com/odelaneau/shapeit4 |
| Flye | Kolmogorov et al.26 | https://github.com/fenderglass/Flye |
| Shasta | Shafin et al.27 | https://github.com/chanzuckerberg/shasta |
| HTSlib | Bonfield et al.71 | https://github.com/samtools/htslib |
| FreeBayes | Garrison and Marth72 | https://github.com/freebayes/freebayes |
| Strelka2 | Kim et al.73 | https://github.com/Illumina/strelka |
| pycoMeth | Snajder et al.44 | https://github.com/PMBio/pycoMeth (https://doi.org/10.5281/zenodo.6637645) |
| Nanopolish | Simpson et al.74 | https://github.com/jts/nanopolish |
| Alfred | Rausch et al.75 | https://github.com/tobiasrausch/alfred |
| NanoVar | Tham et al.76 | https://github.com/cytham/nanovar |
| SvABA | Wala et al.77 | https://github.com/walaj/svaba |
| Visor | Bolognini et al.78 | https://github.com/davidebolo1993/VISOR |
| Circle-seq analysis scripts | Koche et al.25 | https://github.com/henssen-lab/circle-enrich-filter (https://doi.org/10.5281/zenodo.7542388) |
| IGV | Robinson et al.28 | https://github.com/igvteam/igv/ |
| Lorax | This paper | https://github.com/tobiasrausch/lorax (https://doi.org/10.5281/zenodo.7541542) |
| Rayas | This paper | https://github.com/tobiasrausch/rayas (https://doi.org/10.5281/zenodo.7541623) |
| Wally | This paper | https://github.com/tobiasrausch/wally (https://doi.org/10.5281/zenodo.7541485) |
| Analysis scripts | This paper | https://github.com/PMBio/mb-nanopore-2022/ (https://doi.org/10.5281/zenodo.7543715) |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Marc Jan Bonder (bonder.m.j@gmail.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Patient cohort
All biological samples included in this study were obtained after receiving written informed consent in accordance with the Declaration of Helsinki and approval from the respective institutional review boards. The medulloblastoma patient was a male patient aged 8 years at diagnosis. The liposarcoma patient P1 was a male patient aged 55 years and the liposarcoma patient P2 was a female patient aged 65 years. For details on donor characteristics in the Pan-Cancer Analysis of Whole Genomes (PCAWG) consortium, see Table S1 of the main PCAWG paper.2
Method details
Patient material, DNA extraction and short-read whole-genome sequencing
Medulloblastoma samples used for bulk sequencing had a tumor cell content confirmed by neuropathological evaluation of the hematoxylin and eosin stainings. DNA was extracted from frozen tissue and from blood using Qiagen Allprep and Qiagen Blood and tissue kits, respectively. Purified DNA was quantified using the Qubit Broad Range double-stranded DNA assay (Life Technologies, Carlsbad, CA, USA). Genomic DNA was sheared using an S2 Ultrasonicator (Covaris, Woburn, MA, USA). Short-read whole-genome sequencing and library preparations for tumors and matched germline control were performed according to the manufacturer’s instructions (Illumina, San Diego, CA, USA). The quality of the libraries was assessed using a Bioanalyzer (Agilent, Stockport, UK). Sequencing was performed using the Illumina X Ten platform.
DNA methylation array data
Medulloblastoma samples were analyzed using Illumina Infinium HumanMethylation450 BeadChip (450k) arrays or Methylation BeadChip (EPIC) arrays according to the manufacturer’s instructions.
RNA sequencing
RNA was extracted from frozen tissue using Qiagen Allprep kits. RNA quality was assessed using a Bioanalyzer (Agilent, Stockport, UK). Short-read RNA sequencing and library preparations for tumors were performed according to the manufacturer’s instructions (Illumina, San Diego, CA, USA). The quality of the libraries was assessed using a Bioanalyzer (Agilent, Stockport, UK). Sequencing was performed using the Illumina HiSeq 2000.
Fluorescence in situ hybridization (FISH)
Nick translation was carried out for BAC clone RP11 651L9 (chromosome 17) and centromere 17. FISH was performed on metaphase spreads from patient-derived xenograft models or tumor tissue using fluorescein isothiocyanate-labeled probes and rhodamine-labeled probes. Pre-treatment of slides, hybridization, post-hybridization processing and signal detection were performed as described previously.79 Samples showing sufficient FISH efficiency (>90% nuclei with signals) were evaluated. Signals were scored in, at least, 100 non-overlapping metaphases or nuclei. Metaphase FISH for verifying clone-mapping position was performed using peripheral blood cell cultures of healthy donors as outlined previously.79
Long-read library preparation and nanopore sequencing
DNA was quantified using Qubit (Thermo Fisher) and fragment size assessed using FEMTOPulse (Agilent). Libraries were prepared using SQK LSK-109 (Oxford Nanopore) following the manufacturer’s protocol and sequenced on the PromethION for the medulloblastoma patient samples and on the GridION for the patient-derived xenograft and liposarcoma samples (Oxford Nanopore). For the library preparation of the liposarcoma validation samples, 1 ug of high molecular weight DNA was used as input. A tight sample fragment size distribution was assessed by a high-sensitivity pulsed-field capillary electrophoresis fragment analyser (FEMTO-Pulse, Agilent, Santa Clara, CA). DNA was end-repaired and dA-tailed using NEB DNA Ultra II module for 1h at 20°C and 10min 65°C followed by a bead cleanup using SPRI beads (B23319, Beckman Coulter) with an extended sample-SPRI bead incubation time of 30min and 10min elution. The ligation sequencing gDNA kit (SQK-LSK109; Oxford Nanopore Technologies) was used for a 20min adapter ligation. A final SPRI bead cleanup was performed, and DNA was eluting for 20min at 37°C in 15 μl Elution Buffer (Oxford Nanopore Technologies). The final sequencing library was prepared by mixing 50 fmol of adaptor-ligated-library with 37.5 μl sequencing buffer (Oxford Nanopore Technologies) and no loading beads. The library was sequenced using an R9.4.1 flow cell (Oxford Nanopore Technologies) on the GridION. The sequencing run was stopped after 22h, flow cell was washed using the Flow cell wash kit XL (EXP_WSH004-XL, Oxford Nanopore Technologies) and then the library was reloaded.
Circle-seq sequencing and data analysis
For the primary tumor and relapse samples, bulk Circle-seq data was generated and processed as previously described.25 In short, circular DNA enrichment was performed by exonuclease digestion of linear DNA for 5 days at 37°C. In all cases, 1 μg of total DNA was treated with 20 units of Plasmid-Safe DNase (10 units/μl, Epicentre) in 1× Plasmid-Safe reaction buffer (Epicentre) and 1mM ATP.80 After each 24 hour incubation, the enzymatic reaction was supplemented with 20 units of Plasmid-Safe ATP dependent DNase (10 units/μl, Epicentre) and 4 μL of 25 mM ATP. After 5 days of enzymatic digestion, the exonuclease was heat-inactivated by incubating at 70°C for 30 min. The isolated circular DNA was amplified by rolling circle amplification using the REPLI-g mini Kit (Qiagen) and following the manufacturer’s instructions for a starting volume of 10μl of exonuclease-treated DNA. Further, the amplified circular DNA was purified using a 1.7× volumetric ratio of AMPure XP beads (Beckman Coulter). Libraries for Illumina next-generation sequencing were prepared using NEBNext Ultra II FS DNA Library Prep kit (New England Biolabs) according to the manufacturer’s instruction, and sequenced on an Illumina Miseq instrument with 2×75 bp paired-end reads. The raw reads were adapter- and quality-trimmed with trimGalore81 and aligned against a joint reference genome built from hs37d5 and mm10 using bwa mem 0.7.1782 with standard parameters. PCR and optical duplicates were removed with Picard v.2.25.0.83 Further, our internal pipeline was applied to detect circularised genomic regions. The approach uses the overlap of outward-facing split-reads and genomic segments amplified over background (Homer v.4.11 findpeaks)84 to find circularised genomic regions. To select true circles over noise a z-Test score was computed by comparing the distributions of reads spanning the edges of putative circular regions against background, defined as non-circle-enriched regions with similar length and nucleotide composition. For both, CS11-17 assembly and TI thread the genomic regions overlap was computed against Circle-seq calls, followed by manual inspection in IGV.28 To enable the comparison, Circle-seq call coordinates were mapped to GRCh38/hg38 using the UCSC genome browser functionality liftover.85
Short-read alignment, variant calling and copy-number segmentation
Paired-end, short-read FASTQ files (2x151bp) were aligned to the GRCh38 reference genome using the alternate contig-aware bwakit.68 Alignments were sorted and indexed using samtools86 and quality-controlled with Alfred.75 The median coverage of the blood (control), primary tumor and relapse sample were 48x, 45x and 47x, respectively. The insert size ranged from 373bp to 406bp for the three samples.
Single-nucleotide variants (SNVs) and short insertions and deletions (InDels) were called using FreeBayes72 and Strelka2.73 For germline variants we used a consensus approach and only retained polymorphisms supported by FreeBayes and Strelka for subsequent haplotyping. The integration of these two short-read germline call sets on GRCh38 yielded 3,790,471 bi-allelic SNVs and 568,168 bi-allelic insertion and deletions. Bcftools was used to normalize and left-align indels. Copy-number segmentation employed Delly’s cnv mode69 with the GRCh38 mappability map and the DNAcopy87 package of the Bioconductor project (Figure S2). Structural variants were called using Delly,69 Manta88 and SvABA77 in a paired tumor-normal fashion to distinguish germline and somatic SVs. Command-line tools were installed using bioconda.67
Long-read alignment and variant calling
Long reads from Nanopore sequencing were basecalled with guppy version 6.1.7 using the high accuracy model for PromethION (r9.4.1_450bps_hac_prom). Resulting FASTQ files were aligned to the human reference genome (GRCh38) using minimap266 using the ‘--ax map-ont’ option and otherwise default parameters. The long-read coverage was 15x for the blood run, 29x for the primary tumor, and 19x for the relapse sample. The N50 read length was 15,600bp, 21,800bp and 10,800bp for the original blood, primary tumor and relapse runs, respectively. A fourth run was generated, with primary and blood sample multiplexed, which only yielded 6x coverage with an N50 read length of 8,230bp (Table S1, Figure S38). The estimated sequencing error rate of the aligned data using Alfred’s qc mode75 was estimated to be 5.4% for the blood sample and 4.5%-4.6% for the tumor samples.
Structural variants (SVs) from the long-read data were called using Nanovar,76 Sniffles70 and Delly.69 Consensus germline SVs were filtered using a stringent reciprocal overlap of 80% and a maximum breakpoint offset of 50bp, yielding 7,952 deletions and 8,185 insertions, which is lower compared to recent studies using long-reads12,13 likely because of our relatively low germline coverage of only 15x (Figure S39). For somatic SVs we followed a more lenient union approach of short-read SV calls (delly) and long-read SV calls (sniffles and delly) to not miss any interesting variants and only required absence of an SV in the matched control and a minimum support of 2 reads in the tumor, followed by manual inspection of somatic SVs in IGV28 and a newly developed alignment visualization tool, called wally, which enables a fast batch alignment plotting of SVs in a paired tumor-normal split-view.
Nanopore methylation calling
Read-level CpG methylation likelihood ratios were estimated using nanopolish74 version 0.14.0. Methylation rates were computed from binarized methylation calls thresholded at absolute log-likelihood ratio of 2.0 and compared to methylation rates observed in 450k arrays.
Haplotype-phasing of short variants
We used a three-stage approach to phase bi-allelic heterozygous SNVs and InDels present in our consensus call set from FreeBayes and Strelka. In brief, the first stage uses read-based phasing of the long-read data to generate initial haplotype blocks, these are concatenated using population phasing in the second step and finally, remaining switch errors are corrected using shifted allelic ratios in the matched tumor. The procedure is illustrated in Figure S1 where initial phased blocks are colored red and blue that are then extended using statistical phasing and corrected based on the matched tumor genome.
For read-based phasing we used WhatsHap21 with the ‘--indel’ option and the aligned long-read data. The WhatsHap output VCF was indexed using HTSlib.71 WhatsHap determines phased sets which are groups of heterozygous genotypes at which the phase has been inferred using long reads. These phased sets are specified in the PS field of the VCF/BCF file format.89 With the SHAPEIT4 algorithm23 and the phased blocks from WhatsHap we then carried out population phasing using the 1000 Genomes haplotype reference panel.22,90 We used the ‘--use-PS 0.0001’ option to define the expected error rate in the phased sets. The statistically phased VCF files were then augmented for each variant with the matched tumor B-allele frequencies to correct remaining switch errors in regions of unequal haplotype ratio in the tumor sample. As a result of statistical phasing and the use of a haplotype reference panel the statistically phased VCF files are restricted to high-quality variants present in the panel. We therefore used this phased VCF file as a haplotype scaffold to drop in additional variants present in our donor using WhatsHap and the long-read aligned data. Overall, our haplotype-phasing approach phased 2,642,137 bi-allelic heterozygous variants (2,214,532 SNVs and 360,226 InDels) at a median read length of approximately 5kbp which allowed us to study almost the entire mappable genome, 91.13% for the primary tumor and 89.85% for the relapse, in a haplotype-resolved manner. To split alignment files by haplotype we employed Alfred75 using the phased VCF and the unphased alignment as input.
De novo assembly of the primary tumor
We applied two de novo assembly methods, Shasta27 v0.10.0 with the Nanopore config and Flye26 v2.9 with an estimated read error rate of 4.5%, a genome size of 2.9G and the nano-hq option. Due to the relatively low coverage of 30x for de novo assembly, the Shasta assembly contained 7,069 contigs with a longest contig of 52Mbp and an N50 of 3.99Mbp. The Flye assembly contained 2,382 contigs with a longest contig of 109Mbp and an N50 of 22.78Mbp. Both assemblers generated contigs confirming the TI thread in Figure 2 (Figure S10) but failed to assemble the entire CS11-17 structure, possibly because these assemblers compute a so-called squashed assembly of both haplotypes. Nevertheless, multiple contigs appear to confirm individual junctions of the CS11-17 structure and they tile the entire targeted assembly of CS11-17 (Figure S40).
Targeted assembly of complex DNA rearrangements
To enable targeted assembly of complex SVs, we used our haplotype scaffold and the integrated map of somatic structural variants and copy-number alterations. We first applied delly’s cnv mode and the somatic SV calls to identify amplicons on chromosome 11 and chromosome 17 that are inter-connected by split-reads and that have approximately the same total copy-number. We then developed a targeted method to assemble these high copy-number regions by selecting reads that either bridge at least two amplicons or are part of the amplified haplotype based on the depth observed for each germline allele. We implemented the method in our long-read analysis toolbox for cancer genomics, termed lorax, and the tool requires as input the phased germline variants in VCF/BCF format, a set of amplicon regions in BED format and the input tumor BAM file. The method then screens the BAM file for split-reads connecting at least two amplicons and it annotates the haplotype support based on all phased, heterozygous variants covered by the read sequence. Each read is then assigned to either haplotype 1 or haplotype 2 based on the observed variants. The total allelic depth across all reads in the respective amplicon region determines the amplified haplotype which is retained for further analysis. We discard all reads that have confident alignments outside the amplicon boundaries to deplete reads from contaminating normal cells occurring on the same haplotype background or sub-clonal reads from different rearrangement structures. User-defined parameters control the precision of amplicon boundaries (default 1kbp), the minimum required clipping length of split reads (default 100bp) and the minimum mapping quality (default 10). A final pass through the BAM file extracts the sequences of all selected reads, which are then assembled using Shasta.27 Lorax also re-estimates the amplicon boundaries based on the observed read clipping patterns which was used to iteratively refine the input amplicon regions. We trimmed the assembly at repetitive ends that lacked a unique alignment to the reference. The final contigs were aligned back to the reference genome using minimap266 to infer alignment coordinates and breakpoints.
Discovery of TI threads using short and long-reads
To discover complex templated DNA rearrangements using short-reads we devised a graph-based algorithm, called rayas, that uses matched tumor-normal cancer genomics sequencing data. The algorithm parses the tumor and normal BAM file to compute a sample-specific coverage and split-read profile at single-nucleotide resolution. Rayas uses soft- and hard-clips and records the positions where these splits occur. The coverage profile is used to determine the average genome-wide coverage, its standard deviation and to normalize for overall coverage differences between tumor and normal. Using a minimum seed window size (default 100bp) rayas then scans the coverage profile for putative SV breakpoints, always screening two adjacent windows for unexpected coverage increases when entering a TI source segment or unexpected coverage decreases when leaving a TI source segment. Command-line parameters control the minimum number of split-reads required at these SV breakpoints and the required magnitude of the coverage increase or decrease. The matched control is processed simultaneously to account for potential mapping artifacts, i.e. regions where both the tumor and the control show unexpected coverage and split-read patterns which are subsequently filtered out. Once all candidate segments have been identified, rayas re-uses the identified split-reads to connect segments and builds a graph with representing a TI source segment and being an edge from to with representing the split-read support. Using the connected components of , rayas filters out singletons (i.e. segments lacking confident split-read support) as well as connected subgraphs ) with and where all nodes of are nearby in the genome with the definition of nearby depending on a user-defined threshold (by default 10kbp). All remaining connected components are written to a BED file with a unique component id. For each component, all genomic segments and edges are outputted and the results can be visualized as a graph (Figure S23). Using this approach we identified two TI threads in the primary tumor. In addition, a single additional putative instance of this pattern was detected in the Illumina data of the relapse but not in the ONT data from the same sample; this putative event showed much lower split-read support (5 compared to >>100 for the primary tumor TI threads) and an unexpected density of variant calls, suggesting that it may be caused by a mapping artifact or a collapsed repeat rather than a TI thread. A simple threshold for the minimum split-read support (i.e., node out-degree in the rearrangement graph) removes such false positives, indicating excellent sensitivity and specificity of rayas using illumina data, further confirmed by additional simulation experiments using Visor78 (Figure S41). For the PCAWG data, we filtered for connected components with at least one segment with a total copy-number greater than 10, a node degree greater than 50 and evidence of at least one direct self-concatenation supported by at least 3 split-reads, as these features were characteristic of the TI threads found in the medulloblastoma.
The algorithm implemented in lorax for detecting TIs with long reads uses the same discovery approach as rayas, but then scans the original alignment data to extract long reads that span multiple TIs. These reads can be selectively assembled, inspected through self-alignments or back-aligned to the source sequence segments as shown in Figure 2. Simulation experiments of the TI thread discovery show very good sensitivity and specificity even at low coverage (Figure S42). An important distinguishing feature of lorax compared to short-read reconstruction methods is that the tool does not greedily collapse each connected component of the rearrangement graph into a single TI thread reconstruction but instead collects all supporting long reads for a targeted local assembly that can emit multiple contigs in case of TI thread heterogeneity. Our method thus leverages the long sequencing length to separate alleles or tear apart multiple distinct integrations as we show for the primary liposarcoma sample P2 (Figures S27 and S43).
The visualization of long read alignments spanning dozens to hundreds of breakpoint junctions employed minimap2,66 MUMmer,91 custom R scripts and a newly developed tool, called wally, that enables the plotting of long read mappings as chained alignments (Figures 1D and 2F) or augmented dot plots that show pairwise matches as well as alignments widely distributed across the genome (Figure S27).
TI thread simulation experiments and benchmarking
We benchmarked rayas and lorax on simulated data using the SV simulator Visor.78 With Visor, we implanted TI threads in chromosome 18 and then evaluated a range of calling parameters, technologies (Illumina, ONT and PacBio) and coverage thresholds. With default parameters, lorax and rayas show close to 100% specificity at the expense of missing some (10%-30%) source segments of TI threads (Figures S41 and S42). For lorax, there is a high chance of rescuing missed source segments in the subsequent local assembly stage because long reads usually span multiple source segments and we therefore decided to favor specificity over sensitivity in our default parameter selection.
In terms of memory and runtime, rayas required on average 85.2 minutes for analyzing a paired tumor-normal cancer genome from the PCAWG study2 at less than 16G RAM for all samples. Lorax used 37 minutes and 3.3G RAM for the discovery of TI source segments and the identification of connected components, 17 minutes to extract TI supporting reads (at <100MB RAM) and less than 5 minutes to assemble the small set of reads with Shasta for the 30x long read data of the primary tumor.
SV breakpoint junction analysis
We applied Delly-maze69 from the docker container smei/maze to the breakpoint junctions of the TI thread in Figure 2, which uses MUMmer92 and LAST93 for sequence alignment. We summarized the micro-insertion and micro-homology length for each breakpoint and aggregate the results as a histogram (Figure S16).
Large CNVs that lacked adjacent SVs were inspected manually with IGV.28 On chromosome 3 of the primary tumor the p-arm is monosomic and the q-arm trisomic but we did not identify any large SV near or inside the centromere of chromosome 3 even with the T2T assembly as the reference genome. Similarly, the large chromosome 2 deletion could not be fully resolved with SVs because the left-most breakpoint at chr2:57,101,210bp is likely an inter-chromosomal translocation to the chromosome 20 centromere but due to the repetitiveness of the underlying sequence neither delly nor sniffles identified this SV reliably. The right-most breakpoint of the chromosome 2 deletion at chr2:89,753,994 is next to a region masked with Ns in GRCh38.
Short-read complex SV analysis
To confirm TI thread breakpoint junctions using short-reads, we computed SV contigs using SvABA v1.1.3.77 For maximum sensitivity, we used SvABA jointly on both tumor samples with the blood sample as the matched control. We aligned all SvABA SV contigs to GRCh38 and the Shasta assembly to compare the contiguity of short-read SV contigs compared to long-read derived SV contigs. All short-read SV contigs supported at most 3 SV breakpoint junctions and as expected, contigs provided only a very limited view into TI thread complexity (Figure S12).
We further applied two popular higher-level short-read cancer genome reconstruction methods, namely RCK94 and Linx,95 to evaluate the robustness and advantages of our long read approach. RCK requires input in the form of segmented allele-specific copy numbers and novel genomic adjacencies. The somatic SVs of Manta88 were used as novel adjacencies and the allele-specific segment copy numbers were computed using HATCHet96 – with both Manta and HaTCHet being the recommended input methods for RCK. However, HaTCHet, perhaps not unexpectedly because of the nature of TI threads, failed to identify templated insertion source segments at high-copy number because a significant number of these (short) segments do not even have a single (phased) germline SNP. Manta reported a multitude of somatic SVs connecting the templated insertion source segments, e.g. 20 somatic SVs for the region chr4:168,398,000-168,399,000 and 18 for the region chr7:7,805,000-7,806,000 in Figure 2B. We assume that the lack of a distinct copy-number segment matching these templated insertion source sequence regions and the high number of somatic SVs caused RCK to infer huge segments on chr4 with non-plausible copy-number values (Figure S44) – suggesting that extremely complex rearrangement patterns such as TI threads cause artifacts when using RCK with default settings.
Similar to RCK, Linx also requires genomic adjacencies and allele-specific copy-numbers as input which we computed using Gridss and Purple by means of running the gridss/gridss-purple-linx docker container using GRCh38 as the reference bundle. For the CS11-17 assembly with larger segments, Purple confirmed our estimate of a median total copy-number of 3.88 with a major copy-number of 2.7. The estimate for contig 2 which likely belongs to the CS11-17 assembly was a total copy-number of 3.77 and a major copy-number of 2.97. Linx also clustered the segments belonging to the CS11-17 structure into a single complex event, including segments overlapping contig 2. The chaining algorithm, however, failed to predict the entire derivative structure and outputted 14 independent chains for this cluster. Chain 1 was the longest that shared 11 out of 15 (73%) segments of contig 1 (Figure S45). Like HaTCHet, Purple failed to identify the total and major copy-number for some segments involved in the TI thread. However, due to Linx’ heuristic approach with several rules and clustering routines, the algorithm still managed to cluster all SVs related to the TI thread together in a giant complex event with 97 chains and 827 SVs together with many additional SVs from the massive chromothripsis event involving chr4, chr5 and chr7 (among others). Chain 2 best overlapped the TI thread presented in Figure 2 but greatly underestimated the true number of junctions with only 43 compared to 231 estimated from the Shasta assembled TI thread contig (Figure S46) – suggesting clear limitations of Linx and short-reads in general with respect to the characterization of such complex rearrangement structures.
Telomere analysis of derivative chromosomal segments
As part of our long-read analysis toolbox for cancer genomics, termed lorax, we also developed a method that identifies telomeric motifs, repeats of TTAGGG, TGAGGG, TCAGGG, TTGGGG or their reverse complement, in error-prone ONT reads and applied this method to the long read data of the primary tumor and the relapse sample. As suggested previously,97 we start by precomputing all possible strand-specific 18-mer telomere motifs, scan all long-reads for exact motif matches and count their occurence. We then search for distal non-telomeric alignments of these reads and intersect reads that show both a telomeric repeat and a unique alignment outside a telomere region of a minimum length of 1kbp. We use the control genome to filter out likely mapping artifacts due to incomplete reference sequences by masking alignments from the control genome that show both a telomeric repeat and a unique alignment outside a telomere region. In case of mapping ambiguities, we used the CHM13 telomere-to-telomere (CHM T2T) assembly41 as an alternative reference sequence. The method to detect telomere fusions is implemented in our long-read alignment toolkit lorax as a new sub-command. For the matched illumina data, we apply a window-based search (default 1kbp) that counts reads with a telomeric motif based on the mapping location of the read (or its mate if the read is unmapped). If both read1 and read2 are unmapped the sequencing pair is discarded. We filter out all windows that are discovered in the matched control (blood) and retain in the tumor only windows with at least 5 supporting paired-ends. The short-read method is implemented in the alfred toolkit75 as a new sub-command, called ‘alfred telmotif’.
Differential methylation testing
In order to find genomic regions with differential methylation between samples, we used the software package pycoMeth.44 PycoMeth aggregates methylation likelihood ratios reported by Nanopolish over predefined regions, computes a read-level methylation rate from thresholded log-likelihood ratios (threshold 2.0) and then performs a Wilcoxon rank-sum test (for 2-sample comparison) or Kruskal Wallis test (for more than two samples) for methylation rates across samples. P-values were then adjusted for multiple testing using independent hypothesis weighting,98 using a weight based on the variance of methylation rates, and the Benjamini-Hochberg method.99 Regions with FDR<=0.05 are reported as DMRs. Candidate regions for differential methylation testing are selected based on two different segmentation methods: 1) sequence segmentation and 2) methylome segmentation. Sequence segmentation uses pycoMeth's CGI Finder module, which determines CpG islands based on local CG-density. For methylome segmentation pycoMeth Meth_Seg, a de novo methylome segmentation method which implements a bayesian changepoint-detection algorithm, is used to determine regions with consistent methylation rate from the read-level methylation predictions. To achieve one consistent segmentation for all tumor analyses, pycoMeth Meth_Seg was called together on both primary and relapse data, with haplotype information stored in the input MetH5 file. The window-size parameters and max-segments parameter were set to 600 and 16 respectively, as recommended in the pycoMeth benchmark. To assign reads to haplotypes we used WhatsHap’s haplotag command and the three-stage phased blood variants. This haplotype assignment was used as the read-group parameter in pycoMeth, allowing it to consider ASM in the methylome segmentation.
We investigated differentially methylated regions between the primary tumor and the relapse sample by applying pycoMeth Meth_Comp using both candidate region approaches with the parameter using the parameter —hypothesis bs_diff in order to test for difference in read-level methylation rate per segment. In pycoMeth, differential methylation calling was then performed between haplotypes within each sample, in order to determine regions with ASM. For further analyses, DMRs were filtered by an effect size threshold of 0.5 abs methylation rate difference. Differentially methylated regions were then mapped to genes based on their proximity to a TSS, that is they were labeled as promoter methylation if a region was in the range 2,000bps upstream to 500bps downstream from the any transcript’s TSS, or if it overlapped with an enhancer active in Cerebellum as annotated by EnhancerAtlas 2.0.100 Enhancers were then linked to the nearest gene, if the gene is closer than 30kbps. Since detection power in the relapse sample was lower, due to lower read-depth, we investigated whether ASM effects found in primary tumor could be found in relapse as well by applying the same 0.5 absolute methylation rate difference threshold.
RNA alignment and expression quantification
Gene-expression quantification was performed in line with the GTEx standards. In short, we (re)processed the RAW expression data by first aligning the reads to the human reference genome, build 38, using STAR in two step mapping per sample. The mapping was performed in two modes. One for the allele specific expression, using a custom reference genome, replacing the homozygous SNP variants with the relevant genotype of the sample, and supplying a VCF with heterozygous variants when mapping in STAR, used for allele specific expression and gene fusion detection. Second, for the differential expression and splicing analyses we remapped the samples to the standard genome. Gene information was taken from ENSEMBL (v101) and gene-expression quantification was performed using RNASeQC, in line with the GTEx consortium expression quantification. Using LeafCutter101 we quantified splicing across the two samples, as well as a cerebellum reference dataset (SRP151960).102
Reference RNA expression datasets and differential expression
For comparative expression analysis we leverage data from the ALS consortium (SRP151960)102 and GTEx cerebellum expression data. The data from the ALS consortium were reprocessed as done for the two medulloblastoma samples, see above, and the GTEx data103 was used as is. This data was leveraged both for direct comparison of expression levels, and for correction of the gene expression levels.
The first five principal components (PCs) were calculated on the combined ALS and GTEx dataset. The medulloblastoma samples were projected into this same PC space, using the rotation information, and the first five PCs were regressed out from the expression levels of all samples, medulloblastoma, GTEx and ALS. Next we used a Z-score transformation on both the raw and corrected expression of the reference samples and placed the two medulloblastoma samples in these distributions. Given that there are still major differences between the samples and studies, in terms of age, disease and batch, we only use the two samples in a comparative setting. The reference data is used to test for concordance of effects with and without correction. For the differential expression analysis we used the log TPM values and checked concordance in Z-scores.
Allele specific expression and allele specific copy number estimation
ASE on the primary tumor and relapse samples was called from the RNA sequencing data using WASP59 and the phased germline variants, using the approach described in the WASP paper.59 In order to verify whether ASE was driven by DNA copy number amplification or depletion in one haplotype, we estimate allele specific DNA copy number ratio using GATK CollectAllelicCounts104 on the same variants used to identify ASE.
Gene fusion and validation using DNA long reads
Potential gene fusions were detected from RNA sequencing data using Arriba105 (V2.0.0). The SVs called from both short and long read data were used to inform Arriba, and we included the provided blacklist, other settings were left at defaults. After identification of the gene fusion pairs we set out to validate these using the long read DNA data. First, we check for individual read support from ONT reads with chimeric alignments mapping to both genes. Fusion pairs involving long intergenic non-coding RNA genes, which are characterized by long introns of on average 10kbps length,106 or fusion containing large intronic insertions, however often do not have individual genomic reads spanning exons of both genes. In order to additionally validate such fusions with large insertions, for which no single ONT read spans the fusion pairs, we devised a graph-based method to suggest the most plausible gene fusion reconstruction. We construct a graph with nodes representing each base pair position in the reference and edges representing neighboring basepairs. Structural variations, both inter- and intrachromosomal, were then represented as additional edges in the graph, creating shortcuts between the locations on the side of the genomic breakpoint connected by the structural variation). A gene fusion pair was then explained by determining the shortest path between the two fusion partners in the graph using Dijkstra’s algorithm for shortest paths.107 Edges which crossed the exons of a gene not involved in the fusion were removed for the purpose of finding the shortest path. Fusion pairs were classified as either validated by individual read support, explainable using the graph algorithm, or both (high confidence read support).
Acknowledgments
We thank Frauke Devens, Kim Judge, Vladimir Benes, as well as DKFZ and EMBL IT and sequencing core facilities for excellent technical support. The present contribution is supported by the Helmholtz Association under the joint research school HIDSS4Health - Helmholtz Information and Data Science School for Health. We thank Peter Lichter for discussions and Tom Mitchell for sharing long-read sequencing data from a renal cell carcinoma sample for comparison. A.E. received funding from the DFG (project number 460595631) and from the Wilhelm Sander Foundation (project number 2020.115.1). J.O.K. received funding from the BMBF (031L0184C) and from the NIH (1R01HG010169-01 and 2U24HG007497-05).
Author contributions
E.B., O.S., A.E., and J.O.K. designed the study. A.L. and R.S. performed long-read base calling and alignment. R.S. performed methylation calling and differential methylation analysis. T.R. implemented phasing, (targeted) assembly workflows, germline and somatic variant discovery, and complex structural variant calling. R.S. and M.J.B. performed RNA alignment and expression quantification and performed subsequent expression analyses. M.S. performed FISH and established xenograft models for metaphase spreads. T.R., R.S., M.J.B., A.E., and J.O.K. analyzed complex mutation patterns and targeted assemblies. M.G. and A.G.H. performed Circle-seq and analyzed the data. S.F. provided validation samples and helped with the clinical interpretation. L.V. prepared ONT libraries. R.S. implemented the gene fusion validation. T.R. and J.O.K. performed TI analysis and interpretation in PCAWG. R.S., T.R., and M.J.B. prepared the main display items, with additional contributions from A.E. and J.O.K. T.R., R.S., M.J.B., A.E., and J.O.K. wrote the manuscript, with input from all other authors.
Declaration of interests
E.B. is a paid consultant and shareholder of ONT. A.L. has received financial support from ONT for consumables during the course of the project and is currently an employee of ONT. O.S. is a paid consultant of Insitro, Inc.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: March 22, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2023.100281.
Contributor Information
Marc Jan Bonder, Email: bonder.m.j@gmail.com.
Aurelie Ernst, Email: a.ernst@dkfz-heidelberg.de.
Jan O. Korbel, Email: jan.korbel@embl.de.
Supplemental information
For reference the ALS data (RSP151960) is also shown, related to section "functional annotation of the TI threads and telomere SVs,” related to Figures 2 and S35.
Data and code availability
-
•
Sequencing data generated in this project: (1) the primary sample (blood, primary tumor and relapse), as well as a PDX derived from this sample and the Liposarcoma replication data, has been deposited at the European Genome-phenome Archive (EGA), and accession numbers are listed in the key resources table.
-
•
All original code has been deposited at GitHub, archived at Zenodo and is publicly available as of the date of publication. GitHub URLs and DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Priestley P., Baber J., Lolkema M.P., Steeghs N., de Bruijn E., Shale C., Duyvesteyn K., Haidari S., van Hoeck A., Onstenk W., et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575:210–216. doi: 10.1038/s41586-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y., Roberts N.D., Wala J.A., Shapira O., Schumacher S.E., Kumar K., Khurana E., Waszak S., Korbel J.O., Haber J.E., et al. Patterns of somatic structural variation in human cancer genomes. Nature. 2020;578:112–121. doi: 10.1038/s41586-019-1913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gröbner S.N., Worst B.C., Weischenfeldt J., Buchhalter I., Kleinheinz K., Rudneva V.A., Johann P.D., Balasubramanian G.P., Segura-Wang M., Brabetz S., et al. The landscape of genomic alterations across childhood cancers. Nature. 2018;555:321–327. doi: 10.1038/nature25480. [DOI] [PubMed] [Google Scholar]
- 5.Ho S.S., Urban A.E., Mills R.E. Structural variation in the sequencing era. Nat. Rev. Genet. 2020;21:171–189. doi: 10.1038/s41576-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L.B., Martin S., Wedge D.C., et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hadi K., Yao X., Behr J.M., Deshpande A., Xanthopoulakis C., Tian H., Kudman S., Rosiene J., Darmofal M., DeRose J., et al. Distinct classes of complex structural variation uncovered across thousands of cancer genome graphs. Cell. 2020;183:197–210.e32. doi: 10.1016/j.cell.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakamoto Y., Sereewattanawoot S., Suzuki A. A new era of long-read sequencing for cancer genomics. J. Hum. Genet. 2020;65:3–10. doi: 10.1038/s10038-019-0658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakamoto Y., Zaha S., Suzuki Y., Seki M., Suzuki A. Application of long-read sequencing to the detection of structural variants in human cancer genomes. Comput. Struct. Biotechnol. J. 2021;19:4207–4216. doi: 10.1016/j.csbj.2021.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mantere T., Kersten S., Hoischen A. Long-read sequencing emerging in medical genetics. Front. Genet. 2019;10:426. doi: 10.3389/fgene.2019.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nattestad M., Goodwin S., Ng K., Baslan T., Sedlazeck F.J., Rescheneder P., Garvin T., Fang H., Gurtowski J., Hutton E., et al. Complex rearrangements and oncogene amplifications revealed by long-read DNA and RNA sequencing of a breast cancer cell line. Genome Res. 2018;28:1126–1135. doi: 10.1101/gr.231100.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaisson M.J.P., Sanders A.D., Zhao X., Malhotra A., Porubsky D., Rausch T., Gardner E.J., Rodriguez O.L., Guo L., Collins R.L., et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 2019;10:1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebert P., Audano P.A., Zhu Q., Rodriguez-Martin B., Porubsky D., Bonder M.J., Sulovari A., Ebler J., Zhou W., Serra Mari R., et al. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science. 2021;372:eabf7117. doi: 10.1126/science.abf7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porubsky D., Höps W., Ashraf H., Hsieh P., Rodriguez-Martin B., Yilmaz F., Ebler J., Hallast P., Maria Maggiolini F.A., Harvey W.T., et al. Recurrent inversion polymorphisms in humans associate with genetic instability and genomic disorders. Cell. 2022;185:1986–2005.e26. doi: 10.1016/j.cell.2022.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laszlo A.H., Derrington I.M., Brinkerhoff H., Langford K.W., Nova I.C., Samson J.M., Bartlett J.J., Pavlenok M., Gundlach J.H. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc. Natl. Acad. Sci. USA. 2013;110:18904–18909. doi: 10.1073/pnas.1310240110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rausch T., Jones D.T.W., Zapatka M., Stütz A.M., Zichner T., Weischenfeldt J., Jäger N., Remke M., Shih D., Northcott P.A., et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waszak S.M., Northcott P.A., Buchhalter I., Robinson G.W., Sutter C., Groebner S., Grund K.B., Brugières L., Jones D.T.W., Pajtler K.W., et al. Spectrum and prevalence of genetic predisposition in medulloblastoma: a retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol. 2018;19:785–798. doi: 10.1016/S1470-2045(18)30242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A., et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voronina N., Wong J.K.L., Hübschmann D., Hlevnjak M., Uhrig S., Heilig C.E., Horak P., Kreutzfeldt S., Mock A., Stenzinger A., et al. The landscape of chromothripsis across adult cancer types. Nat. Commun. 2020;11:2320. doi: 10.1038/s41467-020-16134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simovic M., Bolkestein M., Moustafa M., Wong J.K.L., Körber V., Benedetto S., Khalid U., Schreiber H.S., Jugold M., Korshunov A., et al. Carbon ion radiotherapy eradicates medulloblastomas with chromothripsis in an orthotopic Li-Fraumeni patient-derived mouse model. Neuro Oncol. 2021;23:2028–2041. doi: 10.1093/neuonc/noab127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patterson M., Marschall T., Pisanti N., van Iersel L., Stougie L., Klau G.W., Schönhuth A. WhatsHap: weighted haplotype assembly for future-generation sequencing reads. J. Comput. Biol. 2015;22:498–509. doi: 10.1089/cmb.2014.0157. [DOI] [PubMed] [Google Scholar]
- 22.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaneau O., Zagury J.-F., Robinson M.R., Marchini J.L., Dermitzakis E.T. Accurate, scalable and integrative haplotype estimation. Nat. Commun. 2019;10:5436. doi: 10.1038/s41467-019-13225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujimoto A., Wong J.H., Yoshii Y., Akiyama S., Tanaka A., Yagi H., Shigemizu D., Nakagawa H., Mizokami M., Shimada M. Whole-genome sequencing with long reads reveals complex structure and origin of structural variation in human genetic variations and somatic mutations in cancer. Genome Med. 2021;13:65. doi: 10.1186/s13073-021-00883-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koche R.P., Rodriguez-Fos E., Helmsauer K., Burkert M., MacArthur I.C., Maag J., Chamorro R., Munoz-Perez N., Puiggròs M., Dorado Garcia H., et al. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nat. Genet. 2020;52:29–34. doi: 10.1038/s41588-019-0547-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolmogorov M., Yuan J., Lin Y., Pevzner P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 27.Shafin K., Pesout T., Lorig-Roach R., Haukness M., Olsen H.E., Bosworth C., Armstrong J., Tigyi K., Maurer N., Koren S., et al. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat. Biotechnol. 2020;38:1044–1053. doi: 10.1038/s41587-020-0503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bignell G.R., Santarius T., Pole J.C.M., Butler A.P., Perry J., Pleasance E., Greenman C., Menzies A., Taylor S., Edkins S., et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–1303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umbreit N.T., Zhang C.-Z., Lynch L.D., Blaine L.J., Cheng A.M., Tourdot R., Sun L., Almubarak H.F., Judge K., Mitchell T.J., et al. Mechanisms generating cancer genome complexity from a single cell division error. Science. 2020;368:eaba0712. doi: 10.1126/science.aba0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., Choi P.S., Francis J.M., Imielinski M., Watanabe H., Cherniack A.D., Meyerson M. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat. Genet. 2016;48:176–182. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weischenfeldt J., Dubash T., Drainas A.P., Mardin B.R., Chen Y., Stütz A.M., Waszak S.M., Bosco G., Halvorsen A.R., Raeder B., et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 2017;49:65–74. doi: 10.1038/ng.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Micci F., Teixeira M.R., Bjerkehagen B., Heim S. Characterization of supernumerary rings and giant marker chromosomes in well-differentiated lipomatous tumors by a combination of G-banding, CGH, M-FISH, and chromosome- and locus-specific FISH. Cytogenet. Genome Res. 2002;97:13–19. doi: 10.1159/000064038. [DOI] [PubMed] [Google Scholar]
- 34.Mandahl N., Magnusson L., Nilsson J., Viklund B., Arbajian E., von Steyern F.V., Isaksson A., Mertens F. Scattered genomic amplification in dedifferentiated liposarcoma. Mol. Cytogenet. 2017;10:25. doi: 10.1186/s13039-017-0325-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosswog C., Bartenhagen C., Welte A., Kahlert Y., Hemstedt N., Lorenz W., Cartolano M., Ackermann S., Perner S., Vogel W., et al. Chromothripsis followed by circular recombination drives oncogene amplification in human cancer. Nat. Genet. 2021;53:1673–1685. doi: 10.1038/s41588-021-00951-7. [DOI] [PubMed] [Google Scholar]
- 36.Horak P., Klink B., Heining C., Gröschel S., Hutter B., Fröhlich M., Uhrig S., Hübschmann D., Schlesner M., Eils R., et al. Precision oncology based on omics data: the NCT Heidelberg experience. Int. J. Cancer. 2017;141:877–886. doi: 10.1002/ijc.30828. [DOI] [PubMed] [Google Scholar]
- 37.Keung E.Z., Somaiah N. Overview of liposarcomas and their genomic landscape. J. Transl. Genet. Genom. 2019;3:8. doi: 10.20517/jtgg.2019.03. [DOI] [Google Scholar]
- 38.Maciejowski J., Li Y., Bosco N., Campbell P.J., de Lange T. Chromothripsis and kataegis induced by telomere crisis. Cell. 2015;163:1641–1654. doi: 10.1016/j.cell.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ernst A., Jones D.T.W., Maass K.K., Rode A., Deeg K.I., Jebaraj B.M.C., Korshunov A., Hovestadt V., Tainsky M.A., Pajtler K.W., et al. Telomere dysfunction and chromothripsis. Int. J. Cancer. 2016;138:2905–2914. doi: 10.1002/ijc.30033. [DOI] [PubMed] [Google Scholar]
- 40.Sieverling L., Hong C., Koser S.D., Ginsbach P., Kleinheinz K., Hutter B., Braun D.M., Cortés-Ciriano I., Xi R., Kabbe R., et al. Genomic footprints of activated telomere maintenance mechanisms in cancer. Nat. Commun. 2020;11:733. doi: 10.1038/s41467-019-13824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nurk S., Koren S., Rhie A., Rautiainen M., Bzikadze A.V., Mikheenko A., Vollger M.R., Altemose N., Uralsky L., Gershman A., et al. The complete sequence of a human genome. Science. 2022;376:44–53. doi: 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liddiard K., Grimstead J.W., Cleal K., Evans A., Baird D.M. Tracking telomere fusions through crisis reveals conflict between DNA transcription and the DNA damage response. NAR Cancer. 2021;3:zcaa044. doi: 10.1093/narcan/zcaa044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tabori U., Nanda S., Druker H., Lees J., Malkin D. Younger age of cancer initiation is associated with shorter telomere length in Li-Fraumeni syndrome. Cancer Res. 2007;67:1415–1418. doi: 10.1158/0008-5472.CAN-06-3682. [DOI] [PubMed] [Google Scholar]
- 44.Snajder R., Leger A., Stegle O., Bonder M.J. pycoMeth: a toolbox for differential methylation testing from Nanopore methylation calls. bioRxiv. 2022 doi: 10.1101/2022.02.16.480699. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Northcott P.A., Buchhalter I., Morrissy A.S., Hovestadt V., Weischenfeldt J., Ehrenberger T., Gröbner S., Segura-Wang M., Zichner T., Rudneva V.A., et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547:311–317. doi: 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bacolod M.D., Lin S.M., Johnson S.P., Bullock N.S., Colvin M., Bigner D.D., Friedman H.S. The gene expression profiles of medulloblastoma cell lines resistant to preactivated cyclophosphamide. Curr. Cancer Drug Targets. 2008;8:172–179. doi: 10.2174/156800908784293631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du W., Gao A., Herman J.G., Wang L., Zhang L., Jiao S., Guo M. Methylation of NRN1 is a novel synthetic lethal marker of PI3K-Akt-mTOR and ATR inhibitors in esophageal cancer. Cancer Sci. 2021;112:2870–2883. doi: 10.1111/cas.14917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pritchard J.I., Olson J.M. Methylation of PTCH1, the Patched-1 gene, in a panel of primary medulloblastomas. Cancer Genet. Cytogenet. 2008;180:47–50. doi: 10.1016/j.cancergencyto.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newell-Price J., Clark A.J., King P. DNA methylation and silencing of gene expression. Trends Endocrinol. Metab. 2000;11:142–148. doi: 10.1016/s1043-2760(00)00248-4. [DOI] [PubMed] [Google Scholar]
- 50.Yamagishi H., Maeda J., Hu T., McAnally J., Conway S.J., Kume T., Meyers E.N., Yamagishi C., Srivastava D. Tbx1 is regulated by tissue-specific forkhead proteins through a common Sonic hedgehog-responsive enhancer. Genes Dev. 2003;17:269–281. doi: 10.1101/gad.1048903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skowron P., Farooq H., Cavalli F.M.G., Morrissy A.S., Ly M., Hendrikse L.D., Wang E.Y., Djambazian H., Zhu H., Mungall K.L., et al. The transcriptional landscape of Shh medulloblastoma. Nat. Commun. 2021;12:1749. doi: 10.1038/s41467-021-21883-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang S., Dai Z., Li W., Wang R., Huang D. Aberrant promoter methylation reduced the expression of protocadherin 17 in nasopharyngeal cancer. Biochem. Cell. Biol. 2019;97:364–368. doi: 10.1139/bcb-2017-0343. [DOI] [PubMed] [Google Scholar]
- 53.Baranova I., Kovarikova H., Laco J., Dvorak O., Sedlakova I., Palicka V., Chmelarova M. Aberrant methylation of PCDH17 gene in high-grade serous ovarian carcinoma. Cancer Biomark. 2018;23:125–133. doi: 10.3233/CBM-181493. [DOI] [PubMed] [Google Scholar]
- 54.Byzia E., Soloch N., Bodnar M., Szaumkessel M., Kiwerska K., Kostrzewska-Poczekaj M., Jarmuz-Szymczak M., Szylberg L., Wierzbicka M., Bartochowska A., et al. Recurrent transcriptional loss of the PCDH17 tumor suppressor in laryngeal squamous cell carcinoma is partially mediated by aberrant promoter DNA methylation. Mol. Carcinog. 2018;57:878–885. doi: 10.1002/mc.22808. [DOI] [PubMed] [Google Scholar]
- 55.Lin Y.-L., Wang Y.-P., Li H.-Z., Zhang X. Aberrant promoter methylation of PCDH17 (protocadherin 17) in serum and its clinical significance in renal cell carcinoma. Med. Sci. Monit. 2017;23:3318–3323. doi: 10.12659/MSM.902077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uyen T.N., Sakashita K., Al-Kzayer L.F.Y., Nakazawa Y., Kurata T., Koike K. Aberrant methylation of protocadherin 17 and its prognostic value in pediatric acute lymphoblastic leukemia. Pediatr. Blood Cancer. 2017;64:e26259. doi: 10.1002/pbc.26259. [DOI] [PubMed] [Google Scholar]
- 57.Meaburn E.L., Schalkwyk L.C., Mill J. Allele-specific methylation in the human genome: implications for genetic studies of complex disease. Epigenetics. 2010;5:578–582. doi: 10.4161/epi.5.7.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Do C., Dumont E.L.P., Salas M., Castano A., Mujahed H., Maldonado L., Singh A., DaSilva-Arnold S.C., Bhagat G., Lehman S., et al. Allele-specific DNA methylation is increased in cancers and its dense mapping in normal plus neoplastic cells increases the yield of disease-associated regulatory SNPs. Genome Biol. 2020;21:153. doi: 10.1186/s13059-020-02059-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van de Geijn B., McVicker G., Gilad Y., Pritchard J.K. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods. 2015;12:1061–1063. doi: 10.1038/nmeth.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Northcott P.A., Shih D.J.H., Peacock J., Garzia L., Morrissy A.S., Zichner T., Stütz A.M., Korshunov A., Reimand J., Schumacher S.E., et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bertrand K.C., Faria C.C., Skowron P., Luck A., Garzia L., Wu X., Agnihotri S., Smith C.A., Taylor M.D., Mack S.C., Rutka J.T. A functional genomics approach to identify pathways of drug resistance in medulloblastoma. Acta Neuropathol. Commun. 2018;6:146. doi: 10.1186/s40478-018-0652-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stevenson B.W., Gorman M.A., Koach J., Cheung B.B., Marshall G.M., Parker M.W., Holien J.K. A structural view of PA2G4 isoforms with opposing functions in cancer. J. Biol. Chem. 2020;295:16100–16112. doi: 10.1074/jbc.REV120.014293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin L.-L., Liu Z.-Z., Tian J.-Z., Zhang X., Zhang Y., Yang M., Zhong H.-C., Fang W., Wei R.-X., Hu C. Integrated analysis of nine prognostic RNA-binding proteins in soft tissue sarcoma. Front. Oncol. 2021;11:633024. doi: 10.3389/fonc.2021.633024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korbel J.O., Campbell P.J. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 65.Cortés-Ciriano I., Lee J.J.-K., Xi R., Jain D., Jung Y.L., Yang L., Gordenin D., Klimczak L.J., Zhang C.-Z., Pellman D.S., et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat. Genet. 2020;52:331–341. doi: 10.1038/s41588-019-0576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grüning B., Dale R., Sjödin A., Chapman B.A., Rowe J., Tomkins-Tinch C.H., Valieris R., Köster J., Bioconda Team Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat. Methods. 2018;15:475–476. doi: 10.1038/s41592-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rausch T., Zichner T., Schlattl A., Stütz A.M., Benes V., Korbel J.O. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sedlazeck F.J., Rescheneder P., Smolka M., Fang H., Nattestad M., von Haeseler A., Schatz M.C. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods. 2018;15:461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bonfield J.K., Marshall J., Danecek P., Li H., Ohan V., Whitwham A., Keane T., Davies R.M. HTSlib: C library for reading/writing high-throughput sequencing data. GigaScience. 2021;10:giab007. doi: 10.1093/gigascience/giab007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 2012 doi: 10.48550/arXiv.1207.3907. Preprint at. [DOI] [Google Scholar]
- 73.Kim S., Scheffler K., Halpern A.L., Bekritsky M.A., Noh E., Källberg M., Chen X., Kim Y., Beyter D., Krusche P., Saunders C.T. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods. 2018;15:591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 74.Simpson J.T., Workman R.E., Zuzarte P.C., David M., Dursi L.J., Timp W. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods. 2017;14:407–410. doi: 10.1038/nmeth.4184. [DOI] [PubMed] [Google Scholar]
- 75.Rausch T., Hsi-Yang Fritz M., Korbel J.O., Benes V. Alfred: interactive multi-sample BAM alignment statistics, feature counting and feature annotation for long- and short-read sequencing. Bioinformatics. 2019;35:2489–2491. doi: 10.1093/bioinformatics/bty1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tham C.Y., Tirado-Magallanes R., Goh Y., Fullwood M.J., Koh B.T.H., Wang W., Ng C.H., Chng W.J., Thiery A., Tenen D.G., Benoukraf T. NanoVar: accurate characterization of patients’ genomic structural variants using low-depth nanopore sequencing. Genome Biol. 2020;21:56. doi: 10.1186/s13059-020-01968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wala J.A., Bandopadhayay P., Greenwald N.F., O’Rourke R., Sharpe T., Stewart C., Schumacher S., Li Y., Weischenfeldt J., Yao X., et al. SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Res. 2018;28:581–591. doi: 10.1101/gr.221028.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bolognini D., Sanders A., Korbel J.O., Magi A., Benes V., Rausch T. VISOR: a versatile haplotype-aware structural variant simulator for short- and long-read sequencing. Bioinformatics. 2020;36:1267–1269. doi: 10.1093/bioinformatics/btz719. [DOI] [PubMed] [Google Scholar]
- 79.Lichter P., Tang C.J., Call K., Hermanson G., Evans G.A., Housman D., Ward D.C. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- 80.Henssen A., MacArthur I., Koche R., Dorado-García H. 2019. Purification and Sequencing of Large Circular DNA from Human Cells. [DOI] [Google Scholar]
- 81.Krueger F., James F., Ewels P., Afyounian E., Schuster-Boeckler B. 2021. FelixKrueger/TrimGalore: v0.6.7 - DOI via Zenodo. [DOI] [Google Scholar]
- 82.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 doi: 10.48550/arXiv.1303.3997. Preprint at. [DOI] [Google Scholar]
- 83.Picard. http://broadinstitute.github.io/picard/
- 84.Duttke S.H., Chang M.W., Heinz S., Benner C. Identification and dynamic quantification of regulatory elements using total RNA. Genome Res. 2019;29:1836–1846. doi: 10.1101/gr.253492.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., et al. The UCSC genome browser database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Venkatraman E.S., Olshen A.B. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 88.Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Källberg M., Cox A.J., Kruglyak S., Saunders C.T. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 89.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.1000 Genomes Project Consortium. Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marçais G., Delcher A.L., Phillippy A.M., Coston R., Salzberg S.L., Zimin A. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 2018;14:e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kiełbasa S.M., Wan R., Sato K., Horton P., Frith M.C. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aganezov S., Raphael B.J. Reconstruction of clone- and haplotype-specific cancer genome karyotypes from bulk tumor samples. Genome Res. 2020;30:1274–1290. doi: 10.1101/gr.256701.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shale C., Cameron D.L., Baber J., Wong M., Cowley M.J., Papenfuss A.T., Cuppen E., Priestley P. Unscrambling cancer genomes via integrated analysis of structural variation and copy number. Cell Genom. 2022;2:100112. doi: 10.1016/j.xgen.2022.100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zaccaria S., Raphael B.J. Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data. Nat. Commun. 2020;11:4301. doi: 10.1038/s41467-020-17967-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Behr J.M., Yao X., Hadi K., Tian H., Deshpande A., Rosiene J., de Lange T., Imieliński M. Loose ends in cancer genome structure. bioRxiv. 2021 doi: 10.1101/2021.05.26.445837. Preprint at. [DOI] [Google Scholar]
- 98.Ignatiadis N., Klaus B., Zaugg J.B., Huber W. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods. 2016;13:577–580. doi: 10.1038/nmeth.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 100.Gao T., Qian J. EnhancerAtlas 2.0: an updated resource with enhancer annotation in 586 tissue/cell types across nine species. Nucleic Acids Res. 2020;48:D58–D64. doi: 10.1093/nar/gkz980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li Y.I., Knowles D.A., Humphrey J., Barbeira A.N., Dickinson S.P., Im H.K., Pritchard J.K. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 2018;50:151–158. doi: 10.1038/s41588-017-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Conlon E.G., Fagegaltier D., Agius P., Davis-Porada J., Gregory J., Hubbard I., Kang K., Kim D., New York Genome Center ALS Consortium. Phatnani H., et al. Unexpected similarities between C9ORF72 and sporadic forms of ALS/FTD suggest a common disease mechanism. Elife. 2018;7:e37754. doi: 10.7554/eLife.37754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Uhrig S., Ellermann J., Walther T., Burkhardt P., Fröhlich M., Hutter B., Toprak U.H., Neumann O., Stenzinger A., Scholl C., et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res. 2021;31:448–460. doi: 10.1101/gr.257246.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chernikova D., Managadze D., Glazko G.V., Makalowski W., Rogozin I.B. Conservation of the exon-intron structure of long intergenic non-coding RNA genes in eutherian mammals. Life. 2016;6:27. doi: 10.3390/life6030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dijkstra E.W. A note on two problems in connexion with graphs. Numer. Math. 1959;1:269–271. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For reference the ALS data (RSP151960) is also shown, related to section "functional annotation of the TI threads and telomere SVs,” related to Figures 2 and S35.
Data Availability Statement
-
•
Sequencing data generated in this project: (1) the primary sample (blood, primary tumor and relapse), as well as a PDX derived from this sample and the Liposarcoma replication data, has been deposited at the European Genome-phenome Archive (EGA), and accession numbers are listed in the key resources table.
-
•
All original code has been deposited at GitHub, archived at Zenodo and is publicly available as of the date of publication. GitHub URLs and DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.