Abstract
Recent studies report the first structures of two CALHM family members, describing unexpected oligomeric architecture and providing insights into the mechanism of function of CALHM channels.
The calcium homeostasis modulator (CALHM) gene family, known previously as the FAM26 gene family, consists of six members that share 20–50% sequence similarity but lack similarities to other proteins. CALHMs 1 and 3 have been identified as pore-forming subunits of plasma membrane ion channels and have been implicated in Alzheimer’s disease, neuronal excitability and taste perception. Understanding how CALHMs function as ion channels and how they contribute to physiology has been hampered by a lack of structural information. Two recent structural reports1,2 reveal remarkable and unexpected features of CALHMs, including an undecameric structure of CALHM2 in both hemichannel and gap-junction channel conformations and a different, octameric assembly of CALHM1. Equally importantly, these studies provide insights into possible mechanisms of channel assembly and gating.
The founding member, CALHM1, was identified in a bioinformatics search for genes expressed in the brain that are linked to a propensity for Alzheimer’s disease3. CALHM1 is a pore-forming subunit of a Ca2+-permeable ion channel that is activated by membrane depolarization, as well as by removal of extracellular Ca2+ (hence the naming of the gene family)4. Sensitivity to extracellular Ca2+ was found to play a role in brain cortical neuron excitability5, although the role of CALHM1 in Alzheimer’s disease is unclear. CALHM1 discriminates poorly among small ions and allows permeation of large molecules, including ATP, consistent with a pore diameter of 14 Å6,7. The large pore size is comparable to those of connexins and innexins, which form gap junctions in vertebrates and insects, respectively, and to those of other ATP-permeable plasma membrane channels, including pannexins and the related leucine-rich-repeat-containing proteins LRRC8s8. It was suggested that CALHM1 has four transmembrane domains (TMDs), with cytoplasmic N and C termini, and that it has a hexameric quaternary structure6. These features, as well as others predicted by secondary-structure analyses, including the presence of a short N-terminal helix preceding the first TMD, are also similar to those found in the other proteins with comparable pore sizes. Accordingly, it was suggested that CALHM1 was related to the connexin and pannexin, innexin and LRRC8 channel families by convergent evolution6.
The two studies, by Choi et al.1 and by Syrjanen et al.2, have now yielded structural insights with surprising results. Both groups report cryo-EM structures of human CALHM2, whereas Syrjanen et al. also report the structure of chicken CALHM1. The structures solved in the absence of divalent cations are likely open-channel conformations. The protomers have similar structures in the two CALHMs. As predicted, each consists of four TMDs, with cytoplasmic N and C termini. The four TMDs are arranged counterclockwise when viewed from the extracellular side, and TM1 is only loosely associated with TM3. TM2, 3 and 4 are aligned approximately in a plane. This distinguishes CALHM proteins from connexins, innexins and LRRC8s, in which the helices are arranged in a clockwise orientation and form a compact bundle. The CALHM1 channel structure revealed an unexpected octameric assembly, similar to innexins (Fig. 1a). Remarkably, CALHM2 was captured as an undecamer (Fig. 1b). Understanding how two members of the same protein family could assemble into channels with distinct numbers of subunits stems from the analysis of the C-terminal tails2. A chimeric CALHM1 channel with its TM4–linker–C-terminal sequences replaced with those from CALHM2 adopted an undecameric structure similar to that of CALHM2. Thus, the TM4–linker and C-terminal helix interactions are strong determinants of the oligomeric state of these CALHM channels. The linker sequence in CALHM1 is similar to that of CALHM3, which is compatible with the observation that CALHM1 and CALHM3 can hetero-oligomerize to form a functional CALHM1/3 ion channel9. Syrjanen et al. suggest that CALHMs 4, 5 and 6, with linker sequences similar to those of CALHM2, may form undecameric assemblies as well.
Fig. 1 |. Structures of CALHM1 and CALHM2.
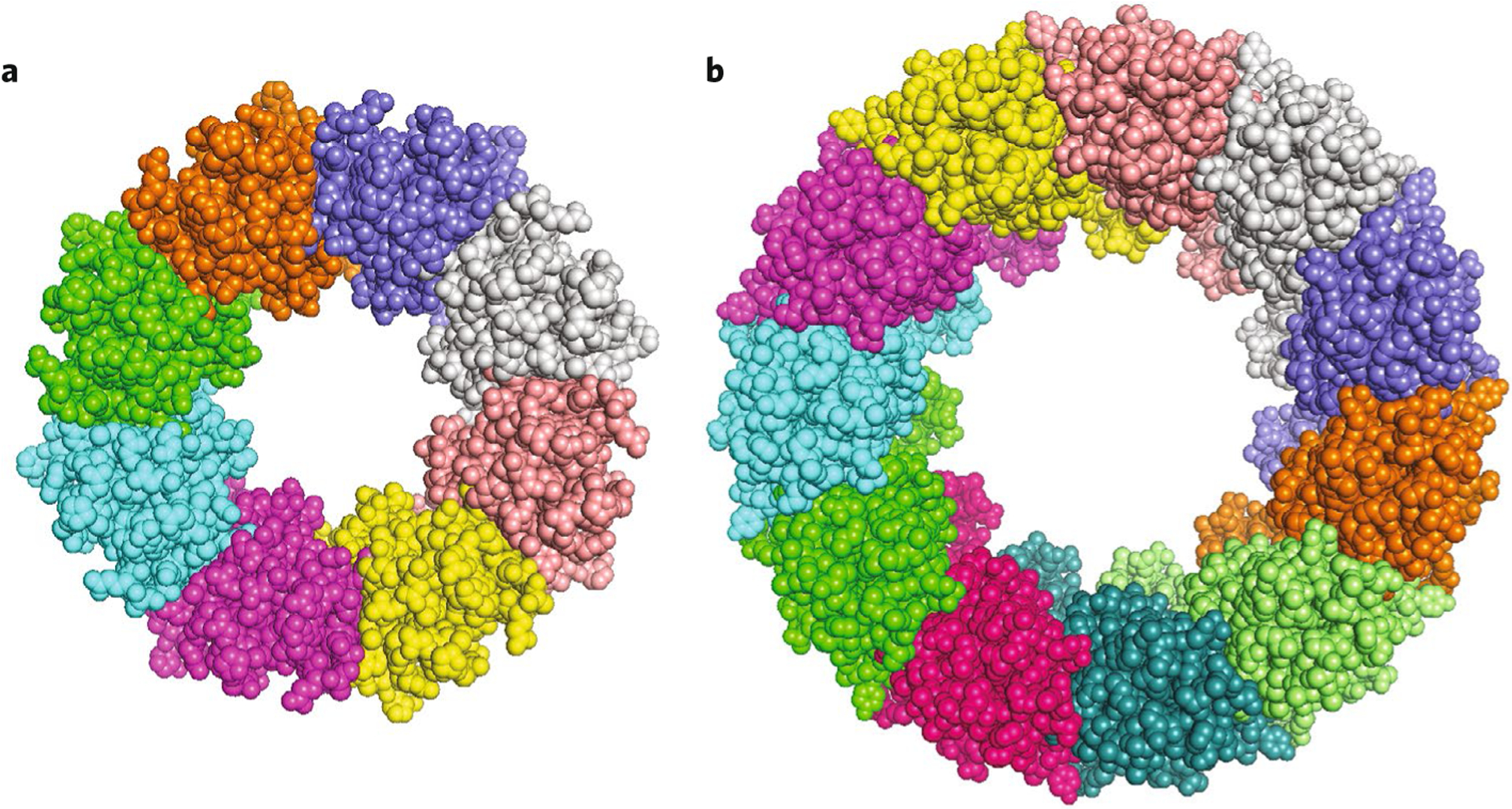
a, Chicken CALHM1(PDB 6VAM). b, human CALHM2 (PDB 6VAK). Both structures are viewed along the pore axis from the extracellular side.
The measured pore diameter of CALHM1 was ~19.5 Å. Considering poorly resolved densities, this is close to the 14 Å diameter previously determined by electrophysiological and dye permeation studies6. In contrast, the pore diameter was larger in CALHM2, ranging from ~50 to 60 Å in both studies1,2. In all the structures, an unresolved density present in the pore, spanning the N-terminal helix and TM1, was attributed to the N terminus, suggesting that these elements are highly flexible. Thus the functional pore diameter in an open channel may be smaller than observed. A structure of CALHM2 bound to ruthenium red (RuR, a known CALHM1 channel inhibitor), reported by Choi et al., revealed that, rather than blocking the pore as expected, RuR occupied the position of TM1, which was displaced (‘lifted’) by nearly 60° towards the pore axis, reducing the pore diameter by 27 Å (Fig. 2). Choi et al. suggest that normal channel gating may involve such a vertical displacement of TM1 to enable the N terminus to physically occlude the pore. Such a displacement in CALHM1, with its smaller pore diameter, might indeed be expected to block the pore. Additional structural and electrophysiological studies will be required to test this model. However, attributing this model of channel gating to CALHM1 requires caution, as the mechanism was proposed on the basis of CALHM2 structures, and CALHM2 may1 or may not2,9 be an ion channel. Syrjanen et al. obtained the CALHM2 structure in lipid nanodiscs. They observed extra density in the pore that they attribute to lipids. Molecular dynamics simulations suggested that it was energetically feasible for the pore to be occupied by a lipid bilayer, which, if true, would prevent ion translocation. Thus the role of CALHM2 as an ion channel remains to be determined.
Fig. 2 |. Displacment of TM1 by RuR in CALHM2.
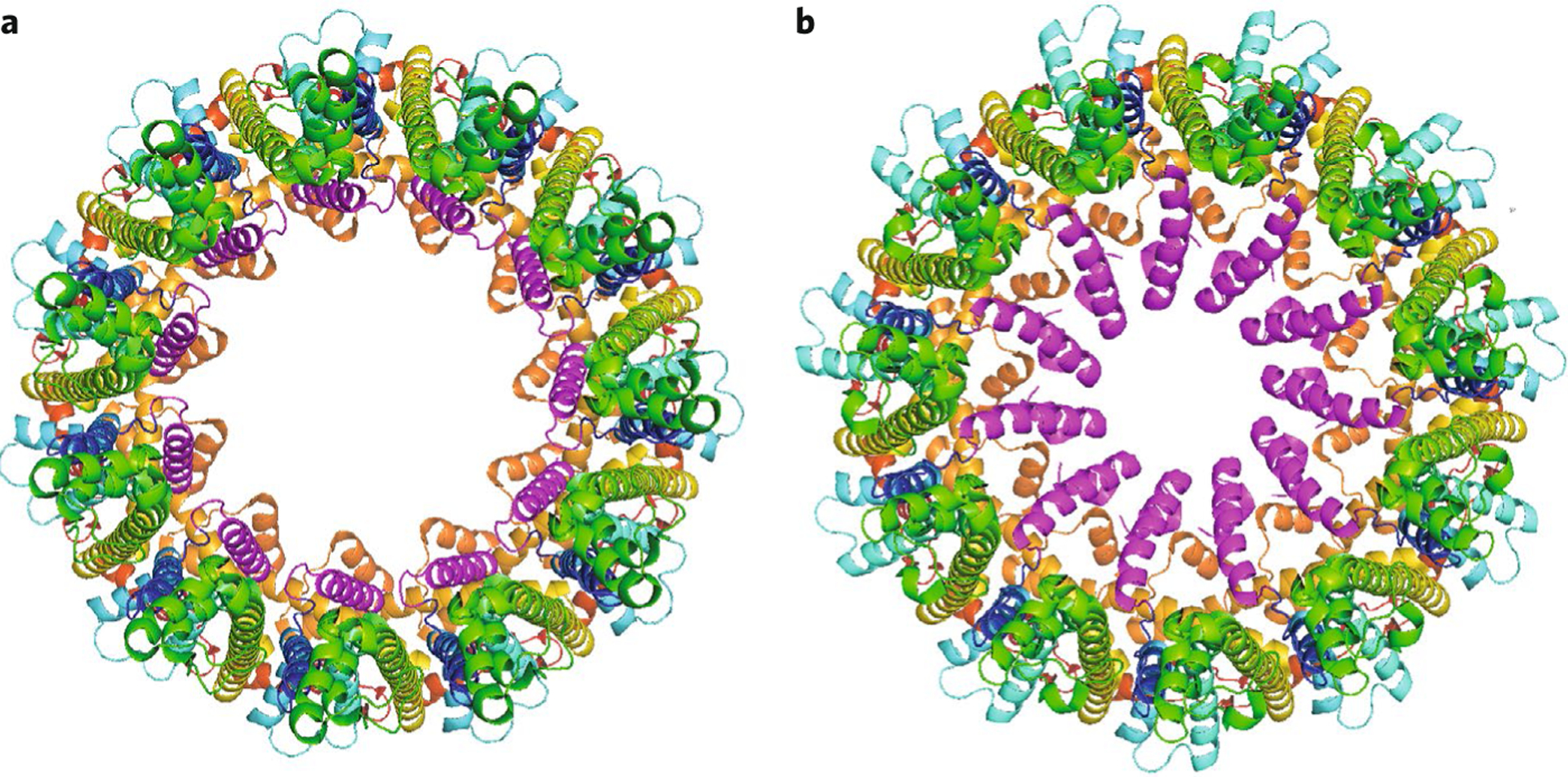
CALHM2 in the absence (PDB 6VAK) (a) and presence (PDB 6UIW) (b) of bound RuR (RuR molecule not shown). TM1 helix is shown in magenta in the vertical (a) and lifted (b) states. Structures are viewed along the pore axis from the extracellular side.
Both groups observed CALHM2 hemichannels, but also gap-junction complexes, in which the extracellular regions interact to form a head-to-head 22-mer pore. It was previously determined that CALHM1 did not form functional gap junctions, possibly because it is glycosylated6. In contrast, Choi et al. found CALHM2 to be unglycosylated. In the CALHM2 gap-junction structure captured by Choi et al., TM1 was in the ‘lifted’ state, even in the absence of RuR. In contrast, the analogous structure captured by Syrjanen et al. shows TM1 in the vertical conformation, similar to that observed in the hemichannel, even though the structure was determined in the presence of Ca2+, which might be expected to promote a closed-channel conformation. On the basis of the similarity of TM1 in their hemichannel and gap-junction structures, Syrjanen et al. suggest that Ca2+ may regulate CALHM channel activity without inducing large structural rearrangements. However, there are several caveats regarding this conclusion, such as the open conformation and the lack of visible Ca2+ ions in the gap-junction structure, and observations in CALHM2 may not translate into mechanisms that operate in the bona fide ion channels of the CALHM family.
Whereas these studies provide important structural insights into the architectures of CALHM family proteins, many questions remain unanswered. The kinetics of voltage activation of CALHM1/3 channels are reminiscent of those in bona fide voltagegated ion channels9. However, unlike those channels, CALHM channels lack an obvious voltage-sensing domain in these structures. Furthermore, it has remained unclear how extracellular Ca2+ interacts with the channel to cooperate with plasma membrane voltage to gate the channel. The structures reported by Syrjanen et al. and Choi et al. do not provide obvious insights. The Caenorhabditis elegans homolog CLHM-1, which has only 16% sequence identity to CALHM1, has permeation, gating properties and regulation that are similar to hCALHM110, which suggests that the CALHM gene family represents a family of ion channels. But the distinct architecture of CALHM2, with the possibility that its pore might be lipid occupied or that it could form gap junctions, now suggests other possibilities. These and other questions remain to be answered by future studies.
Footnotes
Competing interests
The author declares no competing interests.
References
- 1.Choi W, Clemente N, Sun W, Du J & Lu W Nature 576, 163–167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Syrjanen JL et al. Nat. Struct. Mol. Biol 27, 150–159 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dreses-Werringloer U et al. Cell 133, 1149–1161 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma Z, Tanis JE, Taruno A & Foskett JK Pflugers Arch. 468, 395–403 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Z et al. Proc. Natl Acad. Sci. USA 109, E1963–E1971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siebert AP et al. J. Biol. Chem 288, 6140–6153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taruno A et al. Nature 495, 223–226 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taruno A, Matsumoto I, Ma Z, Marambaud P & Foskett JK Bioessays 35, 1111–1118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Z et al. Neuron 98, 547–561.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanis JE et al. J. Neurosci 33, 12275–12286 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]


