Abstract
Rationale
Alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV) is linked to heterozygous mutations in the FOXF1 (Forkhead Box F1) gene, a key transcriptional regulator of pulmonary vascular development. There are no effective treatments for ACDMPV other than lung transplant, and new pharmacological agents activating FOXF1 signaling are urgently needed.
Objectives
Identify-small molecule compounds that stimulate FOXF1 signaling.
Methods
We used mass spectrometry, immunoprecipitation, and the in vitro ubiquitination assay to identify TanFe (transcellular activator of nuclear FOXF1 expression), a small-molecule compound from the nitrile group, which stabilizes the FOXF1 protein in the cell. The efficacy of TanFe was tested in mouse models of ACDMPV and acute lung injury and in human vascular organoids derived from induced pluripotent stem cells of a patient with ACDMPV.
Measurements and Main Results
We identified HECTD1 as an E3 ubiquitin ligase involved in ubiquitination and degradation of the FOXF1 protein. The TanFe compound disrupted FOXF1–HECTD1 protein–protein interactions and decreased ubiquitination of the FOXF1 protein in pulmonary endothelial cells in vitro. TanFe increased protein concentrations of FOXF1 and its target genes Flk1, Flt1, and Cdh5 in LPS-injured mouse lungs, decreasing endothelial permeability and inhibiting lung inflammation. Treatment of pregnant mice with TanFe increased FOXF1 protein concentrations in lungs of Foxf1+/− embryos, stimulated neonatal lung angiogenesis, and completely prevented the mortality of Foxf1+/− mice after birth. TanFe increased angiogenesis in human vascular organoids derived from induced pluripotent stem cells of a patient with ACDMPV with FOXF1 deletion.
Conclusions
TanFe is a novel activator of FOXF1, providing a new therapeutic candidate for treatment of ACDMPV and other neonatal pulmonary vascular diseases.
Keywords: alveolar capillary dysplasia, neonatal pulmonary disease, FOXF1, pulmonary endothelium, pulmonary angiogenesis
At a Glance Commentary
Scientific Knowledge on the Subject
Alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV) is linked to heterozygous mutations in the FOXF1 (Forkhead Box F1) gene, a key transcriptional regulator of pulmonary vascular development. There are no effective treatments for ACDMPV other than lung transplant, and new pharmacological agents activating FOXF1 signaling are urgently needed. Although pharmacological activation of FOXF1 has been suggested to be of interest for future ACDMPV therapies, general difficulties in targeting transcription factors prevented commercial development of FOXF1-activating small-molecule compounds for clinical use.
What This Study Adds to the Field
We identified a novel small-molecule compound, TanFe (transcellular activator of nuclear FOXF1 expression), which specifically increases FOXF1 protein concentrations in pulmonary endothelial cells in vitro and in vivo by interfering with FOXF1 degradation through disruption of protein–protein interactions between FOXF1 and HECTD1 E3 ubiquitin ligase. Treatment of pregnant mice with TanFe stimulated lung angiogenesis in Foxf1+/− embryos and completely prevented the mortality of Foxf1+/− mice after birth. TanFe increased angiogenesis in FOXF1-deficient pulmonary endothelial cells in vitro and in human vascular organoids derived from induced pluripotent stem cells of a patient with ACDMPV with FOXF1 haploinsufficiency. Our studies support the feasibility of developing novel FOXF1-activating small-molecule compounds, such as TanFe, for treatment of patients with ACDMPV and other pulmonary disorders associated with pulmonary vascular disease.
Alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV) is a severe congenital disorder that causes respiratory failure and requires lung transplantation early in life (1–3). Lack of prenatal genetic diagnosis, early onset of ACDMPV, and rapidly progressing respiratory insufficiency greatly complicate clinical care for newborns and infants with ACDMPV, often leading to mortality before lung transplantation becomes an option (1). There is an unmet need for new therapeutic approaches for ACDMPV, which is associated with pulmonary hypertension, reduced alveolar capillary density, lung hypoplasia, and misaligned pulmonary veins (4). A subset of ACDMPV cases is characterized by late onset of the disease and a mosaic pattern of vascular abnormalities in the lung tissue (5, 6). Patients with delayed presentation of ACDMPV are primary candidates for lung transplantation. ACDMPV has been recently linked to mutations in the FOXF1 (Forkhead Box F1) gene locus, which include heterozygous macrodeletions, microdeletions, and point mutations (3, 4). Although recent studies showed the efficacy of nanoparticle-mediated gene therapy (7, 8) and cell therapy with endothelial progenitor cells in mouse ACDMPV models (9, 10), potential use of these therapies in human ACDMPV is unclear.
FOXF1 is a transcription factor that is expressed in pulmonary endothelial cells (ECs), fibroblasts, and their mesenchymal progenitors (2, 11–13). Published studies with mouse genetic models demonstrate that FOXF1 is essential for lung embryonic development (14, 15), lung regeneration after partial pneumonectomy (16), and microvascular repair after acute lung injury (17–19). FOXF1 promotes embryonic and postnatal development of alveolar capillaries by regulating genes critical for Vascular Endothelial Growth Factor (VEGF), BMP9, and PDGF signaling pathways (2, 20–22). Foxf1−/− mice exhibit the embryonic lethal phenotype before the initiation of lung development because of the lack of vasculature in the yolk sac (23). Foxf1 haploinsufficiency (Foxf1+/−) or heterozygous loss-of-function S52F Foxf1 mutation in mice (Foxf1WT/S52F) cause alveolar capillary dysplasia and high mortality rates (3, 24), recapitulating human ACDMPV (2). Although pharmacological activation of FOXF1 has been suggested to be of interest for future ACDMPV therapies (25, 26), general difficulties in targeting transcription factors prevented commercial development of FOXF1-activating small molecule compounds. If available, FOXF1 activators may provide new therapeutic opportunities for treatment of patients with ACDMPV and other pulmonary disorders associated with diminished FOXF1 signaling, such as bronchopulmonary dysplasia (9), acute lung injury (17), and pulmonary arterial hypertension (7).
Herein, we have challenged the concept that transcription factors are “undruggable” therapeutic targets by identifying the small-molecule compound TanFe (transcellular activator of nuclear FOXF1 expression), which stabilizes the FOXF1 protein by inhibiting its degradation. TanFe improves angiogenesis in Foxf1+/− mice and vascular organoids derived from a patient with ACDMPV. Our study supports the feasibility of developing novel FOXF1-activating small-molecule compounds for treatment of ACDMPV.
Materials and Methods
Ethics Statement
Study materials, analytic methods, and experimental data from this manuscript will be made available upon request to the corresponding author for purposes of reproducing the results or replicating the procedures. All animal studies were approved by the Institutional Animal Care and Use Committee of the Cincinnati Children’s Hospital. Consent for use of human tissues was overseen by the Washington University Human Research Protection Office, Boston University Institutional Review Board, and the Cincinnati Children’s Hospital Medical Center Institutional Review Board.
Mouse Strains and Treatment with TanFe
The generation of Foxf1+/− mice has been previously described (27). Eight- to 10-week-old C57Bl/6 mice were purchased from Jackson Labs. The high-throughput screen for FOX-domain–interacting small-molecule compounds was performed using the chemical library of the Genome Research Institute of the University of Cincinnati (28) as described in the online supplement. The twelve best FOXF1-activating small-molecule compounds were further screened by Western blot, identifying the TanFe compound (4- (4- chlorophenyl)- 2- {[2- oxo- 2- (thiophen- 2- yl)ethyl]sulfanyl}- 6- (thiophen- 2- yl)pyridine- 3- carbonitrile; empirical formula, C22H13ClN2OS3; molecular weight, 453.01). For these studies, TanFe was synthesized by TimTec, LLC (catalog number ST4005046) with 95% purity as determined by HPLC. LPS-mediated lung injury, TanFe treatments, and measurements of endothelial permeability in adult mice are described in the online supplement. For the embryonic studies, Foxf1+/− males were bred with wild-type (WT) females, and TanFe was administered intraperitoneally (10 mg/kg of body weight) to pregnant mice on Gestational Days E11.5, E13.5, and E15.5.
Immunoprecipitation and Mass Spectrometry
Immunoprecipitation of FOXF1 was performed from fetal mouse lung endothelial Mouse Fetal Lung Mesenchyme-91 U cells expressing His-Flag–tagged mouse FOXF1 (HF-FOXF1) (29) that were incubated with 20 μM of the compound TanFe or vehicle (Dimethyl Sulfoxide). Purification of proteins interacting with FOXF1, mass spectrometry, and mass spectrometry analysis were performed as described previously (29, 30) and in the online supplement.
Western Blotting and the Ubiquitination Assay
Western blot analysis was performed as described previously (20, 31, 32) using protein extract from lung tissue, MFLM-91 U, and Human Embryonic Kidney (HEK) 293T cells. The antibodies are provided in Table E1 in the online supplement. Chromatin purification was performed as described previously (3). The in vitro ubiquitination assay was performed using the ubiquitination kit from Enzo Life Sciences (catalog number BML-UW9920) according to the manufacturer’s recommendations.
Immunostaining
Lungs were fixed, embedded into paraffin blocks, sectioned, and stained with hematoxylin and eosin or used for immunohistochemistry as described previously (12, 33, 34). Information about primary antibodies is provided in Table E1 and References 35–37. Antibody–antigen complexes were detected using biotinylated secondary antibody, avidin-biotin-horseradish peroxidase complex, and 3,3′-diaminobenzidine substrate (all from Vector Lab) (13, 38, 39). Sections were counterstained with nuclear fast red. Immunofluorescent staining was performed as described previously (40–42).
RNA Preparation, Quantitative RT-PCR, Cell Transfection, and Dual Luciferase Assay
RNeasy micro plus kit (Qiagen) was used to prepare total RNA. Quantitative real-time RT-PCR was performed using a StepOnePlus Real-Time PCR system (Thermo Fisher) as described (43, 44). Samples were amplified using inventoried TaqMan primers for mouse and human FOXF1 mRNA (45). Reactions were analyzed in triplicate, and expression levels were normalized to β-Actin mRNA. Expression plasmids containing HA-tagged Hectd1 cDNA (HA-HECTD1) full length and HA-HECTD1 C2579G mutant were gifted by Irene Zohn, Children’s National Medical Center (46). Transient transfection is described in the online supplement. Dual luciferase (LUC) assay was performed using the standard protocol from Promega as described previously (12, 47). FOXF1-LUC reporter plasmid was previously described (20, 48).
Matrigel Angiogenesis Assay, Generation of Human Induced Pluripotent Stem Cells and Vascular Organoids
Human endothelial Human Umbilical Vein Endothelial Cells (HUVECs) were used for the Matrigel angiogenesis assay as described previously (21). Human induced pluripotent stem cells (iPSCs) were generated by reprogramming dermal fibroblasts obtained from a patient with ACDMPV with genomic loss of one FOXF1 allele as described in the online supplement. Clinical information about the patient with ACDMPV is provided in the online supplement. iPSCs were differentiated into vascular organoids using a previously described protocol (49) with several modifications (online supplement). Vascular organoids were fixed, cryopreserved, sectioned, and stained as described in the online supplement.
Statistical Analysis
One-way ANOVA and Student’s t test were used to determine statistical significance. P values < 0.05 were considered statistically significant. Multiple means were compared using one-way ANOVA with the post hoc Tukey’s test. For datasets with n < 6, a nonparametric Mann-Whitney U test was used to determine statistical significance. Values are presented as mean ± SD.
Results
Identification of Novel FOXF1-Activating Small-Molecule Compound TanFe
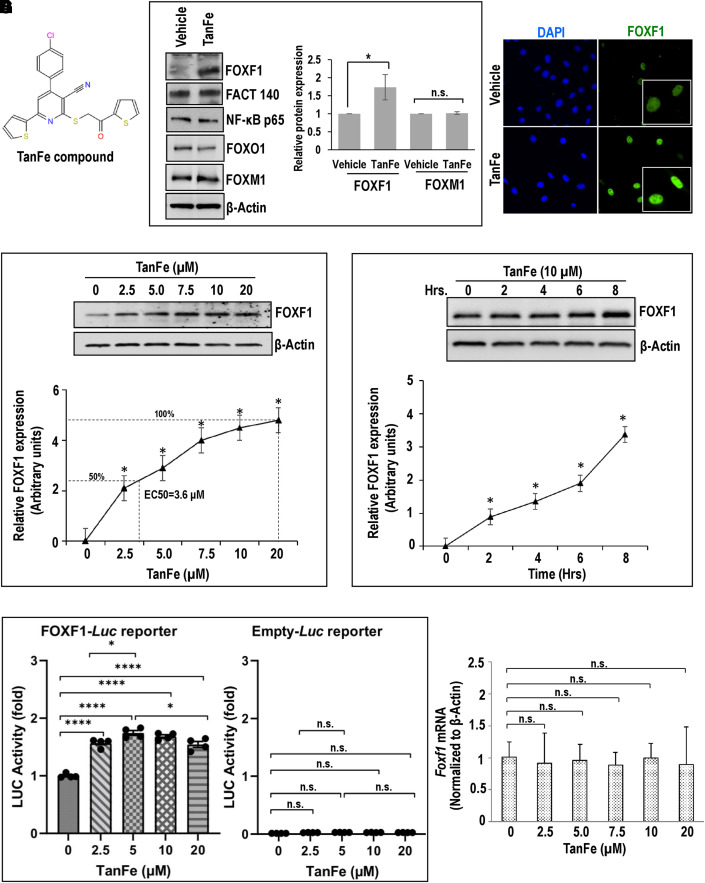
We used small-molecule compounds from the chemical library of the Genome Research Institute, University of Cincinnati (28) for Western blot screening to identify the nitrile compound TanFe (Figure 1A), which increases FOXF1 protein concentrations in mouse embryonic lung endothelial MFLM-91 U cells without affecting other transcription factors, such as FACT 140, NF-κB p65 (nuclear factor-κB p65), FOXO1, and FOXM1 (Figure 1B). After TanFe treatment, increased FOXF1 immunoreactivity was localized to cell nuclei (Figure 1C). TanFe increased the abundance of FOXF1 protein in a dose-dependent manner, with Half maximal effective concentration (EC50) = 3.6 μM as determined by Western blot (Figure 1D). The TanFe effect was detected after 2 hours in cell culture and gradually increased in the 8-hour treatment period (Figure 1E). Consistent with increased FOXF1 protein amounts in TanFe-treated ECs, the FOXF1 transcriptional activity was increased as demonstrated by transient transfection experiments with FOXF1-specific LUC reporter plasmid (Figure 1F). TanFe did not change Foxf1 mRNA in lung ECs (Figure 1G). Thus, TanFe increases FOXF1 protein amounts without altering Foxf1 mRNA.
Figure 1.
The TanFe (transcellular activator of nuclear FOXF1 expression) small-molecule compound increases FOXF1 (Forkhead Box F1) protein amounts in vitro. (A) Chemical structure of the TanFe compound. (B) Western blots show amounts of endogenous FOXF1 protein and other transcription factors in TanFe-treated fetal lung endothelial Mouse Fetal Lung Mesenchyme-91 U (MFLM-91U) cells. Cells were treated with either vehicle alone (control) or 20 μM of TanFe and harvested 24 hours after the treatment. Protein samples were pooled from five independent cell cultures (left panels). Three independent Western blots were quantified and normalized to β-Actin (right graph). (C) TanFe increases nuclear FOXF1 fluorescence. MFLM-91 U endothelial cells were fixed and stained for FOXF1 (green). DAPI was used to visualize cell nuclei (blue). Inserts show high-magnification images of FOXF1-stained cells. (D and E) Western blots show the dose response and the time course of TanFe treatment in MFLM-91 U cells (n = 3 independent experiments). Half maximal effective concentration (EC50) for TanFe is 3.6 μM (dotted lines). (F) Luciferase (Luc) assay shows increased FOXF1 transcriptional activity in TanFe-treated MFLM-91 U cells. Cells were transfected with either FOXF1-specific LUC reporter plasmid (FOXF1-LUC) or Empty-LUC plasmid (control) and treated with TanFe for 24 hours (n = 5 for each group). (G) TanFe treatment does not change Foxf1 mRNA. Quantitative RT-PCR was used to measure Foxf1 mRNA in MFLM-91 U cells after treatment with different concentrations of TanFe (n = 3). Expression levels were normalized to β-Actin (Actb mRNA). *P < 0.05 and ****P < 0.0001. FACT 140 = facilitates chromatin transcription complex subunit SPT16 -140 kDa; NF-κB = nuclear factor-κB; n.s. = not significant.
TanFe Increases FOXF1 Protein Concentrations In Vivo
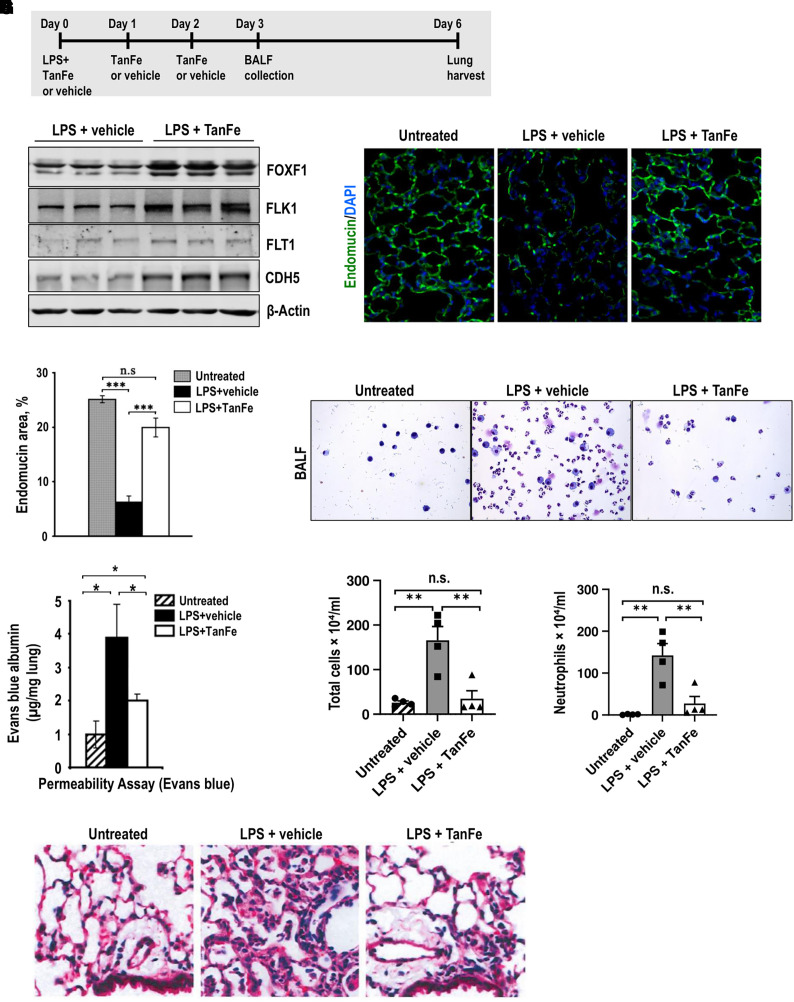
The ability of TanFe to increase FOXF1 in the lung tissue in vivo was tested using an LPS-mediated lung injury model. To induce lung injury, LPS was given as a single intratracheal administration to WT C57Bl/6 mice (Figure 2A). The mice were treated on Days 0, 1, and 2 with either TanFe or vehicle (control) through intraperitoneal injections (Figure 2A). Six days after LPS injury, the mice were harvested. The Western blot of lung homogenates demonstrated that TanFe treatment increased FOXF1 protein amounts in LPS-injured lungs compared with vehicle-treated control mice (Figure 2B). Elevated FOXF1 concentrations were associated with increased expression of FLK1 and FLT1 (Figure 2B), both of which are known transcriptional targets for FOXF1 (21). Thus, TanFe increases protein concentrations of FOXF1 in LPS-injured lungs.
Figure 2.
TanFe (transcellular activator of nuclear FOXF1 expression) increases FOXF1 (Forkhead Box F1) protein amounts in vivo. (A) Schematic shows treatment of C57Bl/6 mice with LPS and TanFe. LPS was administered intratracheally on Day 0. TanFe or vehicle was injected intraperitoneally on Days 0, 1, and 2 (5 mg/kg body weight). (B) TanFe increases FOXF1 protein amounts in LPS-injured mouse lungs. Total lung protein was prepared 6 days after LPS injury and analyzed by Western blot for FOXF1, FLK1, FLT1, CDH5 (VE cadherin), and β-Actin. Samples were obtained from individual mice (n = 3 mice per group). (C and D) Immunostaining for endomucin shows that TanFe increases the capillary density in alveolar regions of LPS-treated mice. Mice were harvested on Day 6 after LPS injury and lung paraffin sections were stained for endomucin (green). Slides were counterstained with DAPI (blue). Ten random slides were used to quantitate the staining (n = 3 mice per group). Magnification is ×400. (E) Endothelial permeability assay shows that TanFe protects endothelial barrier function after LPS lung injury. Evans blue dye was injected intraperitoneally, and lung tissue was collected 4 hours after injection (n = 5 mice in each group). Before the lung harvest, vasculature was perfused with saline to remove intravascular Evans blue dye. (F–H) TanFe inhibits accumulation of inflammatory cells in BAL fluid (BALF) of LPS-injured mice. BALF was collected on Day 3 after LPS lung injury and analyzed for total number of cells and for neutrophil counts (n = 5 mice in each group). (I) Hematoxylin and eosin staining shows that TanFe decreases lung inflammation in the alveolar region of LPS-injured mice. Mice were harvested on Day 6 after LPS injury (n = 3 mice per group). TanFe or vehicle was given on Days 0, 1, and 2. Magnification is ×400. *P < 0.05, **P < 0.01, and ***P < 0.001. n.s. = not significant.
Because increasing FOXF1 expression in transgenic mice improved endothelial barrier function and decreased lung inflammation after injury (17), we tested if TanFe has similar effects in LPS-injured mice. After LPS injury, TanFe improved endothelial coverage in alveoli as shown by immunostaining for endomucin (Figures 2C and 2D) and increased protein concentrations of vascular endothelial cadherin (VE cadherin or, CDH5) (Figure 2B), a critical structural component of EC adherens junctions (17). Consistent with increased CDH5 expression in total lung lysates, endothelial permeability in LPS-injured mice decreased after TanFe treatment (Figure 2E). Compared with vehicle-treated control mice, TanFe reduced the total number of inflammatory cells in BAL fluid (BALF) of LPS-injured mice (Figures 2F and 2G). The number of neutrophils in BALF was decreased (Figure 2H). Histological evaluation of hematoxylin and eosin–stained lung sections showed that TanFe decreased lung inflammation and improved the lung injury score after LPS injury (Figures 2I and E1A–E1C). Thus, TanFe increases FOXF1 protein concentrations in LPS-injured lungs, improving microvascular integrity and decreasing lung inflammation.
TanFe Disrupts Protein–Protein Interactions of FOXF1 with HECTD1 E3 Ubiquitin Ligase
Because TanFe increases FOXF1 protein concentrations without altering Foxf1 mRNA, we tested if TanFe regulates FOXF1 protein stability by inhibiting its degradation. Therefore, we focused on FOXF1 binding to other proteins, prioritizing the interactions that are affected by TanFe. Endothelial MFLM-91 U cells stably expressing HF-FOXF1 (29) were treated with either TanFe or vehicle. Next, we used a two-step affinity purification coupled with mass spectrometry to identify proteins physically bound to FOXF1 (Figure E2A). HF-FOXF1 was found to bind numerous proteins involved in protein ubiquitination and degradation (Table E2). TanFe treatment disrupted the binding of FOXF1 with E3 ligases HECTD1, SHPRH, UNKL isoform 1, and NEURL1B; ubiquitin carboxyl-terminal hydrolases 31, 35, and 2; and the ubiquitin thioesterase Zrab1 but did not influence FOXF1 interactions with HERC2, RBBP6, UBE3A (Ubiquitin protein ligase E3A), Arkadia, RNF181, and Mdm2 (Table E2). Protein–protein interactions of FOXF1 with RNF213, Midline-1, and UNKL isoform X2 were increased after TanFe treatment (Table E2).
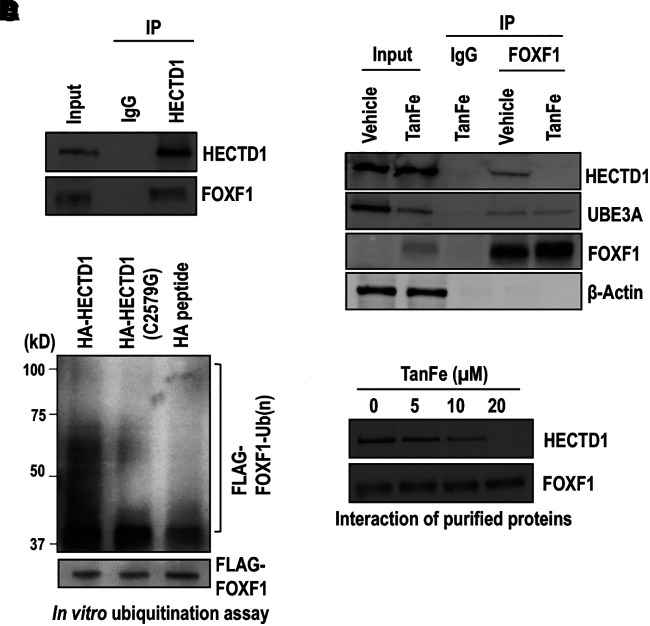
Because the HECTD1-FOXF1 binding was the strongest among protein–protein interactions disrupted by TanFe (Table E2), we prioritized the HECTD1 E3 ligase for our biochemical studies. In vehicle-treated (control) cells, seven HECTD1 unique peptides were identified in the FOXF1 immunoprecipitate (IP) (Table E3 and Figure E2B). In contrast, the HECTD1 peptides were undetectable in FOXF1 IP from TanFe-treated cells (Table E2). To validate the mass spectrometry findings, we used the HECTD1 antibody to immunoprecipitate the endogenous HECTD1 protein from EC lysates (Figure 3A). FOXF1 was detected in HECTD1 IP but not in IgG control IP (Figure 3A). The FOXF1–HECTD1 protein–protein interactions were also confirmed through reciprocal IP experiments, in which FOXF1 was immunoprecipitated from the cell lysates followed by detection of the HECTD1 protein by Western blot (Figure 3B). The TanFe compound completely abolished FOXF1–HECTD1 interactions (Figure 3B). Although TanFe decreased protein amounts of UBE3A in the input, TanFe had no effect on interactions between FOXF1 and UBE3A (Figure 3B), findings consistent with mass spectrometry data (Table E2). Thus, FOXF1 physically binds to HECTD1 E3 ubiquitin ligase, and this binding is disrupted by the TanFe compound.
Figure 3.
TanFe (transcellular activator of nuclear FOXF1 expression) disrupts FOXF1 (Forkhead Box F1)–HECTD1 protein–protein interactions. (A) Immunoprecipitation (IP) shows that HECTD1 interacts with endogenous FOXF1 protein. IP was performed with cell lysates from Mouse Fetal Lung Mesenchyme-91 U (MFLM-91U) cells using the HECTD1 antibody or IgG control. Protein samples were pooled from five independent cell cultures. (B) IP shows that FOXF1 interacts with HECTD1 protein. IP was performed with cell lysates from MFLM-91 U cells stably expressing His-Flag–tagged mouse FOXF1. Cells were treated with 20 μM of TanFe or vehicle (Dimethyl sulfoxide) for 24 hours. FOXF1-interacting proteins were purified using anti-Flag M2 agarose beads and Talon metal affinity resin and were analyzed by Western blot. (C) The in vitro ubiquitination assay shows that HECTD1 ubiquitinates the FOXF1 protein. HA-HECTD1 or HA-HECTD1 C2579G mutants were expressed in Human Embryonic Kidney (HEK) 293T cells, purified using HA magnetic beads, and eluted using HA peptide. The ubiquitination assay was performed in the presence of FLAG-FOXF1 immobilized magnetic beads and the purified HA-HECTD1 proteins. Binding of biotinylated Ub to FLAG-FOXF1 protein was detected by Western blot using Horseradish peroxidase (HRP) streptavidin. (D) Western blot shows that increasing TanFe concentrations interfere with binding of purified HECTD1 and FOXF1 proteins. Endogenous HECTD1 was immunoprecipitated from MFLM-91 U cells and incubated with the purified FLAG-FOXF1 protein immobilized on agarose beads. Protein samples were pooled from three cell cultures. Ub = ubiquitin; UBE3A = Ubiquitin protein ligase E3A.
TanFe Decreases FOXF1 Ubiquitination by HECTD1 E3 Ubiquitin Ligase
To test whether HECTD1 directly ubiquitinates the FOXF1 protein, we transfected mouse Human influenza hemagglutinin (HA)-tagged Hectd1 cDNA (HA-HECTD1) into HEK 293T cells and purified HA-HECTD1 protein using IP. The in vitro ubiquitination assay showed that the purified HA-HECTD1 E3 ligase directly ubiquitinated the purified FLAG-FOXF1 protein (Figures 3C, E3A, and E3B). Neither HA peptide nor C2579G HECTD1 mutant (containing inactivating mutation in the HECTD1 catalytic domain) was capable of ubiquitinating the FOXF1 protein (Figure 3C), indicating that the HECTD1 catalytic domain is required for FOXF1 ubiquitination. Furthermore, the TanFe compound disrupted the binding between purified HECTD1 and FOXF1 proteins in a dose-dependent manner (Figure 3D). Thus, TanFe prevents FOXF1 ubiquitination by directly interfering with FOXF1–HECTD1 protein–protein interactions.
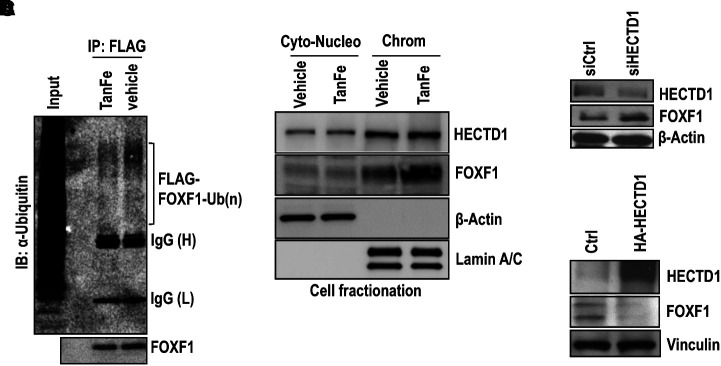
TanFe treatment decreased FOXF1 ubiquitination in EC lysates (Figure 4A) and increased amounts of active, chromatin-bound FOXF1 protein (Figure 4B), findings consistent with increased FOXF1 transcriptional activity in TanFe-treated ECs (Figure 1F). TanFe did not change HECTD1 protein amounts (Figure 4B). Consistent with HECTD1 requirements for FOXF1 degradation, siRNA-mediated inhibition of HECTD1 increased FOXF1 protein amounts (Figure 4C), whereas overexpression of HECTD1 decreased FOXF1 amounts (Figure 4D). Altogether, the TanFe compound increases FOXF1 protein concentrations by preventing FOXF1 ubiquitination through the HECTD1-dependent mechanism.
Figure 4.
TanFe (transcellular activator of nuclear FOXF1 expression) inhibits FOXF1 (Forkhead Box F1) ubiquitination. (A) TanFe decreases ubiquitination of the FOXF1 protein. IP was performed in protein extracts from Mouse Fetal Lung Mesenchyme-91 U (MFLM-91U) cells stably expressing His-Flag–tagged FOXF1 (HF-FOXF1) using the FLAG antibody followed by Western blot with ubiquitin antibodies. TanFe treatment (20 μM) was performed for 24 hours. Equal protein loading is shown by Western blot for FOXF1 (n = 3 pooled cell cultures). (B) Chromatin fractionation shows that TanFe increases FOXF1 amounts in the chromatin fraction. TanFe does not change HECTD1 amounts in cell fractions. Chromatin fractionation was performed in MFLM-91 U cells treated with 20 μM of TanFe for 24 hours. Cell lysates were pooled from three independent cell cultures. (C) Western blot shows that inhibition of HECTD1 by siRNA (siHECTD1) increases FOXF1 protein amounts in MFLM-91 U cells. siCtrl is scrambled siRNA (n = 3 pooled cell cultures). (D) Overexpression of HECTD1 decreases FOXF1 expression. Cells were transfected with the HA-HECTD1 expression vector, and the Western blot was performed 48 hours after transfection (n = 3 pooled cell cultures). Chrom = chromatin; Ctrl = control; Cyto-Nucleo = cytoplasmic and nuclear extract; IB = Immunoblot; IP = immunoprecipitation.
TanFe Increases Lung Angiogenesis and Prevents Mortality in FOXF1-Deficient Mice
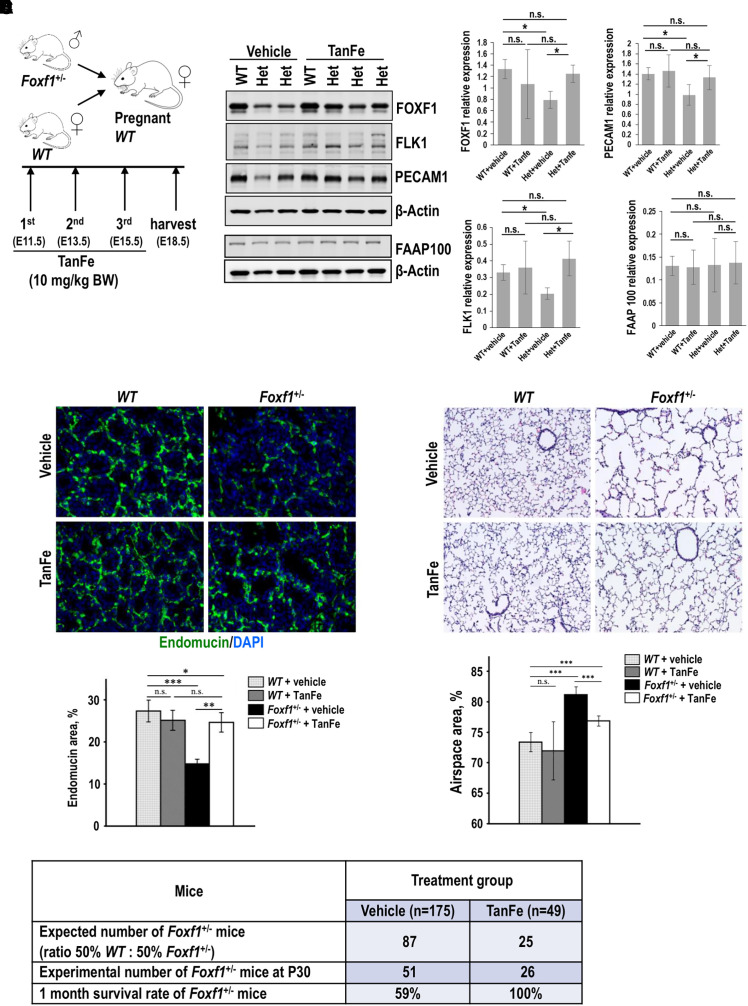
To test whether TanFe increases FOXF1 protein concentrations during embryonic lung development in vivo, we bred WT mice with Foxf1+/− mice, the latter of which exhibit alveolar capillary dysplasia (27) similar to patients with ACDMPV with the loss of one FOXF1 allele (4). Pregnant WT females were intraperitoneally injected with either TanFe or vehicle on Embryonic Days 11.5 (E11.5), E13.5, and E15.5, and embryos were harvested at E18.5 (Figure 5A). Western blot of total lung lysate demonstrated that the TanFe compound increases FOXF1 protein in Foxf1+/− lungs compared with vehicle-treated Foxf1+/− controls (Figure 5B). Consistent with increased expression of FOXF1, TanFe-treated Foxf1+/− lungs exhibited increased expression of endothelial proteins FLK1 and PECAM1, but FAAP100 expression was unchanged (Figure 5B).
Figure 5.
TanFe (transcellular activator of nuclear FOXF1 expression) increases lung angiogenesis and prevents mortality in Foxf1+/− (Forkhead Box F1) mice. (A) Schematic shows a breeding strategy and TanFe treatments of pregnant mice on Embryonic Days E11.5, E13.5, and E15.5 (10 mg/kg body weight). (B) TanFe increases FOXF1 protein amounts in lungs of Foxf1+/− embryos. Immunoblots show the concentrations of FOXF1, FLK1, PECAM-1, FAAP100, and β-Actin in lung extracts from E18.5 embryos treated with either TanFe or vehicle. Protein lysates from three WT lungs were pooled together before the analysis. Foxf1+/− lungs (Het) were analyzed individually (left panels). Quantification of two independent Western blots shows that TanFe does not change FAAP100 protein concentrations but increases FOXF1, FLK1, and PECAM1 in Foxf1+/− embryos collected at E18.5 to P1 (n = 3–6 embryos in each group) (right graphs). (C) TanFe increases capillary density in Foxf1+/− lungs. Images show endomucin staining (green) of E18.5 lungs after TanFe treatment. DAPI (blue) was used to stain cell nuclei. Magnification is ×400. Quantification of endomucin staining was performed using ImageJ software in 10 random images from three mouse lungs in each group. (D) TanFe increases survival of Foxf1+/− mice after birth. Mice were treated with TanFe (10 mg/kg body weight) or vehicle on Embryonic Days E11.5, E13.5, and E15.5. Survival rates of vehicle-treated Foxf1+/− mice (n = 175) and TanFe-treated Foxf1+/− mice (n = 49) were determined at Postnatal Day 30 (P30). (E) TanFe improves alveolarization in Foxf1+/− mice after birth. Hematoxylin and eosin staining shows that TanFe decreases alveolar simplification in Foxf1+/− mice at P35 (n = 7–13 in each group). Magnification is ×100. *P < 0.05, **P < 0.01, and ***P < 0.001. n.s. = not significant; WT = wild-type.
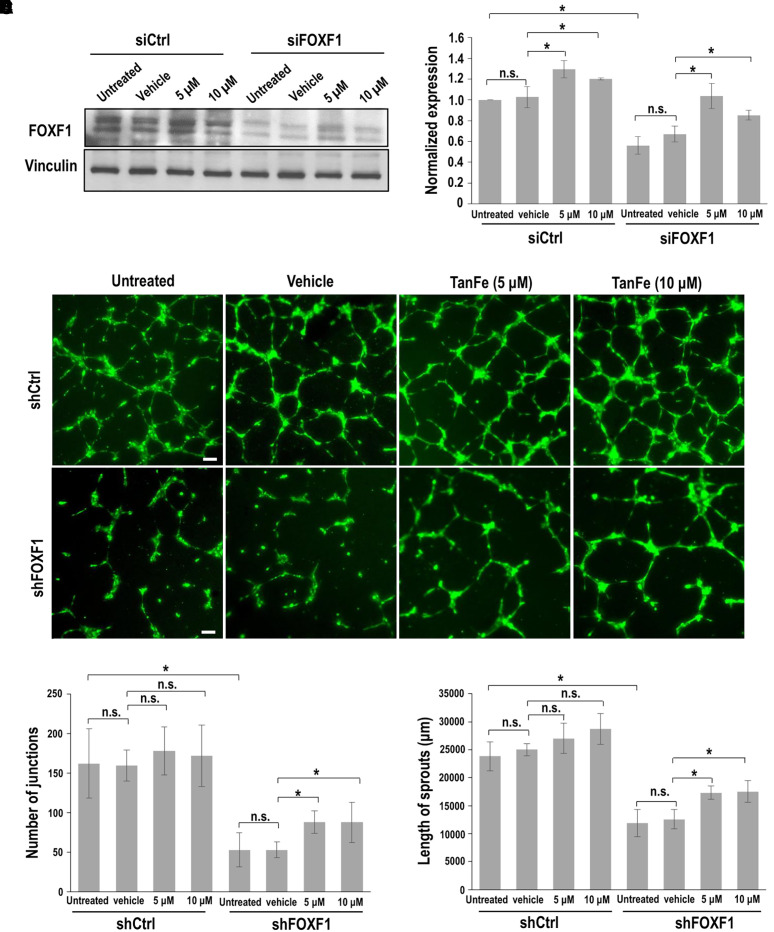
The capillary density was higher in Foxf1+/− mice treated with TanFe than in lungs of vehicle-treated Foxf1+/− embryos (Figures 5C, E4A, and E4B). In colocalization experiments for Ki-67 and endomucin, the number of proliferative ECs was increased in TanFe-treated Foxf1+/− and WT E18.5 embryonic lungs (Figures E5A and E5B), a finding consistent with FOXF1 requirements for endothelial proliferation during lung development and regeneration (16, 21). To model Foxf1 haploinsufficiency in ECs in vitro, we used FOXF1-specific siRNA (siFOXF1) to inhibit approximately 50% of FOXF1 concentrations in lung ECs (Figures 6A and 6B). TanFe increased FOXF1 protein in siFOXF1-transfected ECs (Figures 6A and 6B) and improved angiogenesis in FOXF1-deficient ECs in Matrigel (Figures 6C–6E). TanFe did not influence angiogenesis in control ECs in vitro (Figures 6C–6E) or in WT lungs during embryogenesis (Figure 5C). Thus, TanFe stimulates angiogenesis in FOXF1-deficient ECs in vitro and in vivo.
Figure 6.
TanFe (transcellular activator of nuclear FOXF1 expression) increases angiogenesis in FOXF1 (Forkhead Box F1)-deficient endothelial cells in vitro. (A) Western blot shows that TanFe treatment increases FOXF1 protein concentrations but does not affect Vinculin in FOXF1-deficient endothelial Mouse Fetal Lung Mesenchyme-91 U (MFLM-91U) cells. Inhibition of FOXF1 was achieved by transient transfection with FOXF1-specific siRNA (siFOXF1). siRNA against a nontargeted RNA sequence was used as a control (siCtrl). (B) Quantification of FOXF1 protein concentrations was performed using densitometric analysis of Western blot images and normalized to Vinculin (n = 3). (C–E) In vitro angiogenesis assay shows that TanFe increases the formation of endothelial sprouts in endothelial Human umbilical vein endothelial cells (HUVECs) infected with shFOXF1 lentivirus. Lentiviral particles with shRNA against a nontargeted RNA sequence were used as a control (shCtrl). Scale bars, 100 μm. The complexity of the vascular network in Matrigel was quantitated by measurements of sprout length and counts of the sprout junctions (n = 4 for each group). *P < 0.05. n.s. = not significant.
To test whether TanFe treatment during embryogenesis influences the mortality of Foxf1+/− mice after birth, the survival rates in Foxf1+/− mice were assessed between postnatal day 1 (P1) and P30. Although the 1-month survival rate in vehicle-treated Foxf1+/− mice was 59% (n = 175), the survival of TanFe-treated Foxf1+/− mice was increased to 100% (n = 49) (Figure 5D). Consistent with increased survival of TanFe-treated Foxf1+/− mice, TanFe improved alveolarization and increased the capillary density in Foxf1+/− lung tissues at P35 (Figures 5E and E6). Altogether, TanFe increases EC angiogenesis, improves alveolarization, increases the alveolar capillary density, and prevents mortality in Foxf1+/− mice.
TanFe Increases Angiogenesis in Human Vascular Organoids Formed by iPSCs Derived from a Patient with ACDMPV
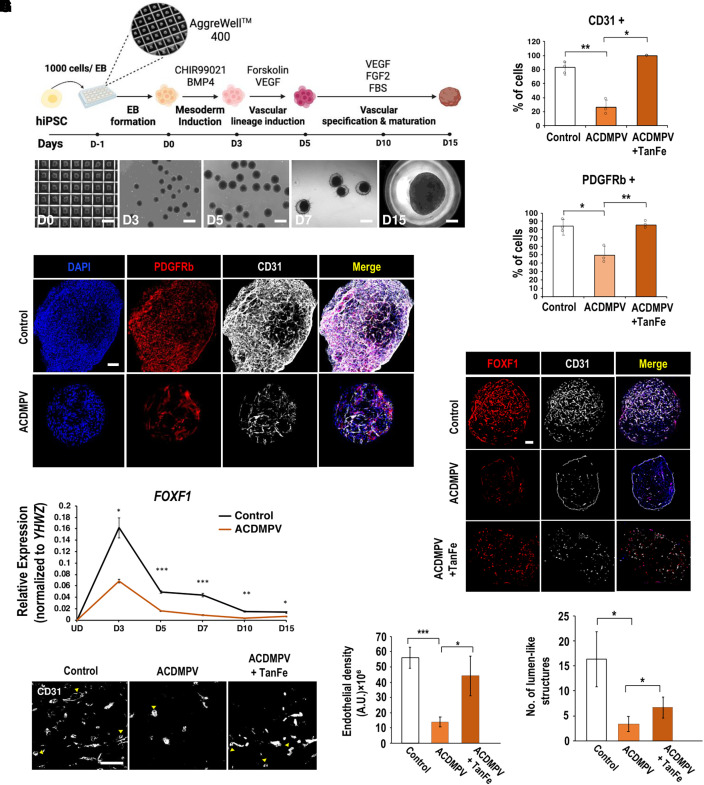
To determine whether TanFe is capable of increasing angiogenesis in human ACDMPV organoids, we produced iPSCs by reprogramming dermal fibroblasts of a patient with ACDMPV with FOXF1 haploinsufficiency. Clinical and genetic information of the patient with ACDMPV as well as reprogramming methods are provided in the online supplement. The resulting iPSC line (clone FOXF1.1, hereafter “ACDMPV iPSC”) exhibited normal karyotype (Figure E7), and its pluripotency was confirmed by staining for pluripotency markers (Figure E8). Vascular organoids were generated from either normal control or ACDMPV iPSCs using a previously established protocol (49). To generate mesodermal vascular progenitors, iPSCs were treated with GSK-3 inhibitor CHIR99021 and BMP4 (Figure 7A). VEGF and Forskolin were used for vascular lineage induction, whereas the treatment with VEGF, FGF2, and fetal bovine serum stimulated further vascular differentiation and maturation (Figure 7A). Consistent with previous studies (49), immunostaining for CD31 (PECAM-1) and PDGFRβ showed an abundance of endothelial cells and pericytes in vascular organoids formed by control iPSCs (Figure 7B). The vascular network in ACDMPV organoids was poorly developed, as demonstrated by decreased percentages of endothelial cells and pericytes (Figures 7B and 7C) and reduced expression of PECAM1 mRNA (Figure E9A). Thus, control iPSCs are more efficient in forming the organoid vascular network than ACDMPV iPSCs.
Figure 7.
TanFe (transcellular activator of nuclear FOXF1 expression) increases angiogenesis in human vascular organoids (VOs) derived from a patient with ACDMPV. (A) Schematic diagram shows the differentiation protocol used to generate VOs from human induced pluripotent stem cells (hiPSCs). Brightfield images show morphological changes undergone by hiPSCs during VO differentiation. Scale bars for Days D0–D7, 500 μm. Scale bar for D15, 250 μm. (B) Images show immunostaining of whole-mount D15 VOs for CD31 (gray, endothelial cells) and PDGFRb (red, mural cells/pericytes). Nuclei were stained with DAPI. Scale bar, 200 μm. (C) TanFe treatment increases the percentages of CD31+ and PDGFRb+ cells in vascular organoids at D15 (n = 3 in each group). (D) Time course gene expression profile of FOXF1 during D3–D15 VO differentiation from human iPSCs. (E) Images show immunostaining of D15 VO sections for CD31 (gray) and FOXF1 (red). Scale bar, 200 μm. (F) High-magnification images of CD31-stained sections show lumen-like structures in D15 VOs (arrowheads). Scale bar, 50 μm. (G) Quantification of the endothelial density within VOs (n = 3). (H) Quantification of the number of lumen-like structures within VOs (n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001. ACDMPV = alveolar capillary dysplasia with misalignment of pulmonary veins; BMP4 = bone morphogenetic protein 4; EB = embryoid body; FBS = fetal bovine serum; FGF2 = fibroblast growth factor 2; UD = undifferentiated human iPSCs; VEGF = vascular endothelial growth factor; YHWZ = tyrosine 3/tryptophan 5 -monooxygenase activation protein.
Although FOXF1 expression in vascular organoids was increased after the vascular lineage induction, FOXF1 concentrations were lower in ACDMPV organoids than in control organoids (Figure 7D). TanFe treatment increased FOXF1 staining in ACDMPV organoids (Figures 7E and E9B) and improved the vascular network as demonstrated by quantification of endothelial density and numbers of lumen-like endothelial structures in the organoids (Figures 7F–7H). TanFe had no effect on vascular density in organoids derived from control iPSCs (Figures E9C and E9D). Altogether, the TanFe compound increases angiogenesis in vascular organoids derived from ACDMPV iPSCs.
Discussion
Published studies demonstrate that lung endothelial cells depend on expression of FOXF1 transcription factor to undergo angiogenesis during lung development (9, 10). Heterozygous deletions or loss-of-function point mutations in the FOXF1 gene locus are linked to ACDMPV (3, 4), a severe congenital disorder associated with maldevelopment of alveolar capillaries in neonates and infants (1). Because FOXF1 is required for development of the alveolar microvasculature in the human lung, pharmacological stabilization of the FOXF1 protein can provide new therapeutic opportunities for patients with ACDMPV by increasing FOXF1 protein concentrations. In this study, we identified the TanFe small-molecule compound, which increases FOXF1 concentrations in pulmonary endothelial cells by preventing HECTD1-mediated ubiquitination of the FOXF1 protein. Treatment of pregnant mice carrying the Foxf1+/− embryos with TanFe compound stimulated neonatal lung angiogenesis and prevented mortality of Foxf1+/− mice after birth. We also found that the TanFe compound is capable of increasing angiogenesis in human vascular organoids formed by iPSCs derived from a patient with ACDMPV. Our studies suggest that TanFe can be considered for stabilization of the WT (normal) FOXF1 protein in human ACDMPV. However, extensive pharmacologic and toxicity studies must be performed to consider any clinical use of the TanFe compound or its derivatives. Another limitation is that the majority of infants with ACDMPV are not recognized until after birth. Improvements in prenatal FOXF1 gene testing are needed to identify subjects with ACDMPV for early intervention.
The TanFe compound appears to directly disrupt FOXF1–HECTD1 interactions as supported by our experiments with purified FOXF1 and HECTD1 proteins under cell-free conditions. Because HECTD1 ubiquitinates FOXF1, it is possible that an increased FOXF1 protein concentration in TanFe-treated cells is a direct consequence of reduced FOXF1 protein degradation. The lack of changes in Foxf1 mRNA suggests that TanFe influences neither Foxf1 mRNA transcription nor its stability. Interestingly, the TanFe compound had no effect on interactions between FOXF1 and UBE3A and several other proteins involved in the ubiquitination system. Furthermore, TanFe did not disrupt the interactions of FOXF1 with MDM2 and MYCBP2 (RBBP6), both of which are known to interact with HECTD1 (50). Although the importance of FOXF1 interactions with UBE3A, MDM2, and MYCBP2 E3 ligases is unclear, our data support the selectivity of TanFe in disrupting the FOXF1–HECTD1 protein complex. The TanFe–FOXF1–HECTD1 cocrystal structure could clarify the exact molecular mechanisms by which the TanFe compound disrupts FOXF1–HECTD1 protein–protein interactions. Because TanFe impacted the interactions between FOXF1 and other ubiquitin E3 ligases, TanFe can have off-target effects in addition to disrupting the FOXF1–HECTD1 binding.
Published studies demonstrated that increasing FOXF1 concentrations in transgenic mice improves endothelial barrier function and decreases lung inflammation after lung injury (17, 18). Therefore, the effects of TanFe on LPS lung injury responses are not surprising. Because FOXF1 transcriptionally activates various genes critical for structural integrity of endothelial adherens junctions (17), it is likely that TanFe-mediated improvement of endothelial barrier function contributes to reduced numbers of neutrophils in BALF of LPS-injured mice. Interestingly, endothelial cells in ACDMPV organoids were mostly located in the peripheral regions. TanFe treatment decreased peripheral endothelial staining and increased the formation of vascular networks inside of ACDMPV organoids, consistent with the critical role of FOXF1 in EC migration and angiogenesis (9, 10). TanFe increased alveolar capillary density in Foxf1+/− embryos, but its effect in WT embryos was modest. It is possible that Foxf1+/− lungs respond better to TanFe treatment because the FOXF1 deficiency creates the vascular-deficient environment sensitive to proangiogenic stimulation.
In summary, the TanFe compound increases FOXF1 protein concentrations by interfering with FOXF1 binding to HECTD1 E3 ubiquitin ligase. Our studies support the feasibility of developing FOXF1-stabilizing small-molecule compounds for potential therapies in human ACDMPV and other neonatal pulmonary vascular diseases.
Acknowledgments
Acknowledgment
The authors thank Dr. Irene Zohn from Children’s National Medical Center for providing expression plasmids containing HA-HECTD1 full length and HA-HECTD1 C2579G mutant.
Footnotes
Supported by NHLBI grants HL141174 (V.V.K.), HL149631 (V.V.K.), HL152973 (V.V.K. and T.V.K.), HL158659 (T.V.K.), U01TR001810 (D.N.K), and N0 175N92020C00005 (D.N.K.).
Author contributions: V.V.K., A.P., and L.C. designed the study. A.P., L.C., V.U., A.A.R., Y.Z., and A.B.A. performed experiments. M.E.S. and A.G.Z. conducted mass spectrometry analysis. N.M.P. and M.G. performed organoid experiments. J.A.W. performed clinical care. D.N.K. provided critical reagents. V.V.K., A.P., T.R.K., and T.V.K. wrote the manuscript with input from all authors.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202207-1332OC on December 8, 2022
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Bishop NB, Stankiewicz P, Steinhorn RH. Alveolar capillary dysplasia. Am J Respir Crit Care Med . 2011;184:172–179. doi: 10.1164/rccm.201010-1697CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Whitsett JA, Kalin TV, Xu Y, Kalinichenko VV. Building and regenerating the lung cell by cell. Physiol Rev . 2019;99:513–554. doi: 10.1152/physrev.00001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pradhan A, Dunn A, Ustiyan V, Bolte C, Wang G, Whitsett JA, et al. The S52F FOXF1 mutation inhibits STAT3 signaling and causes alveolar capillary dysplasia. Am J Respir Crit Care Med . 2019;200:1045–1056. doi: 10.1164/rccm.201810-1897OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dharmadhikari AV, Szafranski P, Kalinichenko VV, Stankiewicz P. Genomic and epigenetic complexity of the FOXF1 locus in 16q24.1: implications for development and disease. Curr Genomics . 2015;16:107–116. doi: 10.2174/1389202916666150122223252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Edwards JJ, Murali C, Pogoriler J, Frank DB, Handler SS, Deardorff MA, et al. Histopathologic and genetic features of alveolar capillary dysplasia with atypical late presentation and prolonged survival. J Pediatr . 2019;210:214–219.e2. doi: 10.1016/j.jpeds.2019.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Towe CT, White FV, Grady RM, Sweet SC, Eghtesady P, Wegner DJ, et al. Infants with atypical presentations of alveolar capillary dysplasia with misalignment of the pulmonary veins who underwent bilateral lung transplantation. J Pediatr . 2018;194:158–164.e1. doi: 10.1016/j.jpeds.2017.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun F, Wang G, Pradhan A, Xu K, Gomez-Arroyo J, Zhang Y, et al. Nanoparticle delivery of STAT3 alleviates pulmonary hypertension in a mouse model of alveolar capillary dysplasia. Circulation . 2021;144:539–555. doi: 10.1161/CIRCULATIONAHA.121.053980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deng Z, Kalin GT, Shi D, Kalinichenko VV. Nanoparticle delivery systems with cell-specific targeting for pulmonary diseases. Am J Respir Cell Mol Biol . 2021;64:292–307. doi: 10.1165/rcmb.2020-0306TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ren X, Ustiyan V, Guo M, Wang G, Bolte C, Zhang Y, et al. Postnatal alveologenesis depends on FOXF1 signaling in c-KIT+ endothelial progenitor cells. Am J Respir Crit Care Med . 2019;200:1164–1176. doi: 10.1164/rccm.201812-2312OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang G, Wen B, Ren X, Li E, Zhang Y, Guo M, et al. Generation of pulmonary endothelial progenitor cells for cell-based therapy using interspecies mouse-rat chimeras. Am J Respir Crit Care Med . 2021;204:326–338. doi: 10.1164/rccm.202003-0758OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Black M, Milewski D, Le T, Ren X, Xu Y, Kalinichenko VV, et al. FOXF1 inhibits pulmonary fibrosis by preventing CDH2-CDH11 cadherin switch in myofibroblasts. Cell Rep . 2018;23:442–458. doi: 10.1016/j.celrep.2018.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim IM, Zhou Y, Ramakrishna S, Hughes DE, Solway J, Costa RH, et al. Functional characterization of evolutionarily conserved DNA regions in forkhead box f1 gene locus. J Biol Chem . 2005;280:37908–37916. doi: 10.1074/jbc.M506531200. [DOI] [PubMed] [Google Scholar]
- 13. Kalinichenko VV, Gusarova GA, Shin B, Costa RH. The forkhead box F1 transcription factor is expressed in brain and head mesenchyme during mouse embryonic development. Gene Expr Patterns . 2003;3:153–158. doi: 10.1016/s1567-133x(03)00010-3. [DOI] [PubMed] [Google Scholar]
- 14. Bolte C, Whitsett JA, Kalin TV, Kalinichenko VV. Transcription factors regulating embryonic development of pulmonary vasculature. Adv Anat Embryol Cell Biol . 2018;228:1–20. doi: 10.1007/978-3-319-68483-3_1. [DOI] [PubMed] [Google Scholar]
- 15. Ustiyan V, Bolte C, Zhang Y, Han L, Xu Y, Yutzey KE, et al. FOXF1 transcription factor promotes lung morphogenesis by inducing cellular proliferation in fetal lung mesenchyme. Dev Biol . 2018;443:50–63. doi: 10.1016/j.ydbio.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bolte C, Flood HM, Ren X, Jagannathan S, Barski A, Kalin TV, et al. FOXF1 transcription factor promotes lung regeneration after partial pneumonectomy. Sci Rep . 2017;7:10690. doi: 10.1038/s41598-017-11175-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cai Y, Bolte C, Le T, Goda C, Xu Y, Kalin TV, et al. FOXF1 maintains endothelial barrier function and prevents edema after lung injury. Sci Signal . 2016;9:ra40. doi: 10.1126/scisignal.aad1899. [DOI] [PubMed] [Google Scholar]
- 18. Kalinichenko VV, Zhou Y, Shin B, Stolz DB, Watkins SC, Whitsett JA, et al. Wild-type levels of the mouse Forkhead Box f1 gene are essential for lung repair. Am J Physiol Lung Cell Mol Physiol . 2002;282:L1253–L1265. doi: 10.1152/ajplung.00463.2001. [DOI] [PubMed] [Google Scholar]
- 19. Kalin TV, Meliton L, Meliton AY, Zhu X, Whitsett JA, Kalinichenko VV. Pulmonary mastocytosis and enhanced lung inflammation in mice heterozygous null for the Foxf1 gene. Am J Respir Cell Mol Biol . 2008;39:390–399. doi: 10.1165/rcmb.2008-0044OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bolte C, Ren X, Tomley T, Ustiyan V, Pradhan A, Hoggatt A, et al. Forkhead box F2 regulation of platelet-derived growth factor and myocardin/serum response factor signaling is essential for intestinal development. J Biol Chem . 2015;290:7563–7575. doi: 10.1074/jbc.M114.609487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ren X, Ustiyan V, Pradhan A, Cai Y, Havrilak JA, Bolte CS, et al. FOXF1 transcription factor is required for formation of embryonic vasculature by regulating VEGF signaling in endothelial cells. Circ Res . 2014;115:709–720. doi: 10.1161/CIRCRESAHA.115.304382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang G, Wen B, Deng Z, Zhang Y, Kolesnichenko OA, Ustiyan V, et al. Endothelial progenitor cells stimulate neonatal lung angiogenesis through FOXF1-mediated activation of BMP9/ACVRL1 signaling. Nat Commun . 2022;13:2080. doi: 10.1038/s41467-022-29746-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mahlapuu M, Ormestad M, Enerbäck S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development . 2001;128:155–166. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- 24. Kalinichenko VV, Lim L, Stolz DB, Shin B, Rausa FM, Clark J, et al. Defects in pulmonary vasculature and perinatal lung hemorrhage in mice heterozygous null for the Forkhead Box f1 transcription factor. Dev Biol . 2001;235:489–506. doi: 10.1006/dbio.2001.0322. [DOI] [PubMed] [Google Scholar]
- 25. Kolesnichenko OA, Whitsett JA, Kalin TV, Kalinichenko VV. Therapeutic potential of endothelial progenitor cells in pulmonary diseases. Am J Respir Cell Mol Biol . 2021;65:473–488. doi: 10.1165/rcmb.2021-0152TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bolte C, Kalin TV, Kalinichenko VV. Molecular, cellular, and bioengineering approaches to stimulate lung regeneration after injury. Semin Cell Dev Biol . 2020;100:101–108. doi: 10.1016/j.semcdb.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 27. Sen P, Dharmadhikari AV, Majewski T, Mohammad MA, Kalin TV, Zabielska J, et al. Comparative analyses of lung transcriptomes in patients with alveolar capillary dysplasia with misalignment of pulmonary veins and in foxf1 heterozygous knockout mice. PLoS One . 2014;9:e94390. doi: 10.1371/journal.pone.0094390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun L, Ren X, Wang IC, Pradhan A, Zhang Y, Flood HM, et al. The FOXM1 inhibitor RCM-1 suppresses goblet cell metaplasia and prevents IL-13 and STAT6 signaling in allergen-exposed mice. Sci Signal . 2017;10:eaai8583. doi: 10.1126/scisignal.aai8583. [DOI] [PubMed] [Google Scholar]
- 29. Pradhan A, Ustiyan V, Zhang Y, Kalin TV, Kalinichenko VV. Forkhead transcription factor FoxF1 interacts with Fanconi anemia protein complexes to promote DNA damage response. Oncotarget . 2016;7:1912–1926. doi: 10.18632/oncotarget.6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ziady AG, Sokolow A, Shank S, Corey D, Myers R, Plafker S, et al. Interaction with CREB binding protein modulates the activities of Nrf2 and NF-κB in cystic fibrosis airway epithelial cells. Am J Physiol Lung Cell Mol Physiol . 2012;302:L1221–L1231. doi: 10.1152/ajplung.00156.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Milewski D, Pradhan A, Wang X, Cai Y, Le T, Turpin B, et al. FoxF1 and FoxF2 transcription factors synergistically promote rhabdomyosarcoma carcinogenesis by repressing transcription of p21Cip1 CDK inhibitor. Oncogene . 2017;36:850–862. doi: 10.1038/onc.2016.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xia H, Ren X, Bolte CS, Ustiyan V, Zhang Y, Shah TA, et al. Foxm1 regulates resolution of hyperoxic lung injury in newborns. Am J Respir Cell Mol Biol . 2015;52:611–621. doi: 10.1165/rcmb.2014-0091OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li E, Ustiyan V, Wen B, Kalin GT, Whitsett JA, Kalin TV, et al. Blastocyst complementation reveals that NKX2-1 establishes the proximal-peripheral boundary of the airway epithelium. Dev Dyn . 2021;250:1001–1020. doi: 10.1002/dvdy.298. [DOI] [PubMed] [Google Scholar]
- 34. Ren X, Zhang Y, Snyder J, Cross ER, Shah TA, Kalin TV, et al. Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Mol Cell Biol . 2010;30:5381–5393. doi: 10.1128/MCB.00876-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wen B, Li E, Ustiyan V, Wang G, Guo M, Na CL, et al. In vivo generation of lung and thyroid tissues from embryonic stem cells using blastocyst complementation. Am J Respir Crit Care Med . 2021;203:471–483. doi: 10.1164/rccm.201909-1836OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang IC, Zhang Y, Snyder J, Sutherland MJ, Burhans MS, Shannon JM, et al. Increased expression of FoxM1 transcription factor in respiratory epithelium inhibits lung sacculation and causes Clara cell hyperplasia. Dev Biol . 2010;347:301–314. doi: 10.1016/j.ydbio.2010.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Flood HM, Bolte C, Dasgupta N, Sharma A, Zhang Y, Gandhi CR, et al. The Forkhead box F1 transcription factor inhibits collagen deposition and accumulation of myofibroblasts during liver fibrosis. Biol Open . 2019;8:bio039800. doi: 10.1242/bio.039800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang X, Bhattacharyya D, Dennewitz MB, Kalinichenko VV, Zhou Y, Lepe R, et al. Rapid hepatocyte nuclear translocation of the Forkhead Box M1B (FoxM1B) transcription factor caused a transient increase in size of regenerating transgenic hepatocytes. Gene Expr . 2003;11:149–162. doi: 10.3727/000000003108749044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wen B, Wang G, Li E, Kolesnichenko OA, Tu Z, Divanovic S, et al. In vivo generation of bone marrow from embryonic stem cells in interspecies chimeras. eLife . 2022;11:e74018. doi: 10.7554/eLife.74018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ustiyan V, Zhang Y, Perl AK, Whitsett JA, Kalin TV, Kalinichenko VV. β-catenin and Kras/Foxm1 signaling pathway are critical to restrict Sox9 in basal cells during pulmonary branching morphogenesis. Dev Dyn . 2016;245:590–604. doi: 10.1002/dvdy.24393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ustiyan V, Wert SE, Ikegami M, Wang IC, Kalin TV, Whitsett JA, et al. Foxm1 transcription factor is critical for proliferation and differentiation of Clara cells during development of conducting airways. Dev Biol . 2012;370:198–212. doi: 10.1016/j.ydbio.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bolte C, Zhang Y, York A, Kalin TV, Schultz Jel J, Molkentin JD, et al. Postnatal ablation of Foxm1 from cardiomyocytes causes late onset cardiac hypertrophy and fibrosis without exacerbating pressure overload-induced cardiac remodeling. PLoS One . 2012;7:e48713. doi: 10.1371/journal.pone.0048713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Milewski D, Balli D, Ustiyan V, Le T, Dienemann H, Warth A, et al. FOXM1 activates AGR2 and causes progression of lung adenomas into invasive mucinous adenocarcinomas. PLoS Genet . 2017;13:e1007097. doi: 10.1371/journal.pgen.1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bolte C, Zhang Y, Wang IC, Kalin TV, Molkentin JD, Kalinichenko VV. Expression of Foxm1 transcription factor in cardiomyocytes is required for myocardial development. PLoS One . 2011;6:e22217. doi: 10.1371/journal.pone.0022217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bolte C, Ustiyan V, Ren X, Dunn AW, Pradhan A, Wang G, et al. Nanoparticle delivery of proangiogenic transcription factors into the neonatal circulation inhibits alveolar simplification caused by hyperoxia. Am J Respir Crit Care Med . 2020;202:100–111. doi: 10.1164/rccm.201906-1232OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sarkar AA, Zohn IE. Hectd1 regulates intracellular localization and secretion of Hsp90 to control cellular behavior of the cranial mesenchyme. J Cell Biol . 2012;196:789–800. doi: 10.1083/jcb.201105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Milewski D, Shukla S, Gryder BE, Pradhan A, Donovan J, Sudha P, et al. FOXF1 is required for the oncogenic properties of PAX3-FOXO1 in rhabdomyosarcoma. Oncogene . 2021;40:2182–2199. doi: 10.1038/s41388-021-01694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hoggatt AM, Kim JR, Ustiyan V, Ren X, Kalin TV, Kalinichenko VV, et al. The transcription factor Foxf1 binds to serum response factor and myocardin to regulate gene transcription in visceral smooth muscle cells. J Biol Chem . 2013;288:28477–28487. doi: 10.1074/jbc.M113.478974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wimmer RA, Leopoldi A, Aichinger M, Kerjaschki D, Penninger JM. Generation of blood vessel organoids from human pluripotent stem cells. Nat Protoc . 2019;14:3082–3100. doi: 10.1038/s41596-019-0213-z. [DOI] [PubMed] [Google Scholar]
- 50. Bento CF, Ashkenazi A, Jimenez-Sanchez M, Rubinsztein DC. The Parkinson’s disease-associated genes ATP13A2 and SYT11 regulate autophagy via a common pathway. Nat Commun . 2016;7:11803. doi: 10.1038/ncomms11803. [DOI] [PMC free article] [PubMed] [Google Scholar]