Abstract
Objective:
Electronic Health Record (EHR) based phenotyping is a crucial yet challenging problem in the biomedical field. Though clinicians typically determine patient-level diagnoses via manual chart review, the sheer volume and heterogeneity of EHR data renders such tasks challenging, time-consuming, and prohibitively expensive, thus leading to a scarcity of clinical annotations in EHRs. Weakly supervised learning algorithms have been successfully applied to various EHR phenotyping problems, due to their ability to leverage information from large quantities of unlabeled samples to better inform predictions based on a far smaller number of patients. However, most weakly supervised methods are subject to the challenge to choose the right cutoff value to generate an optimal classifier. Furthermore, since they only utilize the most informative features (i.e., main ICD and NLP counts) they may fail for episodic phenotypes that cannot be consistently detected via ICD and NLP data. In this paper, we propose a label-efficient, weakly semi-supervised deep learning algorithm for EHR phenotyping (WSS-DL), which overcomes the limitations above.
Materials and Methods:
WSS-DL classifies patient-level disease status through a series of learning stages: 1) generating silver standard labels, 2) deriving enhanced-silver-standard labels by fitting a weakly supervised deep learning model to data with silver standard labels as outcomes and high dimensional EHR features as input, and 3) obtaining the final prediction score and classifier by fitting a supervised learning model to data with a minimal number of gold standard labels as the outcome, and the enhanced-silverstandard labels and a minimal set of most informative EHR features as the input. To assess the generalizability of WSS-DL across different phenotypes and medical institutions, we apply WSS-DL to classify a total of 17 diseases, including both acute and chronic conditions, using EHR data from three healthcare systems. Additionally, we determine the minimum quantity of training labels required by WSSDL to outperform existing supervised and semi-supervised phenotyping methods.
Results:
The proposed method, in combining the strengths of deep learning and weakly semi-supervised learning, successfully leverages the crucial phenotyping information contained in EHR features from unlabeled samples. Indeed, the deep learning model’s ability to handle high-dimensional EHR features allows it to generate strong phenotype status predictions from silver standard labels. These predictions, in turn, provide highly effective features in the final logistic regression stage, leading to high phenotyping accuracy in notably small subsets of labeled data (e.g. n = 40 labeled samples).
Conclusion:
Our method’s high performance in EHR datasets with very small numbers of labels indicates its potential value in aiding doctors to diagnose rare diseases as well as conditions susceptible to misdiagnosis.
Keywords: deep learning, EHR phenotyping, label efficient, silver-standard labels, weakly supervised
1. Introduction
Electronic Health Records (EHRs) are digital records that contain comprehensive, longitudinal patient information, such as administrative and billing data, demographic data, laboratory results, and medical notes written by clinicians during medical visits 1,2. The increasing adoption of EHRs for clinical care has also increased their usage as a resource for epidemiological and biomedical studies 2. One fundamental task in using EHRs for clinical research is to accurately annotate phenotypes (i.e., the presence or absence of disease conditions for individual patients). However, the sheer volume and heterogeneity of EHR data renders such tasks challenging and time-consuming. Typical phenotyping algorithms that predict phenotypes from diverse EHR features can be stratified into (1) rule-based algorithms and (2) supervised machine learning algorithms 2–6, which all require intensive human efforts. Although widely used in the literature, rule-based algorithms are challenging to scale across multiple conditions due to the need for iteratively devising and validating rules. On the other hand, supervised algorithms trained on a subset of manually annotated gold-standard labels necessitate laborious chart review. Furthermore, rule-based or supervised algorithms, by relying exclusively on a small subset of features or a small subset of labeled patients, cannot fully leverage information contained in high dimensional and large sample EHR data. This motivates the need for developing high-throughput phenotyping algorithms capable of handling large datasets with complex structure in an automated fashion, to enhance the speed and efficiency of the clinical workflow and ultimately improve the quality of patient care using only a minimal number of gold-standard labels.
As such, fully unsupervised methods and weakly supervised methods, which require no clinical annotations, have been proposed. Studies for multi-phenotyping applications often utilize raw surrogate features – variables in the EHR dataset that are most predictive of phenotype status (e.g., patient-level counts of ICD codes or NLP mentions in the medical notes for the target phenotype)– as proxies for the true gold standard labels. However, for many diseases, the raw main ICD code is often too imprecise to be reliably used for prediction and can hamper the power of the downstream association study22–26. For example, it was reported in Tedeschi et al. (2018)26 that a published billing code algorithm had very low positive predictive value (PPV) (18%) for the pseudogout phenotype in an academic medical center EHR dataset. To efficiently and accurately assign disease status for patients without intensive human labor, recent efforts have been devoted to the class of weakly supervised methods, which train the supervised classifier using silver-standard labels derived from the most informative EHR features. For example, the “anchor and learn” algorithm trains a regularized supervised classifier using the silver-standard labels derived from “anchor” features 7. The XPRESS algorithm fits a regularized logistic regression on silver-standard labels derived based on the presence or absence of at least one ICD code for the phenotype of interest8. The PheNorm method 9 and High-Throughput Multimodal Automated Phenotyping (MAP) 10 predict patient-level probabilities of positive disease status from counts of ICD codes and NLP mentions for the target phenotype, variables highly predictive of phenotype status. Specifically, MAP first fits Poisson and log-normal mixture models to the ICD and NLP features, as either or both models may provide a good approximation to the observed data, then calculates the posterior probabilities of having the phenotype given the feature information from each fitted mixture model. The final MAP algorithm assigns the predicted probability of having the phenotype as the average of predicted probabilities of all the above mixture models. Wang et al. (2019) propose a weakly supervised deep learning NLP-based algorithm for clinical text classification 11. Ahuja et al. (2019) propose a surrogate-guided ensemble Latent Dirichlet Allocation (sureLDA) method 12. Wagholikar et al. train logistic regression and random forest algorithms on silver standard labels created using a polar labeling algorithm, designed to use the distribution of main ICD code counts to inform the patient phenotype prediction 6. PheVis combines diagnosis codes together with medical concepts extracted from medical notes to provide an interpretable parametric predictor of the occurrence probability for a given medical condition at each visit 13.
While being scalable and retraining good phenotyping performance, the weakly supervised methods mentioned above can still be improved in the following ways. First, most of the weakly supervised methods are subject to the challenge to choose the right cutoff value to generate an optimal classifier. Second, only the most informative features (i.e., main ICD and NLP counts) are utilized leaving the vast amount of moderate informative features unused, which may lead to bad performance for episodic phenotypes (e.g., pseudogout) that cannot be consistently detected via ICD and NLP data. Episodic phenotypes, by definition, are conditions that only occur periodically, being marked by the incidence of specific clinical events (e.g. heart attacks for heart failure, week-long periods of sudden and intense joint inflammation for pseudogout). While ICD and NLP count for chronic conditions such as Type 2 Diabetes Mellitus or Ulcerative Colitis often increase over time, due to a continual need for medical visits to monitor a patient’s condition, those for episodic phenotypes tend to fluctuate, meaning that a patient’s number of ICD codes and NLP mentions at a given point in time may fail to adequately represent their true episodic disease status. For instance, following medical visits during a recovery period for an episodic condition, a patient may have no ICD codes recorded in the EHR system, in the same manner as another patient with no prior history of this disease. Furthermore, due to computing intensity, NLP may not be conducted on all patients for a large cohort, in which case, methods depending on NLP counts are not feasible.
Deep learning methods have been highly effective in EHR phenotyping tasks as they can extract highly complex, latent features in high-dimensional datasets, unlike many other methods 8,11,14–19. In this paper, we propose a weakly semi-supervised deep learning algorithm (WSS-DL) that accurately annotates disease status by mainly using silver-standard labels and unlabeled data, with the help of a very small number of gold-standard labels, to further improve the phenotyping performance beyond that of weakly supervised methods. To address the first limitation, we leverage a very small set of gold-standard labels to help us guide the model fitting. To address the second limitation, we leverage the strengths of the deep learning model to fully utilize the vast information contained in EHRs. To address the third limitation, we only use silver-standard labels as initial values where missingness is allowed. It is worth mentioning that the proposed method is label efficient, in that it only requires a very small set of gold standard labels. The WSS-DL method is designed as follows. First, silver-standard labels are derived from one or multiple surrogate outcome variable(s) in an unsupervised fashion. Second, enhanced-silver-standard labels are derived via a deep learning algorithm, using the silver-standard labels from the first step as the outcome, a very small set of gold standard labels to guide the learning task, and a set of high dimensional EHR variables as input features. Finally, a logistic regression model is applied to predict the gold standard labels using the enhanced-silver-standard labels and a minimal set of selected EHR features.
The paper is structured as follows. In Section 2, we describe the pipeline for the proposed algorithm, as well as the training strategies and evaluation metrics. Section 3 covers experimental results obtained from applying WSS-DL to a total of 17 phenotypes from Massachusetts General Brigham (MGB), an independent cohort of pseudogout patients from MGB (MGB-Pseudogout), and the Boston Children’s Hospital (BCH) EHRs. We discuss the results and implications of WSS-DL’s performance on phenotyping in various EHR-based scenarios in Section 4, and we conclude this paper in Section 5.
2. Material and methods
2.1. Weakly semi-supervised algorithm
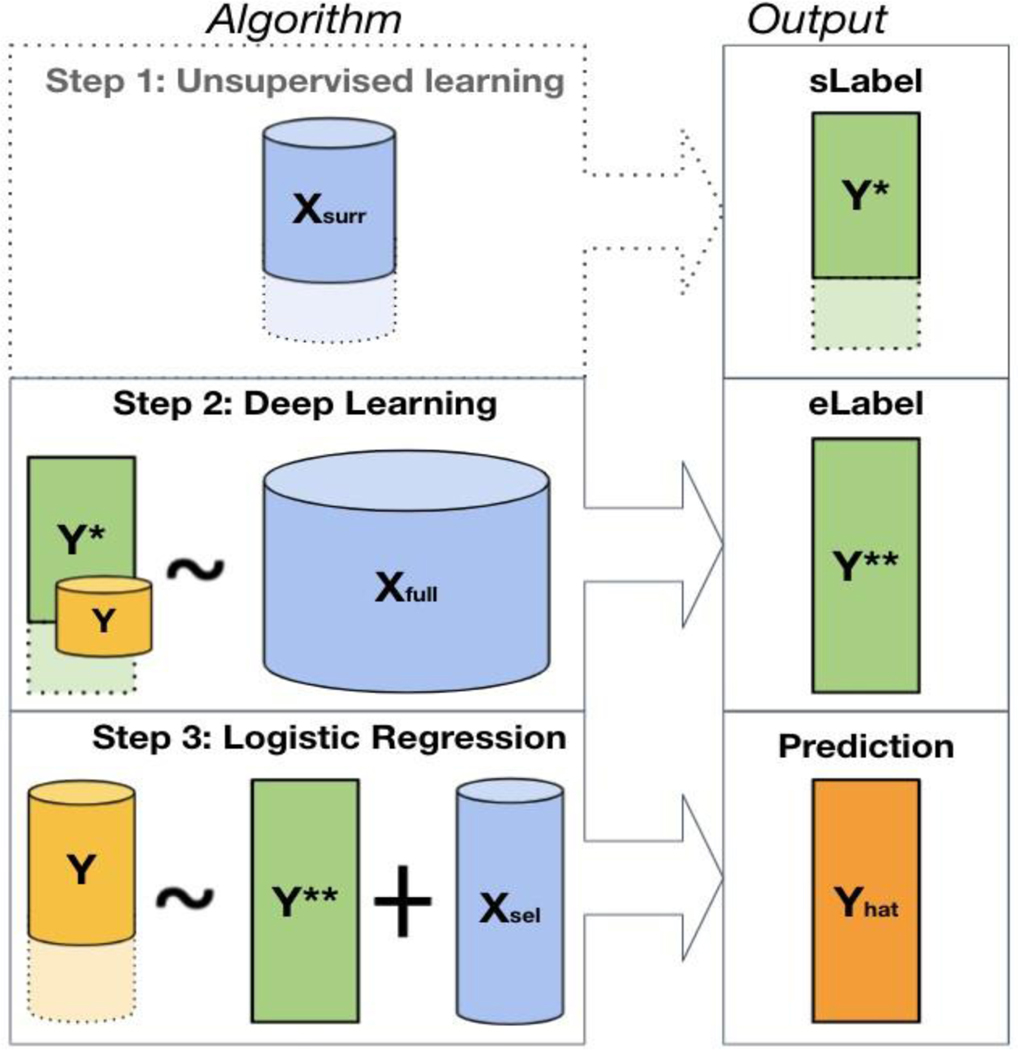
As shown in Figure 1, the proposed weakly semi-supervised algorithm for predicting patient-level disease status consists of three steps: fully unsupervised, semi-supervised, and fully supervised. In the unsupervised stage, we obtain the silver-standard labels for the phenotype of interest by applying a fully unsupervised algorithm to either one or multiple main surrogate variable(s) known to be most predictive of the outcome, based on subject-matter knowledge. This step may be omitted when is a viable surrogate that can be directly used as . In the semi-supervised stage, we train a deep learning algorithm to obtain enhanced-silver-standard labels , with silver-standard labels as the outcome, the collection of all EHR covariates as input features, and with the inclusion of a very small number of labels from to assist in guiding the learning process. Finally, in the fully supervised stage, we obtain the final prediction score and classifier via logistic regression, with a very small number of labels as the outcome, and the enhanced-silver-standard labels and a minimal set of most informative features as the input covariates.
Figure 1.
The WSS-DL method pipeline. Step 1: creation of silver standard labels from using the MAP algorithm; this step may be omitted when is a viable surrogate that can be directly used as . Step 2: creation of the enhanced-silver-standard label using neural network, with as the outcome, as input feature, and a subset of for fine-tuning. Step 3: final prediction of using logistic regression, with the enhanced-silver-standard labels and a minimal set of informative features as input features. The different sizes of represented in steps 2 and 3 indicate that a mere subset of is included for algorithm fine-tuning in Step 2, and the full vector in Step 3 for the final patient-level phenotype classification.
In this work, let represent the binary patient phenotype of interest obtained via chart review, which is only available in a small subset of patients with sample size ; let be the collection of silver standard labels used as a proxy for ; denote a single or multiple surrogate features that are most predictive of , which is available on all patients or a large subset of patients with sample size ; denote the high-dimensional EHR features for the phenotype on all patients with sample size ; and denote a minimal set of most informative features available on all patients with sample size . It is worth mentioning that 1) contains the most informative features that are available on a large amount of patients but may not be available on all patients (e.g. the main NLP mentions in clinical notes when NLP is conducted on only a subset of patients; a lab test conducted on only a subset of patients; patient-reported outcomes that can be combined with EHR data but are only available in a subset of patients); 2) contains all informative features that are available on all patients, including both the stronger and the weaker features, such as a vast amount of ICD codes, laboratory tests, medication codes, procedure codes, healthcare utilization (a variable quantifying how frequent a patient utilize the healthcare system, such as the total number of all ICD codes) and prediction scores from algorithms in the literature; 3) contains only the minimal set of informative features that are available on all patients, such as the MAP-derived feature based on the main ICD code when the ICD code counts are provided for all patients. It is worth mentioning that in many cases, is a subset of .
Unsupervised stage – silver standard label generation
Due to the scarcity of gold standard labels in the EHR, it is necessary to leverage other covariates, measured in a larger subset of patients in the dataset, that are highly predictive of the outcome. In particular, we may use these features to generate silver standard labels, to guide our algorithm in its phenotyping task. For instance, when contains the vectors of counts for the main ICD code and NLP mentions in patient-level clinical notes for the phenotype of interest, we may obtain as the output of an unsupervised phenotyping algorithm, such as MAP8 or PheNorm7. These algorithms predict the patient-level probabilities of positive disease status from the surrogate variables in a fully unsupervised fashion, adjusted for healthcare utilization. In our experiments, we use MAP to generate silver standard labels for most phenotypes. The MAP algorithm predicts the patient-level probabilities of positive disease status by fitting Gaussian and Poisson mixture models to adjusting for the healthcare utilization. In other situations when is a single covariate that is already a viable candidate for from a clinical perspective (e.g., a binary covariate for patient reported outcomes), the first unsupervised step may be omitted.
Due to the very small number of patients with true annotations , inclusion of is necessary. The main contribution of the first stage lies in the fact that is available for most patients, which thus allows for effectively leveraging high dimensional features in the large number of unlabeled samples.
Semi-supervised stage – creation of enhanced silver standard labels
Though alone may already be highly predictive of , it may fail to be representative of phenotype status in the full study population, namely in scenarios where (used to compute ) is not available in all patients. Furthermore, there are some phenotypes in which the correlation between and is not especially strong (e.g. for more common diseases or medical conditions in which misdiagnosis often occurs): in such cases, may contain substantial levels of random noise limiting its predictive power. As such, it is beneficial to leverage the remaining covariates , which may contain additional information supplementary to that in for determining patient-level disease status. To summarize this relationship between and disease status, we train a deep-learning algorithm to learn from , with the inclusion of a very small to guide the learning process.
The deep learning algorithm consists of a neural network with two separate classification layers: one that learns from the labeled observations in the EHR dataset and another that learns from the remaining unlabeled observations. We note that the number of samples with entries for is typically large enough to effectively run a deep learning model with little concern for overfitting. The interdependence between both learning tasks is manifested through the fact that both classifier layers receive the same transformed features as input, being connected to the same core network. We choose to train two separate classifiers, one for each label type, as opposed to training a single one to learn an imputed label vector, with the present values in and values of in the corresponding missing entries, for the following reason. Training a neural network to exclusively learn from on the full EHR dataset may yield predictions with limited accuracy, if has significant noise or is limited in accuracy. Indeed, we have included in the training process to guide the feature learning and alleviate the detrimental effects of low-quality silver-standard labels .
The shared feature extractor network is a feed-forward multilayer perceptron with two hidden (fully connected) layers separated by a single dropout layer, included to overcome potential overfitting. The hidden layers contain 30 and 3 nodes respectively, and the dropout layer involves dropout with 20 % probability. Our model is trained to optimize the joint loss function:
Where and are the total number of labeled and unlabeled training samples, respectively, is the sigmoid function, is the mean cross-entropy, is the vector of gold standard labels, is the vector of silver-standard labels, is the vector of predictions from the gold classification layer, and is the vector of predictions from the silver classification layer.
The expression is the standard cross-entropy loss function. The hyperparameter determines the relative weighting of the gold and silver losses in the global objective function. Following our hyper-parameter selection procedure, we choose to emphasize the importance of the known gold standard labels, which alleviate the influence of potential noise in the silver standard labels and improve the phenotyping ability of the algorithm as a whole. The model is optimized using an Adam optimizer, with learning rate 0.0005 and momentum decay 0.5.
After the training process, we generate predictions as enhanced silver standard labels learned by the network, which not only summarize the additional features in but further describe their relationship to the true disease status, via and.
Fully supervised stage – prediction of gold-standard labels
Finally, we predict the patient-level disease status from and using a very small number of labels where is available (e.g..). Specifically, letting , we conduct a logistic regression:
where is the logit link function. The final prediction is calculated as . We note that the low dimensional matrix efficiently summarizes the key information amidst the vast amount of information contained in the EHR.
The logistic regression method, by design, learns optimal weights for and that represent their relative degrees of association with . As such, it can be said to indirectly balance the influence of and the remaining features in in predicting , on a patient-level basis, without discarding information from either component of the EHR data.
1.2. Data and Evaluation Metrics
We evaluate the performance of WSS-DL using real-world EHR data from Massachusetts General Brigham (MGB), an independent cohort of pseudogout patients from MGB (MGB-Pseudogout), and Boston Children’s Hospital (BCH).
Datasets
MGB:
The MGB Biobank contains linked EHR and genetic data anchored by two large tertiary care hospitals between 1990 and 2015: Brigham and Women’s Hospital and Massachusetts General Hospital in Boston. In the biobank, a total of 17815 had codified data, NLP data, and genetic data. Gold standard labels for a small set of patients were curated for a total of fifteen phenotypes from various clinical categories – respiratory chronic conditions (Asthma, Chronic Obstructive Pulmonary Disease [COPD]), cerebral diseases (Depression, Schizophrenia [SCZ]), neurological disorders (Epilepsy, Multiple Sclerosis [MS]), vascular diseases (Hypertension [HTN], Ischemic Stroke [Stroke], Coronary Artery Disease [CAD]), musculoskeletal conditions (Rheumatoid Arthritis [RA]), digestive disorders (Ulcerative Colitis [UC], Crohn’s Disease [CD]), cancers (Breast Cancer[BrCa]), and diabetic conditions (Type 1 and Type 2 Diabetes Mellitus[T1DM,T2DM]). The additional clinical features consist of 5509 ICD codes, Logical Observation Identifiers Names and Codes (LOINC) codes for labs and Prescription for Electronic Drug Information Exchange (RxNorm) codes for medications. Silver standard labels are obtained from MAP, run using the main ICD and main NLP variables for the target phenotype (refer to step 1 in Figure 1). We remove all features with sparsity ≥ 95%, leaving 813 additional covariates for Stroke, MS, CAD, CD, RA, UC, CD, T2DM and 870 additional covariates for Asthma, COPD, Depression, SCZ, Epilepsy, HTN, BrCa, and T1DM.
MGB-Pseudogout:
The MGB-Pseudogout data derive from a cohort of Pseudogout (also known as acute calcium pyrophosphate crystal arthritis) patients from the Partners HealthCare Research Patient Data Repository (RPDR), containing EHRs from patients throughout 1991–2017. In particular, Partners’ RPDR includes EHR data from 5.5 million patients from BWH and MGH, as well as their affiliated community hospitals, community health centers, and primary care practices. The MGB-Pseudogout dataset’s additional EHR features consist of a vector of main ICD counts, 52 ICD code, LOINC code, and CUI counts, and 6 indicators for lab measurements known to be correlated with Pseudogout. Due to the episodic nature of pseudogout, the ICD and NLP covariates may not reliably represent patient disease status, and thus would yield MAP probabilities with low predictive power. As such, we define the silver standard labels in this dataset based on two binary lab covariates known to effectively identify pseudogout cases: one that indicates whether joint fluid was tested for the presence of crystals, and another that indicates whether calcium pyrophosphate crystals were found in joint fluid. We set the patient’s silver standard label to 1 if these crystals were found in the joint fluid, 0 if not, and −1 if no crystal lab measurement is available. (The indicator for the presence of patient crystal lab measurements is retained as a feature in the dataset). Gold standard labels were determined by rheumatologist manual review of EHR records for the diagnosis of pseudogout.
BCH:
Data were collected from 2012–2020, and the main phenotype of interest is Pediatric Acute Respiratory Disease Syndrome (PARDS). The BCH PARDS dataset contains 13,814 patients, a vector of main ICD counts, and 261 additional ICD code, LOINC code, RxNorm code and CUI counts for various diseases, medications, and labs. Here, unlike in the MGB and MGB-Pseudogout datasets, EHR entries were recorded at the visit-level, as opposed to the patient level, meaning that patients with more than one EHR entry would be included multiple times in the dataset. The prevalence of gold standard labels is notably lower than that of the MGB datasets (1.9 %, n = 100). The highest visit-level P/F ratios (corresponding to the ratio of arterial partial pressure of oxygen to fraction of inspired oxygen delivered) are used as silver standard labels , as this biomarker was found to be highly predictive of PARDS status. Silver standard labels are available only in the subset of 2201 patients in whom an arterial blood gas was analyzed; among these patients 44 also had gold standard labels.
For each phenotype of interest, we retain only patients satisfying a clinically established filter criterion. For the MGB data, this corresponds to having at least one ICD code for the phenotype of interest. Patients in the MGB-Pseudogout dataset were retained if they had at least 1 pseudogout-related ICD code, at least 2 CUI counts for pseudogout, or at least 2 CUI counts for chondrocalcinosis. In the BCH data, patients were selected from three intensive care units if their pGUESS27 score (output probability of the NLP-based algorithm used to determine the likelihood of positive phenotype status) outperformed 0.58. Specifically, we used the pGUESS method as proposed in Cai et al. (2022)27 to define the BCH cohort based on the NLP concepts obtained by Narrative Information Linear Extraction (NILE). The NILE system was normalized against the Systematized Nomenclature of Medicine-Clinical Terms (SNOMED-CT) reference thesaurus. SNOMED-CTs were then mapped to concept unique identifiers from the UMLS. The performance of NILE has been evaluated on MGB and VA cohorts in our previous phenotyping studies as shown in Yu et al. (2013)28 and Cai et al. (2022)27. More details on the pGUESS algorithm can be found in Cai et al. (2022)27. Omitting patients from the analysis that fail to establish the given filtering criteria for the phenotype will therefore increase its prevalence in the dataset, thus boosting the algorithm’s positive predictive value, as is necessary in designing an effective EHR phenotyping algorithm2. In addition, we apply a log transformation to all count data variables. Sample sizes, numbers of labeled patient EHRs, and phenotype prevalences for all filtered datasets are indicated in Table 1.
Table 1.
Summary of EHR datasets by phenotype. The filter positive set was defined as the set of patients passing the clinically established filter criterion as defined in the Method Section. Labeled set was defined as the small set of patients with manually curated gold-standard labels, which was randomly sampled from filter positive set. The prevalence is the proportion of subjects with positive phenotype status among those for whom labels are provided.
| EHR Platform | Phenotype | Sample Size of Filter Positive Patients | No. of Labeled Samples (%) | Prevalence (%) |
|---|---|---|---|---|
| MGB | Asthma | 7289 | 183 (2.5 %) | 47.5 (%) |
| Breast Cancer | 2002 | 94 (4.7 %) | 77.6 | |
| COPD | 3021 | 153 (5.1 %) | 43.1 | |
| Depression | 10189 | 252 (2.5 %) | 54.8 | |
| Epilepsy | 2225 | 117 (5.3 %) | 47.9 | |
| Hypertension | 19853 | 390 (2.0 %) | 79.0 | |
| SCZ | 456 | 108 (23.7 %) | 17.6 | |
| T1DM | 2111 | 128 (6.1 %) | 16.4 | |
| RA | 987 | 153 (15.5%) | 36.6 | |
| CAD | 3793 | 186 (4.9 %) | 37.1 | |
| CD | 519 | 136 (26.2 %) | 53.7 | |
| UC | 476 | 126 (26.5 %) | 49.2 | |
| T2DM | 3460 | 280 (8.1 %) | 35.7 | |
| MS | 136 | 101 (74.3 %) | 52.5 | |
| Stroke | 2052 | 128 ( 6.2 %) | 36.7 | |
|
| ||||
| BWH | Pseudogout | 12035 | 365 (3.0 %) | 21.9 |
|
| ||||
| BCH | ARDS | 2201 | 44 (1.9 %) | 40.9 |
Comparison to benchmark algorithms
We compare the proposed algorithm to five baseline methods: ICD, XPRESS, PheCAP, DAPS20, and NN. ICD predicts the gold standard labels in the dataset based on the raw data in alone. XPRESS performs a LASSO-penalized linear regression of on . Although PheCAP 21 is defined as a semi-supervised method, its learning task is fully-supervised, involving the full labeled dataset to generate phenotyping predictions. As such, we may consider it as a supervised counterpart to our WSS-DL phenotyping algorithm. PheCAP here predicts Y from the features in transformed by a set of orthogonal basis vectors. DAPS, being a semi-supervised deep learning-based algorithm, does not include any silver standard labels. Furthermore, unlike WSS-DL, it does not separate from in its phenotyping task. Rather, it directly applies a denoising autoencoder to (, ). Its hidden layer is then extracted and fed into a random forest algorithm, which returns predicted probabilities . NN merely corresponds to the intermediate output of WSS-DL from the deep learning stage of the algorithm.
Evaluation metrics
We evaluate the predictive performance of our algorithm based on accuracy measures of in predicting , including AUROC, F-score metrics, PPV and NPV. For each dataset, the reported F-score, PPV and NPV are obtained based on the chosen cutoff between positive and negative predictions that yields the highest F-score. To avoid overfitting and to correct for randomness in sampling, we conduct 5-fold cross-validation with 100 bootstrap replicates.
Minimum quantity of labels required by WSS-DL
We wish to determine the minimum quantity of training labels beyond which WSS-DL can outperform the fully supervised PheCAP, implemented on the full training set. For this, we run our algorithm using four different quantities of training data. This gives us four variants of WSS-DL: WSS-DL (20), WSS-DL (40), WSS-DL (60), and WSS-DL (80), with the numbers denoting the number of gold-standard labels included for training. Each variant of WSS−DL (n = 20, 40, 60 and 80) is specifically implemented as follows: in the deep learning stage, we sample gold standard labels exclusively from the subset of training labeled data with n samples. In the fully supervised stage, we train the logistic regression model on this same n-sample subset of training data and validate it on the held-out testing labeled subset. We choose n=20, 40, 60 and 80 to represent very small, relatively small, medium and large sample sizes of the training data. Quantities rather than percentages are used because of the variation in total sample size across different phenotypes. For comparison to WSS-DL, we run ICD and XPRESS using the full training and test subsets. For fair comparison, we also run PheCAP and DAPS with n=20, 40, 60 and 80 labels used for training. All data pre-processing, logistic regression, ICD, XPRESS, and PheCAP are implemented in R, version 3.6.1. The deep learning stage of our algorithm and DAPS are run in Python, versions 3.7.4 and 3.9.1, respectively, using a NVIDIA Tesla K80 GPU.
3. RESULTS
3.1. MGB datasets
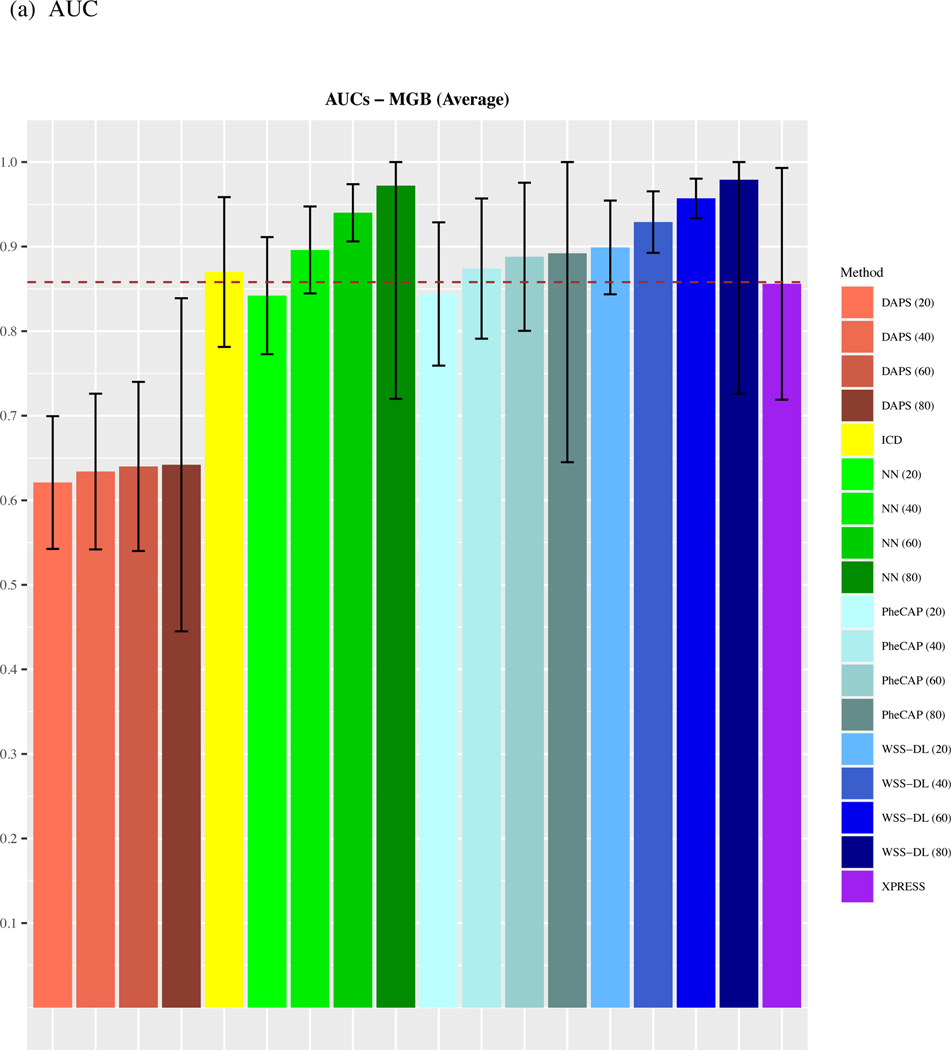
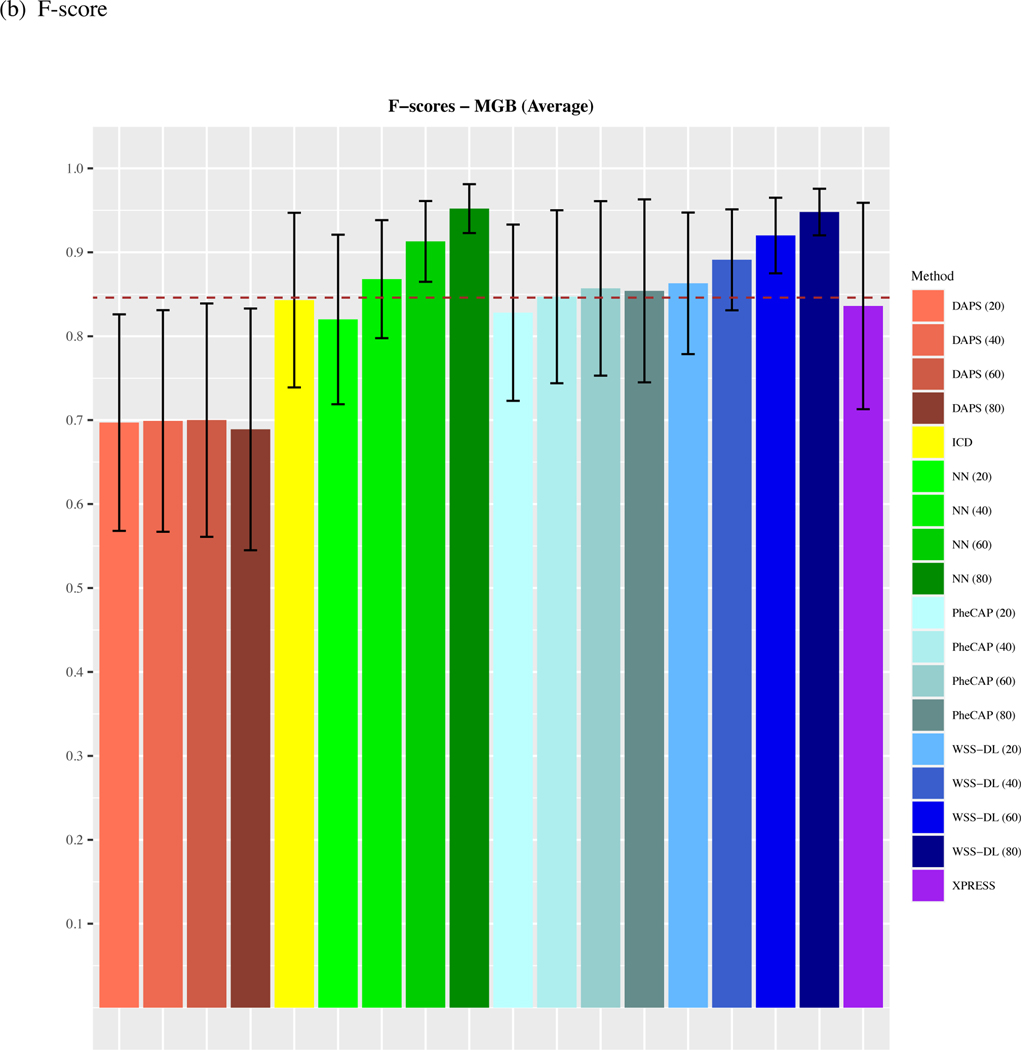
We observe that on average, across all 15 phenotypes from the MGB Biobank, WSS-DL outperformed all benchmark methods when at least n=20 of labeled samples were included for training (Figure 2). In terms of AUC, WSS-DL with n=20 labeled samples (AUC=0.91) outperformed ICD (AUC=0.87), PheCAP with n=80 (AUC=0.894), XPRESS (AUC=0.856), DAPS with n=80 (AUC=0.644), and NN with n=20 (AUC=0.842) respectively. In terms of F-score, WSS-DL with n=20 (F-score=0.863) outperformed ICD (F-score=0.843), XPRESS (F-score=0.836), DAPS with n=80 (F-score=0.699) and NN with n=20 (F-score=0.82), and yielded results only slightly lower than PheCAP’s with n=80 (F-score=0.86). Further details on specific AUCs and F-scores for each phenotype and method are provided in Figure S1 Supplementary Materials. Figures S2 and S3 in the supplementary materials contain PPVs and NPVs for each method.
Figure 2.
Comparison of averaged (a) AUCs and (b) F-scores based on ICD count, XPRESS, PheCAP (n=20 to 80), DAPS (n=20 to 80), and WSS-DL (n=20 to 80) in predicting 15 disease phenotypes using data from MGB biobank. The dash line refers to the performance of the silver-standard label.
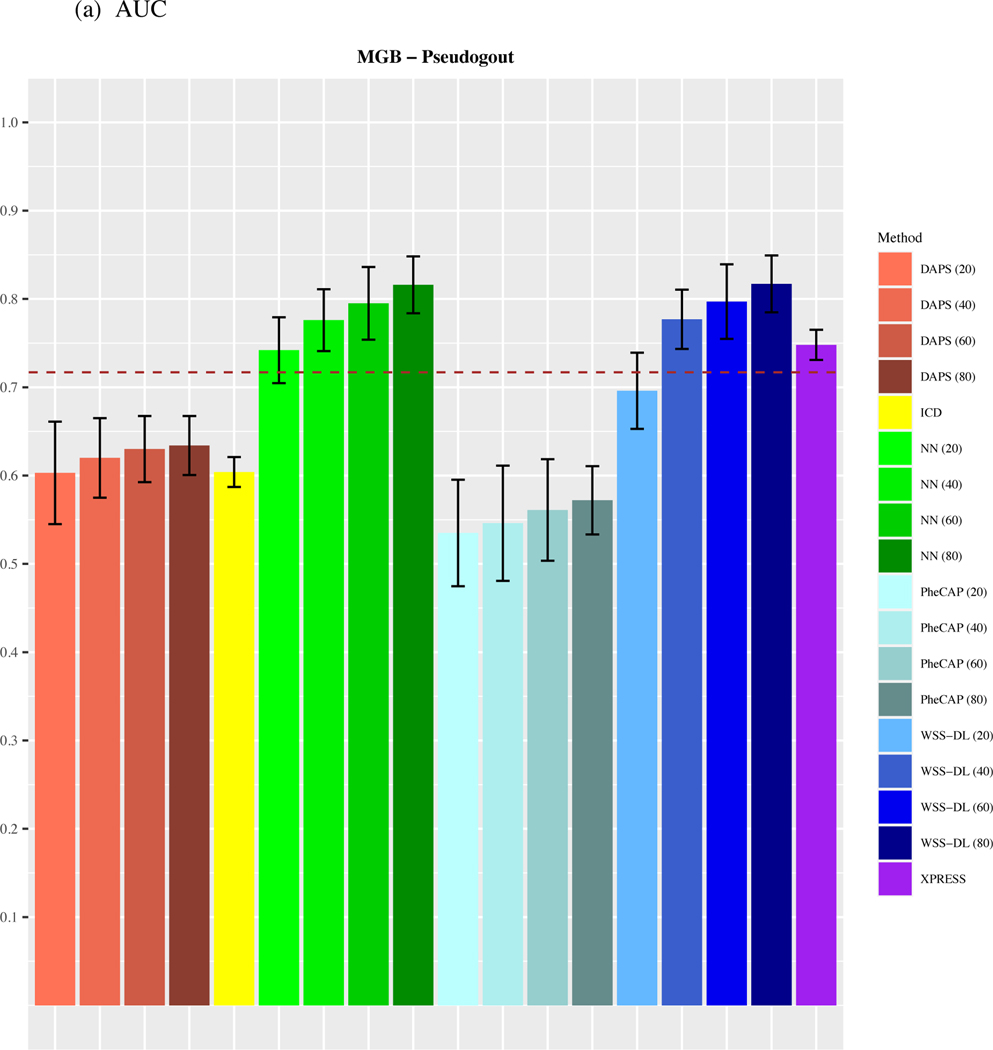
3.2. MGB-Pseudogout dataset
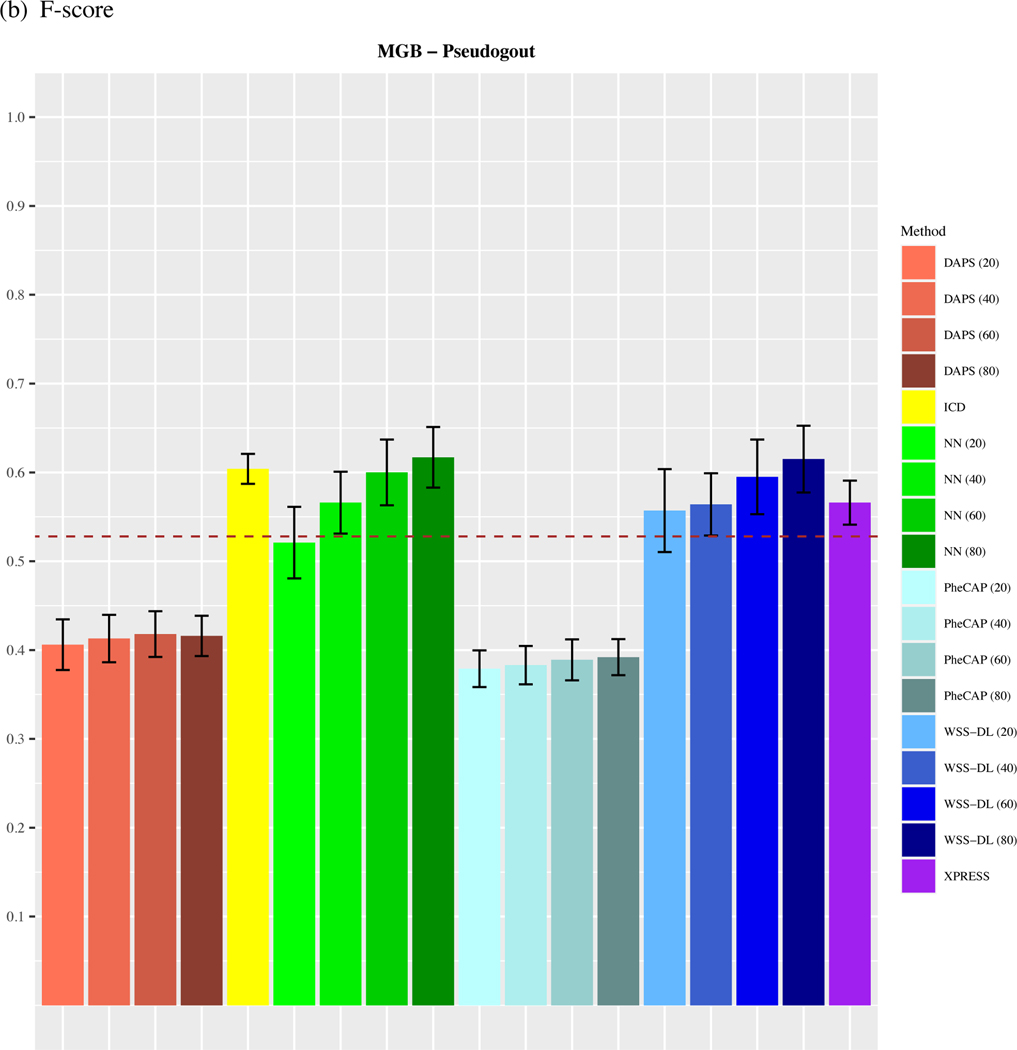
As shown in Figure 3, WSS-DL most notably outperformed all other methods in the Pseudogout dataset, yielding higher accuracy in AUC and in F-score when at least n=40 training labels were included. In terms of AUC, WSS-DL with n=40 labeled samples (AUC=0.777) outperformed ICD (AUC=0.604), PheCAP with n=80 (AUC=0.572), XPRESS (AUC=0.748), DAPS with n=80 (AUC=0.634) and NN with n=40 (AUC=0.776). In terms of F-score, WSS-DL with n=40 (F-score=0.564) outperformed ICD (F-score=0.407), PheCAP with n=80 (F-score=0.392), XPRESS (F-score=0.572), and DAPS with n=80 (F-score=0.416), respectively. Figures S4 and S5 in the supplementary materials contain PPVs and NPVs for each method.
Figure 3.
Comparison of (a) AUCs and (b) F-scores based on ICD count, XPRESS, PheCAP (n=20 to 80), DAPS (n=20 to 80), and WSS-DL (n=20 to 80) in predicting pseudogout using data from MGB EHR. The dash line refers to the performance of the silver-standard label.
3.3. BCH dataset
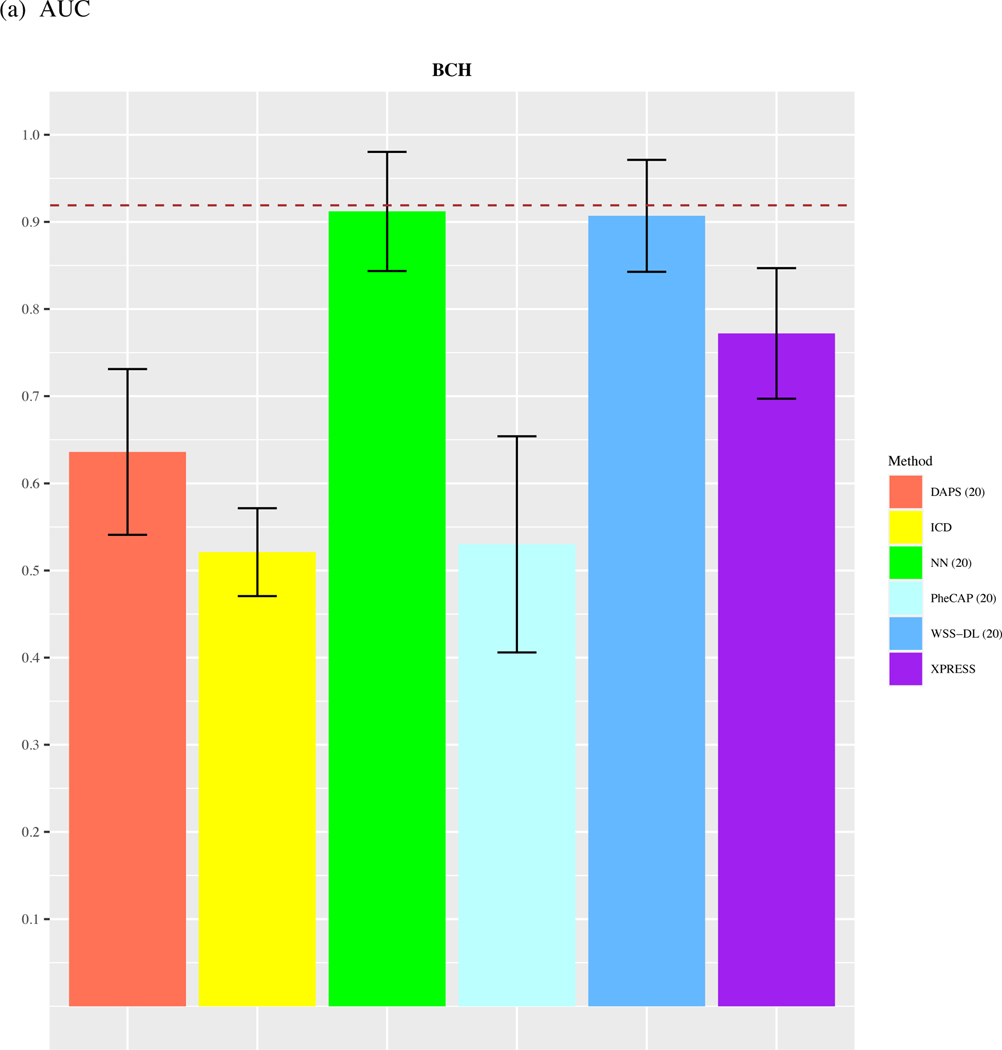
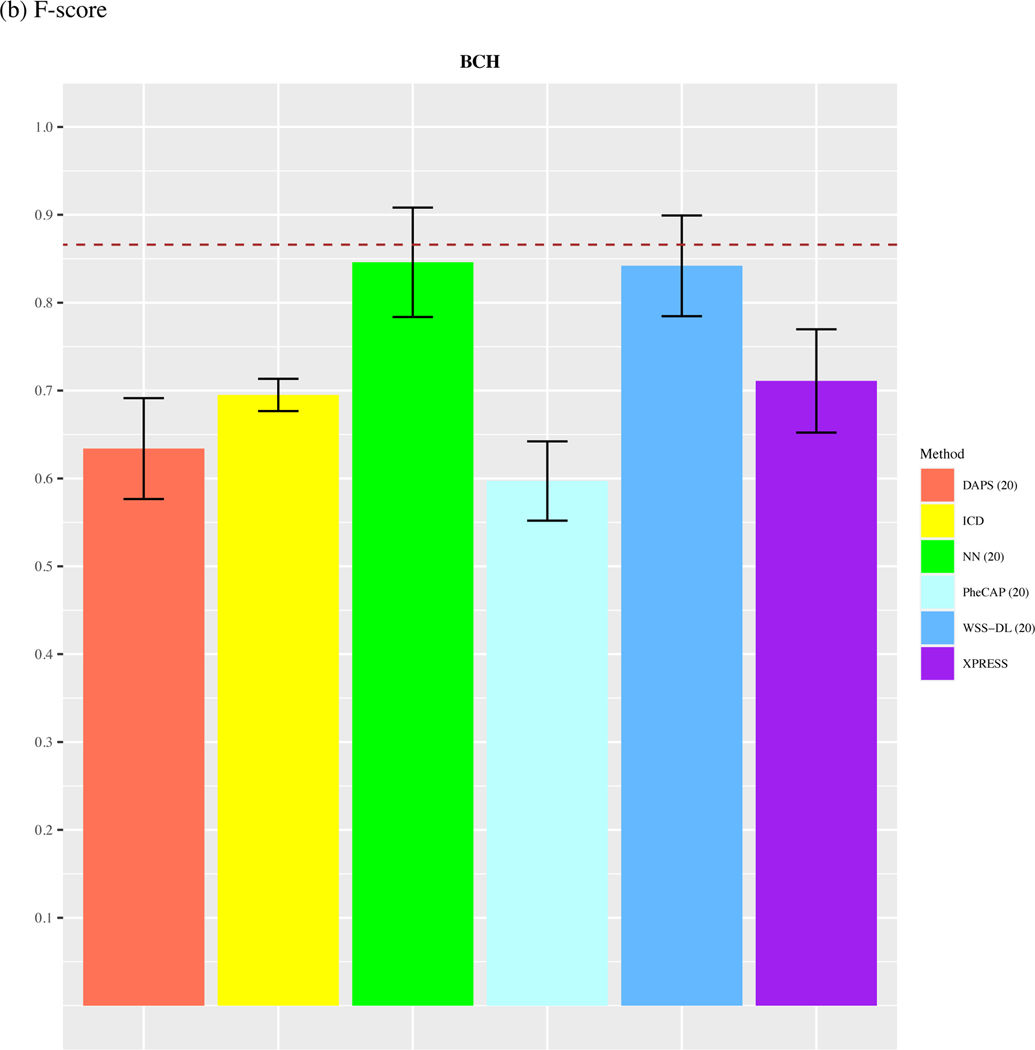
In the ARDS dataset, since the total number of available labels is 44, we only compare the performance of methods with n=20. As shown in Figure 4, we observed that WSS-DL with n=20 (AUC=0.91) outperformed ICD (AUC=0.521), PheCAP with n=20 (AUC=0.53), XPRESS (AUC=0.772), DAPS with n=20 (AUC=0.636), and performed equally well with NN with n=20 (AUC=0.91). In terms of F-score, WSS-DL with n=20 (F-score=0.842) outperformed ICD (F-score=0.693), XPRESS (F-score=0.711), DAPS with n=20 (F-score=0.634), PheCAP with n=20 (0.597), and performed equally well to NN with n=20 (F-score=0.846). Figures S6 and S7 in the supplementary materials contain PPVs and NPVs for each method.
Figure 4.
Comparison of (a) AUCs and (b) F-scores based on ICD count, XPRESS, PheCAP (n=20), DAPS (n=20), and WSS-DL (n) in predicting ARDS using data from BCH. The dash line refers to the performance of the silver-standard label.
4. DISCUSSION
In this paper, we have presented an algorithm that is able to effectively predict patient-level disease status from EHRs, using both the strengths of deep learning and weakly supervised learning. The principal objective is to learn patient level phenotype status from surrogate labels – in some cases generated from all samples in an unsupervised fashion – and a small set of provided gold standard labels, by leveraging and summarizing a large number of features without the need for manual feature engineering or feature selection. In our analyses, we found that the benefits of WSS-DL are most apparent in EHR datasets with moderate to small proportions of labeled data (n=20, or 40), thus confirming that the unlabeled samples contain additional information that is invaluable to determining the patient-level phenotype status. In particular, this information, contained in the additional EHR features, is effectively leveraged by the neural network in its feature-learning task. Additionally, diseases with low baseline ICD accuracies exhibit remarkable improvement in WSS-DL’s performance, even when a very small subset of labels is used for training. Hence, WSS-DL is most beneficial for moderate to large datasets with a small proportion of labeled samples. This suggests that, in practice, WSS-DL may be especially valuable for diagnosing rare diseases. It may also be useful in phenotypes that are difficult to detect in clinical practice, and thus for which there are very few clinical annotations. Indeed, the extent to which WSS-DL outperforms PheCAP highlights the value of including unlabeled samples to assist in the phenotyping task.
The proposed WSS-DL outperforms the other methods based only on main ICD and main NLP for classifying episodic phenotypes. Episodic phenotypes are life-long conditions, for which there is no cure, and some days are better than others. Due to the nature of episodic phenotypes, patients experience periods of fluctuating good health and ill health. As such, the disease information may not be fully captured in their main diagnosis code or main disease mentions in the medical notes. Rather, that information may be captured from other covariates, such as medication prescriptions and laboratory measures. In this situation, approaches only based on the main ICD and main NLP covariates may not reliably represent patients’ disease status, and thus would yield MAP probabilities with low predictive power. On the other hand, the proposed WSS-DL improves the classification accuracy by leveraging more clinical features. For example, the AUCs of MAP in classifying ARDS, Pseudogout and COPD are only 0.52, 0.59 and 0.64, respectively, while those of the WSS-DL are 0.91, 0.82 and 0.91, respectively. Please note that episodic phenotype annotation is just one example of a scenario where MAP fails. In general, WSS-DL performs better than MAP when the main ICD and main NLP are not sufficiently informative.
The limited performance of XPRESS in the episodic phenotypes ARDS, Pseudogout, COPD, and Stroke datasets can be attributed to the low performance of the features in themselves in predicting patient phenotype status 𝑌. Indeed, XPRESS models the relationship between and , a proxy for . As such, it indirectly quantifies how effectively the features in predict . We note that in each case, the low performance of in predicting 𝑌 is reflected in the remarkably low performance of PheCAP. In the Stroke dataset, the low quality of also contributes to the low performance of XPRESS. We note that low quality of and limited predictive power of tend to be inherent to episodic phenotypes, in that patient medical history cannot always help predict the manifestation of these phenotypes. The strong performance of WSS-DL for these episodic phenotypes, by contrast, suggests that its neural network component effectively captured latent patterns in , undetected by most clinicians and existing phenotyping algorithms (such as XPRESS and PheCAP), that are truly predictive of .
Though both WSS-DL and DAPS are semi-supervised deep learning-based algorithms, their main difference lies in the method used to summarize the EHR data in their deep learning stages. While WSS-DL uses a neural network to learn silver standard labels from the remaining non-surrogate EHR variables , DAPS’s denoising autoencoder performs dimensionality reduction on the full set of EHR features (, ) and thus does not isolate the main surrogate from . As such, DAPS does not leverage the valuable relationship between and the remaining EHR features , which in itself is central to determining patient-level phenotype status. The deep learning stage in WSS-DL involves learning silver-standard labels, a target known to be predictive of patient-level unobserved phenotype status. In DAPS, the objective is to extract the main factors of variation in the EHR features, which then are used for phenotype prediction. The notable difference in performance between WSS-DL and DAPS suggests that the current EHR data features learned by the denoising autoencoder are not very informative. This not only implies that the EHR data may be far more complex in structure than the simulation data used in the authors’ original experiments for DAPS24, but also highlights the value of silver standard labels in patient level EHR-based phenotyping. Since we observe that WSS-DL is able to predict phenotype status well in the labeled samples, with the help of the data-learning process in the silver-standard label creation and deep-learning stage, we may infer that the phenotype status predicted by WSS-DL for unlabeled samples is also reliable.
WSS-DL, though already highly effective, may be further improved if its strong benefits could extend beyond rare or sparsely labeled phenotypes. Indeed, diseases with increasing prevalence in the population, such as metabolic and vascular conditions, will greatly increase the number of medical visits and thus the demand for clinical annotations. As such, maximal accuracy in predicting these conditions is crucial. Another limitation of WSS-DL lies in the fact that its phenotyping accuracy strongly depends on the quality of the selected features included in its logistic regression model. Indeed, WSS-DL’s phenotyping accuracy significantly exceeds that of the probability-type silver standard labels (e.g., MAP probabilities) or discrete-value-type silver standard labels (e.g., {−1,0,1} as in the Pseudogout dataset), but it does not hold when using continuous P/F ratios silver standard labels in the PARDS dataset. This may be attributable to the fact that the ICD codes for PARDS are low in quality, thus decreasing the predictive power of the ICD-derived MAP probabilities included in the final logistic regression step. Probabilities derived from surrogate features directly quantify the likelihood of positive phenotype status, unlike P/F ratios which represent a patient-level clinical measure. We note that the silver standard labels created for Pseudogout also act as direct proxies for phenotype status, with −1 entries merely indicating missingness. Furthermore, the usage of continuous silver standard labels, in conjunction with the binary gold standard labels, may require two different types of loss functions in the deep learning stage to accommodate continuous silver standard and binary gold standard labels, thus complexifying the representation of the neural network’s learned features and potentially limiting its optimization and learning tasks. This may be a point of concern, in EHR datasets where the most predictive surrogate labels are continuous and main ICD codes are poor in quality or main NLP counts are not provided, meaning MAP probabilities generated from these ICD and NLP counts would not be effective silver standard labels. These problems may potentially be addressed with the usage of a more sophisticated neural network architecture in the deep learning stage. Potential models include 1) a network that adaptively adjusts the contribution of silver standard labels, giving more importance to those with lower prediction uncertainty or 2) a network that performs multitask learning, to jointly learn silver and gold standard labels. Such a network would possibly be able to extract further information from unlabeled samples, thus boosting the performance of WSS-DL even in EHR datasets with higher proportions of labeled samples (and thus less unlabeled samples). Additionally, it may be capable of balancing the optimization of two different label types (e.g. labels in the form of large continuous values vs. probabilities) more effectively.
5. CONCLUSION
By leveraging a small number of gold-standard labels and a large quantity of unlabeled data, the WSS-DL algorithm can successfully predict patient-level disease status and yield significant improvement compared with existing unsupervised approaches, while improving the efficiency and reducing the need for human annotation. We validated the WSS-DL algorithm in the MGB and BCH systems. The WSS-DL algorithm performed well at both institutions, attaining higher classification accuracy than all other competing methods.
Supplementary Material
FUNDING
This work was funded and supported by the Harvard Department of Biostatistics, the Harvard Department of Biomedical Informatics, Boston (Jamaica Plain) Veterans’ Health Affairs, and the National Science Foundation (research Graduate Fellowship Research Program). Dr. Tedeschi receives support from the National Institutes of Health (K23 AR075070, L30 AR070514).
Footnotes
SUPPLEMENTARY MATERIALS
Additional results including phenotype-specific AUCs and F-scores, averaged PPVs and NPVs, and phenotype-specific PPVs and NPVs are available in the Supplementary Materials.
CONFLICT OF INTEREST STATEMENT
None declared.
Reference
- 1.Ambinder EP Electronic health records. J. Oncol. Pract 1, 57–63 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liao KP et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ 350, h1885 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll RJ, Eyler AE & Denny JC Intelligent use and clinical benefits of electronic health records in rheumatoid arthritis. Expert Review of Clinical Immunology vol. 11 329–337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figueroa RL, Zeng-Treitler Q, Kandula S.& Ngo LH Predicting sample size required for classification performance. BMC Med. Inform. Decis. Mak 12, 8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng Y, Wang F, Zhang P.& Hu J.Risk Prediction with Electronic Health Records: A Deep Learning Approach. Proceedings of the 2016 SIAM International Conference on Data Mining (2016) doi: 10.1137/1.9781611974348.49. [DOI] [Google Scholar]
- 6.Wagholikar KB, Estiri H, Murphy M.& Murphy SN Polar labeling: silver standard algorithm for training disease classifiers. Bioinformatics 36, 3200–3206 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halpern Y, Horng S, Choi Y.& Sontag D.Electronic medical record phenotyping using the anchor and learn framework. Journal of the American Medical Informatics Association vol. 23 731–740 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agarwal V.et al. Learning statistical models of phenotypes using noisy labeled training data. J. Am. Med. Inform. Assoc 23, 1166–1173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu S.et al. Enabling phenotypic big data with PheNorm. J. Am. Med. Inform. Assoc 25, 54–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao KP et al. High-throughput multimodal automated phenotyping (MAP) with application to PheWAS. J. Am. Med. Inform. Assoc 26, 1255–1262 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y.et al. A clinical text classification paradigm using weak supervision and deep representation. BMC Med. Inform. Decis. Mak 19, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahuja Y.et al. sureLDA: A multidisease automated phenotyping method for the electronic health record. J. Am. Med. Inform. Assoc 27, 1235–1243 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Automatic phenotyping of electronical health record: PheVis algorithm. J. Biomed. Inform 117, 103746 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Krizhevsky A, Sutskever I.& Hinton GE ImageNet classification with deep convolutional neural networks. Communications of the ACM vol. 60 84–90 (2017). [Google Scholar]
- 15.Oakden-Rayner L.Exploring Large-scale Public Medical Image Datasets. Acad. Radiol 27, 106–112 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Hu Y-H, Lin W-C, Tsai C-F, Ke S-W & Chen C-W An efficient data preprocessing approach for large scale medical data mining. Technol. Health Care 23, 153–160 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Yan K, Wang X, Lu L.& Summers RM DeepLesion: automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J Med Imaging (Bellingham) 5, 036501 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jagannatha AN & Yu H.Bidirectional RNN for Medical Event Detection in Electronic Health Records. Proc Conf 2016, 473–482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nogues I.et al. Automatic Lymph Node Cluster Segmentation Using Holistically-Nested Neural Networks and Structured Optimization in CT Images. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016 388–397 (2016) doi: 10.1007/978-3-319-46723-8_45. [DOI] [Google Scholar]
- 20.Beaulieu-Jones BK, Greene CS & Pooled Resource Open-Access ALS Clinical Trials Consortium. Semi-supervised learning of the electronic health record for phenotype stratification. J. Biomed. Inform 64, 168–178 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y.et al. High-throughput phenotyping with electronic medical record data using a common semi-supervised approach (PheCAP). Nat. Protoc 14, 3426–3444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benesch C Witter DM Wilder AL et al. Inaccuracy of the International Classification of Diseases (ICD-9-CM) in identifying the diagnosis of ischemic cerebrovascular disease. Neurology.1997;49:660–4. [DOI] [PubMed] [Google Scholar]
- 23.Birman-Deych E Waterman AD Yan Y et al. . Accuracy of ICD-9-CM codes for identifying cardiovascular and stroke risk factors. Med Care.2005;43:480–85. [DOI] [PubMed] [Google Scholar]
- 24.White RH Garcia M Sadeghi B et al. . Evaluation of the predictive value of ICD-9-CM coded administrative data for venous thromboembolism in the United States. Thromb Res.2010;126:61–67. [DOI] [PubMed] [Google Scholar]
- 25.Zhan C Battles J Chiang Y-P et al. . The validity of ICD-9-CM codes in identifying postoperative deep vein thrombosis and pulmonary embolism. Jt Comm J Qual Patient Saf.2007;33:326–31. [DOI] [PubMed] [Google Scholar]
- 26.Tedeschi SK, Solomon DH, Liao KP. Pseudogout among Patients Fulfilling a Billing Code Algorithm for Calcium Pyrophosphate Deposition Disease. Rheumatology international 2018;38:1083–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai Tianrun, He Zeling, Hong Chuan, Zhang Yichi, Ho Yuk-Lam, Honerlaw Jacqueline, Geva Alon, Vidul Ayakulangara Panickan Amanda King, David R Gagnon Michael Gaziano, Cho Kelly, Katherine Liao Tianxi Cai, 2022. Scalable Relevance Ranking Algorithm via Semantic Similarity Assessment Improves Efficiency of Medical Chart Review (manuscript under revision) [DOI] [PubMed] [Google Scholar]
- 28.Yu S, Cai T.and Cai T, 2013. NILE: fast natural language processing for electronic health records. arXiv preprint arXiv:1311.6063. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.