Abstract
Background
The incubation period of SARS-CoV-2 has been estimated for the known variants of concern. However, differences in study designs and settings make comparing variants difficult. We aimed to estimate the incubation period for each variant of concern compared with the historical strain within a unique and large study to identify individual factors and circumstances associated with its duration.
Methods
In this case series analysis, we included participants (aged ≥18 years) of the ComCor case-control study in France who had a SARS-CoV-2 diagnosis between Oct 27, 2020, and Feb 4, 2022. Eligible participants were those who had the historical strain or a variant of concern during a single encounter with a known index case who was symptomatic and for whom the incubation period could be established, those who reported doing a reverse-transcription-PCR (RT-PCR) test, and those who were symptomatic by study completion. Sociodemographic and clinical characteristics, exposure information, circumstances of infection, and COVID-19 vaccination details were obtained via an online questionnaire, and variants were established through variant typing after RT-PCR testing or by matching the time that a positive test was reported with the predominance of a specific variant. We used multivariable linear regression to identify factors associated with the duration of the incubation period (defined as the number of days from contact with the index case to symptom onset).
Findings
20 413 participants were eligible for inclusion in this study. Mean incubation period varied across variants: 4·96 days (95% CI 4·90–5·02) for alpha (B.1.1.7), 5·18 days (4·93–5·43) for beta (B.1.351) and gamma (P.1), 4·43 days (4·36–4·49) for delta (B.1.617.2), and 3·61 days (3·55–3·68) for omicron (B.1.1.529) compared with 4·61 days (4·56–4·66) for the historical strain. Participants with omicron had a shorter incubation period than participants with the historical strain (–0·9 days, 95% CI –1·0 to –0·7). The incubation period increased with age (participants aged ≥70 years had an incubation period 0·4 days [0·2 to 0·6] longer than participants aged 18–29 years), in female participants (by 0·1 days, 0·0 to 0·2), and in those who wore a mask during contact with the index case (by 0·2 days, 0·1 to 0·4), and was reduced in those for whom the index case was symptomatic (–0·1 days, –0·2 to –0·1). These data were robust to sensitivity analyses correcting for an over-reporting of incubation periods of 7 days.
Interpretation
SARS-CoV-2 incubation period is notably reduced in omicron cases compared with all other variants of concern, in young people, after transmission from a symptomatic index case, after transmission to a maskless secondary case, and (to a lesser extent) in men. These findings can inform future COVID-19 contact-tracing strategies and modelling.
Funding
Institut Pasteur, the French National Agency for AIDS Research–Emerging Infectious Diseases, Fondation de France, the INCEPTION project, and the Integrative Biology of Emerging Infectious Diseases project.
Introduction
Assessing the incubation period of SARS-CoV-2 throughout emergences of variants of concern is crucial to establish duration of quarantine, advise testing strategy of contacts, enable contract tracing, and inform epidemic modelling. Initial estimates based on meta-analyses of the incubation period of SARS-CoV-2, before the emergence of variants of concern, ranged between 5·1 and 6·5 days.1, 2, 3, 4 The incubation period has been shown to differ between variants of concern, with a notably reduced incubation for people with the delta (B.1.617.2) variant.5, 6, 7 Cluster investigations published in 2021 report an even shorter incubation period in people with the omicron (B.1.1.529) variant.8, 9
The effect of individual factors, such as age, sex, COVID-19 vaccine status, or underlying conditions, on the length of the incubation period remains largely unestablished. The incubation period is reported to vary with age, with some studies finding an increase in incubation with age. However, a study based on contact-tracing conducted in China found people aged 41–60 years had a shorter incubation period than younger and older people.10, 11, 12 External factors, such as the environment of transmission, the nature of contact with the index case, or the viral load of the index case, might also contribute (possibly through the viral inoculum, which has been shown to affect secondary transmission risk),13, 14 but there is no evidence about how these external factors might affect duration of the incubation period.
Research in context.
Evidence before this study
Iterative assessment of the SARS-CoV-2 incubation period is necessary throughout emergences of variants of concern to adapt mitigation strategies and inform epidemic modelling. A previous meta-analysis of 99 studies last updated in January, 2021, estimated that the mean incubation period was 6·38 days. We searched PubMed without any language restrictions for studies published between Jan 1, 2021, and March 23, 2022, using the keywords “COVID-19” AND “incubation”. We included epidemiological studies reporting an estimate of the mean or median SARS-CoV-2 incubation period with a minimum sample size of ten. The search retrieved 670 results, of which 16 reports of 13 studies (with sample sizes between ten and 1676 participants) were eligible for inclusion. Six studies had data on variants of concern (four studies on the delta [B.1.617.2] variant, one on the alpha [B.1.1.7] variant, and one on the beta [B.1.351] variant). Most studies were rated as good or fair in the quality assessment. Four studies investigated the effects of age or initial viral load on duration of the incubation period. We did not find any studies on the effects of other individual factors, such as sex, vaccine status, mask wearing at the time of contact with an index case, symptomatic status of the index case, or setting where transmission occurred.
Added value of this study
We analysed data from a nationwide study conducted in France that used consistent methods throughout the emergence of multiple variants of concern, allowing comparison between variants. Our study provides evidence of a reduced incubation period for the omicron (B.1.1.529) variant of concern compared with all previously predominant variants while adjusting for individual factors (eg, age and vaccine status). We found evidence for a reduced incubation period in younger people and in situations of high viral inoculum (ie, not wearing a mask during contact or when the index case was symptomatic).
Implications of all the available evidence
Evidence of a notably reduced SARS-CoV-2 incubation period of the currently globally predominant omicron variant compared with the historical strain and other variants of concern has important implications for adapted contact-tracing strategies, which should aim to reduce the time from symptom onset to contact tracing as much as possible to achieve containment. The evidence can also inform infectious disease modelling, which is crucial in estimations of the upcoming effects of COVID-19 on health-care systems in many countries. Although evidence indicates an effect of age on the incubation period, with a shorter incubation period in younger people, the effects of vaccine status and mask wearing at the time of contact with the index case warrant further investigation.
To assess the relative contribution of factors affecting the SARS-CoV-2 incubation period, data documenting incubation periods for a large number of cases, reported over long time periods, and with a stable study design would ideally be used. However, this approach is rarely possible. Meta-analyses have mostly relied on studies with relatively small sample sizes and different study designs. The few studies with more than 1000 participants often date back to the early months of the COVID-19 pandemic.15, 16
In this study, we analysed a dataset documenting incubation periods of SARS-CoV-2 among participants of the large ComCor study, a nationwide case-control study conducted in France for 16 months (October, 2020, to February, 2022). We aimed to characterise the determinants of the SARS-CoV-2 incubation period, including variants of concern, individual factors, and circumstances of infection.
Methods
Study design and participants
The methods of the ComCor study have been described elsewhere.6, 17, 18 Briefly, the ComCor study is a nationwide case-control study of adults aged 18 years or older with a SARS-CoV-2 diagnosis (cases) identified via the Caisse Nationale de l'Assurance Maladie (National Health Insurance Fund) database, which stores all diagnoses of SARS-CoV-2 in France, and control participants (ie, individuals who had never tested positive for SARS-CoV-2 up to February, 2021, when eligibility was broadened to include all adults without ongoing SARS-CoV-2 infection), selected from a representative panel of the French population by Ipsos, a market and opinion research company. Participants receive information about the study online before completing a questionnaire.
For this case series analysis of the ComCor study, we only analysed people who had SARS-CoV-2 after a single encounter with a known index case (ie, cases, not controls) between Oct 27, 2020, and Feb 4, 2022. We excluded participants who tested positive for SARS-CoV-2 after intrahousehold transmission (they were assumed to have had repeated contact with the index case, preventing the incubation period from being established), had an incubation period that could not be defined (including those who were asymptomatic by study completion), had an incubation period longer than 15 days,1, 3, 19 did not report doing a reverse-transcription-PCR (RT-PCR) test, or had an unidentified or non-variant-of-concern non-historical strain of SARS-CoV-2 and tested positive outside periods of large predominance of one strain.
For cases included in our analysis, the incubation period was defined as the number of days from contact with the index case to symptom onset.
We obtained informed consent from all participants. The ComCor study received ethical approval from Comité de Protection des Personnes Sud Ouest et Outre Mer (the Committee for the Protection of Persons South West and Overseas) on Sept 21, 2020. The data protection authority Commission Nationale de l'Informatique et des Libertés (the National Commission on Informatics and Liberty) authorised the processing of ComCor study data on Oct 21, 2020. The ComCor study is registered with ClinicalTrials.gov (NCT04607941).
Procedures
Sociodemographic characteristics, exposure information, circumstances of infection if the index case was identified, testing (ie, RT-PCR or supervised rapid antigen test, not self-tests), COVID-19 vaccination details, smoking status, information about underlying health conditions (eg, diabetes, chronic respiratory disease, hypertension, or coronary artery disease), and BMI were obtained via the online questionnaire between Oct 27, 2020, and Feb 4, 2022 (appendix p 12). Sex data were self-reported, with the options “Female” or “Male” provided.
Variants were established either through variant typing after RT-PCR testing, or by matching the time that a positive test was reported with the predominance of a specific variant at that time. Variant typing was implemented in France by Santé Publique France for most positive RT-PCR tests from January, 2021, onwards. Individual test data were provided by participants if available. Details of mutations used to identify the alpha (B.1.1.7), beta (B.1.351), gamma (P.1), and delta (B.1.617.2) variants via RT-PCR screening are available elsewhere.6, 18 This strategy did not allow for differentiation between the beta and the gamma variants. However, FLASH sequencing studies conducted once per week or once every 2 weeks since January, 2021, on a random sample of positive SARS-CoV-2 tests in France showed that the beta variant largely predominated among tests with a positive result for beta or gamma after screening (eg, on March 2, 2021, beta represented 5·6% of positive samples and gamma represented 0·1%).20 Between Dec 1, 2021, and Jan 16, 2022, we defined cases included in the study with tests showing omicron (B.1.1.529)-specific screening targets (ie, 69–70del, Lys417Asn, Ser371Leu and Ser373Pro, or Gln493Arg), or without the Glu484Lys or the Leu425Arg mutations, as omicron cases. According to FLASH studies, the B.1.640 variant, which has neither the Glu484Lys or the Leu452Arg mutations, represented less than 1% of positive samples in December, 2021, and January, 2022. The routine variant typing strategy did not allow for differentiation between the BA.1 and BA.2 omicron subvariants. The FLASH studies found that BA.1 was largely predominant during the study period; in France, it represented 99·7% of positive samples on Jan 3, 2022, and 90·9% of samples on Jan 31, 2022.21 Because of the absence of variant typing before early 2021 and the reduced proportion of samples typed in periods of high incidence—particularly during the omicron wave in the winter of 2021–22 or in periods of large predominance of one strain—we assumed that people with an unestablished variant had the predominant strain: historical strain for tests conducted before Jan 7, 2021; delta variant for tests conducted between Sept 1, 2021, and Nov 30, 2021; and omicron variant for tests conducted between Jan 17, 2022, and Feb 4, 2022. A predominance period was not defined for the alpha, beta, and gamma variants as they circulated mostly concomitantly during the first 6 months of 2021.22
Statistical analysis
We used ordinary least-squares multivariable linear regression to estimate associations with the incubation period, with adjustment for the covariables of age, sex, underlying conditions (ie, diabetes, hypertension, chronic respiratory disease, coronary artery disease, or underweight defined as a BMI <18·5 kg/m2), smoking status, COVID-19 vaccine status (ie, number of injections and time elapsed since last injection in three categories: <90 days, 90–179 days, and ≥180 days), and characteristics of the index case and onward transmission (ie, symptoms of the index case, duration of the interaction, outdoor or indoor setting, and mask wearing by the participant and the person identified as the index case) in all eligible cases included in this study. During the study period, vaccines available in France were mRNA vaccines (ie, BNT162b2 [Pfizer–BioNTech] and mRNA-1273 [Moderna]) and viral vector vaccines (ie, ChAdOx1 nCoV-19 [AstraZeneca] and Ad26.CoV2.S [Johnson & Johnson]). We applied robust standard errors to account for possible heteroscedasticity. Because of the large sample size, we considered the model to be too robust to violate the normality of residuals' distribution.
We estimated the probability distribution of the true incubation period after correction for probable over-reporting at 7 days due to self-reported data. This calculation was done by estimating the probability of reporting 7 days if the true incubation period was 6 or 8 days (p1) or 5 or 9 days (p2). Poisson, negative-binomial, log-normal, and gamma distributions were considered. We considered one scenario in which p2 was estimated and one scenario in which p2=0. We estimated parameters of each distribution jointly with p1 and p2 using a Monte Carlo Markov Chains algorithm. The deviance information criterion (DIC) was used for model selection (appendix p 12).
We did several sensitivity analyses to test the validity of our results. The first aimed to examine the effect of over-reporting incubation periods of 7 days in the analysis of factors associated with the incubation period. In this analysis, we randomly reassigned 7-day incubation periods to a number of days between 5 days and 9 days using probabilities derived from the distribution retained after model selection on DIC, and did the linear regression analysis. This process was repeated 15 times, and the coefficients and 95% CIs of the 15 regression analyses were averaged for comparison with the initial linear regression analysis results. The second sensitivity analysis aimed to compare the results of a Poisson regression with those of the normal linear regression as the dependent variable, the incubation period, takes only discrete values from 1 to 15. The third sensitivity analysis consisted of restricting the analysis to participants whose index cases were confirmed by a positive SARS-CoV-2 test.
The questionnaire did not allow advancement to the next question unless the previous questions were answered. Therefore, we had no missing data for any items except one. We analysed participants with missing information regarding date of last vaccine dose in a separate category.
Statistical analyses were done with Stata (version 16.0) and R (version 4.2.1).
Role of the funding source
The funders had no role in the study design, collection, analysis and interpretation of data, writing of the report, or decision to submit for publication.
Results
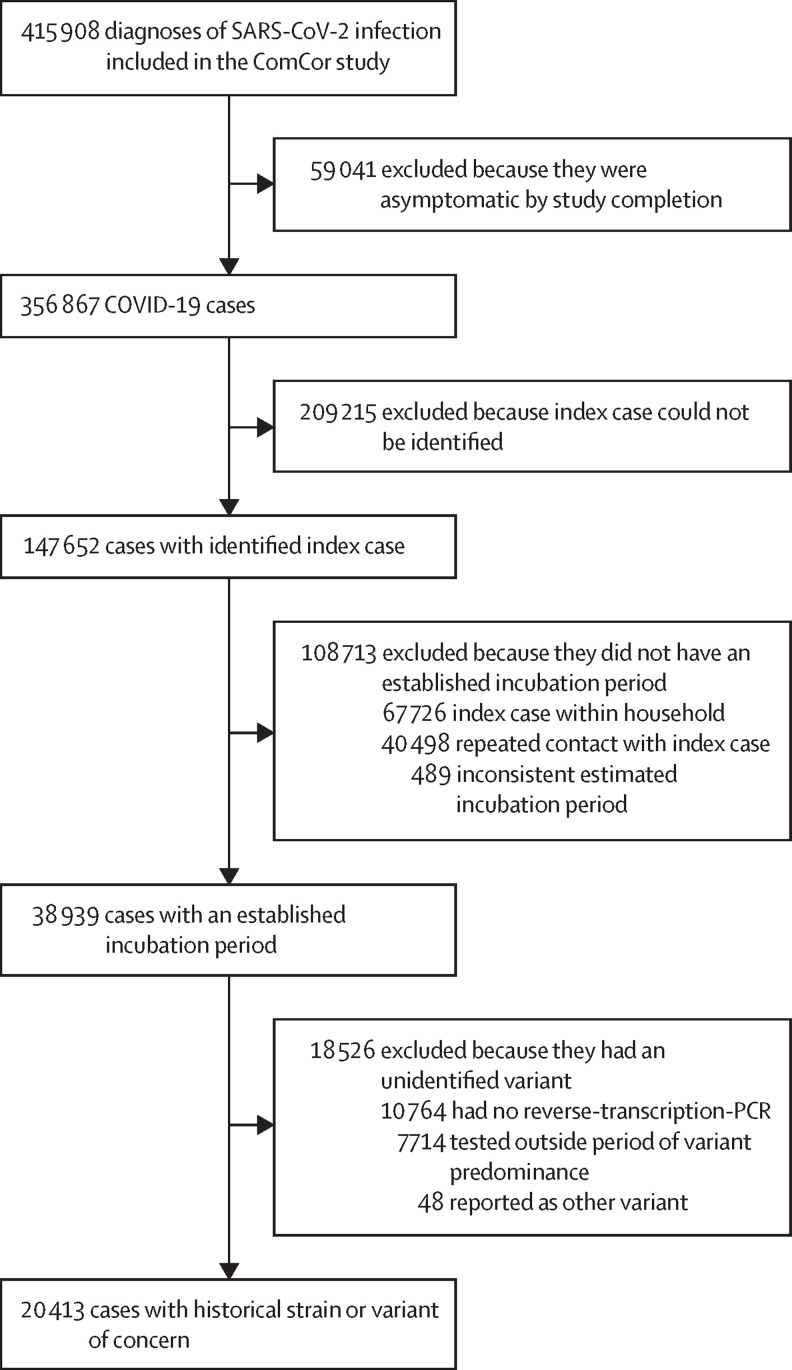
356 867 COVID-19 cases were included in the ComCor study (figure 1 ) between Oct 27, 2020, and Feb 4, 2022. Among these participants, the incubation period could be defined in 38 939 (10·9%) participants who had a single encounter with the index case outside the household. After excluding participants who did not report an RT-PCR and participants with an undetermined or non-variant-of-concern, non-historical strain who tested outside periods of large predominance of one strain, we included 20 413 participants who had the historical strain, alpha variant, beta variant, gamma variant, delta variant, or omicron variant. Variant was established through variant typing on RT-PCR for 13 665 participants and by matching with the predominant strain at the time of the test for 6748 participants.
Figure 1.
Selection of SARS-CoV-2 cases
The ComCor case-control study was conducted in France between October, 2020, and February, 2022.
The study population (n=20 413) was predominantly women and young (ie, aged ≤49 years), with few participants older than 70 years (table 1 ). The most frequent underlying conditions were chronic respiratory disease and hypertension. Approximately a third of participants reported that the index case was symptomatic at the time of contact. A large majority of participants reported a positive test in the index case. Most often, neither the participant or the index case wore a mask during their interaction (table 1). Further demographic description across different variants is provided in the appendix (p 2).
Table 1.
Demographic data for participants with COVID-19 with identified incubation period and variant
| Participants (n=20 413) | ||
|---|---|---|
| Health-care worker | 2766 (13·6%) | |
| Sex | ||
| Female | 13 760 (67·4%) | |
| Male | 6653 (32·6%) | |
| Variant | ||
| Historical | 7539 (36·9%) | |
| Alpha | 5133 (25·1%) | |
| Beta or gamma | 453 (2·2%) | |
| Delta | 4606 (22·6%) | |
| Omicron | 2682 (13·1%) | |
| Age, years | ||
| 18–29 | 4269 (20·9%) | |
| 30–39 | 5526 (27·1%) | |
| 40–49 | 4235 (20·7%) | |
| 50–59 | 3436 (16·8%) | |
| 60–69 | 2228 (10·9%) | |
| ≥70 | 719 (3·5%) | |
| Smoking status | ||
| Non-smoker, no nicotine substitution | 16 465 (80·7%) | |
| Non-smoker, nicotine substitution | 169 (0·8%) | |
| Non-smoker, electronic cigarette only | 801 (3·9%) | |
| <10 cigarettes per day | 1537 (7·5%) | |
| 10–20 cigarettes per day | 1019 (5·0%) | |
| >20 cigarettes per day | 422 (2·1%) | |
| Underlying conditions | ||
| Chronic respiratory disease | 1666 (8·2%) | |
| Hypertension | 1657 (8·1%) | |
| Underweight | 667 (3·3%) | |
| Diabetes | 459 (2·2%) | |
| Coronary artery disease | 165 (0·8%) | |
| COVID-19 vaccine status | ||
| Unvaccinated | 13 868 (67·9%) | |
| One dose, <90 days since last injection | 354 (1·7%) | |
| One dose, 90–179 days since last injection | 143 (0·7%) | |
| One dose, ≥180 days since last injection | 97 (0·5%) | |
| Two doses, <90 days since last injection | 642 (3·1%) | |
| Two doses, 90–179 days since last injection | 3266 (16·0%) | |
| Two doses, ≥180 days since last injection | 942 (4·6%) | |
| Three doses, <90 days since last injection | 903 (4·4%) | |
| Three doses, 90–179 days since last injection | 42 (0·2%) | |
| Three doses, ≥180 days since last injection | 16 (0·1%) | |
| Four doses, <90 days since last injection | 3 (<0·1%) | |
| Undated last dose of vaccine | 137 (0·7%) | |
| Previous SARS-CoV-2 infection | ||
| No | 19 744 (96·7%) | |
| Yes, virologically or serologically confirmed | 512 (2·5%) | |
| Yes, diagnosed on clinical evaluation only | 157 (0·8%) | |
| Symptoms in index case | 6819 (33·4%) | |
| Positive test for index case | 18 005 (88·2%) | |
| Mask wearing | ||
| Neither participant or index case | 15 911 (77·9%) | |
| Index case only | 495 (2·4%) | |
| Participant only | 1694 (8·3%) | |
| Both participant and index case | 2313 (11·3%) | |
| Setting of transmission | ||
| Indoors with closed windows | 15 117 (74·1%) | |
| Indoors with open windows | 4036 (19·8%) | |
| Outdoors | 1260 (6·2%) | |
Data are n (%). COVID-19 cases included in the ComCor case-control study between October, 2020, and February, 2022.
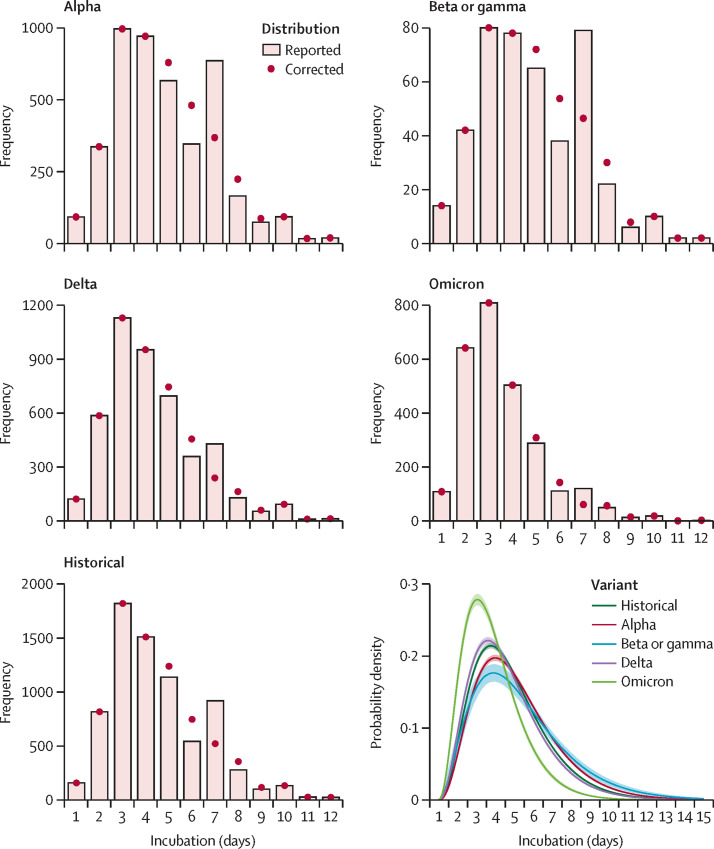
Overall, mean incubation time was 4·54 days (95% CI 4·51–4·57) and varied across different variants of concern (table 2 ). Compared with the historical strain (4·61 days, 4·56–4·66), the alpha (4·96 days, 4·90–5·02) and beta or gamma (5·18 days, 4·93–5·43) variants of concern were associated with a longer incubation period, whereas participants infected by the delta (4·43 days, 4·36–4·49) and omicron (3·61 days, 3·55–3·68) variants of concern had shorter incubation periods (non-parametric Kruskal–Wallis test p=0·0001).
Table 2.
SARS-CoV-2 incubation period by variant, observed and corrected for day 5 to day 9
| N |
Observed, days |
Correction for days 5 to 9, days |
|||
|---|---|---|---|---|---|
| Mean (95% CI) | SD | Mean (95% credible interval) | SD | ||
| Historical | 7539 | 4·61 (4·56–4·66) | 2·24 | 4·57 (4·52–4·62) | 2·22 |
| Alpha | 5133 | 4·96 (4·90–5·02) | 2·32 | 4·92 (4·85–4·98) | 2·31 |
| Beta or gamma | 453 | 5·18 (4·93–5·43) | 2·72 | 5·15 (4·90–5·40) | 2·72 |
| Delta | 4606 | 4·43 (4·36–4·49) | 2·19 | 4·40 (4·33–4·46) | 2·17 |
| Omicron | 2682 | 3·61 (3·55–3·68) | 1·82 | 3·58 (3·52–3·65) | 1·77 |
| Total | 20 413 | 4·54 (4·51–4·57) | 2·25 | 4·50 (4·47–4·53) | 2·23 |
A gamma distribution was fitted to estimate probability of rounding the estimation to 7 days if the underlying incubation period was 6 days or 8 days, or 5 days or 9 days. The included variants are alpha (B.1.1.7), beta (B.1.351), gamma (P.1), delta (B.1.617.2), and omicron (B.1.1.529).
The gamma distribution corrected for p1 and p2 was selected on the basis of deviance information criterion (appendix p 3). The distribution of the incubation period by variant, both observed and corrected for the 7-day effect (applying p1 and p2), and the gamma distribution corrected for p1 and p2 by variant are shown (figure 2 ; appendix p 3). After correction for the 7-day effect, estimates of mean incubation period were shorter overall (4·50 days, 4·47–4·53) and for each variant than before correction for the 7-day effect (table 2).
Figure 2.
SARS-CoV-2 incubation period by variant responsible for infection, observed and corrected for day 5 to day 9 of incubation
Estimation obtained from 20 413 COVID-19 cases included in the ComCor case-control study between October, 2020, and February, 2022, with an identified incubation period and variant. Gamma distribution parameters and probabilities of rounding to the nearest week (ie, 7 days) if the incubation period was 6 days or 8 days (probability p1) or 5 days or 9 days (probability p2) were jointly estimated by fitting the model to data with a Monte Carlo Markov Chains algorithm. Probabilities p1 and p2 were applied to establish corrected distributions. The included variants are alpha (B.1.1.7), beta (B.1.351), gamma (P.1), delta (B.1.617.2), and omicron (B.1.1.529).
In the multivariable linear regression analysis, the incubation period was shorter in people with omicron and was longer in people with the alpha or beta or gamma variants than in people with the historical strain after adjustment for age, sex, smoking status, COVID-19 vaccine status, and setting of transmission (table 3 ). The incubation period increased with increasing age, with the oldest participants (age ≥70 years) having an incubation period 0·4 days (0·2 to 0·6) longer on average than the youngest participants (age 18–29 years). The incubation period was reduced if the index case had symptoms of COVID-19 at the time of the contact, but was moderately increased in settings where either the participant or both the participant and the index case wore a mask. Female participants had a longer incubation period than male participants. Smoking was associated with an increased incubation period, with an effect increasing with daily cigarette count; the same effect was not observed with nicotine substitution.
Table 3.
Factors and their association with duration (days) of SARS-CoV-2 incubation period in univariable and multivariable linear regression models
| Univariable analysis | Multivariable analysis | |
|---|---|---|
| Sex | ||
| Male | 0 (ref) | 0 (ref) |
| Female | 0·04 (−0·03 to 0·1) | 0·1 (0·02 to 0·2) |
| Variant | ||
| Historical strain | 0 (ref) | 0 (ref) |
| Alpha | 0·3 (0·3 to 0·4) | 0·3 (0·3 to 0·4) |
| Beta or gamma | 0·6 (0·3 to 0·8) | 0·6 (0·3 to 0·8) |
| Delta | −0·2 (−0·3 to −0·1) | −0·1 (−0·2 to 0·03) |
| Omicron | −1·0 (−1·1 to −0·9) | −0·9 (−1·0 to −0·7) |
| Age, years | ||
| 18–29 | 0 (ref) | 0 (ref) |
| 30–39 | 0·1 (−0·01 to 0·2) | 0·1 (0·00 to 0·2) |
| 40–49 | 0·2 (0·1 to 0·3) | 0·2 (0·1 to 0·3) |
| 50–59 | 0·2 (0·1 to 0·3) | 0·2 (0·1 to 0·3) |
| 60–69 | 0·4 (0·3 to 0·5) | 0·4 (0·3 to 0·5) |
| ≥70 | 0·3 (0·1 to 0·5) | 0·4 (0·2 to 0·6) |
| Smoking status | ||
| Non-smoker, no nicotine substitution | 0 (ref) | 0 (ref) |
| Non-smoker, nicotine substitution | −0·2 (−0·5 to 0·2) | −0·2 (−0·5 to 0·2) |
| Non-smoker, electronic cigarette only | 0·1 (−0·1 to 0·3) | 0·2 (0·00 to 0·3) |
| <10 cigarettes per day | 0·1 (−0·04 to 0·2) | 0·1 (0·02 to 0·3) |
| 10–20 cigarettes per day | 0·2 (0·02 to 0·3) | 0·2 (0·1 to 0·4) |
| >20 cigarettes per day | 0·3 (0·1 to 0·5) | 0·4 (0·1 to 0·6) |
| COVID-19 vaccine status | ||
| Unvaccinated | 0 (ref) | 0 (ref) |
| One dose, <90 days since last injection | −0·2 (−0·5 to 0·02) | −0·2 (−0·4 to 0·1) |
| One dose, 90–179 days since last injection | −0·9 (−1·3 to −0·6) | −0·3 (−0·7 to 0·1) |
| One dose, ≥180 days since last injection | −1·0 (−1·4 to −0·6) | −0·5 (−0·9 to −0·1) |
| Two doses, <90 days since last injection | −0·2 (−0·4 to −0·1) | 0·1 (−0·1 to 0·3) |
| Two doses, 90–179 days since last injection | −0·6 (−0·7 to −0·5) | −0·1 (−0·3 to −0·03) |
| Two doses, ≥180 days since last injection | −0·7 (−0·8 to −0·5) | −0·2 (−0·4 to −0·05) |
| Three doses, <90 days since last injection | −0·8 (−0·1 to −0·7) | 0·01 (−0·2 to 0·2) |
| Three doses, 90–179 days since last injection | −0·8 (−1·5 to −0·2) | −0·3 (−0·9 to 0·3) |
| Three doses, ≥180 days since last injection | −1·2 (−2·3 to −0·1) | −0·6 (−1·3 to 0·1) |
| Four doses, <90 days since last injection | −0·4 (−2·3 to 2·1) | 0·5 (−2·3 to 3·3) |
| Undated last dose of vaccine | −0·2 (−0·6 to 0·2) | 0·2 (−0·3 to 0·7) |
| Previous SARS-CoV-2 infection | ||
| No | 0 (ref) | 0 (ref) |
| Yes, virologically or serologically confirmed | −0·5 (−0·7 to −0·3) | 0·01 (−0·2 to 0·2) |
| Yes, diagnosed on clinical evaluation only | −0·1 (−0·5 to 0·2) | −0·02 (−0·4 to 0·3) |
| Symptoms in index case | ||
| No | 0 (ref) | 0 (ref) |
| Yes | −0·1 (−0·2 to −0·1) | −0·1 (−0·2 to −0·1) |
| Mask wearing | ||
| Neither participant or index case | 0 (ref) | 0 (ref) |
| Index case only | 0·1 (−0·2 to 0·3) | −0·01 (−0·2 to 0·2) |
| Participant only | 0·3 (0·2 to 0·5) | 0·2 (0·1 to 0·4) |
| Both participant and index case | 0·2 (0·1 to 0·3) | 0·1 (0·03 to 0·2) |
| Setting of transmission | ||
| Indoors with closed windows | 0 (ref) | .. |
| Indoors with open windows | 0·00 (−0·1 to 0·1) | .. |
| Outdoors | 0·1 (−0·1 to 0·2) | .. |
| Underlying conditions | ||
| Chronic respiratory disease | 0·1 (−0·1 to 0·2) | .. |
| Hypertension | 0·2 (0·04 to 0·3) | .. |
| Underweight | −0·2 (−0·4 to −0·1) | .. |
| Diabetes | 0·2 (−0·1 to 0·4) | .. |
| Coronary artery disease | 0·03 (−0·3 to 0·4) | .. |
Data are coefficent (95% CI). The multivariable linear regression model was adjusted for all variables in the table. Underlying conditions and setting of transmission were not included in the multivariable model due to the absence of statistical significance. Robust SEs were applied to account for potential heteroscedasticity.
In an alternative regression model, we analysed the effect of the type of interaction environment (eg, friends, family, or sports activity) on the incubation period. We identified a shorter incubation period in participants infected within a group of friends than in people infected in a family environment (ie, the index case was a family member; –0·2 days, –0·3 to –0·1; appendix pp 4–5).
We did several sensitivity analyses to test the validity of our results. Regarding the effect of redistributing the observed incubation periods of 7 days over days 5 to 9 using probabilities derived from the distribution corrected for p1 and p2, differences between the initial estimates and the redistributed estimates were minimal (appendix p 6). In the sensitivity analyses comparing a Poisson regression model with a linear regression model for the multivariable analysis, the Poisson regression model gave identical results to the linear regression model, expressing the difference in incubation periods between the reference category and comparison category as a ratio of number of days rather than as a difference in number of days (appendix pp 7–9). As a difference in number of days provides a more readily interpretable comparison between the two groups than a ratio, we favoured the linear regression for the presentation of our results. Finally, the analysis restricted to the 18 005 (88·2%) of 20 413 participants for whom the person identified as index case had a positive test provided similar results for the mean (4·52 days [SD 2·22], 95% CI 4·49–4·55) of the incubation period. Results of the univariable and multivariable linear regression analyses in these participants were consistent with those of the main analysis (appendix pp 10–11).
Discussion
To our knowledge, our analysis is the largest study to date to evaluate the incubation period of SARS-CoV-2 with consistent data collection methods used throughout the emergence of consecutive variants of concern. We found a reduced incubation period for the omicron variant, in younger people, in men (to a lesser extent), in participants who did not smoke, in participants not wearing a mask when transmission occurred, or if the index case was symptomatic. These results remained after adjusting for several individual factors, including COVID-19 vaccine status.
The association of the incubation period with mask wearing and symptomatic status of the index case leads to important questions about the role of viral inoculum in the natural history of SARS-CoV-2 infection, considering mask wearing in individuals exposed to SARS-CoV-2 is associated with reduced viral inoculum23, 24 and symptomatic status is associated with increased viral inoculum.13 Increased viral load in index cases has been documented to increase the risk of secondary transmission13, 14 but not the severity of secondary cases,25 whereas the effect on incubation period remains largely undocumented. A study of a series of clusters of COVID-19 found evidence of a reduced incubation period in secondary cases who had an increased viral load, but high viral load in these participants was not associated with high viral load in the index cases.13 We did not have any further details on the type of masks worn during the contact or on the quality of mask wearing. The increased incubation period observed with smoking and electronic cigarette use in our study could be related to reduced viral inoculum if confounded by improved ventilation habits among smokers.
Our findings about the omicron variant having a shorter incubation period than other variants are consistent with available data on an omicron outbreak in a restaurant in Norway and on a cluster in Nebraska, USA, for which the median incubation period was 3 days.8, 9 However, sample sizes in these studies were small and recruitment of participants among clusters might have biased the estimation, as suggested by our findings showing that the incubation period is reduced when transmission occurs among a group of friends. The shortening of the incubation period of the delta variant compared with the historical strain was less substantial than previously estimated in a Chinese contact-tracing study.7 However, that study was conducted on the wild-type virus, whereas the historical strain in France contained the Asp614Gly mutation, possibly affecting comparability.26 A human challenge study conducted in previously uninfected and unvaccinated healthy young adults showed COVID-19 symptom onset from 2 days after inoculation in some infected participants, with an increase in symptom score until 4 days after inoculation.27 This relatively short incubation period compared with our findings, although coherent with a reduced incubation period in younger populations, could be partly explained by the prospective and protocol-driven recording of symptoms in the human challenge study.
Our study found an association between age and incubation period, with increased incubation periods in the oldest individuals (aged ≥60 years), which is consistent with previous findings. Tan and colleagues10 found an incubation period of 8 days for people aged 70 years or older compared with an incubation period of 5 days for people younger than 70 years, and Li and colleagues12 reported a positive correlation between age and duration of the incubation period. We did not find, as Cheng and colleagues11 did in a population of 218 participants in China in 2020, a reduced incubation period for people aged 41–60 years. Different methods and a smaller sample size might explain those discrepancies.
One of the limitations of our study is that we obtained data from self-reported questionnaires. We restricted our analysis to incubation periods within a realistic range (up to 15 days) and most (88·2%) participants reported a positive SARS-CoV-2 test in the person identified as the index case. Results were almost identical after restricting the analysis to people who reported a positive test in the index case (appendix pp 10–11). The identification of the index case might have been erroneous for some of the participants due to the possibility of asymptomatic transmission or exposure to multiple sources of infection. Nonetheless, we believe this limitation affected the computation of the incubation period for only a small number of participants. The over-representation of incubation periods of 7 days is probably a consequence of questionnaire design, as many participants who had an incubation period between 5 days and 9 days might have biased their answer to 7 days (ie, 1 week). This bias seemed more prevalent among participants infected with the alpha, beta, or gamma variant than in those infected with the delta or omicron variant. This discrepancy was most probably a consequence of an increased underlying mean incubation period in participants infected with the alpha, beta, or gamma variant, leading to an increased number of participants likely to round their answer to 1 week. We found no significant differences among populations infected with different variants that could otherwise explain differences in self-reporting quality, although we cannot rule out association with an unmeasured confounder. We developed models of distribution of the incubation period to consider this bias and obtained a slightly lower corrected mean than the observed distribution. We noted only minor changes in the estimates of the linear regression model for the factors associated with the incubation period in the analysis correcting for this bias (appendix p 6).
During the omicron wave between December, 2021, and February, 2022, the proportion of undiagnosed infections probably increased because of the increase in incidence, leading to a high burden on testing platforms, and because of an increased number of infections leading to no symptoms or mild symptoms.28 This increase affected recruitment for our study during that period, and it is difficult to speculate how the impact on recruitment might have affected our findings with regard to the incubation period and its determinants. However, we found no particular differences in the populations across variants (appendix p 2), except for a young age (<40 years) in people infected with the omicron variant.
The over-representation of women (67%) in our study compared with the proportion of women in the French Screening Information System database of SARS-CoV-2-positive cases during the study period (53%) is similar to figures found earlier in the ComCor study6, 17 and might be related to the increased willingness of women to respond to a health survey compared with men, or to the high proportion (14%) of health-care workers in the study population. This over-representation could affect generalisation of our results to under-represented groups (eg, men older than 70 years).
Our variant-typing strategy did not include all cases and could not differentiate between the beta and gamma variants, or between the BA.1 and BA.2 subvariants of the omicron variant. However, evidence from sequencing surveillance in France shows that beta and BA.1 were generally more prominent than gamma and BA.2 during the study period.21
Future research is required to understand the mechanisms underlying differences of duration of incubation period depending on sex, age, and mask wearing during transmission. As predominant strains of SARS-CoV-2 keep changing globally, iterative assessments of incubation period and its associated factors will be necessary.
Our results indicate that the SARS-CoV-2 incubation period is notably shorter with the omicron variant, as well as in younger participants and settings of high viral inoculum. These data should help inform SARS-CoV-2 contact-tracing strategies and modelling. Particular efforts should be made to reduce the number of days from symptom onset to contact tracing and isolation in a context of global predominance of the omicron variant of concern.
Data sharing
The participant data (with identifiers) of this study are available from the French National Health Insurance Fund and from Ipsos. Restrictions apply to the availability of these data, which were used under authorised agreement for this study by the French data protection authority Commission Nationale de l'Informatique et des Libertés (CNIL; the French National Commission on Informatics and Liberty). Access to these data would therefore require previous authorisation by the CNIL. The study protocol and informed consent form will be made available (in French) upon request. The data will be available as soon as access is granted by the CNIL and for the duration authorised by the CNIL, which will entirely determine the beginning and end date of availability for authorised researchers.
Declaration of interests
FC receives consulting fees from Sanofi. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
This study was funded by the Pasteur Institute, Research and Action Targeting Emerging Infectious Diseases, and the French National Agency for AIDS Research–Emerging Infectious Diseases. AF's laboratory receives support from the Labex Integrative Biology of Emerging Infectious Diseases ComCor Project (ANR-10-LABX-62-IBEID) and the INCEPTION project (PIA/ANR-16-CONV-0005) for studies of emerging viruses. SG is funded by the INCEPTION programme's Investment for the Future (ANR-16-CONV-0005). TCh is funded by Fondation de France Alliance Tous unis contre le virus (the Foundation of France United Against the Virus).
Contributors
AF, SG, TCh, LS, FO, CD, FC, SC, and AM designed the study. SG, TCh, LS, AF, and AM developed the study questionnaire. FO, CD, AL, and SM managed the data collection online. OC, CvP, and TCh oversaw the adherence of the study to the regulatory requirements. TCh and LS oversaw the collection of data and maintained the database. SG, TCo, SC, and AF did the statistical analysis. SG, TCo, SC, and AF drafted the first version of the manuscript. To comply with the data processing authorisation granted by the Commission Nationale de l'Informatique et des Libertés (the French National Commission on Informatics and Liberty), SG, TCh, LS, and AF had full access to the data reported in this study. SG and AF accessed and verified the data. All authors reviewed and approved the final manuscript and had final responsibility for the decision to submit for publication.
Supplementary Material
References
- 1.McAloon C, Collins Á, Hunt K, et al. Incubation period of COVID-19: a rapid systematic review and meta-analysis of observational research. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2020-039652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alene M, Yismaw L, Assemie MA, Ketema DB, Gietaneh W, Birhan TY. Serial interval and incubation period of COVID-19: a systematic review and meta-analysis. BMC Infect Dis. 2021;21:257. doi: 10.1186/s12879-021-05950-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elias C, Sekri A, Leblanc P, Cucherat M, Vanhems P. The incubation period of COVID-19: a meta-analysis. Int J Infect Dis. 2021;104:708–710. doi: 10.1016/j.ijid.2021.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xin H, Wong JY, Murphy C, et al. The incubation period distribution of coronavirus disease 2019: a systematic review and meta-analysis. Clin Infect Dis. 2021;73:2344–2352. doi: 10.1093/cid/ciab501. [DOI] [PubMed] [Google Scholar]
- 5.Homma Y, Katsuta T, Oka H, et al. The incubation period of the SARS-CoV-2 B1.1.7 variant is shorter than that of other strains. J Infect. 2021;83:e15–e17. doi: 10.1016/j.jinf.2021.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grant R, Charmet T, Schaeffer L, et al. Impact of SARS-CoV-2 delta variant on incubation, transmission settings and vaccine effectiveness: results from a nationwide case-control study in France. Lancet Reg Health Eur. 2022;13 doi: 10.1016/j.lanepe.2021.100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Chen R, Hu F, et al. Transmission, viral kinetics and clinical characteristics of the emergent SARS-CoV-2 delta VOC in Guangzhou, China. EClinicalMedicine. 2021;40 doi: 10.1016/j.eclinm.2021.101129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandal LT, MacDonald E, Veneti L, et al. Outbreak caused by the SARS-CoV-2 omicron variant in Norway, November to December 2021. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.50.2101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansen L. Investigation of a SARS-CoV-2 B.1.1.529 (omicron) variant cluster—Nebraska, November–December 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1782–1784. doi: 10.15585/mmwr.mm705152e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan WYT, Wong LY, Leo YS, Toh MPHS. Does incubation period of COVID-19 vary with age? A study of epidemiologically linked cases in Singapore. Epidemiol Infect. 2020;148:e197. doi: 10.1017/S0950268820001995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng C, Zhang D, Dang D, et al. The incubation period of COVID-19: a global meta-analysis of 53 studies and a Chinese observation study of 11 545 patients. Infect Dis Poverty. 2021;10:119. doi: 10.1186/s40249-021-00901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Han Z-G, Qin P-Z, et al. Transmission and containment of the SARS-CoV-2 delta variant of concern in Guangzhou, China: a population-based study. PLoS Negl Trop Dis. 2022;16 doi: 10.1371/journal.pntd.0010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marks M, Millat-Martinez P, Ouchi D, et al. Transmission of COVID-19 in 282 clusters in Catalonia, Spain: a cohort study. Lancet Infect Dis. 2021;21:629–636. doi: 10.1016/S1473-3099(20)30985-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerami C, Popkin-Hall ZR, Rapp T, et al. Household transmission of SARS-CoV-2 in the United States: living density, viral load, and disproportionate impact on communities of color. Clin Infect Dis. 2022;74:1776–1785. doi: 10.1093/cid/ciab701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng Y, You C, Liu Y, Qin J, Zhou XH. Estimation of incubation period and generation time based on observed length-biased epidemic cohort with censoring for COVID-19 outbreak in China. Biometrics. 2021;77:929–941. doi: 10.1111/biom.13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nie X, Fan L, Mu G, et al. Epidemiological characteristics and incubation period of 7015 confirmed cases with coronavirus disease 2019 outside Hubei province in China. J Infect Dis. 2020;222:26–33. doi: 10.1093/infdis/jiaa211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galmiche S, Charmet T, Schaeffer L, et al. Exposures associated with SARS-CoV-2 infection in France: a nationwide online case-control study. Lancet Reg Health Eur. 2021;7 doi: 10.1016/j.lanepe.2021.100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charmet T, Schaeffer L, Grant R, et al. Impact of original, B.1.1.7, and B.1.351/P.1 SARS-CoV-2 lineages on vaccine effectiveness of two doses of COVID-19 mRNA vaccines: results from a nationwide case-control study in France. Lancet Reg Health Eur. 2021;8 doi: 10.1016/j.lanepe.2021.100171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Public Health France COVID-19: epidemiological update of 18 March 2021. 2021. https://www.santepubliquefrance.fr/maladies-et-traumatismes/maladies-et-infections-respiratoires/infection-a-coronavirus/documents/bulletin-national/covid-19-point-epidemiologique-du-18-mars-2021 (in French).
- 21.Public Health France Coronavirus: key figures and evolution of COVID-19 in France and around the world. https://www.santepubliquefrance.fr/dossiers/coronavirus-covid-19/coronavirus-chiffres-cles-et-evolution-de-la-covid-19-en-france-et-dans-le-monde (in French).
- 22.Gaymard A, Bosetti P, Feri A, et al. Early assessment of diffusion and possible expansion of SARS-CoV-2 lineage 20I/501Y.V1 (B.1.1.7, variant of concern 202012/01) in France, January to March 2021. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.9.2100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morais FG, Sakano VK, de Lima LN, et al. Filtration efficiency of a large set of COVID-19 face masks commonly used in Brazil. Aerosol Sci Technol. 2021;55:1028–1041. [Google Scholar]
- 24.Bałazy A, Toivola M, Adhikari A, Sivasubramani SK, Reponen T, Grinshpun SA. Do N95 respirators provide 95% protection level against airborne viruses, and how adequate are surgical masks? Am J Infect Control. 2006;34:51–57. doi: 10.1016/j.ajic.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 25.Trunfio M, Longo BM, Alladio F, et al. On the SARS-CoV-2 “variolation hypothesis”: no association between viral load of index cases and COVID-19 severity of secondary cases. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.646679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Killingley B, Mann AJ, Kalinova M, et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat Med. 2022;28:1031–1041. doi: 10.1038/s41591-022-01780-9. [DOI] [PubMed] [Google Scholar]
- 28.Yu W, Guo Y, Zhang S, Kong Y, Shen Z, Zhang J. Proportion of asymptomatic infection and nonsevere disease caused by SARS-CoV-2 omicron variant: a systematic review and analysis. J Med Virol. 2022;94:5790–5801. doi: 10.1002/jmv.28066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The participant data (with identifiers) of this study are available from the French National Health Insurance Fund and from Ipsos. Restrictions apply to the availability of these data, which were used under authorised agreement for this study by the French data protection authority Commission Nationale de l'Informatique et des Libertés (CNIL; the French National Commission on Informatics and Liberty). Access to these data would therefore require previous authorisation by the CNIL. The study protocol and informed consent form will be made available (in French) upon request. The data will be available as soon as access is granted by the CNIL and for the duration authorised by the CNIL, which will entirely determine the beginning and end date of availability for authorised researchers.