ABSTRACT
We show that the zebrafish maternal-effect mutation too much information (tmi) corresponds to zebrafish prc1-like (prc1l), which encodes a member of the MAP65/Ase1/PRC1 family of microtubule-associated proteins. Embryos from tmi homozygous mutant mothers display cytokinesis defects in meiotic and mitotic divisions in the early embryo, indicating that Prc1l has a role in midbody formation during cell division at the egg-to-embryo transition. Unexpectedly, maternal Prc1l function is also essential for the reorganization of vegetal pole microtubules required for the segregation of dorsal determinants. Whereas Prc1 is widely regarded to crosslink microtubules in an antiparallel conformation, our studies provide evidence for an additional function of Prc1l in the bundling of parallel microtubules in the vegetal cortex of the early embryo during cortical rotation and prior to mitotic cycling. These findings highlight common yet distinct aspects of microtubule reorganization that occur during the egg-to-embryo transition, driven by maternal product for the midbody component Prc1l and required for embryonic cell division and pattern formation.
Keywords: Zebrafish, Prc1, Cytokinesis, Microtubule reorganization, Midbody, Dorsal determinant
Summary: Maternal Prc1l is required for microtubule reorganization involved in cell division and the segregation of dorsal determinants in the early zebrafish embryo.
INTRODUCTION
Development prior to zygotic genome activation in animal embryos, including the model system Danio rerio, relies on inherited maternal factors (Pelegri, 2003; Abrams and Mullins, 2009), many of which are RNAs that become translated products after egg activation (Sheets et al., 2017). The process of translation produces a protein supply that is sufficient for the fast-growing embryo, but also generates a temporal lag in product availability. Because of this lag, the earliest events in embryonic development must be driven by ready-to-function maternal factors, which include already-made proteins present in the egg. Because eggs are arrested at meiosis and are transcriptionally quiescent (Svoboda et al., 2017), such maternal products must be synthesized prior to the completion of oocyte maturation. These same maternal products, present in the egg for the early embryo, are also available for use at earlier stages through oocyte maturation and meiotic resumption during egg activation. The temporal usage of such early maternal factors indicates a process in which the egg-to-embryo transition is not a sharp boundary occurring at egg activation, but instead spans the completion of meiosis, egg activation and fertilization, and the earliest embryonic mitotic cycles (Courtois et al., 2012).
A key event that occurs during meiosis after egg activation and in the early embryo is cell division, a process that relies on extensive cytoskeletal remodeling. Early embryonic cells are unusually large and additionally involve patterning determinants (Lindeman and Pelegri, 2010; Wühr et al., 2010), conditions that require unique processes within these cells such as dynamic cytoskeletal reorganization. In the early embryo, cytoskeletal changes are particularly evident, not only in the spindle apparatus during mitosis, but also during furrow formation and maturation (Jesuthasan, 1998; Wühr et al., 2010; Eno and Pelegri, 2018). At furrow induction, ends of anaphase astral microtubules deliver signals to the cleavage plane to induce furrow formation (Rappaport, 1996; Yabe et al., 2009; Nair et al., 2013). These microtubules transition to form the furrow microtubule array (FMA), an array of tubules parallel to each other and perpendicular to the furrow along the length of the furrow (Danilchik et al., 1998; Jesuthasan, 1998). During furrow maturation, FMA tubules continue to undergo increasing bundling as they appear to migrate and form a compact structure at each of the furrow distal ends (Pelegri et al., 1999; Eno et al., 2018). The FMA during furrow maturation in the zebrafish has been proposed to be analogous to midbody formation in smaller cell types (Eno et al., 2018), with the FMA exhibiting features similar to those of midbodies, such as microtubule bundling, coupling to the exocytosis of internal membrane vesicles and eventual microtubule disassembly (Jesuthasan, 1998; Pelegri et al., 1999; Danilchik et al., 2003; Eno et al., 2018). Factors known to participate in the formation or regulation of the mammalian midbody, such as components of the chromosomal passenger complex and the centralspindlin complex, are also associated with the FMA (Chen et al., 2002; Yabe et al., 2009; Nair et al., 2013).
As the egg initiates meiotic resumption and the zygote embryonic cell cycles, other processes essential for embryonic development also ensue. In fish and amphibians, a key event is the off-center displacement of dorsal determinants from the vegetal pole, where they are located during oogenesis, towards their site of action at the future dorsal side of the embryo (reviewed by Blum et al., 2014; Welch and Pelegri, 2017). As in the case of cytokinesis, this symmetry-breaking event also depends on the reorganization of microtubules, this time at the vegetal cortex (Elinson and Rowning, 1988; Houliston and Elinson, 1991a,b; Jesuthasan and Strähle, 1997; Marrari et al., 2003, 2004; Tran et al., 2012; Ge et al., 2014). Such vegetal cortex microtubules are thought to undergo rapid polymerization after egg activation, with the resulting microtubules bundling as parallel arrays that run along cortical arcs towards the presumptive dorsal site and which mediate the directed movement of dorsal determinants (Houliston and Elinson, 1991a,b; Schroeder and Gard, 1992; Marrari et al., 2003; Tran et al., 2012; Olson et al., 2015).
Here, we describe a zebrafish maternal-effect mutation too much information (tmi), originally identified as essential for cytokinesis in the early embryo. We determine that tmi encodes Protein regulator of cytokinesis 1-like (Prc1l), a protein that is part of the MAP65/Ase1/PRC1 family of microtubule-associated proteins known to control the formation of the midzone and the completion of cytokinesis. We find that zebrafish Prc1l functions in cytokinesis during meiosis and early embryonic mitoses, specifically in microtubule reorganization involved in spindle formation and FMA restructuring. We additionally uncover an unexpected role for Prc1l in the reorganization of the microtubule cytoskeleton at the vegetal pole, required for the asymmetric segregation of dorsal determinants in the early embryo. Our results show that Prc1l functions in microtubule bundling during both FMA and vegetal cortex microtubule reorganization, revealing common factors driving contemporaneous processes essential for cell division and pattern formation.
RESULTS
A maternal-effect mutation in tmi affects cytokinesis in the early embryo
Females homozygous for the tmip4anu mutation develop into viable, phenotypically wild-type adults; however, 100% of embryos from such females (henceforth referred to as tmi mutant embryos) display cell division and cytokinesis defects that result in the formation of an acellular embryo (Dosch et al., 2004; Nair et al., 2013; Abrams et al., 2020; Fig. 1). Processes characteristic of egg activation, such as chorion expansion and ooplasmic streaming leading to the lifting of the blastodisc, which occur during the first 30 min post-fertilization (mpf), appeared normal in tmi mutant embryos (Fig. 1A,E). Differences between wild-type and mutant embryos began to appear upon initiation of cell division. At approximately 45 mpf, wild-type embryos had completed the first cell cycle with a clearly evident furrow leading to a two-cell embryo (Fig. 1A), and cleavage furrows corresponding to subsequent cell cycles occurred every 15 min thereafter (Fig. 1B-D). In contrast, tmi mutant embryos at similar stages exhibited furrows that either failed to form or did not undergo furrow deepening (Fig. 1E-H). Labeling to detect DNA and α-tubulin during early stages of tmi mutant development (Fig. 1I,J) showed an apparently normal mitotic progression, as assessed by the number of nuclei, and confirmed furrow initiation, as suggested by the appearance of a microtubule exclusion zone at furrow sites. However, tmi mutant furrows did not accumulate membrane markers such as β-catenin, a component of the cell adhesion junction present in mature furrows of the early zebrafish embryo (Fig. 1K,L), indicative of defects in furrow completion.
Fig. 1.
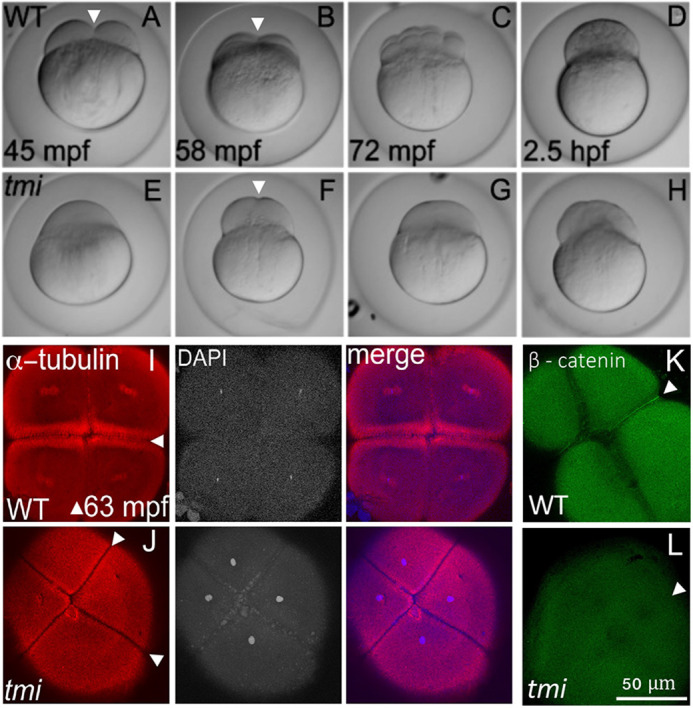
tmi mutant embryos exhibit defects in cytokinesis. (A-H) Developmental time course of wild type (WT; A-D) and tmi mutant (E-H) embryos. In wild type, furrow formation (arrowheads in A,B) and cell division proceeds to generate a blastula. In tmi mutants, incipient furrows form (arrowhead in F) but do not fully contract during cell division, resulting in acellular embryos. (I-L) Immunolabeling of fixed embryos. (I) In wild type, labeling of microtubules (α-tubulin) and DNA (DAPI) show formation of furrows (arrowheads), including reorganization of the microtubular apparatus characteristic of maturing furrows (the FMA, arrowheads in I) as well as nuclear division. (J) In tmi mutants, furrows appear to initiate development as assessed by zones of microtubule exclusion (arrowheads), although the FMA does not fully form, exhibiting a reduction in microtubule enrichment. (K,L) Wild-type embryos (K) show accumulation of β-catenin in mature furrows (arrowhead), but tmi embryos (L) lack β-catenin accumulation (arrowhead). All phenotypes are 100% penetrant with 7-15 embryos per condition in I-L (see Materials and Methods). In spite of acellular phenotype, tmi mutant embryos appear fertilized, judged by the presence of sperm-dependent centrosomes (see Fig. 3 and Dekens et al., 2003; Lindeman and Pelegri, 2012; Yabe et al., 2007) and incipient furrows dividing cells into stereotypically placed, equally sized masses (F,J).
At later stages in mutant embryos, nuclei-like structures appeared to accumulate in the absence of membrane formation, with these structures showing aberrant positioning and apparent clumping into unevenly sized large clusters (Nair et al., 2013; see also Fig. S2). These large DNA aggregates are similar to those observed in embryos mutant for other genes and which lack membranes (Pelegri et al., 1999; Nair et al., 2013; Eno et al., 2016), and are likely formed through the aberrant attraction of chromatin by neighboring asters in the absence of cellular membranes (Tanaka et al., 2005; Shrestha and Draviam, 2013). The observed defects resulted in lysis of 100% of tmi mutant embryos by 6 h post-fertilization (hpf; Nair et al., 2013), similar to other acellular mutant embryos (Nair et al., 2013; Eno et al., 2016).
tmi encodes Prc1-like, a maternally expressed protein of the Microtubule-associated protein (MAP65/ASE1) family
Segregation analysis of the tmip4anua mutant allele using simple sequence length polymorphisms (SSLPs) mapped the mutation to a ∼20 Mb cM region in linkage group 21 (see Materials and Methods). Among the candidate genes in the region, zgc:86764 contained a potential causative mutation resulting in the conversion of a conserved leucine at position 221 into a stop codon (Fig. 2A,B; Fig. S1A). zgc:86764 corresponds to prc1-like (protein regulator of cytokinesis 1-like), belonging to a family of microtubule-associated proteins that includes MAP65 in Arabidopsis, Ase1 in yeast and PRC1 in mammals, characterized by the presence of the MAP65/Ase1/PRC1 homology domain, which is involved in microtubule binding (Jiang et al., 1998; Schuyler et al., 2003; Smertenko et al., 2006) and required for spindle midzone assembly during cytokinesis (Mollinari et al., 2002; Eggert et al., 2006; Zhu et al., 2006). Wild-type prc1l encodes a 580 amino acid protein and the mutated tmi allele codes for a severely truncated protein that lacks the C-terminal 350 amino acids, including the majority of the MAP65/Ase1/PRC1 domain, and is therefore likely a functional null.
Fig. 2.
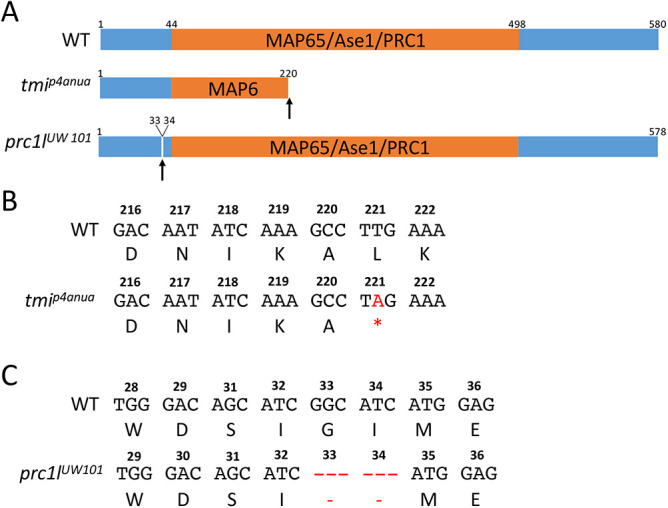
Wild-type and mutant zebrafish Prc1l. (A) Top: Diagram of the predicted wild-type protein, depicting the conserved MAP65/Ase1/PRC1 domain. Middle: The lesion in the ENU-induced maternal-effect allele tmip4anua results in a truncation of the protein deleting a majority of this domain. Bottom: The CRISPR-Cas9-induced allele prc1lUW101 results in the deletion of two conserved amino acids (see Fig. S1B and Fig. S10) N-terminal to the conserved MAP65/Ase1/PRC1 domain. (B,C) Nucleotide sequence and translated amino acids showing the creation of a premature stop codon in tmip4anua (B) and the two-amino acid deletion in prc1lUW101 (C).
To validate the gene assignment of the originally isolated maternal-effect mutation tmi as prc1l, we used CRISPR-Cas9 to induce independent mutations at the prc1l locus, using a guide RNA targeted to a conserved region of the protein upstream of the MAP65/Ase1/PRC1 domain (see Materials and Methods). We isolated a six base pair deletion allele, prc1uw101, that removed two conserved amino acids (Fig. 2A,C, Fig. S1B). Female fish transheterozygous for the tmip4anua and prc1uw101 alleles exhibited an acellular maternal-effect phenotype indistinguishable from that in embryos from tmip4anua homozygous mothers (Fig. S2), indicating non-complementation between the two alleles and corroborating that tmi corresponds to prc1l.
prc1l is one of three prc1 gene paralogs in the zebrafish: prc1a, prc1b and prc1l, with prc1a on linkage group 25 and prc1b on linkage group 7 (Figs S3 and S4). Analysis of the RNA-seq mRNA baseline database available via the Expression Atlas (an ELIXIR database service; White et al., 2017) showed high maternal prc1l RNA levels early in embryonic development that become significantly reduced at the end of the blastula stage period (dome stage, 4.33 hpf) (Fig. S3A). Whole-mount in situ hybridization showed that prc1l RNA is evenly distributed in one- to four-cell wild-type embryos, with similar distribution in tmi mutants (Fig. S3B). In contrast to prc1l, prc1a was expressed during early development through the gastrula period up to the early segmentation period (10.33 hpf), whereas prc1b showed low levels of expression throughout early development (Fig. S3A). Phylogenetic analysis comparing Prc1 family genes in vertebrate species revealed that zebrafish prc1a and prc1b are more closely related to a canonical Prc1 present throughout vertebrate species, whereas prc1l falls within a cluster of Prc1l-related genes present only in fish and amphibian lineages (Fig. S4). Moreover, the root of this prc1l gene cluster appears to have originated at a time that predates various branches within the prc1a and prc1b clusters, which correspond to speciation events within vertebrates. These data indicate that prc1l is an ancient gene duplicate that specifically acts at the egg-to-embryo transition.
Prc1l function is required for microtubule reorganization in embryonic cell divisions
Given the known role for Prc1 family proteins on midzone formation, we visualized spindles in early embryos using antibodies against α-tubulin and γ-tubulin to label microtubules and centrosomes, respectively, and DAPI to label DNA. In early wild-type embryos during metaphase for the first cell cycle (∼45 mpf), chromosomes were aligned at the metaphase plate and metaphase astral microtubules emanated from the centrosomes at each spindle pole (Fig. 3A; 14/14; Jesuthasan, 1998; Wühr et al., 2010, 2011). At this time, mutant embryos showed defects in spindle formation, exhibiting instead shorter and wider spindles that were often asymmetric, with a pole larger than the other (Fig. 3B, two detectable spindle poles as ascertained by γ-tubulin localization, 7/20; Fig. 3C, one detectable spindle pole, 13/20). Later, at around 60 mpf, chromosomes in wild-type embryos began movement towards the poles characteristic of anaphase (Fig. 3D, 15/15; Jesuthasan and Strähle, 1997; Wühr et al., 2010, 2011), whereas in tmi mutants spindle morphology remained abnormal, continuing to exhibit spindle pole asymmetries and additionally showing mis-segregated chromosomes (Fig. 3E, 23/23 with abnormal spindle morphology, 17/23 with mis-segregated chromosomes). Thus, prc1l is essential for proper spindle structure and reorganization during the early blastomere divisions.
Fig. 3.
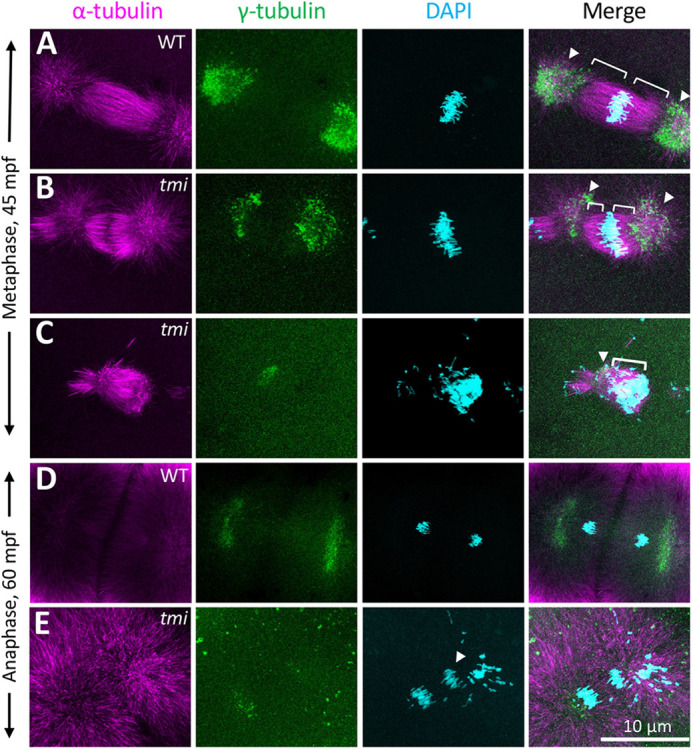
Spindle formation defects in the early mitotic divisions in tmi mutants. (A-C) At metaphase, wild-type asters are highly symmetric (A). In tmi mutants, spindles are often asymmetric (B) or unipolar with chromosomes clustering at one end of the structure (C). Brackets and arrowheads highlight kinetochore microtubule and centrosomes, respectively. (D,E) During anaphase, tmi mutants exhibit chromosomes that fail to segregate (E, arrowhead), which was not observed in wild type (D). Analysis is from embryos at the two- to eight-cell stages (0.751-0.25 hpf), with the specific cell cycle stage assigned by chromosome and spindle morphology.
We also analyzed the effect of the tmi mutation on microtubule reorganization at the furrow. During furrow maturation in wild-type embryos, microtubules of the FMA from opposite sides of the furrow typically exhibit apparent sites of contacts, which also coincide with sites of increased bundling of microtubules at both sides of the furrow (Fig. 4A; Jesuthasan, 1998; Wühr et al., 2010, 2011). Indeed, sites of microtubule bundling at both sides of the furrow at early stages of furrow formation precisely aligned along the length of the furrow (Fig. 4A,A′,E; Fig. S5), consistent with microtubules from both sides of the furrows interacting at specific sites of bundling along the furrow. In contrast, in tmi mutants at the same developmental stage, FMA tubules from both sides of the furrow failed to exhibit the bundling observed in wild type or to contact each other along the furrow (Fig. 4B,B′,F; Fig. S5). Moreover, FMA tips abutting the furrow were tightly apposed in wild type (width of zone of microtubule exclusion: 1.2 µm, s.d.=0.296, n=7), whereas these were spaced further apart in tmi mutants (width of zone of microtubule exclusion: 7.9 µm, s.d.=1.615, n=7). At later stages of furrow maturation, when FMA bundles are undergoing their characteristic movement towards the distal ends of the furrow (Fig. 4C,C′, 11/11), this process was defective in tmi mutants (Fig. 4D,D′, 0/23 with distal enrichment).
Fig. 4.
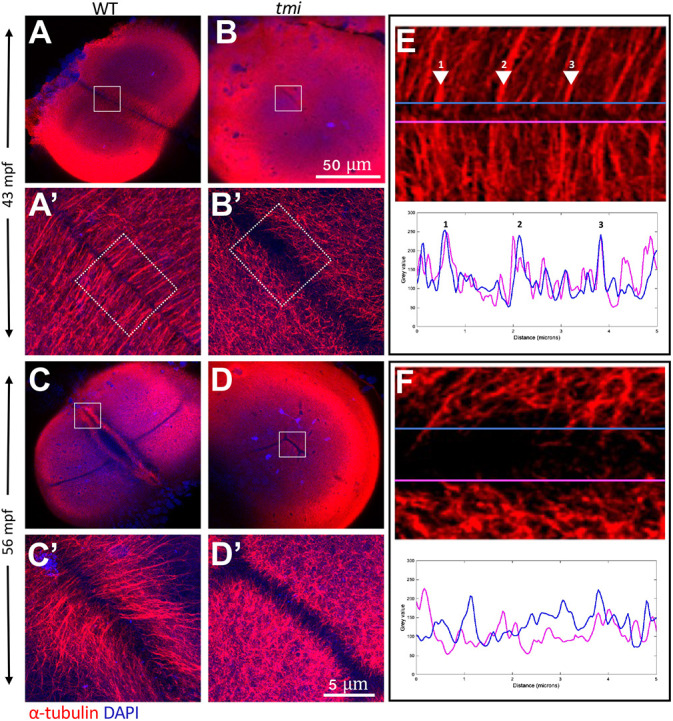
Aberrant reorganization and clustering in the furrow microtubule array. Immunolabeling of fixed embryos to detect microtubules. (A,B) During early stages of furrow formation, the FMA forms along the length of the furrow, consisting of tubule bundles parallel to each other and perpendicular to the furrow (A,A′). tmi mutants fail to show a clearly organized FMA and exhibit a reduction in bundling (B,B′; see also Fig. S5). (C-D′) Upon furrow maturation, tubules of the FMA reorganize, becoming enriched to the distal ends of the furrow and acquiring a tilted angle pointing distally (C,C′), whereas in tmi mutants microtubules fail to reorganize in this manner (D,D′). (E,F) Line scans of FMA bundles along both sides of the furrow show a strong spatial concordance of microtubule bundles in wild type (E, representing the boxed area in A′; numbers correspond to major sites of bundling), consistent with interconnections between bundles across the furrow. Such spatial concordance is absent in tmi mutants (F, corresponding to the boxed area in B′). Images are representative of nine ROIs, each spanning the length of the furrow for each condition (see Fig. S5 and Materials and Methods). Boxed areas in A-D are shown at higher magnification in A′-D′, respectively.
Thus, maternal Prc1l function is essential for spindle formation as well as for the bundling and interconnection of microtubules of the FMA along the furrow.
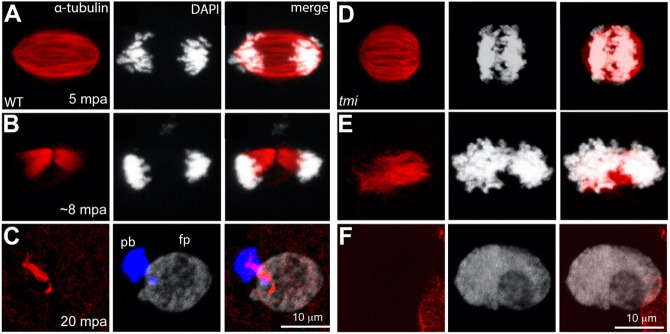
Requirement for Prc1l function during meiosis
Previous studies in zebrafish and other systems have shown that factors involved in early embryonic mitotic cytokinesis also cause cell division defects during the asymmetric cytokinesis that occurs during meiosis (Courtois et al., 2012). We therefore tested for potential meiosis defects in oocytes of tmi mutant females through labeling of α-tubulin and DNA. In eggs from control wild-type females at 5 min post-water activation (mpa), in the absence of sperm, an elongated meiotic spindle with distinct sister chromatids sets could be observed (Fig. 5A, 5/5). In these wild-type water-activated eggs at 8 mpa, this structure transitioned into an incipient midbody flanked by DNA that had undergone segregation to the spindle poles (Fig. 5B, 9/9). By 20 mpa and after polar body cytokinesis completion, each DNA mass exhibits characteristic behaviors, with the female pronucleus acquiring a decondensed appearance and the polar body a highly condensed morphology (Dekens et al., 2003; Lindeman et al., 2012; Nair et al., 2013), with both masses remaining at this stage attached to the meiotic midbody (Fig. 5C, 7/7). In water-activated eggs from homozygous tmi females, a meiotic spindle was visible, although this spindle had a short and tubby appearance (Fig. 5D, 3/3). By 8 mpa in tmi mutants, the spindle had not resolved into a midbody-like structure, with sister chromatids unable to undergo segregation to the spindle poles (Fig. 5E, 4/4). Failure of meiotic cytokinesis in tmi mutants is suggested by each set of segregating chromatids forming adjoining nuclear masses where, as opposed to wild type, neither DNA mass underwent the condensation characteristic of a fully formed polar body (at 8-12 mpf; tmi: 0/5 with a condensed-DNA mass; wild type: 5/5 with one condensed mass; see also Fig. 7A,D). Eventually, by 20 mpa both chromatid sets appeared to collapse into a single DNA mass (Fig. 5F, 3/3) predicted to have a diploid maternal chromosomal component, consistent with a greater nuclear surface area compared with wild type before and after pronuclear fusion (Fig. S6). This diploid maternal ‘pronucleus’ appeared to undergo normal fusion with the sperm-derived haploid pronucleus, as judged by the appearance in tmi mutants of a single nuclear mass at the time corresponding to pronuclear fusion (Fig. S6), with the resulting presumed triploid zygotic nucleus participating in mitoses after fertilization. Triploid embryos can be artificially induced in zebrafish and develop into viable embryos and adults (Kavumpurath and Pandian, 2008; Bazaz et al., 2020). tmi mutant embryos, presumably triploid, appeared to be generated similarly after the failure of polar body extrusion, but failed to cellularize and lyse.
Fig. 5.
Meiotic spindle defects in tmi mutants. (A-C) In wild-type eggs, spindles for the second meiotic division exhibit an elongated shape (A) that resolves into midbody-like structures (B) and becomes highly bundled during abscission (C). During the last stages of cytokinesis, the midbody becomes asymmetric with a longer region adjacent to the polar body (pb; left DNA mass in C, which becomes condensed), and a shorter region adjacent to the female pronucleus (fp; right DNA mass in C, which undergoes decondensation). (D-F) In eggs from tmi mutant females, spindles appear shorter with less-defined poles (D) and fail to form a well-defined midbody (E; see also Fig. 7B). Subsequently, a single DNA mass is observed (F), likely after failure of cytokinesis for meiosis II followed by fusion. In all panels, DNA is represented in white, except the polar body in C, which has been color-coded in blue in the appropriate z-stack sections to better highlight this structure.
Fig. 7.
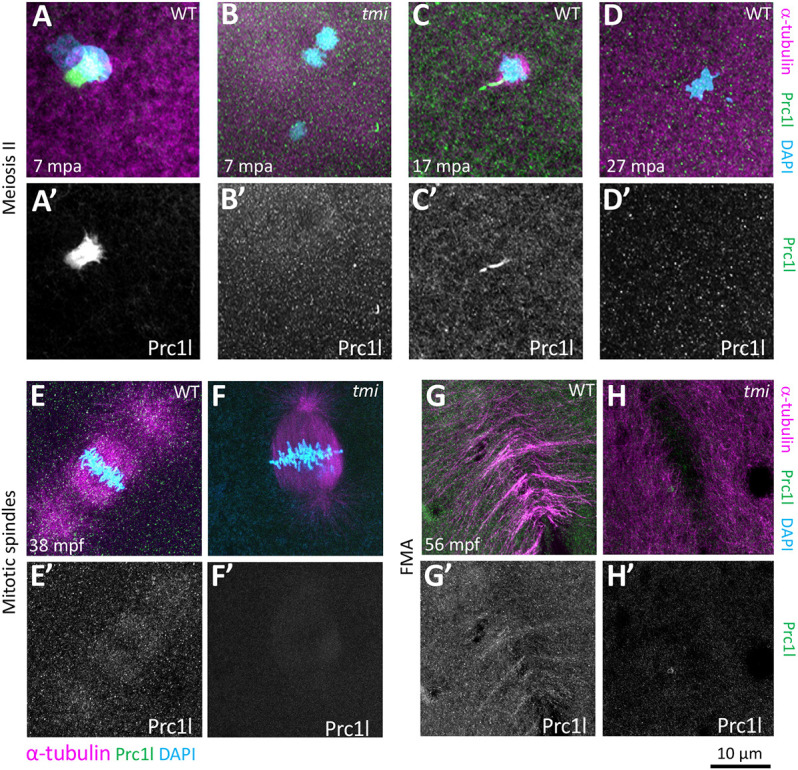
Localization of Prc1l protein to microtubule structures involved in cytokinesis at the animal pole. (A-D′) Prc1l protein localizes to bundling midbody microtubules during meiosis II, (A,A′,C,C′) and becomes delocalized after polar body extrusion (D,D′). tmi mutants do not show antibody labeling (B,B′), as expected from antibody specificity (see also G,H and Fig. S9A). (E-H′) During embryonic mitotic divisions, Prc1l protein localizes to the microtubule apparatus (E,E′) and the FMA (G,G′), with the expected absence of label in tmi mutant embryos (F,F′,H,H′).
Thus, similar to the embryonic divisions, tmi mutants exhibit defects in spindle and midbody organization during meiosis.
Prc1l is required for vegetal microtubule reorganization involved in the segregation of dorsal determinants
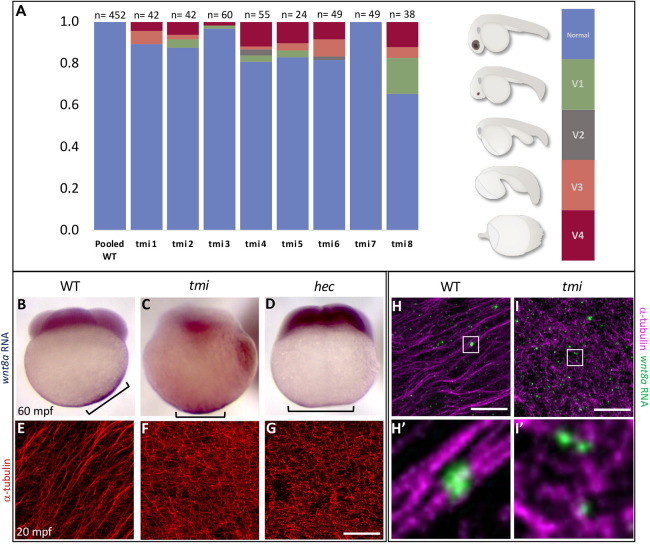
We noticed that a significant proportion of embryos from mothers heterozygous for the tmip4anua mutant allele (genotypically tmip4anua/+) displayed a range of ventralized phenotypes (Fig. 6A, Fig. S7) similar to those described for maternal-effect mutations causing axis induction defects, such as ichabod (Kelly et al., 2000; Bellipanni et al., 2006), tokkaibi (Nojima et al., 2010) and hecate (grip2a) (Ge et al., 2014). This unexpected observation suggested a potential role for Prc1l function in dorsoventral patterning. We reasoned that the observed partially penetrant defects in embryos from mothers heterozygous for a tmi mutant allele may be caused by haploinsufficiency for maternal Prc1l product, and that a full Prc1l functional reduction might exhibit a fully penetrant axis induction phenotype. However, the cell division defect in embryos from homozygous tmi mutant females precludes the cellularization of embryos, and therefore the assessment of axis induction through morphological landmarks. Instead, we tested for defects in tmi embryos in the relocalization of dorsal determinants, a key step leading to axis specification.
Fig. 6.
tmi mutants exhibit defects in the segregation of dorsal determinants. (A) Females heterozygous for the maternal-effect tmi mutation consistently produce a fraction of embryos with ventralization phenotypes in 24 h embryos (V1-V4; ventralization series as in Kishimoto et al., 1997), with V4 the most severe, radially symmetric phenotype (see Fig. S7 for live images). (B-D) wnt8a RNA, originally localized to the vegetal pole of the egg, undergoes a lateral shift towards the future dorsal axis in wild-type embryos (B), a behavior absent in embryos from tmi (C) and hec (D) homozygous mutant mothers. Brackets indicate observed extent of wnt8a RNA at the cortex. (E-G) Microtubules at the vegetal cortex develop into a parallel array of microtubule bundles aligned toward the future dorsal axis (E) (Jesuthasan and Strähle, 1997; Tran et al., 2012; Ge et al., 2014). Vegetal microtubule bundling and alignment is defective in embryos from females homozygous mutant for tmi (F) and hecate (G). (H,I) Labeling of wnt8a RNA-containing particles and microtubules at the vegetal cortex of 20 mpf embryos, showing wnt8a RNA localizing to microtubules in both wild type and tmi mutants, despite different microtubule reorganization. H and I are 2D maximum projections of confocal z-stacks (see Movies 1 and 2 to view individual z-slices), and H′ and I′ show magnified views from individual z-slices. Quantification of individual z-slices (see Materials and Methods; Fig. S8) shows no statistically significant difference in the percentage of wnt8a RNA particles that co-occur with microtubules (wild type: 94.4%, n=10 ROIs, 5 embryos; tmi: 93.0%; n=10 ROIs; 5 embryos; P=0.472; unpaired Student's t-test).
Previous studies have determined that zebrafish wnt8a maternal RNA is initially localized to the vegetal pole of the mature oocyte and undergoes a lateral shift after fertilization (Lu et al., 2011), which reflects a symmetry-breaking event that presages axis induction. We used whole-mount in situ hybridization to test whether the dorsally directed lateral shift of wnt8a RNA was affected in tmi mutant embryos. As expected, wild-type embryos experienced an off-center shift of wnt8a mRNA toward the presumed dorsal region of the early embryo, observable at 30 and 60 mpf (Fig. 6B, 18/18 with shift). tmi mutant embryos at these same stages, however, failed to exhibit an off-center movement of wnt8a RNA, which remained instead symmetrically localized at the base of the vegetal pole (Fig. 6C, 0/28 with shift). This RNA redistribution phenotype is similar to that of embryos from hecate mutant mothers (Fig. 6D, 0/24 with shift; Ge et al., 2014).
The off-center shift of wnt8a RNA in zebrafish, reminiscent of cortical rotation in amphibians (Houston, 2012), takes place minutes after fertilization and depends on the formation of a parallel array of microtubule bundles at the vegetal pole of the embryo (Jesuthasan and Strähle, 1997; Tran et al., 2012; Ge et al., 2014; Welch and Pelegri, 2015). As previously reported, vegetal cortex microtubules in wild-type zebrafish embryos begin to undergo bundling and alignment around 14 mpf, become fully bundled and aligned by 20 mpf, and start to dissociate at approximately 26 mpf (Jesuthasan and Strähle, 1997; Tran et al., 2012). At 20 mpf, when vegetal cortex microtubules were bundled and aligned in wild type (Fig. 6E, 10/10 with microtubule arrays), these microtubules had not become aligned into parallel bundles in tmi mutants, but instead appeared as a disorganized branched meshwork that extended throughout the vegetal cortex (Fig. 6F, 0/12 with arrays), a phenotype again similar to that of hecate mutants (Fig. 6G, 0/10 with arrays; Ge et al., 2014). Co-labeling experiments showed that wnt8a RNA-containing particles localize to aligned microtubule tracks in wild type (Fig. 6H), but also to disorganized cortical microtubules in tmi mutants (Fig. 6I) in similar proportions (wild type=94.4%; tmi=93.0%; P=0.472; Fig. S8, Movies 1 and 2), suggesting that the failure to exhibit an off-center shift for this RNA is due to defective microtubule reorganization and not a lack of RNA particle–microtubule association. Thus, Prc1l function is essential for the relocalization of dorsal determinants, likely through a role in the reorganization of vegetal cortex microtubules into aligned parallel bundles.
Localization of Prc1l protein and requirement for microtubule bundling and alignment
To determine the subcellular localization of Prc1l, we developed antibodies against amino acids 59-102 of the predicted 580-amino acid Prc1l protein. Western blot analysis showed that the antibodies recognized a band of the expected size (67 kD) in wild-type embryos that was absent in embryos from females homozygous for the tmip4anua mutation (Fig. S9A).
In wild-type embryos immediately after fertilization, Prc1 protein was associated with remnants of the spindle apparatus for meiosis II that began to undergo bundling (Fig. 7A,A′, 7 mpf, 4/4) and reorganize into compact midbodies (Fig. 7C,C′, 17 mpa, 5/5). Prc1 localization to meiotic midbodies ceased coinciding with the disassembly of the midbody upon completion of polar body extrusion (Fig. 7D,D′, 27 mpa, 3/3). In dividing blastomeres of the early embryo, Prc1l protein was observed to localize both at the metaphase spindle (Fig. 7E,E′, 10/10) and at sites of bundling of the FMA ends along the forming furrow (Fig. 7G,G′, 13/13). Specificity of the antibody labeling was further supported by the absence of label in tmi mutants in spindles during meiosis (Fig. 7B,B′, 0/5 with label, compare with A,A′) and embryonic mitosis (Fig. 7F,F′, 0/5 with label; compare with Fig. 7E,E′) and the FMA (Fig. 7H,H′, 0/5 with label, compare with Fig. 7G,G′).
Given the unexpected role for Prc1 function in the reorganization of vegetal cortex microtubules, we also investigated Prc1l protein localization in these microtubules during the first cell cycle interphase (15 mpf). Immunolocalization studies within this region revealed that Prc1l protein was localized along parallel tracks of bundled microtubules (Fig. 8A,A′, 17/17). Interestingly, Prc1l protein along these tracks exhibited a pattern of repetitive enrichments, with an accumulation every 14-28 μm and the most common distance between enrichments at about 19 μm (average=18.9 µm, s.d.=4.19; Fig. 8B, Fig. S9B,B′). Colocalization control images, in which the color labeling channels have been rotated by 90°, show no repetitive pattern of Prc1l protein accumulation (Fig. 8C, Fig. S9C,C′).
Fig. 8.
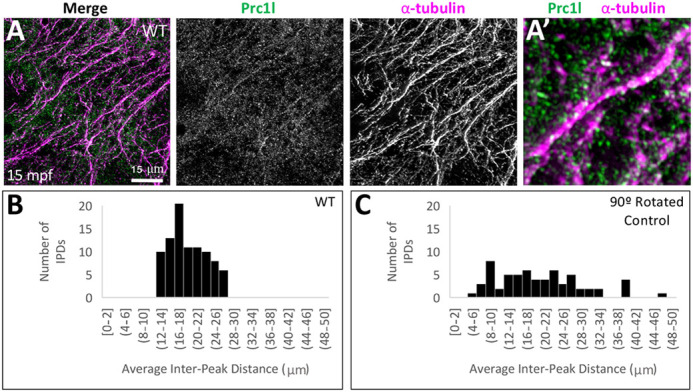
Localization of Prc1l protein to aligned vegetal microtubule arrays at the vegetal pole. (A,A′) Array of aligned bundles of vegetal cortical microtubules and localization of Prc1l protein along bundles in wild type (A, magnification in A′). (B,C) Distribution of distances between sites of Prc1l localization along microtubule tracks, showing a restricted distribution range in wild type (B) and random distribution in a rotated-channel control in wild type (C). The difference between distributions is statistically significant according to a Kolmogorov–Smirnov test (P=0.002; see Fig. S9B,C).
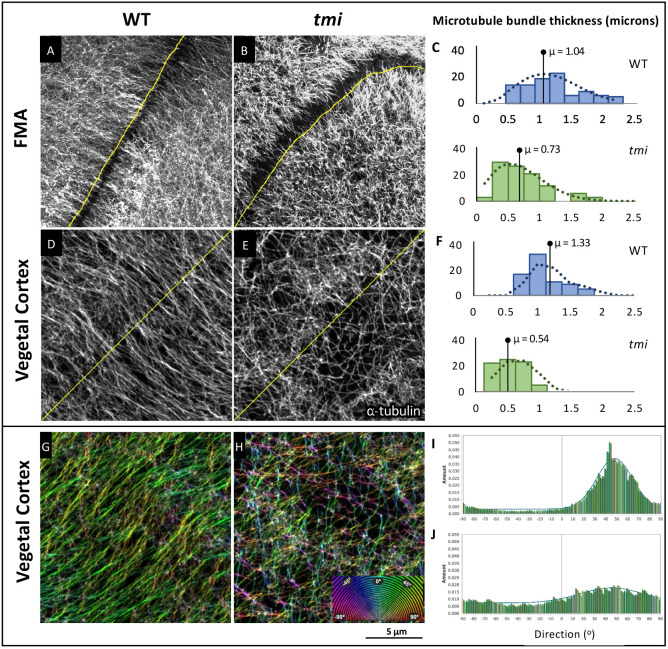
The localization pattern of Prc1l at microtubules in both the FMA and the vegetal cortex, as well as defects in microtubule networks in tmi mutant embryos, suggests a role for this factor in microtubule bundling, Consistent with such a role, quantification showed that the average thickness of microtubule tracks is significantly greater in wild type (1.04 µm) compared with tmi mutants (0.73 µm) at the FMA tips at the time at which they are expected to undergo interdigitation (Fig. 9A-C; P-value≤0.001). Similarly, the average thickness of microtubule tracks was significantly greater in wild type (1.33 µm) than in tmi mutants (0.54 µm) at the vegetal cortex at a time of microtubule track formation and alignment (Fig. 9D-F; P≤0.001). Additionally, analysis of the angles of orientation of vegetal cortex microtubules showed that microtubule orientation falls within a narrow range of approximately 40° in wild type (Fig. 9G,I), but exhibits largely random orientation across a 180° span in tmi mutants (Fig. 9H,J). Thus, tmi function is required for microtubule bundling at both the FMA and vegetal cortex, and the precise alignment of vegetal cortex microtubule tracks necessary for the relocalization of dorsal determinants.
Fig. 9.
Prc1l functions in microtubule bundling and orientation. (A-C) FMA microtubules. (A) In wild type during furrow formation, microtubules coalesce in thick bundles along the FMA. (B) Microtubules along the FMA in tmi mutant embryos exhibit lack of bundling. (C) Quantification of the average width of the microtubules and microtubule bundles along the FMA in wild-type (blue, n=30) and mutant (green, n=30) embryos at the four-cell stage (56-63 mpf). (D-F) Vegetal cortex microtubules. (D) Microtubule bundles at the vegetal cortex of wild-type embryos. (E) Microtubules at the vegetal cortex of tmi mutant embryos show reduced bundling. (F) Quantification of the average width of vegetal cortex microtubules and microtubule bundles in wild type (blue, n=30) and mutants (green, n=30) at 20 mpf, showing decreased thickness in mutants. Statistical analysis in C,F was carried out using the Mann–Whitney test. (G-J) Microtubule track directionality analysis of vegetal cortex microtubules (RGB 180° spectral color coding). Microtubule tracks exhibit a cohesive orientation in wild type (single-color coding in G, n=6), and a disorientated network in tmi embryos (multi-color coding in H, n=6). Microtubule track angle distribution at 20 mpf (shown in I,J for representative single ROIs) shows regularity in wild type (I) but not mutants (J). The difference between distributions is statistically significant according to a Kolmogorov–Smirnov test (P<0.009).
Previous studies have shown that microtubule bundling at the midzone involves crosslinking between anti-parallel microtubules (Mollinari et al., 2002; Zhu and Jiang, 2005; Bieling et al., 2010; Subramanian et al., 2010; Kellogg et al., 2016). Indeed, our studies show a role for Prc1l in the bundling of microtubule tips during cytokinesis for both meiosis II and the early embryonic mitoses, which are expected to be arranged in an anti-parallel orientation (Danilchik et al., 2003; Otegui et al., 2005; Fededa and Gerlich, 2012). However, the localization and bundling function of Prc1l protein on vegetal cortex microtubule tracks, known to be arranged in a parallel conformation (Houliston and Elinson, 1991a; Schroeder and Gard, 1992; Marrari et al., 2003; Tran et al., 2012; Olson et al., 2015; Wijeratne and Subramanian, 2018), was unexpected. The vegetal cortex microtubule bundling defect in tmi mutants indicates that, in addition to its ability to mediate antiparallel microtubule intercalation required for midzone and midbody formation, Prc1l functions in the bundling of parallel microtubules. Despite divergence of the Prc1l protein sequence compared with that of zebrafish Prc1a and Prc1b and human PRC1 (Fig. S10), protein modeling did not reveal obvious differences in the predicted three-dimensional structure for these products (Fig. S11). Rather, this apparent differential activity may rely on additional regulatory interactions.
Thus, Prc1l protein is present in the egg and early embryo in spindles and in areas of active microtubule bundling during cytokinesis, as well as in aligned vegetal cortex microtubules required for the relocalization of dorsal determinants.
DISCUSSION
We show that the zebrafish maternal-effect mutation tmi corresponds to prc1-like, a gene belonging to the MAP65/Ase1/PRC1 family of microtubule-associated proteins. Prc1l is essential for meiosis, mitosis and cytokinesis in the early embryo, and unexpectedly also plays a role in the vegetal microtubule reorganization and bundling necessary for the relocalization of dorsal determinants.
The maternal-effect gene tmi corresponds to a prc1 gene duplicate with an essential function at the egg-to-embryo transition
Meiosis and mitosis of early embryos depend critically on the associated steps of cytokinesis. Females homozygous for mutations in tmi produce embryos that exhibit shallow membrane indentations corresponding to incipient furrows, but which nevertheless fail to undergo full membrane contraction or adhesive septum formation (Dosch et al., 2004; Nair et al., 2013). We find that tmi mutant eggs also exhibit defects in cytokinesis for meiosis, exhibiting a lack of polar body extrusion. Thus, tmi function is required for the progression of cytokinesis in both meiotic and early mitotic cell divisions.
A positional cloning approach finds that the maternal-effect tmi mutation is associated with an early truncation of the Prc1l protein. Given the inherent challenge to rescue early developmental defects caused by mutations in maternal-effect genes (Yabe et al., 2009; Lindeman and Pelegri, 2012; Nair et al., 2013), we corroborated gene assignment through non-complementation to newly generated CRISPR/Cas9 pcr1l alleles, an approach used in other studies in zebrafish (Eno et al., 2016) and other model systems (Ching et al., 2010; Chao et al., 2019).
Phylogenetic analysis indicates that prc1l is a duplicated copy of a canonical prc1 gene that pre-dates the split between amphibian and fish lineages, with expression analysis suggesting that the gene is exclusively maternally expressed. This is in contrast to two other prc1 genes in zebrafish, prc1a and prc1b, which exhibit both maternal and zygotic expression. Thus, prc1l corresponds to a duplicated prc1 gene that appears to have acquired a specialized function at the egg-to-embryo transition early in vertebrate evolution, and which has been maintained in amphibians and fish lineages. Prc1l function encompasses both meiosis and early mitotic divisions, consistent with a need to use previously translated products for events associated with egg activation and the earliest embryonic divisions. Thus, as previously found in the mouse embryo (Courtois et al., 2012), a functional transition at the egg-to-embryo transition does not necessarily coincide with the rapid processes of egg activation and fertilization, but appears to be a longer-lasting period, lasting from meiosis to the early embryonic cellular divisions.
Prc1l acts in microtubule reorganization during meiosis and early embryonic mitosis
In many in vivo and in vitro systems, PRC1 is known to localize and bind microtubules at the spindle midzone, where it mediates antiparallel microtubule overlap, and later to the transient midbody (Mollinari et al., 2002; Zhu and Jiang, 2005; Bieling et al., 2010; Subramanian et al., 2010, 2013; Kellogg et al., 2016). In zebrafish, we find that in the absence of Prc1l function, eggs undergoing meiosis as well as embryonic blastomeres undergoing mitosis exhibit defects in the microtubule apparatus. Spindles in tmi mutants exhibit a reduced length during meiosis and a lack of the symmetry during early embryonic mitosis (depicted for mitosis in Fig. 10, center). These results are consistent with recent studies using Xenopus extracts that show the maternal Prc1l ortholog (named Prc1E in that study), together with the kinesin Kif4A, is required to regulate the organization of spindle asters and the extent of interdigitation in the early Xenopus embryo (Nguyen et al., 2018).
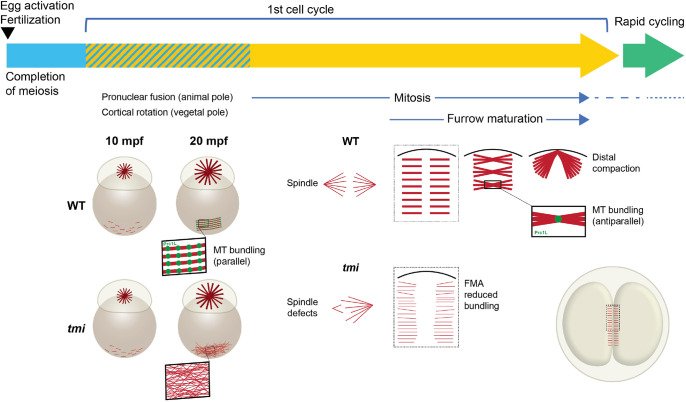
Fig. 10.
Model highlighting multiple roles for Prc1l at the egg-to-embryo transition. After egg activation and completion of meiosis, the embryo undergoes a first longer cycle, which is followed by rapid mitotic cycles. In wild type, the first cycle includes an interphase period (striped section) with microtubule growth both at the animal pole, where it forms a sperm aster involved in pronuclear fusion, and at the vegetal pole, where microtubules coalesce into tracks involved in a cortical rotation-like process that generates an early dorsoventral asymmetry. Prc1l is required for the bundling of vegetal cortex microtubules. During the mitotic cycles, Prc1l function is required for microtubule interdigitation at the spindle midzone and the FMA, the latter reorganizing as multiple midbody equivalents that gradually coalesce at the distal end of the furrow. Prc1l appears to promote bundling of parallel microtubules during interphase but antiparallel microtubules during mitosis. Prc1l is also required for midbody formation during polar body extrusion (not depicted). Overview embryo images on the left are side views, and on the lower right an animal view, with boxes indicating cytoskeletal changes presented in greater detail.
Cytokinesis during meiosis and mitosis requires the formation of a midbody (Agromayor and Martin-Serrano, 2013; Mierzwa and Gerlich, 2014). During meiosis, the midbody can be observed to form between the female pronucleus and the forming polar body, a process dependent on factors known to be involved in midbody reorganization, such as the chromosomal passenger complex (Nair et al., 2013). Spindles in tmi mutant oocytes are unable to form midbodies during meiosis, demonstrating a role for Prc1l in this process.
In the large embryonic cells of the early zebrafish embryo, FMA tubules undergo localized bundling at their plus ends along the furrow. These bundled microtubule ends progressively coalesce into a large, compact structure at each of the two distal ends of the furrow (Jesuthasan, 1998; Pelegri et al., 1999) through what appears to be a modified process of midbody formation (Eno et al., 2018). In tmi mutants, FMA microtubules fail to undergo bundling and subsequent distal enrichment. We find that in wild type, when the FMA forms as a parallel array, Prc1l-dependent bundling and crosslinking initially occur at multiple sites along the furrow, each apparently representing a minimal midbody-like structure. Moreover, these sites of coordinated microtubule bundling appear enriched with Prc1l protein. Previous studies in Xenopus have also identified localization of the Xenopus Prc1l interacting partner Kif4A at the interdigitating tips of the FMA in this organism (Nguyen et al., 2018). Together with previous work (Nguyen et al., 2018), our studies support the notion that Prc1l functions in the crosslinking of antiparallel microtubules across the cleavage plane in the early embryo (Fig. 10, right), similar to the proposed role for this factor at the midzone and midbodies in smaller cell types (Bieling et al., 2010; Agromayor and Martin-Serrano, 2013; Mierzwa and Gerlich, 2014).
As the FMA undergoes reorganization during furrow maturation, these minimal midbodies migrate and coalesce to form large microtubule masses present at the distal ends of the mature furrow. The directional process that mediates the migration of these bundled ends depends on an intracellular calcium wave along the forming furrow and polarized F-actin dynamics (Miranda-Rodríguez et al., 2017; Eno et al., 2018). This alternative mechanism for midbody coalescence at the outward edge of the furrow may reflect an adaptation to cytokinesis completion in large embryonic cells. Additionally, in zebrafish, FMA tubules are also associated with segregating germ plasm (Pelegri et al., 1999; Knaut et al., 2000), a maternal determinant of germ cell fate (Knaut et al., 2000; Hashimoto et al., 2004; Bontems et al., 2009; reviewed by Eno and Pelegri, 2016).
Thus, the defects in microtubule-based structures observed in tmi mutant embryos are consistent with the role for zebrafish Pcr1l in the crosslinking of antiparallel microtubules, which promotes microtubule bundling and reorganization during meiosis and the early embryonic mitoses.
Function of Prc1l in microtubule reorganization during the segregation of dorsal determinants
tmi mutant embryos are acellular and lyse at 4-6 hpf, coincident with the initiation of epiboly in wild type. This acellular phenotype and subsequent lysis precludes assessment of the consequences of tmi functional reduction in downstream developmental processes. However, we fortuitously found a partially penetrant apparent axis-induction phenotype associated with maternal haploinsufficiency for tmi. Indeed, molecular landmarks show that complete loss of Prc1l function results in the absence of the off-center shift characteristic of vegetally localized RNAs, known to be essential for the specification of the embryonic dorsal axis. In wild type, cortical microtubules at the vegetal pole become aligned into parallel arrays, with microtubules becoming visibly thicker as they bundle and align as parallel arrays (Jesuthasan and Strähle, 1997; Tran et al., 2012; Ge et al., 2014) (Fig. 10, left). In tmi mutants, vegetal cortical microtubules exhibit reduced bundling and alignment, indicating a role for Prc1l in these processes.
Vegetal cortex microtubule reorganization, together with microtubule-based motors, is known to mediate cortical rotation, a movement of the outer embryonic cortex with respect to the embryonic core (reviewed by Houston, 2012; Welch and Pelegri, 2017) originally identified in amphibians (Gerhart et al., 1989; Houliston and Elinson, 1991a,b; Schroeder and Gard, 1992) and later zebrafish (Tran et al., 2012). Moving such embryonic structures likely requires large forces, which may be facilitated by microtubule bundling. The movement of the cortex itself is thought to cause further microtubule alignment, with microtubule-based movement, bundling and alignment acting as an amplifying feed-forward loop (Olson et al., 2015).
Symmetry-breaking leading to axis specification is thought to additionally involve the directional, off-center transport of RNA-containing particles through the action of microtubule-based motors (reviewed by Houston, 2012). Although microtubule alignment is likely key for directed transport, it is unclear what the role of microtubule bundling is in this process. Bundling may provide structural support, but may additionally facilitate long-range particle movement, for example by allowing productive switching of motors at the end of processive runs (Ali et al., 2008).
A role for PRC1 in the bundling of parallel microtubules
Visualization of the Prc1l protein shows that it localizes to microtubules of the vegetal cortical array, consistent with a role in microtubule bundling. Remarkably, Prc1l protein is found to be enriched along vegetal microtubule tracks in an apparently repeated pattern, with enrichments at regular intervals of about 19 µm. Previous studies have shown vegetal microtubule tracks as bundles of aligned parallel microtubules, with their plus ends oriented towards the prospective dorsal region (reviewed by Houston, 2012). Thus, the localization and function of Prc1l protein is unlikely to be caused by crosslinking of antiparallel microtubules, as occurs at the spindle midzone and midbody, but is likely instead related to the bundling of parallel microtubules.
Bundling of arrays of microtubules reminiscent of parallel tracks at the vegetal cortex has been previously observed after overexpression of mammalian PRC1 in HeLa cells (Mollinari et al., 2002). In these studies, overexpressed PRC1 could be observed during interphase to both promote and localize to cytoplasmic arrays of long microtubule bundles. Subsequently, as the cells entered mitosis, overexpressed PRC1 redistributes to the spindle midzone (Mollinari et al., 2002). The cell cycle-dependent effects and localization of overexpressed mammalian PRC1 in mammalian cell lines are therefore remarkably similar to that which occurs for endogenous levels of Prc1l in the early zebrafish embryo. In the latter case, the early embryo undergoes a long first cell cycle with an interphase that includes pronuclear congression and fusion, followed by a period of fast mitotic cell cycles that lack interphase (Kane and Kimmel, 1993; see also Tsai et al., 2014). In the early zebrafish, Prc1l-dependent cortical microtubule bundles form during interphase prior to mitotic cycling, and Prc1l-dependent spindle midzone and FMA microtubule interdigitation occur during the subsequent embryonic mitotic cycles. In smaller dividing cells, PRC1 levels are typically low during interphase and rise only during mitosis (Jiang et al., 1998). It may be that high levels of maternally inherited Prc1l protein present in the egg, and active in the vegetal pole during the first cell cycle interphase, result in the bundling of parallel microtubules, similar to ectopically expressed PRC1 in interphase of smaller dividing cells. The signals that modulate the function of Prc1l in the bundling of vegetal cortex microtubules remain unknown but may depend on cell cycle-dependent activity of PRC1 regulators (Jiang et al., 1998; Mollinari et al., 2002), and/or differential distribution of such factors. Our studies show for the first time a role for endogenous levels of a PRC1-related product in the bundling of parallel microtubules.
Previous studies suggest a mechanism by which microtubule growth originates from local nucleation in multiple locations of the vegetal cortex (Houliston and Elinson, 1991a,b; Schroeder and Gard, 1992; Marrari et al., 2003; Tran et al., 2012; Olson et al., 2015). Our findings suggest that, following this nucleation, Prc1l-mediated microtubule bundling generates and/or amplifies an initial microtubule alignment bias, thus stabilizing microtubule tracks and amplifying the rotation of the vegetal cortex. In zebrafish, loss of function for the maternal Kinesin-1 Kif5ba results in vegetal cortex microtubule alignment defects (Campbell et al., 2015), and Prc1 has been shown to interact with a subset of Kinesin-family motors (Kurasawa et al., 2004; Zhu and Jiang, 2005; Gruneberg et al., 2006; Fu et al., 2009; Bieling et al., 2010; Subramanian et al., 2013; Vitre et al., 2014; Nguyen et al., 2018). Thus, zebrafish maternal Kif5Ba and Prc1l may act in concert to mediate vegetal cortex microtubule alignment and bundling.
Additionally, microtubule growth in the vegetal pole in zebrafish (Jesuthasan and Strähle, 1997; Tran et al., 2012) occurs at a time concomitant with growth of the sperm-derived monoaster at the animal pole (Dekens et al., 2003; Lindeman and Pelegri, 2012), a temporal coincidence that also occurs in Xenopus (Houliston and Elinson, 1991a,b; Schroeder and Gard, 1992). This suggests embryo-wide signals promote microtubule growth during that early developmental time. Our studies suggest that following such microtubule growth, microtubules growing in the vegetal pole are additionally modified through Prc1l-dependent bundling to produce parallel microtubule tracks involved in the relocalization of dorsal determinants. Further research will be required to improve our understanding of the common signals as well as specific factors involved in these processes.
In summary, we show that a maternal-specific copy of prc1l has a role at the egg-to-embryo transition, spanning meiosis and early embryonic mitoses, in key processes involving microtubule reorganization: at the spindle and midbody during cell division, and for vegetal microtubule array reorganization required for the segregation of dorsal determinants.
MATERIALS AND METHODS
Ethics statement
All animals were handled in strict accordance with good animal practice as defined by the relevant national and/or local animal welfare bodies, and all animal work was approved by the School of Medicine and Public Health Institutional Animal Care and Use Committee at the University of Wisconsin–Madison (assurance number A3369-01).
Fish maintenance and genetic lines
Fish stocks were raised and maintained under standard conditions at 26.5°C. tmi was originally isolated from an N-ethyl-N-nitrosourea (ENU)-induced mutagenesis screen (Dosch et al., 2004; Nair et al., 2013; Abrams et al., 2020). Homozygous tmi mutant fish were identified phenotyping, or by using the locked nucleic acid (LNA) endpoint genotyping method, which employs two fluorescent probes: HEX for identification of the wild-type allele, and FAM for identification of the mutant allele. Mutant embryos were obtained by pairing homozygous mutant females with male siblings. Wild-type controls were obtained from the AB line. Embryos were collected and allowed to develop in E3 embryonic medium until the desired stage, at which point they were fixed using 4% paraformaldehyde. All maternal-effect phenotypes were 100% penetrant with respect to early embryonic lethality (no cellularization followed by lysis) and across multiple tested generations, with experiments analyzing at least three different clutches each from wild-type and mutant females.
For LNA genotyping, fish were anesthetized with 0.014% tricaine and the tail fin was clipped using a razor blade and placed in 100 μl of 50 mM NaOH. Tissue lysates were incubated at 95°C for 20 min then cooled to 4°C for approximately 5 min. One-tenth of the volume of Tris-HCl pH 8.0 was added to neutralize the lysate, and it was centrifuged at 13,000 rpm (14,000 g) for 2 min. A typical 10 μl endpoint genotyping reaction contained 5 μl primetime gene expression master mix (IDT), 1 μl of 10 μM forward and reverse primer mix, 0.5 μl, FAM, 0.5 μl HEX, 10-25 ng of genomic DNA, and ddH2O up to 10 μl.
The primers for LNA genotyping of tmi were: (5′→3′): forward: CTGCTCACGCATGACAATATC; reverse: AGTTGATAGAGAGTAGAGACACTTT. LNA probes were: HEX: /5HEX/AGC+C+T+T+GAA+A+CT/3IABkFQ/; FAM: /56-FAM/AGC+C+T+A+GAA+A+CT/3IABkFQ/.
Cloning and sequence analysis
Homozygous tmi mutant males were crossed to WIK females to raise F1 fish, which were then in-crossed to obtain the F2 mapping generation. Embryos from F2 females were screened for the syncytial phenotype at 2-4 hpf. Genomic DNA from F2 females that produced syncytial mutant clutches was analyzed for segregation of a pan-genomic panel of 244 SSLP markers (Nair et al., 2013). This analysis found linkage of tmi to markers z25665, z65461 and z9233 in linkage group 21, with recombination frequencies of 3/268, 0/132 and 21/52, respectively (varying numbers of total reflect available polymorphic markers within analyzed crosses). The genetically defined region included seven candidate genes encoding proteins with predicted function in cytoskeletal dynamics and/or cell division: Sfswap, FYN binding protein, MOZART, Mitotic spindle organizing protein 2B, Smarcb1b and two novel proteins Zgc:193801 and Zgc:86764 present in this interval. Full-length cDNAs for each of the seven candidate genes were cloned from mature eggs of wild-type and tmi homozygous females, sequenced to identify a potential lesion, and only zgc:86764 contained a non-synonymous mutation.
Complementation test using CRISPR-Cas9
A CRISPR design web tool, CHOPCHOP, was used to identify a guide RNA site, GTGGACATATGGGACAGCATCGG, immediately upstream of the first predicted protein domain of the Prc1l protein, as identified by Ensembl. The guide RNA DNA template was produced using to two oligonucleotides. One oligonucleotide included a T7 promoter, the 20-base target site sequence from ChopChop without the PAM site, and a complementary region to the constant oligonucleotide. The constant oligonucleotide contained the reverse complement of the tracrRNA tail, allowing for interaction between the guide RNA and Cas9 protein. The two oligos were annealed and filled in using T4 DNA polymerase (Gagnon et al., 2014). The guide RNA was synthesized using the MEGAshortscript T7 Transcription Kit (AM1334, Thermo Fisher Scientific) and purified using an ethanol/ammonium acetate protocol. The concentration of the guide RNA was measured using a NanoDrop spectrophotometer, and its integrity was checked by gel electrophoresis. Single-use aliquots of guide RNAs were stored at −80°C.
To create the CRISPR-Cas9 mutant zebrafish line, we injected a mixture of guide RNA (200 pg/nl total final concentration) and Cas9 protein (PNA Bio; 400 pg/nl final concentration) into one-cell-stage AB embryos. To confirm Cas9 activity, a subset of the injected embryos was collected at 24 hpf, DNA was extracted from these embryos and a 95 bp fragment across the guide RNA site was amplified by PCR (forward: TGAGATCAATCATGCGATGG; reverse: TTTTTACAGTTTGCATTCTCTCCA) and resolved on a 2.5% agarose gel. If Cas9-mediated genetic edits were created in the somatic cells, the population of DNA that contains a variety of INDELS generates a smear, confirming Cas9 activity (Moravec and Pelegri, 2019). Once the prc1l F0-injected fish reached sexual maturity, offspring carriers of insertions and deletions (INDELs) were identified using fin clip DNA and the above method for INDEL identification, and the induced mutations were sequenced. The genotype of tmip4anua/prc1luw101 females was confirmed by testing for each allele independently. The prc1luw10 line is available upon request.
In situ hybridization
Whole-mount in situ hybridization was carried out to detect RNAs of interest using digoxygenin-labeled antisense RNA probes (Thisse and Thisse, 2008). To generate the prc1l antisense probe, the T7 promoter sequence was attached to a 20 bp antisense sequence of prc1l cDNA by PCR. Subsequently, the PCR product was used as a template for in vitro transcription using T7 polymerase and labeling the transcribed RNA with digoxygenin-conjugated ribonucleotides. A sense control probe was also designed by attaching the T7 promoter sequence to the 20 bp sense cDNA sequence. These sequences were also used as primers for RT-PCR. The wnt8a probe was a gift from the Lekven lab (Lekven et al., 2001), and generated by linearizing with Ap1I. For a visible label, the digoxygenin was detected with alkaline phosphatase-conjugated anti-digoxygenin antibodies and the chromogenic substrates NBT and BCIP. Embryos exhibiting reduced or no label (e.g. 4-8% of the embryos in Fig. 6B-D) were excluded from the scoring.
Western blotting
Approximately 200-500 embryos were collected from wild-type and tmi mutant fish. Embryos were lysed in RIPA buffer using a 22G syringe. Lysates were centrifuges at 13,000 rpm (14,000 g) for 5 min at 4°C to collect debris. Protein concentration was determined and 150 or 300 µg was loaded onto precast 4-15% TGX gels (Bio-Rad) and blotted onto PVDF membranes for 1 h at 100 V at 4°C. Membranes were blocked in 5% milk and blotted with 1:200 mouse-anti Prc1l (amino acids 59-102, Abmart) and 1:5000 anti-mouse HRP (31430, Thermo Fisher Scientific). Membranes were developed using SuperSignal West Pico (Thermo Fisher Scientific).
Immunolabeling
Embryos from wild-type and tmi mutant females were chemically dechorionated with a pronase solution (∼2 mg/ml in E3 embryonic medium) and forceps, and fixed at the appropriate time points in cytoskeleton-preserving fixative (4% paraformaldehyde, 0.25% glutaraldehyde, 5 mM EGTA, 0.2% Triton X-100 in ddH2O) overnight at 4°C. Fixed embryos were washed with PBS, permeabilized in methanol overnight at −20°C, rehydrated in a methanol:PBS series, and treated with 0.5 mg/ml sodium borohydrate 30 min at room temperature to inactivate remaining glutaraldehyde. Antibodies against Prc1l were derived from three separate peptides to amino acids 59-102 of Prc1l (Abmart, antibodies available upon request), used as unpurified samples and validated by lack of labeling in tmi mutants by whole-mount immunofluorescence and western analysis. Primary antibodies used for immunolabeling of fixed embryos were as follows: rabbit anti-β-catenin (1:1000, C2206, Sigma-Aldrich), mouse anti-Prc1l (amino acids 59-102; 1:400; Abmart), mouse anti-α-Tubulin (1:2500, T5168, Sigma-Aldrich), rat anti-α-Tubulin (1:1000, ab6161, Abcam), mouse anti-γ-tubulin (1:2000, T6557, Sigma-Aldrich). Confocal microscopy images were obtained using a Zeiss LSM 510 for fixed images or Zeiss LSM 780 (for high magnification, including super-resolution microscopy) and processed with Fiji (Schindelin et al., 2012).
For quantification of the interval between Prc1l protein on vegetal cortex microtubules, 30 regions of interest (ROIs) were randomly placed on the image in Fiji using an adapted version of the macro ‘RandomSamplePerimeterMethod’ (see also https://blog.bham.ac.uk/intellimic/g-landini-software/). Using the freehand line tool, a line was drawn along the longest microtubule in each ROI. The plot profile tool was then used to assess detection of Prc1l protein aggregate localization to the microtubule. Peaks of colocalized fluorescence were defined as an increase above a gray value of 100. On the plot profiles, the interpeak distances (IPDs) were measured between the apex of each peak to determine the distance (µm) between protein aggregates. Data presented are averages of IPDs representing the collective IPDs within each ROI. Technical controls were obtained by taking measurements from wild-type images with the green channel (detecting Prc1l protein aggregates) rotated 90° clockwise. To assess relative dispersion of data, the coefficient of variation (CV) was calculated with the ‘CVTEST’ macro on Microsoft Excel using the average IPDs from three images for each condition (90 average IPDs per condition).
Fluorescence in situ hybridization with microtubule immunolabeling
Embryos were fixed with a paraformaldehyde:glutaraldehyde fixative (see the ‘Immunolabeling’ section) and first processed to detect wnt8a RNA using fluorescence in situ hybridization and digoxygenin-labeled antisense wnt8a RNA probe as described in the ‘In situ hybridization’ section, except using Tyramide Signal Amplification kit (PerkinElmer) for fluorescence detection. Subsequently, labeled embryos were processed for microtubule immunolabeling (mouse anti-α-Tubulin, 1:2500, T5168, Sigma-Aldrich) (Hansen and Pelegri, 2021) as above. After labeling, embryos were manually dissected using fine forceps to separate the vegetal hemisphere of each embryo from the animal hemisphere. The vegetal sections were then mounted flat with the cut side towards the slide and imaged using a Zeiss LSM780 confocal microscope (63× objective). Quantification of wnt8a RNA particle association with microtubules was performed using Fiji (Schindelin et al., 2012) (Fig. S6). For analysis, two representative 20 µm×20 µm ROIs per embryo were selected from single focal planes (z-slices) from confocal micrographs of five wild-type and five tmi/prc1l embryos. All detectable wnt8a RNA particles in each ROI were selected, counted and outlined using the ‘Make Binary’ and ‘Analyze Particles’ functions and the resulting outlines were overlaid on the corresponding microtubule channel of the ROI. Each outlined particle area was visually inspected to determine whether or not microtubule labeling was also observed within the boundaries of the outlined RNA particle in the same focal plane. The percentage of wnt8a RNA particles that overlapped with microtubules for each group (wild type versus tmi) was compared using an unpaired Student's t-test.
Microtubule analysis
Images were taken of microtubules in early two-cell wild-type and tmi embryos at the FMA and vegetal cortex. Images were processed by generating z-projected images (representing 6.5-8.0 µm in depth) in Fiji and subsequently used to analyze microtubule bundling alignment, thickness and orientation.
Using FMA images, line segment ROIs were placed at the tips of the FMA on each side and along the length of the furrow and analyzed using the ‘Plot Profile’ function. Each plot was then processed using the ‘Find Peaks’ plugin to identify the x-coordinates of peak apices. Peaks were defined by a minimum gray value amplitude (y-coordinate; ai) threshold, whereby the minimum amplitude of a peak was set to one standard deviation of the plotted gray value data. Peaks that had two maxima (x-coordinates) without reaching the minimum amplitude between the apices (observed in cases of spatially restricted detanglement on an otherwise continuous microtubule bundle), were considered a single microtubule bundle. For each embryo, maxima data from the described plots were compiled into two categories, right- and left-sided coordinates (XR, XL), corresponding to respective sides of the furrow. The difference of the right- and the left-side values [XR−XL] was calculated as an interpeak distance (di), which, if below a threshold of 0.3 µm, was used as an indication of microtubule alignment. Single alignment of microtubule tracks on opposite sides of the furrow was determined based on whether right- or left-sided value had >1 contrary value (i.e. peak maxima) within the 0.3 µm threshold. Once a di of two peaks was considered aligned, the next two contrary peaks were analyzed for alignment. Upon analysis of all peak profiles, the values for each sample group were used to generate a contrasting histogram that demonstrates the frequency of interpeak distances for each sample group.
A microtubule thickness analysis was performed to assess microtubule bundling at the FMA and vegetal cortex. The analysis at the FMA was conducted using the plot profiles from the alignment analysis. Subsequently, additional plot profiles were generated using images representing the vegetal cortex. ROIs were drawn diagonally across these images and plots were created using the ‘Plot Profile’ function as described above. The thickness of the microtubule bundles was quantified using the width of the plot peaks (i.e. the distance between peak minima) from the plots of both the FMA and vegetal cortex.
Orientation was qualified for microtubule tracks at both the vegetal cortex and FMA. The ‘OrientationJ Analysis’ plugin was applied to each image to encode microtubule bundle track orientation with colors based on a 180° plane (Püspöki et al., 2016). Quantitative values were assigned to each track in an image based on its angle. To assess microtubule orientation synchronicity, the angularity values for each sample group were collated and plotted onto histograms. The distribution spread of these histograms was then used to evaluate the synchronicity of microtubule track orientations.
Sequence and protein structure analysis
The DNA and amino acid sequences for the full protein alignment of zebrafish Prc1a, Prc1b and Prc1l and human PRC1 were collected from Ensembl (http://www.ensembl.org/index.html). Sequence alignment for full proteins and mutations was carried out with Clustal Omega (Madeira et al., 2019). The highlighted domains for zebrafish Prc1l protein were labeled according to previous analysis of human PRC1 (Subramanian et al., 2010). Gene phylogenetic trees were carried out using Phylogeny.fr (Dereeper et al., 2008; Dereeper et al., 2010).
Three-dimensional protein structures of zebrafish Prc1a, Prc1b and Prc1l were predicted using the Phyre2 web server (Kelley et al., 2015). The output secondary structures were color-coded using a rainbow gradient to distinguish the N terminus and the C terminus (red to blue, respectively). All predictive protein structures were constructed from a homologous protein folding template, c4l6yB, provided by the RCSB Protein Data Bank (PDB) (https://www.rcsb.org/).
Supplementary Material
Acknowledgements
We thank Drs Mary Mullins (Perelman School of Medicine at the University of Pennsylvania) and Arne Lekven (University of Houston) for generously providing the tmi mutation and wnt8a probes, respectively. We also thank past and current members of the Pelegri lab for their contributions to our research, particularly our animal husbandry staff for the care of the aquatic facility. Microscopy was performed at the Newcomb Imaging Center, Department of Botany at the University of Wisconsin–Madison.
Footnotes
Author contributions
Conceptualization: S.N., E.L.W., F.P.; Methodology: S.N., E.L.W., C.E.M., R.L.T., C.L.H., F.P.; Validation: S.N., E.L.W., C.E.M., R.L.T., C.L.H.; Formal analysis: S.N., E.L.W., C.E.M., R.L.T., C.L.H.; Investigation: S.N., E.L.W., C.E.M., C.L.H.; Data curation: R.L.T.; Writing - original draft: S.N., E.L.W.; Writing - review & editing: C.E.M., R.L.T., C.L.H., F.P.; Visualization: S.N., E.L.W., C.E.M., R.L.T., C.L.H., F.P.; Supervision: F.P.; Funding acquisition: F.P.
Funding
This work was supported by the National Institute of General Medical Sciences (R01 GM065303) and the University of Wisconsin–Madison. Deposited in PMC for release after 12 months.
Data availability
All relevant data can be found within the article and its supplementary information.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.200564
References
- Abrams, E. W. and Mullins, M. C. (2009). Early zebrafish development: it's in the maternal genes. Curr. Opin. Genet. Dev. 19, 396-403. 10.1016/j.gde.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrams, E. W., Fuentes, R., Marlow, F. L., Kobayashi, M., Zhang, H., Lu, S., Kapp, L., Joseph, S. R., Kugath, A., Gupta, T.et al. (2020). Molecular genetics of maternally-controlled cell divisions. PLoS Genet. 16, e1008652. 10.1371/journal.pgen.1008652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agromayor, M. and Martin-Serrano, J. (2013). Knowing when to cut and run: mechanisms that control cytokinetic abscission. Trends Cell Biol. 23, 433-441. 10.1016/j.tcb.2013.04.006 [DOI] [PubMed] [Google Scholar]
- Ali, M. Y., Lu, H., Bookwalter, C. S., Warshaw, D. M. and Trybus, K. M. (2008). Myosin V and Kinesin act as tethers to enhance each others’ processivity. Proc. Natl. Acad. Sci. USA 105, 4691-4696. 10.1073/pnas.0711531105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazaz, A. I., Ahmad, I., Nafath-Ul-Arab, T. H., Shah, O. A., Asimi, B. A., Bhat, Z., Yousuf, S. H., Baba, N., Razak, and Fatima, A. (2020). A review of induction of triploidy in fish using heat, pressure and cold shock treatments. J. Entol. Zool. Studies 8, 381-385. [Google Scholar]
- Bellipanni, G., Varga, M., Maegawa, S., Imai, Y., Kelly, C., Myers, A. P., Chu, F., Talbot, W. S. and Weinberg, E. S. (2006). Essential and opposing roles of zebrafish β-catenins in the formation of dorsal axial structures and neuroectoderm. Development 133, 1299-1309. 10.1242/dev.02295 [DOI] [PubMed] [Google Scholar]
- Bieling, P., Telley, I. A. and Surrey, T. (2010). A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell 142, 420-432. 10.1016/j.cell.2010.06.033 [DOI] [PubMed] [Google Scholar]
- Blum, M., Schweickert, A., Vick, P., Wright, C. V. E. and Danilchik, M. V. (2014). Symmetry breakage in the vertebrate embryo: when does it happen and how does it work? Dev. Biol. 393, 109-123. 10.1016/j.ydbio.2014.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontems, F., Stein, A., Marlow, F., Lyautey, J., Gupta, T., Mullins, M. C. and Dosch, R. (2009). Bucky ball organizes germ plasm assembly in zebrafish. Curr. Biol. 19, 414-422. 10.1016/j.cub.2009.01.038 [DOI] [PubMed] [Google Scholar]
- Campbell, P. D., Heim, A. E., Smith, M. Z. and Marlow, F. L. (2015). Kinesin-1 interacts with Bucky ball to form germ cells and is required to pattern the zebrafish body axis. Development 142, 2996-3008. 10.1242/dev.124586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, T., Liu, Z., Zhang, Y., Zhang, L., Huang, R., He, L., Gu, Y., Chen, Z., Zheng, Q., Shi, L.et al. (2019). Precise and rapid validation of candidate gene by allele specific knockout with CRISPR/Cas9 in wild mice. Front. Genet. 10, 124. 10.3389/fgene.2019.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.-C., Zhou, Y. and Detrich, H. W.III. (2002). Zebrafish mitotic kinesin-like protein 1 (Mklp1) functions in embryonic cytokinesis. Physiol. Genom. 8, 51-66. 10.1152/physiolgenomics.00042.2001 [DOI] [PubMed] [Google Scholar]
- Ching, Y.-H., Munroe, R. J., Moran, J. L., Barker, A. K., Mauceli, E., Fennell, T., Dipalma, F., Lindblad-Toh, K., Abcunas, L. M., Gilmour, J. F.et al. (2010). High resolution mapping and positional cloning of ENU-induced mutations in gthe Rw region of mouse chromosome 5. BMC Genet. 11, 106. 10.1186/1471-2156-11-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois, A., Schuh, M., Ellenberg, J. and Hiiragi, T. (2012). The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J. Cell Biol. 198, 357-370. 10.1083/jcb.201202135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilchik, M. V., Funk, W. C., Brown, E. E. and Larkin, K. (1998). Requirement for microtubules in new membrane formation during cytokinesis of Xenopus embryos. Dev. Biol. 194, 47-60. 10.1006/dbio.1997.8815 [DOI] [PubMed] [Google Scholar]
- Danilchik, M. V., Bedrick, S. D., Brown, E. E. and Ray, K. (2003). Furrow microtubules and localized exocytosis in cleaving Xenopus laevis embryos. J. Cell Sci. 116, 273-283. 10.1242/jcs.00217 [DOI] [PubMed] [Google Scholar]
- Dekens, M. P. S., Pelegri, F. J., Maischein, H.-M. and Nüsslein-Volhard, C. (2003). The maternal-effect gene futile cycle is essential for pronuclear congression and mitotic spindle assembly in the zebrafish zygote. Development 130, 3907-3916. 10.1242/dev.00606 [DOI] [PubMed] [Google Scholar]
- Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chenevet, F., Dufayard, J.-F., Guindon, S., Lefort, V., Lescot, M.et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nuc. Acid Res. 36, W465-W469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper, A., Audic, S., Claverie, J.-M. and Blanc, G. (2010). BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10, 8. 10.1186/1471-2148-10-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosch, R., Wagner, D. S., Mintzer, K. A., Runke, G., Wiemelt, A. P. and Mullins, M. C. (2004). Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Dev. Cell 6, 771-780. 10.1016/j.devcel.2004.05.002 [DOI] [PubMed] [Google Scholar]
- Eggert, U. S., Mitchison, T. J. and Field, C. M. (2006). Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 75, 543-566. 10.1146/annurev.biochem.74.082803.133425 [DOI] [PubMed] [Google Scholar]
- Elinson, R. P. and Rowning, B. (1988). A transient array of parallel microtubules in frog eggs: potential tracks for a cytoplasmic rotation that specifies the dorso-ventral axis. Dev. Biol. 128, 185-197. 10.1016/0012-1606(88)90281-3 [DOI] [PubMed] [Google Scholar]
- Eno, C. and Pelegri, F. (2016). Germ cell determinant transmission, segregation and function in the zebrafish embryo. In Insights from animal reproduction (ed. Carreira R. P.), pp. 115-142. Rijeka, Croatia: InTech. [Google Scholar]
- Eno, C. and Pelegri, F. (2018). Modulation of F-actin dynamics by maternal Mid1ip1L controls germ plasm aggregation and furrow recruitment in the zebrafish embryo. Development 145, dev156596. 10.1242/dev.156596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eno, C., Solanki, B. and Pelegri, F. (2016). aura (mid1ip1l) regulates the cytoskeleton at the zebrafish egg-to-embryo transition. Development 143, 1585-1599. 10.1242/dev.130591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eno, C., Gomez, T., Slusarski, D. C. and Pelegri, F. (2018). Slow calcium waves mediate furrow microtubule reorganization and germ plasm compaction in the early zebrafish embryo. Development 145, dev156604. 10.1242/dev.156604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fededa, J. P. and Gerlich, D. W. (2012). Molecular control of animal cell cytokinesis. Nature Cell Biol. 14, 440-447. 10.1038/ncb2482 [DOI] [PubMed] [Google Scholar]
- Fu, C., Ward, J. J., Loiodice, I., Velve-Casquillas, G., Nedelec, F. J. and Tran, P. T. (2009). Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev. Cell 17, 257-267. 10.1016/j.devcel.2009.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon, J. A., Valen, E., Thyme, S. B., Huang, P., Ahkmetova, L., Pauli, A., Montague, T. G., Zimmerman, S., Richter, C. and Schier, A. F. (2014). Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9, e106396. 10.1371/journal.pone.0098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, X., Grotjahn, D., Welch, E., Lyman-Gingerich, J., Holguin, C., Dimitrova, E., Abrams, E. W., Gupta, T., Marlow, F. L., Yabe, T.et al. (2014). Hecate/Grip2a acts to reorganize the cytoskeleton in the symmetry-breaking event of embryonic axis induction. PLoS Genet. 10, e1004422. 10.1371/journal.pgen.1004422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart, J., Danilchik, M., Doniach, T., Roberts, S., Rowning, B. and Stewart, R. (1989). Cortical rotation of the Xenopus egg: consequences for the anteroposterior pattern of embryonic dorsal development. Development 107 Suppl., 37-51. 10.1242/dev.107.Supplement.37 [DOI] [PubMed] [Google Scholar]
- Gruneberg, U., Neef, R., Li, X., Chan, E. H. Y., Chalamalasetty, R. B., Nigg, E. A. and Barr, F. A. (2006). KIF14 and citron kinase act together to promote efficient cytokinesis. J. Cell Biol. 172, 363-372. 10.1083/jcb.200511061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, C. L. and Pelegri, F. (2021). Methods for visualization of RNA and cytoskeletal elements in the early zebrafish embryo. Meth. Mol. Biol. 2218, 219-244. 10.1007/978-1-0716-0970-5_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, Y., Maegawa, S., Nagai, T., Yamaha, E., Suzuki, H., Yasuda, K. and Inoue, K. (2004). Localized maternal factors are required for zebrafish germ cell formation. Dev. Biol. 268, 152-161. 10.1016/j.ydbio.2003.12.013 [DOI] [PubMed] [Google Scholar]
- Houliston, E. and Elinson, R. P. (1991a). Evidence for the involvement of microtubules, ER, and kinesin in the cortical rotation of fertilized frog eggs. J. Cell Biol. 114, 1017-1028. 10.1083/jcb.114.5.1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houliston, E. and Elinson, R. P. (1991b). Patterns of microtubule polymerization relating to cortical rotation in Xenopus laevis eggs. Development 112, 107-117. 10.1242/dev.112.1.107 [DOI] [PubMed] [Google Scholar]
- Houston, D. W. (2012). Cortical rotation and messenger RNA localization in Xenopus axis formation. WIREs Dev. Biol. 1, 371-388. 10.1002/wdev.29 [DOI] [PubMed] [Google Scholar]
- Jesuthasan, S. (1998). Furrow-associated microtubule arrays are required for the cohesion of zebrafish blastomeres following cytokinesis. J. Cell Sci. 111, 3695-3703. 10.1242/jcs.111.24.3695 [DOI] [PubMed] [Google Scholar]
- Jesuthasan, S. and Strähle, U. (1997). Dynamic microtubules and specification of the zebrafish embryonic axis. Curr. Biol. 7, 31-42. 10.1016/S0960-9822(06)00025-X [DOI] [PubMed] [Google Scholar]
- Jiang, W. J., Jimenez, G., Wells, N. J., Hope, T. J., Wahl, G. M., Hunter, T. and Fukunaga, R. (1998). PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol. Cell 2, 877-885. 10.1016/S1097-2765(00)80302-0 [DOI] [PubMed] [Google Scholar]
- Kane, D. A. and Kimmel, C. B. (1993). The zebrafish midblastula transition. Development 119, 447-456. 10.1242/dev.119.2.447 [DOI] [PubMed] [Google Scholar]
- Kavumpurath, S. and Pandian, T. J. (2008). Induction of triploidy in the zebrafish, Brachydanio rerio (Hamilton). Aquaculture Res. 21, 299-306. 10.1111/j.1365-2109.1990.tb00468.x [DOI] [Google Scholar]
- Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. and Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nature Prot. 10, 845-858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, E. H., Howes, S., Ti, S.-C., Ramírez-Aportela, E., Kapoor, T. M., Chacón, P. and Nogales, E. (2016). Near-atomic cryo-EM structure of PRC1 bound to the microtubule. Proc. Natl. Acad. Sci. USA 113, 9430-9439. 10.1073/pnas.1609903113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, C., Chin, A. J., Leatherman, J. L., Kozlowski, D. J. and Weinberg, E. S. (2000). Maternally controlled β-catenin-mediated signaling is required for organizer formation in the zebrafish. Development 127, 3899-3911. 10.1242/dev.127.18.3899 [DOI] [PubMed] [Google Scholar]
- Kishimoto, Y., Lee, K. H., Zon, L., Hammerschmidt, M. and Schulte-Merker, S. (1997). The molecular nature of zebrafish swirl: BMP2 function is essential during early dorsoventral patterning. Development 124, 4457-4466. 10.1242/dev.124.22.4457 [DOI] [PubMed] [Google Scholar]
- Knaut, H., Pelegri, F., Bohmann, K., Schwarz, H. and Nüsslein-Volhard, C. (2000). Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J. Cell Biol. 149, 875-888. 10.1083/jcb.149.4.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasawa, Y., Earnshaw, W. C., Mochizuki, Y., Dohmae, N. and Todokoro, K. (2004). Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 23, 3237-3248. 10.1038/sj.emboj.7600347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekven, A. C., Thorpe, C. J., Waxman, J. S. and Moon, R. T. (2001). Zebrafish wnt8 encodes two wnt8 proteins on a bicistronic transcript and is required for mesoderm and neurectoderm patterning. Dev. Cell 1, 103-114. 10.1016/S1534-5807(01)00007-7 [DOI] [PubMed] [Google Scholar]
- Lindeman, R. E. and Pelegri, F. (2010). Vertebrate maternal-effect genes: insights into fertilization, early cleavage divisions, and germ cell determinant localization from studies in the zebrafish. Mol. Rep. Dev. 77, 299-313. 10.1002/mrd.21128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman, R. E. and Pelegri, F. (2012). Localized products of futile cycle/lrmp promote centrosome-nucleus attachment in the zebrafish zygote. Curr. Biol. 22, 843-851. 10.1016/j.cub.2012.03.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, F.-I., Thisse, C. and Thisse, B. (2011). Identification and mechanism of regulation of the zebrafish dorsal determinant. Proc. Natl. Acad. Sci. USA 108, 15876-15880. 10.1073/pnas.1106801108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira, F., Park, Y., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., Basutkar, P., Tivey, A. R. N., Potter, S. C., Finn, R. D.et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nuc. Acids Res. 47, W636-W641. 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrari, Y., Clarke, E. J., Rouvière, C. and Houliston, E. (2003). Analysis of microtubule movement on isolated Xenopus egg cortices provides evidence that the cortical rotation involves dynein as well as Kinesin Related Proteins and is regulated by local microtubule polymerisation. Dev. Biol. 257, 55-70. 10.1016/S0012-1606(03)00057-5 [DOI] [PubMed] [Google Scholar]
- Marrari, Y., Rouvière, C. and Houliston, E. (2004). Complementary roles for dynein and kinesins in the Xenopus egg cortical rotation. Dev. Biol. 271, 38-48. 10.1016/j.ydbio.2004.03.018 [DOI] [PubMed] [Google Scholar]
- Mierzwa, B. and Gerlich, D. W. (2014). Cytokinetic abscission: molecular mechanisms and temporal control. Dev. Cell 31, 525-538. 10.1016/j.devcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Miranda-Rodríguez, J. R., Salas-Vidal, E., Lomelí, H., Zurita, M. and Schnabel, D. (2017). RhoA/ROCK pathway activity is essential for the correct localization of the germ plasm mRNAs in zebrafish embryos. Dev. Biol. 421, 27-42. 10.1016/j.ydbio.2016.11.002 [DOI] [PubMed] [Google Scholar]
- Mollinari, C., Kleman, J.-P., Jiang, W., Schoehn, G., Hunter, T. and Margolis, R. L. (2002). PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157, 1175-1186. 10.1083/jcb.200111052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravec, C. E. and Pelegri, F. (2019). An accessible protocol for the generation of CRISPR-Cas9 knockouts using INDELS in zebrafish. Methods Mol. Biol. 1920, 377-392. 10.1007/978-1-4939-9009-2_23 [DOI] [PubMed] [Google Scholar]
- Nair, S., Marlow, F., Abrams, E., Kapp, L., Mullins, M. C. and Pelegri, F. (2013). The chromosomal passenger protein Birc5b organizes microfilaments and germ plasm in the zebrafish embryo. PLoS Genet. 9, e1003448. 10.1371/journal.pgen.1003448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, P. A., Field, C. M. and Mitchison, T. J. (2018). Prc1E and Kif4A control microtubule organization within and between large Xenopus egg asters. Mol. Biol. Cell 29, 304-316. 10.1091/mbc.E17-09-0540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima, H., Rothhämel, S., Shimizu, T., Kim, C.-H., Yonemura, S., Marlow, F. L. and Hibi, M. (2010). Syntabulin, a motor protein linker, controls dorsal determination. Development 137, 923-933. 10.1242/dev.046425 [DOI] [PubMed] [Google Scholar]
- Olson, D. J., Oh, D. and Houston, D. W. (2015). The dynamics of plus end polarization and microtubule assembly during Xenopus cortical rotation. Dev. Biol. 401, 249-263. 10.1016/j.ydbio.2015.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui, M. S., Verbrugghe, K. J. and Skop, A. R. (2005). Midbodies and phragmoplasts: analogous structures involved in cytokinesis. Trends Cell Biol. 15, 404-413. 10.1016/j.tcb.2005.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegri, F. (2003). Maternal factors in zebrafish development. Dev. Dyn. 228, 535-554. 10.1002/dvdy.10390 [DOI] [PubMed] [Google Scholar]
- Pelegri, F., Knaut, H., Maischein, H.-M., Schulte-Merker, S. and Nüsslein-Volhard, C. (1999). A mutation in the zebrafish maternal-effect gene nebel affects furrow formation and vasa RNA localization. Curr. Biol. 9, 1431-1440. 10.1016/S0960-9822(00)80112-8 [DOI] [PubMed] [Google Scholar]
- Püspöki, Z., Storath, M., Sage, D. and Unser, M. (2016). Transforms and operators for directional bioimage analysis: a survey. Adv. Anat. Embryol. Cell Biol. 219, 69-93. 10.1007/978-3-319-28549-8_3 [DOI] [PubMed] [Google Scholar]
- Rappaport, R. (1996). Cytokinesis in animal cells. Cambridge: Cambridge University Press. [Google Scholar]
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B.et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Meth. 9, 676-682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, M. M. and Gard, D. L. (1992). Organization and regulation of cortical microtubules during the first cell cycle of Xenopus eggs. Development 114, 699-709. 10.1242/dev.114.3.699 [DOI] [PubMed] [Google Scholar]
- Schuyler, S. C., Liu, J. Y. and Pellman, D. (2003). The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. J. Cell Biol. 160, 517-528. 10.1083/jcb.200210021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets, M. D., Fox, C. A., Dowdle, M. E., Blaser, S. I., Chung, A. and Park, S. (2017). Controlling the messenger: regulated translation of maternal mRNAs in Xenopus laevis development. Adv. Exp. Med. Biol. 953, 49-82. 10.1007/978-3-319-46095-6_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha, R. L. and Draviam, V. M. (2013). Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK. Curr. Biol. 23, 1514-1526. 10.1016/j.cub.2013.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko, A. P., Chang, H.-Y., Sonobe, S., Fenyk, S. I., Weingartner, M., Bögre, L. and Hussey, P. J. (2006). Control of the AtMAP65-1 interaction with microtubules through the cell cycle. J. Cell Sci. 119, 3227-3237. 10.1242/jcs.03051 [DOI] [PubMed] [Google Scholar]
- Subramanian, R., Wilson-Kubalek, E. M., Arthur, C. P., Bick, M. J., Campbell, E. A., Darst, S. A., Milligan, R. A. and Kapoor, T. M. (2010). Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell 142, 433-443. 10.1016/j.cell.2010.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, R., Ti, S.-C., Tan, L., Darst, S. A. and Kapoor, T. M. (2013). Marking and measuring single microtubules by PRC1 and Kinesin-4. Cell 154, 377-390. 10.1016/j.cell.2013.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda, P., Fulka, H. and Malik, R. (2017). Clearance of parental products. Adv. Exp. Med. Biol. 953, 489-535. 10.1007/978-3-319-46095-6_10 [DOI] [PubMed] [Google Scholar]
- Tanaka, K., Mukae, N., Dewar, H., Van Breugel, M., James, E. K., Prescott, A. R., Antony, C. and Tanaka, T. U. (2005). Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 434, 987-994. 10.1038/nature03483 [DOI] [PubMed] [Google Scholar]
- Thisse, C. and Thisse, B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Methods 3, 59-69. 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Tran, L. D., Hino, H., Quach, H., Lim, S., Shindo, A., Mimori-Kiyosue, Y., Mione, M., Ueno, N., Winkler, C., Hibi, M.et al. (2012). Dynamic microtubules at the vegetal cortex predict the embryonic axis in zebrafish. Development 139, 3644-3652. 10.1242/dev.082362 [DOI] [PubMed] [Google Scholar]
- Tsai, T. Y.-C., Theriot, J. A. and Ferrell, J. E.Jr. (2014). Changes in oscillatory dynamics in the cell cycle of early Xenopus laevis embryos. PLoS Biol. 12, e1001788. 10.1371/journal.pbio.1001788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre, B., Gudimchuk, N., Borda, R., Kim, Y., Heuser, J. E., Cleveland, D. W. and Grishchuk, E. L. (2014). Kinetochore-microtubule attachment throughout mitosis potentiated by the elongated stalk of the kinetohore kinesin CENP-E. Mol. Biol. Cell 25, 2272-2281. 10.1091/mbc.e14-01-0698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, E. and Pelegri, F. (2015). Cortical depth and differential transport of vegetally localized dorsal and germ line determinants in the zebrafish embryo. Bioarchitecture 5, 13-26. 10.1080/19490992.2015.1080891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, E. L. and Pelegri, F. (2017). Reorganization of vegetal cortex microtubules and its role in axis induction in the early vertebrate embryo. In Cytoskeleton - Structure, Dynamics, Function and Disease. (ed. Jimenez-Lopez J. C.), pp. 3-31. Rijeka, Croatia: InTech. [Google Scholar]
- White, R. J., Collins, J. E., Sealy, I. M., Wali, N., Dooley, C. M., Digby, Z., Stemple, D. L., Murphy, D. N., Billis, K., Hourlier, T.et al. (2017). A high-resolution mRNA expression time course of embryonic development in zebrafish. eLife 6, e30860. 10.7554/eLife.30860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne, S. and Subramanian, R. (2018). Geometry of antiparallel microtubule bundles regulates relative sliding and stalling by PRC1 and Kif4A. eLife 7, e32595. 10.7554/eLife.32595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr, M., Tan, E. S., Parker, S. K., Detrich, H. W., III and Mitchison, T. J. (2010). A model for cleavage plane determination in early amphibian and fish embryos. Curr. Biol. 20, 2040-2045. 10.1016/j.cub.2010.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr, M., Obholzer, N. D., Megason, S. G., Detrich, H. W., III and Mitchison, T. J. (2011). Live imaging of the cytoskeleton in early cleavage-stage zebrafish embryos. Methods Cell Biol. 101, 1-18. 10.1016/B978-0-12-387036-0.00001-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe, T., Ge, X. and Pelegri, F. (2007). The zebrafish maternal-effect gene cellular atoll encodes the centriolar component sas-6 and defects in its paternal function promote whole genome duplication. Dev. Biol. 312, 44-60. 10.1016/j.ydbio.2007.08.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe, T., Ge, X., Lindeman, R., Nair, S., Runke, G., Mullins, M. C. and Pelegri, F. (2009). The maternal-effect gene cellular island encodes Aurora B kinase and is essential for furrow formation in the early zebrafish embryo. PLoS Genet. 5, e1000518. 10.1371/journal.pgen.1000518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, C. and Jiang, W. (2005). Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc. Natl. Acad. Sci. USA 102, 343-348. 10.1073/pnas.0408438102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, C., Lau, E., Schwarzenbacher, R., Bossy-Wetzel, E. and Jiang, W. (2006). Spatiotemporal control of spindle midzone formation by PRC1 in human cells. Proc. Natl. Acad. Sci. USA 103, 6196-6201. 10.1073/pnas.0506926103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.