Abstract
The placenta is a fast-evolving organ with large morphological and histological differences across eutherians, but the genetic changes driving placental evolution have not been fully elucidated. Transposable elements, through their capacity to quickly generate genetic variation and affect host gene regulation, may have helped to define species-specific trophoblast gene expression programs. Here we assess the contribution of transposable elements to human trophoblast gene expression as enhancers or promoters. Using epigenomic data from primary human trophoblast and trophoblast stem-cell lines, we identified multiple endogenous retrovirus families with regulatory potential that lie close to genes with preferential expression in trophoblast. These largely primate-specific elements are associated with inter-species gene expression differences and are bound by transcription factors with key roles in placental development. Using genetic editing, we demonstrate that several elements act as transcriptional enhancers of important placental genes, such as CSF1R and PSG5. We also identify an LTR10A element that regulates ENG expression, affecting secretion of soluble endoglin, with potential implications for preeclampsia. Our data show that transposons have made important contributions to human trophoblast gene regulation, and suggest that their activity may affect pregnancy outcomes.
Subject terms: Transcriptional regulatory elements, Epigenomics
Here the authors investigate the contribution of transposable elements to regulation of gene expression in human trophoblasts. Amongst other examples, they identify an LTR10A element with potential implications for preeclampsia.
Main
The success of human pregnancy depends on the healthy development and function of the placenta. Following implantation, fetally derived trophoblast cells invade maternal tissues interstitially, remodeling uterine spiral arteries well into the myometrium1. Aberrations to this process result in serious pregnancy complications that cause maternal and fetal morbidity and mortality, including recurrent pregnancy loss, fetal growth restriction, preterm birth and preeclampsia in the case of too little invasion, or disorders of the placenta accreta spectrum where invasion is too extensive2. However, the genetic determinants of these disorders remain unclear, as genome-wide association studies have revealed very few candidates, with the notable exception of FLT1 in preeclampsia3.
Placental development and structure displays wide variation across eutherian species, even within the primate order4. Notably, the deep interstitial trophoblast invasion observed in humans is unique to great apes5. Placentas differ in cellular composition, histological arrangement and gross morphology, as well as many molecular aspects, all of which shape interactions between conceptus and mother, and their outcomes. This striking variation reflects the myriad selective pressures associated with the feto-maternal conflicts that fuel fast evolution of this organ6. Yet, the genetic drivers of placental evolution remain to be fully elucidated.
Transposable elements (TEs) are one important yet understudied source of genetic variation. These abundant repetitive elements, which include endogenous retroviruses (ERVs), have made major contributions to human evolution, helping to shape both the coding and regulatory (noncoding) landscape of the genome. Akin to the variable and species-specific development and structure of the placenta, TEs are highly species-specific, making them putative drivers of placental evolution. Indeed, multiple genes with key roles in placentation have been derived from TEs7, most prominently the syncytin genes, whose products mediate cell–cell fusion to generate a syncytialized trophoblast layer that directly contacts the maternal blood8. Additionally, the noncoding portions of TEs (for example, the long terminal repeats (LTRs) in ERVs) have the ability to regulate gene transcription, and through this action contribute to human embryonic development9–11, innate immunity12, the development of cancer13 and the evolution of the feto-maternal interface, among others14,15. TEs can recruit host transcription factors, often in a highly tissue-specific manner, and gain epigenetic hallmarks of gene regulatory activity, acting as transcriptional promoters or distal enhancer elements14–16. We and others have previously shown that in mouse trophoblast stem cells, several ERV families can act as major enhancers of gene expression17,18. In humans, several examples of TE-encoded placenta-specific promoters have been uncovered, such as those driving expression of CYP19A1, NOS3 and PTN genes19. A fascinating example of a human placental TE-derived enhancer has also been described that affects gestational length when inserted into the mouse genome20. More recently, the Macfarlan laboratory has identified a group of putative lineage-specific placental enhancers that are derived from ERVs21. However, as the placenta is a heterogeneous tissue, it remains unclear whether all of these ERVs are active in trophoblast cells, and a genetic demonstration of their regulatory action is lacking.
In this Article, we identify ERV families that exhibit hallmarks of gene regulatory activity in human trophoblast. We show that these ERVs bind transcription factors required for placental development and lie close to genes with preferential trophoblast expression in a species-specific manner. Using genetic editing, we show examples of ERVs that act as gene enhancers in trophoblast, including an LTR10A element within the ENG gene that regulates the secretion of soluble ENG protein by the syncytiotrophoblast (SynT), which is both a marker for, and contributor to, the pathogenesis of preeclampsia.
Results
ERVs with regulatory potential in human trophoblast
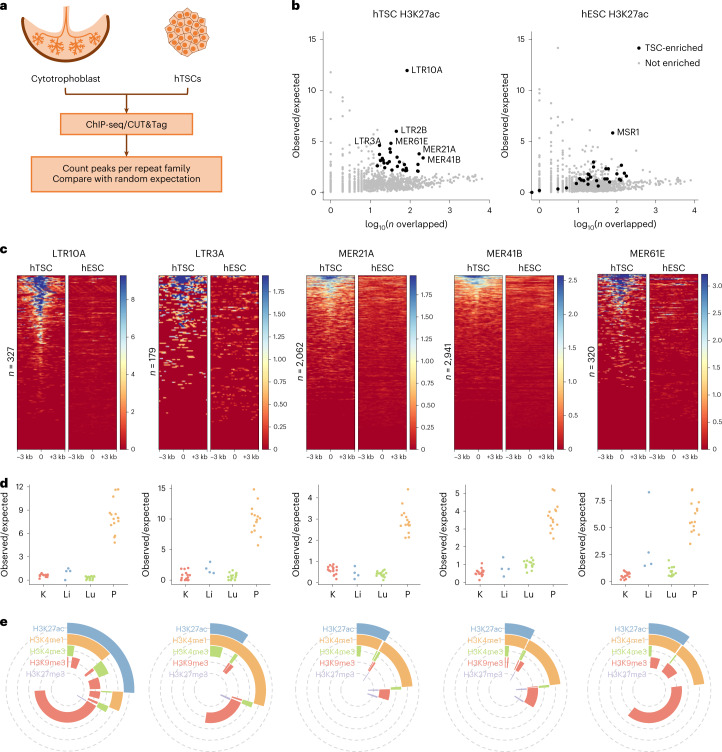
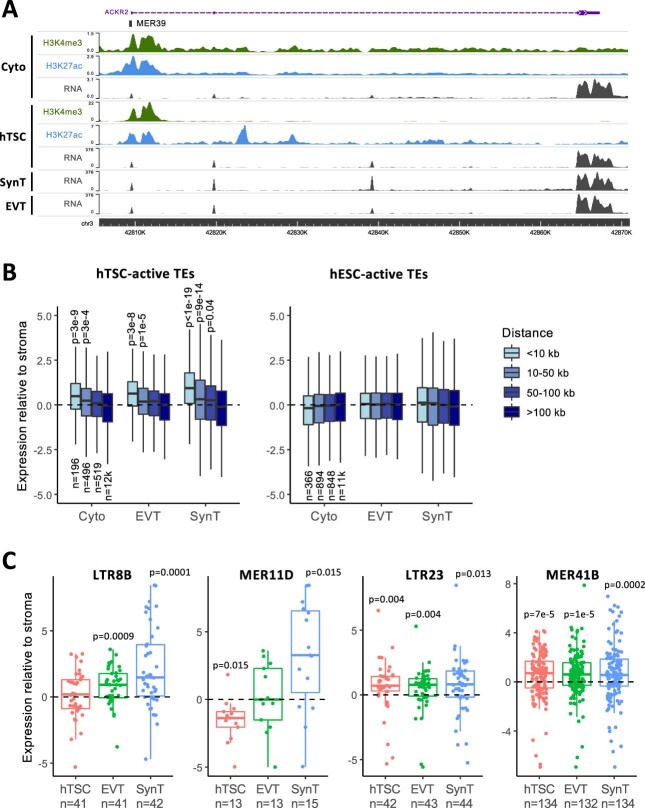
To identify interspersed repetitive elements bearing hallmarks of activating regulatory potential in human trophoblast, we performed H3K27ac profiling using either chromatin immunoprecipitation sequencing (ChIP-seq) or CUT&Tag (cleavage under targets and tagmentation)22. We analyzed our previously published data from primary human cytotrophoblast23, as well as newly generated data from cytotrophoblast-like human trophoblast stem cells (hTSCs)24, which can be differentiated in vitro and allow for easy genetic manipulation (Fig. 1a). Using the RepeatMasker annotation, we determined the frequency of H3K27ac peaks per repeat family and compared it with random controls using a permutation test (Fig. 1a). This revealed 29 repeat families enriched for H3K27ac peaks in both primary cytotrophoblast and hTSCs, the vast majority of which were primate-specific ERVs (Fig. 1b, Extended Data Fig. 1a and Supplementary Table 1). For comparison, we performed the same analysis on published H3K27ac CUT&Tag data from human embryonic stem cells (hESCs)22. Most hTSC-enriched repeats displayed little to no enrichment in hESCs (Fig. 1b), despite the fact that hESCs make use of a large set of TEs for gene-regulatory purposes10,25. A more detailed analysis of H3K27ac signals across each of the enriched families confirmed the asymmetry between hTSCs and hESCs (Fig. 1c). Human ERV subfamily H (HERVH)-associated LTRs present an interesting case, with LTR7C elements H3K27ac-enriched in hTSCs, whereas the related LTR7 family is H3K27ac-enriched in hESCs (Extended Data Fig. 1b). Each of the LTR7 subfamilies displays a unique combination of transcription-factor-binding motifs26, which may underlie their cell-type-specific expression.
Fig. 1. Gene regulatory signatures of ERVs in human trophoblast.
a, Primary cytotrophoblast and hTSCs were profiled for H3K27ac to identify repeat families with putative gene regulatory potential. b, Enrichment for H3K27ac peaks for each repeat family in hTSCs or hESCs. Families with significant enrichment in hTSCs are highlighted. c, H3K27ac profiles of a subset of hTSC-enriched ERV families in hTSCs and hESCs. Each line represents an element in that family. d, Enrichment for DNase hypersensitive sites in the same ERV families, in the kidney (K), liver (Li), lung (Lu) and placenta (P). Each datapoint represents a different ENCODE dataset. e, Proportion of elements from the same ERV families overlapping particular combinations of histone modifications. The data availability statement provides details of source data for b and c.
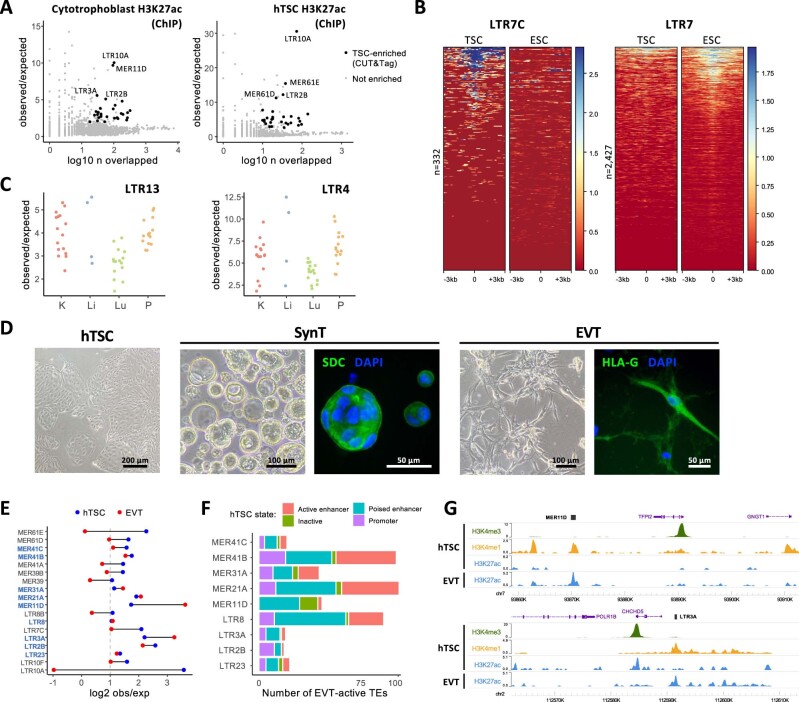
Extended Data Fig. 1. ERV regulatory signatures in undifferentiated and differentiated cells.
A, Enrichment for H3K27ac peaks for each repeat family in ChIP-seq data from primary cytotrophoblast (n = 2) or hTSCs (n = 1). Families with significant enrichment in CUT&Tag data from hTSCs (Fig. 1b) are highlighted. B, H3K27ac profiles of LTR7C and LTR7 families in hTSCs and hESCs. Each line represents an element in that family. C, Enrichment for Dnase hypersensitive sites in LTR13 and LTR4 families, in the kidney (K), liver (Li), lung (Lu) and placenta (P). Each datapoint represents a different ENCODE dataset. D, Representative phase contrast and immunofluorescence images for hTSCs and hTSC-derived SynT and EVT. Expression of the SynT marker SDC or EVT marker HLAG is shown (green), with DAPI counterstain (blue). E, Enrichment for H3K27ac peaks in hTSC or EVT for all hTSC-enriched ERV families. Families that are also significantly enriched in EVT are highlighted in blue. F, Number of H3K27ac-marked ERVs in EVT, divided by their chromatin state in hTSCs: active enhancer (H3K4me1 + H3K27ac), poised enhancer (H3K4me1 alone), promoter (H3K4me3) or inactive (none of the three marks). G, Genome browser snapshots showing examples of ERVs that are in a poised state in hTSCs and become active in EVT. Please see the data availability statement for details on source data for panels A, C, E and F.
To further validate our findings and narrow down the list of candidate regulatory repeat families, we also analyzed placental DNAse-seq data from ENCODE. When compared with the data from liver, lung and kidney, most hTSC-associated families displayed tissue-specific enrichment of elements with accessible chromatin (Fig. 1d; non-tissue-specific examples are provided in Extended Data Fig. 1c). Based on stringent criteria (Methods), we focused on 18 placenta-specific candidate regulatory repeat families, all of which were ERV-associated LTRs (Supplementary Table 1). This included families with known examples of elements bearing promoter activity in the placenta: LTR10A (NOS3 gene), LTR2B (PTN gene), MER39 (PRL gene), MER39B (ENTPD1 gene) and MER21A (HSD17B1 and CYP19A1 genes)19.
We then characterized in more detail the H3K27ac-marked elements from each of these families by performing CUT&Tag for H3K4me1, H3K4me3, H3K9me3 and H3K27me3 in hTSCs. This revealed that a large proportion of H3K27ac-marked elements were also marked by H3K4me1 (median 72%, range 44–85%), a signature of active enhancers (Fig. 1e). Only a small proportion (median 3%, range 0–20%) was marked by H3K4me3, a signature of active promoters (Fig. 1e). There was also a large fraction of elements that were marked by H3K4me1 alone (median 71% of all H3K4me1 elements, range 27–81%), which is normally associated with poised enhancers (Fig. 1e), raising the possibility that this group of elements becomes active following the differentiation of hTSCs. To test this, we differentiated hTSCs into extravillous trophoblast (EVT; Extended Data Fig. 1d) and performed CUT&Tag for H3K27ac. Half of the hTSC-active ERV families remained H3K27ac-enriched in EVT, whereas others were specific to the stem cell state (for example, LTR10A and MER61E; Extended Data Fig. 1e). In line with our hypothesis, a large proportion of ERVs that are active in EVT were in a poised enhancer state in hTSCs (Extended Data Fig. 1f,g). It is possible that other poised ERV enhancers become active upon differentiation into SynT (Extended Data Fig. 1d), but the multinucleated nature of these cells seemingly interfered with our CUT&Tag attempts.
Our analyses suggest that a large number of ERVs (nearly all of which are primate-specific) may act as gene regulatory elements in human trophoblast, showing dynamic changes during differentiation.
hTSC-active ERVs bind key placental transcription factors
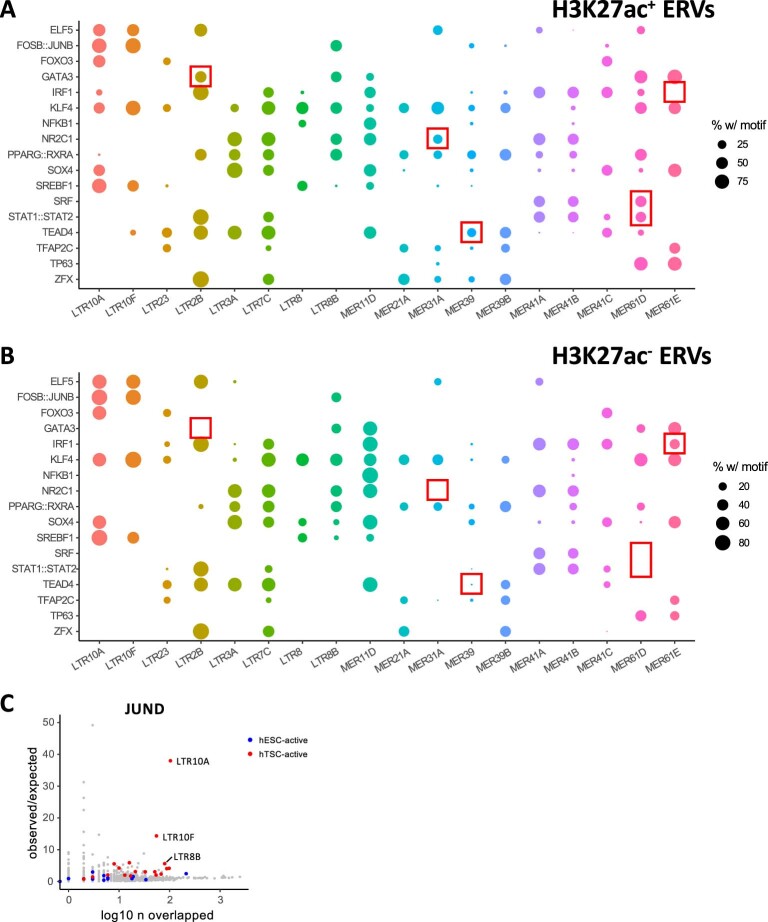
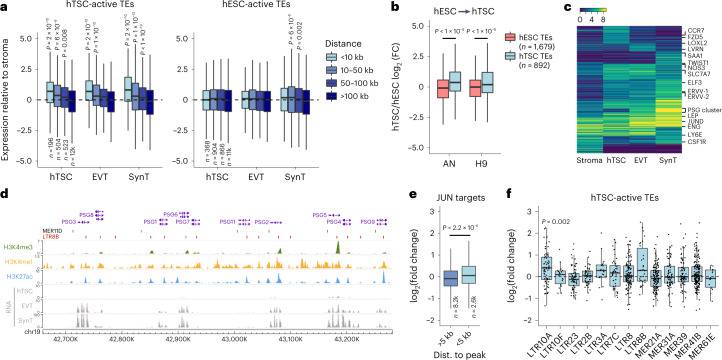
Enhancers function to regulate gene expression through the binding of transcription factors. We therefore identified transcription-factor binding motifs that were enriched within each hTSC-active (that is, marked by H3K27ac) ERV family, and focused on a selection of transcription factors that are expressed in trophoblast (Methods). Reassuringly, we identified previously described motifs on MER41B for signal transducer and activator of transcription (STAT) proteins and serum response factor (SRF; Extended Data Fig. 2a)12,21. We also uncovered a large collection of motifs for transcription factors with known roles in trophoblast development. Multiple families bore motifs for key factors involved in the maintenance of the stem cell state, such as ELF5, GATA3, TFAP2C, TP63 and TEAD4 (Fig. 2a and Extended Data Fig. 2a). Additional transcription factors with known roles in placental development and/or physiology included JUN/FOS27,28, PPARG/RXRA29 and FOXO330. Most of these motifs were also present in elements negative for H3K27ac (Extended Data Fig. 2b), suggesting that there are additional genetic or epigenetic determinants of their activity. There were nonetheless exceptions where motifs were only found in active elements, such as GATA3 in LTR2B elements and SRF in MER61D elements (Extended Data Fig. 2b).
Extended Data Fig. 2. Transcription factor binding motifs for all hTSC-active ERV families.
A, Proportion of H3K27ac-marked elements from all analyzed ERV families bearing motifs for the transcription factors on the y axis. B, As in B, but for H3K27ac-negative elements. Particularly striking examples of differential motif enrichment between H3K27ac+ and H3K27ac- ERVs are highlighted. C, Repeat family-wide enrichment for peaks from JUND CUT&Tag data on hTSCs. H3K27ac-enriched families in hTSCs or hESCs are highlighted. Please see the data availability statement for details on source data.
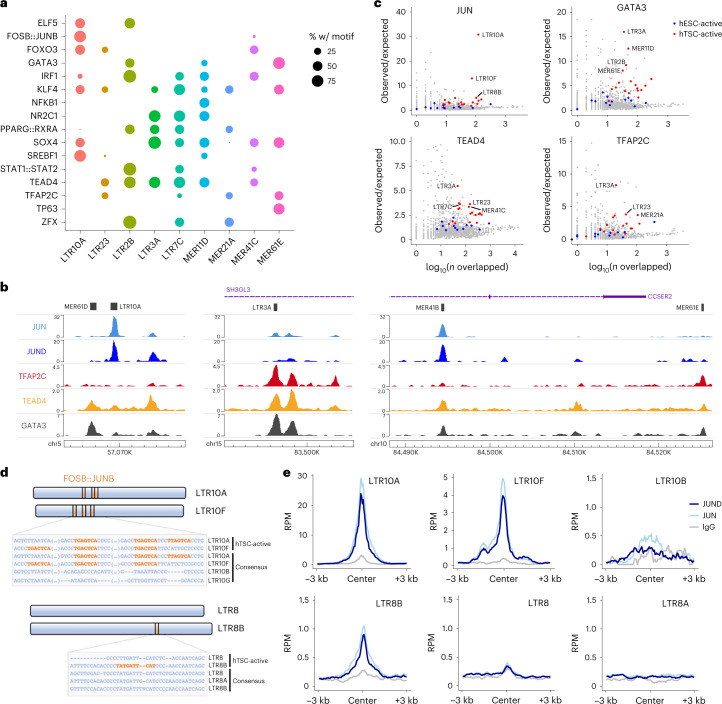
Fig. 2. Transcription-factor repertoire at hTSC-active ERVs.
a, Proportion of H3K27ac-marked elements from selected ERV families bearing motifs for the transcription factors on the y axis. b, Genome browser snapshots of transcription-factor CUT&Tag/RUN data, showing examples of enrichment over ERVs. c, Repeat family-wide enrichment for peaks from transcription-factor CUT&Tag/RUN data on hTSCs. H3K27ac-enriched families in hTSCs or hESCs are highlighted. d, Schematic and alignment of LTR10 and LTR8 subfamilies showing the presence of FOSB::JUNB (that is, AP-1) motifs in the genome-wide consensus sequence and/or in a consensus of the H3K27ac-marked elements. e, Mean CUT&Tag profiles for JUN and JUND over LTR10 and LTR8 subfamilies. See the data availability statement for details on source data for a, c and e.
To validate the binding of some of these transcription factors to hTSC-active ERVs, we performed CUT&Tag or CUT&RUN31 for JUN, JUND, GATA3, TEAD4 and TFAP2C (Fig. 2b). We applied the same peak enrichment pipeline as described above (Fig. 1a) and found that several of these families were enriched for one or more of the evaluated transcription factors (Fig. 2c and Extended Data Fig. 2c). In contrast, TE families active specifically in hESCs showed little to no enrichment of these factors (Fig. 2c and Extended Data Fig. 2c). LTR10A and LTR10F elements were strongly enriched for JUN binding, as predicted from our motif analysis. Similarly, binding to motif-bearing ERVs was confirmed for GATA3 (for example, LTR2B, MER11D and MER61E), TEAD4 (for example, LTR3A, LTR7C and MER41C) and TFAP2C (for example, LTR23 and MER21A). We also found instances of enriched transcription-factor binding to families that seemingly do not bear the corresponding motif (for example, GATA3 and TFAP2C binding at LTR3A elements). This could reflect limitations of motif-finding approaches and/or suggest indirect transcription-factor recruitment—for example, TEAD4 may recruit TFAP2C32.
Given the striking enrichment for JUN and JUND binding over LTR10A/F elements, we further explored the corresponding motifs in these families, as well as in LTR8B. JUN and JUND are two subunits of the activator protein-1 (AP-1) complex, which can heterodimerize with the FOS family of transcription factors. AP-1 plays important roles in cell proliferation and survival, and has been implicated in the regulation of trophoblast differentiation and invasion27,28. Both LTR10A and LTR10F active elements contained three AP-1 motifs that were also present in the family-wide consensus sequence, whereas other LTR10-related families lacked any such motifs (Fig. 2d). In strict correspondence with this, binding was observed for LTR10A/F, but not other LTR10 families (Fig. 2e). JUN/JUND enrichment at LTR10A elements may be substantially more pronounced than for LTR10F due to differences in motif arrangement, cooperative binding with other transcription factors, or chromatin environment. In the case of LTR8B, only active elements contained one AP-1 motif, suggesting divergence of a subset of elements after retroviral endogenization (Fig. 2d). CUT&Tag profiles confirmed that JUN/JUND bound LTR8B only, and not other LTR8-related families (Fig. 2e).
These results show that ERV families bearing regulatory potential in human trophoblast are bound by multiple transcription factors that play important gene regulatory roles in trophoblast.
Genes near active ERVs display biased trophoblast expression
To assess the potential of hTSC-active ERVs to drive trophoblast gene expression, we first asked whether some functioned as gene promoters. We performed de novo transcriptome assembly on our primary cytotrophoblast data23 and extracted transcripts for which an ERV from the selected families overlapped the transcriptional start site. In line with the relatively small proportion of H3K4me3-containing elements (Fig. 1e), we identified few ERVs with apparent promoter activity (Supplementary Table 2), many of which had been previously reported (promoters for CYP19A133, PTN34, PRL35 and MID136). Another notable gene was ACKR2, which encodes a chemokine scavenger whose expression in trophoblast is driven by a MER39 element (Extended Data Fig. 3a), and deficiency of which leads to placental defects and pre/neonatal mortality in mice37. Most other transcripts associated with ERV promoters were lowly expressed in cytotrophoblast (Supplementary Table 2).
Extended Data Fig. 3. Associations between hTSC-active ERVs and gene expression.
A, Genome browser snapshot showing an example of an ERV-derived gene promoter in primary cytotrophoblast. B, Gene expression in primary cytotrophoblast, EVT and SynT relative to placental stroma. Genes are grouped based on their distance to the nearest H3K27ac-marked TE in hTSCs. P values are for the difference to the ‘>100 kb’ group based on ANOVA and Tukey post-hoc test. c) Expression in hTSCs and hTSC-derived EVT and SynT relative to primary placental stroma for genes within 50 kb of an H3K27ac-marked ERV of the indicated family. Genes are grouped based on their distance to the nearest H3K27ac-marked TE in hTSCs. P values are from two-sided Wilcoxon tests comparing each distribution to 0, with multiple comparisons correction. Boxplots show median center, 25th and 75th percentile box bounds, and 1.5× IQR whisker limits. Please see the data availability statement for details on source data for panels B and C.
We next sought to evaluate the regulatory potential of candidate ERV-derived enhancers. Using published RNA-seq data24, we asked whether the distance to active ERVs correlated with gene expression in trophoblast cells, by comparing it to the expression in placental stroma (non-trophoblast tissue). We found a strong association between gene–ERV distance and preferential expression in both undifferentiated and differentiated trophoblast (Fig. 3a and Extended Data Fig. 3b). In contrast, genes proximal to H3K27ac-marked ERVs in hESCs displayed no preferential expression in trophoblast (Fig. 3a and Extended Data Fig. 3b). We also analyzed RNA-seq data from transdifferentiation experiments of hESCs into hTSC-like cells38, which showed that genes near hTSC-active ERVs displayed higher expression upon transdifferentiation than those close to hESC-active ERVs (Fig. 3b).
Fig. 3. Expression of genes close to regulatory ERVs.
a, Gene expression in hTSCs and hTSC-derived EVT and SynT relative to primary placental stroma. Genes are grouped based on their distance to the nearest H3K27ac-marked TE in hTSCs. The number of genes (n) in each set is shown. P values are for the difference in comparison to the ‘>100-kb’ group based on the ANOVA and Tukey post-hoc test. b, Gene expression in hESC-transdifferentiated hTSCs relative to their respective parental hESC lines (AN or H9). Expression ratios are shown for genes within 50 kb of either hESC- or hTSC-active (H3K27ac-marked) TEs. P values are from two-sided Wilcoxon tests with multiple comparisons correction. FC, fold change. c, Expression patterns of variably expressed genes across stroma and trophoblast cells. d, Genome browser snapshot of the human PSG cluster, highlighting the overlap between H3K27ac peaks and MER11D or LTR8B elements. e, FC in gene expression after treatment of hTSCs with JNK inhibitor SP600125, with genes divided according to their distance to the nearest JUN CUT&Tag peak. The number of genes in each set is shown; the P value is from a two-sided Wilcoxon test. f, FC in expression after SP600125 treatment for genes within 100 kb of an H3K27ac-marked ERV from selected families. The P value is from a two-sided Wilcoxon test comparing each distribution to 0, with multiple comparisons correction. The boxplots show median center, 25th and 75th percentile box bounds, and 1.5× interquartile range whisker limits. See the data availability for details of source data for a–c, e and f.
Most genes proximal to hTSC-active ERVs were expressed in all three trophoblast cell types analyzed, with smaller subsets displaying expression in a single trophoblast cell type (Fig. 3c). A relatively large fraction showed increased expression upon differentiation into SynT, including a cluster harboring several genes encoding for pregnancy-specific glycoproteins (PSGs), which are highly expressed in SynT. PSGs are the most abundant conceptus-derived proteins circulating in the maternal blood39. Their function in pregnancy remains elusive, although they have been associated with immune responses to pregnancy, and low levels of circulating PSGs are linked to recurrent pregnancy loss, fetal growth restriction and preeclampsia40–42. Within the PSG cluster, virtually every H3K27ac peak overlaps either a MER11D or LTR8B element (Fig. 3d), which presumably were already present in the ancestral PSG gene before its duplication. These two ERV families are associated with SynT-biased gene expression, mostly due to the PSG cluster (Extended Data Fig. 3c). This is in contrast with other families that are associated with genes with biased expression across all three trophoblast cell types analyzed (Extended Data Fig. 3c).
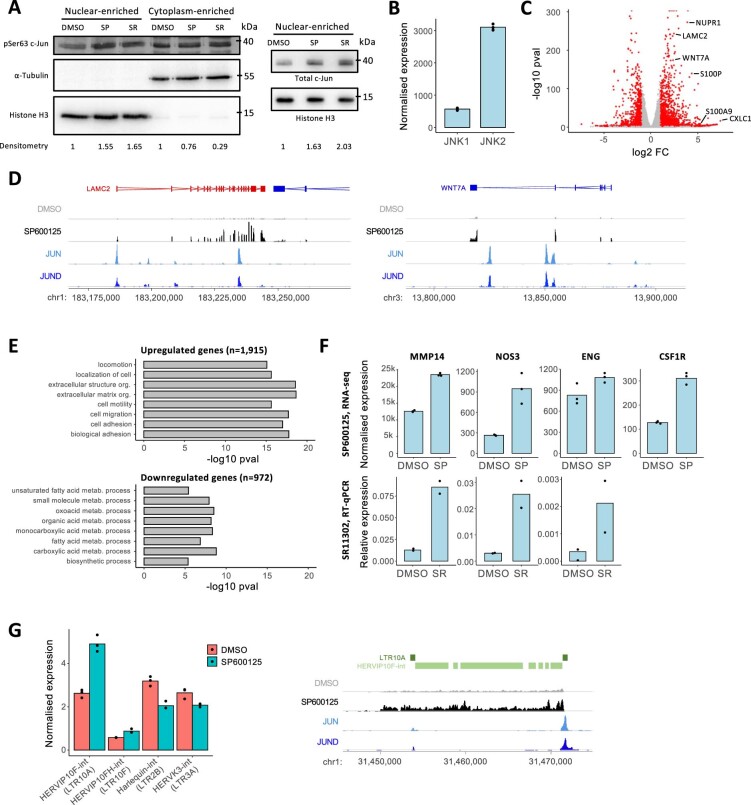
We then leveraged information from our motif analysis to interfere with ERV regulatory activity. Given the enrichment in JUN/JUND binding at LTR10A, LTR10F and LTR8B elements, we treated hTSCs with SP600125, an inhibitor of c-Jun N-terminal kinases (JNKs). Unexpectedly, this led to increased levels of phosphorylated c-Jun in the nucleus, and the same result was obtained with a second AP-1 inhibitor (Extended Data Fig. 4a). JNK signaling is known to have cell-type specific effects43, partly due to the opposing roles of JNK1 and JNK244. JNK2 deficiency increases c-Jun phosphorylation and stability44, potentially explaining our results, as JNK2 is highly expressed in hTSCs (Extended Data Fig. 4b). Irrespective of the mechanism, higher levels of phosphorylated c-Jun are predicted to increase expression of AP-1 target genes. Indeed, RNA-seq of SP600125-treated hTSCs revealed that genes proximal to JUN binding sites were on average upregulated (Fig. 3e and Extended Data Fig. 4d). A large number of upregulated genes are implicated in cell migration (Extended Data Fig. 4c–e), which is in line with observations that SP600125 increases trophoblast cell migration27. Finally, we found that LTR10A target genes were upregulated upon SP600125 treatment (Fig. 3f), including NOS3 (also observed with a second AP-1 inhibitor; Extended Data Fig. 4f), whose placenta-specific gene expression is driven by an LTR10A-derived promoter45. Although the majority of LTR10F and LTR8B target genes were also upregulated (Fig. 3f), their low number precluded robust statistical analysis. More prominent effects on LTR10A target genes were expected based on the stronger binding of JUN/JUND to this family (Fig. 2e). Nonetheless, expression of all PSG genes was increased by at least twofold, suggesting that JUN regulates this cluster via LTR8B elements. We also found that SP600125 led to upregulation of proviral LTR10A elements, but less so for LTR10F (we found no proviral LTR8B elements), and not for other H3K27ac-enriched families (Extended Data Fig. 4g), providing more direct evidence that AP-1 supports the regulatory activity of LTR10A elements.
Extended Data Fig. 4. Effects of SP600125 treatment in hTSCs.
A, Western blot of phosphorylated c-Jun (pSer63 c-Jun) and total c-Jun in enriched nuclear or cytoplasmic fractions of cells treated with the AP-1 inhibitors SP600125 (‘SP’) or SR11302 (‘SR’). Densitometry values normalized to the respective loading controls are shown below. Full blot images are in Supplementary Figure 7. B, Expression of JNK1 (MAPK8) and JNK2 (MAPK9) in hTSCs (data from RNA-seq). C, Volcano plot of RNA-seq data from cells treated with SP600125. Some genes implicated in cell migration are highlighted. P values are from the DESeq2 R package. D, Genome browser snapshots showing examples of genes highlighted in C, together with CUT&Tag data for JUN and JUND. E Gene ontology biological processes enriched terms for genes up- or downregulated after treatment of hTSCs with SP600125. P values are from the topGO R package. F, Expression of MMP14 (a known AP-1 target), NOS3, ENG and CSF1R in hTSCs treated with SP600125 (data from RNA-seq) and/or SR11302 (see data from RT–qPCR). G, Expression of internal ERV fragments driven by LTRs of H3K27ac-enriched families (in brackets) upon SP600125 treatment. An example of LTR10A-driven transcription of a proviral HERVIP10-F locus is shown on the right. Please see the data availability statement for details on source data for panels B, C and E–G.
ERVs are associated with species-specific gene expression
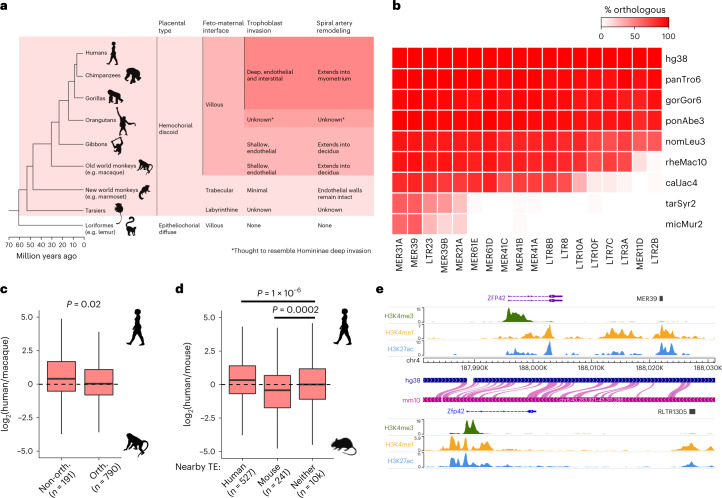
Primate evolution has involved dramatic divergence in placental phenotypes, including differences in the cellular arrangement of the feto-maternal interface and the extent of trophoblast invasion into the maternal decidua (Fig. 4a)46. Great apes display a unique and deep form of trophoblast invasion47. The integration of ERVs with regulatory capacity in trophoblast may have helped to fuel such fast placental evolution across primates. To test this, we first identified ERVs in non-human primates that are orthologous to human elements from each selected family. This showed a wide spread of inter-species differences that are in accordance with the evolutionary age of the selected families (Fig. 4b). We then used published RNA-seq data from rhesus macaque TSCs (macTSCs), which were recently derived using the same culture conditions as for hTSCs48. We found that one-to-one orthologous genes close to human-specific ERVs displayed, on average, higher expression in hTSCs than in macTSCs, when compared to genes close to conserved ERVs (Fig. 4c). The majority of non-orthologous elements were from the LTR2B family (as expected from its near absence in macaque) and included the previously characterized placenta-specific promoter of PTN34. Additional LTR2B-associated genes with human-specific expression included KCNE3, which encodes an estrogen-receptor-regulated potassium channel49, and STOML2, which regulates trophoblast proliferation and invasion50.
Fig. 4. Association of ERVs to species-specific gene expression.
a, Primate phylogeny highlighting cross-species differences in placental morphology and invasion4,47. b, Proportion of human ERVs from each of the selected families that contain orthologous elements in the non-human primates highlighted in a. c, Expression difference between human and macTSCs for genes within 100 kb of an H3K27ac-marked ERV from hTSC-active families, depending on whether there is an orthologous element in macaque. The P value is from a two-sided Wilcoxon test. d, Expression difference between human and mouse TSCs for genes within 50 kb of an H3K27ac-marked ERV in human, mouse (RLTR13D5 and RLTR13B families) or neither species. P values are from an ANOVA with Tukey post-hoc test. e, Genome browser snapshot showing a putative example of convergent evolution between mouse and human, wherein different enhancer-like ERVs lie downstream of the ZFP42/Zfp42 gene. Boxplots show median center, 25th and 75th percentile box bounds, and 1.5× interquartile range whisker limits. See the data availability statement for details of the source data for b–d.
We and others have previously shown that Mus-specific ERVs also act as distal enhancers in mouse TSCs (mTSCs)17,18. We therefore extended our comparative expression analysis by asking whether human- and/or mouse-specific ERVs were associated with increased gene expression in the respective species. Indeed, genes with active ERVs nearby in human but not in mouse displayed higher expression in hTSCs, whereas those close to active mouse ERVs had higher expression in mTSCs (Fig. 4d). Genes with mouse-specific expression included a component of the fibroblast growth factor signaling pathway (Fgfbp1), which maintains the stem cell state in mouse but not in human trophoblast, and other mTSC markers (Duox, Duox2 and Nr0b1)51. Conversely, human-specific ERVs were associated with expression of a Wnt signaling receptor (FZD5), a pathway that is important for hTSC derivation24, and MMP14, which is important for trophoblast invasion52. We also considered potential cases of convergent evolution, whereby the same gene may be regulated by different ERVs in human and mouse. Of the 12 genes that were close to active ERVs in both mouse and human, ten were expressed in mTSCs and hTSCs, including Zfp42/ZFP42 (Fig. 4e and Supplementary Table 3). Despite being a well-known marker of ESCs, Zfp42 is also expressed in mouse trophoblast, especially in early embryos53,54, where it regulates the expression of some imprinted genes53. This raises the possibility that different ERVs have convergently been co-opted to maintain the expression of Zfp42/ZFP42 in mouse and human trophoblast.
These analyses suggest that some of the putative regulatory ERVs that we identified help to drive species-specific expression of genes that are important for trophoblast development and function.
ERVs regulate genes involved in human trophoblast function
Previous observations have shown that epigenetic markers are not predictive of gene regulatory activity18. To test whether ERVs can act as enhancers in vivo, we utilized CRISPR to genetically excise a subset of candidate regions, and then measured nearby gene expression. Because the efficiency of growing clonal hTSCs from single cells was extremely low, we employed a population-wide lentiviral approach that we had previously used55, achieving an average of 49% deletion efficiency across different targets and experiments (Supplementary Table 4).
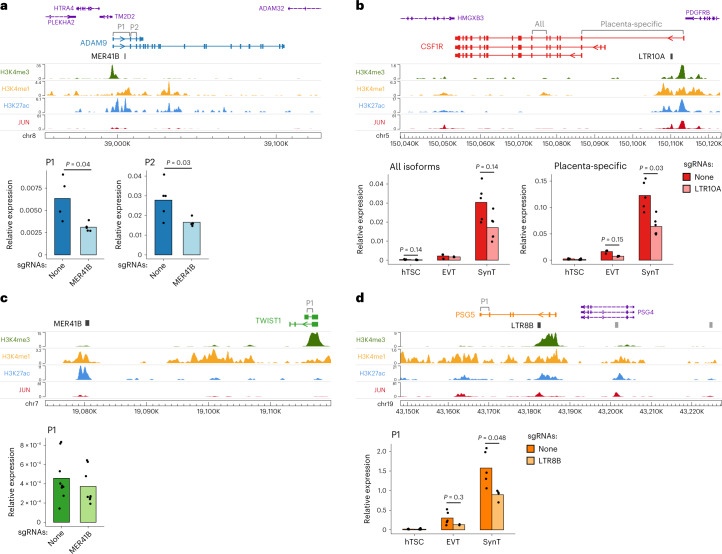
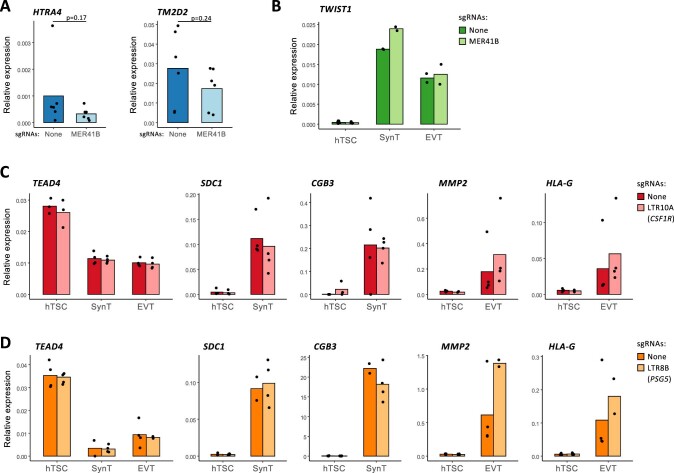
We first excised an enhancer-like MER41B element that is located in the first intron of the ADAM9 gene (Fig. 5a), which encodes for a metalloproteinase. Genetic variants of ADAM9 are implicated in preeclampsia56, and its known substrates play roles in inflammation, angiogenesis, cellular migration and proliferation57. Two independent MER41B excisions were derived, resulting in a 1.7–2-fold decrease in ADAM9 expression in hTSCs compared to no-sgRNA controls (Fig. 5a). The MER41B LTR is also 8 kb upstream of TM2D2 and 16 kb upstream of HTRA4, a placenta-specific serine peptidase that is upregulated in early-onset preeclampsia58,59. Expression of these genes was low and remained largely unchanged following MER41B excision (Extended Data Fig. 5a).
Fig. 5. Genetic excision of hTSC-active ERVs.
a, Genome browser snapshot of the ADAM9 locus containing an enhancer-like MER41B element, and ADAM9 expression (using primer pairs highlighted on the gene annotation) in hTSC populations treated with lentiviral CRISPR constructs carrying either no sgRNAs (n = 4) or sgRNAs that excise the MER41B element (n = 5 from two sgRNA sets). b, As in a, but for CSF1R and its intronic LTR10A elements. CSF1R expression was measured in hTSCs (n = 5 from one sgRNA set) and hTSC-derived EVT (n = 3) and SynT (n = 5 no sgRNA, n = 6 LTR10A sgRNAs). c, As in a, but for TWIST1 and a downstream MER41B element (n = 9 no sgRNA, n = 8 MER41B sgRNAs; from three infections using one sgRNA set). d, As in a, but for PSG5 and its intronic LTR8B elements. PSG5 expression was measured in hTSCs (n = 3 no sgRNA, n = 6 LTR8B sgRNAs from two sgRNA sets) and hTSC-derived EVT (n = 5 no sgRNA, n = 2 LTR8B sgRNAs) and SynT (n = 5 no sgRNA, n = 4 LTR8B sgRNAs). P values throughout are from two-sided Wilcoxon tests with multiple comparisons correction. See the data availability statement for details of the source data for RT–qPCR.
Extended Data Fig. 5. Effects of ERV genetic excisions on other genes.
A, RT–qPCR data for HTRA4 and TM2D2 in hTSC populations treated with lentiviral CRISPR constructs carrying either no sgRNAs or sgRNAs that excise the MER41B element highlighted in Fig. 5a (n = 6). B, TWIST1 expression in EVT and SynT derived from hTSC populations with no sgRNAs or sgRNAs that excise the MER41B element highlighted in Fig. 5c. C, Expression of stem cell (TEAD4), SynT (SDC1, CGB3) and EVT (MMP2, HLAG) markers in cell populations with no sgRNAs or sgRNAs that excise the LTR10A element highlighted in Fig. 5b. D, As in C, but for the LTR8B element in Fig. 5d. No significant differences were detected in any of the data after two-sided Wilcoxon tests with multiple comparisons correction. Please see the data availability statement for details on source data.
The CSF1R gene has a placenta-specific promoter60, downstream of which lies an enhancer-like LTR10A element (Fig. 5b). Both CSF1 and CSF1R expression increase in the placenta during pregnancy61. CSF1 signaling via CSF1R promotes the growth, proliferation and migration of trophoblasts in humans and mice62–64, and high CSF1 levels are correlated with preeclampsia development65. Expression of CSF1R was low in undifferentiated hTSCs, but increased following differentiation to EVT, and was highest in SynT-differentiated hTSCs, particularly for the placenta-specific CSF1R variant (Fig. 5b). CSF1R expression also increased upon SP600125 treatment, suggesting it is regulated by AP-1 (Extended Data Fig. 4f). An excision of the LTR10A was derived in hTSCs, with CSF1R expression of the placenta-specific variant being reduced by around twofold in both EVT and SynT cell pools (and to a lesser extend in hTSCs), compared to no-sgRNA controls (Fig. 5b). These differences were not caused by an impairment in trophoblast differentiation efficiency, as judged by the expression of key marker genes (Extended Data Fig. 5c).
We excised a second MER41B element with enhancer-like chromatin conformation (Fig. 5c). The TWIST1, FERD3L and HDAC9 genes lie in the vicinity of this LTR, but only TWIST1 is expressed in hTSCs. TWIST1 regulates the syncytialization of trophoblast66,67 and promotes epithelial to mesenchymal transition—a key process in EVT differentiation68. We measured the expression of TWIST1 in MER41B excision hTSC pools and found its expression to be unchanged (Fig. 5c). Because TWIST1 is important for trophoblast differentiation, we also differentiated hTSCs to EVT and SynT, resulting in a 25–41-fold increase in TWIST1 expression, but there was no difference in TWIST1 expression in excision versus no-sgRNA differentiated cells (Extended Data Fig. 5b). This particular MER41B element is therefore either a redundant enhancer or does not regulate TWIST1, highlighting the importance of these genetic experiments.
As previously mentioned, the PSG cluster on chromosome 19 includes MER11D and LTR8B elements at each tandemly repeated gene locus, all featuring enhancer-like chromatin features in hTSCs (Fig. 3d). We excised an LTR8B element within the second intron of one of the most highly expressed PSG in humans, PSG5 (Fig. 5d)69, deriving two independent excisions. PSG5 expression was low in undifferentiated hTSCs and remained unchanged in LTR8B excision pools compared to no-sgRNA controls (Fig. 5d). However, PSG5 expression was increased following differentiation to EVT and SynT and was reduced in LTR8B excision pools compared to no-sgRNA controls in both trophoblast types (Fig. 5d), whereas differentiation efficiency was unaffected by the excision (Extended Data Fig. 5d). The fact that the enhancer activities of this LTR8B-PSG5 element and the LTR10A-CSF1R element are most strongly expressed after differentiation supports the notion that some hTSC-active ERVs also play roles in differentiated trophoblast, as suggested by our epigenomic and transcriptomic analyses above.
An LTR10A-derived enhancer promotes ENG expression
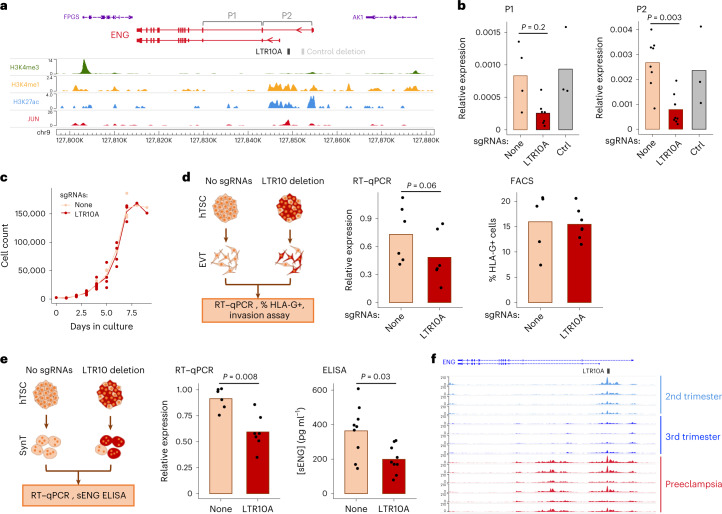
We were particularly interested in an enhancer-like LTR10A element within the first intron of the endoglin gene (ENG/CD105; Fig. 6a). ENG is a transforming growth factor-beta (TGF-β) 1 and 3 co-receptor, with both membrane-bound and soluble cleavage variants, highly expressed in the endothelium and SynT, and involved in the pathogenesis of preeclampsia70. The serum levels of soluble ENG (sENG) are strongly correlated with the severity of preeclampsia71. Membrane-bound ENG is also expressed in villous cell columns in the first trimester of pregnancy, regulating trophoblast differentiation to EVT72,73.
Fig. 6. Impact of an LTR10A element within the ENG gene.
a, Genome browser snapshot of the ENG locus containing an enhancer-like LTR10A element in its first intron. b, ENG expression (using the primer pairs highlighted in a) in hTSC populations treated with lentiviral CRISPR constructs carrying no sgRNAs (n = 4 P1, n = 8 P2), sgRNAs that excise the LTR10A element (n = 7 P1, n = 9 P2 from three sgRNA sets) or sgRNAs that excise a control region highlighted in a (n = 3). c, Growth curves for hTSC populations carrying no sgRNAs or LTR10A-excising sgRNAS. d, The same hTSC populations were differentiated to EVT, and assays were performed for ENG RNA levels (n = 6) and HLAG cell surface expression (n = 5 no sgRNAs, n = 7 LTR10A sgRNAs). e, The same hTSC populations were differentiated to SynT, and assays were performed for ENG RNA levels (n = 6 no sgRNAs, n = 7 LTR10A sgRNAs) and secretion of soluble ENG protein (n = 9). f, Genome browser snapshot of the ENG locus with cytotrophoblast H3K27ac ChIP-seq data from second- or third-trimester uncomplicated pregnancies, or third-trimester severe preeclampsia placentas. P values throughout are from two-sided Wilcoxon tests with multiple comparisons correction where relevant. See the data availability statement for details of the source data for b–e.
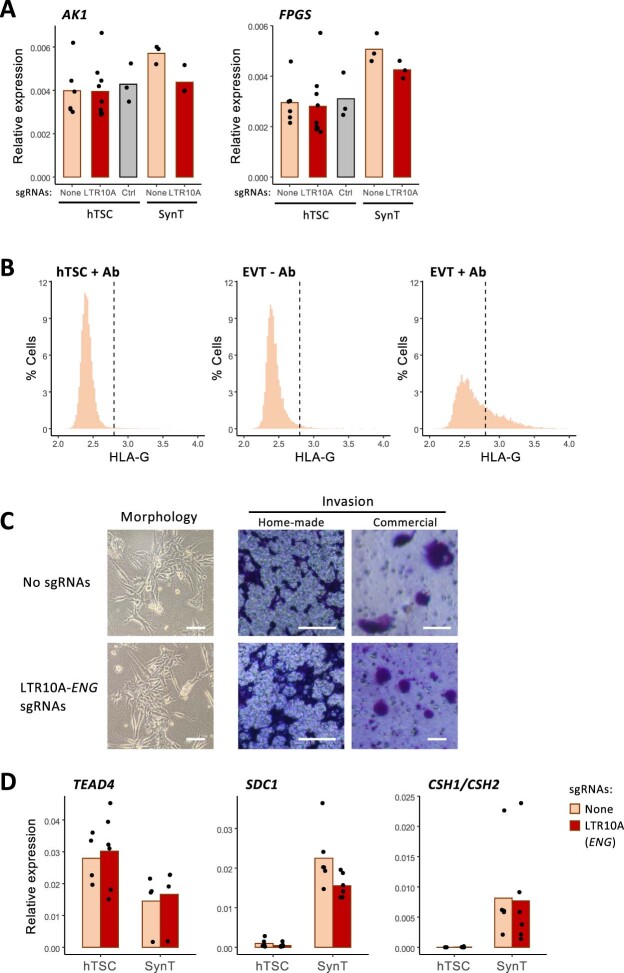
We derived three independent excisions of the ENG LTR10A in hTSCs, which resulted in a striking decrease in ENG expression compared to no-sgRNA controls (Fig. 6b). Expression of neighboring genes (AK1 and FPGS) did not change (Extended Data Fig. 6a). To confirm that loss of ENG expression was strictly associated with deletion of the LTR10A element, and not transcriptional interference by the CRISPR–Cas9 machinery, we also deleted a control region upstream of the putative LTR10A enhancer that was devoid of active histone modification marks in hTSCs (Fig. 6a). Reassuringly, deletion of this region had no effect on ENG expression (Fig. 6b). In line with the role of AP-1 in regulating LTR10A regulatory activity, increased c-Jun phosphorylation led to ENG upregulation (Extended Data Fig. 4f). To assess the phenotypic impact of LTR10A deletion, we first measured cell proliferation in hTSCs, finding no difference when compared to no-sgRNA controls (Fig. 6c). Differentiation to EVT was not affected by LTR10A deletion, as measured by the percentage of human leukocyte antigen G (HLAG)-positive cells (Fig. 6d and Extended Data Fig. 6b). LTR10A-deleted EVT were also morphologically similar to control EVT and retained the capacity to invade Matrigel in a transwell assay (Extended Data Fig. 6c), although technical variation precluded a more quantitative assessment of the extent of invasion. Notably, ENG expression remained lower (albeit variable) in LTR10A-deleted EVT, when compared with no-sgRNA controls (Fig. 6d). Differentiation into SynT was also unaffected by deletion of the LTR10A element (Extended Data Fig. 6d). Despite a large rise in ENG levels following SynT differentiation, LTR10A-deleted cells expressed less ENG than no-sgRNA controls (Fig. 6e). We therefore tested whether sENG protein levels were impacted by the LTR10A enhancer by performing enzyme-linked immunosorbent assays (ELISA) in the media of SynT cultures. We detected sENG at concentrations varying from ~100 pg ml−1 to 600 pg ml−1 and observed a significant decrease in sENG protein levels in LTR10A-ENG excised SynT cultures when compared with no-sgRNA control (Fig. 6e).
Extended Data Fig. 6. Transcriptional and phenotypic effects of LTR10A-ENG deletion.
A, AK1 and FPGS expression in hTSC or hTSC-derived SynT populations carrying lentiviral CRISPR constructs with no sgRNAs, sgRNAs that excise the LTR10A element highlighted in Fig. 6a, or sgRNAs that excise a control region also highlighted in Fig. 6a. B, Representative FACS profiles for HLAG immunolabelling in hTSCs and hTSC-derived EVTs. The middle panel is a no-antibody control. C, Images of cells in culture (to assess morphology) or crystal violet-stained cells after an invasion assay using Matrigel-coated chambers (either home-made chambers or from a commercial provider). Images representative from six independent experiments. Scale bars are 100 µm. D, Expression of stem cell (TEAD4) and SynT (SDC1, CSH1/CSH2) in cell populations with no sgRNAs or sgRNAs that excise the LTR10A-ENG element. No significant differences were detected in any of the qRT–PCR data after Wilcoxon tests with multiple comparisons correction. Please see the data availability statement for details on source data for panels A, B and D.
These experiments raise the possibility that deregulation of the ENG-LTR10A element may be associated with elevated levels of ENG in preeclampsia. We leveraged recently published H3K27ac ChIP-seq data from cytotrophoblast isolated from preeclamptic or uncomplicated pregnancies74. In uncomplicated pregnancies, there was clear H3K27ac enrichment over the LTR10A-ENG element in second-trimester placentas, which decreased in third-trimester placentas (Fig. 6f). However, in third-trimester placentas from severe preeclampsia, H3K27ac levels remained high over the LTR10A-ENG enhancer (Fig. 6f), suggesting that this ERV may help to maintain ENG expression unduly high in preeclampsia.
These data show that the LTR10A-ENG element acts as an enhancer in human trophoblast, regulating ENG expression and sENG protein production, with potential implications for preeclampsia.
Discussion
We have identified multiple ERV families that are enriched for a chromatin signature of cis regulatory elements in human trophoblast, most resembling enhancers. Our stringent criteria ensured that these regulatory profiles reflect what is observed in vivo and are not driven by non-trophoblast cell types present in the placenta. Indeed, we find that the activity of these families is largely trophoblast-specific, with several playing important roles in both undifferentiated and differentiated trophoblast cell types. Our genetic editing experiments have demonstrated that at least a subset of the ERVs we identified act as bona fide enhancers of genes, with important roles in placentation. Notably, the fact that we were limited to performing CRISPR on a population scale implies that the effects we observed are actually an underestimation of the true importance of those ERVs to gene expression.
When we compared our selected families with those identified by the Macfarlan laboratory as being enriched for lineage-specific placental enhancers, we found multiple ERV families in common, including MER21A, MER41B, LTR8 and MER3921. On the other hand, the study by Sun et al.21 study did not list other ERV families identified here, most prominently LTR10A, which has a strong regulatory signature and copies of which we demonstrated act as enhancers of important placental genes. Conversely, we find no evidence of regulatory activity from the MaLR group of ERV families identified by Sun and colleagues. One possible reason for these discrepancies is that Sun et al. used whole placental explants, leading to a mixed epigenomic profile from multiple cell types.
We also noted that a number of our trophoblast-active ERV families were recently identified as active in several cancers, including LTR10A, LTR10F, LTR2B and MER11D75. Multiple parallels have previously been drawn between trophoblast and cancer cells, including an epigenetic landscape that is strikingly different from other differentiated cell types76. The combination of an arguably permissive chromatin conformation and shared signaling pathways77 may make the co-option of ERVs for both placental development/function and cancer a frequent occurrence. For example, the Hippo signaling pathway, acting through YAP and TEAD4, plays key roles in both tumorigenesis and placental development78. The activation of LTR10A elements in both contexts is also seemingly driven by a shared signaling pathway, that is, MAPK/AP-175.
Several specific elements identified here suggested a potential important role for ERVs in placental evolution, including the multiple LTR8B and MER11D copies associated with the PSG gene cluster. The PSG cluster is semi-conserved in primates, with 6 to 24 genes found in Old World monkeys, one to seven genes in New World monkeys and none in more distantly related primates such as lemurs, suggesting that the presence of PSG may correlate with hemochorial placentae, given that lemurs have an epitheliochorial placenta79. Notably, PSG clusters are also present in mice, which also have a hemochorial placenta, and this region was expanded independently in mice and primates, suggesting convergent evolution80. Our results suggest that the integration of LTR8B elements ahead of PSG cluster expansion in humans was an important step that contributed to high trophoblast expression of these genes. MER11D elements may play a similar role and, together with LTR8B, be responsible for much of the transcriptional regulation of this important locus. Similarly, MER61D/E retrotransposition may have played a key role in setting the TP63 binding landscape, which in human trophoblast supports cell proliferation and prevents cellular differentiation in trophoblast81. TP63 belongs to the same family of transcription factors as TP53, sharing many of its binding sites82. It was previously shown that MER61 elements expanded the TP53 binding network in primates, and a number of copies have been exapted to mediate cellular stress responses in lymphoblastoid cells83.
Other examples suggest a role of ERVs in coordinating the expression of genes from the same pathway. MER21A elements were previously shown to act as promoters of the steroidogenesis pathway genes CYP19A1 and HSD17B1, implicating these LTRs in the regulation of steroidogenesis in human trophoblast. We also noted that both NOS3 and ENG bear LTR10A elements as major transcriptional regulators. Interestingly, the contribution of sENG to vascular pathology in preeclampsia is partially due to effects on NOS3; in concert with FLT1, ENG reduces placental angiogenesis and vasodilation of maternal spiral arteries, and increases vessel permeability70,84.
Our results implicate regulatory ERVs in pregnancy complications, prompting the need for further investigation of their functional impact on pregnancy outcomes. These ERVs may bear genetic variants that are difficult to investigate due to their repetitive nature, and that may affect their regulatory activity in the placenta. Notably, structural variants in LTR10A/F elements (in the form of variable number tandem repeats), some potentially contributing to cancer, were recently described75. Research into the effects of such variants in the placenta will benefit from more complex models that can assess the impact of regulatory ERVs on cell–cell interactions, such as those between the conceptus and the mother, that make pregnancy so unique. Such experiments will be greatly supported by the recent development of placental and endometrial organoids, as well as platforms that support the study of trophoblast invasion85,86.
Methods
Tissue culture
hTSCs were cultured according to ref. 24, with modifications as outlined in ref. 87. Briefly, ~1 × 105 hTSCs were seeded onto six-well plates coated with either 5–10 µg ml−1 collagen IV (10376931, Fisher Scientific) or 0.5 µg ml−1 iMatrix 511 (NP892-011, Generon). The basal TS medium comprised DMEM/F12 (Gibco) supplemented with 1% KnockOut serum replacement (KSR; Life Technologies Ltd Invitrogen Division), 0.5% penicillin-streptomycin (Pen-Strep, Gibco), 0.15% BSA (A9205, Sigma-Aldrich), 1% ITS-X supplement (Fisher Scientific) and 200 µM l-ascorbic acid (Sigma-Aldrich). Complete medium contained 2.5 μM Rho-associated protein kinase (ROCK) inhibitor Y27632, TGF-B inhibitors A83-01 (5 μM), CHIR99021 (2 μM) (all StemMACS) and epidermal growth factor (50 ng ml−1, E9644, Sigma-Aldrich), with 0.8 mM histone deacetylase inhibitor valproic acid (PHR1061, Sigma-Aldrich). The hTSCs were passaged using TrypLE Express (Fisher Scientific) and split 1:3 to 1:5 every 2–3 days. All cells were cultured at 37 °C in 5% CO2 and 20% O2. For JNK inhibition experiments, hTSCs were seeded onto 12-well or six-well plates and treated with either 10 µM SP600125 (Cambridge Bioscience) for 2–4 days or 20 µM SR11302 (Cambridge Bioscience) for 1–2 days. Control wells were treated with DMSO. Changes in the expression of diagnostic genes were consistent across different cell-collection time points.
Differentiation to EVT and SynT
Differentiation was performed as outlined in ref. 24. For EVT, hTSCs were seeded in a six-well plate coated with 1 μg ml−1 Col IV at a density of 0.75 × 105 cells per well and cultured in 2 ml of EVT medium: DMEM/F12 supplemented with 0.1 mM 2-mercaptoethanol, 0.5% Pen-Strep, 0.3% BSA, 1% ITS-X, 100 ng ml−1 human neuregulin-1 (NRG1; Cell Signalling), 7.5 μM A83-01, 2.5 μM ROCK inhibitor Y27632 and 4% KSR. Cells were suspended in the medium, and Matrigel (LDEV-free, 354234, Scientific Laboratory Supplies) was added to a final concentration of 2%. On day 3, the medium was replaced with EVT medium without NRG1, and Matrigel was added to a final concentration of 0.5%. On day 6, the medium was replaced with EVT medium without NRG1 and KSR, and Matrigel was added to a final concentration of 0.5%, with cells grown for a further two days. Differentiation efficiency was measured using fluorescence-activated cell sorting (FACS) following staining with an allophycocyanin (APC) conjugated antibody to HLAG (APC anti-human HLAG antibody clone: 87G, BioLegend UK); in unmodified hTSCs, this was consistently between 40 and 70%, and in lentivirus-infected CRISPR excision lines and controls, between 10 and 30%. SynT (three-dimensional) differentiation was carried out by seeding 2.5 × 105 hTSCs in uncoated six-well plates, cultured in 2 ml of SynT medium: DMEM/F12 supplemented with 0.1 mM 2-mercaptoethanol, 0.5% Pen-Strep, 0.3% BSA, 1% ITS-X, 2.5 μM ROCK inhibitor Y27632, 50 ng ml−1 epidermal growth factor, 2 μM forskolin (Cambridge Bioscience) and 4% KSR. A 2-ml volume of fresh ST(3D) medium was added on day 3. Cells were passed through a 40-μm mesh strainer, and cells remaining on the strainer were collected and analyzed. Differentiation was assessed using quantitative polymerase chain reaction with reverse transcription (RT–qPCR) and immunostaining (the primers used are listed in Supplementary Table 5).
FACS
Cells were single-cell-dissociated using TrypLE Express and washed in FACS buffer (PBS supplemented with 4% KSR). The cells were then resuspended in 500 μl of fresh FACS buffer, passed through a 70-µm cell strainer, and GFP+ (green fluorescent protein) cells collected and replated (lentiviral infections). To sort EVT-differentiated cells, 400 μl of the cell suspension was incubated with APC-HLAG for 15 min in the fridge, then 100 μl was used as an unstained control. Following antibody incubation, the cells were washed twice with FACS buffer, resuspended in fresh FACS buffer, and passed through a 70-μm cell strainer. Flow cytometry was performed using a FACS Aria II system, and the data were analyzed with FlowJo software.
Immunostaining
Five thousand hTSCs per well were grown on collagen-IV-coated glass coverslips in a 24-well plate, and the differentiation protocol to EVT followed. On day 8 of EVT differentiation, the medium was removed and the cells washed with PBS three times, before fixing in 4% paraformaldehyde for 10 min then staining. Differentiated SynT cells were centrifuged briefly (300g, 1 min), resuspended gently in 500 μl PBS + 4% KSR, and added dropwise to poly-l-lysine-coated glass coverslips. Once SynT cells had gathered on the coverslip, the PBS + KSR was removed and cells were fixed in 4% paraformaldehyde for 10 min. Both fixed EVT and SynT cells were permeabilized with 0.1% Triton X in blocking buffer (1% BSA (wt/vol) 2% FCS (vol/vol)) for 5 min, followed by 1 h of incubation in blocking buffer. Incubation with the primary antibody (SDC-1 (1:200, CD138 mouse anti-human, phycoerythrin, Clone: MI15, Fisher Scientific, UK) for SynT and HLAG (1:100, APC anti-human HLAG antibody clone: 87G, BioLegend UK) for EVT) diluted in blocking buffer was performed at 4 °C overnight. The secondary goat anti-mouse IgG, Alexa Fluor Plus 488 secondary antibody (cat. no. A32723, Thermo Fisher Scientific) was added at a 1:250 dilution for 1 h at room temperature. 4′,6-Diamidino-2-phenylindole (300 nM) was used as a nuclear stain. Images were collected using a Leica DM4000 epifluorescence microscope.
Western blot
Cells were washed with ice-cold PBS, lysed with ice-cold ‘Triton lysis buffer’ (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, supplemented with protease inhibitor cocktail, PMSF and sodium orthovanadate) and pelleted. Supernatants were labeled as ‘Triton-soluble fractions’, which are primarily cytoplasmic. Pellets were washed with ice-cold PBS and lysed with 2% sodium dodecyl sulfate (SDS), followed by sonication (these were labeled as ‘Triton-insoluble fractions’, which are primarily nuclear). Protein concentrations were assessed using a Pierce BCA protein assay kit (Thermo Fisher Scientific). Samples were separated by SDS–polyacrylamide gel electrophoresis (PAGE) and transferred into nitrocellulose membranes. Membranes were incubated with 5% skimmed milk in PBS-T for 30 min at room temperature, followed by overnight incubation with the following antibodies at +4 °C: anti pSer63 c-Jun (Cell Signalling, cat. no. 2361, 1:1,000) and anti c-Jun (Cell Signalling, cat. no. 9165, 1:1,000). After washing with PBS-T, membranes were incubated with peroxidase-conjugated anti-rabbit IgG (Sigma-Aldrich, A6154, 1:10,000) for 1 h at room temperature, washed with PBS-T and exposed to ECL reagent (Sigma-Aldrich, cat no. WBKLS0500). Signals were visualized using X-ray film and a film processor (pSer63 c-Jun) or using a ChemiDoc MP Imaging System (Bio-Rad). Membranes were re-incubated with anti α-tubulin (Sigma-Aldrich, T9026, 1:20,000) or anti H3 (Abcam, ab1791, 1:10,000) for 1 h at room temperature, washed, and then incubated for 1 h with peroxidase-conjugated anti-mouse IgG (Sigma-Aldrich, A0168, 1:10,000) or anti-rabbit IgG, respectively, followed by visualization using ChemiDoc.
ChIP-seq
hTSCs were dissociated using TrypLE Express, pelleted, and washed twice with PBS. The cell pellets were fixed with 1% formaldehyde for 12 min in PBS, followed by quenching with glycine (final concentration of 0.125 M) and washing. Chromatin was sonicated using a Bioruptor Pico device (Diagenode) to an average size of 200–700 bp. Immunoprecipitation was performed using 10 μg of chromatin and 2.5 μg of human antibody (H3K4me3 (RRID:AB_2616052, Diagenode C15410003), H3K4me1 (RRID:AB_306847, Abcam ab8895) and H3K27ac (RRID:AB_2637079, Diagenode C15410196)). Final DNA purification was performed using a GeneJET PCR purification kit (Thermo Scientific, K0701) and eluted in 80 μl of elution buffer. ChIP-seq libraries were prepared from 1 to 5 ng eluted DNA using an NEBNext Ultra II DNA library Prep Kit (New England Biolabs) with 12 cycles of library amplification.
CUT&Tag
CUT&Tag was carried out as in ref. 22. A total of 100,000 hTS or 50,000 EvT cells per antibody were collected fresh using TrypLE Express and centrifuged for 3 min at 600g at room temperature. Cells were washed twice in 1.5 ml of wash buffer (20 mM HEPES pH 7.5; 150 mM NaCl; 0.5 mM spermidine; 1× protease inhibitor cocktail; Roche 11836170001 (PIC)) by gentle pipetting. BioMagPlus concanavalin A-coated magnetic beads (Generon) were activated by washing and resuspension in binding buffer (20 mM HEPES pH 7.5, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2), then 10 µl of activated beads were added per sample and incubated at room temperature for 15 min. A magnet stand was used to isolate bead–cell complexes (henceforth ‘cells’), the supernatant was removed, and the cells were resuspended in 50–100 µl of antibody buffer (20 mM HEPES pH 7.5; 150 mM NaCl; 0.5 mM spermidine; 1× PIC; 0.05% digitonin, 2 mM EDTA, 0.1% BSA) and a 1:50 dilution of primary antibody (H3K27Ac (39034), active motif, H3K9me3 (C15410193) and H3K27me3 (C15410195), Diagenode; c-Jun (60A8)–9165T, and JunD (D17G2) – 5000S, Cell Signalling; GATA3 sc-268 and TFAP2C sc-12762, Santa Cruz; TEAD4 CSB-PA618010LA01HU, Stratech; negative control rabbit IgG, sc-2027, Santa Cruz). Primary antibody incubation was performed on a Nutator shaker for 2 h at room temperature or overnight at 4 °C. Primary antibody was removed and secondary antibody (guinea pig anti-rabbit IgG (heavy and light chain) ABIN101961) diluted 1:50 in 50–100 µl of Dig-Wash buffer, and added to the cells for 30 min incubation at room temperature. The supernatant was removed and cells were washed three times in Dig-Wash buffer. Protein A-Tn5 transposase fusion protein (pA-Tn5) loaded with Illumina NEXTERA adapters (a kind gift from the Henikoff laboratory, via the Madapura laboratory, diluted 1:250, or Epicypher CUTANA pAG-Tn5, diluted 1:10) was prepared in Dig-300 buffer (0.05% digitonin, 20 mM HEPES, pH 7.5, 300 mM NaCl, 0.5 mM spermidine, 1× PIC) and 50–100 µl was added to the cells with gentle vortexing, followed by incubation at room temperature for 1 h. Cells were washed three times in 800 μl of Dig-300 buffer to remove unbound pA-Tn5. Next, the cells were resuspended in 50–100 µl tagmentation buffer (10 mM MgCl2 in Dig-300 buffer) and incubated at 37 °C for 1 h. To stop tagmentation, 2.25 µl of 0.5 M EDTA, 2.75 µl of 10% SDS and 0.5 µl of 20 mg ml−1 Proteinase K was added to the sample and incubated overnight at 37 °C. To extract the DNA, 300 µl phenol:chloroform:isoamyl alcohol (PCI 25:24:1, vol/vol; Sigma-Aldrich) was added to the sample and vortexed. Samples were added to five PRIME Phase Lock Gel light tubes and centrifuged for 3 min at 16,000g. The samples were washed in chloroform, centrifuged for 3 min at 16,000g, and supernatant was added to 100% ethanol, chilled on ice and centrifuged at 16,000g for 15 min. Pellets were washed in 100% ethanol and allowed to air-dry, followed by resuspension in 30 μl of 10 mM Tris-HCl pH 8 1 mM EDTA containing 1/400 RNAse A. Libraries were indexed and amplified, using 21 µl of DNA per sample and adding 25 µl NEBNext HiFi 2× PCR Master mix + 2 µl Universal i5 primer (10 µM) + 2 µl uniquely barcoded i7 primers (10 µM) in 0.2-ml PCR tube strips, using a different barcode for each sample. Cycling parameters were 72 °C for 5 min, 98 °C for 30 s, then 12 cycles of 98 °C for 10 s, 63 °C for 10 s, followed by 72 °C for 1 min, followed by purification with 1X Agencourt AMPure XP beads as per the manufacturer’s instructions (Beckman Coulter).
Cleavage under targets and release using nuclease
CUT&RUN was carried out as in ref. 31. A total of 500,000 hTSCs were washed and resuspended in wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine, plus PIC). Following concanavalin A bead preparation (as for CUT&Tag), the cells were incubated with 50 μl of bead slurry for 15 min and the supernatant removed. Cell–bead complexes (hereafter, ‘cells’) were incubated overnight with antibody buffer (as for CUT&Tag) and primary antibodies (H3K27Ac (39034), Active Motif, H3K9me3 (C15410193) and H3K27me3 (C15410195), Diagenode; c-Jun (60A8) – 9165T and JunD (D17G2) – 5000S, Cell Signalling; GATA3 sc-268 and TFAP2C sc-12762, Santa Cruz; TEAD4 CSB-PA618010LA01HU, Stratech and negative control rabbit IgG, sc-2027, Santa Cruz). Primary antibody was removed and the cells were washed three times in Dig-Wash buffer and incubated with 1:200 pA-MNase (a gift from the Hurd laboratory) for 1 h at 4 °C. The cells were washed three times in Dig-Wash buffer, followed by digestion (adding 2 μl 100 mM CaCl2 to 100 μl of sample) for 30 min at 0 °C, and the reaction stopped in STOP buffer (170 mM NaCl, 20 mM EGTA, 0.05% digitonin, 100 μg ml−1 RNAse A, 50 μg ml−1 glycogen) for 30 min at 37 °C. DNA was extracted using phenol/chloroform, as for CUT&Tag, and barcoded libraries constructed with the NEBNext Ultra II DNA Library Prep Kit for Illumina, using 12 cycles of PCR, followed by purification with 1X Agencourt AMPure XP beads.
RNA isolation and RT–qPCR
Total RNA was extracted with the AllPrep DNA/RNA Mini Kit (Qiagen) and treated with a TURBO DNA-free Kit (Ambion, AM1907) to remove contaminating genomic DNA. RNA (200 ng) was retrotranscribed using Revertaid reverse transcriptase (Thermo Scientific, EP0441), and cDNA was diluted 1/50 for qPCRs using KAPA SYBR FAST (Sigma-Aldrich, KK4610). RT–qPCR was carried out on a Roche LC480 for 40–45 cycles.
RNA-seq
Before library construction, 10–100 ng of total RNA was treated with the NEBNext rRNA Depletion Kit. Library construction was performed with the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina, according to the manufacturer’s protocol. RNA concentration and integrity were assessed using a Bioanalyzer 2100 system (Agilent Technologies). All hTSC samples had an RNA integrity number equivalent (RINe) value of >9.
Guide RNA cloning into CRISPR–Cas9 plasmids
For CRISPR/Cas9 deletion of LTRs, sgRNA oligonucleotides (Integrated DNA Technologies) were designed to target upstream and downstream of the LTRs of interest, using Benchling (https://benchling.com) and annealed. Either the upstream or the downstream guide was cloned into plasmid LRG (Lenti_sgRNA_EFS_GFP; deposited by C. Vakoc (Addgene 65656), which expresses GFP. The other guide was cloned into lentiCRISPR v2, deposited by F. Zhang (Addgene 52961). Clones were verified by Sanger sequencing (Source Bioscience). Guide sequences are listed in Supplementary Table 6.
Lentivirus-mediated hTSC transduction and selection
Lentivirus was produced in 293T cells by quadruple transfection with CRISPR/Cas9 delivery vectors (see above) and the packaging plasmids psPAX2, (deposited by D. Trono, Addgene 12260) and pMD2.G (deposited by D. Trono, Addgene 12259) using FuGENE HD transfection reagent (Promega E2311). Viral supernatant was collected at 48 h and again at 72 h post transfection, pooled, filtered through 0.45 μm and either used fresh or aliquoted and stored at −80 °C. hTSCs were transduced with lentiviral supernatant supplemented with 4 µg ml−1 polybrene for 6 h. Supernatant from lentivirus transfected with psPAX2, pMDG.2, empty LRG and empty lentiCRISPR v2 (that is, no gRNAs) was used alongside each new hTSC infection as a no-sgRNA control. At 48 h after transduction, GFP+ cells were sorted on a FACS Aria II system and replated onto 6- or 12-well plates depending on the cell number. At 24–48 h after sorting, cells were treated with puromycin sulfate for 48 h. DNA and RNA were extracted from the resultant cell populations and genotyped for the presence of excisions. Successful excision pools were further analyzed for the percent of excised alleles and for the expression of genes nearby the targeted LTRs using qPCR and RT–qPCR. The number of independent RT–qPCR replicates used is visible on the respective plots—these were derived using different sgRNA sets or independent infections (as detailed in the figure legends and Supplementary Table 4), and/or from RNA collections at different passage numbers. Genotyping and RT–qPCR primers are listed in Supplementary Table 5.
Endoglin ELISA
hTSC pools containing the LTR10A excision at ENG were seeded in parallel with no-sgRNA controls at 2.5 × 105 cells per well of a six-well plate and differentiated to SynT, as described above. On day 6, medium was collected and stored at −80 °C. SynT cells were snap-frozen for RT–qPCR analysis of differentiation markers and ENG expression. Media aliquots were frozen and subjected to ENG ELISA analysis with the human endoglin ELISA kit (Sigma-Aldrich, RAB0171) according to the manufacturer’s instructions. Absorbance was measured at 450 nm.
Transwell invasion assay
hTSC pools containing the LTR10A excision at ENG were seeded in parallel with no-sgRNA controls at 2.5 × 105 cells per well of a collagen-IV-coated six-well plate and differentiated to EVT as described above. On days 7–9 of differentiation, cells were treated with TrypLE and counted. A subset (~5%) of cells were tested for differentiation efficiency by HLAG staining and FACS to ascertain the proportion of HLAG+ cells, as described above. Invasion assays were carried out on either inserts coated with 75 μl of Matrigel Basement Membrane Matrix (Fisher Scientific, cat. no. 11573620) diluted 1:1 with serum-free EVT medium; or commercial Matrigel pre-coated inserts (BioCoat Matrigel Invasion Chamber, VWR, cat. no. 734-1047), rehydrated by the addition of 500 μl of serum-free D6 EVT medium in both upper and lower chambers for >1 h at 37 °C/5% CO2, both utilizing polycarbonate Transwell inserts (8.0-μm-diameter pores, 6.5-mm-diameter, Costar cat. no. CLS3464-48EA). The remaining cells (9,000–80,000, depending on the experiment) were resuspended in 500 μl of serum-free D6 EVT medium and plated in the upper transwell insert chamber in duplicate. The lower chamber was filled with 500 μl of medium supplemented with 20% KSR. Cells were left to invade through the Matrigel for 48 or 72 h at 37 °C/5% CO2. Non-migrated cells were removed from the upper chamber using a cotton bud, while cells that had migrated were fixed using 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet solution for 15 min. Images were taken using a light microscope at ×10 magnification.
Primary sequencing data processing
High-throughput sequencing reads (2 × 150-bp format; NovaSeq 6000) were trimmed using Trim_galore, with default settings. Mapping was done with either Bowtie288 (for CUT&Tag, CUT&RUN and ChIP-seq; default settings) or Hisat289 (for RNA-seq; with --no-softclip) to the reference genome of the species of origin: hg38, mm10 or rheMac10. Reads with MAPQ below 2 were discarded. Bigwig files were generated using deepTools90 (bamCoverage tool with --binSize 200 --normalizeUsing CPM). Peak detection was performed using either MACS291 (with -q 0.05 --broad) or SEACR92 (with ‘norm’ and ‘relaxed’ options). RNA-seq gene raw counts or reads per kilobase per million values were extracted using the RNA-seq pipeline in Seqmonk. DESeq293 (default parameters) was used to perform differential expression analysis. SQuIRE94 was used to measure ERV family-wide expression from RNA-seq data.
Peak enrichment at repeat families
For a given ChIP-seq/CUT&Tag/CUT&RUN experiment, the number of peaks overlapping each RepeatMasker-annotated repeat family was compared with overlap frequencies across 1,000 random controls (shuffled peaks, avoiding unmappable regions of the genome), yielding enrichment values and associated P values. Significantly enriched repeat families had P < 0.05, >2-fold enrichment, and at least ten copies overlapped by peaks. Families were further selected by only keeping those for which >80% of ENCODE placental DNase-seq samples had an enrichment above twofold, and a median enrichment difference larger than 2 when compared to liver, lung and kidney ENCODE DNase-seq data.
Transcription factor motif analysis
Motifs enriched at active TE families were identified using the AME tool of the MEME suite95 (default parameters) and the 2020 JASPAR vertebrate database. Relevant motifs were selected based on the expression of the respective transcription factors in hTSCs (>1 log2(FPKM)), followed by clustering to find redundant motifs and further selection based on literature searches. The FIMO tool was then used to extract motif locations and frequencies (default parameters).
Human RNA-seq analysis
TE-derived promoters were identified by performing transcriptome assembly on primary cytotrophoblast data using Stringtie (with --rf and guided by the Gencode v38 annotation)96, and intersecting the transcription start sites of multi-exonic transcripts with hTSC-active TE families. To evaluate putative enhancer effects, gene expression differences were determined based on the distance to the nearest H3K27ac-marked TE. JNK target genes were determined based on their distance to the nearest JUN binding peak. For all analyses, log2 fold differences were calculated using only genes that passed a minimal expression threshold (variable depending on dataset and normalization strategy) in at least one of the two samples compared. Gene ontology analysis of differentially expressed genes after JNK inhibition was performed using topGO.
Comparative analysis
Human TE orthologs were identified in non-human primates by performing a reciprocal liftOver. Human TSC RNA-seq data were merged to either mouse TSC or rhesus macTSCs data based on one-to-one gene orthologs. For the human–mouse comparison, information about the proximity to hTSC-active TEs (from the selected families) and mTSC-active TEs (from the RLTR13D5 and RLTR13B families) was used to separate genes into different groups. For the human–macaque comparison, information about whether the nearest hTSC-active TE had a macaque ortholog was used.
Statistical analyses
Wilcoxon tests with Benjamini-Hochberg correction for multiple comparisons or analysis of variance (ANOVA) followed by the Tukey post-hoc test were used, as specified in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this Article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41594-023-00960-6.
Supplementary information
Supplementary tables 1–7.
Acknowledgements
We thank J. Kropp Schmidt and T. Golos for sharing their macaque TSC RNA-seq data, S. Henikoff (Fred Hutchinson Cancer Research Center), P. Madapura (QMUL), H. Wan (QMUL) and P. Hurd (QMUL) for reagents, and S. Papa (Univ. Leeds) for insights into the complexities of JNK activity. We also thank G. Warnes for his gracious assistance with FACS. This work was supported by the BBSRC (research grant no. BB/T000031/1, awarded to M.B. and J.F.), Barts Charity (MRC0297, awarded to M.B. and J.F.) and The Leakey Foundation (awarded to J.F.). This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement InvADeRS no. 841172 (to J.F.).
Extended data
Source data
Unprocessed western blots.
Author contributions
J.M.F. and M.R.B. designed the study and wrote the manuscript. J.M.F. performed cell culture, epigenomic, transcriptomic and genetic editing experiments in human cells. S.M.A. performed epigenomic experiments in mouse TSCs. H.O. and T.A. provided hTSCs and assisted in their culture. E.M.J. and M.P.C. performed immunofluorescence experiments. B.A., R.M.L. and J.K.C. isolated and cultured primary human cytotrophoblast. T.M. performed western blots and invasion assays. M.R.B. and J.M.F. performed bioinformatic analyses.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Atma Ivancevic, Johan Jakobsson and Todd Macfarlan for their contribution to the peer review of this work. Primary Handling Editor: Carolina Perdigoto, in collaboration with the Nature Structural & Molecular Biology team. Peer reviewer reports are available.
Data availability
CUT&Tag, CUT&RUN, ChIP-seq and RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus under accession no. GSE200763. Details of other datasets used can be found in Supplementary Table 7. The GitHub repository https://github.com/MBrancoLab/Frost_2022_hTroph contains the following source data: (1) TE family enrichments for chromatin features (‘Peak_enrichment’ folder), (2) motif frequencies (‘Transcription_factors/FIMO’), (3) JUN/JUND binding profiles (‘Transcription_factors/AP1_profiles’), (4) processed RNA-seq data (‘RNA-seq’ and ‘Comparative_analysis’), (5) TE orthology (‘Active_families/orthologues’) and (6) RT–qPCR, growth curves, FACS and ELISA (‘Assays’). A Readme file is included describing how each figure was generated. Source data are provided with this paper.
Code availability
All code associated with the manuscript is available in the GitHub repository https://github.com/MBrancoLab/Frost_2022_hTroph.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jennifer M. Frost, Email: j.frost@qmul.ac.uk
Miguel R. Branco, Email: m.branco@qmul.ac.uk
Extended data
is available for this paper at 10.1038/s41594-023-00960-6.
Supplementary information
The online version contains supplementary material available at 10.1038/s41594-023-00960-6.
References
- 1.Turco MY, Moffett A. Development of the human placenta. Development. 2019;146:dev163428. doi: 10.1242/dev.163428. [DOI] [PubMed] [Google Scholar]
- 2.Brosens I, Pijnenborg R, Vercruysse L, Romero R. The ‘Great Obstetrical Syndromes’ are associated with disorders of deep placentation. Am. J. Obstet. Gynecol. 2011;204:193–201. doi: 10.1016/j.ajog.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGinnis R, et al. Variants in the fetal genome near FLT1 are associated with risk of preeclampsia. Nat. Genet. 2017;49:1255–1260. doi: 10.1038/ng.3895. [DOI] [PubMed] [Google Scholar]
- 4.Carter AM, Enders AC. Comparative aspects of trophoblast development and placentation. Reprod. Biol. Endocrinol. 2004;2:46. doi: 10.1186/1477-7827-2-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter AM. Unique aspects of human placentation. Int. J. Mol. Sci. 2021;22:8099. doi: 10.3390/ijms22158099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuong EB, Tong W, Hoekstra HE. Maternal-fetal conflict: rapidly evolving proteins in the rodent placenta. Mol. Biol. Evol. 2010;27:1221–1225. doi: 10.1093/molbev/msq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imakawa K, Nakagawa S, Miyazawa T. Baton pass hypothesis: successive incorporation of unconserved endogenous retroviral genes for placentation during mammalian evolution. Genes Cells. 2015;20:771–788. doi: 10.1111/gtc.12278. [DOI] [PubMed] [Google Scholar]
- 8.Lavialle C, et al. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuentes DR, Swigut T, Wysocka J. Systematic perturbation of retroviral LTRs reveals widespread long-range effects on human gene regulation. eLife. 2018;7:e35989. doi: 10.7554/eLife.35989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pontis J, et al. Hominoid-Specific transposable elements and KZFPs facilitate human embryonic genome activation and control transcription in naive human ESCs. Cell Stem Cell. 2019;24:724–735. doi: 10.1016/j.stem.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pontis J, et al. Primate-specific transposable elements shape transcriptional networks during human development. Nat. Commun. 2022;13:7178. doi: 10.1038/s41467-022-34800-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang HS, et al. Transposable elements drive widespread expression of oncogenes in human cancers. Nat. Genet. 2019;51:611–617. doi: 10.1038/s41588-019-0373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 2017;18:71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fueyo R, Judd J, Feschotte C, Wysocka J. Roles of transposable elements in the regulation of mammalian transcription. Nat. Rev. Mol. Cell Biol. 2022;23:481–497. doi: 10.1038/s41580-022-00457-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sundaram V, et al. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014;24:1963–1976. doi: 10.1101/gr.168872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chuong EB, Rumi MAK, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 2013;45:325–329. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Todd CD, Deniz O, Taylor D, Branco MR. Functional evaluation of transposable elements as enhancers in mouse embryonic and trophoblast stem cells. eLife. 2019;8:e44344. doi: 10.7554/eLife.44344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 20.Dunn-Fletcher CE, et al. Anthropoid primate-specific retroviral element THE1B controls expression of CRH in placenta and alters gestation length. PLoS Biol. 2018;16:e2006337. doi: 10.1371/journal.pbio.2006337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun, M. et al. Endogenous retroviruses drive lineage-specific regulatory evolution across primate and rodent placentae. Mol. Biol. Evol.10.1093/molbev/msab223 (2021). [DOI] [PMC free article] [PubMed]
- 22.Kaya-Okur HS, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019;10:1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashley B, et al. Placental uptake and metabolism of 25(OH)vitamin D determine its activity within the fetoplacental unit. eLife. 2022;11:e71094. doi: 10.7554/eLife.71094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okae H, et al. Derivation of human trophoblast stem cells. Cell Stem Cell. 2018;22:50–63. doi: 10.1016/j.stem.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 26.Carter T, et al. Mosaic cis-regulatory evolution drives transcriptional partitioning of HERVH endogenous retrovirus in the human embryo. eLife. 2022;11:e76257. doi: 10.7554/eLife.76257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He G, et al. p57KIP2‑mediated inhibition of human trophoblast apoptosis and promotion of invasion in vitro. Int. J. Mol. Med. 2019;44:281–290. doi: 10.3892/ijmm.2019.4175. [DOI] [PubMed] [Google Scholar]
- 28.Kubota K, Kent LN, Rumi MAK, Roby KF, Soares MJ. Dynamic regulation of AP-1 transcriptional complexes directs trophoblast differentiation. Mol. Cell. Biol. 2015;35:3163–3177. doi: 10.1128/MCB.00118-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadam L, Kohan-Ghadr HR, Drewlo S. The balancing act—PPAR-γ’s roles at the maternal-fetal interface. Syst. Biol. Reprod. Med. 2015;61:65–71. doi: 10.3109/19396368.2014.991881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, et al. Potential role of FoxO3a in the regulation of trophoblast development and pregnancy complications. J. Cell. Mol. Med. 2021;25:4363–4372. doi: 10.1111/jcmm.16499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skene PJ, Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife. 2017;6:e21856. doi: 10.7554/eLife.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chi F, Sharpley MS, Nagaraj R, Roy SS, Banerjee U. Glycolysis-independent glucose metabolism distinguishes TE from ICM fate during mammalian embryogenesis. Dev. Cell. 2020;53:9–26. doi: 10.1016/j.devcel.2020.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Lagemaat LN, Landry J-R, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Schulte AM, et al. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ-line insertion of an endogenous retrovirus. Proc. Natl Acad. Sci. USA. 1996;93:14759–14764. doi: 10.1073/pnas.93.25.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emera D, Wagner GP. Transformation of a transposon into a derived prolactin promoter with function during human pregnancy. Proc. Natl Acad. Sci. USA. 2012;109:11246–11251. doi: 10.1073/pnas.1118566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landry J-R, Rouhi A, Medstrand P, Mager DL. The Opitz syndrome gene Mid1 is transcribed from a human endogenous retroviral promoter. Mol. Biol. Evol. 2002;19:1934–1942. doi: 10.1093/oxfordjournals.molbev.a004017. [DOI] [PubMed] [Google Scholar]
- 37.Teoh PJ, et al. Atypical chemokine receptor ACKR2 mediates chemokine scavenging by primary human trophoblasts and can regulate fetal growth, placental structure and neonatal mortality in mice. J. Immunol. 2014;193:5218–5228. doi: 10.4049/jimmunol.1401096. [DOI] [PubMed] [Google Scholar]
- 38.Dong C, et al. Derivation of trophoblast stem cells from naïve human pluripotent stem cells. eLife. 2020;9:e52504. doi: 10.7554/eLife.52504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin TM, Halbert SP, Spellacy WN. Measurement of pregnancy-associated plasma proteins during human gestation. J. Clin. Invest. 1974;54:576–582. doi: 10.1172/JCI107794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnold LL, et al. Pregnancy-specific glycoprotein gene expression in recurrent aborters: a potential correlation to interleukin-10 expression. Am. J. Reprod. Immunol. 1999;41:174–182. doi: 10.1111/j.1600-0897.1999.tb00530.x. [DOI] [PubMed] [Google Scholar]
- 41.Karg NJ, Csaba IF, Than GN, Arany AA, Szabó DG. The prognosis of the possible foetal and placental complications during delivery by measuring maternal serum levels of pregnancy-specific β-1-glycoprotein (SP1) Arch. Gynecol. 1981;231:69–73. doi: 10.1007/BF02110026. [DOI] [PubMed] [Google Scholar]
- 42.Towler CM, Horne CH, Jandial V, Campbell DM, MacGillivray I. Plasma levels of pregnancy-specific β 1-glycoprotein in complicated pregnancies. Br. J. Obstet. Gynaecol. 1977;84:258–263. doi: 10.1111/j.1471-0528.1977.tb12573.x. [DOI] [PubMed] [Google Scholar]
- 43.Bubici C, Papa S. JNK signalling in cancer: in need of new, smarter therapeutic targets: JNKs in cancer. Br. J. Pharmacol. 2014;171:24–37. doi: 10.1111/bph.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabapathy K, et al. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol. Cell. 2004;15:713–725. doi: 10.1016/j.molcel.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 45.Huh J-W, Ha H-S, Kim D-S, Kim H-S. Placenta-restricted expression of LTR-derived NOS3. Placenta. 2008;29:602–608. doi: 10.1016/j.placenta.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 46.Crosley EJ, Elliot MG, Christians JK, Crespi BJ. Placental invasion, preeclampsia risk and adaptive molecular evolution at the origin of the great apes: evidence from genome-wide analyses. Placenta. 2013;34:127–132. doi: 10.1016/j.placenta.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 47.Carter AM, Pijnenborg R. Evolution of invasive placentation with special reference to non-human primates. Best Pract. Res. Clin. Obstet. Gynaecol. 2011;25:249–257. doi: 10.1016/j.bpobgyn.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt JK, et al. Placenta-derived macaque trophoblast stem cells: differentiation to syncytiotrophoblasts and extravillous trophoblasts reveals phenotypic reprogramming. Sci. Rep. 2020;10:19159. doi: 10.1038/s41598-020-76313-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo Y, Kumar P, Mendelson CR. Estrogen-related receptor γ (ERRγ) regulates oxygen-dependent expression of voltage-gated potassium (K+) channels and tissue kallikrein during human trophoblast differentiation. Mol. Endocrinol. 2013;27:940–952. doi: 10.1210/me.2013-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, et al. Stomatin-like protein 2 (SLP2) regulates the proliferation and invasion of trophoblast cells by modulating mitochondrial functions. Placenta. 2020;100:13–23. doi: 10.1016/j.placenta.2020.07.008. [DOI] [PubMed] [Google Scholar]
- 51.Kuales G, Weiss M, Sedelmeier O, Pfeifer D, Arnold SJ. A resource for the transcriptional signature of bona fide trophoblast stem cells and analysis of their embryonic persistence. Stem Cells Int. 2015;2015:218518. doi: 10.1155/2015/218518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H, et al. Leptin-promoted human extravillous trophoblast invasion is MMP14 dependent and requires the cross talk between Notch1 and PI3K/Akt signaling. Biol. Reprod. 2014;90:78. doi: 10.1095/biolreprod.113.114876. [DOI] [PubMed] [Google Scholar]
- 53.Kim JD, et al. Rex1/Zfp42 as an epigenetic regulator for genomic imprinting. Hum. Mol. Genet. 2011;20:1353–1362. doi: 10.1093/hmg/ddr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers MB, Hosler BA, Gudas LJ. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development. 1991;113:815–824. doi: 10.1242/dev.113.3.815. [DOI] [PubMed] [Google Scholar]
- 55.Deniz Ö, et al. Endogenous retroviruses are a source of enhancers with oncogenic potential in acute myeloid leukaemia. Nat. Commun. 2020;11:3506. doi: 10.1038/s41467-020-17206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmed SIY, Ibrahim ME, Khalil EAG. High altitude and pre-eclampsia: adaptation or protection. Med. Hypotheses. 2017;104:128–132. doi: 10.1016/j.mehy.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 57.Chou C-W, Huang Y-K, Kuo T-T, Liu J-P, Sher Y-P. An overview of ADAM9: structure, activation and regulation in human diseases. Int. J. Mol. Sci. 2020;21:E7790. doi: 10.3390/ijms21207790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inagaki A, et al. Upregulation of HtrA4 in the placentas of patients with severe pre-eclampsia. Placenta. 2012;33:919–926. doi: 10.1016/j.placenta.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 59.Singh H, et al. Human HtrA4 expression is restricted to the placenta, is significantly up-regulated in early-onset preeclampsia, and high levels of HtrA4 cause endothelial dysfunction. J. Clin. Endocrinol. Metab. 2015;100:E936–E945. doi: 10.1210/jc.2014-3969. [DOI] [PubMed] [Google Scholar]
- 60.Visvader J, Verma IM. Differential transcription of exon 1 of the human c-fms gene in placental trophoblasts and monocytes. Mol. Cell. Biol. 1989;9:1336–1341. doi: 10.1128/mcb.9.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kauma SW, Aukerman SL, Eierman D, Turner T. Colony-stimulating factor-1 and c-fms expression in human endometrial tissues and placenta during the menstrual cycle and early pregnancy. J. Clin. Endocrinol. Metab. 1991;73:746–751. doi: 10.1210/jcem-73-4-746. [DOI] [PubMed] [Google Scholar]
- 62.Ahmad SF, et al. Targeting colony stimulating factor-1 receptor signalling to treat ectopic pregnancy. Sci. Rep. 2020;10:15638. doi: 10.1038/s41598-020-72785-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamilton GS, Lysiak JJ, Watson AJ, Lala PK. Effects of colony stimulating factor-1 on human extravillous trophoblast growth and invasion. J. Endocrinol. 1998;159:69–77. doi: 10.1677/joe.0.1590069. [DOI] [PubMed] [Google Scholar]
- 64.Pollard JW, et al. Apparent role of the macrophage growth factor, CSF-1, in placental development. Nature. 1987;330:484–486. doi: 10.1038/330484a0. [DOI] [PubMed] [Google Scholar]
- 65.Hayashi M, Ohkura T, Inaba N. Elevation of serum macrophage colony-stimulating factor before the clinical manifestations of preeclampsia. Am. J. Obstet. Gynecol. 2003;189:1356–1360. doi: 10.1067/S0002-9378(03)00674-4. [DOI] [PubMed] [Google Scholar]
- 66.Lu X, et al. Twist1 is involved in trophoblast syncytialization by regulating GCM1. Placenta. 2016;39:45–54. doi: 10.1016/j.placenta.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 67.Ng YH, Zhu H, Leung PCK. Twist regulates cadherin-mediated differentiation and fusion of human trophoblastic cells. J. Clin. Endocrinol. Metab. 2011;96:3881–3890. doi: 10.1210/jc.2010-2714. [DOI] [PubMed] [Google Scholar]
- 68.Qin Q, Xu Y, He T, Qin C, Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell. Res. 2012;22:90–106. doi: 10.1038/cr.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Camolotto S, et al. Expression and transcriptional regulation of individual pregnancy-specific glycoprotein genes in differentiating trophoblast cells. Placenta. 2010;31:312–319. doi: 10.1016/j.placenta.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 70.Venkatesha S, et al. Soluble endoglin contributes to the pathogenesis of preeclampsia. Nat. Med. 2006;12:642–649. doi: 10.1038/nm1429. [DOI] [PubMed] [Google Scholar]
- 71.Leaños-Miranda A, et al. Soluble endoglin as a marker for preeclampsia, its severity, and the occurrence of adverse outcomes. Hypertension. 2019;74:991–997. doi: 10.1161/HYPERTENSIONAHA.119.13348. [DOI] [PubMed] [Google Scholar]
- 72.Caniggia I, Taylor CV, Ritchie JW, Lye SJ, Letarte M. Endoglin regulates trophoblast differentiation along the invasive pathway in human placental villous explants. Endocrinology. 1997;138:4977–4988. doi: 10.1210/endo.138.11.5475. [DOI] [PubMed] [Google Scholar]
- 73.Mano Y, et al. The loss of endoglin promotes the invasion of extravillous trophoblasts. Endocrinology. 2011;152:4386–4394. doi: 10.1210/en.2011-1088. [DOI] [PubMed] [Google Scholar]
- 74.Zhang B, et al. Human placental cytotrophoblast epigenome dynamics over gestation and alterations in placental disease. Dev. Cell. 2021;56:1238–1252. doi: 10.1016/j.devcel.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ivancevic, A., Simpson, D. M. & Chuong, E. B. Endogenous retroviruses mediate transcriptional rewiring in response to oncogenic signaling in colorectal cancer. Preprint at https://www.biorxiv.org/content/10.1101/2021.10.28.466196v2 (2021). [DOI] [PMC free article] [PubMed]
- 76.Smith ZD, et al. Epigenetic restriction of extraembryonic lineages mirrors the somatic transition to cancer. Nature. 2017;549:543–547. doi: 10.1038/nature23891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Costanzo V, Bardelli A, Siena S, Abrignani S. Exploring the links between cancer and placenta development. Open Biol. 2018;8:180081. doi: 10.1098/rsob.180081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meinhardt G, et al. Pivotal role of the transcriptional co-activator YAP in trophoblast stemness of the developing human placenta. Proc. Natl Acad. Sci. USA. 2020;117:13562–13570. doi: 10.1073/pnas.2002630117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zimmermann W, Kammerer R. The immune-modulating pregnancy-specific glycoproteins evolve rapidly and their presence correlates with hemochorial placentation in primates. BMC Genomics. 2021;22:128. doi: 10.1186/s12864-021-07413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rudert F, Zimmermann W, Thompson JA. Intra- and interspecies analyses of the carcinoembryonic antigen (CEA) gene family reveal independent evolution in primates and rodents. J. Mol. Evol. 1989;29:126–134. doi: 10.1007/BF02100111. [DOI] [PubMed] [Google Scholar]
- 81.Li Y, Moretto-Zita M, Leon-Garcia S, Parast MM. p63 inhibits extravillous trophoblast migration and maintains cells in a cytotrophoblast stem cell-like state. Am. J. Pathol. 2014;184:3332–3343. doi: 10.1016/j.ajpath.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Riege K, et al. Dissecting the DNA binding landscape and gene regulatory network of p63 and p53. eLife. 2020;9:e63266. doi: 10.7554/eLife.63266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Su D, et al. Interactions of chromatin context, binding site sequence content, and sequence evolution in stress-induced p53 occupancy and transactivation. PLoS Genet. 2015;11:e1004885. doi: 10.1371/journal.pgen.1004885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seligman SP, Buyon JP, Clancy RM, Young BK, Abramson SB. The role of nitric oxide in the pathogenesis of preeclampsia. Am. J. Obstet. Gynecol. 1994;171:944–948. doi: 10.1016/S0002-9378(94)70064-8. [DOI] [PubMed] [Google Scholar]
- 85.Abbas Y, Turco MY, Burton GJ, Moffett A. Investigation of human trophoblast invasion in vitro. Hum. Reprod. Update. 2020;26:501–513. doi: 10.1093/humupd/dmaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ojosnegros S, Seriola A, Godeau AL, Veiga A. Embryo implantation in the laboratory: an update on current techniques. Hum. Reprod. Update. 2021;27:501–530. doi: 10.1093/humupd/dmaa054. [DOI] [PubMed] [Google Scholar]
- 87.Takahashi S, et al. Loss of p57KIP2 expression confers resistance to contact inhibition in human androgenetic trophoblast stem cells. Proc. Natl Acad. Sci. USA. 2019;116:26606–26613. doi: 10.1073/pnas.1916019116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meers MP, Tenenbaum D, Henikoff S. Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenet. Chromatin. 2019;12:42. doi: 10.1186/s13072-019-0287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang WR, Ardeljan D, Pacyna CN, Payer LM, Burns KH. SQuIRE reveals locus-specific regulation of interspersed repeat expression. Nucleic Acids Res. 2019;47:e27. doi: 10.1093/nar/gky1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME Suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables 1–7.
Data Availability Statement
CUT&Tag, CUT&RUN, ChIP-seq and RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus under accession no. GSE200763. Details of other datasets used can be found in Supplementary Table 7. The GitHub repository https://github.com/MBrancoLab/Frost_2022_hTroph contains the following source data: (1) TE family enrichments for chromatin features (‘Peak_enrichment’ folder), (2) motif frequencies (‘Transcription_factors/FIMO’), (3) JUN/JUND binding profiles (‘Transcription_factors/AP1_profiles’), (4) processed RNA-seq data (‘RNA-seq’ and ‘Comparative_analysis’), (5) TE orthology (‘Active_families/orthologues’) and (6) RT–qPCR, growth curves, FACS and ELISA (‘Assays’). A Readme file is included describing how each figure was generated. Source data are provided with this paper.
All code associated with the manuscript is available in the GitHub repository https://github.com/MBrancoLab/Frost_2022_hTroph.