Abstract
Human Angiotensin-Converting Enzyme 2 (hACE2) is the major receptor enabling host cell invasion by SARS-CoV-2 via interaction with Spike. The murine ACE2 does not interact efficiently with SARS-CoV-2 Spike and therefore the laboratory mouse strains are not permissive to SARS-CoV-2 replication. Here, we generated new hACE2 transgenic mice, which harbor the hACE2 gene under the human keratin 18 promoter, in “HHD-DR1” background. HHD-DR1 mice are fully devoid of murine Major Histocompatibility Complex (MHC) molecules of class-I and -II and express only MHC molecules from Human Leukocyte Antigen (HLA) HLA 02.01, DRA01.01, DRB1.01.01 alleles, widely expressed in human populations. We selected three transgenic strains, with various hACE2 mRNA expression levels and distinctive profiles of lung and/or brain permissiveness to SARS-CoV-2 replication. These new hACE2 transgenic strains display high permissiveness to the replication of SARS-CoV-2 Omicron sub-variants, while the previously available B6.K18-ACE22Prlmn/JAX mice have been reported to be poorly susceptible to infection with Omicron. As a first application, one of these MHC- and ACE2-humanized strains was successfully used to show the efficacy of a lentiviral-based COVID-19 vaccine.
Keywords: Transgenesis, COVID-19 animal model, SARS-CoV-2, Omicron variant, HLA
1. Introduction
With the persistence of the coronavirus disease (COVID)-19 pandemic, the research for second-generation vaccines and drugs remains an important public health issue worldwide and therefore new research tools are still indispensable. Human Angiotensin-Converting Enzyme 2 (hACE2) is the major receptor enabling host cell invasion by SARS-CoV-2 via interaction with Spike glycoprotein [1]. The murine ACE2 ortholog does not interact properly with Spike and therefore the conventional laboratory mouse strains are not permissive to SARS-CoV-2 replication. C57BL/6 mice transgenic for hACE2 (B6.K18-ACE22Prlmn/JAX) [2] are available at JAX Laboratories. We also generated a new C57BL/6 transgenic strain carrying hACE2 gene under the human keratin 18 (K18) promoter, namely “B6.K18-hACE2IP-THV”. Compared to B6.K18-ACE22Prlmn/JAX mice, the B6.K18-hACE2IP-THV strain has distinctive characteristics, including higher hACE2 mRNA expression and strong permissiveness to viral replication in the brain, in addition to the lung [3, 4]. In the present study, we generated three new K18-hACE2 transgenic mice in the “HHD-DR1” genetic background. HHD-DR1 mice are fully devoid of murine Major Histocompatibility Complex (MHC) molecules of class-I (MHC-I) and -II (MHC-II) and express only MHC-I and -II molecules from Human Leukocyte Antigen (HLA) alleles, i.e., HLA 02.01, DRA01.01, DRB1.01.01, widely expressed in human populations, notably in Caucasian, Asian and African populations. More precisely, HLA 02.01 is expressed by 50% of Caucasian, 50% of Asian and 35% of African population, while DR1 is expressed by 20% of Caucasian, 10% of Asian and 8% of African populations. Therefore, in the HHD-DR1 hACE2 transgenic mice, antigen presentation and cognate interactions between MHC-I and -II molecules and repertoire of T-Cell Receptors for antigen (TCR) reproduce key molecular aspects of antigen presentation in humans.
HHD-DR1 mice are triple transgenic for HLA 02.01, DRA01.01, DRB1.01.01 and quintuple knocked-out for murine β2-microglobulin (β2m), H-2Db, I-Aα b, I-Aβ b and I-Eβ b. Now homozygote for all these loci, HHD-DR1 mice resulted from: (i) an initial crossing of H2Db KO mice with mice Cre Lox-deleted of a complete 80 kb-long genomic region, encompassing the genes encoding I-Aα b, I-Aβ b, I-Eβ b MHC-II molecules [5], to select mice having linked the MHC deleted region and the H-2Db KO locus by crossing over, (ii) a subsequent crossing to the first generation HHD mice, transgenic for a fusion of HLA A2.01 and human β2m single chain and double KO for murine β2m and H-2Db [6], followed by (iii) a final crossing to another HHD strain, double transgenic for HLA DRA01.01, DRB1.01.01 mice [7, 8]. Therefore, any possibility of epitope presentation by the H-2Db heavy chain alone [[9], [10], [11]] or by a putative HLA DRA.01.01 - I-Eβ b hybrid MHC-II molecule is fully discarded in HHD-DR1 mice [12, 13].
The HHD-DR1 hACE2 transgenic strains generated in the present study are: (i) permissive to SARS-CoV-2 replication, either in the lung or in the lung and brain, (ii) express high amounts of hACE2 mRNA, comparable to those detected in the B6.K18-hACE2IP-THV strain [3], and (iii) have marked permissiveness to SARS-CoV-2 Omicron replication, in addition to previous SARS-CoV-2 variants. This last feature also strongly distinguishes HHD-DR1 hACE2 mice from the B6.K18-ACE22Prlmn/JAX mice [2], which have been reported to have scarce permissiveness to SARS-CoV-2 Omicron replication [14]. As a first application, in one of the established MHC- and ACE2-humanized strains, we demonstrated the strong efficacy of a lentiviral vector-based COVID-19 vaccine candidate, the mode of action of which is largely dependent on T-cell immunity [3, 4, 15, 16].
2. Materials and Methods
2.1. Mice
HHD-DR1 adult males and 3-week-old female mice were purchased from Charles River (Les Oncins, Saint Germain- Nuelles, France) and housed in ventilated cages under specific pathogen-free conditions at the Institut Pasteur animal facilities. B6CBAF1 females were purchased from Janvier Labs (Janvier Labs, Le Genest-Saint-Isle). B6.K18-hACE2IP-THV mice [3, 4] were bred and housed in animal facilities of Institut Pasteur. All procedures were performed in accordance with the European and French guidelines (Directive 86/609/CEE and Decree 87-848 of 19 October 1987) subsequent to approval by the Institut Pasteur Safety, Animal Care and Use Committee, protocol agreement delivered by local ethical committee (CETEA #DAP20007, CETEA #DAP200058) and Ministry of High Education and Research APAFIS#31068-2021041613059523 v1, APAFIS#24627-2020031117362508 v1, APAFIS#28755-2020122110238379 v1.
2.2. hACE-2 transgenesis in HHD-DR1 mice
The pK18-hACE2 plasmid, purchased from Addgene (Watertown, MA), was double digested with Hpa1 and Xba1 enzymes, and the 6.8 kb DNA fragment generated was purified with Qiaex II gel extraction kit (Qiagen). Female HHD-DR1 mice were injected with pregnant mare serum gonadotropin and human chorionic gonadotropin with a 46h interval, and then mated with HHD-DR1 males. The fertilized one-cell embryos were collected from the oviducts. Then, 2 ng/μl of the purified transgene were microinjected into the pronuclei of HHD-DR1 zygotes according to standard protocols [17]. The microinjected eggs were then transferred into pseudo-pregnant B6CBAF1 foster mothers (Janvier Labs).
The lentiviral vector used for the alternative transgenesis method has been previously described [3]. Briefly, the human K18 promoter (GenBank: AF179904.1 nucleotide 90–2,579) was PCR amplified from A549 cells [18, 19]. The K18 promoter is completed in 3’ with a modified “i6x7” intron from human K18 gene (GenBank: AF179904.1 nucleotide 2,988–3,740) which was synthesized by Genscript. As a result, the K18 IP-ThV lentiviral pFLAP plasmid includes the human K18 promoter and the i6x7 intron at 5’, and the transgene is also flanked with the wildtype WPRE element at 3’ (Fig. S2). To facilitate the construction, a ClaI restriction site was introduced between the promoter and the intron. The resulted segment was inserted into a pFLAP lentiviral plasmid between the MluI and BamHI sites. In the case of K18-hACE2 pFLAP plasmid, hACE2 cDNA was introduced between the BamHI and XhoI sites by restriction/ligation. A high titer (≈ 109 TU/ml) of integrative lentiviral K18-hACE2 IP-ThV construct was used for transgenesis by means of subzonal microinjection under the pellucida of fertilized HHD-DR1 eggs, followed by transplantation of the microinjected eggs into pseudo-pregnant B6CBAF1 females. Lentiviral vector allows particularly efficient transfer of the transgene into the nuclei of the fertilized eggs [20].
The progeny was studied for integration of hACE2 gene by using hACE2-forward: 5’-TCC TAA CCA GCC CCC TGT T-3’ and hACE2-reverse: 5’-TGA CAA TGC CAA CCA CTA TCA CT-3’ primers in PCR, applied on genomic DNA prepared from toe clipping, as described previously [3]. PCR-identified hACE2 + founders were backcrossed to HHD-DR1 mice in order to preserve the state of homozygosity for HLA 02.01, DRα01.01, DRβ01.01 humanized loci and for quintuple KO for β2m, H-2Db, I-Aα b, I-Aβ b and I-Eα b murine loci.
It is noteworthy that the female founder F0#9 gave birth to only one hACE2 + male, in the progeny of which only females were hACE2 +, indicating that the hACE2 transgene is on the X chromosome. Therefore, in the resulted strain, the hACE2 transgene insertion is not lethal since males are in good state. The transition to homozygosity will be easy as it can be determined by a simple progeny test with which a hACE2 + male is "pseudo homozygous", and if crossed with heterozygote females, will give rise to 50% of homozygote females. In addition, whether male or female, the founder F0#9 descendance will express only one functional hACE2 allele. In fact, in females this will result from methylation of the hACE2 allele on the second X chromosome.
2.3. SARS-CoV-2 inoculation and measurement of viral RNA contents
The hACE2 + offspring were inoculated i.n., with 0.3 × 105 Median Tissue Culture Infectious Dose (TCID)50 – contained in 20 μl – of the Delta/2021/I7.2 200 [21], Omicron BA.1 [22] or Omicron BA.5 SARS-CoV-2 clinical isolate. The Delta/2021/I7.2 200 is a clinical isolate of the B.1.617.2 Delta variant, originated from an individual with COVID-19 who had returned to France from India in 2021 [21]. The SARS-CoV-2 Omicron Pango lineage BA.1 has been isolated in Belgium from a traveler returning from Egypt in 2022 [22]. The SARS-CoV-2 Omicron BA.5 (hCoV-19/France/BRE-IPP34319/2022 strain) has been isolated by the National Reference Centre for Respiratory Viruses (Institut Pasteur) from a human sample provided by Laboratoire Alliance Anabio, (Melesse, France). For i.n. inoculation, mice were anesthetized by i.p. injection of Ketamine (Imalgene, 80 mg/kg) and Xylazine (Rompun, 5 mg/kg). Animals were then housed in an isolator in BioSafety Level 3 animal facilities of Institut Pasteur. Viral RNA contents were determined in the organs by quantification of total or sub-genomic RNA from ECoV-2 by quantitative real-time (qRT)-PCR. The Esg RT-PCR measures only active viral replication [15, [23], [24], [25]]. In the context of vaccine efficacy assays, we previously compared head-to-head the viral content quantification by plaque-forming unit (PFU) and Esg qRT-PCR [15]. We observed that, in vaccinated animals, the PFU approach widely underestimated the amounts of cultivable SARS-CoV-2 viral particles because of the high activity of anti-Spike neutralizing antibodies present in the organ homogenates which are incubated with the Vero cells in vitro in the PFU assay. Based on this fact and also the lack of additional information provided by the PFU method, we opted for the Esg qRT-PCR approach.
2.4. qRT-PCR for Inflammation study
The qRT-PCR quantification of inflammatory mediators in the lungs and brain was performed as detailed elsewhere [15] on total RNA extracted by TRIzol reagent (Invitrogen) and stored at −80°C. The RNA quality was first assessed using a Bioanalyzer 2100 (Agilent Technologies). The RNA Integrity Number (RIN) was 7.5 — 10.0. RNA samples were quantitated using a NanoDrop Spectrophotometer (Thermo Scientific NanoDrop). One μg of each RNA was used per qRT-PCR sample in 96 wells plate.
2.5. ELISPOT assay and T-cell epitope mapping
Splenocytes from individual mice were homogenized and filtered through 70 μm-pore filters and centrifuged at 300g for 10 min. Cells were treated with Red Blood Cell Lysing Buffer (Sigma) and resuspended at 2.5 × 107 cells/mL in complete α-MEM medium containing 10% heat-inactivated FBS, 100 U/mL penicillin and 100 μg/ml streptomycin, 1 × 10-4 M non-essential amino-acids, 10 mM Hepes, 1 mM sodium pyruvate and 5 × 10-5 M of β-mercapto-ethanol. For each mouse, 100 μl of cell suspension (2.5 × 105 splenocyte/well) were stimulated overnight (∼16h) with 100 μl of SARS-CoV-2 matrix peptide pools (2 μg/ml of each peptide, Mimotopes, UK), SARS-CoV-2 individual peptides (2 μg/ml) or blank control (complete medium with dimethylsulfoxide) onto sterile nitrocellulose MultiScreenIP 96-well plates (Millipore, Bedford, MA) coated with capture antibodies against mouse IFN-γ. Panels of 253 15-mer peptides spanning the prototype Spike were organized into 32 pools. The overlapping peptide pools were assigned to a 2D matrix in which each peptide was represented in 2 different peptide pools. In this way, the intersection of two positive pools identifies a potentially positive peptide [26]. Fifteen-mer candidate peptides at the intersections of the positive matrix-pools were tested individually for confirmation. The results were expressed as IFN-γ spot forming units (SFU) per 106 splenocytes and were determined using on an ELISpot analyzer (ImmunoSpot®, CTL, Germany). To quantify antigen-specific responses, mean spots of the control wells were subtracted from the positive wells.
2.6. Intracellular cytokine staining
Splenocytes were plated at 4 × 106 cells/well in 24-well plates and co-cultured during 6h in the presence of 10 μg/ml of appropriate peptide, 1 μg/ml of anti-CD28 (clone 37.51) and 1 μg/ml of anti-CD49d (clone 9C10-MFR4.B) mAbs (BD Biosciences). During the last 3h of incubation, a mixture of Golgi Plug and Golgi Stop (BD Biosciences) were added. Cells were then collected, washed and stained for 25 min at 4°C with a mixture of Near IR Live/Dead (Invitrogen), FcγII/III receptor blocking anti-CD16/CD32 (clone 2.4G2), PerCP-Cy5.5-anti-CD3ε (clone 145-2C11), PE-Cy7-anti-CD4 (clone RM4-5) and BV711-anti-CD8 (clone 53-6.7) mAbs (BD Biosciences or eBioscience). Cells were washed in FACS buffer, permeabilized in Cytofix/Cytoperm kit (BD Bioscience), washed twice with PermWash 1X buffer and incubated with a mixture of BV421-anti-IL-2 (clone JES6-5H4), FITC-anti-TNF-α (MP6-XT22), and APC-anti-IFN-γ (clone XMG1.2) mAbs (BD Biosciences), during 25 min at 4°C. Cells were then washed in PermWash and then in FACS buffer and fixed with Cytofix (BD Biosciences) overnight at 4°C. Samples were acquired in an Attune NxT cytometer system (Invitrogen) and data were analyzed using FlowJo software (Treestar, OR, USA).
2.7. Vaccine assay
Methods used for the measurement of Spike-specific antibody and T-cell responses were described elsewhere [15]. Mice were immunized (i.m. or i.n.) with 1 × 108 TU of LV::SBeta-2P, contained in 50 μl for i.m. injection and in 20 μl for i.n injection. Mice were inoculated i.n. with 0.3 × 105 TCID50 of the Delta variant of SARS-CoV-2 clinical isolate and housed in filtered cages in an isolator in biosafety level 3 animal facilities. The organs recovered from the infected animals were manipulated according to the approved standard procedures of these facilities in a Bio-Safety Level 3 laboratory.
2.8. Lung histopathology
Histopathological study was performed on the left lung lobes, fixed in formalin for 7 days, then transferred in ethanol and embedded in paraffin. Five μm-thick sections were stained with hematoxylin and eosin. Lesions were scored on images acquired in a double-blinded manner on an Axioscan Z1 Zeiss slide scanner, using the Zen 2 blue edition software.
2.9. Statistical analyses
Statistical significances were determined by use of two-tailed unpaired t test or one-way Anova. When indicated data were subjected to Pearson correlation coefficient analyses. All statistical analyses were performed by use of Prism GraphPad v9.
2.10. Data availability
The published article includes all datasets generated and analyzed during this study. All plasmids, lentiviral vectors and mouse strains generated in this study will be available under material transfer agreement for research use. Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author Laleh Majlessi (laleh.majlessi@pasteur.fr).
3. Results
3.1. hACE-2 transgenesis in HHD-DR1 mice
Two technologically distinct transgenesis methods were performed by: (i) pronuclear DNA micro-injection of a 6.8-kb Hpal I-XbaI fragment from the pK18-hACE2 vector [2] into the fertilized HHD-DR1 eggs, or (ii) sub-zonal micro-injection of the integrative LV::pK18-hACE2 lentiviral vector under the zona pellucida of fertilized HHD-DR1 eggs. With the former approach, the DNA fragment must be directly injected into the pronuclei of the zygotes and usually multiple head-to-tail tandem copies of the transgene are inserted into a unique site of the host genome [27]. With the second technology, the nuclear transfer of the genetic material from the zona pellucida to the nuclei of fertilized HHD-DR1 eggs is assured by cell invasion followed by retro-transcription of the lentiviral recombinant RNA into double stranded proviral DNA and very efficient nuclear import of the transgene into the host nuclei. The strong transduction capacity of lentiviral vector is related to a three-stranded DNA structure, known as a "DNA flap", which is formed following retro-transcription of RNA into double stranded proviral DNA [28]. In net opposition to the first approach, with the lentiviral technology, the transgene is not in tandem and is generally integrated at numerous potential integration sites [29].
The progenies were studied for integration of hACE2 gene. The hACE2 + F0#8 male, F0#9 female and F0#11 female founders were identified in the DNA fragment-based transgenesis. The hACE2 + F30# male and F0#47 male founders were identified in the lentiviral vector-based transgenesis (Table S1). To stabilize the progenies, each founder was backcrossed to HHD-DR1 non-hACE2 transgenic mice to preserve the homozygous status for: (i) HLA 02.01, DRA01.01 and DRB1.01.01 humanized loci, and (ii) KO genes encoding for murine β2m, H-2Db, I-Aα b, I-Aβ b and I-Eα b.
3.2. Permissiveness of the hACE2+ offspring to SARS-CoV-2 replication
First, the hACE2 + offspring of the founders generated by the DNA fragment-based transgenesis were evaluated for their permissiveness to SARS-CoV-2 replication. Individuals from the previously described B6.K18-hACE2IP-THV strain [3, 4] were also included as positive controls. Mice were inoculated intranasally (i.n.) with 0.3 × 105 TCID50 of a Delta SARS-CoV-2 clinical isolate [21]. Four days post-inoculation (dpi), viral loads were determined in the lung and brain by ECoV-2 (E)- or sub-genomic E (Esg)-specific quantitative real-time (qRT)-PCR. It is well established that the Esg qRT-PCR is an indicator of only active viral replication [24, 25, 30]. We previously established the good correlation between viral contents determined by PFU and Esg qRT-PCR methods in non-immune mice and hamsters. [3, 15]. The Esg qRT-PCR method is also technically easier to perform, especially in brain homogenates, which are very rich in fat material and difficult to culture in PFU assay.
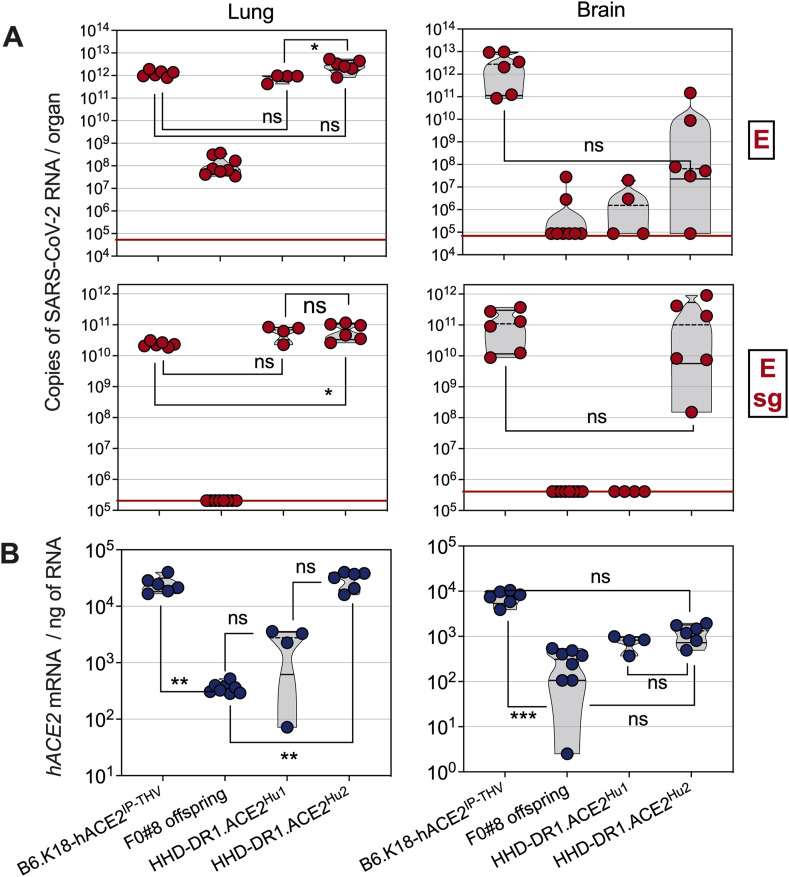
The progeny of the founder F0#8 was not permissive to SARS-CoV-2 replication (Fig. 1 A). In contrast, the progeny of the founder F0#9, hereafter named "HHD-DR1.ACE2Hu1", displayed a notable lung – but not brain – permissiveness to SARS-CoV-2 replication. The progeny of the founder F0#11, hereafter named “HHD-DR1.ACE2Hu2”, was not only markedly permissive to SARS-CoV-2 replication in their lung (Fig. 1A, top) but also displayed significant Esg RNA contents in their brain (Fig. 1A, bottom). The viral RNA contents in the lungs of HHD-DR1.ACE2Hu1 and HHD-DR1.ACE2Hu2 mice were statistically comparable to those of the previously described B6.K18-hACE2IP-THV strain. The main characteristics of these strains are recapitulated in the Table S1. Comparative assessment of hACE2 mRNA transcription showed that the non-permissive F0#8 offspring had detectable but the lowest hACE2 mRNA expression level in the lung and brain (Fig. 1B). Such transcription levels were obviously not sufficient to allow productive SARS-CoV-2 infection. Analysis of the hACE2 mRNA transcription levels in both lung and brain established the following hierarchy: B6.K18-hACE2IP-THV = HHD-DR1.ACE2Hu2 > HHD-DR1.ACE2Hu1 > F0#8 offspring (Fig. 1B).
Fig. 1.
Permissiveness of HHD-DR1.ACE2Hu1 and HHD-DR1.ACE2Hu2 transgenic mice to SARS-CoV-2 replication. hACE2+ offspring from the founders #8, #9 (HHD-DR1.ACE2Hu1) or #11 (HHD-DR1.ACE2Hu2), resulting from a conventional DNA-based transgenesis, or B6.K18-hACE2IP-THV positive control mice were inoculated i.n. with 0.3 × 105 TCID50/mouse of SARS-CoV-2 Delta variant. A. Lung and brain viral RNA contents were determined by E- (top) or sub-genomic Esg- (bottom) specific qRT-PCR at 4 dpi. Red lines indicate the qRT-PCR detection limits. B. Quantification of hACE-2 mRNA in the lung and brain of the same mice. Statistical significance was evaluated by one-way Anova (*= p < 0.05, **= p < 0.01, ***= p < 0.001).
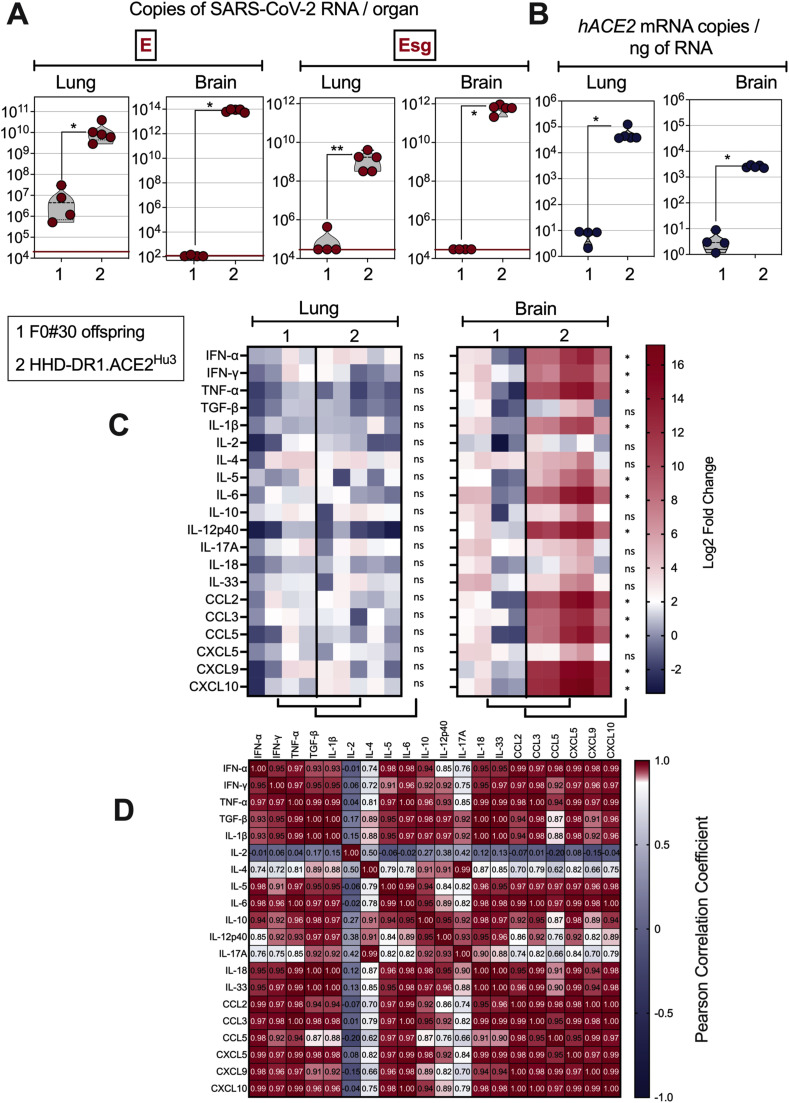
In the lentiviral vector-based transgenesis, the progeny of the founder F0#30 was not permissive to SARS-CoV-2 Delta replication, whereas the progeny of the founder F0#47, hereafter named "HHD-DR1.ACE2Hu3", displayed strong lung and brain permissiveness to SARS-CoV-2 Delta replication (Fig. 2 A). Comparative assessment of the hACE2 mRNA transcription levels in the lung and brain (Fig. 2B) of the F0#30 offspring and HHD-DR1.ACE2Hu3 mice established a positive correlation between the hACE2 transgene expression levels and the permissiveness to viral replication. Therefore, based on the transgenesis method, hACE2 expression levels and permissiveness of the organs to SARS-CoV-2 replication, we defined three distinct humanized HHD-DR1.ACE2Hu strains (Table S1).
Fig. 2.
Permissiveness of HHD-DR1.ACE2Hu3 transgenic mice to SARS-CoV-2 replication and inflammatory status of their lungs and brain after SARS-CoV-2 inoculation. hACE2+ offspring from the founders #30 or #47 (HHD-DR1.ACE2Hu3), resulting from a lentiviral vector-based transgenesis, were inoculated i.n. with 0.3 × 105 TCID50/mouse of SARS-CoV-2 Delta variant. A. Lung and brain viral RNA contents were determined by E-specific or sub-genomic Esg-specific qRT-PCR at 4 dpi. B. Quantification of hACE-2 mRNA in the lung and brain of the same mice. C. Heatmaps represent log2 fold change in cytokine and chemokine mRNA expression in the lungs or brain of the same mice (n = 4-5/group). Data were normalized versus untreated controls. Statistical significance was evaluated by Mann-Whitney test (ns = not significant, * = p < 0.05, ** = p < 0.01). D. Pearson correlation coefficient of the analytes studied in the brain of SARS-CoV-2 Delta-inoculated HHD-DR1.ACE2Hu3 mice.
HHD-DR1.ACE2Hu3 mice, which had the highest viral loads notably in the brain, were further characterized for their infection-mediated inflammation after inoculation with SARS-CoV-2 Delta. At 4 dpi, as evaluated by qRT-PCR study of 20 pro- or anti-inflammatory analytes, applied to RNA extracted from total lung homogenate, no sizable modification of the inflammatory transcriptome was observed in the lungs of the permissive HHD-DR1.ACE2Hu3 mice, compared to the non-permissive F0#30 offspring (Fig. 2C, left). In net contrast, at this time point, the brain of the permissive HHD-DR1.ACE2Hu3 mice displayed a marked inflammatory status, compared to the non-permissive F0#30 offspring (Fig. 2C, right). This brain inflammation was characterized by statistically significant upregulation of IFN-α, IFN-γ, TNF-α, IL-1β, IL-5, IL-6, IL-12p40, CCL2, CCL3, CCL5, CXCL9 and CXCL10 (Fig. 2C, right), in a correlative manner (Fig. 3 D). No correlation was observed between IL-2 and IL-4 expression and the former.
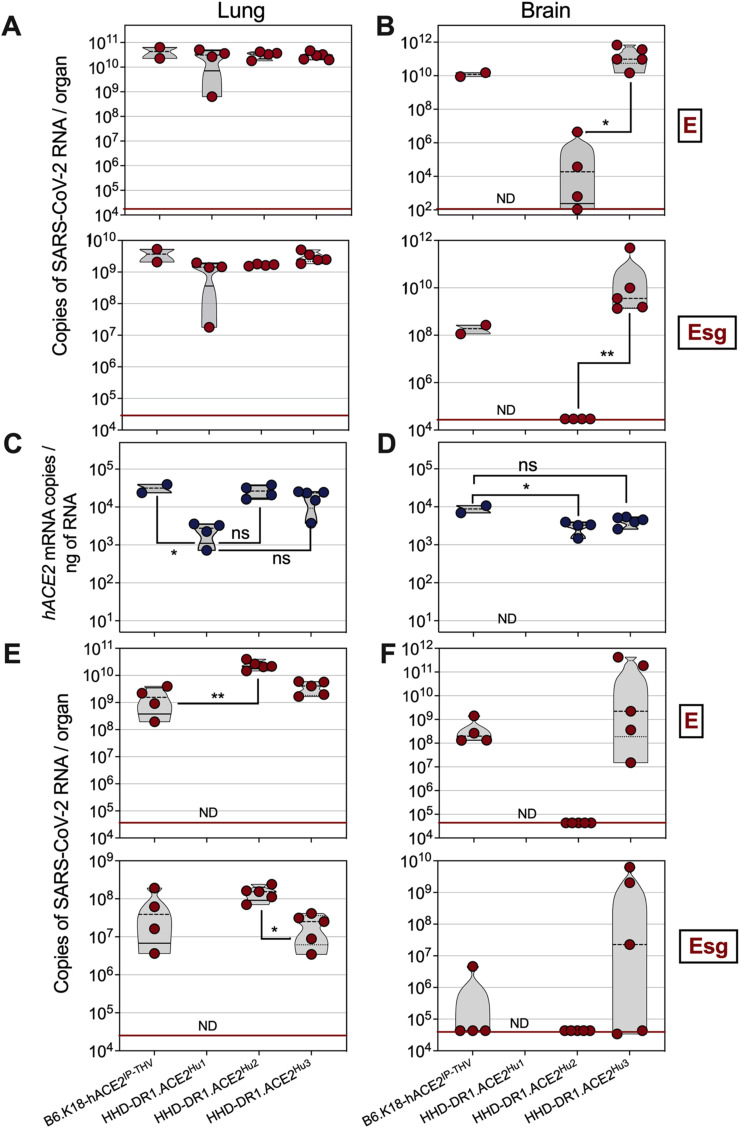
Fig. 3.
Permissiveness of HHD-DR1.ACE2Hu1/2/3 mice to replication of SARS-CoV-2 Omicron BA.1 or BA.5 sub-variants. HHD-DR1.ACE2Hu1/2/3 mice or B6.K18-hACE2IP-THV positive control mice were inoculated i.n. with 0.3 × 105 TCID50/mouse of SARS-CoV-2 Omicron BA.1 or BA.5. A. Lung and B. brain viral RNA contents were determined by E-specific (top) or sub-genomic Esg-specific (bottom) qRT-PCR at 4 dpi with Omicron BA.1. C-D. Comparative quantification of hACE-2 mRNA in the lung and brain of individual B6.K18-hACE2IP-THV or HHD-DR1.ACE2Hu1/2/3 transgenic mice. E-F. Lung and brain viral RNA contents were determined by E-specific (top) or sub-genomic Esg-specific (bottom) qRT-PCR at 4 dpi with Omicron BA.5. Statistical significance was evaluated by one-way Anova (*= p < 0.05, **= p < 0.01). ND = Not determined.
3.3. Permissiveness of HHD-DR1.ACE2 mice to replication of SARS-CoV-2 Omicron sub-variants
The previously available B6.K18-ACE22Prlmn/JAX transgenic mice are much less permissive to Omicron replication than to the other previously emerged SARS-CoV-2 variants [14]. In net contrast, the B6.K18-hACE2IP-THV mice, that we previously generated [3], allow the replication of Omicron as efficiently as other SARS-CoV-2 variants like Delta [4]. Correlatively, B6.K18-hACE2IP-THV mice express significantly higher amounts of hACE2 mRNA in lungs and brain than B6.K18-ACE22Prlmn/JAX [3].
Here, we first evaluated the permissiveness of the three HHD-DR1.ACE2Hu1/2/3 lineages to SARS-CoV-2 Omicron BA.1 sub-variant. As determined at 4 dpi, by SARS-CoV-2 E- or Esg-specific qRT-PCR, the lungs of all three HHD-DR1.ACE2Hu1/2/3 strains were strongly permissive to the replication of Omicron BA.1 (Fig. 3A). The brain viral RNA content of HHD-DR1.ACE2Hu1 was not studied here, as we previously determined the brain unpermissiveness in this strain to SARS-CoV-2 replication (Fig. 1A). In HHD-DR1.ACE2Hu2 mice, the brain E viral RNA content was relatively weak and no brain Esg viral RNA was detected (Fig. 3B). In contrast, substantial Omicron BA.1 replication was detected in the brain of HHD-DR1.ACE2Hu3 mice which, like the previously described B6.K18-hACE2IP-THV mice, resulted from lentiviral vector-based transgenesis (Fig. 3B).
Analysis of hACE2 mRNA expression levels established the following hierarchy: HHD-DR1.ACE2Hu3 = HHD-DR1.ACE2Hu2 > HHD-DR1.ACE2Hu1 in the lungs (Fig. 3C). In the brain, we observed a tendency to a higher hACE2 mRNA expression in HHD-DR1.ACE2Hu3 compared to HHD-DR1.ACE2Hu2. Although this trend did not reach significance, it was associated with a significantly higher permissiveness of HHD-DR1.ACE2Hu3 mice to Omicron BA.1 mice (Fig. 3D).
Like the B6.K18-hACE2IP-THV mice, the HHD-DR1.ACE2Hu2 and HHD-DR1.ACE2Hu3 mice were also strongly permissive to replication of Omicron BA.5 (Fig. 3E). In contrast to the brains of HHD-DR1.ACE2Hu2 mice, which were not permissive to replication of Omicron BA.5, the brain of 3 out of 5 HHD-DR1.ACE2Hu3 mice had high cerebral Omicron BA.5 loads (Fig. 3F).
Therefore, permissiveness to the two Omicron sub-variants tested was observed in both lungs and brains of the MHC-humanized mice, reaching levels as high or even higher than in B6.K18-hACE2IP-THV mice.
3.4. Use of HHD-DR1.ACE2Hu1 to evaluate the efficacy of a lentiviral vector-based COVID-19 vaccine candidate
To conduct a vaccine efficacy evaluation in one of the HHD-DR1.ACE2Hu strains, we first studied, in HHD-DR1 mice, the immunogenicity of the lentiviral vector-based COVID-19 vaccine candidate, namely "LV::SBeta-2P" [3, 4, 15, 16]. LV::SBeta-2P encodes the full length sequence of Spike from the SARS-CoV-2 Beta variant, stabilized by K986P and V987P substitutions in the S2 domain [4].
As we previously established that an intramuscular (i.m.) prime followed by an i.n. boost was the most efficacious scheme to achieve anti-SARS-CoV-2 protection, HHD-DR1 mice were primed (i.m.) with 1 × 108 Transduction Units (TU) of LV::SBeta-2P at wk 0 and then boosted (i.n.) with the same amount of this vaccine at wk 8. Control mice received an empty LV vector (Ctrl LV). The LV::SBeta-2P-immunized mice mounted high titers of serum anti-Spike IgG, albeit weaker than those detected in the conventional C57BL/6 mice (Fig. S2) (see Discussion).
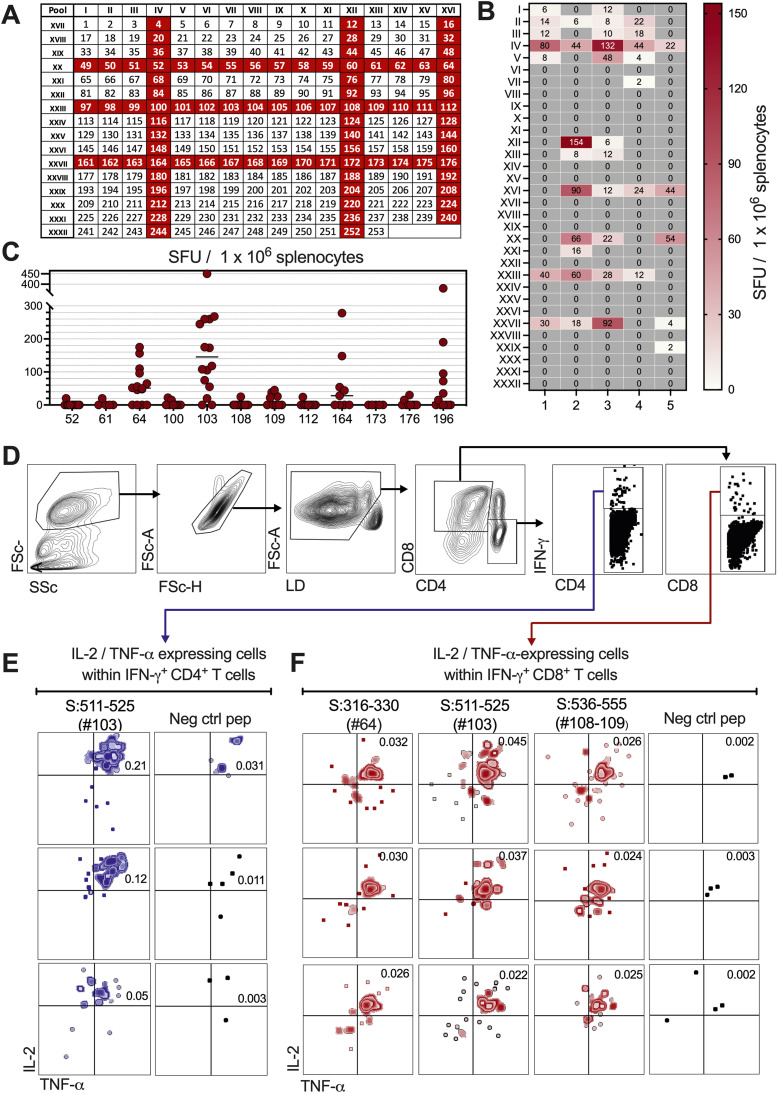
T-cell immunogenicity of LV::SBeta-2P in HHD-DR1 mice was studied by IFN-γ ELISPOT epitope mapping of Spike, based on a 2D-peptide pool matrix [26], using splenocytes (Fig. 4 A, B). To do so, panels of 253 15-mers spanning the full-length Spike protein were organized into 32 pools, assigned to a 2D matrix in which each peptide was represented in two distinct pools. In this approach, the intersection of two immunogenic pools predicts presence of a potentially positive epitope. By analyzing 5 individual LV::SBeta-2P-vaccinated HHD-DR1 mice, we identified the positive IV, XII, XVI, XX, XXIII and XXVII peptide pools (Fig. 4A, B). The intersection peptides, as well as S:511-525 (#103, VVLSFELLHAPATVC) and S:976-990 (#196, VLNDILSRLDKVEAE), previously identified in humans by others [31, 32], were then individually tested in IFN-γ ELISPOT (Fig. 4C). This assay clearly confirmed the immune-recognition of S:316-330 (#64, SNFRVQPTESIVRFP), S:511-525 (#103, VVLSFELLHAPATVC), S:536-555 (#108 and #109, NKCVNFNFNGLTGTGVLTES), S:816-830 (#164, SFIEDLLFNKVTLAD), and S:976-990 (#196, VLNDILSRLDKVEAE) by T cells from HHD-DR1 vaccinated mice. Interestingly, none of these identified T-cell epitopes overlapped with regions where mutations occurred in the Omicron Spike. This suggests that LV::SBeta-2P-induced T cells will recognize Omicron-infected host cells as efficiently as those infected with the ancestral virus.
Fig. 4.
Spike T-cell epitope mapping in HHD-DR1 mice vaccinated with LV::SBeta-2P. HHD-DR1 mice were primed (i.m.) at wk 0 and boosted (i.n.) at wk 8 with a control empty lentiviral vector (Ctrl LV) or LV::SBeta-2P. A. Panels of 253 15-mers spanning the Spike protein, were organized into 32 pools (I to XXXII) containing no more than 16 individual peptides. The peptide pools were assigned to a 2D matrix system in which each peptide was represented in two different pools. The intersection of two positive pools identified potentially positive peptides. B. Splenocytes from HHD-DR1 mice (n = 5) immunized with LV::SBeta-2P or a Ctrl LV were harvested at wk 2 post boost and were analyzed in an IFN-γ ELISPOT after stimulation with each of the 32 distinct I to XXXII pools. Heat map representing the number of IFN-γ-producing cells per million of splenocytes responding to each peptide pool. For each peptide pool, the background generated by the splenocytes from Ctrl LV-injected mice was subtracted. C. Intersection or previously identified peptides were tested individually in ELISPOT. The threshold of positivity was defined by a twofold increase in the number of spots upon stimulation with the peptide, compared to the average number of spots for the same splenocyte sample in the medium alone. D-F. The positive peptides identified were used in CD4 and CD8 surface staining and IFN-γ, IL-2, and TNF-α ICS and cytometric analysis to determine the T-subset specific to the corresponding epitopes. D. Gating strategy, E-F. IL-2/TNF-α expressing cells within the IFN-γ+ CD4+(E) or CD8+(F) T cells after stimulation with the indicated Spike-derived peptides or a negative control peptide. The numbers in the upper right quadrant of the dot plots indicate the percentage of the IL-2+ TNF-α+ IFN-γ+ population within the CD4+(E) or CD8+(F) T cells. See also Fig. S3.
Intracellular cytokine staining (ICS) and cytometric analysis performed with splenocytes from LV::SBeta-2P-vaccinated HHD-DR1 mice, stimulated in vitro with these identified peptides, showed that: (i) S:511-525 (#103, VVLSFELLHAPATVC) was recognized by CD4+ T cells and thus restricted by DRA01.01 + DRB1.01.01 MHC-II molecule (Fig. 4D, E, and Fig. S3). In parallel, S:316-330 (#64, SNFRVQPTESIVRFP), S:511-525 (#103, VVLSFELLHAPATVC) and S:536-555 (#108 and #109, NKCVNFNFNGLTGTGVLTES) were recognized by CD8+ T cells and thus contain epitopes restricted by HLA 02.01 MHC-I molecule (Fig. 4D, F, and Fig. S3). Note that S:511-525 (#103, VVLSFELLHAPATVC) contains both MHC-I- and -II-restricted epitopes. Other peptides detected positive in the ELISPOT (Fig. 4C) were not identifiable by ICS, the most likely because of the weaker sensitivity of ICS compared to ELISPOT. Further analysis of these immunogenic regions by the SYFPEITHI software (http://www.syfpeithi.de/bin/MHCServer.dll/EpitopePrediction.htm) also indicated the presence of high-scoring predicted T-cell epitopes in all these identified regions (Table 2S).
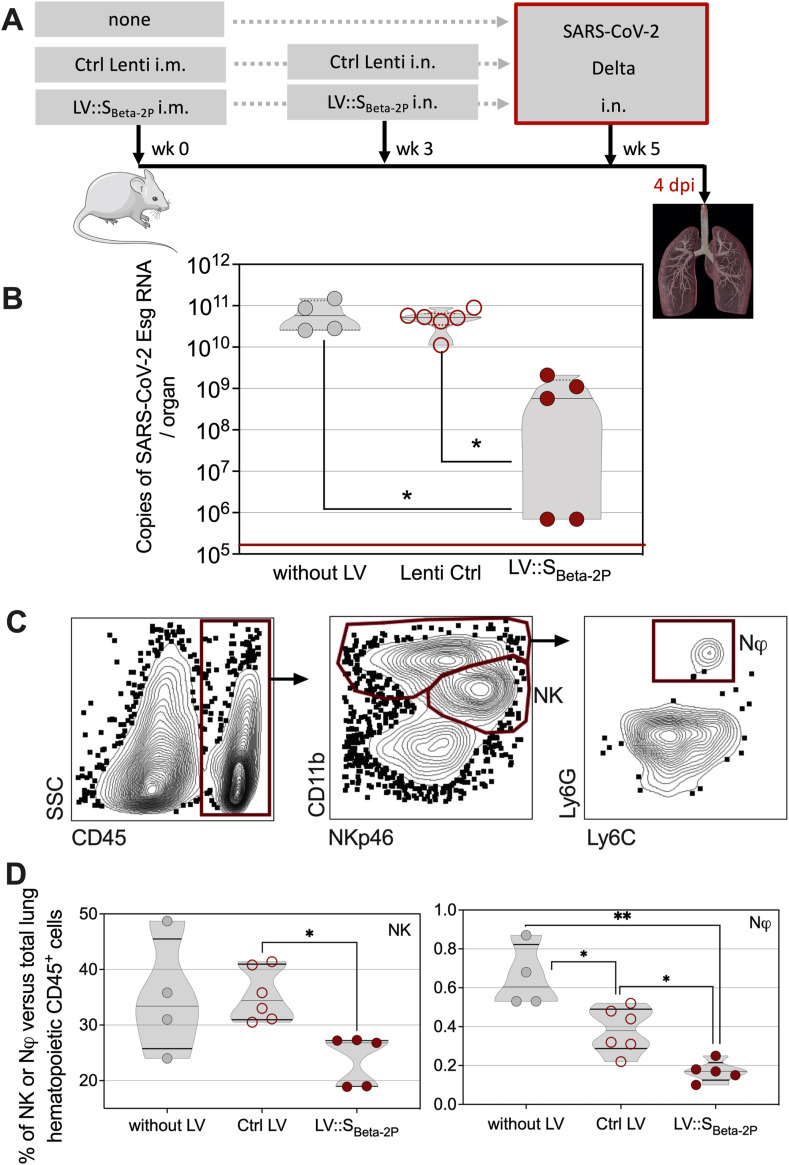
Given the emergency of making research tools available to the community in the context of the pandemic, the transgenic mouse colony that most quickly gave us enough age- and sex-matched animals, i.e., HHD-DR1.ACE2Hu1 mice, was used to perform a protection experiment. HHD-DR1.ACE2Hu1 mice were primed i.m. at wk 0 and boosted i.n. at wk 3 with LV::SBeta-2P, as detailed above [3, 4, 15, 16] or Ctrl LV, and were then challenged i.n. at wk 5 with 0.3 × 105 TCID50 of SARS-CoV-2 Delta variant. LV::SBeta-2P prime-boost vaccination of HHD-DR1.ACE2Hu1 mice led to statistically significant protection against SARS-CoV-2 replication in the lungs, as determined at 4 dpi (Fig. 5 A, B). At this time point, cytometric analysis of the lung innate immune cell subsets detected significantly lower percentages of CD11bint NKp46+ Natural Killer (NK) cells and CD11b+ Ly6G+ neutrophils among the CD45+ cells in the vaccinated HHD-DR1.ACE2Hu1 mice, than in the control animals which received Ctrl LV (Fig. 5C, D). NK and neutrophils have been associated with enhanced lung inflammation and poor COVID-19 outcome and their decrease is a biomarker of good prognosis in humans [33, 34]. The reason why the percentages of lung neutrophils in unprotected Ctrl LV-injected mice were reduced compared with unvaccinated, unprotected controls is not known and requires further investigation.
Fig. 5.
Use of HHD-DR1.ACE2Hu1 mice to evaluate the protective capacity of LV::SBeta-2P vaccine candidate against SARS-CoV-2. A. Timeline of vaccination and SARS-CoV-2 challenge. B. HHD-DR1.ACE2Hu1 mice were untreated or primed (i.m.) at wk 0 and boosted (i.n.) at wk 3 with 1 × 108 TU/mouse of LV::SBeta-2P or Ctrl LV, before i.n. challenge at wk 5 with 0.3 × 105 TCID50 of SARS-CoV-2 Delta variant. Lung viral Esg-specific RNA contents was evaluated by qRT-PCR at 4 dpi. Red lines indicate the detection limits. C. Cytometric detection of neutrophils or NK cells in the lungs of untreated, Ctrl-Lenti-, or LV::SBeta-2P-vaccinated and challenged mice at 4 dpi. D. Percentages of neutrophils or NK cells in the lungs of untreated, Ctrl LV-, or LV::SBeta-2P-vaccinated and challenged mice at 4 dpi. Percentages were calculated versus total lung live CD45+ cells. Statistical significance was evaluated by one-way Anova test (*= p < 0.05, **= p < 0.01).
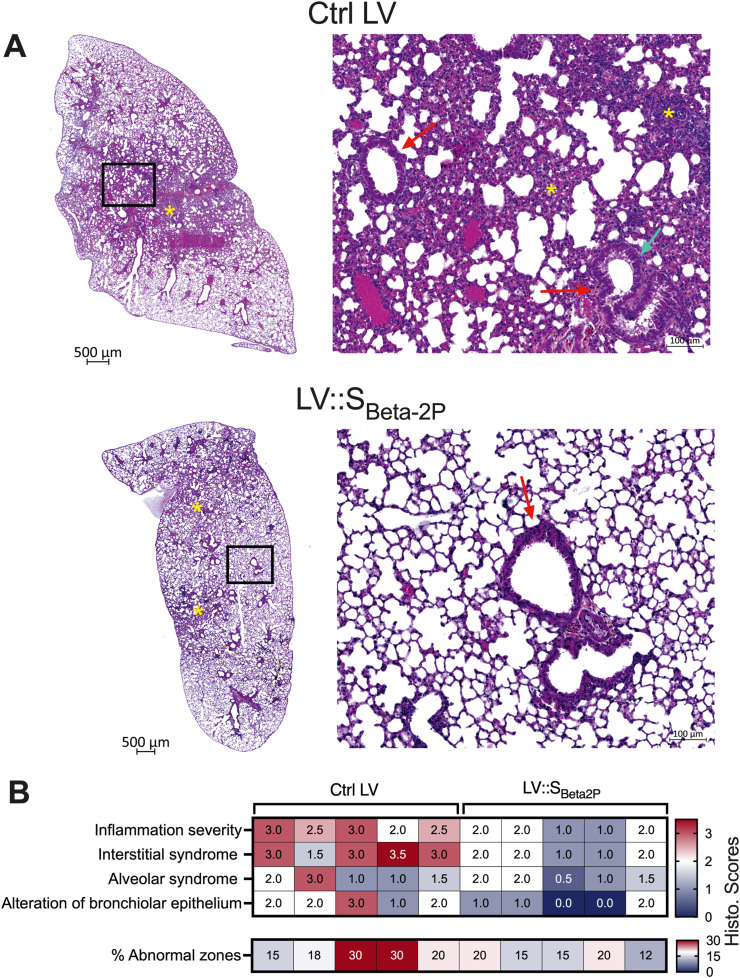
Histological examination of SARS-CoV-2-inoculated mice pre-treated with Ctrl LV revealed a mild to moderate inflammation of the lung, with zones of interstitial infiltration sometimes accompanied by alveolar exudates (Fig. 6 A). Although the bronchiolar epithelium integrity was preserved, degenerative lesions of epithelium cells could be seen in the inflammatory areas. The histological scores are recapitulated in the heatmap (Fig. 6B). In vaccinated HHD-DR1.ACE2Hu1 mice, the alveolo-interstitial syndrome was more limited in size, less severe (minimal to mild), and accompanied by discreet alterations of the bronchiolar epithelium.
Fig. 6.
Lung histopathology of unvaccinated or LV::SBeta-2P-vaccinated HHD-DR1.ACE2Hu1 mice after inoculation with SARS-CoV-2. Mice are those detailed in the Fig. 5. A. Examples of haematoxylin-eosin saffron staining of whole-lung sections (left, scale bar: 500 μm) and close-up views (right, scale bar: 100 μm), at 4 dpi. Stars mark areas of minimal (bottom) or mild-to-moderate (top) infiltration. In the blow-up panels, red arrows indicate bronchiolar epithelia. While in the bottom blow-up the epithelium is subnormal, in the top panel epithelial degenerative lesions can be seen, e.g., perinuclear clear spaces (green arrow), associated with cells remnants and proteinaceous material in the bronchiolar lumen. B. Heatmap representing the histological scores for: (i) inflammation seriousness, (ii) interstitial syndrome, and (iii) alveolar syndrome, (iv) alteration of bronchial epithelium, and (v) percent of abnormal lung zones.
Therefore, virological, immunological and histopathological criteria showed that HHD-DR1.ACE2Hu mice can be used to evaluate protective potential of COVID-19 vaccine candidates.
4. Discussion
We generated three new transgenic mice expressing: (i) hACE2 under the human K18 promoter, and (ii) only human – but not murine – MHC-I and -II molecules from HLA 02.01, DRA01.01, DRB1.01.01 alleles, widely expressed in human populations. The three strains, namely “HHD-DR1.ACE2Hu1/2/3”, are permissive to SARS-CoV-2 replication, yet each displays distinctive characteristics in terms of lung- and/or brain-specific permissiveness to viral replication, and their site-specific hACE2 mRNA transcription levels. Similar to B6.K18-hACE2IP-THV strain [3], one particularity of the HHD-DR1.ACE2Hu1/2/3 strains is their marked permissiveness to the replication of Omicron sub-variant(s). The high hACE2 mRNA transcription notably in the HHD-DR1.ACE2Hu3 generated by lentiviral based transgenesis, is correlated with their permissiveness to SARS-CoV-2 Omicron replication. The reasons why the two independent lentiviral-based transgeneses, which gave rise to B6.K18-hACE2IP-THV and HHD-DR1.ACE2Hu3 strains, led to high lung and brain transgene expression are not clear. It is plausible that the lentiviral insertion sites in the host chromosome(s) lead to more accessible and more active transcription of the transgene. In addition, the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE), present in the genetic material delivered by the lentiviral vector, enhances the transgene expression and transgenic mRNA export from the host nuclei [35], which is not the case for the transgene inserted by the conventional, DNA fragment-based transgenesis.
Quantitative comparison of hACE2 transcript levels in hACE2 transgenic mice with physiological levels of this transcript in humans is very important but also very challenging. The tissue expression pattern and level of ACE2 mRNA has been established and compared in 31 tissues [36, 37]. Small intestine, testis, kidney, heart, thyroid, and adipose tissue have the highest ACE2 expression levels, lung, colon, liver, bladder, and adrenal gland have medium ACE2 expression levels, while blood, spleen, bone marrow, brain, blood vessels, and muscle display the lowest ACE2 expression levels [37]. These amounts were determined as transcripts per kilobase of exon pattern per million mapped reads, by the RNA-Seq approach [36], and therefore very difficult to compare to the hACE2 mRNA levels we determined in transgenic mice by qRT-PCR.
Among all SARS-CoV-2 variants of concern, Omicron harbors the highest number, i.e., 37 mutations in its Spike glycoprotein [[38], [39], [40]]. Among these mutations, 15 are located inside the Receptor-Binding Domain (RBD). Inside the Omicron RBD, the 96 amino acid-long Receptor-Binding Motif (RBM) carries 10 mutations, i.e., N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y and Y505H, among which, G446S, E484A, Q493R, G496S, Q498R and Y505H are unique to Omicron and are believed to reinforce by threefold the binding of its Spike to hACE2, compared to the Spike of ancestral or Delta SARS-CoV-2. This strengthened interaction occurs via increased electrostatic and hydrophobic interactions, and formation of ACE2 salt bridge and ACE2 hydrogen bond, improving the potential of Omicron to invade the host cells [[38], [39], [40]]. On the other hand, the Omicron Spike protein is less efficiently cleaved into S1 and S2 subunits, reducing its fusogenic potential and thereby its pathogenicity [41, 42]. Other mutations elsewhere in the genome of Omicron may also be responsible its reduced pathogenic potential. Despite the increased affinity of Omicron RBM interaction with hACE2, replication of Omicron variants in the upper and lower respiratory airways of B6.K18-ACE22Prlmn/JAX transgenic mice is 3-to-5 log10 lower compared to the earlier SARS-CoV-2 variants [14]. The notable permissiveness of HHD-DR1.ACE2Hu1/2/3 strains to Omicron, is probably largely explained by their high expression of hACE2 transgene transcription levels, because SARS-CoV-2 enters the host cells mainly via hACE2, although alternative routes of entry such as the transmembrane protease serine 2 (TMPRSS2) or the cathepsin-dependent endocytic pathway have been reported [43]. However, these are unlikely to be different in the B6.K18-ACE22Prlmn/JAX, B6.K18-ACE2IP-THV and HHD-DR1.ACE2Hu1/2/3 mouse strains, which harbor a common C57BL/6 genetic background.
As a first application of these strains, we demonstrated in HHD-DR1.ACE2Hu1 mice the efficacy of a lentiviral vector-based COVID-19 vaccine candidate. This vaccine induced anti-Spike serum IgG, CD4+ and CD8+ T cells [3, 4, 15, 16]. In the LV::SBeta-2P-immunized HHD-DR1 mice, the titers of anti-Spike IgG were lower than in their LV::SBeta-2P-immunized wild type C57BL/6 counterparts. This may be linked to the absence of free murine β2m in HHD-DR1 mice, which harbor only the human β2m covalently linked to the HLA 02.01. β2m contributes to the formation of “neonatal Fc receptor” (FcRn) for IgG [44]. In the absence of association with β2m, the FcRn heavy chain remains sequestered and unfunctional in endoplasmic reticulum. Increased clearance of IgG in β2m KO mice has suggested that FcRn protects IgG from degradation, and is thus critical in maintaining IgG levels in the circulation [45]. In a non-mutually-exclusive manner, it is also possible that the size of the T-cell compartment in HHD-DR1 mice is reduced compared to wild-type mice. In this case, the CD4+ T-cell helper function may be attenuated, resulting in weaker antibody responses. Spike epitope mapping in LV::SBeta-2P-immunized HHD-DR1 mice allowed for the identification of both MHC-I- and II-restricted immunogenic Spike regions for humans. Furthermore, LV::SBeta-2P-vaccinated HHD-DR1Hu1 mice were protected from a challenge by SARS-CoV-2, as evidenced by virological, immunological and histopathological criteria, validating the HHD-DR1Hu preclinical model for immunogenicity and vaccine evaluation investigations.
HHD-DR1.ACE2Hu1/2/3 murine strains provide new pre-clinical research tools for SARS-CoV-2 infection and research and development of new COVID-19 drugs or vaccines. These strains will notably pave the way for: (i) identification of SARS-CoV-2-derived epitopes recognized by human T cells, (ii) investigation of T-cell immunogenicity of new-generation COVID-19 vaccines, and (iii) evaluation of protective potential of new-generation COVID-19 vaccine, in small rodent models, in which the MHC-I and -II molecules are from widely expressed HLA alleles in human populations. In addition, since coronaviruses can infect the brain [[46], [47], [48]], the HHD-DR1.ACE2Hu2/3 strains provide also valuable pre-clinical models for investigation of immune protection of the central nervous system against SARS-CoV-2.
Funding sources
This work was supported by Institut Pasteur and TheraVectys.
Declaration of competing interest
PC is the founder and CSO of TheraVectys. FLC, PA, BV, IF, FM, AN, KN and FA are employees of TheraVectys. LM has a consultancy activity for TheraVectys. Other authors declare no competing interests.
Funding
MB, PC, Study concept and design: MB, PC, FL, FLV, LM, acquisition of data: FLC, PA, MB, BV, IF, KN, LM, lung histopathology: FG, DH, transgenesis experiments and breeding: SC, DC, YS, FLV, construction and production of lentiviral vector for transgenesis: FM, AN, CB, FA, analysis and interpretation of data: PC, FL, FLV, LM, drafting of the manuscript: LM.
Declaration of Competing Interest
PC is the founder and CSO of TheraVectys. FLC, PA, BV, IF, FM, AN, KN and FA are employees of TheraVectys. LM has a consultancy activity for TheraVectys. Other authors declare no competing interests.
Acknowledgments
The authors are grateful to Magali Tichit and Johan Bedel for excellent technical assistance in preparing histological sections. The SARS-CoV2 variant Delta/2021/I7.2 200 was supplied by the Virus and Immunity Unit (Institut Pasteur, Paris, France) headed by Dr. Olivier Schwartz. The SARS-CoV-2 Omicron BA.1 variant was initially supplied by the Virus and Immunity Unit (Institut Pasteur, Paris, France) headed by Dr. Olivier Schwartz, and was provided to our lab by Matthieu Prot and Dr. Etienne Simon-Lorière (G5 Evolutionary Genomics of RNA Viruses, Institut Pasteur, Paris, France). The strain hCoV-19/France/BRE-IPP34319/2022 (Omicron BA.5) was supplied by the National Reference Centre for Respiratory Viruses hosted by Institut Pasteur (Paris, France) and headed by Pr. Sylvie van der Werf. The human sample from which strain hCoV-19/France/BRE-IPP34319/2022 was isolated have been provided by Dr. Marque-Juillet from Laboratoire Alliance Anabio, in Melesse.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micinf.2023.105142.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ku M.W., Authie P., Bourgine M., Anna F., Noirat A., Moncoq F., et al. Brain cross-protection against SARS-CoV-2 variants by a lentiviral vaccine in new transgenic mice. EMBO Mol Med. 2021;13 doi: 10.15252/emmm.202114459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vesin B., Lopez J., Noirat A., Authie P., Fert I., Le Chevalier F., et al. An intranasal lentiviral booster reinforces the waning mRNA vaccine-induced SARS-CoV-2 immunity that it targets to lung mucosa. Mol Ther. 2022;30:2984–2997. doi: 10.1016/j.ymthe.2022.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madsen L., Labrecque N., Engberg J., Dierich A., Svejgaard A., Benoist C., et al. Mice lacking all conventional MHC class II genes. Proc Natl Acad Sci U S A. 1999;96:10338–10343. doi: 10.1073/pnas.96.18.10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pascolo S., Bervas N., Ure J.M., Smith A.G., Lemonnier F.A., Perarnau B. HLA-A2.1-restricted education and cytolytic activity of CD8(+) T lymphocytes from beta2 microglobulin (beta2m) HLA-A2.1 monochain transgenic H-2Db beta2m double knockout mice. J Exp Med. 1997;185:2043–2051. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altmann D.M., Douek D.C., Frater A.J., Hetherington C.M., Inoko H., Elliott J.I. The T cell response of HLA-DR transgenic mice to human myelin basic protein and other antigens in the presence and absence of human CD4. J Exp Med. 1995;181:867–875. doi: 10.1084/jem.181.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pajot A., Michel M.L., Fazilleau N., Pancre V., Auriault C., Ojcius D.M., et al. A mouse model of human adaptive immune functions: HLA-A2.1-/HLA-DR1-transgenic H-2 class I-/class II-knockout mice. Eur J Immunol. 2004;34:3060–3069. doi: 10.1002/eji.200425463. [DOI] [PubMed] [Google Scholar]
- 9.Allen H., Fraser J., Flyer D., Calvin S., Flavell R. Beta 2-microglobulin is not required for cell surface expression of the murine class I histocompatibility antigen H-2Db or of a truncated H-2Db. Proc Natl Acad Sci U S A. 1986;83:7447–7451. doi: 10.1073/pnas.83.19.7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potter T.A., Boyer C., Verhulst A.M., Golstein P., Rajan T.V. Expression of H-2Db on the cell surface in the absence of detectable beta 2 microglobulin. J Exp Med. 1984;160:317–322. doi: 10.1084/jem.160.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potter T.A., Zeff R.A., Schmitt-Verhulst A.M., Rajan T.V. Molecular analysis of an EL4 cell line that expresses H-2Db but not H-2Kb or beta 2-microglobulin. Proc Natl Acad Sci U S A. 1985;82:2950–2954. doi: 10.1073/pnas.82.9.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bix M., Raulet D. Functionally conformed free class I heavy chains exist on the surface of beta 2 microglobulin negative cells. J Exp Med. 1992;176:829–834. doi: 10.1084/jem.176.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrance S.K., Karlsson L., Price J., Quaranta V., Ron Y., Sprent J., et al. Transgenic HLA-DR alpha faithfully reconstitutes IE-controlled immune functions and induces cross-tolerance to E alpha in E alpha 0 mutant mice. Cell. 1989;58:583–594. doi: 10.1016/0092-8674(89)90439-x. [DOI] [PubMed] [Google Scholar]
- 14.Halfmann P.J., Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ku M.W., Bourgine M., Authie P., Lopez J., Nemirov K., Moncoq F., et al. Intranasal vaccination with a lentiviral vector protects against SARS-CoV-2 in preclinical animal models. Cell Host Microbe. 2021;29:236–249 e6. doi: 10.1016/j.chom.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majlessi L., Charneau P. [An anti-Covid-19 lentiviral vaccine candidate that can be administered by the nasal route] Med Sci (Paris) 2021;37:1172–1175. doi: 10.1051/medsci/2021173. [DOI] [PubMed] [Google Scholar]
- 17.Gordon J.W., Ruddle F.H. Integration and stable germ line transmission of genes injected into mouse pronuclei. Science. 1981;214:1244–1246. doi: 10.1126/science.6272397. [DOI] [PubMed] [Google Scholar]
- 18.Chow Y.H., O'Brodovich H., Plumb J., Wen Y., Sohn K.J., Lu Z., et al. Development of an epithelium-specific expression cassette with human DNA regulatory elements for transgene expression in lung airways. Proc Natl Acad Sci U S A. 1997;94:14695–14700. doi: 10.1073/pnas.94.26.14695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koehler D.R., Chow Y.H., Plumb J., Wen Y., Rafii B., Belcastro R., et al. A human epithelium-specific vector optimized in rat pneumocytes for lung gene therapy. Pediatr Res. 2000;48:184–190. doi: 10.1203/00006450-200008000-00011. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa T., Hoogenraad C.C. Lentiviral transgenesis. Methods Mol Biol. 2011;693:117–142. doi: 10.1007/978-1-60761-974-1_8. [DOI] [PubMed] [Google Scholar]
- 21.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 22.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 23.Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. May 2020 doi: 10.1126/science.abc4776. PMID: 32434946. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tostanoski L.H., Wegmann F., Martinot A.J., Loos C., McMahan K., Mercado N.B., et al. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat Med. 2020;26:1694–1700. doi: 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmeister B., Kiecker F., Tesfa L., Volk H.D., Picker L.J., Kern F. Mapping T cell epitopes by flow cytometry. Methods. 2003;29:270–281. doi: 10.1016/s1046-2023(02)00349-3. [DOI] [PubMed] [Google Scholar]
- 27.Haruyama N., Cho A., Kulkarni A.B. Overview: engineering transgenic constructs and mice. Curr Protoc Cell Biol. 2009 doi: 10.1002/0471143030.cb1910s42. Chapter 19:Unit 19 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zennou V., Petit C., Guetard D., Nerhbass U., Montagnier L., Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 29.Dussaud S., Pardanaud-Glavieux C., Sauty-Colace C., Ravassard P. Lentiviral Mediated Production of Transgenic Mice: A Simple and Highly Efficient Method for Direct Study of Founders. J Vis Exp. 2018 doi: 10.3791/57609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habel J.R., Nguyen T.H.O., van de Sandt C.E., Juno J.A., Chaurasia P., Wragg K., et al. Suboptimal SARS-CoV-2-specific CD8(+) T cell response associated with the prominent HLA-A*02:01 phenotype. Proc Natl Acad Sci U S A. 2020;117:24384–24391. doi: 10.1073/pnas.2015486117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavalcante-Silva L.H.A., Carvalho D.C.M., Lima E.A., Galvao J., da Silva J.S.F., Sales-Neto J.M., et al. Neutrophils and COVID-19: The road so far. Int Immunopharmacol. 2021;90 doi: 10.1016/j.intimp.2020.107233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masselli E., Vaccarezza M., Carubbi C., Pozzi G., Presta V., Mirandola P., et al. NK cells: A double edge sword against SARS-CoV-2. Adv Biol Regul. 2020;77 doi: 10.1016/j.jbior.2020.100737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zufferey R., Donello J.E., Trono D., Hope T.J. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.https://www.proteinatlas.org/ENSG00000130234-ACE2.
- 37.Li M.Y., Li L., Zhang Y., Wang X.S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty. 2020;9:45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar R., Murugan N.A., Srivastava V. Improved Binding Affinity of Omicron's Spike Protein for the Human Angiotensin-Converting Enzyme 2 Receptor Is the Key behind Its Increased Virulence. Int J Mol Sci. 2022;23 doi: 10.3390/ijms23063409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki R., Yamasoba D., Kimura I., Wang L., Kishimoto M., Ito J., et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 2022;603:700–705. doi: 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Z., Zhou J., Tian M., Huang M., Liu S., Xie Y., et al. Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape. Nat Commun. 2022;13:4958. doi: 10.1038/s41467-022-32665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito A., Irie T., Suzuki R., Maemura T., Nasser H., Uriu K., et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2022;602:300–306. doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balint G., Voros-Horvath B., Szechenyi A. Omicron: increased transmissibility and decreased pathogenicity. Signal Transduct Target Ther. 2022;7:151. doi: 10.1038/s41392-022-01009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu X., Peng J., Raychowdhury R., Nakajima A., Lencer W.I., Blumberg R.S. The heavy chain of neonatal Fc receptor for IgG is sequestered in endoplasmic reticulum by forming oligomers in the absence of beta2-microglobulin association. Biochem J. 2002;367:703–714. doi: 10.1042/BJ20020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Israel E.J., Wilsker D.F., Hayes K.C., Schoenfeld D., Simister N.E. Increased clearance of IgG in mice that lack beta 2-microglobulin: possible protective role of FcRn. Immunology. 1996;89:573–578. doi: 10.1046/j.1365-2567.1996.d01-775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aghagoli G., Gallo Marin B., Katchur N.J., Chaves-Sell F., Asaad W.F., Murphy S.A. Neurological Involvement in COVID-19 and Potential Mechanisms: A Review. Neurocrit Care. 2020 doi: 10.1007/s12028-020-01049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ali Awan H., Najmuddin Diwan M., Aamir A., Ali M., Di Giannantonio M., Ullah I., et al. SARS-CoV-2 and the Brain: What Do We Know about the Causality of 'Cognitive COVID? J Clin Med. 2021:10. doi: 10.3390/jcm10153441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bourgonje A.R., Abdulle A.E., Timens W., Hillebrands J.L., Navis G.J., Gordijn S.J., et al. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19) J Pathol. 2020;251:228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated and analyzed during this study. All plasmids, lentiviral vectors and mouse strains generated in this study will be available under material transfer agreement for research use. Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author Laleh Majlessi (laleh.majlessi@pasteur.fr).