Abstract
The presence of antibiotic resistance has increased the urgency for more effective treatments of bacterial infections. Biofilm formation has complicated this issue as biofilm bacteria become tolerant to antibiotics due to environmental factors such as nutrient deprivation and adhesion. In septic arthritis, a disease with an 11% mortality rate, bacteria in synovial fluid organize into floating, protein-rich, bacterial aggregates (mm-cm) that display depressed metabolism and antibiotic tolerance. In this study, Staphylococcus aureus (S. aureus), which is the most common pathogen in septic arthritis, was tested against different inhibitors that modulate bacterial surface protein availability and that should decrease bacterial aggregation. One of these, berberine, a quaternary ammonium compound, was found to reduce bacterial counts by 3–7 logs in human synovial fluid (aggregating medium) with no effect in tryptic soy broth (TSB, non-aggregating). Unlike traditional antibiotics, the bactericidal activity of berberine appeared to be independent of bacterial metabolism. To elucidate the mechanism, we used synovial fluid fractionation, targeted MRSA transposon insertion mutants, dyes to assess changes in membrane potential (DiSC3(5)) and membrane permeability (propidium iodide (PI)), colony counting, and fluorescence spectroscopy. We showed that berberine's activity was dependent on an alkaline pH and berberine killed both methicillin-sensitive S. aureus and MRSA in alkaline media (pH 8.5–9.0; p < 0.0001 vs. same pH controls). Under these alkaline conditions, berberine localized to S. aureus where berberine was isolated in cytoplasmic (∼95%) and DNA (∼5%) fractions. Importantly, berberine increased bacterial cell membrane permeability, and disrupted the proton motive force, suggesting a mechanism whereby it may be able to synergize with other antibacterial compounds under less harsh conditions. We suggest that berberine, which is cheap and readily available, can be made into an effective treatment.
Keywords: Staphylococcus aureus, Berberine, Septic arthritis, Synovial fluid
1. Introduction
The antibiotic age was ushered in by Alexander Fleming's 1928 discovery of penicillin, causing an almost 75% decrease in infectious disease mortality rates in the United States from 1937 to 1952 [1,2]. However, the utility of these antimicrobials was hampered by bacterial antibiotic resistance [3] and the tolerance of bacteria and their biofilms to antibiotic treatments [4], effectively narrowing the antibiotic's therapeutic window. This antibiotic tolerance arose from several causes, including decreased bacterial metabolism, encasement in a biofilm slime, and ultimately evasion of immune surveillance, the critical partner of any clinical antimicrobial therapy. In the joint, bacteria cluster to form micro- and macroscopic proteinaceous bacterial aggregates in synovial fluid, the viscous fluid that lubricates joints; these aggregates have characteristics of floating biofilms [5,6]. We have shown that Staphylococcus aureus (S. aureus) forms antibiotic tolerant aggregates in both human and equine synovial fluid, even in the presence of prophylactic antibiotics [7]. The antibiotic tolerance of these aggregates is thought to play a major role in septic arthritis, a disease with an 11% mortality rate and which is caused by S. aureus in 40–50% of cases [8]. New, effective strategies to overcome the antibiotic tolerance of bacterial aggregates are therefore crucial.
Bacterial aggregation is dependent on interaction with serum proteins, where fibrin[ogen] and albumin play important roles [9]. These interactions are mediated by Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMM) [10], and we previously showed that the MSCRAMM clumping factor A (ClfA) has a major role in aggregation [5]. We reasoned that antibiotic sensitivity could be restored if we could prevent bacterial surface display of ClfA and other MSCRAMMs, a process mediated by the transpeptidase Sortase A (SrtA) [11].
Among other putative SrtA inhibitors, we examined berberine [12], a quaternary ammonium compound extracted from plants in the Berberidaceae family [13]. Berberine, an over-the-counter supplement, has been used for diarrheal diseases [14], inflammation and autoimmune diseases [15,16], and metabolic syndrome [17,18]. Berberine's antimicrobial activity has been associated with modulation of SrtA [12] and its binding to phenol soluble modulins (PSMs) [19]; both would decrease virulence. Importantly, berberine's mammalian cell toxicity was reported to be minimal [20].
In this manuscript, we hypothesized that preventing display of MSCRAMMs responsible for aggregation would result in increased S. aureus antibiotic sensitivity and perhaps even be antimicrobial. We first examined srtA mutants and inhibitors of SrtA. Upon finding that the putative SrtA inhibitor berberine was antimicrobial, we asked whether a synovial fluid component responsible for aggregation was necessary for the antibacterial effect of berberine. Furthermore, using the innate fluorescence of berberine, we asked if localization to and into the target bacteria was required. Finally, using propidium iodide, which only fluoresces when localized to DNA, we asked if berberine caused increased membrane permeability and if this was accompanied by an alteration in proton motive force as detected by localization of DiSC3(5), an indicator of membrane potential [21].
2. Methods
Study approval. Collection of equine synovial fluid was approved by the IACUC of the University of Pennsylvania, in accordance with ARRIVE guidelines. Human synovial fluid and other surgical waste were obtained from therapeutically necessary joint aspirations or operations. This material, designated as “waste” (no identifiers), was retrieved, and designated as “not human research” by the Thomas Jefferson University Office of Human Research Protections, in keeping with the revised Federal Policy for the Protection of Human Subjects (revised Common Rule, 2018).
Bacteria and reagents.S. aureus ATCC®25923™, and community-acquired methicillin-resistant S. aureus (CA-MRSA) LAC/USA300 (gift from Dr. Michael Otto, NIH [5]) were cultured in tryptic soy broth (TSB; Becton-Dickinson, Sparks, MD), 37 °C, 180 rpm. USA300 transposon insertion mutants for sortase A (srtA) (NR48329), and norA (NR47577) (Nebraska Transposon Bank) [22] were cultured in TSB containing 5 μg/ml erythromycin (Fisher, Waltham, MA). Overnight cultures were sub-cultured for 2 h and diluted in phosphate buffered saline, pH 7.4 (PBS; MP Biomedicals, Irvine, CA) by comparison with a 0.5 McFarland standard. Experiments were performed in 24-well TrueLine cell culture plates (MedSupply Partners, Atlanta, GA) unless otherwise stated.
Measurement of S. aureus aggregation. One hundred μL of 1 × 108 colony forming units/mL (CFU/mL) of (1) S. aureus 25923, (2) USA300, (3) USA300 srtA mutant were incubated with 900 μL of TSB, a synthetic “pseudo-synovial fluid” (pSynF) [9], or human synovial fluid for 90 min, 37 °C, 180 rpm. Potential aggregation inhibitors: 100 μM putrescine dihydrochloride (1 M stock in dH2O) (MP Biomedicals, Irvine, CA), 50 mM calcium chloride (0.5 M stock in dH2O) (CaCl2; Sigma, St. Louis, MO). Compounds were added concomitantly with bacteria using the above conditions. Cultures were serially diluted and plated on 3M™ Petrifilm™ aerobic films (3 M, Saint Paul, MN).
Trypsin dispersal of bacteriaS. aureus 25923 (50 μL of 1 × 108 CFU/mL) were pelleted for 4 min, 10,000×g and dispersed by incubating with 0.25% trypsin, 2.21 mM ethylene diamine tetraacetic acid (EDTA, Corning, Corning, NY) for 20 min, 37 °C, 180 rpm. Samples were pelleted again for 4 min, 10,000×g, resuspended in PBS, serially diluted, and plated.
Berberine in synovial fluid.S. aureus 25923, USA300, USA300 srtA mutant, or USA300 norA mutant (all 50 μL of 1 × 108 CFU/mL) were incubated in 450 μL of TSB, human, or equine synovial fluid in the presence of 0, 10, 20, or 30 μg/mL berberine chloride (15 mg/mL stock in dimethylsulfoxide (DMSO) (Fisher, Waltham, MA)). Harvested samples were pelleted for 4 min, 10,000×g, dispersed with trypsin, serially diluted, and plated.
Fractionating synovial fluid. Human synovial fluid (500 μL) was filtered through a 10 kDa cutoff, 0.5 mL Pierce™ PES Protein Concentrator (Sigma, St. Louis, MO) for 60 min in 10 min intervals at 10,000×g and separated into filtrate (<10 kDa) and retentate (>10 kDa) fractions (reconstituted in 500 μL TSB or Dulbecco's Modified Eagle Medium (DMEM; Corning, Corning, NY)). S. aureus 25923 (50 μL of 1 × 108 CFU/mL) were incubated in 450 μL filtrate or retentate fractions with 0 or 30 μg/mL berberine for 3 h, 37 °C, 180 rpm. Samples were pelleted for 4 min, 10,000×g, dispersed with trypsin, serially diluted, and plated.
DMEM conditions.S. aureus 25923 (50 μL of 1 × 108 CFU/mL) were incubated in 450 μL DMEM, custom DMEM formulations (US Biological, Salem, MA), PBS pH 7.4 with 0 or 30 μg/mL berberine or 0 or 3.7 mg/mL sodium bicarbonate (Fisher, Waltham, MA) for 3 h, 37 °C, 180 rpm. Samples were pelleted for 4 min, 10,000×g, dispersed with trypsin, serially diluted, and plated.
pH dependence.S. aureus 25923 (50 μL of 1 × 108 CFU/mL) were incubated in 450 μL PBS (pH adjusted to 6.0–9.0 using 1 M HCl or NaOH) or equine synovial fluid in the presence of 0, 10, 20, or 30 μg/mL berberine or 0 or 30 μg/mL berberine ± 10X phosphate buffer (PB) (100X stock in dH2O) for 3 h, 37 °C, 180 rpm. PB stock (100X): 14.4 g Na2HPO4 (Alfa Aesar, Haverhill, MA), 2.4 g KH2PO4 (Fisher, Waltham, MA), 100 mL dH2O, pH adjusted to 7.0 with 1 M HCl or NaOH). Samples were pelleted for 4 min, 10,000×g, dispersed with trypsin, serially diluted, and plated.
Berberine localization with membrane potential.DiSC3(5) (3,3-Dipropylthiadicarbocyanine iodide) fluorescence. Buffer solutions (pH 6.0, 7.0, 8.0, 8.5, 9.0) composed of 0.15 M citric acid monohydrate (Fisher, Waltham, MA) and 0.15 M disodium phosphate (Alfa Aesar, Haverhill, MA) (CAP) were prepared. S. aureus 25923 (50 μL of 1 × 108 CFU/mL) were incubated in CAP buffers pH 6.0, 7.0, 8.0, 9.0 in a ThermoScientific™ 96-well black/optical bottom plate (Fisher, Waltham, MA) with 1 μM DiSC3(5) (100 μM stock in DMSO) (TCI, Portland, OR) or PBS and incubated in the dark for 1 h, 37 °C, 180 rpm. A parallel setup was prepared with 20 μl PBS in place of S. aureus to serve as a control. Fluorescence (λex/em, 620/685 nm) was measured using an Infinite® M1000 plate reader (Tecan.com), excitation/emission bandwidth = 5 nm. Percentage fluorescence for each pH level was calculated as follows: (fluorescence of bacteria sample/fluorescence of no bacteria sample) x 100. Berberine fluorescence. S. aureus 25923 (50 μL of 1 × 109 CFU/mL) were incubated in PBS pH 7.0 or pH 9.0 (adjusted using 1 M HCl or NaOH) with 30 μg/mL berberine for 1 h, 37 °C, 180 rpm. A parallel setup was prepared without berberine to serve as a control. Samples were pelleted for 4 min, 10,000×g, washed once with PBS, and resuspended in 500 μL PBS. One hundred μL of each sample was transferred to a 96-well black/optical bottom plate and fluorescence (λex/em, 355/517 nm) and optical density (λ = 600 nm) was measured using an Infinite® M1000 plate reader, excitation/emission bandwidth = 5 nm, bandwidth = 5 nm. Fluorescence was divided by the optical density to normalize the data.
NorA.Berberine fluorescence. Fifty μL of 1 × 108 CFU/mL USA300 or USA 300 norA mutant were incubated in 450 μL PBS with 30 μg/mL berberine for 1 h, 37 °C, 180 rpm. Samples were pelleted for 4 min, 10,000×g, washed once with PBS, and resuspended in 500 μL PBS. One hundred μL of each sample was transferred to a 96-well black/optical bottom plate and fluorescence (λex/em, 355/517 nm) and optical density (λ = 600 nm) was measured using an Infinite® M1000 plate reader, excitation/emission bandwidth = 5 nm. Fluorescence was divided by the optical density to normalize the data. NorA inhibition. S. aureus 25923 (50 μL of 1 × 108 CFU/mL) were incubated in 450 μL PBS with 30 μg/mL berberine and/or 20 μg/mL reserpine (10 mg/mL stock in DMSO, for this and all reserpine experiments) (Alfa Aesar, Haverhill, MA) for 3 h, 37 °C, 180 rpm. Samples were pelleted for 4 min, 10,000×g, dispersed with trypsin, serially diluted, and plated.
Berberine DNA binding. Five mL of an overnight S. aureus 25923 culture was pelleted for 10 min, 4700×g, and resuspended in 1 mL DMEM (Fisher, Waltham, MA) with carbonate-bicarbonate buffer added to maintain a pH of 9.0–9.5 (1 capsule/100 mL) (Sigma, St. Louis, MO). Two μL of the berberine stock solution, 3 μL of SYTO 9 LIVE/DEAD™ BacLight™ Bacterial Viability Kit (Invitrogen, Waltham, MA), or 100 μL of Alamar Blue™ Cell Viability Reagent (Invitrogen, Waltham, MA) was added to each sample. A parallel setup was prepared without treatment. All samples were incubated for 1 h, 37 °C, 180 rpm. Samples were washed 2x with PBS and resuspended in 1 mL PBS. One hundred μL was transferred to a 96-well black plate. Fluorescence was measured (berberine: λex/em 355/517 nm; SYTO 9: λex/em 483/503 nm; Alamar blue: λex/em 540/590 nm) using a Spark® plate reader (Tecan.com), excitation/emission bandwidth = 5 nm. Readings were multiplied by 10 to account for the entire resuspended volume of 1 mL. DNA was extracted by cell lysis as follows: Samples were pelleted for 4 min, 10,000×g, and resuspended in 350 μl PBS. Cells were lysed using zirconia beads from the Ribopure™ RNA Purification Kit (Invitrogen, Carlsbad, CA), following manufacturer's instructions. The lysate was separated from the zirconia beads by centrifugation for 5 min, 14,000×g, 4 °C and brought to ∼600 μl using PBS. Proteins were precipitated with 200 μL of Protein Precipitation Solution from the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI), and the supernatant was isolated using manufacturer's instructions. DNA was then precipitated, washed, and resuspended using 200 μL (instead of 100 μL) DNA Rehydration Solution from the Wizard® Genomic DNA Purification Kit, following manufacturer's instructions. Samples were vortexed and vigorously pipetted to facilitate resuspension. Each sample (200 μL) was transferred to a 96-well black/optical bottom plate and fluorescence was measured (λex/em, 355/517 nm) using a Spark® plate reader, excitation/emission bandwidth = 5 nm.
Propidium iodide fluorescence.S. aureus 25923 (1 × 108 CFU/mL) were incubated in CAP buffers pH 7.0 or 8.5 in 96-well black/optical bottom plates with 10 μM propidium iodide (100 μM stock in dH2O) (PI; Alfa Aesar, Haverhill, MA). The suspension was incubated at 37 °C, 180 rpm, and fluorescence (λex/em, 535/617 nm) was measured at 10 and 20 min, excitation/emission bandwidth = 5 nm. Thirty μg/mL berberine, or 0 or 10 μg/ml daptomycin (100 μg/ml stock in PBS) with 0 or 1.25 mM CaCl2 (12.5 mM stock in dH2O), respectively, was then added to each sample. A parallel setup was prepared without any S. aureus. Samples were incubated at 37 °C, 180 rpm, and fluorescence (λex/em, 535/617 nm) was measured at 10, 20, 30, 40, 50, 60, 120, 180, and 240 min.
Concentration dependence of berberine's effect on proton motive force.S. aureus 25923 (1 × 108 CFU/mL) was incubated in a buffer of 250 mM sucrose, 5 mM MgSO4, and 10 mM KH2PO4, brought to pH 7.0 with NaOH, and sterilized by filtering through a 0.2 μm nylon syringe filter. DiSC3(5) (1 μM) was added, followed by 0.3, 3.0, 30, or 300 μg/mL berberine. Controls included S. aureus 25923 (1 × 108 CFU/mL) incubated with 1 μM of DiSC3(5) in the presence of 10 μg/mL Daptomycin in PBS, 1% DMSO, or 5% DMSO. All samples were incubated in a 96-well black/optical bottom plate for 45 min, 37 °C. Fluorescence (λex/em 620/680 nm) was measured using a Spark® plate reader.
Statistics. All results were assessed for normality using the Kolmogorov-Smirnov test. Normally distributed data was analyzed using ordinary one-way ANOVA with the Dunnett's correction. Non-normally distributed data was analyzed using the non-parametric ANOVA Kruskal-Wallis test with the Dunn-Sidak correction or the Mann-Whitney U test set to a two-sided hypothesis test. In some cases, due to the presence of 0's or 1's in the data, the less powerful U or in some cases t-tests were used. In those cases, the comparisons were between independent sets and advice was taken from a statistician. All analysis was performed using MATLAB R2019a (MathWorks, Natick, MA) and GraphPad Prism 9.1.2 (GraphPad, La Jolla, CA). All graphs were generated using GraphPad Prism 9.1.2 or Excel for Microsoft 365 (Microsoft, Albuquerque, NM).
3. Results
3.1. S. aureus aggregation is decreased via inhibition of clumping factor A (ClfA)
S. aureus and other bacteria aggregate in synovial fluid [5,6], resulting in a range of aggregate sizes, including visible aggregates. By scanning electron microscopy (SEM), these aggregates are encased in a matrix as would be found in floating biofilms [5] (Fig. 1A). Previously we showed that fibrin[ogen] was critical for bacterial aggregation in synovial fluid [9] and that aggregation was dependent on Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMMs) [5]. Because of aggregation, the apparent number of colonies is decreased when colony forming units are directly counted (shown with a synthetic synovial fluid, pSynF [9], Fig. 1B). However, if the bacterial culture is digested with trypsin at harvest and prior to plating, bacterial numbers are no longer significantly different from those measured in tryptic soy broth (TSB). A cartoon (Fig. 1C) illustrates how aggregation results in a net decrease in colony forming units (CFU), where protease digestion can restore bacterial number showing that viability is not affected. Using the decrease in apparent counts as an indication of aggregation, we characterized aggregation in three fluids: TSB, the synthetic synovial fluid pSynF that contains fibrin [ogen], albumin, and hyaluronic acid [9], and human synovial fluid. Using TSB, a slight increase in bacterial number (∼2X) was measured after 90 min, consistent with bacterial proliferation, whereas both the synthetic pSynF and synovial fluid showed decreased CFU/mL (∼2 and ∼1 log, respectively), consistent with aggregation (Fig. 1D). To test if aggregation depended on fibrin[ogen] cross-linking, we used the diamine putrescine which competes with the cross-linking site on fibrin[ogen] [23]. After incubation for 90 min in the presence of putrescine, no change in recovered S. aureus CFU/mL was measured in TSB (Fig. 1E). Bacteria in synovial fluid showed the usual aggregation, resulting in lower CFU/mL at 90 min. Because putrescine did not change CFU/mL, fibrin[ogen] cross-linking via the coagulation cascade is unlikely to be central to bacterial aggregation.
Fig. 1.
S. aureus aggregation in different media. (A) Scanning electron micrograph of S. aureus aggregate formed in human synovial fluid showing its ∼1 mm size and the structure of the surface, with Staphylococci visible through the matrix on the higher magnification. (B) Comparison of CFU/mL recovered after incubation of bacteria in TSB or a synthetic pSynF without and with trypsin dispersion (n = 12; simple one-way ANOVA with Šídák's multiple comparisons test) (C) Cartoon: changes in bacterial counts are used to indicate aggregation (CFU = colony forming units/mL). (D)S. aureus (SA) aggregation in tryptic soy broth (TSB), the synthetic synovial fluid pSynF, and synovial fluid (SynF); n = 12 for each point. Each media set was analyzed as independent pairs by a two-tailed Mann-Whitney U test. The effect of (E) putrescine and (F) calcium (Ca2+) on S. aureus aggregation in TSB, pSynF, and synovial fluid. n = 9–12; both using two-tailed Student's t-test. ns = not significant; *≤0.05; ****p ≤ 0.0001.
We then tested S. aureus aggregation during Ca2+ inhibition of ClfA. ClfA is one of the main cell wall-anchored adhesins involved in aggregation [24]. In TSB, we observed a slight increase in CFU/mL in the presence of 50 mM Ca2+ (∼25% increase, Fig. 1F). In the synthetic pSynF, 50 mM Ca2+ resulted in a 2–3 log increase in CFU/mL. In human synovial fluid, the presence of 50 mM Ca2+ resulted in an ∼0.5 log increase in CFU/mL. Overall, this experiment suggested that inhibition of ClfA resulted in fewer bacteria clustered in aggregates.
3.2. Mutation of sortase A (SrtA) alters aggregation
We had previously reasoned that Sortase A (SrtA), the enzyme that anchors ClfA (among other proteins) to the S. aureus cell wall [10,11], would be critical for aggregation. We switched to methicillin resistant S. aureus (MRSA) USA300 which we had previously used in our aggregation studies [5,25] to enable us to test well-characterized USA300 mutants. In TSB, USA300 and a USA300 srtA mutant [22] showed increased CFU/mL after a 90 min incubation (Fig. 2A, ∼87% and ∼60% increase, respectively), consistent with bacterial proliferation. In the synthetic pSynF, USA300 showed an ∼2 log decrease in CFU/mL after 90 min indicating aggregation, while the USA300 srtA mutant showed no significant change in counts, consistent with attenuated aggregation and some proliferation (Fig. 2B). In human synovial fluid, USA300 CFU/mL decreased by ∼80% due to aggregation; this decrease was not significant for the USA300 srtA mutant (∼25% decrease, Fig. 2C). Together, these data supported a role for SrtA-anchored proteins in bacterial aggregation.
Fig. 2.
Sortase A (SrtA) and S. aureus aggregation. USA300 and USA300 srtA mutant aggregation in (A) tryptic soy broth (TSB), (B) the synthetic pSynF, and (C) human synovial fluid (SynF); n = 12 for each condition. Columns were analyzed as independent pairs by a two-tailed Mann-Whitney U test. Effects of (D) morin and (E) phenylvinylsulfone on S. aureus survival in TSB and equine synovial fluid (EqSynF); n = 9 for each condition. The effect of purported SrtA inhibitor berberine (BBR) on S. aureus in (F) TSB and equine synovial fluid (n = 15) or (G) TSB and human synovial fluid (n = 9 for each condition). Comparisons for panel D were performed using a One-way ANOVA followed by Dunnett's multiple comparisons test relative to the “0” dose control. Panel E, F, and G were analyzed by a Kruskal-Wallis ANOVA using Dunn's multiple comparisons test. ns = not significant, *p < 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
We then tested the effects of three putative SrtA inhibitors. Because ascribing decreased CFU/mL to aggregation would not be possible if toxicity was present, we first tested bacterial survival. The inhibitors morin [26] (Fig. 2D) and phenyl vinyl sulfone [27] (Fig. 2E) showed some mild toxicity in TSB at concentrations of >10 μg/mL. In equine synovial fluid, neither inhibitor showed toxicity as evidenced by no decrease in counts after aggregate dispersal. The putative SrtA inhibitor berberine (BBR) [12] only showed mild toxicity above 30 μg/mL in TSB (Fig. 2F). In contrast, in equine synovial fluid, berberine caused significant bacterial killing both at 20 μg/mL and 30 μg/mL (∼3 logs, p < 0.0001). In human synovial fluid (Fig. 2G), berberine at 20 μg/mL and 30 μg/mL resulted in large decreases in S. aureus CFU/mL (∼3 logs, p < 0.0001; ∼6 logs, p < 0.0001, respectively). Because berberine appeared to have bactericidal activity in synovial fluid (both human and equine synovial fluid, but not the synthetic pSynF, Supplementary Fig. S1), we further investigated its use as an antimicrobial against synovial fluid aggregates. Notably, berberine exhibited bactericidal effects that were not observed with the USA300 srtA mutant or with the SrtA inhibitors morin and phenyl vinyl sulfone.
3.3. Berberine is bactericidal in alkaline pH
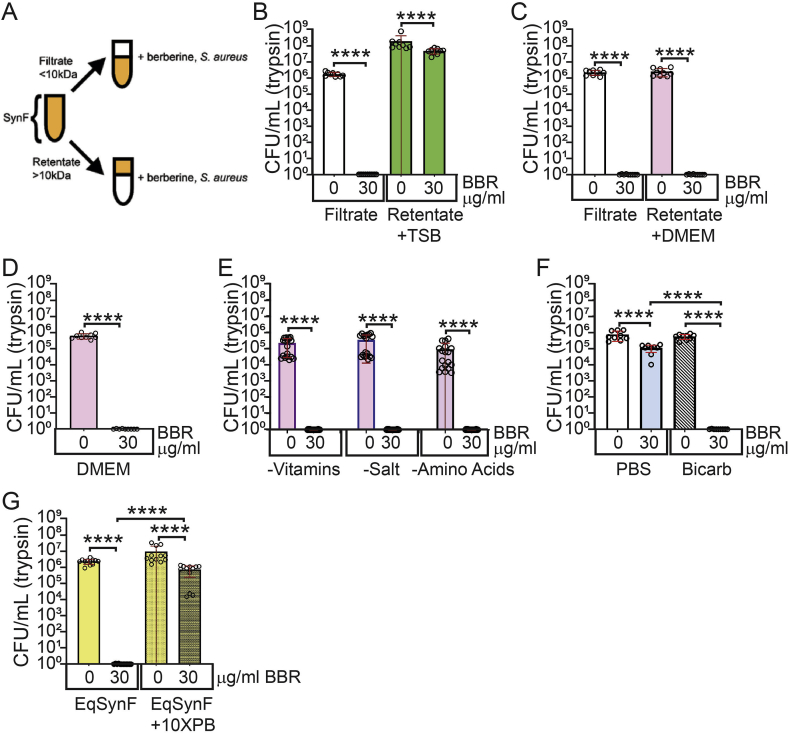
To localize the host factor facilitating berberine's anti-staphylococcal effect, we fractionated synovial fluid into a filtrate (<10 kDa) and a retentate (>10 kDa) (Fig. 3A). In the filtrate, 30 μg/mL berberine decreased S. aureus CFU/mL by 6–7 logs. When the retentate was restored to its original volume with TSB, 30 μg/mL berberine showed only a small, albeit significant (p < 0.0001) effect (Fig. 3B). However, when the retentate was restored with Dulbecco's Modified Eagle Medium (DMEM), 30 μg/mL berberine caused a 6–7 log decrease in CFU/mL (p < 0.0001, Fig. 3C). Thus, berberine's bactericidal effects were observed when S. aureus was cultured in retentate reconstituted with DMEM, but not with retentate reconstituted with TSB.
Fig. 3.
pH dependence of the antibacterial effect of berberine (BBR). (A) Cartoon: fractionation of synovial fluid (SynF) into a filtrate (<10 kDa) and a retentate (>10 kDa). S. aureus treated with berberine in filtrate and retentate dissolved with (B) Tryptic soy broth (TSB) and (C) Dulbecco's Modified Eagle Medium (DMEM); n = 9 for each condition. S. aureus treated with berberine in (D) DMEM (n = 9 for each condition), (E) different formulations of DMEM (n = 18 for each condition), and (F) bicarbonate (n = 9 for each condition). (G)S. aureus treated with berberine in equine synovial fluid (EqSynF) and equine synovial fluid buffered to pH 7.0 (EqSynF + 10XPB); n = 12 for each condition. All pairs were compared using two-tailed Mann Whitney U test. ****p ≤ 0.0001.
We then asked if DMEM alone would support berberine's bactericidal activity. In DMEM, 30 μg/mL berberine resulted in a 5–6 log decrease in S. aureus CFU/mL (Fig. 3D). To determine component(s) in DMEM that were fostering berberine's activity, we used DMEM that lacked (i) amino acids and glucose, (ii) phenol red, pyruvate and vitamins, or (iii) inorganic salts. In all three DMEM formulations, berberine eradicated bacteria so that no CFU/mL were recovered (Fig. 3E; ∼6-log decrease, p < 0.0001). Because all DMEM formulations contained sodium bicarbonate to buffer the medium, we tested berberine in sodium bicarbonate buffer alone. This combination caused a 5–6 log reduction in S. aureus CFU/mL (Fig. 3F).
Sodium bicarbonate buffers blood, synovial fluid, and cell culture media. As is apparent in cell culture medium, sodium bicarbonate-buffered solutions become more alkaline in vitro over time. We therefore asked if berberine's bactericidal activity was pH dependent. Synovial fluid pH was measured at ∼8.5 in the berberine/S. aureus experiment (Fig. 3G). When synovial fluid was forced to remain at pH 7 by addition of 10X phosphate buffer (PB), berberine caused at most a 1 log decrease in S. aureus CFU/mL which was not statistically significant. Collectively, these data showed berberine antimicrobial activity was dependent on pH such that by pH 8.5–9.0, a potent anti-Staphylococcal effect was measured.
3.4. Berberine is associated with S. aureus
We next used berberine's innate fluorescence to probe its activity against S. aureus. We first asked if berberine associated with S. aureus and if any association increased in alkaline compared to neutral media (Fig. 4A). At pH 7, treatment of S. aureus with 30 μg/mL berberine caused slightly increased fluorescence of pelleted bacteria; at pH 9, bacterial fluorescence increased markedly (∼2–3 fold).
Fig. 4.
Mechanisms of berberine's (BBR's) bactericidal effect. I. (A) Berberine internalization into S. aureus as measured by fluorescence as a function of pH (n = 18). Comparisons were done using the Kruskal-Wallis test with the Dunn-Sidak correction. (B) Fluorescence of S. aureus lysate with no exposure and after exposure to berberine (n = 6; two-tailed Mann-Whitney U test). (C) Fluorescence that localizes to the supernatant vs the DNA fraction in berberine-loaded S. aureus (n = 4; two-tailed Mann-Whitney U test). (D) Propidium iodide fluorescence following addition of Daptomycin to S. aureus. (E) Propidium iodide fluorescence following exposure of S. aureus to berberine. Comparisons for D and E were done using the Kruskal-Wallis test with the Dunn-Sidak correction. ns = not significant; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001****p ≤ 0.0001.
Because berberine association with S. aureus increased as the bactericidal effect increased, we asked if berberine internalization and subsequent DNA binding, as suggested by others [28], could account for its bactericidal activity. DNA extracted from S. aureus and then exposed to 30 μg/mL berberine in alkaline media exhibited ∼3X greater fluorescence than the innate fluorescence of the control (Fig. 4B). To test DNA binding, berberine-loaded bacteria were lysed and separated into a DNA fraction and a soluble fraction. In this setting, DNA localization was minor (∼5%) compared to the total berberine contained within the S. aureus (Fig. 4C), suggesting that DNA-berberine interactions are not a major mechanism. For the negative control alamar blue, the DNA:cytoplasm ratio was ∼0.03%; for the positive control SYTO 9, ∼8% (Supplementary Fig. 2).
We next asked if berberine caused membrane destabilization, either through pore formation or through more non-specific destabilization. We first tracked the effects of Daptomycin, an antibiotic with known pore forming capabilities. In the presence of Daptomycin, propidium iodide fluorescence steadily increased as a function of time (Fig. 4D). When berberine and propidium iodide were added to S. aureus in pH 8.5 media, a slight but statistically significant increase in propidium iodide fluorescence was measured over the first 2 h (Fig. 4E). However, the propidium iodide fluorescence only began to increase more rapidly after 60 min, suggesting a mechanism that was distinct from the pore formation caused by Daptomycin.
3.5. Berberine affects membrane potential
While berberine localized to S. aureus, there were only mild effects on either DNA binding or membrane permeability. We next asked if perhaps S. aureus was altering drug efflux. We chose to first examine NorA, an S. aureus multidrug efflux pump that is well-characterized and exports dyes and quaternary ammonium compounds [29] (berberine is a quaternary amine [30], Fig. 5A). If berberine was internalized into S. aureus, an USA300 norA mutant should exhibit attenuated export and result in increased retention of the fluorescent berberine. After treatment with berberine in phosphate buffered saline (PBS), pH 7.4, the USA300 norA mutant exhibited ∼5X greater berberine fluorescence/localization than that of USA300 (Fig. 5B). Furthermore, treatment with reserpine, a NorA inhibitor (Fig. 5C), increased the bactericidal effect of 30 μg/mL berberine in pH 7.4 PBS by 2–3 logs. Berberine or reserpine alone resulted in <1 log reduction in bacterial counts (Fig. 5C). Collectively, these results support an interplay between NorA efflux of berberine and its facilitated localization at alkaline pH.
Fig. 5.
Mechanisms of berberine's (BBR's) bactericidal effect II. (A) Berberine structure. (B) Berberine localization to S. aureus as measured by fluorescence in a USA300 norA mutant (n = 18 for each point; unpaired t-test). (C) Berberine in combination with the NorA inhibitor reserpine effects on bacterial survival (right; n = 12 for each; Kruskal-Wallis ANOVA with Dunn's multiple comparisons test). (D) Cartoon: alkalinization reduces the proton gradient and increases the membrane potential across Gram-positive bacterial cell membranes (created with BioRender.com). (E) The effect of pH on DiSC3(5) localization as measured by fluorescence (n = 18 for each point; Kruskal-Wallis ANOVA with Dunn's multiple comparisons test). (F) The bactericidal effect of berberine with increasing pH (n = 9 for each; Kruskal-Wallis ANOVA with Dunn's multiple comparisons test). (G) The effect of increasing berberine concentration on DiSC3(5) localization as measured by fluorescence (n = 24 for each; two-tailed Mann-Whitney U test for independent comparisons with control). ns = not significant; *p ≤ 0.05, **p ≤ 0.01, ****p ≤ 0.0001.
Efflux pumps and extracellular pH both interact with membrane potential to ultimately alter the electron transport chain. As electrons are shuttled down the electron transport chain, protons (H+) are transported extracellularly before passing back through the membrane-bound ATPase to generate ATP [31] (Fig. 5D). We thus reasoned that berberine in the alkaline medium would reduce the proton gradient while increasing membrane potential (ΔΨ). This increase in ΔΨ would exert an attractive force on positivecharges, such as the quaternary amine in berberine.
To test this, we asked if ΔΨ of S. aureus increased as the medium became more alkaline, causing localization of berberine within S. aureus. We used DiSC3(5), a dye that associates with bacteria as a function of increasing ΔΨ, but due to self-quenching, yields an inverse correlation between ΔΨ and DiSC3(5) fluorescence [21]. With increasing pH, DiSC3(5) fluorescence decreased, showing an ∼50% decrease in fluorescence at pH 9.0 (Fig. 5E). This increased permeability paralleled the bactericidal effect of berberine. Specifically, no significant decrease in CFU/mL (after trypsin dispersal) was seen in PBS buffered to pH 6.0, 6.5, and 7.0. Beginning at pH 7.5, berberine caused a pH-dependent decrease in S. aureus number, such that by pH 9.0, a 5–6 log decrease in S. aureus CFU/mL was measured (Fig. 5F).
These data suggested that berberine could act by dissipating ΔΨ. We asked if increasing concentrations of berberine in pH 7.0 media could increase release of DiSC3(5) localized to S. aureus (Fig. 5F). Dye fluorescence was unchanged with 0.3–30 μg/mL berberine. However, 300 μg/mL berberine resulted in a 4X increase in fluorescence after 45 min, where again, fluorescence is inversely related to DiSC3(5) concentration. Neither 1% nor 10% DMSO, which was used as the solvent for berberine, resulted in a comparable level of DiSC3(5) fluorescence. The positive control, 10 μg/mL Daptomycin, showed dye fluorescence comparable to that measured at low concentrations of berberine. Thus, membrane destabilization appeared to factor into berberine's bactericidal effects, perhaps amplified by the pH dependence of membrane potential and the activity of NorA and other efflux pumps.
4. Discussion
Bacterial aggregates exhibit tolerance to antibiotics that are similar to that of bacterial biofilms [[5], [6], [7],9,25,32,33] so that successful treatment of septic arthritis requires addressing the reservoir of S. aureus aggregates that form in synovial fluid [34]. The findings of this study demonstrated that berberine, a compound used for treatment of diarrheal diseases [14], inflammation [15,16,35], diabetes [36], and infections [[37], [38], [39], [40]], possesses a pH-dependent, metabolism-independent antibacterial effect that can be effective against these aggregated, antibiotic-tolerant bacteria. This is to our knowledge the first study describing such an effect in the context of antibiotic tolerant bacteria in synovial fluid.
We have shown in previous work that aggregate dispersal can increase the antibiotic sensitivity of S. aureus in synovial fluid [5,6]. Therefore, in this manuscript, we initially sought to prevent bacterial aggregation to increase antibiotic susceptibility. We initially used the synthetic synovial fluid pSynF, a synovial fluid mimic our group developed to study bacterial aggregation and antibiotic susceptibility [9]. S. aureus showed greater aggregation in pSynF compared to synovial fluid. In both fluids, the competitive transglutaminase inhibitor putrescine [41] did not alter aggregation and indicated fibrin[ogen] crosslinking was unlikely to be the cause of aggregation. The difference in aggregation between the two fluids could be related to their relative calcium content and calcium's ability to inhibit clumping factor A (ClfA) binding to serum proteins [24]. The serum concentration of ionized calcium is 1.1–1.3 mM, which would also be found in synovial fluid due to the equilibration of low molecular weight molecules between serum and synovial fluid [[42], [43], [44]]; no calcium is added to pSynF [9]. This interpretation is supported by the inhibition of S. aureus aggregation by Ca2+ in the synthetic pSynF, but only a minimal inhibition in synovial fluid. Overall, the calcium data supported the importance of Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMMs) in aggregation,
In keeping with the critical role of the cell surface MSCRAMMs, we focused on the bacterial transpeptidase sortase A (SrtA), which mediates protein anchoring to the bacterial surface. Indeed, a USA300 srtA mutant resulted in less aggregation in synovial fluid and the synthetic pSynF. When we tested toxicity of SrtA inhibitors, we surprisingly found that berberine [12] caused bacterial death in synovial fluid but not in TSB. Based on our results, we asked if synovial fluid possessed a component that could potentiate berberine. Based on recapitulation of the activity in Dulbecco's Modified Eagle Medium (DMEM), we identified alkaline pH as the factor that potentiates the bactericidal effect of berberine. As we attempted to clarify the bactericidal mechanism, we also questioned berberine's effects on aggregation—any effects were minor compared to the antibacterial activity. These data integrate with previous observations that alkaline pH can decrease the minimum inhibitory concentration of berberine against S. aureus [45].
In the joint, the absence of a synovial basement membrane allows near equal distribution of small molecular weight molecules [46] between serum and synovial fluid [[42], [43], [44]] so that bicarbonate will be present in synovial fluid. In vivo, this bicarbonate allows retention of physiological pH. Synovial fluid stored in vitro showed a drift towards higher pH, presumably because of bicarbonate breakdown. Bicarbonate in solution combines with H2O to form OH− and carbonic acid, which breaks down into H2O and CO2. In an open system, CO2 diffuses out, increasing production of OH− and alkalinization of the media. This alkaline pH is critical to berberine's bactericidal effect, as further evidenced by decreased effects in equine synovial fluid buffered to ∼pH 7.0.
Although we noted that berberine binds to bacterial DNA, membrane destabilization and collapsing of the proton motive force ΔΨ appears to explain its bactericidal activity. Membrane destabilization measured using propidium iodide fluorescence showed an ∼2 h delay for berberine to exert its bactericidal effect; the membrane pore-forming antibiotic Daptomycin took effect within 20 min. This difference in activation time could be due to berberine destabilizing the membrane in a non-specific manner that is both time- and concentration-dependent or more likely due to the collapse of the proton motive force ΔΨ.
In keeping with this effect, the USA300 norA mutant accumulated more berberine than wild type at neutral pH. Use of the NorA inhibitor reserpine [45] also augmented the bactericidal effect of berberine at neutral pH. Thus, berberine's bactericidal effect appears to be tied to the alkaline environment, which reduces the activity of 4 of 5 multidrug efflux pump families in S. aureus [47]. In keeping with these effects, we also measured disruption of the electron transport chain (ETC) and the proton motive force (PMF) in bacteria.
In bacteria, the PMF is maintained by offsetting any change in proton gradient by a counteracting change in ΔΨ [48]. Alkalinization of the media would reduce the proton gradient and therefore induce a reactionary increase in ΔΨ, as we observed through a decrease in DiSC3(5) fluorescence. This increase in ΔΨ would exert an attractive force on H+ and berberine, which is also cationic, causing localization to S. aureus, which we detected as increasing berberine fluorescence under alkaline media conditions.
The ability of berberine treatment to reduce the bacterial burden in synovial fluid through a mechanism that appears largely independent of bacterial metabolism shows promise for clinical applicability, with the caveat that it requires inducing the infected environment to become alkaline. Inclusion of other frequent causative pathogens that can aggregate in synovial fluid, such as coagulase negative Staphylococcus and Escherichia coli [6], will be important to assess the scope of berberine's coverage. Additionally, the bactericidal efficacy of berberine needs to be tested against mature bacterial aggregates in synovial fluid, aggregates that have shown high levels of tolerance to standard first-line antibiotics. Ultimately, an in vivo model will be necessary to evaluate the ability of berberine to eradicate bacteria inside the joint and the effect of induction of an alkaline environment on joint physiology.
Finally, the presence of antibiotic resistance has increased the urgency for more effective treatments for bacterial infections. Biofilm formation has complicated this issue in that bacteria species can become tolerant to antibiotics through environmental factors rather than genetic alterations. Synovial fluid provides the necessary conditions for bacteria to quickly form aggregates that are essentially floating biofilms. In this manuscript, we have taken the first steps to show that berberine, which is cheap and readily available, can be effective against these antibiotic tolerant bacteria.
Funding
Research reported in this publication was supported by the National Institutes of Health (NIH) under award numbers R01 AR069119, R01 AR072513, and RO1 AR076941 (NH and TS) and T32AR052273 (NZ). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
CRediT authorship contribution statement
Neil Zhao: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. Selin Isguven: Conceptualization, Data curation, Methodology, Writing – review & editing. Rachel Evans: Conceptualization, Data curation, Methodology, Writing – review & editing. Thomas P. Schaer: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing. Noreen J. Hickok: Conceptualization, Data curation, Formal analysis, Funding acquisition, Resources, Supervision, Visualization, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no conflicts of interest associated with the research reported in this manuscript.
Acknowledgements
We thank Catherine Gurr for help in data visualization and Matthew Sherman for statistical consultations.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2023.100117.
Appendix A. Supplementary data
The following is the supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Armstrong G.L., Conn L.A., Pinner R.W. Trends in infectious disease mortality in the United States during the 20th century. JAMA, J Am Med Assoc. 1999;281(1):61–66. doi: 10.1001/jama.281.1.61. [DOI] [PubMed] [Google Scholar]
- 2.Spellberg B., Blaser M., Guidos R.J., Boucher H.W., Bradley J.S., et al. Combating antimicrobial resistance: policy recommendations to save lives. Clin Infect Dis. 2011;52(Suppl 5):S397–S428. doi: 10.1093/cid/cir153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson T. The staggering death toll of drug-resistant bacteria. Nature. 2022 doi: 10.1038/d41586-022-00228-x. PMID: 35102288. [DOI] [PubMed] [Google Scholar]
- 4.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 5.Dastgheyb S., Parvizi J., Shapiro I.M., Hickok N.J., Otto M. Effect of biofilms on recalcitrance of staphylococcal joint infection to antibiotic treatment. J Infect Dis. 2015;211(4):641–650. doi: 10.1093/infdis/jiu514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbertie J.M., Schnabel L.V., Hickok N.J., Jacob M.E., Conlon B.P., Shapiro I.M., Parvizi J., Schaer T.P. Equine or porcine synovial fluid as a novel ex vivo model for the study of bacterial free-floating biofilms that form in human joint infections. PLoS One. 2019;14(8) doi: 10.1371/journal.pone.0221012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dastgheyb S.S., Hammoud S., Ketonis C., Liu A.Y., Fitzgerald K., Parvizi J., Purtill J., Ciccotti M., Shapiro I.M., Otto M., Hickok N.J. Staphylococcal persistence due to biofilm formation in synovial fluid containing prophylactic cefazolin. Antimicrob Agents Chemother. 2015;59(4):2122–2128. doi: 10.1128/AAC.04579-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathews C.J., Weston V.C., Jones A., Field M., Coakley G. Bacterial septic arthritis in adults. Lancet. 2010;375(9717):846–855. doi: 10.1016/S0140-6736(09)61595-6. [DOI] [PubMed] [Google Scholar]
- 9.Knott S., Curry D., Zhao N., Metgud P., Dastgheyb S., Purtill C., et al. Staphylococcus aureus floating biofilm formation and phenotype in synovial fluid depends on albumin, fibrinogen, and hyaluronic acid. Front Microbiol. 2021;12:70-82 doi: 10.3389/fmicb.2021.655873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster T.J. Surface proteins of Staphylococcus aureus. Microbiol Spectr. 2019 Jul;7(4) doi: 10.1128/microbiolspec.GPP3-0046-2018. PMID: 31267926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marraffini L.A., DeDent A.C., Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70(1):192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S.-H., Shin D.-S., Oh M.-N., Chung S.-C., Lee J.-S., Oh K.-B. Inhibition of the bacterial surface protein anchoring transpeptidase sortase by isoquinoline alkaloids. Biosci Biotechnol Biochem. 2004;68(2):421–424. doi: 10.1271/bbb.68.421. [DOI] [PubMed] [Google Scholar]
- 13.Neag M.A., Mocan A., Echeverría J., Pop R.M., Bocsan C.I., Crişan G., et al. Berberine: botanical occurrence, traditional uses, extraction methods, and relevance in cardiovascular, metabolic, hepatic, and renal disorders. Front Pharmacol. 2018 Aug 21;9:557. doi: 10.3389/fphar.2018.00557. PMID: 30186157; PMCID: PMC6111450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C., Tao C., Liu Z., Lu M., Pan Q., Zheng L., Li Q., Song Z., Fichna J. A randomized clinical trial of berberine hydrochloride in patients with diarrhea-predominant irritable bowel syndrome. Phytother Res. 2015;29(11):1822–1827. doi: 10.1002/ptr.5475. [DOI] [PubMed] [Google Scholar]
- 15.Tew X.N., Xin Lau N.J., Chellappan D.K., Madheswaran T., Zeeshan F., Tambuwala M.M., Aljabali A.A., Balusamy S.R., Perumalsamy H., Gupta G., Oliver B.G., Hsu A., Wark P., Reddy K., Wadhwa R., Hansbro P.M., Dua K. Immunological axis of berberine in managing inflammation underlying chronic respiratory inflammatory diseases. Chem Biol Interact. 2020;317 doi: 10.1016/j.cbi.2020.108947. [DOI] [PubMed] [Google Scholar]
- 16.Shen P., Jiao Y., Miao L., Chen J.-H., Momtazi-Borojeni A.A. Immunomodulatory effects of berberine on the inflamed joint reveal new therapeutic targets for rheumatoid arthritis management. J Cell Mol Med. 2020;24(21):12234–12245. doi: 10.1111/jcmm.15803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chi L., Peng L., Pan N., Hu X., Zhang Y. The anti-atherogenic effects of berberine on foam cell formation are mediated through the upregulation of sirtuin 1. Int J Mol Med. 2014;34(4):1087–1093. doi: 10.3892/ijmm.2014.1868. [DOI] [PubMed] [Google Scholar]
- 18.Yang J., Yin J., Gao H., Xu L., Wang Y., Xu L., et al. Berberine improves insulin sensitivity by inhibiting fat store and adjusting adipokines profile in human preadipocytes and metabolic syndrome patients. Evid Based Compl Alternat Med. 2012 doi: 10.1155/2012/363845. 2012 Epub 2012 Mar 8. PMID: 22474499; PMCID: PMC3310165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu M., Zhang M.-b., Liu Y.-c., Kang J.-r., Chu Z.-y., Yin K.-l., Ding L.-y., Ding R., Xiao R.-x., Yin Y.-n., Liu X.-y., Wang Y.-d. Role of berberine in the treatment of methicillin-resistant Staphylococcus aureus infections. Sci Rep. 2016 Apr 22;6 doi: 10.1038/srep24748. PMID: 27103062; PMCID: PMC4840435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., He P., Hou Y., Chen S., Xiao Z., Zhan J., Luo D., Gu M., Lin D. Berberine inhibits the interleukin-1 beta-induced inflammatory response via MAPK downregulation in rat articular chondrocytes. Drug Dev Res. 2019;80(5):637–645. doi: 10.1002/ddr.21541. [DOI] [PubMed] [Google Scholar]
- 21.Te Winkel J.D., Gray D.A., Seistrup K.H., Hamoen L.W., Strahl H. Analysis of antimicrobial-triggered membrane depolarization using voltage sensitive dyes. Front Cell Dev Biol. 2016;4:29. doi: 10.3389/fcell.2016.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fey P.D., Endres J.L., Yajjala V.K., Widhelm T.J., Boissy R.J., Bose J.L., Bayles K.W. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio. 2013;4(1) doi: 10.1128/mBio.00537-12. e00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dolynchuk K.N., Bendor-Samuel R., Bowness J.M. Effect of putrescine on tissue transglutaminase activity in wounds: decreased breaking strength and increased matrix fucoprotein solubility. Plast Reconstr Surg. 1994;93(3):567–573. [PubMed] [Google Scholar]
- 24.O'Connell D.P., Nanavaty T., McDevitt D., Gurusiddappa S., Höök M., Foster T.J. The fibrinogen-binding MSCRAMM (clumping factor) of Staphylococcus aureus has a Ca2+-dependent inhibitory site. J Biol Chem. 1998;273(12):6821–6829. doi: 10.1074/jbc.273.12.6821. [DOI] [PubMed] [Google Scholar]
- 25.Dastgheyb S.S., Villaruz A.E., Le K.Y., Tan V.Y., Duong A.C., Chatterjee S.S., Cheung G.Y., Joo H.S., Hickok N.J., Otto M. Role of phenol-soluble modulins in formation of Staphylococcus aureus biofilms in synovial fluid. Infect Immun. 2015;83(7):2966–2975. doi: 10.1128/IAI.00394-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang P., Hu P., Zhou S.Y., Li Q., Chen W.M. Morin inhibits sortase A and subsequent biofilm formation in Streptococcus mutans. Curr Microbiol. 2014;68(1):47–52. doi: 10.1007/s00284-013-0439-x. [DOI] [PubMed] [Google Scholar]
- 27.Frankel B.A., Bentley M., Kruger R.G., McCafferty D.G. Vinyl sulfones: inhibitors of SrtA, a transpeptidase required for cell wall protein anchoring and virulence in Staphylococcus aureus. J Am Chem Soc. 2004;126(11):3404–3405. doi: 10.1021/ja0390294. [DOI] [PubMed] [Google Scholar]
- 28.Bhadra K., Maiti M., Kumar G.S. Berberine-DNA complexation: new insights into the cooperative binding and energetic aspects. Biochim Biophys Acta. 2008;1780(9):1054–1061. doi: 10.1016/j.bbagen.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Monteiro K.L.C., de Aquino T.M., Mendonça Junior F.J.B. An update on Staphylococcus aureus NorA efflux pump inhibitors. Curr Top Med Chem. 2020;20(24):2168–2185. doi: 10.2174/1568026620666200704135837. [DOI] [PubMed] [Google Scholar]
- 30.Cicero A.F., Baggioni A. Berberine and its role in chronic disease. Adv Exp Med Biol. 2016;928:27–45. doi: 10.1007/978-3-319-41334-1_2. [DOI] [PubMed] [Google Scholar]
- 31.Kashket E.R. The proton motive force in bacteria: a critical assessment of methods. Annu Rev Microbiol. 1985;39:219–242. doi: 10.1146/annurev.mi.39.100185.001251. [DOI] [PubMed] [Google Scholar]
- 32.Crosby H.A., Kwiecinski J., Horswill A.R. Staphylococcus aureus aggregation and coagulation mechanisms, and their function in host-pathogen interactions. Adv Appl Microbiol. 2016;96:1–41. doi: 10.1016/bs.aambs.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pestrak M.J., Gupta T.T., Dusane D.H., Guzior D.V., Staats A., Harro J., Horswill A.R., Stoodley P. Investigation of synovial fluid induced Staphylococcus aureus aggregate development and its impact on surface attachment and biofilm formation. PLoS One. 2020;15(4) doi: 10.1371/journal.pone.0231791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz E.M., McLaren A.C., Sculco T.P., Brause B., Bostrom M., Kates S.L., et al. Adjuvant antibiotic-loaded bone cement: Concerns with current use and research to make it work. J Orthop Res. 2021;39(2):227–239. doi: 10.1002/jor.24616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu P.-f., Chen W.-p., Tang J.-l., Bao J.-p., Wu L.-d. Protective effects of berberine in an experimental rat osteoarthritis model. Phytother Res. 2011;25(6):878–885. doi: 10.1002/ptr.3359. [DOI] [PubMed] [Google Scholar]
- 36.Dong H., Wang N., Zhao L., Lu F. Berberine in the treatment of type 2 diabetes mellitus: a systemic review and meta-analysis. Evid Based Compl Alternat Med. 2012 doi: 10.1155/2012/591654. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu H.-H., Kim K.-J., Cha J.-D., Kim H.-K., Lee Y.-E., Choi N.-Y., You Y.-O. Antimicrobial activity of berberine alone and in combination with ampicillin or oxacillin against methicillin-resistant Staphylococcus aureus. J Med Food. 2005;8(4):454–461. doi: 10.1089/jmf.2005.8.454. [DOI] [PubMed] [Google Scholar]
- 38.Xie Q., Johnson B.R., Wenckus C.S., Fayad M.I., Wu C.D. Efficacy of berberine, an antimicrobial plant alkaloid, as an endodontic irrigant against a mixed-culture biofilm in an in vitro tooth model. J Endod. 2012;38(8):1114–1117. doi: 10.1016/j.joen.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 39.Zorić N., Kosalec I., Tomić S., Bobnjarić I., Jug M., Vlainić T., Vlainić J. Membrane of Candida albicans as a target of berberine. BMC Compl Alternative Med. 2017;17(1):268. doi: 10.1186/s12906-017-1773-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X., Yao X., Zhu Z.a., Tang T., Dai K., Sadovskaya I., Flahaut S., Jabbouri S. Effect of berberine on Staphylococcus epidermidis biofilm formation. Int J Antimicrob Agents. 2009;34(1):60–66. doi: 10.1016/j.ijantimicag.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 41.Siegel M., Khosla C. Transglutaminase 2 inhibitors and their therapeutic role in disease states. Pharmacol Ther. 2007;115(2):232–245. doi: 10.1016/j.pharmthera.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kushner I., Somerville J.A. Permeability of human synovial membrane to plasma proteins. Relationship to molecular size and inflammation. Arthritis Rheum. 1971;14(5):560–570. doi: 10.1002/art.1780140503. [DOI] [PubMed] [Google Scholar]
- 43.Cajori F.A., Pemberton R., Stile E. The chemical composition of synovial fluid in cases of joint effusion. J Biol Chem. 1928;76:471–480. [Google Scholar]
- 44.Fremont-Smith F., Dailey E. Studies in the distribution of chloride and protein between plasma and synovial fluid. J Biol Chem. 1926;70:779–784. [Google Scholar]
- 45.Hsieh P.C., Siegel S.A., Rogers B., Davis D., Lewis K. Bacteria lacking a multidrug pump: a sensitive tool for drug discovery. Proc Natl Acad Sci USA. 1998;95(12):6602–6606. doi: 10.1073/pnas.95.12.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ropes M.W., Bennett G.A., Bauer W. The origin and nature of normal synovial fluid. J Clin Investig. 1939;18(3):351–372. doi: 10.1172/JCI101050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lekshmi M., Ammini P., Adjei J., Sanford L.M., Shrestha U., Kumar S., Varela M.F. Modulation of antimicrobial efflux pumps of the major facilitator superfamily in Staphylococcus aureus. AIMS Microbiol. 2018;4(1):1–18. doi: 10.3934/microbiol.2018.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bakker E.P., Mangerich W.E. Interconversion of components of the bacterial proton motive force by electrogenic potassium transport. J Bacteriol. 1981;147(3):820–826. doi: 10.1128/jb.147.3.820-826.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.