Highlights
-
•
The core group of the gut microbiome generally never gets affected by any of the environmental factors.
-
•
Phenotypic variations have been correlated with the structural composition of the intestinal microbiome.
-
•
The microbiome present in fish produces a particular antibody in their gills in response to the pathogenic microbiome called ‘immunoglobulin IgT’, the function of which was discovered in the recent past.
-
•
Several genes have been discovered that govern different physiological functions in the gut tract and these genes have been found to be regulated by the presence or absence of microbes in the gut system.
-
•
The mechanisms and the key molecules that are involved in this complex process have also been identified. However, further research is required on host-microbiome interactions in relation to the genomes.
Keywords: Microbiome, Fish, Shellfish, Host, Interaction, Mechanism
Abstract
The importance of the gut microbiome in the management of various physiological activities including healthy growth and performance of fish and shellfish is now widely considered and being studied in detail for potential applications in aquaculture farming and the future growth of the fish industry. The gut microbiome in all animals including fish is associated with a number of beneficial functions for the host, such as stimulating optimal gastrointestinal development, producing and supplying vitamins to the host, and improving the host's nutrient uptake by providing additional enzymatic activities. Besides nutrient uptake, the gut microbiome is involved in strengthening the immune system and maintaining mucosal tolerance, enhancing the host's resilience against infectious diseases, and the production of anticarcinogenic and anti-inflammatory compounds. Because of its significant role, the gut microbiome is very often considered an “extra organ,” as it plays a key role in intestinal development and regulation of other physiological functions. Recent studies suggest that the gut microbiome is involved in energy homeostasis by regulating feeding, digestive and metabolic processes, as well as the immune response. Consequently, deciphering gut microbiome dynamics in cultured fish and shellfish species will play an indispensable role in promoting animal health and aquaculture productivity. It is mentioned that the microbiome community available in the gut tract, particularly in the intestine acts as an innovative source of natural product discovery. The microbial communities that are associated with several marine organisms are the source of natural products with a diverse array of biological activities and as of today, more than 1000 new compounds have been reported from such microbial species. Exploration of such new ingredients from microbial species would create more opportunities for the development of the bio-pharma/aquaculture industries. Considering the important role of the microbiome in the whole life span of fish and shellfish, it is necessary to understand the interaction process between the host and microbial community. However, information pertaining to host-microbiome interaction, particularly at the cellular level, gene expression, metabolic pathways, and immunomodulation mechanisms, the available literature is scanty. It has been reported that there are three ways of interaction involving the host-microbe-environment operates to maintain homeostasis in the fish and shellfish gut i.e. host intrinsic factors, the environment that shapes the gut microbiome composition, and the core microbial community present in the gut system itself has equal influence on the host biology. In the present review, efforts have been made to collect comprehensive information on various aspects of host-microbiome interaction, particularly on the immune system and health maintenance, management of diseases, nutrient uptake, digestion and absorption, gene expression, and metabolism in fish and shellfish.
1. Introduction
The microbiome communities comprising several micro-organisms like protists, yeasts, Viruses, Bacteria, and Archaea are generally found not only in the digestive tract the fish and shellfish but these communities also inhabit the different parts of the body particularly in the skin, gills, and muscular tissues. The population density and composition of microbiome communities are generally affected by a number of environmental factors, so also the effects of different seasons, host genetic factors, sex/age, and diet are seen prominently. Despite this, the microbiome contains core components of the microbes, especially within the digestive tract, which are well adapted to the selection pressures associated with the host species, and as such these core microbes are commonly found in individuals of the same species even when reared in different locations or conditions [1,2]. The different microbial species present in the gastrointestinal tract and other body parts play a vital role in maintaining a homeostatic physiological environment in the body and protecting it from harmful pathogens. The most important functions of differentiation of the digestive tract, its morphology, immunomodulation, and nutrient absorption always depend on the type of microbial composition present in the fish and shellfish and this has been proved experimentally [2,3]. The mechanisms involved in these actions have been partially worked out and various molecules that are important in the complicated processes of host-microbe have also been identified to some extent. Sharma and Thaiss [4] in their review while discussing the host-microbiome interactions in single-cell reported that, microbes play an important role in modulating host physiology, health, and disease management in most organisms. Further, it is mentioned that due to technological advancement in single-cell analysis particularly in the area of genomics, transcriptomics, and spatial resolution, our understanding of the knowledge complexities involved in single organisms has enhanced to large extent.
It is now well known that host-microbiome interaction plays a very significant role in host development, immunity, metabolisms, and pathways, but it is also a fact that the information pertaining to host-microbiome interactions is limited as far as fish and shellfish are concerned. In the recent past, some efforts have been made to investigate host-microbiome interaction with respect to different physiological functions in fish and shellfish. Perez et al. [5] carried out studies on host-microbiome interactions in hybrid fish, derived by crossing fish from herbivorous and carnivorous dietary adaptations. It has been reported that there was no difference in the growth pattern during the early developmental stages of both groups of fish and the microbial composition also did not show much difference. However, significant changes have been observed in the microbiome composition during later stages when there was a change in dietary adaptations. Further analysis showed that there was the dominance of microbiome species that are associated with metabolism and growth in both groups of fish. In addition, it was also observed that the differentially expressed homologous genes in the intestine associated with cell growth, immunity, and the metabolism was related to the dominant gut microbiome species. These findings indicate that host genetics-gut microbiome interactions contribute to dietary adaptation in hybrid fish. Studies on the fish gut microbiome and their interactions with mucosal tissue have been carried out by Merrifield and Rodiles [3]. Similarly, Nerea Arias Jayo et al. [6] investigated host-microbiome interactions in adult zebrafish in response to a diet rich in high saturated fat supplemented with fish oil. As it is well known that diet is a very important factor that affects the host's health, hence interaction of microbial species with the host's nutrition becomes a vital study. Nikouli et al. [7] while working on the gut microbiome composition of five sympatrically farmed marine fish reported that the intestinal microbiome species act as a second genome of the animals controlling various vital functions of the body. Further, it is mentioned that the colonization of the fish gut microbiome community depends on both intrinsic and extrinsic factors including feeding habits. Simon et al. (2019) while emphasizing the importance of the holobiont concept reported that the interaction between hosts and their associated microbial communities is necessary to know the biological entity of an organism involving the host and its inherited microbiome community which together constitute a holobiont concept.
There are a number of reports mentioning the functional role of the gut microbiome species and their interaction with hosts both in fish and shellfish. The role of the gut microbiome in the production of digestive enzymes and the digestion process has been reported by Ray et al. [8]. Similarly, the production of vitamins, short-chain fatty acids, biofilm formation, and iron metabolism have been investigated in the gut microbiome of freshwater and marine fish by a number of researchers [3,[9], [10], [11]]. While studying the host-microbiome interaction, Li et al. [2] carried out an in-depth analysis of growth performance in the hybrid fish in relation to the composition of the microbiome species in the gut system. There are few studies assessing the phenotypic variations in fish and shellfish and their correlations with the microbiome present in the gut system. Mushegian et al. [12] worked on the genome of the gut microbiome species to investigate whether the genes of the microbiome are involved in the selection preference of the core microbiome. A number of workers have also investigated the role of the host-gut microbiome interaction in several fish in the management of different types of stress responses [13,14].
As far as shellfish are concerned particularly the penaeid shrimp, the information available on host-microbiome interaction is scanty. A number of workers have reported the presence of harmful pathogens in different body parts of penaeid shrimp and their catastrophic effects on animal health and the quality of the harvest during shrimp farming activities. Frequent reports of outbreaks of bacterial and viral diseases and mass mortality continued to be a major challenge for the shrimp aquaculture industry [15,1]. Awareness of host-microbiome interaction and its importance has been realized in recent years after understanding the microbiome benefits for improving overall physiological activities leading to a better immune system and healthy growth of organisms. Therefore, it is also necessary to know how various factors both extrinsic and intrinsic can shape the gut microbiota of shrimp is crucial for the future utilization of the gut microbiome as a tool for controlling overall health and quality. Chaiyapechara et al. [15] while studying host-microbiome interaction in shrimp P. monodon carried out investigations on intestinal microbial species and their transcriptomics profile analysis at different salinity levels. It was reported that the shrimp acclimatized at higher salinities showed that the Proteobacteria was the dominant phyla in the gut microbiome composition followed by other bacterial groups i.e. Bacteroidetes, Planctomycetes, and Firmicutes. The most abundant genus was Vibrio belonging to the Harveyi clade. Further, it has been mentioned that in higher salinities genes involved with stress and immune responses were differentially expressed and there was also a close relationship between the abundance of pathogenic Vibrio and the expression of genes related to innate immunity. Similar work and observations were made by a number of other researchers while investing the effect of salinity on gut microbial composition in shrimp [[16], [17], [18], [19]]. Holt et al. [20] carried out studies on the role of the gut microbiome in the regulation of shrimp health and disease management. In their review, they discussed that manipulating intestinal microbes using beneficial microbial supplementation, there can be a positive effect on the growth and survival of shrimp. Several workers have reported that the alternate method to prevent disease occurrence and improve shrimp health is through modulations of the gut microbiome to stimulate the growth of beneficial bacteria. Gut microbiome species can play important roles in the health and immune response of farm animals [21], and commercially important aquatic animals (Tarnecki et al., 2017). There are several factors like culture environments, growth, and developmental stages, and health status that can influence gut microbial composition in shrimp, and a strategic research approach is required to understand this complex interaction [16,18,19,22]. Uengwetwanit et al. [23] while studying the growth performance of shrimp P. monodon and its correlation with gut microbiome composition, transcriptome, and metabolites reported that there was a relative abundance of bacteria like Brevibacillus, Fusibacter, and Spongiimonas in the majority of shrimp gut. Further, it was observed shrimp with different growth performances had different intestinal transcriptomic profiles and immune-related genes. Duan et al. [[24], [25], [26]] studied microbiome and transcriptome profiles in shrimp in relation to probiotic feeding and it has been reported that the correlation between microbiota and host gene expression was significant as far as immunity, digestion, and apoptosis processes are concerned. Few workers while studying the gut microbial composition of different penaeid shrimp emphasized the importance of the host-microbiome interaction directly or indirectly to reveal the mechanisms involved in various physiological activities [[27], [28], [29], [30]].
In the present review, efforts have been made to generate comprehensive and updated information on various aspects of host-microbiome interaction in commercially important finfish and shellfish. Such attempts will be quite useful for developing effective technic and tools of the microbiome for growing healthy and quality organisms in captive conditions.
2. Holobiont concept and its importance
In an organism whatever physiological changes take place during its entire life span and shape the body, are due to the resultant of the complex interactions between the combined expression of the host and its associated microbial communities, leading to the popularization of notions defined as holobiont and the hologenome [[31], [32], [33]]. A host and its microbiome community thus constitute a holobiont environment. The importance of holobiont has been highlighted in the recent past by a number of workers in most organisms including the shrimp and research in this particular niche area has become an imperative across various fields of life [[34], [35], [36]]. The holobiont term is now widely used in different contexts and applies to virtually all animals including human beings, with current research focusing mainly on human, animal, and plant holobionts. The term hologenome was introduced by Rosenberg and Rosenberg [36] to describe the sum of the host genome and associated microbial genomes. Many researchers do not consider the holobiont as a new phenomenon and earlier the same is compared to the term symbiosis [37]. However, in recent times due to a thorough understanding of the ubiquitous nature of the host-associated microbes and their significant role in host biology, the holobiont environment in the animal body has gained a lot of importance. Further, the availability of advanced molecular tools and NGS technologies have helped us in revealing the vital role in the recognition of microbes as key inhabitants in the host, and as players in biological and evolutionary processes. As host-associated microbes cannot be cultivable outside their host, environmental genomics approaches have been successfully applied to unravel the diversity and roles of microbes in organisms and in all ecosystems, whether terrestrial, marine or aquatic [38]. In the holobiont environment assembly of microbial species in the host and the mechanism that is being used to maintain such core microbial group in the gut system is an important issue for investigation. It has been reported that several environmental factors and host genetic factors have a vital role to play in determining microbial assembly [39]. Some workers have reported that the stability of microbes is determined by the transmission pattern of microbiome species in the body of the host. There is a wide range of transmission modes for microbes along a continuum from vertical to horizontal and environmental acquisition, but this important determinant of host-microbiome interactions is unknown in many systems [38]. Rosenberg and Rosenberg [40] suggested the concept of hologenome for determining the holobiont environment of the host and holobionts are considered super-organisms. Further, it is mentioned that host-microbiome interactions take place with mutual understanding and cooperation of the host and the members of the holobiont microbiome species [[32], [41], [42]].
Dittami et al. [43] reported a novel perspective model on the holobiont of marine habitat organisms. According to them, there are two important factors that govern the holobiont environment. One is the intervention of several environmental factors that determines the holobiont composition of the host and the second one is the impact and roles of different partners involved in this complex system. It is further mentioned that microbiome transmission into the host takes place in two ways i.e. vertical transmission and horizontal transmission and sometimes there can be the influence of both vertical and horizontal transmission. Vertical transmission is the maternal transmission of core microbiome species into the host and horizontal is through the environment [44,45]. Therefore, identifying the factors that shape the holobiont composition of the host and understanding their evolution also becomes very much relevant for marine organisms to decide their specificity for microbiome species preference in their body system [46,47]. Liwinski & Elinav (2020), mentioned that even the immune system of animals plays an important role in the preference of microbiome species as the animal needs to produce specific antimicrobial peptides to prevent pathogenic infection. It is also suggested that to have in-depth knowledge of the complex system of the holobiont environment, molecular interaction studies between the microbiome present in the environment and the host are very much needed [43].
3. Host-microbiome interaction in fish
While discussing the importance of holobiont research, the question remains what functions are carried out by the microbes to the host organisms resulting in phenotypic and fitness effects. So also the mechanism involved in host-microbiome interaction becomes a vital issue in understanding the functions of holobionts (Simon et al., 2019). Though there are several reports regarding the gut microbial composition of different finfish and shellfish, information pertaining to the functional properties of the microbes is scanty and poorly understood. Many earlier studies mentioned that gut microbes are capable of producing various digestive enzymes that have led to the conclusion that these communities are likely to contribute to the hosts’ digestive function [8]. There are also reports that fish gut microbes produce vitamin B12 and this has been proved experimentally [9,10]. Production of short-chain fatty acids by digesting dietary fiber with the help of gut microbes has been reported in herbivorous fish species [48]. The short-chain fatty acids contribute considerably to the host energy requirements of herbivorous fish species and in addition, also help to reduce pH in the gut system which may not be a favorable environment for certain harmful pathogens thus protecting the fish from bacterial infections [3]. The concept of the presence of core microbiome species in the gut system of fish and shellfish has gained a lot of importance because the core microbiomes have certain definite functions to perform in the body and this particular group of microbes generally never gets affected by any of the extrinsic or intrinsic factors. Although this theory of the presence of a core microbiome in the gut tract is rather speculative because there will be some microbial species that break this rule. This has been observed in some mammalian groups of animals including humans. However, this is also true in the case of fish and other animals of aquaculture species remains to be a matter of investigation. Xing et al., [11] while working on the microbiome composition of turbot fish reported some interesting observations in the context of bacterial abundance and functional analysis using metagenomics combined with 16S rRNA gene sequence analyses. They found that genes present in the Vibrio genus in the digestive tract of turbot were more abundant and performed various functions like biofilm formation, iron acquisition, and metabolism when compared with other fish like striped bass where such activities were low suggesting that differences in metabolic potential may occur in marine and freshwater fish microbiomes. Li et al. [2] while working on microbial communities and host-microbial interaction in hybrid fish-derived by crossing herbivorous Megalobrama amblycephala (♀) × carnivorous Culter alburnus (♂) in response to the dietary adaptations, mentioned that the host-microbiome interactions play vital roles host development, immunity, metabolism, and behavior. In two groups of these fishes when fed one with herbivorous diet and another with a carnivorous diet, there was no much difference in the growth rate during the early developmental stages of both the groups, and a large of number bacterial communities were observed in the gut system and the dominant groups were Acinetobacter, Gemmobacter, Microbacterium, Vibrio, and Aeromonas. The abundance of other bacterial groups like Firmicutes, Actinobacteria, and Chloroflexi was also observed which can be correlated with host growth level. Further, it was mentioned by using Spearman's correlation analysis they could establish that the differentially expressed homologous genes in the gut system of these fishes associated with cell growth, immunity, and metabolism was related to the dominant group of microbiome present. These results indicated that host genetics-gut microbiota interactions contribute to dietary adaptation in hybrid fish. Therefore, we need in-depth knowledge of the core microbiome in the gut system of several finfish and shellfish and their metagenomic functions to draw definite conclusions on such aspects.
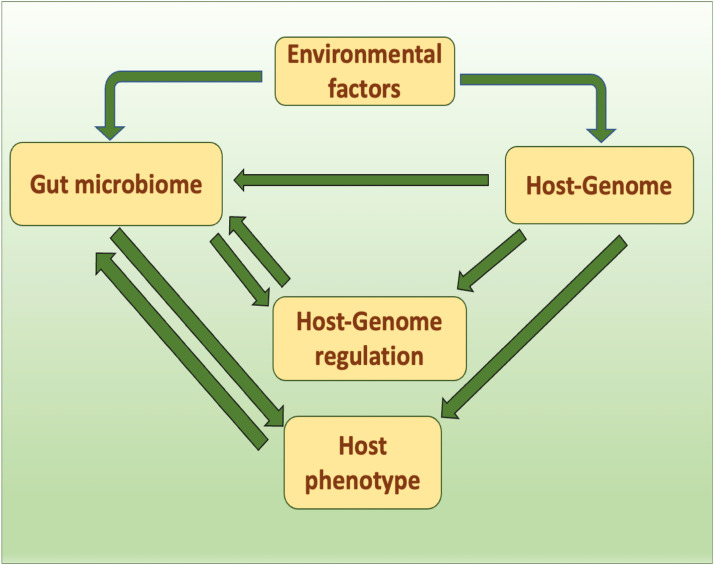
In order to understand the host-microbiome interaction, a number of workers suggested carrying out studies on the production of phenotypic traits as a basis and its correlation with the microbiomes present in the gut system of fish or shellfish. Phenotypic variations in the animals have been correlated with the structural composition of the intestinal microbiome as the microbiome species present in the gut system produce a number of biochemical compounds including short chain fatty acids, vitamins, and amino acids, and cooperate with the host intestinal immune system to inhibit the invasion of exogenous pathogenic microorganisms. The animal's host genome determines the selection of specific microbiome species and completes the construction of the microbiome community in the gut system (Fig. 1). Therefore it is mentioned that host genes are the designers of the structural composition of the gut microbiome [12]. Host genetics-gut microbiome interactions contribute to food digestion, physical development, and environmental adaptation [2,49]. While working on host-microbiome interaction in zebrafish, Robinson et al. [50] mentioned that there are several intra and extra-host factors that determine bacterial selection and adaptations in the digestive tract. Based on these findings by several other workers also, it was inferred that intestinal microbes play a very significant role in genetic evolution and growth and development not only in fish and shellfish but also in other animals [51]. There is also evidence that the gut microbiome has a great influence on brain function including stress responses and behavior further it is reported that the brain receives the signals through the gut microbiome through various pathways [52]. Including the production of microbial metabolites and peptides, immune activation, and activation of the vagus nerve in the gut itself. Research has shown that in fish, as in mammals, the intestinal microbiome community affects the hypothalamic-pituitary-adrenal axis and the management of all the stress responses they execute effectively through this mechanism [13,14]. Similarly in fish (gilthead sea bream) also, it has been reported that the intensity of the growth performance always depends on the presence of specific bacterial species and their abundance in the gut system. Zhang et al. [53] also mentioned that bacterial species with different abundances in the intestine of fish showed distinct growth rates performance. Even in the genetic selective breeding of fish, it is observed that these fishes have the distinct presence of microbial species in their intestinal tract [54]. In many organisms including fish and shellfish, the intestine serves as a multifunctional organ involved in the process of nutrient uptake, pathogen recognition, and regulating intestinal microbiome composition. Several workers have reported that due to the multifunctional role of the intestine, structural changes occur in the histology of the intestine [55,9]. The influence of gut microbiome flora on various host functions and structural changes in the gastrointestinal tract has also been reported earlier by a number of workers [56]. However, each type of physiological activity and its correlation with the gut microbiome structure have to be worked out in detail at the genetic expression and molecular levels. The mechanism between the host and its microbiome community interaction is given in Fig. 2.
Fig. 1.
Host-microbiome interaction and phenotypic expression possible pathway.
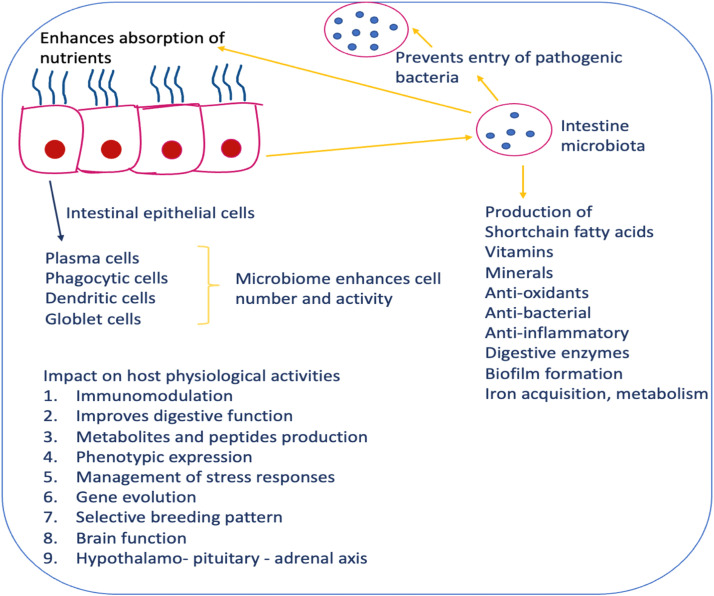
Fig. 2.
Host-microbiome interaction possible mechanisms.
3.1. Microbiome associated with other body parts and their interaction
The microbiome community associated with skin and other tissues and their interaction with the host and mechanism involved is a matter of thorough investigation in both fish and shellfish. There is a good amount of information available with regard to microbes associated with the skin of fish species. It is well known that the mucous layer of the epithelium of the skin provides a mechanical and chemical protective barrier against pathogenic organisms and it has been reported that this mucous contains antibacterial agents like immunoglobulins, defensins, lysozyme, lectin-like agglutinins, and a variety of antimicrobial peptides that provides a broad spectrum of antimicrobial activity. In addition, due to its viscous nature, mucus can bind and trap microbes and the high mucus secretion rates can effectively remove microbes from the epidermal surfaces (Perez et al. 2010). It is mentioned that mucus which contains several antibacterial compounds are generally metabolized by the mucus-dwelling bacteria in a mutualistic relationship between the fish and cutaneous microbiota. The microbes present in the skin interfere with pathogenic bacteria by antagonistic activity and competition for adhesion sites [[57], [58], [59]]. Depending upon the abundance of cutaneous bacteria and externally invaded bacteria present in the mucous and their interaction with the host will decide the health issues caused by the pathogens in fish. Further insight and knowledge revealing how the mechanism works among these three components i. e. skin bacteria, invading bacteria in the mucous and the host is a matter of future investigation.
Guivier et al. (2019) worked on the taxonomic and functional microbes associated with the skin, gills, and gut tissues of the fish Parachondrostoma toxostoma collected from the wild and basin environment and were compared. It was observed that the microbes dwelling in the skin in the wild population of fish showed diverse and variable bacterial community composition than the gut microbiome group. These findings indicated that the different factors actually determine the bacterial composition communities for the specific tissues. Further, it is reported that by using the Picrust tool, they analyzed taxonomic variation of the microbiota composition in the fish populations’ from river basins and found that the enrichment of microbes in different tissues particularly in the gut system could be correlated with different metabolic pathways. In particular, carbohydrate metabolism was more represented and xenobiotic biodegradation pathways were less represented in the gut microbiota. These findings indicate that environmental features are very important for determining the microbial composition and potential prediction of functional capacities of the microbiome in fish. A similar type of work on comparative analysis of the microbial composition associated with the gut and gill tissues of filter-feeding fish, (Hypophthalmichthys molitrix and Hypophthalmichthys nobilis) residing in different habitats has been carried out by Kuang et al. [60]. The main aim of this study was to find out whether the surrounding sediment and water of the habitat ecosystem influence the fish microbial community in an unfed aquaculture system. The study revealed that there was a significant variation in the microbial composition in both the fish and microbial diversity was significantly higher in the habitat samples than in the fish host samples and was significantly higher in the gills than in the gut. The gill and gut possessed unique core microbial groups and predictive functions in comparison to the surrounding environment. These findings should improve our understanding of the composition, diversity, and function of aquatic host microbiome and their associations with habitat [60].
Considerable work has been carried out on the microbiome associated with gills in fish and shellfish belonging to marine and freshwater environments and it is mentioned that the gills support a high population of a wide range of bacterial species [[60], [61], [62], [63], [64]]. The presence of non-pathogenic microbes with antimicrobial properties on the mucosal surface of the gills has also been reported by a number of workers [64]. It is observed that due to the continuous flow of water current over the gills, habitation and colonization of microbes are restricted, but the microbes that have been identified on gill tissues have been reported to be typical of the same order or higher than those present on the skin and of a lower order than those reported in the gut tract [65]. The presence of aerobic bacteria was more dominant in the gill filaments of several fish like Atlantic salmon, turbot Scophthalmus maximus, striped bass, Nile tilapia Oreochromis niloticus, brown trout, pike, Atlantic mackerel, and rainbow trout. Several studies have also reported that the microbial diversity of fish gills is lower than that of fish skin [66,67]. In a cultivable variety of fish in captive conditions, it has been observed that the microbial composition of gill tissue is almost similar to surrounding waters [[65], [68], [69]]. There are also reports mentioning of similarity of microbes of gill tissues with that the microbes found in the skin and gut tract [70]. The bacterial components identified in the gill tissues include Proteobacteria (Pseudomonadales, Enterobacteriales, Aeromonadales, Vibrionales, Aeromonadales, Rhizobiales, Burkholderiales, Pasteurellales, Caulobacterales, Xanthomonadales), Actinobacteria (Actinomycetales), Firmicutes (Bacillales, Lactobacillales, Erysipelotrichales), and Bacteroidetes (Flavobacteriales, Bacteriodales). Several workers have suggested that there are several factors like environmental stress, water quality, pollutants and contaminants, nutritional deficiencies, overcrowding, trauma, parasitism, or primary viral infections that could affect the microbial diversity of the gills and skin [71].
While discussing the mechanism involved between the gill microbiome and the microbes that is dwelling in the fish body, Zhang et al. [72] and Salinas et al. [73] made a breakthrough and revealed that the microbiome present in the fish produces a particular antibody in their gills in response to the pathogenic microbiome and microbes present in the mucosal layers of the gills also produce the same antibody called ‘immunoglobulin IgT’, the function of which was discovered by Zhang et al. [72]. It has been reported that immunoglobulin IgT is the primary immunoglobulin produced in response to the pathogenic microbiome in fish guts and skin and prevents any damaging effects of harmful pathogens on the fish. As the gills are respiratory organs having mucosal surfaces, the researchers wanted to see if similar immune defense mechanisms were present there. They observed that in the rainbow trout IgT was more abundant in the mucus of the gills, though they also noticed the presence of other immunoglobulins like IgD and IgM. By examining the mitochondria of the gills it was observed that IgT was the primary antibody coating bacteria in the gills. In order to confirm the role of IgT in response to pathogens in the gills, the fish were exposed to WSSV and it was observed that the IgT was found to be more abundant in the mucus of the gills. Fish that survived infection also had a significant increase in IgT-producing B cells in their gills, an additional sign that the IgT response was key to fighting the pathogens. These results indicated that there is a possibility of developing cheaper fish vaccines so-called ‘bath vaccines’ that are simply dropped in the water and the same will be absorbed by gills and skin very effectively for the protection of fish from infectious diseases. Legrand et al. [74] while working on skin and gill microbes in yellowtail kingfish reported that the microbial composition and health status of the gut have control over the assemblage of the microbes of skin and gills. It is mentioned that understanding the skin and gill microbial assemblages and the factors that control this assemblage may provide useful insights into the dynamics of fish host-microbial relationships, and may also reveal underlying changes in the health status of the host. When fish are infected with pathogenic microbes and lose their microbial diversity in the gut tract, its impact will be also reflected in the microbial composition of the skin and gills. It is further mentioned that by looking into the microbiome composition of the skin and gills one can assess the health status of the host. However, future research is necessary for understanding the underlying functional contribution and connectivity of these mucosal microbiomes across the skin, gills, and gut and their interplay with the host during changes in health.
There are published reports regarding fish microbiome-mucosa interaction and considerable work has been carried out covering these aspects. Besides several functions of the gut microbial species, it has been recognized that the microbiome plays a significant role in mucosal barrier function to prevent the entry of pathogens into the host. In this context, the work carried out by Bates et al., [75] and Rawls et al., [76] on zebrafish has shown that when zebrafish larvae were reared in germ-free environments with no microbial interactions, the development of the larvae was not proper and in such animals, the gut tract fails to differentiate, enteroendocrine cells and goblet cells in intestinal layers could not be differentiated, cells lack brush border, there was reduced intestinal alkaline phosphatase (IAP) activity, reduced epithelial cell turnover rates, immature patterns of glycans on the enterocytes was seen and a loss of epidermal integrity was also observed. These characteristics ultimately lead to a failure of the intestine to uptake protein macromolecules. Rawls et al. [77] found that there are 212 genes that govern different functions like immunity, nutrition, metabolism, and other physiological activities present in the gut tract of zebrafish and these genes were regulated by the presence or absence of the microbes in the gut system. Further, these authors have also investigated host-microbe cross-talk at the mucosal interface that determines the nature of the subsequent host-microbe relationship. The mechanisms and the key molecules that are involved in this complex process have been identified. However, further research is required to improve our understanding of host-microbe interactions not only at the mucosal interface but other physiological functions.
Perez et al. [5] carried out work on the mechanism of the host-microbiome interaction within the teleost fish gastrointestinal tract. It is mentioned that the microbes present in the gastrointestinal tract have direct implications on the health of the fish and the mucosal surfaces in the intestine are the main sites of the interaction between environmental bacteria and host microbes. The gut-associated lymphoid tissues are capable of discriminating pathogenic bacteria and commensal microorganisms and colonization of normal bacteria in the intestinal mucosal surfaces has a positive effect on immune regulatory functions. However, any disturbance in the immune system due to dysbiosis in the microbial composition in the intestinal tract may lead to the development of diseases. In such situations, the use of probiotics has been suggested to restore the normal microbiota to regain health by preventing diseases. A number of workers have assessed the microbiome composition of the gastrointestinal tract of several marine and freshwater fishes and reported the presence of bacteria from the members of the genera Aeromonas, Alcaligenes, Alteromonas, Carnobacterium, Flavobacterium, Micrococcus, Moraxella, Pseudomonas, and Vibrio that constitute the predominant intestinal microbiome group of marine fish species and in contrast, the intestinal microbial composition of freshwater fish species found to be dominated by members of the genera Acinetobacter, Aeromonas, Flavobacterium, Lactococcus, and Pseudomonas, representatives of the family Enterobacteriaceae, and obligate anaerobic bacteria of the genera, Bacteroides, Clostridium, and Fusobacterium [78,79]. Pamer [80] while working on immune responses to commensal and environmental microbes mentioned that gut microbes are considered an ‘extra organ’ of the host playing a vital role in immunomodulatory regulation in response to pathogenic microbes. In fact, the mucosal surfaces of the gastrointestinal tract are the main sites in which environmental microorganisms and antigens interact with the host, through intensive cross-talks [81]. Therefore, the first line of defense is provided by the mucus layer over the epithelial cells which secretes various antimicrobial substances including complement components, mucins, enzymes, piscidins, and defensins [82,83]. The gut-associated lymphoid tissue in the gastrointestinal tract develops the mechanism to distinguish pathogenic bacteria and commensal microbes and determine the responses of immune regulation depending on the intensity of the pathogenic bacterial population. The lymphocyte cells present in the lymphoid tissue play a vital role against harmful foreign pathogens and here also immunoglobulin A is most abundantly produced and protects the host. It has been discovered that the commensal bacteria that are present in the intestinal mucosa are responsible for the development of gut-associated lymphoid tissue and in the absence of some of these bacteria called luminal bacteria, immunoglobulin A is not produced. Some workers have reported that intestinal epithelial cells are capable of recognizing ingredients of microbes through the expression of the pattern of receptors and these receptors are defined as Toll-free receptors [84,85]. These Toll-like receptors actually regulate the immune cells that initiate or enhance the immune responses and the system.
4. Host-microbiome interaction in shellfish
In shellfish also the gut microbial community interacts directly with planktonic microbes available in aquatic ecosystems and therefore, characterization of the gut microbiome community of aquatic organisms is a priority to understand host-microorganism interactions and the corresponding relationship with the surrounding microbes. Additionally, the mechanisms underlying host-microbiome interactions are pivotal in our understanding of the functioning of holobionts. Considerable information is available on host-microbiome interaction in soft corals and sponges and it has been reported that the bioactive compounds which are produced in such organisms are regulated by the microbes present in the host's gut tract and their symbiotic relationship [86,87]. While working on sponges, Pita and colleagues [88] emphasized how key functions, provided by holobionts to ecosystem functioning can be affected by changes in microbial composition. By analyzing metagenomics and host transcriptomic datasets, it is possible to analyze both the host and its microbiome interaction and the mechanism involved in the process. Zhang and Sun [89] while working on host-microbiome interaction in response to the immune system in four different species of shrimp reported that there was a huge variation in the core gut microbial composition. These changes in the core gut composition and structure were due to changes in the environmental factors that led to infection with the WSSV virus. However, the alterations of the core microbe group among different shrimp showed the same trend and are related to immune function in the prediction of its metabolic function potential. Further, it is mentioned that when the metabolic analysis was done, nine metabolites possibly produced from the gut microbes were significantly up-regulated after viral infection, and these metabolites had antiviral properties. These findings indicate that these metabolites are responsible for maintaining the immune homeostasis of the host and the function of the gut microbes in protecting the shrimp from pathogens.
Holt et al. [20] carried out work on the role of the gut microbiome in the regulation of shrimp (L. vannamei, P. monodon) health and disease and also on how the gut microbial composition changes in response to different shrimp pathogens. While discussing these issues, they have highlighted the vital role of the gut microbiome in controlling a number of key physiological processes including digestion and immunity. Therefore, it is suggested that instead of using antibiotics for controlling diseases and health management in shrimp, the alternate method is the manipulation of the gut microbiome. Microbiome supplementation has also been demonstrated with positive effects on the growth and survival of several different commercial species, including shrimp. Chaiyapechara et al. [15] studied the gut microbiome of shrimp P. monodon and also transcriptome analysis of the microbes in response to different variations in the salinity levels. They observed that different salinities of the rearing environment of the shrimp have a significant influence on certain species of gut microbiome. Acclimatized Shrimp from 20 ppt, when transferred to 10 and 30 ppt for a period of 10 days, showed a relatively similar microbial composition with the dominance of Proteobacteria (83.4%), Bacteroidetes (8.1%), Planctomycetes (3.2%), Verrucomicrobia (2.5%), and Firmicutes (1.5%). The most abundant genus was found to be Vibrio and its level was lower at 10 ppt and higher at 30 ppt. Other dominant genera in the gut tract not affected by the change in salinity were Pseudoaltermonas and Tenacibaculum. When the microbes of the surrounding water were analyzed simultaneously at different salinity levels, there were significant changes in the microbiome composition. For assessing host-microbiome interaction, transcriptome analysis was carried out for the gut microbiome composition in response to different salinity levels, and it was observed that genes involved with stress and immune responses were differentially expressed and there was also a significant correlation between pathogenic vibrio and genes relating to innate immunity. The results indicated the influence of water salinity on both the gut microbiome and transcriptome of shrimp. It remains to be proven whether the host shrimp, under different rearing salinity, actively controls the microbes via its expressed genes or passively reacts to the changing microbiome. The interaction between the microbiome in the gut of aquatic animals and rearing water remains to be further explored. Several other workers also performed similar experiments to find out the influence of rearing water microbial community on the shrimp, particularly at the early developmental stages of P. monodon, and L. vannamei and it has been reported that the microbe structure between animals and rearing water showed significant differences [16,22]. However further research is required on these aspects for a better understanding of the microbiome and the host-microbiome interaction, which will help to support the sustainability of aquaculture production.
Wen-Fang Dai et al. [90] reported starvation stress effects on host gut microbiome interaction in shrimp in relation to digestion and immune activities. The results indicated that in starved shrimp digestion activities were lower than the normal shrimp, while immune activities were higher. These changes in the digestive enzymes have been correlated with the structural changes in the gut microbiome of the shrimp. It was also observed that there was a significant decrease in the functional pathways involved in carbohydrate and protein metabolism, so also in glycan biosynthesis, lipid and enzyme metabolism. These results suggest that under the influence of harmful pathogens, there is an increase in the susceptibility of starved shrimp leading to the chances of more infection of the pathogenic bacteria and further it also reflects a novel insight into the host-microbiome interaction mechanism of the gut microbes in response to starvation stress. The host-microbiome interaction has also been correlated with the selective breeding of fish and shellfish by a few workers. Fan & Li [91] while working on the shrimp L. vannamei reported that the growth performance and weight gain always depend on the structural composition of the gut microbes and further the breeding activity of shrimp has been correlated with gut microbial species.
5. Gut-microbiome and its impact on host health
The concept of the microbiome is increasingly recognized for its major role in host health, and an important field of application of holobiont research that deals with the prevention and therapy of diseases based on treatments restoring altered microbes. A Number of researchers have suggested use of healthy microbes or modulation of microbes in the gut tract of fish and shellfish by using probiotics, prebiotics, and synbiotics or by developing a biofloc system, for controlling the spread of diseases among cultivable organisms and promoting healthy growth in captive condition during aquaculture farming [1,[92], [93], [94]]. In recent studies, it has been reported that the interactions between the gut microbiome and the host are an integral part of the development, maintenance, and effective functionality of the intestinal mucosa and gut-associated lymphoid tissues. The microbes present in the gut tract of the fish also provide protection against harmful pathogens in the gastrointestinal tract and aid the host's digestive function via the production of exogenous digestive enzymes and vitamins. The use of probiotics can stimulate immune responses, enhance growth performance, feed utilization, digestive enzyme activities, antioxidant enzyme activities, gene expression, disease resistance, larval survival, gut morphology, modulate GI microbes and mediate stress responses. Regarding the use of prebiotics, synbiotics, and biofloc technology applications for improving the immune status and the quality of fish production, the literature available is scanty. Additionally, the probiotic and prebiotic mechanisms which mediate host benefits at the mucosal interface are poorly understood. Hence, future studies should focus on these interactions to provide a better understanding of how to extract the full potential of biotic applications to promote immune function among cultivable species of aquaculture importance [1,20].
6. Manipulation of the gut microbiome using supplementary diet
It has been now well established that any dysbiosis in the microbiome composition in the gastrointestinal tract of fish or shellfish creates a negative impact on the regulatory functions of the immune system and other physiological processes leading to increased chances of infection and spread of diseases. In earlier studies also, it was shown that the gut microbes have close relationship with gene expression patterns, specifically its involvement with stimulation of epithelial proliferation, an increase of nutrient metabolism, and innate immune responses. Therefore, in recent times much attention has been focused on the role of probiotics in the induction or restoration of a disturbed microbiome to its normal beneficial composition (Rawl et al. 2007, [95]). The use of probiotics as an alternative to antibiotics in aquaculture farming is gaining a lot of importance and has become now common practice [96]. Several microbial resources have been identified for the preparation of probiotics and among them, Gram-positive and Gram-negative bacteria, bacteriophages, microalgae, and yeasts have all been tested as potential probiotics in fish [97]. Other sources are LAB Bacillus, Lactococcus, Shewanella, and Aeromonas genera are being used for the preparation of probiotics [[98], [99], [100]]. In a review, Carnevali et al. [101] gave very exhaustive information on the use of probiotics in teleosts fish for the manipulation of the gut microbial composition and thereby enhancing the growth rate and modulation of immune status in fish [[18], [102], [103], [104]]. There are indications that feeding with probiotics for 2 weeks, resulted in a higher survival rate of rainbow trout even when they were challenged with Aeromonas salmonicida. In addition, it was also observed there was a correlation between colonization of the intestinal microbiome community with the probiotic strains used, and also increase in innate immune responses by more phagocytic activity and alternative complement pathway activity [57,105]. The use of several other probiotics prepared from different strains of bacteria (L. mesenteroides and Lactobacillus plantarum) also showed positive effects from protection against infectious pathogens in rainbow trout [106]. It has been also reported that probiotics made out of multistrain species of bacteria have been proven to have synergistic beneficial effects on the host health, although the underlying mechanisms remain unclear. However, in higher vertebrates, the use of multistrain probiotics has shown more positive effects like greater survival, growth, viability, or adhesion to mucosal surfaces of one species in the presence of another species, production of different enzymes, etc. [107,108]. There are also pieces of evidence indicating that probiotic feeding helps in activating the anaerobic microbiota of the gastrointestinal tract in fish which produces a number of vitamins, and a variety of digestive enzymes [109,110]. The use of probiotics for the growth of the aquaculture industry particularly, in the management of outbreaks of diseases, nutritional supplements, and immunomodulation has been highlighted by several workers in the recent past [[111], [112], [113], [114], [115], [116], [117]]. Several strains of microbial species like Lactobacillus, Enterococcus, Bacillus, Aeromonas, Alteromonas, Arthrobacter, Bifidobacterium, Clostridium, Microbacterium, Paenibacillus, Phaeobacter, Pseudoalteromonas, Pseudomonas, Rhodosporidium, Roseobacter, Streptomyces and Vibrio have been mentioned for the preparation of probiotics. In shellfish particularly in shrimp, Leptopenaeus vannamei, Restrepo et al. [118] developed a probiotic from Vibrio diabolicus bacterium for preventing the spread of infection of acute hepatopancreatic necrosis disease (AHPND) virus. In this study, it was shown that gastrointestinal microbiome composition was totally different in healthy shrimp compared to the infected ones with AHPND. There are also reports mentioning the impact of probiotic feeding on the modulation of gastrointestinal microbial composition in shrimp. Rajeev et al. [92] while discussing the impact of a healthy microbiome in shrimp aquaculture, gave a detailed account of the usage of probiotics for accelerating the quality and sustainable growth of the shrimp industry and also for the effective management of preventing the spread of diseases in the shrimp farming sector. Holt et al. (2020) while discussing the role of gut microbiome in preventing the diseases and health management in shrimp, emphasized that manipulating the gut microbiome composition using probiotics was quite beneficial for improving the host's health. Similar observations were made by number of other researchers who also demonstrated that supplemental bacteria in the form of probiotics can directly affect and eliminate harmful pathogens in shrimp, P. monodon and L. vannamei ([119,120]; García [121]; Mazón-Suástegui et al. [122]). In the shrimp L. vannamei a probiotic preparation with a combination of bacterial species like Streptomyces and Bacillus was found to be very effective in producing antibacterial gut bacteria (Mazón-Suástegui et al. 2019). The use of probiotics made from the lactic acid bacteria, Lactobacillus Plantarum was also found to be very effective in preventing the infection caused by Vibrio harveyi in the shrimp P. monodon and L. vannamei. [123,124]. Successful use of several probiotics made from different strains of bacterial species and their impact on modulation of the gut microbiome composition, immunity improvement, improvement in the digestive enzymes, survival rate, and hastening of the growth process in the number of shrimp species has been reported by a number of researchers ([[125], [126], [127], [128], [129]], Riya et al. 2020). Now many probiotics with different combinations are available in the market for their use in the shrimp aquaculture of industry, however, it is also a fact that the application of general combinations of bacterial strains in probiotics may not be beneficial to the host [[130], [131], [132]].
A number of workers also suggested the use of prebiotic supplementation in place of probiotic strains for developing a beneficial microbiome in the gastrointestinal tract in shrimp. Zhang et al. [133] used prebiotics like mannan oligosaccharides in the shrimp diet and it has been reported that the use of this particular prebiotic significantly improved the growth rate and increase the weight of the shrimp, besides, there was also an increase in the length of intestinal microvilli which accounted for creating more mucosal surface area in the intestinal tract for effective nutrient absorption. The use of other prebiotics like oligosaccharides isolated from grain, fruits, and vegetables and also inulin when tested in different shrimp species, showed positive effects in developing a beneficial microbiome community in the gut tract particularly at the early developmental stages while rearing the shrimp in captive conditions [134]. Some of the prebiotics used in the fish farming sector are fructose oligosaccharides, short-chain fructose-oligosaccharides, oligo-fructose, mannan oligosaccharides, trans-galacto-oligosaccharides, inulin, galacto oligosaccharides, Xylo oligosaccharides, arabinoxylo-oligosaccharides and isomalto-oligosaccharides [135]. Some workers also suggested the use of a combination of pro and prebiotics which is termed synbiotics can also stimulate immune responses against viral pathogens [136]. While working on the fish microbiome Seyed Hossein et al. [137] mentioned that microbial feed additives including synbiotics when fed in the form of a diet showed very positive effects in producing antioxidant enzymes and also improvement in the immune system. Su et al. [138] while discussing antibiotic-resistance genes in shrimp aquaculture reported that, the antibiotic-resistant genes in adult shrimp have been found to be more abundant than the juvenile shrimp and these genes are transferred from gastrointestinal microbes into the culture environment and this kind of horizontal transfer of gene can spread resistance between microbes in the culture systems [139,140]. However, further research is needed to establish this fact. In the literature, the information available is scanty on how probiotics influence on developing beneficial microbiome in the gastrointestinal tract of fish and shellfish and also on the mechanism involved in the replacement of beneficial microbes in place of non-beneficial microbes and further, the interaction between the host and the replaced microbiome. Some workers have reported that after feeding the specific probiotics, the beneficial microbial species colonize and increase their density in the gut tract with result that the non-beneficial microbes either slowly get eliminated or interfere with harmful pathogens, producing positive effects [92,141].
Considerable work has been carried out on the modulation of the gut microbiome of fish and shrimp in captive conditions by using the technology of biofloc. It is believed that different types of probiotics and other health-promoting bioactive compounds within the biofloc like vitamin C, poly-β-hydroxybutyrate, and carotenoids help in improving immunity to disease resistance in many cultivable organisms [[142], [143], [144], [145]]. It has been reported that the inclusion of dried biofloc in the diet of the shrimp L. vannamei and P. monodon has led to developing greater resistance to the infection of Vibrio harveyi and V. parahaemolyticus, compared to those fed a biofloc-free control diet ([146,147]). Kheti et al., [148] while working on the effect of the use of biofloc in the diet of freshwater fish, Labeo rohita reported that the fish developed greater resistance to the virus Edwardsiella tarda when compared with the normal fish. Qiao et al. [149] while working on the goldfish, Carassius aurata also observed that when poly-β-hydroxybutyrate was obtained from biofloc and used in the diet of fish, they found that the gene expression for immunity was upregulated reducing the infection of herpes virus and the bacillus species of the gut microbial composition increased in their density in the gut tract of the fish. Indeed, the consumption of poly-β-hydroxybutyrate produced by microbes has similarly shown immunological benefits in various other aquatic animals (e.g., [[150], [151], [152]]). Luo et al., [153] while working on tilapia fish, Oreochromis niloticus observed that fish did not show much difference in the microbial community even after feeding with biofloc containing poly-β-hydroxybutyrate as a carbon source. However, the implications of poly-β-hydroxybutyrate as a carbon source on immunity/disease resistance have not yet been investigated.
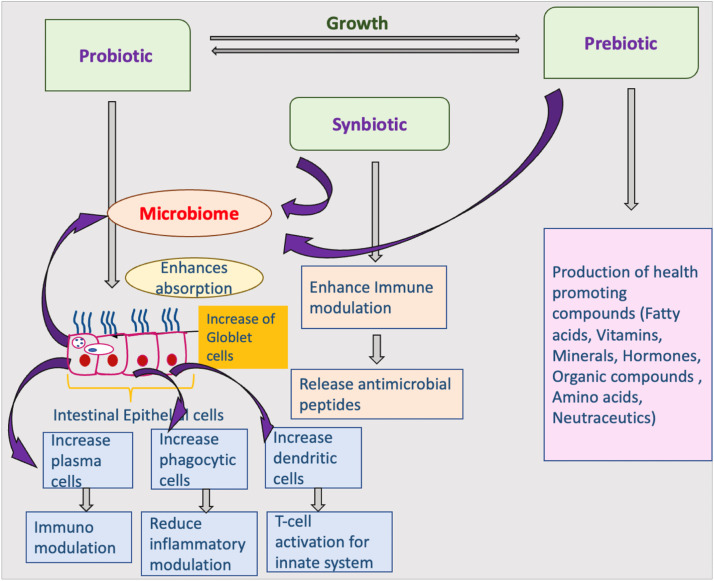
It has been mentioned that the presence of biofloc is quite beneficial in stimulating the innate immunity in shrimp [[154], [155], [156], [157], [158]] and hence the shrimp grown in the biofloc environment were found to be more resistant to pathogensshrimp . The effect of biofloc on shrimp's innate immunity has been proposed to act through ingestion of biofloc microflora [114,159] and micro, P. monodon and L. vannamei bes present in the biofloc play a vital role in stimulating immunity. However, the mechanism involved in the interaction of the host and biofloc microflora is not clear and requires fuvannamei rther investigation. A recent study demonstrated that gut bacterial communities of biofloc-grown shrimp were not identical to those of shrimp cultivated in clear seawater [155]. While previous microbiome studies have primarily focused on some prime factors shifting bacterial community in the shrimp gastrointestinal tract (Chen et al. 2017; [18,27,160,161]), functional aspects of the gut microbiome on shrimp health remain largely uncharacterized. Although biofloc has been shown to alter the shrimp gut microbiome and promote innate immunity in the host, however, there is no direct evidence and these findings were accumulated from different investigations (Carodona et al. 2016). Tepaamorndech et al. [162] while dealing with metagenomic studies in the biofloc microbes and their impact on the gut microbiome and immunity in white shrimp, Leptopenaeus vannamei observed that the biofloc system has a greater impact on the bacterial population and contained vibrios as the dominant member in the shrimp. The presence of biofloc significantly promoted an abundance of Vibrio and suppressed Photobacterium density in the shrimp gut. It was also observed that genes involved in the digestive enzymes were upregulated in biofloc-grown shrimp. In addition, shrimp also showed an elevation in hemocyte number and antioxidant activity, which possibly contributed to the high survival rate when they were cultured in captive conditions. In this regard, some work has been carried out on the quantity of nutrient absorption from the biofloc system and it was observed that herbivorous animals are better in absorption than carnivorous ones [163,164]. Fig. 3 describes the possible mechanism between the host-microbiome interactions in relation to supplementary diets like probiotics, prebiotics and synbiotics.
Fig. 3.
Host-microbiome interaction mechanism in relation to probiotics, probiotics, and synbiotics.
7. Host-microbiome interaction and aquatic environment
Several workers have reported the importance of the gut microbiome in controlling various physiological functions including growth, breeding and immune system. Outbreak of diseases in cultivable organisms has been correlated with the composition of the microbial community in the surrounding aquatic environment [1,165]. Number of workers have also pointed out that changes in the microbial community in the surrounding environment have a greater impact on the host gut microbiome assembly and also on the mechanisms involved in the host's microbiome interaction with invasive microbiota [[166], [167], [168]]. It is a fact that a number of intrinsic and extrinsic factors play a vital role in determining the composition and structure of the gut microbiota of fish and shellfish and in most other cultivable organisms. The interaction between the environmental factors and the gut microbiome has been reported to be the most complex one [169] and therefore it is crucial to understand the mechanism involved between these two modes of identity. The intrinsic factors that affect the composition of the gut microbiome are host genetics, developmental stage, physiological conditions, and starvation and extrinsic factors include diet factors and environmental factors. The interaction between the surrounding water environment and the intestinal microbes is expected to be two-way directional. Chen et al. [170] in their recent review paper reported the driving mechanism involved in the interaction between the gut microbiome of fish and the microbiome present in the surrounding water ecosystem. They observed that the interaction between the fish gut microbiome and the environment has a vital role in microbial adaptation to the environment and also composition and functions which further determines the health status of the fish. Similar observations have also been made by other researchers working on cultivable aquatic organisms and it was emphasized that the assembly pattern of the gut microbiome in aquatic animals is usually a deterministic process [[171], [172], [173]]. The composition of the microbiome in the water column of the aquaculture system always depends on the physico-chemical parameters like temperature, salinity, pH, chemical oxygen demand, total nitrogen, phosphorus, carbon, and inorganic nitrogen [165,174] and any disturbance in these parameters alters the structural composition of the microbiome community of the ecosystem that will further influence the profile of gut microbiome composition of fish and shellfish. In several cultivable species like tilapia larvae [175], pearl oyster, Pinctada fucata [[176], [177]], silver carp (Hypophthalmichthys molitrix), and bighead carp (H. nobilis) [178], it has been observed that the gut microbiome always gets influenced by the surrounding aquatic microbial ecology. The microbiome present in the surrounding ecosystem plays an important role in providing essential ecosystem services and energy to these systems regulating the gut health of the organisms [170,174]. Hou et al., [179] mentioned that the microbiome present in the aquatic ecosystem performs various functions like the productivity of the ecosystem, water quality control, controlling the nutrient cycle, and defense against pathogenic microorganisms. For this purpose, it is necessary to understand how this microbiome community functions at the genome and cellular levels in the environment and also in animals in response to many abrasive factors in the aquatic environment. It has been noticed that the gut microbiome of tilapia fish, Oreochromis niloticus was found to be almost similar to the microbes present in the surrounding environment [180] whereas in the case of shrimp L. vannamei the gut microbes was found to be more closely related to the microbes present in the sediment [17,181,182]. All these studies indicated that the water and sediment of the aquatic ecosystem are the primary sources of deriving the microbiome resources in the environment and also in the gut tract of fish and shellfish [170]. Though such a relationship of the gut microbes and the microbes present in the surrounding aquatic ecosystems has been established by a number of workers, however, when the proportion of pathogenic microbes increases, the outbreak of diseases has been observed with severe damage to the aquaculture yield. Therefore, there is a need to understand the underlying regulatory mechanisms of such pathogenic bacteria present in sediment and water with that of the microbes present in the gut system of the cultivable organisms [165,183]. Li et al. [2] already pointed out that the microbes present in the surrounding environment plays a very significant role in shaping the gut microbiome composition and structure and also the functionality controlling the various physiological activities in organisms. Any dysbiosis in the environmental factors has been shown to affect bacterial growth and this has been worked in several fish and shellfish wherein the gut microbiome composition was affected by thermal stress as well as low and high pH levels. A number of studies have shown that the use of antibiotic treatment caused dysbiosis of the fish gut microbial composition leading to a reduction of the growth of probiotic bacteria and even eradicating normal bacteria [[184], [185], [186]]. It has also been shown that gut commensal bacteria may stimulate intestinal cells directly through interbacterial signaling, producing an immune response leading to microbiome dysbiosis (Perez et al., 2010, [[59], [187]]). There are several other contaminants/pollutants in the surrounding environment which may also cause dysbiosis not only in the environment but also in animals living in such habitats.
In order to understand the evolutionary basis of the symbiotic relationship between animal hosts and indigenous microbes, several workers carried out studies on the gut microbiome of the host and symbionts. Hentschel et al. [188] studied the molecular mechanism of symbiosis and pathogens to find out whether there is any difference in the manifestation of bacteria-host interaction with that of symbionts and pathogen interaction. From their findings, it was concluded that the underlying strategies of bacteria-host interaction are remarkably similar in pathogens and symbionts. Their study was based on the genetic variability protocols in both symbionts and pathogens. Identical observations were also reported by Emie et al. [189] while working on the microbiome of clownfish and their symbiotic host anemone. Kim et al. (2021) investigated the gut microbiome of 227 individual fishes. From their studies, it was concluded that the gut microbial community was more strongly shaped by host habitat adaptations rather than host taxonomy or trophic level. Though several studies have been conducted on the gut microbiome of different fishes, the information pertaining to the correlation of the composition of the microbiome with that of the host habitat is not adequate.
8. Summary and conclusions
The concept of hologenome and holobiont has gained a lot of significance in the life of an organism because the development and shaping of the whole body is the result of the physiological interaction between the host and its microbiome community available in the body. Any dysbiosis in the composition of the gut microbiome due to any abrasive factor may lead the physiological disturbance and affect the health of an organism. Though this kind of theory has been accepted by the scientific community by and large, however, there is a need to undertake further research in this niche area of science to build up its strength. In the holobiont environment assembly of microbial species in the host and the mechanism that is being used to maintain a core group of the microbiome in the gut system is an important issue for future investigation. Though a large amount of literature is available on the gut microbiome composition of fish and shellfish, the information is scanty on the functional aspects of microbial species. The concept of the presence of core microbiome species in the gut system of fish and shellfish has gained a lot of importance because the core microbiomes have certain definite functions to perform in the body and generally never get affected by any of the extrinsic or intrinsic factors. However, we need in-depth knowledge of the core microbiome in the gut system of several finfish and shellfish and their metagenomic functions to draw definite conclusions. In order to understand the host-microbiome interaction, a number of workers suggested carrying out studies on the production of phenotypic traits as a basis and its correlation with the microbiomes present in the gut system of fish or shellfish. Phenotypic variations have been correlated with the structural composition of the intestinal microbiome. However further research is needed on the animal's genome that determines the selection of specific microbiome species and shapes the composition of the microbiome community in the gut system. There is also evidence that the gut microbiome has a great influence on brain function including stress responses and behavior. Further, it is reported that the brain receives the signals through the gut microbiome through various pathways which need in-depth studies. The host-microbiome interaction has also been correlated with the selective breeding and also breeding activity of fish and shellfish by a few workers but again the available literature is scanty and we need in-depth knowledge and research in this particular area. The microbiome associated with skin and other tissues and their interaction with the host and mechanism involved is a matter of thorough investigation in both fish and shellfish. There is a good amount of information available with regard to microbiota associated with the skin of fish species. Depending upon the abundance of cutaneous bacteria and externally invaded bacteria present in the mucous and their interaction with the host will decide the health issues caused by the pathogens. Further insight and knowledge revealing how the mechanism works among these three components i. e. skin bacteria, invading bacteria in the mucous and the host is a matter of future investigation.
While discussing the mechanism involved between the gill microbes and the microbes that are dwelling in the fish body, a breakthrough has been made and it has been revealed that the microbiome present in the fish produces a particular antibody in their gills in response to the pathogenic microbiome and microbes present in the mucosal layers of the gills also produce the same antibody called ‘immunoglobulin IgT’, the function of which was discovered. Several genes have been discovered that govern different functions like immunity, nutrition, metabolism, and other physiological activities present in the gut tract and these genes have been found to be regulated by the presence or absence of the microbes in the gut system. The mechanisms and the key molecules that are involved in this complex process have also been identified. However, further research is required to improve our understanding of host-microbe interactions in relation to the genomes not only at the mucosal interface but other physiological functions in several other organisms.
Considerable work has been carried out on the modulation of the microbial communities in the intestine through dietary administration of probiotics, prebiotics, or synbiotics and also by using biofloc technology to improve microbial metabolite production, the immune signaling pathways, and the host defense mechanisms against pathogens. It has been now well established that any dysbiosis in the microbiome composition in the gastrointestinal tract of fish or shellfish creates a negative impact on the regulatory functions of the immune system and other physiological processes leading to increased chances of infection and spread of diseases. Therefore, in recent times much attention has been focused on the role of microbiome use as a supplementary diet not only in the induction or restoration of a disturbed microbiota to its normal beneficial composition but to accelerate the healthy growth of animals in captive conditions. However, we need to discover more and more sources of such microbial species for their use in aquaculture farming.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
The authors express their gratitude and thanks to the Chancellor and the Secretary, Shri Ankushrao Kadam, Mahatma Gandhi Mission University (MGMU), Aurangabad, Maharashtra, India, for his incessant encouragement and generous cooperation in providing all the facilities for writing this review.
Animal welfare statement
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required as this is a review article with no original research data
Data availability statement
The data supporting the findings of this study are available from the corresponding author upon request.
Authors contribution
A D Diwan has articulated the idea of writing this article and prepared the draft of the manuscript. S N Harke was responsible for the collection of relevant literature and helping in the articulation of the manuscript. Archana N Panche has helped in the collection of the data and also in the analysis of the interpretation in the discussion part of the manuscript. She also assisted me in designing the figures
Data availability
No data was used for the research described in the article.
References
- 1.Diwan A.D., Harke S.N., Gopalkrishna Panche, Archana N. Aquaculture industry prospective from gut microbiome of fish and shellfish: an overview. J. Anim. Physiol. Anim. Nutr. 2021;106:441–469. doi: 10.1111/jpn.13619. [DOI] [PubMed] [Google Scholar]
- 2.Li W., Zhou Z., Li H., Wang S., Ren L., Hu J., et al. Successional changes of microbial communities and host-microbiota interactions contribute to dietary adaptation in allodiploid hybrid fish. Microb. Ecol. 2022 doi: 10.1007/s00248-022-01993-y. [DOI] [PubMed] [Google Scholar]
- 3.Merrifield D.L., Rodiles A. The fish microbiome and its interactions with mucosal tissues. Mucosal Health Aquacult. 2015:273–295. doi: 10.1016/b978-0-12-417186-2.00010-8. [DOI] [Google Scholar]
- 4.Sharma P.V., Thaiss C.A. Host-microbiome interactions in the era of single-cell biology. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.569070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pérez T., Balcázar J.L., Ruiz-Zarzuela I., Halaihel N., Vendrell D., de Blas Iú, Zquiz J.L.M. Host–microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol. 2010;3(4):355–360. doi: 10.1038/mi.2010.12. [DOI] [PubMed] [Google Scholar]
- 6.Arias-Jayo Nerea, Abecia Leticia, Luis José, Itziar Lavín, Arranz Tueros Sara, Ramírez Andoni, Miguel García, Pardo Angel. Host-microbiome interactions in response to a high-saturated fat diet and fish-oil supplementation in zebrafish adult. J. Funct. Foods. 2019;60 [Google Scholar]
- 7.Nikouli E., Meziti A., Smeti E., et al. Gut microbiota of five sympatrically farmed marine fish species in the Aegean Sea. Microb. Ecol. 2021;81:460–470. doi: 10.1007/s00248-020-01580-z. 2021. [DOI] [PubMed] [Google Scholar]
- 8.Ray A.K., Ghosh K., Ringø E. Enzyme-producing bacteria isolated from fish gut: a review. Aquacult. Nutr. 2012;18:465–492. [Google Scholar]
- 9.Romero J., Ringø E., Merrifield D.L. In: Aquaculture Nutrition: Gut Health, Probiotics and Prebiotics. Merrifield D.L., Ringø E., editors. Wiley; London: 2014. The gut microbiota of fish; pp. 75–100. Eds. [Google Scholar]
- 10.Tsuchiya C., Sakata T., Sugita H. Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett. Appl. Microbiol. 2008;46(1):43–48. doi: 10.1111/j.1472-765X.2007.02258.x. [DOI] [PubMed] [Google Scholar]
- 11.Xing M., Hou Z., Yuan J., Liu Y., Qu Y., Liu B. Taxonomic and functional metagenomic profiling of gastrointestinal tract microbiome of the farmed adult turbot (Scophthalmus maximus) FEMS Microbiol. Ecol. 2013;86(3):432–443. doi: 10.1111/1574-6941.12174. [DOI] [PubMed] [Google Scholar]
- 12.Mushegian A.A., Arbore R., Walser J.C., Ebert D. Environmental sources of bacteria and genetic variation in behavior influence host-associated microbiota. Appl. Environ. Microbiol. 2019;85 doi: 10.1128/AEM.01547-18. e01547-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui X., Zhang Q., Zhang Q., Zhang Y., Chen H., Liu G., et al. Research progress of the gut microbiome in hybrid fish. Microorganisms. 2022;10:891. doi: 10.3390/microorganisms10050891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohanta L., Das B.C., Patri M. Microbial communities modulating brain functioning and behaviors in zebrafish: a mechanistic approach. Microb. Pathog. 2020;145 doi: 10.1016/j.micpath.2020.104251. [DOI] [PubMed] [Google Scholar]
- 15.Chaiyapechara S., Tanaporn U., Sopacha A., Phimsucha B., Metavee P., Waraporn J., Siriporn T., Nitsara K., Wanilada R. Understanding the host-microbe-environment interactions: intestinal microbiota and transcriptome of black tiger shrimp Penaeus monodon at different salinity levels. Aquaculture. 2021;546 doi: 10.1016/j.aquaculture.2021.737371. [DOI] [Google Scholar]
- 16.Angthong P., Uengwetwanit T., Arayamethakorn S., Chaitongsakul P., Karoonuthaisiri N., Rungrassamee W. Bacterial analysis in the early developmental stages of the black tiger shrimp (Penaeus monodon) Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-61559-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan L., Wang Z., Chen M., Qu Y., Li J., Zhou A., Zou J. Microbiota comparison of Pacific white shrimp intestine and sediment at freshwater and marine cultured environment. Sci. Total Environ. 2019;657:1194–1204. doi: 10.1016/j. [DOI] [PubMed] [Google Scholar]
- 18.Huang Z., Li X., Wang L., Shao Z. Changes in the intestinal bacterial community during the growth of white shrimp, Litopenaeus vannamei. Aquac. Res. 2016;47:1737–1746. [Google Scholar]
- 19.Rungrassamee W., Klanchui A., Chaiyapechara S., Maibunkaew S., Tangphatsornruang S., Jiravanichpaisal P., Karoonuthaisiri N. Bacterial population in intestines of the black Tiger shrimp (Penaeus monodon) under different growth stages. PLoS One. 2013;8 doi: 10.1371/journal.pone.0060802. e60802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holt C.C., David B., Stentiford G.D., Giezen M. Understanding the role of the shrimp gut microbiome in health and disease. J. Invertebr. Pathol. 2020;4(1):1–16. doi: 10.1016/j.jip.2020.107387. [DOI] [PubMed] [Google Scholar]
- 21.Ma T., Suzuki Y., Guan L.L. Dissect the mode of action of probiotics in affecting host-microbial interactions and immunity in food producing animals. Vet. Immunol. Immuno Pathol. 2018;205:35–48. doi: 10.1016/j.vetimm.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Zheng Y., et al. Bacterial community associated with healthy and diseased Pacific white shrimp (Litopenaeus vannamei) larvae and rearing water across different growth stages rearing of shrimp larvae. Front. Microbiol. 2017;8:1362. doi: 10.3389/fmicb.2017.01362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uengwetwanit T., Uawisetwathana U., Arayamethakorn S., Khudet J., Chaiyapechara S., Karoonuthaisiri N., Rungrassamee W. Multi-omics analysis to examine microbiota, host gene expression and metabolites in the intestine of black tiger shrimp (Penaeus monodon) with different growth performance. PeerJ. 2020;8 doi: 10.7717/peerj.9646. e9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan Y., et al. Impairment of the intestine barrier function in Litopenaeus vannamei exposed to ammonia and nitrite stress. Fish Shellfish Immunol. 2018;78:279–288. doi: 10.1016/j.fsi.2018.04.050. [DOI] [PubMed] [Google Scholar]
- 25.Duan Y., et al. Changes in the intestine barrier function of Litopenaeus vannamei in response to pH stress. Fish Shellfish Immunol. 2019;88:142–149. doi: 10.1016/j.fsi.2019.02.047. [DOI] [PubMed] [Google Scholar]
- 26.Duan Y., et al. Transcriptomic and microbiota response on Litopenaeus vannamei intestine subjected to acute sulphide exposure. Fish Shellfish Immunol. 2019;88:335–343. doi: 10.1016/j.fsi.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 27.Granados F.C., Lopez-Zavala A.A., Gallardo-Becerra L., Mendoza-Vargas A., Sanchez F., Vichido R., Brieba L.G., Viana M.T., Sotelo-Mundo R.R., OchoaLeyva A. Microbiome of Pacific White leg shrimp reveals differential bacterial community composition between wild, aquacultured and AHPND/EMS outbreak conditions. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-11805-w. (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imaizumi K., Tinwongger S., Kondo H., Hirono I. Analysis of microbiota in the stomach and midgut of two penaeid shrimps during probiotic feeding. Sci. Rep. 2021;11(1):9936. doi: 10.1038/s41598-021-89415-w. May 11; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai Wen-Fang, Zhang Jin-Jie, Qiu Qiong-Fen, Chen Jiong, Yang Wen, Ni Sui, Xiong Jin-Bo. Starvation stress affects the interplay among shrimp gut microbiota, digestion and immune activities. Fish Shellfish Immunol. 2018;80:191–199. doi: 10.1016/j.fsi.2018.05.040. Epub 2018 May 24. PMID: 29803665. [DOI] [PubMed] [Google Scholar]
- 30.Zoqratt M.Z.H.M., Eng W.H., Thai B.T., Austin C.M., Gan H.M. Microbiome analysis of Pacific white shrimp gut and rearing water from Malaysia and Vietnam: implications for aquaculture research and management. PeerJ. 2018;6 doi: 10.7717/peerj.5826. e5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bordenstein S.R., Theis K.R. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 2015;13(8) doi: 10.1371/journal.pbio.1002226. e1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foster K.R., Schluter J., Coyte K.Z., Rakoff-Nahoum S. The evolution of the host microbiome as an ecosystem on a leash. Nature. 2017;548:43. doi: 10.1038/nature23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McFall-Ngai M., Hadfield M.G., Bosch T.C.G., Carey H.V., Domazet-Lošo T., Douglas A.E., Dubilier N., Eberl G., Fukami T., Gilbert S.F., et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. 2013;110(9):3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margulis L. (1991) Symbiosis as a source of evolutionary innovation: speciation and morphogenesis. In: Cambridge MA MLFR, editor. Symbiogenesis and Symbionticism: MIT Press, 1–14.
- 35.Rosenberg E., Koren O., Reshef L., Efrony R., Zilber-Rosenberg I. The role of microorganisms in coral health, disease, and evolution. Nat. Rev. Microbiol. 2007;5:355. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- 36.Rosenberg Z.I., Rosenberg E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 2008;32(5):723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 37.Egerton F.N. History of ecological sciences, part 52: symbiosis studies. Bull. Ecol. Soc. Am. 2015;96(1):80–139. [Google Scholar]
- 38.Simon J.C., Julian R.M., Christophe M., Marc A.S. Host-microbiota interactions: from holobiont theory to analysis. Microbiome. 2019;7:5. doi: 10.1186/s40168-019-0619-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brener-Raffalli K., Clerissi C., Vidal-Dupiol J., Adjeroud M., Bonhomme F., Pratlong M., Aurelle D., Mitta G., Toulza E. Thermal regime and host clade, rather than geography, drive Symbiodinium and bacterial assemblages in the scleractinian coral Pocillopora damicornis sensu lato. Microbiome. 2018;6(1):39. doi: 10.1186/s40168-018-0423-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenberg E., Zilber-Rosenberg I. The hologenome concept of evolution after 10 years. Microbiome. 2018;6(1):78. doi: 10.1186/s40168-018-0457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Douglas A.E., Werren J.H. Holes in the hologenome: why host-microbe symbioses are not holobionts. MBio. 2016;7(2) doi: 10.1128/mBio.02099-15. e02099–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moran N.A., Sloan D.B. The Hologenome concept: helpful or hollow? PLoS Biol. 2015;13(12) doi: 10.1371/journal.pbio.1002311. e1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dittami S.M., Arboleda E., Auguet J.-.C., Bigalke A., Briand E., Cárdenas P., Cardini U., Decelle J., Engelen A.H., Eveillard D., Gachon C.M.M., Griffiths S.M., Harder T., Kayal E., Kazamia E., Lallier F.H., Medina M., Marzinelli E.M., Morganti T.M., Núñez Pons L., Prado S., Pintado J., Saha M., Selosse M.-.A., Skillings D., Stock W., Sunagawa S., Toulza E., Vorobev A., Leblanc C., Not F. A community perspective on the concept of marine holobionts: current status, challenges, and future directions. PeerJ. 2021;9:1–34. doi: 10.7717/peerj.10911. e10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bright M., Bulgheresi S. A complex journey: transmission of microbial symbionts. Nat. Rev. Microbiol. 2010;8:218–230. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Funkhouser L.J., Bordenstein S.R. Mom knows best: the universality of maternal microbial transmission. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001631. e1001631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kazamia E., Helliwell K.E., Purton S., Smith A.G. How mutualisms arise in phytoplankton communities: building eco-evolutionary principles for aquatic microbes. Ecol. Lett. 2016;19:810–822. doi: 10.1111/ele.12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollock F.J., McMinds R., Smith S., Bourne D.G., Willis B.L., Medina M., Thurber R.V., Zaneveld J.R. Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 2018;9:4921. doi: 10.1038/s41467-018-07275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mountfort Douglas O., Campbell J., Clements Kendall D. Hindgut fermentation in three species of marine herbivorous fish. Society. 2002;68(3):1374–1380. doi: 10.1128/AEM.68.3.1374-1380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macke E., Callens M., De Meester L., Decaestecker E. Host-genotype dependent gut microbiota drives zooplankton tolerance to toxic cyanobacteria. Nat. Commun. 2017;8:1608. doi: 10.1038/s41467-017-01714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson C.D., Klein H.S., Murphy K.D., Parthasarathy R., Guillemin K., Bohannan B.J.M. Experimental bacterial adaptation to the zebrafish gut reveals a primary role for immigration. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2006893. e2006893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Redfern L.K., Jayasundara N., Singleton D.R., Di Giulio R.T., Carlson J., Sumner S.J., et al. The role of gut microbial community and metabolomic shifts in adaptive resistance of Atlantic killifish (Fundulus heteroclitus) to polycyclic aromatic hydrocarbons. Sci. Total Environ. 2021;776 doi: 10.1016/j.scitotenv.2021.145955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye L., Amberg J., Chapman D., Gaikowski M., Liu W.T. Erratum: fish gut microbiota analysis 639 differentiates physiology and behavior of invasive Asian carp and indigenous American fish. ISME J. 2016;10 doi: 10.1038/ismej.2016.71. 2076-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y., Wen B., David M.A., Gao J.-.Z., Chen Z.Z. Comparative analysis of intestinal microbiota of discus fish (Symphysodon haraldi) with different growth rates. Aquaculture. 2021;540 [Google Scholar]
- 54.Piazzon M.C., Naya-Català F., Perera E., Palenzuela O., Sitjà-Bobadilla A., Pérez-Sánchez J. Genetic selection for growth drives differences in intestinal microbiota composition and 559 parasite disease resistance in gilthead sea bream. Microbiome. 2020;8:168. doi: 10.1186/s40168-020-00922-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin S.A.M., Carola E.D., Elzbieta K. Review Transcriptomic responses in the fish intestine. Dev. Compar. Immunol. 2016;64:103–117. doi: 10.1016/j.dci.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 56.Esser D., Janina L., Georgios M., Michael S., Lena B., Daniela P., Jay B., Malte C., Rühlemanne Katherin B., Cornelia J., Felix S. Functions of the microbiota for the physiology of animal metaorganisms. J. Innate Immun. 2019;11:393–404. doi: 10.1159/000495115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Balcazar J.L., Vendrell D., de Blas I., Ruiz-Zarzuela I., Muzquiz J.L., Girones O. Characterization of probiotic properties of lactic acid bacteria isolated from intestinal microbiota of fish. Aquaculture. 2008;278:188–191. doi: 10.1016/j.aquaculture.2008.03.014. [DOI] [Google Scholar]
- 58.Olafsen J. Interactions between fish larvae and bacteria in marine aquaculture. Aquaculture. 2001;200(1–2):223–247. [Google Scholar]
- 59.Pérez-Sánchez T., Balcázar J.L., Merrifield D.L., Carnevali O., Gioacchini G., de Blas I., Ruiz-Zarzuela I. Expression of immune-related genes in rainbow trout (Oncorhynchus mykiss) induced by probiotic bacteria during Lactococcus garvieae infection. Fish Shellfish Immunol. 2011;31:196–201. doi: 10.1016/j.fsi.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 60.Kuang T., He Anyou, Lin Yifei, Huang Xiande, LiLiu Lei Zhou. Comparative analysis of microbial communities associated with the gill, gut, and habitat of two filter-feeding fish. Aquacult. Rep. 2020;18:1–11. doi: 10.1016/j.aqrep.2020.100501. [DOI] [Google Scholar]
- 61.Brown R.M., Gregory D.W., Irene S. Analysis of the gut and gill microbiome of resistant and susceptible lines of rainbow trout (Oncorhynchus mykiss) Fish Shellfish Immunol. 2019;86:497–506. doi: 10.1016/j.fsi.2018.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guivier E., Nicolas P., Rémi C., André G. Microbiota associated with the skin, gills, and gut of the fish Parachondrostoma toxostoma from the Rhône basin. Freshw. Biol. 2019;65(3):446–459. doi: 10.1111/fwb.13437. [DOI] [Google Scholar]
- 63.Nedoluha P.C., Westhoff D. Microbiological analysis of Stripped bass (Morone saxatilis) grown in flow-through tanks. Food Protect. 1995;58:1363–1368. doi: 10.4315/0362-028X-58.12.1363. [DOI] [PubMed] [Google Scholar]
- 64.Ringø E., Holzapfel W. Identification and characterization of carnobacteria associated with the gills of Atlantic salmon (Salmo salar L.) Syst. Appl. M. 2000;23(4):523–527. doi: 10.1016/s0723-2020(00)80026-0. [DOI] [PubMed] [Google Scholar]
- 65.Mudarris M., Austin B. Quantitative and qualitative studies of the bacterial microflora of turbot, Scophthalmus maximus L., gills. J. Fish Biol. 1988;32:223–229. [Google Scholar]
- 66.Kapetanovic D., Kurtovic B., Vardic I., Valic D., Teskeredzic Z., Teskeredzic E. Preliminary studies on bacterial diversity of cultured Bluefin tuna Thunnus thynnus from the Adriatic Sea. Aquacult. Res. 2006;37:1265–1266. [Google Scholar]
- 67.Wang W., Zhou Z., He S., Liu Y., Cao Y., Shi P., Yao B., Ringø E. Identification of the adherent microbiota on the gills and skin of poly-cultured gibel carp (Carassius auratus gibelio) and blunt nose black bream (Megalobrama amblycephala) Aquacult. Res. 2010;41 e72–e83. [Google Scholar]
- 68.Al-Harbi A., Uddin N. Bacterial diversity of tilapia (Oreochromis niloticus) cultured in brackish water in Saudi Arabia. Aquaculture. 2005;250:566–572. [Google Scholar]
- 69.Nedoluha P.C., Westhoff D. Microbiological analysis of striped bass (Morone saxatilis) grown in a three aquaculture system. Food Microbiol. 1997;14:255–264. [Google Scholar]
- 70.Wang C.Z., Lin G.R., Yan T., Zheng Z.P., Chen B., Sun F.L. The cellular community in the intestine of the shrimp Penaeus penicillatus and its culture environments. Fish. Sci. 2014;80(5):1001–1007. doi: 10.1007/s12562-014-0765-3. [DOI] [Google Scholar]
- 71.Cahill M.M. Bacterial flora of fishes: a review. Microb. Ecol. 1990;19(1):21–41. doi: 10.1007/BF02015051. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Y.A., Salinas I., Li J., et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 2010;11:827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salinas I., Zhang Y.A., Sunyer J.O. Mucosal immunoglobulins and B cells of teleost fish. Dev. Compar. Immunol. 2011;35:1346–1365. doi: 10.1016/j.dci.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Legrand T.P.R.A., Catalano S.R., Wos-Oxley M.L., Stephens F., Landos M., Bansemer M.S., Stone D.A.J., Qin J.G., Oxley A.P.A. The inner workings of the outer surface: skin and gill microbiota as indicators of changing gut health in yellowtail kingfish. Front. Microbiol. 2018;8:2664. doi: 10.3389/fmicb.2017.02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bates J.M., Mittge E., Kuhlman J., Baden K.N., Cheesman S.E., Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 2006;297(2):374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 76.Rawls J.F., Mahowald M.A., Goodman A.L., Trent C.M., Gordon J.I. In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7622–7627. doi: 10.1073/pnas.0702386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rawls J.F., Samuel B.S., Gordon J.I. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl Acad. Sci. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Austin B. The bacterial microflora of fish. Sci. World J. 2006;6:931–945. doi: 10.1100/tsw.2006.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gómez G.D., Balcazar J.L. A review on the interactions between gut microbiota and innate immunity of fish. FEMS Immunol. Med. Microbiol. 2008;52:145–154. doi: 10.1111/j.1574-695X.2007.00343.x. [DOI] [PubMed] [Google Scholar]
- 80.Pamer E.G. Immune responses to commensal and environmental microbes. Nat. Immunol. 2007;8:1173–1178. doi: 10.1038/ni1526. [DOI] [PubMed] [Google Scholar]
- 81.Montalto M., D’ Onofrio F., Gallo A., Cazzato A, Gabardine G. Intestinal microbiota and its functions. Dig. Liver Dis. Suppl. 2009;3:30–34. [Google Scholar]
- 82.Silphaduang U., Colorni A., Noga E.J. Evidence for widespread distribution of piscidins antimicrobial peptides in teleost fish. Dis. Aquat. Org. 2006;72:241–252. doi: 10.3354/dao072241. [DOI] [PubMed] [Google Scholar]
- 83.Zou J., Mercier C., Koussounadis A., Secombes C. Discovery of multiple beta-defensins like homologues in teleost fish. Mol. Immunol. 2007;44:638–647. doi: 10.1016/j.molimm.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 84.Fasano A., Shea-Donohue Mechanisms of disease: the role of intestinal barrier function in the pathogenesis of gastrointestinal autoimmune diseases. Nat. Clin. Pract. Gastroenterol. Hepatol. 2005;2:416–422. doi: 10.1038/ncpgasthep0259. [DOI] [PubMed] [Google Scholar]
- 85.Hormannsperger G., Haller D. Molecular crosstalk of probiotic bacteria with the intestinal immune system: clinical relevance in the context of inflammatory bowel disease. Int. J. Med. Microbiol. 2010;300:63–73. doi: 10.1016/j.ijmm.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 86.Sorek M., Schnytzer Y., Ben-Asher H.W., Caspi V.C., Chen C.S., Miller D.J., Levy O. Setting the pace: host rhythmic behaviour and gene expression patterns in the facultatively symbiotic cnidarian Aiptasia are determined largely by Symbiodinium. Microbiome. 2018;6(1):83. doi: 10.1186/s40168-018-0465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van de Water J.A.J.M., Allemand D., Ferrier-Pagès C. Host-microbe interactions in octocoral holobionts-recent advances and perspectives. Microbiome. 2018;6(1):64. doi: 10.1186/s40168-018-0431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pita L., Rix L., Slaby B.M., Franke A., Hentschel U. The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome. 2018;6(1):46. doi: 10.1186/s40168-018-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang S., Sun X. Core gut microbiota of shrimp function as a regulator to maintain immune homeostasis in response to WSSV. Infection. Microbiol Spectr. 2022;27(2) doi: 10.1128/spectrum.02465-21. 10e0246521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dai W., et al. Integrating molecular and ecological approaches to identify potential polymicrobial pathogens over a shrimp disease progression. Appl. Microbiol. Biotechnol. 2018;102:3755–3764. doi: 10.1007/s00253-018-8891-y. [DOI] [PubMed] [Google Scholar]
- 91.Fan L., Li Q. Characteristics of intestinal microbiota in the Pacific white shrimp Litopenaeus vannamei differing growth performances in the marine cultured environment. Rev. Aquacult. 2019;505:450–461. [Google Scholar]
- 92.Rajeev R., Adithya K.K., Kiran G.S., Joseph S. Healthy microbiome: a key to successful and sustainable shrimp aquaculture. Rev. Aquacult. 2020;13:238–258. doi: 10.1111/raq.12471. [DOI] [Google Scholar]
- 93.Van Doan H., Hoseinifar S.H., Ringø E., Angeles Esteban M., Dadar M., Dawood M.A., et al. Host-associated probiotics: a key factor in sustainable aquaculture. Rev. Fish. Sci. Aquacult. 2020;28(1):16–42. [Google Scholar]
- 94.Wang Y.C., Hu S.Y., Chiu C.S., Liu C.H. Multiple-strain probiotics appear to be more effective in improving the growth performance and health status of white shrimp, Litopenaeus vannamei, than single probiotic strains. Fish Shellfish Immunol. 2019;84:1050–1058. doi: 10.1016/j.fsi.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 95.Egerton S., Culloty S., Whooley J., Stanton C., Ross R.P. The gut microbiota of marine fish. Front. Microbiol. 2018;9:873. doi: 10.3389/fmicb.2018.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abelli L., Randelli E., Carnevali O., Picchietti S. Stimulation of gut immune system by early administration of probiotic strains in Dicentrarchus labrax and Sparus aurata. Ann. N. Y. Acad. Sci. 2009;1163(1):340–342. doi: 10.1111/j.1749-6632.2008.03670.x. [DOI] [PubMed] [Google Scholar]
- 97.Akhter N., Wu B., Memon A.M., Mohsin M. Probiotics and prebiotics associated with aquaculture: a review. Fish Shellfish Immunol. 2015;45:733–774. doi: 10.1016/j.fsi.2015.05.038. [DOI] [PubMed] [Google Scholar]
- 98.Burr G., Delbert G., Steven R. Microbial ecology of the gastrointestinal tract of fish and the potential application of prebiotics and probiotics in finfish aquaculture. J. World Aquac. Soc. 2005;36:425–436. doi: 10.1111/j.1749-7345.2005. tb00390.x. [DOI] [Google Scholar]
- 99.Hagi T., Tanaka D., Iwamura Y., Hoshino T. Diversity and seasonal changes in lactic acid bacteria in the intestinal tract of cultured freshwater fish. Aquaculture. 2004;234:335–346. doi: 10.1016/j.Aquaculture2004.01.018. [DOI] [Google Scholar]
- 100.Merrifield, D.L., & Carnevali, O. (2014). Probiotic modulation of the gut microbiota of fish. In D. Merrifield, & E. Ringø (Eds.), Aquaculture Nutrition: Gut Health, Probiotics and Prebiotics (pp. 185–222). John Wiley & Sons, Ltd.
- 101.Carnevali O., Maradonna F., Gioacchini G. Integrated control of fish metabolism, wellbeing and reproduction: the role of probiotic. Aquaculture. 2017;472:144–155. doi: 10.1016/aquaculture. [DOI] [Google Scholar]
- 102.Balcázar J.L., de Blas I., Ruiz-Zarzuela I., Vendrell D., Giron E.S.O., Muzquiz J.L. Enhancement of the immune response and protection induced by probiotic lactic acid bacteria against furunculosis in rainbow trout (Oncorhynchus mykiss) FEMS Immunol. Med. Microbiol. 2007;51:185–193. doi: 10.1111/j.1574-695X.2007.00294.x. [DOI] [PubMed] [Google Scholar]
- 103.Cordero H., Guardiola F.A., Tapia-Paniagua S.T., Cuesta A., Meseguer J., Balebona M.C., Moriñigo M.Á., Esteban M.Á. Modulation of immunity and gut microbiota after dietary administration of alginate encapsulated Shewanella putrefaciens Pdp11 to gilthead sea bream (Sparus aurata L.) Fish Shellfish Immunol. 2015;45:608–618. doi: 10.1016/j.fsi.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 104.Lobo C., Moreno-Ventas X., Tapia-Paniagua S., Rodrıguez C., Morinigo M.A., de La Banda I.G. Dietary probiotic supplementation (Shewanella putrefaciens) modulates gut microbiota and promotes growth and condition in Senegalese sole larviculture. Fish Physiol. Biochem. 2014;40:295–309. doi: 10.1007/s10695-013-9844-0. [DOI] [PubMed] [Google Scholar]
- 105.Kwak J.K., Park S.W., Koo J.G., Cho M.G., Buchholz R., Goetz P. Enhancement of the non-specific defense activities in carp (Cyprinus carpio) and flounder (Paralichthys olivaces) by oral administration of Schizophyllan. Acta. Biotechnol. 2003;23:359–371. [Google Scholar]
- 106.Vendrell D., Balcazar J.L., de Blas I., Ruiz-Zarzuela I., Giron E.S.O., M ú zquiz J.L. Protection of rainbow trout (Oncorhynchus mykiss) from lactococcosis by probiotic bacteria. Comp. Immunol. Microbiol. Infect. Dis. 2008;31:337–345. doi: 10.1016/j.cimid.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 107.Gatesoupe F.J. Updating the importance of lactic acid bacteria in fish farming: natural occurrence and probiotic treatments. J. Mol. Microbiol. Biotechnol. 2008;14:107–114. doi: 10.1159/000106089. [DOI] [PubMed] [Google Scholar]
- 108.Kim D.H., Austin B. Innate immune responses in rainbow trout (Oncorhynchus mykiss, Walbaum) induced by probiotics. Fish Shellfish Immunol. 2006;21:513–524. doi: 10.1016/j.fsi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 109.Aly S.M., Ahmed Y.A.G., Ghareeb A.A.A., Mohamed M.F. Studies on Bacillus subtilis and Lactobacillus acidophilus, as potential probiotics, on the immune response and resistance of Tilapia nilotica (Oreochromis niloticus) to challenge infections. Fish Shellfish Immunol. 2008;25:128–136. doi: 10.1016/j.fsi.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 110.Ramirez R.F., Dixon B.A. Enzyme production by obligate intestinal anaerobic bacteria isolated from Oscars (Astronotus ocellatus), angelfish (Pterophyllum scalare) and southern flounder (Paralichthys lethostigma) Aquaculture. 2003;227:417–426. [Google Scholar]
- 111.Ayisi C.L., Apraku A., Afriyie G. A review of probiotics, prebiotics, and symbiotic in crab: present research, problems, and future perspectives. J. Shellfish Res. 2017;36(3):799–806. doi: 10.2983/35.036.0329. [DOI] [Google Scholar]
- 112.Hoseinifar S.H., Sun Y.Z., Wang A., Zhou Z. Probiotic as means of diseases control in aquaculture, a review of current knowledge and future perspectives. Front. Microbiol. 2018;9:2429. doi: 10.3389/fmib.2018.02429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hoseinifar, S.H., van Dadar, M., Doan, H., & Harikrishnan, R. (2019). Feed additives impacts on shellfish microbiota, health, and development. In N. Derome (Ed.), Microbial Communities in Aquaculture Ecosystems (pp. 143–163). Springer Nature.
- 114.Ninawe A.S., Selvin J. Probiotics in shrimp aquaculture: avenues and challenges. Crit. Rev. Microbiol. 2009;35:43–66. doi: 10.1080/10408410802667202. [DOI] [PubMed] [Google Scholar]
- 115.Ringø E. Probiotics in shellfish aquaculture. Aquacult. Fish. 2020;5(1):1–27. doi: 10.1016/j.aaf.2019.12.001. [DOI] [Google Scholar]
- 116.Shefat S.H.T. Use of probiotics in shrimp aquaculture in Bangladesh. Acta Sci. Microbiol. 2018;1(11):20–27. [Google Scholar]
- 117.Soltani M., Ghosh K., Hoseinifar S., Kumar V., Lymbery A.J., Roy S. Genus Bacillus, promising probiotics in aquaculture: aquatic animal origin, bio-active components, bioremediation and efficacy in fish and shellfish. Rev. Fish. Sci. Aquacult. 2019;27(3):331–379. doi: 10.1080/23308249.2019. [DOI] [Google Scholar]
- 118.Restrepo L., Cristóbal D.B., Leandro B., Irma B., Jenny R., Bonny B., Alejandro R. Microbial community characterization of shrimp survivors to AHPND challenge test treated with an effective shrimp probiotic (Vibrio diabolicus) Microbiome. 2021;9:88. doi: 10.1186/s40168-021-01043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Augustine D., Jacob J.C., Philip R. Exclusion of Vibrio spp. by an antagonistic marine actinomycete Streptomyces rubrolavendulae M56. Aquac. Res. 2016;47(9):2951–2960. [Google Scholar]
- 120.Das S., Ward L.R., Burke C. Screening of marine Streptomyces spp. for potential use as probiotics in aquaculture. Aquaculture. 2010;305(1–4):32–41. [Google Scholar]
- 121.García Bernal M., et al. Probiotic effect of Streptomyces strains alone or in combination with Bacillus and Lactobacillus in juveniles of the white shrimp Litopenaeus vannamei. Aquacult. Int. 2017;25:927–939. [Google Scholar]
- 122.Mazon-Suastegui J.M., et al. Effect of Streptomyces probiotics on the gut microbiota of Litopenaeus vannamei challenged with Vibrio parahaemolyticus. Microbiol. Open. 2019;9(2) doi: 10.1002/mbo3.967. e967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kongnum K., Hongpattarakere T. Effect of Lactobacillus plantarum isolated from digestive tract of wild shrimp on growth and survival of white shrimp (Litopenaeus vannamei) challenged with Vibrio harveyi. Fish Shellfish Immunol. 2012;32:170–177. doi: 10.1016/j.fsi.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 124.Rungrassamee W., et al. Bacterial dynamics in intestines of the black tiger shrimp and the Pacific white shrimp during Vibrio harveyi exposure. J. Invertebr. Pathol. 2016;133:12–19. doi: 10.1016/j.jip.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 125.Du Y., Wang B., Jiang K., Wang M., Zhou S., Liu M., et al. Exploring the influence of the surface proteins on probiotic effects performed by Lactobacillus pentosus HC-2 using transcriptome analysis in Litopenaeus vannamei. Fish Shellfish Immunol. 2019;87:853–870. doi: 10.1016/j.fsi.2019.02.027. [DOI] [PubMed] [Google Scholar]
- 126.Foysal M.J., Fotedar R., Siddik M.A., Tay A. Lactobacillus acidophilus and L. plantarum improve health status, modulate gut microbiota and innate immune response of marron (Cherax cainii) Sci. Rep. 2020;10(1):1–3. doi: 10.1038/s41598-020-62655-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Panigrahi A., Das R.R., Sivakumar M.R., Saravanan M.R., Saranya C., Sudheer N.S., et al. Bio-augmentation of heterotrophic bacteria in biofloc system improves growth, survival, and immunity of Indian white shrimp Penaeus indicus. Fish Shellfish Immunol. 2020;98:477–487. doi: 10.1016/j.fsi.2020.01.021. [DOI] [PubMed] [Google Scholar]
- 128.Swain S.M., Singh C., Arul V. Inhibitory activity of probiotics Streptococcus phocae PI80 and Enterococcus faecium MC13 against Vibriosis in shrimp Penaeus monodon. World J. Microbiol. Biotechnol. 2009;25:697–703. doi: 10.1007/s11274-008-9939-4. [DOI] [Google Scholar]
- 129.Zuo Z.H., Shang B.J., Shao Y.C., Li W.Y., Sun J.S. Screening of intestinal probiotics and the effects of feeding probiotics on the growth, immune, digestive enzyme activity and intestinal flora of Litopenaeus vannamei. Fish Shellfish Immunol. 2018;86:160–168. doi: 10.1016/j.fsi.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 130.Landsman A., St-Pierre B., Rosales-Leija M., Brown M., Gibbons W. Impact of aquaculture practices on intestinal bacterial profiles of Pacific White leg shrimp Litopenaeus vannamei. Microorganisms. 2019;7:93. doi: 10.3390/microorganisms7040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu J., et al. Strain-specific changes in the gut microbiota profiles of the white shrimp Litopenaeus vannamei in response to cold stress. Aquaculture. 2019;503:357–366. [Google Scholar]
- 132.Liu Z., et al. Effects of a commercial microbial agent on the bacterial communities in shrimp culture system. Front. Microbiol. 2018;9:1–10. doi: 10.3389/fmicb.2018.02430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang J., et al. Effects of dietary mannan oligosaccharide on growth performance, gut morphology and stress tolerance of juvenile Pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 2012;33(4):1027–1032. doi: 10.1016/j.fsi.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 134.Hoseinifar S.H., Zare P., Kolangi Miandare H. ‘The effects of different routes of inulin administration on gut microbiota and survival rate of Indian white shrimp post-larvae (Fenneropenaeus indicus)’. Vet. Res. Forum: Int. Q. J. 2015;6(4):331–335. [PMC free article] [PubMed] [Google Scholar]
- 135.Ringø E., Zhou, J., Vecino L.G., Wadsworth S., Romero J., Krogdahl Å., Olsen R.E., Dimitroglou A., Foey A., Davies S., Owen M., Lauzon H.L., Martinsen L.L., De Schryver P., Bossier P. (2016) Effect of dietary components on the gut microbiota of aquatic animals. A never-ending story? Aquac. Nutr., 10.1111/anu.12346. [DOI]
- 136.Li J., Tan B., Mai K. Dietary probiotic Bacillus OJ and isomalto oligosaccharides influence the intestine microbial populations, immune responses and resistance to white spot syndrome virus in shrimp (Litopenaeus vannamei) Aquaculture. 2009;291:35–40. doi: 10.1016/j.aquaculture.2009. 03.005. [DOI] [Google Scholar]
- 137.Seyed Hossein H., Samira Y., Hien V.D., Ghasem A., Giorgia G., Francesca M., Oliana C. Oxidative stress and antioxidant defense in fish: the implications of probiotic, prebiotic, and synbiotics. Rev. Fish. Sci. Aquacult. 2020;29(2):198–217. doi: 10.1080/23308249.2020.1795616. [DOI] [Google Scholar]
- 138.Su H., et al. Persistence and spatial variation of antibiotic resistance genes and bacterial populations change in reared shrimp in South China. Environ. Int. 2018;119:327–333. doi: 10.1016/j.envint.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 139.Tomova A., et al. Antimicrobial resistance genes in marine bacteria and human uropathogenic Escherichia coli from a region of intensive aquaculture. Environ. Microbiol. Rep. 2015;7(5):803–809. doi: 10.1111/1758-2229.12327. [DOI] [PubMed] [Google Scholar]
- 140.Zeng S., Hou D., Liu J., Ji P., Weng S., He J., et al. Antibiotic supplement in feed can perturb the intestinal microbial composition and function in Pacific white shrimp. Appl. Microbiol. Biotechnol. 2019;103:3111–3122. doi: 10.1007/s00253-019-09671-9. [DOI] [PubMed] [Google Scholar]
- 141.Sanders M.E. Impact of probiotics on colonizing microbiota of the gut. J. Clin. Gastroenterol. 2011;5:115–119. doi: 10.1097/MCG.0b013e318227414a. [DOI] [PubMed] [Google Scholar]
- 142.Ju Z.Y., Forster I., Conquest L., Dominy W., Kuo W.C., David Horgen F. Determination of microbial community structures of shrimp floc cultures by biomarkers and analysis of floc amino acid profiles. Aquac. Res. 2008;39:118–133. 10.1111/j.1365-2109.2007.01856.x [Google Scholar]
- 143.Kumar V., Roy S., Behera B.K., Swain H.S., Das B.K. Biofloc microbiome with bioremediation and health benefits. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.741164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Qiao F., Liu Y.K., Sun Y.H., Wang X.D., Chen K., Li T.Y., et al. Influence of different dietary carbohydrate sources on the growth and intestinal microbiota of Litopenaeus vannamei at low salinity. Aquac. Nutr. 2017;23:444–452. [Google Scholar]
- 145.Ruan W., Engevik M.A., Spinler J.K., Versalovic J. Healthy human gastrointestinal microbiome: composition and function after a decade of exploration. Dig. Dis. Sci. 2020;65:695–705. doi: 10.1007/s10620-020-06118-4. [DOI] [PubMed] [Google Scholar]
- 146.Lee C., Kim S., Lim S., Lee K. Supplemental effects of biofloc powder on growth performance, innate immunity, and disease resistance of Pacific white shrimp Litopenaeus vannamei. Fish. Aquat. Sci. 2017;20:1–7. doi: 10.1186/s41240-017-0059-7. [DOI] [Google Scholar]
- 147.Promthale P., Pongtippatee P., Withyachumnarnkul B., Wongprasert K. Bioflocs substituted fishmeal feed stimulates immune response and protects shrimp from Vibrio parahaemolyticus infection. Fish Shellfish Immunol. 2019;93:1067–1075. doi: 10.1016/j.fsi.2019.07.084. [DOI] [PubMed] [Google Scholar]
- 148.Kheti B., Kamilya D., Choudhury J., Parhi J., Debbarma M., Singh S.T. Dietary microbial floc potentiates immune response, immune relevant gene expression and disease resistance in rohu, Labeo rohita (Hamilton, 1822) fingerlings. Aquaculture. 2017;468:501–507. 10.1016/j.aquaculture [Google Scholar]
- 149.Qiao F.Y.K., Liu Y.H., Sun X.D., Wang K., Chen T.Y., Li E.C., Zhang M.L. Influence of different dietary carbohydrate sources on the growth and intestinal microbiota of Litopenaeus vannamei at low salinity. Aquac. Nutr. 2017;23:444–452. [Google Scholar]
- 150.Schryver P., De Crab R., Defoirdt T., Boon N., Verstraete W. The basics of bio-flocs technology: the added value for aquaculture. Aquaculture. 2008;277 doi: 10.1016/j.aquaculture. 3–4125–137. [DOI] [Google Scholar]
- 151.Duan Y., Dong H., Wang Y., Li H., Liu Q., Zhang Y., et al. Intestine oxidative stress and immune response to sulphate stress in Pacific white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 2017;63:201–207. doi: 10.1016/j.fsi.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 152.Suguna P., Binuramesh C., Abirami P., Saranya V., Poornima K., Rajeswari V., Shenbagarathai R. Immunostimulation by poly-β hydroxybutyrate–hydroxyl valerate (PHB–HV) from Bacillus thuringiensis in Oreochromis mossambicus. Fish Shellfish Immunol. 2014;36:90–97. doi: 10.1016/j.fsi.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 153.Luo G., Zhang N., Cai S., Tan H., Liu Z. Nitrogen dynamics, bacterial community composition and biofloc quality in biofloc-based systems cultured Oreochromis niloticus with poly-β-hydroxybutyric and polycaprolactone as external carbohydrates. Aquaculture. 2017;479:732–741. [Google Scholar]
- 154.Cardona E., Saulnier D., Lorgeoux B., Chim L., Gueguen Y. Rearing effect of biofloc on antioxidant and antimicrobial transcriptional response in Litopenaeus stylirostris shrimp facing an experimental sub-lethal hydrogen peroxide stress. Fish Shellfish Immunol. 2015;45(2):933–939. doi: 10.1016/j.fsi.2015.05.041. [DOI] [PubMed] [Google Scholar]
- 155.Cardona E., Gueguen Y., Magré K., et al. Bacterial community characterization of water and intestine of the shrimp Litopenaeus stylirostris in a biofloc system. BMC Microbiol. 2016;16:157. doi: 10.1186/s12866-016-0770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ekasari J., Hanif Azhar M, Surawidjaja E.H., Nuryati S., De Schryver P., Bossier P. Immune response J. and disease resistance of shrimp fed biofloc grown on different carbon sources. Fish Shellfish Immunol. 2014;41(2):332–339. doi: 10.1016/j.fsi.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 157.Kim S.K., Pang Z., Seo H.C., Cho Y.R., Samocha T., Jang I.K. Effect of bioflocs on growth and immune activity of Pacific white shrimp, Litopenaeus vannamei post larvae. Aquacult. Res. 2014;45(2):362–371. doi: 10.1111/are.12319. [DOI] [Google Scholar]
- 158.Xu W.J., Pan L.Q. Enhancement of immune response and antioxidant status of Litopenaeus vannamei juvenile in biofloc-based culture tanks manipulating high C/N ratio of feed input. Aquaculture. 2013 doi: 10.1016/j.aquaculture.2013.07.017. 412–413, 117–124. [DOI] [Google Scholar]
- 159.Smith V.J., Brown J.H., Hauton C. Immunostimulation in crustaceans: does it really protect against infection? Fish Shellfish Immunol. 2003;15(1):71–90. doi: 10.1016/S1050-4648(02)00140-7. [DOI] [PubMed] [Google Scholar]
- 160.Yu W., Wu J.H., Zhang J., Yang W., Chen J., Xiong J. A meta-analysis reveals universal gut bacterial signatures for diagnosing the incidence of shrimp disease. FEMS Microbiol. Ecol. 2018;94(10) doi: 10.1093/femsec/fiy147. [DOI] [PubMed] [Google Scholar]
- 161.Xiong J. Progress in the gut microbiota in exploring shrimp disease pathogenesis and incidence. Appl. Microbiol. Biotechnol. 2018;102(17):7343–7350. doi: 10.1007/s00253-018-9199-7. [DOI] [PubMed] [Google Scholar]
- 162.Tepaamorndech S., Nookaew I., Shawn M.H., Santiyanont P., Phromson M., Chantarasakha K., Mhuantong W., Plengvidhya V., Visessanguan W. Metagenomics in bioflocs and their effects on gut microbiome and immune responses in Pacific white shrimp. Fish Shellfish Immunol. 2020;106:733–741. doi: 10.1016/j.fsi.2020.08.042. [DOI] [PubMed] [Google Scholar]
- 163.Dauda A.B., Romano N., Ebrahimi M., Teh J.C., Ajadi A., Chong C.M., et al. Influence of carbon/nitrogen ratios on biofloc production and biochemical composition and subsequent effects on the growth, physiological status and disease resistance of African catfish (Clarias gariepinus) cultured in glycerol-based biofloc systems. Aquaculture. 2018;483:120–130. 10.1016/j.aquaculture [Google Scholar]
- 164.Zhao Z., Xu Q., Luo L., Wang C., Li J., Wang L. Effect of feed C/N ratio promoted bioflocs on water quality and production performance of bottom and filter feeder carp in minimum-water exchanged pond polyculture system. Aquaculture. 2014;434:442–448. doi: 10.1016/j.aquaculture.2014.09.006. [DOI] [Google Scholar]
- 165.Sun F., et al. Insights into the intestinal microbiota of several aquatic organisms and association with the surrounding environment. Aquaculture. 2019;507:196–202. [Google Scholar]
- 166.Akbar S., et al. Understanding host-microbiome-environment interactions: insights from Daphnia as a model organism. Sci. Total Environ. 2022;808 doi: 10.1016/j.scitotenv.2021.152093. [DOI] [PubMed] [Google Scholar]
- 167.Xiao F., Zhu W., Yu Y., He Z., Wu B., Wang C., Yan Q. Host development overwhelms environmental dispersal in governing the ecological succession of zebrafish gut microbiota. Biofilms Microbiomes. 2021;7(1):1–12. doi: 10.1038/s41522-020-00176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yan Q., et al. Environmental filtering decreases with fish development for the assembly of gut microbiota. Environ. Microbiol. 2016;18:4739–4754. doi: 10.1111/1462-2920.13365. [DOI] [PubMed] [Google Scholar]
- 169.Ding X., et al. The impact of aquaculture system on the microbiome and gut metabolome of juvenile Chinese soft-shell turtle (Pelodiscus sinensis) IMeta. 2022;1 doi: 10.1002/imt2.17. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen C.Z., Ping L., Ling L., Zhi-Hua L. Exploring the interactions between the gut microbiome and the shifting surrounding aquatic environment in fisheries and aquaculture: a review. Environ. Res. 2022;214(4) doi: 10.1016/j.envres.2022.114202. [DOI] [PubMed] [Google Scholar]
- 171.Khurana H., et al. Gut milieu shapes the bacterial communities of invasive silver carp. Genomics. 2021;113:815–826. doi: 10.1016/j.ygeno.2021.01.013. [DOI] [PubMed] [Google Scholar]
- 172.Zhou L., et al. Deep insight into bacterial community characterization and relationship in the pond water, sediment and the gut of shrimp (Penaeus japonicus) Aquaculture. 2021;539 [Google Scholar]
- 173.Zhu J., et al. Robust host source tracking building on the divergent and nonstochastic assembly of gut microbiomes in wild and farmed large yellow croaker. Microbiome. 2022;10:1–15. doi: 10.1186/s40168-021-01214-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sehnal L., Brammer-Robbins E., Wormington A.M., Blaha L., Bisesi J., Larkin I., Martyniuk C.J., Simonin M., Adamovsky O. Microbiome composition and function in aquatic vertebrates: small organisms making big impacts on aquatic animal health. Front. Microbiol. 2021;12:358. doi: 10.3389/fmicb.2021.567408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Giatsis C., Sipkema D., Smidt H., Heilig H., Benvenuti G., Verreth J., Verdegem M. The impact of rearing environment on the development of gut microbiota in tilapia larvae. Sci. Rep. 2015;5(1):1–15. doi: 10.1038/srep18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zheng D., Liwinski T., Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zheng Z., et al. Microbiota diversity in pearl oyster Pinctada fucata martensii intestine and its aquaculture environment. Front. Mar. Sci. 2021;8:271. [Google Scholar]
- 178.Meng L.-.J., et al. Comparative analysis of bacterial communities of water and intestines of silver carp (Hypophthalmichthys molitrix) and bighead carp (H. nobilis) reared in aquaculture pond systems. Aquaculture. 2021;534 [Google Scholar]
- 179.Hou D., et al. Comparative analysis of the bacterial community compositions of the shrimp intestine, surrounding water and sediment. J. Appl. Microbiol. 2018;125:792–799. doi: 10.1016/j.jff.2019.103416. [DOI] [PubMed] [Google Scholar]
- 180.Del'Duca A., et al. Bacterial community of pond's water, sediment and in the guts of tilapia (Oreochromis niloticus) juveniles characterized by fluorescent in situ hybridization technique. Aquacult. Res. 2015;46:707–715. [Google Scholar]
- 181.Huang Z., Zeng S., Xiong J., Hou D., Zhou R., Xing C. Microecological Koch's postulates reveal that intestinal microbiota dysbiosis contributes to shrimp white faces syndrome. Microbiome. 2020;8(1):1–13. doi: 10.1186/s40168-020-00802-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Huang F., et al. Microbiota assemblages of water, sediment, and intestine and their associations with environmental factors and shrimp physiological health. Appl. Microbiol. Biotechnol. 2018;102:8585–8598. doi: 10.1007/s00253-018-9229-5. [DOI] [PubMed] [Google Scholar]
- 183.Kim P.S., et al. Host habitat is the major determinant of the gut microbiome of fish. Microbiome. 2021;9:1–16. doi: 10.1186/s40168-021-01113-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Almeida A.R., et al. The impact of antibiotic exposure in water and zebrafish gut microbiomes: a 16S rRNA gene-based metagenomic analysis. Ecotoxicol. Environ. Saf. 2019;186 doi: 10.1016/j.ecoenv.2019.109771. [DOI] [PubMed] [Google Scholar]
- 185.Jia J., et al. Characterization of tetracycline effects on microbial community, antibiotic resistance genes and antibiotic resistance of Aeromonas spp. in gut of goldfish Carassius auratus Linnaeus. Ecotoxicol. Environ. Saf. 2020;191 doi: 10.1016/j.ecoenv.2020.110182. [DOI] [PubMed] [Google Scholar]
- 186.Zhou L., et al. Environmental concentrations of antibiotics impair zebrafish gut health. Environ. Pollut. 2018;235:245–254. doi: 10.1016/j.envpol.2017.12.073. [DOI] [PubMed] [Google Scholar]
- 187.Pérez-Sánchez T., Balcázar J.L., García Y., Halaihel N., Vendrell D., de Blas I., Merrifield D.L., Ruiz-Zarzuela I. Identification and characterization of lactic acid bacteria isolated from rainbow trout Oncorhynchus mykiss (Walbaum) with inhibitory activity against Lactococcus garvieae. J. Fish Dis. 2011;34:499–507. doi: 10.1111/j.1365-2761.2011.01260.x. [DOI] [PubMed] [Google Scholar]
- 188.Hentschel U., Michael S., Jörg H. Common molecular mechanisms of symbiosis and pathogenesis. Trend Microbiol. 2000;226(8):5. doi: 10.1016/s0966-842x(00)01758-3. PII: S0966-842X (00)01758-3. [DOI] [PubMed] [Google Scholar]
- 189.Émie A.G., Sylvain F.É., Bouslama S., Derome N. Microbiomes of clownfish and their symbiotic host anemone converge before their first physical contact. Microbiome. 2021;9:109. doi: 10.1186/s40168-021-01058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.
The data supporting the findings of this study are available from the corresponding author upon request.