Abstract
Rearranged renal cell carcinomas (RCC) are rare types of kidney cancer. The clinicopathological features of rearranged RCC require further validation. The pathological diagnosis usually depends on immunohistochemistry and molecular analysis. This study aimed to explore the expression features of anti-TFE3, TFEB, and ALK in different renal entities. In addition, we collected thirty-six TFE3-rearranged RCC, two TFEB-altered RCC, and one ALK-rearranged RCC to explore their clinicopathological features. We observed that TFE3 can sometimes be weakly expressed in non-TFE3-rearranged RCC. TFE3-rearranged RCC usually exhibited strong TFE3 expression. However, clear cell RCC and FH-deficient RCC also displayed strong TFE3 expression. TFEB also can be weakly expressed in clear cell RCC. However, ALK IHC showed a relatively high specificity and was negative for all non-ALK-rearranged RCC. The ALK-rearranged RCC was analyzed using next generation sequencing to explore gene alterations, and we identified a novel gene partner, SLIT1. ALK-rearranged RCC appears to have eosinophilic cytoplasm. Tumor cells with clear cytoplasm may exclude this diagnosis. Psammomatous bodies (22/38) and pattern multiplicity (35/38) were observed in more than half of the patients. In conclusion, weak TFE3 expression did not indicate TFE3 rearrangement. Strong TFE3 expression had a higher value for indicating TFE3-rearranged RCC, although other entities can also exhibit a strong pattern. Young age combined with morphological features (psammomatous calcification and pattern multiplicity) may indicate the diagnosis of rearranged RCC.
Keywords: TFE3, TFEB, ALK, Immunohistochemistry, Fluorescence in situ hybridization, DNA-seq, Rearranged renal cell carcinoma
1. Introduction
Rearranged renal cell carcinomas, including TFE3-rearranged renal cell carcinoma (TFE3-RCC), TFEB-altered renal cell carcinoma (TFEB-RCC), and ALK-rearranged renal cell carcinoma (ALK-RCC), account for a relatively low proportion of renal cancers. TFE3-RCC and TFEB-RCC were classified as microphthalmia-associated transcription factor (MiT) family translocation RCC according to the 2016 WHO classification [1], and termed TFE3 rearranged renal cell carcinoma and TFEB altered renal cell carcinoma in the 2022 WHO classification [2]. ALK-RCC is another recently determined novel entity characterized by ALK fusion [3], which has limited case reports and requires more work to extend its clinicopathological features [[4], [5], [6], [7]]. An early study identified six RCCs with TFE3 rearrangement from 632 RCCs and demonstrated that the cancer-specific survival of TFE3-RCC was worse than that of papillary renal cell carcinoma (PRCC) and was not different from that of clear cell renal cell carcinoma (CCRCC) [8]. It is well established that other rearranged renal cancers can exhibit aggressive behavior [5,9]. Rearranged RCCs may benefit from medicines targeting their rearranged genes [10,11]. Therefore, it is important to distinguish rearranged RCCs from non-rearranged RCC. However, rearranged RCCs can exhibit a morphology similar to that of renal cancers without rearrangement. The diagnosis of rearranged RCCs is mainly based on the relatively specific morphology and presence of staining for TFE3, TFEB, or ALK and is further supported by fluorescence in situ hybridization (FISH) or gene analysis. However, limited specificity of TFE3 IHC has been reported [12]. Another study demonstrated that TFE3 protein expression in renal cell carcinoma predicts a poor outcome [13]. Whether TFEB or ALK is expressed in other renal cell neoplasms remains unclear. Whether rearranged RCCs share similar clinicopathological features among cases remains unclear because of the limited number of reported cases. In this study, we aimed to evaluate the expression of TFE3, TFEB, and ALK in different renal entities, and introduce 36 TFE3-RCC, one ALK-RCC with a novel gene fusion partner, SLIT1, and two TFEB-RCC with detailed clinicopathological features to extend our understanding of rearranged RCC.
2. Materials and methods
2.1. Case selection and clinicopathological assessment
This study was conducted with the approval of the Department of Pathology, Ruijin Hospital The datasets of renal cell tumors using TFE3 immunohistochemistry (IHC), with/without TFEB and ALK were retrieved, and 1678 patients were identified. Thirty-six TFE3-RCC, one ALK-RCC, and two TFEB-RCC were retrieved, and slides were reviewed by two experienced urological pathologists. Clinicopathological data, including age, sex, size, and laterality, were obtained from the datasets. Microscopic features, including tumor cell traits, architectural patterns, foamy macrophages, invasion, necrosis, psammomatous calcification, and encapsulation were re-evaluated. Follow-up duration was determined from the time of diagnosis to the most recent contact.
2.2. Immunohistochemistry and FISH analysis
IHC and FISH were performed on 4-μm sections using antibodies TFE3 (ZSGB-BIO; OTI1H6, prediluted), TFEB (ABCAM; ab2636, 1:100), ALK (ZSGB-BIO; 1A4, prediluted), CK7 (DAKO; OV-TL 12/30, prediluted), CD117 (DAKO; polyclonal, 1:500), Vimentin (DAKO; V9, prediluted), CD10 (DAKO; 56C6, prediluted), CK20 (DAKO; Ks20.8, prediluted), CA9 (MXB; RAB-0615; 1:100), HMB45 (DAKO; GA05261; prediluted), Melan A (DAKO; IR63361; prediluted), Cathepsin K (ABCAM; ab37259; 1:100), PAX8 (MXB; EP298, prediluted), INI-1 (ZSGB-BIO; ZA-0696; prediluted), FH (ABCAM; ab233394; 1:100), SDHB (ZSGB-BIO; ZM-0162; prediluted), and probes for chromosomes 3p (LBP, China), 7, 17, and Y (GPMEDICAL, China); and TFE3 and TFEB break-apart (LBP, China). On IHC, proteins expressed in <10% of tumor cells were considered negative (−). Focal (10%–50% tumor cells) or weak expression was noted. Diffuse and strong expression was considered positive (+). Focal, weak, or diffuse and strong expression were defined as positive in the statistical description. Positive expression of TFE3 or TFEB was defined as their positive nuclear reactivity in the neoplastic cells. Cytoplasmic and membranous expression was not considered positive for TFE3 or TFEB. Positive cytoplasmic, membranous, or nuclear reactivity for ALK was interpreted as positive. For FISH assessment, three or more signals in ≥10% of the tumor cells were identified as trisomy 7 or 17 chromosomes. One signal in ≥70% or 40% of tumor cells was viewed as a loss of chromosome Y or 3p. TFE3 and TFEB break-apart cut off values were determined to be 10%. Chromosomal or gene alterations were evaluated as positive, otherwise negative.
2.3. Next-generation sequencing (NGS)
Considering the extremely rare reports of ALK-RCC, available sections were obtained from the patient with ALK-RCC for further gene analysis. The QIAamp DNA FFPE Tissue Kit (Qiagen, USA) was used to extract DNA, and the Qubit assay (BGI, China) was used to measure the DNA concentration. Each step was performed in accordance with the manufacturer's instructions. DNA samples were utilized for pan-cancer laboratory gene testing (BGI, China), an NGS assay including 688 cancer-associated gene target sequencing panels (Supplementary Table 1).
3. Results
3.1. TFE3, TFEB, ALK IHC and FISH assessment
TFE3 IHC was performed on 1677 renal tumors, and the expression levels are shown in Table 1. A total of 228 renal tumors showed focal/weak staining and 38 tumors showed strong staining. TFE3 was often strongly expressed (30/36) in TFE3-RCC. Only few TFE3-RCC exhibited weak TFE3 expression (6/36). Except for TFE3-RCC showing positivity for TFE3, TFE3 can be focally or weakly expressed in various renal entities. TFE3 was weakly expressed in TFEB-RCC and ALK-RCC (Fig. 1A). TFE3 was observed to be positive in more than 10% of CCRCC and PRCC (Fig. 1B). In addition, TFE3 expression can be positive in chromophobe RCC, clear cell papillary renal cell tumor (CCPRCT), renal oncocytoma (RO), collecting duct carcinoma (CDC), papillary renal neoplasm with reverse polarity (PRNRP), and FH-deficient RCC. However, TFE3 IHC was limited to weak or focal expression in these renal entities. Of these, only five CCRCC and one FH-deficient RCC exhibited strong nuclear TFE3 expression (Fig. 1C), similar to most TFE3-RCC (Fig. 1D). However, TFE3 FISH was performed on these six patients and yielded negative results. The diagnosis of CCRCC was based on classic morphology with absence of papillary architecture, immunohistochemical panel (positive CA9 and CD10 and negative CK7), and 3p loss. The diagnosis of FH-deficient RCC was based on FH IHC loss. Melan A and HMB45 were performed on tumors with TFE3 expression, and were all negative in non-TFE3-RCC tumors. TFE3 FISH was employed on 72 patients. Positive results were obtained in 34 patients. In the remaining cases, most were confirmed as CCRCC, depending on classic histology, and chromosome 3p loss if necessary. We observed diffuse and strong TFE3 expression in several tumors without positive FISH results. Cathepsin K, HMB45 and/or Melan A were expressed in two tumors, and we still diagnosed the patients with TFE3-RCC. Three cases without melanocytic marker expression were diagnosed with RCC-NOS.
Table 1.
TFE3 protein expression in renal cancers.
| negative | focal/weak positive | positive | |
|---|---|---|---|
| CCRCC (n = 1288) | 1107 (85.9%) | 176 (13.7%) | 5 (0.4%) |
| PRCC (n = 109) | 92 (84.4%) | 17 (15.6%) | 0 |
| CHRCC (n = 95) | 88 (92.6%) | 7 (7.4%) | 0 |
| CCPRCT (n = 27) | 26 (96.3%) | 1 (3.7%) | 0 |
| RO (n = 48) | 45 (93.7%) | 3 (6.3%) | 0 |
| TFE3-RCC (n = 35) | 0 | 6 (17.1%) | 29 (82.9%) |
| TFEB-RCC (n = 2) | 0 | 2 | 0 |
| ALK-RCC (n = 1) | 0 | 1 | 0 |
| RCC-FMS (n = 3) | 3 | 0 | 0 |
| CDC (n = 9) | 6 (66.7%) | 3 (33.3%) | 0 |
| PRNRP (n = 7) | 6 (85.7%) | 1 (14.3%) | 0 |
| INI1-RCC (n = 3) | 3 | 0 | 0 |
| FH-RCC (n = 7) | 2 (28.6%) | 4 (57.1%) | 1 (14.3%) |
| BHPRCC (n = 1) | 1 | 0 | 0 |
| RCC-NOS (n = 42) | 32 | 7 | 3 |
CCRCC, clear cell renal cell carcinoma; PRCC, papillary renal cell carcinoma; CHRCC, chromophobe renal cell carcinoma; CCPRCT, clear cell papillary renal cell tumor; RO, renal oncocytoma; RCC-FMS, RCC with fibromyomatous stroma; CDC, collecting duct carcinoma; PRNRP, papillary renal neoplasm with reverse polarity; BHPRCC, biphasic hyalinizing psammomatous renal cell carcinoma. RCC-NOS, renal cell carcinoma, not otherwise specified.
Fig. 1.
TFE3 can be expressed in different renal entities. For example, TFE3 was weakly expressed in ALK-RCC (A) and papillary RCC (B). FH-deficient RCC exhibited strong TFE3 expression (FH IHC loss in tumor cells is shown in the upper right) (C). TFE3-RCC exhibited strong TFE3 expression (D).
Anti-TFEB was performed on 557 tumors, and weak or focal nuclear staining of TFEB was observed in 6 patients. Similar to the findings of TFE3 IHC, CCRCC with low or high WHO/ISUP grades can also partially express TFEB. FISH was performed in 16 patients and no TFEB break-apart was identified. Strongly positive for TFEB was observed in two patients and further FISH revealed TFEB break-apart in 70% and 73% of the tumor cells, respectively.
Anti-ALK was performed on 146 tumors with a morphology indicative of ALK-RCC. A total of 145 tumors were negative for ALK and were finally diagnosed as non-ALK-RCC. No focal or weak staining was observed. Only one showed diffuse cytoplasmic ALK positivity. ALK IHC was performed in 100 CCRCC and 20 CCPRCT, all of which were negative.
3.2. TFE3-rearranged renal cell carcinoma
Detailed clinical, microscopic, immunohistochemical, and molecular results for each patient are shown in Table 2 (and Supplementary Tables 2 and 3). The clinicopathological features of the TFE3-RCC patients are summarized in Table 3. TFE3-RCC showed no clear laterality or sex predominance. Tumor size varied between 1.0 and 10 cm. Follow-up data were obtained for 33 patients, and the follow-up duration ranged from 7 to 56 months. Six patients with TFE3-RCC had metastases to the lymph node, bone, or abdominal cavity, of which four died of the disease. TFE3-RCC showed heterogeneous histology among the cases. Classic biphasic tumor cell populations were observed in five tumors (Fig. 2A). A combination of papillary, tubular, solid, and cystic architectures can be observed in TFE3-RCC (Fig. 2B–F). Heterogeneity was also observed in the individual cases. Clear cells displayed alveolar patterns, and eosinophilic cells exhibited papillary architecture (Fig. 2E). Neoplastic cells forming the same architecture also can have diverse cellular morphologies. Papillae were covered by clear or eosinophilic cells with low or high WHO/ISUP grade. Psammomatous calcification was observed in 60% (21/35) of the patients. Foamy macrophages were found in 11% (4/35) of the patients. Necrosis, invasion, and sarcomatoid differentiation were observed in 23% (8/35), 11% (4/35), and 6% (2/35) of the patients, respectively. CK7 was negative or weakly expressed in the tumor cells (3/35). Melan A and HMB45 were positive in 41% (11/27) and 36% (13/36) of patients, respectively. Cathepsin K was positive in 60% (21/35) of patients. CD10 (33/33) was mostly positive, whereas CA9 (3/18) was usually negative in tumor cells. In patients positive for TFE3 FISH, the break-apart rates ranged from 14% to 91%. Chromosomal alterations have been assessed in several patients. 3p (1/9) or Y (1/6) loss and gain of chromosomes 7 (3/14) or 17 (3/14) were observed.
Table 2.
IHC and molecular results of rearranged RCC.
| Patient | TFE3 | TFEB | ALK | Melan A | HMB45 | Cathepsin K | CK7 | CA9 | CD10 | molecular comment |
|---|---|---|---|---|---|---|---|---|---|---|
| TFE3-RCC | ||||||||||
| 1 | + | – | – | NP | – | + | – | NP | + | TFE3 break-apart:23.57% negative for 7, 17, and Y |
| 2 | + | – | – | – | – | – | F+ | – | + | TFE3 break-apart:43% negative for TFEB |
| 3* | + | – | – | – | – | NP | – | NP | + | TFE3 break-apart:59% |
| 4 | + | – | – | – | – | – | – | – | + | TFE3 break-apart:23% |
| 5 | + | – | – | + | F+ | + | – | – | F+ | TFE3 break-apart:1.5% |
| 6 | + | – | – | – | + | + | – | – | + | TFE3 break-apart:71% negative for 3p, 7 and 17 |
| 7 | + | – | – | NP | – | – | – | F+ | + | TFE3 break-apart:67% negative for 3p |
| 8 | W+ | – | – | – | – | – | – | – | + | TFE3 break-apart:30% |
| 9 | + | – | – | – | – | – | F+ | – | + | TFE3 break-apart:59.3% |
| 10 | + | – | – | NP | – | + | – | NP | NP | TFE3 break-apart: 63% negative for 3p, 7, 17, and Y |
| 11 | + | – | – | F+ | – | + | – | – | + | TFE3 break-apart: 30.67% positive for 7 negative for 17 and Y |
| 12 | + | – | – | – | + | + | – | – | + | TFE3 break-apart:83% negative for 3p, 7, and 17 |
| 13 | F+ | – | – | + | F+ | + | – | NP | + | TFE3 break-apart:49% |
| 14 | F+ | – | – | NP | – | + | – | – | + | TFE3 break-apart:75.52% negative for 7, 17, and Y |
| 15 | + | – | – | + | – | + | NP | – | F+ | TFE3 break-apart:2% negative for 7 and 17 |
| 16 | + | – | – | NP | – | + | – | – | + | TFE3 break-apart:85% positive for 3p |
| 17 | + | – | – | – | + | + | – | – | + | TFE3 break-apart:14% |
| 18 | + | – | – | – | + | – | – | – | + | TFE3 break-apart:88.81% |
| 19 | W+ | – | – | NP | – | + | – | NP | + | TFE3 break-apart:60.26% positive for 7, 17 |
| 20 | + | – | – | – | – | – | F+ | F+ | + | TFE3 break-apart:50% negative for 7 and 17 |
| 21 | + | – | – | – | – | – | – | – | + | TFE3 break-apart:30.48% |
| 22 | + | – | – | + | + | + | – | NP | + | TFE3 break-apart:53% |
| 23 | + | – | – | – | F+ | + | – | + | + | TFE3 break-apart:30.19% positive for 7, 17, and Y negative for 3p |
| 24 | + | – | – | NP | – | + | – | NP | NP | TFE3 break-apart:43.09% positive for 17 negative for 7 |
| 25 | + | – | – | NP | F+ | + | – | NP | + | TFE3 break-apart:15.28% |
| 26 | W+ | – | – | NP | – | – | – | NP | NP | TFE3 break-apart:90% |
| 27 | + | – | – | + | – | – | – | – | F+ | TFE3 break−apart:31.67% |
| 28 | + | – | – | F+ | – | – | – | F+ | + | TFE3 break-apart:68.46% negative for 3p, 7, and 17 |
| 29 | W+ | – | – | – | – | + | – | NP | + | TFE3 break-apart:66.67% negative for 3p and TFEB |
| 30 | + | – | – | F+ | – | – | – | NP | F+ | TFE3 break-apart:42.1% negative for 7 and 17 |
| 31 | + | – | – | – | + | – | – | F+ | + | TFE3 break-apart:81% |
| 32 | + | – | – | + | + | – | – | – | + | TFE3 break-apart:50% |
| 33 | + | – | – | F+ | F+ | + | – | F+ | + | TFE3 break-apart:91% negative for 7, 17, and Y |
| 34 | + | – | – | – | – | + | – | – | + | TFE3 break-apart: 60.9% |
| 35 | + | – | – | – | + | + | – | – | + | TFE3 break-apart: 69% negative for 3p |
| 36 | + | – | – | + | – | + | – | NP | F+ | TFE3 break-apart: 70% negative for TFEB |
| TFEB-RCC | ||||||||||
| 1 | W+ | + | – | + | F+ | + | – | – | – | TFEB break-apart:73% negative for TFE3 |
| 2 | W+ | + | – | + | – | + | – | – | – | TFEB break-apart:70% negative for TFE3 |
| ALK-RCC | ||||||||||
| 1 | W+ | – | + | – | – | + | + | – | F+ | DNA-seq: SLIT1::ALK negative for 7, 17, and Y |
F, focal; NP, not performed; W, weak.
Patient 3 did not undergo the nephrectomy and IHC and FISH were performed on the metastasis.
Table 3.
Clinicopathologic characteristics of TFE3-RCC.
| Characteristics | TFE3-RCC |
|---|---|
| age at diagnosis (y) | 37.6 ± 13.9 |
| sex | |
| male | 15 |
| female | 21 |
| laterality | |
| left | 14 |
| right | 22 |
| tumor size (cm) | 4.4 ± 2.4 |
| follow-up | |
| recurrence or metastasis | 6 |
| death | 4 |
| biphasic morphology | 5 |
| cytoplasm | |
| only clear cytoplasm | 0 |
| only eosinophilic cytoplasm | 3 |
| mixed | 32 |
| architecture | |
| papillary | 30 |
| nest/solid | 30 |
| cystic | 7 |
| tubular | 2 |
| sarcomatoid | 2 |
| lymphovascular or perinephric invasion | 4 |
| necrosis | 8 |
| foamy macrophages | 4 |
| psammomatous calcification | 21 |
| Immunohistochemistry (-/FW+/+) | |
| TFE3 | 0/6/30 |
| Melan A | 16/4/7 |
| HMB45 | 23/5/8 |
| Cathepsin K | 14/0/21 |
| CD10 | 0/5/28 |
| CA9 | 15/2/1 |
| CK7 | 32/3/0 |
| FISH analysis | |
| chromosome 3p loss | 1/9 |
| chromosome 7 gain | 3/14 |
| chromosome 17 gain | 3/14 |
| chromosome Y loss | 1/6 |
| TFE3 break-apart | 34/36 |
| TFEB break-apart | 0/3 |
Fig. 2.
TFE3-rearranged renal cell carcinoma. The tumor exhibited a biphasic phenotype. Large epithelioid cells displayed nest patterns, and small cell clustered within the nests (A). Tubulopapillary architecture and psammomatous calcification were observed (B, C). Eosinophilic tumor cells exhibited solid and cystic patterns (D). Alveolar (left), cystic (middle), and papillary (right) patterns were observed (E). Tumor cells with eosinophilic cytoplasm abutted directly against normal renal tissue, and lymphocyte aggregation was observed (F).
3.3. TFEB-altered renal cell carcinoma
The patients confirmed to have TFEB-RCC were female at 26 and 24 years of age, with no clinical symptoms. Both cases revealed classic biphasic morphology in most areas (Fig. 3A and B). Tumor cells with abundant clear or eosinophilic cytoplasm displayed nest architecture. A cystic growth pattern was also observed (Fig. 3C). Foamy macrophages, psammomatous calcification, necrosis, or invasion were not observed. The tumor cells were positive for TFEB (Fig. 3D). CK7, CA9, and CD10 levels were negative. HMB45, Melan A, and Cathepsin K were positive, and TFE3 was weakly expressed in tumor cells. However, TFE3 FISH did not reveal any break apart.
Fig. 3.
TFEB-altered renal cell carcinoma. The tumor exhibited a biphasic phenotype. Large epithelioid cells displayed nest patterns, and small cell clustered within the nests (A and B). The tumor cells displayed a cystic architecture (C). Strong nuclear reactivity for TFEB was observed (D).
3.4. ALK-rearranged renal cell carcinoma
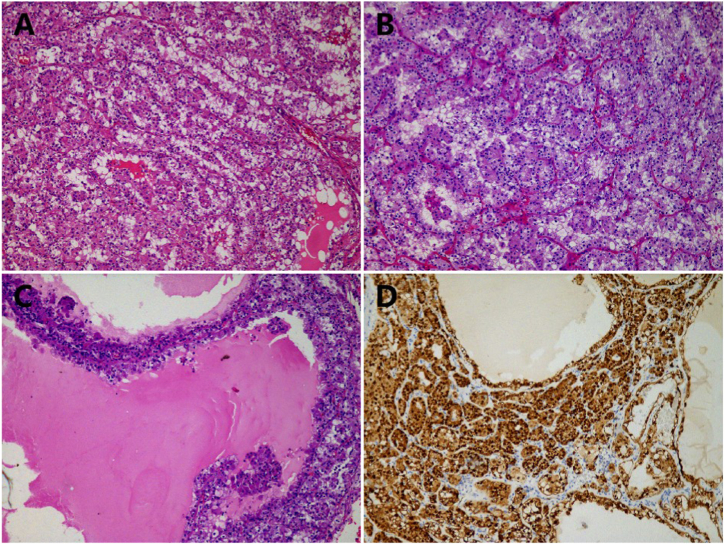
A solitary mass in the left kidney was occasionally found in a 34-year-old male and he underwent a partial nephrectomy. No sickle cell traits were observed. No other confirmed diseases with ALK rearrangements were found. The tumor had a solid-cystic appearance with the largest diameter of 3.5 cm. Histologically, tumor cells with eosinophilic cytoplasm and large nuclei, but inconspicuous nucleoli, abutted directly against normal renal tissue (Fig. 4A). Tumor cells displayed solid and papillary architectural patterns and stromal fibrosis can be observed (Fig. 4B–D). In focal areas, reverse polarity was found, similar to papillary renal neoplasm with reverse polarity (PRNRP) (Fig. 4B). Tumor cells with clear cytoplasm were not observed. Calcification was observed in the stroma. Mucus was not seen. No adverse events, such as sarcomatoid differentiation, necrosis, or lymphovascular or perinephric invasion occurred. CK7, PAX8, and Vimentin were diffusely expressed in the tumor cells (Fig. 4E). CD117, CK20, and CA9 levels were negative. INI1, FH, and SDHB were retained. ALK staining was diffusely positive in tumor cells (Fig. 4F). TFE3 showed a weak immunoreactivity. TFEB and melanocytic markers, HMB45 and Melan A, were negative. Unexpectedly, Cathepsin K was diffusely positive in tumor cells (Supplementary Fig. 1). In the PRNRP-like areas, GATA3 was diffusely positive. Ki67 index was 5%. FISH showed no chromosomal alterations in 7, 17, or Y. Next-generation sequencing was performed in the patient with diffuse ALK expression. Missense mutation in MST1 and a novel ALK (exon 19) gene fusion partner, SLIT1 (exon 36), were identified (Supplementary Fig. 2). No KRAS mutation was observed.
Fig. 4.
ALK-rearranged renal cell carcinoma. Tumor cells with eosinophilic cytoplasm and large nuclei, but inconspicuous nucleoli, abutted directly against normal renal tissue (A). Papillary (B), stromal fibrosis (C), and solid growth patterns (D) were observed. CK7 (E) and ALK (F) were diffusely positive in tumor cells.
4. Discussion
Novel renal entities with specific morphological, immunohistochemical, and genetic features have been investigated [3]. Rearranged renal cancers are still uncommon, and further studies are needed to explore their clinicopathological features. In the current study, we performed TFE3, TFEB, and ALK IHC in a relatively large series of renal entities to explore whether expression patterns varied between rearranged renal cell carcinoma and other renal tumors. In addition, thirty-six TFE3-RCC, two TFEB-RCC, and one ALK-RCC were included to explore the clinicopathological features of rearranged renal cell carcinomas.
TFE3, TFEB, and ALK protein expression are useful tools for screening rearrangements. Limited specificity has been validated for TFE3 IHC in TFE3-RCC [12]. CCRCC also exhibits strong TFE3 expression [14]. Another study demonstrated that RCC patients with TFE3 protein expression had worse outcomes than those without [13]. A previous study demonstrated that TFE3 affects the viability of CCRCC cells, and in CCRCC, TFE3 can elevate PD-L1 expression, resulting in immune evasion and resistance [15]. In another study, the authors discovered that TFEB did not affect the proliferation or migration of CCRCC but could mediate resistance to mTOR inhibition by regulating PD-L1 [16]. In the current study, anti-TFE3 was employed in 1677 renal tumors. We found that a large proportion of patients with high-grade RCC indeed expressed TFE3. TFE3 can be expressed in low-grade CCRCC, even in CCPRCT and PRNRP, which have relatively favorable prognoses [17,18]. Whether they can predict poor outcomes requires further investigation. In non-TFE3-RCC tumors, TFE3 IHC was usually limited to weak or focal expression. TFE3-RCC often exhibited diffuse and strong TFE3 expression. In this study, one FH-deficient RCC exhibited strong TFE3 expression. However, TFE3 FISH yielded a negative result. Loss of FH and diffuse 2SC expression indicated the diagnosis of FH-deficient RCC. Five CCRCC showed strong TFE3 expression. We validated the diagnoses based on 3p loss and negative TFE3 FISH results. ALK and TFEB IHC analyses were performed in a relatively large number of patients. Similar to TFE3, TFEB can also be weakly or focally expressed in some CCRCC. However, we did not observe TFEB expression in other renal entities. Regarding ALK IHC, no focal or weak expression was observed, indicating relatively high specificity for ALK IHC.
TFE3-RCC is more common than the other two rearranged RCCs. Melan A, HMB45, cathepsin K staining, and other biomarkers have been investigated as useful markers for indicating TFE3-RCC [19,20]. Break-apart fluorescence in situ hybridization (FISH) with higher sensitivity has been recommended [21]. Different fusion partners have been reported to exhibit different morphologies [22]. A multicenter study reported that Melan A and HMB45 were viewed as most preferred IHC assays for diagnosing of TFE3-RCC by most pathologists (>85%) [19]. However, a review including 397 TFE3-rearranged renal cell carcinomas demonstrated low positive rates for Melan A (19%) and HMB45 (17%) [23]. In this study, both Melan A and HMB45 antibodies showed high specificity, but limited sensitivity. Melan A and HMB45 were positive in 41% (11/27) and 36% (13/36) of patients, respectively. Similar to the review [23], CD10 was positive and CA9 and CK7 were mostly negative in TFE3-RCC. The FISH break-apart rates varied between 14% and 91%. False-negative results have been reported because of paracentric inversions involving Xp11 [22]. We observed three cases with diffuse TFE3 and melanocytic marker expression but without TFE3 break-apart, of which one displayed biphasic morphology. The patients were still diagnosed with TFE3-RCC based on the IHC panel (positive PAX8, TFE3, CD10, Melan A, HMB45, Cathepsin K, and negative CK7, CA9) [24]. Lymph nodes, bones and abdominal cavities were favorable sites for metastasis. In the current study, six patients showed metastasis and four died of the disease. Loss of chromosome 3p is the characteristic feature of CCRCC. However, in the current study, one TFE3-RCC patient with negative CA9, had both 3p loss and TFE3 break apart. Chromosomal alterations of 7, 17, and Y are viewed as molecular alterations of PRCC. Other studies have demonstrated that chromosomal alterations can also be detected in tubulocystic carcinoma or FH-deficient RCC [25,26]. We observed that alterations of chromosomes 7, 17, and Y can also occur in TFE3-RCC.
TFEB gene rearrangement or amplification were both named as “TFEB-altered renal cell carcinoma”. Based on the previous literature, TFEB-RCC is less common and behaves less aggressively than TFE3-RCC [27]. Recent studies have demonstrated that TFEB with amplification exhibits a more aggressive behavior than TFEB with rearrangement [9,[28], [29], [30], [31], [32], [33]]. A previous study found that the classic biphasic morphology was only observed in TFEB with fusions [28]. Another study further validated that typical biphasic tumor cells were only observed in MALAT1-TFEB fusion by reviewing 31 cases of TFEB rearrangement [34]. Necrosis was validated as the only morphological feature associated with poor outcomes of TFEB-RCC [34]. In our study, both patients displayed typical biphasic morphology. We did not find necrosis or other adverse events, such as sarcomatoid differentiation or lymphovascular invasion, which may be associated with poor outcomes. In the current study, the follow-up duration was limited (6 and 41 months), and both patients had benign follow-ups.
ALK-RCC is a rare renal entity, with a limited number of reported cases. Different gene fusion partners, including VCL, TPM3, EML4, STRN, CLIP1, and KIF5B have been reported [5]. In the current study, we present an ALK-RCC with a novel fusion partner, SLIT1, that has not been previously reported. Similar to other rearranged RCC, ALK-RCC displays pattern multiplicity. They can exhibit solid, papillary, tubular, and sarcomatoid architectures [5]. In this study, the tumor cells displayed papillary, cystic, and solid patterns, and the neoplastic cells had eosinophilic cytoplasm, with large nuclei and inconspicuous nucleoli. Together with previous reports, ALK-RCC appears to have eosinophilic cytoplasm [[4], [5], [6], [7]]. Tumor cells with clear cytoplasm may exclude this diagnosis. Whether this method can be used for differential diagnoses requires further validation. We also found that ALK-RCC can display papillary architecture with reverse polarity, similar to PRNRP. A recent study reported eight cases of ALK-RCC and GATA3 was expressed in only one case with weak immunoreactivity [5]. Interestingly, GATA3 was diffusely expressed in the present study. However, ALK-RCC showed nuclear pseudostratification in most areas, whereas PRNRP had a lower chance of exhibiting this morphology. KRAS mutations are viewed as early events in PRNRP [35]. No KRAS mutation was detected in this patient. ALK-RCC may exhibit sickle cell traits. However, in this study, the patient with ALK-RCC had no blood abnormalities. ALK-RCC may exhibit metastasis or even death [5,6]. In this study, the follow-up duration was limited (15 months) and the patient was disease-free. Whether malignant behavior may be validated by a longer follow-up period. Based on pertinent literature, we identified IHC markers similar to those of ALK-RCC. CK7 was diffusely expressed, whereas CD117 and CK20 were not. Unexpectedly, diffuse positivity for Cathepsin K was observed in this study, which was unreported before. IHC was performed on two different blocks and both showed positive results. Only one ALK-RCC was identified in this study, indicating its extremely rare incidence. Therefore, it is likely to be a low-yield method for screening for ALK rearrangements. However, it remains meaningful to identify ALK-RCC because targeted medicine, such as alectinib, a potent and specific ALK kinase inhibitor, has proven its efficacy in three patients with EML4-ALK fusion [11]. In the current study, the patient did not receive any treatment after the nephrectomy. Therefore, we cannot evaluate whether ALK inhibitors are effective in ALK-RCC.
TFE3-RCC is sometimes misdiagnosed as CCRCC or PRCC. CCRCC may also exhibit heterogeneity within the cases. However, CCRCC usually has clear cytoplasm with low-grade and eosinophilic cytoplasm with high-grade nuclei [36]. In addition, it is uncommon for CCRCC to display a papillary architecture [37]. In PRCC, it is rare for classical PRCC to have clear or eosinophilic cells within the same tumor. It is also rare for PRCC to display nest or solid patterns. In the current study, we found that rearranged RCC often displayed papillary architecture mixed with nest or solid architecture. Tumor cells in TFE3-rearranged RCC can have clear cytoplasm with high-grade nuclei or eosinophilic cytoplasm with inconspicuous nucleoli. In the current study, we introduced 5 CCRCC with strong TFE3 expression, which was a rare event for CCRCC. The diagnosis of such tumors should be issued based on combination of classic morphology (nest or alveolar architecture with absence of papillary architecture), IHC panel (positive CA9 and CD10 and negative CK7), and molecular results (3p loss and intact TFE3). In such circumstances, CA9 is a useful IHC marker to differentiate CCRCC with strong TFE3 expression between TFE3-rearranged RCC. CA9 is mostly negative in rearranged RCC, including those with cryptic fusion, and is usually positive in CCRCC [23]. These findings can aid in excluding the diagnosis of CCRCC or PRCC. ALK-RCC also exhibits pattern multiplicity. Therefore, in the work up for a high grade RCC with multiple patterns and cytoplasmic eosinophilia, ALK-RCC is also one of the differential diagnoses to be ruled out along with TFE3-RCC and TFEB-RCC, among many others.
Our study had some limitations. A related review reported that TFE3 IHC has variability in antibody performance or when automated [23]. In this study, TFE3 and TFEB were expressed in a number of non-TFE3/TFEB-RCC and we only used one clone of TFE3/TFEB. We did not compare with different clones to validate whether the results varied based on the clone used. In addition, most patients with rearranged RCC were confirmed using FISH. Next-generation sequencing was not performed to further explore the gene fusion partners. The available follow-up data on rearranged RCC were limited: 55% (18/33) were <24 months.
In conclusion, rearranged renal cell carcinomas are rare renal cell tumors that are relatively common in young patients. They usually exhibit pattern multiplicity (nest/solid and papillary growth patterns). Except for pattern multiplicity, TFE3-RCC and TFEB-RCC showed heterogeneity within cases. Tumor cells with clear or eosinophilic cytoplasm were found. However, ALK-RCC appears to have only eosinophilic, but not clear tumor cells. Weak expression of TFE3 can be observed across many renal entities and strong expression has more favorable value indicating TFE3-RCC, although it can be detected in few CCRCC and FH-deficient RCC. Compared with TFE3 IHC, TFEB and ALK IHC exhibited a relatively high specificity. However, unless suggested by young age, mixed growth patterns, heterogeneous cell traits, or eosinophilic tumor cells, routine screening for TFEB or ALK IHC is not recommended because of its low incidence.
Author contribution statement
Yang Liu: Performed the experiments; Wrote the paper.
Xiangyun Li; Yue Fan; Luting Zhou: Performed the experiments.
Haimin Xu; Lei Dong: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Yijin Gu: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Xiaoqun Yang; Chaofu Wang: Conceived and designed the experiments.
Funding
This study was supported by the National Natural Science Foundation of China (81903050), National Natural Science Foundation of China (82002667), and Fund Cooperation Project of Shanghai Municipal Science and Technology Commission (20Z11900303).
Data availability statement
Data will be made available on request.
Declaration of interest’s statement
The authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e15159.
Contributor Information
Lei Dong, Email: dl11968@rjh.com.cn.
Luting Zhou, Email: zhouluting@163.com.
Xiaoqun Yang, Email: yxq11964@rjh.com.cn.
Chaofu Wang, Email: wangchaofu@126.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.International Agency for Research on Cancer (IARC) and H. Moch . World Health Organization Classification of Tumours. fourth ed. International Agency for Research on Cancer; Lyon: 2016. WHO classification of tumours of the urinary system and male genital organs; p. 356. [Google Scholar]
- 2.Moch H., et al. The 2022 world Health Organization classification of tumours of the urinary system and male genital organs-Part A: renal, penile, and testicular tumours. Eur. Urol. 2022;82(5):458–468. doi: 10.1016/j.eururo.2022.06.016. [DOI] [PubMed] [Google Scholar]
- 3.Trpkov K., et al. Novel, emerging and provisional renal entities: the Genitourinary Pathology Society (GUPS) update on renal neoplasia. Mod. Pathol. 2021;34(6):1167–1184. doi: 10.1038/s41379-021-00737-6. [DOI] [PubMed] [Google Scholar]
- 4.Gorczynski A., et al. ALK-rearranged renal cell carcinomas in Polish population. Pathol. Res. Pract. 2019;215(12) doi: 10.1016/j.prp.2019.152669. [DOI] [PubMed] [Google Scholar]
- 5.Kuroda N., et al. ALK rearranged renal cell carcinoma (ALK-RCC): a multi-institutional study of twelve cases with identification of novel partner genes CLIP1, KIF5B and KIAA1217. Mod. Pathol. 2020;33(12):2564–2579. doi: 10.1038/s41379-020-0578-0. [DOI] [PubMed] [Google Scholar]
- 6.Sukov W.R., et al. ALK alterations in adult renal cell carcinoma: frequency, clinicopathologic features and outcome in a large series of consecutively treated patients. Mod. Pathol. 2012;25(11):1516–1525. doi: 10.1038/modpathol.2012.107. [DOI] [PubMed] [Google Scholar]
- 7.Yu W., et al. Genetic analysis and clinicopathological features of ALK-rearranged renal cell carcinoma in a large series of resected Chinese renal cell carcinoma patients and literature review. Histopathology. 2017;71(1):53–62. doi: 10.1111/his.13185. [DOI] [PubMed] [Google Scholar]
- 8.Sukov W.R., et al. TFE3 rearrangements in adult renal cell carcinoma: clinical and pathologic features with outcome in a large series of consecutively treated patients. Am. J. Surg. Pathol. 2012;36(5):663–670. doi: 10.1097/PAS.0b013e31824dd972. [DOI] [PubMed] [Google Scholar]
- 9.Argani P., et al. TFEB-Amplified renal cell carcinomas: an aggressive molecular subset demonstrating variable melanocytic marker expression and morphologic heterogeneity. Am. J. Surg. Pathol. 2016;40(11):1484–1495. doi: 10.1097/PAS.0000000000000720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damayanti N.P., et al. Therapeutic targeting of TFE3/IRS-1/PI3K/mTOR Axis in translocation renal cell carcinoma. Clin. Cancer Res. 2018;24(23):5977–5989. doi: 10.1158/1078-0432.CCR-18-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pal S.K., et al. Responses to alectinib in ALK-rearranged papillary renal cell carcinoma. Eur. Urol. 2018;74(1):124–128. doi: 10.1016/j.eururo.2018.03.032. [DOI] [PubMed] [Google Scholar]
- 12.Sharain R.F., et al. Immunohistochemistry for TFE3 lacks specificity and sensitivity in the diagnosis of TFE3-rearranged neoplasms: a comparative, 2-laboratory study. Hum. Pathol. 2019;87:65–74. doi: 10.1016/j.humpath.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Lee H.J., et al. TFE3 translocation and protein expression in renal cell carcinoma are correlated with poor prognosis. Histopathology. 2018;73(5):758–766. doi: 10.1111/his.13700. [DOI] [PubMed] [Google Scholar]
- 14.Yang B., et al. Xp11 translocation renal cell carcinoma and clear cell renal cell carcinoma with TFE3 strong positive immunostaining: morphology, immunohistochemistry, and FISH analysis. Mod. Pathol. 2019;32(10):1521–1535. doi: 10.1038/s41379-019-0283-z. [DOI] [PubMed] [Google Scholar]
- 15.Guo X., et al. TFE3-PD-L1 axis is pivotal for sunitinib resistance in clear cell renal cell carcinoma. J. Cell Mol. Med. 2020;24(24):14441–14452. doi: 10.1111/jcmm.16066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C., et al. TFEB mediates immune evasion and resistance to mTOR inhibition of renal cell carcinoma via induction of PD-L1. Clin. Cancer Res. 2019;25(22):6827–6838. doi: 10.1158/1078-0432.CCR-19-0733. [DOI] [PubMed] [Google Scholar]
- 17.Weng S., et al. The clinicopathologic and molecular landscape of clear cell papillary renal cell carcinoma: implications in diagnosis and management. Eur. Urol. 2021;79(4):468–477. doi: 10.1016/j.eururo.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y., et al. Papillary renal neoplasm with reverse polarity with a favorable prognosis should be separated from papillary renal cell carcinoma. Hum. Pathol. 2022;127:78–85. doi: 10.1016/j.humpath.2022.06.016. [DOI] [PubMed] [Google Scholar]
- 19.Akgul M., et al. Diagnostic approach in TFE3-rearranged renal cell carcinoma: a multi-institutional international survey. J. Clin. Pathol. 2021;74(5):291–299. doi: 10.1136/jclinpath-2020-207372. [DOI] [PubMed] [Google Scholar]
- 20.Wang X.M., et al. TRIM63 is a sensitive and specific biomarker for MiT family aberration-associated renal cell carcinoma. Mod. Pathol. 2021;34(8):1596–1607. doi: 10.1038/s41379-021-00803-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao Q., et al. TFE3 break-apart FISH has a higher sensitivity for Xp11.2 translocation-associated renal cell carcinoma compared with TFE3 or cathepsin K immunohistochemical staining alone: expanding the morphologic spectrum. Am. J. Surg. Pathol. 2013;37(6):804–815. doi: 10.1097/PAS.0b013e31827e17cb. [DOI] [PubMed] [Google Scholar]
- 22.Argani P. Translocation carcinomas of the kidney. Genes Chromosomes Cancer. 2022;61(5):219–227. doi: 10.1002/gcc.23007. [DOI] [PubMed] [Google Scholar]
- 23.Tretiakova M.S. Chameleon TFE3-translocation RCC and how gene partners can change morphology: accurate diagnosis using contemporary modalities. Adv. Anat. Pathol. 2022;29(3):131–140. doi: 10.1097/PAP.0000000000000332. [DOI] [PubMed] [Google Scholar]
- 24.Calio A., et al. TFE3 and TFEB-rearranged renal cell carcinomas: an immunohistochemical panel to differentiate from common renal cell neoplasms. Virchows Arch. 2022;481(6):877–891. doi: 10.1007/s00428-022-03380-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarungbam J., et al. Tubulocystic renal cell carcinoma: a distinct clinicopathologic entity with a characteristic genomic profile. Mod. Pathol. 2019;32(5):701–709. doi: 10.1038/s41379-018-0185-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y., et al. GATA3 aids in distinguishing fumarate hydratase-deficient renal cell carcinoma from papillary renal cell carcinoma. Ann. Diagn. Pathol. 2022;60 doi: 10.1016/j.anndiagpath.2022.152007. [DOI] [PubMed] [Google Scholar]
- 27.Calio A., et al. TFEB rearranged renal cell carcinoma. A clinicopathologic and molecular study of 13 cases. Tumors harboring MALAT1-TFEB, ACTB-TFEB, and the novel NEAT1-TFEB translocations constantly express PDL1. Mod. Pathol. 2021;34(4):842–850. doi: 10.1038/s41379-020-00713-6. [DOI] [PubMed] [Google Scholar]
- 28.Gupta S., et al. TFEB expression profiling in renal cell carcinomas: clinicopathologic correlations. Am. J. Surg. Pathol. 2019;43(11):1445–1461. doi: 10.1097/PAS.0000000000001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendel L., et al. Comprehensive study of three novel cases of TFEB-amplified renal cell carcinoma and review of the literature: evidence for a specific entity with poor outcome. Genes Chromosomes Cancer. 2018;57(3):99–113. doi: 10.1002/gcc.22513. [DOI] [PubMed] [Google Scholar]
- 30.Gupta S., et al. TFEB-VEGFA (6p21.1) co-amplified renal cell carcinoma: a distinct entity with potential implications for clinical management. Mod. Pathol. 2017;30(7):998–1012. doi: 10.1038/modpathol.2017.24. [DOI] [PubMed] [Google Scholar]
- 31.Kammerer-Jacquet S.F., et al. Comprehensive study of nine novel cases of TFEB-amplified renal cell carcinoma: an aggressive tumour with frequent PDL1 expression. Histopathology. 2022;81(2):228–238. doi: 10.1111/his.14683. [DOI] [PubMed] [Google Scholar]
- 32.Calio A., et al. VEGFA amplification/increased gene copy number and VEGFA mRNA expression in renal cell carcinoma with TFEB gene alterations. Mod. Pathol. 2019;32(2):258–268. doi: 10.1038/s41379-018-0128-1. [DOI] [PubMed] [Google Scholar]
- 33.Skala S.L., et al. Detection of 6 TFEB-amplified renal cell carcinomas and 25 renal cell carcinomas with MITF translocations: systematic morphologic analysis of 85 cases evaluated by clinical TFE3 and TFEB FISH assays. Mod. Pathol. 2018;31(1):179–197. doi: 10.1038/modpathol.2017.99. [DOI] [PubMed] [Google Scholar]
- 34.Xia Q.Y., et al. Clinicopathologic and molecular analysis of the TFEB fusion variant reveals new members of TFEB translocation renal cell carcinomas (RCCs): expanding the genomic spectrum. Am. J. Surg. Pathol. 2020;44(4):477–489. doi: 10.1097/PAS.0000000000001408. [DOI] [PubMed] [Google Scholar]
- 35.Al-Obaidy K.I., et al. Recurrent KRAS mutations are early events in the development of papillary renal neoplasm with reverse polarity. Mod. Pathol. 2022;35(9):1279–1286. doi: 10.1038/s41379-022-01018-6. [DOI] [PubMed] [Google Scholar]
- 36.Nilsson H., et al. Features of increased malignancy in eosinophilic clear cell renal cell carcinoma. J. Pathol. 2020;252(4):384–397. doi: 10.1002/path.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alaghehbandan R., et al. Papillary pattern in clear cell renal cell carcinoma: clinicopathologic, morphologic, immunohistochemical and molecular genetic analysis of 23 cases. Ann. Diagn. Pathol. 2019;38:80–86. doi: 10.1016/j.anndiagpath.2018.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.