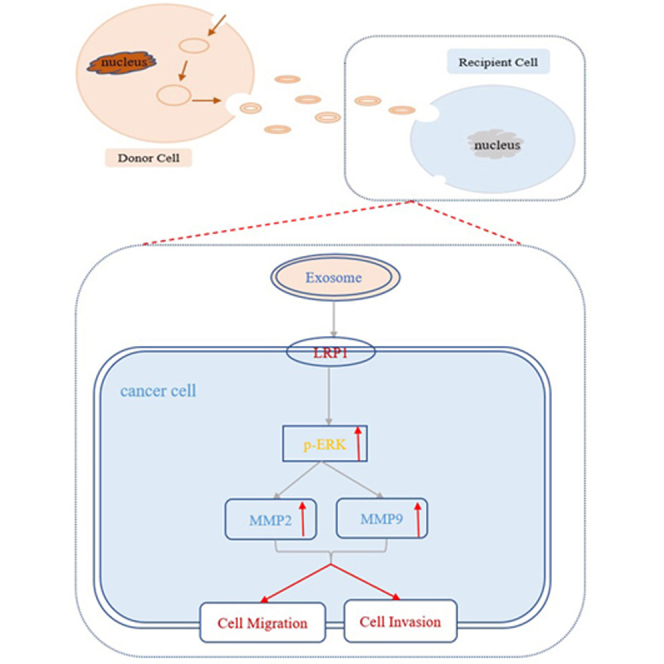
Abstract
Ovarian cancer is a gynecological tumor with extremely high mortality and poor prognosis. Exosomes derived from tumor cells contain abundant proteins that may influence tumor metastasis. The purpose of our study was to explore the proteomic profile of serum exosomes from epithelial ovarian cancer (EOC) patients and to find potential diagnostic markers for EOC. We obtained purified exosomes from serum using ultracentrifugation. Migration assay was used to evaluate the effects of exosomes on the migration of EOC cells. Proteomic profile of serum exosomes was analyzed by liquid chromatogram-tandem mass spectrometry. The levels of low-density lipoprotein receptor-related protein 1 (LRP1) in serum and serum exosomes were determined by enzyme-linked immunosorbent assay. Western blot and Immunohistochemistry were used to determine the level of LRP1 in tissues. Moreover, we performed small-interfering RNA-mediated knockdown of LRP1 in EOC cells to obtain SI-LRP1-Exos and SI-NC-Exos. The detailed mechanisms by which exosomal LRP1 affected the migration of EOC cells in vitro and in vivo were also explored. We found that serum exosomes from EOC patients contributed to the migration of EOC cells. The level of serum exosomal LRP1 of EOC patients was significantly upregulated compared with that of healthy volunteers, which was consistent with the result of enzyme-linked immunosorbent assay. We found that exosomal LRP1 regulated the expression of MMP2 and MMP9 through ERK signaling pathway and affected the migration of EOC cells in vitro and in vivo. Therefore, we propose that exosomal LRP1 contributes to the migration of EOC and may act as an important diagnostic and prognostic biomarker of EOC.
Keywords: epithelial ovarian cancer, exosome, proteomics, low-density lipoprotein receptor-related protein 1, migration
Graphical abstract
The levels of low-density lipoprotein receptor-related protein 1 were increased in exosomes from serum of epithelial ovarian cancer patients, exosomal low-density lipoprotein receptor-related protein 1 affected the migration of epithelial ovarian cancer cells through p-ERK/MMP2/MMP9 signaling pathway.
Highlights
-
•
Ovarian cancer is the deadliest gynecological malignancy.
-
•
Exosomes play a key role in mediating cancer metastasis.
-
•
Proteomic analysis of exosomes from serum.
-
•
Exosomal LRP1 may act as a marker of epithelial ovarian cancer.
In Brief
Ovarian cancer is the most lethal malignant gynecological tumor, and the clinical diagnosis and treatment of OC have always been difficult. Cancer cell–derived exosomes play a key role in mediating cancer progression. We performed proteomics analysis of serum exosomes to discover biomarkers for epithelial ovarian cancer.
Ovarian cancer (OC) is a malignant tumor that occurs in the female reproductive system (1). However, the lethality rate of OC patients is higher than that of cervical cancer and endometrial cancer. OC is reported to be the 7th most common cancer in women worldwide and the 10th most common cancer among women in China (2). According to a report, OC affecting more than 240,000 women worldwide every year (3). Most patients were diagnosed with advanced OC with distant metastases, and the 5-year relative survival rates of metastatic patients were less than 30% (4). The mortality rate of OC patients has shown a significant upward trend in China (5). The clinical diagnosis and treatment of OC have always been difficult, and it is an urgent clinical task to explore the mechanism of OC progression and search for diagnostic biomarkers. Epithelial ovarian cancer (EOC) is the most common pathological type (approximately 90%) (6). Therefore, this research focuses on EOC.
Exosomes are vesicles with diameters of 30 to 100 nm that are secreted by most cells and naturally exist in blood, urine, and ascites (7). The special lipid bilayer structure of exosomes can protect the contents from damage, so their biological activities are stable. Exosomes carry a variety of biological molecules derived from different originating cells, including proteins, RNA and DNA, lipids, and metabolites. Exosomes are considered as important mediators of intercellular communication. Metastasis, a key process involved in tumor progression, is not only a major challenge for cancer treatment but also a main cause of cancer death. Numerous studies have reported that exosomes in body fluids contain proteins that cause the tumorigenesis and metastasis of cancer, including endometrial cancer (8), breast cancer (9), gastric cancer (10), and oral squamous cell carcinoma (11). The changes of exosomal proteins are related to the pathological process of many cancers, and exosomal proteins are recommended as diagnostic and prognostic indicators of many cancers (12).
Low-density lipoprotein receptor-related protein 1 (LRP1) is a large endocytosis receptor (13). LRP1 is reported to be the most multifunctional member of the low-density lipoprotein receptor (LDL-R) family (14). Previous studies have found that LRP1 may influence multiple physiological processes by acting as an endocytic receptor (15) and regulating cellular signaling (16, 17). The protein level of LRP1 was upregulated in cancer cells, which may facilitate the growth and metastasis of cancer cells by affecting the expression of vascular endothelial growth factor (18). The dysregulation of Notch signaling pathway is closely correlated with the occurrence of various cancers, and researchers found that LRP1 was a key protein in the protein–protein interaction (PPI) network map of Notch pathway through bioinformatics analysis (19). The roles of LRP1 in the progression of EOC have not been well characterized.
Research showed that increased levels of serum exosomes derived from OC patients than that of those derived from people with benign disease or no disease (20). Analysis of the biological functions of exosomes from EOC may be of great significance in exploring their role in tumor metastasis. At present, there are few proteomics studies on serum exosomes from EOC patients. We found that serum exosomes from EOC patients contributed to the migration of EOC cells. Then, we performed proteomic to explore the protein profiles of purified exosomes from the serum. Proteomics analysis proved that the level of exosomal LRP1 was increased in serum from EOC patients than that from healthy volunteers. Furthermore, we proved that exosomal LRP1 affected the migration of EOC through the p-ERK/MMP2/MMP9 pathway in vitro and in vivo.
Experimental Procedures
Clinical Samples
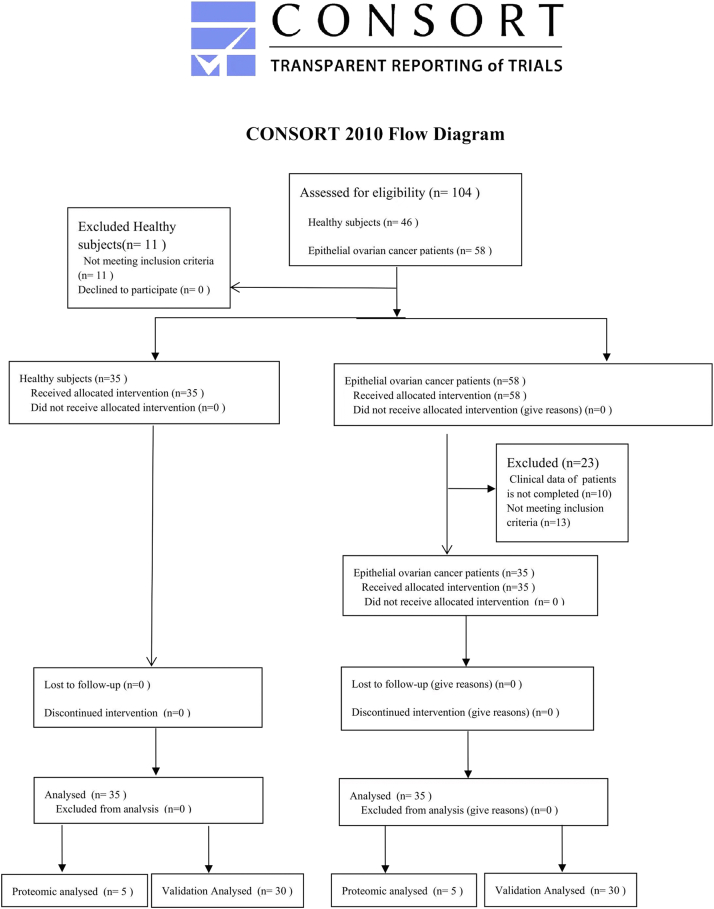
Venous blood samples of five EOC patients (n = 5, mean age: 57 ± 4.64) and five age-matched healthy volunteers (n = 5, mean age: 53.8 ± 3.35) from Xuzhou Central Hospital were collected for serum exosome isolation. Serum of 30 healthy volunteers (n = 30, mean age: 53.4 ± 6.52) and 30 EOC patients (n = 30, mean age: 55.6 ± 9.38) were also collected for the verification of specific biomarkers screened from the proteomics analysis. Blood samples were centrifuged at 3000g for 15 min at 4 °C. The separated serum was stored at −80 °C for subsequent experiments. Tissues from six ovarian cancer patients and six benign disease patients were collected undergoing surgery at Xuzhou Central Hospital. All tissues were stored at −80 °C until analyzed. Detailed information for this study population was presented in supplemental Tables S1–S3. Detailed information, inclusion criteria, and exclusion criteria for this study population have been described in the flow diagram (supplemental Fig. S1). The protocol was approved by the Ethics Committee of Xuzhou Central Hospital. Written informed consent was obtained from each subject before the study.
Cell Culture
The human EOC cell lines SKOV3 and HO8910 were kindly provided by Cancer Institute of Xuzhou Medical University. The cells were cultured in 1640 medium (Gibco) containing 10% fetal bovine serum (Clark) and 1% penicillin streptomycin at 37 °C and 5% CO2 in an incubator.
Isolation of Exosomes
To perform proteomic analysis, purified exosomes were obtained from serum by ultracentrifugation. Firstly, 3 ml serum was diluted to 10 ml with phosphate-buffered saline (1× PBS). Then, diluted samples were centrifuged at 500g for 10 min and at 2000g for 15 min (4 °C). Next, the supernatants were centrifuged at 12,000g for 30 min at 4 °C and filtered with a 0.22 μm syringe filter (Millipore) to remove large vesicles. The pellets were then ultracentrifuged twice at 110,000g for 70 min at 4 °C to obtain exosomes (Optima L-100XP, 90 Ti 11U1942 rotor, Beckman). The exosomes were resuspended in 100 μl 1× PBS for subsequent analysis. For verification, the Exosome Isolation Kit (H-Wayen, China, EIQ3-01001) was used to obtain exosomes from the serum of individual subjects following the manufacturer's instructions. Exosomes were obtained from cell supernatants by using Exosome Isolation Kit (H-Wayen, China, EIQ3-04001). The specific steps were carried out according to the instructions.
Transmission Electron Microscopy Analysis
Transmission electron microscopy (TEM) was used to observe the morphology of negatively stained exosomes. First, a copper grid was placed on the purified exosome droplet. Next, the copper grid was placed onto 3% glutaraldehyde droplet. Then, the copper grid was transferred to water droplets to wash the remaining fixative ten times. Finally, the exosomes on the copper grid were stained with 4% uranyl acetate. At the end of all the above steps, filter paper was used to remove the excess fluid. Morphologies of exosomes were observed using a TEM (Tecnai G2 Spirit Twin).
Dynamic Light Scattering Analysis
A laser nanoparticle size/potentiometer instrument (380ZLS, PSS) was used to analyze the size distribution of exosomes. The purified sample was added to the instrument to analyze the diameter of exosomes.
Western Blot Analysis
Total proteins were lysed with ice-cold RIPA buffer, and the insoluble materials were removed. BCA method was used to determine total protein concentration of samples. The specific steps of Western blot were consistent with our previous study (21). The main antibodies were CD9 (Affinity), CD63 (Bioworld), TSG101 (Bioworld), LRP1 (Abmart), ERK (Proteintech), p-ERK (Proteintech), MMP2 (Proteintech), MMP9 (Proteintech) and GAPDH (Bioworld).
Exosome Labeling
The purified exosomes were stained with 1,10-dioctadecyl 3,3,30,30- tetramethylindocarbocyanine perchlorate (Beyotime) and incubated at 37 °C for 20 min. SKOV3 and HO8910 cells were incubated with Dil-labeled exosomes for 24 h. Then, cells were fixed with 4% paraformaldehyde for 10 min and stained with DAPI for 5 min. Finally, the cells were imaged under a fluorescence microscope (OLYMPUS).
Wound Healing Assay
SKOV3 and HO8910 cells were seeded into 6-well plates and cultured to 90% confluence. The monolayers of cells were scratched with a micropipette tip to form a wound on the plate. PBS was used to remove floating cells. Serum-free medium containing exosomes (50 μg) was added to the chambers. SKOV3 and HO8910 cells were allowed to migrate into the wound area for 24 h. Pictures of wound at 0 h and 24 h were observed using a microscope (OLYMPUS).
Cell Migration Assay
SKOV3 cells (5 × 104) and HO8910 cells (8 × 104) were seeded into the upper chambers. Six hundred microliter of complete medium was added to the lower chambers. Serum-free medium with exosomes (50 μg) and PBS (50 ul) were added into the upper chambers. After incubation for 24 h, all chambers were washed with PBS. Cells were fixed in 4% paraformaldehyde for 15 min. Then, we use 0.1% crystal violet to stain cells. Unmigrated cells were wiped off with cotton swab. Migrated cells in random fields of view were observed under an inverted microscope (OLYMPUS).
Reverse Transcriptase—Quantitative PCR
Following the manufacturer’s protocol, total RNA was extracted using TRIZOL (Invitrogen). The PrimeScript RT Reagent Kit (TaKaRa) was used to perform reverse transcription. The reaction was conducted according to the following protocol: 95 °C for 30 s, 55 cycles at 95 °C for 10 s and 60 °C for 30 s. The primers for LRP1 and β-actin were purchased from Sheng Gong Biological Company (Shanghai). All sequences of specific primers were shown in supplemental Table S4. Gene expression was analyzed according to 2−ΔΔCq relative quantification method using Light Cycle 96 software version 1.1 (Roche).
Immunohistochemistry
Immunohistochemistry (IHC) assay was performed to detect the protein expression in tissues. The detailed steps of IHC followed by our previous study. The main primary antibodies used in IHC are as follows:LRP1 (Abmart, 1:200), MMP2 (Proteintech, 1:150), MMP9 (Proteintech, 1:200).
Mass Spectrometry of Exosomes
Exosomes obtained from the serum of healthy volunteers (N-Exos, n = 5) and EOC patients (EOC-Exos, n = 5) were used to investigate the protein profile. Protein concentrations of exosomes were determined with a BCA protein detection kit (Beyotime). First, exosomes were lysed with 8 M urea to obtain protein solution. The protein solution was reduced with 100 mM TCEP for 60 min at 56 °C, then the solution was alkylated with 200 mM CAA for 30 min at room temperature in the dark. Then, 50 mM NH3HCO3 was added to the solution to dilute the urea concentration. Finally, trypsin (1 μg/μl) was added for digestion overnight at 37 °C (50:1, protein to trypsin). The polypeptide was desalted using C18E SPE columns (Welchrom) and dried under vacuum. Peptides were analyzed using an LTQ-Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher) coupled to an EASY-nLC 1200 (Thermo Fisher). Chromatographic gradients were 95 min with a flow rate of 300 nl/min. Buffer A: 0.1% formic acid in water and buffer B: 0.1% formic acid in acetonitrile. Chromatographic gradients set as follows: 0 to 5 s, 3 to 5% Buffer B; 5 s-40 min 5 s, 5 to 15% Buffer B; 40 min 5 S-74 min 55 s, 15 to 28% Buffer B; 74 min 55 S-86 min 55 s, 28 to 38% Buffer B; 86 min 55 s-87 min, 38 to 100% Buffer B; 87 min to 95 min, 100% Buffer B. The mass spectrometer was operated in positive ionization mode with nanospray voltage set at 2.2 kV and source temperature at 300 °C. The acquisition was performed in data-dependent acquisition mode, and full MS scans with one micro scans at resolution of 60,000 were acquired over a mass range of m/z 350 to 1500 with detection in the Orbitrap mass analyzer. The resolution of secondary mass spectrometry is 15,000, the fragmentation mode is HCD. Fragment ion spectra were produced via high-energy collision dissociation fragmentation at a normalized collision energy of 30%, and they were acquired in the ion trap mass analyzer. AGC was set to 50,000 and a maximum injection time of 30 ms were used. All data were acquired in Xcalibur software, version 3. MaxQuant software (1.6.15.0) search engine was used to quantify proteins using MS/MS data. Differentially expressed proteins (DEPs) were screened with Perseus software (1.6.0.7) for subsequent bioinformatics analysis.
Mass Spectrometry Data Analysis
All acquired spectra were analyzed with MaxQuant (1.6.15.0) using default settings. The data were searched against Swiss-Prot human database (downloaded July 7, 2020). Search criteria included carbamidomethylation of cysteine as a fixed modification, methionine oxidation and N-terminal protein acetylation as variable modifications, and trypsin/P with two missed cleavages as the enzyme. Mass tolerance was set at 4.5 ppm for precursor ions and 20 ppm for fragment ions. Peptide spectrum match and protein FDR were set at 0.01. For DDA peptide quantification, match between runs was performed with a matching time window of 0.7 min and an alignment window of 20 min. MS1 summed isotope intensities were used. For protein quantification, all peptide MS1 intensities for a given protein were summed to yield the total protein intensity. The MaxQuant output file was exported as an Excel file and was included in the supplemental Table S5. MaxLFQ method was used for proteome quantification. All required files from the MaxQuant analysis output have been uploaded to MS-Viewer (https://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer), the search key for the saved dataset is oohebltixe.
Enzyme-Linked Immunosorbent Assay
An enzyme-linked immunosorbent assay (ELISA) kit purchased from Lanpaibio was used to measure the level of LRP1 in serum and serum exosomes. All samples were diluted with diluent reagent (1:5) before analysis. Absorbance (A) was read at 450 nm.
Experiment on Animals
Animal study was approved by the Animal Ethics Committee of Xuzhou Medical University. Eighteen female BALB/c nude mice were purchased from Wei Tong Li Hua. HO8910 cells (2 × 107 cells/mouse) were injected subcutaneously into the armpit of nude mice to establish a xenograft model. After the tumors were formed, the 18 mice were randomly divided into three groups: PBS, SI-NC-Exos, and SI-LRP1-Exos. Exosomes (5 μg) were injected into the tumor every other day for 2 weeks. The long side (L) and the short side (W) were measured every 3 days, the tumor volume V = LW2/2.
Experimental Design and Statistical Rationale
To explore the proteomic profile of serum exosomes from EOC patients and find potential diagnostic markers for EOC. Exosomes isolated from five biological replicate of each experimental group for proteomic analysis. To assess the statistical significance of protein fold changes in mass spectrometry analysis, log2-fold change ratios were averaged across the biological replicates, and p-values were calculated using Student’s t test. For all cell experiments, three biological replicates were applied. For all animal experiments, six biological replicates were applied. Statistical analysis was performed using SPSS software (20.0), and all experimental data were expressed as the mean ± SEM. Differences between two groups were tested by Student’s t test. Comparison among three groups were performed using one-way analysis of variance. Differences were considered significant if p-value <0.05.
Results
Exosomes From Serum of EOC Patients Promote EOC Cells Migrate in Vitro
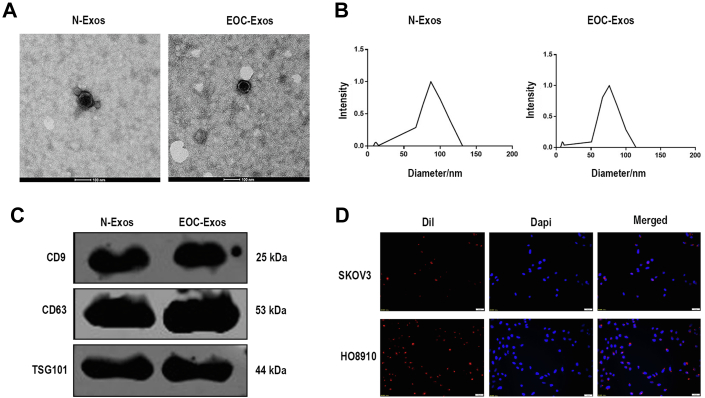
Purified serum exosomes from healthy volunteers (N-Exos) and EOC patients (EOC-Exos) were collected by ultracentrifugation. The TEM analysis showed that the morphologies of the exosomes were typical dish-like vesicles (Fig. 1A). We next performed dynamic light scattering analysis to observe the size of exosomes, results revealed the pellets with diameters ranging from 30 to 100 nm (Fig. 1B). The data confirmed that there was no significant difference between EOC-Exos and N-Exos in shape and size. In addition, Western blot showed that CD9, CD63, and TSG101, typical exosomal markers, were well expressed in exosomes from serum (Fig. 1C). In conclusion, our results confirmed that we have successfully isolated exosomes from serum. To confirm whether serum exosomes could be accepted by EOC cells, we stained exosomes with Dil to track the main location of exosomes in cells. We found that red fluorescently labeled exosomes could enter SKOV3 and HO8910 cells after incubation for 24 h (Fig. 1D).
Fig. 1.
Characterization of serum exosomes.A, TEM analysis of exosomes. B, DLS analysis of exosomes. C, Western blot analysis of exosomal markers. D, representative fluorescent photos of EOC cells (SKOV3 and HO8910) incubated with Dil-labeled exosomes (Dil in red and DAPI in blue). DLS, dynamic light scattering; EOC, epithelial ovarian cancer; EOC-Exos: exosomes from EOC patients; N-Exos: exosomes from healthy volunteers.
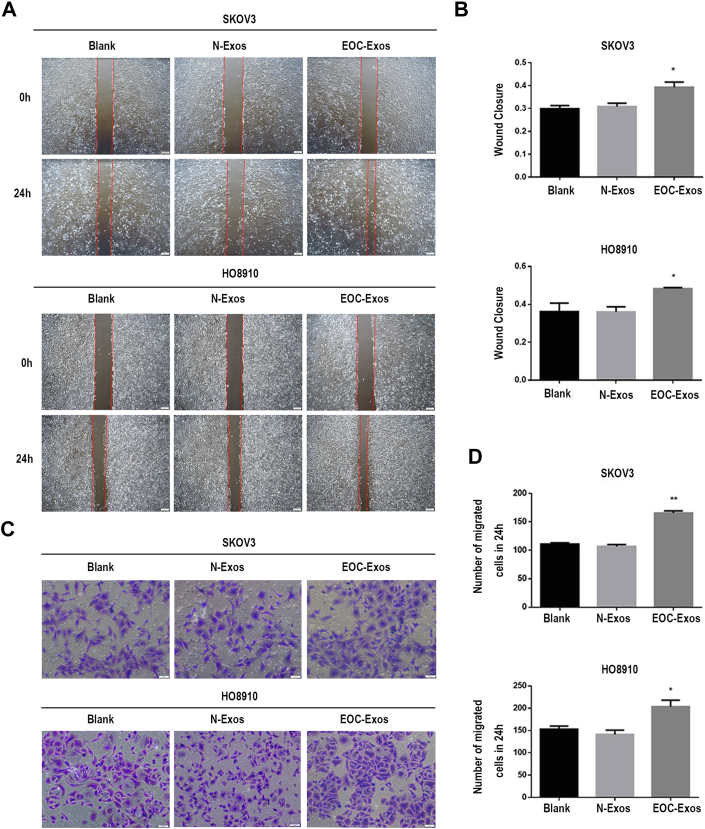
To gain insight into the effects of N-Exos and EOC-Exos on the migration of EOC, we performed wound healing and transwell assay to examine the migratory abilities of EOC cells. Figure 2, A and B demonstrated that the migratory abilities of EOC cells treated with EOC-Exos were significantly higher than that treated with N-Exos. In addition, EOC-Exos significantly increased the migratory abilities of both SKOV3 and HO8910 cells compared to N-Exos (Fig. 2, C and D). Cell counting kit-8 analysis showed that there was no significant difference in the proliferation abilities of cells treated with exosomes from different groups (supplemental Fig. S2A). Taken together, these findings showed that serum exosomes from EOC patients may promote the migration of EOC cells, which were consistent with previously published studies (22).
Fig. 2.
Serum exosomes from EOC patients promote EOC cell migration.A and B, the migratory abilities of SKOV3 and HO8910 cells incubated with 1× PBS or exosomes were determined by wound healing assay. C and D: the migratory abilities of SKOV3 and HO8910 cells incubated with 1× PBS or exosomes were observed by migration assay. Data are expressed as the mean ± SEM; n = 3, ∗p < 0.05, ∗∗p < 0.01, compared to the N-Exos group (Blank: negative control; N-Exos: exosomes from healthy volunteers; EOC-Exos: exosomes from EOC patients). EOC, epithelial ovarian cancer.
Quantitative Mass Spectral Analysis of Exosomes From Serum
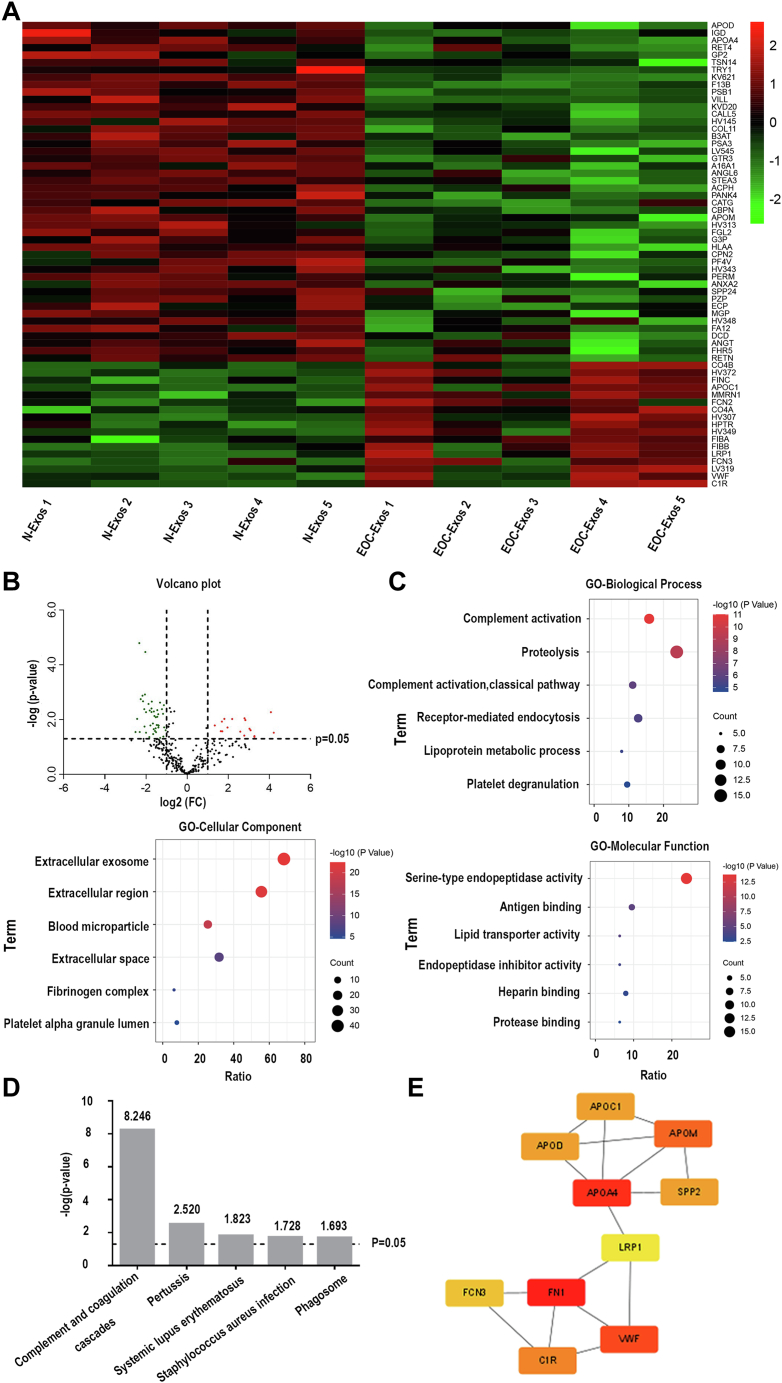
Three hundred eight proteins were identified in research groups, and 63 proteins were selected as DEPs between the two groups (fold change >2 or fold change <1/2 and p-value <0.05). As can be seen from Figure 3A, the DEPs were depicted in a heatmap, including 17 upregulated proteins and 46 downregulated proteins. Detailed information about the DEPs were shown in supplemental Table S6. Furthermore, we performed a volcano plot to display all quantified proteins (Fig. 3B).
Fig. 3.
Bioinformatics analysis of proteomic results.A, heatmap of DEPs. B, volcano plot of all quantified proteins. C, GO analysis of DEPs. The top six items are listed. D, KEGG pathway enrichment analysis of DEPs. Top five items are listed. E, top ten key proteins in the PPI network constructed by the STRING. DEPs, differentially expressed proteins; EOC, epithelial ovarian cancer; EOC-Exos: exosomes from EOC patients; GO, gene ontology; N-Exos: exosomes from healthy volunteers; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein–protein interaction.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Analysis of DEPs
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analysis of exosomal DEPs were performed using DAVID (https://david.ncifcrf.gov/) database. The top six items in cellular components, biological processes, and molecular functions were listed in Figure 3C. When referring to cell components, these DEPs were mainly enriched in extracellular exosomes. In the biological process category, exosomal DEPs were mainly related to complement activation, mediating endocytosis, and lipoprotein metabolism. The abnormal regulation of signal transduction may be related to the metastasis of cancer. Molecular function analysis showed that DEPs were involved in lipid transport and activity antigen binding. As shown in Figure 3D, Kyoto Encyclopedia of Genes and Genomes pathway analysis showed that serum exosomal DEPs were enriched in complement activation pathway.
Interaction Networks of DEPs
To gain further insight into the potential PPIs among 63 DEPs, the STRING (https://string-db.org) database was used for performing a PPI network. As shown in supplemental Fig. S3, the 29 nodes that mapped in PPI network represent 29 exosomal proteins. Subsequently, the PPI network was visualized in Cytoscape software (3.9.0). By using the MCC method in plugin cytoHubba, the ten high-scoring hub proteins (FN1, APOA4, VWF, APOM, C1R, APOC1, SPP2, APOD, FCN3, and LRP1) were selected from the comprehensive PPI network (Fig. 3E).
Exosomal LRP1 Was Associated With EOC Progression
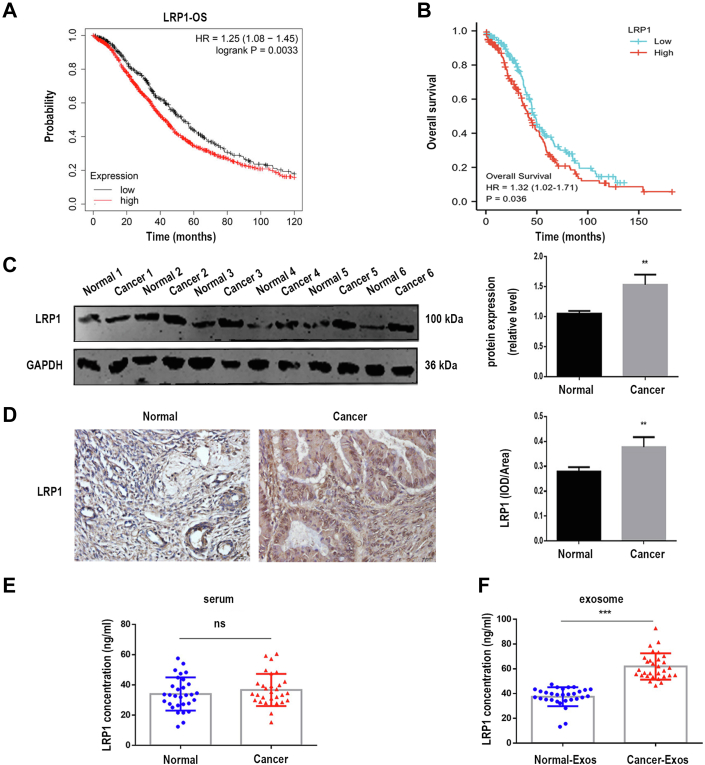
We used online Kaplan–Meier plotter (http://kmplot.com) and The Cancer Genome Atlas (https://genome-cancer.ucsc.edu) database to search for potential prognostic markers of EOC among the ten hub proteins. Surprisingly, we found that only LRP1 was related to EOC prognosis, and high expression of LRP1 lead to shorter overall survival of EOC patients (Fig. 4, A and B). These results implied that LRP1 might be a prognostic marker for EOC, and LRP1 was selected as the key protein. The expression of LRP1 in ovarian tissue was analyzed by Western blot and Immunohistochemistry. Figure 4C showed that increased LRP1 level was found in ovarian tissues of EOC patients compared with normal volunteers. IHC results also revealed that LRP1 was upregulated in EOC tissues (Fig. 4D). In an ELISA test, there were no differences in the levels of LRP1 in serum between healthy volunteers and EOC patients (Fig. 4E), while the levels of exosomal LRP1 were upregulated in EOC patients compared with healthy volunteers (Fig. 4F).
Fig. 4.
Exosomal LRP1 was associated with EOC progression.A, K-M Plotter was used to analysis the role of LRP1 in EOC prognosis. B, high expression of LRP1 was connected with shorter OS of EOC patients. C, Western blot analysis of LRP1 expression in ovarian tissues from EOC patients and benign disease (Normal: normal ovarian tissue from benign disease patients, n = 6; Cancer: ovarian tissue from EOC patients, n = 6). D, IHC results of LRP1 in EOC tissue sections (normal: n = 6; cancer: n = 50). Original magnification: 40× . E and F, ELISA revealed LRP1 concentration in serum and serum exosomes from healthy volunteers and EOC patients (normal: n = 30, cancer: n = 30). Data are expressed as the mean ± SEM; ∗∗p < 0.01, ∗∗∗p < 0.001, compared to the Normal group. EOC, epithelial ovarian cancer; LRP1, low-density lipoprotein receptor-related protein 1; OS, overall survival.
Exosomal LRP1 May Affect the Migration of EOC via the p-ERK/MMP2/MMP9 Signaling Pathway
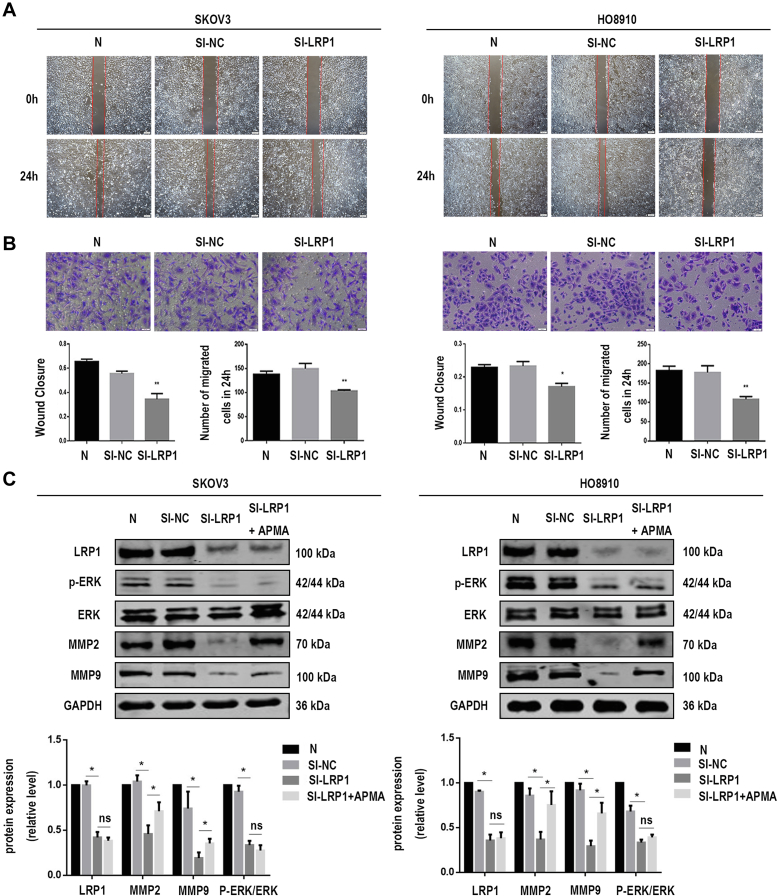
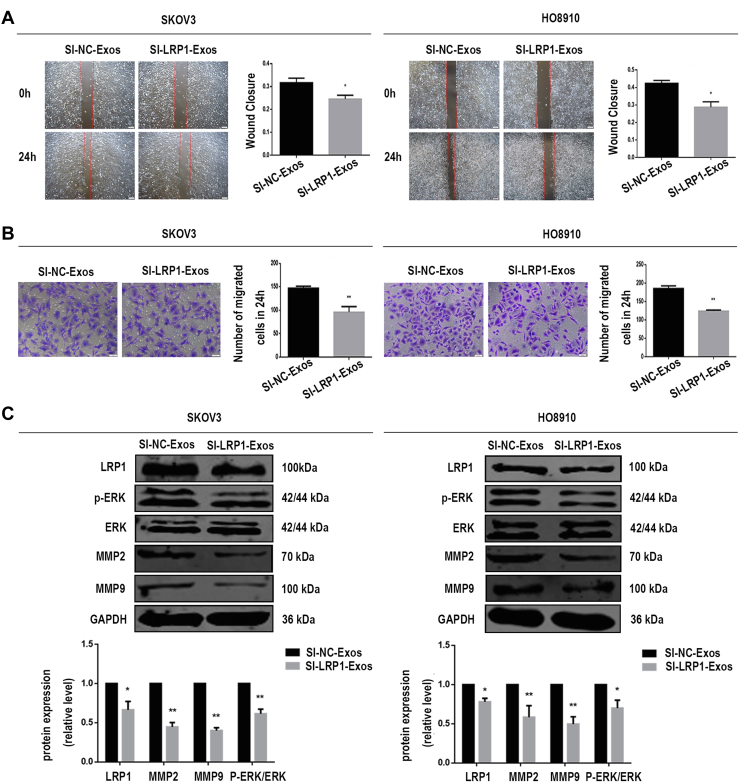
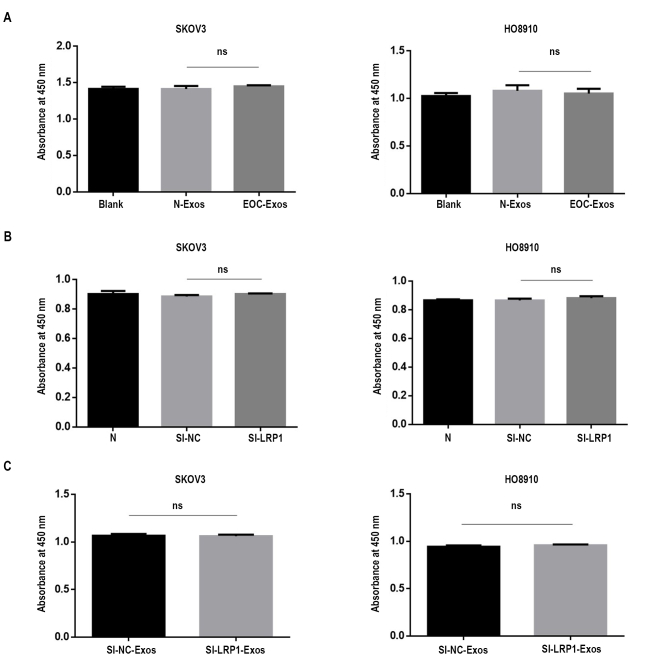
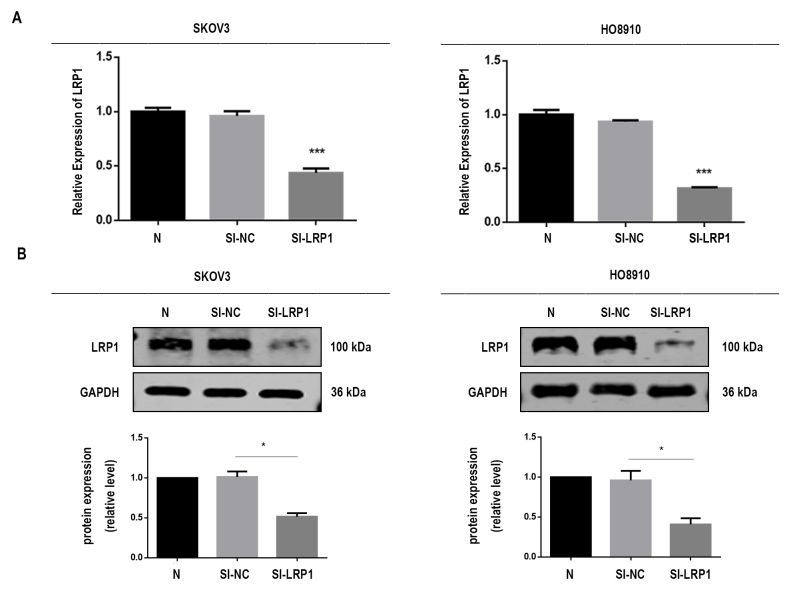
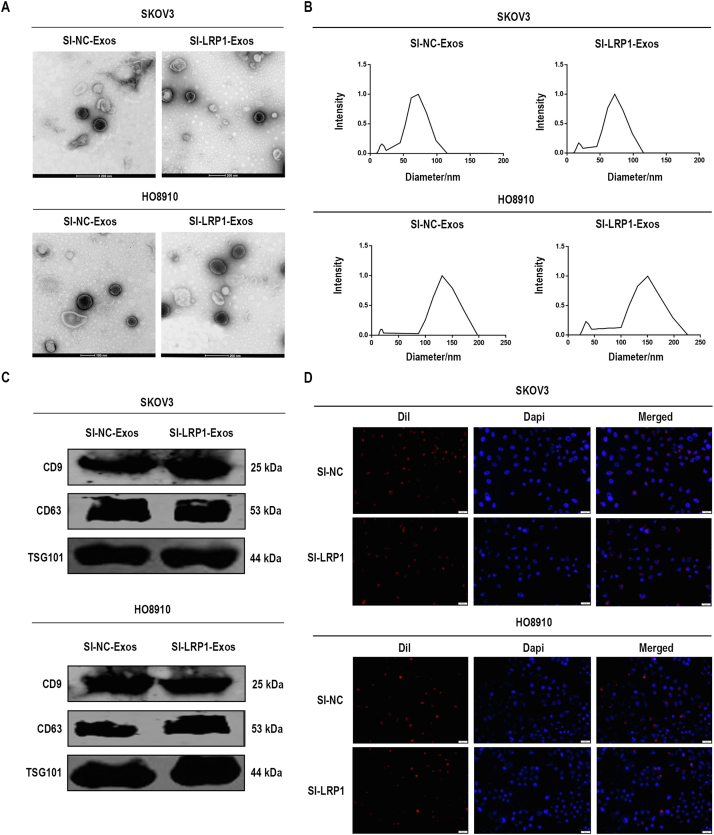
To address the biological role of exosomal LRP1 in migration, we knocked down LRP1 expression by using siRNA in SKOV3 and HO8910 cells to obtain SI-NC-Exos and SI-LRP1-Exos from the culture supernatants. RT–qPCR analysis and Western blot analysis confirmed LRP1 was downregulated in EOC cells (supplemental Fig. S4). As showed in supplemental Fig. S5, the characterization of exosomes, including the morphology (supplemental Fig. S5, A and B), markers (supplemental Fig. S5C), and penetration (supplemental Fig. S5D) of exosomes were not altered by LRP1 knockdown. Next, we explored the effects of LRP1 and exosomal LRP1 on the migration of EOC cells. It is noteworthy that LRP1 knockdown reduced migratory ability in SKOV3 and HO8910 cells (Fig. 5, A and B). Meanwhile, increased migration was observed in SKOV3 and HO8910 cells after treated with SI-NC-Exos compared with those treated with SI-LRP1-Exos (Fig. 6, A and B). The proliferation assay was performed to confirm cells are migratory and not proliferative. As shown in supplemental Fig. S2B, the proliferation abilities of EOC cells have not been altered by the regulation of LRP1. Moreover, no significant change was observed in the proliferation of EOC cells treated with SI-NC-Exos and SI-LRP1-Exos (supplemental Fig. S2C).
Fig. 5.
LRP1 contributes to EOC cell migration.A, the migratory abilities of transfected SKOV3 and HO8910 cells were determined by wound healing assay. B, the migratory abilities of transfected SKOV3 and HO8910 cells were determined by migration assay. C, the protein levels of LRP1, MMP2, MMP9, GAPDH, ERK, and p-ERK in SKOV3 and HO8910 cells after LRP1 downregulation. Data are expressed as the mean ± SEM; n = 3, ∗p < 0.05, ∗∗p < 0.01 compared to the SI-LRP1 group. EOC, epithelial ovarian cancer; LRP1, low-density lipoprotein receptor-related protein 1.
Fig. 6.
Exosomal LRP1 affects EOC cell migration through the p-ERK/MMP2/MMP9 signaling pathway.A, the migratory abilities of SKOV3 and HO8910 cells incubated with exosomes were determined by wound healing assay. B, the migratory abilities of SKOV3 and HO8910 cells incubated with exosomes were determined by migration assay. C, after incubated with exosomes (SI-NC-Exos, SI-LRP1-Exos), the protein levels of LRP1, MMP2, MMP9, GAPDH, ERK, and p-ERK in EOC cells. Data are expressed as the mean ± SEM; n = 3, ∗p < 0.05, ∗∗p < 0.01, compared to the SI-NC-Exos group (SI-NC-Exos: exosomes secreted by EOC cells transfected with SI-NC; SI-LRP1-Exos: exosomes secreted by EOC cells transfected with SI-LRP1). EOC, epithelial ovarian cancer; LRP1, low-density lipoprotein receptor-related protein 1.
The mechanism by which exosomal LRP1 affected the migration of EOC cells was also explored. According to previous reports, LRP1 regulated the expression of MMP2 and MMP9 through ERK signaling pathways. Matrix metalloproteinases (MMPs), especially MMP2 and MMP9, were dysregulated during the invasion and metastasis of various tumors. Moreover, MMPs can be activated in vitro by multiple mechanisms such as treatment with organomercurials. It has been suggested that organomercurials such as 4-aminophenylmercuric acetate is an organic mercury activator for MMPs. We found that the levels of MMP2, MMP9, and p-ERK were decreased in EOC cells with LRP1 downregulation (Fig. 5C). Meanwhile, MMPs agonist 4-aminophenylmercuric acetate rescued the expression of MMP2 and MMP9 after the knockdown of LRP1 in our study (Fig. 5C). A shown in Figure 6C, the protein levels of LRP1, MMP2, MMP9, and p-ERK in EOC cells incubated with SI-NC-Exos were significantly higher than those treated with SI-LRP1-Exos. Both secreted LRP1 and exosomal LRP1 affected the migration of EOC cells through p-ERK/MMP2/MMP9 signaling pathway. Therefore, we concluded that the potential mechanism that exosomal LRP1 affected EOC migration may be similar to secreted LRP1.
Exosomal LRP1 Facilitated Tumor Migration in Vivo
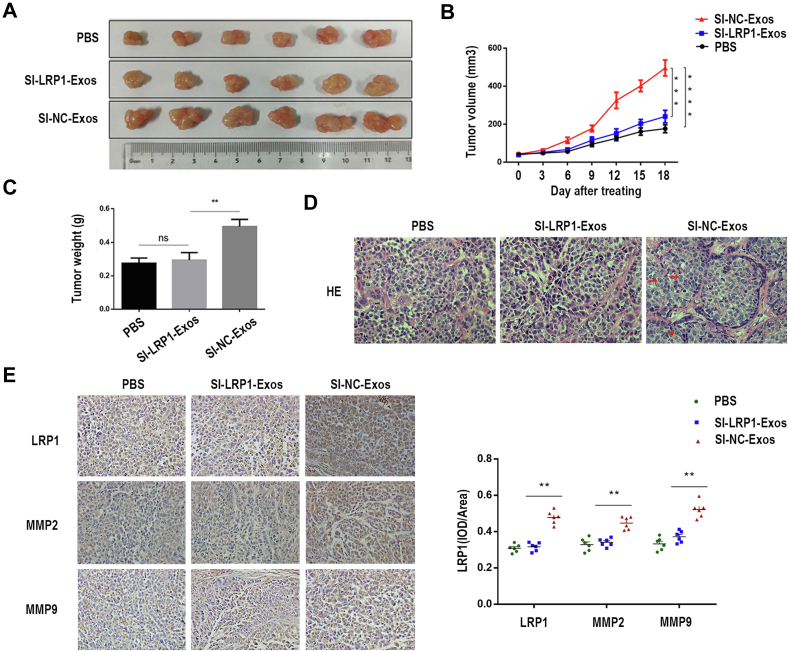
In order to identify the effects of exosomal LRP1 on the migration of EOC in vivo, we performed a xenograft model of EOC in nude mice. Images of tumors of all nude mice were shown in Figure 7A. As shown in Figure 7, B and C, mice treated with SI-NC-Exos had larger and heavier tumors. Compared with the SI-LRP1-Exos, the tumor cells treated with SI-NC-Exos showed diffuse infiltrating growth, no specific structural characteristics, and obvious cell atypia (Fig. 7D). Furthermore, higher levels of LRP1, MMP2, and MMP9 were in tumor tissue treated with SI-NC-Exos (Fig. 7E). These findings suggested that exosomal LRP1 could facilitate the migration of EOC cells in vivo.
Fig. 7.
The role of exosomal LRP1 in tumor migration in vivo.A, images of tumor tissue. B, tumor growth curve. C, statistical map of tumor weight. D, HE staining of tumor tissues. Original magnification: 40× . E, representative images of IHC staining show the levels of LRP1, MMP2, and MMP9 in tumors. Data are expressed as the mean ± SEM; n = 6, ∗∗p < 0.01, compared to the SI-NC-Exos group (SI-NC-Exos: exosomes secreted by EOC cells transfected with SI-NC; SI-LRP1-Exos: exosomes secreted by EOC cells transfected with SI-LRP1). EOC, epithelial ovarian cancer; LRP1, low-density lipoprotein receptor-related protein 1.
Discussion
It has been reported that the lack of early specific symptoms of patients is an important reason for the poor prognosis and high mortality of EOC (23). The early detection of EOC genesis is the key to improving the survival and prognosis of EOC patients. Vaginal ultrasound examination and serum CA125 examination are recognized diagnostic technologies for OC, and they cannot detect OC at an early stage due to their poor sensitivity and specificity (24). The tumor microenvironment provides important conditions for tumor growth and metastasis. Exosomes secreted by cancer cells are important players in the formation of tumor microenvironment, which may facilitate cell proliferation and metastasis (25), drug resistance (26), tumor angiogenesis (27), and the formation of premetastatic niches (28). Detecting protein profiles of serum exosomes from EOC patients might provide a new perspective for EOC diagnosis and prognosis.
Accumulated results have demonstrated exosomes derived from tumor cells could promote cancer metastasis. We also found that serum exosomes from EOC patients promoted EOC cells migration (Fig. 2). As far as we know, there were few studies about the systematic proteomic analysis of serum exosomes from EOC patients. We performed proteomic analysis to explore proteins of serum exosomes, and the DEPs were shown in Figure 3, A and B. PPI networks play a key role in regulating cell signaling. The top ten most pivotal proteins were selected among DEPs. Bioinformatics analysis indicated that high level of LRP1 may predict a poor prognosis of EOC patients. Then, LRP1 was selected as a candidate protein for further studies.
LRP1 is involved in the receptor-mediated endocytosis pathway as shown in GO-BP analysis (Fig. 3C). The preprotein encoded by LRP1 is enzymatically hydrolyzed to form a mature receptor, which takes part in intracellular signal transduction and lipid homeostasis. The level of LRP1 was dysregulated in many tumors. Elevated levels of LRP1 were observed in serum exosomes from EOC patients by proteomics analysis and ELISA in our study (Fig. 4F). LRP1 may promote tumor development and tumor cell migration, especially in epithelial malignant tumors. It had been reported that the mechanism of LRP1 affecting the migratory and invasive ability of glioblastoma cells was that LRP1 regulated the levels of MMP2 and MMP9 through p-ERK (29). Langlois (30) reported that LRP1 contributed to malignant invasion of cancer cells by activating ERK and inhibiting JNK. At present, the mechanism of exosomal LRP1 affecting the migration of EOC has not been reported in detail. In this study, we provided evidence that secreted LRP1, and exosomal LRP1 affected EOC cell migration in vitro (Figs. 5, A and B and 6, A and B). As shown in Figures 5C and 6C, we proved that both LRP1 and exosomal LRP1 affected EOC cell migration via the p-ERK/MMP2/MMP9 signaling pathway, which indicated that exosomal LRP1 was likely to utilize a molecular mechanism similar to that of secreted LRP1 for regulating cancer migration. Exosomes are important mediators of LRP1 transfer between cells. Exosomal LRP1 can be stably transported to recipient cells due to the advantage of lipid bilayer structure. However, the detailed mechanism of exosomal LRP1 delivery to EOC cells remains unknown.
A previous study performed proteomic analysis of purified exosomes from NCI-H838 cells, and they showed that LRP1 may be a potential biomarker for lung cancer (31). Similarly, exosomal CD91 (LRP1) was identified as one of the candidate markers of lung adenocarcinoma by mass spectrometric quantification (32). In our study, high levels of LRP1 may lead to shorter overall survival of EOC patients. Hence, our research confirmed that circulating exosomal LRP1 may be a promising diagnostic marker of EOC.
Above all, our results showed that the levels of LRP1 were increased in exosomes from serum of EOC patients. In addition, exosomal LRP1 affected the migration of EOC cells through p-ERK/MMP2/MMP9 signaling pathway. Based on this study, we proposed exosomal LRP1 from serum might be a promising diagnostic and prognostic factor for EOC for the first time. However, further research should be conducted to reveal the potential value of exosomal LRP1 due to the limited sample size of this study.
Data Availability
All data described in the manuscript are contained within the manuscript and the supplemental data. All raw mass spectrometry data have been uploaded to the ProteomeXchange Consortium via the iProX repository with the dataset identifier PXD039347.
The dataset can be accessed using the following URL: https://www.iprox.cn/page/PSV023.html;?url=1676444019997O7Xe; Password: 93HU.
Supplemental Data
This article contains supplemental data.
Conflicts of interest
All authors declare no competing interest.
Acknowledgments
We thank all volunteers in this study for their cooperation and the support of gynecologists in Xuzhou Central Hospital.
Funding and additional information
This work was supported by the Natural Science Foundation of China [No. 82173883]; the Science and Technology Foundation of Xuzhou [No. KC21010]; the Natural Science Foundation of the Jiangsu Higher Education Institutions of China [No. 18KJA350002]; the Natural Science Foundation of Jiangsu Province [No. BK20181470]; the Provincial Commission of Health and Family Planning in Jiangsu Province [No. H2017079]; the Science and Technology Planning Project of Jiangsu Province [No. BE2019636].
Author contributions
X. Z. supervision; X. Z. conceptualization; B. Z. funding acquisition; B. Z. project administration; W. Z. and J. M. writing-original draft; W. Z., J. M., and Z. C. methodology; W. Z. writing-review and editing; H. Z. and Z. C. data curation; H. Z., X. G., and J. X. software; H. Z. visualization; Q. W. and L. C. resources; Q. W. investigation; X. G. validation; J. X. formal analysis.
Contributor Information
Bei Zhang, Email: bettyzhang10@163.com.
Xueyan Zhou, Email: zxy851107@xzhmu.edu.cn.
Supplementary Data
Supplemental Figure S1.
Flow diagram of inclusion criteria and exclusion criteria for this study population.
Supplemental Figure S2.
The proliferation abilities of SKOV3 and HO8910 cells. A, the proliferation abilities of SKOV3 and HO8910 incubated with 1× PBS or serum exosomes. B, the proliferation abilities of transfected SKOV3 and HO8910 cells. C, the proliferation abilities of SKOV3 and HO8910 cells incubated with exosomes.
Supplemental Figure S3.
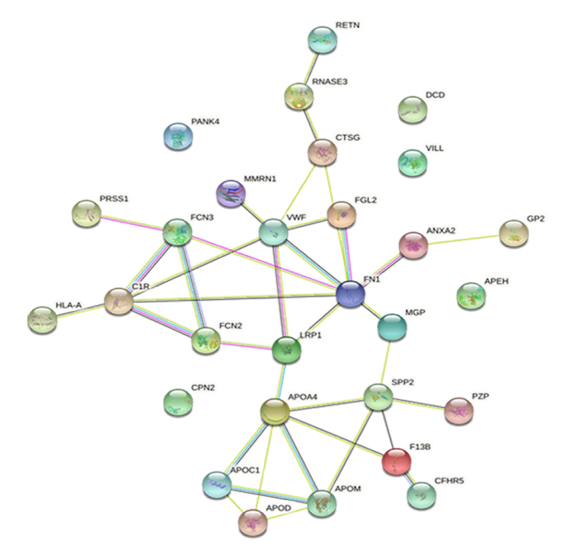
Protein–protein interaction network of DEPs.
Supplemental Figure S4.
The mRNA and protein levels of LRP1 in transfected SKOV3 and HO8910 cells. A, mRNA levels of LRP1. B, the protein levels of LRP1.
Supplemental Figure S5.
Characterization of exosomes from transfected SKOV3 and HO8910 cells. A, TEM analysis of exosomes. B, DLS analysis of exosomes. C, Western blot analysis of exosomal markers. D, representative fluorescent photos of SKOV3 and HO8910 cells incubated with Dil-labeled exosomes (Dil in red and DAPI in blue). DLS, dynamic light scattering; TEM, transmission electron microscopy.
References
- 1.Li X., Wang X. The emerging roles and therapeutic potential of exosomes in epithelial ovarian cancer. Mol. Cancer. 2017;16:92. doi: 10.1186/s12943-017-0659-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reid B.M., Permuth J.B., Sellers T.A. Epidemiology of ovarian cancer: a review. Cancer Biol. Med. 2017;14:9–32. doi: 10.20892/j.issn.2095-3941.2016.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Webb P.M., Jordan S.J. Epidemiology of epithelial ovarian cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2017;41:3–14. doi: 10.1016/j.bpobgyn.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 5.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., et al. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 6.Torre L.A., Trabert B., DeSantis C.E., Miller K.D., Samimi G., Runowicz C.D., et al. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018;68:284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang M.K., Wong A.S. Exosomes: emerging biomarkers and targets for ovarian cancer. Cancer Lett. 2015;367:26–33. doi: 10.1016/j.canlet.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 8.Song Y., Wang M., Tong H., Tan Y., Hu X., Wang K., et al. Plasma exosomes from endometrial cancer patients contain LGALS3BP to promote endometrial cancer progression. Oncogene. 2021;40:633–646. doi: 10.1038/s41388-020-01555-x. [DOI] [PubMed] [Google Scholar]
- 9.Maji S., Chaudhary P., Akopova I., Nguyen P.M., Hare R.J., Gryczynski I., et al. Exosomal annexin II promotes angiogenesis and breast cancer metastasis. Mol. Cancer Res. 2017;15:93–105. doi: 10.1158/1541-7786.MCR-16-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu H., Yang H., Zhang X., Wang B., Mao J., Li X., et al. Exosomal TRIM3 is a novel marker and therapy target for gastric cancer. J. Exp. Clin. Cancer Res. 2018;37:162. doi: 10.1186/s13046-018-0825-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao M., Zhang J., Chen W., Chen W. M1-like tumor-associated macrophages activated by exosome-transferred THBS1 promote malignant migration in oral squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2018;37:143. doi: 10.1186/s13046-018-0815-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W., Li C., Zhou T., Liu X., Liu X., Li X., et al. Role of exosomal proteins in cancer diagnosis. Mol. Cancer. 2017;16:145. doi: 10.1186/s12943-017-0706-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herz J., Hamann U., Rogne S., Myklebost O., Gausepohl H., Stanley K.K. Surface location and high affinity for calcium of a 500-KD liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 1988;7:4119–4127. doi: 10.1002/j.1460-2075.1988.tb03306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herz J., Strickland D.K. Lrp: a multifunctional scavenger and signaling receptor. J. Clin. Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang H.S., Kim J., Lee H.J., Kwon B.M., Lee D.K., Hong S.H. LRP1-dependent pepsin clearance induced by 2'-hydroxycinnamaldehyde attenuates breast cancer cell invasion. Int. J. Biochem. Cell Biol. 2014;53:15–23. doi: 10.1016/j.biocel.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 16.Muratoglu S.C., Mikhailenko I., Newton C., Migliorini M., Strickland D.K. Low density lipoprotein receptor-related protein 1 (LRP1) forms a signaling complex with platelet-derived growth factor receptor-beta in endosomes and regulates activation of the MAPK pathway. J. Biol. Chem. 2010;285:14308–14317. doi: 10.1074/jbc.M109.046672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mantuano E., Lam M.S., Gonias S.L. LRP1 assembles unique co-receptor systems to initiate cell signaling in response to tissue-type plasminogen activator and myelin-associated glycoprotein. J. Biol. Chem. 2013;288:34009–34018. doi: 10.1074/jbc.M113.509133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montel V., Gaultier A., Lester R.D., Campana W.M., Gonias S.L. The low-density lipoprotein receptor-related protein regulates cancer cell survival and metastasis development. Cancer Res. 2007;67:9817–9824. doi: 10.1158/0008-5472.CAN-07-0683. [DOI] [PubMed] [Google Scholar]
- 19.Bian W., Tang M., Jiang H., Xu W., Hao W., Sui Y., et al. Low-density-lipoprotein-receptor-related protein 1 mediates notch pathway activation. Dev. Cell. 2021;56:2902–2919 e2908. doi: 10.1016/j.devcel.2021.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Taylor D.D., Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 21.Zhou X., Xu Y., Yin D., Zhao F., Hao Z., Zhong Y., et al. Type 2 diabetes mellitus facilitates endometrial hyperplasia progression by activating the proliferative function of mucin o-glycosylating enzyme GALNT2. Biomed. Pharmacother. 2020;131:110764. doi: 10.1016/j.biopha.2020.110764. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura K., Sawada K., Kobayashi M., Miyamoto M., Shimizu A., Yamamoto M., et al. Role of the exosome in ovarian cancer progression and its potential as a therapeutic target. Cancers (Basel) 2019;11:1147. doi: 10.3390/cancers11081147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coleman R.L., Monk B.J., Sood A.K., Herzog T.J. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat. Rev. Clin. Oncol. 2013;10:211–224. doi: 10.1038/nrclinonc.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W., Ou X., Wu X. Proteomics profiling of plasma exosomes in epithelial ovarian cancer: a potential role in the coagulation cascade, diagnosis and prognosis. Int. J. Oncol. 2019;54:1719–1733. doi: 10.3892/ijo.2019.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu J., Liu B., Wang Z., Wang D., Ni H., Zhang L., et al. Exosomes from nicotine-stimulated macrophages accelerate atherosclerosis through miR-21-3p/PTEN-mediated VSMC migration and proliferation. Theranostics. 2019;9:6901–6919. doi: 10.7150/thno.37357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milman N., Ginini L., Gil Z. Exosomes and their role in tumorigenesis and anticancer drug resistance. Drug Resist. Updat. 2019;45:1–12. doi: 10.1016/j.drup.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Ludwig N., Yerneni S.S., Azambuja J.H., Gillespie D.G., Menshikova E.V., Jackson E.K., et al. Tumor-derived exosomes promote angiogenesis via adenosine A2B receptor signaling. Angiogenesis. 2020;23:599–610. doi: 10.1007/s10456-020-09728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng W., Dean D.C., Hornicek F.J., Shi H., Duan Z. Exosomes promote pre-metastatic niche formation in ovarian cancer. Mol. Cancer. 2019;18:124. doi: 10.1186/s12943-019-1049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song H., Li Y., Lee J., Schwartz A.L., Bu G. Low-density lipoprotein receptor-related protein 1 promotes cancer cell migration and invasion by inducing the expression of matrix metalloproteinases 2 and 9. Cancer Res. 2009;69:879–886. doi: 10.1158/0008-5472.CAN-08-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langlois B., Perrot G., Schneider C., Henriet P., Emonard H., Martiny L., et al. LRP-1 promotes cancer cell invasion by supporting ERK and inhibiting JNK signaling pathways. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan D., Chen J., Feng C., Wu W., Wang Y., Tong J., et al. Preferential localization of MUC1 glycoprotein in exosomes secreted by non-small cell lung carcinoma cells. Int. J. Mol. Sci. 2019;20:323. doi: 10.3390/ijms20020323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueda K., Ishikawa N., Tatsuguchi A., Saichi N., Fujii R., Nakagawa H. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci. Rep. 2014;4:6232. doi: 10.1038/srep06232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in the manuscript are contained within the manuscript and the supplemental data. All raw mass spectrometry data have been uploaded to the ProteomeXchange Consortium via the iProX repository with the dataset identifier PXD039347.
The dataset can be accessed using the following URL: https://www.iprox.cn/page/PSV023.html;?url=1676444019997O7Xe; Password: 93HU.