SUMMARY
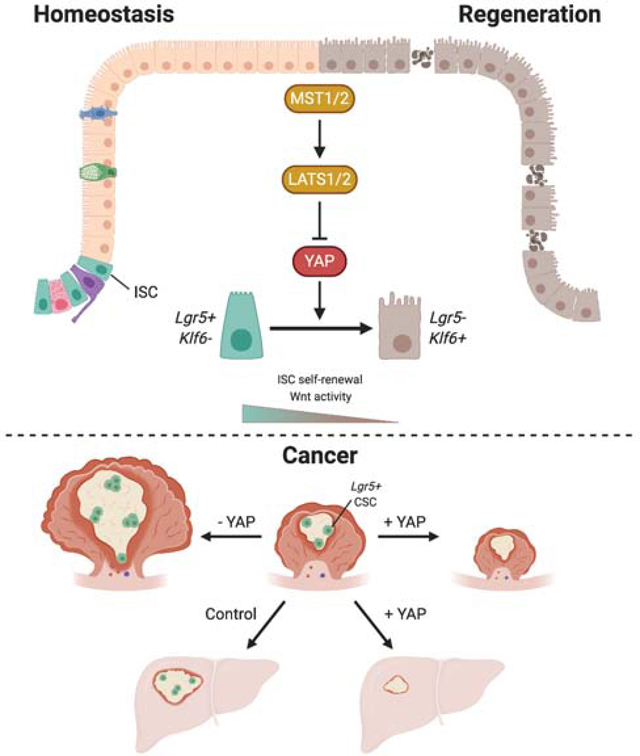
While the Hippo transcriptional coactivator YAP is considered oncogenic in many tissues, its roles in intestinal homeostasis and colorectal cancer (CRC) remain controversial. Here, we demonstrate that the Hippo kinases LATS1/2 and MST1/2, which inhibit YAP activity, are required for maintaining Wnt signaling and canonical stem cell function. Hippo inhibition induces a distinct epithelial cell state marked by low Wnt signaling, a wound healing response, and transcription factor Klf6 expression. Notably, loss of LATS1/2 or overexpression of YAP is sufficient to reprogram Lgr5+ cancer stem cells to this state and thereby suppress tumor growth in organoids, patient-derived xenografts, and mouse models of primary and metastatic CRC. Finally, we demonstrate that genetic deletion of YAP and its paralog TAZ promotes the growth of these tumors. Collectively, our results establish the role of YAP as a tumor suppressor in the adult colon and implicate Hippo kinases as therapeutic vulnerabilities in colorectal malignancies.
eTOC Blurb
Hippo inhibition reprograms intestinal stem cells to a wound healing-like cell state that can be induced to suppress tumorigenesis in human and mouse metastatic colorectal cancer.
Graphical Abstract
INTRODUCTION
The gut epithelium is organized in a monolayer of cells that carries out essential absorptive, barrier, and endocrine functions (Peterson & Artis, 2014). Cellular turnover is sustained by highly proliferative Lgr5+ stem cells found at the bottom of intestinal and colonic crypts (Barker et al., 2007), and the presence of an active Wnt signaling pathway acts as a defining property for these stem cells. Wnt agonists are secreted by neighboring cells, forming the niche, and as cells move up the crypt, they undergo differentiation (Gehart & Clevers, 2019). Thus, regulated control of this pathway is essential for maintaining normal crypt structure and function.
The transcriptional co-activator YAP, which is negatively regulated by the Hippo kinases MST1/2 and LATS1/2, plays an important role in maintaining intestinal homeostasis. It has been shown to mediate epithelial repair following injury (Cai et al., 2010; Gregorieff et al., 2015; Yui et al., 2018), but the molecular events that follow YAP activation remain controversial. We previously have shown that YAP overexpression in the intestine leads to Wnt inhibition and crypt degeneration (Barry et al., 2013), but subsequent reports on the effects of YAP on Wnt signaling have been contradictory (Azzolin et al., 2014; Cai et al., 2015; Gregorieff et al., 2015). YAP is also known to induce a transcriptional program that generally is associated with promoting stemness and proliferation (Fu et al., 2017) and has been shown to function as an oncogene in a number of tissues, including pancreas, liver, and skin. Its role in colorectal cancer (CRC) however is debated (Hong et al., 2016).
The most common event that triggers formation of colon tumors is the acquisition of mutations that constitutively activates Wnt signaling, which is thought to be the first step in the sequence that leads to malignant transformation of colon epithelial cells (Fearon & Vogelstein, 1990; Siegel et al., 2014). The growth of colon tumors is driven by cancer stem cells that are labeled by Lgr5 as in healthy tissue (Merlos-Suárez et al., 2011; Schepers et al., 2012). As targeting the Wnt pathway for the treatment of CRC has been historically challenging, the recent discovery that depletion of Lgr5+ cancer stem cells can arrest tumor growth and prevent metastasis in preclinical models has opened the door for developing strategies to specifically target this cell population (De Sousa E Melo et al., 2017; Shimokawa et al., 2017). Thus, crosstalk between the Hippo and Wnt signaling pathways could serve as a potential therapeutic window for which treatment of CRC could be developed.
Here, we use multiple genetic models to uncover a role for YAP in reprogramming gut epithelial cells into a state distinct from those found in steady-state intestine. This state is characterized molecularly by a wound healing signature, expression of Krüppel-like factor 6 (Klf6), and low levels of Wnt signaling. Using mouse and human models of CRC, we show that YAP is capable of reprogramming cancer stem cells into a low Wnt, non-proliferative state, which leads to tumor regression in primary and metastatic disease. In addition, deletion of YAP favors growth of focally induced colonic tumors, further supporting the notion that YAP acts as a tumor suppressor in the colon and that targeting the Hippo kinases represents a novel therapeutic approach for combatting CRC.
RESULTS
YAP Reprograms Intestinal Epithelial Cells into a Wound Healing State
Our previous data show that overexpression of an active version of YAP in the mouse intestine leads to Wnt inhibition and degeneration of intestinal crypts (Barry et al., 2013). To explore fully the cellular and molecular consequences of YAP dysregulation in the intestinal mucosa, we studied additional models of YAP activation via deletion of its upstream negative regulatory kinases LATS1/2 and MST1/2. First, we used a Lrig1-CreERT2 driver to delete the LATS1/2 kinases in mouse intestinal stem cells (ISCs) (hereafter referred to as Lats1/2 cKO) and transcriptionally profiled single cells isolated from the small intestine of these mice seven days after one dose of tamoxifen injection (Figure S1A) (Powell et al., 2012). Unsupervised clustering followed by uniform manifold approximation and projection (UMAP) representation of the single-cell gene expression profiles reveals a distinct clustering of the Lats1/2 knockout cells from the control cells (Figure S1B) (McInnes et al., 2018). While the cells in the controls could be identified by markers of normal intestinal cell states, the Lats1/2 knockout cells could not be assigned to any known intestinal cell type (Figures 1A and S1C-D). The Lats1/2 knockout cells lack expression of characteristic markers of normal intestinal states but still belong to the intestinal lineage as seen by expression of Villin and Cdx2 (Figure S1D), thus defining a distinct cell type not present in the homeostatic intestinal epithelium. Upregulated genes in Lats1/2 knockout cells relative to control cells are overrepresented in gene ontology (GO) terms involved in wound healing and actin reorganization, which have been associated with YAP activation (Cai et al., 2010; Dupont et al., 2011; Zhao et al., 2007, 2011, 2012), and those downregulated are overrepresented in terms related to intestinal absorption and villus organization (Figure 1B and Table S1). Gene set enrichment analysis (GSEA) confirms that the Lats1/2 knockout cells are highly enriched for two previously published YAP gene signatures and reveals that they are negatively enriched for intestinal β-catenin targets compared to the stem cells (Figures S1E-F and 1C) (Barry et al., 2013; Fevr et al., 2007; Gregorieff et al., 2015). Upregulated genes include known targets of YAP, such as AmotL2, and fetal marker Ly6a/Sca1, which has been identified to label a cell state induced after damage or infection (Figure 1D) (Nusse et al., 2018; Yui et al., 2018). Interestingly, these cells are also marked by high expression of the transcription factor Klf6, which has been characterized as an immediateearly gene in response to injury and implicated to be a tumor suppressor in CRC (Inuzuka et al., 1999; Kojima et al., 2000; Ratziu et al., 1998; Reeves et al., 2004). These findings altogether suggest a role for YAP in molecularly reprogramming epithelial cells into a distinct transcriptional state that is not present in steady-state intestine.
Figure 1: YAP Reprograms Intestinal Epithelial Cells into a Wound Healing State.
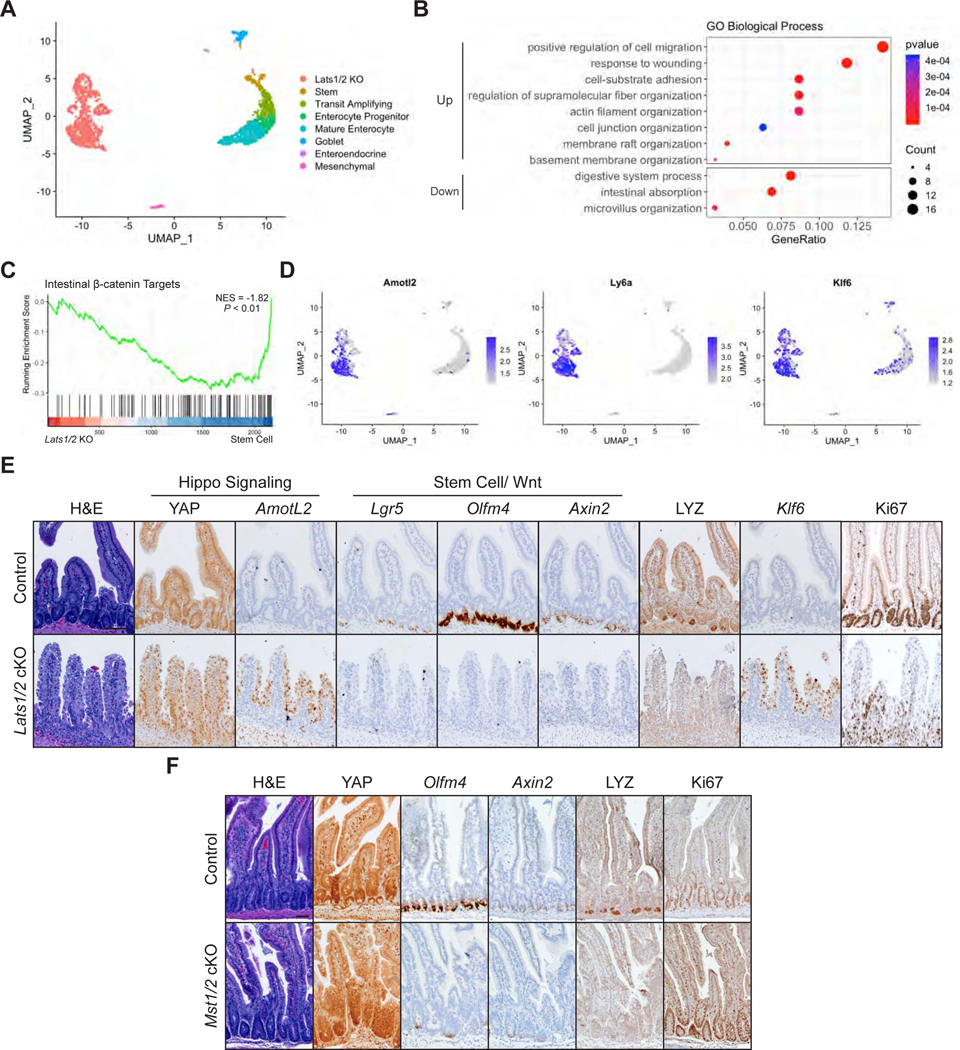
(A) UMAP representation of single cell transcriptome profiles of intestinal epithelial cells from 2 Lats1fl/fl Lats2fl/fl (control) and 2 Lrig1-CreERT2 Lats1fl/fl Lats2fl/fl (Lats1/2 cKO) animals 7 days after tamoxifen induction. Clusters are annotated by cell type based on expression of known marker genes.
(B) GO enrichment analysis of upregulated and downregulated genes in Lats1/2 KO cells compared to control cells, excluding the mesenchymal population.
(C) GSEA of intestinal β-catenin targets in Lats1/2 KO cells compared to the stem cells.
(D) Normalized expression of the indicated genes overlaid on the UMAP plots.
(E) Histological analysis of control and Lats1/2 cKO duodenal samples 7 days after tamoxifen induction: H&E stains, IHC for YAP, LYZ, and Ki67, and RNA-ISH for Amotl2, Lgr5, Olfm4, Axin2, and Klf6. Scale bar, 100 μm.
(F) Histological analysis of Mst1−/− Mst2fl/fl (control) and Vil-Cre Mst1−/− Mst2fl/fl (Mst1/2 cKO) samples: IHC for YAP, LYZ, and Ki67 and RNA-ISH for Olfm4 and Axin2. Scale bar, 50 μm.
To validate these results, we performed immunohistochemistry analysis of the intestine from Lats1/2 cKO mice seven days post tamoxifen treatment. This analysis reveals an accumulation of YAP in the nucleus and expression of the YAP target gene AmotL2 (Figure 1E). Consistent with the single-cell RNA-sequencing (scRNA-seq) data, characteristic markers of normal intestinal cell states are not present, such as those associated with ISCs (Lgr5 and Olfm4), secretory cells (LYZ), and targets of the Wnt signaling pathway (Axin2). Lats1/2 knockout cells retain their proliferative capacity as observed by staining with Ki67 and show marked increase in Klf6 levels, corroborating our scRNA-seq findings of YAP-mediated induction of a distinct Klf6+ cell state.
This loss of typical intestinal cellular organization was confirmed in additional models for activation of endogenous YAP through knockout of the MST1/2 kinases in Mst1−/− Mst2fl/fl mice via an intestinal specific, developmental Villin-Cre driver and the adult-inducible Villin-CreERT2 driver (hereafter referred to as Mst1/2 cKO) (El Marjou et al., 2004; Madison et al., 2002; Zhou et al., 2011). In both models, we observed loss of ISC markers and Wnt targets accompanied by YAP activation (Figures 1F and S1G). Altogether, our results point to a requirement for Hippo kinases in maintaining Wnt signaling in ISCs and suggest that persistent YAP activation cannot sustain normal homeostasis.
Activation of YAP Leads to Loss of Canonical Stem Cell Properties
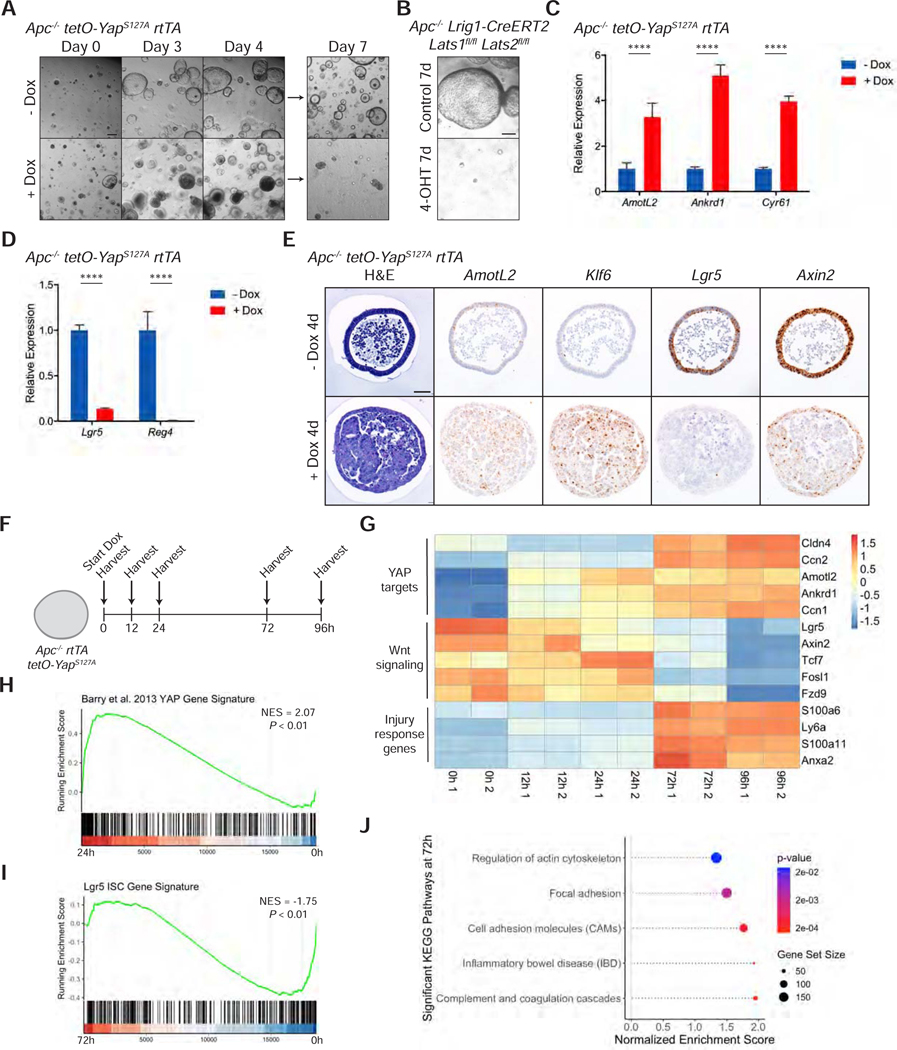
As YAP-expressing cells are transcriptionally distinct from normal ISCs, we examined their capacity to form intestinal organoids. To test the effect of YAP activation in organoids, we generated a colon organoid line that would allow for inducible overexpression of YAPS127A, a transcriptionally active mutant form of YAP (hereafter referred to as tetO-YAP rtTA) (Figure S2A) (Yimlamai et al., 2014; Zhao et al., 2007). Induction of YAP expression by doxycycline for 48 hours causes the organoids to lose single cell, columnar organization and prevents them from growing or forming spheroids upon passage (Figure 2A). Lats1/2 cKO colon organoids upon 4-OHT treatment also exhibit a similar phenotype of not being able to grow or form 3D structures (Figure 2B), which is not due to 4-OHT toxicity (Figure S2B). In both contexts of YAP activation, we observed upregulation of mRNA levels of YAP targets and downregulation of ISC and secretory cell markers and the Wnt target Axin2 (Figures 2C-D and S2C). Histological analysis confirms these results and reveals a progressive loss of the monolayer structure, upregulation of Klf6 expression, and proliferation arrest (Figure 2E). These results collectively suggest that this Wnt-low, Klf6-high state induced by YAP is incompatible with organoid growth in vitro.
Figure 2: Activation of YAP Leads to Loss of Canonical Stem Cell Properties.
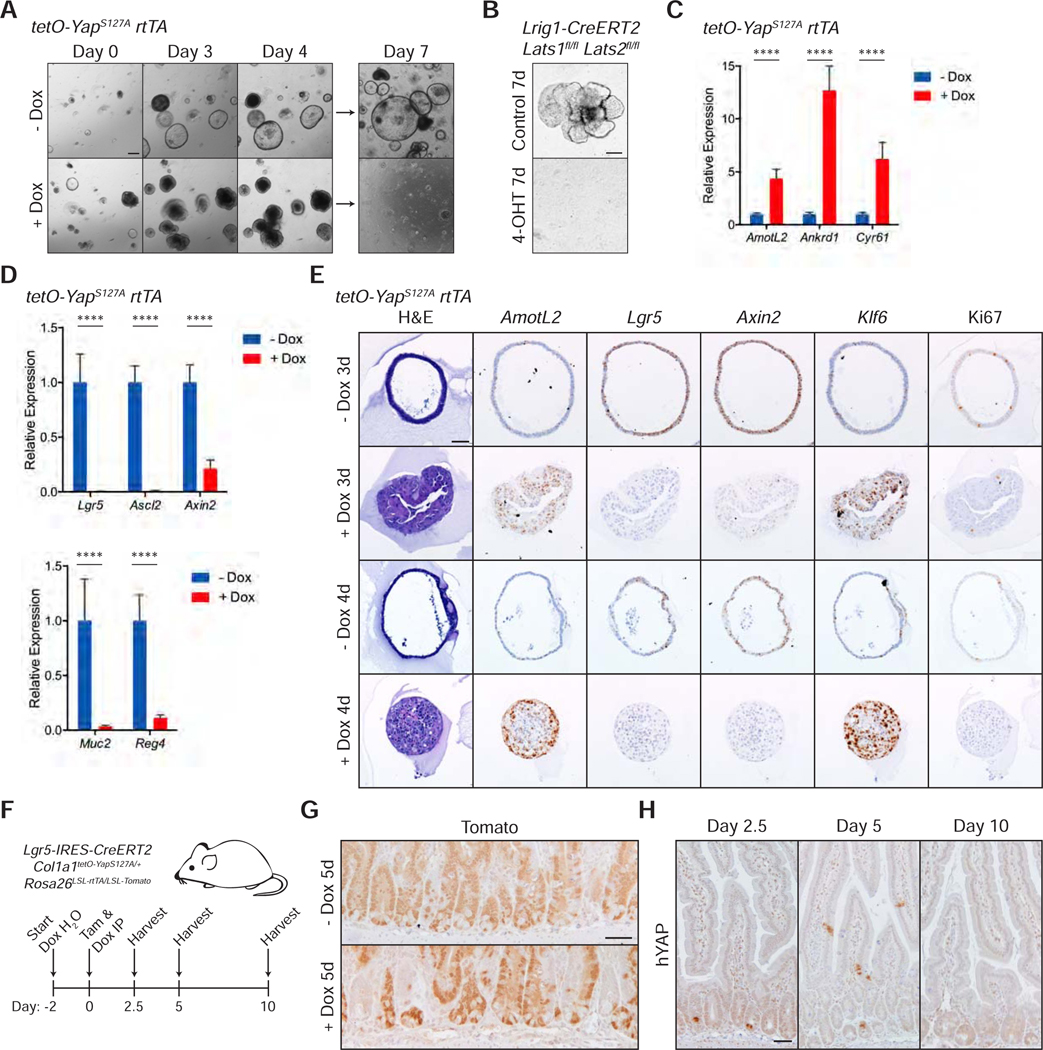
(A) Brightfield images of tetO-YAPS127A rtTA organoids on the indicated days in the presence or absence doxycycline. The organoids were split on day 4. Scale bar, 200 μm.
(B) Brightfield images of Lrig1-CreERT2 Lats1fl/fl Lats2fl/fl organoids 7 days after plating in the presence of 4-OHT for 24 hours. Scale bar, 200 μm.
(C-D) RT-qPCR analysis of the indicated genes in tetO-YAPS127A rtTA organoids 2 days after induction with doxycycline. Data are represented as mean ± SEM; n = 3 biological and 2 technical replicates. ****P < 0.001.
(E) Histological analysis of tetO-YAPS127A rtTA organoids 3 and 4 days after induction with doxycycline: H&E stains, IHC for YAP and Ki67, and RNA-ISH for AmotL2, Lgr5, Axin2, and Klf6. Scale bar, 50 μm.
(F) Schematic of Lgr5 lineage tracing, including timeline of doxycycline administration, doxycycline and tamoxifen injections, and tissue collection.
(G) IHC for Tomato in the small intestine of Lgr5-IRES-CreERT2 Col1a1tetO-YapS127A/+ Rosa26LSL-rtTA/LSL-Tomato mice 5 days after tamoxifen injection with or without administration of doxycycline. Scale bar, 50 μm.
(H) IHC for human YAP in the small intestine of Lgr5-IRES-CreERT2 Col1a1tetO-YapS127A/+ Rosa26LSL-rtTA/LSL-Tomato mice on the indicated days after tamoxifen and doxycycline administration. Scale bar, 50 μm.
See also Figure S2.
To determine whether these YAP-expressing cells have functional stem cell activity in vivo, we leveraged lineage tracing in Lgr5-CreERT2 knock-in mice to trace Lgr5+ ISCs with and without activation of YAP (Barker et al., 2007; Huch et al., 2013). We induced YAP activation and tdTomato expression by intraperitoneal injection of tamoxifen and doxycycline in Lgr5-IRES-CreERT2 Col1a1tetO-YapS127A/+ Rosa26LSL-rtTA/LSL-Tomato mice (Figure 2F). Compared to the homogeneous Tomato labeling in uninduced crypts 5 days post induction, crypts with YAP-activated ISCs exhibit mosaic Tomato labeling in the small intestine (Figure 2G), indicating that YAP expression in ISCs negatively affects their ability to contribute to intestinal lineages. Indeed, these YAP-activated ISCs are pushed out of intestinal crypts and villi over the course of 10 days (Figure 2H), suggesting that these mutant cells are quickly replaced by wildtype ISCs. Similarly, lineage tracing in Lgr5-CreERT2 Lats1fl/fl Lats2fl/fl Rosa26mT/mG mice reveals progressive loss of GFP+ ISCs at 2 weeks and 1 month after induction (Figure S2D). Altogether, these models suggest that YAP activation within ISCs leads to loss of normal stem cell function in vivo.
A YAP Gene Signature is Activated during Mucosal Healing
The activation of YAP previously has been shown to be required for mucosal regeneration in different models of intestinal and colonic injury (Cai et al., 2010; Gregorieff et al., 2015). Thus, we investigated whether such physiological activation of YAP via injury induces reprogramming of intestinal epithelial cells to this Wnt-low, Klf6+ state that we described above. To characterize epithelial cells during wound healing, we induced a punch lesion in mice by colonoscopy and collected samples from injured animals after six days (Seno et al., 2009). Histological analysis reveals an increased accumulation of YAP in the nucleus of epithelial cells at sites of wound healing and concomitant upregulation of Klf6 and the YAP target AmotL2 when compared to uninjured controls (Figure 3A). Intriguingly, these areas are also depleted of Lgr5+ ISCs and Reg4+ deep crypt secretory (DCS) cells, suggesting that physiological regeneration activates a transcriptional state similar to that which we observed in Lats1/2 cKO and Mst1/2 cKO animals.
Figure 3: A YAP Gene Signature is Activated during Mucosal Healing.
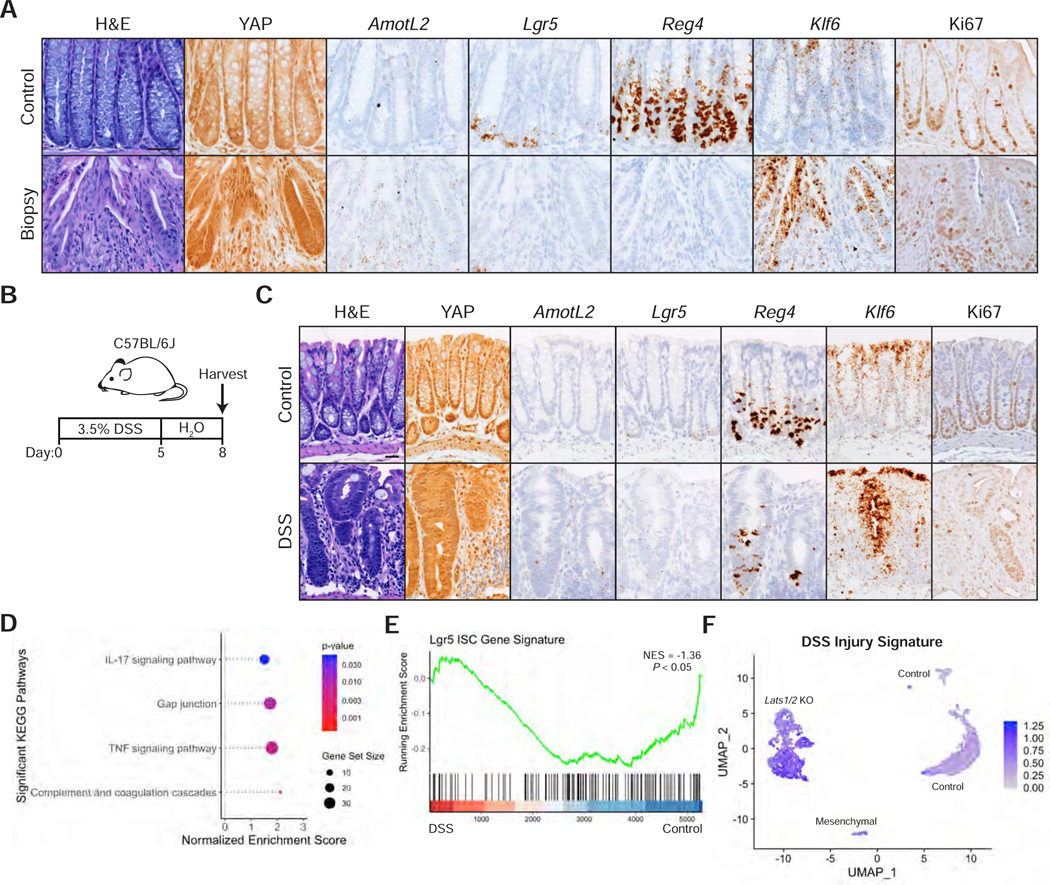
(A) Histological analysis of colon samples from mice 6 days after inducing a punch biopsy: H&E stains, IHC for YAP and Ki67, and RNA-ISH for AmotL2, Lgr5, Reg4, and Klf6. Scale bar, 50 μm.
(B) Schematic of colonic injury and regeneration by dextran sulfate sodium (DSS), including timeline of DSS administration and tissue collection.
(C) Histological analysis of colon samples from mice after a 5-day DSS treatment and 3-day recovery: H&E stains, IHC for YAP and Ki67, and RNA-ISH for AmotL2, Lgr5, Reg4, and Klf6. Scale bar, 25 μm.
(D-E) GSEA of KEGG pathways (D) and a Lgr5 ISC gene signature (E) in the DSS sample compared to control.
(F) Expression of the DSS injury signature, characterized by genes upregulated at least 2-fold in stem cells, progenitors, goblet cells, or colonocytes in DSS versus control samples, overlaid onto the UMAP of Lats1/2 cKO scRNA-seq.
In a second model of colonic injury, we induced acute inflammation in adult mice by supplementing drinking water with 3.5% dextran sulfate sodium (DSS) for five days (Figure 3B), which leads to a loss of colonic crypts and immune cell infiltration (Figure S3A). Three days after DSS washout, crypts in inflamed areas of the colon show marked activation of YAP, increased Klf6 expression, and loss of Lgr5 and Reg4 markers (Figure 3C). These data altogether suggest that YAP activation is a general and specific response to injury in the colon during regeneration.
To characterize the molecular changes in the colonic epithelium during regeneration, we performed scRNA-seq on epithelial cells isolated from DSS-treated and control mice (Figure S3B). UMAP representation of the transcriptomic profiles reveals that the DSS-treated cells, despite being distinct from control cells (Figure S3C), maintain some features of normal colonic cells and could be assigned to known colonic cell types by marker expression (Figures S3D-F). As reported previously (Yui et al., 2018), the DSS sample presents with upregulation of Ly6a/Sca1 transcript levels (Figure S3G). Differentially expressed genes between DSS and control are positively enriched for wound healing response, cell-cell contact pathways, and YAP gene signatures and negatively enriched for a previously published ISC gene signature (Figures 3D-E, S3H-I, and Table S2) (Barry et al., 2013; Gregorieff et al., 2015; Muñoz et al., 2012). Remarkably, this DSS injury signature is highly enriched in the transcriptomes of Lats1/2 knockout cells (Figure 3F). Collectively, these data suggest that the cellular state induced by knockout of Lats1/2 or activation of YAP likely represents the physiological equivalent of a transiently repairing cell state.
YAP can Reprogram Lgr5+ Stem Cells in the Presence of Constitutively Active Wnt Signaling
The fact that YAP remodels stem cells in vitro even in the presence of exogenous Wnt signals prompted us to analyze the effect of YAP in the context of constitutively active Wnt. To assess the role of YAP in this process, we introduced an Apc mutation via CRISPR/Cas9 into tetO-YAP rtTA colon organoids (Figure S4A). Strikingly, Apc−/− organoids lose their characteristic spheroid structure as early as three days after doxycycline induction and could not grow after passage (Figure 4A). Lrig1-CreERT2-mediated deletion of Lats1/2 in Apc mutant organoids results in a similar growth inhibition upon plating (Figure 4B). As in Apc wildtype organoids, YAP targets and Klf6 are upregulated along with concomitant downregulation of ISC, DCS, and Wnt markers (Figures 4C-E and S4B). These results show that YAP is able to suppress Wnt and the colonic stem cell program even in the presence of constitutive Wnt signaling caused by the deletion of Apc.
Figure 4: YAP can Reprogram Lgr5+ Stem Cells in the Presence of Constitutively Active Wnt Signaling.
(A) Brightfield images of Apc−/− tetO-YAPS127A rtTA organoids on the indicated days in the presence or absence of doxycycline. The organoids were split on day 4. Scale bar, 200 μm.
(B) Brightfield images of Apc−/− Lrig1-CreERT2 Lats1/2fl/fl organoids 7 days after plating in the presence of 4-OHT for 24 hours. Scale bar, 200 μm.
(C-D) RT-qPCR analysis of the indicated genes in Apc−/− tetO-YAPS127A rtTA organoids 1.5 days after induction with doxycycline. Data are represented as mean ± SEM; n = 3 biological and 3 technical replicates. ****P < 0.0001.
(E) Histological analysis of Apc−/− tetO-YAPS127A rtTA organoids 4 days after induction with doxycycline: H&E stains and RNA-ISH for AmotL2, Klf6, Lgr5, and Axin2. Scale bar, 50 μm.
(F) Schematic of RNA-seq time course analysis in Apc−/− tetO-YAPS127A rtTA organoids.
(G) Heatmap of YAP downstream target genes, genes involved in Wnt signaling, and injury response genes over the time course of doxycycline induction in Apc−/− tetO-YAPS127A rtTA organoids.
(H) GSEA of a YAP gene signature from Barry et al., 2013 in Apc−/− tetO-YAPS127A rtTA organoids after 24 hours on doxycycline compared to 0 hours
(I-J) GSEA of a Lgr5 ISC gene signature (I) and KEGG pathways (J) in Apc−/− tetO-YAPS127A rtTA organoids after 72 hours on doxycycline compared to 0 hours.
To determine whether this reprogramming is mediated through YAP’s transcriptional or cytoplasmic function, we generated Apc−/− CAGs-rtTA3 organoids, carrying inducible expression of either YAP5SA or YAP5SA/S94A (see STAR Methods) (Premsrirut et al., 2011). YAP5SA, which cannot be phosphorylated by LATS1/2, is therefore nuclear and constitutively active while YAPS94A cannot bind TEAD and is unable to activate transcription of target genes (Zhao et al., 2007, 2008). Only YAP5SA expression is able to arrest organoid growth (Figures S4C-D), suggesting that binding to TEAD is required for YAP to prevent growth of Apc−/− organoids. We next determined whether YAP’s transcription activation domain, which mediates recruitment of transcription machinery (Yagi et al., 1999), is required to mediate this phenotype. Expression of HA-tagged deletion versions of YAP5SA in Apc−/− CAGs-rtTA3 organoids reveals that only fulllength YAP is able to induce proliferation arrest despite comparable levels of expression among the variants (Figures S4E-G). Collectively, these data suggest that YAP causes the loss of stem cell properties through TEAD-dependent transcriptional activity.
To gain further insight into the molecular changes underlying this change in cell fate, we performed RNA-seq analysis at sequential time points in Apc−/− tetO-Yap rtTA organoids upon YAP activation (Figure 4F). This analysis reveals an overall distinct transcriptional state of treated cells (Figure S4H), suggesting a global transcriptional reprogramming. This data also validate the immediate activation of YAP targets, substantiated by the enrichment of the Hippo signaling pathway and two published YAP gene signatures by GSEA (Figures 4G-H and S4I-J), and the downregulation of Wnt and ISC gene signatures, which predominantly occurs between 24 and 72 hours (Figures 4G, 4I, and S4J) (Barry et al., 2013; Gregorieff et al., 2015; Muñoz et al., 2012). Interestingly, Wnt nuclear co-factors are not downregulated over the course of YAP activation with the exception of Tcf7, which is also a target of the Wnt pathway (Figure S4K), suggesting that downregulation of Wnt signaling upon YAP activation may be due to posttranslational modifications or changes in protein interaction partners of these factors unrelated to transcript levels. Additionally, injury signature genes, such as Anxa2, Ly6a/Sca1, S100A6, and S100A11, are concomitantly upregulated, and pathways related to cell-cell contact, actin cytoskeleton, and response to injury are significantly enriched at 72 hours (Figures 4G and 4J). These enriched pathways are also recapitulated in GO overrepresentation analysis and GSEA of the transcription factors differentially expressed at 72 hours (Table S3 and Figures S4L-M), underscoring their role in mediating this cell fate change. Altogether, these findings substantiate the transcriptional activity of YAP in driving a complete reprogramming of intestinal cells even in the context of an Apc deletion.
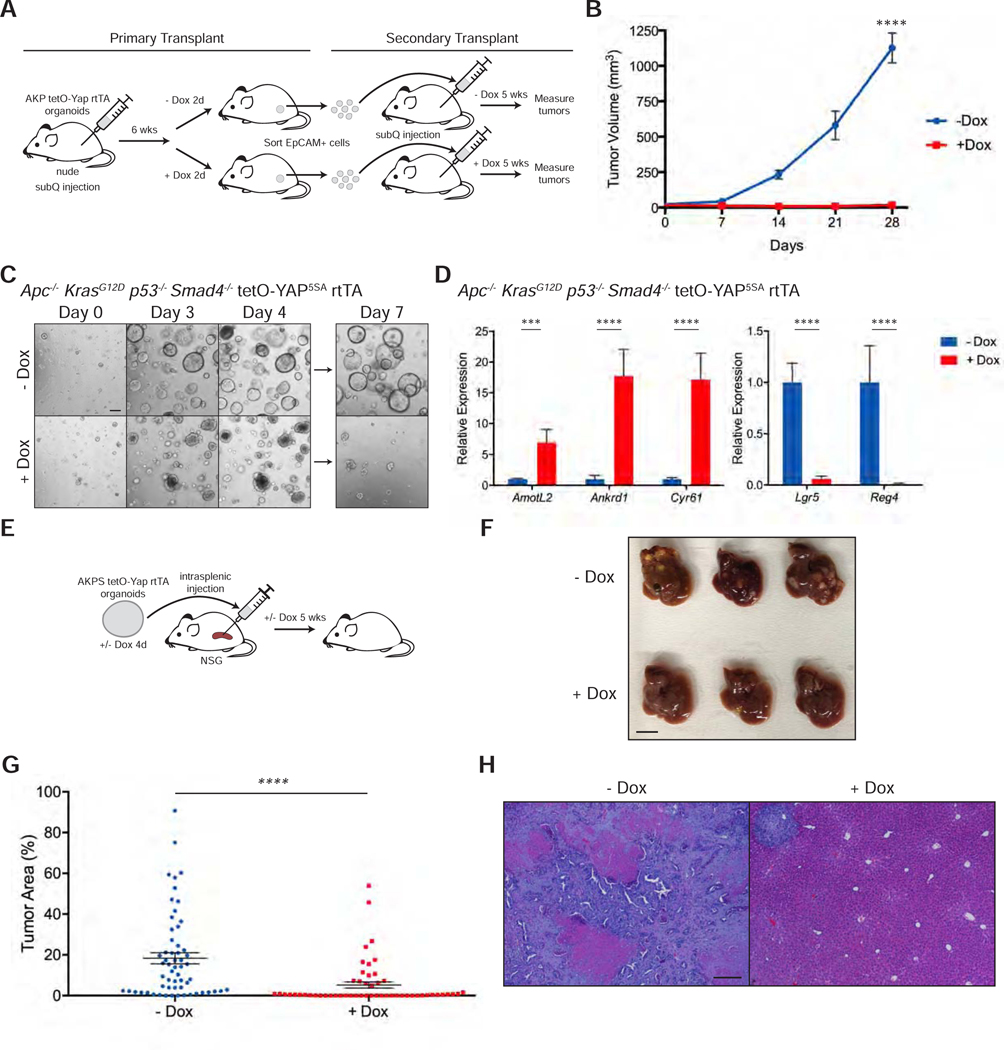
Activation of YAP Induces Loss of Cancer Stem Cells and Tumor Regression
We next tested whether YAP expression could also induce similar molecular changes in colon organoids carrying mutations associated with malignant colon cancer disease. In order to do so, we generated Apc−/− KrasG12D p53−/− (AKP) cancer organoids with inducible YAP5SA overexpression (Figure S5A). Overexpression of YAP5SA by addition of doxycycline to the culture media leads to death of AKP organoids by seven days after induction (Figure 5A). As in wild-type and Apc−/− organoids, ISC marker Lgr5 and DCS marker Reg4 are downregulated after doxycycline induction (Figure 5B). Similar to other contexts, RNA-seq analysis of these organoids at 72 hours post doxycycline reveals significant enrichment of previously published YAP gene signatures and pathways involving cell-cell contact, actin cytoskeleton, and response to injury and negative enrichment of those related intestinal absorption and the Lgr5 ISC gene signature (Figures 5C-E and S5B) (Barry et al., 2013; Gregorieff et al., 2015; Muñoz et al., 2012), confirming our previous findings that YAP is reprogramming these cancer cells to a low Wnt, wound healing-like state.
Figure 5: Activation of YAP Induces Loss of Cancer Stem Cells and Tumor Regression.
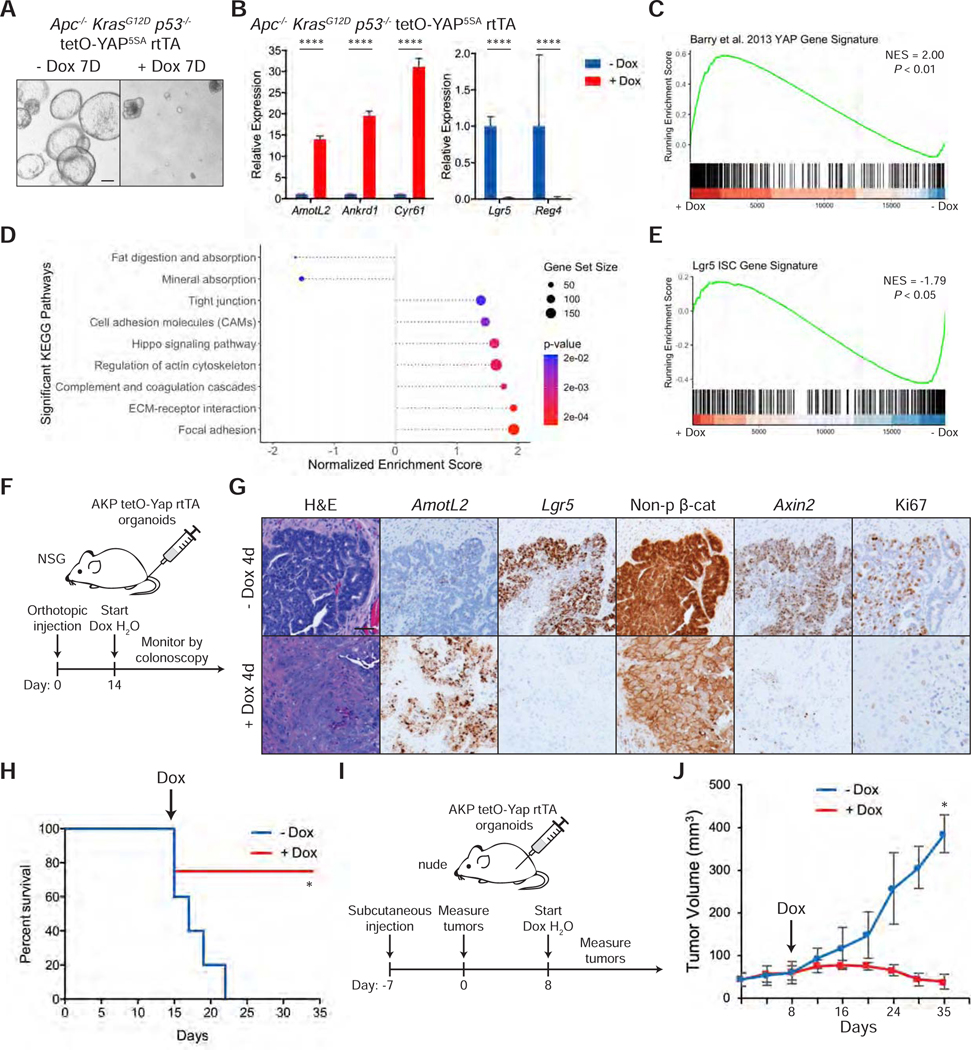
(A) Brightfield images of Apc−/− KrasG12D p53−/− tetO-YAP5SA rtTA organoids after 7 days in the presence or absence of doxycycline. The organoids were split on day 4. Scale bar, 200 μm.
(B) RT-qPCR analysis of the indicated genes in Apc−/− KrasG12D p53−/− tetO-YAP5SA rtTA organoids 3 days after induction with doxycycline. Data are represented as mean ± SEM; n = 3 biological and 2 technical replicates. ****P < 0.0001.
(C-E) GSEA of a YAP gene signature from Barry et al., 2013 (C), KEGG pathways (D), and a Lgr5 ISC gene signature (E) in Apc−/− KrasG12D p53−/− tetO-YAP5SA rtTA organoids 3 days after induction with doxycycline.
(F) Schematic of timeline for orthotopic injection of Apc−/− KrasG12D p53−/− tetO-YAP5SA rtTA organoids in the colons of NSG mice and doxycycline administration.
(G) Histological analysis of orthotopic tumors from Apc−/− KrasG12D p53−/− tetO-YAP5SA rtTA organoids 4 days after doxycycline administration: H&E stains, IHC for non-phosphorylated (active) β-catenin and Ki67, and RNA-ISH for AmotL2, Lgr5, and Axin2. Scale bar, 50 μm.
(H) Kaplan-Meier survival curve of mice injected orthotopically with Apc−/− KrasG12D p53−/− tetO-YAP5SA rtTA organoids. Doxycycline treatment was started 14 days after injection as indicated with the arrow. - dox: n = 5, + dox: n = 4. *P < 0.05.
(I) Schematic of timeline for subcutaneous injection of Apc−/− KrasG12D p53−/− tetO-YAP5SA rtTA organoids in the flanks of nude mice, tumor measurements, and doxycycline administration.
(J) Growth curve of subcutaneous tumors from Apc−/− KrasG12D p53−/− tetO-YAP5SA rtTA organoids injected in the flanks of nude mice. Measurements were started one week after injection (shown as day 0 on the plot). The arrow indicates the day doxycycline treatment was started. Data are represented as mean ± SD. *P < 0.05.
See also Figure S5.
To test if YAP is capable of reprogramming AKP cells in vivo, where the cells are exposed to additional niche factors, we injected AKP organoids in the colon of NSG mice (Figure 5F). Two weeks after injection, YAP5SA overexpression was induced by adding doxycycline to drinking water. Strikingly, histological analysis shows that tumor cells 4 days post doxycycline present with a different morphology, with an enlarged cytoplasm and loss of the characteristic columnar shape (Figure 5G). Expression of Lgr5 is completely lost, and β-catenin is dislocated from the nucleus to the cytoplasm (Figures 5G and S5C). Further supporting the notion that these cells lack active Wnt signaling, expression of Axin2 is greatly reduced. These YAP-overexpressing cells are also not proliferative, and as a consequence, YAP induction by doxycycline confers a survival advantage in these transplants (Figure 5H). To quantify tumor growth, we injected AKP organoids in the flanks of nude mice and confirmed that YAP-overexpressing cells in this model undergo the same phenotypic and molecular changes as cells growing in the colon (Figure 5I and S5D). Remarkably, YAP overexpression induces loss of cellularity and tumor regression (Figures S5E and 5J).
We also validated the role of YAP in human CRC as engineered inducible YAP5SA expression leads to the reduction in growth of a patient-derived CRC organoid line (Figure S5F). This is accompanied by increased YAP target expression and strong downregulation of ISC and Wnt markers LGR5 and AXIN2 (Figure S5G). Additionally, expression of YAP in established human CRC xenografts leads to a statistically significant reduction in growth compared to their uninduced counterparts (Figure S5H). Taken together, our results indicate that YAP activation in CRC organoids leads to molecular reprogramming and proliferation arrest.
YAP Suppresses Cancer Stemness in Primary and Metastatic Tumor Growth
We next tested the ability of YAP to suppress two critical features of malignant growth: tumor initiating capacity and metastatic potential. To measure tumor propagating capacity, we performed secondary transplantation of AKP colon organoids with and without YAP activation. After allowing the tumors to grow in mice for 6 weeks, we induced them with doxycycline for 2 days and isolated the EpCAM+ cells via FACS (Figures 6A and S6A). As before, we observed an increase in nuclear YAP and the YAP target AmotL2 and a decrease in ISC and Wnt markers Lgr5 and Axin2 after only 2 days (Figure S6B). Equal numbers of uninduced and induced EpCAM+ tumor cells were transplanted subcutaneously into nude mice. While uninduced tumor cells grow dramatically over the course of 4 weeks, cells from doxycycline-induced tumors have virtually no capacity to reinitiate tumor growth (Figures 6B and S6C). Similarly, AKP organoids induced with doxycycline in culture for 4 days also exhibit a lack of growth following transplant compared to uninduced controls (Figures S6D-F), suggesting that YAP activation abolishes the tumor propagating potential of triple mutant AKP colon cancer cells.
Figure 6: YAP Suppresses Cancer Stemness in Primary and Metastatic Growth.
(A) Schematic of primary and secondary transplantation of Apc−/− KrasG12D p53−/− tetO-YAP5SA rtTA organoid cells into nude mice, including timeline of subcutaneous injections, doxycycline administration, and tumor isolation.
(B) Growth curve of subcutaneous tumors from Apc−/− KrasG12D p53−/− tetO-YAP5SA rtTA organoid cells following secondary transplantation with or without doxycycline treatment. Measurements were started one week after injection (shown as day 0 on the plot). Data are represented as mean ± SEM; - dox: n = 10 tumors; + dox: n = 6 tumors. ****P < 0.0001.
(C) Brightfield images of Apc−/− KrasG12D p53−/− Smad4−/− tetO-YAP5SA rtTA organoids on the indicated days in the presence or absence of doxycycline. The organoids were split on day 4. Scale bar, 200 μm.
(D) RT-qPCR analysis of the indicated genes in Apc−/− KrasG12D p53−/− Smad4−/− tetO-YAP5SA rtTA organoids 2 days after induction with doxycycline. Data are represented as mean ± SEM; n = 3 biological and 2 technical replicates. ***P < 0.001 and ****P < 0.0001.
(E) Schematic of intrasplenic injection of uninduced and doxycycline-induced Apc−/− KrasG12D p53−/− Smad4−/− tetO-YAP5SA rtTA organoids into NSG mice.
(F) Gross morphology of representative livers of NSG mice injected intrasplenically with Apc−/− KrasG12D p53−/− Smad4−/− tetO-YAP5SA rtTA organoids induced with or without doxycycline for 4 days in culture. Scale bar, 10 mm. </p/> (G) Quantification of tumor area per liver piece from Figure 6F. Dot plot is represented as mean ± SEM; - dox: n = 9–10 liver pieces per mouse, n = 6 mice; + dox: n = 9–10 liver pieces per mouse, n = 5 mice. ****P < 0.0001.
(H) Representative H&E staining of livers from Figure 6F. Scale bar, 200 μm.
See also Figure S6.
To test the effect of YAP on metastatic potential, we engineered an additional Smad4 mutation via CRISPR/Cas9 in AKP colon organoids to enhance their metastatic capacity (Figures S6G-H) (Fumagalli et al., 2017; Miyaki et al., 1999; Zhang et al., 2010). Strikingly, even in the presence of a fourth mutation, AKPS organoids with engineered YAP5SA expression are not able to reform organoids by day 7 (Figure 6C). Similarly, these organoids also downregulate ISC and DCS markers as early as day 2 (Figure 6D). To test their metastatic potential in vivo, we injected control and doxycycline-induced AKPS organoids in the spleens of NSG mice (Figures 6E), which leads to formation of metastases in the liver (Fujii et al., 2016; O’Rourke et al., 2017). Five weeks post injection, we found that the livers of NSG mice injected with YAP-expressing AKPS organoids have a drastic and significant reduction in tumor burden (Figures 6F-G). Histological analysis of these livers reveals small metastatic foci compared to the extensive metastatic burden in livers with uninduced organoids (Figure 6H), suggesting YAP-expressing cancer cells are less able to seed and establish metastases in the liver.
To characterize the role of YAP on established liver metastases, we allowed AKPS organoids to grow in the livers of NSG mice for 5 weeks before administering doxycycline for 10 days. YAP activation has a drastic effect on the morphology of established metastatic tumors with obvious and large areas of necrosis (Figure S6I). Additionally, we observed the near complete elimination of Lgr5+ ISCs and a significant reduction in their proliferative index (Figures S6J-K). Altogether, our data point to the therapeutic role of activating YAP in preventing both the establishment and growth of liver metastases.
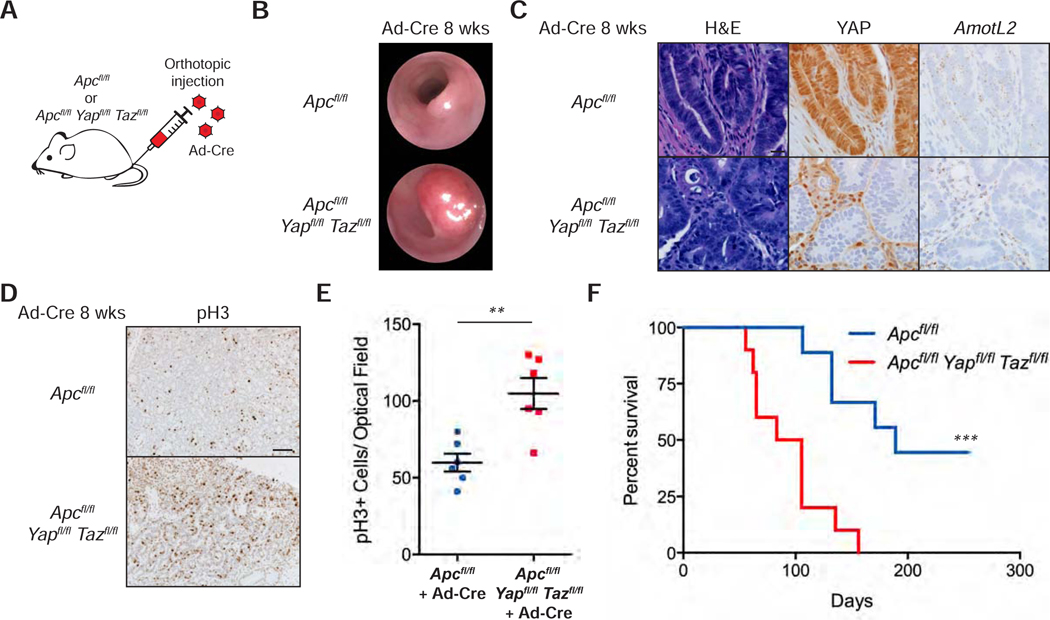
Deletion of YAP/TAZ Increases Colon Tumor Growth
The data presented in this manuscript relating to YAP overactivation speak to a tumor suppressive role for YAP in CRC. However, other work has reported that YAP is required for intestinal and colonic tumor growth either in cell lines or in a mouse model of familial adenomatous polyposis (FAP), where one copy of Apc is mutated, and numerous tumors appear upon stochastic loss of the second copy (Cai et al., 2015; Gregorieff et al., 2015; Konsavage et al., 2012; Rosenbluh et al., 2012; Zhou et al., 2011). These experimental settings use a developmental deletion of YAP/TAZ and predominantly look at tumor formation in the small intestine. To test unbiasedly the effects of Yap and Taz knockout on the growth of discrete Apc-mutant tumors in the adult colon in vivo, we injected adenovirus expressing Cre recombinase in the colonic mucosa of Apcfl/fl and Apcfl/fl Yapfl/fl Tazfl/fl mice (Figure 7A) (Colnot et al., 2004; Schlegelmilch et al., 2011; Xin et al., 2013). While all mice at eight-weeks post injection develop tumors, Apcfl/fl Yapfl/fl Tazfl/fl mice present with higher tumor burden (Figures 7B and S7A-B). We observed YAP deletion efficiency ranging from 40 to 100% in the tumors from Apcfl/fl Yapfl/fl Tazfl/fl mice and downregulation of the YAP target AmotL2 in the YAP-deleted cells (Figures S7C and 7C). Furthermore, Yap/Taz knockout tumors have a significantly higher number of proliferating cells by phospho-histone H3 staining (Figures 7D-E), explaining the difference in tumor burden. Although these adenomas rarely become malignant, they grow until they obstruct the colonic lumen of the mice at which point the animals have to be euthanized. Taking this into account, we observed a significant decrease in survival in animals bearing Yap/Taz deleted tumors compared to their control counterparts (Figure 7F). Collectively, these results show that Yap/Taz are not required for the growth of genetically defined colon tumors driven by Wnt but are rather inhibitory towards their growth.
Figure 7: Deletion of YAP/TAZ Increases Colon Tumor Growth.
(A) Schematic of orthotopic injection of adenovirus expressing Cre recombinase (Ad-Cre) into mice of the indicated genotypes.
(B) Representative colonoscopy images of colon tumors 8 weeks after orthotopic injection of Ad-Cre in mice of the indicated genotypes.
(C) Histological analysis of Ad-Cre injected Apcfl/fl and Apcfl/fl Yapfl/fl Tazfl/fl colonic tumors: H&E stains, YAP IHC, and AmotL2 RNA-ISH. Scale bar, 20 μm.
(D) IHC for phospho-histone H3 (pH3) in Ad-Cre injected Apcfl/fl and Apcfl/fl Yapfl/fl Tazfl/fl colonic tumors. Scale bar, 100 μm.
(E) Quantification of pH3+ cells per optical field in Ad-Cre injected Apcfl/fl and Apcfl/fl Yapfl/fl Tazfl/fl colonic tumors. Dot plot is represented as mean ± SEM; n = 2 measurements per mouse, n = 3 mice per group. **P < 0.01.
(F) Kaplan-Meier survival curve for mice of the indicated genotypes injected with Ad-Cre in the colonic mucosa. Apcfl/fl: n = 9 mice; Apcfl/fl Yapfl/fl Tazfl/fl: n = 10 mice. ***P < 0.001.
See also Figure S7.
We also tested the role of YAP/TAZ in sporadic tumor formation by inducing tumors in Villin-Cre Yapfl/fl Tazfl/fl mice and their Yapfl/fl Tazfl/fl littermates as controls with six weekly intraperitoneal injections of 10 mg/kg azoxymethane (AOM) carcinogen (Figure S7D) (Neufert et al., 2007). These conditional mouse knockouts lack YAP in the entire intestinal epithelium (Figure S7E). Colonoscopy imaging shows formation of tumors in both wildtype and Yap/Taz knockout mice 1 month after the last AOM injection (Figure S7F). Eight weeks after the last dose, we observed a higher tumor burden in the colons of Yap/Taz knockout mice than in their control littermates (Figure S7G-H). In this case, all epithelial cells of the tumors are fully deleted for Yap/Taz as are the cells of origin (Figure S7I). Altogether, our results not only establish YAP/TAZ as a bona fide tumor suppressor in Apc mutant colon tumors but also implicate YAP activation as a viable therapeutic strategy in this cellular context.
DISCUSSION
YAP is known to play an important role during mucosal healing and has been proposed to remodel stem cells, activate expression of a fetal program, or expand quiescent stem cells upon injury (Ayyaz et al., 2019; Gregorieff et al., 2015; Yui et al., 2018). Using scRNA-seq, we elucidate a role for YAP in reprogramming colon cells into a cell type marked by high Klf6 expression that is unique to the repairing epithelium and distinct from all cell types present during homeostasis. This state is characterized by the expression of injury-associated genes and a low Wnt signature, which confirms our earlier finding that YAP inhibits Wnt signaling in the intestine. Our work is consonant with a recent scRNA-seq study identifying a stem cell type enriched in YAP target genes (Ayyaz et al., 2019). In contrast to this work, our data suggest that this YAP-driven cell state does not exist in homeostasis, and it instead arises from the reprogramming of traditional ISCs. Remarkably, YAP antagonizes stemness and inhibits Wnt even in the presence of multiple oncogenic mutations, rendering Lgr5+ cancer stem cells non-proliferative and inducing tumor regression in mouse and patient-derived models of primary and metastatic CRC. These data collectively suggest that this YAP-dependent regenerative program can be reactivated during CRC progression and be dominant over Wnt-driven outputs.
Interestingly, growth of certain CRC cell lines requires YAP, an observation that supports the idea of YAP as an oncogene in the colon as seen in other tissues (Konsavage et al., 2012; Rosenbluh et al., 2012; Zhou et al., 2011). However, it is possible that YAP dependence is a feature of in vitro culture and does not reflect physiological requirements in vivo. For instance, while YAP deletion has no effect on intestinal homeostasis in vivo (Cai et al., 2010), in vitro growth in intestinal organoids is dependent on YAP (Azzolin et al., 2014; Gregorieff et al., 2015; Serra et al., 2019). Our data show that in vivo growth of Apc knockout adenomas is not only independent of YAP but is also favored upon YAP/TAZ depletion. Moreover, activation of YAP in vivo and in vitro hampers cancer growth by suppressing Wnt and antagonizing the stem cell state. Our observations are supported by recent data demonstrating transient stages of YAP activation during organoid formation, where YAP activity is negatively correlated with the presence of Lgr5+ cells (Serra et al., 2019). A few studies have also proposed an oncogenic role for YAP based on data from a mouse model of FAP, where deletion of YAP reduces the number of tumors that appear as a consequence of a germline mutation in one of two Apc alleles (Cai et al., 2015; Gregorieff et al., 2015). However, as tumor outcome in this model depends on the loss of the second Apc allele (Haigis & Dove, 2003; Haines et al., 2005), it is possible that these results could reflect a role of YAP in mitotic recombination as opposed to tumor growth per se. Additionally, these experiments have been done in the context of developmental YAP deletions and might not be informative of what would occur in an adult mutation. Therefore, the use of an acute adult and colon-specific manipulation of YAP as done here represents the most relevant model to study the role of YAP in adenoma growth. Additionally, the appearance of sporadic colon tumors through the action of a mutagen does not require YAP, ruling out the role for YAP in tumor initiation. Altogether, our data using models of both YAP activation and depletion are consistent and strongly suggest that YAP functions as a tumor suppressor in the colon.
Multiple strategies are being developed to target solid tumor stem cells. Our study suggests that reprogramming of these cells into a regenerative state could be harnessed as a novel therapeutic approach. In this context, the cell state induced by YAP, marked by high Klf6 expression during colonic regeneration, can be induced in CRC. Given that YAP is negatively regulated by the upstream kinases LATS1/2 and MST1/2, our results indicate that Hippo kinase inhibitors could serve as potential therapeutic agents to combat CRC.
Limitations of the Study
Here, we demonstrate that YAP activation leads to the reprogramming of colonic cells, which may serve as a therapeutic vulnerability in the treatment of CRC. We leveraged multiple genetic murine models to inactivate the Hippo signaling pathway at different levels of pathway regulation, including Mst1/2 deletion, Lats1/2 deletion, and YAP activation. Further work needs to be done to translate our findings to clinically tractable modalities, such as small molecule inhibitors of these Hippo kinases. In addition, we primarily rely on the deletion of the Apc tumor suppressor as an initial driver of CRC in many of our murine models of the disease. It remains to be seen whether the therapeutic activation of YAP may translate in the context of other cancer driving mutations.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Fernando Camargo (Fernando.Camargo@childrens.harvard.edu).
Materials Availability
Plasmids generated in this current study are available upon request.
Data and Code Availability
The datasets generated in this study are available in the NCBI GEO database under GSE152376. Code to reproduce the analyses of the single-cell and bulk RNA-seq datasets can be found at https://github.com/cheungpriscilla/CellStemCell_2020.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All animal protocols and procedures were approved by the respective local animal institutional committees. All mice used for this study were on a C57BL/6J background unless indicated otherwise.
Models of YAP activation:
Lats1fl/fl Lats2fl/fl (controls) and their Lrig1-CreERT2 Lats1fl/fl Lats2fl/fl littermates were injected intraperitoneally with 2 mg of tamoxifen and harvested on the days indicated for single cell RNA sequencing and histology experiments. The intestine and colon of Villin-Cre Mst1−/− Mst2fl/fl mice and their Mst1−/− Mst2fl/fl littermates were collected at the indicated ages for histological analysis. Villin-CreERT2 Mst1−/− Mst2fl/fl mice and their Mst1−/− Mst2fl/fl littermates were injected intraperitoneally with 2 mg of tamoxifen and harvested 1 month after for histology experiments.
Lineage tracing experiments:
2–4 months old Lgr5-IRES-CreERT2 Col1a1tetO-YapS127A/+ Rosa26 LSL-rtTA/LSL-Tomato mice were administered doxycycline (2 mg/ml with 0.5% sucrose) in drinking water ad libitum two days prior to intraperitoneal injection with 3 mg of tamoxifen and 25 μg/g doxycycline and were harvested on the indicated days for histology experiments (Lgr5-IRES-CreERT2 mice were a kind gift of Meritxell Huch). 2-month old Lgr5-CreERT2 Lats1fl/fl Lats2fl/fl Rosa26mT/mG mice were injected intraperitoneally with 120 mg/kg tamoxifen for two consecutive days and harvested on the indicated days for immunofluorescence.
Organoid transplantation experiments:
Both male and female 2–3 months old nude mice were used for flank injections, and male and female 2–4 months old NSG mice were used for colon orthotopic and intrasplenic injections. Mice were administered doxycycline (2 mg/ml with 0.5% sucrose) in drinking water ad libitum either immediately upon transplantation or starting on the days indicated as specified in the figure legends.
Colon tumorigenesis:
Both male and female Apcfl/fl and Apcfl/fl Yapfl/fl Tazfl/fl mice at 6–8 weeks old were used for injection of adenovirus expressing Cre recombinase. AOM-induced carcinogenesis was performed in Villin-Cre Yapfl/fl Tazfl/fl animals and their Yapfl/fl Tazfl/fl littermates as controls.
Experimental Colitis
Colitis was induced in 2-month old female C57BL/6 mice by administration of 3.5% DSS in drinking water for 5 days followed by normal water for 3 days.
Azoxymethane-Induced Tumorigenesis
A stock solution of azoxymethane (AOM) was prepared at a concentration of 1 mg/mL. Colon tumorigenesis was induced by intraperitoneal injection of 10 mg/kg of AOM once weekly for six consecutive weeks into 8-week old mice of the indicated genotypes. Mice were harvested 2 months after the last injection for analysis.
Mouse Colon Organoid Isolation and Culturing
For colon epithelial cell isolation for organoid culture, a 2 cm piece of distal colon was extracted from 2-month old mice and incubated in 4 mM EDTA in PBS with mild agitation for 45 min at 4°C. The colon pieces were cut open, and colon crypts were scraped from the tissue with a coverslip. After washing with cold PBS and centrifugation at 1000 g for 5 min, crypts were resuspended in Matrigel and plated in 24-well plates (50 μL Matrigel/well). The Matrigel was allowed to solidify for 15–30 min in a 37°C incubator. 500 μL of organoid culture media was then overlaid onto the Matrigel and changed every 2–3 days. The organoid cultures were maintained at 37°C in fully humidified chambers containing 5% CO2.
Colon organoids were prepared from animals with the following genotypes: Col1a1tetO-YapS127A/+ Rosa26LSL-rtTA/+, Lrig1-CreERT2 Lats1fl/fl Lats2fl/fl, Apcfl/fl KrasLSL-G12D p53fl/fl, and Rosa26CAGs-rtTA3. Organoids were grown in conditioned media containing Wnt3a, R-Spondin, and noggin, generated as described previously from L-WRN cells (Miyoshi & Stappenbeck, 2013) and diluted 1:1 with Advanced DMEM/F-12 containing 1x N-2, 1x B-27, 1% Pen/Strep, and 2 mM L-glutamine. The growth media also contained 50 ng/mL EGF, 0.1 mg/mL primocin, 1 μM N-acety-L-cysteine, 10 mM HEPES, and 10 μM Y-27632 (added only upon passaging). Induction of Lats1/2 knockout was performed by addition of 100 nM 4-OHT overnight whereas Yap overexpression was induced with 1 μg/mL doxycycline in the media.
To generate tetO-YapS127A rtTA organoids, organoids derived from Col1a1tetO-YapS127A/+ Rosa26LSL-rtTA/+ mice were transduced with a lentivirus expressing Cre recombinase (generated from Cre-IRES-PuroR, a gift from Darrell Kotton) to activate rtTA expression (Somers et al., 2010). Three days after infection, organoids were selected by adding 1 μg/mL puromycin.
Knockout of Apc was performed by transfecting or transducing a plasmid or lentivirus, respectively, expressing Cas9 and a sgRNA against Apc (Apc sgRNA cloned into lentiCRISPR, a gift from Feng Zhang) (Schwank et al., 2013; Shalem et al., 2014). For organoids derived from Col1a1tetO-YapS127A/+ Rosa26LSL-rtTA/+ mice, the lentivirus additionally expressed Cre recombinase to activate rtTA (Apc sgRNA cloned into pSECC, a gift from Tyler Jacks) (Sanchez-Rivera et al., 2014). Three days after infection, organoids were selected by removing Wnt and R-Spondin from the media. These organoids were grown in the same media as wild-type organoids but did not contain conditioned media and were supplemented with 100 ng/mL noggin. Apc−/− CAGs-rtTA3 organoids were infected with lentiviruses carrying inducible expression of mutant versions of YAP. Three days after infection, organoids were selected by adding 1 μg/mL puromycin for 3 days.
Apcfl/fl KrasLSL-G12D p53fl/fl organoids were infected with an adenovirus expressing Cre recombinase and GFP (Cat#VVC-U of Iowa-1174) and selected 3 days after infection for 1 week by adding 1 μM EGFR inhibitor gefitinib and 5 μM nutlin-3 to organoid media without Wnt, R-Spondin, or EGF. Growth media was the same as for wildtype organoids but did not contain conditioned media or EGF and was supplemented with 100 ng/mL noggin. Organoids with inducible Yap overexpression were obtained by infecting with a lentivirus overexpressing Yap in the pLVX-Tight-Puro system (Clonetech Cat#632162) followed by selection with 1 μg/mL puromycin (starting 3 days after infection) and subsequent infection with a lentivirus expressing rtTA (generated from pLenti CMV rtTA3 Blast (w756–1), a gift from Eric Campeau) and selection with 10 μg/mL blasticidin. We isolated 20 clones for which we tested YAP overexpression by western blot and used the clone that displayed higher levels of YAP expression for all the experiments shown in this manuscript.
Knockout of Smad4 was performed by transfecting AKP organoids with a plasmid expressing Cas9 and a sgRNA against Smad4 (Smad4 sgRNA cloned into lentiCRISPR, a gift from Feng Zhang) (Shalem et al., 2014; Weber et al., 2015). The organoids were selected 3 days after transfection by removing noggin from the culture media for AKP organoids and supplementing with 10 ng/ml TGF-β (Matano et al., 2015). Selected clones were maintained in the same culture media as AKP organoids but without noggin.
Human Colorectal Cancer Organoids
The collection of fresh human CRC specimens and the generation of organoid lines was described previously (Roper et al., 2017). The line used in this study is derived from a moderately differentiated liver metastasis from a MSS rectal adenocarcinoma and carries mutations in KRAS, TP53, and PTCH1. Human CRC organoids were grown in Matrigel and maintained in the same culture media as for AKP mouse organoids.
Murine Colonoscopy and Mucosal Injection
Mouse colonoscopy was performed on anesthetized mice using Storz equipment. Injections were performed as described previously (Roper et al., 2017). We used a custom-made flexible steel needle, which was introduced through the working channel of the colonoscope. The injection was performed under observation by a very gentle mucosal penetration with the open side of the bevel heading up in a flat angle. A volume of 50–100 μL of virus or organoid solution was then injected into the colonic lamina propria. The viral titer of adenovirus expressing Cre recombinase (Cat#VVC-U of Iowa-5) was 10^9 pfu/ml with 3 injections per mouse at different locations.
Apc−/− KrasG12D p53−/− tetO-YAP5SA rtTA organoids were prepared for infection by mechanically resuspending the Matrigel and growth media with a pipette. They were dissociated by passing them 3 times through a syringe with a 25G needle followed by a cold PBS wash. They were resuspended in PBS containing 10% Matrigel and kept on ice before injection. Typically, one well of a 24-well plate containing 50 μL of Matrigel and grown to confluency (~100,000 cells) was resuspended in a volume of 250 μL for injection.
Subcutaneous Transplantation of Organoids
Apc−/− KrasG12D p53−/− and human organoids expressing tetO-YAP5SA and rtTA were injected subcutaneously in the flanks of nude mice. The organoids were prepared as described for colon injections but were resuspended in Matrigel diluted 1:1 in DMEM. Each flank was injected with organoids coming from three wells of a 24-well plate (~300,000 cells) resuspended in a volume of 100 μL. For secondary transplantation, 18,000 sorted EpCAM+ Apc−/− KrasG12D p53−/− tetO-YAP5SA rtTA tumor cells were transplanted into the flanks of nude mice.
Intrasplenic Injection of Organoids
Apc−/− KrasG12D p53−/− Smad4−/− tetO-YAP5SA rtTA organoids were prepared as single cells following a 20 min incubation in dispase supplemented with 10 μM Y-27632 at 37°C, subsequent incubation in 0.25% trypsin with 10 μM Y-27632 at 37°C for 10 min, and enzyme inactivation with Advanced DMEM/F12 (+1% Pen/Strep, 2 mM L-glutamine, 20% FBS). The cells were mechanically dissociated with a 25G needle, centrifuged, and resuspended in cold Advanced DMEM/F12. Mice were anesthetized with isoflurane, and under sterile conditions, the abdomens were depilated and sterilized. A small flank incision was made to expose the tip of the spleen, which was gently withdrawn from the abdomen with sterile tweezers and held in place with a sterile clamp by the fat tissue attached to the spleen. The bottom third of the spleen was loosely ligated using a non-absorbable Vicryl coated suture, and 300,000 cells in a 100 μL volume were injected with a 25G needle underneath the splenic capsule past the ligation. After needle withdrawal, the spleen was ligated to close the wound, and the incision was closed with sutures.
Lentivirus Production
Lentiviruses were generated in 293X cells by transfecting backbone and packaging plasmids using TransIT reagent following manufacturer’s instructions. 293X cells were maintained in DMEM (+10% FBS, 2 mM L-glutamine, 1% Pen/Strep). The supernatant containing the virus was collected 48 and 60–72 hours after transfection, concentrated by ultracentrifugation at 16,000 rpm for 90 min at 4°C, and resuspended in the remaining supernatant.
METHOD DETAILS
Organoid Transfection and Infection
2–3 days prior to transfection or infection, organoids were treated with 10 mM nicotinamide. Organoids were washed with PBS and treated with dispase to dissolve the Matrigel. They were passaged through a 25G syringe three times, spun down for 5 min at 1000 g and resuspended in 450 μL of culture media. The cell suspension was transferred to a 48-well plate, and 10 μL of a lentivirus solution (titer approximately 10^7 pfu/mL) or 50 μL of lipofectamine-DNA complex were added to the well. Cells were spinoculated for 1 hr at 30°C at 600 g. After a 4–6 h recovery at 37°C, the cells were resuspended, centrifuged in an Eppendorf tube, resuspended in 50 μL Matrigel, and cultured as described above.
Histology
Intestine, colon, and tumor samples were fixed overnight in 10% formalin, washed with PBS and 70% ethanol, and embedded in paraffin. Organoids grown in Matrigel were fixed in 4% paraformaldehyde/PBS, washed extensively with PBS and encapsulated in Histogel prior to embedding in paraffin. Livers were fixed 4% paraformaldehyde/PBS for 36–48 hours and cut into 9–10 pieces before embedding in paraffin. The paraffin blocks were cut into 5 μm sections using a microtome.
For immunohistochemistry, antigen retrieval for most epitopes was performed with citric-acid based pH 6.0 Antigen Unmasking Solution at 95°C in a pressure cooker for 1 hr. For epitopes requiring high pH, antigen retrieval was performed with tris-based pH 9.0 Antigen Unmasking Solution at 37°C for 10 min. The slides were blocked with 0.3% hydrogen peroxide and subsequently 2.5% donkey serum in PBS and incubated with primary antibody in blocking buffer overnight at 4°C. After washing with PBS three times, slides were incubated with biotinylated secondary antibodies in PBS for 1 hr. The signal was amplified with VectaStain ABC Reagent and developed with DAB. Harris modified hematoxylin was used to counterstain nuclei and subsequently slides were washed, dehydrated, and mounted using VectaMount. The list of antibodies used has been provided in the key resource table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-YAP (clone D8H1X) | Cell Signaling Technology | Cat#14074; RRID: AB_2650491 |
| Rabbit polyclonal anti-human Lysozyme | Dako | Cat#A0099; RRID: AB_2341230 |
| Rabbit monoclonal anti-Ki67 (clone SP6) | GeneTex | Cat#GTX16667; RRID: AB_422351 |
| Rat monoclonal anti-mouse CD44 | BD Biosciences | Cat#550538; RRID: AB_393732 |
| Goat polyclonal anti-mCherry (tdTomato) | Sicgen | Cat#AB0040-200; RRID: AB_2333092 |
| Rat monoclonal anti-Ki67 (clone SolA15) | Invitrogen | Cat#14-5698-82; RRID: AB_10854564 |
| Rabbit monoclonal non-phospho (active) β-catenin (Ser33/37/Thr41) (clone D13A1) | Cell Signaling Technology | Cat#8814; RRID: AB_11127203 |
| Rabbit monoclonal anti-phospho-histone H3 (Ser10) (clone E173) | Millipore | Cat#04-1093; RRID: AB_1977262 |
| Rabbit monoclonal anti-HA-tag (clone C29F4) | Cell Signaling Technology | Cat#3724; RRID: AB_1549585 |
| Rabbit monoclonal anti-GAPDH (clone 14C10) (HRP conjugated) | Cell Signaling Technology | Cat#3683; RRID: AB_1642205 |
| APC rat monoclonal anti-mouse CD326 (Ep-CAM) | BioLegend | Cat#118214; RRID: AB_1134102 |
| APC/Cy7 rat monoclonal anti-mouse TER-119 | BioLegend | Cat#116223; RRID: AB_2137788 |
| APC/Cy7 rat anti-mouse CD45 (clone 30-F11) | BD Biosciences | Cat#557659; RRID: AB_396774 |
| APC/Cy7 rat anti-CD11b (clone M1/70) | BD Biosciences | Cat#557657; RRID: AB_396772 |
| Biotinylated goat anti-rabbit IgG | Vector Laboratories | Cat#BA-1000; RRID: AB_2313606 |
| Biotinylated goat anti-rat IgG | Vector Laboratories | Cat#BA-9400; RRID: AB_2336202 |
| Donkey anti-rabbit IgG, Alexa Fluor 488 | Thermo Fisher Scientific | Cat#A-21206; RRID: AB_2535792 |
| Donkey anti-rat IgG, Alexa Fluor 594 | Thermo Fisher Scientific | Cat#A-21209; RRID: AB_2535795 |
| Bacterial and Virus Strains | ||
| Ad5CMVCre-eGFP | University of Iowa - Viral Vector Core | Cat#VVC-U of Iowa-1174 |
| Ad5CMVCre | University of Iowa - Viral Vector Core | Cat#VVC-U of Iowa-5 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma-Aldrich | Cat#T5648 |
| Corn oil | Fisher Scientific | Cat#s93205 |
| Doxycycline hyclate | Sigma-Aldrich | Cat#D9891 |
| Dextran sulfate sodium salt (36,000-50,000 M.Wt.) | MP Biomedicals | Cat#0216011080 |
| Azoxymethane | Sigma-Aldrich | Cat#A5486 |
| Matrigel | Corning | Cat#CB-40234 |
| Advanced Dulbecco’s modified Eagle medium/Ham’s F-12 | Gibco | Cat#12634028 |
| Tet system approved FBS, US-sourced | Clontech | Cat#631101 |
| Fetal bovine serum (FBS) | Gibco | Cat#26140079 |
| L-glutamine (200 mM) | Gibco | Cat#25030164 |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat#15140163 |
| N-2 supplement (100x) | Gibco | Cat#17502048 |
| B-27 supplement (50x), minus vitamin A | Gibco | Cat#12587010 |
| HEPES 1 M sterile solution pH 7.3 | Amresco | Cat#J848 |
| Recombinant murine EGF | Peprotech | Cat#315-09 |
| Primocin | InvivoGen | Cat#ant-pm-1 |
| N-acetyl-L-cysteine | Sigma-Aldrich | Cat#9165 |
| Y-27632 dihydrochloride | ApexBio | Cat#A3008 |
| Recombinant murine noggin | Peprotech | Cat#250-38 |
| Nicotinamide | Sigma-Aldrich | Cat#N0636 |
| (Z)-4-Hydroxytamoxifen | Sigma-Aldrich | Cat#H7904 |
| Recombinant human TGF-β1 (HEK293 derived) | Peprotech | Cat#100-21 |
| Blasticidin (solution) | InvivoGen | Cat#ant-bl-1 |
| Puromycin dihydrochloride | Gibco | Cat#A1113802 |
| Gefitinib | ApexBio | Cat#A8219 |
| Nutlin-3 | ApexBio | Cat#A4228 |
| TransIT-293 transfection reagent | Mirus | Cat#MIR 2705 |
| Dispase (5 U/mL) | STEMCELL Technologies | Cat#07913 |
| Trypsin-EDTA (0.25%), phenol red | Gibco | Cat#25200114 |
| Collagenase/Dispase | Roche | Cat#10269638001 |
| Collagenase, type IV | Gibco | Cat#17104019 |
| HBSS (10X), no calcium, no magnesium, no phenol red | Gibco | Cat#14185052 |
| EDTA 0.5 M sterile solution pH 8.0 | Amresco | Cat#E177 |
| Paraformaldehyde aqueous solution 16% | Electron microscopy sciences | Cat#15711 |
| HistoGel specimen processing gel | Thermo Fisher Scientific | Cat#NC9150318 |
| Antigen unmasking solution, citric acid based | Vector Laboratories | Cat#H-3300 |
| Antigen unmasking solution, tris based | Vector Laboratories | Cat#H-3301 |
| Donkey serum | Sigma-Aldrich | Cat#D9663 |
| Vectastain Elite ABC HRP reagent, R.T.U. | Vector Laboratories | Cat#PK-7100 |
| Hematoxylin solution, Harris modified | Sigma-Aldrich | Cat#HHS32 |
| Eosin Y solution, alcoholic, with phloxine | Sigma-Aldrich | Cat#HT110332 |
| VectaMount permanent mounting medium | Vector Laboratories | Cat#H-5000 |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Invitrogen | Cat#D1306 |
| ProLong Gold antifade mountant with DAPI | Invitrogen | Cat#P36935 |
| TRIzol reagent | Invitrogen | Cat#15596018 |
| Fast SYBR Green master mix | Applied Biosystems | Cat#4385617 |
| Critical Commercial Assays | ||
| InDrop single-cell RNA seq kit | 1CellBio | N/A |
| DAB peroxidase (HRP) substrate kit (with nickel), 3,3’-diaminobenzidine | Vector Laboratories | Cat#SK-4100 |
| RNAscope 2.5 HD Reagent Kit-BROWN | Advanced Cell Diagnostics | Cat#322300 |
| RNAscope probe-Mm-AmotL2 | Advanced Cell Diagnostics | Cat#515181 |
| RNAscope probe-Mm-Lgr5 | Advanced Cell Diagnostics | Cat#312171 |
| RNAscope probe-Mm-Olfm4 | Advanced Cell Diagnostics | Cat#311831 |
| RNAscope probe-Mm-Axin2 | Advanced Cell Diagnostics | Cat#400331 |
| RNAscope probe-Mm-Reg4 | Advanced Cell Diagnostics | Cat#409601 |
| RNAscope probe-Mm-Klf6 | Advanced Cell Diagnostics | Cat#426901 |
| iScript cDNA synthesis kit | Bio-Rad | Cat#1708891BUN |
| NucleoSpin RNA XS | Takara Bio | Cat#740902.50 |
| TruSeq RNA Library Preparation Kit v2, Set A | Illumina | Cat#RS-122-2001 |
| Deposited Data | ||
| Raw and processed data | This paper | GEO: GSE152376 |
| Experimental Models: Cell Lines | ||
| Mouse: Col1a1tetO-YapS127A/+ Rosa26LSL-rtTA/+ organoid | This paper | N/A |
| Mouse: Lrig1-CreERT2 Lats1fl/fl Lats2fl/fl organoid | This paper | N/A |
| Mouse: Apc−/− Col1a1tetO-YapS127A/+ Rosa26LSL-rtTA/+ organoid | This paper | N/A |
| Mouse: Apc−/− Lrig1-CreERT2 Lats1fl/fl Lats2fl/fl organoid | This paper | N/A |
| Mouse: Apc−/− KrasG12D p53−/− tetO-Yap5SA rtTA organoid | This paper | N/A |
| Mouse: Apc−/− CAGs-rtTA3 organoid | This paper | N/A |
| Mouse: L-WRN | ATCC | Cat#CRL-3276; RRID: CVCL_DA06 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Lrig1-CreERT2: Lrig1tm1.1(cre/ERT2)Rjc/J | The Jackson Laboratory | JAX: 018418; RRID: IMSR_JAX:018418 |
| Mouse: Lats1fl/fl: Lats1tm1.1Jfm/RjoJ | The Jackson Laboratory | JAX: 024941; RRID: IMSR_JAX:024941 |
| Mouse: Lats2fl/fl: Lats2tm1.1Jfm/RjoJ | The Jackson Laboratory | JAX: 025428; RRID: IMSR_JAX:025428 |
| Mouse: Villin-Cre: B6.Cg-Tg(Vil1-cre)997Gum/J | The Jackson Laboratory | JAX: 004586; RRID: IMSR_JAX:004586 |
| Mouse: Villin-CreERT2: B6.Cg-Tg(Vil1-cre/ERT2)23Syr/J | The Jackson Laboratory | JAX: 020282; RRID: IMSR_JAX:020282 |
| Mouse: Mst1−/− Mst2fl/fl | Zhou et al., 2011 | N/A |
| Mouse: Col1a1tetO-YapS127A/+; Rosa26LSL-rtTA/+ | Yimlamai et al., 2014 | N/A |
| Mouse: Lgr5-IRES-CreERT2 | Huch et al., 2013 | N/A |
| Mouse: Rosa26LSL-Tomato: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | The Jackson Laboratory | JAX: 007909; RRID: IMSR_JAX:007909 |
| Mouse: Lgr5-CreERT2: B6.129P2-Lgr5tm1(cre/ERT2)Cle/J | The Jackson Laboratory | JAX: 008875; RRID: IMSR_JAX:008875 |
| Mouse: Rosa26mTmG: B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | The Jackson Laboratory | JAX: 007676; RRID: IMSR_JAX:007676 |
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664; RRID: IMSR_JAX:000664 |
| Mouse: CAGs-rtTA3: B6N.FVB(Cg)-Tg(CAG-rtTA3)4288Slowe/J | The Jackson Laboratory | JAX: 016532; RRID: IMSR_JAX:016532 |
| Mouse: Apcfl/fl | Colnot et al., 2004 | N/A |
| Mouse: KrasLSL-G12D: B6.129S4-Krastm4Tyj/J | The Jackson Laboratory | JAX: 008179; RRID: IMSR_JAX:008179 |
| Mouse: p53fl/fl: B6.129P2-Trp53tm1Brn/J | The Jackson Laboratory | JAX: 008462; RRID: IMSR_JAX:008462 |
| Mouse: NSG: NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ | The Jackson Laboratory | JAX: 005557; RRID: IMSR_JAX:005557 |
| Mouse: nude: J:NU | The Jackson Laboratory | JAX: 007850; RRID: IMSR_JAX:007850 |
| Mouse: Yapfl/fl | Schlegelmilch et al., 2011 | N/A |
| Mouse: Tazfl/fl | Xin et al., 2013 | N/A |
| Oligonucleotides | ||
| Primers for RT-qPCR, see Table S4 | This paper | N/A |
| sgRNA: mouse Apc GTCTGCCATCCCTTCACGTT | Schwank et al., Cell Stem Cell 2013 | N/A |
| sgRNA: mouse Smad4 GACAACCCGCTCATAGTGATA | Weber et al., 2015 | N/A |
| Recombinant DNA | ||
| Cre-IRES-PuroR | Somers et al., 2010 | Addgene Plasmid #30205; RRID: Addgene_30205 |
| pLenti CMV rtTA3 Blast (w756-1) | Addgene | Addgene Plasmid #26429; RRID: Addgene_26429 |
| lentiCRISPR-sgApc | This paper | N/A |
| pSECC-sgApc | This paper | N/A |
| lentiCRISPR-sgSmad4 | This paper | N/A |
| pLVX-Tight-YAP5SA | This paper | N/A |
| pLVX-Tight-YAP5SA/S94A | This paper | N/A |
| pLVX-Tight-HA-YAP5SA(full length) | This paper | N/A |
| pLVX-Tight-HA-YAP5SA(amino acids 1-170) | This paper | N/A |
| pLVX-Tight-HA-YAP5SA(amino acids 1-285) | This paper | N/A |
| Software and Algorithms | ||
| rCASC | Alessandri et al., Gigascience 2019 | https://kendomaniac.github.io/rCASC/index.html |
| Pipeline for processing inDrops sequencing data | Zilionis et al., 2017 | https://github.com/indrops/indrops |
| Bowtie v1.1.1.1 | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/index.shtml |
| Samtools v1.3.1 | Li et al., 2009 | http://samtools.sourceforge.net/ |
| RSEM v1.3.0 | Li & Dewey, 2011 | https://deweylab.github.io/RSEM/ |
| Salmon v0.14.1 | Patro et al., 2017 | https://combine-lab.github.io/salmon/; RRID: SCR_017036 |
| RStudio v1.1.383 | Rstudio, Inc. | https://rstudio.com/; RRID: SCR_000432 |
| R v3.6.2 | R Foundation for Statistical Computing | https://www.r-project.org/; RRID: SCR_001905 |
| Seurat v3.1.3 | Butler et al., 2018 | https://satijalab.org/sseura/; RRID: SCR_016341 |
| sctransform v0.2.1 | Hafemeister & Satija, 2019 | https://satijalab.org/seurat/v3.1/sctransform_vignette.html |
| tximport v1.14.0 | Soneson et al., 2015 | https://github.com/mimikelo/tximport; RRID: SCR_016752 |
| DESeq2 v1.26.0 | Love et al., 2014 | https://github.com/mimikelo/DESeq2; RRID: SCR_015687 |
| fgsea v1.12.0 | Sergushichev, 2016 | https://github.com/ctlab/fgsea |
| clusterProfiler v3.14.3 | Yu et al., 2012 | https://github.com/YYuLa-SMU/clusterProfiler; RRID: SCR_016884 |
| Code for analyses of single-cell and bulk RNA-seq datasets | This paper | https://github.com/cheungpriscilla/CellStemCell_2020 |
| GraphPad Prism6 | GraphPad | https://www.graphpag.com/scientific-software/prism/; RRID: SCR_002798 |
| ImageJ v1.51m9 | Schneider et al., 2012 | https://imagej.nih.gog/ij/; RRID: SCR_003070 |
| Fiji v2.0.0-rc-69/1.52p | Schindelin et al., 2012 | https://imagej.net/Fiji; RRID: SCR_00228 |
| FlowJo | FlowJo | https://www.flowjo.coc/; RRID: SCR_008520 |
| BioRender | BioRender | https://biorender.com/ |
| Other | ||
For immunofluorescence, following antigen retrieval, blocking, and primary antibody incubation, slides were incubated with appropriate fluorescent antibodies and 1 μg/ml DAPI for 1 hour and then mounted with Prolong Gold antifade with DAPI.
For RNA in situ hybridization, we used the RNAScope Brown HD 2.5 kit according to the manufacturer’s instructions with the probes listed in the key resource table.
IHC and RNAScope images were taken using a Zeiss Axio Scope. IF images were obtained on a Zeiss AxioObserver Z1.
RNA Isolation and RT-qPCR
RNA from organoids was extracted using Trizol reagent. cDNA was obtained using the cDNA Synthesis Kit and diluted 1:40 for RT-qPCR if starting from 500 ng of RNA. RT-qPCR was performed on a One Step plus Sequence Detection System (Applied Biosystems) using Fast SYBR® Green Master Mix (Life Technologies), and gene expression data was quantified using the DeltaDeltaCt method and normalized to Gapdh/GADPH. The list of primers used for RT-qPCR has been provided in the key resources table.
Protein Extraction and Western Blot Analyses
Organoids were washed in PBS, and the Matrigel was dissolved with dispase. After washing with PBS, the cells were lysed with RIPA buffer and quantified using Bradford Reagent. For western blot, 10 μg of total protein were loaded into each well of an SDS-PAGE gel. The list of antibodies used can be found in the key resources table.
Fluorescence-Activated Cell Sorting
For isolation of intestinal and colonic epithelial cells for fluorescence-activated cell sorting (FACS), a piece of intestine was extracted from mice and incubated in 4 mM EDTA in PBS with gentle rocking for 40 min at 4°C. The small intestine was shaken to dissociate epithelial cells whereas the colon was cut open and scraped with a coverslip to dissociate the crypts. The smooth muscle layer was removed, and the remaining supernatant was centrifuged and incubated in 3 mg/mL collagenase/dispase in PBS for 6 min at 37°C with repeated up and down pipetting. The single cells were then placed on ice, centrifuged, filtered through 70 μm strainers, and stained in 2% FBS/PBS with fluorescent conjugated EpCAM, CD45, and TER119 antibodies diluted 1:100 for 30 min at 4°C.
For isolation of tumor cells for FACS and subsequent secondary transplantation, tumors were chopped into small pieces in 300 U/ml collagenase IV in digestion buffer (1x HBSS, 10 mM HEPES, 1.25 mM CaCl2*2H2O, and 4 mM MgCl2*6H2O in MilliQ H2O) and subsequently incubated at 37°C for 30 min with occasional pipetting. The tumor pieces were then centrifuged and incubated in 0.25% trypsin at 37°C for 30 min with occasional pipetting followed by an addition of FBS to a 5% final concentration. The tumors cells were washed with cold PBS twice, filtered through 70 μm strainers, and stained in 2% FBS/PBS with fluorescent conjugated EpCAM, CD45, TER119, and CD11b antibodies diluted 1:100 for 30 min at 4°C.
All sorting was performed on a BD FACSAria II, using a 100 μm nozzle, and FlowJo (Tree Star) software was used for all flow cytometry analysis. The following combinations were used to isolate each of the respective populations: intestinal/colonic epithelial cells: EpCAM+CD45-TER119-; tumor epithelial cells: EpCAM+CD45-TER119-CD11b-. For all sorts, 4’, 6-diamidino-2-phenylindole (DAPI) was used to eliminate dead cells.
Single-cell RNA Sequencing and Analysis
InDrop-v2 (1CellBio) encapsulation protocol was performed following manufacturer’s instructions found at the company’s website. 5,000 cells were encapsulated per library and condition. Whole-transcriptome libraries were prepared as previously described (Zilionis et al., 2017) and sequenced on an Illumina NextSeq 500 using paired-end 75 cycles v2 kits (Read 1: 36 cycles; Index Read: 6 cycles; Read 2: 50 cycles). The fastq to counts conversion was performed using the indropIndex and the indropCounts functions, which are part of the rCASC framework (Alessandrì et al., 2019). The indropCounts function, which invokes the inDrop pipeline v20170126 (https://github.com/indrops/indrops), was used with the following options: M=10, U=2, D=400, and low.complexity.mask=“False” and the following packages: Bowtie v1.1.1.1, Samtools v1.3.1, RSEM v1.3.0, and Java v1.8.0 (Langmead et al., 2009; B. Li & Dewey, 2011; H. Li et al., 2009).
To visualize the single cell data, we used Seurat (v3.1.3 with default parameters except where indicated) (Butler et al., 2018). In both datasets, cells that had more than 15% of the total UMIs in mitochondrial genes were filtered out, and those with a log10(number of genes per UMI) greater than 0.8 were kept. We also set a minimum threshold of UMIs/cell and genes/cell: 600 UMIs/cell and 250 genes/cell in both the control and Lats1/2 cKO samples and 300 UMIs/cell and 300 genes/cell in the colon control and DSS sample. For each experiment, the number of cells were down sampled to the sample with the fewest cells to obtain an equal number of cells per condition for downstream analysis. Next, sctransform was used to normalize UMI counts and find variable features for dimensionality reduction (Hafemeister & Satija, 2019). Dimensionality reduction was performed using RunPCA to generate points for embedding, which were then used to construct UMAP plots (RunUMAP) and find neighbors for clustering (FindNeighbors). For all relevant functions, 40 principal components were used. FindClusters (resolution=1.4) was used to run the Louvain clustering algorithm.
For differential expression analysis, the bimod test (McDavid et al., 2013) implemented in Seurat was used to find the differentially expressed genes between the Lats1/2 KO cells and control cells, excluding the mesenchymal population, or stem cells in the Lats1/2 cKO scRNA-seq and between the same cell type in the DSS versus control in the DSS scRNA-seq. For the Lats1/2 cKO scRNA-seq, gene ontology enrichment analysis was performed on genes with an adjusted p-value < 0.05 and a log2FC > 1 or < −1 in the Lats1/2 knockout cells versus control using enrichGO in the clusterProfiler R package to identify significant GO biological processes overrepresented (Yu et al., 2012). For both datasets, gene set enrichment analysis (GSEA) was performed on the ranked average log fold change of the differentially expressed genes using the fgsea R package to identify significant KEGG pathways enriched (Sergushichev, 2016). The GSEA function in clusterProfiler was used to perform GSEA of previously published gene signatures. The intestinal β-catenin signature is derived from the supplementary table of downregulated genes in intestinal crypt cells upon deletion of β-catenin (Fevr et al., 2007). The Lgr5 ISC gene signature is derived from the “mRNA stem cell signature” in the supplementary table of Muñoz et al., 2012. For the YAP gene signatures, the Barry et al., 2013 signature is derived from the supplementary table of genes upregulated upon transgenic YAP overexpression in mouse crypts whereas the Gregorieff et al., 2015 signature is derived from the supplementary table of YAP activated genes with a log2 fold change < −1. To generate the DSS injury signature, we kept all genes with adjusted p-value < 0.05 and log2FC > 1 and plotted this injury signature onto the UMAP of control and Lats1/2 cKO cells by taking the average of the normalized expression values of the signature genes for each cell and plotting the log2 + 1 of these values.
Bulk RNA Sequencing and Analysis
RNA was isolated from organoids using the NucleoSpin RNA XS kit. The libraries for the RNA-seq analysis were prepared with 100 ng of RNA using the TruSeq RNA Library Preparation Kit v2 (Illumina) according to the manufacturer’s protocol. All of the libraries were sequenced on an Illumina HiSeq 4000 using paired end 150 cycles kits at Novogene. Raw sequencing reads were aligned to a reference transcriptome generated from the Ensembl v98 database with Salmon v0.14.1 using options “--seqBias --useVBOpt --gcBias --numBootstraps 30 --validateMappings” (Patro et al., 2017). Length-scaled transcripts per million were acquired using the tximport function, and log2 fold changes and false discovery rates were determined by DESeq2 in R (Love et al., 2014; Soneson et al., 2015). Shrunken log2 fold changes were determined with DESeq2, which were used to rank genes for GSEA of significant KEGG pathways by fgsea or of previously published gene signatures using the GSEA function (Kanehisa, 2000). In cases where there were duplicate gene symbols or Entrez IDs, the more significant gene was kept for the ranking. To identify the transcription factors differentially expressed, the differentially expressed genes at 72 hours with adjusted p-value < 0.05 and absolute log2 fold change > 1 were overlapped with the CIS-BP v2.00 database of mouse transcription factors (Weirauch et al., 2014). enrichGO in the clusterProfiler R package was used to identify significant GO biological processes overrepresented (Yu et al., 2012).
Image Analysis
Quantification of tumor area was performed in ImageJ (Schneider et al., 2012). TIFF files were imported into ImageJ, and the scale for each image was set. Each liver piece was thresholded manually, which was used to calculate its area. The freehand selection tool was used to circle each tumor and calculate the area, which was summed for each liver piece to calculate the percent tumor burden.
For quantification of RNA-ISH images, digital morphometric analysis was performed using the Trainable Weka Segmentation (TWS) plugin in Fiji (Arganda-Carreras et al., 2017; Schindelin et al., 2012). TIFF files were imported into Fiji, and the TWS plugin was trained to produce a classifier, segmenting images into areas of background, nuclei, and probe. The same trained classifier was applied to all images, providing a percentage of positive staining for each image.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data represented are expressed as mean ± standard error of mean unless otherwise specified. Data were analyzed using Prism Software 6.0 (GraphPad). Technical and biological replicates are specified for each experiment in the figure legends. P-values were determined by a two-tailed t test with Welch’s correction unless otherwise indicated whereas p-values for the Kaplan-Meier survival curve were determined with the log rank test. The p-values are presented as follows: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Material
Table S4. Primers for RT-qPCR, Related to STAR Methods
Table S1. Differentially Expressed Genes from Lats1/2 cKO scRNA-seq Dataset, Related to Figure 1
Table S2. Differentially Expressed Genes between DSS and Control by Cell Type, Related to Figure 3
Table S3. Differentially Expressed Transcription Factors in Apc KO YAP Overexpressing Organoids at 72h vs 0h, Related to Figure 4
Highlights.
Hippo signaling inhibition reprograms Lgr5+ ISCs to a Klf6+ wound healing cell state
Klf6+ cells are characterized by low Wnt activity and reduced stemness
YAP activation suppresses tumor growth in human and mouse metastatic CRC
YAP/TAZ deletion increases colon tumor growth
ACKNOWLEDGMENTS
We are grateful to all the members of the Camargo lab for insightful discussion and support. We thank Jonathan D. Lee, Sachin Patel, Brian Pepe-Mooney, and Lucia Suarez-Lopez for providing technical assistance; Ronald Mathieu for help with FACS; and Maxine McGredy and Bing Shui for contributions that were not included in the final version of this manuscript. The graphical abstract was created with BioRender. This work was supported in part by a Landry Cancer Biology Consortium Research Fellowship and NIH F31CA235893 and T32GM007226-41 to P.C. and a HFSP (LT000845/2014-L) and T32 fellowships to J.X. Further support was provided by NIH grants R01 DK099559 and R01 AR064036 (F.D.C.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alessandrì L, Cordero F, Beccuti M, Arigoni M, Olivero M, Romano G, Rabellino S, Licheri N, De Libero G, Pace L, & Calogero RA (2019). RCASC: Reproducible classification analysis of single-cell sequencing data. GigaScience. 10.1093/gigascience/giz105 [DOI] [PMC free article] [PubMed]
- Arganda-Carreras I, Kaynig V, Rueden C, Eliceiri KW, Schindelin J, Cardona A, & Seung HS (2017). Trainable Weka Segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics. 10.1093/bioinformatics/btx180 [DOI] [PubMed]
- Ayyaz A, Kumar S, Sangiorgi B, Ghoshal B, Gosio J, Ouladan S, Fink M, Barutcu S, Trcka D, Shen J, Chan K, Wrana JL, & Gregorieff A. (2019). Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature. 10.1038/s41586-019-1154-y [DOI] [PubMed]
- Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, Guzzardo V, Fassina A, Cordenonsi M, & Piccolo S. (2014). YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell, 158(1), 157–170. 10.1016/j.cell.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Barker N, Van Es JH, Kuipers J, Kujala P, Van Den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, & Clevers H. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature, 449(7165), 1003–1007. 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- Barry ER, Morikawa T, Butler BL, Shrestha K, De La Rosa R, Yan KS, Fuchs CS, Magness ST, Smits R, Ogino S, Kuo CJ, & Camargo FD (2013). Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature, 493(7430), 106–110. 10.1038/nature11693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, & Satija R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed]
- Cai J, Maitra A, Anders RA, Taketo MM, & Pan D. (2015). β-catenin destruction complex-independent regulation of Hippo-YAP signaling by APC in intestinal tumorigenesis. Genes and Development, 29(14), 1493–1506. 10.1101/gad.264515.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhang N, Zheng Y, De Wilde RF, Maitra A, & Pan D. (2010). The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes and Development, 24(21), 2383–2388. 10.1101/gad.1978810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colnot S, Niwa-Kawakita M, Hamard G, Godard C, Le Plenier S, Houbron C, Romagnolo B, Berrebi D, Giovannini M, & Perret C. (2004). Colorectal cancers in a new mouse model of familial adenomatous polyposis: Influence of genetic and environmental modifiers. Laboratory Investigation, 84(12), 1619–1630. 10.1038/labinvest.3700180 [DOI] [PubMed] [Google Scholar]
- De Sousa E Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, Anderson JE, Storm EE, Modrusan Z, Koeppen H, Dijkgraaf GJP, Piskol R, De Sauvage FJ, Sousa F. De Antonina V, Harnoss JM, Kljavin N, Hoeck JD, Hung J, … Sauvage F. J. De. (2017). A distinct role for Lgr5 + stem cells in primary and metastatic colon cancer. Nature, 543(7647), 676–680. 10.1038/nature21713 [DOI] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, & Piccolo S. (2011). Role of YAP/TAZ in mechanotransduction. Nature, 474(7350), 179–184. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- El Marjou F, Janssen KP, Chang BHJ, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, & Robine S. (2004). Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 10.1002/gene.20042 [DOI] [PubMed]
- Fearon ER, & Vogelstein B. (1990). A genetic model for colorectal tumorigenesis. Cell, 61(5), 759–767. 10.1016/0092-8674(90)90186-I [DOI] [PubMed] [Google Scholar]
- Fevr T, Robine S, Louvard D, & Huelsken J. (2007). Wnt/ -Catenin Is Essential for Intestinal Homeostasis and Maintenance of Intestinal Stem Cells. Molecular and Cellular Biology. 10.1128/mcb.01034-07 [DOI] [PMC free article] [PubMed]
- Fu V, Plouffe SW, & Guan KL (2017). The Hippo pathway in organ development, homeostasis, and regeneration. In Current Opinion in Cell Biology (Vol. 49). 10.1016/j.ceb.2017.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, Ohta Y, Toshimitsu K, Nakazato Y, Kawasaki K, Uraoka T, Watanabe T, Kanai T, & Sato T. (2016). A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell. 10.1016/j.stem.2016.04.003 [DOI] [PubMed]
- Fumagalli A, Drost J, Suijkerbuijk SJE, Van Boxtel R, & De J. (2017). Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. 1–19. 10.1073/pnas.1701219114 [DOI] [PMC free article] [PubMed]
- Gehart H, & Clevers H. (2019). Tales from the crypt: new insights into intestinal stem cells. Nature Reviews Gastroenterology and Hepatology, 16(1), 19–34. 10.1038/s41575-018-0081-y [DOI] [PubMed] [Google Scholar]
- Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, & Wrana JL (2015). Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer. Nature, 526(7575), 715–718. 10.1038/nature15382 [DOI] [PubMed] [Google Scholar]
- Hafemeister C, & Satija R. (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biology. 10.1186/s13059-019-1874-1 [DOI] [PMC free article] [PubMed]
- Haigis KM, & Dove WF (2003). A Robertsonian translocation suppresses a somatic recombination pathway to loss of heterozygosity. Nature Genetics. 10.1038/ng1055 [DOI] [PubMed]
- Haines J, Johnson V, Pack K, Suraweera N, Slijepcevic P, Cabuy E, Coster M, Ilyas M, Wilding J, Sieber O, Bodmer W, Tomlinson I, & Silver A. (2005). Genetic basis of variation in adenoma multiplicity in ApcMin/+ Mom1S mice. Proceedings of the National Academy of Sciences, 102(8), 2868–2873. 10.1073/pnas.0500039102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong AW, Meng Z, & Guan KL (2016). The Hippo pathway in intestinal regeneration and disease. In Nature Reviews Gastroenterology and Hepatology. 10.1038/nrgastro.2016.59 [DOI] [PMC free article] [PubMed]
- Huch M, Dorrell C, Boj SF, Van Es JH, Li VSW, Van De Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, Haft A, Vries RG, Grompe M, & Clevers H. (2013). In vitro expansion of single Lgr5 + liver stem cells induced by Wnt-driven regeneration. Nature. 10.1038/nature11826 [DOI] [PMC free article] [PubMed]
- Inuzuka H, Wakao H, Masuho Y, Muramatsu M. aki, Tojo H, & Nanbu-Wakao R. (1999). cDNA cloning and expression analysis of mouse zf9, a Kruppel-like transcription factor gene that is induced by adipogenic hormonal stimulation in 3T3-L1 cells. Biochimica et Biophysica Acta - Gene Structure and Expression. 10.1016/S0167-4781(99)00161-X [DOI] [PubMed]
- Kanehisa M. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed]
- Kojima S, Hayashi S, Shimokado K, Suzuki Y, Shimada J, Crippa MP, & Friedman SL (2000). Transcriptional activation of urokinase by the Kruppel-like factor Zf9/COPEB activates latent TGF-β1 in vascular endothelial cells. Blood. [PubMed]
- Konsavage WM, Kyler SL, Rennoll SA, Jin G, & Yochum GS (2012). Wnt/β-catenin signaling regulates yes-associated protein (YAP) gene expression in colorectal carcinoma cells. Journal of Biological Chemistry. 10.1074/jbc.M111.327767 [DOI] [PMC free article] [PubMed]
- Langmead B, Trapnell C, Pop M, & Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed]
- Li B, & Dewey CN (2011). RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, & Durbin R. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed]
- Love MI, Huber W, & Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, & Gumucio DL (2002). cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. Journal of Biological Chemistry. 10.1074/jbc.M204935200 [DOI] [PubMed]
- Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, & Sato T. (2015). Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nature Medicine, 21(3), 256–262. 10.1038/nm.3802 [DOI] [PubMed] [Google Scholar]
- McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, Roederer M, & Gottardo R. (2013). Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics. 10.1093/bioinformatics/bts714 [DOI] [PMC free article] [PubMed]
- McInnes L, Healy J, Saul N, & Großberger L. (2018). UMAP: Uniform Manifold Approximation and Projection. Journal of Open Source Software. 10.21105/joss.00861 [DOI]
- Merlos-Suárez A, Barriga FM, Jung P, Iglesias M, Céspedes MV, Rossell D, Sevillano M, Hernando-Momblona X, Da Silva-Diz V, Muñoz P, Clevers H, Sancho E, Mangues R, & Batlle E. (2011). The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell, 8(5), 511–524. 10.1016/j.stem.2011.02.020 [DOI] [PubMed] [Google Scholar]
- Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, Hishima T, Koike M, Shitara N, Iwama T, Utsunomiya J, Kuroki T, & Mori T. (1999). Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 10.1038/sj.onc.1202642 [DOI] [PubMed]
- Miyoshi H, & Stappenbeck TS (2013). In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nature Protocols, 8(12), 2471–2482. 10.1038/nprot.2013.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz J, Stange DE, Schepers AG, Van De Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, Myant K, Versteeg R, Sansom OJ, Van Es JH, Barker N, Van Oudenaarden A, Mohammed S, Heck AJR, & Clevers H. (2012). The Lgr5 intestinal stem cell signature: Robust expression of proposed quiescent ′ +4′ cell markers. EMBO Journal. 10.1038/emboj.2012.166 [DOI] [PMC free article] [PubMed]
- Neufert C, Becker C, & Neurath MF (2007). An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nature Protocols. 10.1038/nprot.2007.279 [DOI] [PubMed]
- Nusse YM, Savage AK, Marangoni P, Rosendahl-Huber AKM, Landman TA, De Sauvage FJ, Locksley RM, & Klein OD (2018). Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature. 10.1038/s41586-018-0257-1 [DOI] [PMC free article] [PubMed]
- O’Rourke KP, Loizou E, Livshits G, Schatoff EM, Baslan T, Manchado E, Simon J, Romesser PB, Leach B, Han T, Pauli C, Beltran H, Rubin MA, Dow LE, & Lowe SW (2017). Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nature Biotechnology. 10.1038/nbt.3837 [DOI] [PMC free article] [PubMed]
- Patro R, Duggal G, Love MI, Irizarry RA, & Kingsford C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nature Methods. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed]
- Peterson LW, & Artis D. (2014). Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. In Nature Reviews Immunology. 10.1038/nri3608 [DOI] [PubMed]
- Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, Guo Y, Shyr Y, Aronow BJ, Haigis KM, Franklin JL, & Coffey RJ (2012). The pan-ErbB negative regulator lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 10.1016/j.cell.2012.02.042 [DOI] [PMC free article] [PubMed]
- Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, Scuoppo C, Zuber J, Dickins RA, Kogan SC, Shroyer KR, Sordella R, Hannon GJ, & Lowe SW (2011). A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell, 145(1), 145–158. 10.1016/j.cell.2011.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratziu V, Lalazar A, Wong L, Dang Q, Collins C, Shaulian E, Jensen S, & Friedman SL (1998). Zf9, a Kruppel-like transcription factor up-regulated in vivo during early hepatic fibrosis. Proceedings of the National Academy of Sciences of the United States of America. 10.1073/pnas.95.16.9500 [DOI] [PMC free article] [PubMed]
- Reeves HL, Narla G, Ogunbiyi O, Haq AI, Katz A, Benzeno S, Hod E, Harpaz N, Goldberg S, Tal-Kremer S, Eng FJ, Arthur MJP, Martignetti JA, & Friedman SL (2004). Kruppel-Like Factor 6 (KLF6) Is a Tumor-Suppressor Gene Frequently Inactivated in Colorectal Cancer. Gastroenterology. 10.1053/j.gastro.2004.01.005 [DOI] [PubMed]
- Roper J, Tammela T, Cetinbas NM, Akkad A, Roghanian A, Rickelt S, Almeqdadi M, Wu K, Oberli MA, Sánchez-Rivera F, Park YK, Liang X, Eng G, Taylor MS, Azimi R, Kedrin D, Neupane R, Beyaz S, Sicinska ET, … Yilmaz ÖH (2017). In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nature Biotechnology. 10.1038/nbt.3836 [DOI] [PMC free article] [PubMed]
- Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, Shao DD, Schumacher SE, Weir BA, Vazquez F, Cowley GS, Root DE, Mesirov JP, Beroukhim R, Kuo CJ, … Hahn WC (2012). β-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell, 151(7), 1457–1473. 10.1016/j.cell.2012.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, Joshi NS, Subbaraj L, Bronson RT, Xue W, & Jacks T. (2014). Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 10.1038/nature13906 [DOI] [PMC free article] [PubMed]
- Schepers AG, Snippert HJ, Stange DE, Van Den Born M, Van Es JH, Van De Wetering M, Clevers H, & Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, and Clevers H. (2012). Lineage Tracing Reveals Lgr5+ Stem Cell Activity in Mouse Intestinal Adenomas. Science, 337(6095), 730–735. 10.1126/science.1224676 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, & Cardona A. (2012). Fiji: An open-source platform for biological-image analysis. In Nature Methods. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, & Camargo FD (2011). Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell, 144(5), 782–795. 10.1016/j.cell.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, & Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. In Nature Methods. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed]
- Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, Van Der Ent CK, Nieuwenhuis EES, Beekman JM, & Clevers H. (2013). Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 10.1016/j.stem.2013.11.002 [DOI] [PubMed]
- Seno H, Miyoshi H, Brown SL, Geske MJ, Colonna M, & Stappenbeck TS (2009). Efficient colonic mucosal wound repair requires Trem2 signaling. Proceedings of the National Academy of Sciences of the United States of America. 10.1073/pnas.0803343106 [DOI] [PMC free article] [PubMed]
- Sergushichev AA (2016). An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. BioRxiv. 10.1101/060012 [DOI]
- Serra D, Mayr U, Boni A, Lukonin I, Rempfler M, Challet Meylan L, Stadler MB, Strnad P, Papasaikas P, Vischi D, Waldt A, Roma G, & Liberali P. (2019). Self-organization and symmetry breaking in intestinal organoid development. Nature. 10.1038/s41586-019-1146-y [DOI] [PMC free article] [PubMed]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, & Zhang F. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science, 343(6166), 84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa M, Ohta Y, Nishikori S, Matano M, Takano A, Fujii M, Date S, Sugimoto S, Kanai T, & Sato T. (2017). Visualization and targeting of LGR5 + human colon cancer stem cells. Nature, 1–21. 10.1038/nature22081 [DOI] [PubMed]
- Siegel R, DeSantis C, & Jemal A. (2014). Colorectal cancer statistics, 2014. CA: A Cancer Journal for Clinicians. 10.3322/caac.21220 [DOI] [PubMed]
- Somers A, Jean JC, Sommer CA, Omari A, Ford CC, Mills JA, Ying L, Sommer AG, Jean JM, Smith BW, Lafyatis R, Demierre MF, Weiss DJ, French DL, Gadue P, Murphy GJ, Mostoslavsky G, & Kotton DN (2010). Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 10.1002/stem.495 [DOI] [PMC free article] [PubMed]
- Soneson C, Love MI, & Robinson MD (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed]
- Weber J, Öllinger R, Friedrich M, Ehmer U, Barenboim M, Steiger K, Heid I, Mueller S, Maresch R, Engleitner T, Gross N, Geumann U, Fu B, Segler A, Yuan D, Lange S, Strong A, Rosa JD La Esposito I, … Rad R. (2015). CRISPR/Cas9 somatic multiplex-mutagenesis for high-Throughput functional cancer genomics in mice. Proceedings of the National Academy of Sciences of the United States of America. 10.1073/pnas.1512392112 [DOI] [PMC free article] [PubMed]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, Zheng H, Goity A, van Bakel H, Lozano JC, Galli M, Lewsey MG, Huang E, Mukherjee T, Chen X, … Hughes TR (2014). Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 10.1016/j.cell.2014.08.009 [DOI] [PMC free article] [PubMed]
- Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, Richardson JA, Sadek HA, Bassel-Duby R, & Olson EN (2013). Hippo pathway effector Yap promotes cardiac regeneration. Proceedings of the National Academy of Sciences, 110(34), 13839–13844. 10.1073/pnas.1313192110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, & Ito Y. (1999). A WW domain-containing Yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO Journal. 10.1093/emboj/18.9.2551 [DOI] [PMC free article] [PubMed]
- Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, Shrestha K, Cahan P, Stanger BZ, & Camargo FD (2014). Hippo pathway activity influences liver cell fate. Cell, 157(6), 1324–1338. 10.1016/j.cell.2014.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, & He QY (2012). ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS A Journal of Integrative Biology. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed]
- Yui S, Azzolin L, Maimets M, Pedersen MT, Fordham RP, Hansen SL, Larsen HL, Guiu J, Alves MRP, Rundsten CF, Johansen JV, Li Y, Madsen CD, Nakamur T., Watanab M., Nielse OH., Schweige PJ., Piccol S., & Jensen KB (2018). YAP/TAZ-Dependent Reprogramming of Colonic Epithelium Links ECM Remodeling to Tissue Regeneration. Cell Stem Cell, 22(1), 35–49.e7. 10.1016/j.stem.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Halder SK, Kashikar ND, Cho YJ, Datta A, Gorden DL, & Datta PK (2010). Antimetastatic Role of Smad4 Signaling in Colorectal Cancer. Gastroenterology. 10.1053/j.gastro.2009.11.004 [DOI] [PMC free article] [PubMed]
- Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, & Guan KL (2011). Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes and Development, 25(1), 51–63. 10.1101/gad.2000111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Wang L, Wang CY, Yu J, & Guan KL (2012). Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes and Development, 26(1), 54–68. 10.1101/gad.173435.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu JJ, Li L, Li W, Li S, Yu JJ, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, & Guan KL (2008). TEAD mediates YAP-dependent gene induction and growth control. Genes and Development, 22(14), 1962–1971. 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Zhao B, Wei X, Wei X, Li W, Li W, Udan RS, Udan RS, Yang Q, Yang Q, Kim J, Kim J, Xie J, Xie J, Ikenoue T, Ikenoue T, Yu J, Yu J, Li LL, … Guan KLK-LK-L (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes & Development, 21(21), 2747–2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Zhang Y, Wu H, Barryg E, Yin YY, Lawrence E, Dawson D, Willis JE, Markowitz SD, Camargo FD, Avruch J, Barry E, Yin YY, Lawrence E, Dawson D, Willis JE, Markowitz SD, Camargo FD, & Avruch J. (2011). Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proceedings of the National Academy of Sciences of the United States of America, 108(49), 1312–1320. 10.1073/pnas.1110428108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM, & Mazutis L. (2017). Single-cell barcoding and sequencing using droplet microfluidics. Nature Protocols, 12(1), 44–73. 10.1038/nprot.2016.154 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4. Primers for RT-qPCR, Related to STAR Methods
Table S1. Differentially Expressed Genes from Lats1/2 cKO scRNA-seq Dataset, Related to Figure 1
Table S2. Differentially Expressed Genes between DSS and Control by Cell Type, Related to Figure 3
Table S3. Differentially Expressed Transcription Factors in Apc KO YAP Overexpressing Organoids at 72h vs 0h, Related to Figure 4
Data Availability Statement
The datasets generated in this study are available in the NCBI GEO database under GSE152376. Code to reproduce the analyses of the single-cell and bulk RNA-seq datasets can be found at https://github.com/cheungpriscilla/CellStemCell_2020.