Abstract
Class C radical SAM methyltransferases catalyze a diverse array of difficult chemical transformations in the biosynthesis of a range of compounds of biomedical importance. Phylogenetic analysis suggests that all of these enzymes are related to “CpdH” (formerly “HemN”) and “HemW”, proteins with essential roles in anaerobic heme biosynthesis and heme transport, respectively. These functions are essential to anaerobic metabolism in Escherichia coli. Interestingly, evolution has come full circle, and the divergence of this protein sequence/fold has resulted in the class C radical SAM methyltransferases. Several pathogenic organisms have further adapted this fold to catalyze the anaerobic degradation of heme. In this review, we summarize what is known about the mechanism of anaerobic heme degradation and the evolutionary implications.
Keywords: radical SAM enzyme, methyltransferase, heme degradation, enteric pathogens
Introduction
Very few cofactors or coenzymes have as prestigious a role in the evolution of life on this planet as that of iron–sulfur (Fe–S) clusters and porphyrins. One notable exception would be the highly reactive adenosyl radical that is produced by a single-electron reduction of the coenzyme S-adenosylmethionine (SAM). Moreover, it is clear that SAM preceded the emergence of the porphyrin-derived and the more complicated adenosylcobalamin or coenzyme B12.1,2 Other cofactors that contain the adenosyl moiety include ATP, NAD(P), FAD, and coenzyme A. The connection to an early RNA world is unavoidable when the catalytic adenosine base in rRNA is also considered. In fact, phylogenic analysis and comparative biochemistry suggest that the reduction of ribose to deoxyribose was one of the outcomes of corrinoid synthesis, a pathway that stops just short of porphyrin formation.3 It was Eschenmoser that made the case for the non-enzymatic formation of simple corrinoids.4 When these observations are further considered in light of the energetically favorable reactions underlying the in vitro self-assembly potential of porphyrins, the ubiquitous nature of porphyrin cofactors, such as heme, becomes apparent. Finally, considering all of these factors and the availability/solubility of iron throughout Earth’s history, it is clear why Nature capitalized on the utility of heme, especially as atmospheric oxygen levels increased. In this review, we consider the mechanism of two class C radical SAM methyltransferases (RSMTs), ChuW and HutW, enzymes involved in the anaerobic degradation of heme.
RSMTs in Heme Biosynthesis and Degradation
The evolution of mammals, humans in particular, has provided another ecological niche for microbes, specifically organisms found in the gut microbiome. It is from within this competitive environment that enteric pathogens have evolved to take advantage of these essential cofactors/coenzymes (Fe–S clusters and SAM) to cannibalize heme, the largest source of iron in the human diet.5 The degradation of heme under aerobic conditions has been well studied;6 however, only recently has a mechanism for the anaerobic catabolism of heme been identified. The enzymes ChuW and ChuY have been characterized in at least two pathogenic strains of Escherichia coli.7−9 The available evidence indicates that ChuW is a radical SAM methyltransferase responsible for opening the porphyrin ring of heme under anaerobic conditions,7,10 whereas ChuY is an NADPH-dependent reductase that reduces the tetrapyrrole product of ChuW.8 Interestingly, Vibrio cholera, another enteric pathogen that colonizes the tight mucosal layer of the intestine, can also utilize heme as an iron source. Similarly, V. cholera also expresses an enzyme, HutW (39% sequence identity with ChuW), that is capable of opening the porphyrin ring under anaerobic conditions.11 However, in contrast, V. cholera does not contain a ChuY homologue.
A careful phylogenetic analysis, based on the primary structure of E. coli CpdH (formerly HemN) and 828 class C RSMTs, by Wilkens et al. allowed for the discrimination of enzymes involved in menaquinone biosynthesis as well as a number of other functionalities.12 On a phylogenetic basis, YtkT and Jaw5 are more closely related to CpdH (Figure 1) and are on a distinct branch with ChuW and HutW.12 YtkT and Jaw5 have critical roles in the biosynthesis of the natural products yatakemycin and jawsamycin, respectively. Similar to ChuW/HutW, the mechanism of YtkT/Jaw5 involves the methylation of a sp2-hybridized carbon atom via the generation of a methylene radical. It should be distinguished that YtkT/Jaw5 are responsible for cyclopropane ring formation, and the methylated intermediates that have been identified are due to the capture of nonproductive side reactions.13−15 Regardless, based on the crystal structure of CpdH and numerous experimental observations, insight into the evolution of function from anaerobic heme biosynthesis to anaerobic heme degradation can be gained.
Figure 1.
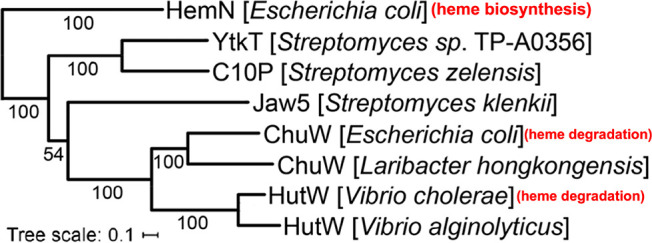
Phylogenetic analysis of eight CpdH-like class C RSMTs, with E. coli CpdH (formerly “HemN”) included as the outlier. Adapted with permission from ref (12). Copyright 2021 Society for Applied Microbiology.
Mechanism of ChuW and HutW
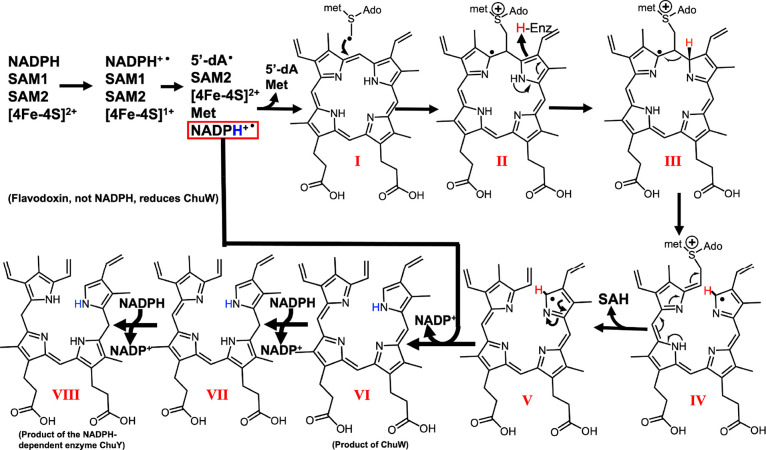
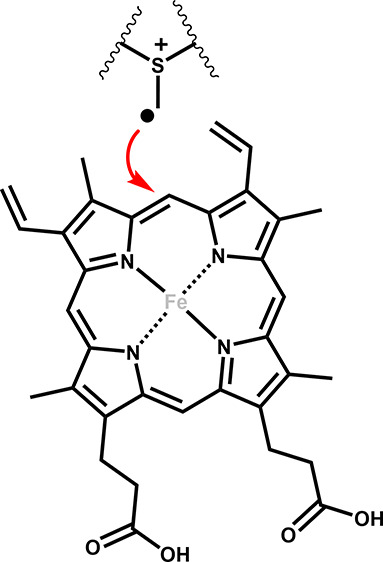
Both ChuW and HutW initiate catalysis through reductive cleavage of SAM, yielding the 5′-deoxyadenosyl radical (5′-dA•). The 5′-dA• rapidly abstracts a hydrogen atom from a second SAM molecule to yield a methylene radical. Interestingly, observation of the methylene radical attack was first observed in YtkT and NosN.16 The methylene radical represents a point of divergence in the evolution of the mechanism. Specifically, for CpdH, the methylene radical functions as part of a “hydrogen relay” and simply serves to abstract a hydrogen atom from the substrate, coproporphyrinogen III.17 Ji et al. demonstrated that this methylene radical could add to the porphyrin substrate through the identification of a SAM–porphyrin adduct,17 an important observation in light of the function of class C RSMTs. In the case of the latter, the methylene radical is added to a double bond, resulting in a methyl transfer reaction and, in many cases, further radical-catalyzed chemical rearrangements.18 For both ChuW and HutW, this includes the β-scission of a carbon–carbon bond, resulting in opening of the porphyrin ring along with the liberation of the iron atom.10,11
As Figure 2 shows, the iron atom does not participate in the reaction, and in fact, several metal-free porphyrins have been shown to be acceptable substrates for ChuW as well as HutW.10,11 It is also important to point out that the electron source as well as the tetrapyrrole product produced by ChuW is different from what has been observed for HutW. ChuW utilizes the E. coli flavodoxin (EcFldA)/ferredoxin (flavodoxin):NADP+ oxidoreductase (EcFpr)/NADPH system to reduce the [4Fe–4S] cluster, and the resulting tetrapyrrole still has a high degree of aromaticity (Figure 2, molecule VI).11 Interestingly, characterization of HutW revealed that NADPH could be utilized directly as the electron source.11
Figure 2.
Mechanism proposed for the class C RSMT HutW incorporating the observation that NADPH can be used as a direct source of electrons. The electron donor for ChuW is reduced flavodoxin, and compound VI is the proposed product of the ChuW reaction. In fact, in the organism E. coli O157:H7, the reduction of molecule VI has been shown to require the NADPH-dependent enzyme ChuY.8 Adapted with permission from ref (19). Copyright 2022 Elsevier.
The observation that NADPH can be used as an electron donor for HutW when SAM is utilized to initiate the reaction is significant. Moreover, this is surprising because the [4Fe–4S]2+ cluster will accept a single electron while NADPH transfers a hydride. When considering this, it is important to remember that all radical SAM enzymes must quench a radical intermediate at some step in the mechanism. This is consistent with the mechanism proposed in Figure 2 and could be facilitated by the porphyrin substrate and/or the tetrapyrrole itself. In fact, direct electron transfer from NADPH to heme has been observed for P450nor.20,21 Specifically, heme facilitates a hydride transfer reaction in this system. The proposal that HutW utilizes a similar mechanism is consistent with the requirement of two electrons and a proton in order to get from heme to molecule VI (Figure 2).7,11 How the peptide environment of all radical SAM enzymes controls these reactive intermediates remains a universally important question.
Structure Predictions
The radical SAM domain of all class C RSMTs is predicted to be highly conserved, as has been found to be the case for the majority of radical SAM enzymes,22,23 and consists of a full or partial, triose-phosphate isomerase mutase (TIM)-barrel protein fold. In addition to the radical SAM domain, class C RSMTs contain a “CpdH-like” domain.24 There is considerably more sequence diversity in the latter, and it stands to reason that subtle differences in the structural architecture of this domain account for the substrate specificity as well as the deviation in function of the second molecule of SAM. Specifically, the role of the second SAM molecule has evolved from functioning as a hydrogen relay, in the case of CpdH, to formation of a methylene radical, in the case of class C RSMTs. Furthermore, a comparison of the mechanism of CpdH and ChuW/HutW is particularly useful because the substrate, in both cases, is a tetrapyrrole macrocycle.
While no structural information is available for any class C RSMT, including ChuW and HutW, an interesting development has arisen in computational space through the application of “deep learning” to protein structure prediction.25 Application of this software provides some new insight into ChuW and HutW structure. Consistent with current knowledge, Alphafold2 predicts a similar radical SAM domain containing a partial TIM barrel fold for ChuW (Figure 3). In contrast, significant differences in structure are found in the “CpdH-like” domain.
Figure 3.
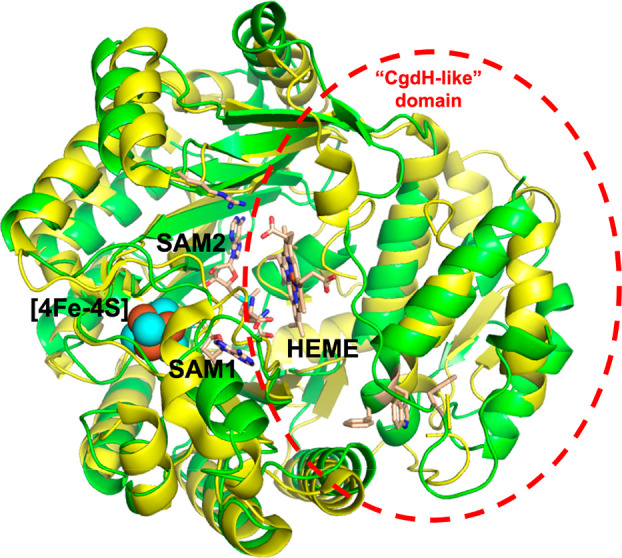
Cartoon representation showing a structural overlay of the CpdH (yellow) model (PDB ID 1OLT) with a model for ChuW (green) produced by Alphafold2. The [4Fe–4S] cluster (spheres), SAM1, and SAM2 (stick representation) of the CpdH model are also shown. Based on the Alphafold2 and binding modes of the [4Fe–4S] cluster, SAM1, and SAM2, the heme binding mode was predicted using Autodock.
The models produced by Alphafold2 and PHYRE2 for ChuW both accommodate the [4Fe–4S] cluster as well as the two SAM binding modes observed for CpdH. Manually placing the [4Fe–4S] cluster and both SAM molecules into the Alphafold2 model, predicted for ChuW, serves as a reasonable starting point to dock heme and provide clues to the mechanism (Figure 3). Interestingly, the predicted models (PHYRE2 versus Alphafold2) for ChuW are not identical and alignment of these models results in a root-mean-square deviation (rmsd) for the backbone carbon atoms of 2.9 Å. Regardless, alignment of the Alphafold2 model with the X-ray model for CpdH (Figure 3), results in a RMSD of 2.1 Å for the backbone carbon atoms. Only the Alphafold2 model produced a reasonable binding pocket for heme. Interestingly, application of Autodock VINA26 placed the bridging α-meso-carbon atom of heme within 4 Å of the SAM2 methyl group (Figure 3). Several regions containing residues conserved in ChuW and HutW are also within a few angstroms of the porphyrin ring. For example, R311 (R342 in HutW) is predicted to be capable of forming a salt bridge with the heme propionate side chain (Figure 4). There is a group of conserved hydrophobic residues from both the N and C termini that cluster together and appear to form a “tether” that may play an important role in stabilizing the overall TIM barrel fold of the heme-degrading enzymes (Figure 4).
Figure 4.
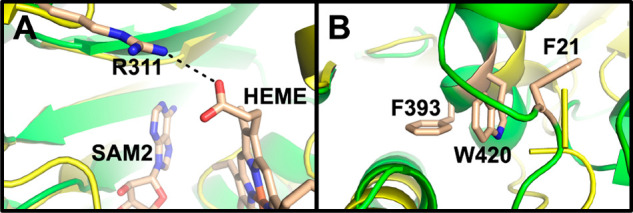
Close up views of the model presented in Figure 3 showing the relative locations of heme, SAM2, the conserved amino acids R311 in the active site (A) as well as conserved amino acids in the “hydrophobic tether” (B). Numbering is based on ChuW.
Similar to the observations for ChuW discussed above, models produced by PHYRE2 as well as Alphafold2 for HutW can be aligned with each other or the CpdH structure and result in RMSDs of 2.9 and 3.0 Å2, respectively. However, there is a notable problem with the HutW model generated by Alphafold2. Specifically, the loop region containing H31 (a potential heme ligand) and the glycine-rich NAD(P) binding motif (328GCGAGGNMGG337) is modeled within the active site cleft.
Genetic Organization
In addition to the functional differences reported for the enzymes ChuW and HutW, notable differences in the heme uptake and degradation genes are observed. Specifically, both of the enteric pathogens, E. coli O157:H7 and V. cholerae, express the “W” genes from a three gene operon that is associated with another larger operon involved in heme uptake and degradation.27,28 Both operons are controlled by the ferric uptake regulator (fur), and the larger operon contains genes for heme uptake and transfer into the cytoplasm. A major difference in the large operon is that V. cholerae has an additional gene that is homologous to the hemophore receptor HasR.29 Another notable difference is that there is no ChuS homologue in V. cholerae. ChuS (PhuS in P. aeruginosa) has been shown to function in heme storage/transport as well as aerobic heme degradation.10,30,31 It should be emphasized that, compared with the enterohemorrhagic E. coli, V. cholerae has additional mechanisms for obtaining iron and seems more adept at incorporating external heme into its own heme proteins.27,32−34
In regard to the smaller three gene operon, the “W” and “X” genes are the first two genes in both organisms. However, the third gene in V. cholerae is the hutZ gene that substitutes for the chuY gene found in E. coli. Moreover, the protein products, HutZ and ChuY, have been shown to have distinctly different structures and functions.8,28,35 Specifically, HutZ has been shown to be a peroxide-dependent heme oxygenase and not a reductase like ChuY.35 In fact, ChuY has been shown to catalyze the NADPH-dependent reduction of the tetrapyrrole product of ChuW. The phenotype reported for the ChuY deletion strain suggests that accumulation of the tetrapyrrole product, from the ChuW reaction, is toxic.9 This observation is consistent with ChuY functioning as a tetrapyrrole reductase, similar to biliverdin reductase. As described in the proceeding section, HutW has been shown to utilize NADPH as an electron source when SAM is utilized to initiate turnover.11 HutW also produces a tetrapyrrole that is in a more reduced state (Figure 2, molecule VII) when compared to the product of ChuW (Figure 2, molecule VI). In contrast, ChuW relies on the E. coli flavodoxin (EcFldA)/ferredoxin (flavodoxin):NADP+ oxidoreductase (EcFpr)/NADPH system. Taken together, it makes sense that HutW can utilize NADPH to catalyze the opening of the porphyrin ring as well as the reduction of the tetrapyrrole product. This suggests a distinct evolution of function and perhaps additional structural features, including a binding site for NADPH.
Conclusions
Efforts to obtain structural information are ongoing, but the models and observations discussed herein provide testable hypotheses for future work. The investigation is warranted given the available evidence, suggesting that the mechanism of methylene radical generation is common to all class C radical SAM enzymes. As has been reviewed by others,12 understanding the mechanism of these enzymes has tremendous potential for metabolic engineering as well as contributing to the improvement of human health and disease outcomes.
Acknowledgments
The authors would like to thank Dr. Amy Medlock and Dr. Harry Dailey for their critical reviews during preparation of this manuscript.
Author Contributions
† L.G.M. and M.B. contributed equally to this work.
Funding from the National Institute of General Medical Sciences (Grant No. R01GM124203) to W.N.L. is gratefully acknowledged.
The authors declare no competing financial interest.
References
- Frey P. A. Lysine 2,3-aminomutase: is adenosylmethionine a poor man’s adenosylcobalamin?. FASEB J. 1993, 7, 662–670. 10.1096/fasebj.7.8.8500691. [DOI] [PubMed] [Google Scholar]
- Wang S. C.; Frey P. A. S-adenosylmethionine as an oxidant: the radical SAM superfamily. Trends Biochem. Sci. 2007, 32, 101–110. 10.1016/j.tibs.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Lazcano A. Planetary change and biochemical adaptation: molecular evolution of corrinoid and heme biosyntheses. Hematology 2012, 17 (Suppl 1), S7–10. 10.1179/102453312X13336169155015. [DOI] [PubMed] [Google Scholar]
- Eschenmoser A. Vitamin-B12 - Experiments Concerning the Origin of Its Molecular-Structure. Angew. Chem., Int. Ed. Engl. 1988, 27, 5–39. 10.1002/anie.198800051. [DOI] [Google Scholar]
- Carpenter C. E.; Mahoney A. W. Contributions of heme and nonheme iron to human nutrition. Crit. Rev. Food Sci. Nutr. 1992, 31, 333–367. 10.1080/10408399209527576. [DOI] [PubMed] [Google Scholar]
- Wilks A.; Ikeda-Saito M. Heme utilization by pathogenic bacteria: not all pathways lead to biliverdin. Acc. Chem. Res. 2014, 47, 2291–2298. 10.1021/ar500028n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMattina J. W.; Nix D. B.; Lanzilotta W. N. Radical new paradigm for heme degradation in Escherichia coli O157:H7. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 12138–12143. 10.1073/pnas.1603209113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMattina J. W.; et al. Anaerobic Heme Degradation: ChuY Is an Anaerobilin Reductase That Exhibits Kinetic Cooperativity. Biochemistry 2017, 56, 845–855. 10.1021/acs.biochem.6b01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.; et al. Structural and functional study of ChuY from Escherichia coli strain CFT073. Biochem. Biophys. Res. Commun. 2017, 482, 1176–1182. 10.1016/j.bbrc.2016.12.008. [DOI] [PubMed] [Google Scholar]
- Mathew L. G.; Beattie N. R.; Pritchett C.; Lanzilotta W. N. New Insight into the Mechanism of Anaerobic Heme Degradation. Biochemistry 2019, 58, 4641–4654. 10.1021/acs.biochem.9b00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimberry M.; Toma M. A.; Hines K. M.; Lanzilotta W. N. HutW from Vibrio cholerae Is an Anaerobic Heme-Degrading Enzyme with Unique Functional Properties. Biochemistry 2021, 60, 699–710. 10.1021/acs.biochem.0c00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkens D.; Meusinger R.; Hein S.; Simon J. Sequence analysis and specificity of distinct types of menaquinone methyltransferases indicate the widespread potential of methylmenaquinone production in bacteria and archaea. Environ. Microbiol. 2021, 23, 1407–1421. 10.1111/1462-2920.15344. [DOI] [PubMed] [Google Scholar]
- Huang W.; et al. Characterization of yatakemycin gene cluster revealing a radical S-adenosylmethionine dependent methyltransferase and highlighting spirocyclopropane biosynthesis. J. Am. Chem. Soc. 2012, 134, 8831–8840. 10.1021/ja211098r. [DOI] [PubMed] [Google Scholar]
- Ding W.; et al. The Catalytic Mechanism of the Class C Radical S-Adenosylmethionine Methyltransferase NosN. Angew. Chem., Int. Ed. 2017, 56, 3857–3861. 10.1002/anie.201609948. [DOI] [PubMed] [Google Scholar]
- LaMattina J. W.; et al. NosN, a Radical S-Adenosylmethionine Methylase, Catalyzes Both C1 Transfer and Formation of the Ester Linkage of the Side-Ring System during the Biosynthesis of Nosiheptide. J. Am. Chem. Soc. 2017, 139, 17438–17445. 10.1021/jacs.7b08492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.; LaMattina J. W.; Marshall R.; Booker S. J. Capturing Intermediates in the Reaction Catalyzed by NosN, a Class C Radical S-Adenosylmethionine Methylase Involved in the Biosynthesis of the Nosiheptide Side-Ring System. J. Am. Chem. Soc. 2019, 141, 5788–5797. 10.1021/jacs.8b13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X.; et al. Revisiting the Mechanism of the Anaerobic Coproporphyrinogen III Oxidase HemN. Angew. Chem., Int. Ed. 2019, 58, 6235–6238. 10.1002/anie.201814708. [DOI] [PubMed] [Google Scholar]
- Jin W. B.; Wu S.; Xu Y. F.; Yuan H.; Tang G. L. Recent advances in HemN-like radical S-adenosyl-l-methionine enzyme-catalyzed reactions. Nat. Prod. Rep. 2020, 37, 17–28. 10.1039/C9NP00032A. [DOI] [PubMed] [Google Scholar]
- Brimberry M. A.; Mathew L.; Lanzilotta W. Making and breaking carbon-carbon bonds in class C radical SAM methyltransferases. J. Inorg. Biochem. 2022, 226, 111636. 10.1016/j.jinorgbio.2021.111636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima R.; et al. Structural evidence for direct hydride transfer from NADH to cytochrome P450nor. J. Mol. Biol. 2004, 342, 207–217. 10.1016/j.jmb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Kramos B.; Menyhard D. K.; Olah J. Direct hydride shift mechanism and stereoselectivity of P450nor confirmed by QM/MM calculations. J. Phys. Chem. B 2012, 116, 872–885. 10.1021/jp2080918. [DOI] [PubMed] [Google Scholar]
- Nicolet Y. Structure–function relationships of radical SAM enzymes. Nature Catalysis 2020, 3, 337–350. 10.1038/s41929-020-0448-7. [DOI] [Google Scholar]
- Dowling D. P.; Vey J. L.; Croft A. K.; Drennan C. L. Structural diversity in the AdoMet radical enzyme superfamily. Biochim. Biophys. Acta, Proteins Proteomics 2012, 1824, 1178–1195. 10.1016/j.bbapap.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerle M. R.; Schwalm E. L.; Booker S. J. Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation. J. Biol. Chem. 2015, 290, 3995–4002. 10.1074/jbc.R114.607044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A. G.; Payne S. M. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 1997, 23, 825–833. 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- Wyckoff E. E.; Schmitt M.; Wilks A.; Payne S. M. HutZ is required for efficient heme utilization in Vibrio cholerae. J. Bacteriol. 2004, 186, 4142–4151. 10.1128/JB.186.13.4142-4151.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey A. R.; Payne S. M. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 2001, 42, 835–849. 10.1046/j.1365-2958.2001.02683.x. [DOI] [PubMed] [Google Scholar]
- Suits M. D.; Jaffer N.; Jia Z. Structure of the Escherichia coli O157:H7 heme oxygenase ChuS in complex with heme and enzymatic inactivation by mutation of the heme coordinating residue His-193. J. Biol. Chem. 2006, 281, 36776–36782. 10.1074/jbc.M607684200. [DOI] [PubMed] [Google Scholar]
- Kaur A. P.; Lansky I. B.; Wilks A. The role of the cytoplasmic heme-binding protein (PhuS) of Pseudomonas aeruginosa in intracellular heme trafficking and iron homeostasis. J. Biol. Chem. 2009, 284, 56–66. 10.1074/jbc.M806068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley S. M.; et al. Comparison of the heme iron utilization systems of pathogenic Vibrios. J. Bacteriol. 1999, 181, 3594–3598. 10.1128/JB.181.11.3594-3598.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhino D. A.; Wyckoff E. E.; Henderson D. P.; Wrona T. J.; Payne S. M. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 1998, 29, 1493–1507. 10.1046/j.1365-2958.1998.01034.x. [DOI] [PubMed] [Google Scholar]
- Peng E. D.; Wyckoff E. E.; Mey A. R.; Fisher C. R.; Payne S. M. Nonredundant Roles of Iron Acquisition Systems in Vibrio cholerae. Infect. Immun. 2016, 84, 511–523. 10.1128/IAI.01301-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida T.; Sekine Y.; Matsui T.; Ikeda-Saito M.; Ishimori K. A heme degradation enzyme, HutZ, from Vibrio cholerae. Chem. Commun. (Cambridge, U. K.) 2012, 48, 6741–6743. 10.1039/c2cc31147j. [DOI] [PubMed] [Google Scholar]