Zhang et al. identify the lysosomal Cl−/H+ antiporter CLH-6/ClC-7 as an important factor for protecting lysosome integrity. They reveal that CLH-6 maintains the luminal chloride levels required for cathepsin activity, thus facilitating substrate digestion to preserve lysosomal membrane stability.
Abstract
Lysosomal integrity is vital for cell homeostasis, but the underlying mechanisms are poorly understood. Here, we identify CLH-6, the C. elegans ortholog of the lysosomal Cl−/H+ antiporter ClC-7, as an important factor for protecting lysosomal integrity. Loss of CLH-6 affects lysosomal degradation, causing cargo accumulation and membrane rupture. Reducing cargo delivery or increasing CPL-1/cathepsin L or CPR-2/cathepsin B expression suppresses these lysosomal defects. Inactivation of CPL-1 or CPR-2, like CLH-6 inactivation, affects cargo digestion and causes lysosomal membrane rupture. Thus, loss of CLH-6 impairs cargo degradation, leading to membrane damage of lysosomes. In clh-6(lf) mutants, lysosomes are acidified as in wild type but contain lower chloride levels, and cathepsin B and L activities are significantly reduced. Cl− binds to CPL-1 and CPR-2 in vitro, and Cl− supplementation increases lysosomal cathepsin B and L activities. Altogether, these findings suggest that CLH-6 maintains the luminal chloride levels required for cathepsin activity, thus facilitating substrate digestion to protect lysosomal membrane integrity.
Introduction
Lysosomes degrade extra- and intra-cellular macromolecules, recycle catabolites, and serve as signaling hubs to maintain cell and tissue homeostasis. Lysosomes are also involved in secretion, plasma membrane repair, immune responses, and many other physiological or pathological processes (Ballabio and Bonifacino, 2020). Dysfunction of lysosomes is associated with metabolic disorders, neurodegenerative diseases, and cancer (Appelqvist et al., 2013; Bajaj et al., 2019).
Lysosomes are acidic membrane-bound organelles that contain ∼60 soluble acid hydrolases and hundreds of integral and peripheral membrane proteins. The 7–10 nm limiting membrane of lysosomes distinguishes the acidic lumen from the cytosol to protect other cellular constituents from unwanted degradation (Saftig et al., 2010). On the other hand, lysosomes are susceptible to membrane damage by various stressors such as endocytosed and phagocytosed membrane-damaging materials, invading pathogens, reactive oxygen species, and changes in lysosomal lipid composition (Boyle and Randow, 2013; Cantuti-Castelvetri et al., 2018; Gómez-Sintes et al., 2016; Papadopoulos et al., 2020; Papadopoulos and Meyer, 2017). Partial lysosomal membrane permeabilization induces regulated cell death via cathepsin release, while massive rupture of lysosomes causes hydrolysis of cytosolic contents and generalized cytoplasmic acidification, ultimately leading to death of the cell (Serrano-Puebla and Boya, 2016). The integrity of lysosomes is vital for cell homeostasis and viability, but the mechanisms by which lysosomal membrane stability is maintained are not well understood. The lysosomal membrane contains abundant and highly glycosylated membrane proteins such as LAMPs (lysosomal-associated membrane proteins) and LIMPs (lysosomal integral membrane proteins). They may form a continuous carbohydrate layer called the glycocalyx lining the luminal leaflet to prevent membrane damage by the hydrolytic enzymes (Fehrenbacher et al., 2008; Fukuda, 1991; Neiss, 1984; Rudnik and Damme, 2021). We found previously that SCAV-3/LIMP-2 plays an essential role in preserving lysosomal membrane integrity, and the insulin/IGF-1 pathway modulates lysosome integrity to regulate longevity (Li et al., 2016). In addition to glycosylated membrane proteins, Hsp70 is found to stabilize lysosomes and thus promote survival of transformed cells (Nylandsted et al., 2004). The protection effect is achieved by binding of Hsp70 to the anionic phospholipid bis(monoacylglycero) phosphate (BMP), which in turn facilitates activity of acid sphingomyelinase (Kirkegaard et al., 2010). How increased acid sphingomyelinase activity promotes lysosomal stability is unclear. Stabilization of lysosomal membranes ensures cargo degradation, but it remains to be determined whether and how substrate digestion affects lysosomal membrane stability. Cathepsins are the most abundant hydrolases in lysosomes. They are released upon lysosomal membrane permeabilization to trigger cell death, but their role in lysosomal integrity has not been characterized.
Lysosomal hydrolases require an acidic lumen for optimal function. The vacuolar-type ATPase (V-ATPase) pumps protons across the lysosomal membrane coupled with ATP hydrolysis to establish and maintain the acidity in the lumen (Forgac, 2007; Ohkuma et al., 1982). The vectorial translocation of protons across the membrane generates an inside-positive transmembrane potential that, if not compensated for, would rapidly shut down the activity of V-ATPase and hamper further entry of protons. Therefore, parallel transport of counter ions—influx of anions and/or efflux of cations—is needed to dissipate the mounting voltage and maintain the pH gradient (DiCiccio and Steinberg, 2011; Mindell, 2012; Stauber and Jentsch, 2013). As the most abundant anion in the lysosomal lumen, chloride is thought to act as a major neutralizing counter ion for efficient operation of V-ATPase (Deriy et al., 2009; Di et al., 2006; Graves et al., 2008). Nevertheless, replacement of cytosolic Cl− with impermeant anions has no effect on proton pumping in macrophages, while a monovalent cation flux is sufficient to sustain normal lysosome acidification in the absence of Cl− (Steinberg et al., 2010). The CLC family protein ClC-7 transports chloride into the lysosomal lumen. It acts as a Cl−/H+ antiporter to mediate coupled movement of Cl− and H+ in opposite directions (Brandt and Jentsch, 1995; Graves et al., 2008; Leisle et al., 2011). ClC-7 is ubiquitously expressed and enriched in the ruffled border of bone-resorbing osteoclasts where it acts in concert with the V-ATPase to acidify the resorption lacuna (Kornak et al., 2001). ClC-7 forms a complex with Ostm1, an ancillary β-subunit that stabilizes ClC-7 protein and modulates its transport activity (Lange et al., 2006; Leisle et al., 2011; Schrecker et al., 2020; Zhang et al., 2020). Consistent with this, loss of CLCN7 or OSTM1 causes severe osteopetrosis in mice and human (Chalhoub et al., 2003; Kornak et al., 2001). Moreover, CLCN7−/− and gl mice, which bear a mutation in OSTM1, display severe lysosomal storage defects and widespread neurodegeneration (Kasper et al., 2005; Kornak et al., 2001; Lange et al., 2006; Pressey et al., 2010). Of note, the lysosomal chloride level is lower in CLCN7-deficient fibroblasts, but the acidity of lysosomes is unaffected in CLCN7-deficient fibroblasts, neurons, and macrophages (Kasper et al., 2005; Steinberg et al., 2010; Wartosch et al., 2009; Weinert et al., 2010). In line with this, chloride reduction correlates with reduced degradation capacity of lysosomes in Caenorhabditis elegans coelomocytes and cultured mammalian cells independent of acidity regulation (Chakraborty et al., 2017). The mechanism by which ClC-7 and luminal Cl− regulates lysosomal function beyond pH regulation remains to be addressed.
In this study, we identify CLH-6, the C. elegans ortholog of ClC-7, as an important factor required for the protection of lysosomal integrity. Our data suggest that CLH-6 maintains the luminal chloride levels required for optimal cathepsin function and thus facilitates substrate digestion to protect lysosomal membrane stability.
Results
Loss of clh-6 affects cargo degradation and membrane integrity of lysosomes
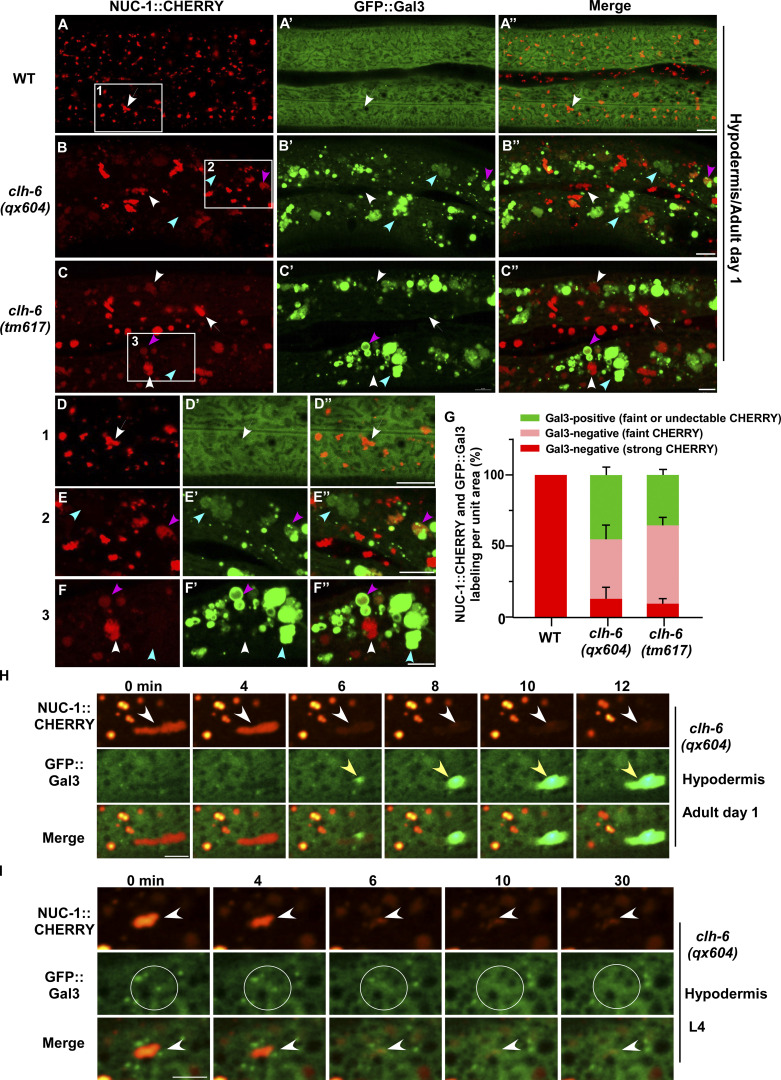
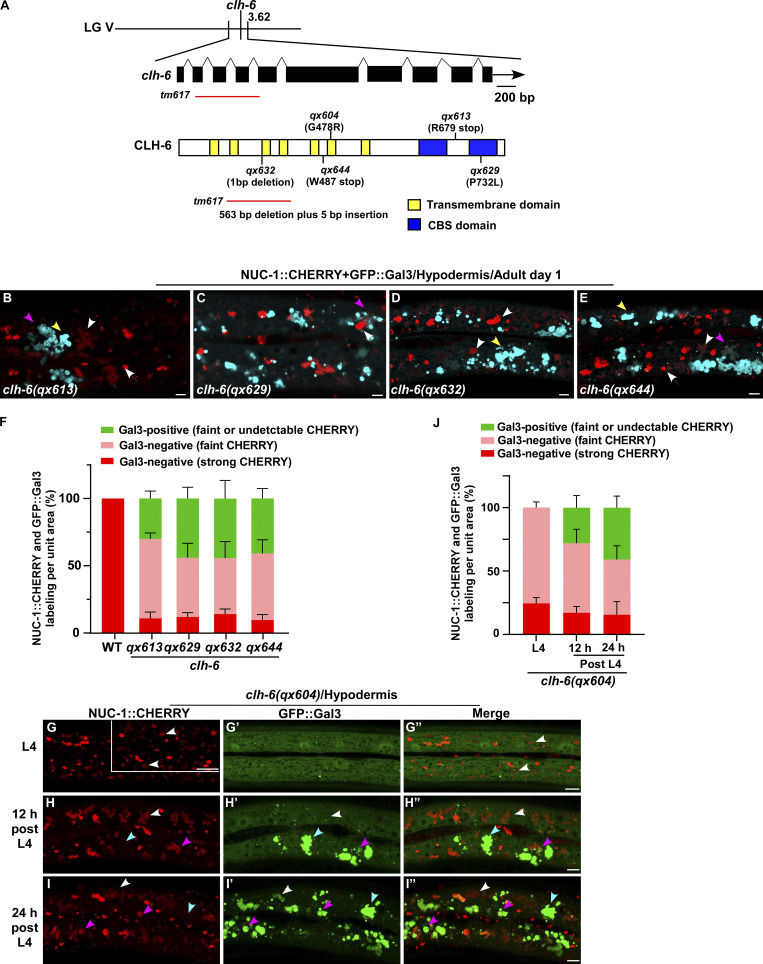
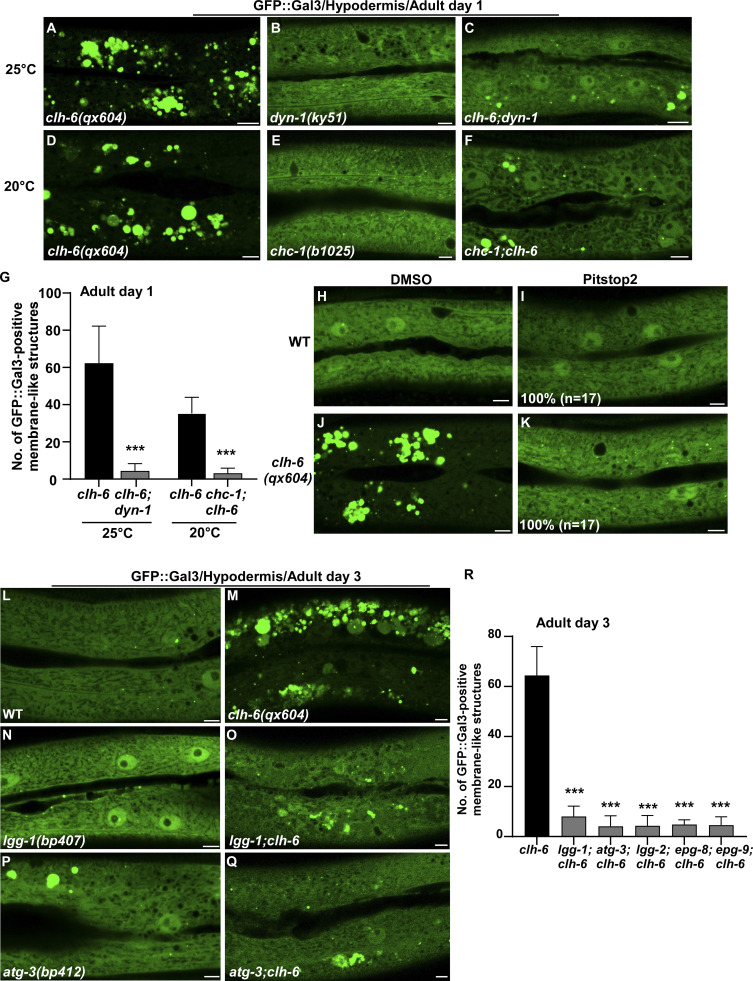
We co-expressed the lysosomal DNase II NUC-1 (NUC-1::CHERRY) and the endomembrane damage reporter Galectin-3 (GFP::Gal3), and performed genetic screens for mutants that affect lysosomal membrane integrity (Guo et al., 2010; Maejima et al., 2013). From the screen, we isolated five recessive mutations, which caused accumulation of high levels of Gal3-positive structures in the hypodermis (Fig. 1, A–B′′; and Fig. S1, A–E). All five mutations affect the gene clh-6, which encodes a C. elegans ortholog of the human chloride transporter ClC-7 (Fig. S1 A and Fig. S2 A). CLH-6 and ClC-7 both contain multiple transmembrane spans and two cystathionine beta-synthase (CBS) domains (Fig. S1 A and Fig. S2 A; Dutzler et al., 2002; Schrecker et al., 2020; Zhang et al., 2020). They exhibit 64% sequence similarity and 47% sequence identity (Fig. S2 A). The clh-6 mutant alleles carry nonsense or missense mutations at various places throughout the gene (Fig. S1 A). Moreover, we obtained tm617, a deletion mutant of clh-6. It carries a 563-bp deletion and 5-bp insertion that removes exons 2–4 and causes premature termination of the protein (Fig. S1 A). clh-6(tm617) worms accumulated high levels of Gal3-positive structures, as observed for the other clh-6 alleles (Fig. 1, C–C′′). We used qx604 and tm617 alleles in later experiments.
Figure 1.
clh-6 mutants accumulate damaged lysosomes. (A–G) Confocal fluorescence images of the hypodermis in wild-type (A–A′′), clh-6(qx604) (B–B′′), and clh-6(tm617) (C–C′′) adults co-expressing NUC-1::CHERRY and GFP::Gal3. The boxed region in A–C is magnified in D–F. Intact lysosomes contain strong NUC-1::CHERRY fluorescence and are not labeled by GFP::Gal3 (white arrows). Damaged lysosomes have either faint NUC-1::CHERRY fluorescence with GFP::Gal3 labeling (purple arrowheads) or without GFP::Gal3 labeling (white arrowheads), or strong GFP::Gal3 fluorescence with undetectable NUC-1::CHERRY (blue arrowheads). Quantification is shown in G. At least 10 animals were scored in each strain and data are shown as mean ± SD. (H and I) Time-lapse images of lysosomes in clh-6(qx604) expressing GFP::Gal3 and NUC-1::CHERRY. “0” min represents the time point before NUC-1::CHERRY release (white arrowheads) and initial appearance of GFP::Gal3 (yellow arrowheads). Circles indicate the region where NUC-1::CHERRY leakage but not GFP::Gal3 enrichment was observed. Scale bars: 5 µm.
Figure S1.
clh-6 mutants contain damaged lysosomes. (A) Cloning of clh-6. The clh-6 gene structure is shown with filled boxes representing exons and thin lines indicating introns. The arrow delineates the direction of transcription. A schematic diagram showing the organization of the CLH-6 protein is shown below the transcript. The mutation sites identified in all clh-6 alleles are indicated. Yellow boxes indicate transmembrane domains and blue boxes designate CBS domains. (B–J) Confocal fluorescence images of the hypodermis in the indicated strains co-expressing GFP::Gal3 and NUC-1::CHERRY. Intact lysosomes contain strong NUC-1::CHERRY fluorescence and are not labeled by GFP::Gal3 (white arrows). Damaged lysosomes have either faint NUC-1::CHERRY fluorescence with GFP::Gal3 labeling (purple arrowheads) or without GFP::Gal3 labeling (white arrowheads), or strong GFP::Gal3 fluorescence with undetectable NUC-1::CHERRY (yellow arrowheads). Quantification analyses are shown in F and J. At least 10 animals were scored in each strain and data are shown as mean ± SD. Scale bars: 5 µm.
Figure S2.
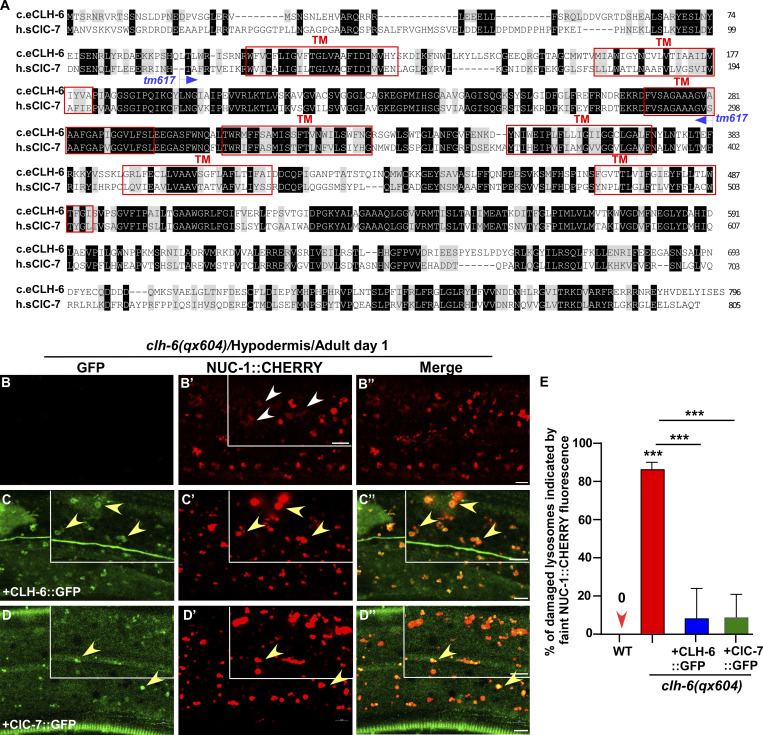
CLH-6 is homologous to human ClC-7. (A) Sequence alignment of C. elegans (c.e) CLH-6 and human (h.s) ClC-7. Identical residues are shaded in black and similar ones in gray. Red boxes indicate the transmembrane (TM) domains in CLH-6 predicted by uniProt. The region deleted in the tm617 allele is indicated. (B–D′′) Confocal fluorescence images of the hypodermis in clh-6(qx604) mutants carrying NUC-1::CHERRY without (B–B′′) or with expression of CLH-6::GFP (C–C′′) or ClC-7::GFP (D–D′′). Both CLH-6::GFP and ClC-7::GFP colocalize with NUC-1::CHERRY (yellow arrowheads). (E) The percentage of damaged lysosomes indicated by faint NUC-1::CHERRY fluorescence (white arrowheads) is quantified in E. Unpaired two-tailed Student’s t test was performed to compare mutant datasets with wild type or datasets that are linked by lines. At least 10 animals were scored in each strain and data are shown as mean ± SD. ***P < 0.0001. All other points had P > 0.5. Scale bars: 5 µm.
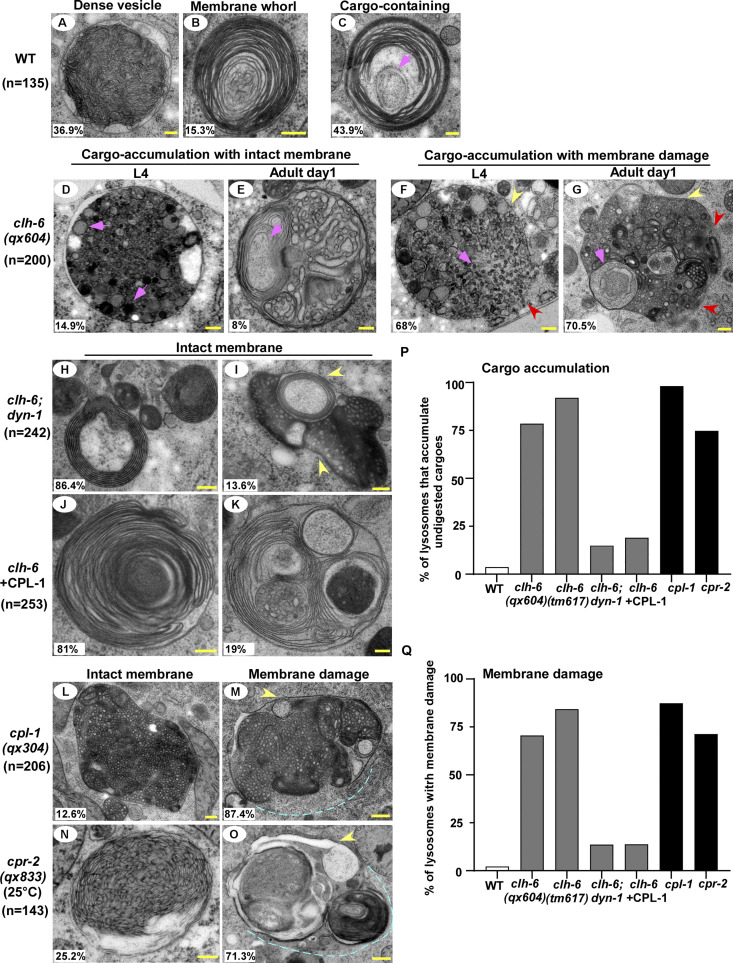
In wild-type worms, lysosomes contained bright NUC-1::CHERRY fluorescence, while GFP::Gal3 was diffuse in the cytosol (Fig. 1, A–A′′ and D–D′′). In clh-6 mutants, NUC-1::CHERRY fluorescence was greatly weakened, and GFP::Gal3 signals accumulated extensively as vesicular or irregular membrane-like structures (Fig. 1, B–C′′; and Fig. S1, B–E). The GFP::Gal3 speckles contained either faint or undetectable NUC-1::CHERRY, whereas lysosomes with bright NUC-1::CHERRY fluorescence lacked Gal3 (Fig. 1, B–C′′ and E–G; and Fig. S1, B–F). We performed time-lapse analyses in clh-6(qx604) worms and observed a big reduction of NUC-1::CHERRY fluorescence followed by appearance of GFP::Gal3, which gradually expanded and became enriched (Fig. 1 H). These data suggest that loss of clh-6 affects lysosome membrane integrity, causing leakage of the luminal hydrolase NUC-1 and gradual enrichment of cytosolic Gal3, probably through its binding with the luminal glyco-conjugates exposed upon lysosomal membrane damage. Of note, GFP::Gal3 did not appear on all lysosomes following NUC-1::CHERRY leakage. In some cases, as shown in Fig. 1 I, the lysosomal NUC-1::CHERRY signal was quickly reduced, and GFP::Gal3 did not appear or become enriched on the leaking lysosome. In line with this, >40% of leaking lysosomes indicated by faint NUC-1::CHERRY fluorescence were not associated with GFP::Gal3 signals in clh-6 adults (Fig. 1, B–C′′, F–F′′, and G [white arrowheads]; and Fig. S1, B–F). Moreover, weakened NUC-1::CHERRY fluorescence was observed in clh-6 mutants at the fourth larval stage (L4), but Gal3 accumulation was not readily seen until adult stages (Fig. S1, G–J). Galectin-3 is known to detect large but not small holes on the lysosomal membrane (Skowyra et al., 2018). These data suggest that clh-6 mutants contain lysosomes that are damaged to different extents and the lysosomal damage becomes more severe at the adult stages than in larvae. We performed transmission electron microscopy (TEM) analyses to further examine lysosomal damage in clh-6 mutants. In wild type, hypodermal lysosomes appeared as spherical membrane-enclosed vesicles with electron-dense contents, membrane whorls, or few cargoes (Fig. 2, A–C). In clh-6 mutants, about 80% of lysosomes in the hypodermis were filled with granular contents including intraluminal vesicles resembling those in multivesicular bodies, autophagosomes, and vesicles with dense or lucent contents (Fig. 2, D–G and P). In addition to accumulating undigested cargoes, membrane damage was observed in 68% and 71–84% of lysosomes in clh-6 mutants at L4 and adult day 1 stages, respectively (Fig. 2, F, G, and Q). These data indicate that loss of clh-6 affects lysosome function and integrity, causing cargo accumulation and membrane damage.
Figure 2.
Loss of clh-6 causes rupture of lysosome membranes. (A–O) Transmission electron micrographs of lysosomes in the hypodermis of wild type (A–C), clh-6(qx604) (D–G), clh-6;dyn-1 (H and I), clh-6 with CPL-1 overexpression (J and K), cpl-1(qx304) (L and M), and cpr-2(qx833) (N and O). Yellow arrowheads indicate lysosomal membranes, while red arrowheads and blue dashed lines designate areas that have no detectable membranes. Cargoes and granules are indicated by pink arrowheads. The percentage of lysosomes with the representative pattern is quantified and shown at the lower left corner in each panel. (P and Q) The percentage of lysosomes that accumulate undigested cargoes (P) or with membrane damage (Q) is quantified in the indicated stains. Scale bars: 200 nm.
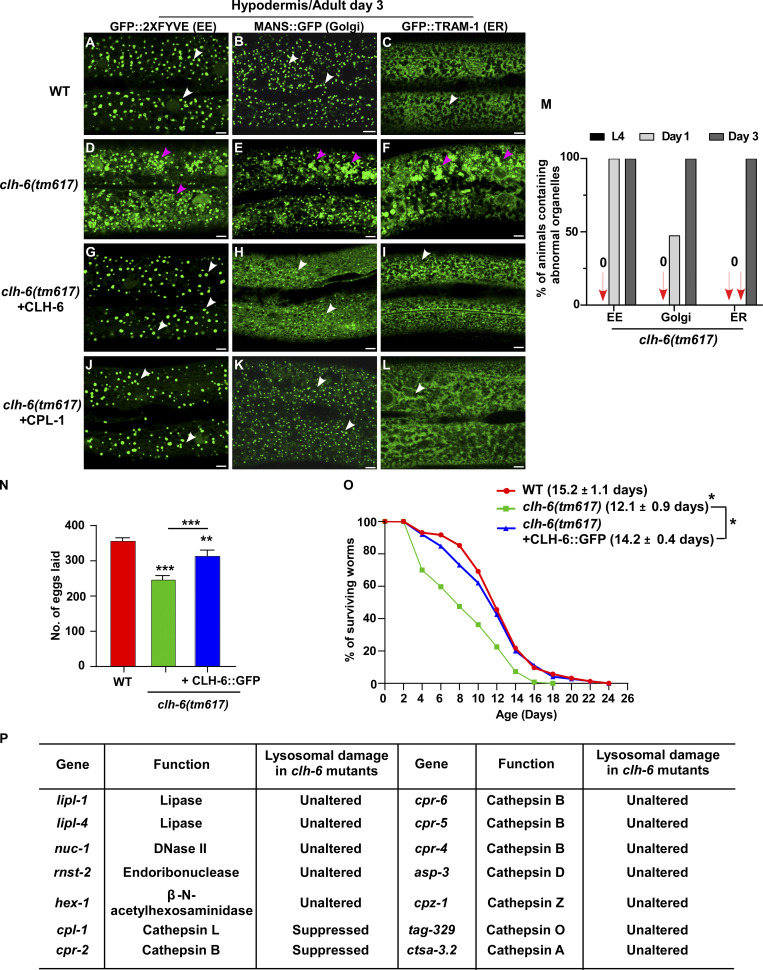
In addition to lysosomes, patterns of endosomes, Golgi, and ER in the hypodermis were also affected in clh-6 mutants. The punctate pattern of early endosomes and Golgi was altered, causing aggregation of the Golgi protein MANS::GFP and the endosomal marker GFP::2xFYVE (Fig. S3, A, B, D, and E). Moreover, the ER reporter GFP::TRAM-1 lost its network pattern and became aggregated in clh-6(tm617) worms (Fig. S3, C and F). The abnormal pattern of endosomes, Golgi, and ER in clh-6 mutants was observed only in the hypodermis at adult stages when lysosomal damage is severe, which suggests that lysosomal rupture and the subsequent leakage of hydrolases may lead to abnormalities of these organelles (Fig. S3 M). In line with this, expression of CLH-6 rescued the lysosomal integrity defects and restored the morphology of endosomes, Golgi, and ER in clh-6 mutants (Fig. S2, B–C′′ and E; and Fig. S3, G–I). clh-6 mutants produced significantly fewer progeny and had a shortened lifespan compared to wild type, which suggests that CLH-6 function is important for worm fertility and longevity (Fig. S3, N and O).
Figure S3.
Loss of clh-6 affects endosome, ER and Golgi patterns. (A–L) Confocal fluorescence images of the hypodermis in the indicated strains expressing different organellar markers. The punctate pattern of early endosomes (EE) and Golgi and the network pattern of ER (white arrows) are disrupted in clh-6(tm617) mutants (purple arrowheads) and restored by overexpression of CLH-6 and CPL-1. (M) The abnormal pattern of early endosomes, Golgi, and ER was quantified in clh-6(tm617) mutants at larval and adult stages. At least 30 animals were scored for each organellar marker in each stage. (N and O) Brood size and lifespan analyses were performed in the indicated strains. >15 and >100 worms were quantified in brood size and lifespan analyses, respectively. At least three independent experiments were performed, and data are shown as mean ± SD. Unpaired two-tailed Student’s t test was performed to compare mutant datasets with wild type or datasets that are linked by lines. *P < 0.05, **P < 0.001, ***P < 0.0001. Scale bars: 5 µm. (P) Lysosomal hydrolases that were tested by overexpression in clh-6(tm617) mutants. Lysosomal damage was examined by comparing accumulation of Gal3-positive structures in clh-6(tm617) mutants without and with the expression of each lysosomal hydrolase.
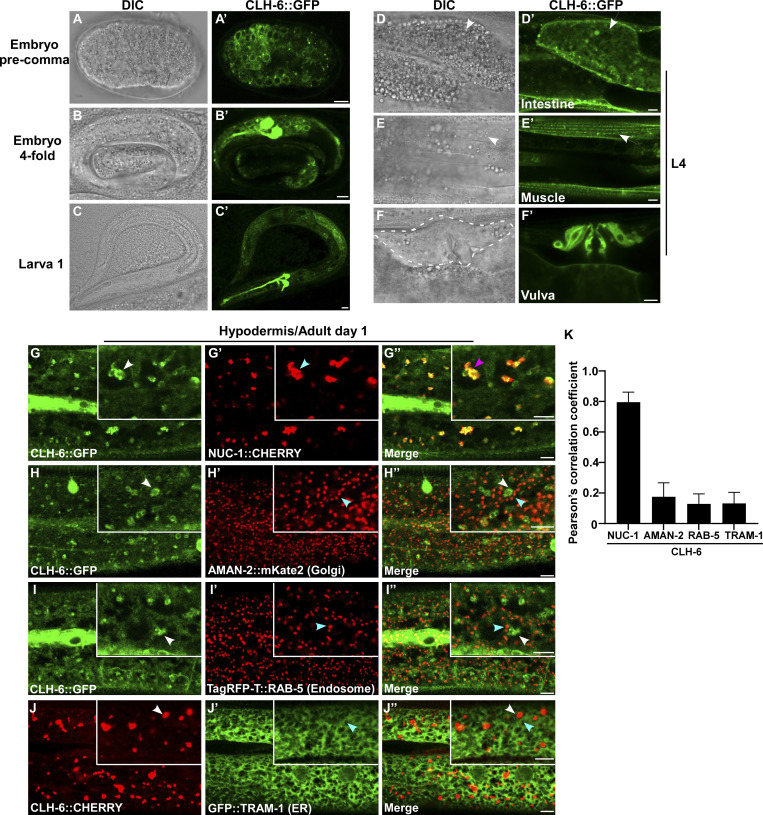
CLH-6 is widely expressed and localizes to lysosomes
We generated a CLH-6::GFP reporter driven by its own promoter, which efficiently rescued the lysosomal integrity defect in clh-6 mutants (Fig. S2, B–C′′ and E). CLH-6::GFP is expressed throughout embryonic, larval, and adult stages in various tissues including hypodermis, intestine, body wall muscle, and vulva region (Fig. 3, A–F′). CLH-6 co-localized with NUC-1::CHERRY, but not reporters of Golgi, ER, or early endosomes (Fig. 3, G–K). This suggests that CLH-6 localizes to lysosomes. We expressed human ClC-7 in worms and found that it localized to lysosomes and showed rescue activity to the same extent as CLH-6 (Fig. S2, D and E). This suggests that CLH-6 and ClC-7 play a conserved role in the maintenance of lysosome integrity.
Figure 3.
CLH-6 is widely expressed and localizes to lysosomes. (A–F′) DIC and confocal fluorescence images of wild-type worms expressing CLH-6::GFP at different stages (A–C′) and in different tissues at the L4 stage (D–F′). White arrows indicate intestine and body wall muscle. The vulva region is surrounded by the dashed line. (G–K) Confocal fluorescence images of the hypodermis in wild-type adults co-expressing CLH-6::GFP or CLH-6::CHERRY with different organellar markers including NUC-1::CHERRY (lysosome, G–G′′), AMAN-2::mKate2 (Golgi, H–H′′), TagRFP-T::RAB-5 (early endosome, I–I′′), GFP::TRAM-1 (ER, J-J′′). White and blue arrowheads indicate structures labeled by CLH-6 and organellar markers, respectively, and the purple arrowheads indicates co-localization of CLH-6 and NUC-1::CHERRY. Quantifications are shown in K. At least 10 animals were scored in each strain, and data are shown as mean ± SD. Scale bars: 5 µm.
Inhibiting cargo delivery suppresses lysosomal damage in clh-6 mutants
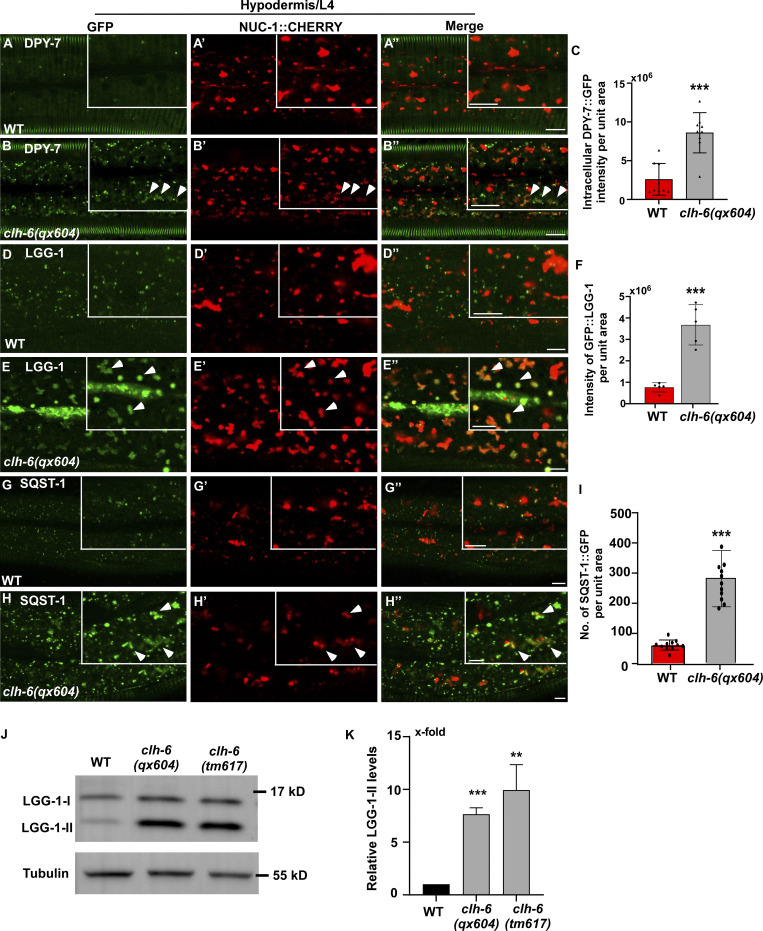
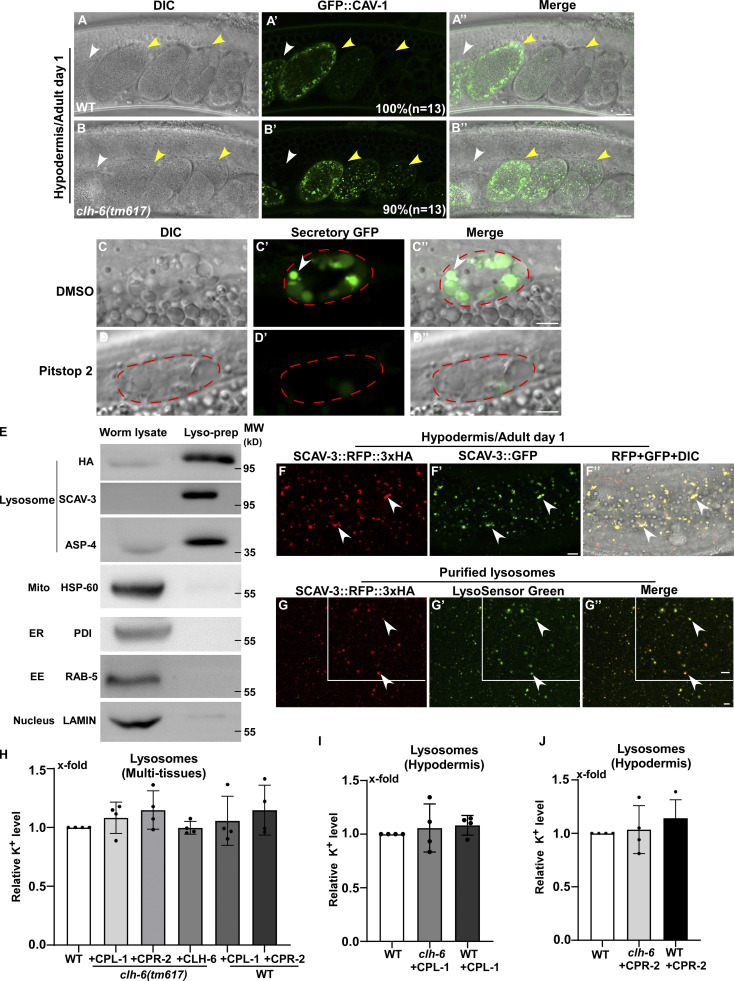
The TEM analyses revealed that loss of clh-6 causes accumulation of undigested cargoes in lysosomes. We further examined endocytic and autophagic cargo degradation in clh-6 mutants. DPY-7 collagen localizes to the annular furrows of the C. elegans cuticle (McMahon et al., 2003). It is turned over via endocytosis followed by degradation in lysosomes (Miao et al., 2020). In wild type, DPY-7::GFP appeared on the cuticle as circumferential bands with very few GFP puncta present in the cytosol of hypodermal cells (Fig. 4, A–A′′). In clh-6(qx604) mutants, DPY-7::GFP accumulated intracellularly at a significantly higher level than in wild type (Fig. 4, B and C). We also examined the cell surface protein CAV-1, which is delivered to lysosomes through the endocytic pathway and degraded shortly after fertilization. CAV-1 was degraded in wild-type but persisted in clh-6 mutant embryos (Fig. S4, A–B′′). LGG-1 and SQST-1, the C. elegans homologs of human LC3 and p62, respectively, associate with autophagic structures and/or substrates and are degraded through autophagy (Tian et al., 2010). Both LGG-1 and SQST-1 accumulated at significantly higher levels in clh-6 mutants than in wild type (Fig. 4, D–I). Moreover, clh-6 mutants accumulated LGG-1-II (the PE-conjugated form of LGG-1), consistent with defects in autophagic cargo degradation (Fig. 4, J and K). These data are consistent with the TEM analyses and suggest that endocytic and autophagic cargo degradation is defective in clh-6 mutants.
Figure 4.
Loss of clh-6 affects degradation of endocytic and autophagic cargoes. (A–I) Confocal fluorescence images of the hypodermis in wild-type (A–A′′, D–D′′, G–G′′) and clh-6(qx604) (B–B′′, E–E′′, H–H′′) adults expressing NUC-1::CHERRY and DPY-7::GFP (A–B′′), GFP::LGG-1 (D–E′′) or SQST-1::GFP (G–H′′). Arrowheads indicate DPY-7–, LGG-1–, and SQST-1–positive structures that contain faint NUC-1::CHERRY fluorescence. Quantification analyses are shown in C, F, and I. At least 15 animals were scored in each strain. (J and K) Western blot analysis of LGG-1-I and LGG-1-II (lipid-conjugated form) in wild type, clh-6(qx604), and clh-6(tm617). LGG-1 accumulation was quantified and normalized to onefold in wild type (K). At least three independent experiments were performed. In C, F, I, and K, data are shown as mean ± SD. Student’s two-tailed unpaired t test was performed to compare mutant datasets with wild type. **P < 0.001, ***P < 0.0001. Scale bars: 5 µm. Source data are available for this figure: SourceData F4.
Figure S4.
Loss of clh-6 affects endocytic cargo degradation. (A–B′′) DIC, confocal fluoresence, and merged images of wild-type (A, A', and A'') and clh-6(tm617) (B, B', and B'') adults expressing CAV-1::GFP. White arrowheads indicate the spermatheca; yellow arrowheads indicate fertilized and dividing embryos. The percentage of worms with the representative pattern is quantified and shown at the lower right corner in each panel. 13 animals (n) were scored in each strain. (C–D′′) DIC and confocal fluorescence images of a coelomocyte in wild-type adults expressing GFP secreted from body wall muscle cells (Secretory GFP). Worms were treated with DMSO (C–C′′) or Pitstop 2 (D–D′′). GFP is endocytosed into the coelomocyte (outlined by the dashed line) in DMSO- but not Pitstop 2–treated worms. (E) The purity of lysosomes obtained by LysoIP was examined by Western blot using antibodies that detect the lysosomal reporter SCAV-3::RFP::3xHA (anti-HA) and recognize proteins in lysosomes (anti-SCAV-3, anti-ASP-4), mitochondria (anti-HSP-60), ER (anti-PDI), endosomes (anti-RAB-5) or nuclei (anti-LAMIN). 1% of the whole worm lysate and 2% of the purified lysosomal prep were loaded. EE, early endosomes. (F–F′′) DIC and confocal fluorescence images of the hypodermis in wild type expressing the lysosomal markers SCAV-3::RFP::3xHA and SCAV-3::GFP. The RFP signal overlaps well with SCAV-3::GFP (white arrowheads). (G–G′′) The integrity of lysosomes purified by LysoIP was examined by LysoSensor green staining. SCAV-3::RFP::3xHA-positive lysosomes are well stained by LysoSensor green (white arrowheads). Scale bars in A–D′′ and F–G′′ represent 5 µm. (H–J) The potassium concentration in lysosomes purified from multi-tissues (H) or the hypodermis of C. elegans (I and J) was determined as described in the Materials and methods. Relative potassium levels are presented. At least four independent experiments were performed, and data are shown as mean ± SD. Unpaired two-tailed Student’s t test was performed to compare mutant datasets with wild type. All points had P > 0.5. Source data are available for this figure: SourceData FS4.
The failure in cargo degradation may lead to membrane damage of lysosomes in clh-6 mutants. We tested this hypothesis by inhibiting cargo delivery to lysosomes. dyn-1(ky51), a temperature-sensitive mutation of dyn-1, and chc-1(b1025) mutation affect the essential endocytosis regulators dynamin and clathrin, respectively. Both mutations caused significant reduction of Gal3-positivie structures in clh-6 mutants (Fig. 5, A–G). Moreover, the proportion of lysosomes that contained undigested cargoes and/or membrane damage was greatly reduced in clh-6;dyn-1 double mutants revealed by TEM (Fig. 2, H, I, P, and Q). In addition, we inhibited endocytosis using Pitstop 2, a potent inhibitor of clathrin-dependent and clathrin-independent endocytosis (Dutta et al., 2012). Pitstop 2 treatment blocked endocytosis in coelomocytes and suppressed appearance of Gal3-positive structures in clh-6 mutants (Fig. 5, H–K; and Fig. S4, C–D′′) Furthermore, autophagy-defective mutations greatly reduced the number of Gal3-positive structures in clh-6 mutants (Fig. 5, L–R). Altogether, these data suggest that inhibiting endocytic or autophagic cargo delivery suppresses lysosomal damage in clh-6 mutants, and they support the hypothesis that impaired cargo digestion leads to membrane damage of clh-6 lysosomes.
Figure 5.
Blocking endocytosis or autophagy suppresses lysosome integrity defects in clh-6 mutants. (A–R) Confocal fluorescence images of the hypodermis in the indicated strains expressing GFP::Gal3 without treatment (A–F and L–Q), or with DMSO treatment (H and J), or with Pitstop 2 treatment (I and K). The experiments using the temperature-sensitive dyn-1(ky51) allele were performed at 25°C, which is the non-permissive temperature of the mutant allele. The average number of GFP::Gal3-positive membrane-like structures in the indicated strains is quantified in G and R. Scale bars: 5 µm. In I and K, 17 animals (n) were scored in each strain. In G and R, data are shown as mean ± SD. Unpaired two-tailed Student’s t test was performed to compare double mutant datasets with clh-6(qx604) worms. 10 animals were scored in each strain. ***P < 0.0001.
CPL-1 and CPR-2 activity is important for preserving lysosomal membrane integrity
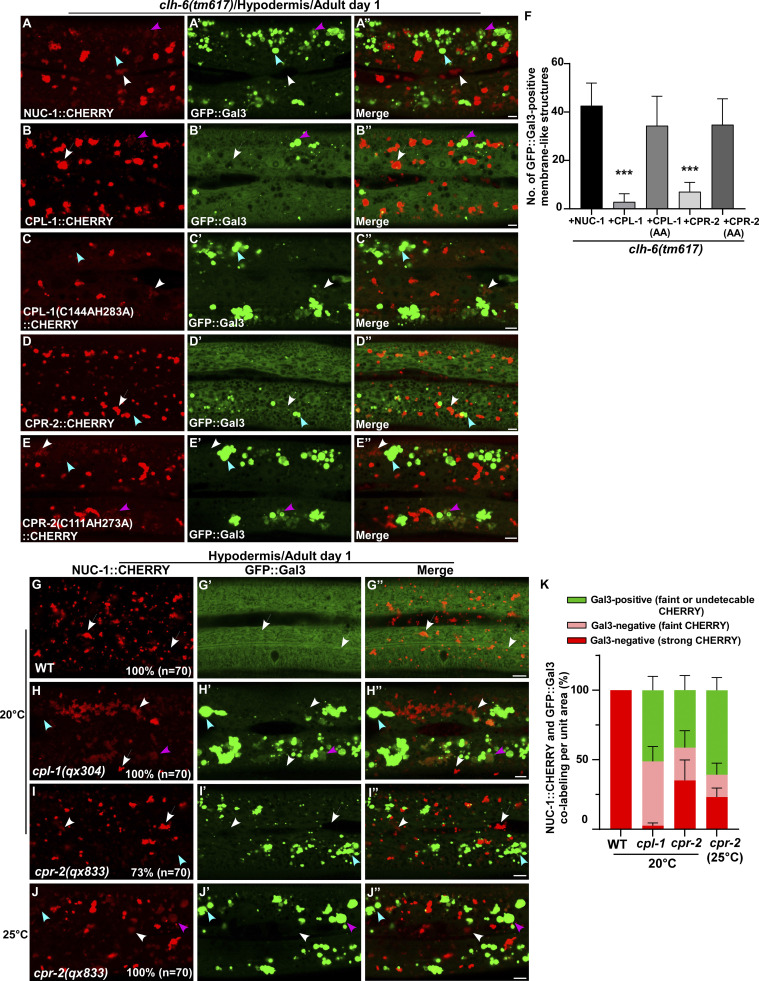
We expressed various lysosomal hydrolases (DNase, RNase, lipase, and cathepsin) to facilitate cargo digestion in clh-6 mutants and examined whether lysosomal membrane integrity is restored (Fig. S3 P). Overexpression of CPL-1 and CPR-2, orthologs of cathepsin L and B, respectively, but not other hydrolases that we tested, suppressed the lysosomal integrity defects in clh-6 mutants (Fig. S3 P). Expression of wild-type but not catalytically inactive CPL-1 or CPR-2 caused significant reduction of Gal3-positive structures in clh-6(tm617) mutants, which suggests that peptidase activity is essential for damage suppression by the two cathepsins (Fig. 6, A–F). CPL-1 overexpression significantly suppressed both cargo accumulation and membrane damage of lysosomes in clh-6 mutants and restored the morphology of endosomes, ER, and Golgi (Fig. 2, J, K, P, and Q; and Fig. S3, J–L). These data suggest that elevated expression of CPL-1 and CPR-2 promotes cargo degradation to protect lysosome membrane integrity in clh-6 mutants. Moreover, loss of cpl-1 or cpr-2 function affects lysosome integrity. In cpl-1(qx304) mutants, NUC-1::CHERRY fluorescence was significantly weakened, while GFP::Gal3 accumulated extensively (Fig. 6, G–H′′ and K). TEM analyses indicated that both cargo digestion and lysosomal membrane integrity were severely affected in cpl-1(qx304) mutants (Fig. 2, L, M, P, and Q). These phenotypes resembled the ones in clh-6 mutants. In cpr-2(qx833) mutants, NUC-1::CHERRY leakage and Gal3 accumulation were observed at 20°C and became more evident at 25°C when cellular stress is increased (Fig. 6, I–K). By TEM, we found that over 70% of cpr-2 lysosomes accumulated undigested cargoes and exhibited membrane damage (Fig. 2, N–Q). Altogether, these data suggest that the functions of CPL-1 and CPR-2 are important for cargo degradation and lysosome membrane stability.
Figure 6.
CPL-1 and CPR-2 overexpression suppresses accumulation of damaged lysosomes in clh-6 mutants. (A–E′′) Confocal fluorescence images of the hypodermis in clh-6(tm617) co-expressing GFP::Gal3 and NUC-1::CHERRY (A–A′′), wild-type CPL-1::CHERRY (B–B′′), catalytically inactive CPL-1::CHERRY (C–C′′), wild-type CPR-2::CHERRY (D–D′′), or catalytically inactive CPR-2::CHERRY (E–E′′). CPL-1::CHERRY and CPR-2::CHERRY exhibit a vesicular pattern like NUC-1::CHERRY. (G–J′′) Confocal fluorescence images of the hypodermis in the indicated strains co-expressing NUC-1::CHERRY and GFP::Gal3. In A–E′′ and G–J′′, intact lysosomes contain strong NUC-1::CHERRY fluorescence and are not labeled by GFP::Gal3 (white arrows), while damaged lysosomes have either faint NUC-1::CHERRY fluorescence with GFP::Gal3 labeling (purple arrowheads) or without GFP::Gal3 labeling (white arrowheads), or strong GFP::Gal3 fluorescence with undetectable NUC-1::CHERRY (blue arrowheads). (F and K) Quantification analyses are shown in F and K. At least 10 animals were scored in each strain and data are shown as mean ± SD. In F, unpaired two-tailed Student’s t test was performed to compare datasets from CPL-1– or CPR-2–expressing worms with the NUC-1–expressing strain. ***P < 0.0001. All other points had P > 0.5. Scale bars: 5 µm.
Loss of clh-6 affects chloride levels and cathepsin B and L activity in lysosomes
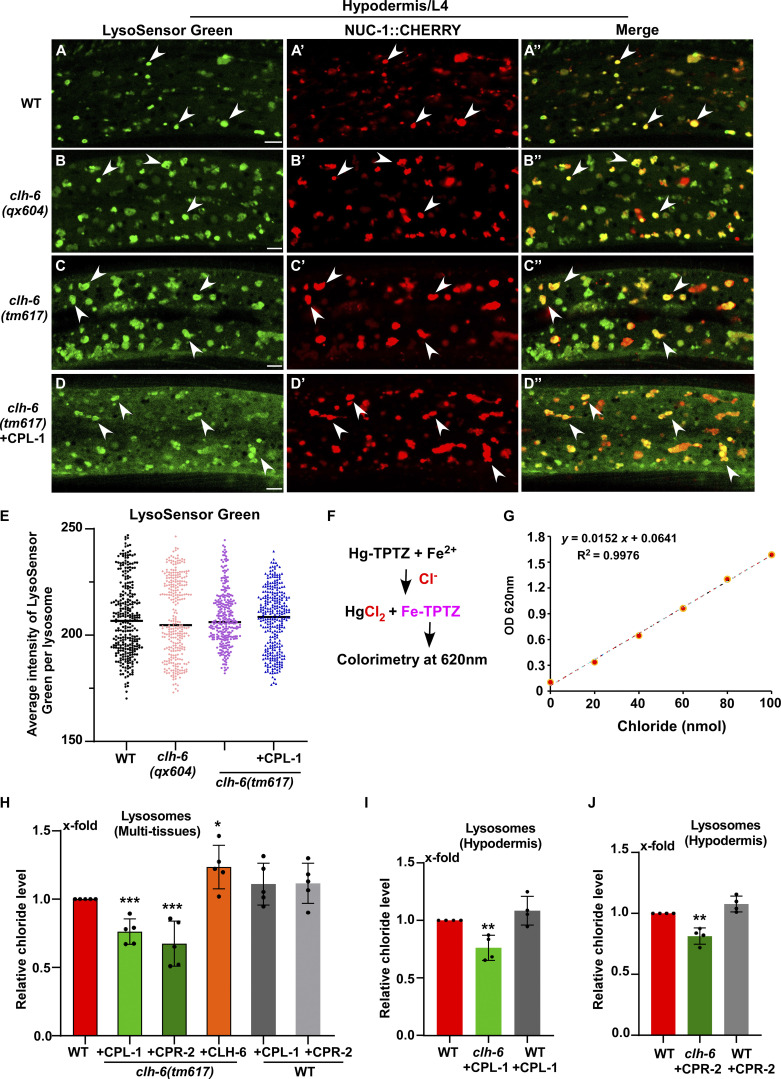
We found that lysosomes indicated by bright NUC-1::CHERRY fluorescence were well stained by LysoSensor green in wild type and in clh-6 mutants without or with overexpression of CPL-1, which blocks lysosomal damage (Fig. 7, A–E). This is consistent with the previous report and suggests that lysosomal acidity is unaffected in clh-6 mutants (Chakraborty et al., 2017).
Figure 7.
Lysosome chloride level, but not acidity, is affected in clh-6 mutants. (A–D′′) Confocal fluorescence images of the hypodermis in the indicated strains expressing NUC-1::CHERRY and stained by LysoSensor green. Intact lysosomes indicated by strong NUC-1::CHERRY fluorescence were scored and they were well stained by LysoSensor green (white arrowheads). (E) The mean intensity of LysoSensor green per lysosome is quantified in E. 300 lysosomes in 6–7 worms were scored in each strain. Scale bars: 5 µm. (F–J) Chloride concentration is determined by a competition reaction (F). The standard curve (G) and lysosomal chloride concentration in multi-tissues (H) and hypodermis (I and J) were determined as described in the Materials and methods. Relative chloride levels are presented. At least four independent experiments were performed. Data are shown as mean ± SD. In E and H–J, unpaired two-tailed Student’s t test was performed to compare mutant datasets with wild type. **P < 0.001, *P < 0.05, ***P < 0.0001. All other points had P > 0.5.
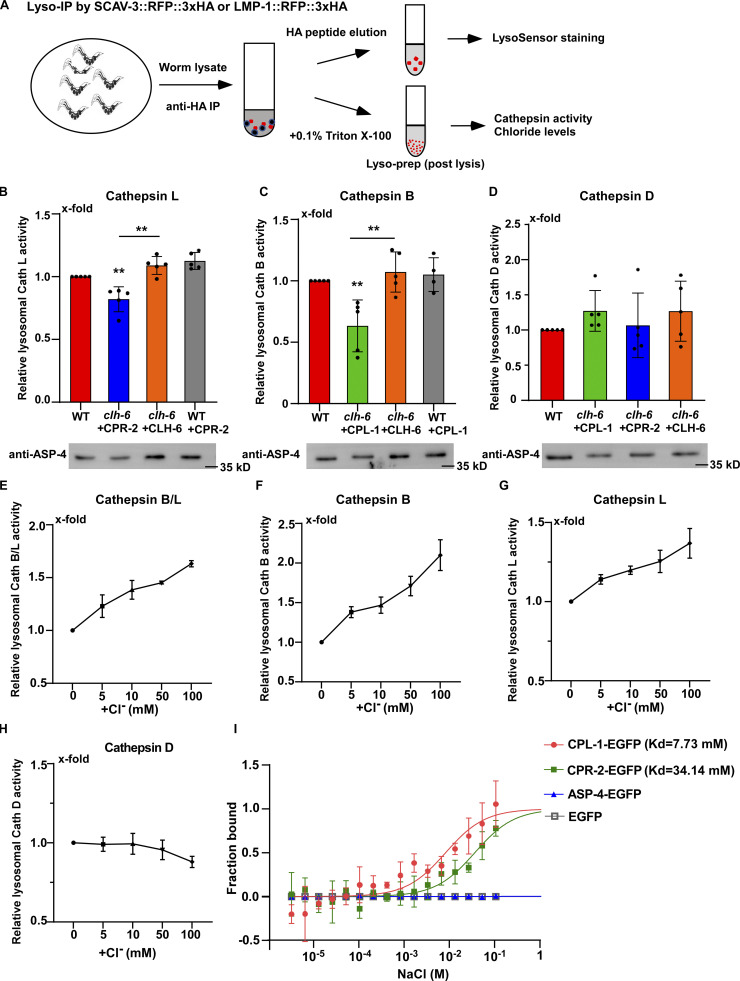
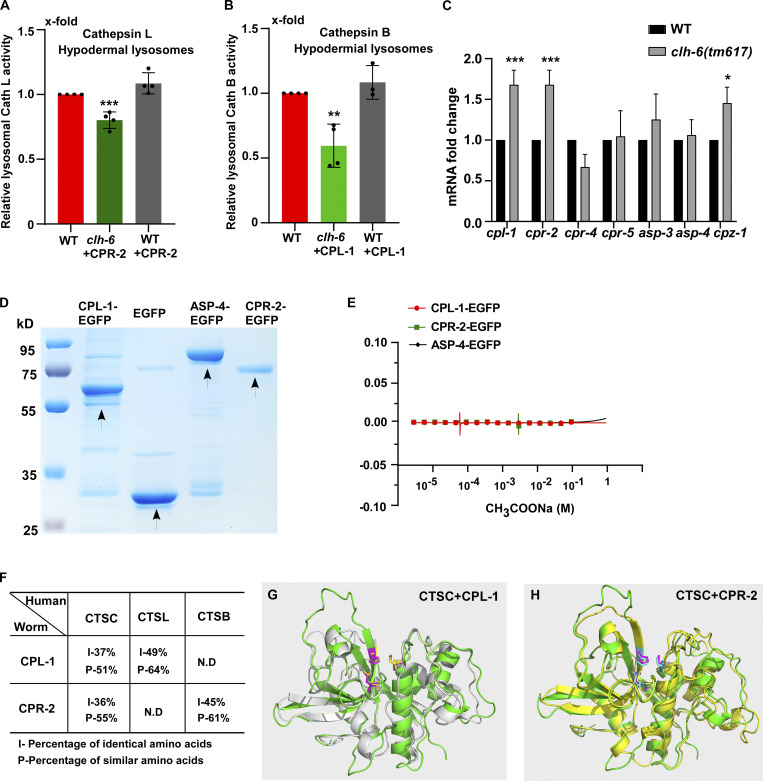
We performed LysoIP experiments using a tagged marker based on the lysosomal membrane protein SCAV-3 (SCAV-3::RFP::3xHA) or LMP-1 (LMP-1::RFP::3xHA) to isolate C. elegans lysosomes by affinity purification (Fig. 8 A and Fig. S4 E; Yu et al., 2022 Preprint). In these experiments, CPL-1 or CPR-2 overexpression was included in clh-6 mutants to suppress lysosomal damage (clh-6 + CPL-1, clh-6 + CPR-2), and in wild type to serve as a control (WT + CPL-1, WT + CPR-2). We found that the chloride concentration, determined by a competition reaction, was significantly lower in lysosomes purified either from multiple tissues including hypodermal cells or specifically from the hypodermis in clh-6+CPL-1 and clh-6+CPR-2 worms than in wild type (WT, WT + CPL-1, WT + CPR-2; Fig. 7, F–J). Overexpression of CLH-6 restored lysosomal chloride levels in clh-6 mutants (Fig. 7 H). By contrast, lysosomal potassium levels were unaffected in clh-6 mutants (Fig. S4, H–J). These data are consistent with the previous report (Chakraborty et al., 2017) and suggest that CLH-6 is important for maintaining chloride levels in lysosomes. We next examined whether loss of clh-6 affects substrate cleavage by CPL-1/cathepsin L and CPR-2/cathepsin B. We found that cathepsin L activity was significantly lower in lysosomes purified from clh-6 + CPR-2 worms than from wild type (WT, WT + CPR-2; Fig. 8 B and Fig. S5 A). Similarly, lysosomes purified from clh-6 + CPL-1 worms exhibited lower cathepsin B activity than in wild type (WT, WT + CPL-1; Fig. 8 C and Fig. S5 B). Cathepsin D activity was unaltered in clh-6 lysosomes compared to wild type (Fig. 8 D). Expression of CLH-6 restored cathepsin L and B activity in clh-6 mutants (Fig. 8, B and C). The expression levels of cpl-1 and cpr-2 were increased in clh-6 mutants, probably due to a feedback response, and delivery of CPL-1 and CPR-2 to lysosomes was unaffected (Fig. 6, B and D; and Fig. S5 C). This raises the possibility that CLH-6 may regulate CPL-1 and CPR-2 activity via chloride. In support of this, supplements of Cl− led to increased activity of cathepsin L and B, but not cathepsin D, in lysosomes in a concentration-dependent manner (Fig. 8, E–H). By microscale thermophoresis (MST) assay, we found that recombinant CPL-1 and CPR-2 proteins bound with Cl− but not Na+, while ASP-4/cathepsin D had no ion binding activity (Fig. 8 I and Fig. S5, D and E).
Figure 8.
Loss of clh-6 affects lysosomal cathepsin B and L activity. (A) Schematic illustration of the lysosome purification experiment by LysoIP. (B–D) Relative activity of cathepsin L (B), cathepsin B (C), and cathepsin D (D) in purified lysosomes from the indicated strains. The amount of lysosomes in each strain was determined by Western blot with an anti-ASP-4 antibody. Relative cathepsin activity was quantified as described in the Materials and methods and normalized to onefold in wild type. At least five independent experiments were performed. Data are shown as mean ± SD. Student’s two-tailed unpaired t test was performed to compare mutant datasets with wild type or datasets linked by lines. **P < 0.001, all other points had P > 0.5. (E–H) Chloride supplements increase the activity of cathepsin B and L, but not cathepsin D, in lysosomes purified from wild-type worms. Relative cathepsin activity was quantified as described in the Materials and methods and normalized to onefold in the sample without NaCl addition. NaCl provides Cl− and Na-gluconate was used to substitute Na+ and Cl− in the low NaCl solutions. At least three independent experiments were performed. Data are shown as mean ± SD. (I) Chloride binds to recombinant CPL-1-EGFP and CPR-2-EGFP, but not ASP-4-EGFP or EGFP, in MST assays. At least three independent experiments were performed. Data are shown as mean ± SD. Source data are available for this figure: SourceData F8.
Figure S5.
CPL-1 and CPR-2 share sequence and structural similarity with human cathepsin C. (A and B) Relative activity of cathepsin L (A) and cathepsin B (B) in lysosomes purified from hypodermal cells in the indicated strains. The amount of lysosomes in each strain was determined by Western blot with an anti-ASP-4 antibody. Relative cathepsin activity was quantified as described in the Materials and methods and normalized to onefold in wild type. At least four independent experiments were performed. (C) Quantitative RT-PCR analyses of cathepsin genes in wild type and clh-6(tm617) at adult day 1. At least three independent experiments were performed. In A–C, data are shown as mean ± SD. Student’s two-tailed unpaired t test (A and B) or two-way ANOVA followed by Bonferroni post-test (C) was performed to compare CPR-2– or CPL-1–overexpression datasets with wild type (A and B) or mutant datasets with wild type (C). *P < 0.05, **P < 0.001, ***P < 0.0001; all other points had P > 0.5. (D) Coomassie blue staining of CPL-1-EGFP, CPR-2-EGFP, ASP-4-EGFP, and EGFP purified from insect cells by two-step affinity purification. (E) Recombinant CPL-1-EGFP, CPR-2-EGFP, and ASP-4-EGFP proteins do not bind to CH3COONa in MST assays. At least three independent experiments were performed. (F) CPL-1 and CPR-2, the C. elegans orthologs of cathepsin L (CTSL) and B (CTSB), respectively, also share sequence similarity with human cathepsin C (CTSC). (G and H) Superimposition of the predicated tertiary structures of C. elegans CPL-1 (gray), with human cathepsin C (CTSC, green; G), and C. elegans CPR-2 (yellow) with human CTSC (green; H). The tertiary structures are predicated by AlphaFold. Key active site residues are highlighted in magenta (CTSC), yellow (CPL-1), and blue (CPR-2). Source data are available for this figure: SourceData FS5.
Discussion
It remains unclear how the Cl−/H+ antiporter ClC-7 and luminal chloride regulate lysosome activity. In this study, we identify CLH-6, the C. elegans ortholog of ClC-7, as an important factor for protecting lysosome integrity. Expression of human ClC-7 efficiently rescued the lysosome integrity defect in clh-6 mutants, which suggests that CLH-6 and ClC-7 play a conserved role in preserving lysosomal membrane stability. Our data suggest that CLH-6 maintains luminal chloride levels to facilitate substrate digestion by cathepsins, and thus protects lysosomal membrane integrity. We show that chloride binds to CPL-1 and CPR-2 and promotes cathepsin L and B activity in lysosomes.
CLH-6/ClC-7 facilitates substrate digestion to protect lysosome membrane integrity
The integrity of lysosomal membranes is essential for maintaining the function of lysosomes. However, it remains poorly understood how lysosomes protect the stability of their membranes while accomplishing essential functions. We showed previously that the lysosomal membrane glycoproteins SCAV-3/LIMP-2 and LMP-1/LAMP1 protect membrane integrity probably by forming the glycocalyx at the luminal leaflet (Li et al., 2016). Here we found that loss of CLH-6 causes defects in both cargo degradation and membrane integrity. Reducing cargo delivery or increasing CPL-1 and CPR-2 expression suppresses cargo accumulation and restores membrane integrity of lysosomes in clh-6 mutants. Loss of CPL-1 or CPR-2 impairs cargo degradation and causes rupture of lysosomal membranes as in clh-6(lf) mutants. These data suggest that optimal substrate digestion by cathepsins is important for stabilizing lysosomal membranes. It is conceivable that undigested or incompletely digested cathepsin substrates and/or their derivatives become harmful to the membrane if they persist in the lysosomal lumen. Like in scav-3 mutants, loss of clh-6, cpl-1, or cpr-2 causes lysosomal damage in the hypodermis but not in the intestine or body wall muscle cells (Li et al., 2016). High demands for lysosomal cargo degradation and/or differentially regulated stress resistance may be responsible for the high occurrence of lysosomal rupture in the hypodermis.
Of note, CLCN7 or OSTM1 deficiency causes lysosomal storage phenotypes consistent with those in clh-6(lf) mutants. CLCN7−/− and OSTM1 deficient mice display severe lysosomal storage defects in the central nervous system with features of neuronal ceroid lipofuscinosis, a lysosomal storage disease, and exhibit widespread neurodegeneration (Kasper et al., 2005; Kornak et al., 2001; Lange et al., 2006; Pressey et al., 2010). CLCN7 deletion in renal proximal tubular cells causes defects in lysosomal degradation of endocytosed proteins (Wartosch et al., 2009). It is possible that CLCN7 deficiency affects lysosomal membrane integrity, and rupture of lysosomes may contribute to the massive loss of neurons in CLCN7−/− mice (Kasper et al., 2005). Like clh-6(lf), inactivation of C. elegans ostm1 causes decreased chloride levels and reduced degradation capacity of lysosomes in coelomocytes (Chakraborty et al., 2017; Gee et al., 2017), but whether loss of ostm1 function impairs lysosome degradation in other cell types or affects lysosome integrity awaits further investigation. Mutations in lysosomal hydrolases cause lysosomal storage disorders, which are characterized by intralysosomal accumulation of specific undegraded substrates (Platt et al., 2012; Segatori, 2014). Future study is needed to determine whether membrane stability is generally affected by deficiency of lysosomal hydrolases, or whether the lysosomal membrane is particularly vulnerable to loss of certain types of degradative enzymes.
Luminal chloride modulates cathepsin activity in lysosomes
Cl− is the most abundant anion in the lysosomal lumen, but whether and how it directly regulates lysosomal degradation activity remains unclear. Consistent with previous reports, we found that the lysosomal chloride level is reduced in clh-6 mutants, while acidity of lysosomes is unaffected (Chakraborty et al., 2017; Weinert et al., 2010). Our data suggest that luminal Cl−, maintained by CLH-6, is important for cathepsin L and B activity. Loss of clh-6 causes reduced lysosomal cathepsin L and B activity, which is restored by CLH-6 expression. Expression of the cpl-1 and cpr-2 genes is specifically increased in clh-6 mutants, probably due to a feedback response to impaired cathepsin activity. Cl− binds to CPL-1 and CPR-2 in vitro and its supplementation increases lysosomal cathepsin B and L activity in a concentration-dependent manner. Cl− is known to bind and activate human cathepsin C (Cigic and Pain, 1999; McDonald et al., 1966). CPL-1 and CPR-2 exhibit a Cl−-binding activity with equilibrium dissociation constants (Kd) of about 7 and 34 mM, respectively, consistent with the millimolar range of cathepsin C activation by Cl− (Cigic and Pain, 1999). Lysosomes contain the highest Cl− levels (∼75–108 mM) among all the endocytic compartments (Chakraborty et al., 2017; Saha et al., 2015; Stauber and Jentsch, 2013). Binding with Cl− at a millimolar range may ensure that CPL-1 and CPR-2 activity is modulated only under high Cl− conditions, such as in the lysosomal lumen. Cathepsin C is a cysteine-type aminodipeptidase with no clear orthologs present in worms. CPL-1 and CPR-2 are both cysteine-type peptidases that share sequence similarity with cathepsin C (Fig. S5 F). Interestingly, the tertiary structures of CPL-1 and CPR-2, predicted by AlphaFold, closely resemble that of cathepsin C (Fig. S5, G and H). More in-depth biochemical and structural studies are needed to understand whether CPL-1 and CPR-2 activity is modulated by Cl− in a similar manner as human cathepsin C. While CPL-1 is the only cathepsin L ortholog, worms have a divergent multigene family of cathepsin B–like proteins with 11 members. It is possible that multiple cathepsin B proteases are modulated by Cl−, which would explain the less severe lysosomal defects in cpr-2 mutants than in clh-6 and cpl-1 mutants.
Our data suggest that chloride modulates cathepsin L and B activity, which is important for substrate digestion and stabilization of lysosomal membranes. Thus, optimal activity of lysosomal hydrolases requires both acidic pH and luminal Cl−. Future investigations should address whether additional cysteine cathepsins or other types of hydrolases are also modulated by Cl−, and whether acidic pH and luminal Cl− coordinate to regulate hydrolase activity as suggested for cathepsin C (Cigic and Pain, 1999).
Materials and methods
C. elegans strains
Strains of C. elegans were cultured and maintained using standard protocols. The N2 Bristol strain was used as the wild-type strain except for polymorphism mapping in which Hawaiian strain CB4856 was used. The following strains were used in this work: Linkage group I (LG I): epg-8(bp251); LG II: lgg-1(bp407); LG III: cup-5(bp510), chc-1(b1025); LG IV: atg-3(bp412), epg-9(bp320), lgg-2(tm6474); LG V: clh-6(tm617, qx632, qx644, qx604, qx613, qx629), cpr-2(qx833), cpl-1(qx304); LG X: dyn-1(ky51).
The reporter strains used in this study are listed below. Transgenic animals carrying extrachromosomal arrays (qxEx) were generated using standard microinjection methods, and genome-integrated arrays (qxIs) were obtained by γ-ray irradiation to achieve stable expression from arrays with low copy numbers (Evans and Hunter, 2005). Single copy insertion worms (qxSi) were generated by CRISPR/Cas9 (Paix et al., 2014).
qxIs257(Pced-1NUC-1::CHERRY), qxIs582(Psemo-1sfGFP::Gal3), qxIs285(Psemo-1GFP::2xFYVE), qxIs439(Psemo-1GFP::TRAM-1), qxIs684(Psemo-1MANS::GFP), qxIs430(Pscav-3SCAV-3::GFP), qxIs915(Pclh-6CLH-6::GFP), qxIs767(Psemo-1CPL-1::CHERRY), qxIs769(Psemo-1CPR-2::CHERRY), qxIs882(Psemo-1SQST-1::GFP), qxIs712(Pdpy-7DPY-7::GFP), qxIs864(Pcol-12LMP-1::RFP::3xHA), qxSi13(Plgg-1GFP::LGG-1), qxSi36(Pscav-3SCAV-3::RFP::3xHA), qxSi121(Pcol-19SCAV-3::RFP::3xHA), qxEx8869(Pclh-6CLH-6::GFP), qxSi104(Pcol-12AMAN-2::mKate2), qxEx9446(Pdpy-7TagRFP-T::RAB-5), qxEx8574(Phyp-7CLH-6::CHERRY), qxEx9256, qxEx9257(Pclh-6CLCN7::GFP), qxEx9376[Psemo-1CPL-1(C144A,H283A)::CHERRY], qxEx9301[Psemo-1CPR-2(C111A,H273A)::CHERRY]. We obtained pwIs281(Ppie-1CAV-1::GFP) from Dr. B. Grant (Rutgers University, USA).
Isolation, mapping, and cloning of clh-6
Worms carrying sfGFP::Gal3 were mutagenized using Ethylmethanesulfonate (EMS, M0880; Sigma-Aldrich). Young adult worms at the F3 generation were examined for accumulation of sfGFP::Gal3-positive membrane-like structures. From a screen that covered ∼20,000 haploid genomes, 44 mutants were obtained. The recessive mutations qx604, qx613, qx629, qx632, and qx644 failed to complement one another in causing sfGFP::Gal3 accumulation, which suggests that they affect the same gene. qx604 was mapped to the left arm of linkage group III by single nucleotide polymorphism mapping. Whole-genome sequencing and transformation rescue experiments led to identification of a DNA fragment containing the clh-6 gene, which fully rescued the qx604 defect. The sequence of the clh-6 gene was determined in all clh-6 alleles. qx604 and qx644 contain G to A mutations that result in substitution of Gly 478 with Arg and a premature stop codon after Leu 486, respectively. qx613 and qx629 contain C to T mutations that cause a premature stop codon after Asn 678 and substitution of Pro 732 with Leu, respectively. qx632 contains a 1-bp deletion that causes a frameshift, leading to early termination of protein after only the first 294 amino acids. The tm617 allele contains a 563-bp deletion and a 5-bp insertion that removes most of the fifth and sixth exons and generates an early stop codon, resulting in a truncated protein containing only the first 164 amino acids. All mutants were backcrossed with N2 animals at least six times before further analyses.
Microscopy and imaging analysis
Differential interference contrast (DIC) images and confocal microscopy images were taken with an inverted laser scanning confocal microscope (LSM 880; Carl Zeiss) with 488 (emission filter BP 503-530) and 543 (emission filter BP 560-615) lasers. Images were processed and viewed using LSM Image Browser and ZEN software (Carl Zeiss). All images were taken at 20°C.
Time-lapse recording using spinning-disk microscopy
C. elegans larvae or adults were mounted on agar pads in M9 buffer with 5 mM levamisole. Fluorescence images were captured using a 100× objective (CFIPlan Apochromat Lambda; NA 1.45; Nikon) with immersion oil (type NF) on an inverted fluorescence microscope (Eclipse Ti-E; Nikon) with a spinning-disk confocal scanner unit (UltraView, PerkinElmer) with 488 [emission filter 525 (W50)] and 561 [dual-band emission filter445 (W60)] lasers. To follow dynamic changes in worms co-expressing sfGFP::Gal3 and NUC-1::CHERRY, images were captured every 30 s for 1–3 h. The collected images were viewed and analyzed using Velocity software (PerkinElmer).
Quantification of damaged lysosomes
To examine lysosome damage, fluorescence images of the hypodermis in each strain were captured using an inverted confocal microscope (LSM 880; Carl Zeiss) with equal exposure time. Intact lysosomes contain strong NUC-1::CHERRY fluorescence and are not labeled by GFP::Gal3. Leaking or damaged lysosomes have either faint NUC-1::CHERRY fluorescence with or without labeling of GFP::Gal3, or strong GFP::Gal3 fluorescence with undetectable NUC-1::CHERRY. The proportion of intact and damaged lysosomes within the unit area (1,000 μm2) was quantified by dividing the area of intact or damaged lysosomes by the total area of lysosomes. The number of damaged lysosomes was quantified by scoring the number of sfGFP::Gal3-positive membrane-like structures within the unit area (1,000 μm2). At least 10 animals were scored in each strain at each stage.
TEM analyses
C. elegans larvae (L4 stage) or adults (day 1 of adulthood) were rapidly frozen using a high-pressure freezer (HPM100; Leica Biosystems). Freeze substitution was performed in anhydrous acetone containing 1% osmium tetroxide. The samples were kept sequentially at −90°C for 72 h, −60°C for 10 h, and −30°C for 10 h and were finally brought to 0°C for 1 h in a freeze-substitution unit (EM AFS2; Leica Biosystems). The samples were washed three times (15 min each time) in fresh anhydrous acetone and were gradually infiltrated with Embed-812 resin in the following steps: resin/acetone 1:3 for 3 h, 1:1 for 5 h, 3:1 overnight, and 100% resin for 48 h. Samples were then kept overnight and embedded at 60°C for 48 h. The fixed samples were cut into 70 nm sections with a microtome EM UC6 (Leica Biosystems) and electron stained with uranyl acetate and lead citrate. Sections were observed with a HT7700 (Hitachi) operating at 80 kV.
LysoSensor staining
>50 L4-staged larvae were soaked in 100 μl LysoSensor Green solution (1:200 dilution by M9; Invitrogen) for 30 min at 20°C in the dark. Worms were then transferred to fresh OP50-seeded NGM plates and allowed to recover at 20°C for 1 h in the dark before examination by fluorescence microscopy.
Pitstop 2 treatment
NGM plates were supplemented with 1 ml of concentrated stocks (1 mM) of Pitstop 2 (SML1169; Sigma-Aldrich), then spotted with Escherichia coli strain OP50. L4 larvae of each genotype were cultured on the Pitstop 2–supplemented plates for about 12 h, after which the lysosome integrity was examined. Worms cultured on DMSO-supplemented plates to the similar stage (12 h post L4) were used as control. DMSO or Pitstop 2–supplemented plates were kept in the dark during the whole process.
Lifespan assay
Lifespan assays were performed at 20°C as described previously (Hansen et al., 2005). Briefly, worms at the L4 stage (day 0, >100) were placed on NGM plates seeded with OP50, with 10 worms per plate. The animals were transferred to new plates when their progeny grew up to the L3 stage and dead animals were counted every 2 d. Animals that crawled off the plate, exploded, bagged, or became contaminated were discarded.
Examination of brood size
To examine brood size, >25 L4 staged worms were placed on NGM plates seeded with E. coli OP50, with five worms per plate. The animals were transferred to new plates every 8 h until they were no longer laying eggs. All eggs and larvae were quantified in each plate. At least three independent experiments were performed.
Mutagenesis and generation of knock-in worms using CRISPR/Cas9
The cpr-2(qx833) mutation was obtained by CRISPR/Cas9 editing as described before (Paix et al., 2014). In brief, guide RNA (sgRNA) sites for cpr-2 (sgRNA1: 5′-CTTAGTTGATAATAGGACAC-3′, sgRNA2: 5′-GACACAGGCAGAGCTCAGTC-3′) were cloned into pDD162 vector (pDD162-Peft-3::Cas9-Pu6::sgRNA), which expresses the Cas9 protein and sgRNA. Repair templates containing desired editing sequences and homologous arms with targeted region were used to achieve site-directed editing. dpy-10 was used as a selection marker for co-conversion events in the F1 progeny as reported previously. Dpy or roller F1 worms were picked and screened for recombination by restriction enzyme digestion. Positive candidates were confirmed by sequencing. Single-copy insertion of the SCAV-3::RFP::3xHA reporter was generated as described before (Takayanagi-Kiya et al., 2016). In brief, the Pscav-3SCAV-3::RFP::3xHA 3′UTR and Pcol-19SCAV-3::RFP::3xHA 3′UTR DNA cassettes were cloned into a donor vector containing LG II–targeted homology arms and a hygromycin resistance gene as the selection marker. Injected worms were screened on hygromycin plates for array insertion. RFP signals were also used to exclude candidates with extrachromosomal arrays which were also hygromycin-resistant. The positive recombinants were sequenced to confirm correct insertion. All strains obtained were outcrossed with the N2 wild-type strain for four times before further analysis.
Lysosome purification
The lysosomal membrane protein SCAV-3::RFP::3xHA or LMP-1::RFP::3xHA was expressed in multiple tissues driven by the scav-3 promoter or specifically in hypodermal cells controlled by either the col-19 or col-12 promoter. Lysosomes from multiple tissues or the hypodermis of C. elegans were then purified by LysoIP using anti-HA magnetic beads (Thermo Fisher Scientific) as described previously (Savini et al., 2022). Briefly, strains carrying the lysosomal marker protein SCAV-3::RFP::3xHA or LMP-1::RFP::3xHA were used for LysoIP experiments. 500 μl to 1 ml adult worms were collected for each strain, then washed three times in M9 and once in KPBS buffer (136 mM KCl, 10 mM KH2PO4, pH 7.25), followed by Dounce homogenization on ice. The resulting worm lysates were centrifuged at 1,000 g for 3 min at 4°C to remove debris. The resulting supernatants were incubated with 50 μl of anti-HA magnetic beads for 1 h at 4°C and transferred to a magnetic stand followed by washing with cold KPBS for at least four times. The resulting lysosome-bound magnetic beads were washed by chloride-free buffer (25 mmol/liter Hepes, 150 mmol/liter N-Methyl-D-glucamine [NMDG], pH 7.2, adjusted with Methanesulfonic acid [MSA]) and lysosomes were either eluted by 3xHA peptide for LysoSensor staining or treated with 0.1% Triton X-100 on beads for measurement of chloride concentration and cathepsin activity. The purity of lysosomes was examined by antibodies that detect different intracellular organelles as shown in Fig. S4 E. The integrity of lysosomes was examined by LysoSensor staining and the lysosome amounts were determined by anti-HA and anti-ASP-4 antibodies.
Measurement of chloride and potassium concentration and cathepsin activity
The lysosome-bound magnetic beads were incubated with chloride- and potassium-free buffer containing 0.1% Triton X-100 (VWR International) for 10 min on ice, vortexed intermittently for four times, and centrifuged at 15,000 rpm for 15 min at 4°C. The resulting supernatant was transferred to new tubes for measurement of chloride or potassium concentration and subjected to Western blot to examine lysosome amounts using anti-ASP-4 antibody. The standard curve and concentration of chloride in lysosomal samples were determined by a colorimetric chloride assay kit according to the manufacturer’s instructions (MAK023; Sigma-Aldrich). The concentration of potassium in lysosomal samples was determined by a colorimetric potassium assay kit according to the manufacturer’s instructions (BC2775; Solabio).
To measure cathepsin activity, the lysosome-bound magnetic beads were treated with 0.1% Triton X-100 as described above and resuspended with the supernatant after centrifugation. The resulting lysosomal prep was used to examine cathepsin activity, and to determine the lysosome amount by western blot using anti-ASP-4 antibody. To measure cathepsin activity, 10 μl lysosomal prep was added into 200 μl reaction buffer (0.1 M sodium citrate, pH 5.3, 1 mM DTT) containing each cathepsin substrate at a final concentration of 10 μM (cathepsin B: Z-Arg-Arg-AMC, Sigma-Aldrich; cathepsin L: Ac-FR-AFC Abcam; cathepsin D Bz-Arg-Gly-Phe-Phe-Pro-4MeOβNA, Sigma-Aldrich; cathepsin B/L: Z-Phe-Arg-AMC, Glpbio). DTT was omitted and pH was adjusted to 4.0 in sodium citrate buffer for measurement of cathepsin D and B/L activity, respectively. For chloride supplementation experiments, NaCl was included in sodium citrate buffer at different concentrations as shown in Fig. 8, E–H. In these experiments, the ionic strength and osmolarity were maintained at constant levels by adding an appropriate amount of Na-gluconate when the concentration of NaCl was below 100 mM. The reaction was performed at 37°C for 1–2 h and the magnetic beads were collected by a magnetic stand and discarded. The cathepsin activity was measured in the supernatant in a fluorescence plate reader (EnSpire, PerkinElmer) at 460 nm (cathepsin B, B/L), 505 nm (cathepsin L), and 425 nm (cathepsin D). Negative control wells contained reaction buffer without or with the cathepsin substrate. Each sample was assayed in duplicates or triplicates in a 96-well flat-bottom ELISA Plate (Sangon Biotech).
To compare chloride concentration and cathepsin activity in different strains, both the colorimetric or fluorometric reading and the lysosome amount determined by anti-ASP-4 antibody (ImageJ, National Institutes of Health) in wild type were normalized to “onefold” and used as the denominator to get the fold changes in mutant strains. The ratio of normalized colorimetric/fluorometric reading vs. lysosome amount is presented as relative chloride level and cathepsin activity in each sample. Three independent experiments were performed and quantified.
Immunoblotting
To examine protein levels of LGG-1, ∼100 adult day 1 worms of each genotype were lysed by three rounds of freeze–thaw in 25 μl 1xSDS loading buffer followed by boiling at 100°C for 10 min. The resulting whole-worm lysate samples were resolved by SDS-PAGE and blotted with anti-LGG-1 antibody (rat; 1:1,000; prepared by the Antibody Center of the National Institute of Biological Sciences, Beijing, China) and anti-α-tubulin antibody (mouse; 1:5,000; Sigma-Aldrich). The ratio of LGG-1-II vs. tubulin in wild type was normalized to “1” fold and used as the denominator to get the fold changes in clh-6 mutant strains. Three independent experiments were performed and quantified in each strain. Purified lysosomal preps were boiled in 25 μl 1xSDS loading buffer at 100°C for 10 min, and blotted with anti-HA (rabbit; 1:1,000; Abcam), anti-SCAV-3 (rat; 1:1,000; prepared by the Antibody Center of the National Institute of Biological Sciences, Beijing, China), anti-ASP-4 (rat; 1:1,000; prepared by the Antibody Center of the Institute of Genetics and Developmental Biology, Chinese Academy of Science), anti-Hsp-60 (mouse; 1:1,000; DSHB), anti-RAB-5 (rat; 1:1,000; prepared by the Antibody Center of the National Institute of Biological Sciences, Beijing, China), anti-LAMIN (rabbit; 1:1,000; ABclonal), or anti-PDI (rabbit; 1:1,000; kindly provided by Chi-chen Wang’s lab, Insitute of Biophysics CAS, Beijing, China) antibodies. The secondary antibodies used are goat anti-rat IgG (H + L), goat anti-rabbit IgG (H + L), and goat anti-mouse IgG (H + L) conjugated with HRP (1:10,000; Jackson ImmunoResearch). The signals were detected using ECL chemiluminescence reagent (RPN2232; Cytiva) and captured by ChemiDoc system (5300; Clinx). The intensity of the protein band was quantified by ImageJ (National Institutes of Health).
Real-time quantitative PCR
Worms were synchronized and cultured at 20°C to adult day 1. Total RNA was extracted from ∼20 μl worm pellets at each stage using Trizol (Invitrogen) and reverse transcribed by a PrimeScript RT Reagent Kit (Takara). The reverse transcription products (cDNA) were diluted to 10 ng/ml, and 1 μl of the diluted cDNA was used as the template for quantitative PCR in a 20 μl reaction mixture. For quantitative RT-PCR, FastStart Universal SYBR Green Master (Roche) was used on an Applied Biosystems QuantStudio 3 Flex real-time PCR system (Life Technologies). The gene act-1 was used as the internal reference. At least three independent experiments were performed with three replications each time.
Recombinant proteins
Full-length cDNA of cpr-2 and asp-4, and cDNA of cpl-1, which encodes the mature form of CPL-1(120-337), were cloned into pFastBac1-EGFP with a His-Strep tag. The recombinant proteins were produced in insect sf9 cells (1.8–2.3 × 106, >95% viability) in IB905 medium (Yishengke). CPL-1(120-337) was purified from cell lysates using Ni-NTA resin (Qiagen) followed by Strep-Tactin (IBA). CPR-2 and ASP-4 proteins were purified from culture medium by Ni2+-Sepharose 6 Fast Flow (GE Healthcare) affinity chromatography followed by size-exclusion chromatography (Superdex-200; 26/60; GE Healthcare).
MST assay
To examine chloride binding, recombinant proteins (200 nmol/liter) were incubated with NaCl or CH3COONa at different concentrations in a 20-μl reaction mixture and loaded into NT.115 standard coated capillaries (NanoTemper Technologies). The buffer used in the MST assays was free of chloride (25 mmol/liter Hepes, 150 mmol/liter NMDG, pH 7.2 adjusted with MSA). MST measurements were performed at 25°C with 20% excitation power and medium MST power. All experiments were repeated three times for each measurement. Data analyses were performed using NanoTemper software (NanoTemper).
Plasmid construction
To generate Pclh-6CLH-6::GFP, a genomic DNA fragment containing both the promoter and gene sequence of clh-6 was amplified using primers PQQZ237/388 and was seamlessly assembled with pPD49.26-GFP through Bam HI and Nhe I sites. To generate Pclh-6ClC-7::GFP, firstly the promoter sequence of clh-6 was amplified by primers PQQZ388/373 and ligated into pPD49.26-GFP through Bam HI and Nhe I sites, followed by ligation with the CLCN7 cDNA amplified by primers PQQZ489/490 through NheI site. To construct Psemo-1CPL-1::CHERRY and Psemo-1CPR-2::CHERRY, genomic DNA fragments containing the cpl-1 and cpr-2 genes were amplified by primers PQQ318/319 and PQQZ322/323, respectively, followed by seamless assembly with Psemo-1CHERRY through the Nhe I site. To build Psemo-1CPL-1C144A,H283A::CHERRY and Psemo-1CPR-2C111A,H273A::CHERRY, site-directed mutagenesis was performed to introduce the corresponding mutations into Psemo-1CPL-1::CHERRY and Psemo-1 CPR-2::CHERRY using primers PQQZ487/488, PLRC204/205 and PQQZ497/498, PQQZ499/500, respectively. To build Psemo-1 CPR-4::CHERRY, Psemo-1 CPR-5::CHERRY, Psemo-1 CPR-6::CHERRY, Psemo-1 ASP-3::CHERRY, Psemo-1 CPZ-1::CHERRY and Psemo-1 TAG-329::CHERRY, corresponding genomic DNA fragments were amplified by primers PQQZ363/364, PQQZ365/366, PQQZ343/344, PQQZ367/368, PQQZ335/336, PQQZ337/338, respectively and ligated into Psemo-1CHERRY through the Nhe I site. CTSA-3.2 was amplified by PQQZ6/7 and ligated into 49.26-Psemo-1 through Kpn I and Nhe I sites. To generate PFastBac1-CPL-1(120–337)-EGFP and PFastBac1-CPR-2-EGFP, corresponding cDNA fragments were amplified using primers PQQZ493/494 and PQQZ491/492, respectively, CPL-1(120–337) and CPR-2 were ligated into pFastBac1-EGFP vector through Bam H I and Kpn I sites. Primers are listed in Table 1.
Table 1.
Primers used for plasmid construction
| Primers | Sequences (5′-3′) |
|---|---|
| PQQZ318 | 5′-TTTCAGGAGGACCCTTGGCTAGCATGAACCGATTCATTCTTCTGG-3′ |
| PQQZ319 | 5′-TCATGGTACCGTCGACGCTAGCGACCAATGGATAACTGGCCTTG-3′ |
| PQQZ237 | 5′-ATACCATGGTACCGTCGACGCTAGCAGATTCAGAAATATAAAGTTCA-3′ |
| PQQZ388 | 5′-CTGCAGGTCGACTCTAGAGGATCCTCATTTCAATTGTAAAGCAACCAA-3′ |
| PQQZ373 | 5′-ATACCATGGTACCGTCGACGCTAGCCTGAAATGTTGAAATAAATCC-3′ |
| PQQZ322 | 5′-CATTTTCAGGAGGACCCTTGGCTAGCATGGTAAGTTTTTCATTTAACCAA-3′ |
| PQQZ323 | 5′-GAGACCATGGTACCGTCGACGCTAGCTCTCGGCAATCCAGCGACAATACG-3′ |
| PLRC204 | 5′-GCTCATCCGAAGAGCTCGACGCCGGAGTGCTTC-3′ |
| PLRC205 | 5′-GAAGCACTCCGGCGTCGAGCTCTTCGGATGAGC-3′ |
| PQQZ497 | 5′-GGATCGGCCTGGGCTTTCTCAACAGCTGAGGTGATTT-3′ |
| PQQZ498 | 5′-AGCCCAGGCCGATCCGCAGTTGGACTGCTCGCGGA-3′ |
| PQQZ499 | 5′-GGAGCCGCTGTAAAGCTAATTGGCT-3′ |
| PQQZ500 | 5′-GGCTCCTCCCTTTGATCTACCGGCG-3′ |
| PQQZ487 | 5′-AATGTGCGGATCGGCCTGGGCCTT-3′ |
| PQQZ488 | 5′-TCCGCACATTCCTTGGTTCTTGAC-3′ |
| PQQZ489 | 5′-CATTTTCAGGAGGACCCTTGGCTAGCATGGCCAACGTCTCTAAGAAGGTG-3′ |
| PQQZ490 | 5′-GAGACCATGGTACCGTCGACGCTAGCCGTCTGGGCCAGCGAGAGCTCC-3′ |
| PQQZ491 | 5′-ACCGTCCCACCATCGGGCGCGGATCCATGAACCTCATCCTTCTTTCTTCA-3′ |
| PQQZ492 | 5′-AACAGAACTTCCAGAAGCTTGGTACCTCTCGGCAATCCAGCGACAATACG-3′ |
| PQQZ493 | 5′-ACCGTCCCACCATCGGGCGCGGATCCATGGTCCCAGATGAGGTTGACTGGCG-3′ |
| PQQZ494 | 5′-AACAGAACTTCCAGAAGCTTGGTACCGACCAATGGATAACTGGCCTTGGTG-3′ |
| PQQZ495 | 5′-ACCGTCCCACCATCGGGCGCGGATCCATGAACCGCTGTATTCTTTTG-3′ |
| PQQZ496 | 5′-AACAGAACTTCCAGAAGCTTGGTACCTTCGTCGTCTTGTTCCATGGATTC-3′ |
| PQQZ363 | 5′-CATTTTCAGGAGGACCCTTGGCTAGCATGGTAAGTGGTTGTTTTAGCG-3′ |
| PQQZ364 | 5′-GAGACCATGGTACCGTCGACGCTAGCGACTTTTGGGACTCCTCCGACA-3′ |
| PQQZ365 | 5′-CATTTTCAGGAGGACCCTTGGCTAGCATGTGGAAGCTCTCCGCTATTC-3′ |
| PQQZ366 | 5′-GAGACCATGGTACCGTCGACGCTAGCGTTGTGACGAGCCAAGTCTGGA-3′ |
| PQQZ343 | 5′-CATTTTCAGGAGGACCCTTGGCTAGCATGGTAGGCTTTTGAACTTTGAA-3′ |
| PQQZ344 | 5′-GAGACCATGGTACCGTCGACGCTAGCGTAGTTGTCATCGTAGACGTGGC-3′ |
| PQQZ367 | 5′-CATTTTCAGGAGGACCCTTGGCTAGCATGTCGGGCCGCGTTTTCCTTC-3′ |
| PQQZ368 | 5′-GAGACCATGGTACCGTCGACGCTAGCTTTTCCGGTTCTAGAGGTGGCA-3′ |
| PQQZ335 | 5′-CATTTTCAGGAGGACCCTTGGCTAGCATGCGAACTTTTGTGCTCCTTCTT-3′ |
| PQQZ336 | 5′-GAGACCATGGTACCGTCGACGCTAGCAACAATCGGATCAGCCCAAACACA-3′ |
| PQQZ337 | 5′-CATTTTCAGGAGGACCCTTGGCTAGCATGGCGTCTCTTCTGGCGCTCTT-3′ |
| PQQZ338 | 5′-GAGACCATGGTACCGTCGACGCTAGCTTCAGGGATTCTTGCACCATATC-3′ |
| PQQZ6 | 5′-AGAACATTTTCAGGAGGACCCTTGGCTAGCATGTGGTGGACGTCATTAGT-3′ |
| PQQZ7 | 5′-GCGGAGCTCAGATATCAATACCATGGTACCTCAAACGGGCGAGGTGTAGT-3′ |
Statistical analysis
The SD was used as the y-axis error bar for bar charts plotted from the mean value of the data. Data derived from different genetic backgrounds were compared by Student’s two-tailed unpaired t test or two-way ANOVA followed by Bonferroni post-test, as indicated in the figure legends. Data distribution was assumed to be normal but this was not formally tested. Data were considered statistically different at P < 0.05. P < 0.05 is indicated with single asterisks, P < 0.001 with double asterisks, and P < 0.0001 with triple asterisks.
Online supplemental material
Fig. S1 shows that clh-6 mutants contain damaged lysosomes. Fig. S2 shows that CLH-6 is homologous to human ClC-7. Fig. S3 shows that loss of clh-6 affects endosome, ER, and Golgi patterns. Fig. S4 shows that loss of clh-6 affects endocytic cargo degradation. Fig. S5 shows that CPL-1 and CPR-2 share sequence and structural similarity with human cathepsin C.
Supplementary Material
is the source file for Fig. 4.
is the source file for Fig. 8.
is the source file for Fig. S4.
is the source file for Fig. S5.
Acknowledgments
We thank Drs. B. Grant (Rutgers University, New Brunswick-Piscataway, NJ, USA), Meng Wang (Janelia Research Campus, Howard Hughes Medical Institute, Ashburn, VA, USA), and S. Mitani (Tokyo Women’s Medical University, Tokyo, Japan) for strains, Min Wang (Xiaochen Wang’s lab, Institute of Biophysics) for technical assistance, and Dr. Isabel Hanson for editing services. Some strains were from Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40OD010440).
This work was supported by grants from the National Natural Science Foundation of China (92254303, 32130028, 32293202) and the National Key R&D Program of China (2021YFA1300303) to X. Wang.
Author contribution: X. Wang conceived and supervised the research. Q. Zhang performed most of the experiments and Y. Li performed some genetic analyses. Y. Jian and M. Li performed EM analyses. X. Wang, Q. Zhang, and Y. Li wrote the manuscript.
Data availability
Strains and reagents generated in this study would be made available upon reasonable request.
References
- Appelqvist, H., Wäster P., Kågedal K., and Öllinger K.. 2013. The lysosome: From waste bag to potential therapeutic target. J. Mol. Cell Biol. 5:214–226. 10.1093/jmcb/mjt022 [DOI] [PubMed] [Google Scholar]
- Bajaj, L., Lotfi P., Pal R., Ronza A.D., Sharma J., and Sardiello M.. 2019. Lysosome biogenesis in health and disease. J. Neurochem. 148:573–589. 10.1111/jnc.14564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio, A., and Bonifacino J.S.. 2020. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 21:101–118. 10.1038/s41580-019-0185-4 [DOI] [PubMed] [Google Scholar]
- Boyle, K.B., and Randow F.. 2013. The role of ‘eat-me’ signals and autophagy cargo receptors in innate immunity. Curr. Opin. Microbiol. 16:339–348. 10.1016/j.mib.2013.03.010 [DOI] [PubMed] [Google Scholar]
- Brandt, S., and Jentsch T.J.. 1995. ClC-6 and ClC-7 are two novel broadly expressed members of the CLC chloride channel family. FEBS Lett. 377:15–20. 10.1016/0014-5793(95)01298-2 [DOI] [PubMed] [Google Scholar]
- Cantuti-Castelvetri, L., Fitzner D., Bosch-Queralt M., Weil M.T., Su M., Sen P., Ruhwedel T., Mitkovski M., Trendelenburg G., Lütjohann D., et al. 2018. Defective cholesterol clearance limits remyelination in the aged central nervous system. Science. 359:684–688. 10.1126/science.aan4183 [DOI] [PubMed] [Google Scholar]
- Chakraborty, K., Leung K., and Krishnan Y.. 2017. High lumenal chloride in the lysosome is critical for lysosome function. Elife. 6:e28862. 10.7554/eLife.28862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub, N., Benachenhou N., Rajapurohitam V., Pata M., Ferron M., Frattini A., Villa A., and Vacher J.. 2003. Grey-lethal mutation induces severe malignant autosomal recessive osteopetrosis in mouse and human. Nat. Med. 9:399–406. 10.1038/nm842 [DOI] [PubMed] [Google Scholar]
- Cigic, B., and Pain R.H.. 1999. Location of the binding site for chloride ion activation of cathepsin C. Eur. J. Biochem. 264:944–951. 10.1046/j.1432-1327.1999.00697.x [DOI] [PubMed] [Google Scholar]
- Deriy, L.V., Gomez E.A., Zhang G., Beacham D.W., Hopson J.A., Gallan A.J., Shevchenko P.D., Bindokas V.P., and Nelson D.J.. 2009. Disease-causing mutations in the cystic fibrosis transmembrane conductance regulator determine the functional responses of alveolar macrophages. J. Biol. Chem. 284:35926–35938. 10.1074/jbc.M109.057372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di, A., Brown M.E., Deriy L.V., Li C., Szeto F.L., Chen Y., Huang P., Tong J., Naren A.P., Bindokas V., et al. 2006. CFTR regulates phagosome acidification in macrophages and alters bactericidal activity. Nat. Cell Biol. 8:933–944. 10.1038/ncb1456 [DOI] [PubMed] [Google Scholar]
- DiCiccio, J.E., and Steinberg B.E.. 2011. Lysosomal pH and analysis of the counter ion pathways that support acidification. J. Gen. Physiol. 137:385–390. 10.1085/jgp.201110596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, D., Williamson C.D., Cole N.B., and Donaldson J.G.. 2012. Pitstop 2 is a potent inhibitor of clathrin-independent endocytosis. PLoS One. 7:e45799. 10.1371/journal.pone.0045799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutzler, R., Campbell E.B., Cadene M., Chait B.T., and MacKinnon R.. 2002. X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of anion selectivity. Nature. 415:287–294. 10.1038/415287a [DOI] [PubMed] [Google Scholar]
- Evans, T.C., and Hunter C.P.. 2005. Translational Control of Maternal RNAs. In WormBook. WormBook. pp. 1–11. doi: 10.1895/wormbook.1.34.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbacher, N., Bastholm L., Kirkegaard-Sørensen T., Rafn B., Bøttzauw T., Nielsen C., Weber E., Shirasawa S., Kallunki T., and Jäättelä M.. 2008. Sensitization to the lysosomal cell death pathway by oncogene-induced down-regulation of lysosome-associated membrane proteins 1 and 2. Cancer Res. 68:6623–6633. 10.1158/0008-5472.CAN-08-0463 [DOI] [PubMed] [Google Scholar]
- Forgac, M. 2007. Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8:917–929. 10.1038/nrm2272 [DOI] [PubMed] [Google Scholar]
- Fukuda, M. 1991. Lysosomal membrane glycoproteins. Structure, biosynthesis, and intracellular trafficking. J. Biol. Chem. 266:21327–21330. 10.1016/S0021-9258(18)54636-6 [DOI] [PubMed] [Google Scholar]
- Gee, K., Zamora D., Horm T., George L., Upchurch C., Randall J., Weaver C., Sanford C., Miller A., Hernandez S., et al. 2017. Regulators of lysosome function and dynamics in Caenorhabditis elegans. G3. 7:991–1000. 10.1534/g3.116.037515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Sintes, R., Ledesma M.D., and Boya P.. 2016. Lysosomal cell death mechanisms in aging. Ageing Res. Rev. 32:150–168. 10.1016/j.arr.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Graves, A.R., Curran P.K., Smith C.L., and Mindell J.A.. 2008. The Cl-/H+ antiporter ClC-7 is the primary chloride permeation pathway in lysosomes. Nature. 453:788–792. 10.1038/nature06907 [DOI] [PubMed] [Google Scholar]
- Guo, P., Hu T., Zhang J., Jiang S., and Wang X.. 2010. Sequential action of Caenorhabditis elegans Rab GTPases regulates phagolysosome formation during apoptotic cell degradation. Proc. Natl. Acad. Sci. USA. 107:18016–18021. 10.1073/pnas.1008946107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, M., Hsu A.L., Dillin A., and Kenyon C.. 2005. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 1:119–128. 10.1371/journal.pgen.0010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper, D., Planells-Cases R., Fuhrmann J.C., Scheel O., Zeitz O., Ruether K., Schmitt A., Poët M., Steinfeld R., Schweizer M., et al. 2005. Loss of the chloride channel ClC-7 leads to lysosomal storage disease and neurodegeneration. EMBO J. 24:1079–1091. 10.1038/sj.emboj.7600576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard, T., Roth A.G., Petersen N.H., Mahalka A.K., Olsen O.D., Moilanen I., Zylicz A., Knudsen J., Sandhoff K., Arenz C., et al. 2010. Hsp70 stabilizes lysosomes and reverts Niemann-Pick disease-associated lysosomal pathology. Nature. 463:549–553. 10.1038/nature08710 [DOI] [PubMed] [Google Scholar]
- Kornak, U., Kasper D., Bösl M.R., Kaiser E., Schweizer M., Schulz A., Friedrich W., Delling G., and Jentsch T.J.. 2001. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell. 104:205–215. 10.1016/S0092-8674(01)00206-9 [DOI] [PubMed] [Google Scholar]
- Lange, P.F., Wartosch L., Jentsch T.J., and Fuhrmann J.C.. 2006. ClC-7 requires Ostm1 as a beta-subunit to support bone resorption and lysosomal function. Nature. 440:220–223. 10.1038/nature04535 [DOI] [PubMed] [Google Scholar]
- Leisle, L., Ludwig C.F., Wagner F.A., Jentsch T.J., and Stauber T.. 2011. ClC-7 is a slowly voltage-gated 2Cl(-)/1H(+)-exchanger and requires Ostm1 for transport activity. EMBO J. 30:2140–2152. 10.1038/emboj.2011.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Chen B., Zou W., Wang X., Wu Y., Zhao D., Sun Y., Liu Y., Chen L., Miao L., et al. 2016. The lysosomal membrane protein SCAV-3 maintains lysosome integrity and adult longevity. J. Cell Biol. 215:167–185. 10.1083/jcb.201602090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima, I., Takahashi A., Omori H., Kimura T., Takabatake Y., Saitoh T., Yamamoto A., Hamasaki M., Noda T., Isaka Y., and Yoshimori T.. 2013. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32:2336–2347. 10.1038/emboj.2013.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, J.K., Reilly T.J., Zeitman B.B., and Ellis S.. 1966. Cathepsin C: A chloride-requiring enzyme. Biochem. Biophys. Res. Commun. 24:771–775. 10.1016/0006-291X(66)90392-5 [DOI] [PubMed] [Google Scholar]
- McMahon, L., Muriel J.M., Roberts B., Quinn M., and Johnstone I.L.. 2003. Two sets of interacting collagens form functionally distinct substructures within a Caenorhabditis elegans extracellular matrix. Mol. Biol. Cell. 14:1366–1378. 10.1091/mbc.e02-08-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, R., Li M., Zhang Q., Yang C., and Wang X.. 2020. An ECM-to-nucleus signaling pathway activates lysosomes for C. elegans larval development. Dev. Cell. 52:21–37.e5. 10.1016/j.devcel.2019.10.020 [DOI] [PubMed] [Google Scholar]
- Mindell, J.A. 2012. Lysosomal acidification mechanisms. Annu. Rev. Physiol. 74:69–86. 10.1146/annurev-physiol-012110-142317 [DOI] [PubMed] [Google Scholar]
- Neiss, W.F. 1984. A coat of glycoconjugates on the inner surface of the lysosomal membrane in the rat kidney. Histochemistry. 80:603–608. 10.1007/BF02400979 [DOI] [PubMed] [Google Scholar]
- Nylandsted, J., Gyrd-Hansen M., Danielewicz A., Fehrenbacher N., Lademann U., Høyer-Hansen M., Weber E., Multhoff G., Rohde M., and Jäättelä M.. 2004. Heat shock protein 70 promotes cell survival by inhibiting lysosomal membrane permeabilization. J. Exp. Med. 200:425–435. 10.1084/jem.20040531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuma, S., Moriyama Y., and Takano T.. 1982. Identification and characterization of a proton pump on lysosomes by fluorescein-isothiocyanate-dextran fluorescence. Proc. Natl. Acad. Sci. USA. 79:2758–2762. 10.1073/pnas.79.9.2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix, A., Wang Y., Smith H.E., Lee C.Y., Calidas D., Lu T., Smith J., Schmidt H., Krause M.W., and Seydoux G.. 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics. 198:1347–1356. 10.1534/genetics.114.170423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos, C., Kravic B., and Meyer H.. 2020. Repair or lysophagy: Dealing with damaged lysosomes. J. Mol. Biol. 432:231–239. 10.1016/j.jmb.2019.08.010 [DOI] [PubMed] [Google Scholar]
- Papadopoulos, C., and Meyer H.. 2017. Detection and clearance of damaged lysosomes by the endo-lysosomal damage response and Lysophagy. Curr. Biol. 27:R1330–R1341. 10.1016/j.cub.2017.11.012 [DOI] [PubMed] [Google Scholar]
- Platt, F.M., Boland B., and van der Spoel A.C.. 2012. The cell biology of disease: Lysosomal storage disorders: The cellular impact of lysosomal dysfunction. J. Cell Biol. 199:723–734. 10.1083/jcb.201208152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressey, S.N., O’Donnell K.J., Stauber T., Fuhrmann J.C., Tyynelä J., Jentsch T.J., and Cooper J.D.. 2010. Distinct neuropathologic phenotypes after disrupting the chloride transport proteins ClC-6 or ClC-7/Ostm1. J. Neuropathol. Exp. Neurol. 69:1228–1246. 10.1097/NEN.0b013e3181ffe742 [DOI] [PubMed] [Google Scholar]
- Rudnik, S., and Damme M.. 2021. The lysosomal membrane-export of metabolites and beyond. FEBS J. 288:4168–4182. 10.1111/febs.15602 [DOI] [PubMed] [Google Scholar]
- Saftig, P., Schröder B., and Blanz J.. 2010. Lysosomal membrane proteins: Life between acid and neutral conditions. Biochem. Soc. Trans. 38:1420–1423. 10.1042/BST0381420 [DOI] [PubMed] [Google Scholar]
- Saha, S., Prakash V., Halder S., Chakraborty K., and Krishnan Y.. 2015. A pH-independent DNA nanodevice for quantifying chloride transport in organelles of living cells. Nat. Nanotechnol. 10:645–651. 10.1038/nnano.2015.130 [DOI] [PubMed] [Google Scholar]
- Savini, M., Folick A., Lee Y.T., Jin F., Cuevas A., Tillman M.C., Duffy J.D., Zhao Q., Neve I.A., Hu P.W., et al. 2022. Lysosome lipid signalling from the periphery to neurons regulates longevity. Nat. Cell Biol. 24:906–916. 10.1038/s41556-022-00926-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrecker, M., Korobenko J., and Hite R.K.. 2020. Cryo-EM structure of the lysosomal chloride-proton exchanger CLC-7 in complex with OSTM1. Elife. 9:e59555. 10.7554/eLife.59555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segatori, L. 2014. Impairment of homeostasis in lysosomal storage disorders. IUBMB Life. 66:472–477. 10.1002/iub.1288 [DOI] [PubMed] [Google Scholar]
- Serrano-Puebla, A., and Boya P.. 2016. Lysosomal membrane permeabilization in cell death: New evidence and implications for health and disease. Ann. N. Y. Acad. Sci. 1371:30–44. 10.1111/nyas.12966 [DOI] [PubMed] [Google Scholar]
- Skowyra, M.L., Schlesinger P.H., Naismith T.V., and Hanson P.I.. 2018. Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science. 360:360. 10.1126/science.aar5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber, T., and Jentsch T.J.. 2013. Chloride in vesicular trafficking and function. Annu. Rev. Physiol. 75:453–477. 10.1146/annurev-physiol-030212-183702 [DOI] [PubMed] [Google Scholar]
- Steinberg, B.E., Huynh K.K., Brodovitch A., Jabs S., Stauber T., Jentsch T.J., and Grinstein S.. 2010. A cation counterflux supports lysosomal acidification. J. Cell Biol. 189:1171–1186. 10.1083/jcb.200911083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi-Kiya, S., Zhou K., and Jin Y.. 2016. Release-Dependent Feedback Inhibition by a Presynaptically Localized Ligand-Gated Anion Channel. eLife. 5:e21734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Y., Li Z., Hu W., Ren H., Tian E., Zhao Y., Lu Q., Huang X., Yang P., Li X., et al. 2010. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 141:1042–1055. 10.1016/j.cell.2010.04.034 [DOI] [PubMed] [Google Scholar]
- Wartosch, L., Fuhrmann J.C., Schweizer M., Stauber T., and Jentsch T.J.. 2009. Lysosomal degradation of endocytosed proteins depends on the chloride transport protein ClC-7. FASEB J. 23:4056–4068. 10.1096/fj.09-130880 [DOI] [PubMed] [Google Scholar]
- Weinert, S., Jabs S., Supanchart C., Schweizer M., Gimber N., Richter M., Rademann J., Stauber T., Kornak U., and Jentsch T.J.. 2010. Lysosomal pathology and osteopetrosis upon loss of H+-driven lysosomal Cl− accumulation. Science. 328:1401–1403. 10.1126/science.1188072 [DOI] [PubMed] [Google Scholar]
- Yu, Y., Gao S.M., Guan Y., Hu P.-W., Zhang Q., Liu J., Jing B., Zhao Q., Sabatini D.M., Abu-Remaileh M., et al. 2022. Organelle proteomic profiling reveals lysosomal heterogeneity in association with longevity. bioRxiv. (Preprint posted October 16, 2022). 10.1101/2022.10.16.512400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Liu Y., Zhang B., Zhou J., Li T., Liu Z., Li Y., and Yang M.. 2020. Molecular insights into the human CLC-7/Ostm1 transporter. Sci. Adv. 6:eabb4747. 10.1126/sciadv.abb4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is the source file for Fig. 4.
is the source file for Fig. 8.
is the source file for Fig. S4.
is the source file for Fig. S5.
Data Availability Statement
Strains and reagents generated in this study would be made available upon reasonable request.