Expression of N-terminal truncated cMyBPC is sufficient to modulate systolic function in vivo, demonstrating that these domains have an important functional role. Understanding of region-specific function of cMyBPC can be leveraged to manipulate various aspects of cardiac function.
Abstract
Cardiac myosin binding protein C (cMyBPC) is an 11-domain sarcomeric protein (C0–C10) integral to cardiac muscle regulation. In vitro studies have demonstrated potential functional roles for regions beyond the N-terminus. However, the in vivo contributions of these domains are mostly unknown. Therefore, we examined the in vivo consequences of expression of N-terminal truncated cMyBPC (C3C10). Neonatal cMyBPC−/− mice were injected with AAV9-full length (FL), C3C10 cMyBPC, or saline, and echocardiography was performed 6 wk after injection. We then isolated skinned myocardium from virus-treated hearts and performed mechanical experiments. Our results show that expression of C3C10 cMyBPC in cMyBPC−/− mice resulted in a 28% increase in systolic ejection fraction compared to saline-injected cMyBPC−/− mice and a 25% decrease in left ventricle mass-to-body weight ratio. However, unlike expression of FL cMyBPC, there was no prolongation of ejection time compared to saline-injected mice. In vitro mechanical experiments demonstrated that functional improvements in cMyBPC−/− mice expressing C3C10 were primarily due to a 35% reduction in the rate of cross-bridge recruitment at submaximal Ca2+ concentrations when compared to hearts from saline-injected cMyBPC−/− mice. However, unlike the expression of FL cMyBPC, there was no change in the rate of cross-bridge detachment when compared to saline-injected mice. Our data demonstrate that regions of cMyBPC beyond the N-terminus are important for in vivo cardiac function, and have divergent effects on cross-bridge behavior. Elucidating the molecular mechanisms of cMyBPC region-specific function could allow for development of targeted approaches to manipulate specific aspects of cardiac contractile function.
Introduction
Cardiac myosin binding protein C (cMyBPC) is a multidomain sarcomeric protein that is integral to the regulation of cardiac muscle. It is made up of 11 sequential domains, C0 through C10, joined by interdomain linkers. The linker between domains C1 and C2 is termed the motif (or M) domain, which contains several phosphorylatable serines that are particularly important in modulating the function of cMyBPC (Gautel et al., 1995; Gresham et al., 2014; Mamidi et al., 2017). The importance of cMyBPC in regulating cardiac function is highlighted by the fact that deficiency or lack of cMyBPC results in significant cardiac dysfunction. Mutations that result in decreased expression of cMyBPC or mutated cMyBPC that is incorporated into sarcomeres can lead to hypertrophic cardiomyopathy (HCM) in humans (Alfares et al., 2015; Jacques et al., 2008; Suay-Corredera and Alegre-Cebollada, 2022). Mice genetically engineered to lack cMyBPC similarly display a hypertrophic cardiomyopathy phenotype with significant cardiac dysfunction (Harris et al., 2002). The dysfunction is a result of the loss of functional cMyBPC itself, rather than a side effect of the resultant hypertrophy, as indicated by conditional knock-down of cMyBPC by Giles et al. (2019). Cardiac dysfunction in cMyBPC−/− mice is characterized by hypercontractile cross-bridge kinetics at the fiber level, which translates to truncated ejection time at the organ level (Harris et al., 2002; Stelzer et al., 2006; Tong et al., 2008; Mamidi et al., 2014; Gresham and Stelzer, 2016; Li et al., 2018).
The N-terminal portion of cMyBPC consisting of domains C0 through C2 has been extensively studied and is known to modulate contractile function via its M-domain phosphorylation sites (Heling et al., 2020; Moss et al., 2015; Suay-Corredera and Alegre-Cebollada, 2022). These N-terminal domains (NTDs) are known to activate thin filaments while inhibiting thick filaments in heart muscle (Kampourakis et al., 2014; Mun et al., 2014). Previous studies have shown that AAV9-driven cardiac expression of the cMyBPC fragment consisting of domains C0 through C2 (C0C2) is sufficient to ameliorate systolic and diastolic dysfunction in cMyBPC−/− mice (Li et al., 2020). Improved cardiac function in this case was attributable to normalization of hypercontractile cross-bridge kinetics, which led to prolonged ejection time and increased ejection fraction in vivo (Li et al., 2020).
In contrast to the N-terminus, relatively little is known about the functional contributions made by the remainder of cMyBPC. Domains C8 through C10 at the C-terminus are known to anchor the protein to the thick filament through binding with titin and light meromyosin (Okagaki et al., 1993; Freiburg and Gautel, 1996; Gilbert et al., 1999; Miyamoto et al., 1999). Recent studies have revealed high-affinity interaction sites for myosin S1 and S2 in central regions of cMyBPC (Ponnam and Kampourakis, 2022). These in vitro studies using cMyBPC lacking the N-terminal domains (i.e., Δ-C0C2) demonstrate that the Δ-C0C2 fragment binds to myosin S1 with similar affinity to full-length cMyBPC, which is an order of magnitude higher than the affinity of C0C2 itself for myosin S1 (Ponnam and Kampourakis, 2022). Interest in central domains of cMyBPC has been further fueled by recent findings demonstrating sites for post-translational modification outside of the N-terminus, which may impact molecular structure and function (Doh et al., 2022a; Doh et al., 2022b).
Investigations of cMyBPC that lacks N-terminal domains have indicated that there may be functional relevance to the remainder of the protein beyond anchoring it to the thick filament. However, the in vivo roles of central and C-terminal domains and how they contribute to cardiac function remain largely unknown. Therefore, in this study, we performed in vivo gene transfer to express C3C10 cMyBPC lacking the N-terminus in the heart and determined its effects on whole organ and myofilament function. We present evidence that C3C10 reconstitution by gene transfer into cMyBPC−/− mice improves in vivo cardiac function, which is reflected by parallel improvement in cardiac morphology, and mechanistically distinct from functional improvements mediated by reconstitution with full length cMyBPC.
Materials and methods
Transgenic animals
cMyBPC−/− mice were generated previously (Harris et al., 2002) on a 129SV background. These mice are known to develop a severe HCM-related phenotype shortly after birth that remains stable over the course of their lifespan.
AAV9 production and administration
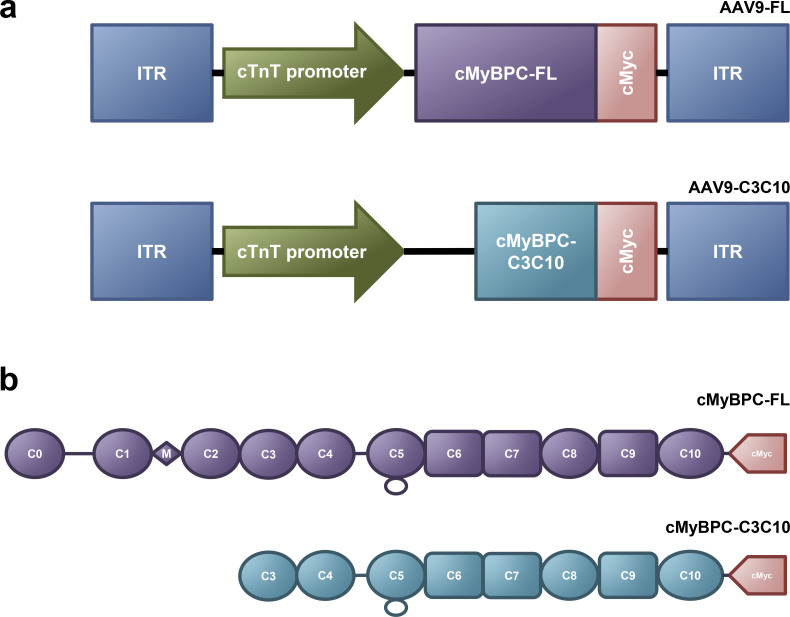
AAV9-pseudotyped vectors expressing FL cMyBPC and C3C10 cMyBPC were designed under the control of a truncated chicken cardiac troponin T (cTnT) promoter (Prasad et al., 2011). The FL vector contained the rabbit β-globin poly-A sequence, while the C3C10 vector contained the bovine growth hormone poly-A sequence. The C3C10 vector contained the woodchuck hepatitis virus post-transcriptional regulatory element. Vectors were produced by the Penn Vector Core at the University of Pennsylvania as described previously (Lock et al., 2010). Schematic representations of the vectors are displayed in Fig. 1.
Figure 1.
Vector design and construct maps. (a) Schematic of AAV9 vectors containing cMyBPC-FL and –C3C10 with C-terminal c-Myc tag under the control of a truncated chicken cardiac troponin T promoter. (b) Schematics of full-length (FL) cMyBPC and truncated cMyBPC consisting of domains C3 through C10 (C3C10), including the C-terminal c-Myc tag used to identify exogenously expressed cMyBPC.
Within the first 36 h following birth, neonatal pups were anesthetized by placing them on ice. Pups were kept on ice for a maximum of 5 min. 2 × 1011 genome copies (GC)/mouse were delivered by injection into the temporal facial vein using a 33 G needle. Pups weighed ∼1 g at birth, making this dose effectively 2 × 1014 GC/kg. Injections were carried out under standard biosafety level 1 (BSL 1) and animal biosafety level 1 (ABSL 1) conditions. Following injection, pups were placed in soiled bedding from their home cage on heat support. Pups were returned to their home cage once recovered.
In vivo cardiac function
Noninvasive transthoracic echocardiography was performed on 6-wk-old mice as previously described (Li et al., 2018) using a Vevo 3100 ultrasonography 40 MHz transducer (Visual Sonics) to acquire 2-D B mode, M mode, and Doppler images of the left ventricle. Mice were anesthetized by 1–2% isoflurane during the procedure, and heart rate was managed using heat support.
Images were analyzed using Vevo Lab 3.1.1 (Visual Sonics). Left ventricular (LV) ejection fraction (EF), anterior and posterior wall thickness, chamber diameter, and LV mass were measured over 3–4 cardiac cycles from midventricular short axis M-mode images. AET was measured from apical four chamber pulsed wave Doppler images with the marquee placed over the region of peak inflow and outflow. Speckle tracking echocardiography-based longitudinal strain parameters were measured from parasternal long-axis B mode video using the semi-automated Vevo Strain module of Vevo Lab.
Western blot analysis of myocardial samples
Myocardial protein samples were prepared from hearts that had been excised from mice anesthetized with 4% isoflurane and immediately flash-frozen in liquid nitrogen. Hearts were stored at −80°C. Frozen ventricular tissues were thawed in relaxing solution with protease and phosphatase inhibitors (cOmplete ULTRA EDTA-free and PhosStop tablets, MilliporeSigma) at 4°C and mechanically homogenized. Homogenates were skinned with 1% Triton-X100 (Thermo Fisher Scientific) for 15 min at room temperature, then centrifuged for 5 min at 10,000 rcf. The supernatant was discarded, and the pellet consisting of the myofibril fraction was resuspended in relaxing solution with protease and phosphatase inhibitors. Protein concentrations of the resuspended myofibril fractions were determined by BCA colorimetric assay (Pierce BCA Protein Assay Kit, Thermo Fisher Scientific). Samples were prepared by diluting to 1 µg/μl protein, reducing with Laemmli Buffer, and heating to 95°C for 10 min. 10 µg of solubilized myofibrils were loaded and electrophoretically separated using TruPAGE precast 4–12% gels (MilliporeSigma) or hand-cast 17% gels at 180 V for 45 min. Proteins were transferred to PVDF membranes at 0.2 A for 2.5 h at 4°C. Protein loading was evaluated using No-Stain Protein Labeling Reagent (catalog A44449; Invitrogen). Western blot analysis was conducted using 1:200 mouse anti-myc primary antibodies (catalog sc-40; Santa Cruz Biotechnology Inc,) or 1:1,000 mouse anti-cMyBPC antibodies (catalog sc-137237; Santa Cruz Biotechnology Inc.), followed by 1:5,000 HRP-conjugated mouse IgG κ binding protein (catalog sc-516102; Santa Cruz Biotechnology Inc.).
Immunohistochemistry (IHC) staining and fluorescence imaging
Immunofluorescent characterization of construct localization within myofibrils was performed as previously described (Doh et al., 2019). In brief: tissue sections from the left ventricular midwall were thawed in relaxing solution at 4°C and then transferred to rigor buffer for homogenization. Myofibrils were skinned with 1% Triton X-100 (Thermo Fisher Scientific) for 30 min at room temperature. Myofibrils were centrifuged at 1,500 rcf for 2 min, transferred to fresh solution, and resuspended four times. Final resuspension was done using rigor buffer containing 1 mg/ml bovine serum albumin to prevent clumping. 25 μl of fresh skinned myofibrils was dispensed onto each of two 15-mm-diameter circles on fluorescent antibody microscope slides. Myofibrils were allowed to dry partially, washed three times with 150 μl of PBS, and fixed with 35 μl of 4% paraformaldehyde. After washing again, slides were blocked for 30 min at room temperature with 50 μl of 5% goat serum. Primary antibodies were diluted in 5% goat serum and incubated on the slides for 1 h at room temperature. Slides were washed, then secondary antibodies diluted in 5% goat serum were added and incubated for 30 min at room temperature. After removing the secondary antibodies and washing the slides again, coverslips were mounted using ProLong Gold Antifade Mountant (Invitrogen). Slides were allowed to cure for at least 48 h before imaging with an Olympus Fluoview-1000 confocal microscope using appropriate excitation and emission wavelengths.
Primary antibodies were as follows. From Santa Cruz Biotechnology Inc.: mouse anti-myc 1:50 (catalog; sc-40). From MilliporeSigma: rabbit anti-ACTN2 1:50 (catalog SAB4503474).
Secondary antibodies were as follows. From Invitrogen: goat anti-mouse Alexa Fluor 488 1:500 (catalog A-21121); goat anti-rabbit Texas red 1:500 (catalog T2767).
Preparation of skinned myocardial and Ca2+ solutions for steady state and cross-bridge kinetic experiments
Skinned myocardial samples were prepared as previously described (Doh et al., 2019; Mamidi et al., 2021). In brief, frozen ventricular tissue was homogenized in relaxing solution and chemically skinned by incubating in 1% Triton X-100 (Thermo Fisher Scientific) for 1 h.
Ca2+ solutions were prepared as previously described (Li et al., 2020). First, all solutions were prepared by admixing the following reagents: 14.5 mM creatine phosphate, 7 mM EGTA, and 20 mM imidazole. Then, 65.45 mM KCl, 7.01 mM CaCl2, 5.27 mM MgCl2, and 4.81 mM ATP were added to the solution to make a maximal activating solution (pCa 4.5; pCa = −log [Ca2+]free), whereas 72.45 mM KCl, 0.02 mM CaCl2, 5.42 mM MgCl2, and 4.76 mM ATP were added to the solution to make relaxing solution (pCa 9.0). The pH and ionic strength of the solution were set to 7.0 and 180 mM, respectively. A range of pCa solutions (pCa 6.2 to 5.6) containing varying amounts of [Ca2+]free were then prepared by mixing appropriate volumes of pCa 9.0 and 4.5 stock solutions.
Measurement of steady state and dynamic contractile properties of the skinned myocardial
Skinned multicellular ventricular samples were incubated in a relaxing solution, and myocardial preparations ∼100 µm in width and 400 µm in length were selected for the study. The myocardial preparations were securely fixed between a motor arm (model 315C; Aurora Scientific Inc.) and a force transducer (model 403 A; Aurora Scientific Inc.), as previously described (Mamidi et al., 2016). Changes in motor arm positions and force transducer signals were sampled at 2.0 kHz using custom-built SL control software. All the experiments were performed at room temperature (∼24 ± 1°C).
At the beginning of each experiment, the ventricular preparation’s sarcomere length (SL) was stretched to ≈2.1 µm (Stelzer et al., 2006). Force–pCa relationships were characterized by measuring the forces generated by skinned myocardial preparations in a series of pCa solutions that produced submaximal to maximal forces. The apparent cooperativity of force generation was estimated from the steepness of the Hill plot transformation of the force–pCa relationships. The force–pCa data were fit using the equation where P is the force generated at a given pCa, Po is the force generated at maximal Ca2+ activation, nH is the Hill coefficient, and k is the pCa needed to elicit half-maximal force (pCa50).
The stretch-activation properties were analyzed as previously described (Doh et al., 2019; Mamidi et al., 2021; Stelzer et al., 2006). The forces of the skinned myocardial preparations were activated in a pCa 6.1 solution which generated steady-state forces of ≈30% of maximal Ca2+-activated force. Once the myocardial preparations reached a steady-state force, they were subjected to a sudden 2% stretch of their initial muscle length (ML), held for 8 s, and returned to their initial ML.
The stretch activation responses of the cardiac muscle stretch activation have been described in detail elsewhere (Li et al., 2020; Mamidi et al., 2016). In brief, the muscle shows an instantaneous rise in the force once a sudden 2% stretch in ML is produced. This is due to a sudden strain of elastic elements within the strongly bound cross-bridges (phase 1). Then the force decreases rapidly (phase 2) due to the detachment of strained cross-bridges (krel), which are subsequently recruited to a force generating state (kdf; phase 3). Krel and kdf were estimated by fitting a single exponential to the time course of force decay using the equation: where “a” is the amplitude of the single exponential phase and k is the rate constant of the force decay or force redevelopment.
Statistics
All data are reported as mean ± SEM. For quantifying Western blots, one-tailed unpaired t tests were used to compare expression between two groups. For echo parameters, one-way analysis of variance (ANOVA) was performed followed by post hoc Tukey’s multiple comparison test to test the significance of the mean values of multiple groups. For mechanical studies, data from multiple myocardial preparations from each heart were analyzed in a hierarchical, nested manner using one-way repeated measured ANOVA followed by Tukey’s multiple comparisons test. R version 4.2.1 via RStudio version 2022.07.2 + 576 and GraphPad Prism 9.4.1 were used to perform statistical analysis. Statistical significance was set at P < 0.05.
Study approval
All procedures involving animal care and handling, including viral injections and BSL-1/ABSL-1 practices, were reviewed and approved by the Case Western Reserve University Animal Care and Use Committee.
Results
Expression of AAV9-derived constructs
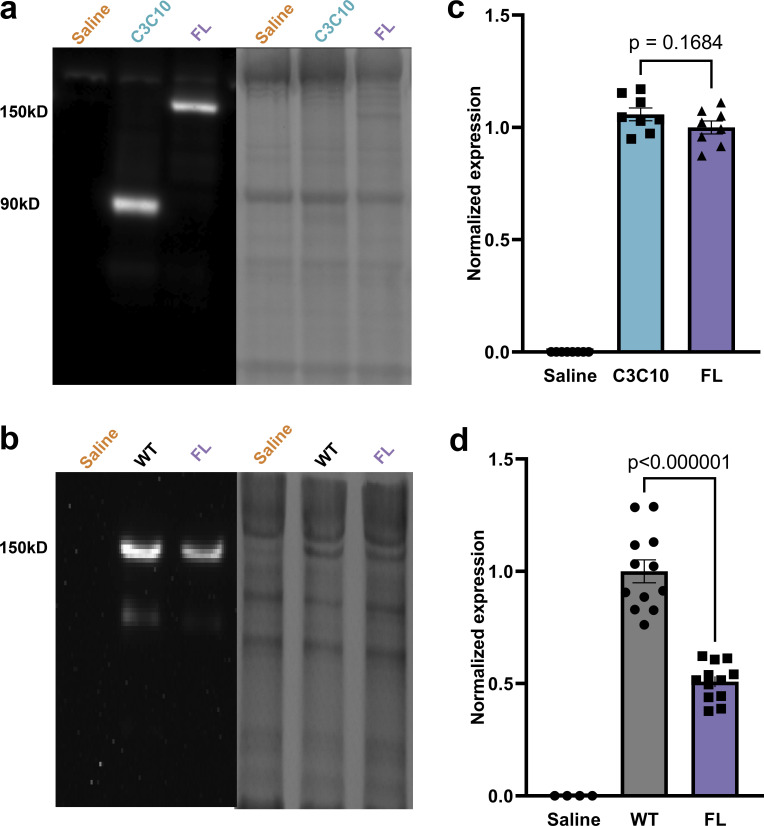
To determine the effectiveness of AAV9 gene transfer, the expression of cMyc-tagged AAV9-derived constructs was quantified in myocardial samples taken from AAV9-FL−, AAV9-C3C10−, and saline-injected cMyBPC−/− mice at 6 wk post injection. The absence of cMyc-containing constructs in saline-treated hearts was confirmed by Western blot (Fig. 2). Overall expression of cMyc-tagged protein was normalized to the average expression of cMyc-tagged FL in FL-treated hearts. There was no statistically significant difference in the average content of cMyc-tagged protein in FL- and C3C10-treated hearts.
Figure 2.
Expression of AAV9-delivered constructs. (a) Expression of AAV9-FL and –C3C10 as determined by immunoblotting (left) against the c-Myc epitope, normalized to total protein loading (right). (b) Expression of endogenously produced cMyBPC (WT) versus AAV9-FL as determined by immunoblotting (left) against the N-terminus of cMyBPC, normalized to total protein loading (right). (c) Plot of mean ± SEM c-Myc expression normalized to the average c-Myc expression of the FL group, for n = 8 hearts per group. (d) Plot of mean ± SEM cMyBPC expression normalized to the average cMyBPC expression of the WT group. n = 12 hearts per group. Comparisons between C3C10 and FL, and WT and FL, made by one-tailed unpaired t test.
The goal of our study was to understand differences in the function of FL cMyBPC and cMyBPC lacking the N-terminal domains. A tolerability study revealed that expression of the C3C10 construct was limited by dose-dependent mortality in AAV9-C3C10–injected mice. As such, AAV9-FL was dosed so that expression of FL and C3C10 were equivalent. In this way, differences between groups are attributable to differences between the constructs, rather than differences in protein abundance. When dosed to match expression of C3C10 at the maximum tolerable level of AAV9-C3C10, expression of AAV9-FL was found to be 50.81 ± 2.41% as compared to endogenously produced WT cMyBPC (P < 0.000001; Fig. 2).
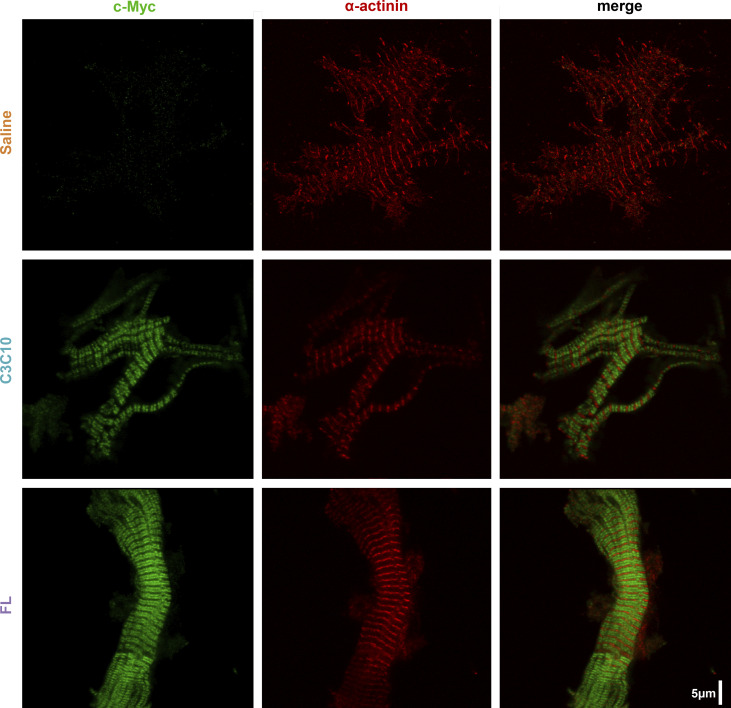
Incorporation of constructs into sarcomeres
The localization of AAV9-delivered constructs within the sarcomere was characterized by IHC. In mammalian-striated muscle, cMyBPC is known to be localized to the C zone within the A band of sarcomeres in a characteristic doublet pattern formed by adjacent C zones separated on one side by the Z disc and on the other side by the M line. Appropriately, AAV9-FL- and AAV9-C3C10-treated myocardium showed incorporation of cMyc-tagged constructs restricted to doublets between each Z disc. Doublets were absent in myocardium from saline-treated animals. (Fig. 3)
Figure 3.
Incorporation of constructs into sarcomeres. Representative IHC of c-Myc- and α-actinin-stained myofibrils from respective hearts showing incorporation of AAV9-derived constructs in doublets between z-lines.
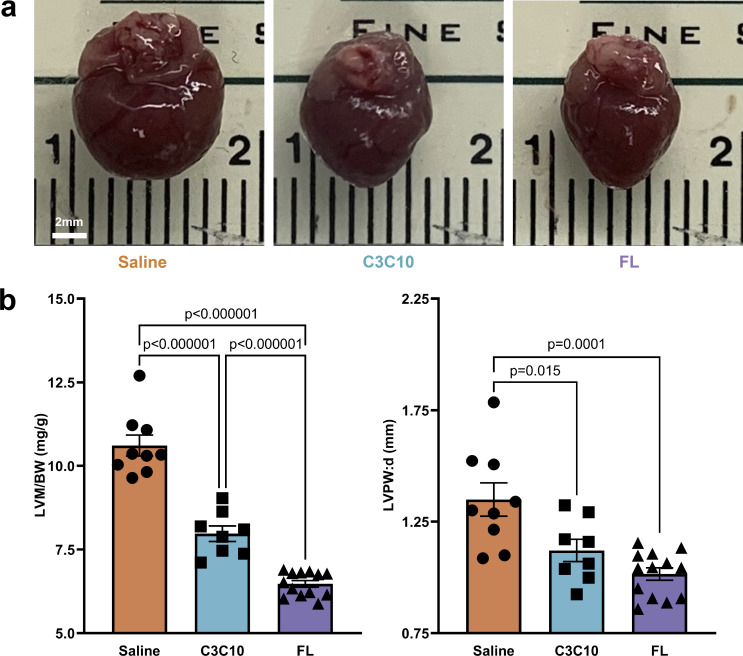
Effect of AAV9-derived constructs on cardiac morphology
Cardiac morphology was evaluated 6 wk after FL or C3C10 gene transfer. cMyBPC−/− mice are known to develop cardiac hypertrophy shortly after birth (Harris et al., 2002; de Lange et al., 2013). As expected, hearts from saline-treated cMyBPC−/− mice exhibited severe cardiomegaly, while hearts from FL-treated mice were smaller and exhibited a normalized overall morphology. Hearts from cMyBPC−/− mice treated with C3C10 were intermediate in size and morphology, indicating morphologic improvement in comparison to saline-treated hearts.
Differences in apparent size of hearts between groups were borne out by comparing ratios of left ventricle mass to body weight (LVM/BW). Saline-treated mice had a significantly higher LVM/BW ratio at 6 wk (10.61 ± 0.31) than FL-treated mice (6.48 ± 0.10), while C3C10-treated mice had an intermediate ratio (7.97 ± 0.23), P < 0.000001 (Fig. 4 and Table 1).
Figure 4.
Morphology normalized by C3C10. (a) Representative images of hearts from mice injected with saline or AAV9-FL or –C3C10. (b) Plots of mean ± SEM LVM/BW and LVPW:d. Group comparison made by one-way ANOVA with post-hoc Tukey’s multiple comparisons test.
Table 1.
n indicates number of biological replicates, i.e., number of mice
| Saline (n = 9) | C3C10 (n = 8) | FL (n = 13) | |
|---|---|---|---|
| Age (wk) | 6.32 ± 0.08 | 6.34 ± 0.05 | 6.25 ± 0.03 |
| HR (BPM) | 451 ± 2 | 448 ± 2 | 449 ± 2 |
| BW (g) | 17.44 ± 0.38 | 17.75 ± 0.59 | 16.31 ± 0.52 |
| LVM (mg) | 184.92 ± 5.93a, P < 0.000001 | 141.35 ± 6.05 aP < 0.000001, bP < 0.000001 |
105.57 ± 3.77, P < 0.000001 |
| LVM/BW (mg/g) | 10.61 ± 0.31a, P < 0.000001 | 7.97 ± 0.23 aP < 0.000001, bP < 0.000001 |
6.48 ± 0.10b, P < 0.000001 |
| LVAW:d (mm) | 1.19 ± 0.04a, P = 0.0046 | 1.065 ± 0.07 | 0.998 ± 0.03b, P = 0.0046 |
| LVPW:d (mm) | 1.35 ± 0.08a, P = 0.00010 | 1.12 ± 0.05b, P = 0.015 | 1.02 ± 0.03b, P = 0.00010 |
| Diameter:d (mm) | 4.05 ± 0.13a, P = 0.00024 | 3.93 ± 0.06a, P = 0.0056 | 3.50 ± 0.07b, P = 0.00024 |
| FS (%) | 15.97 ± 0.54a, P < 0.000001 | 21.15 ± 0.73b, P = 0.00006 | 22.91 ± 0.64b, P < 0.000001 |
| EF (%) | 34.02 ± 1.01a, P < 0.000001 | 43.51 ± 1.27b, P = 0.000033 | 46.9 ± 1.12b, P < 0.000001 |
| GLS (%) | −8.52 ± 0.65a, P < 0.000001 | −11.07 ± 0.26 aP = 0.019, bP = 0.0052 |
−13.06 ± 0.43b, P < 0.000001 |
| AET (ms) | 32.18 ± 1.08a, P = 0.000023 | 35.00 ± 0.96a, P = 0.0014 | 41.62 ± 1.08b, P = 0.000023 |
| rLSR (s−1) | 3.51 ± 0.34a, P = 0.00070 | 4.32 ± 0.52a, P = 0.019 | 6.35 ± 0.52b, P = 0.00070 |
Values are expressed as mean ± SEM., analyzed by one-way ANOVA with post hoc Tukey’s multiple comparisons test. HR, heart rate; BW, body weight; LVM, left ventricle mass; LVAW:d, left ventricle anterior wall thickness in diastole; LVPW:d, left ventricle posterior wall thickness in diastole; Diameter:d, left ventricle diameter in diastole; FS, fractional shortening; EF, ejection fraction; GLS, global longitudinal strain; AET, aortic ejection time; rLSR, reverse longitudinal strain rate.
Significantly different compared to FL.
Significantly different compared to saline.
Echocardiographic evaluation provided further evidence for reduction of cardiac hypertrophy following gene transfer of FL and C3C10 cMyBPC. Hearts from FL- and C3C10-treated mice had decreased LV posterior wall thickness as measured in diastole (LVPW:d) when compared to hearts from saline-treated mice (P = 0.00010 and P = 0.015, respectively; Fig. 4 and Table 1). Measurement of LV anterior wall thickness in diastole (LVAW:d) followed a similar trend, though differences between the C3C10-treated mice and either control did not reach significance (P = 0.49 vs. FL and 0.12 vs. saline; Table 1). In contrast, chamber diameter as measured in diastole (D:d) was decreased in hearts from FL-treated mice (P = 0.0056), but not different between hearts from C3C10- and saline-treated mice (P = 0.60; Table 1).
Effect of AAV9-driven cMyBPC expression on in vivo cardiac function
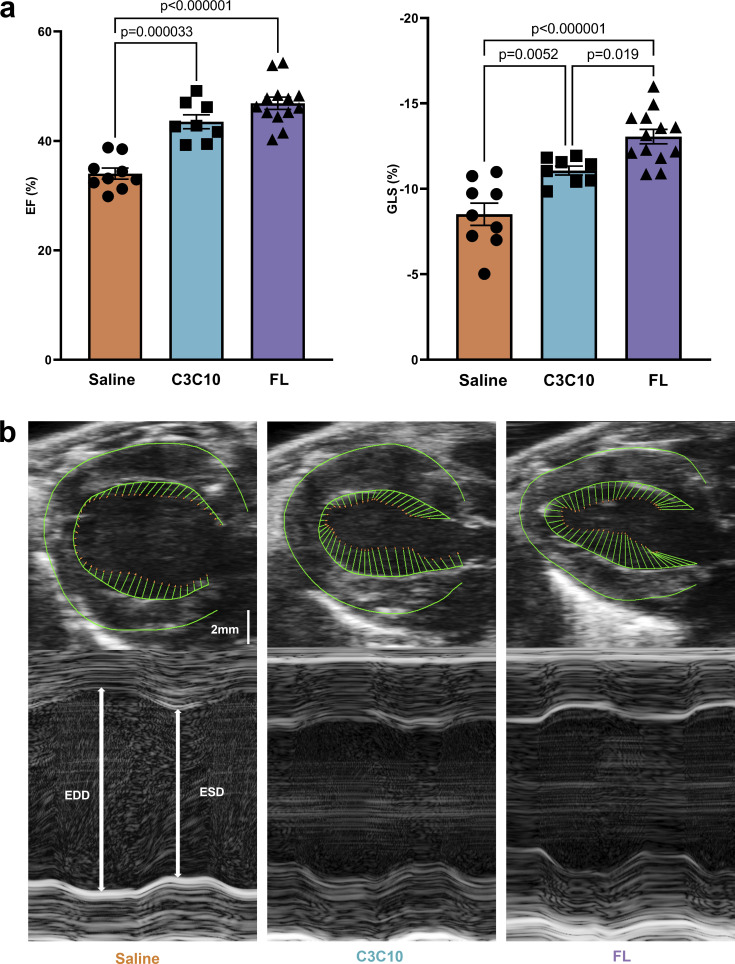
In vivo cardiac function was also evaluated via echocardiography. Saline-treated cMyBPC−/− mice displayed severely depressed systolic function evidenced by low EF (34.02 ± 1.01%). The EF was significantly improved to 46.87 ± 1.11% (P < 0.000001) in cMyBPC−/− mice following FL gene transfer. cMyBPC−/− mice expressing C3C10 cMyBPC also displayed a significant improvement in ejection fraction compared to saline treated cMyBPC−/− mice (43.51 ± 1.27%, P = 0.000033), which was not statistically different from that of FL cMyBPC-expressing mice (P = 0.12; Fig. 5 and Table 1).
Figure 5.
Contractile function improved by C3C10. (a) Plots of mean ± SEM EF (left) and GLS (right). Group comparison made by one-way ANOVA with post-hoc Tukey’s multiple comparisons test. (b) Representative PSLAX B-mode images (top) with vectors indicating longitudinal strain along the endocardial border. Representative SAX M-mode images (bottom) indicating end diastolic diameter (EDD) and end systolic diameter (ESD).
In vivo function was further characterized by performing strain analysis on echocardiograms using speckle tracking software. Global longitudinal strain (GLS), which is a sensitive indicator of systolic function (Cho et al., 2009; Park et al., 2018), significantly improved in mice expressing FL cMyBPC (−13.06 ± 0.43%) compared with saline-treated cMBPC−/− mice (−8.52 ± 0.6%, P < 0.000001). cMyBPC−/− mice expressing C3C10 cMyBPC had a mean GLS of −11.07 ± 0.26%, which was intermediate and statistically distinct from both the saline- and FL cMyBPC-treated groups (P = 0.0052 and 0.019, respectively; Fig. 5 and Table 1).
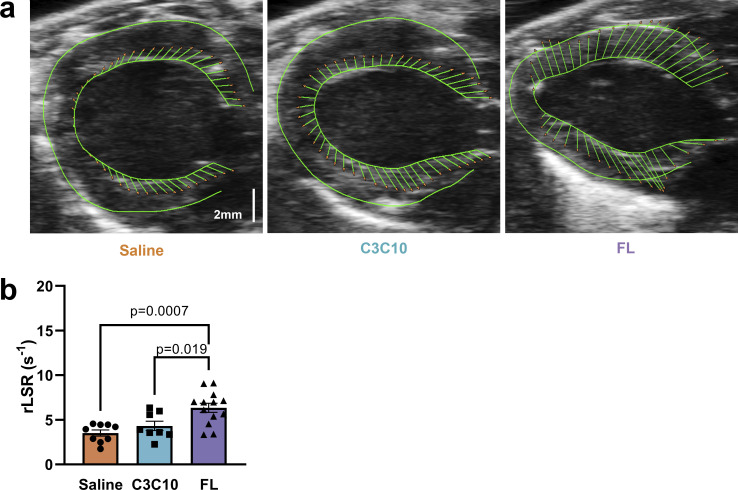
Reverse longitudinal strain rate (rLSR) is an indicator of diastolic function that reflects the peak rate of strain development during early LV filling (Schnelle et al., 2018). Diastolic dysfunction in saline-treated cMyBPC−/− mice was indicated by depressed rLSR of 3.50 ± 0.34 s−1. Relative improvement in diastolic function in the FL-treated group was indicated by an increased rLSR of 6.35 ± 0.52 s−1 (P = 0.00070; Fig. 6 and Table 1). The C3C10-treated group had an average rLSR of 4.31 ± 0.51 s−1, which was not statistically different from the saline-treated group (P = 0.54).
Figure 6.
Diastolic impairment in the absence of NTDs. (a) Representative para-sternal long axis (PSLAX) B-mode images with vectors indicating longitudinal strain along the endocardial border. (b) Plot of mean ± SEM rLSR. Group comparison made by one-way ANOVA with post-hoc Tukey’s multiple comparisons test.
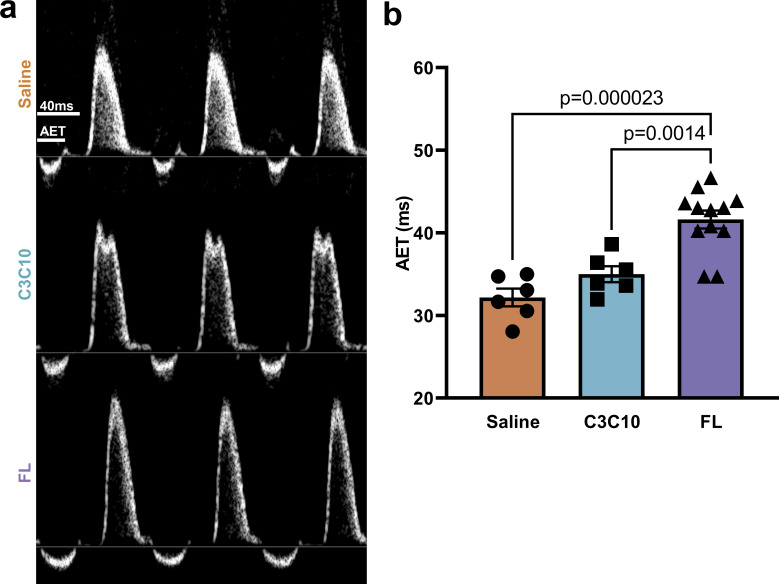
Abbreviated systolic ejection is the hallmark of cMyBPC ablation (Nagayama et al., 2007; Tong et al., 2008; Gresham and Stelzer, 2016; Li et al., 2020), and accordingly, aortic ejection time (AET) in saline-treated cMyBPC−/− mice was 32.18 ± 1.08 ms. In FL-treated mice, AET was significantly prolonged to 41.62 ± 1.08 ms (P = 0.000023); however, mice expressing C3C10 cMyBPC had an AET of 35.00 ± 0.96 ms, which was not statistically different from the saline-treated group (P = 0.30; Fig. 7 and Table 1).
Figure 7.
Truncated ejection in the absence of NTDs. (a) Representative pulsed wave Doppler traces depicting the velocity of blood flow through a region of interest positioned at the area of peak inflow during diastole. Velocities above the baseline represent ventricular inflow, and below baseline represent ventricular outflow. (b) AET is indicated, and plotted as mean ± SEM. Group comparison made by one-way ANOVA with post-hoc Tukey’s multiple comparisons test.
Effect of FL and C3C10 cMyBPC on myofilament Ca2+ sensitivity (pCa50) and cooperativity of force development (nH)
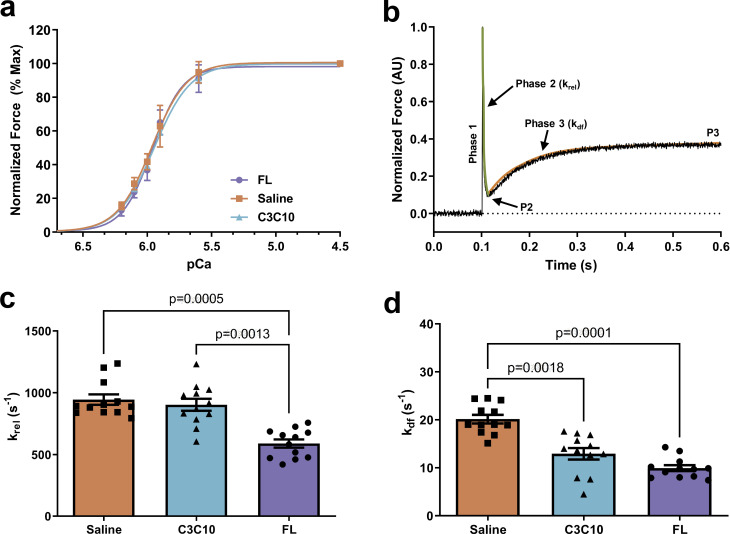
Myocardial preparations were sequentially bathed in pCa 9.0 (relaxing solution), pCa 4.5 (maximal activating solution), and Ca2+ solutions containing increasing Ca2+ levels (pCa 6.2–5.6) to yield submaximal forces. All three experimental groups (Saline, C3C10, and FL) exhibited similar steady-state force generation and cooperativity of force as demonstrated in the force–pCa relationship (Fig. 8 and Table 2). The pCa50 did not show differences among the groups. The Ca2+-independent force (Fmin) was analyzed at pCa 9.0 and did not alter among the groups (Table 2). The maximal Ca2+-activated force (Fmax) was measured at pCa 4.5 and did not show statistically significant differences between groups (Table 2).
Figure 8.
Effect of FL and C3C10 cMyBPC on myofilament pCa50 and cooperativity of force development (nH), and effect of FL and C3C10 cMyBPC on dynamic stretch activation parameters. (a) Force–pCa relationship of FL, C3C10, and saline-treated myocardium analyzed by plotting normalized forces generated at a range of pCa. No significant differences were observed between the groups (b). Representative force trace in response to a sudden 2% stretch in ML in an isometrically contracting FL myocardium preparation at an SL of 2.1 µm. The green line represents the rate of cross-bridge detachment (krel) and the orange line represents the rate of cross-bridge recruitment (kdf). (c) The rate of cross-bridge detachment, krel. (d) The rate of cross-bridge recruitment, kdf. Values are expressed as mean ± SEM. Data from n = 4 hearts per group with three fiber preparations per heart was analyzed in a hierarchical, nested manner using one-way repeated measures ANOVA followed by Tukey’s multiple comparisons test.
Table 2.
Steady-state and dynamic contractile parameters measured in chemically skinned myocardial preparations from cMyBPC−/− mice treated with saline, AAV9-FL, and AAV9-C3C10
| Saline | C3C10 | FL | |
|---|---|---|---|
| Fmin (mN/mm2) | 0.71 ± 0.08 | 0.60 ± 0.09 | 0.55 ± 0.09 |
| Fmax (mN/mm2) | 11.35 ± 0.72 | 10.73 ± 0.74 | 8.48 ± 0.74 |
| nH | 3.5 ± 0.3 | 3.1 ± 0.3 | 3.8 ± 0.3 |
| pCa50 | 5.97 ± 0.02 | 5.93 ± 0.01 | 5.94 ± 0.02 |
| krel (s−1) | 945 ± 147a, P = 0.0005 | 903 ± 168a, P = 0.0013 | 590 ± 115b, P = 0.0005 |
| kdf (s−1) | 20 ± 3a, P = 0.0001 | 13 ± 4b, P = 0.0018 | 10 ± 2b, P = 0.0001 |
Values are expressed as mean ± SEM. n = 4 hearts with three preparations per heart, analyzed by one-way repeated measures ANOVA with post hoc Tukey’s multiple comparisons test. Fmin, Ca2+-independent force analyzed at pCa9.0; Fmax, Ca2+-activated maximal force measured at pCa 4.5; nH, Hill coefficient of the force–pCa relationship; pCa50, pCa required for the generation of half-maximal force; krel, rate of cross-bridge detachment; kdf, rate of cross-bridge recruitment.
Significantly different compared to FL.
Significantly different compared to saline.
Effect of FL and C3C10 cMyBPC on dynamic stretch activation parameters
The rate of stretch-induced cross-bridge detachment (krel) and cross-bridge recruitment (kdf) was analyzed by fitting a single exponential of the rapid force decay in phase 2 and force reuptake at the end of phase 2 to the point of a plateau region of phase 3 (P3), respectively (Fig. 8). The skinned myocardial preparations isolated from Saline and C3C10 hearts exhibited significantly accelerated krel rates when compared to the FL group (P = 0.0005 and P = 0.0013, respectively; Fig. 8 and Table 2). The rate of kdf of the Saline group showed a significantly higher rate when compared to the FL (P = 0.0001), whereas kdf in the C3C10 group was not significantly different from that of FL (P = 0.15; Fig. 8 and Table 2).
Discussion
We performed in vivo gene transfer of N-terminal truncated cMyBPC in cMyBPC null mice to investigate the effects of cMyBPC domains beyond the well-studied N-terminus on cardiac function. While it is established that NTDs play a vital role in modulating myocardial force generation and myofilament function (Heling et al., 2020; Moss et al., 2015; Suay-Corredera and Alegre-Cebollada, 2022), our data demonstrate an important role for cMyBPC domains beyond C0C2 that modulate in vivo contractile function. In vivo gene transfer of C3C10 cMyBPC in cMyBPC−/− hearts improved systolic cardiac function compared to untreated cMyBPC−/− hearts, with corresponding improvements in wall thickness and overall left ventricle mass. Mechanical studies in skinned myocardium isolated from hearts expressing C3C10 cMyBPC support a distinct mechanism for the mode of regulation compared to expression of NTD-containing full-length cMyBPC.
Neonatal gene transfer of C3C10 cMyBPC in cMyBPC−/− mice resulted in systolic functional improvements evidenced by increased EF and global longitudinal strain (GLS; Fig. 5 and Table 1). EF in cMyBPC−/− mice expressing C3C10 cMyBPC was normalized to the same degree as in cMyBPC−/− mice expressing FL cMyBPC (Fig. 5 and Table 1). GLS, which is a more sensitive measure of systolic performance, was improved in cMyBPC−/− mice expressing C3C10 when compared to saline-treated cMyBPC−/− mice, however, not to the full extent as the improvement seen in mice expressing FL cMyBPC (Fig. 5 and Table 1). These functional improvements in systolic function were sufficient to improve cardiac morphology as evidenced by improvements in markers of hypertrophy, like LVPW:d and the ratio of LVM/BW, compared to saline injected cMyBPC−/− mice (Fig. 4 and Table 1). Whereas mice expressing C3C10 had reduced wall thickness compared to saline-injected cMyBPC−/− mice, LV chamber diameter was not different (Table 1). This indicates that morphologic improvement in cMyBPC−/− mice expressing C3C10 did not reach the extent of improvement in mice expressing FL, as the geometry of the chamber failed to normalize. This agrees with the extent of systolic functional improvement measured by EF and GLS: while EF was not statistically different between cMyBPC−/− mice expressing FL and those expressing C3C10, GLS was improved to a greater extent in the FL group as compared to the C3C10 group. It is established that GLS is a more sensitive measure of cardiac dysfunction in failing hearts (Cho et al., 2009; Park et al., 2018); therefore, more subtle differences in systolic function between mice expressing C3C10 cMyBPC and FL cMyBPC could be detected.
There is substantial evidence that cMyBPC is important for prolonging late systolic ejection and thereby increasing cardiac output (Nagayama et al., 2007; Tong et al., 2008; Gresham and Stelzer, 2016; Li et al., 2020). Our data suggests that late systolic prolongation is mainly due to the NTDs of cMyBPC (i.e., C0C2), because AET (a measure of systolic ejection time) in cMyBPC−/− mice expressing C3C10 did not significantly differ from saline-treated cMyBPC−/− mice (Fig. 7 and Table 1). It has been proposed that the N-terminus of cMyBPC prolongs ejection via enhanced actin binding in late systole when calcium levels have already waned, by maintaining cooperative cross-activation of the thin filament (Herron et al., 2006; Razumova et al., 2006; Mun et al., 2014; Kampourakis et al., 2014; Inchingolo et al., 2019). Furthermore, our data are consistent with previous studies that have shown that in vivo altered N-terminal behavior modulates ejection time (van Dijk et al., 2018; Li et al., 2020).
In addition to systolic function, it has been increasingly recognized that cMyBPC also plays a role in modulating diastolic function (Rosas et al., 2015; Gresham and Stelzer, 2016) through variable phosphorylation levels, which alters rates of cross-bridge cycling and detachment (Stelzer et al., 2006; Gresham et al., 2014; Mamidi et al., 2021). It is also possible that the presence of cMyBPC NTDs improves diastolic function by improving cardiac mechanical function. Previously, we showed (Li et al., 2020) that in vivo expression of C0C2 cMyBPC via gene transfer in cMyBPC−/− mice resulted in improved systolic function as evidenced by increased GLS and normalized AET, as well as improved diastolic function as evidenced by improved rLSR. In the present study, cMyBPC−/− mice expressing C3C10 cMyBPC (i.e., cMyBPC lacking NTDs) showed improved GLS compared to saline-injected cMyBPC−/− mice, but rLSR remained similarly impaired (Fig. 6). Importantly, AET remained shortened in cMyBPC−/− mice expressing C3C10 cMyBPC to the same extent as saline-injected cMyBPC−/− mice, whereas it was prolonged in mice expressing FL cMyBPC (which contains NTDs).
In the heart, diastolic function and systolic function are interdependent. In vitro, the maximum lengthening velocity of cardiac muscle fibers is dependent on the extent of fiber shortening (Tamiya et al., 1979). This is reflected in vivo by the fact that augmentation of cardiac contractility (and thus the extent of fiber shortening reached at the end of systole) during exercise results in corresponding enhancement of ventricular relaxation (Nonogi et al., 1988). Contraction during systole creates a compressive force on sarcomere “springs” as they shorten below their resting slack length, storing potential energy which is released as recoil during diastole (Udelson et al., 1990; Opdahl et al., 2009; Opdahl et al., 2012). The restoring force stored during systole is critical for early diastolic recoil and ventricular filling (Nonogi et al., 1988). The giant sarcomeric protein titin is considered the main determinant of cardiomyocyte passive tension responsible for the spring-like behavior of sarcomeres (Granzier et al., 2002; Granzier and Labeit, 2004; LeWinter et al., 2007; Tonino et al., 2017). Thus, increased fiber shortening during systole contributes to increased lengthening during diastole (Opdahl et al., 2009). Prolongation of ejection time by cMyBPC NTDs results in decreased end systolic volume as a result of greater fiber shortening, thereby enhancing cardiac systolic function (Li et al., 2020). Therefore, the absence of NTDs in the hearts expressing C3C10 cMyBPC and corresponding truncation of ejection time may explain the lack of improvement in diastolic function in these mice.
The molecular hallmark of the absence of cMyBPC in the sarcomere on the fiber level is a dramatic acceleration of the stretch activation including the rates of cross-bridge cycling (krel) and recruitment (kdf; Stelzer et al., 2006). Re-expression of full-length cMyBPC in the cMyBPC-null heart has been shown to slow stretch activation cross-bridge kinetics as demonstrated by a normalization of krel and kdf (Merkulov et al., 2012; Li et al., 2018; Li et al., 2020), which underlie improved in vivo function because of reduced ATP utilization, improved mechanical efficiency, systolic performance, and coupling between systolic ejection and diastolic filling. Previous studies have demonstrated that NTDs also impact both the rate of cross-bridge recruitment as well as the rate of detachment, albeit to a lesser extent compared with full length cMyBPC (Li et al., 2020). This difference between the effects of full length cMyBPC and the effects of cMyBPC NTDs alone suggests that the remaining domains could also play a role in modulating cross-bridge kinetics. In this study, we present evidence that the C3C10 construct slows the rate of cross-bridge recruitment without impacting the rate of cross-bridge cycling. Although there was no effect on krel, C3C10 cMyBPC expression significantly slowed kdf compared to saline injected cMyBPC−/− hearts, which was similar to the effect of expression of FL cMyBPC (Fig. 8 and Table 2). Cooperative cross-bridge recruitment modulates myocardial force generation in early systole (Moss et al., 2015), thus, slowed kdf may underlie the in vivo functional improvement in C3C10 cMyBPC hearts, especially in systolic parameters such as EF and GLS.
The rate of cross-bridge recruitment is, in part, dependent on the pool of myosin heads available for recruitment to the thin filament. One factor that could impact myosin recruitment is the ratio of myosin heads in the super-relaxed and disordered-relaxed states (SRX and DRX, respectively), which are biochemically defined by the rate of ATP hydrolysis by the myosin ATPase. The rate of ATP hydrolysis is dramatically slowed in the SRX compared to the DRX (Stewart et al. 2010; Hooijman et al., 2011). Cardiac myosin heads in the SRX may represent a cardioprotective mechanism for decreasing the metabolic load on the heart (Hooijman et al., 2011), and/or may represent a reserve population that can be called upon to increase contractility (McNamara et al., 2015). In humans, HCM-causing mutations in cMyBPC are associated with a decreased population of myosin heads in the SRX (McNamara et al., 2017), suggesting that stabilization of the SRX may be a key function of cMyBPC. One possibility is that the central domains of cMyBPC could modulate the SRX by stabilizing the auto-inhibitory binding of myosin heads to each other and the thick filament backbone (termed the interacting heads motif or IHM; Nag et al., 2017; Rahmanseresht et al., 2021). Indeed, recent work by Ponnam and Kampourakis (2022) has shown that the sites with the highest affinity for myosin S1 are in the central domains of cMyBPC, which are suggested to be able to crosslink the two myosin heads of a given myosin molecule and thus stabilize its folded state, which is hypothesized to be the structural basis behind the SRX (Hooijman et al., 2011). Region-specific modulation of the SRX is an area of active investigation. Lynch et al. (2021) found that cMyBPC lacking N-terminal domains did not affect the proportion of myosin heads in the SRX state. Interestingly, a recent study using the same mouse model (Nelson et al., 2023) indicated that the absence of cMyBPC N-terminal domains resulted in an SRX landscape that was similar to that in myofibrils from cMyBPC-null mice. These discrepant results could be explained by differences in experimental methodology including tension applied to myofibrils that could impact the SRX state (Nelson et al., 2023). Our model shows that N-terminal truncated C3C10 cMyBPC is capable of slowing the rate of cross-bridge recruitment, but factors other than SRX are known to mediate the impact of cMyBPC on myosin recruitment as well, including phosphorylation of the M-domain (Mamidi et al., 2016). We have shown previously that NTDs alone can decrease the rate of cross-bridge recruitment (Li et al., 2020). As it stands, the mechanism by which C3C10 cMyBPC results in a decreased rate of cross-bridge recruitment and the role that potential modulation of the SRX plays therein remains to be determined and warrants further study.
The rate of cross-bridge detachment is thought to be modulated by interaction of the NTDs of cMyBPC with actin (Razumova et al., 2006; Walcott et al., 2015). In vivo, the rate of cross-bridge detachment is closely correlated with ejection time (Gresham and Stelzer, 2016; van Dijk et al., 2018) because prolonged activation of the thin filament delays cross-bridge cooperative deactivation and slows the rate of force relaxation (McDonald 2011). A drag-activation-competition model has been proposed to describe the multifactorial mechanism by which NTDs may alter the rate of cross-bridge detachment and thus the longevity of cross-bridges (described in detail in Walcott et al., 2015). In short: NTDs can bind to actin and displace tropomyosin, thus activating the thin filament and uncovering myosin binding sites. This is most relevant at low calcium, when thin filament activation is disfavored by calcium-dependent troponin regulation. However, binding of NTDs to actin creates steric interference and renders some actin binding sites unavailable to myosin. The significance of this effect is dependent on the concentration of NTDs, with increasing concentration leading to increased competition for binding sites. Finally, simultaneous binding of NTDs to the thick and thin filament imposes a viscous drag on filament sliding, slowing the rate. Given that the C3C10 construct lacks NTDs, it is not surprising that incorporation of C3C10 into the sarcomere did not impact the rate of cross-bridge detachment following rapid stretch, and correspondingly, ejection time remained truncated in vivo (Fig. 7). Therefore, our findings are in agreement with previous work demonstrating the necessity of NTDs in regulating the rate of cross-bridge detachment (Li et al., 2020).
Absence or paucity of cMyBPC in the sarcomere can result in HCM and potentially severe cardiac dysfunction in humans. It is of clinical interest to understand the specific functions of the different regions of cMyBPC that make its absence so unfavorable. The function of the N-terminus has been well characterized, and in this study, we shed further light on the function of the remainder of the protein. In this cMyBPC−/− model of a severely compromised hypercontractile heart, suppression of myosin activation alone by the C3C10 portion of cMyBPC yielded improvements in cardiac morphology and function in vivo. This suggests that there is therapeutic potential in targeting specific elements of sarcomere function. Understanding the function of different regions of cMyBPC, and therefore the implications of mutations in these different regions, will allow for better tailoring of therapy. Additionally, understanding of different regions of cMyBPC could illuminate new therapeutic targets directed at specific elements of the cross-bridge cycle.
Acknowledgments
Henk L. Granzier served as editor.
All experiments were performed at the Department of Physiology and Biophysics, Case Western Reserve University, Cleveland, OH.
This work was supported by the National Institutes of Health (grants R01-HL 146676, R01 HL114770, R01 HL153236, HIH GTRP RSA 2050).
Author contributions: K.L. Dominic and J.E. Stelzer contributed to the conception and design of the experiments. K.L. Dominic, J.B. Holmes, and J.E. Stelzer contributed to data analysis and data interpretation. K.L. Dominic, J. Choi, and J.E. Stelzer participated in writing and revising the manuscript. K.L. Dominic, J. Choi, J.B. Holmes, M. Singh, M.J. Majcher, and J.E. Stelzer participated in performing the experiments and data collection. All authors approved the final version of the manuscript.
Footnotes
This work is part of a special issue on Myofilament Function 2022.
Data availability
All data supporting this paper will be made available from the authors upon request.
Data availability
All data supporting this paper will be made available from the authors upon request.
References
- Alfares, A.A., Kelly, M.A., McDermott, G., Funke, B.H., Lebo, M.S., Baxter, S.B., Shen, J., McLaughlin, H.M., Clark, E.H., Babb, L.J., et al. 2015. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: Expanded panels offer limited additional sensitivity. Genet. med. 17:880–888. 10.1038/gim.2014.205 [DOI] [PubMed] [Google Scholar]
- Cho, G.Y., Marwick T.H., Kim H.S., Kim M.K., Hong K.S., and Oh D.J.. 2009. Global 2-dimensional strain as a new prognosticator in patients with heart failure. J. Am. Coll. Cardiol. 54:618–624. 10.1016/j.jacc.2009.04.061 [DOI] [PubMed] [Google Scholar]
- de Lange, W.J., Grimes A.C., Hegge L.F., and Ralphe J.C.. 2013. Ablation of cardiac myosin-binding protein-C accelerates contractile kinetics in engineered cardiac tissue. J. Gen. Physiol. 141:73–84. 10.1085/jgp.201210837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doh, C.Y., Li J., Mamidi R., and Stelzer J.E.. 2019. The HCM-causing Y235S cMyBPC mutation accelerates contractile function by altering C1 domain structure. Biochim. Biophys. Acta Mol. Basis Dis. 1865:661–677. 10.1016/j.bbadis.2019.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doh, C.Y., Bharambe N., Holmes J.B., Dominic K.L., Swanberg C.E., Mamidi R., Chen Y., Bandyopadhyay S., Ramachandran R., and Stelzer J.E.. 2022a. Molecular characterization of linker and loop-mediated structural modulation and hinge motion in the C4-C5 domains of cMyBPC. J. Struct. Biol. 214:107856. 10.1016/j.jsb.2022.107856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doh, C.Y., Dominic K.L., Swanberg C.E., Bharambe N., Willard B.B., Li L., Ramachandran R., and Stelzer J.E.. 2022b. Identification of phosphorylation and other post-translational modifications in the central C4C5 domains of murine cardiac myosin binding protein C. ACS Omega. 7:14189–14202. 10.1021/acsomega.2c00799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiburg, A., and Gautel M.. 1996. A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur. J. Biochem. 235:317–323. 10.1111/j.1432-1033.1996.00317.x [DOI] [PubMed] [Google Scholar]
- Gautel, M., Zuffardi O., Freiburg A., and Labeit S.. 1995. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: A modulator of cardiac contraction?. EMBO J. 14:1952–1960. 10.1002/j.1460-2075.1995.tb07187.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, R., Cohen J.A., Pardo S., Basu A., and Fischman D.A.. 1999. Identification of the A-band localization domain of myosin binding proteins C and H (MyBP-C, MyBP-H) in skeletal muscle. J. Cell Sci. 112:69–79. 10.1242/jcs.112.1.69 [DOI] [PubMed] [Google Scholar]
- Giles, J., Patel J.R., Miller A., Iverson E., Fitzsimons D., and Moss R.L.. 2019. Recovery of left ventricular function following in vivo reexpression of cardiac myosin binding protein C. J. Gen. Physiol. 151:77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzier, H.L., and Labeit S.. 2004. The giant protein titin: A major player in myocardial mechanics, signaling, and disease. Circ. Res. 94:284–295. 10.1161/01.RES.0000117769.88862.F8 [DOI] [PubMed] [Google Scholar]
- Granzier, H., Labeit D., Wu Y., and Labeit S.. 2002. Titin as a modular spring: Emerging mechanisms for elasticity control by titin in cardiac physiology and pathophysiology. J. Muscle Res. Cell Motil. 23:457–471. 10.1023/a:1023458406346 [DOI] [PubMed] [Google Scholar]
- Gresham, K.S., and Stelzer J.E.. 2016. The contributions of cardiac myosin binding protein C and troponin I phosphorylation to β-adrenergic enhancement of in vivo cardiac function. J. Physiol. 594:669–686. 10.1113/JP270959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham, K.S., Mamidi R., and Stelzer J.E.. 2014. The contribution of cardiac myosin binding protein-c Ser282 phosphorylation to the rate of force generation and in vivo cardiac contractility. J. Physiol. 592:3747–3765. 10.1113/jphysiol.2014.276022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S.P., Bartley C.R., Hacker T.A., McDonald K.S., Douglas P.S., Greaser M.L., Powers P.A., and Moss R.L.. 2002. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ. Res. 90:594–601. 10.1161/01.res.0000012222.70819.64 [DOI] [PubMed] [Google Scholar]
- Heling, L.W.H.J., Geeves M.A., and Kad N.M.. 2020. MyBP-C: One protein to govern them all. J. Muscle Res. Cell Motil. 41:91–101. 10.1007/s10974-019-09567-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron, T.J., Rostkova E., Kunst G., Chaturvedi R., Gautel M., and Kentish J.C.. 2006. Activation of myocardial contraction by the N-terminal domains of myosin binding protein-C. Circ. Res. 98:1290–1298. 10.1161/01.RES.0000222059.54917.ef [DOI] [PubMed] [Google Scholar]
- Hooijman, P., Stewart M.A., and Cooke R.. 2011. A new state of cardiac myosin with very slow ATP turnover: A potential cardioprotective mechanism in the heart. Biophys. J. 100:1969–1976. 10.1016/j.bpj.2011.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inchingolo, A.V., Previs S.B., Previs M.J., Warshaw D.M., and Kad N.M.. 2019. Revealing the mechanism of how cardiac myosin-binding protein C N-terminal fragments sensitize thin filaments for myosin binding. Proc. Natl. Acad. Sci. USA. 116:6828–6835. 10.1073/pnas.1816480116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques, A., Hoskins A.C., Kentish J.C., and Marston S.B.. 2008. From genotype to phenotype: A longitudinal study of a patient with hypertrophic cardiomyopathy due to a mutation in the MYBPC3 gene. J. Muscle Res. Cell Motil. 29:239–246. 10.1007/s10974-009-9174-0 [DOI] [PubMed] [Google Scholar]
- Kampourakis, T., Yan Z., Gautel M., Sun Y.B., and Irving M.. 2014. Myosin binding protein-C activates thin filaments and inhibits thick filaments in heart muscle cells. Proc. Natl. Acad. Sci. USA. 111:18763–18768. 10.1073/pnas.1413922112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, T.L., IV, Kumar M., McNamara J.W., Kuster D.W.D., Sivaguru M., Singh R.R., Previs M.J., Lee K.H., Kuffel G., Zilliox M.J., et al. 2021. Amino terminus of cardiac myosin binding protein-C regulates cardiac contractility. J. Mol. Cell. Cardiol. 156:33–44. 10.1016/j.yjmcc.2021.03.00 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeWinter, M.M., Wu, Y., Labeit, S., & Granzier, H. (2007). Cardiac titin: Structure, functions and role in disease. Clinica chimica acta. 375:1–9. 10.1016/j.cca.2006.06.035 [DOI] [PubMed] [Google Scholar]
- Li, J., Gresham K.S., Mamidi R., Doh C.Y., Wan X., Deschenes I., and Stelzer J.E.. 2018. Sarcomere-based genetic enhancement of systolic cardiac function in a murine model of dilated cardiomyopathy. Int. J. Cardiol. 273:168–176. 10.1016/j.ijcard.2018.09.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Mamidi R., Doh C.Y., Holmes J.B., Bharambe N., Ramachandran R., and Stelzer J.E.. 2020. AAV9 gene transfer of cMyBPC N-terminal domains ameliorates cardiomyopathy in cMyBPC-deficient mice. JCI Insight. 5:e130182. 10.1172/jci.insight.130182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock, M., Alvira M., Vandenberghe L.H., Samanta A., Toelen J., Debyser Z., and Wilson J.M.. 2010. Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum. Gene Ther. 21:1259–1271. 10.1089/hum.2010.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidi R., Gresham K.S., Stelzer J.E.. 2014. Length-dependent changes in contractile dynamics are blunted due to cardiac myosin binding protein-C ablation. Front Physiol. 5:461. 10.3389/fphys.2014.00461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidi, R., Gresham K.S., Verma S., and Stelzer J.E.. 2016. Cardiac myosin binding protein-C phosphorylation modulates myofilament length-dependent activation. Front. Physiol. 7:38. 10.3389/fphys.2016.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidi R., Gresham K.S., Li J., Stelzer J.E.. 2017. Cardiac myosin binding protein-C Ser302 phosphorylation regulates cardiac β-adrenergic reserve. Sci Adv. 3:e1602445. 10.1126/sciadv.1602445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidi, R., Holmes J.B., Doh C.Y., Dominic K.L., Madugula N., and Stelzer J.E.. 2021. cMyBPC phosphorylation modulates the effect of omecamtiv mecarbil on myocardial force generation. J. Gen. Physiol. 153:e202012816. 10.1085/jgp.202012816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, K.S. 2011. The interdependence of Ca2+ activation, sarcomere length, and power output in the heart. Pflugers Arch. 462:61–67. 10.1007/s00424-011-0949-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, J.W., Li A., Dos Remedios C.G., and Cooke R.. 2015. The role of super-relaxed myosin in skeletal and cardiac muscle. Biophys. Rev. 7:5–14. 10.1007/s12551-014-0151-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, J.W., Li A., Lal S., Bos J.M., Harris S.P., van der Velden J., Ackerman M.J., Cooke R., and Dos Remedios C.G.. 2017. MYBPC3 mutations are associated with a reduced super-relaxed state in patients with hypertrophic cardiomyopathy. PLoS One. 12:e0180064. 10.1371/journal.pone.0180064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkulov, S., Chen X., Chandler M.P., and Stelzer J.E.. 2012. In vivo cardiac myosin binding protein C gene transfer rescues myofilament contractile dysfunction in cardiac myosin binding protein C null mice. Circ. Heart Fail. 5:635–644. 10.1161/CIRCHEARTFAILURE.112.968941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, C.A., Fischman D.A., and Reinach F.C.. 1999. The interface between MyBP-C and myosin: Site-directed mutagenesis of the CX myosin-binding domain of MyBP-C. J. Muscle Res. Cell Motil. 20:703–715. 10.1023/a:1005513312939 [DOI] [PubMed] [Google Scholar]
- Moss, R.L., Fitzsimons D.P., and Ralphe J.C.. 2015. Cardiac MyBP-C regulates the rate and force of contraction in mammalian myocardium. Circ. Res. 116:183–192. 10.1161/CIRCRESAHA.116.300561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun, J.Y., Previs M.J., Yu H.Y., Gulick J., Tobacman L.S., Beck Previs S., Robbins J., Warshaw D.M., and Craig R.. 2014. Myosin-binding protein C displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc. Natl. Acad. Sci. USA. 111:2170–2175. 10.1073/pnas.1316001111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, S., Trivedi D.V., Sarkar S.S., Adhikari A.S., Sunitha M.S., Sutton S., Ruppel K.M., and Spudich J.A.. 2017. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat. Struct. Mol. Biol. 24:525–533. 10.1038/nsmb.3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayama, T., Takimoto E., Sadayappan S., Mudd J.O., Seidman J.G., Robbins J., and Kass D.A.. 2007. Control of in vivo left ventricular [correction] contraction/relaxation kinetics by myosin binding protein C: protein kinase A phosphorylation dependent and independent regulation. Circulation. 116:2399–2408. 10.1161/CIRCULATIONAHA.107.706523 [DOI] [PubMed] [Google Scholar]
- Nelson, S., Beck-Previs S., Sadayappan S., Tong C., and Warshaw D.M.. 2023. Myosin-binding protein C stabilizes, but is not the sole determinant of SRX myosin in cardiac muscle. J. Gen. Physiol. 155:e202213276. 10.1085/jgp.202213276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogi, H., Hess O.M., Ritter M., and Krayenbuehl H.P.. 1988. Diastolic properties of the normal left ventricle during supine exercise. Br. Heart J. 60:30–38. 10.1136/hrt.60.1.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki, T., Weber F.E., Fischman D.A., Vaughan K.T., Mikawa T., and Reinach F.C.. 1993. The major myosin-binding domain of skeletal muscle MyBP-C (C protein) resides in the COOH-terminal, immunoglobulin C2 motif. J. Cell Biol. 123:619–626. 10.1083/jcb.123.3.619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdahl, A., Remme E.W., Helle-Valle T., Lyseggen E., Vartdal T., Pettersen E., Edvardsen T., and Smiseth O.A.. 2009. Determinants of left ventricular early-diastolic lengthening velocity: Independent contributions from left ventricular relaxation, restoring forces, and lengthening load. Circulation. 119:2578–2586. 10.1161/CIRCULATIONAHA.108.791681 [DOI] [PubMed] [Google Scholar]
- Opdahl, A., Remme E.W., Helle-Valle T., Edvardsen T., and Smiseth O.A.. 2012. Myocardial relaxation, restoring forces, and early-diastolic load are independent determinants of left ventricular untwisting rate. Circulation. 126:1441–1451. 10.1161/CIRCULATIONAHA.111.080861 [DOI] [PubMed] [Google Scholar]
- Park, J.J., Park J.B., Park J.H., and Cho G.Y.. 2018. Global longitudinal strain to predict mortality in patients with acute heart failure. J. Am. Coll. Cardiol. 71:1947–1957. 10.1016/j.jacc.2018.02.064 [DOI] [PubMed] [Google Scholar]
- Ponnam, S., and Kampourakis T.. 2022. Microscale thermophoresis suggests a new model of regulation of cardiac myosin function via interaction with cardiac myosin-binding protein C. J. Biol. Chem. 298:101485. 10.1016/j.jbc.2021.101485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, K.M., Xu Y., Yang Z., Acton S.T., and French B.A.. 2011. Robust cardiomyocyte-specific gene expression following systemic injection of AAV: In vivo gene delivery follows a Poisson distribution. Gene Ther. 18:43–52. 10.1038/gt.2010.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmanseresht, S., Lee K.H., O’Leary T.S., McNamara J.W., Sadayappan S., Robbins J., Warshaw D.M., Craig R., and Previs M.J.. 2021. The N terminus of myosin-binding protein C extends toward actin filaments in intact cardiac muscle. J. Gen. Physiol. 153:e202012726. 10.1085/jgp.202012726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razumova, M.V., Shaffer J.F., Tu A.Y., Flint G.V., Regnier M., and Harris S.P.. 2006. Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay: Evidence for long-lived cross-bridges. J. Biol. Chem. 281:35846–35854. 10.1074/jbc.M606949200 [DOI] [PubMed] [Google Scholar]
- Rosas, P.C., Liu Y., Abdalla M.I., Thomas C.M., Kidwell D.T., Dusio G.F., Mukhopadhyay D., Kumar R., Baker K.M., Mitchell B.M., et al. 2015. Phosphorylation of cardiac myosin-binding protein-C is a critical mediator of diastolic function. Circ. Heart Fail. 8:582–594. 10.1161/CIRCHEARTFAILURE.114.001550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnelle, M., Catibog N., Zhang M., Nabeebaccus A.A., Anderson G., Richards D.A., Sawyer G., Zhang X., Toischer K., Hasenfuss G., et al. 2018. Echocardiographic evaluation of diastolic function in mouse models of heart disease. J. Mol. Cell. Cardiol. 114:20–28. 10.1016/j.yjmcc.2017.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer, J.E., Dunning S.B., and Moss R.L.. 2006. Ablation of cardiac myosin-binding protein-C accelerates stretch activation in murine skinned myocardium. Circ. Res. 98:1212–1218. 10.1161/01.RES.0000219863.94390.ce [DOI] [PubMed] [Google Scholar]
- Stewart, M.A., Franks-Skiba K., Chen S., and Cooke R.. 2010. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc. Natl. Acad. Sci. USA. 107:430–435. 10.1073/pnas.0909468107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suay-Corredera, C., and Alegre-Cebollada J.. 2022. The mechanics of the heart: Zooming in on hypertrophic cardiomyopathy and cMyBP-C. FEBS Lett. 596:703–746. 10.1002/1873-3468.14301 [DOI] [PubMed] [Google Scholar]
- Tamiya, K., Sugawara M., and Sakurai Y.. 1979. Maximum lengthening velocity during isotonic relaxation at preload in canine papillary muscle. Am. J. Physiol. 237:H83–H89. 10.1152/ajpheart.1979.237.1.H83 [DOI] [PubMed] [Google Scholar]
- Tong, C.W., Stelzer J.E., Greaser M.L., Powers P.A., and Moss R.L.. 2008. Acceleration of crossbridge kinetics by protein kinase A phosphorylation of cardiac myosin binding protein C modulates cardiac function. Circ. Res. 103:974–982. 10.1161/CIRCRESAHA.108.177683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonino, P., Kiss B., Strom J., Methawasin M., Smith J.E. III, Kolb J., Labeit S., and Granzier H.. 2017. The giant protein titin regulates the length of the striated muscle thick filament. Nat. Commun. 8:1041. 10.1038/s41467-017-01144-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udelson, J.E., Bacharach S.L., Cannon R.O. III, and Bonow R.O.. 1990. Minimum left ventricular pressure during beta-adrenergic stimulation in human subjects. Evidence for elastic recoil and diastolic “suction” in the normal heart. Circulation. 82:1174–1182. 10.1161/01.cir.82.4.1174 [DOI] [PubMed] [Google Scholar]
- van Dijk, S.J., Kooiker K.B., Napierski N.C., Touma K.D., Mazzalupo S., and Harris S.P.. 2018. Point mutations in the tri-helix bundle of the M-domain of cardiac myosin binding protein-C influence systolic duration and delay cardiac relaxation. J. Mol. Cell. Cardiol. 119:116–124. 10.1016/j.yjmcc.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walcott, S., Docken S., and Harris S.P.. 2015. Effects of cardiac Myosin binding protein-C on actin motility are explained with a drag-activation-competition model. Biophys. J. 108:10–13. 10.1016/j.bpj.2014.11.1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting this paper will be made available from the authors upon request.
All data supporting this paper will be made available from the authors upon request.