Abstract
Accurate intraoperative diagnosis is essential for providing safe and effective care during brain tumor surgery. Our standard-of-care diagnostic methods are time, resource, and labor intensive, which restricts access to optimal surgical treatments. To address these limitations, we propose an alternative workflow that combines stimulated Raman histology (SRH), a rapid optical imaging method, with deep learning-based automated interpretation of SRH images for intraoperative brain tumor diagnosis and real-time surgical decision support. Here, we present OpenSRH, the first public dataset of clinical SRH images from 300+ brain tumors patients and 1300+ unique whole slide optical images. OpenSRH contains data from the most common brain tumors diagnoses, full pathologic annotations, whole slide tumor segmentations, raw and processed optical imaging data for end-to-end model development and validation. We provide a framework for patch-based whole slide SRH classification and inference using weak (i.e. patient-level) diagnostic labels. Finally, we benchmark two computer vision tasks: multiclass histologic brain tumor classification and patch-based contrastive representation learning. We hope OpenSRH will facilitate the clinical translation of rapid optical imaging and real-time ML-based surgical decision support in order to improve the access, safety, and efficacy of cancer surgery in the era of precision medicine. Dataset access, code, and benchmarks are available at https://opensrh.mlins.org.
1. Introduction
The optimal surgical management of brain tumors varies widely depending on the underlying pathologic diagnosis [1]. Surgical goals range from needle biopsies (e.g. primary central nervous system lymphoma [2]) to supramaximal resections (e.g. diffuse gliomas [3]). A major obstacle to the precision care of brain tumor patients is that the pathologic diagnosis is usually unknown at the time of surgery. For other tumor types, such as breast or lung cancer, diagnostic biopsies are obtained prior to definitive surgical management, which provides essential clinical information used to inform the goals of surgery. Routine diagnostic biopsies in neuro-oncology are not feasible due to high surgical morbidity and the potential for permanent neurologic injury. Consequently, the importance of intraoperative pathologic diagnosis in brain tumor surgery has been recognized for nearly a century[4].
Unfortunately, our current intraoperative pathologic techniques are time, resource, and labor intensive [7, 8]. Conventional diagnostic methods, including frozen sectioning and cytologic preparations, require an extensive pathology infrastructure for tissue processing and specimen analysis by a board-certified neuropathologist [9]. While the conventional pathology workflow with board certified neuropathologist interpretation has a diagnostic accuracy between 86 – 96% [5], the pathologist workforce in the US declined in absolute and population-adjusted numbers by nearly 20% between 2007–2017 [10]. This decline has unevenly affected neuropathology, with a 40% fellowship vacancy rate and it is projected to worsen [11]. The number of medical centers performing brain tumor surgery outnumbers board-certified neuropathologists, reducing patient access to expert intraoperative consultation and, consequently, optimal surgical management.
An ideal system for surgical specimen analysis and intraoperative tumor diagnosis would be accessible, fast, standardized, and accurate. An intraoperative pathology system requires, at minimum, (1) a data/image acquisition modality and (2) a diagnostic interpretation. Conventional intraoperative pathology uses light microscopy interpreted by a neuro-pathologist (>20–30 minutes). We propose an innovative diagnosis system that uses a rapid (2–3 minutes, >10× speedup), label-free optical histology method, called stimulated Raman histology (SRH), combined with deep learning-based interpretation of fresh, unprocessed surgical specimens. We have previously demonstrated the feasibility of large-scale clinical SRH imaging [5] and the use of deep neural networks for SRH image classification of brain tumor patients [6, 12, 13]. These studies demonstrate the potential for AI-based diagnosis and interpretation of SRH images to better inform brain tumor surgery and provide personalized surgical goals in the era of precision medicine.
Here, we seek to facilitate this area of active research by releasing OpenSRH, a collection of intraoperative SRH data, including raw SRH acquisition data, processed high-resolution image patches for model development, virtually-stained whole slide images, semantic segmentation of tumor regions, and full intraoperative diagnostic annotations. OpenSRH is the first and only publicly available dataset of any human cancer imaged using optical histology. We release the OpenSRH dataset with the intention to foster translational AI research within the field of precision oncology. The main contributions of this work are:
OpenSRH dataset: We curate and open-source the largest dataset of intraoperative SRH images with pathologic annotations to facilitate the development of innovative machine learning solutions to improve brain tumor surgery.
Classification benchmarks: We benchmark performance for patch-based histologic brain tumor classification across multiple tumor types, computer vision architectures, and transfer learning methods.
Contrastive representation learning benchmarks: We evaluate both self-supervised and weakly supervised patch contrastive learning methods for SRH representation learning. Contrastive learning methods are evaluated using linear evaluation protocols and benchmarked as a model pretraining strategy.
2. Background
Stimulated Raman Histology
SRH is based on Raman scattering. Raman scattering occurs when incident photons on a media either gain or lose energy when scattered (i.e. inelastic scattering), shifting the frequency/wavenumber of the scattered photons. This Raman shift can be measured to characterize the biochemical composition of both inorganic and organic materials using narrow-band laser excitation and a spectrometer [14]. A major limitation of using spontaneous Raman scattering for biochemical analysis is that the Raman effect is weak compared to elastic scattering. Therefore, long acquisition times (> 30 minutes) and spectral averaging are required to obtain representative biochemical spectra. Stimulated Raman scattering (SRS) microscopy was discovered in 2008 as a highly sensitive, label-free biomedical imaging method [15]. Rather than acquiring broad-band spectra, SRS microscopy uses a second laser excitation source to achieve non-linear amplification of narrow-band Raman spectral regions that correspond to specific molecular vibrational modes (see Figure 1). SRS images can then be generated at specific narrow-band Raman wavenumbers. Translational research led to the development of a fiber-laser-based SRS imaging system that could be used at the patient’s bedside to generate rapid histologic images of fresh surgical specimens, called SRH [5, 16, 17]. A major advantage of SRH over other histologic imaging methods is that image contrast is generated by the intrinsic biochemical properties of the tissue only and does not require any tissue processing, staining, dyeing, or labelling (i.e. label-free).
Figure 1:
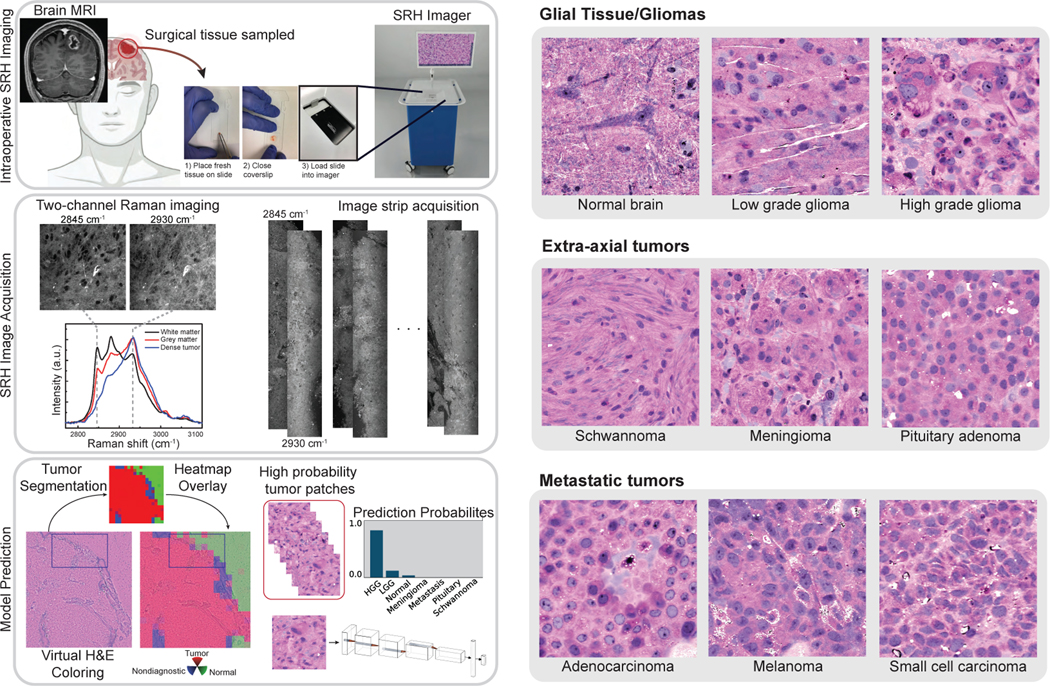
Left, A patient with a newly diagnosed brain lesion undergoes a surgery for tissue diagnosis and/or tumor resection. The tumor specimen is sampled from the patient’s tumor and directly loaded into a premade, disposable microscope slide. The specimen is placed into the SRH imager for rapid optical imaging. SRH images are acquired sequentially as strips at two Raman shifts, 2845 cm−1, and 2930 cm−1. The size and number of strips to be acquired are set by the operator who defines the desired image size. Standard image sizes range from 1–5mm2 and image acquisition time ranges from 30 seconds to 3 minutes. The strips are edge clipped, field flattened and co-registered to generate whole slide SRH images. Images can be colored using a custom virtual H&E colorscheme for pathologic review [5]. The whole slide image is divided into non-overlapping 300×300 pixel patches and each patch undergoes a feedforward pass through a previously trained tumor segmentation model [6] to segment the patches into tumor regions, normal brain, and nondiagnostic regions. The tumor patches are then used for both training and inference to predict the patient’s brain tumor diagnosis. Right, Examples of virtually-colored SRH images from brain tumor diagnoses included in OpenSRH. We include a diversity of tumor diagnoses that cover the most common brain tumor subtypes.
ML applications for SRH
Unlike conventional intraoperative histology with light microscopy, SRH provides high-resolution digital images that can be used directly for downstream ML tasks. Whole slide image digitization of frozen or paraffin-embedded tissue is slow and memory intensive, presenting a major bottleneck for its routine use in intraoperative histology, and clinical medicine in general [18]. Previous studies showed that SRH plus shallow ML models can be used to detect and quantify tumor infiltration in adult and pediatric fresh surgical specimens [5, 19, 20, 21]. We subsequently demonstrated that SRH combined with convolutional neural network architectures can be used for intraoperative diagnostic decision support [6, 12, 13]. These preliminary studies, while demonstrating the feasibility of applying deep architectures to SRH, did not include rigorous hyperparameter tuning, explicit representation learning, or ablation studies to optimize model performance. Moreover, all previous studies required manual annotations, including dense patch-level annotations [6], for model training.
3. Related Work
To date, no SRH datasets are publicly available. The work most directly related to OpenSRH comes from digital and computational pathology research. The Cancer Genome Atlas (TCGA) and The Cancer Imaging Atlas (TCIA) include a large repository of digitized histopathology slides processed using hematoxylin and eosin (H&E) staining. Many studies have used this dataset for image classification tasks across several cancer types, including, but not limited to, lung [22], gastrointestinal [23], prostate [24, 25], brain [26], and pan-cancer studies [23, 27, 28]. Another related histopathology dataset comes from the CAMELYON16 research challenge [29]. The challenge is to detect lymph node metastases in women with breast cancer. Digital pathology remains an active area of research in precision oncology. However, ML applications in digital pathology are mainly applied to postoperative tissue assessment and do not play a major role in informing cancer surgery.
One application of SRH is the detection of tumor infiltration in real-time to improve the extent of tumor resection and reduce residual tumor burden. Real-time SRH-based tumor delineation has been studied in sinonasal/skull base cancers [13, 30, 31] and diffuse gliomas [20, 32]. OpenSRH provides the necessary dataset to explore this topic for multiple brain tumor types, including metastatic tumors and extra-axial tumors, such as meningiomas (Figure 1).
Overall Need
High-quality, public, biomedical datasets with expert annotations are rare. Moreover, the clinical significance of some existing datasets is unclear due to the lack of a roadmap for clinical translation [33]. We believe that OpenSRH has the potential to address a currently unmet clinical need of improving cancer surgery in order to advance precision oncology, both in the US and globally [7]. OpenSRH can address a pressing and significant clinical problem, while having high translational potential because, as previously mentioned, an ideal system for intraoperative tumor specimen evaluation should be:
Accessible: SRH imaging systems are FDA-approved and commercially available for intraoperative imaging
Fast: imaging acquisition time and time-to-diagnosis is 10× faster than the current standard-of-care H&E histology
Standardized: SRH image acquisition is invariant to patient demographic features, clinical workforce, and geographic location
Accurate: preliminary results [6, 13] and diagnostic performance benchmarks (see Figure 4) are on par with the pathologist-based interpretation of H&E histology
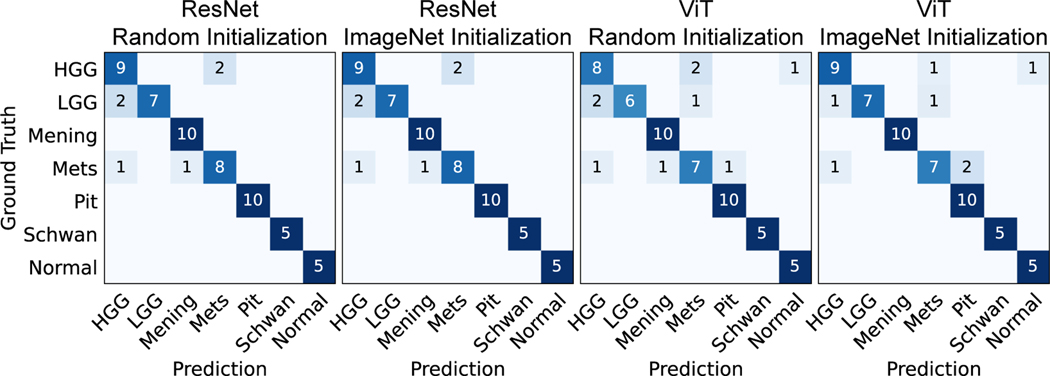
Figure 4:
Patient-level confusion matrices for the four different training strategies on the validation set. Most of the errors occurred in the HGG, LGG, and metastasis classes. Only seed 1 is shown here, other seeds are included in Appendix D. Mening, meningioma; Mets, metastasis; Pit, pituitary adenoma; Schwan, schwannoma.
4. Data Description
Patient population
Patients were consecutively and prospectively enrolled for intraoperative SRH imaging. This study was approved by Institutional Review Board (HUM00083059). Informed consent was obtained for each patient prior to SRH imaging and approved the use of tumor specimens for research and development. All patient health information (PHI) are removed from all OpenSRH data. The inclusion criteria are (1) patients with planned brain tumor or epilepsy surgery at Michigan Medicine (UM), (2) subjects or durable power of attorney able to give informed consent, and (3) subjects in whom there was additional specimen beyond what was needed for routine clinical diagnosis.
SRH imaging
Intraoperative SRH imaging and data processing workflow can be found in Figure 1. A small tumor specimen (3×3 mm3) is placed into a premade microscope slide, which is then loaded into the commercially available NIO Imaging System (Invenio Imaging, Inc.) for SRH imaging. The tissue is then excited with a dual-wavelength fiber laser source, which provides spectral access to Raman shifts in the range of 2800 cm−1 to 3130 cm−1. SRH images are acquired at two Raman shifts: 2845 cm−1 highlights lipid-rich regions and 2930 cm−1 highlights DNA and protein-rich regions [6]. The images are acquired sequentially as 0.5 mm wide strips, stitched together and the two image channels are co-registered to generate the final whole slide image. The co-registration between the two image channels is performed using discrete Fourier transform. A virtual H&E colorscheme [5] can be applied to SRH images for clinician review, but is not used for model development.
Image preprocessing
Image processing starts by applying a sliding window over the two-channel image to generate 300×300 pixel non-overlapping patches. A third channel is obtained by performing a pixel-wise subtraction from the two registered channels (2930cm−1 − 2845cm−1), which highlights the nuclear contrast and cellular density of the tissue [19]. The third channel is concatenated depth-wise to generate a final three-channel patch for model training and inference. Each patch then undergoes a feedforward pass through a pretrained segmentation model to classify the patch into tumor, normal brain, or non-diagnostic tissue [12]. The model was trained using manually labelled patches. The segmentation prediction for each patch is released as part of the OpenSRH dataset.
Dataset breakdown
OpenSRH consists of 307 patients. A total of 304 patients underwent intraoperative SRH imaging and three patients had postmortem specimen collection. We strategically selected the most common brain tumor types to be included in OpenSRH. The included brain tumor diagnoses cover more than 90% of all newly diagnosed brain tumors in the US [34]. OpenSRH includes a diversity of brain tumor types, including primary brain tumors (high-grade gliomas, low-grade gliomas), secondary brain tumors (metastases), and extra-axial tumors (meningiomas, schwannomas, pituitary adenomas). A panel of patch samples is included in Figure 1. The dataset is randomly divided into training (247 patients) and validation set (60 patients, about 20%). Figure 2 shows a histogram of the number of patches and slides per patient, and Figure 3 shows the distribution of the number of patients, slides, and patches per class in the training and validation set. Technical details of the data release and companion source code are in Appendix A.
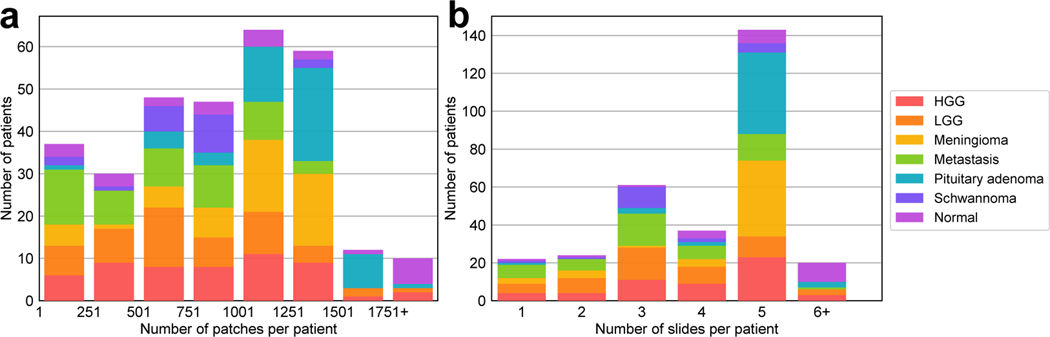
Figure 2:
Histogram for the number of patches and slides per patient. a shows the total number of patches varies across the patients and, b shows most patients have 5 slides. The difference between these two distributions may be caused by specimen size, non-diagnostic regions, surgeon preference, etc. HGG, high grade glioma; LGG, low grade glioma.
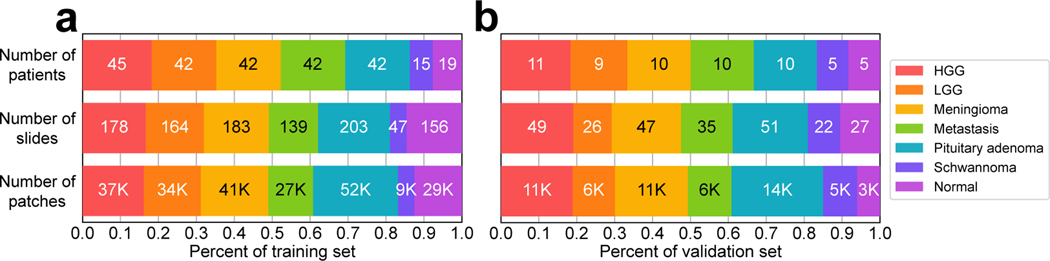
Figure 3:
Bar chart for the number of patients, slides and patches for each diagnostic class. Validation set was randomly selected and contains approximately 20% of patients in OpenSRH (60/307 patients). Training and validation sets have approximately equivalent class distributions.
5. Histologic brain tumor classification benchmarks
In this section, we present the results of the baseline multiclass brain tumor classification task. We aim to benchmark the results for common training strategies. We investigate the value of transfer learning/pretraining from natural image datasets, specifically ImageNet [35], for improving classification performance. The value of transfer learning for SRH [6] and medical imaging, in general [36], remains an active area of research. We selected representative models from the two most competitive computer vision architectures: convolutional neural networks (ResNet50 [37]) and vision transformers (ViT-S[38, 39]). These architectures were selected because they contain a similar number of parameters (~23.5 million for ResNet50, ~21.7 million for ViT-S).
5.1. Training protocol
ResNet50
In the ResNet50 architecture, we changed the output dimension of the last layer to 7 for our experiments. We used a batch size 96 and trained on 300×300 images with horizontal and vertical flipping of probability 0.5 as augmentations. We used categorical cross-entropy loss and AdamW optimizer [40] with β1 =0.9, β2 =0.999, and a weight decay of 0.001. The initial learning rate was 0.001, with a step scheduler with a decay rate γ = 0.5 every epoch. We trained for 20 epochs with two Nvidia RTX 2080Ti GPUs. Training wall time was ~9.5 hours for each experiment.
ViT-S
ViT training protocol was adjusted based on previously published results[38, 39]. In addition to the augmentations in the ResNet50 protocol, we also resized the image to 224×224 to fit the standard ViT-S model and ImageNet pretrained weights [41]. We used AdamW as the optimizer, with the same parameters in ResNet50. The initial learning rate was 1E-4, with a cosine learning rate scheduler. First 20% of training steps were set as the linear warm-up stage to increase training stability. We trained 20 epochs with a batch size of 256 for 9 hours using the same GPU resources mentioned above. Detailed training parameters are included in Appendix C.
5.2. Prediction aggregation and benchmark metrics
Patch-level predictions from the same whole slide or patient need to be aggregated to generate a slide- or patient-level prediction. We aggregated patch-level logits after softmax using average pooling to compute slide and patient-level prediction. We preferred using average pooling over hard patch voting to retain the full patch-level model predictions during slide- or patient-level inference. Model performance evaluation metrics include top-1 accuracy, mean class accuracy (MCA), and mean average precision (MAP). Additional classification metrics including top-2 accuracy and false negative rate (tumor vs. normal) can be found in Appendix D.
5.3. Experimental results
Patch- and patient-level results can be found in Table 1. Patient-level metrics are generally higher than patch-level metrics. Patch-level prediction errors can be mitigated through the average pooling aggregation function. In our preliminary benchmark, ResNet50 achieved overall better performance than ViT-S (e.g., by 7.2 patch accuracy and 5.6 patient accuracy). A potential reason is due to ViT requiring large-scale image datasets on the scale of ImageNet21K or JFT300M [38] to overcome low inductive bias. Insufficient pretraining is known to result in worse performance compared to convolutional neural networks (CNNs). We did observe improved patch-level performance when using ImageNet pretraining (2.1 for ResNet50 and 6.5 for ViT-S at patch accuracy). In general, pretraining was more beneficial to ViT than ResNet50. We believe vision transformers may outperform CNNs with data efficient pretraining. Figure 4 summarizes the patient-level confusion matrix. Both models had similar diagnostic errors differentiating HGG and LGG, a known challenging diagnostic task for pathologists and computer vision models [5]. Metastatic tumors have diverse histologic features (see Figure 1) that can result in diagnostic errors across multiple classes [6]. From the confusion matrices in figure 4, it is important to note that we can also observe some false negatives in the model prediction (tumor vs. normal). Additional metrics on false negative rate for these experiments are also included in appendix D.
Table 1:
Classification benchmarks for ResNet50 and ViT-S. Pretrain refers to the pretraining strategy. Each experiment included three random initial seeds. Mean value and standard deviation (in parentheses) for each metric are reported here. The full table including false negative rates and slide-level metrics can be found in the Appendix D.
| Backbone | Pretrain | Patch | Patient | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Accuracy | MCA | MAP | Accuracy | MCA | MAP | ||
| ResNet50 | Random | 84.4 (0.4) | 83.8 (0.5) | 89.5 (0.5) | 90.0 (0.0) | 91.4 (0.0) | 92.8 (0.2) |
| ResNet50 | ImageNet | 86.5 (0.4) | 85.6 (0.3) | 91.2 (0.3) | 88.9 (0.8) | 90.5 (0.6) | 94.0 (0.1) |
| ViT-S | Random | 77.2 (0.5) | 76.8 (0.8) | 82.3 (0.5) | 85.0 (1.4) | 87.2 (1.1) | 93.2 (0.4) |
| ViT-S | ImageNet | 83.7 (0.5) | 82.7 (0.9) | 88.8 (0.1) | 88.9 (0.8) | 90.5 (0.6) | 93.9 (0.4) |
6. Contrastive representation learning benchmark
Our previous studies demonstrate that contrastive representation learning is well suited for patch-based representation learning [42]. The focus for this section is to investigate the effectiveness of contrastive learning strategies for OpenSRH. We used both unsupervised contrastive learning (SimCLR [43]) and supervised contrastive learning (SupCon [44]) on ResNet50 and ViT-S architectures. The contrastive loss for SimCLR aims to solve the pretext task of instance discrimination. The model is trained to identify two augmented positive pairs of the same image from other images in a minibatch. SupCon loss has the similar training objective but aims at optimizing class discrimination. All images from the same class are treated as positive instances and other images are negative instances. Our general contrastive learning workflow uses SimCLR and SupCon as a representation learning strategy on our dataset followed with a linear evaluation protocol. We compared contrastive representation learning methods with ImageNet pretraining.
6.1. Training and evaluation protocol
Training protocol For both SimCLR and SupCon methods, we use the same protocol except for the loss function. We applied ResNet50 and ViT-S with a linear projection head to project the image representation to a low dimensional hypersphere (128 for ResNet50, 24 for ViT-S) to compute the contrastive loss. The data augmentation strategy followed [43]: a composition of multiple augmentations including flipping, color jittering, and Gaussian blur (for details, see Appendix C). We use AdamW optimizer for both models and same parameters as in Section 5.1. Different learning rates (1E-3 for ResNet50 and 5E-4 for ViT-S) were adopted for each model. We trained using a batch size 224 for ResNet50 and 512 for ViT-S for 40 epochs. We use linear warmup for the first 10% epochs and cosine decay scheduler for ViT only. Detailed protocols are included in Appendix C.
Evaluation protocol To evaluate the learned image representations, we followed a standard linear evaluation protocol [43, 45, 46, 47], where a linear classifier is trained on top of the frozen pretrained backbone. We consider the same evaluation metrics and aggregation function as in Section 5.2. Apart from the classification metrics, we performed qualitative evaluation of our learned representations through t-distributed stochastic neighbor embedding (tSNE) [48] in Figure 5. Additional fine-tuning protocols and results are included in Appendix F.
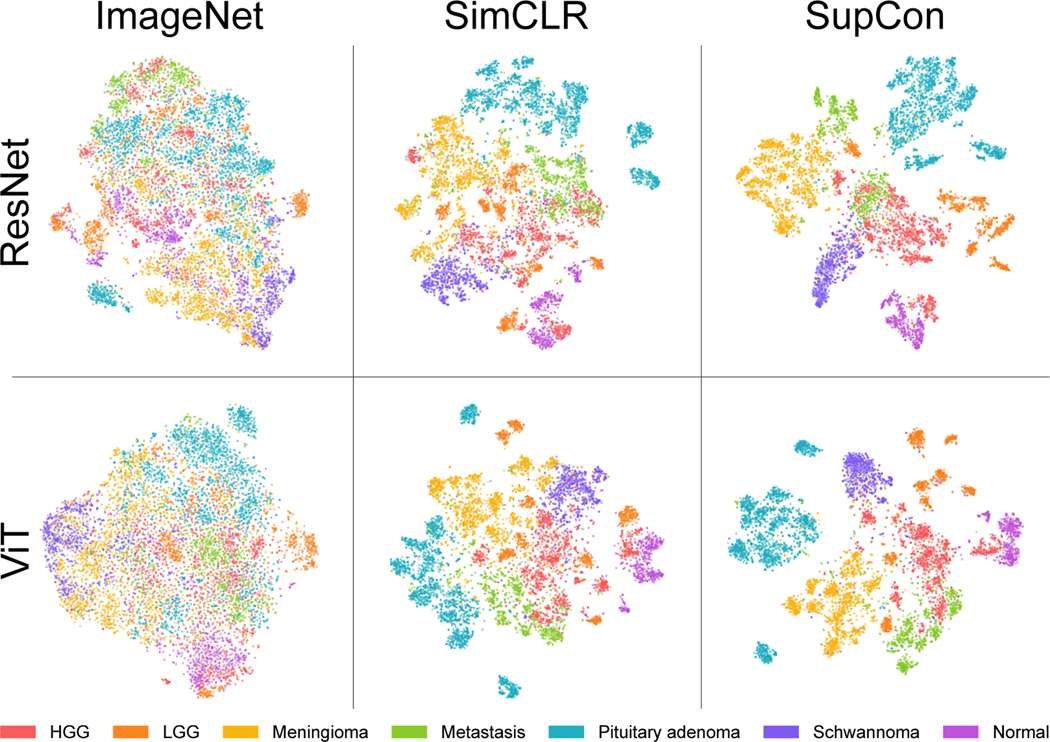
Figure 5:
Patch-level SRH representations of validation images. Latent feature vectors are colored by tumor class labels. ImageNet pretraining fails to represent discriminative SRH image features. SimCLR shows discernable SRH feature learning capabilities, while improved class separation can be learned with SupCon. Tumor classes with similar SRH histologic features tend to show similar feature representations, such as HGG/LGG and HGG/Metastasis. A single seed is shown here. Figure best viewed in color.
6.2. Experimental results
Results of linear evaluations could be found in Table 2. Self-supervised representation learning with SimCLR was able to achieve a patient-level accuracy of 85.6 for ResNet50 and 78.3 for ViT-S. These results demonstrate improvement over our previous self-supervised classification performance [49]. Self-supervised contrastive representation learning using OpenSRH outperforms ImageNet transfer learning for patch-based metrics (e.g., by 3.2 for ResNet50 and 2.3 for ViT-S). These results emphasize the large domain gap between natural images and SRH optical images [50, 36]. Similar to other computer vision tasks, optimal representation learning can be achieved with additional supervision. SupCon outperforms both ImageNet pretraining and SimCLR in patch-based metrics. Linear evaluation showed an overall increase of 4.4 and 8.4 in patient-level accuracy between SupCon and SimCLR for ResNet50 and Vit-S, respectively. Interestingly, the patient-level metrics for pretrained ViT-S were prominently high, while the patch-level metrics were comparatively worse. We believe these results may be due to a simple soft voting aggregation of patch-level predictions. This opens the question for better (learnable) aggregation functions for SRH images. The tSNE plot in Figure 5 was consistent with our patch-based evaluation metrics for both models, where SupCon showed more discrete image representations.
Table 2:
Linear evaluation protocol results for contrastive representation learning. Each experiment included three random initial seeds. Mean value and standard deviation (in parentheses) for each metric are reported here. The full table including false negative rates and slide-level metrics can be found in the Appendix E.
| Backbone | Methods | Patch | Patient | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Accuracy | MCA | MAP | Accuracy | MCA | MAP | ||
| ResNet50 | ImageNet | 68.3 (0.0) | 67.9 (0.0) | 72.9 (0.1) | 80.0 (0.0) | 82.9 (0.0) | 88.8 (0.1) |
| ResNet50 | SimCLR | 79.1 (0.4) | 78.9 (0.4) | 84.2 (0.6) | 83.9 (1.0) | 86.1 (0.9) | 92.4 (0.1) |
| ResNet50 | SupCon | 87.5 (0.3) | 86.8 (0.3) | 91.5 (0.5) | 90.0 (0.0) | 91.4 (0.1) | 94.6 (0.5) |
| ViT-S | ImageNet | 71.8 (0.1) | 71.1 (0.1) | 77.1 (0.1) | 88.3 (0.0) | 89.8 (0.0) | 93.9 (0.0) |
| ViT-S | SimCLR | 76.8 (0.5) | 76.3 (0.5) | 82.5 (0.3) | 80.0 (1.7) | 83.0 (1.3) | 92.3 (0.0) |
| ViT-S | SupCon | 81.4 (0.2) | 80.2 (0.3) | 85.6 (0.5) | 87.8 (1.0) | 89.4 (0.7) | 94.0 (0.4) |
7. Limitations, Open Questions, and Ethical Consequences
OpenSRH contains data collected from a single institution. While SRH imagers have standardized settings, different operating room workflows, tumor sampling strategies, and surgeons may produce SRH data distribution shifts. Moreover, while our OpenSRH does contain the most common brain tumor types, rare tumor classes are not included. This is a limitation because rare tumor diagnosis is one of the contexts in which ML-based diagnostic decision support can be most beneficial to clinicians. We intend to include multicenter data with additional tumor rare classes in future releases of OpenSRH.
There are many open questions for the machine learning community that can be explored through OpenSRH. The most important questions are given below:
Domain adaptation. Many domain adaptation literature uses datasets that have very small domain gaps such as MNIST [51], SVHN[52], Office 31[53], or datasets that are artificially crafted or generated, such as Adaptiope [54] or DomainNet [55]. OpenSRH can be combined with existing H&E dataset such as TCGA, to create a large-scale benchmark for domain adaptation, with intrinsic pathologic features captured using different imaging modalities.
Multiple instance learning. Besides our patch-based classification workflow, multiple instance learning may be a good strategy for histopathology analysis [56]. By removing patch labels in OpenSRH, histologic classification becomes a generic multiple instance learning task. A model can learn to select the important patches with only slide-level labels. Our current patch-level annotation can be used as a ground truth to interpret instance-level predictions from multiple instance learning paradigms.
Aggregation of patch-level predictions. We have relied on individual patch-level predictions and average pooling as a general method for whole slide inference. However, this strategy is limited because it does not account for discriminative heterogeneity within whole slide images. Expectation maximization [57], clustering [58, 28], attention [56, 59], and other multiple instance learning strategies [60] have been proposed as learnable aggregation functions, but questions related to scalability, training efficiency, and data domain differences remain open.
Self-supervised learning. Self-supervised learning and contrastive learning methods have been explored using histology images, but the effectiveness of different augmentation strategies has not been studied. Our preliminary experiments indicate that augmentations used for natural image self-supervised representation learning are sub-optimal. It remains an open question whether domain specific augmentation would improve self-supervised learning performance.
Data efficient training. It is known that ViTs require large image datasets on the scale of ImageNet21K or JFT300M, and insufficient pretraining can result in inferior performance [38]. Acquiring these large supervised datasets is currently infeasible in medical imaging. In addition to low inductive bias, ViTs demonstrate better interpretability compared to CNNs, and their clinical adoption can improve reliability and physician’s trust in AI-assisted diagnosis. By demonstrating a performance gap between CNNs and ViTs, our OpenSRH benchmarks encourage research in data efficient training of ViTs suited for medical imaging.
Lastly, we have ensured that all patients consented to release a portion of their tumor for research. There is minimal additional risk for patients because samples are collected from tumors removed as part of the standard patient care, and their personal health information is protected in OpenSRH. OpenSRH is released to promote translational AI research. The dataset, algorithms and benchmarks discussed in the paper are for research purposes only.
8. Conclusion
In this work, we introduce OpenSRH, an intraoperative brain tumor dataset of SRH, a rapid, label-free, optical imaging method. OpenSRH contains both raw SRH acquisition data and processed high-resolution image patches for model development using diagnostic annotations from expert neuropathologists. OpenSRH is the first and only publicly available dataset of human cancers imaged using optical histology. We benchmark classification performance for histologic brain diagnosis across the most common brain tumor types. We also provide benchmarks for self-supervised and weakly supervised contrastive representation learning. We release the OpenSRH dataset with the intention to promote translational AI research within the field of precision oncology and optimize the surgical management of human cancers.
Supplementary Material
Checklist.
- For all authors...
- Do the main claims made in the abstract and introduction accurately reflect the paper’s contributions and scope? [Yes]
- Did you describe the limitations of your work? [Yes] The limitations of OpenSRH are described in section 7.
- Did you discuss any potential negative societal impacts of your work? [Yes] Potential negative societal impacts are described in section 7.
- Have you read the ethics review guidelines and ensured that your paper conforms to them? [Yes]
- If you are including theoretical results...
- Did you state the full set of assumptions of all theoretical results? [N/A]
- Did you include complete proofs of all theoretical results? [N/A]
- If you ran experiments (e.g. for benchmarks)...
- Did you include the code, data, and instructions needed to reproduce the main experimental results (either in the supplemental material or as a URL)? [Yes] All code, data, and instructions are available on https://opensrh.mlins.org.
- Did you specify all the training details (e.g., data splits, hyperparameters, how they were chosen)? [Yes] Detailed training protocol is listed in appendix C, and data split information is described in section 4 and appendix A.
- Did you report error bars (e.g., with respect to the random seed after running experiments multiple times)? [Yes] All experiments are repeated with 3 random seeds and error bars are reported in all tables.
- Did you include the total amount of compute and the type of resources used (e.g., type of GPUs, internal cluster, or cloud provider)? [Yes] Details of the compute resources and time to produce the OpenSRH benchmarks are described in appendix C.
- If you are using existing assets (e.g., code, data, models) or curating/releasing new assets...
- If your work uses existing assets, did you cite the creators? [Yes] Our benchmark implementation uses existing open source framework. More details are described in appendices A and C.
- Did you mention the license of the assets? [Yes] All licenses of these frameworks are included in the THIRD_PARTY file in the root level of our repository.
- Did you include any new assets either in the supplemental material or as a URL? [Yes] The OpenSRH dataset and benchmark source code can be accessed on https://opensrh.mlins.org.
- Did you discuss whether and how consent was obtained from people whose data you’re using/curating? [Yes] Informed consent was obtained for each patient in OpenSRH. Details are described in section 4.
- Did you discuss whether the data you are using/curating contains personally identifiable information or offensive content? [Yes] All personally identifiable information are removed before data release and benchmark training. Details are described in section 4.
- If you used crowdsourcing or conducted research with human subjects...
- Did you include the full text of instructions given to participants and screenshots, if applicable? [N/A]
- Did you describe any potential participant risks, with links to Institutional Review Board (IRB) approvals, if applicable? [Yes] This study was approved by Institutional Review Board (HUM00083059), and more details are described in section 4. Potential participant risk is minimal, and described in section 7.
- Did you include the estimated hourly wage paid to participants and the total amount spent on participant compensation? [N/A]
Acknowledgements, Disclosure of Funding and Competing Interests
We would like to thank Karen Eddy, Lin Wang, Andrea Marshall and Katherine Lee for their support in data collection and processing.
This work was supported by grants NIH R01CA226527, NIH/NIGMS T32GM141746, NIH K12 NS080223, Cook Family Brain Tumor Research Fund, Mark Trauner Brain Research Fund: Zenkel Family Foundation, and Ian’s Friends Foundation.
Research reported in this publication was also supported by the Investigators Awards grant program of Precision Health at the University of Michigan.
This research was also supported in part through computational resources and services provided by Advanced Research Computing (ARC), a division of Information and Technology Services (ITS) at the University of Michigan, Ann Arbor.
Footnotes
Competing interests: C.W.F. is an employee and shareholder of Invenio Imaging, Inc., a company developing SRH microscopy systems. D.A.O. is an advisor and shareholder of Invenio Imaging, Inc, and T.C.H. is a shareholder of Invenio Imaging, Inc.
References
- [1].David N Louis Arie Perry, Wesseling Pieter, Brat Daniel J, Cree Ian A, Dominique Figarella-Branger Cynthia Hawkins, Ng HK, Pfister Stefan M, Reifenberger Guido, Soffietti Riccardo, Deimling Andreas von, and Ellison David W. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro. Oncol, June 2021. [DOI] [PMC free article] [PubMed]
- [2].Scott Brian J, Vanja C Douglas Tarik Tihan, Rubenstein James L, and Josephson S Andrew. A systematic approach to the diagnosis of suspected central nervous system lymphoma. JAMA Neurol., 70(3):311–319, March 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Di Long, Ashish H Shah Anil Mahavadi, Daniel G Eichberg Raghuram Reddy, Sanjurjo Alexander D, Morell Alexis A, Lu Victor M, Ampie Leonel, Luther Evan M, Komotar Ricardo J, and Ivan Michael E. Radical supramaximal resection for newly diagnosed left-sided eloquent glioblastoma: safety and improved survival over gross-total resection. J. Neurosurg, pages 1–8, May 2022. [DOI] [PubMed]
- [4].Eisenhardt Land Cushing H. Diagnosis of intracranial tumors by supravital technique. Am. J. Pathol, 6(5):541–552.7, September 1930. [PMC free article] [PubMed] [Google Scholar]
- [5].Daniel A Orringer Balaji Pandian, Niknafs Yashar S, Todd C Hollon Julianne Boyle, Lewis Spencer, Garrard Mia, Hervey-Jumper Shawn L, Garton Hugh J L, Maher Cormac O, Heth Jason A, Sagher Oren, Wilkinson D Andrew, Snuderl Matija, Venneti Sriram, Ramkissoon Shakti H, McFadden Kathryn A, Fisher-Hubbard Amanda, Lieberman Andrew P, Johnson Timothy D, Xie X Sunney, Trautman Jay K, Freudiger Christian W, and Camelo-Piragua Sandra. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated raman scattering microscopy. Nat Biomed Eng, 1, February 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Todd C Hollon Balaji Pandian, Urias Esteban, Save Akshay V, Adapa Arjun R, Srinivasan Sudharsan, Jairath Neil K, Farooq Zia, Marie Tamara, Al-Holou Wajd N, Eddy Karen, Heth Jason A, Khalsa Siri Sahib S, Conway Kyle, Sagher Oren, Jeffrey N Bruce Peter Canoll, Christian W Freudiger Sandra Camelo-Piragua, Lee Honglak, and Orringer Daniel A. Rapid, label-free detection of diffuse glioma recurrence using intraoperative stimulated raman histology and deep neural networks. Neuro. Oncol, July 2020. [DOI] [PMC free article] [PubMed]
- [7].Sullivan Richard, Alatise Olusegun Isaac, Anderson Benjamin O, Audisio Riccardo, Autier Philippe, Aggarwal Ajay, Balch Charles, Brennan Murray F, Dare Anna, D’Cruz Anil, Eggermont Alexander M M, Fleming Kenneth, Gueye Serigne Magueye, Hagander Lars, Herrera Cristian A, Hampus Holmer, Ilbawi André M, Jarnheimer Anton, Ji Jia-Fu, Kingham T Peter, Liberman Jonathan, Andrew JM Leather, Meara John G, Mukhopadhyay Swagoto, Murthy Shilpa S, Omar Sherif, Parham Groesbeck P, Pramesh CS, Robert Riviello, Rodin Danielle, Santini Luiz, Shailesh V Shrikhande Mark Shrime, Thomas Robert, Tsunoda Audrey T, Cornelis van de Velde, Umberto Veronesi, Dehannathparambil Kottarathil Vijaykumar David Watters, Wang Shan, Wu Yi-Long, Zeiton Moez, and Arnie Purushotham. Global cancer surgery: delivering safe, affordable, and timely cancer surgery. Lancet Oncol., 16(11):1193–1224, September 2015. [DOI] [PubMed] [Google Scholar]
- [8].Cheah Phaik-Leng, Looi Lai Meng, and Susan Horton. Cost analysis of operating an anatomic pathology laboratory in a Middle-Income country. Am. J. Clin. Pathol, 149(1):1–7, January 2018. [DOI] [PubMed] [Google Scholar]
- [9].Somerset Hilary Lynchand Kleinschmidt-DeMasters Bette Kay. Approach to the intraoperative consultation for neurosurgical specimens. Adv. Anat. Pathol, 18(6):446–449, November 2011. [DOI] [PubMed] [Google Scholar]
- [10].Metter David M, Colgan Terence J, Leung Stanley T, Timmons Charles F, and Park Jason Y. Trends in the US and canadian pathologist workforces from 2007 to 2017. JAMA Netw Open, 2(5):e194337, May 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Stanley J Robboy Sally Weintraub, Horvath Andrew E, Bradden W Jensen C Alexander Bruce, Fody Edward P, Crawford James M, Clark Jimmy R, Cantor-Weinberg Julie, Joshi Megha G, Cohen Michael B, Prystowsky Michael B, Bean Sarah M, Gupta Saurabh, Powell Suzanne Z, Speights VO Jr, Gross David J, and Black-Schaffer W Stephen. Pathologist workforce in the united states: I. development of a predictive model to examine factors influencing supply. Arch. Pathol. Lab. Med, 137(12):1723–1732, December 2013. [DOI] [PubMed] [Google Scholar]
- [12].Todd C Hollon Balaji Pandian, Arjun R Adapa Esteban Urias, Save Akshay V, Khalsa Siri Sahib S, Eichberg Daniel G, D’Amico Randy S, Farooq Zia U, Spencer Lewis, Petridis Petros D, Marie Tamara, Shah Ashish H, Garton Hugh J L, Maher Cormac O, Heth Jason A, McKean Erin L, Sullivan Stephen E, Hervey-Jumper Shawn L, Patil Parag G, Thompson B Gregory, Sagher Oren, McKhann Guy M 2nd, Komotar Ricardo J, Ivan Michael E, Snuderl Matija, Otten Marc L, Johnson Timothy D, Sisti Michael B, Bruce Jeffrey N, Muraszko Karin M, Trautman Jay, Christian W Freudiger Peter Canoll, Lee Honglak, Camelo-Piragua Sandra, and Orringer Daniel A. Near real-time intraoperative brain tumor diagnosis using stimulated raman histology and deep neural networks. Nat. Med, January 2020. [DOI] [PMC free article] [PubMed]
- [13].Jiang Cheng, Bhattacharya Abhishek, Linzey Joseph R, Rushikesh S Joshi Sung Jik Cha, Srinivasan Sudharsan, Alber Daniel, Kondepudi Akhil, Urias Esteban, Pandian Balaji, Al-Holou Wajd N, Sullivan Stephen E, Thompson B Gregory, Heth Jason A, Freudiger Christian W, Khalsa Siri Sahib S, Pacione Donato R, Golfinos John G, Camelo-Piragua Sandra, Orringer Daniel A, Lee Honglak, and Hollon Todd C. Rapid automated analysis of skull base tumor specimens using intraoperative optical imaging and artificial intelligence. Neurosurgery, March 2022. [DOI] [PMC free article] [PubMed]
- [14].Hollon Todd, Lewis Spencer, Freudiger Christian W, Xie X Sunney, and Orringer Daniel A. Improving the accuracy of brain tumor surgery via raman-based technology. Neurosurg. Focus, 40(3):E9, March 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Christian W Freudiger Wei Min, Brian G Saar Sijia Lu, Gary R Holtom Chengwei He, Tsai Jason C, Kang Jing X, and Xie X Sunney. Label-free biomedical imaging with high sensitivity by stimulated raman scattering microscopy. Science, 322(5909):1857–1861, December 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Christian W Freudiger Wenlong Yang, Gary R Holtom Nasser Peyghambarian, Xie X Sunney, and Kieu Khanh Q. Stimulated raman scattering microscopy with a robust fibre laser source. Nat. Photonics, 8(2):153–159, February 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Di Long, Daniel G Eichberg Kevin Huang, Shah Ashish H, Jamshidi Aria M, Luther Evan M, Lu Victor M, Komotar Ricardo J, Ivan Michael E, and Gultekin Sakir H. Stimulated raman histology for rapid intraoperative diagnosis of gliomas. World Neurosurg., 150:e135–e143, June 2021. [DOI] [PubMed] [Google Scholar]
- [18].Farahani Navid, Parwani Anil V, and Pantanowitz Liron. Whole slide imaging in pathology: advantages, limitations, and emerging perspectives. Pathol. Lab. Med. Int, 7:23–33, June 2015. [Google Scholar]
- [19].Ji Minbiao, Orringer Daniel A, Christian W Freudiger Shakti Ramkissoon, Liu Xiaohui, Lau Darryl, Alexandra J Golby Isaiah Norton, Hayashi Marika, Nathalie YR Agar, Young Geoffrey S, Spino Cathie, Santagata Sandro, Camelo-Piragua Sandra, Ligon Keith L, Oren Sagher, and X Sunney Xie. Rapid, label-free detection of brain tumors with stimulated raman scattering microscopy. Sci. Transl. Med, 5(201):201ra119, September 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ji Minbiao, Lewis Spencer, Camelo-Piragua Sandra, Shakti H Ramkissoon Matija Snuderl, Venneti Sriram, Amanda Fisher-Hubbard Mia Garrard, Fu Dan, Wang Anthony C, Heth Jason A, Maher Cormac O, Sanai Nader, Johnson Timothy D, Freudiger Christian W, Oren Sagher, Xiaoliang Sunney Xie, and Orringer Daniel A. Detection of human brain tumor infiltration with quantitative stimulated raman scattering microscopy. Sci. Transl. Med, 7(309):309ra163, October 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Todd C Hollon Spencer Lewis, Pandian Balaji, Niknafs Yashar S, Mia R Garrard Hugh Garton, Cormac O Maher Kathryn McFadden, Snuderl Matija, Andrew P Lieberman Karin Muraszko, Camelo-Piragua Sandra, and Orringer Daniel A. Rapid intraoperative diagnosis of pediatric brain tumors using stimulated raman histology. Cancer Res., 78(1):278–289, January 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Coudray Nicolas, Paolo Santiago Ocampo Theodore Sakellaropoulos, Narula Navneet, Snuderl Matija, Fenyö David, Moreira Andre L, Razavian Narges, and Tsirigos Aristotelis. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med, 24(10):1559–1567, October 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jakob Nikolas Kather, Heij Lara R, Heike I Grabsch Chiara Loeffler, Echle Amelie, Hannah Sophie Muti Jeremias Krause, Niehues Jan M, Sommer Kai A J, Bankhead Peter, Kooreman Loes F S, Schulte Jefree J, Cipriani Nicole A, Buelow Roman D, Peter Boor, Nadi-Na Ortiz-Brüchle, Hanby Andrew M, Valerie Speirs, Sara Kochanny, Akash Patnaik, Andrew Srisuwananukorn, Hermann Brenner, Hoffmeister Michael, Piet A van den Brandt, Dirk Jäger, Christian Trautwein, Pearson Alexander T, and Tom Luedde. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat Cancer, 1(8):789–799, August 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nagpal Kunal, Foote Davis, Liu Yun, Po-Hsuan Cameron Chen Ellery Wulczyn, Tan Fraser, Olson Niels, Jenny L Smith Arash Mohtashamian, Wren James H, Greg S Corrado Robert MacDonald, Peng Lily H, Amin Mahul B, Evans Andrew J, Sangoi Ankur R, Mermel Craig H, Hipp Jason D, and Stumpe Martin C. Development and validation of a deep learning algorithm for improving gleason scoring of prostate cancer. NPJ Digit Med, 2:48, June 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Po-Hsuan Cameron Chen Krishna Gadepalli, Robert MacDonald Yun Liu, Kadowaki Shiro, Nagpal Kunal, Kohlberger Timo, Dean Jeffrey, Corrado Greg S, Hipp Jason D, Mermel Craig H, and Stumpe Martin C. An augmented reality microscope with real-time artificial intelligence integration for cancer diagnosis. Nat. Med, 25(9):1453–1457, September 2019. [DOI] [PubMed] [Google Scholar]
- [26].Mobadersany Pooya, Yousefi Safoora, Amgad Mohamed, Gutman David A, Barnholtz-Sloan Jill S, José E Velázquez Vega, Brat Daniel J, and Cooper Lee A D. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. U. S. A, 115(13):E2970–E2979, March 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fu Yu, Alexander W Jung Ramon Viñas Torne, Gonzalez Santiago, Harald Vöhringer Artem Shmatko, Lucy R Yates Mercedes Jimenez-Linan, Moore Luiza, and Gerstung Moritz. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat Cancer, 1(8):800–810, August 2020. [DOI] [PubMed] [Google Scholar]
- [28].Lu Ming Y, Chen Tiffany Y, Williamson Drew F K, Melissa Zhao, Maha Shady, Jana Lipkova, and Faisal Mahmood. AI-based pathology predicts origins for cancers of unknown primary. Nature, 594(7861):106–110, June 2021. [DOI] [PubMed] [Google Scholar]
- [29].Babak Ehteshami Bejnordi Mitko Veta, Paul Johannes van Diest, Bram van Ginneken, Karssemeijer Nico, Litjens Geert, Jeroen AWM van der Laak, the CAMELYON16 Consortium, Meyke Hermsen, Quirine F Manson, <Balkenhol Maschenka, Geessink Oscar, Stathonikos Nikolaos, Marcory Crf van Dijk, Peter Bult, Francisco Beca, Andrew H Beck, Dayong Wang, Aditya Khosla, Rishab Gargeya, Humayun Irshad, Zhong Aoxiao, Dou Qi, Li Quanzheng, Chen Hao, Lin Huang-Jing, Heng Pheng-Ann, Christian Haß Elia Bruni, Wong Quincy, Halici Ugur, Mustafa Ümit Öner, Rengul Cetin-Atalay Matt Berseth, Khvatkov Vitali, Vylegzhanin Alexei, Kraus Oren, Shaban Muhammad, Rajpoot Nasir, Awan Ruqayya, Sirinukunwattana Korsuk, Qaiser Talha, Tsang Yee-Wah, Tellez David, Annuscheit Jonas, Hufnagl Peter, Valkonen Mira, Kartasalo Kimmo, Latonen Leena, Ruusuvuori Pekka, Liimatainen Kaisa, Albarqouni Shadi, Mungal Bharti, George Ami, Demirci Stefanie, Navab Nassir, Watanabe Seiryo, Seno Shigeto, Takenaka Yoichi, Matsuda Hideo, Hady Ahmady Phoulady, Kovalev Vassili, Kalinovsky Alexander, Liauchuk Vitali, Bueno Gloria, M Milagro Fernandez-Carrobles Ismael Serrano, Deniz Oscar, Racoceanu Daniel, and Rui Venâncio. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA, 318(22):2199–2210, December 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Conall WR Fitzgerald, Dogan Snjezana, Rabih Bou-Nassif Tim Mclean, Woods Robbie, Jennifer R Cracchiolo Ian Ganly, Tabar Viviane, and Cohen Marc A. Stimulated raman histology for rapid IntraOperative diagnosis of sinonasal and skull base tumors. Laryngoscope, May 2022. [DOI] [PMC free article] [PubMed]
- [31].Hoesli Rebecca C, Orringer Daniel A, McHugh Jonathan B, and Spector Matthew E. Coherent raman scattering microscopy for evaluation of head and neck carcinoma. Otolaryngol. Head Neck Surg, 157(3):448–453, September 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pekmezci Melike, Ramin A Morshed Pranathi Chunduru, Pandian Balaji, Young Jacob, Javier E VillanuevaMeyer Tarik Tihan, Sloan Emily A, Aghi Manish K, Molinaro Annette M, Berger Mitchel S, and Hervey-Jumper Shawn L. Detection of glioma infiltration at the tumor margin using quantitative stimulated raman scattering histology. Sci. Rep, 11(1):1–11, June 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wiens Jenna, Saria Suchi, Sendak Mark, Ghassemi Marzyeh, Vincent X Liu Finale Doshi-Velez, Jung Kenneth, Heller Katherine, Kale David, Saeed Mohammed, Pilar N Ossorio Sonoo Thadaney-Israni, and Goldenberg Anna. Do no harm: a roadmap for responsible machine learning for health care. Nat. Med, 25(9):1337–1340, September 2019. [DOI] [PubMed] [Google Scholar]
- [34].Quinn T Ostrom Nirav Patil, Cioffi Gino, Waite Kristin, Kruchko Carol, and Jill S Barnholtz-Sloan. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the united states in 2013–2017. Neuro. Oncol, 22(12 Suppl 2):iv1–iv96, October 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].J Deng W Dong, R Socher, L Li, Kai Li, and Li Fei-Fei. ImageNet: A large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition, pages 248–255, June 2009. [Google Scholar]
- [36].Raghu Maithra, Zhang Chiyuan, Kleinberg Jon, and Bengio Samy. Transfusion: Understanding transfer learning for medical imaging. February 2019.
- [37].K He X Zhang, Ren S, and Sun J. Deep residual learning for image recognition. Proc. IAPR Int. Conf. Pattern Recogn, 2016.
- [38].Dosovitskiy Alexey, Beyer Lucas, Kolesnikov Alexander, Weissenborn Dirk, Zhai Xiaohua, Unterthiner Thomas, Dehghani Mostafa, Minderer Matthias, Heigold Georg, Gelly Sylvain, Uszkoreit Jakob, and Houlsby Neil. An image is worth 16×16 words: Transformers for image recognition at scale. September 2020.
- [39].Steiner Andreas, Kolesnikov Alexander, Zhai Xiaohua, Wightman Ross, Uszkoreit Jakob, and Beyer Lucas. How to train your ViT? data, augmentation, and regularization in vision transformers. June 2021.
- [40].Loshchilov Ilya and Hutter Frank. Decoupled weight decay regularization. November 2017.
- [41].Wightman Ross. Pytorch image models, 2019.
- [42].Jiang Cheng, Bhattacharya Abhishek, Linzey Joseph, Joshi Rushikesh, Sung Jik Cha Sudharsan Srinivasan, Alber Daniel, Kondepudi Akhil, Urias Esteban, Pandian Balaji, Wajd Al-Holou Steve Sullivan, B Gregory Thompson Jason Heth, Freudiger Chris, Khalsa Siri, Pacione Donato, John G Golfinos Sandra Camelo-Piragua, Daniel A Orringer Honglak Lee, and Hollon Todd. Contrastive representation learning for rapid intraoperative diagnosis of skull base tumors imaged using stimulated raman histology. August 2021.
- [43].Chen Ting, Kornblith Simon, Norouzi Mohammad, and Hinton Geoffrey. A simple framework for contrastive learning of visual representations. February 2020.
- [44].Khosla Prannay, Teterwak Piotr, Wang Chen, Sarna Aaron, Tian Yonglong, Isola Phillip, Maschinot Aaron, Liu Ce, and Krishnan Dilip. Supervised contrastive learning. April 2020.
- [45].Zhang Chiyuan, Bengio Samy, Hardt Moritz, Recht Benjamin, and Vinyals Oriol. Understanding deep learning requires rethinking generalization. November 2016.
- [46].Aaron van den Oord, Yazhe Li, and Oriol Vinyals. Representation learning with contrastive predictive coding. July 2018.
- [47].Kolesnikov Alexander, Zhai Xiaohua, and Beyer Lucas. Revisiting self-supervised visual representation learning. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pages 1920–1929. openaccess.thecvf.com, 2019. [Google Scholar]
- [48].Laurens van der Maaten and Geoffrey Hinton. Visualizing data using t-SNE. J. Mach. Learn. Res, 9(86):2579–2605, 2008. [Google Scholar]
- [49].Jiang Cheng, Bhattacharya Abhishek, Linzey Joseph, Joshi Rushikesh, Sung Jik Cha Sudharsan Srinivasan, Alber Daniel, Kondepudi Akhil, Urias Esteban, Pandian Balaji, Wajd Al-Holou Steve Sullivan, B Gregory Thompson Jason Heth, Freudiger Chris, Khalsa Siri, Pacione Donato, John G Golfinos Sandra Camelo-Piragua, Daniel A Orringer Honglak Lee, and Hollon Todd. Contrastive representation learning for rapid intraoperative diagnosis of skull base tumors imaged using stimulated raman histology. August 2021.
- [50].A S Razavian H Azizpour, Sullivan J, and Carlsson S. CNN features Off-the-Shelf: An astounding baseline for recognition. In 2014 IEEE Conference on Computer Vision and Pattern Recognition Workshops, pages 512–519, June 2014. [Google Scholar]
- [51].Y Lecun L Bottou, Bengio Y, and Haffner P. Gradient-based learning applied to document recognition. Proc. IEEE, 86(11):2278–2324, November 1998. [Google Scholar]
- [52].Netzer Yuval, Wang Tao, Coates Adam, Bissacco Alessandro, Wu Bo, and Ng Andrew Y. Reading digits in natural images with unsupervised feature learning. 2011.
- [53].Saenko Kate, Kulis Brian, Fritz Mario, and Darrell Trevor. Adapting visual category models to new domains. In Computer Vision – ECCV 2010, pages 213–226. Springer; Berlin Heidelberg, 2010. [Google Scholar]
- [54].Ringwald and Stiefelhagen. Adaptiope: A modern benchmark for unsupervised domain adaptation. Proceedings of the IEEE/CVF, 2021.
- [55].Peng Can, Zhao Kun, Wiliem Arnold, Zhang Teng, Hobson Peter, Jennings Anthony, and Lovell Brian C. To what extent does downsampling, compression, and data scarcity impact renal image analysis? In 2019 Digital Image Computing: Techniques and Applications (DICTA). IEEE, December 2019.
- [56].Ilse Maximilian, Tomczak Jakub, and Welling Max. Attention-based deep multiple instance learning. In Jennifer Dyand Andreas Krause, editors, Proceedings of the 35th International Conference on Machine Learning, volume 80 of Proceedings of Machine Learning Research, pages 2127–2136. PMLR, 2018. [Google Scholar]
- [57].L Hou D Samaras, Kurc TM, Gao Y, Davis JE, and Saltz JH. Patch-Based convolutional neural network for whole slide tissue image classification. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pages 2424–2433, June 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lu Ming Y, Williamson Drew F K, Chen Tiffany Y, Chen Richard J, Barbieri Matteo, and Mahmood Faisal. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat Biomed Eng, 5(6):555–570, June 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Shao Zhuchen, Bian Hao, Chen Yang, Wang Yifeng, Zhang Jian, Ji Xiangyang, and Zhang Yongbing. TransMIL: Transformer based correlated multiple instance learning for whole slide image classification. May 2021.
- [60].Campanella Gabriele, Matthew G Hanna Luke Geneslaw, Miraflor Allen, Vitor Werneck Krauss Silva, Klaus J Busam, Edi Brogi, Reuter Victor E, Klimstra David S, and Fuchs Thomas J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med, 25(8):1301–1309, August 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Matěj Týč and Christoph Gohlke. imreg_dft. https://github.com/matejak/imreg_dft, 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.