Abstract
Ischemic cardiomyopathy (ICM) is a prominent form of heart failure but the molecular mechanisms underlying ICM remain relatively understudied due to marked phenotypic heterogeneity. Alterations in post-translational modifications (PTMs) and isoform switches in sarcomeric proteins play important roles in cardiac pathophysiology. Thus, it is essential to define sarcomeric proteoform landscape to better understand ICM. Herein, we have implemented a top-down liquid chromatography (LC)-mass spectrometry (MS)-based proteomics method for the identification and quantification of sarcomeric proteoforms in the myocardia of donors without heart diseases (n = 16) compared to end-stage ICM patients (n = 16). Importantly, quantification of post-translational modifications (PTMs) and expression reveal significant changes in various sarcomeric proteins extracted from ICM tissues. Changes include altered phosphorylation and expression of cardiac troponin I (cTnI) and enigma homolog 2 (ENH2) as well as an increase in muscle LIM protein (MLP) and calsarcin-1 (Cal-1) phosphorylation in ICM hearts. Our results imply that the contractile apparatus of the sarcomere is severely dysregulated during ICM. Thus, this is the first study to uncover significant molecular changes to multiple sarcomeric proteins in the LV myocardia of the end-stage ICM patients using LC-MS-based top-down proteomics. Raw data are available via the PRIDE repository with identifier PXD038066.
Keywords: ischemic cardiomyopathy, human heart proteomics, quantitative proteomics, top-down proteomics, myofilament proteins, Z-disk proteins, sarcomere
For TOC Only
Statement: All images used in the TOC graphic are original and 100% created by the authors. Our TOC is now in compliance with the Journal of Proteome Research guidelines.
Introduction
Heart failure (HF) is the leading cause of death worldwide and produces immense economic costs.1–4 Ischemic cardiomyopathy (ICM) is a prominent form of HF wherein left ventricular (LV) systolic dysfunction reduces blood flow to the heart, leading to oxygen deprivation and myocardial hypoxia.5–7 However, the molecular mechanisms underlying ICM remain relatively understudied due to marked phenotypic heterogeneity.
The sarcomere, the basic contractile unit of the heart, contains the structural building blocks for myocardium including actin-based thin and myosin-based thick filaments laterally bordered by the Z-disk.8–10 In the presence of intracellular calcium, conformational changes within the cardiac troponin complex and tropomyosin induce actin-myosin cross-bridge formation to regulate cardiac contraction.11,12 The Z-disk protein network helps cross-link myofilaments into highly organized three-dimensional structures to stabilize the sarcomere.13,14 Alterations in post-translational modifications (PTMs) in sarcomeric proteins such as phosphorylation, oxidation, degradation, and isoform switches play important roles in cardiac pathophysiology. 15–19 Thus, it is essential to quantitatively determine the changes in the PTMs and isoforms of sarcomeric proteins to better understand ICM.
“Top-down” mass spectrometry (MS)-based proteomics is a powerful tool for the analysis and characterization of diverse proteoforms, the myriad number of expressed protein products that arise from polymorphisms, alternative splice variants, and post-translational modifications (PTMs).20–24 Distinct from the “bottom-up” MS approach in which intact proteins are enzymatically digested into peptides,25 top-down MS analyzes intact proteins that can detect and comprehensively characterize intact proteoforms from complex biological samples.26–30 Quantitative analysis of proteoforms using the top-down approach can reveal significant changes in isoform expression and relative abundance of PTMs across various heart disease phenotypes.31,32 We previously detected a decrease in phosphorylation of cardiac troponin I (cTnI) in ischemic human hearts using quantitative top-down MS/MS and affinity purification.33,34 Recently, our lab has developed a highly robust top-down LC-MS/MS method that can simultaneously quantify protein expression and PTM and applied to non-human skeletal and cardiac muscle tissues.35 However, a study characterizing changes in the sarcomeric proteoform landscape of human ICM remains lacking.
Herein, we aimed to uncover sarcomeric proteoform changes in failing heart tissues from patients in end-stage ischemic heart failure and non-failing heart tissues from donors without heart disease to better understand ICM. We have implemented a top-down MS proteomics method for the simultaneous quantification of sarcomeric proteoforms with high reproducibility to reveal the molecular changes protein PTMs and isoforms correlated with LV dysfunction in ICM patients. Our results elucidate significant PTM and expression -level changes in myofilament and Z-disk proteins during ischemic heart failure. Collectively, our results demonstrate the unique advantages of top-down proteomics for defining the sarcomeric proteoform landscape and quantifying protein expression and PTMs in parallel, providing valuable insights into the molecular alterations underlying human ICM.
Experimental Procedures
Chemicals and reagents.
All reagents were purchased from MilliporeSigma (Burlington, MA, USA) unless otherwise noted. Buffers were prepared with HPLC-grade water from Fisher Scientific (Fair Lawn, NJ, USA). Isopropanol and acetonitrile were purchased from Fisher Scientific (Fair Lawn, NJ, USA). Amicon, 0.5 mL cellulose ultra-centrifugal filters with a molecular weight cutoff (MWCO) of 10 kDa were purchased from MilliporeSigma.
Human Cardiac Tissue Collection.
LV myocardium from nonfailing donor hearts with no history of heart diseases were used as control tissues (donor, n = 16). Apex myocardium from failing ICM hearts were collected during left ventricular assist device (LVAD) implantation in which left ventricular apex is removed (ICM, n = 16).36 Donor heart tissue was stored in cardioplegic solution prior to dissection and snap-frozen immediately in liquid nitrogen and stored at −80 °C. ICM heart tissue was stored on dry ice immediately after surgical removal and then stored at −80 °C. Donor and ICM hearts were obtained from the University of Wisconsin (UW)-Madison Organ and Tissue Donation and University of Michigan, respectively. The procedures for the collection of human non-failing donor heart and ICM failing heart tissues were approved by the Institutional Review Board (IRB) of the UW-Madison and University of Michigan, respectively. Available clinical deidentified data including age, gender, cause of death, and medical history are listed for the heart tissues used in this study in Table S1.
Sample Preparation.
Sarcomeric proteins were extracted from human heart tissues using a differential pH-based extraction as previously published.37 First, ~10 mg of heart tissue was quickly homogenized at 4 °C and washed twice in 100 μL of HEPES buffer (25 mM HEPES pH = 7.4, 60 mM NaF, 10 mM L-Methionine, 1 mM DTT, 1 mM PMSF, 1 mM Na3VO4, 1x Protease Inhibitor Cocktail, and 1x Phosphatase Inhibitor Cocktail) with a Teflon pellet pestle. The homogenate was centrifuged at 21,000 g for 30 minutes at 4 °C. The pellets were re-dispersed in 50 μL of freshly prepared trifluoroacetic acid (TFA) buffer (1% TFA pH = 2.0, 1 mM TCEP, 10 mM L-Methionine). The homogenate was centrifuged at 21,000 g for 30 minutes at 4 °C and the supernatant was collected. All samples were normalized to 0.08 mg/mL protein in 0.2% formic acid with 2 mM L-methionine by the Pierce™ BCA assay prior to LC-MS/MS analysis.
Top-down Data Acquisition.
Top-down LC-MS/MS was carried out by using a NanoAcquity ultra-high pressure LC system (Waters) coupled to a high-resolution maXis II quadrupole time-of-flight mass spectrometer (Bruker Daltonics). 5 μL (400 ng) of total protein was injected onto a home-packed PLRP column (PLRP-S) (Agilent Technologies), 10-μm particle size, 500-μm inner diameter, 1,000 Å pore size) using an organic gradient of 5 to 95% mobile phase B (mobile phase A: 0.2% formic acid in water; mobile phase B: 0.2% formic acid in 50:50 acetonitrile:isoproponal) at a flow rate of 12 μL/min and temperature of 35 °C. Column pressure was maintained between 700–1200 psi. Mass spectra were taken at a scan rate of 0.5 Hz over 530–2000 m/z range. A total of three replicate runs were collected for each concentration between 250–1200 ng to establish instrument sensitivity and reproducibility. Samples were randomized during processing and LC-MS/MS analysis to correct for batch effects.38
Data-dependent LC-MS/MS was performed on sarcomeric protein extracts. The three most intense ions in each mass spectrum were selected and fragmented by collision-activated dissociation (CAD) with a scan rate of 2 Hz from 200–3000 m/z. The isolation window for online AutoMS/MS CAD was 10 m/z. The collision DC bias was set from 18 to 35 eV for CAD with nitrogen as the collision gas.
Data Analysis.
LC-MS raw files were processed and analyzed with Compass DataAnalysis 4.3 (Bruker Daltonics) software. All chromatograms were smoothened by the Gauss algorithm with a smoothing width of 2.04 s. Sophisticated Numerical Annotation Procedure (SNAP) peak-picking algorithm (quality factor: 0.4; signal-to-noise ratio (S/N): 3.0) was used to determine the monoisotopic mass of all detected ions. Mass spectra were deconvoluted using the Maximum Entropy algorithm within the DataAnalysis 4.3 software. The resolving power for Maximum Entropy deconvolution was set to 60,000k for proteins that were isotopically resolved.
Protein modifications were quantified based on the ratio of the peak intensity of the proteoform to the summed peak intensities of all proteoforms of the same protein using the deconvoluted mass spectrum. In order to quantify protein expression, the top 3–5 most abundant charge state ions of all proteoforms of the same protein were combined to generate one extracted ion chromatogram (EIC) (Table S2). The area under the curve (AUC) of each EIC was then manually integrated using the DataAnalysis 4.3 software.
Top-down LC-MS/MS data was processed using MASH Native software with the topPIC (v. 1.5.4) algorithm embedded.39,40 The fragment ions in the MS/MS spectra were searched and assigned based on the canonical entries of the Homo sapiens UniProt (UniProtKB) protein database (UP000005640, 20,389 reviewed entries, version December 2022). Monoisotopic masses were used for all proteoform determinations, and all fragment ions were manually validated with a mass tolerance of 20 ppm.
Statistical analysis.
Statistical analysis for group comparison was completed using paired-student t tests to determine the level of statistical significance for the quantification of sarcomere protein modification and expression. All p-values at p < 0.05 were considered significant. All error bars indicated in the figures represent the mean ± standard error of the mean (S.E.M.). Statistical analysis for the coordinated decrease in enigma homolog 2 (ENH2) and cTnI phosphorylation was completed using a Pearson’s correlation. The Pearson correlation coefficient (r) is considered strongly correlated if it is above 0.7.
Results
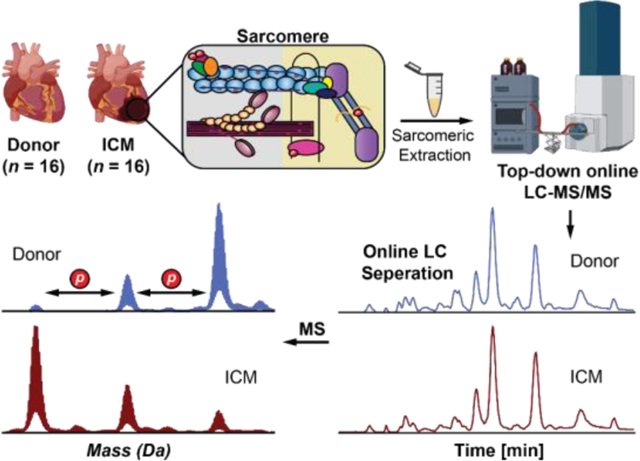
Analysis of sarcomeric proteins in donor versus ICM tissues by online top-down LC-MS/MS.
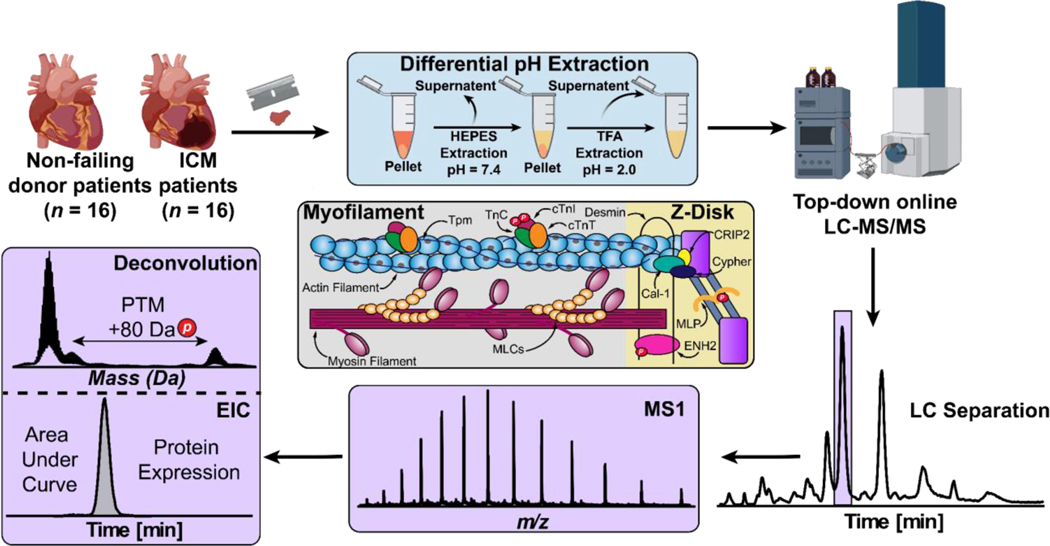
The goal of this project was to reveal the molecular alteractions underlying ICM using top-down MS-based proteomics. To do so, we implemented an online top-down LC-MS/MS method to simultaneously quantify sarcomeric protein expression and modification levels between LV tissue from non-failing donor hearts and left ventricular apex from ICM tissues collected during LVAD surgeries (Figure 1). We have demonstrated that LV and apex tissues in donor hearts (n = 4) have similar sarcomeric proteoform levels (Figure S1), which allows sarcomeric proteoform comparison between donor LV and ICM apex tissues in this study.
Figure 1. Label-free top-down proteomics workflow for the simultaneous quantification of sarcomeric protein expression and modification in ICM.
Sarcomeric proteins were extracted from ~10 mg of failing donor (n = 16) and ICM (n = 16) human cardiac tissues using a differential pH-based extraction. Tissue was first homogenized in HEPES buffer (pH = 7.4) to remove cytosolic proteins. After centrifugation, the remaining pellets were homogenized in TFA buffer (pH = 2.0) to enrich for sarcomeric proteins. Intact proteins (400 ng) were then separated by online reverse-phase LC and MS data (MS1) were acquired using a Bruker maXis II quadrupole-time-of-flight (QTOF) mass spectrometer. All data analysis was performed using Bruker DataAnalysis v. 4.3. Protein modifications were quantified by calculating the relative abundance of the most abundant proteoforms from the deconvoluted spectra. Protein expression was quantified by finding the area under the curve (AUC) of the EIC.
The whole procedure including tissue homogenization, sarcomeric protein extraction, and LC-MS/MS analysis can be performed in less than 3 h, demonstrating a fast and high-throughput method to simultaneously characterize protein expression and PTMs. SDS-PAGE was used to visualize proteins in the HEPES versus TFA extractions (Figure S2). The results showed that our differential pH-based extraction method successfully depletes cytosolic proteins and enriches sarcomeric proteins in donor and ICM cardiac tissues. Our online top-down LC-MS/MS method revealed a diverse sarcomeric proteoform landscape in donor and ICM tissue samples (Figure 2).
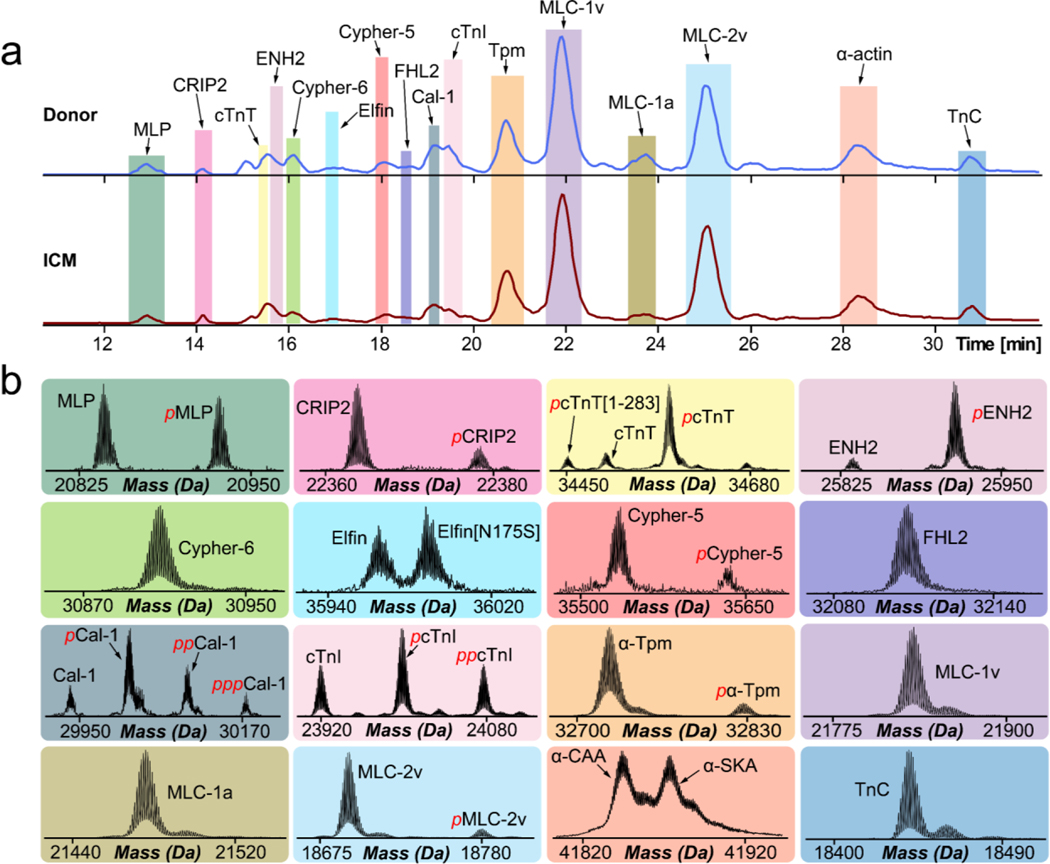
Figure 2. Diverse proteoform landscape detected in donor and ICM human cardiac tissues.
(a) Representative base peak chromatograms (BPC) showing high-resolution separation of intact sarcomeric and Z-disk proteins muscle lim protein (MLP), cysteine-rich protein 2 (CRIP2), cardiac troponin T (cTnT), enigma homolog 2 (ENH2), cypher-6, elfin, cypher-5, four and a half LIM domain protein 2 (FHL2), calsarcin-1, cardiac troponin I (cTnI), tropomyosin (Tpm), ventricular myosin light chain-1 (MLC-1V), atrial myosin light chain-1 (MLC-1a), ventricular myosin light chain-2 (MLC-2v), alpha-actin (α-actin), troponin C (TnC) in non-failing donor (blue trace) and ICM (red trace) tissues procured via left ventricular device surgery. (b) High-resolution deconvoluted mass spectra highlighting the variety of sarcomeric and Z-disk proteoforms observed. Red p and pp denote monophosphorylated and bisphosphorylated proteoforms, respectively.
Myofilament proteins identified include cTnI, cardiac troponin T (cTnT), troponin C (TnC), tropomyosin (Tpm) isoforms, α-actin isoforms, ventricular isoform of MLC-2 (MLC-2v), ventricular isoform of MLC-1 (MLC-1v), and atrial isoform of MLC-1 (MLC-1a). We also detected various Z-disk proteins such as ENH2, muscle LIM protein (MLP), cysteine-rich protein 2 (CRIP2), cypher-5, cypher-6, elfin, calsarcin-1 (Ca1–1), and four and a half LIM domains 2 (FHL2).41,42 Sarcomeric proteins were first identified based on their intact protein mass with a mass error tolerance within 4 ppm. A total of 35 human sarcomeric proteoforms were identified compared to 63 human sarcomeric proteins present in Uniprot. Conceivably, it is difficult to identify large proteins due to the limited fragments obtained from online LC-MS/MS data.26,43,44 A full list of sarcomeric proteoforms identified in donor and ICM tissues can be found in Table S3.
For further protein characterization we employed online LC-MS/MS analysis with CAD fragmentation to generate b- and y-ions. LC-MS/MS was performed on all sarcomeric proteins detected. Fragment assignments were confirmed in MASH Native software to identify the ventricular isoforms of MLC-1 and MLC-2. The method applies for all sarcomeric proteins detected in the analysis. Uniprot databases were used to map the sequences of human MLC-1v and MLC-2v. Removal of N-terminal methionine and addition of N-terminal trimethylation was validated for human MLC-1v and MLC-2v which is in good agreement with previous literature.45 The determination of the N-terminal modifications was further validated by comparison of the theoretical fragment ion masses and the experimental fragment ion masses. Previous studies have reported the localization of mono-phosphorylated MLC-2v to Ser15 or Ser19.46,47 This data indicates that Ser19 is a site of MLC-2v phosphorylation due to sequence informative b and y-ions further demonstrating the power of top-down MS/MS. Total fragmentation of MLC-1v reveals 25 b-ions and 23 y-ions achieving 23% sequence coverage (Figure S3). Similarly, total fragmentation of MLC-2v reveals 14 b-ions and 22 y-ions achieving 21% sequence coverage (Figure S4). Overall, these data reveal how top-down MS/MS can be used to unambiguously sequence intact myofilament proteins for protein identification and characterization.
Simultaneous quantification of sarcomeric protein expression and modification in ICM using online top-down LC-MS/MS method with high reproducibility and linearity.
We next assessed the reproducibility and linearity of our top-down online LC-MS/MS method. To ensure our extraction protocol and LC-MS/MS method was reproducible we performed three extraction replicates using donor tissue sample (Figure S5 and Figure S6). With the same amount of protein loaded for each biological replicate, overlaid base peak chromatograms (BPC) and total ion chromatograms (TIC) show highly similar chromatographic profiles and MS signal intensities demonstrating the comparative reproducibility of our method.
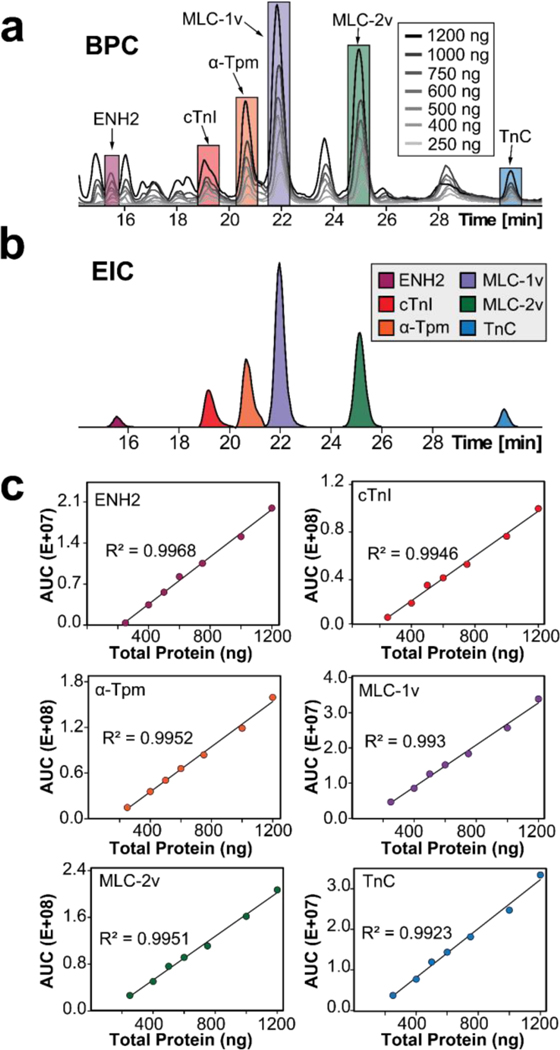
To compare protein expression levels between ICM and donor samples using top-down quantitative analysis, we first evaluated the linearity of our instrument response with 250, 400, 500, 600, 750, 1000, and 1200 ng of total protein from the same donor tissue extract injected in triplicate.35 EICs of representative sarcomeric proteins ENH2, cTnI, α-Tpm, MLC-1v, MLC-2v, and TnC were generated, and a mutual linear range of 250–1200 ng was established by measuring the summed abundance of the AUC of each EIC (Figure 3a, 3b). Injection replicates across each amount of total protein loaded demonstrated excellent reproducibility, sensitivity, and linearity achieving R2 values greater than 0.99. (Figure 3c and Figure S7). As the instrument responds linearly to differing amounts of total protein loaded, this analysis allows us to be confident in quantitating changes in expression level. Therefore, we can also be confident in quantifying changes in protein abundance between samples.
Figure 3. Mutual linear range response determination for key sarcomeric proteins.
(a) Overlaid BPCs of 250, 400, 500, 600, 750, 1000, and 1200 ng of total protein loaded into the LC-MS demonstrating highly similar proteoform profiles. (b) EICs were generated for ENH2, cTnI, α-Tpm, MLC-1v, MLC-2v, and TnC by combining the top 3–5 most abundant charge states of all proteoforms of the same protein. (c) Mutual linear range response determination for key sarcomeric proteins. AUCs of the individual sarcomeric proteins exhibit mutual linear correlation with 250–1200 ng of total protein injected with 3 injection replicates for each point.
Correlated decrease in ENH2 and cTnI expression and modification in ICM tissues.
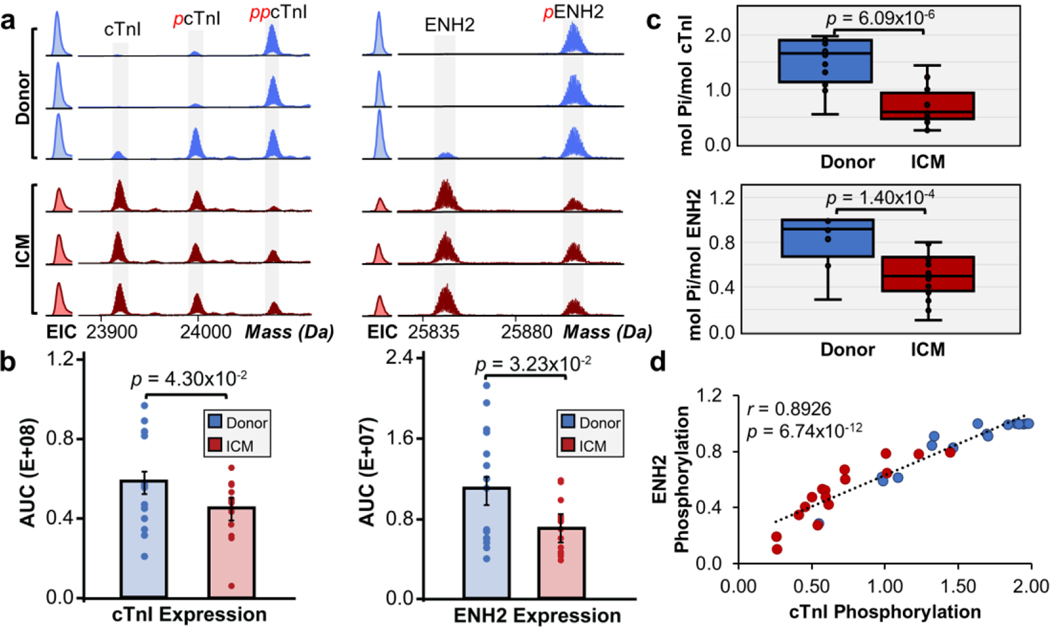
Significant alterations in PTM and expression level changes in ENH2 and cTnI were identified in ICM compared to non-failing donor samples. Three major cTnI proteoforms were observed in the deconvoluted mass spectra for donor and ICM samples including unphosphorylated cTnI, monophosphorylated cTnI (pcTnI), and bisphosphorylated cTnI (ppcTnI) (Figure 4a). Two major ENH2 proteoforms were observed in donor and ICM samples including unphosphorylated ENH2 and monophosphorylated ENH2 (pENH2) (Figure 4a). We then quantified the expression of cTnI and ENH2 and observed decreased expression levels in ICM compared to donor tissues (Figure 4b). Total phosphorylation levels were quantified by calculating the relative abundance of proteoforms as their representative percentages among all detected proteoforms in the deconvoluted mass spectra (Ptotal). Total phosphorylation levels of ENH2 in donor tissues were between 0.29 to 1.00 mol Pi/mol protein and 0.10 to 0.80 mol Pi/mol protein in ICM tissues (Figure 4c). We observed a significant decrease in pENH2, with total phosphorylation of ENH2 decreasing from 85% to 50% in ICM tissues compared to donor tissues. Total phosphorylation levels of cTnI in donor tissues were between 0.55 to 1.98 mol Pi/mol protein and 0.25 to 1.44 mol Pi/mol protein in ICM tissues (Figure 4c). Notably, we also observed a significant decrease in cTnI phosphorylation, with total phosphorylation of cTnI decreasing from 90% to 56% in ICM tissues compared to donor tissues. To determine if the decrease in phosphorylation of ENH2 and cTnI in ICM tissues were dependent on one another, we plotted a linear correlation achieving a strong, positive Pearson correlation coefficient (r = 0.8926) and p < 0.00001 (Figure 4d). These findings agree with a previous study in human hearts with hypertrophic cardiomyopathy (HCM) which implied that the coordinated decrease in ENH2 and cTnI phosphorylation is due to dysregulation of the cAMP-dependent protein kinase A (PKA)-mediated pathway.26 PKA signaling is essential for cardiac function and plays important roles in cardiac contractility, metabolism, and homeostasis. Inactivation of this pathway has previously been associated with impaired cardiac contraction and myocardial ischemia.48,49 β-blockers remain widely used as antihypertensives to manage cardiovascular symptoms such as high blood pressure and irregular arrhythmia. While β-blockers antagonize β-adrenergic receptors in which the PKA-mediated pathway is activated by, the effect of β-blockade on the PKA-mediated pathway remains incompletely understood.50,51 Nonetheless, our previous study investigated the effect of β-blockade on reduced cTnI phosphorylation and found no significant correlation between reduced phosphorylation in cTnI and β-blocker treatments.33 Thus, the decreased PKA-mediated phosphorylation of cTnI and ENH2 in ICM hearts may be independent of β-blocker treatment.
Figure 4. Coordinated decrease in cTnI and ENH2 phosphorylation in ICM.
(a) Representative high-resolution deconvoluted spectra and EICs of cTnI and ENH2 in non-failing donor (blue) compared to ICM (red) tissues. Red p and pp denote monophosphorylated and bisphosphorylated proteoforms, respectively. (b) Quantification of cTnI and ENH2 expression. Groups were considered significantly different by paired-student t tests with p < 0.05. (c) Total protein phosphorylation calculated by mol pi/mol protein for cTnI and ENH2. Box, median and interquartile range (25%, 75%); whiskers, minimum and maximum values. Horizontal lines represent the median of the group. Groups were considered significantly different by paired-student t tests with p < 0.001. (d) Linear correlation between ENH2 and cTnI phosphorylation. The Pearson correlation coefficient (r = 0.8926) considers the groups strongly correlated and statistically significant with p < 0.00001.
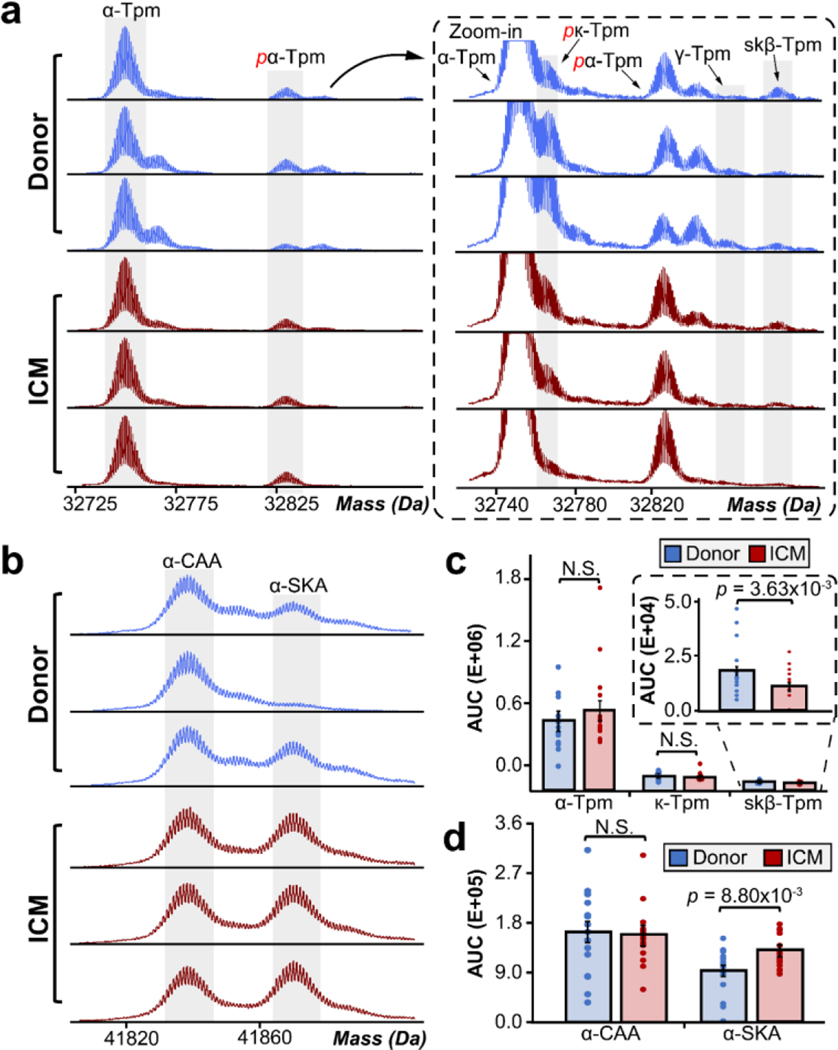
Differential isoform expression of Tpm and α-actin.
Human Tpm is a thin-filament associated protein which is expressed as several isoforms that closely interact with cTnT and α-actin to regulate muscle contraction.52–54 Tpm1 encodes for both α-Tpm and κ-Tpm, whereas Tpm2 and Tpm3 encode for β-Tpm and γ-Tpm, respectively.55,56 Our previous study confirmed the presence of α-Tpm, κ-Tpm, and β-Tpm in the human heart with α-Tpm being the most abundantly expressed isoform.55 The major isoforms of Tpm that we detected in this study were unphosphorylated α-Tpm, monophosphorylated α-Tpm (pα-Tpm), and monophosphorylated κ-Tpm (pκ-Tpm) (Figure 5a). The low abundance isoforms detected were unphosphorylated κ-Tpm, unphosphorylated γ-Tpm, and unphosphorylated skeletal β-Tpm (skβ-Tpm). We quantified the abundance of each isoform relative to the total abundance of all isoforms by finding the AUC for α-Tpm, κ-Tpm, and skβ-Tpm in donor versus ICM tissues (Figure 5c). γ-Tpm was too low in abundance and could not be accurately quantified. We observed a significant decrease in skβ-Tpm expression, with total skβ-Tpm expression decreasing from 2.9% to 1.5% of total Tpm abundance in ICM tissues compared to donor tissues. The difference in isoform expression of α-Tpm and κ-Tpm in donor versus ICM tissues were not significant. The mechanism of how Tpm isoform ratios affect cardiac function remains unclear; however, we also detected a decrease in skβ-Tpm expression in HCM tissues using top-down MS/MS.26 Thus, our data implies that changes in skβ-Tpm isoform expression in the heart may alter cardiac function and impair systolic function in ICM.
Figure 5. Differential isoform expression of tropomyosin and α-actin.
(a) Representative high-resolution deconvoluted spectra of tropomyosin in non-failing donor (blue) compared to ICM (red) tissues. We identified four Tpm isoforms including α-Tpm, β-Tpm, κ-Tpm, and γ-Tpm. Red p denotes monophosphorylated proteoforms. (b) Representative deconvoluted spectra of α-CAA and α-SKA in non-failing donor (blue) compared to ICM (red) tissues. (c) Quantification of Tpm and α-actin isoforms by finding the AUCs of each respective isoform. Groups were considered significantly different by paired-student t tests with p < 0.005.
In cardiac and skeletal muscle, ACTC and ACTA encode for skeletal α-actin (α-SKA) and cardiac α-actin (α-CAA) isoforms, respectively. These two isoforms are co-expressed in myocardium and play roles in sarcomere structure and integrity. Here, we observed both α-actin isoforms in donor and ICM tissues (Figure 5b). We observed a significant increase in α-SKA expression, with total α-SKA expression increasing from 36% to 45% of total α-actin abundance in ICM tissues compared to donor tissues (Figure 5d). Previously, our lab has found a significant increase in the relative abundance of α-SKA in non-failing donor hearts compared to failing dilated cardiomyopathy (DCM) hearts.57 Therefore, these data agree with previous reports that the upregulation of α-SKA expression in failing hearts is a promising biomarker of heart disease.
Increased phosphorylation of full-length cTnT in ICM tissues.
The deconvoluted mass spectra for cTnT showed multiple proteoforms including unphosphorylated cTnT, monophosphorylated cTnT (pcTnT), unphosphorylated truncated cTnT with a lysine cleaved from the C-terminus (cTnT [aa 1–286]), and monophosphorylated truncated cTnT (pcTnT [aa1–286]) (Figure S8a). We observed a significant increase in full-length cTnT phosphorylation, with total cTnT phosphorylation increasing from 79% to 86% ICM tissues compared to donor tissues (Figure S8b). There also appeared to be an increase in total phosphorylation of truncated cTnT [aa 1–286]; however, the change in total cTnT [aa 1–286] phosphorylation between donor and ICM tissues was not significant (Figure S8c). Similar to cTnI, cTnT can be phosphorylated by a variety of kinases and is a choice biomarker for the detection of cardiac injury due to its cardiac specificity.58,59 cTnT mutations are known to be a common cause of many different cardiomyopathies and PTMs play a vital role in cardiac contractility.60–62 Interestingly, we observed a homozygous and heterozygous cTnT polymorphism in four ICM and one donor samples (Figure S9). The polymorphism corresponded to a +28 Da mass shift, indicating a possible alanine (A) to valine (V) point mutation which has previously been reported as a cardiomyopathy variant at amino acid positions 38 or 114.63,64 However, being that this point mutation was also present in one donor tissue sample, it is unlikely this polymorphism to be a potential biomarker for ICM.
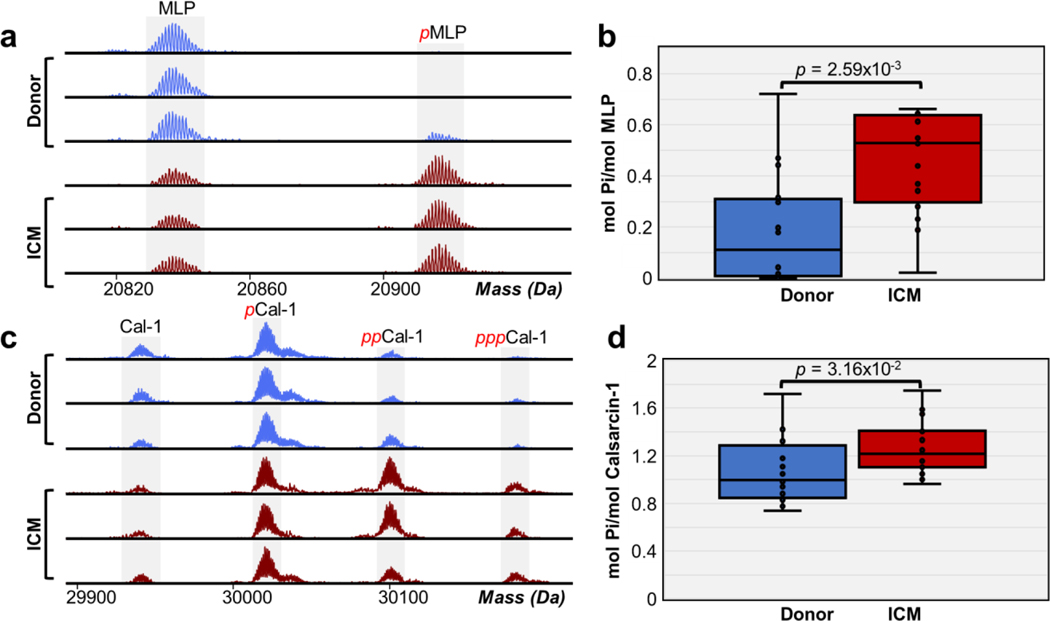
Increased phosphorylation of Z-disk proteins in ICM tissues.
In addition to myofilament proteins, we also identified and quantified Z-disk proteins such as MLP and calsarcin-1. The deconvoluted mass spectra for MLP showed unphosphorylated MLP and monophosphorylated MLP (pMLP) (Figure 6a). The deconvoluted mass spectra for calsarcin-1 revealed multiple phosphorylated proteoforms including unphosphorylated calsarcin-1, monophosphorylated calsarcin-1 (pCal-1), bisphosphorylated calsarcin-1 (ppCal-1), and trisphosphorylated calsarcin-1 (pppCal-1) (Figure 6c). Total phosphorylation levels of MLP in donor tissues were between 0.0 to 0.44 mol Pi/mol protein and 0.02 to 0.66 mol Pi/mol protein in ICM tissues. We observed a significant increase in MLP phosphorylation, with total phosphorylation increasing from 19% to 46% of total MLP abundance in ICM tissues compared to donor tissues (Figure 6b). We also quantified phosphorylation of calsarcin-1 (Figure S10). The total phosphorylation levels of calsarcin-1 in donor tissues were between 0.73 to 1.72 mol Pi/mol protein and 0.96 to 1.75 mol Pi/mol protein in ICM tissues. Similar to MLP, we observed a significant increase in calsarcin-1 phosphorylation, with total phosphorylation of calsarcin-1 increasing from 86% to 90% of total calsarcin-1 abundance in ICM tissues compared to donor tissues (Figure 6d). MLP and calsarcin-1 are known to be involved in severe forms of cardiomyopathy. Mutations in the CSRP3 gene which encodes for MLP have been directly involved in the development of HCM and DCM.65 Calsarcin-1 is a mediator of protein phosphatase activity and deficiency of this protein sensitizes the heart to calcineurin, a protein phosphatase that plays important roles in HCM when dysregulated.66,67 Nevertheless, the molecular mechanisms and effect of PTMs on MLP and calsarcin-1 remain relatively unknown. This is the first study to report increased phosphorylation of MLP and calsarcin-1 in ICM. The total phosphorylation of MLP and calsarcin-1 exhibited a negative linear correlation with a Pearson correlation coefficient r = −0.03, indicating that these two phosphorylation are not correlated which are unlikely to be phosphorylated by the same kinase or closely interact with each other in the z-disk. Despite reports of MLP being significantly down regulated in DCM and ICM68, we did not observe a significant change in MLP expression.
Figure 6. Increase in MLP and calsarcin-1 phosphorylation in ICM.
(a) Representative high-resolution deconvoluted spectra of MLP in non-failing donor (blue) compared to ICM (red) tissues. Red p denotes monophosphorylated MLP. (b) Total protein phosphorylation calculated by mol pi/mol protein for MLP. Box, median and interquartile range (25%, 75%); whiskers, minimum and maximum values. Horizontal lines represent the median of the group. Groups were considered significantly different by paired-student t tests with p < 0.01. (c) Representative high-resolution deconvoluted spectra of calsarcin-1 (Cal-1) in non-failing donor (blue) compared to ICM (red) tissues. Red p, pp, and ppp denotes mono-, bis-, and tris-phosphorylated proteoforms, respectively. (d) Total protein phosphorylation calculated by mol pi/mol protein for calsarcin-1. Box, median and interquartile range (25%, 75%); whiskers, minimum and maximum values. Horizontal lines represent the median of the group. Groups were considered significantly different by paired-student t tests with p < 0.05.
Discussion
Overall, in this study, we have focused on sarcomere subproteome and achieved simultaneous quantification sarcomeric protein expression and PTMs from non-failing donor and failing ICM human cardiac tissues. ICM is a highly heterogenous cardiovascular disease that is characterized by significant alterations to the left ventricle of the heart, impacting systolic function and inducing myocardial hypoxia. While ischemic heart disease is the leading cause of death worldwide, there are only very few proteomics studies that have been performed directly using human ICM samples due to the difficulty in obtaining human heart tissue samples.69,70 Roselló-Lletí et al. analyzed cardiac protein changes in ICM from human LV tissues using 2-dimensional gel electrophoresis followed by in-gel digestion and bottom-up MS, showing that proteins involved in cellular stress response, respiratory chain, and cardiac metabolism were altered.69 More recently, Yi et al. quantitatively analyzed the LV proteome in human ICM using bottom-up LC-MS/MS and observed differentially expressed proteins related to metabolism, immune response, muscle contraction, and signal transduction.70 Nonetheless, bottom-up proteomics suffers from many limitations including the protein interference problem and loss of vital information about PTMs and alternative splice variants due to proteolytic cleave of intact proteins into peptides.22,71 On the other hand, top-down proteomics can directly analyze intact proteins allowing for identification and characterization of proteoforms with full sequence coverage.20–24 Therefore, top-down proteomics is ideally suited for studying human ICM at the proteoform level to reveal molecular changes within the human sarcomeric proteome.
In this study, we have extracted sarcomeric proteins and utilized top-down online LC-MS/MS for simultaneous quantification of many sarcomeric proteoforms in failing human ICM tissues compared to non-failing donor tissues. Specifically, we discovered a significant decrease in cTnI and ENH2 phosphorylation with a significant increase in MLP, Calsarcin-1, and cTnT phosphorylation in failing ICM tissues compared to non-failing donor tissues (Table S4). We also found sarcomeric protein expression levels to be significantly altered in ICM tissues compared to non-failing donor tissues with a significant decrease in cTnI, ENH2, and β-Tpm expression and significant increase in α-SKA expression. Previously, our lab has employed a top-down MS combined with affinity purification of cTnI to characterize cTnI proteoforms in ICM and DCM tissues, which revealed that cTnI phosphorylation was greatly reduced in failing ICM and DCM hearts compared to brain-dead donor hearts, implying that phosphorylation of cTnI is a potential biomarker for chronic heart failure.33 Moreover, in another previous study, Peng et al. generated swine models with acute myocardial infarction (AMI) and identified reduced phosphorylation in cTnI and MLC2 in the myofilaments and ENH2 in the Z-disk.72 We do observe some similar proteoform changes in swine and human ischemia such as reduced phosphorylation of cTnI and ENH2. On the other hand, we have also observed unique differences only in human ischemia but not in swine ischemia such as significant increases in cTnT, calsarcin-1, and MLP phosphorylation, whereas reduced phosphorylation of MLC2 was observed in swine AMI. Additionally, our lab has previously published an extensive study of the sarcomeric proteoform landscape in human HCM.26 We found some similar proteoform changes in ICM and HCM such as decreased phosphorylation of cTnI and ENH2, whereas significant increases in MLP and calsarcin-1 phosphorylation were only observed only in ICM but not in HCM. Since human heart tissue samples in ICM and HCM were obtained from end-stage/late-stage heart failure patients, conceivably they share similarity likely due to the converging pathways and similar molecular processes that leads towards heart failure. Conversely, distinct proteoform changes observed are representative of their respective cardiomyopathies.
One major limitation of this study is the difficulty in detecting the large proteins (>50 kDa) using our online top-down LC-MS/MS approach mainly due to the co-elution of proteins during the 1DLC separation prior to MS/MS analysis and the exponential decrease in the signal-to-noise ratio as protein molecular weight increases.28,43 Moreover, it is difficult to identify large proteins due to the limited fragments from online LC-MS/MS data. Our lab has previously developed a top-down 2D-LC method using offline serial size exclusion chromatography (sSEC) to separate high MW proteins up to 223 kDa and coupling with top-down MS for detection of large proteoforms.43 While the 2DLC sSEC method allows for high-resolution size-based fractionation of large intact proteins from complex protein mixtures, this offline 2DLC method was time consuming and required a large amount of samples. Here we only have a limited amount of ICM tissue samples which were collected during LVAD surgery. Despite these limitations, the major advantages of this online top-down LC-MS/MS method include high reproducibility, superior linearity, and unambiguous quantification of protein PTMs and expression in the sarcomeric subproteome from failing ICM and non-failing donor human cardiac tissues.
Conclusion
We employed a top-down MS proteomics method for the simultaneous quantification of sarcomeric protein expression and PTMs from ICM and donor human cardiac tissues. This study represents the first top-down LC-MS/MS-based proteomics study of human ICM, which uncovers a diverse sarcomeric proteoform landscape and molecular changes associated with end-stage ischemic heart failure. We identified five significant changes in phosphorylation and four significant changes in protein expression in ICM. Notably, we observed a coordinated decrease in the phosphorylation and expression of cTnI and ENH2, implying dysregulation in the PKA-mediated pathway during ischemic HF. Our results also revealed that Z-disk proteins are altered during ICM, as we observed a significant increase in phosphorylation of calsarcin-1 and MLP. Finally, our results showed differential isoform expression of α-SKA and skβ-Tpm in ICM. Overall, our results highlight the importance of characterizing ICM at the proteoform level to reveal molecular changes within the extremely complex cardiac proteome. In-depth understanding of the molecular consequences underlying ICM will provide valuable information on ICM disease progression and possible therapeutic interventions.
Supplementary Material
Supplementary Figure S1. Sarcomeric proteoform levels between left ventricle and apex tissues in donor hearts are comparable.
Supplementary Figure S2. SDS-PAGE of HEPES (H) and TFA (T) extracts from donor and ICM human cardiac tissues.
Supplementary Figure S3. Online LC-MS/MS characterization of MLC-1v.
Supplementary Figure S4. Online LC-MS/MS characterization of MLC-2v.
Supplementary Figure S5. SDS-PAGE of HEPES (H) and TFA (T) from three extraction replicates from donor human cardiac tissue.
Supplementary Figure S6. Extraction replicates showing highly similar and reproducible chromatographic profiles.
Supplementary Figure S7. Mutual linear range response determination for major sarcomeric proteins.
Supplementary Figure S8. Quantification of full-length and truncated cTnT phosphorylation.
Supplementary Figure S9. cTnT point mutation present in ICM samples.
Supplementary Figure S10. Quantification of calsarcin-1 proteoform phosphorylation.
Supplementary Table S1. Available clinical data for donors and ICM patients.
Supplementary Table S2. Generation of extracted ion chromatograms for sarcomeric proteins.
Supplementary Table S3. Sarcomeric proteoforms identified.
Supplementary Table S4. Proteoform changes in ICM.
Acknowledgements
This work was supported by NIH R01 HL109810 (to Y.G.). Y.G. also would like to acknowledge R01 HL096971, R01 GM125085, and S10 OD018475. E.A.C. would like to acknowledge support from the NIH Chemistry-Biology Interface Training Program NIH T32GM008505. T.J.A. would like to acknowledge support from the Training Program in Molecular and Cellular Pharmacology, T32 GM008688-20. J.A.M. and T.Z would like to acknowledge support from the Training Program in Translational Cardiovascular Science, T32 HL007936. K.J.R. would like to acknowledge support from the National Science Foundation Graduate Research Fellowship Program, DGE-1747503 and the Graduate School and the Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison, funded by Wisconsin Alumni Research Foundation.
Abbreviations
- MS
mass spectrometry
- LC
liquid chromatography
- MS/MS
tandem MS
- Q-TOF
quadrupole time of flight
- LV
left ventricle
- LVAD
left ventricular assist device
- ICM
ischemic cardiomyopathy
- HCM
hypertrophic cardiomyopathy
- DCM
dilated cardiomyopathy
- PTM
post-translational modification
- TFA
trifluoroacetic acid
- CAD
collision activated dissociation
- BPC
base peak chromatogram
- TIC
total ion chromatogram
- EIC
extracted ion chromatogram
- AUC
area under curve
- PKA
protein kinase A
- PKC
protein kinase C
- MLP
muscle lim protein
- CRIP2
cysteine-rich protein 2
- cTnT
cardiac troponin T
- ENH2
enigma homolog 2
- FHL2
four and a half LIM domain protein 2
- cTnI
cardiac troponin I
- tpm
tropomyosin
- MLC
myosin light chain
- TnC
troponin C
Footnotes
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD038066.
References
- (1).Virani SS; Alonso A; Aparicio HJ; Benjamin EJ; Bittencourt MS; Callaway CW; Carson AP; Chamberlain AM; Cheng S; Delling FN; et al. Heart Disease and Stroke Statistics-2021 Update: A Report From the American Heart Association. Circulation 2021, 143 (8), E254–E743. [DOI] [PubMed] [Google Scholar]
- (2).Mudd JO; Kass DA Tackling Heart Failure in the Twenty-First Century. Nat. 2008 4517181 2008, 451 (7181), 919–928. [DOI] [PubMed] [Google Scholar]
- (3).Arrigo M; Jessup M; Mullens W; Reza N; Shah AM; Sliwa K; Mebazaa A Acute Heart Failure. Nat. Rev. Dis. Prim 2020 61 2020, 6 (1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ziaeian B; Fonarow GC Epidemiology and Aetiology of Heart Failure. Nat. Rev. Cardiol 2016, 13 (6), 368–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Briceno N; Schuster A; Lumley M; Perera D Ischaemic Cardiomyopathy: Pathophysiology, Assessment and the Role of Revascularisation. Heart 2016, 102 (5), 397–406. [DOI] [PubMed] [Google Scholar]
- (6).Felker GM; Shaw LK; O’Connor CM A Standardized Definition of Ischemic Cardiomyopathy for Use in Clinical Research. J. Am. Coll. Cardiol 2002, 39 (2), 210–218. [DOI] [PubMed] [Google Scholar]
- (7).Bloom MW; Greenberg B; Jaarsma T; Januzzi JL; Lam CSP; Maggioni AP; Trochu JN; Butler J Heart Failure with Reduced Ejection Fraction. Nat. Rev. Dis. Prim 2017, 3, 17058. [DOI] [PubMed] [Google Scholar]
- (8).Yuan C; Solaro RJ Myofilament Proteins: From Cardiac Disorders to Proteomic Changes. Proteomics. Clin. Appl 2008, 2 (6), 788–799. [DOI] [PubMed] [Google Scholar]
- 9.) van der Velden J; Stienen GJM Cardiac Disorders and Pathophysiology of Sarcomeric Proteins. Physiol. Rev 2019, 99 (1), 381–426. [DOI] [PubMed] [Google Scholar]
- (10).Hwang PM; Sykes BD Targeting the Sarcomere to Correct Muscle Function. Nat. Rev. Drug Discov 2015 145 2015, 14 (5), 313–328. [DOI] [PubMed] [Google Scholar]
- (11).Solís C; John Solaro R Novel Insights into Sarcomere Regulatory Systems Control of Cardiac Thin Filament Activation. J. Gen. Physiol 2021, 153 (7), e202012777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Solaro RJ; Henze M; Kobayashi T Integration of Troponin I Phosphorylation With Cardiac Regulatory Networks. Circ. Res 2013, 112 (2), 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Pyle WG; Solaro RJ At the Crossroads of Myocardial Signaling. Circ. Res 2004, 94 (3), 296–305. [DOI] [PubMed] [Google Scholar]
- (14).Wang X; Su H Unraveling Enigma in the Z-Disk. Circ. Res 2010, 107 (3), 321–323. [DOI] [PubMed] [Google Scholar]
- (15).Fert-Bober J; Murray CI; Parker SJ; Van Eyk JE Precision Profiling of the Cardiovascular Post-Translationally Modified Proteome. Circ. Res 2018, 122 (9), 1221–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Marston SB; Walker JW Back to the Future: New Techniques Show That Forgotten Phosphorylation Sites Are Present in Contractile Proteins of the Heart Whilst Intensively Studied Sites Appear to Be Absent. J. Muscle Res. Cell Motil 2009, 30 (3–4), 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ramirez-Correa GA; Jin W; Wang Z; Zhong X; Gao WD; Dias WB; Vecoli C; Hart GW; Murphy AM O-Linked GlcNAc Modification of Cardiac Myofilament Proteins. Circ. Res 2008, 103 (12), 1354–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Solaro RJ; Warren CM; Scruggs SB Why Is It Important to Analyze the Cardiac Sarcomere Subproteome? Expert Rev. Proteomics 2010, 7 (3), 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Lau E; Han Y; Williams DR; Thomas CT; Shrestha R; Wu JC; Lam MPY Splice-Junction-Based Mapping of Alternative Isoforms in the Human Proteome. Cell Rep. 2019, 29 (11), 3751–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Smith LM; Kelleher NL; Proteomics TC for T. D. Proteoform: A Single Term Describing Protein Complexity. Nat. Methods 2013, 10 (3), 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Aebersold R; Agar JN; Amster IJ; Baker MS; Bertozzi CR; Boja ES; Costello CE; Cravatt BF; Fenselau C; Garcia BA; et al. How Many Human Proteoforms Are There? Nat. Chem. Biol 2018, 14 (3), 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Smith LM; Kelleher NL Proteoforms as the next Proteomics Currency. Science 2018, 359 (6380), 1106–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Gregorich ZR; Ge Y Top-down Proteomics in Health and Disease: Challenges and Opportunities. Proteomics 2014, 14 (10), 1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Chen B; Brown KA; Lin Z; Ge Y Top-Down Proteomics: Ready for Prime Time? Anal. Chem 2018, 90 (1), 110–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zhang Y; Fonslow BR; Shan B; Baek MC; Yates JR Protein Analysis by Shotgun/Bottom-up Proteomics. Chem. Rev 2013, 113 (4), 2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Tucholski T; Cai W; Gregorich ZR; Bayne EF; Mitchell SD; McIlwain SJ; de Lange WJ; Wrobbel M; Karp H; Hite Z; et al. Distinct Hypertrophic Cardiomyopathy Genotypes Result in Convergent Sarcomeric Proteoform Profiles Revealed by Top-down Proteomics. Proc. Natl. Acad. Sci. U. S. A 2020, 117 (40), 24691–24700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Tiambeng TN; Roberts DS; Brown KA; Zhu Y; Chen B; Wu Z; Mitchell SD; Guardado-Alvarez TM; Jin S; Ge Y Nanoproteomics Enables Proteoform-Resolved Analysis of Low-Abundance Proteins in Human Serum. Nat. Commun 2020 111 2020, 11 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Melby JA; Roberts DS; Larson EJ; Brown KA; Bayne EF; Jin S; Ge Y Novel Strategies to Address the Challenges in Top-Down Proteomics. J. Am. Soc. Mass Spectrom 2021, 32 (6), 1278–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Smith LM; Agar JN; Chamot-Rooke J; Danis PO; Ge Y; Loo JA; Paša-Toliæ L; Tsybin YO; Kelleher NL The Human Proteoform Project: Defining the Human Proteome. Sci. Adv 2021, 7 (46), 734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Drown BS; Jooß K; Melani RD; Lloyd-Jones C; Camarillo JM; Kelleher NL Mapping the Proteoform Landscape of Five Human Tissues. J. Proteome Res 2022, 21 (5), 1299–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Cai W; Tucholski TM; Gregorich ZR; Ge Y Top-down Proteomics: Technology Advancements and Applications to Heart Diseases. Expert Rev. Proteomics 2016, 13 (8), 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Gregorich ZR; Chang YH; Ge Y Proteomics in Heart Failure: Top-down or Bottom-Up? Pflugers Arch. 2014, 466 (6), 1199–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhang J; Guy MJ; Norman HS; Chen YC; Xu Q; Dong X; Guner H; Wang S; Kohmoto T; Young KH; et al. Top-down Quantitative Proteomics Identified Phosphorylation of Cardiac Troponin I as a Candidate Biomarker for Chronic Heart Failure. J. Proteome Res 2011, 10 (9), 4054–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tiambeng TN; Tucholski T; Wu Z; Zhu Y; Mitchell SD; Roberts DS; Jin Y; Ge Y Analysis of Cardiac Troponin Proteoforms by Top-down Mass Spectrometry. Methods Enzymol. 2019, 626, 347–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Lin Z; Wei L; Cai W; Zhu Y; Tucholski T; Mitchell SD; Guo W; Ford SP; Diffee GM; Ge Y Simultaneous Quantification of Protein Expression and Modifications by Top-down Targeted Proteomics: A Case of the Sarcomeric Subproteome. Mol. Cell. Proteomics 2019, 18 (3), 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Han JJ; Acker MA; Atluri P Left Ventricular Assist Devices. Circulation 2018, 138 (24), 2841–2851. [DOI] [PubMed] [Google Scholar]
- (37).Cai W; Hite ZL; Lyu B; Wu Z; Lin Z; Gregorich ZR; Messer AE; McIlwain SJ; Marston SB; Kohmoto T; et al. Temperature-Sensitive Sarcomeric Protein Post-Translational Modifications Revealed by Top-down Proteomics. J. Mol. Cell. Cardiol 2018, 122, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Nakayasu ES; Gritsenko M; Piehowski PD; Gao Y; Orton DJ; Schepmoes AA; Fillmore TL; Frohnert BI; Rewers M; Krischer JP; et al. Tutorial: Best Practices and Considerations for Mass-Spectrometry-Based Protein Biomarker Discovery and Validation. Nat. Protoc 2021 168 2021, 16 (8), 3737–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Wu Z; Roberts DS; Melby JA; Wenger K; Wetzel M; Gu Y; Ramanathan SG; Bayne EF; Liu X; Sun R; et al. MASH Explorer: A Universal Software Environment for Top-Down Proteomics. J. Proteome Res 2020, 19 (9), 3867–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Kou Q; Xun L; Liu X TopPIC: A Software Tool for Top-down Mass Spectrometry-Based Proteoform Identification and Characterization. Bioinformatics 2016, 32 (22), 3495–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Li A; Ponten F; dos Remedios CG The Interactome of LIM Domain Proteins: The Contributions of LIM Domain Proteins to Heart Failure and Heart Development. Proteomics 2012, 12 (2), 203–225. [DOI] [PubMed] [Google Scholar]
- (42).Wadmore K; Azad AJ; Gehmlich K The Role of Z-Disc Proteins in Myopathy and Cardiomyopathy. Int. J. Mol. Sci 2021, 22 (6), 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Cai W; Tucholski T; Chen B; Alpert AJ; McIlwain S; Kohmoto T; Jin S; Ge Y Top–down Proteomics of Large Proteins up to 223 KDa Enabled by Serial Size Exclusion Chromatography Strategy. Anal. Chem 2017, 89 (10), 5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Compton PD; Zamdborg L; Thomas PM; Kelleher NL On the Scalability and Requirements of Whole Protein Mass Spectrometry. Anal. Chem 2011, 83 (17), 6868–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Gregorich ZR; Cai W; Lin Z; Chen AJ; Peng Y; Kohmoto T; Ge Y Distinct Sequences and Post-Translational Modifications in Cardiac Atrial and Ventricular Myosin Light Chains Revealed by Top-down Mass Spectrometry. J. Mol. Cell. Cardiol 2017, 107, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Scruggs SB; Reisdorph R; Armstrong ML; Warren CM; Reisdorph N; Solaro RJ; Buttrick PM A Novel, In-Solution Separation of Endogenous Cardiac Sarcomeric Proteins and Identification of Distinct Charged Variants of Regulatory Light Chain. Mol. Cell. Proteomics 2010, 9 (9), 1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Chang AN; Chen G; Gerard RD; Kamm KE; Stull JT Cardiac Myosin Is a Substrate for Zipper-Interacting Protein Kinase (ZIPK). J. Biol. Chem 2010, 285 (8), 5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Wang C; Taskinen JH; Segersvärd H; Immonen K; Kosonen R; Tolva JM; Mäyränpää MI; Kovanen PT; Olkkonen VM; Sinisalo J; et al. Alterations of Cardiac Protein Kinases in Cyclic Nucleotide-Dependent Signaling Pathways in Human Ischemic Heart Failure. Front. Cardiovasc. Med 2022, 0, 1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Liu Y; Chen J; Fontes SK; Bautista EN; Cheng Z Physiological and Pathological Roles of Protein Kinase A in the Heart. Cardiovasc. Res 2022, 118 (2), 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Grandi E; Ripplinger CM Antiarrhythmic Mechanisms of Beta Blocker Therapy. Pharmacol. Res 2019, 146, 104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Ong HT β Blockers in Hypertension and Cardiovascular Disease. BMJ Br. Med. J 2007, 334 (7600), 946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Peng Y; Ayaz-Guner S; Yu D; Ge Y Top-down Mass Spectrometry of Cardiac Myofilament Proteins in Health and Disease. PROTEOMICS – Clin. Appl 2014, 8 (7–8), 554–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Peng Y; Yu D; Gregorich Z; Chen X; Beyer AM; Gutterman DD; Ge Y In-Depth Proteomic Analysis of Human Tropomyosin by Top-down Mass Spectrometry. J. Muscle Res. Cell Motil 2013, 34 (3–4), 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Jin Y; Peng Y; Lin Z; Chen YC; Wei L; Hacker TA; Larsson L; Ge Y Comprehensive Analysis of Tropomyosin Isoforms in Skeletal Muscles by Top-down Proteomics. J. Muscle Res. Cell Motil 2016, 37 (1–2), 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Peng Y; Yu D; Gregorich Z; Chen X; Beyer AM; Gutterman DD; Ge Y In-Depth Proteomic Analysis of Human Tropomyosin by Top-down Mass Spectrometry. J. Muscle Res. Cell Motil 2013, 34 (3–4), 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Geeves MA; Hitchcock-DeGregori SE; Gunning PW A Systematic Nomenclature for Mammalian Tropomyosin Isoforms. J. Muscle Res. Cell Motil 2015, 36 (2), 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Chen YC; Ayaz-Guner S; Peng Y; Lane NM; Locher MR; Kohmoto T; Larsson L; Moss RL; Ge Y Effective Top-down LC/MS+ Method for Assessing Actin Isoforms as a Potential Cardiac Disease Marker. Anal. Chem 2015, 87 (16), 8399–8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Garg P; Morris P; Fazlanie AL; Vijayan S; Dancso B; Dastidar AG; Plein S; Mueller C; Haaf P Cardiac Biomarkers of Acute Coronary Syndrome: From History to High-Sensitivity Cardiac Troponin. Intern. Emerg. Med 2017, 12 (2), 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Zhang J; Zhang H; Ayaz-Guner S; Chen YC; Dong X; Xu Q; Ge Y Phosphorylation, but Not Alternative Splicing or Proteolytic Degradation, Is Conserved in Human and Mouse Cardiac Troponin T. Biochemistry 2011, 50 (27), 6081–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Zhang J; Zhang H; Ayaz-Guner S; Chen YC; Dong X; Xu Q; Ge Y Phosphorylation, but Not Alternative Splicing or Proteolytic Degradation, Is Conserved in Human and Mouse Cardiac Troponin T. Biochemistry 2011, 50 (27), 6081–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Perry SV Troponin T: Genetics, Properties and Function. J. Muscle Res. Cell Motil 1998, 19 (6), 575–602. [DOI] [PubMed] [Google Scholar]
- (62).Wei B; Jin JP Troponin T Isoforms and Posttranscriptional Modifications: Evolution, Regulation and Function. Arch. Biochem. Biophys 2011, 505 (2), 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Millat G; Bouvagnet P; Chevalier P; Sebbag L; Dulac A; Dauphin C; Jouk PS; Delrue MA; Thambo JB; Le Metayer P; et al. Clinical and Mutational Spectrum in a Cohort of 105 Unrelated Patients with Dilated Cardiomyopathy. Eur. J. Med. Genet 2011, 54 (6), 570–575. [DOI] [PubMed] [Google Scholar]
- (64).Nakajima-Taniguchi C; Matsui H; Fujio Y; Nagata S; Kishimoto T; Yamauchi-Takihara K Novel Missense Mutation in Cardiac Troponin T Gene Found in Japanese Patient with Hypertrophic Cardiomyopathy. J. Mol. Cell. Cardiol 1997, 29 (2), 839–843. [DOI] [PubMed] [Google Scholar]
- (65).Vafiadaki E; Arvanitis DA; Sanoudou D Muscle Lim Protein: Master Regulator of Cardiac and Skeletal Muscle Function. Gene 2015, 566 (1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Frey N; Richardson JA; Olson EN Calsarcins, a Novel Family of Sarcomeric Calcineurin-Binding Proteins. Proc. Natl. Acad. Sci. U. S. A 2000, 97 (26), 14632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Frank D; Kuhn C; Van Eickels M; Gehring D; Hanselmann C; Lippl S; Will R; Katus HA; Frey N Calsarcin-1 Protects against Angiotensin-II-Induced Cardiac Hypertrophy. Circulation 2007, 116 (22), 2587–2596. [DOI] [PubMed] [Google Scholar]
- (68).Buyandelger B; Ng KE; Miocic S; Piotrowska I; Gunkel S; Ku CH; Knöll R MLP (Muscle LIM Protein) as a Stress Sensor in the Heart. Pflugers Arch. 2011, 462 (1), 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Roselló-Lletí E; Alonso J; Cortés R; Almenar L; Martínez-Dolz L; Sánchez-Lázaro I; Lago F; Azorín I; Juanatey JRG; Portolés M; et al. Cardiac Protein Changes in Ischaemic and Dilated Cardiomyopathy: A Proteomic Study of Human Left Ventricular Tissue. J. Cell. Mol. Med 2012, 16 (10), 2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Yi X; Jiang DS; Feng G; Jiang X jun; Zeng HL An Altered Left Ventricle Protein Profile in Human Ischemic Cardiomyopathy Revealed in Comparative Quantitative Proteomics. Kardiol. Pol 2019, 77 (10), 951–959. [DOI] [PubMed] [Google Scholar]
- (71).Plubell DL; Käll L; Webb-Robertson BJ; Bramer LM; Ives A; Kelleher NL; Smith LM; Montine TJ; Wu CC; Maccoss MJ Putting Humpty Dumpty Back Together Again: What Does Protein Quantification Mean in Bottom-Up Proteomics? J. Proteome Res 2022, 21 (4), 891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Peng Y; Gregorich ZR; Valeja SG; Zhang H; Cai W; Chen YC; Guner H; Chen AJ; Schwahn DJ; Hacker TA; et al. Top-down Proteomics Reveals Concerted Reductions in Myofilament and Z-Disc Protein Phosphorylation after Acute Myocardial Infarction. Mol. Cell. Proteomics 2014, 13 (10), 2752–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Sarcomeric proteoform levels between left ventricle and apex tissues in donor hearts are comparable.
Supplementary Figure S2. SDS-PAGE of HEPES (H) and TFA (T) extracts from donor and ICM human cardiac tissues.
Supplementary Figure S3. Online LC-MS/MS characterization of MLC-1v.
Supplementary Figure S4. Online LC-MS/MS characterization of MLC-2v.
Supplementary Figure S5. SDS-PAGE of HEPES (H) and TFA (T) from three extraction replicates from donor human cardiac tissue.
Supplementary Figure S6. Extraction replicates showing highly similar and reproducible chromatographic profiles.
Supplementary Figure S7. Mutual linear range response determination for major sarcomeric proteins.
Supplementary Figure S8. Quantification of full-length and truncated cTnT phosphorylation.
Supplementary Figure S9. cTnT point mutation present in ICM samples.
Supplementary Figure S10. Quantification of calsarcin-1 proteoform phosphorylation.
Supplementary Table S1. Available clinical data for donors and ICM patients.
Supplementary Table S2. Generation of extracted ion chromatograms for sarcomeric proteins.
Supplementary Table S3. Sarcomeric proteoforms identified.
Supplementary Table S4. Proteoform changes in ICM.