Abstract
Background
There is a limited literature describing the oral microbiome and its diagnostic potential in paediatric inflammatory bowel disease [IBD].
Methods
We examined the dorsum tongue microbiome by V1–V2 sequencing in a cohort of 156 treatment-naïve children diagnosed with IBD compared to 102 healthy control children. Microbiome changes over time following treatment were examined in a subset of patients and associations between IBD diagnosis and dysbiosis were explored.
Results
Analysis of community structure of the microbiome in tongue samples revealed that IBD samples diverged significantly from healthy control samples [PERMANOVA p = 0.0009] and exhibited a reduced abundance of Clostridia in addition to several major oral genera [Veillonella, Prevotella and Fusobacterium species] with an increased abundance of streptococci. This dysbiosis was more marked in patients with severe disease. Higher levels of the potential pathobionts Klebsiella and Pseudomonas spp. were also associated with IBD. In terms of predicted functions, the IBD oral microbiome was potentially more acidogenic and exhibited reduced capacity for B vitamin biosynthesis. We used a machine learning approach to develop a predictive model of IBD which exhibited a mean-prediction AUC [area under the ROC curve] of 0.762. Finally, we examined a subset of 53 patients following 12 months of therapy and could show resolution of oral dysbiosis as demonstrated by a shift towards a healthy community structure and a significant reduction in oral dysbiosis.
Conclusion
Oral dysbiosis found in children with IBD is related to disease severity and resolves over time following successful IBD treatment.
Keywords: Oral microbiome, paediatric, IBD, dysbiosis
1. Introduction
Inflammatory bowel disease [IBD], which includes Crohn’s disease [CD] and ulcerative colitis [UC], is a complex disorder that is partly the result of an inappropriate immune response to the patient’s gut microbiota.1 The exact role of the microbiome in the pathogenesis of IBD is still under investigation but current evidence suggests that the disease involves a strong inflammatory response that may be triggered by acquired infection or alternatively by dysbiotic changes to the host’s own microbiome. In CD, the cause of this disruption in host–microbe homeostasis is unknown, but studies have suggested roles for environmental factors, inherited genetic factors and possibly transit of microorganisms from the oral cavity in the perturbation of the relationship between the mucosal immune system and the gut microbiota.2–4
Within the last decade, microbiome studies have unexpectedly identified increased abundance of common oral taxa in the gut microbiomes of patients with IBD [e.g. Veillonella, Haemophilus and Eikenella spp.] whereas many bacteria with important roles in the generation of short-chain fatty acids [SCFAs] were reduced.5–8 The involvement of oral taxa in the pathogenesis of IBD has been suggested and experiments have shown that inoculation of the oral microbiome from children with IBD in germ-free mice resulted in colonization with Fusobacterium, Veillonella and Klebsiella spp. and the accumulation of inflammatory TH1 cells.9 These results suggest that the oral cavity may be a reservoir for pathobionts, and that transit from the oral cavity to the gut may play a role in inducing inflammation in IBD patients.9
Inflammation in CD most frequently affects the distal ileum and colon but may occur in any part of the gastrointestinal tract including the mouth.10 Oral CD [OCD] is a phenomenon that can occur in patients with intestinal CD and is characterized by a wide variety of disease-specific oral lesions.11 The most frequently described oral lesions in CD patients include swelling of the lips, buccal mucosal swelling or ‘cobble-stoning’, mucogingivitis, deep linear ulceration along the buccal gutters and mucosal tags.11,12 The oral manifestations of IBD are diverse and based on their relationship with CD activity can be classified as specific [e.g. mucosal swelling] and non-specific oral lesions [e.g. angular cheilitis].13 Some studies have suggested that oral manifestations are a good cutaneous marker of IBD activity14 and are useful diagnostic markers.15 Although oral manifestations of CD are well described in the literature, the nature and the extent of dysbiosis in the oral microbiome in IBD patients is still poorly characterized. 16S rRNA gene sequence-based approaches have been used to examine the salivary microbiome in IBD.16–21 Taken together, these sequence-based studies show that oral dysbiosis occurs in IBD, although the exact nature of the dysbiosis characterized differs between studies. This may be due to the relatively small patient cohorts examined [the largest study cited here included 80 IBD patients] and the failure to recruit treatment-naïve patients who have not yet been prescribed disease-modulating medications. In addition, none have exclusively focused on children and do not include longitudinal elements to determine if treatment is associated with reversal of dysbiosis. Despite these limitations, most studies concur that the genera Prevotella, Haemophilus and Veillonella are somehow affected in the oral cavity in IBD, which corresponds to the genera shown to be enriched in the gut microbiota in CD patients.5 Orofacial granulomatosis [OFG], an oral condition that may be related to IBD, has also been investigated which identified a specific sequence type [oligotype] of Streptococcus salivarius with increased abundance in individuals with CD or OFG compared to controls and may have promise as an early biomarker of CD.22
In the current study we characterize the oral microbiome profiles of children with and without IBD, examine associations between dysbiosis and clinical features and explore the impact of IBD treatment on oral dysbiosis over time.
2. Materials and Methods
2.1. Patients
Ireland has a single national centre for paediatric gastroenterology and all Irish children up to age 16 years with IBD are diagnosed and managed at the centre. The DOCHAS study [Determinants and Outcomes of Children and Adolescents with IBD] is a national prospective inception cohort study of paediatric IBD, which commenced in 2012 following research ethics committee approval at thr Children’s Health Ireland at Crumlin, Dublin [GEN/193/11]. Since 2012, 94% of all new patients have been enrolled in DOCHAS, and thus the data are reflective of the national disease cohort. For this sub-study of DOCHAS, patients were eligible for inclusion if they were <16 years old at diagnosis and diagnosed with IBD between April 1, 2014 and October 31, 2018. Patients were excluded from this study if they had been diagnosed outside Ireland, had received induction treatment or antibiotics prior to endoscopy, or had evidence of immunodeficiency or monogenic disease.23
Control subjects included patients who did not have IBD but were either undergoing endoscopy for other clinical indications, or attending the Dublin Dental University Hospital [DDUH] for dental check-ups. Samples recovered from children attending DDUH were recovered following ethical approval from the Joint Hospitals’ Research Ethics Committee, Dublin [015-07 Chairman’s Action17]. Children were excluded if they had taken antibiotics in the previous 2 months or were using steroid inhalers.
Diagnosis of IBD was based on clinical, endoscopic and histological features as per established criteria.23 Disease extent was determined by the performing endoscopist and categorized as per the Paris classification.24 Data were collected using a standardized form at diagnosis: age, gender, ethnicity, family history, environmental background [urban/rural dwelling, home smoking exposure], treatments, clinical and endoscopic activity scores, and presence or absence of mucosal atrophy. Disease activity was assigned per the Pediatric Ulcerative Colitis Activity Index [PUCAI] score and Pediatric Crohn’s Disease Activity Index [PCDAI] as appropriate.25,26 Follow-up data including disease activity, treatments, occurrence of relapse, medication escalation at time of relapse and need for surgery were collected prospectively at 3, 6 and 12 months after diagnosis and yearly thereafter using standardized forms. Patients with CD treated with exclusive enteral nutrition [EEN] were treated exclusively with polymeric formulas to provide 100% of the estimated energy and nutritional requirements calculated following clinical dietetic assessment for 8 weeks of duration, followed by stepwise reintroduction of standard diet over the subsequent 2 weeks. Steroid-free remission was defined as PUCAI or PCDAI ≤ 10 without steroids in the preceding 3 months [excluding the initial 3 months post-diagnosis]. Clinical relapse was defined as an increase in disease activity score plus the need for dose intensification or adjunctive therapy or hospitalization. To investigate the relationship between disease severity and the level of dysbiosis, CD disease activity was defined by PCDAI score as follows: mild ≥10, moderate to severe≥30. To define the microbiome in severe disease, we defined severe CD as PCDAI ≥ 50. For analysis of UC patients, mild disease was classed as PUCAI ≥ 10 and moderate to severe as ≥35. To characterize the microbiome in severe disease, a PUCAI ≥ 65 was used to define severe UC.
2.2. Swabbing protocol
The oral microbiome was sampled using a polyurethane sponge swab [CultureSwab EZ, Becton, Dickinson] to sample the dorsum of the tongue in all participants, and additionally of the buccal mucosa in the initial cohort. Swabs were transferred to −80°C immediately and processed within 6 months. The first 96 children recruited to the study [referred to as the initial cohort] also had a buccal swab taken and, within this group, 73 children also received a full oral examination from a dentist to record decayed, missing and filled teeth [DMFT], the presence of active caries, gingivitis or mucosal ulceration. Swabs were taken on all patients at baseline, while treatment naïve. Swabs were taken at follow-up timepoints from patients undergoing induction treatment with polymeric enteral nutrition for CD at 6 weeks and after 12 months post-diagnosis.
2.3 DNA extraction and sequencing
DNA was extracted from mucosal swabs using the MasterPure Complete DNA/RNA Purification Kit [Epicentre Biotechnologies]. The manufacturer’s extraction protocol was supplemented with an additional incubation step with Ready-Lyse lysozyme [Epicentre Biotechnologies] and a bead disruption step, as described by Amer et al.27 The DNA was resuspended in Tris-EDTA buffer [pH 7.5] and stored at −80°C.
Amplification of the V1–V2 region of the 16S rRNA gene was carried using the KAPA HiFi Hot start system [KapaBiosystems] with the primers 27F-YM and 338R-R [27F-YM: 5ʹTCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGTCAGTCTGTCAGAGTTTGATYMTGGCTCAG; 338R-R: 5ʹGTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTATGGTAATTCATGCTGCCTCCCGTAGRAGT].28 The V1–V2 region was selected on the basis that ~90% of species in the Human Oral Microbiome Database [HOMD] can be correctly identified using these sequences.29 Sample indexing was carried out with the Nextera XT Index Kit [Illumina] and library quantification and purification were carried out according to the Illumina protocol ‘16S Metagenomic Sequence Library Preparation’.30 The size and integrity of indexed amplimers were determined using a Bioanalyzer [Agilent Technologies] and samples were normalized to 4 nM. Samples were combined to generate a pooled library, denatured and combined with PhiX control DNA [5%] and loaded at a concentration of 6 pM. Paired-end sequencing was performed using the Illumina 500-cycle MiSeq reagent kit.
2.4. Data analysis
Bacterial 16S rRNA sequences were processed and filtered using Dada230 with the following parameters: maxN = 0, maxEE = c[2,5] and truncQ = 2.31 The ten terminal bases were trimmed from reverse reads due to the drop in sequence quality. Following error estimation and correction using the Dada2 algorithm, paired reads were merged and chimeras were removed following their identification with the removeBimeraDenovo command. Taxonomy was assigned using the HOMD classifications [eHOMD 16S rRNA RefSeq version 15.1].32 Phyloseq was used to calculate alpha diversity and richness indices for each sample.33 For beta diversity analysis, the Aitchison distance metric was used by applying principal components analysis [PCA] to the centre log-ratio [CLR]-transformed counts using package ‘microbiome’ in R studio. Further visualizations [Iris plots, biplots] and analysis of community structure, including PERMANOVA [permutational multivariate analysis of variance], were conducted using the package ‘MicroViz’ in R studio.34,35 Multivariate analysis was conducted with MaAsLin 2 using CLR-transformed counts and the indicated patient metadata variables as ‘fixed-effects’.35 Further differential analysis was carried out using a combination of methods including DeSeq236 and following normalization to the geometric mean for each taxon and using the check.associations function in ‘Siamcat’, which applies a non-parametric Wilcoxon test on relative abundances.37 All p values were adjusted for multiple comparisons using the Benjamini–Hochberg correction.
2.5. Dysbiosis index
An oral dysbiosis index [ODI] was generated based on the analytical approach used to describe intestinal dysbiosis in paediatric CD by Gevers et al.5 The ODI is the log of [total abundance in organisms increased in IBD] over [total abundance of organisms decreased in IBD] for all samples. Based on empirical analysis of the results from MaAsLin 2, the ratio of Bacilli and Flavobacteriia over Clostridia, Negativicutes, Betaproteobacteria and Fusobacteriia was chosen to generate the dysbiosis index presented here.
2.6. Machine learning
Breiman’s random forest algorithm was applied to the data using the SiamCat package in R studio in order to determine whether oral microbiome profiles could distinguish IBD patients from healthy controls.38 To develop a model that could be cross-validated, the data were split into a training set [80%], which was used to train the machine learning model, and a test set [20%], which was used to make predictions based on the model. We created 100 such random splits for the training and test datasets. The random forest classifier was run on the training set, with 1000 random trees, and the predicted model was then used on the test data set to obtain the area under the ROC curve [AUC].
2.7. Metagenome prediction
PICRUSt 2 was used to predict the metagenome from 16S rRNA community profiles. Sequence variants were included if they appeared more than twice in at least 5% of samples. Data were converted to BIOM format using make.biom in Mothur and PICRUSt 2 was used to generate a table of overall EC number abundances and MetaCyc pathway abundances for each sample.39,40 Statistical analysis of the relative abundances of each data set was carried out using LEfSe.39
3. Results
A total of 225 treatment-naïve children were prospectively swabbed at the gastroenterology centre; 156 children were diagnosed with IBD (94 with CD, 52 with UC and ten IBD unclassified [IBD-U]). The ten children with IBD-U were excluded from analyses of CD and UC accordingly. The remaining 69 children attending the clinic were confirmed not to have IBD and included as controls for this study. An additional 33 non-IBD controls were recruited from children attending the Dublin Dental University Hospital for routine dental check-up, resulting in a total of 102 non-IBD control children recruited for the study. The mean age of controls was 11.2 [+3.9] years and patients was 12.1 [+2.8] and was not significantly different [Mann–Whitney p = 0.06]. Males outnumbered females in both cases and controls [IBD M:F ratio 2.6:1; Control M:F ratio 2:1]. The baseline characteristics for the entire cohort are summarized in Table 1 and full clinical parameters are given in Supplementary Table 1.
Table 1.
Characteristics of the cohort. Phenotype details were assessed according to the Paris classification17
| Patient category | |||||
|---|---|---|---|---|---|
| IBD | CD | UC | Controls | ||
| n | 146 | 94 | 52 | 102 | |
| Mean age [+SD] | 12.1 [2.8] | 11.8 [2.9] | 12.3 [2.8] | 11.2 [3.9] | |
| Male:female ratio | 2.6:1 | 3:1 | 1.8:1 | 2:1 | |
| Paris phenotype, n [%] | |||||
| L1 | — | 6 [6] | E1 | 4 [8] | — |
| L2 | — | 24 [26] | E2 | 8 [15] | — |
| L3 | — | 57 [61] | E3 | 2 [4] | — |
| +L4a | — | 37 [39] | E4 | 35 [67] | — |
| +L4b | — | 14 [15] | S1 | 18 [35] | — |
| B1 | — | 92 [98] | — | — | |
| B2 | — | 2 [2] | |||
| +p | — | 19 [20] | |||
| G1 | — | 11 [11] | |||
| Baseline disease activity [per PCDAI or PUCAI], n [%] | |||||
| Mild | — | 32 [34] | 13 [25] | — | |
| Moderate | — | 26 [28] | 23 [44] | — | |
| Severe | — | 36 [38] | 16 [31] | — | |
IBD, inflammatory bowel disease; UC, ulcerative colitis; CD, Crohn;s disease; PCDAI, Pediatric Crohn’s Disease Activity Index; PUCAI, Pediatric Ulcerative Colitis Activity Index.
3.1. Analysis of the oral microbiome in IBD
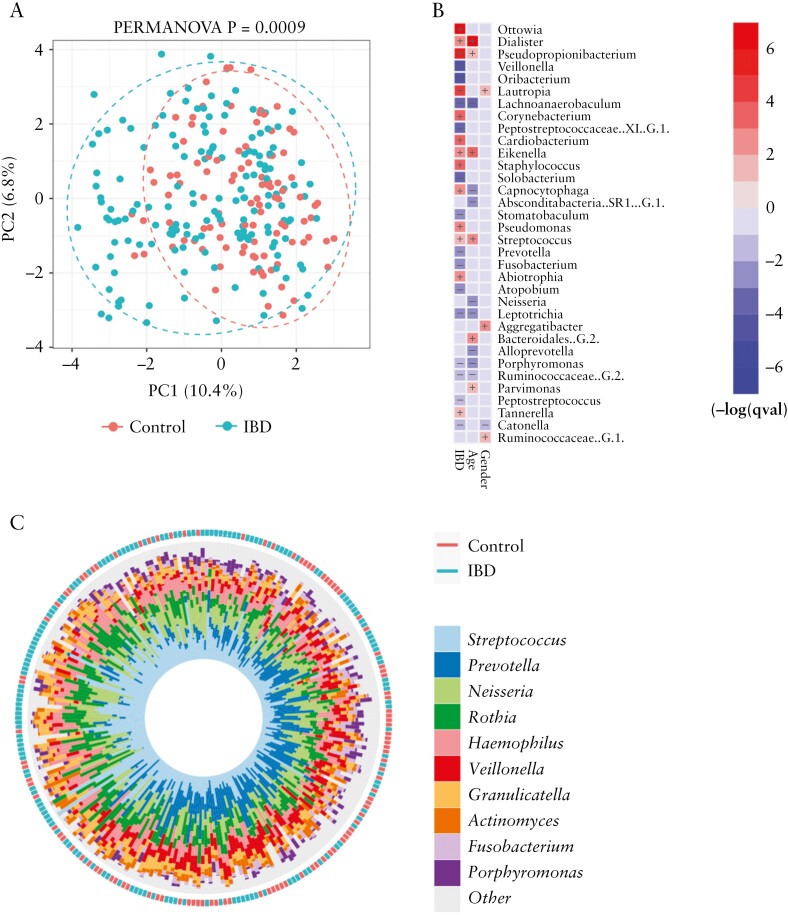
We first determined the optimal site for sampling, given the potential for microbial variance within the oral cavity. The first 96 children sampled [henceforth called the ‘initial cohort’, IBD = 59, non-IBD = 37] had both a dorsal tongue and buccal mucosal swab taken to determine whether buccal or tongue swabs were more effective at identifying oral dysbiosis in paediatric IBD. The V1–V2 region of the 16S rRNA gene was sequenced from all swabs taken from study participants. The buccal and tongue communities of IBD and healthy controls were compared using the Aitchison distance metric. This analysis showed that tongue microbiome communities from healthy children and IBD children exhibited significant divergence [PERMANOVA p = 0.0009; Supplementary Figure 1a] whereas buccal communities were more similar [p = 0.023; Supplementary Figure 1b]. Within this initial cohort, 73 children also underwent a full oral examination and these data were used to determine the impact of oral health on the tongue microbiome. A multivariate analysis was carried out using MaAsLin2 on the initial cohort tongue samples, with fixed effects including oral ulceration, DMFT, active caries and gingivitis. This analysis identified a large number of taxa with differential abundance in children with IBD and showed that these changes were independent of changes associated with oral health [Supplementary Figure 1c]. Gingivitis was associated with increased abundance of species linked to plaque development [Campylobacter gracilis, Prevotella olourum and Dialister invisus], an increasing DMFT score was associated with Streptococcus parasanguinus and Aggregatibacter sp. HMT513 and no taxa could be identified as caries-enriched [Supplementary Figure 1c]. This preliminary analysis indicated that the tongue microbiome of IBD patients had the greatest diagnostic potential and for this reason we concentrated the remaining analysis in the full cohort on DNA recovered from tongue samples.
Analysis of alpha diversity metrics [Chao1, Shannon, Inverse Simpson] in all 258 tongue samples [IBD = 156; controls = 102] did not reveal any significant differences in species richness between healthy controls and the IBD cohort [Supplementary Figure 2a]. Within the IBD cohort, patients with more severe activity, abnormal C-reactive protein [CRP; >10 mg/l] or abnormal albumin [<37 g/l] levels had lower mean alpha diversity values, but these differences were only statistically significant in the case of albumin levels [Supplementary Figure S3c; Shannon p = 0.02; Chao1 p = 0.0023]. Community structure in tongue samples was analysed using PCA of the Aitchison distance metric, which revealed that IBD samples formed a more dispersed group that significantly diverged from healthy control samples along axis 1 [PERMANOVA p = 0.0009; Figure 1a]. Several taxa could be identified as differentially abundant across axis 1 including Corynebacterium, Pseudopropionibacterium and Abiotrophia [IBD-associated] and Peptostreptococcus, Solobacterium and Peptostreptococcaceae [XI][G-1] [Health associated; Supplementary Figure 2b]. Multivariate analysis was also used to identify taxa associated with IBD independently of demographic factors [Figure 1b]. After adjusting for age and gender, a number of genera were found to be enriched or depleted in IBD children [Figure 1b]. Highly significant [padj < 0.05] alterations in abundance were detected for Ottowia, Pseudopropionobacterium, Lautropia, Staphylococcus, Pseudomonas and Corynebacterium species [all increased in IBD; Figure 1b] and Veillonella, Oribacterium, Peptostreptococcaceae [XI] [G-1] and Lachnoanaerobaculum species [reduced in IBD; Figure 1b]. Plots comparing abundance of individual taxa in IBD patients and controls are shown in Supplementary File 2. This analysis also showed that some of the most common oral taxa were also depleted in IBD patients, including Prevotella, Fusobacterium, Leptotrichia and Porphyromonas species, with a general increase in Eikenella and Streptococcus species in IBD [Figure 1b and c]. Additional analysis using a non-parametric Wilcoxon test comparing relative abundances of genera in IBD and control children produced similar results [Supplementary Figure S4]
Figure 1.
[a] Community structure of the tongue microbiome in IBD and non-IBD children analysed by PCA of the Aitchison distance metric. [b] Heatmap summarizing the most significant microbiome associations identified between IBD diagnosis and demographic factors using MaAsLin2. Taxa in red or blue show increased or decreased abundance, respectively, in that category. Colour intensity reflects significance [−log q value]. [c] Iris plot summarizing the relative abundances of the ten most abundance genera in IBD and non-IBD children, generated with MicroViz.
3.2. Oral dysbiosis and disease severity
Next, we classified IBD patients in terms of disease severity [remission, mild–moderate disease or severe disease, see Methods]. Analysis of population structure within the IBD samples showed that clinical disease activity significantly impacted population structure [PERMANOVA p = 0.0089], independent of CRP, serum albumin or any other patient data examined [Supplementary Figure 3].
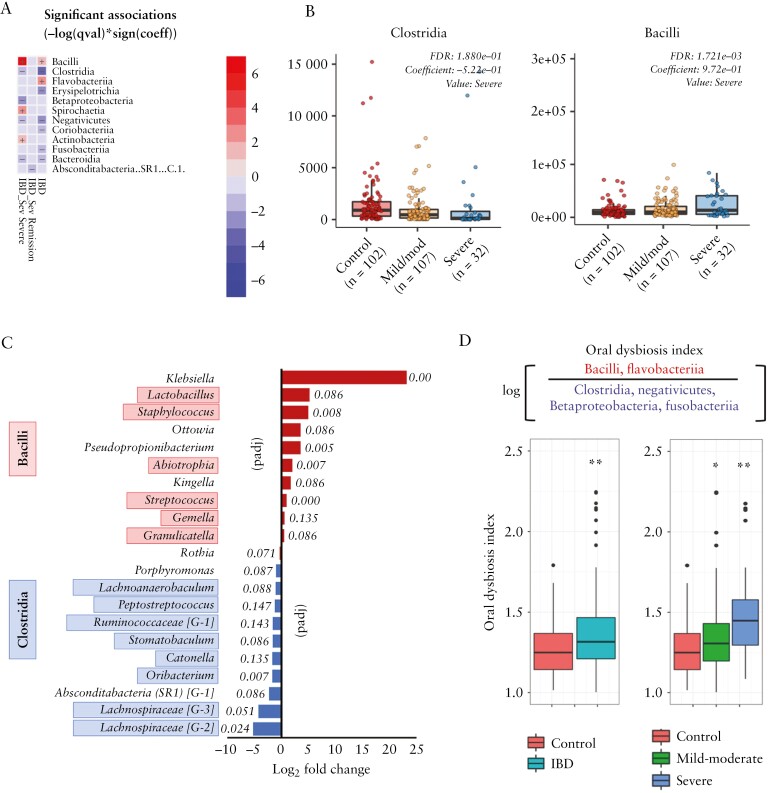
Analysis to identify taxa associated with disease severity found that severe IBD was associated with a change in the composition of the phylum Firmicutes, with increased levels of Bacilli and reduced levels of Clostridia [Figure 2a and b]. At the genus level, severe IBD was associated with increased abundance of genera within the class Bacilli such as Lactobacillus, Streptococcus and Staphylococcus relative to controls, with a concomitant decrease in abundance of taxa within the class Clostridia such as Lachnospiraceae, Oribacterium, Catonella, Stomatobaculum and Ruminococcaceae [G-1] [Figure 2c]. Klebsiella spp. showed the most significant increase in abundance in severe disease relative to controls [Figure 2c], but only 11 children in total [all IBD] were positive for Klebsiella spp. Based on these data, we generated an ODI comparing the log ratio of differentially abundant taxonomic classes [Bacilli + Flavobacteriia/Clostridia + Negativicutes + Betaproteobacteria + Fusobacteriia]. This ODI was significantly greater in IBD patients vs controls [p = 0.0001, Figure 2d] and showed a relationship with disease severity, with a moderately significant increase in mild–moderate disease [p = 0.02] and a highly significant increase in severe disease [p < 0.0001] relative to controls [Figure 2d]. The increasing level of dysbiosis observed with greater disease severity was supported by a global analysis of community structure using Bray–Curtis dissimilarity measures. PERMANOVA of these data showed that communities from children with severe disease were highly divergent from healthy children [padj = 0.001], whereas children with mild to moderate disease harboured communities that were less significantly divergent [padj = 0.025].
Figure 2.
[a] Heatmap summarizing the most significant microbiome associations identified in different categories of IBD severity [see Methods] using MaAsLin2. Taxa in red or blue show increased or decreased abundance, respectively, in that category. Colour intensity reflects significance [−log q value]. [b] Box-plots showing the abundances of Clostridia and Bacilli in non-IBD controls and IBD children with mild to moderate and severe disease. [c] Plot showing the log2 fold-change in abundance of taxa associated with severe IBD [red] or health [blue], identified using DeSeq2. Adjusted p values for each taxon are shown. [d] The oral dysbiosis index. Plots show the index generated for IBD patients vs controls and for IBD patients with mild–moderate disease and severe disease vs controls [*Mann–Whitney p = 0.02; **Kruskall–Walis p = 0.0001].
3.3. Machine learning
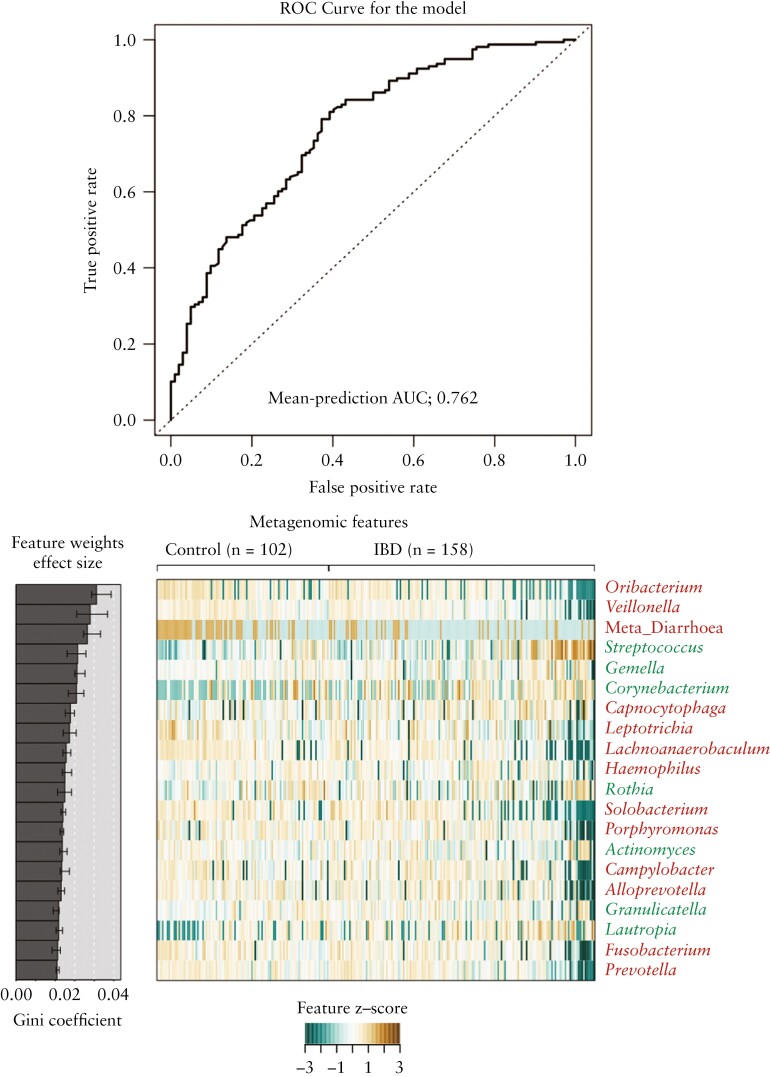
Next, to investigate whether oral dysbiosis in IBD could be diagnostically useful, we used a random forest classifier to determine whether the microbiome profile could effectively classify patients as healthy controls or IBD patients. Based on genus-level profiles, serial receiver operating characteristic [ROC] curves were generated to compare the predictive utility of the different microbial profiles that corresponded with a diagnosis of IBD. Our classifier attained an AUC of 0.738. Incorporation of whether the patient presented with diarrhoea into the model increased the ROC AUC to 0.762 and the Precision recall AUC to 0.834 [Figure 3].
Figure 3.
[a] Evaluation of the performance of the random forest classifier to differentiate IBD patients and control patients based on genus-level microbiome profiles. ROC is based on the tongue microbiome profile, incorporating metadata on whether the patient presented with diarrhoea. The AUC is indicated on the plot. [b] Heatmap showing the 20 most significant features included in the majority of models fitted during cross-validation. Heatmap shows normalized values across all samples. Features are listed based on weight, indicated by the Gini coefficient, as shown in the ‘Feature Weight’ barplot. Taxon names are coloured by Z score with control-associated taxa in red and IBD-associated taxa in green.
3.4. Analysis of CD vs UC
We then analysed samples from CD [n = 94] and UC [n = 52] separately, to assess the extent of oral dysbiosis associated with each condition. Ordination plots of CD and UC samples showed that CD samples diverged from healthy samples to a greater degree than UC samples [Supplementary Figure 5a and b]. CD population structure was significantly different from healthy controls [PERMANOVA p = 0.0009; Supplementary Figure 5a]. A significantly reduced abundance of several major oral taxa was observed in CD patients including Veillonella spp., Prevotella spp., Fusobacterium spp. and Rothia spp. as well as increases in Coynebacterium spp. and Streptococcus spp. [Supplementary Figure S5c]. In UC, population structure exhibited less significant differences from healthy controls [PERMANOVA p = 0.02, Supplementary Figure 5b] and fewer taxa exhibiting significant divergence were identified [Supplementary Figure 5d]. The ODI was able to discriminate severe CD [Supplementary Figure 6a] and moderate-to-severe CD [Supplementary Figure 6b] from controls to a greater extent than in UC patients [Supplementary Figure 6c and d]. Direct comparison of CD and UC populations using PCA could not accurately discriminate these populations [PERMANOVA p = 0.054; Supplementary Figure 7a] although some discriminatory taxa were identified, including specific genera of the class Clostridia (Lachnoanaerobaculum, Stomatobaculum and Ruminococcaceae [G2]) and Pseudopropionabacterium [Supplementary Figure 7b and c]. We did not identify any relationship between Paris classification phenotypes and population structure in CD or UC [data not shown].
3.5. Metagenome prediction
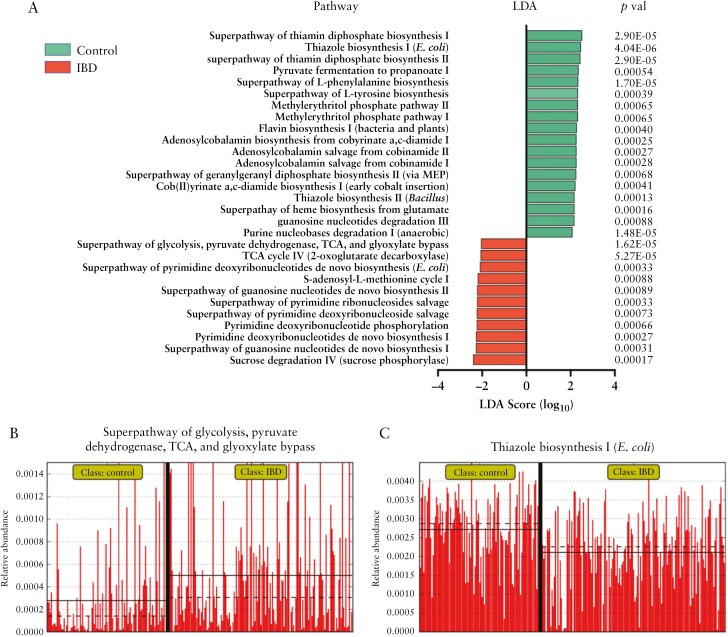
Next we used PiCrust2 to generate metagenome predictions based on the 16S community profiles resulting in predicted EC abundances and predicted MetaCyc pathway abundances. Comparative analysis of pathway abundance in IBD and control communities using LEfSe indicated a higher degree of sugar degradation in IBD samples, including pathways for sucrose [p = 0.00017] and lactose and galactose degradation [p = 0.0037] [Figure 4a]. The metabolism of sugars was also enhanced in IBD samples with greater abundance of the ‘superpathway’ of glycolysis and the TCA [tricarboxylic acid] cycle [p < 0.0001; Figure 4b]. In control populations, we detected greater abundances of pathways involved in B vitamin biosynthesis including thiazole [Figure 4b], thiamine diphosphate [vitamin B1], adenosylcobalamin biosynthesis [vitamin B12] and amino acid metabolism.
Figure 4.
[a] LEfSe plot showing predicted MetaCyc pathways exhibiting significant differences in abundance in control or IBD patients. Differential abundance is indicated by the linear discriminant analysis [LDA] effect size score. Green bars are control associated and red bars IBD associated. Only features with Kruskall–Wallis p < 0.001 are shown. [b] Relative abundance of selected pathways [Thiazole biosynthesis and Superpathway of glycolysis and TCA] in controls and IBD patients.
3.6. Oral dysbiosis resolves following treatment
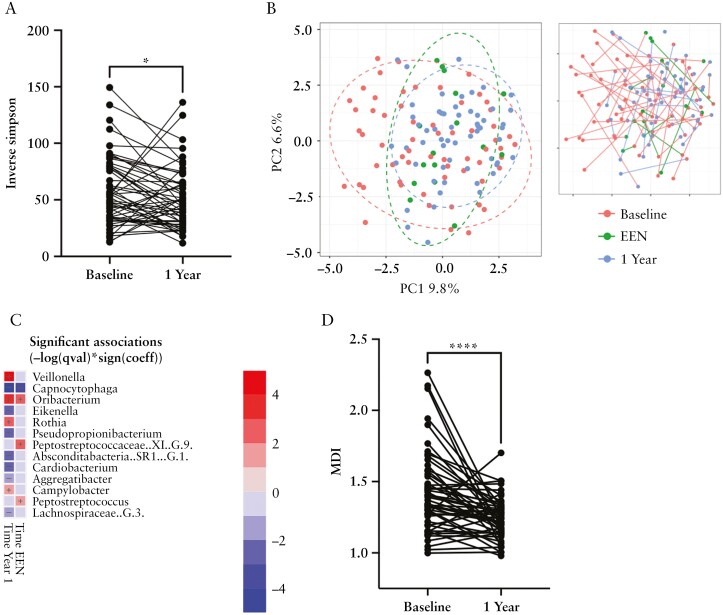
To examine whether IBD treatment alters oral dysbiosis, we next analysed samples from patients following treatment. Paired baseline and 1-year samples were available for 52 patients. The majority of these patients were in remission or had mild activity at 1 year of follow-up [see Table 2 for clinical summary]. We also analysed oral microbiome changes following an 8-week course of EEN in 16 patients with CD. Post-EEN and 1-year follow-up samples showed a tendency towards reduced biodiversity relative to baseline samples [Supplementary Figure 8] and 1-year samples exhibited a significant reduction in Inverse Simpson value when paired with baseline samples [Figure 5a; paired Wilcoxon p = 0.023]. In terms of population structure, PCA plots of the Aitchison distance metric showed that post-EEN and 1-year samples were significantly shifted to the right of axis 1 [PERMANOVA p = 0.003; Figure 5b]. This change in population structure could be in part be attributed to increased abundance of several genera of the class Clostridia, including Stomatobaculum, Peptostreptococcaceae [XI][G-1] and Peptostreptococcus in follow-up samples [Supplementary Figure 8]. Additionally, we observed an increased abundance of the ‘health-associated’ taxa Veillonella spp. and Oribacterium spp. and a reduction the ‘IBD-associated’ taxa Eikenella and Pseudopropionibacterium spp. following treatment [Figure 5c]. Calculation of the MDI for post-EEN and 1-year samples showed a significant reduction in MDI following treatment [Supplementary Figure 8] which was highly significant when paired baseline and 1-year samples were compared [p < 0.001], further indicating the recovery of a normal oral microbiome [Figure 5d].
Table 2.
Treatment details for patients with 1-year follow-up swabs
| IBD category | Treatments | |
|---|---|---|
| CD | UC | |
| n [%] | 36 | 12 |
| Treatments used in first 3 months following diagnosis, n [%] | ||
| EEN | 32 [89] | — |
| 5-ASA | 6 [17] | 11 [92] |
| Steroids | 5 [14] | 5 [42] |
| Antibiotics | 8 [22] | 1 [8] |
| Anti-TNFα drugs | 12 [33] | 3 [25] |
| Immunomodulators | 21 [58] | 1 [8] |
| Treatments at time of 1-year follow-up swab, n [%] | ||
| 5-ASA | 6 [17] | 11 [92] |
| Steroids | — | 2 [17] |
| Antibiotics | — | — |
| Anti-TNFα drugs | 16 [44] | 3 [25] |
| Immunomodulators | 24 [67] | 3 [25] |
| Disease activity at 1-year follow-up swab [per PCDAI or PUCAI], n [%] | ||
| Remission | 27 [75] | 9 [75] |
| Mild | 8 [22] | 2 [17] |
| Moderate | 1 [3] | 1 [8] |
| Severe | — | — |
IBD, inflammatory bowel disease; UC, ulcerative colitis; CD, Crohn;s disease; EEN, exclusive enteral nutrition; 5-ASA, 5-aminosalicylic acid; TNF, tumour necrosis factor; PCDAI, Pediatric Crohn’s Disease Activity Index; PUCAI, Pediatric Ulcerative Colitis Activity Index.
Figure 5.
Analysis of follow-up samples after 6 weeks of EEN therapy [post-EEN] or 12 months therapy [1 Year]. [a] Inverse Simpson values in paired baseline and 1 Year samples; *paired Wilcoxon p = 0.023. [b] Community structure in baseline, post-EEN and 1 Year samples analysed by PCA of the Aitchison distance metric. [c] Heatmap summarizing the most significant microbiome associations identified in post-EEN and 1 Year samples using MaAsLin2. Taxa in red or blue show increased or decreased abundance, respectively, in that category. Colour intensity reflects significance [−log q value]. [d] Oral dysbiosis index in paired baseline and 1 Year samples; ***paired Wilcoxon p < 0.0001.
4. Discussion
This study for the first time describes oral dysbiosis in a relatively large cohort [n = 156] of treatment-naïve children with IBD. One of the strengths of this study is that all patients are treatment naïve as immunosuppressive or nutritional therapies are likely to confound analysis of disease-specific changes to the microbiome. The oral dysbiosis described here is characterized by reduced abundance of many dominant oral taxa including Veillonella, Neisseria, Prevotella, Fusobacterium and Porphyromonas species, with increased abundance of Streptococci. Some of the most significant changes in species abundance occurred within the phylum Firmicutes, with increased levels of Bacilli and reduced levels of Clostridia. This dysbiosis was more pronounced in the tongue compared to the buccal mucosa and multivariate analysis showed that these changes in abundance were independent of age, gender and oral health. Our findings of significantly higher dysbiosis in patients with more severe disease supports the hypothesis that this dysbiosis is intrinsically disease related. Past studies describing oral dysbiosis in IBD have reported discordant findings of relative abundance of various taxa and genera.16,18,20,21,41 This discordance may be due to the fact that previous studies analysed relatively small cohorts of adult patients who are not treatment naïve and often involve samples from different oral niches. Our findings that active IBD treatments impact on oral dysbiosis also have implications for designing future studies in the field.
Uniquely, the current study shows a reduction in the abundance of Clostridia, similar to the gut dysbiosis described in IBD. Oral dysbiosis in IBD is associated with a disturbance in the phylum Firmicutes, characterized by reduced abundance of Clostridia and increased abundance of Bacilli. Reduced levels of Clostridia have been reported in the gut microbiome of treatment-naïve IBD patients and this study confirms this depletion also occurs in the oral cavity. Gevers et al. reported reduced abundance of the families Lachnospiraceae and Ruminococcaceae in the gut microbiomes of IBD children and these taxa were among the most significantly depleted in this study in the oral cavities of those with severe IBD.5 Thus, depletion of the Clostridiales appears to be a fundamental element of microbiome dysbiosis in IBD. The explanation remains speculative, but may be due to the host response to these organisms or due to displacement by pathobionts. In the oral cavity, this displacement may be due to the increased abundance of bacilli including Streptococcus, Staphylococcus and Lactobacillus species.
We also observed reduced relative abundance of some major oral taxa including Veillonella and Fusobacterium species. This was unexpected as studies of the gut microbiome indicate that the abundance of Veillonella and Fusobacterium species is often increased in the gut of IBD patients.5,7 The reduced abundance of oral Veillonella and Fusobacterium species in IBD patients could be due to displacement by streptococci or alternatively due to systemic immune responses which could lead to depletion of these taxa in the oral cavity. Atarashi et al. showed in a mouse model that ectopic colonization of the murine gut with oral Veillonella and Fusobacteria induced strong inflammatory responses.9 Rengarajan et al. also showed significant antibody responses in IBD patients to bacteria normally found in the oral cavity including Gemella, Peptostreptococcus and Streptococcus species.42 It is therefore possible that ectopic colonization of the gut results in a systemic response that may lead to depletion of these taxa in the oral cavity.
A unique finding of our study was the extent of recovery from dysbiosis following therapy for paediatric IBD. Therapy with EEN for 8 weeks led to a reduction in Capnoctyophaga and the recovery of Oribacterium. After 1 year we observed a return towards a healthy population structure and a significant reduction in the dysbiosis index, with recovery of Veillonella and Oribacterium and reduced levels of Eikenella, Pseudopropionobacterium and Capnocytophaga. Although this may not represent a complete reversion of dysbiosis, it is clear that disease resolution results in microbiome recovery, which may be due to resolution of systemic inflammation. Some variation in the patient responses may be related to differences in individual treatment plans or medications. Improved diet and nutrition may also have impacted on the post-treatment microbiome, but diet was not analysed specifically in our study. For example, we observed a strong increase in Rothia spp., which are known to increase with consumption of nitrate-rich foods such as leafy green vegetables.43 Our findings of dysbiosis worsening with disease activity and recovering with treatment and remission suggest that the oral microbiome may be a bellwether of disease status in certain patient groups.
We found moderate ability of the oral microbiome to predict a diagnosis of IBD. The oral cavity is easily accessible and oral swabs are minimally invasive as a diagnostic tool. Incorporating clinical data enhanced the predictive performance. Our findings were broadly in keeping with those described for rectal biopsies and stool samples5,8 and values for saliva samples analysed in a small study of children with IBD [n = 47].21 The oral microbiome has potential as a significant component in a diagnostic predictive tool for IBD, and is worthy of ongoing exploration in this regard. Although tongue swabs were analysed here, Somineni at al. have indicated that saliva may be more effective than mucosal surfaces for measuring oral dysbiosis.21 Metagenomic sequencing data, including fungal and viral sequences, could further improve the accuracy of the models.
Discrimination between CD and UC using oral microbiome data was more difficult. Although PCA showed that dysbiosis was more severe in CD, classifiers used here did not effectively discriminate between the two diagnoses [data not shown].
We detected increased levels of opportunistic pathogens in the oral cavities of IBD patients, including Klebsiella, Pseudomonas and Staphylococcus spp. [Supplementary Figure 9]. Increased levels of Klebsiella have been reported in the gut microbiome of IBD patients and Atarashi et al. have demonstrated in a murine model of IBD that strains of Klebsiella from the saliva of IBD patients are highly inflammatory.9Klebsiella was detected in 11 of our IBD patients [eight CD, three UC] and in none of the control patients [Supplementary Figure 9]. Although only a small proportion were positive for Klebsiella, this organism could be a driver of inflammation in a small subset of patients. Similarly, Pseudomonas spp. were recovered from only six controls and 21 IBD patients [15 CD, six UC; Supplementary Figure 9]. Pseudomonas has also be implicated as a pathobiont in paediatric IBD with a higher proportion of IBD children being PCR positive for Pseudomonas relative to controls.44Pseudomonas colonization was also linked to a mutation in PSMG1 [proteasome assembly chaperone 1 gene], suggesting that a subset of IBD patients may be susceptible to colonization with Pseudomonas.45 Staphylococci were identified in ten controls and 29 IBD patients [21 CD, eight UC; Supplementary Figure S9]. Staphylococci have not been implicated in the pathogenesis of IBD but they can be responsible for complicating infections.46 Overall these data show that pathogens may reside in the oral cavities of these patients and, as has been suggested by others, may represent a reservoir of pathobionts that can transit to the gut, maintaining inflammation and dysbiosis.47
Although our study greatly improves our knowledge of the oral microbiome in paediatric IBD, there are some limitations which temper interpretation of our results but could be addressed in future studies. To properly address the relationship between oral and gut communities, dual sampling of both sites would have been ideal but was not possible for the current study. This may allow identification of oral–gut transit and even identification of specific sequence types that can prosper in both environments. We did not undertake tandem salivary microbiome analysis which would have enabled greater comparisons with some previous oral microbiome studies, as we were keen to sample mucosa-associated flora. Other factors such as diet and oral hygiene could impact on these results. Although only a subset of the patients received a dental examination for oral hygiene and dental caries, we could identify clear signatures of gingivitis and caries in these samples which was unrelated to the IBD-specific dysbiosis characterized subsequently. We found no significant differences in our microbiome analysis between groups in relation to rural vs urban dwelling, household number or exposure to smoking in the home environment [data not shown]. Formal dietary monitoring is challenging and prone to inaccuracy, but adoption of food frequency questionnaires in future studies would enhance the interpretive analysis at different timepoints. Patients were treated at patient–physician discretion rather than per protocol, including choice of EEN brands, but treatments at the centre follow the contemporary ESPGHAN guidelines of the day. Proper functional analysis of these communities will require metagenomic sequencing which may also provide further biomarkers to improve the diagnostic capabilities of the oral microbiome.
5. Conclusion
Oral dysbiosis is a feature of treatment-naïve paediatric IBD which mirrors disease severity and responds to treatment. Variation in microbial community structure showed similar patterns to the gut in IBD. Several pathobionts were found to be enriched in the oral cavities of patients with IBD, suggesting that the oral cavity could be a reservoir for these organisms. The oral microbiome has potential as a non-invasive diagnostic tool and measure of treatment success.
Supplementary Material
Acknowledgments
The authors wish to thank Kathleen McGrath and Elaine Kenny at TrinSeq [St James’s Hospital, Dublin] for assistance and advice with next generation sequencing.
Contributor Information
Khalid Elmaghrawy, School of Dental Science, Trinity College Dublin and Dublin Dental University Hospital, Dublin 2, Republic of Ireland.
Paddy Fleming, School of Dental Science, Trinity College Dublin and Dublin Dental University Hospital, Dublin 2, Republic of Ireland.
Kirsten Fitzgerald, School of Dental Science, Trinity College Dublin and Dublin Dental University Hospital, Dublin 2, Republic of Ireland.
Sarah Cooper, DOCHAS Study, Children’s Health Ireland, Crumlin, Dublin, Republic of Ireland.
Anna Dominik, DOCHAS Study, Children’s Health Ireland, Crumlin, Dublin, Republic of Ireland.
Séamus Hussey, DOCHAS Study, Children’s Health Ireland, Crumlin, Dublin, Republic of Ireland; Department of Paediatrics, University of Medicine and Health Sciences, RCSI, Dublin and University College Dublin, Ireland.
Gary P Moran, School of Dental Science, Trinity College Dublin and Dublin Dental University Hospital, Dublin 2, Republic of Ireland.
Funding
K.E. was supported by a scholarship from the Libyan Ministry of Higher Education and Scientific Research. G.P.M. was supported by a grant from the Irish Health Research Board [Grant no: ILP-POR-2019-030]. S.H. and the DOCHAS study were supported by grants from the National Children’s Research Centre, Dublin [Grant nos: J/15/1 and C/18/2].
Conflict of Interest
The authors can confirm that they have no conflicts of interest to declare.
Author Contributions
G.P.M.: study design, funding, data analysis, manuscript preparation; K.E.: sample preparation, data analysis; P.F.: study design, sample collection, oral health screening; K.F.: sample collection, oral health screening; S.C.: data collection, data analysis; A.D.: sample collection, manuscript preparation; S.H.: study design, data collection, funding, data analysis, manuscript preparation.
Data Availability
All sequence data have been submitted to the NCBI sequence read archive [SRA], submission: SUB11582071.
References
- 1. López RL, Burgos MJG, Gálvez A, Pulido RP.. The human gastrointestinal tract and oral microbiota in inflammatory bowel disease: a state of the science review. APMIS 2016;125:3–10. [DOI] [PubMed] [Google Scholar]
- 2. McGovern DPB, Gardet A, Törkvist L, et al. ; NIDDK IBD Genetics Consortium. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet 2010;42:332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. ; IBDMDB Investigators. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knights D, Silverberg MS, Weersma RK, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med 2014;6:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014;15:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shaw KA, Bertha M, Hofmekler T, et al. Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med 2016;8:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schirmer M, Denson L, Vlamakis H, et al. Compositional and temporal changes in the gut microbiome of pediatric ulcerative colitis patients are linked to disease course. Cell Host Microbe 2018;24:600–10.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang F, Kaplan JL, Gold BD, et al. Detecting microbial dysbiosis associated with pediatric Crohn disease despite the high variability of the gut microbiota. Cell Rep 2016;14:945–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Atarashi K, Suda W, Luo C, et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 2017;358:359–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hussey S, Fleming P, Rowland M, et al. Disease outcome for children who present with oral manifestations of Crohn’s disease. Eur Arch Paediatr Dent 2012;12:167–9. [DOI] [PubMed] [Google Scholar]
- 11. Rowland M, Fleming P, Bourke B.. Looking in the mouth for Crohn’s disease. Inflamm Bowel Dis 2010;16:332–7. [DOI] [PubMed] [Google Scholar]
- 12. Harikishan G, Reddy NR, Prasad H, Anitha S.. Oral Crohn’s disease without intestinal manifestations. J Pharm Bioallied Sci 2012;4:S431–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lankarani KB, Sivandzadeh GR, Hassanpour S.. Oral manifestation in inflammatory bowel disease: a review. World J Gastroenterol 2013;19:8571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lourenço SV, Hussein TP, Bologna SB, Sipahi AM, Nico MMS.. Oral manifestations of inflammatory bowel disease: a review based on the observation of six cases. J Eur Acad Dermatol Venereol 2009;24:204–7. [DOI] [PubMed] [Google Scholar]
- 15. Katsanos KH, Torres J, Roda G, Brygo A, Delaporte E, Colombel J-F.. Review article: non-malignant oral manifestations in inflammatory bowel diseases. Aliment Pharmacol Ther 2015;42:40–60. [DOI] [PubMed] [Google Scholar]
- 16. Said HS, Suda W, Nakagome S, et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res 2014;21:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xun Z, Zhang Q, Xu T, Chen N, Chen F.. Dysbiosis and ecotypes of the salivary microbiome associated with inflammatory bowel diseases and the assistance in diagnosis of diseases using oral bacterial profiles. Front Microbiol 2018;9:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang T, Kayani MR, Hong L, et al. Dynamics of the salivary microbiome during different phases of Crohn’s disease. Front Cell Infect Microbiol 2020;10:544704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qi Y, Zang S, Wei J, et al. High-throughput sequencing provides insights into oral microbiota dysbiosis in association with inflammatory bowel disease. Genomics 2021;113:664–76. [DOI] [PubMed] [Google Scholar]
- 20. Hu S, Mok J, Gowans M, Ong DEH, Hartono JL, Lee JWJ.. Oral microbiome of Crohn’s disease patients with and without oral manifestations. J Crohns Colitis 2022. Doi: 10.1093/ecco-jcc/jjac063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Somineni HK, Weitzner JH, Venkateswaran S, et al. Site- and taxa-specific disease-associated oral microbial structures distinguish inflammatory bowel diseases. Inflamm Bowel Dis 2021;27:1889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goel RM, Prosdocimi EM, Amar A, et al. Streptococcus salivarius: a potential salivary biomarker for orofacial granulomatosis and Crohn’s disease? Inflamm Bowel Dis 2019;25:1367–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levine A, Koletzko S, Turner D, et al. ESPGHAN revised porto criteria for the diagnosis of inflammatory bowel disease in children and adolescents. J Pediatr Gastr Nutr 2014;58:795–806. [DOI] [PubMed] [Google Scholar]
- 24. Levine A, Griffiths A, Markowitz J, et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis 2011;17:1314–21. [DOI] [PubMed] [Google Scholar]
- 25. Hyams JS, Ferry GD, Mandel FS, et al. Development and validation of a pediatric Crohn’s disease activity index. J Pediatr Gastr Nutr 1991;12:439. [PubMed] [Google Scholar]
- 26. Turner D, Otley AR, Mack D, et al. Development, validation, and evaluation of a pediatric ulcerative colitis activity index: a prospective multicenter study. Gastroenterology 2007;133:423–32. [DOI] [PubMed] [Google Scholar]
- 27. Amer A, Galvin S, Healy CM, Moran GP.. The microbiome of potentially malignant oral leukoplakia exhibits enrichment for Fusobacterium, Leptotrichia, Campylobacter, and Rothia species. Front Microbiol 2017;8:2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ.. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microb 2008;74:2461–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Diaz PI, Dupuy AK, Abusleme L, et al. Using high throughput sequencing to explore the biodiversity in oral bacterial communities. Mol Oral Microbiol 2012;27:182–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Illumina. 16S Metagenomic Sequencing Library Preparation, 2013:1–28. https://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html
- 31. Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol 2010;192:5002–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McMurdie PJ, Holmes S.. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barnett D, Arts I, Penders J.. microViz: an R package for microbiome data visualization and statistics. J Open Source Softw 2021;6:3201. [Google Scholar]
- 35. Mallick H, Rahnavard A, McIver LJ, et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput Biol 2021;17:e1009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wirbel J, Zych K, Essex M, et al. Microbiome meta-analysis and cross-disease comparison enabled by the SIAMCAT machine learning toolbox. Genome Biol 2021;22:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Breiman L. Random forests. Mach Learn 2001;45:5–32. [Google Scholar]
- 39. Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Langille MGI, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31:814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Docktor MJ, Paster BJ, Abramowicz S, et al. Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm Bowel Dis 2012;18:935–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rengarajan S, Vivio EE, Parkes M, et al. Dynamic immunoglobulin responses to gut bacteria during inflammatory bowel disease. Gut Microbes 2019;11:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kapil V, Haydar SMA, Pearl V, Lundberg JO, Weitzberg E, Ahluwalia A.. Physiological role for nitrate-reducing oral bacteria in blood pressure control. Free Radical Bio Med 2013;55:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wagner J, Short K, Catto-Smith AG, Cameron DJS, Bishop RF, Kirkwood CD.. Identification and characterisation of Pseudomonas 16S ribosomal DNA from ileal biopsies of children with Crohn’s disease. PLoS One 2008;3:e3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wagner J, Catto-Smith AG, Cameron DJS, Kirkwood CD.. Pseudomonas infection in children with early-onset crohn’s disease: an association with a mutation close to PSMG1. Inflamm Bowel Dis 2012;19:E58–9. [DOI] [PubMed] [Google Scholar]
- 46. Bettenworth D, Nowacki TM, Friedrich A, Becker K, Wessling J, Heidemann J.. Crohn’s disease complicated by intestinal infection with methicillin-resistant Staphylococcus aureus. World J Gastroenterol 2013;19:4418–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Elmaghrawy K, Hussey S, Moran GP.. The oral microbiome in pediatric IBD: a source of pathobionts or biomarkers? Front Pediatr 2021;8:620254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data have been submitted to the NCBI sequence read archive [SRA], submission: SUB11582071.