Abstract
Understanding the exact molecular mechanisms involved in the aetiology of epileptogenic pathologies with or without tumour activity is essential for improving treatment of drug-resistant focal epilepsy. Here, we characterize the landscape of somatic genetic variants in resected brain specimens from 474 individuals with drug-resistant focal epilepsy using deep whole-exome sequencing (>350×) and whole-genome genotyping. Across the exome, we observe a greater number of somatic single-nucleotide variants in low-grade epilepsy-associated tumours (7.92 ± 5.65 single-nucleotide variants) than in brain tissue from malformations of cortical development (6.11 ± 4 single-nucleotide variants) or hippocampal sclerosis (5.1 ± 3.04 single-nucleotide variants). Tumour tissues also had the largest number of likely pathogenic variant carrying cells. low-grade epilepsy-associated tumours had the highest proportion of samples with one or more somatic copy-number variants (24.7%), followed by malformations of cortical development (5.4%) and hippocampal sclerosis (4.1%). Recurring somatic whole chromosome duplications affecting Chromosome 7 (16.8%), chromosome 5 (10.9%), and chromosome 20 (9.9%) were observed among low-grade epilepsy-associated tumours. For germline variant-associated malformations of cortical development genes such as TSC2, DEPDC5 and PTEN, germline single-nucleotide variants were frequently identified within large loss of heterozygosity regions, supporting the recently proposed ‘second hit’ disease mechanism in these genes. We detect somatic variants in 12 established lesional epilepsy genes and demonstrate exome-wide statistical support for three of these in the aetiology of low-grade epilepsy-associated tumours (e.g. BRAF) and malformations of cortical development (e.g. SLC35A2 and MTOR). We also identify novel significant associations for PTPN11 with low-grade epilepsy-associated tumours and NRAS Q61 mutated protein with a complex malformation of cortical development characterized by polymicrogyria and nodular heterotopia. The variants identified in NRAS are known from cancer studies to lead to hyperactivation of NRAS, which can be targeted pharmacologically. We identify large recurrent 1q21–q44 duplication including AKT3 in association with focal cortical dysplasia type 2a with hyaline astrocytic inclusions, another rare and possibly under-recognized brain lesion. The clinical-genetic analyses showed that the numbers of somatic single-nucleotide variant across the exome and the fraction of affected cells were positively correlated with the age at seizure onset and surgery in individuals with low-grade epilepsy-associated tumours. In summary, our comprehensive genetic screen sheds light on the genome-scale landscape of genetic variants in epileptic brain lesions, informs the design of gene panels for clinical diagnostic screening and guides future directions for clinical implementation of epilepsy surgery genetics.
Keywords: epilepsy, malformation of cortical development, low-grade epilepsy-associated tumour, hippocampal sclerosis, genetics
López-Rivera et al. discover differences in genetic architecture across major epileptic brain lesion types. They describe novel somatic chromosomal alterations, identify novel genes and genotype–phenotype associations, and provide support for the role of genetics in the histopathological diagnosis of epileptic lesions.
Introduction
Drug-resistant epilepsies due to focal brain lesions represent a huge health burden and challenge for everyday clinical practice.1 Neurosurgical resection strategies have proven helpful in carefully selected patients, especially for brain lesions visible through MRI and confirmed by histopathology diagnosis.2,3 The most common types of epilepsy-associated brain lesions comprise hippocampal sclerosis (HS),4 low-grade epilepsy-associated developmental brain tumours (LEAT)5,6 such as ganglioglioma (GG) and dysembryoplastic neuroepithelial tumours (DNET), and malformations of cortical development (MCD) such as focal cortical dysplasia (FCD).7 Overall, these three most common categories accounted for >80% of the almost 10 000 patients who underwent epilepsy surgery and were collected in the European Epilepsy Brain Bank.2
Genetic factors have been associated with many common8 and rare epilepsies.9–13 Until recently, specific genes with variants of large effects have mainly been discovered in rare and severe forms of paediatric epilepsy that typically do not show structural abnormalities on MRI.10–13 In the past decade, >15 genes have been associated with somatic variants in epilepsy-associated MCD and LEAT.5,14–16 In contrast and to the best of our knowledge, the burden of somatic variants has not been systematically evaluated yet for HS. For MCD, somatic variants in genes encoding proteins of the canonical PI3K-AKT-MTOR pathway and germline loss-of-function variants—with or without co-occurring somatic losses of heterozygosity—in genes encoding proteins of the GATOR complex, a negative regulator of the PI3K-AKT-MTOR pathway, have been associated with FCD type 2 and hemimegalencephaly (HME).15–18 In addition, the SLC35A2 gene has been associated with the recently discovered mild MCD with oligodendroglial hyperplasia in the epilepsy (MOGHE) disease entity.19,20 For LEAT, somatic variants that affect the RAS-RAF-MAPK pathway (i.e. BRAF and FGFR1) play a major role, with the BRAF variant V600E reported in 18–56% of GG and pathogenic variants in FGFR1 in 58–82% of DNET.5,21,22 There is also emerging evidence for somatic variants in the RAS-RAF-MAPK pathway underlying MCD, with recent case studies reporting variants in KRAS associated with epilepsy-associated tumours and malformations.23–25
The search for actionable treatment targets in drug-resistant focal epilepsies pushed genetic studies of epilepsy-associated brain lesions into an emerging field. Most current studies use ultra-deep targeted sequencing with coverages >1000× to identify low allelic fraction disease-causing single-nucleotide variants (SNV) with high sensitivity and specificity. Due to the high coverage and correlated high sequencing cost, current studies typically prioritized targeted over whole-exome sequencing (WES).16,26 Nevertheless, this approach has been very successful, leading to the discovery of 15 lesional epilepsy-associated genes.5,14–16 However, such candidate gene approaches did not interrogate the mutational signature for a structural lesion across the whole exome or genome, which is a typical study design in cancer research.27,28 As a result, the mutation rate and genetic architecture across epileptic brain lesion categories have not been systematically described. Subsequently, it is unclear whether MCD are single gene or oligogenic somatic disorders and whether somatic variation contributes to the development of HS. In addition, the role of genome-wide postzygotic copy-number variants (CNV) in different epileptic lesions has not been investigated. Recent evidence suggests that low-frequency mosaicism of variants in germline epilepsy-associated ion-channel encoding genes (e.g. SCN8A) can cause epilepsy in rodent models.29 However, it is unclear whether somatic variants in these genes can cause focal epilepsy in humans. It is also unclear whether variants in the 15 recently identified genes5,14–16 represent the most common genetic cause of lesional epilepsy as most previous studies were gene panel candidate studies or small-scale exome-wide studies, underpowered for statistical gene burden analyses across the exome.16,26
Here, we explored the exome-wide somatic variant burden and the genome-wide CNV burden of the three most common categories of epileptogenic brain lesions (i.e. LEAT, MCD and HS). We analysed differential variant burden across lesion categories, statistically identified novel epileptogenic pathology-associated genes, explored the role of driver mutations observed in cancer and refined genotype–phenotype associations. The results of our study will shed light on the genetic architecture for a lot of epileptogenic brain lesions and inform the design of diagnostic genetic tests with reliably high sensitivity and specificity.
Materials and methods
Study cohorts
Snap-frozen surgical brain tissue samples obtained from 474 individuals with epilepsy-associated brain lesions (223 MCD, 154 LEAT and 97 HS) were retrieved from the Cleveland Clinic Epilepsy Center biorepository (n = 154) and European Epilepsy Brain Bank consortium (n = 320). All studies were performed following institutional guidelines and regulations regarding research involving human subjects and approved by the ethics review boards of the Cleveland Clinic and the University of Erlangen, Germany. All patients underwent a comprehensive presurgical evaluation followed by a discussion at a multidisciplinary patient management conference where the surgical strategy was approved. Histopathological review of all resected brain tissue was performed and interpreted by board-certified clinical neuropathologists at the Cleveland Clinic in all patients and followed by a detailed re-review by experienced neuropathologists (I.B. and R.C.) using the International League against Epilepsy (ILAE) classification scheme for FCD,7 HS4 and the 4th edition of the WHO classification of tumours of the central nervous system.30 Characteristics of our study cohorts are detailed in Table 1. Genomic DNA was extracted from fresh-frozen brain tissue for all 474 individuals. The DNeasy Blood and Tissue Kit (Qiagen) was used for DNA extraction from fresh-frozen brain samples according to the manufacturer’s protocol.
Table 1.
Study cohort
| Total (n = 474) | CCF (n = 154) | EEBB (n = 320) | |
|---|---|---|---|
| HS | 97 | 3 | 94 |
| ȃType 1 HS | 52 | 2 | 50 |
| ȃType 2 HS | 6 | 0 | 6 |
| ȃType 3 HS | 2 | 0 | 2 |
| ȃHS (NOS) | 37 | 1 | 36 |
| MCD | 223 | 111 | 112 |
| ȃFCD 1 | 9 | 5 | 4 |
| ȃȃFCD 1a | 8 | 4 | 4 |
| ȃȃFCD 1b | 1 | 1 | 0 |
| MOGHE | 31 | 7 | 24 |
| ȃFCD 2 | 111 | 46 | 65 |
| ȃȃFCD 2a | 45 | 25 | 20 |
| ȃȃFCD 2b | 65 | 21 | 44 |
| ȃȃFCD 2 (NOS) | 1 | 0 | 1 |
| ȃFCD (NOS) | 30 | 30 | 0 |
| ȃHME | 7 | 1 | 6 |
| ȃPMG | 10 | 3 | 7 |
| ȃOther MCD | 25 | 19 | 6 |
| ȃȃmMCD | 22 | 16 | 6 |
| ȃȃNH | 3 | 3 | 0 |
| LEAT | 154 | 40 | 114 |
| ȃGG | 83 | 8 | 75 |
| ȃDNET | 24 | 5 | 19 |
| ȃOther LEAT | 47 | 27 | 20 |
| ȃȃGlioma | 16 | 0 | 16 |
| ȃȃMVNT | 3 | 1 | 2 |
| ȃȃLEAT (NOS) | 26 | 26 | 0 |
| ȃȃMeningioangiomatosis | 2 | 0 | 2 |
The bold highlighted values represent the numbers of individuals with one of the three main classes of epilepsy-associated brain lesions. CCF = Cleveland Clinic Foundation; EEBB = European Epilepsy Brain Bank; mMCD = mild MCD; NOS = not otherwise specified; PMG = polymicrogyria; NH = nodular heterotopia; MVNT = multinodular vacuolated neuronal tumour.
Somatic and germline SNV calling
Deep WES (>350×) of all samples in this study was performed using Agilent SureSelect Human All Exon V7 enrichment and paired-end reads (151 bp) Illumina sequencing. All paired-end FASTQ files were aligned to the GRCh37/hg19 human reference genome, including the hs37d5 decoy sequence using BWA-MEM,31 following GATK (Genome Analysis Toolkit) best practices.32 We used MuTect233 to generate a panel of normals (PoN) from 24 additional surgically resected and sequenced brain samples to exclude sequencing and alignment artefacts. According to GATK best practices guidelines,34 these 24 brain samples were best suited for the PoN on the basis of their technical properties (i.e. sequenced with the same methodology, platform and in the same batch as the study samples). All 24 samples included in the PoN underwent surgery for epilepsy and were histopathologically confirmed as non-lesional or due to an external insult (glial scar or encephalitis, detailed in Supplementary Table 1) and thus had a low likelihood of carrying overgrowth disorder or cancer-driver variants that are predominantly involved in MCD or tumours. We then called somatic SNV, insertions and deletion polymorphisms (indels) using MuTect233 in conjunction with the PoN. We only retained somatic variants with a maximum minor allele frequency (MAF) < 10−5 in the Genome Aggregation Database (gnomAD). Variants within segmental duplication regions or non-diploid regions were removed. Low-quality calls flagged by MuTect2 with ‘t_lod_fstar’, ‘str_contraction’ and ‘triallelic_site’ were removed. Finally, we excluded somatic indels within RepeatMasker35 or simple repeat regions.
We then used MosaicForecast36 to perform read-based phasing and identify high-confidence somatic mosaic variants from all called variants. MosaicForecast is a machine-learning-based method optimized to detect somatic mutations without a matched reference tissue. Briefly, the method trains a random forest model on read-based features from phased variants and has been shown to reliably identify somatic mosaic mutations with variant allelic fractions (VAF) as low as 2%. Therefore, the method has the sensitivity to detect variants present in as few as 4% of cells in a given sample of brain tissue.36,37 After all training and filtration, we identified 217 560 putative somatic mosaic SNV across all samples. We then filtered all exonic or splice site variants with a VAF between 0.02 and 0.40 and an alternative read depth ≥3. Only non-recurring mutations within the cohort were considered, except for variants in 15 genes that have been previously associated with epileptogenic lesions: MTOR, SLC35A2, AKT3, PIK3CA, RHEB, TSC1, TSC2, NPRL2, NPRL3, DEPDC5, PTEN, BRAF, FGFR1, MYB and MYBL1.5,14–16 Last, to ensure that no potentially meaningful variants in established lesional epilepsy genes were mistakenly filtered out, we used the Integrated Genomics Viewer (IGV)38 browser to visually examine all somatic variants in established epileptic lesion genes called by Mutect2-PoN, but judged as potential artefacts by MosaicForecast. However, such variants were not included in the somatic variant enrichment analyses and were only included when reporting the final yield from our screen.
Finally, we used GATK’s HaplotypeCaller (GATK v.4.1.9.0),39 following GATK best practices,40 to identify germline variants in lesional epilepsy-associated genes in negative regulators shown to act through a germline or two-hit mechanism (i.e. TSC1, TSC2, DEPDC5, NPRL2, NPRL3 and PTEN).5,14–16 We also used GATK’s HaplotypeCaller to identify somatic VAF > 0.30) in other lesional epilepsy-associated genes (i.e. MTOR, SLC35A2, AKT3, PIK3CA, RHEB, BRAF, FGFR1, MYB and MYBL1)5,14–16 that MuTect2 with PoN or MosaicForecast may have missed. The resulting variants were filtered for variants with: (i) phred quality score (QUAL) < 30; (ii) genotype quality (GQ) < 99; (iii) sample read depth (DP) ≥ 30; (iv) max DP < 1000; and (v) GATK truth sensitivity tranche >99.5% for SNV and >95% for indels and were not included in the somatic variant enrichment analyses (See ‘Materials and methods’ section ‘Exome-wide statistical identification of genes associated with epileptogenic brain lesions’).
Somatic CNV and CNN-LOH calling
A total of 688 032 single-nucleotide polymorphisms (SNPs) were genotyped for all samples of this study using the Global Screening Array with Multi-disease drop-in (GSA-MD v.1.0; Illumina). The SNP data were used to detect somatic CNV using MoChA. MoChA is available as a bcftools41 extension that uses phased VCF files with B allele frequency (BAF) and log R ratios (LRR) to identify somatic CNV and copy-number neutral loss of heterozygosity (CNN-LOH). MoChA uses a three-state hidden Markov model to capture somatic-CNV-induced deviations in allelic balance (|ΔBAF|) at heterozygous sites. After affine-normalization and GC wave-correction, the BAF and LRR values were transformed as described elsewhere.42 Autosomes and sex chromosomes were separately considered. Before phasing with Eagle v.2,43 we generated a list of variants that were excluded from modelling by both Eagle and MoChA using the following parameters: (i) segmental duplications with low divergence (<2%); (ii) high levels of missingness (>2%); (iii) variants with excess heterozygosity (P < 10−6); and (iv) variants that unexpectedly correlate with sex (P < 10−6). After somatic CNV calling, we removed samples on the basis of the following quality control (QC) parameters: (i) call rates <0.97 and (ii) baf_auto > 0.3. We then removed variants on the basis of the following QC parameters: (i)lod_baf_phase < 10 unless the somatic CNV was larger than 5 Mbp (or 10 Mbp if they span the centromere); (ii) CNV calls flagged as germline CNV; and (iii) possible constitutional duplications with length >10 Mb and either LRR > 0.35 or LRR > 0.2 and |ΔBAF|>0.16 or length <10 Mb and either LRR > 0.2 or LRR > 0.1 and |ΔBAF|>0.1.44 The estimation of the allelic fraction for each somatic CNV or CNN-LOH by MoChA has been detailed elsewhere.45 All somatic CNV and CNN-LOH were examined visually by plotting the LRR, BAF and phased BAF values with MoChA.
Variant annotation and assessment of deleteriousness
We applied different strategies for the identified CNV and SNV to assess the likelihood of a deleterious effect on disease-relevant loci or genes. We used ANNOVAR46 with custom databases to perform variant annotation. The deleteriousness of SNV used to characterize the genetic architecture of the different brain lesion categories was assessed on the basis of two filters: (i) variant type and population frequency; and (ii) predicted variant deleteriousness. The frequency filter was based on the variant not being present at an AF > 10−5 in the gnomAD47 database. From the remaining variants, we selected only variants with a high-confidence prediction to be deleterious using the following criteria: (i) loss-of-function (LoF) variants ranked in the top 1% most deleterious variants in the human genome according to the Combined Annotation Dependent Depletion score (CADD, scaled CADD phred score ≥ 20)48 found in established lesional epilepsy genes5,14–16 or highly LoF-intolerant genes (genes with a gnomAD LoF Observed/Expected Upper bound Fraction LOEUF ≤ 0.35)47; and (ii) missense variants ranked in the top 1% most deleterious variants in the human genome (scaled CADD phred score ≥20)48 in established lesional epilepsy genes or found in missense-constrained sites (MPC score ≥2)49,50 of missense intolerant genes (gnomAD missense Z-score >3.09, corresponding to P < 10−3).47 The supporting aligned reads of all reported variants were visually inspected using the IGV browser.38
Somatic CNV were annotated with triplosensitive,51 haploinsufficient47 and lesional epilepsy genes.5,14–16 Somatic CNV or CNN-LOH that had a size >1 Mbp were considered deleterious. Deleterious CNV that overlapped genes associated with lesional epilepsies (i.e. MTOR, SLC35A2, AKT3, PIK3CA, RHEB, TSC1, TSC2, NPRL2, NPRL3, DEPDC5, PTEN, BRAF, FGFR1, MYB and MYBL1)5,14–16 were considered pathogenic and are reported in Fig. 2C.
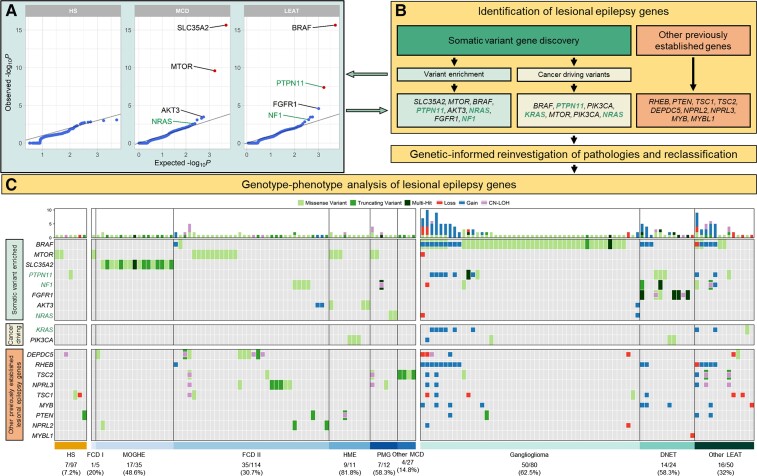
Figure 2.
Characterization and histopathological annotation of genes associated with epileptogenic lesions. (A) Quantile–quantile plots of the gene burden analysis results. Highlighted genes represent identified genes of interest (see ‘Materials and methods’ section). Known and novel genetic associations and candidate genes are labelled with the corresponding gene name (MCD: SLC35A2, MTOR and AKT3 = known causal genes, NRAS = candidate gene; LEAT: BRAF and FGFR1 = known causal genes, NF1 = candidate gene, PTPN11 = significantly enriched gene). Statistically significant somatic variant enrichment was defined by false discovery rate-corrected P-value < 0.1. (B) Flow diagram for identifying genes of interest associated with lesional epilepsies and genotype-phenotype relationships. PTPN11, NF1, NRAS and KRAS were considered novel genetic associations or candidate genes. (C) Somatic mutation profiles for 151 mutation-positive epileptogenic lesions. Each column represents an individual sample with rectangles highlighting identified variants across 19 genes associated with lesional epilepsies. Larger rectangles represent SNV, and smaller squares represent CNV or CNN-LOH. Gain = somatic copy number gain; Loss = somatic copy number loss; PMG = polymicrogyria.
Exome-wide statistical identification of genes associated with epileptogenic brain lesions
To detect genes under evolutionary mutational selection, we used a Poisson-based dN/dS model using dNdScv.52 This model tests the normalized ratio of non-synonymous (missense, nonsense and splicing) over background (synonymous) mutations while correcting for sequence composition and mutational signatures. This method has been shown to reliably identify genes under positive selection in cancer and normal tissues.52 A global false discovery rate adjusted P-value q ≤ 0.1 was used to identify statistically significant non-synonymous variant-enriched genes and an unadjusted P ≤ 0.005 to identify nominally significant non-synonymous variant-enriched genes. Genes that showed a suggestive enrichment of non-synonymous variants were only considered when their biological role and known disease associations matched the associated phenotype. To improve the quality of our results, the reads for all variants in genes identified by dNdScv were visually inspected using the IGV browser38 to assess variant quality. Low-quality variants were flagged as potential artefacts, and genes with only low-quality variants were excluded.
Post hoc genetics-informed histopathological review
In addition to the evaluation on enrolment, we performed an in-depth post hoc histopathological review of all mutation-positive samples in the context of their genetic diagnosis. All slides and formalin-fixed paraffin-embedded tissue blocks were retrieved from the laboratory archives. Following microscopic review of the H&E stained sections, additional immunohistochemical stainings recommended by the ILAE for a comprehensive neuropathologic workup of epilepsy surgery brain tissue were added to the review where necessary.53
Statistical analysis
All statistical analyses and filtering were done using R. We used the Chi-square, Fisher’s exact or Wilcoxon rank-sum test where appropriate. Correlation statistics were generated with Pearson’s correlation.
Data availability
All somatic variant calls are available from the supplementary material. All code is available at https://github.com/LalResearchGroup. Additional data are available from the corresponding author on reasonable request.
Results
Cohort overview
We extracted DNA from snap-frozen brain tissue samples of 474 individuals with drug-resistant epilepsy who underwent epilepsy surgery and were diagnosed with histopathologically confirmed HS, MCD or LEAT (Table 1). HS and LEAT samples primarily originated from the temporal lobe, whereas MCD samples were primarily derived from extratemporal resections and had the largest proportion of multilobular involvement (Table 2). The average age at seizure onset for MCD patients was significantly earlier than that of HS and LEAT (MCD = 6.3 years, HS = 13.73 years, LEAT = 13.14 years; Wilcoxon rank-sum test: MCD versus HS P = 1.17 × 10−7, MCD versus LEAT P = 2.27 × 10−10). Conversely, the average age at surgery for HS patients was significantly later than that of MCD and LEAT (HS = 39.13 years, MCD = 18.48 years, LEAT = 21.82 years; Wilcoxon rank-sum test: HS versus MCD P = 1.02 × 10−21, HS versus LEAT P = 9.59 × 10−15, MCD versus LEAT P = 0.17). Last, individuals with HS had a longer duration of epilepsy before undergoing surgery, followed by individuals with MCD, and with LEAT (HS = 25.81, MCD = 12.24, LEAT = 8.53 years; Wilcoxon rank-sum test: HS versus MCD P = 4.33 × 10−13, HS versus LEAT P = 6.63 × 10−19, MCD versus LEAT P = 1.31 × 10−3).
Table 2.
Clinical characteristics of the study cohort
| HS (n = 97) | MCD (n = 223) | LEAT (n = 154) | |
|---|---|---|---|
| Sex | |||
| ȃMale | 46 (47.4%) | 107 (48%) | 92 (59.7%) |
| ȃFemale | 51 (52.6%) | 116 (52%) | 62 (40.3%) |
| Age of onset (years) | |||
| ȃAverage | 13.73 ± 13.25 | 6.32 ± 8.61 | 13.17 ± 12.98 |
| ȃMedian [IQR] | 9.5 [2, 22] | 3 [0.5, 9.25] | 8.75 [4.25, 18] |
| Age at surgery (years) | |||
| ȃAverage ± SD | 39.13 ± 14.15 | 18.5 ± 14.65 | 21.8 ± 16.09 |
| ȃMedian [IQR] | 40 [27, 48] | 17.86 [5, 28] | 17 [9, 31.75] |
| Duration of epilepsy (years) | |||
| ȃAverage ± SD | 25.81 ± 15.26 | 12.23 ± 10.89 | 8.49 ± 9.57 |
| ȃMedian [IQR] | 23.75 [12.75, 36.5] | 10 [3.5, 17.2] | 5 [2, 11] |
| Lateralization | |||
| ȃRight | 45 (52.3%) | 132 (61.1%) | 83 (54.6%) |
| ȃLeft | 41 (47.7%) | 83 (38.4%) | 69 (45.4%) |
| ȃBoth | 0 | 1 (0.5%) | 0 |
| Localization | |||
| ȃTemporal | 97 (100%) | 48 (21.8%) | 114 (74.5%) |
| ȃȃRight | 45 | 28 | 63 |
| ȃȃLeft | 41 | 18 | 50 |
| ȃȃNot specified | 11 | 2 | 1 |
| ȃFrontal | 0 | 96 (43.6%) | 17 (11.1%) |
| ȃȃRight | 0 | 60 | 8 |
| ȃȃLeft | 0 | 35 | 9 |
| ȃȃNot specified | 0 | 1 | 0 |
| ȃParietal | 0 | 11 (5%) | 8 (5.2%) |
| ȃȃRight | 0 | 5 | 5 |
| ȃȃLeft | 0 | 5 | 3 |
| ȃȃNot specified | 0 | 1 | 0 |
| ȃOccipital | 0 | 6 (2.7%) | 3 (2%) |
| ȃȃRight | 0 | 5 | 1 |
| ȃȃLeft | 0 | 1 | 2 |
| ȃMultiple Lobes | 0 | 59 (26.8%) | 11 (7.2%) |
| ȃȃRight | 0 | 34 | 6 |
| ȃȃLeft | 0 | 24 | 5 |
| ȃȃBoth | 0 | 1 | 0 |
| Follow-up time (months) | |||
| ȃAverage ± SD | 31.88 ± 29.4 | 37.71 ± 34.37 | 27.2 ± 19.28 |
| Surgical outcome at last follow-up | |||
| ȃEngel I | 47 (81%) | 120 (63.8%) | 104 (82.5%) |
| ȃȃEngel II | 9 (15.5%) | 20 (10.6%) | 12 (9.5%) |
| ȃȃEngel III-IV | 2 (3.5%) | 48 (25.6%) | 10 (8%) |
IQR = Interquartile range, SD = standard deviation.
Somatic SNV profiles differ across the major categories of epilepsy-associated brain lesions
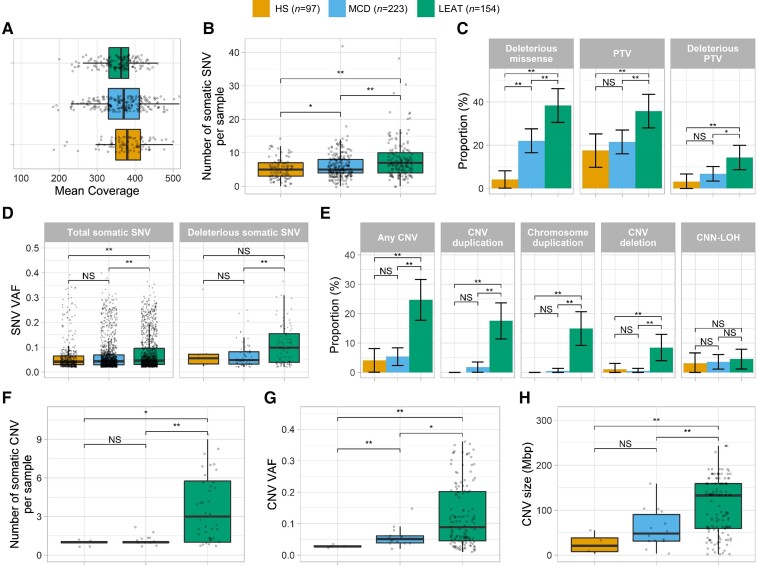
Deep WES achieved an average mean target coverage of 364 × across all 474 samples. DNA quality and experimental performance across pathology groups were similar, with no differences in mean coverage among the three phenotype groups (Fig. 1A). On average, the number of somatic SNV carried per sample was higher in LEAT (7.92 ± 5.65) and MCD (6.11 ± 4) than in HS (5.1 ± 3.04; Fig. 1B). From a total of 3078 somatic SNV, 172 SNV affecting 141 individuals were bioinformatically classified as potentially deleterious. These included 128 missense variants and 44 protein-truncating variants. Overall, a higher proportion of LEAT (48.1%, 74/154) had at least one potentially deleterious SNV, compared to MCD (26.9%, 60/223) and HS (6.2%, 6/97; Fig. 1C). The average VAF of somatic SNV was 1.18-fold higher in LEAT (average VAF = 0.078) than in MCD (average VAF = 0.066) and 1.26-fold higher than in HS (average VAF = 0.062). MCD and HS had similar VAF distributions (Fig. 1D). When considering only potentially deleterious variants, the average VAF in LEAT (average deleterious VAF = 0.108) was 1.63-fold higher than in MCD (average deleterious VAF = 0.066) and 1.22-fold higher than in HS (average deleterious VAF = 0.088; Fig. 1D). Overall, MCD and HS showed similar somatic SNV profiles, although MCD carried a nominally significant higher number of somatic SNV per sample and a larger proportion of samples with at least one deleterious missense SNV more than HS (Fig. 1B and C).
Figure 1.
Different somatic mutation profiles observed across major categories of epilepsy-associated brain lesions. (A) Median coverage is similar across major lesion categories. (B) LEAT and MCD have a higher number of somatic SNV per sample than HS. (C) LEAT have a higher proportion of samples with at least one more deleterious variant than HS and MCD. (D) The average VAF for total somatic SNV is higher in LEAT. Potentially deleterious variants (missense variants with MPC > 2 and truncating variants in LoF-intolerant genes with LOEUF < 0.35) in LEAT have a higher VAF than those in HS and MCD. (E) LEAT have a higher proportion of samples with at least one more CNV than MCD and HS. (F) LEAT samples with somatic CNV have significantly more somatic CNV per sample than MCD/HS. (G) LEAT CNV occur at significantly higher VAF than MCD/HS. (H) LEAT have larger CNV than MCD/HS. Asterisk indicates nominally significant difference (P < 0.05 before multiple-testing correction); double asterisks indicate significant difference (P < 0.05 after multiple-testing correction). NS = not significant; PTV = protein-truncating variant.
Somatic CNV profiles differ across the major categories of epilepsy-associated brain lesions
After QC, we identified a total of 153 large CNV and CNN-LOH across 56 samples: 105 duplications (85 whole chromosome gains), 27 deletions (5 whole chromosome losses) and 21 CNN-LOH. LEAT had the highest proportion of samples with one or more somatic CNV or CNN-LOH (38/154, 24.7%), followed by MCD (12/223, 5.4%) and HS (4/97, 4.1%; Fig. 1E). LEAT were also enriched for CNV duplications and deletions compared to MCD and HS, whereas CNN-LOH were not distributed differently across all three categories. Among LEAT, somatic duplications were the most frequently observed type of CNV, with 71% (27/38) of CNV or CNN-LOH-positive samples carrying one or more somatic duplications. Somatic whole chromosome duplications were the major type of duplications among LEAT (82/101 LEAT gain CNV, 81.2%; Fig. 1E). Conversely, somatic CNN-LOH were predominant among MCD and HS (8/12 CNV-positive MCD, 66.7%; 3/4 CNV-positive HS, 75%).
Most somatic CNV-positive LEAT had multiple somatic CNV (27/38, 71%), whereas only two (2/12, 16.7%) CNV-positive MCD had multiple somatic CNV or CNN-LOH (Fig. 1F). No CNV-positive HS sample carried multiple somatic CNV or CNN-LOH. The highest average VAF of somatic CNV and CNN-LOH was identified in LEAT (average VAF = 0.125), followed by MCD (average VAF = 0.058) and HS (average VAF = 0.028; Fig. 1G). Last, LEAT samples carried larger somatic CNV and CNN-LOH (average size = 108.6 Mbp) than MCD (average size = 59.6 Mbp) and HS (average size = 25.3 Mbp; Fig. 1H). Overall, LEAT showed the most distinct CNV profile; they had a larger proportion of samples with CNV or CNN-LOH, were enriched for duplications and deletions, tended to carry more CNV per sample and had larger CNV affecting a higher number of cells.
The number of somatic SNV and the fraction of mutated tumour cells are associated with age at seizure onset and age at surgery in LEAT
For LEAT, we identified positive correlations between the number of somatic SNV per sample and later age at seizure onset and older age at surgery (Pearson’s correlation; LEAT total somatic SNV and age at surgery: r = 0.26, P = 1.16 × 10−3; LEAT total somatic SNV and age at onset: r = 0.22, P = 9 × 10−3). Similarly, we also identified positive correlations between higher somatic VAF (a proxy marker for the number of mutated cells) and later age at seizure onset and older age at surgery for LEAT (Pearson’s correlation; LEAT somatic VAF and age at onset: r = 0.15, P = 1.61 × 10−7; LEAT somatic VAF and age at surgery: r = 0.11, P = 3.55 × 10−5). This signal was driven primarily by the somatic SNV VAF, with the VAF of the CNV and CNN-LOH having little effect (Pearson’s correlation; LEAT somatic SNV VAF and age at surgery: r = 0.16, P = 1.47 × 10−8; LEAT somatic SNV VAF and age at onset: r = 0.18, P = 1.44 × 10−9; LEAT somatic CNV VAF and age at surgery: r = −0.2, P = 0.0176; LEAT somatic CNV VAF and age at onset: r = 0.01, P = 0.95). We did not identify any similar correlations between genetic architecture and clinical variables for MCD and HS.
Unbiased somatic variant burden analysis confirms previously reported and discovers novel lesional epilepsy-associated genes
We performed the first exome-wide burden analysis for a lesional epilepsy cohort (HS, MCD and LEAT) to identify genes under positive mutational selection with significant enrichment of somatic SNV in any of the three main lesion categories. In line with previous results from candidate gene studies, we identified four genes with significant somatic SNV burden for LEAT and MCD and no genetic associations with HS (Supplementary Table 2 and Fig. 2A). For LEAT, the gene with the most significant exome-wide somatic SNV burden was BRAF, which represents the most well-established previously reported LEAT gene and is primarily associated with GG.5,54 The second gene identified with a significant variant enrichment was PTPN11, which has not been previously associated with LEAT. The PTPN11 gene encodes Protein Tyrosine Phosphatase Non-Receptor Type 11, an upstream regulator of the RAS/MAPK and mTOR signalling pathways. Activating mutations in PTPN11 have been shown to cause Noonan syndrome (a congenital developmental syndrome), play a role in tumorigenesis and affect the development of white matter microstructure in humans.55–57 For MCD, our burden analysis identified SLC35A2 and MTOR, which have been previously reported in multiple candidate gene studies or descriptive exome analysis screens as the two most frequently mutated genes in mild MOGHE and FCD type 2, respectively.14 However, we are the first to use an unbiased statistical approach to show that variants in these genes are a major genetic cause of MCD entities.
Identification of additional lesional epilepsy genes with suggestive statistical support and high biological plausibility
The previous statistical analysis could identify and validate the most frequently mutated lesional epilepsy genes that remained significant after controlling for the false discovery rate (q ≤ 0.1). Next, we investigated variants in genes with suggestive variant enrichment (P < 5 × 10−3) for association with LEAT or MCD by considering additional evidence criteria (see ‘Materials and methods’ section and Supplementary Table 2). We also screened our cohort for variants in 579 high-confidence cancer-driver sites, previously identified in 53 genes (Supplementary Table 3).58 The genes FGFR1, NF1, PIK3CA and KRAS had additional evidence for a role in the LEAT aetiology (Fig. 2A). FGFR1 and NF1 had nominally significant variant enrichment and strong evidence in the literature based on patient reports with similar phenotypes (Supplementary Table 2).5,21,22 We also identified functionally validated cancer-driving variants in PIK3CA and KRAS, both of which are established tumour or growth disorder genes59,60 (Supplementary Table 3). The genes AKT3, NRAS and PIK3CA had nominally significant mutational burden and/or variants in cancer-driving sites and met additional disease-association criteria for a role in the aetiology of MCD (Supplementary Tables 2 and 3). AKT3 and PIK3CA are well-established MCD genes and the identified variants have been molecularly characterized as activating variants.61–63 Additionally, we identified a novel association of the NRAS gene with MCD (Fig. 3A). NRAS is a key regulator of cell proliferation, differentiation and growth as an upstream regulator of the RAS-RAF-MAPK and PI3K-AKT-MTOR pathways.64 The variants driving the enrichment occurred at the previously established NRAS Q61 cancer-driving site (Supplementary Table 3).58,65
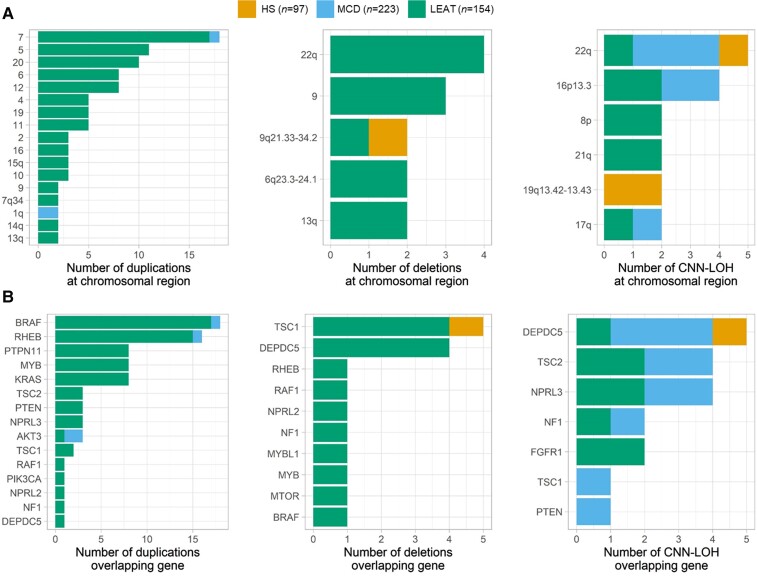
Figure 3.
Recurring somatic CNV and CNN-LOH regions often overlap with lesional epilepsy genes. (A) Recurring regions of overlap among somatic CNV and CNN-LOH. Smaller regions are identified on the basis of included cytobands, CNV or CNN-LOH affecting >80% of a chromosomal arm are identified by the chromosomal arm, and CNV affecting the entire chromosome are identified by the chromosome number. (B) Detected CNV and CNN-LOH often overlap established and newly identified lesional epilepsy genes.
Large somatic chromosomal alterations in lesional epilepsies frequently include established lesional epilepsy-associated genes
We identified several somatic CNV patterns and recurring regions across the lesion categories represented in our cohort. In LEAT specifically, the most frequently recurring duplicated regions were whole chromosome duplications of chromosome 7 (17/101, 16.8%), chromosome 5 (11/101, 10.9%) and chromosome 20 (10/101, 9.9%; Fig. 3A). We also identified a recurring CNV gain of the q-arm of chromosome 1 in our MCD cohort (n = 2). Duplications of chromosome 1q have been previously identified in cases of macrocephaly, megalencephaly, polymicrogyria and FCD.66–68 The most frequently recurring regions of CNN-LOH mapped to the longer q-arm of chromosome 22 (5/21, 23.8%) and chromosome 16p13.3 (4/21, 19%; Fig. 3A). These regions were most prevalent in MCD, but were also identified in HS or LEAT. A single recurring region of CNN-LOH was unique to HS, which mapped to the q-arm of chromosome 19 (19q13.42–13.43).
To pinpoint the putative causative genes, we investigated whether the identified CNV and CNN-LOH overlapped with previously established lesional epilepsy genes. We observed that 68.4% of LEAT, 83.3% of MCD and 50% of HS with a somatic CNV or CNN-LOH carried at least one chromosomal alteration affecting one of the 15 established lesional epilepsy genes5,14–16 (Fig. 3B). The most frequently recurring CNV gain (chromosome 7) overlapped with the most frequently mutated gene in lesional epilepsies (BRAF). Additionally, the two most frequently recurring regions of CNN-LOH included DEPDC5 (22q) or TSC2 (16p13.3), both established lesional epilepsy genes that require a somatic second hit such as a CNN-LOH to cause disease.
Genetics-informed histopathology review reveals novel genotype–phenotype associations
In total, we identified 19 genes of interest either through our somatic variant gene discovery analysis (SLC35A2, MTOR, BRAF, PTPN11, AKT3, NRAS, FGFR1, NF1, PIK3CA and KRAS) or previously reported as genes associated with epileptogenic brain lesions (RHEB, PTEN, TSC1, TSC2, DEPDC5, NPRL2, NPRL3, MYB and MYBL1).5,14–16 Across our entire cohort, we identified 151 samples that carried at least one SNV, CNV or CNN-LOH affecting one of these genes (31.4% of cohort; 7.2% of HS, 31.8% of MCD and 51.9% of LEAT; Fig. 2C and Supplementary Tables 4 and 5). By considering known pathogenic pathways, we identified 74 samples (64 LEAT, 9 MCD and 1 HS) with variants affecting genes in the RAS-RAF-MAPK pathway (FGFR1, PTPN11, KRAS, NRAS, NF1), 64 samples (32 MCD, 27 LEAT and five HS) with variants in genes of the PI3K-AKT-MTOR pathway (PIK3CA, PTEN, AKT3, TSC1, TSC2, RHEB, MTOR) and 31 samples (19 MCD, 11 LEAT and 1 HS) with variants in genes of the GATOR1 complex (DEPDC5, NPRL2 and NPRL3). All mutation-positive samples underwent an in-depth post hoc histopathological review that included their genetic diagnosis (Table 3). The genes with the most variants (SNV, CNV or CNN-LOH) which had primarily homogeneous histopathology included: BRAF (52/67 in GG), SLC35A2 (17/17 in MOGHE), FGFR1 (14/14 in DNET) and MTOR (13/19 in FCD 2b or HME). A detailed summary of the genotype–phenotype analysis for the established LEAT and MCD genes is provided in the Supplementary material and Fig. 2C.
Table 3.
Genetic-positive samples where genetic diagnosis had an impact on histopathological review
| Sample name | Lesion category | Original pathology | Genetic diagnosis | Inheritance (VAF) | Revised pathology | Additional information |
|---|---|---|---|---|---|---|
| MCD_EEBB_10 | MCD | PMG | NRAS (p.Q61R) | Somatic (0.13) | Complex MCD | PMG, NH, FCD 2a and GG |
| MCD_EEBB_65 | MCD | PMG | NRAS (p.Q61K) | Somatic (0.24) | Complex MCD | PMG, NH, FCD 2a and DNET |
| MCD_CCF_29 | MCD | FCD (NOS) | TSC2 (p.C644X) | Germline (0.46) | Cortical tuber | FCD 2b |
| MCD_CCF_55 | MCD | FCD (NOS) | TSC2 (p.F1619S) | Germline (0.46) | Cortical tuber | FCD 2b |
| MCD_CCF_56 | MCD | FCD (NOS) | TSC2 (p.M276Vfs*61) | Germline (0.43) | Cortical tuber | FCD 2b |
| MCD_CCF_27 | MCD | FCD 2b | TSC2 (p.H1135Pfs*33) | Germline (0.45) | Cortical tuber | FCD 2b |
| MCD_CCF_62 | MCD | FCD 2a | Chr1q duplication | Somatic (0.06) | FCD 2a | Hyaline astrocytic inclusions |
| MCD_EEBB_110 | MCD | MOGHE | Chr1q duplication | Somatic (0.08) | FCD 2a | Hyaline astrocytic inclusions |
| MCD_CCF_30 | MCD | mMCD | DEPDC5 (p.V272L) | Germline (0.46) | FCD 2a | – |
| MCD_CCF_80 | MCD | mMCD | DEPDC5 (p.P779A) | Germline (0.49) | FCD 2a | – |
| MCD_EEBB_100 | MCD | mMCD | SLC35A2 (p.Q108X) | Somatic (0.25) | MOGHE | – |
| MCD_CCF_79 | MCD | FCD 1a | SLC35A2 (p.A116E) | Somatic (0.14) | MOGHE | – |
| MCD_EEBB_58 | MCD | FCD 1a | SLC35A2 (p.S308Wfs*106) | Somatic (0.11) | MOGHE | – |
| MCD_EEBB_87 | MCD | FCD 1a | SLC35A2 (p.Q108X) | Somatic (0.09) | MOGHE | – |
NOS = Not otherwise specified; PMG = polymicrogyria; NH = nodular heterotopia.
Major novel findings of our analysis include eight somatic PTPN11 SNV across seven LEAT samples (three in cancer-driving sites) and eight samples with somatic duplications covering PTPN11 (Fig. 3C). Despite a clear association with LEAT, no specific LEAT histopathological subtype was strongly associated with PTPN11 alterations; variants occurred in GG, DNET and three different glioma subtypes (Fig. 3C and Supplementary Tables 4 and 5).
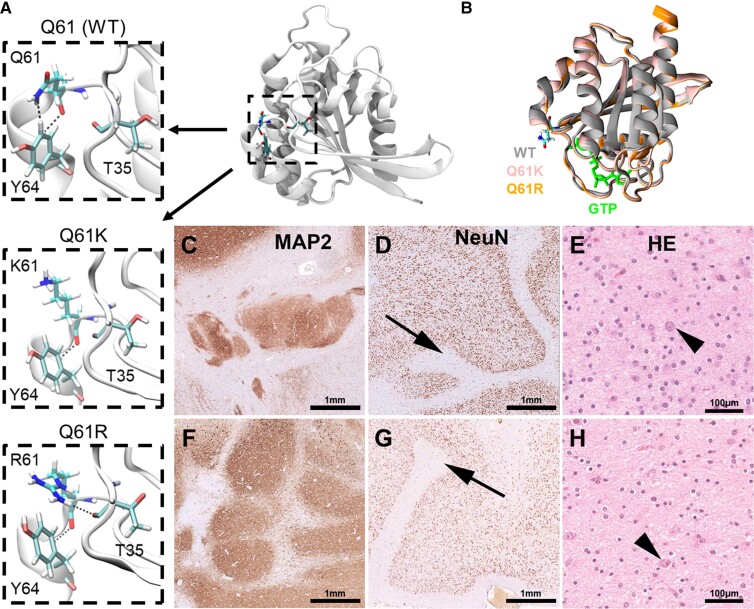
Second, we observed a novel association between polymicrogyria and variants in NRAS, with 17% (2/12) of polymicrogyria samples having an activating variant in NRAS. Both variants (p.Q61K and p.Q61R) have been well recognized in the literature as cancer-driver variants (Fig. 4A).65,69 Conformational changes of the NRAS Q61 mutated protein lead to a prolonged active state (guanosine-5′-triphosphate GTP-bound) compared to the wild-type form, with variants such as Q61R having an increased affinity for GTP, a slower rate of GTP exchange and a lower rate of intrinsic GTP hydrolysis relative to wild-type (Fig. 4B).70 On microscopic re-review, both samples with NRAS somatic variants had a concordant complex MCD phenotype composed of polymicrogyria and nodular heterotopia with tumour-like glio-neuronal growth patterns (Fig. 4C–H).
Figure 4.
Novel complex MCD phenotype is associated with NRAS mutations at Q61. (A) Crystal structure of the wild-type (WT) N-Ras protein. Close-up of the 61st amino acid site highlights changes in intermolecular interactions between different residues at position 61 and nearby positions T35 and Y64. (B) Overlap of the active GTP-bound 3D structures of WT, Q61K and Q61R N-Ras highlighting changes in structure due to the introduced amino acid changes that increase the protein’s affinity to GTP. (C–E) Twenty-year-old female patient with right temporal lobe epilepsy secondary to polymicrogyria seen on MRI. Histopathology examination of the surgical resection sample revealed a complex cortical malformation with nodular heterotopia shown in C, polymicrogyria (arrow in D), and a tumour-like glio-neuronal growth pattern resembling DNET in E. The arrow in E points to a dysplastic neuronal cell element. (F–H) Twenty-five-year-old male patient with left temporal lobe epilepsy and complex malformation seen on MRI. Histopathology examination of the surgical resection sample confirmed a complex cortical malformation with nodular heterotopia shown in F, polymicrogyria (arrow in G) and a tumour-like glio-neuronal growth pattern in H (arrow showing a dysplastic neuronal cell). MAP2 = immunohistochemistry using antibodies directed against microtubule-associated protein 2, NeuN = immunohistochemistry using antibodies directed against the neuronal nucleus epitope, HE = haematoxylin and eosin staining.
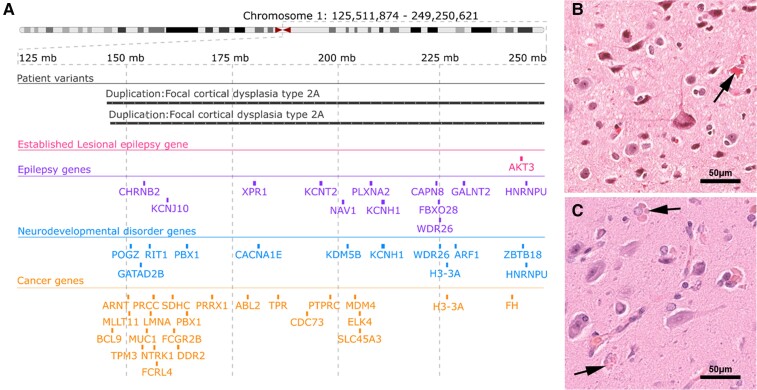
Last, we identified two MCD samples with duplications of the q-arm of chromosome 1 (Table 3). Both samples had similar duplications spanning the 1q21-q44 region, including AKT3 (Fig. 5A). Histopathological review revealed that both samples with the 1q21-q44 duplication, including AKT3, had a concordant phenotype of FCD 2a with hyaline astrocytic inclusions, a rare and seldom reported epileptogenic brain lesion (Fig. 5B and C).71 This is a novel genotype–phenotype association for somatic duplications of chromosome 1q and the first genetic association reported for hyaline astrocytic inclusions.
Figure 5.
Novel association between large chromosome 1q duplications that include AKT3 and type 2 FCD with hyaline astrocytic inclusions. (A) Genomic coordinates of two large patient somatic duplications at 1q21-q44. Both duplications cover the established lesional epilepsy gene AKT3, as well as additional genes of potential interest. Breakpoints of both somatic duplications are similar. (B and C) Histopathology findings were remarkably similar in both cases, showing hyaline astrocytic inclusions (arrows in B and C) next to FCD 2a.
Discussion
We generated deep (>350×) WES data to identify somatic SNV and whole-genome genotyping data to identify somatic CNV across surgically resected epileptogenic brain lesions from 474 individuals with focal drug-resistant epilepsy. While other studies performed targeted sequencing of lesional brain tissue from <100 individuals,16 or WES in <130 individuals,72 our study sample represents the largest cohort of epilepsy-associated brain lesions analysed through deep whole-exome analysis. Using this rich source of data, we demonstrated differential somatic variant profiles across LEAT, MCD and HS. For LEAT and MCD, we confirmed previously reported and identified novel pathology-gene associations and explored the role of genetic information in clinical characteristics and histopathological classification.
We observed a differential somatic variant burden across the three most common epileptogenic pathologies (LEAT, MCD and HS). LEAT showed a higher burden of somatic SNV and CNV than MCD or HS. This finding indicates a proliferative advantage of mutated tumour cells and conclusively shows for the first time that the genetic architecture of epilepsy-associated tumours is different from that underlying MCD or HS. We showed that increasing numbers of somatic SNV and fractions of mutated tumour cells are correlated with later age at seizure onset and older age at surgery in LEAT. This finding is in line with studies that found positive correlations between higher somatic burden and later age at diagnosis for all human genes and all cancer-associated genes across all types of cancer.73 We confirmed previously reported and discovered novel lesional epilepsy-associated genes using a combined statistical genetic and biological approach. Our study confirms that somatic variants affecting 12/15 recently established lesional epilepsy genes5,14–16 represent common genetic aetiologies of lesional epilepsies (29.1%, 138/474 pathogenic variant carriers in our cohort across 12 genes). We did not observe somatic variants in RHEB, MYB and MYBL1, in which previously identified pathogenic variants are primarily structural rearrangements such as gene fusions and CNV.5,14
Our study could not provide evidence for a significant role of somatic pathogenic SNV and CNV in the aetiology of HS. We analysed 97 individuals with HS, and did not identify any genetic association with HS. However, we identified a small subset of patients with somatic variants in recognized lesional epilepsy genes or the newly described PTPN11. Whether these variants are indeed contributing to rare subtypes of HS needs to be clarified in future studies. To the best of our knowledge, this is the first large-scale exome-level investigation of genetic variants in surgically resected HS tissues.
Most CNV (>69%) affected one of the 15 established lesional epilepsy genes.5,14 Microscopic re-review of these samples led to the identification of a novel genotype–phenotype association between duplications at q21-q44 of chromosome 1 that affects AKT3 and many additional genes and a FCD 2a phenotype with hyaline astrocytic inclusions. This is the first report of a genotype–phenotype correlation for this lesion type and expands the spectrum of MCD phenotypes associated with somatic duplications of chromosome 1q. Compared to our study, previously reported 1q duplications were larger in size and observed in individuals with larger lesions such as polymicrogyria.62,67,68 Furthermore, we identified nine cases with large somatic CNN-LOH regions overlapping lesional epilepsy genes with pathogenic mechanisms known follow the ‘two-hit’ model (TSC1, TSC2, DEPDC5, PTEN, FGFR1 and NF1).17,74 For five of these (with the exception of TSC1), we identified an accompanying pathogenic germline SNV in the same gene. This result is in line with previous lesional epilepsy studies that identified CNN-LOH as a somatic second hit for germline variants in DEPDC5, TSC1 and TSC2.17,75 Overall, our findings add to a growing body of evidence that somatic CNV are involved in the aetiology of epilepsy-associated brain lesions.62,76
Our genetic screen identified five LEAT samples carrying a total of six variants in PTPN11, which encodes the Protein Tyrosine Phosphatase Non-Receptor Type 11—a regulator of the RAS-RAF-MAPK signalling pathway. Five of the six PTPN11 variants identified in LEAT have been previously reported in the clinical variant database ClinVar as pathogenic variants associated with Noonan syndrome or various neoplasms (p.A72G, p.E76K, p.D61N, p.D61H, p.E139D, p.T507K), with the final variant (p.A72V) reported as having ‘conflicting interpretations of pathogenicity’ (Supplementary Table 4). Additionally, three identified variants occurred in established cancer-driver hotspots in PTPN11 (p.A72 and p.E76). According to the American College of Medical Genetics and Genomics (ACMG) guidelines,77 this finding provides strong evidence of the pathogenicity of the identified variants (see Supplementary Table 6 for full criteria). Furthermore, our exome-wide somatic variant enrichment analysis provides the first statistical evidence for PTPN11, a ClinGen recognized criterion for gene to disorder association validity, therefore considering PTPN11 as a novel disease-associated gene for LEAT.78 In addition to the evidence provided by the association result, the clinical phenotype of previously reported PTPN11 mutations matches the phenotype of the carriers identified in our study. Heterozygous gain of function mutations in PTPN11 cause ∼50% of all Noonan syndrome cases,79 a germline overgrowth disorder where multiple cases of co-occurring epileptic tumours have been reported.55,80,81 The LEAT samples with somatic PTPN11 SNV had other pathogenic somatic SNV in TSC2, FGFR1, NF1 or BRAF (Supplementary Table 4). This finding suggests a modifier role for PTPN11 in LEAT, as shown for other components of the RAS-RAF-MAPK signalling pathway.82
We also identified an individual with HS who carried a likely pathogenic somatic mutation in PTPN11 (p.S502L; Supplementary Table 6). Our findings are in line with a recent study that reported somatic PTPN11 mutations in two individuals with FCD type 3, another non-LEAT epileptogenic lesion.83 Interestingly, PTPN11 also regulates the PI3K-AKT-MTOR pathway84 and plays a role in the development of human white matter microstructure.56 Therefore, it is possible that mutations in PTPN11 may also cause other epileptogenic lesions beyond LEAT. However, gene-disease validity criteria78 are currently only support associations with Noonan syndrome, metachondromatosis and certain cancer types including, as of this study, LEAT. Confirming and further elucidating the role of PTPN11 in LEAT as well as other epileptic lesions will require more in-depth evaluations of large lesional epilepsy cohorts as well as comprehensive animal model.54,85
We identified three additional candidate genes for lesional epilepsy within the RAS-RAF-MAPK pathway (i.e. NF1, KRAS and NRAS), all of which had moderate levels of evidence for gene-disease association according to ACMG guidelines based on variant burden and concordance between previously reported and patient phenotypes.77 Our gene burden analysis identified an enrichment of somatic NF1 variants in LEAT. Similar to PTPN11, no particular LEAT pathology was associated with NF1 variants, although alterations of NF1 have been previously identified in GG.5,6 Interestingly, we also identified somatic variants in NF1 in four FCD 2 and one complex MCD (Fig. 2C). Although HS and FCD 3a have been described in a subset of germline neurofibromatosis type 1, our study is the first direct report of NF1 variants in bona fide MCD.86,87 We also identified an individual with sporadic meningioangiomatosis and a brain somatic cancer-driving variant in KRAS. Germline meningioangiomatosis is associated with neurofibromatosis type 2, but no causal gene for the sporadic form is known yet.88 Last, we identified two established cancer-driving SNV in NRAS in two individuals with a distinct and histopathologically concordant complex MCD phenotype composed of polymicrogyria and nodular heterotopia with tumour-like glio-neuronal growth patterns (Fig. 4). The identification of potentially pharmacologically targetable NRAS variants89 in such patients may present a promising target for precision medicine when epilepsy surgery cannot achieve successful seizure control.90
When the genetic information was disclosed for histopathology re-review for all 151 individuals carrying somatic variants in known or novel candidate genes for lesional epilepsy, we identified consistent genotype–phenotype associations not previously recognized. These included: (i) a complex MCD pattern of polymicrogyria with nodular heterotopia and low-grade glio-neuronal tumours associated with NRAS mutations (Fig. 4 and Table 3); and (ii) hyaline astrocytic inclusions91 associated with chromosomal 1q gains (Fig. 5 and Table 3). The recently defined association of MOGHE with SLC35A2 was confirmed in our case series and resulted in a histopathology reclassification of four cases previously diagnosed as mild MCD (mMCD) or FCD1a.19 Notwithstanding, mMCD and FCD1a are the most difficult and challenging differential diagnoses of MOGHE.92 There were four cases with a germline mutation in TSC2 leading to the reclassification of FCD2b to cortical tuber. However, the tuberous sclerosis complex syndrome was not documented in the clinical charts at the time of histopathological diagnosis (Table 3). Our findings exemplify the increasing impact of an integrated molecular-histopathology diagnosis also in the field of focal epilepsies, as was suggested previously20 and already introduced successfully into the clinical management of individuals with brain tumours.20,93 Last, we also observed variants in established lesional epilepsy genes, which were identified and confirmed to occur in pathologies that do not match the previously reported associated phenotype (Fig. 2C and Supplementary Table 4). These included an MTOR variant (p.R360W) in an individual with FCD1a, a germline DEPDC5 variant (p.R78P) in an individual with MOGHE, and a BRAF variant (p.I61V) in an individual with FCD2a. On the basis of the available data, we cannot definitively assess whether these variants are benign or pathogenic, thus expanding the phenotypic spectrum associated with these genes. Future larger association studies or molecular functional investigations will have to be conducted to confirm our findings.
It is important to note that our exome-wide SNV and genome-wide CNV screening design casts a wide net to elucidate the genomic architecture of the lesional epilepsies and uncover potential disease-associated genes that are typically not on hypothesis-based gene panels. However, despite a high coverage of 364×, well within the standard for recent high-impact deep-coverage WES studies,94–96 our approach is not well-powered to reliably identify somatic variants carried only by a very small number of cells within a lesion (VAF < 2%). Our study was also limited in distinguishing between somatic VAF and germline variants because of the lack of matched blood samples. Therefore, we implemented upper and lower thresholds for the allelic fraction of somatic variants that may have reduced the total number of identified somatic variants in all downstream analyses. Our variant QC using individuals with non-lesional epilepsies as the PoN may have led to the exclusion of somatic variants associated with epilepsy only. It is unlikely that the individuals with non-lesional epilepsy carried genetic factors involved in over/abnormal growth disorders (i.e. tumours and FCDs). However, we cannot rule out the possibility that our strategy may have excluded some somatic variants involved in HS. Overall, our quality filtering strategy was stringent to overcome the direct need for confirmatory sequencing analyses, but will have likely reduced the number of identified somatic variants. As such, the genetic-positive rate is likely higher than detected in our study and other studies with a diagnostic focus that include ultra-deep-coverage sequencing and secondary validation, such as amplicon sequencing or digital PCR, should be cited for diagnostic yield estimates.
Understanding the exact molecular mechanisms involved in the aetiology of epileptogenic pathologies with or without tumour activity is essential for improving treatment of drug-resistant focal epilepsy. Our study systematically shed light on the genomic landscape of the lesional epilepsies, identified four novel candidate genes for lesional epilepsy (i.e. NRAS, KRAS, NF1 and PTPN11), and observed several potentially pathogenic somatic CNV. More basic and clinical-genetic research is needed to elucidate the potential suitability of post-surgery screenings after somatic variants to inform surgery outcome prediction, patient management and our general understanding of the aetiology of the various pathologies underlining focal epilepsy.
Supplementary Material
Acknowledgements
We wish to thank Anna Ossowski from the WGGC for her expert technical assistance.
Contributor Information
Javier A López-Rivera, Department of Molecular Medicine, Cleveland Clinic Lerner College of Medicine, Case Western Reserve University, Cleveland, OH 44195, USA; Genomic Medicine Institute, Lerner Research Institute, Cleveland Clinic, Cleveland, OH 44195, USA; Charles Shor Epilepsy Center, Neurological Institute, Cleveland Clinic, Cleveland, OH 44195, USA.
Costin Leu, Genomic Medicine Institute, Lerner Research Institute, Cleveland Clinic, Cleveland, OH 44195, USA; Charles Shor Epilepsy Center, Neurological Institute, Cleveland Clinic, Cleveland, OH 44195, USA; Stanley Center for Psychiatric Research, Broad Institute of Harvard and M.I.T., Cambridge, MA 02142, USA; Department of Clinical and Experimental Epilepsy, Institute of Neurology, University College London, London WC1N 3BG, UK.
Marie Macnee, Cologne Center for Genomics (CCG), University of Cologne, Faculty of Medicine and University Hospital Cologne, 50931 Cologne, Germany.
Jean Khoury, Charles Shor Epilepsy Center, Neurological Institute, Cleveland Clinic, Cleveland, OH 44195, USA.
Lucas Hoffmann, Department of Neuropathology, University Hospital Erlangen, Erlangen 91054, Germany.
Roland Coras, Department of Neuropathology, University Hospital Erlangen, Erlangen 91054, Germany.
Katja Kobow, Department of Neuropathology, University Hospital Erlangen, Erlangen 91054, Germany.
Nisha Bhattarai, Genomic Medicine Institute, Lerner Research Institute, Cleveland Clinic, Cleveland, OH 44195, USA; Charles Shor Epilepsy Center, Neurological Institute, Cleveland Clinic, Cleveland, OH 44195, USA.
Eduardo Pérez-Palma, Universidad del Desarrollo, Centro de Genética y Genómica, Facultad de Medicina Clínica Alemana, Santiago 12438, Chile.
Hajo Hamer, Epilepsy Center, University Hospital Erlangen, 91054 Erlangen, Germany.
Sebastian Brandner, Department of Neurosurgery, University Hospital Erlangen, Erlangen 91054, Germany.
Karl Rössler, Department of Neurosurgery, Medical University Vienna, 1090 Vienna, Austria.
Christian G Bien, Department of Epileptology (Krankenhaus Mara), Medical School, Bielefeld University, 33617 Bielefeld, Germany.
Thilo Kalbhenn, Department of Neurosurgery - Epilepsy surgery, Evangelisches Klinikum Bethel, Universitätsklinikum OWL, Bielefeld University, 33617 Bielefeld, Germany.
Tom Pieper, Center for Pediatric Neurology, Neurorehabilitation and Epileptology, Schoen-Clinic, 83569 Vogtareuth, Germany.
Till Hartlieb, Center for Pediatric Neurology, Neurorehabilitation and Epileptology, Schoen-Clinic, 83569 Vogtareuth, Germany; Research Institute ‘Rehabilitation, Transition, Palliation’, PMU Salzburg, 5020 Salzburg, Austria.
Elizabeth Butler, Charles Shor Epilepsy Center, Neurological Institute, Cleveland Clinic, Cleveland, OH 44195, USA.
Giulio Genovese, Stanley Center for Psychiatric Research, Broad Institute of Harvard and M.I.T., Cambridge, MA 02142, USA.
Kerstin Becker, Cologne Center for Genomics (CCG), University of Cologne, Faculty of Medicine and University Hospital Cologne, 50931 Cologne, Germany.
Janine Altmüller, Cologne Center for Genomics (CCG), University of Cologne, Faculty of Medicine and University Hospital Cologne, 50931 Cologne, Germany; Berlin Institute of Health at Charité, Universitätsmedizin Berlin, Core Facility Genomics, 10117 Berlin, Germany; Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC), 13125 Berlin, Germany.
Lisa-Marie Niestroj, Cologne Center for Genomics (CCG), University of Cologne, Faculty of Medicine and University Hospital Cologne, 50931 Cologne, Germany.
Lisa Ferguson, Charles Shor Epilepsy Center, Neurological Institute, Cleveland Clinic, Cleveland, OH 44195, USA.
Robyn M Busch, Genomic Medicine Institute, Lerner Research Institute, Cleveland Clinic, Cleveland, OH 44195, USA; Charles Shor Epilepsy Center, Neurological Institute, Cleveland Clinic, Cleveland, OH 44195, USA.
Peter Nürnberg, Cologne Center for Genomics (CCG), University of Cologne, Faculty of Medicine and University Hospital Cologne, 50931 Cologne, Germany; Center for Molecular Medicine Cologne (CMMC), University of Cologne, Faculty of Medicine and University Hospital Cologne, 50931 Cologne, Germany.
Imad Najm, Charles Shor Epilepsy Center, Neurological Institute, Cleveland Clinic, Cleveland, OH 44195, USA.
Ingmar Blümcke, Charles Shor Epilepsy Center, Neurological Institute, Cleveland Clinic, Cleveland, OH 44195, USA; Department of Neuropathology, University Hospital Erlangen, Erlangen 91054, Germany.
Dennis Lal, Genomic Medicine Institute, Lerner Research Institute, Cleveland Clinic, Cleveland, OH 44195, USA; Charles Shor Epilepsy Center, Neurological Institute, Cleveland Clinic, Cleveland, OH 44195, USA; Stanley Center for Psychiatric Research, Broad Institute of Harvard and M.I.T., Cambridge, MA 02142, USA; Cologne Center for Genomics (CCG), University of Cologne, Faculty of Medicine and University Hospital Cologne, 50931 Cologne, Germany.
Funding
I.B. and P.N. received research funding from the German Research Foundation (DFG, grant agreement numbers BL 421/4-1 and NU 50/13-1). Sequencing was facilitated by the DFG-funded West German Genome Center (WGGC).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Engel J. What can we do for people with drug-resistant epilepsy? The 2016 Wartenberg Lecture. Neurology. 2016;87:2483–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blumcke I, Spreafico R, Haaker G, et al. . Histopathological findings in brain tissue obtained during epilepsy surgery. N Engl J Med. 2017;377:1648–1656. [DOI] [PubMed] [Google Scholar]
- 3. Lamberink HJ, Otte WM, Blümcke I, et al. . Seizure outcome and use of antiepileptic drugs after epilepsy surgery according to histopathological diagnosis: A retrospective multicentre cohort study. Lancet Neurol. 2020;19:748–757. [DOI] [PubMed] [Google Scholar]
- 4. Blümcke I, Thom M, Aronica E, et al. . International consensus classification of hippocampal sclerosis in temporal lobe epilepsy: A task force report from the ILAE commission on diagnostic methods. Epilepsia. 2013;54:1315–1329. [DOI] [PubMed] [Google Scholar]
- 5. Slegers RJ, Blumcke I. Low-grade developmental and epilepsy associated brain tumors: A critical update 2020. Acta Neuropathol Commun. 2020;8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blümcke I, Aronica E, Becker A, et al. . Low-grade epilepsy-associated neuroepithelial tumours—The 2016 WHO classification. Nat Rev Neurol. 2016;12:732–740. [DOI] [PubMed] [Google Scholar]
- 7. Blümcke I, Thom M, Aronica E, et al. . The clinicopathologic spectrum of focal cortical dysplasias: A consensus classification proposed by an ad hoc task force of the ILAE diagnostic methods Commission1. Epilepsia. 2011;52:158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leu C, Stevelink R, Smith AW, et al. . Polygenic burden in focal and generalized epilepsies. Brain. 2019;142:3473–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feng YCA, Howrigan DP, Abbott LE, et al. . Ultra-rare genetic variation in the epilepsies: A whole-exome sequencing study of 17,606 individuals. Am J Hum Genet. 2019;105:267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heyne HO, Singh T, Stamberger H, et al. . De novo variants in neurodevelopmental disorders with epilepsy. Nat Genet. 2018;50:1048–1053. [DOI] [PubMed] [Google Scholar]
- 11. May P, Girard S, Harrer M, et al. . Rare coding variants in genes encoding GABAA receptors in genetic generalised epilepsies: An exome-based case-control study. Lancet Neurol. 2018;17:699–708. [DOI] [PubMed] [Google Scholar]
- 12. Ellis CA, Petrovski S, Berkovic SF. Epilepsy genetics: Clinical impacts and biological insights. Lancet Neurol. 2020;19:93–100. [DOI] [PubMed] [Google Scholar]
- 13. McTague A, Howell KB, Cross JH, Kurian MA, Scheffer IE. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol. 2016;15:304–316. [DOI] [PubMed] [Google Scholar]
- 14. Blumcke I, Budday S, Poduri A, Lal D, Kobow K, Baulac S. Neocortical development and epilepsy: Insights from focal cortical dysplasia and brain tumours. Lancet Neurol. 2021;20:943–955. [DOI] [PubMed] [Google Scholar]
- 15. Baldassari S, Picard F, Verbeek NE, et al. . The landscape of epilepsy-related GATOR1 variants. Genet Med. 2019;21:398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baldassari S, Ribierre T, Marsan E, et al. . Dissecting the genetic basis of focal cortical dysplasia: A large cohort study. Acta Neuropathol. 2019;138:885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ribierre T, Deleuze C, Bacq A, et al. . Second-hit mosaic mutation in mTORC1 repressor DEPDC5 causes focal cortical dysplasia-associated epilepsy. J Clin Invest. 2018;128:2452–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. D’Gama AM, Woodworth MB, Hossain AA, et al. . Somatic mutations activating the mTOR pathway in dorsal telencephalic progenitors cause a continuum of cortical dysplasias. Cell Rep. 2017;21:3754–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonduelle T, Hartlieb T, Baldassari S, et al. . Frequent SLC35A2 brain mosaicism in mild malformation of cortical development with oligodendroglial hyperplasia in epilepsy (MOGHE). Acta Neuropathol Commun. 2021;9:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blümcke I, Coras R, Busch RM, et al. . Toward a better definition of focal cortical dysplasia: An iterative histopathological and genetic agreement trial. Epilepsia. 2021;62:1416–1428. [DOI] [PubMed] [Google Scholar]
- 21. Surrey LF, Jain P, Zhang B, et al. . Genomic analysis of dysembryoplastic neuroepithelial tumor spectrum reveals a diversity of molecular alterations dysregulating the MAPK and PI3K/mTOR pathways. J Neuropathol Exp Neurol. 2019;78:1100–1111. [DOI] [PubMed] [Google Scholar]
- 22. Pekmezci M, Villanueva-Meyer JE, Goode B, et al. . The genetic landscape of ganglioglioma. Acta Neuropathol Commun. 2018;6:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pepi C, de Palma L, Trivisano M, et al. . The role of KRAS mutations in cortical malformation and epilepsy surgery: A novel report of nevus sebaceous syndrome and review of the literature. Brain Sci. 2021;11:793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adachi M, Abe Y, Aoki Y, Matsubara Y. Epilepsy in RAS/MAPK syndrome: Two cases of cardio-facio-cutaneous syndrome with epileptic encephalopathy and a literature review. Seizure. 2012;21:55–60. [DOI] [PubMed] [Google Scholar]
- 25. Chang CA, Perrier R, Kurek KC, et al. . Novel findings and expansion of phenotype in a mosaic RASopathy caused by somatic KRAS variants. Am J Med Genet A. 2021;185:2829–2845. [DOI] [PubMed] [Google Scholar]
- 26. Sim NS, Ko A, Kim WK, et al. . Precise detection of low-level somatic mutation in resected epilepsy brain tissue. Acta Neuropathol. 2019;138:901–912. [DOI] [PubMed] [Google Scholar]
- 27. Campbell PJ, Getz G, Korbel JO, et al. . Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alexandrov LB, Kim J, Haradhvala NJ, et al. . The repertoire of mutational signatures in human cancer. Nature. 2020;578(7793):94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu W, Smolen CE, Hill SF, Meisler MH. Spontaneous seizures and elevated seizure susceptibility in response to somatic mutation of sodium channel Scn8a in the mouse. Hum Mol Genet. 2021;30:902–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 31. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DePristo MA, Banks E, Poplin R, et al. . A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cibulskis K, Lawrence MS, Carter SL, et al. . Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Panel of Normals (PON). GATK . Accessed 22 April 2022. https://gatk.broadinstitute.org/hc/en-us/articles/360035890631-Panel-of-Normals-PON-35
- 35. Smit AFA, Hubley R, Green P. RepeatMasker. http://www.repeatmasker.org. 2015; 2013-2015.
- 36. Dou Y, Kwon M, Rodin RE, et al. . Accurate detection of mosaic variants in sequencing data without matched controls. Nat Biotechnol. 2020;38:314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodin RE, Dou Y, Kwon M, et al. . The landscape of somatic mutation in cerebral cortex of autistic and neurotypical individuals revealed by ultra-deep whole-genome sequencing. Nat Neurosci. 2021;24:176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robinson JT, Thorvaldsdóttir H, Winckler W, et al. . Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Poplin R, Ruano-Rubio V, DePristo MA, et al. . Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. [Preprint] 10.1101/201178 [DOI] [Google Scholar]
- 40. Van der Auwera GA, Carneiro MO, Hartl C, et al. . From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013; 11(1110):11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Danecek P, Bonfield JK, Liddle J, et al. . Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Diskin SJ, Li M, Hou C, et al. . Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36:e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Loh PR, Danecek P, Palamara PF, et al. . Reference-based phasing using the haplotype reference consortium panel. Nat Genet. 2016;48:1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Loh PR, Genovese G, Handsaker RE, et al. . Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature. 2018;559(7714):350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jacobs KB, Yeager M, Zhou W, et al. . Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Karczewski KJ, Francioli LC, Tiao G, et al. . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Samocha KE, Kosmicki JA, Karczewski KJ, et al. . Regional missense constraint improves variant deleteriousness prediction. bioRxiv. [Preprint] doi: 10.1101/148353v1 [DOI] [Google Scholar]
- 50. Traynelis J, Silk M, Wang Q, et al. . Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Res. 2017;27:1715–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Collins RL, Glessner JT, Porcu E, et al. . A cross-disorder dosage sensitivity map of the human genome. Cell. 2022;185:3041–3055.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martincorena I, Raine KM, Gerstung M, et al. . Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171:1029–1041.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Blümcke I, Aronica E, Miyata H, et al. . International recommendation for a comprehensive neuropathologic workup of epilepsy surgery brain tissue: A consensus task force report from the ILAE Commission on Diagnostic Methods. Epilepsia. 2016;57:348–358. [DOI] [PubMed] [Google Scholar]
- 54. Koh HY, Kim SH, Jang J, et al. . BRAF Somatic mutation contributes to intrinsic epileptogenicity in pediatric brain tumors. Nat Med. 2018;24:1662–1668. [DOI] [PubMed] [Google Scholar]
- 55. Siegfried A, Cances C, Denuelle M, et al. . Noonan syndrome, PTPN11 mutations, and brain tumors. A clinical report and review of the literature. Am J Med Genet A. 2017;173:1061–1065. [DOI] [PubMed] [Google Scholar]
- 56. Fattah M, Raman MM, Reiss AL, Green T. PTPN11 mutations in the Ras-MAPK signaling pathway affect human white matter microstructure. Cerebral Cortex. 2021;31:1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Martinelli S, Carta C, Flex E, et al. . Activating PTPN11 mutations play a minor role in pediatric and adult solid tumors. Cancer Genet Cytogenet. 2006;166:124–129. [DOI] [PubMed] [Google Scholar]
- 58. Bailey MH, Tokheim C, Porta-Pardo E, et al. . Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–385.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cook JH, Melloni GEM, Gulhan DC, Park PJ, Haigis KM. The origins and genetic interactions of KRAS mutations are allele- and tissue-specific. Nat Commun. 2021;12:1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Canaud G, Hammill AM, Adams D, Vikkula M, Keppler-Noreuil KM. A review of mechanisms of disease across PIK3CA-related disorders with vascular manifestations. Orphanet J Rare Dis. 2021;16:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee JH, Huynh M, Silhavy JL, et al. . De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44:941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Poduri A, Evrony GD, Cai X, et al. . Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron. 2012;74:41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jansen LA, Mirzaa GM, Ishak GE, et al. . PI3K/AKT Pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain. 2015;138:1613–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Posch C, Moslehi H, Feeney L, et al. . Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo. Proc Natl Acad Sci U S A. 2013;110:4015–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Grill C, Larue L. NRAS, NRAS, which mutation is fairest of them all? J Invest Dermatol. 2016;136:1936–1938. [DOI] [PubMed] [Google Scholar]
- 66. Conti V, Pantaleo M, Barba C, et al. . Focal dysplasia of the cerebral cortex and infantile spasms associated with somatic 1q21.1-q44 duplication including the AKT3 gene. Clin Genet. 2015;88:241–247. [DOI] [PubMed] [Google Scholar]
- 67. Wang D, Zeesman S, Tarnopolsky MA, Nowaczyk MJM. Duplication of AKT3 as a cause of macrocephaly in duplication 1q43q44. Am J Med Genet A. 2013;161:2016–2019. [DOI] [PubMed] [Google Scholar]
- 68. Kobow K, Jabari S, Pieper T, et al. . Mosaic trisomy of chromosome 1q in human brain tissue associates with unilateral polymicrogyria, very early-onset focal epilepsy, and severe developmental delay. Acta Neuropathol. 2020;140:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hélias-Rodzewicz Z, Funck-Brentano E, Terrones N, et al. . Variation of mutant allele frequency in NRAS Q61 mutated melanomas. BMC Dermatol. 2017;17:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Burd CE, Liu W, Huynh MV, et al. . Mutation-specific RAS oncogenicity explains NRAS codon 61 selection in melanoma. Cancer Discov. 2014;4:1418–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hazrati LN, Kleinschmidt-DeMasters BK, Handler MH, et al. . Astrocytic inclusions in epilepsy: Expanding the spectrum of filaminopathies. J Neuropathol Exp Neurol. 2008;67:669–676. [DOI] [PubMed] [Google Scholar]
- 72. Lai D, Gade M, Yang E, et al. . Somatic mutation involving diverse genes leads to a spectrum of focal cortical malformations. medRxiv. [Preprint] doi: 10.1101/2021.12.22.21267563v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Qing T, Mohsen H, Marczyk M, et al. . Germline variant burden in cancer genes correlates with age at diagnosis and somatic mutation burden. Nat Commun. 2020;11:2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Carbonara C, Longa L, Grosso E, et al. . Apparent preferential loss of heterozygosity at TSC2 over TSC1 chromosomal region in tuberous sclerosis hamartomas. Genes Chromosomes Cancer. 1996;15:18–25. [DOI] [PubMed] [Google Scholar]
- 75. Martin KR, Zhou W, Bowman MJ, et al. . The genomic landscape of tuberous sclerosis complex. Nat Commun. 2017;8:15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cai X, Evrony GD, Lehmann HS, et al. . Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 2014;8:1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Richards S, Aziz N, Bale S, et al. . Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. DiStefano MT, Goehringer S, Babb L, et al. . The gene curation coalition: A global effort to harmonize gene–disease evidence resources. Genet Med. 2022;24:1732–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Aoki Y, Niihori T, Inoue Si, Matsubara Y. Recent advances in RASopathies. J Hum Genet. 2016;61:33–39. [DOI] [PubMed] [Google Scholar]
- 80. McWilliams GD, SantaCruz K, Hart B, Clericuzio C. Occurrence of DNET and other brain tumors in Noonan syndrome warrants caution with growth hormone therapy. Am J Med Genet A. 2016;170:195–201. [DOI] [PubMed] [Google Scholar]
- 81. Lodi M, Boccuto L, Carai A, et al. . Low-grade gliomas in patients with Noonan syndrome: Case-based review of the literature. Diagnostics. 2020;10:582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Da R, Wang M, Jiang H, Wang T, Wang W. BRAF AMP frequently co-occurs with IDH1/2, TP53, and ATRX mutations in adult patients with gliomas and is associated with poorer survival than that of patients harboring BRAF V600E. Front Oncol. 2020;10:531968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bedrosian TA, Miller KE, Grischow OE, et al. . Detection of brain somatic variation in epilepsy-associated developmental lesions. Epilepsia. 2022;63:1981–1997. [DOI] [PubMed] [Google Scholar]
- 84. Tajan M, de Rocca Serra A, Valet P, Edouard T, Yart A. SHP2 Sails from physiology to pathology. Eur J Med Genet. 2015;58:509–525. [DOI] [PubMed] [Google Scholar]
- 85. Cases-Cunillera S, van Loo KMJ, Pitsch J, et al. . Heterogeneity and excitability of BRAF V600E-induced tumors is determined by Akt/mTOR-signaling state and Trp53-loss. Neuro Oncol. 2022;24:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bernardo P, Cinalli G, Santoro C. Epilepsy in NF1: A systematic review of the literature. Childs Nerv Syst. 2020;36:2333–2350. [DOI] [PubMed] [Google Scholar]
- 87. Gales J, Prayson RA. Hippocampal sclerosis and associated focal cortical dysplasia-related epilepsy in neurofibromatosis type I. J Clin Neurosci. 2017;37:15–19. [DOI] [PubMed] [Google Scholar]
- 88. Tomkinson C, Lu JQ. Meningioangiomatosis: A review of the variable manifestations and complex pathophysiology. J Neurol Sci. 2018;392:130–136. [DOI] [PubMed] [Google Scholar]
- 89. Qian L, Chen K, Wang C, Chen Z, Meng Z, Wang P. Targeting NRAS-mutant cancers with the selective STK19 kinase inhibitor chelidonine. Clin Cancer Res. 2020;26:3408–3419. [DOI] [PubMed] [Google Scholar]
- 90. D’Gama AM, Poduri A. Precision therapy for epilepsy related to brain malformations. Neurotherapeutics. 2021;18:1548–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hedley-Whyte ET, Goldman JE, Nedergaard M, et al. . Hyaline protoplasmic astrocytopathy of neocortex. J Neuropathol Exp Neurol. 2009;68:136–147. [DOI] [PubMed] [Google Scholar]
- 92. Holthausen H, Coras R, Tang Y, et al. . Multilobar unilateral hypoplasia with emphasis on the posterior quadrant and severe epilepsy in children with FCD ILAE type 1A. Epilepsia. 2022;63:42–60. [DOI] [PubMed] [Google Scholar]
- 93. Louis DN, Perry A, Wesseling P, et al. . The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro Oncol. 2021;23:1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Vosoughi A, Zhang T, Shohdy KS, et al. . Common germline-somatic variant interactions in advanced urothelial cancer. Nat Commun. 2020;11:6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Bailey MH, Meyerson WU, Dursi LJ, et al. . Retrospective evaluation of whole exome and genome mutation calls in 746 cancer samples. Nat Commun. 2020;11:4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Miao D, Margolis CA, Vokes NI, et al. . Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumours. Nat Genet. 2018;50:1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All somatic variant calls are available from the supplementary material. All code is available at https://github.com/LalResearchGroup. Additional data are available from the corresponding author on reasonable request.