Abstract
A mass sea urchin die-off in the Caribbean Sea in the 1980s may have resulted from a single-cell protist called a scuticociliate.
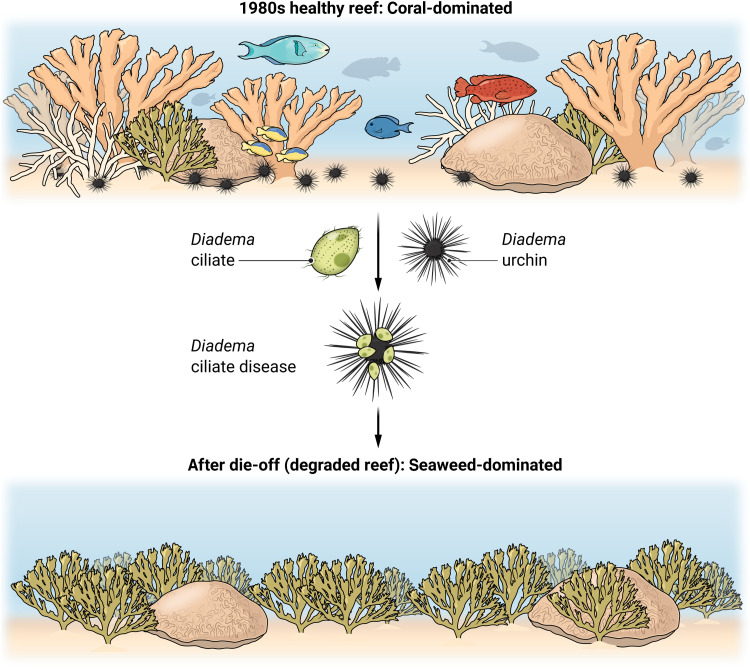
In 1983, the common sea urchin Diadema antillarum, living near the Caribbean opening to the Panama Canal, started dropping their spines and dying. When populations experienced infection, 93 to 100% of urchins died within 2 weeks, and within about 13 months of these first diseased urchins dying in Panama, 98% of Diadema died throughout the entire Caribbean (1). On heavily fished reefs like those in the U.S. Virgin Islands and Jamaica, where fishes that consume urchins or compete with urchins had been overharvested, predisease densities of these urchins were commonly reported as 10 to more than 70/m2, and they functioned as critical herbivores—consuming algae and suppressing algal competition with corals (1). After the Caribbean-wide death of Diadema, macroalgae escaped control, largely replaced corals, and the coral-generated topographic complexity on which many reef organisms depend declined dramatically. In Jamaica, coral cover declined from ~50 to ~3% and macroalgal cover increased from ~4 to ~90% (2). Thus, the disease that killed Diadema appeared to cascade through the ecosystem to fundamentally alter the structure and function of Caribbean coral reefs. Numerous reefs began converting from structurally complex and species-rich systems to flattened reefs with fewer species and compromised ecosystem functions (Fig. 1).
Fig. 1.
Fundamental alterations of Caribbean coral reefs due to the Diadema epidemic. Before Diadema die-off, even heavily fished reefs of the Caribbean were coral-rich, topographically complex, and generated a structure that hosted many species due to abundant Diadema whose grazing suppressed macroalgae and protected corals from algal competition (top). The pathogenic ciliate attacked Diadema sea urchins (center). After die-off, macroalgae escaped control by Diadema, seaweeds replaced corals, and Caribbean reefs lost 3D structure, diversity, and function (bottom). Credit: Austin Fisher/Science Advances
Despite such extensive mortality, the causative agent of the 1980's die-off remained a mystery until now (3). At the time of the outbreak, marine scientists and practitioners were not prepared or equipped to investigate the disease, and its rapid progression meant that little material was left to study after the disease swept through the Caribbean. However, a recent die-off event of the same species, beginning in early 2022 and with a similar ecology to the 1980s event (4), provided a second opportunity to evaluate what may have killed these urchins. During this new event, scientists, community members, and managers seized the opportunity to better investigate the cause. Hewson et al. (3) investigated the current die-off using a holistic set of traditional (e.g., culture-based work and histopathology) and modern (high-throughput sequencing and qPCR) methodologies to identify the culprit as an infectious single-celled protist, a scuticociliate (Fig. 1).
Because ciliates are commonly associated with marine species and not always pathogens, it makes it difficult to conclusively identify an individual ciliate species as the cause of a given disease. In this study, researchers compared specimens from 23 sites around the Northern and Eastern Caribbean that were either notably afflicted by disease or normal. They found that, in almost all cases, the diseased urchins contained evidence of DNA consistent with that of a ciliate pathogen but no evidence of viral, bacterial, or archaeal DNA differentially associated with the disease (i.e., the “usual” groups of microbial pathogens did not seem to be the cause). Further, the ciliate morphology and its DNA were very similar to known pathogenic ciliates isolated and characterized from fish eggs in 2009. Using this information, the researchers created molecular methods (i.e., qPCR) allowing them to track the abundance of the ciliate within and among their specimens. Compared to normal urchins, these molecular methods indicated that ciliates were more abundant in animals with signs of the disease, as well as in asymptomatic animals from sites where the disease had been recorded. Researchers then grew cultures of ciliates, confirmed their identities, and showed that urchins exposed to the ciliate became infected and showed disease symptoms like those seen in the field. These findings strongly support their hypothesis that this ciliate caused the disease that killed these Caribbean sea urchins; they named the disease D. antillarum scuticociliate or DaSc.
Mass mortality events due to infectious diseases, like seastar wasting disease, abalone withering syndrome, morbilliviruses in marine mammals, and numerous other diseases, have been particularly damaging to marine species in many regions of the world (5). Although the causative agents of most marine diseases are unknown, the disease impacts often are obvious. Massive mortality events can fundamentally alter entire ecosystems, and major impacts of disease outbreaks are not uncommon. When diseases affect species that create the structure on which others depend (e.g., corals on coral reefs) or that have strong impact on the community for other reasons, community-wide or even ecosystem-wide effects can occur. For example, the sunflower star Pyconopodia succumbed to seastar wasting disease along the Pacific coast from Canada to Mexico (6). This subtidal predator normally consumed kelp-eating sea urchins, and release from this predator led to a plague of herbivorous sea urchins that overgrazed and suppressed kelp beds upon which many near-shore species depended. A few years after the infamous Diadema die-off, white-band disease that attacked Acropora corals in the Caribbean (7) caused the two species there to go from the most abundant species in the Caribbean to being listed as endangered species in only a couple of decades. It is telling that the names of most marine diseases focus on the species they affect or the symptoms they produce such as seagrass wasting disease, stony coral tissue loss disease (SCTLD), or white-band disease, rather than the pathogens that cause the disease. This occurs because the causative agents are rarely known.
Identifying and managing disease outbreaks in the sea are major challenges. Much of what we do know about marine diseases is limited to commercially important (often aqua-cultured) species like oysters, shrimps, and finfish. However, most marine disease research remains far behind other areas of disease research for several important reasons. First, for marine organisms, we do not have the thousands of years of experience and intimate interaction that we have with humans or terrestrial plants and animals used in agriculture; humans invented and used scuba gear to explore marine habitats for the first time less than 100 years ago. Second, studying disease identity and dynamics in a liquid medium as vast, dynamic, and complex as the ocean poses challenges that terrestrial humans are not well equipped to confront. Third, we lack the basic foundational research on the natural history of most marine microbes, and even many larger marine species, limiting our ability to respond adequately during an outbreak. Finally, we simply do not have systems in place to quickly and effectively respond to new diseases in the sea, even ones that cause catastrophic effects on the entire ecosystem.
Marine ecosystems have not had a Centers for Disease Control or a Department of Agriculture with deep histories of studying diseases nor the organization and infrastructure to quickly mobilize resources when an outbreak occurs. When mystery outbreaks occur among humans (for instance, COVID-19, Legionnaires’ disease, or Four Corners disease) or within domestic or agricultural animals (such as bird flu, chronic wasting disease, or white-nose syndrome), we now often identify the cause within weeks to months. In contrast, for one of the largest epidemics in a marine system and one that killed ~98% of all individuals and produced cascading effects on the entire ecosystem, it has taken 40 years to identify the suspected pathogen. Essentially, the tools, scientific knowledge, and human infrastructure to identify marine diseases are underdeveloped and limit our ability to quickly, accurately, and inexpensively identify and better manage the pathogens causing disease epizootics in the sea. For example, currently unknown pathogens associated with SCTLD are ravaging what remains of the Caribbean’s coral reefs, and despite hundreds of researchers and managers focused on Caribbean coral reefs, no conclusive data link any single or multiple pathogens to the disease (8).
Despite the above challenges, Hewson et al. (3) managed to solve this 40-year-old mystery of a major marine disease. One reason for this new success is that, after some recent marine epidemics, NOAA, state-funded programs, and other coalitions of scientists have begun early detection programs that track specific diseases, like the current program focused on SCTLD (https://cdhc.noaa.gov/coral-disease/characterized-diseases/stony-coral-tissue-loss-disease-sctld/) and the group that initially tracked the 2022 Diadema outbreak (https://www.agrra.org/sea-urchin-die- off/). However, marine disease surveillance and forecasting programs need additional and continuous support (9). In addition, citizen science programs (https://marine.ucsc.edu/data-products/collaborative-monitoring/index.html) that helped track seastar wasting disease can be replicated in other areas around the world so that rapid response teams can be activated quickly and efficiently. With new diagnostic tools like those used by Hewson et al. (3) and with international cooperation and financing to track emerging infectious disease in the sea, we can be more prepared and potentially able to understand and hopefully mitigate marine disease outbreaks in future.
Notice (12 May 2023): Some readers may infer from the Focus that researchers discovered the definitive cause of the Diadema die-off in the 1980s. However, despite the symptoms of the recent Diadema die-off being similar to the one in the early 1980s, one cannot be sure that the cause was the same due to a lack of samples from the early 1980s die-off for rigorous assessment.
References
- 1.H. A. Lessios, The great Diadema antillarum die-off: 30 years later. Ann. Rev. Mar. Sci. 8, 267–283 (2016). [DOI] [PubMed] [Google Scholar]
- 2.T. P. Hughes, Catastrophes, phase shifts, and large-scale degradation of a Caribbean coral reef. Science 265, 1547–1551 (1994). [DOI] [PubMed] [Google Scholar]
- 3.I. Hewson, I. T. Ritchie, J. S. Evans, A. Altera, D. Behringer, E. Bowman, M. Brandt, K. A. Budd, R. A. Camacho, T. O. Cornwell, P. D. Countway, A. Croquer, G. A. Delgado, C. De Rito, E. Duermit-Moreau, R. Francis-Floyd, S. Gittens Jr., L. Henderson, A. Hylkema, C. A. Kellogg, Y. Kiryu, K. A. Kitson-Walters, P. Kramer, J. C. Lang, H. Lessios, L. Liddy, D. Marancik, S. Nimrod, J. T. Patterson, M. Pistor, I. C. Romero, R. Sellares-Blasco, M. L. B. Sevier, W. C. Sharp, M. Souza, A. Valdez-Trinidad, M. van der Laan, B. Vilanova-Cuevas, M. Villalpando, S. D. Von Hoene, M. Warham, T. Wijers, S. M. Williams, T. M. Work, R. P. Yanong, S. Zambrano, A. Zimmermann, M. Breitbart, A Scuticociliate causes mass mortality of Diadema antillarum in the Caribbean Sea. Sci. Adv. 9, eadg3200 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D. R. Levitan, R. M. Best, P. J. Edmunds, Sea urchin mass mortalities 40 y apart further threaten Caribbean coral reefs. Proc. Natl. Acad. Sci. U.S.A. 120, e2218901120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D. C. Behringer, K. D. Lafferty, B. R. Silliman, Marine Disease Ecology (Oxford Univ. Press, 2020). [Google Scholar]
- 6.C. D. Harvell, D. Montecino-Latorre, J. M. Caldwell, J. M. Burt, K. Bosley, A. Keller, S. F. Heron, A. K. Salomon, L. Lee, O. Pontier, C. Pattengill-Semmens, J. K. Gaydos, Disease epidemic and a marine heat wave are associated with the continental-scale collapse of a pivotal predator (Pycnopodia helianthoides). Sci. Adv. 5, eaau7042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.R. E. Aronson, W. F. Precht, White-band disease and the changing face of Caribbean coral reefs. Hydrobiologia 460, 25–38 (2001). [Google Scholar]
- 8.E. M. Muller, C. Sartor, N. I. Alcaraz, R. van Woesik, Spatial epidemiology of the stony-coral-tissue-loss disease in Florida. Front. Mar. Sci. 7, 163 (2020). [Google Scholar]
- 9.J. Maynard, R. van Hooidonk, C. D. Harvell, C. M. Eakin, G. Liu, B. L. Willis, G. J. Williams, M. L. Groner, A. Dobson, S. F. Heron, R. Glenn, K. Reardon, J. D. Shields, Improving marine disease surveillance through sea temperature monitoring, outlooks, and projections. Philos Trans R Soc Lond B Biol. Sci. 371, 20150208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]