Abstract
Intracellular G protein-coupled receptors (GPCRs) can be activated by permeant ligands, which contributes to agonist selectivity. Opioid receptors (ORs) provide a notable example, where opioid drugs rapidly activate ORs in the Golgi apparatus. Our knowledge on intracellular GPCR function remains incomplete, and it is unknown whether OR signaling in plasma membrane (PM) and Golgi apparatus differs. Here, we assess the recruitment of signal transducers to mu- and delta-ORs in both compartments. We find that Golgi ORs couple to Gαi/o probes and are phosphorylated but, unlike PM receptors, do not recruit β-arrestin or a specific Gα probe. Molecular dynamics simulations with OR–transducer complexes in bilayers mimicking PM or Golgi composition reveal that the lipid environment promotes the location-selective coupling. We then show that delta-ORs in PM and Golgi have distinct effects on transcription and protein phosphorylation. The study reveals that the subcellular location defines the signaling effects of opioid drugs.
Signal transduction of drug-activated opioid receptors in the plasma membrane and the Golgi apparatus is location specific.
INTRODUCTION
Cells sense changes in their environment through the actions of G protein-coupled receptors (GPCRs) that detect incoming stimuli and transmit the signal by initiating intracellular responses. GPCR-mediated signal transduction plays critical and ubiquitous roles in normal physiology, and aberrations in GPCR pathways can cause disease, making GPCRs key therapeutic targets (1, 2). Recent methodological advances, including the use of optical biosensors, have allowed investigating GPCR activation and signaling with unprecedented spatial and temporal resolution at the level of individual cells (3–5). This has led to accumulating evidence that some agonists bind and activate GPCRs in intracellular organelles in addition to the plasma membrane (PM) and has established a discrete layer of functional selectivity (6–8). An essential step for understanding the diversity of responses triggered by individual ligands is to gain insights into GPCR signaling at different subcellular locations. It remains largely unknown if GPCRs in internal organelles can couple to the canonical PM signal transduction machinery, or alternatively, exhibit a location-selective interaction profile to drive unique downstream responses. Here, we start to address these gaps by investigating the compartmentalized signaling of Gi/o-coupled opioid receptors (ORs), which are prototypical members of the rhodopsin-like GPCR family.
ORs are the targets of endogenous neuromodulatory neuropeptides and of therapeutically important analgesic opioid drugs, such as morphine and fentanyl. Nanobody-based sensors recently revealed that mu- (MOR), delta- (DOR), and kappa- (KOR) ORs undergo ligand-dependent activation not only in the PM but also in different cellular organelles, including endosomes and the Golgi apparatus (7, 9, 10). Opioid neuropeptides and opioid drugs notably differ in their subcellular activation patterns in that small-molecule drugs uniquely trigger activation of ORs in the Golgi apparatus (7). Opioid drug access to the cell interior is likely achieved by free diffusion of the small, permeant molecules across cellular membranes. Neurons are known to contain ORs in the PM and in intracellular organelles, including the endoplasmic reticulum and Golgi apparatus, under basal conditions (11, 12). Transgenic mice expressing labeled ORs at endogenous levels have revealed varied but often predominant intracellular OR pools across brain regions and neuronal types (13–15). Previously, the importance of the internal biosynthetic ORs has mainly been attributed to the ability to replenish PM receptors (16, 17); however, the recent findings suggest that Golgi-localized ORs may initiate opioid drug-driven responses and have signaling functions.
Elements of the GPCR signaling cascade comprise both spatially confined and diffusing molecules, including transducers, effectors, and second messengers that can act at varying distances relative to the site of ligand–receptor binding. The rapid and dynamic signal propagation makes it difficult to trace back downstream signaling readouts to a precise subcellular site of GPCR activation. Studies that followed rapid receptor-proximal signaling events started to provide location-resolved insights and uncovered Gs-mediated 3',5'-cyclic adenosine monophosphate (cAMP) responses of several GPCRs in endosomes and Golgi apparatus, in addition to the PM (6, 18–21). Other studies restricted GPCR activation or signal initiation to a given subcellular site with pharmacological or light-controlled tools and identified phosphorylation changes, effector activation, and transcriptional changes downstream of intracellular Gs-coupled GPCRs (20, 22–24). However, little is known about Gi/o-coupled receptor signaling at internal organelles.
Here, we probe MOR and DOR signaling with unprecedented spatial resolution by applying unique pharmacological tools to isolate OR activation in the Golgi apparatus from activation in the PM. First, we demonstrate that canonical GPCR-interacting proteins including G protein-based probes, GPCR kinases (GRKs), and β-arrestin show unique engagement profiles with ORs in the Golgi apparatus, revealing location selectivity of specific receptor-proximal coupling events. Given that the lipid composition of the PM and Golgi apparatus varies remarkably, we then test whether differences in membrane lipids can contribute to the observed coupling selectivity. Using molecular dynamics (MD) simulations, we analyze the interactions of two distinct G protein probes with MOR embedded in membranes with a PM- or Golgi-like lipid composition. We find that lipids can directly influence OR coupling and reveal that PM phospholipids interact with a G protein probe to promote a stable OR interaction, while coupling of the same protein is altered in the Golgi-like bilayer. We then measure the downstream signaling effects promoted by PM- or by Golgi-localized DOR activation by unbiased global gene expression and protein phosphorylation analyzes and find that, unlike OR activation in the PM, Golgi-driven DOR activation does not alter gene expression but drives a unique signaling response involving protein phosphorylation.
Together, the study identifies location-selective OR signal transduction, which occurs at the receptor-proximal level and leads to distinct downstream effects at later time points. Our results further reveal that lipids can directly regulate OR–transducer coupling, providing a possible mechanism to achieve location-selective GPCR responses. Given the highly conserved coupling mechanisms across the GPCR family, the spatial differences that we delineate here for ORs may be widely applicable to signal propagation of GPCRs in different cellular locations.
RESULTS
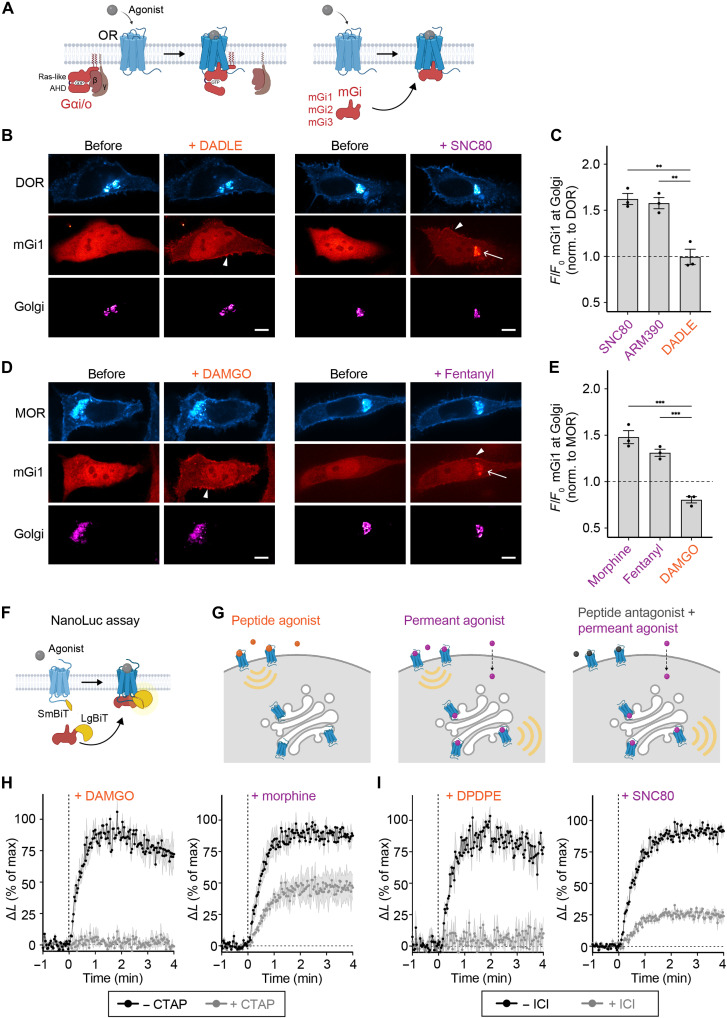
Golgi-localized ORs recruit Gαi/o sensors in response to permeant agonists
Activation of ORs residing in the Golgi apparatus has previously been detected using active state-selective nanobodies that bind to Golgi-localized ORs in response to permeant opioid ligands (7, 9). We aimed to test whether the activated internal receptors engage canonical GPCR transducers including G proteins, GRKs, and β-arrestin. First, we assessed whether Golgi-localized MOR and DOR can interact with G proteins, and for this, we used mini-G (mG) proteins, which are engineered Ras-like guanosine triphosphatase (GTPase) domains of Gα proteins that preserve the molecular contacts formed between the active GPCR and the G protein (Fig. 1A) (25, 26). Since ORs couple to Gi/o, we used mG proteins derived from the Gαi/o subunits. We developed three new mG probes based on Gαi1, Gαi2, and Gαi3, which contain deletions and mutations to remove the Gαi membrane anchor, the α-helical domain, and improve protein stability, based on the previously generated mGo probe (fig. S1A) (27). The mGi1, mGi2, mGi3, and mGo proteins were well expressed in HeLa cells, albeit at different levels (fig. S1, B and C). We coexpressed mRuby2-tagged mGi1 with green fluorescent protein (GFP)-tagged ORs in HeLa cells and tested whether the mGi1 probe, which was diffusely distributed in the cytoplasm at basal state, relocalized to activated receptors in different cellular compartments. mGi1 was recruited to DOR and MOR in the PM after adding peptide agonists (DADLE or DAMGO) or permeant small-molecule agonists (SNC80 or fentanyl), as observed using live cell confocal microscopy (Fig. 1, B and D) and total internal reflection fluorescence microscopy (TIR-FM) (fig. S1, D and E). To assess mGi1 recruitment to the Golgi pool of ORs, we labeled the Golgi apparatus in living cells with the specific marker α-mannosidase II (ManII)-BFP and quantified the mGi1 signal before and after agonist addition. The small-molecule agonists SNC80, ARM390, fentanyl, and morphine strongly induced the recruitment of mGi1 to Golgi-localized DOR or MOR (Fig. 1, C and E, and movies S1 and S2), which was dependent on the ligand concentration (fig. S1F). HeLa cells stably expressing DOR also showed mGi1 binding to DOR in PM and Golgi apparatus (fig. S1G). Peptide-based agonists did not promote mGi1 recruitment to the Golgi apparatus, confirming the inaccessibility of Golgi-residual ORs to peptide ligands (Fig. 1, C and E) (7). We next extended our live cell assays to the other Gi/o-based proteins and coexpressed mGi2, mGi3, or mGo with DOR or MOR. Similar to mGi1, the probes had a cytosolic localization in unstimulated cells, and they were recruited to PM- and Golgi-localized ORs selectively after treatment with permeant agonists (fig. S2, A to C).
Fig. 1. Novel Gαi probes bind ligand-activated MOR and DOR in the Golgi apparatus.
(A) OR coupling to heterotrimeric Gi/o proteins (left) and mGi1,2,3 proteins generated (right). The α-helical domain (AHD) of Gα is deleted and the Ras-like domain was modified, resulting in cytosolic probes that are recruited to the site of OR activation. (B) Confocal images of HeLa cells expressing SEP-DOR (cyan), mGi1-mRuby2 (red), and Golgi marker ManII-BFP (magenta) before and 5 min after adding 10 μM DADLE or SNC80. Scale bars, 10 μm. Arrowheads depict mGi1 at PM, and arrow depicts mGi1 at Golgi. (C) Quantification of mGi1-mRuby2 recruitment to DOR at Golgi upon agonist addition. Ligands at 10 μM. N = 3 with >20 cells, mean ± SD. **P = 0.001 by ordinary one-way analysis of variance (ANOVA). (D) Confocal images of HeLa cells expressing MOR-GFP (cyan), mGi1-mRuby2 (red), and ManII-BFP (magenta) before and 5 min after adding 10 μM DAMGO or morphine. Scale bars, 10 μm. Arrowheads depict mGi1 at PM, and arrow depicts mGi1 at Golgi. (E) Quantification of mGi1-mRuby2 recruitment to MOR at Golgi upon adding 10 μM DAMGO or morphine or 1 μM fentanyl. N = 3 with >20 cells, mean ± SD. ***P = 0.0002 by ordinary one-way ANOVA. (F) NanoLuc assay to measure mGi–ORs interaction. OR-SmBiT and mGi-LgBiT interaction drives NanoLuc complementation. (G) Ligands used to selectively activate ORs in the PM (nonpermeant peptide agonists), in PM and Golgi (permeant agonists), or only in Golgi (combining excess peptide antagonist and permeant agonist). (H) Kinetics of mGi1 recruitment to MOR upon DAMGO (100 nM) or morphine (100 nM) addition, in the absence or presence of CTAP (10 μM). N = 3, mean ± SEM. (I) Kinetics of mGi1 recruitment to DOR upon DPDPE (100 nM) or SNC80 (100 nM) addition, in the absence or presence of ICI 174,864 (100 μM). N = 3, mean ± SEM.
To determine the kinetics of mGi/o recruitment to ORs and measure the Golgi-localized component of the response, we used a split nanoluciferase (NanoLuc)-based complementation assay (Fig. 1F) (28). We fused the intracellular C terminus of DOR and MOR with the small subunit of NanoLuc (SmBiT, 13 amino acids) and the mGi probes with the larger subunit (LgBiT, 158 amino acids). HeLa cells expressing OR-SmBiT and mGi1-LgBiT were incubated with the NanoLuc substrate furimazine, and luminescence kinetics were recorded after adding different ligands or ligand combinations (Fig. 1G). The peptide agonists DAMGO and DPDPE promoted robust mGi1 recruitment to activated MOR and DOR, with a rapid onset of t1/2 (half-time) < 30 s (Fig. 1, H and I, left). The recruitment signal was abolished when cells were preincubated with excess of the peptide antagonists CTAP or ICI 174,864 (ICI) to pharmacologically block PM-localized ORs (Fig. 1, H and I, left). Adding the small-molecule agonists morphine and SNC80 led to engagement of mGi1 with MOR and DOR with a similarly rapid onset (Fig. 1, H and I, right), but the maximal luminescence signal was higher than for the peptide agonists (fig. S2, D and E). When cells were preincubated with 1000-fold molar excess of peptide antagonists, we found that an important fraction of the OR–mGi1 interaction driven by morphine or SNC80 remained (Fig. 1, H and I, right). The results show that MOR and DOR bind mGi1 at intracellular organelles and that the recruitment occurs independently of receptor activation at the PM. Given that the Golgi apparatus is the main site containing intracellular ORs in our experimental system (Fig. 1, B and D, and fig. S2), we conclude that Golgi-localized DOR and MOR are Gi/o protein coupling competent and contribute a substantial fraction of the OR–mGi interaction promoted by permeant small-molecule agonists.
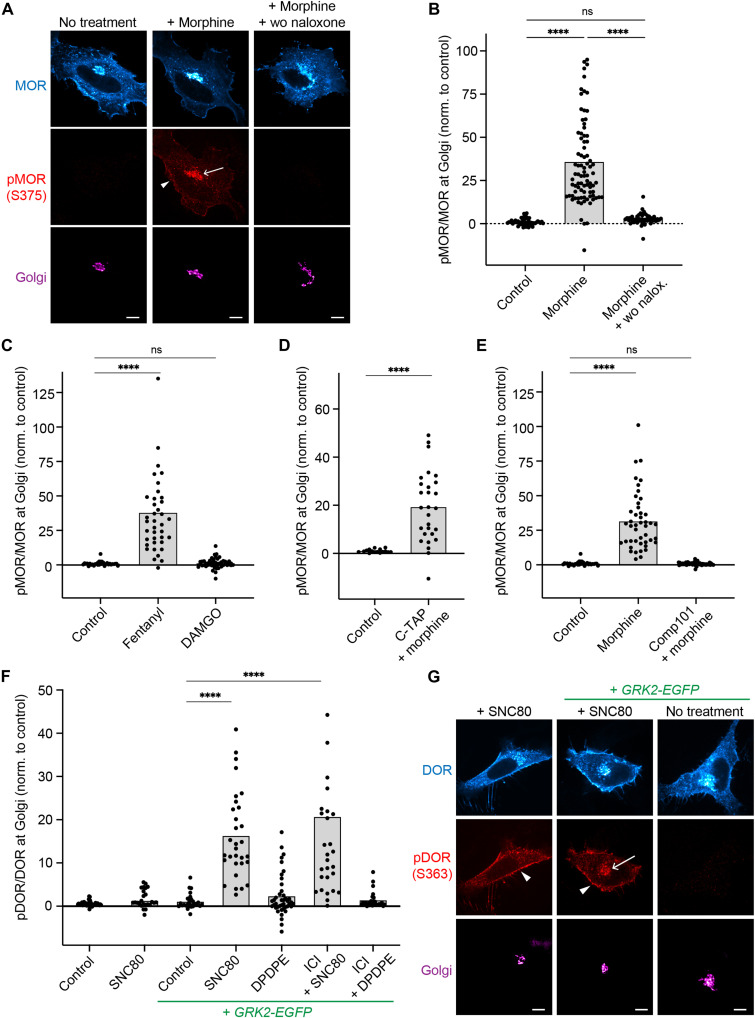
GRK2/3 phosphorylate activated GPCRs in the Golgi apparatus
ORs in the PM are regulated through interaction with and phosphorylation by GRKs, which control downstream responses as they promote β-arrestin binding and concomitant termination of G protein signaling (29). The GRKs 2 and 3 (GRK2/3) primarily mediate agonist-driven OR phosphorylation in serine and threonine residues in the cytoplasmic C-terminal tail (30, 31). We tested whether Golgi-resident ORs are phosphorylated in response to permeant agonists. We used a phosphosite-specific antibody that detects MOR when phosphorylated at serine 375, a primary target site for GRK2/3. In basal conditions, cells showed a low signal of MOR-pS375 in both PM and Golgi apparatus (Fig. 2A). Adding the permeant ligand morphine drove a significant increase in MOR phosphorylation within minutes of agonist application at the PM and also in the Golgi area labeled with ManII-BFP (Fig. 2, A and B). Upon agonist washout and antagonist (naloxone) addition, MOR phosphorylation at the Golgi apparatus was reversed within 10 min, concomitant with the reversal of receptor dephosphorylation at the PM (Fig. 2, A and B). In the continuous presence of morphine, phosphorylation of Golgi-localized MOR persisted for at least 1.5 hours (fig. S3A). Fentanyl also induced phosphorylation of MOR at both PM and Golgi (Fig. 2C and fig. S3B). The peptide agonist DAMGO did not promote phosphorylation of Golgi-resident MOR while driving phosphorylation of PM receptors, showing that local activation is required for MOR phosphorylation at the Golgi apparatus (Fig. 2C and fig. S3B). When cells were preincubated with the peptide antagonist CTAP to block PM receptors, morphine still drove pronounced phosphorylation of Golgi-resident MOR, revealing that regulation of MOR via phosphorylation occurs locally and independently of PM signaling (Fig. 2D). When we pretreated cells with the GRK2/3 inhibitor compound 101 (Cmpd101), no morphine-driven MOR-pS375 signal was detected at the Golgi apparatus, which identified GRK2/3 as the likely kinases that mediate MOR S375 phosphorylation at PM and Golgi (Fig. 2E).
Fig. 2. GRK2/3 phosphorylate activated MOR and DOR in the Golgi apparatus.
(A) Confocal images of HeLa cells expressing FLAG-MOR and ManII-BFP (magenta). Cells were fixed, permeabilized, and immunolabeled with anti-FLAG (cyan) and anti-MOR-pS375 (red) antibodies. Cells treated with 10 μM morphine for 5 min. In the washout condition (wo, right), 10 μM morphine was washed out after 5 min and 100 μM naloxone was added for 10 min. Arrow depicts pMOR at Golgi, and arrowhead depicts pMOR at PM. Scale bar, 10 μm. (B to E) Quantification of pMOR/MOR fluorescence at Golgi (ManII-labeled). Cells transfected and stained as in (A). (C) DAMGO used at 10 μM, fentanyl at 1 μM. (D) Cells pretreated with 10 μM CTAP for 5 min, followed by 100 nM morphine for 5 min. ****P < 0.0001 by Welch’s t test. (E) Cmpd101 (30 μM; GRK2/3 inhibitor) was added 45 min before adding 10 μM morphine for 5 min. N = 3 with >30 cells. ****P < 0.0001 by ordinary one-way ANOVA for (B), (C), and (E). (F) Quantification of pDOR/DOR fluorescence at Golgi. Cells transfected and stained as in (G). Cells treated with 10 μM SNC80 or DADLE. In ICI conditions, 100 μM ICI was added 10 min before treatment with 100 nM SNC80 or DADLE. Conditions with GRK2-EGFP coexpression are labeled. N = 3 with >25 cells. ****P < 0.0001 by ordinary one-way ANOVA. (G) Confocal images of HeLa cells expressing FLAG-DOR and ManII-BFP (magenta). Cells fixed, permeabilized, and immunolabeled with anti-FLAG (cyan) and anti-DOR-pS363 (red) antibodies. Treatment with 10 μM SNC80 for 5 min of cells with or without GRK2-GFP expression. Arrow depicts pDOR at Golgi, and arrowheads depict pDOR at PM. Scale bars, 10 μm. ns, not significant.
We next tested whether the Golgi-resident pool of DOR also underlies regulation by GRK2/3 and probed DOR phosphorylation using a phosphosite-specific antibody against serine 363 in the receptor C-tail. We initially did not detect a significant increase in phosphorylation of Golgi-localized DOR after adding the permeant ligand SNC80, while the signal of phosphorylated DOR in the PM strongly increased (Fig. 2, F and G). We then performed similar immunostainings in cells overexpressing GRK2-GFP. At higher GRK2 levels, a significant phosphorylation of both PM- and Golgi-resident DOR was detected within minutes of SNC80 treatment (Fig. 2, F and G). As for MOR, DOR phosphorylation was independent of PM signaling as preincubation with ICI did not block the Golgi-localized DOR phosphorylation, and phosphorylation persisted for over 1.5 hours (Fig. 2F and fig. S3, C and D). The peptide agonist DPDPE did not drive phosphorylation of DOR in the Golgi apparatus (Fig. 2F).
Last, we tested whether the Golgi pool of the β2-adrenergic receptor (β2AR), a prototypical Gs-coupled GPCR, was also regulated by phosphorylation following their activation. Previous reports indicate that adrenergic receptors in the Golgi apparatus can be activated in HeLa cells upon addition of epinephrine and its derivatives (6). Similar to DOR, endogenous kinase levels did not lead to detectable agonist-driven phosphorylation of β2AR (serines 355/356) in the Golgi area, while PM-localized receptors were phosphorylated (fig. S3, E and F). Yet in cells coexpressing GRK2, a pronounced β2AR-pS355/S356 signal was detected for Golgi-resident β2AR minutes after the agonist isoproterenol was applied (fig. S3, E and F).
The findings show that the Golgi pools of MOR, DOR, and β2AR are regulated via phosphorylation by GRK2/3 upon activation by permeant ligands. The phosphorylation requires local GPCR activation and occurs independently of PM signaling. In HeLa cells, Golgi-localized GRK2/3-dependent phosphorylation occurs at endogenous kinase levels for MOR and is detectable at increased GRK2 levels for DOR and β2AR.
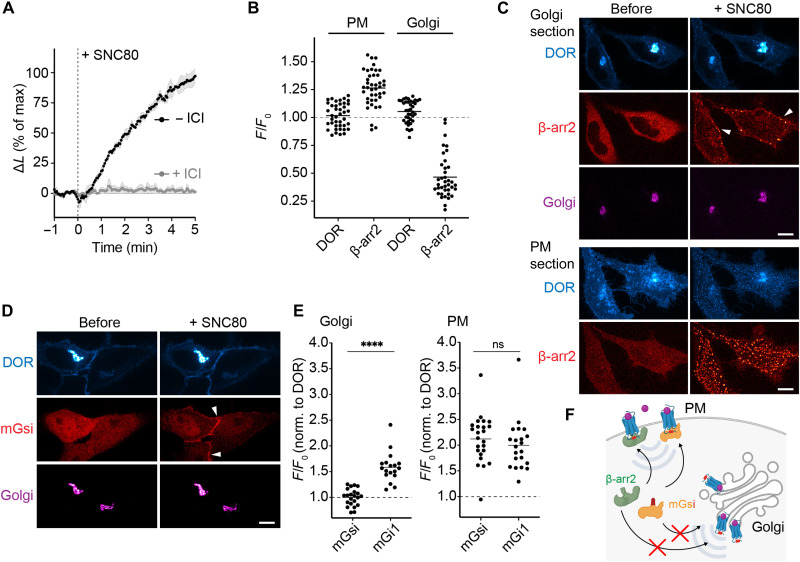
β-arrestin and the mGsi probe show location-selective OR binding
Since the phosphorylated OR C-tail promotes β-arrestin binding at the PM, we tested whether activated and phosphorylated DOR and MOR in the Golgi apparatus also recruit β-arrestin2. Using the NanoLuc complementation assay, we tested binding of β-arrestin2 in cells with elevated GRK2 levels to ensure DOR phosphorylation in both the PM and Golgi. As expected, SNC80 treatment drove strong and rapid β-arrestin2 engagement with DOR (Fig. 3A). However, when PM receptors were blocked with the peptide antagonist ICI, the β-arrestin2–DOR interaction was abolished (Fig. 3A), suggesting a lack of β-arrestin binding to phosphorylated DOR in the Golgi apparatus. We next assessed β-arrestin2 recruitment to DOR and MOR in cells with elevated GRK2 levels using live cell confocal microscopy. We found that OR activation with permeant drugs rapidly recruited β-arrestin2 to receptors in the PM but not in the Golgi apparatus (Fig. 3, B and C, and fig. S4A). The lack in β-arrestin2 recruitment to Golgi-localized OR was also detected at longer time points (45 min and 90 min; fig. S4B). We conclude that ligand-activated and phosphorylated DOR and MOR in the Golgi apparatus do not couple to β-arrestin2.
Fig. 3. Location-selective OR binding to β-arrestin2 and mGsi.
(A) Agonist-induced interaction of β-arrestin2-LgBiT with DOR-SmBiT upon adding 100 nM SNC80 in the absence or presence of 100 μM ICI. Cells coexpress GRK2-GFP. Change in luminescence signal normalized to untreated cells and signal before agonist addition. N = 3, mean ± SEM. (B) Quantification of β-arrestin2-mCherry and DOR at PM or at Golgi before and 5 min after adding 10 μM SNC80. Images of the same cells acquired with PM or Golgi focus, as described in (C). N = 3 with >35 cells, mean ± SD. (C) Confocal images of HeLa cells, expressing SEP-DOR (cyan), β-arrestin2-mCherry (red), ManII-BFP (magenta), and GRK2 (not shown) before and 5 min after adding 10 μM SNC80. Two confocal sections of the same cells are shown. Top: Focus on the Golgi. Bottom: Focus on PM. Arrowheads depict β-arrestin2 at the PM. Scale bars, 10 μm. (D) Confocal images of HeLa cells, expressing DOR-SEP (cyan), mGsi-mRuby2 (red), and ManII-BFP (magenta) before and 5 min after adding 10 μM SNC80. Arrowheads depict mGsi at PM. Scale bar, 10 μm. (E) Quantification of mGsi-mRuby2 and mGi1-mRuby2 recruitment to DOR at Golgi or PM. Left: Probe recruitment to DOR at Golgi as measured by confocal imaging. Right: Probe recruitment to PM as measured with TIR-FM. Same cells were imaged before and 5 min after adding 10 μM SNC80. N = 3 with >15 cells, mean ± SD. ****P < 0.0001 by unpaired t test. (F) Scheme depicting location-selective recruitment of β-arrestin2 (green) and mGsi (yellow). Both proteins are recruited to ligand-activated, phosphorylated (red dots) ORs in the PM but do not bind to phosphorylated ORs in the Golgi apparatus.
The location-dependent binding of β-arrestin to ORs was reminiscent of the recruitment pattern of a specific mG probe, termed mGsi, which had recently been shown to bind to active DOR in the PM, but not in the Golgi apparatus (32). mGsi consists of the Ras-like domain of Gαs but contains selectivity determining residues of Gαi in the C-terminal α5-helix (fig. S4C) and is widely used as a tool to study Gi/o-coupled GPCR activation and GPCR coupling selectivity (33, 34). We hypothesized that the β-arrestin and mGsi engagement with ORs may underlie a shared regulatory mechanism that differs based on the subcellular location and which is distinct to mGi1 that bound active ORs in both PM and Golgi (Fig. 1). We also reasoned that comparing OR binding to mGsi and to mGi1, two proteins that have similar overall conformation and sequence, may enable uncovering molecular mechanisms that promote location-selective OR coupling of one protein but not of the other. First, we confirmed the lack of mGsi binding to Golgi-localized DOR upon small-molecule agonist (SNC80) addition, while PM-localized DOR engaged mGsi (Fig. 3, D and E). We found the same location-selective engagement pattern when we extended our analyses to MOR. The permeant ligand fentanyl drove mGsi recruitment to active MOR in the PM but not in the Golgi apparatus (fig. S4D). To rule out the possibility that the different recruitment patterns of mGsi and mGi1 were caused by differences in the probes’ sensitivities for OR, we quantified the agonist-driven recruitment of mGsi and mGi1 to cell surface ORs using TIR-FM. Both mGsi and mGi1 were recruited to PM-localized DOR or MOR to the same extent and with similar kinetics (Fig. 3E and fig. S4, C and D). The mGsi-specific location bias suggested that the determinants for OR–G protein coupling in the Golgi apparatus comprise structural features outside the Gαi/o C terminus, which is shared between the probes. Furthermore, the similar subcellular recruitment patterns of β-arrestin2 and mGsi (Fig. 3F) suggested a common mechanism and motivated us to mechanistically explore the location-selective GPCR coupling behavior of the mGsi probe.
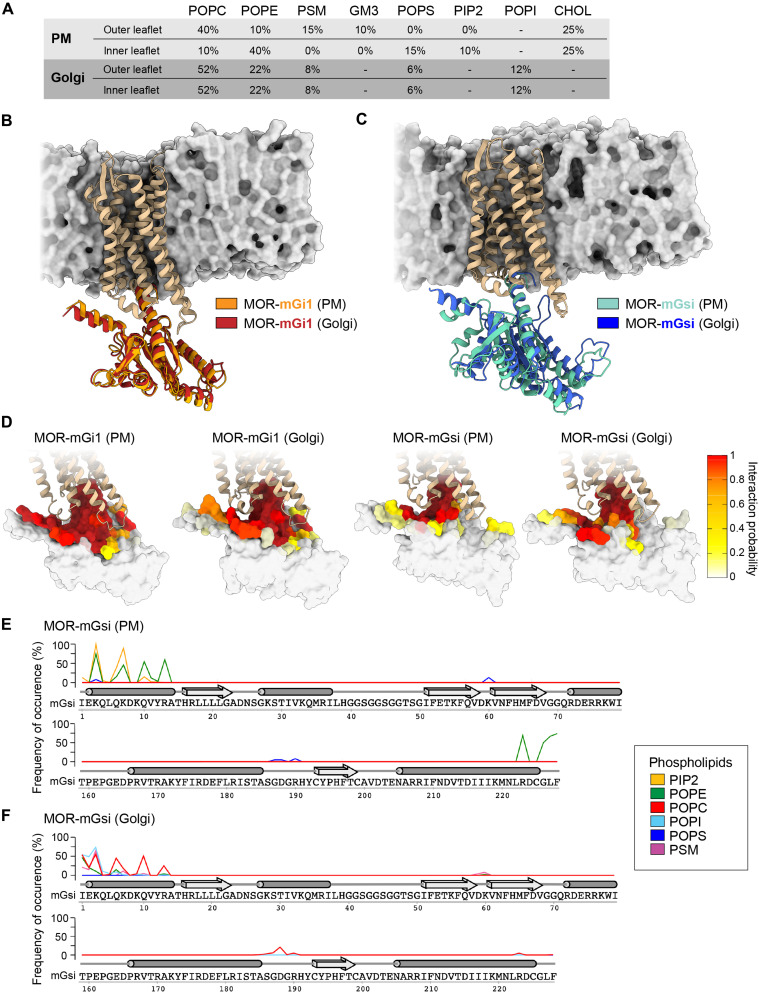
Subcellular membrane composition affects OR coupling
One clear distinction between the PM and the Golgi apparatus is the composition of lipids that make up the membrane and therefore we set out to test whether differences in the local lipid environments contribute to the distinct recognition of ORs by mGsi and mGi1. We used MD simulations to perform a comprehensive molecular assessment of the MOR–mG interactions in model membrane bilayers with PM- or Golgi-like lipid composition and properties (Fig. 4A). We focused on MOR because the high-resolution cryo-electron microscopy insights into MOR–Gi coupling provided a valuable basis for our analyses (35). The membrane models were built on the basis of previous computational studies of realistic membranes, with multiple lipid types and a high degree of compositional complexity (Fig. 4A) (36–39). The initial structures of the MOR–mGsi and MOR–mGi1 complexes were generated using “Alphafold2-multimer,” and each complex was embedded in either membrane model by using the Chemistry at Harvard Macromolecular Mechanics - Graphical User Interface (CHARMM-GUI) (see Materials and Methods). All four final structures underwent 1.0-μs-long classical MD calculations.
Fig. 4. The lipid environment regulates OR–mGsi interaction.
MD simulations carried out with MOR-mGi1 and MOR-mGsi heterodimers in PM and Golgi-like membrane models. (A) Lipid composition of PM and Golgi-like membrane models used. POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; POPE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine; PSM, palmitoyl sphingomyelin; GM3, monosialo dihexosyl ganglioside; POPS, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine; PIP2, 1,2-diacyl-sn-glycero-3-phospho-1-D-myo-inositol 4,5-bisphosphate; POPI, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol; CHOL, cholesterol. (B and C) Superimposition of the most relevant conformations assumed by the MOR-mGi1 and MOR-mGsi heterodimers in PM and Golgi membranes. MOR is tan. (B) mGi1 in the PM environment is orange, and mGi1 in the Golgi environment is red. (C) mGsi in the PM environment is teal, and mGsi in the Golgi environment is blue. Lipid bilayer schematics are based on the Golgi membrane models, displayed through their solvent exposed surface (gray). (D) Frequency of occurrence of contacts between MOR and mGi1 or MOR and mGsi in PM or Golgi membranes. mGi1 and mGsi are represented through their solvent exposed surfaces and colored according to the local interaction probability values, following depicted color gradient. (E and F) Frequencies of occurrence of contacts between mGsi and phospholipids in the PM (E) and the Golgi membrane (F). The α-helices (tubes) and β-sheets (arrows) of mGsi are displayed. The frequency of interaction of each phospholipid species is colored according to the legend. mGsi residues 70 to 159 are omitted since no lipid interactions were detected.
First, we assessed the overall dynamics and conformational stability of the OR–mG complexes. For this, we computed the root mean square deviation (RMSD) of the secondary structure Cα atoms and plotted the RMSD values for both MOR and the mGi1/mGsi probes as a function of the simulation time (fig. S5, A and B). MOR exhibited good conformational stability in all four simulations (MOR bound to mGi1 or mGsi, in PM or Golgi membrane) (average RMSD ~1.0 Å) (fig. S5A); however, the mG probes exhibited a more intricate behavior (fig. S5B). While mGi1 displayed limited conformational flexibility in both membrane environments (average RMSD ~1.0 Å), mGsi engaged a stable binding pose only when MOR was embedded in the PM bilayer. In the Golgi-like environment, a higher average RMSD value for mGsi (~2.2 Å) was observed, suggesting a less favorable MOR–mGsi interaction. A membrane-dependent behavior of MOR–mGsi binding was also detected by cluster analyses of the conformational states visited by the protein complexes during the different MD simulations. We found that MOR–mGi1 heterodimers assumed a similar binding mode in both PM- and Golgi-like membranes (Fig. 4B), while the MOR–mGsi heterodimers exhibited significantly different binding geometries in the two membranes (Fig. 4C).
We then investigated whether distinct interaction networks between the receptor and the mG probes might contribute to the different coupling behaviors of mGi1 and mGsi. We computed the frequency of occurrence of the contacts formed between MOR and mGi1 or MOR and mGsi during the simulations. An overall higher degree of engagement between MOR and mGi1 in comparison to mGsi was observed in both membrane models (Fig. 4D). MOR and mGi1 interacted via an extensive network of salt bridges (fig. S6), mediated by mGi1 residues in the C-terminal helix (H5), the N-terminal helix (HN), and the hns1, s2s3, and h4s6 linkers (domains according to GPCRdb database), closely paralleling the binding of full length Gαi to MOR (35). The MOR–mGi1 interactions were virtually identical in both membrane environments (fig. S6). In particular, the three salt bridges mGi1-K171–MOR-E272, mGi1-E175–MOR-K273, and mGi1-D198–MOR-K273 were able to strongly engage helix 6 in MOR and ease the retention of MOR’s active state. In contrast, MOR–mGsi binding was dominated by hydrophobic interactions and characterized by a lower number of salt bridges that differed according to the membrane environment (Fig. 4D and fig. S7). Beyond the interactions mediated by the mGsi C-terminal helix, the MOR–mGsi complex was held together by three interprotein salt bridges in the PM environment (mGsi-R13–MOR-D179, mGsi-D157–MOR-K271, and mGsi-D215–MOR-K273), yet only one salt bridge was detected in the Golgi-like membrane (mGsi-D215–MOR-K273) (fig. S7). The analyses suggest that MOR binds mGsi with lower affinity than mGi1 due to a reduced interaction network and are consistent with membrane-dependent binding of mGsi to MOR.
To assess whether the membrane composition could influence mGsi–MOR binding, we analyzed the frequency of occurrence of contacts between the mGsi residues and each phospholipid species in the PM- and the Golgi-like bilayers. We found that both the N-terminal and C-terminal helices of mGsi bind to PM lipids, yet different lipid species were involved (Fig. 4E). While both helices engaged with POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine), the positively charged residues in the N-terminal helix specifically bound to PIP2 (phosphatidylinositol-4,5-biphosphate), a highly negatively charged phospholipid that is enriched in the PM. The interactions of mGsi with lipids in the Golgi-like membrane exhibited a different pattern (Fig. 4F). The C-terminal helix of mGsi interacted poorly with lipids and the N-terminal helix also showed a lower level of membrane engagement, mainly via the zwitterionic phospholipid POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine). The findings identify interactions between mGsi and individual phospholipids that may promote MOR–mGsi complex assembly in the PM, but not in the Golgi environment.
Together, the results suggest a lipid-based mechanism for location-selective MOR coupling and provide a molecular explanation for the lack of mGsi binding to Golgi-localized ORs, which is consistent with the subcellular binding pattern we find in living cells (Fig. 3). We conclude that OR coupling to specific proteins may require the interplay between receptor-mediated and phospholipid-mediated contacts in complex assembly.
OR signaling at the PM, but not at the Golgi apparatus, alters gene expression
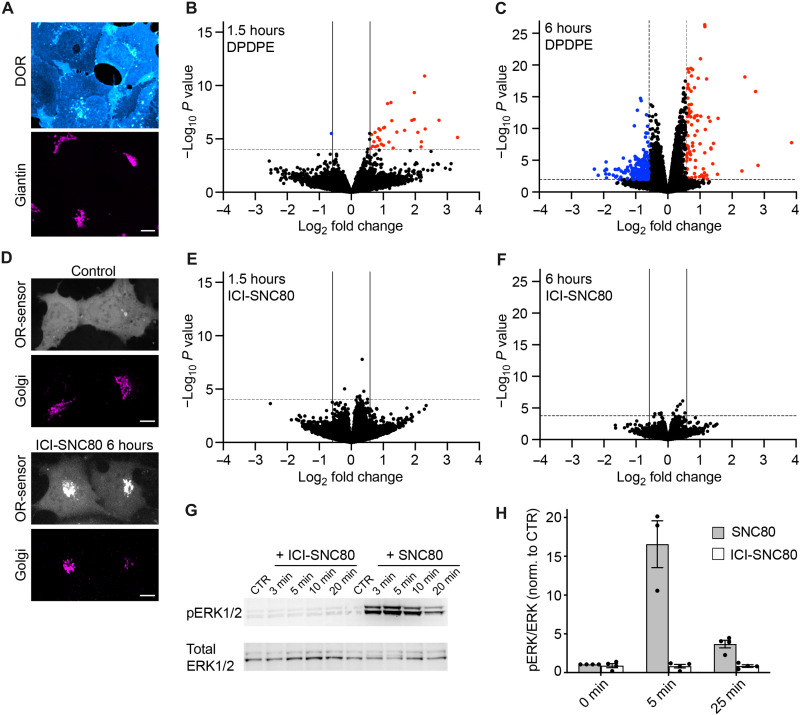
As our results have revealed differences in the molecular interactions between ORs and signaling partners at the PM and the Golgi apparatus, we set out to test whether signals initiated in the different cellular compartments result in distinct downstream effects. GPCRs exert their functions by engaging transducers and effectors that produce second messengers and cause phosphorylation changes, which can give rise to changes in gene expression. We profiled OR-mediated signaling using two parallel approaches, transcriptomics and phosphoproteomics. First, we performed differential gene expression analyses using RNA sequencing (RNA-seq) and compared the transcriptome of control cells and cells treated with agonists for 1.5 or 6 hours. The signaling studies were performed using human embryonic kidney (HEK) 293 cells stably expressing DOR (HEK293-DOR), which contained pronounced receptor pools in both the PM and the Golgi apparatus at steady state (Fig. 5A). To probe whether acute signaling of PM- localized DOR drives robust changes in gene expression, we treated cells with the peptide agonist DPDPE. At 1.5 and 6 hours, significant transcriptional changes were detected, with differential expression of 34 genes at 1.5 hours and 272 genes at 6 hours [fold change (FC) > 1.5 or < –1.5, adjusted P < 0.05] (Fig. 5, B and C; fig. S8, A and B; and table S1). Consistent with previous reports on transcriptional regulation by ORs (40–42), the 1.5-hour hits comprised immediate-early response genes (IER3, EGR1, and EGR3), regulators of mitogenic signaling (DUSPs), Hippo pathway genes (CTGF, ANKRD1, and CYR61), and transcription factors (e.g., MAFF and FOSL1), and we also detected their up-regulation in separate experiments using quantitative reverse transcription PCR (RT-qPCR; fig. S8C). The transcriptional response driven by DOR was distinct to the changes promoted by isoproterenol-activated Gs-coupled β2AR (fig. S8A), showing that gene expression changes can reveal specific downstream signals. We then probed whether activated Golgi-localized ORs initiate transcriptional changes. To restrict DOR signaling to the Golgi area, we again used the peptide antagonist ICI at high concentrations to block PM receptors and subsequently stimulated Golgi-resident ORs by adding the permeant agonist SNC80 for 1.5 or 6 hours (ICI-SNC80 condition). First, we confirmed that ICI treatment alone did not affect cellular gene expression (fig. S8D). We then performed live cell confocal imaging of HEK293-DOR cells expressing the OR-activity sensor EGFP-Nb33 (7), which confirmed that the ICI-SNC80 treatment drove Golgi-localized DOR activation that lasted for 6 hours, while no DOR activation at the PM was detected (Fig. 5D). Notably, RNA-seq-based differential gene expression analyses identified a complete lack of transcriptional changes upon Golgi-restricted DOR activation at 1.5 or 6 hours (FC > 1.5 or < –1.5, adjusted P < 0.05) (Fig. 5, E and F). This was not due to agonist-specific differences, because when we performed RNA-seq analysis of cells treated with SNC80 in the absence of ICI, we detected a pronounced transcriptional response that was identical to the DPDPE effects (fig. S8, A and E, and table S1).
Fig. 5. Golgi-restricted DOR activation does not promote a transcriptional response.
(A) Confocal images of HEK293 cells stably expressing FLAG-DOR (HEK293-DOR). Cells were fixed, permeabilized, and immunolabeled with anti-FLAG (cyan) and anti-giantin (magenta) antibodies. Scale bar, 10 μm. (B and C) Volcano plots of differentially expressed genes in HEK293-DOR cells treated with DPDPE (100 nM) for 1.5 hours (B) or 6 hours (C) versus mock-treated control cells. Results are presented as the mean differential gene expression from three replicates and expressed as log2FC. Genes significantly up-regulated are shown in red [quasi-likelihood F-test (QL F-test), FC > 1.5, adjusted P < 0.05]. Genes significantly down-regulated are shown in blue (QL F-test, FC < –1.5, adjusted P < 0.05). (D) Confocal images of HEK293-DOR cells expressing EGFP-Nb33 (gray) and GalT-DsRed (magenta). Control cells and cells treated for 6 hours with ICI (100 μM, 5 min preincubation) and SNC80 (100 nM). Scale bar, 10 μm. (E and F) Volcano plots of differentially expressed genes in HEK293-DOR cells treated with ICI-SNC80 (ICI = 100 μM, SNC80 = 100 nM) for 1.5 hours (E) or 6 hours (F) versus control cells (100 μM ICI treated). Results are presented as the mean differential gene expression from three replicates and expressed as log2FC. No genes are significantly changed in their expression [criteria as in (B) and (C)]. (G) Immunoblot of ERK1/2 activation upon treatment of HEK293-DOR cells with ICI-SNC80 (100 μM and 100 nM) or SNC80 (100 nM) for indicated times or untreated control (CTR). (H) Quantification of pERK1/2 protein levels normalized to total ERK1/2 in HEK293-DOR cells treated as in (G), for 5 min and 25 min. N = 3, mean ± SEM.
It is well established that GPCRs, including ORs, regulate expression of primary and secondary response genes through activation of mitogen-activated protein kinase (MAPK) modules. Our RNA-seq results suggested that Golgi-localized DOR may not drive MAPK signaling. We probed extracellular signal-regulated kinase 1/2 (ERK1/2) MAPK activity after Golgi-restricted DOR activation (ICI-SNC80) and indeed detected no ERK1/2 phosphorylation (Fig. 5, G and H). SNC80 treatment in the absence of ICI produced increased ERK1/2 activity downstream of OR activation (Fig. 5, G and H), showing that transcription regulators are differentially affected by activation of DOR in the PM and Golgi apparatus.
Activation of ORs in the Golgi apparatus produces unique effects on protein phosphorylation
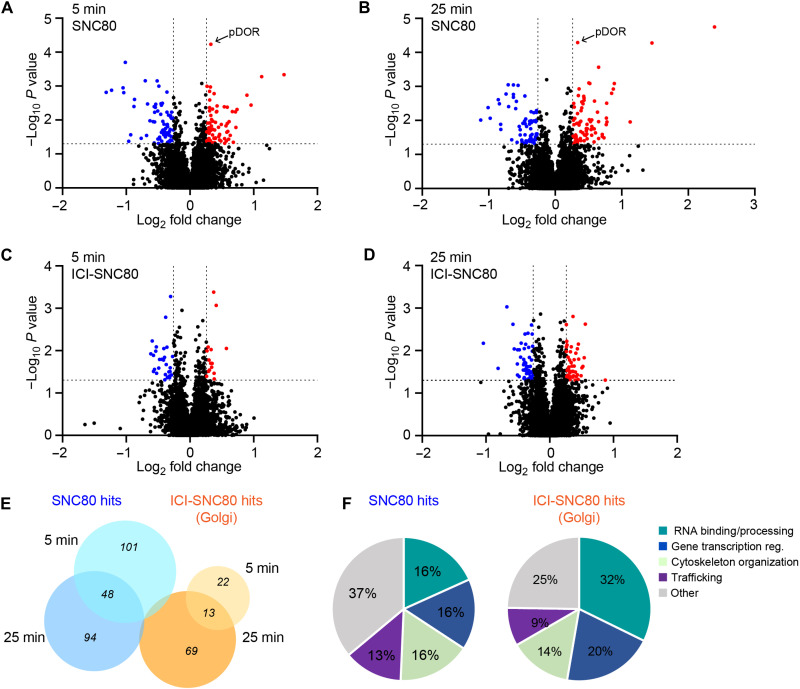
We next turned to mass spectrometry-based analyses of the phosphoproteome as a parallel unbiased and global method for profiling cellular signaling and focused on early time points since GPCR-driven phosphorylation events occur rapidly (43–45). Using liquid chromatography-electrospray ionization-tandem mass spectrometric (LC-ESI-MS/MS) analyses, we captured OR-mediated phosphorylation changes in HEK293-DOR cells treated with SNC80 (PM signaling control) or ICI-SNC80 (Golgi-restricted signaling) for 5 or 25 min. DOR-mediated changes of phosphopeptides were considered significant if a FC > 1.2 or < –1.2 and P < 0.05 was reached, and the corresponding protein level was unchanged. The cutoff was based on the detected FC of a DOR C-terminal peptide, known to be phosphorylated upon OR activation (site: DOR-p363, 1.26 FC) (Fig. 6, A and B). SNC80 treatment induced phosphorylation of 149 and 142 peptides after 5 and 25 min, respectively, with 48 shared phosphopeptides between the time points (Fig. 6, A, B, and E, and table S2). Golgi-restricted DOR activation by ICI-SNC80 induced phosphorylation of 35 and 82 peptides after 5 and 25 min treatment respectively, with 13 shared hits between the time points (Fig. 6, C, D, and E, and table S2). Signaling from the PM and from the Golgi apparatus resulted in largely different phosphorylation effects as only three phosphopeptides were shared between the SNC80 and ICI-SNC80 hits (Fig. 6E and table S2), which is consistent with a unique Golgi-localized OR response.
Fig. 6. DOR activation in the Golgi apparatus promotes unique changes in the phosphoproteome.
(A to D) Volcano plots of phosphopeptides identified by mass spectrometry in HEK293-DOR cells treated with SNC80 (100 nM) for 5 min (A) and 25 min (B) or with ICI-SNC80 (ICI = 100 μM, SNC80 = 100 nM) for 5 min (C) and 25 min (D) relative to control cells. Results are presented as the mean phosphopeptide abundance ratios between treated and mock-treated cells from three replicates and expressed as log2FC. Peptides showing a significantly higher phosphorylation upon treatment are shown in red (ANOVA, FC > 1.2, P < 0.05), and peptides with significantly lower phosphorylation are shown in blue (ANOVA, FC < –1.2, P < 0.05). (E) Venn diagrams showing the overlap between phosphopeptides regulated upon 5 and 25 min of SNC80 treatment or 5 and 25 min of ICI-SNC80 treatment. Three phosphopeptides are shared between the SNC80 and ICI-SNC80 hits (VIM, ATP2B1, and SRRM2). (F) Classification of the modulated phosphoproteins downstream of SNC80 or ICI-SNC80 treatment into functional groups based on Metacore (Clarivate), STRING (v. 11.5), and Pubmed and UniProt curation.
Analyzing the phospho-responses revealed that 40% of regulated proteins downstream of SNC80 treatment have been previously linked to OR function (table S2), which provided a validation of the phosphoproteomic dataset. Notably, the mTOR pathway previously detected upon KOR activation in mice brain (45–47) was also affected downstream of DOR activation by SNC80 [SLC3A2, PRKAA2, MTOR, RPS6KA1, and BRAF, according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway]. In addition, PAK1 and PAK2 were significantly phosphorylated upon SNC80 and previously also identified upon KOR activation in HEK293 cells (48). None of these phosphoproteins were, however, regulated downstream of Golgi-restricted OR activation. Similarly, several endocytic proteins were detected upon SNC80 treatment (PARD3, IGF2R, DNM1, EPS15L1, RAB11FIP1, CBL, and SNX1, according to KEGG pathway), yet they did not change upon ICI-SNC80 addition. While the ERK1/2 kinases did not pass the cutoff criteria used to define hits, an increase in pERK2 (MAPK1) was detected in the SNC80 condition (t = 25 min, FC = 1.18). pERK was not detected after adding ICI-SNC80, consistent with the location-dependent DOR effects on ERK1/2 activity described earlier (Fig. 5, G and H). We then aimed to identify upstream kinases by applying kinase-substrate enrichment analyses (KSEA app), pooling all unique phosphosites regulated by SNC80 or by ICI-SNC80 treatment (fig. S9, A to C, and table S2). In the SNC80 condition, ERK1 and ERK2 were predicted as active kinases with highest confidence, and the cAMP-regulated PKA was predicted to be significantly less active (fig. S9, A and C, and table S2), pinpointing kinases with known roles downstream of OR signaling. No significant kinase activity could be assigned based on the phosphosites identified in the ICI-SNC80 conditions (fig. S9B). We noted that 5 min after Golgi-localized ORs activation, 75% of regulated proteins showed a reduction in their phosphorylation status, which suggests that internal OR activation may predominantly down-regulate a basal kinase tone.
Next, we categorized the phosphoproteins regulated by SNC80 or by ICI-SNC80 treatment according to cellular function (Fig. 6F). Downstream of Golgi-localized OR activation, 32% of hits had roles in RNA binding and processing, and Gene Ontology (GO) term analysis identified significant enrichment of molecular functions related to RNA regulation in the ICI-SNC80 response (table S2). In addition, 20% ICI-SNC80 hits were linked to transcriptional regulation, and several regulated proteins showed functions in cytoskeleton remodeling and membrane trafficking, some of them with established cellular localization at the Golgi apparatus (SNAPIN, ARHGAP21, ALG2, and CAMSAP1/3) (Fig. 6F). The identified hits present candidate proteins that may specifically act downstream of the drug-induced Golgi-localized OR activation wave (table S2).
Together, the unbiased downstream signaling analyses demonstrate that Golgi-localized ORs have a unique signaling profile that shares little overlap with the responses initiated by ORs in the PM. DOR activation at the Golgi apparatus by permeant drugs leads to minimal effects on gene transcription yet has a unique imprint on the phosphoproteome.
DISCUSSION
The present study establishes that activation of ORs in different cellular compartments promotes distinct signal transduction. Focusing on MOR and DOR in the PM and in the Golgi apparatus, we uncover similarities and differences in receptor-proximal protein recruitment and delineate location-specific effects on gene expression and protein phosphorylation. The results convey that intracellular ORs do not merely mimic the actions of PM receptors but that they uniquely drive cellular responses upon activation by permeant drugs. We find that OR coupling can be promoted by transducer–lipid interactions that depend on the local membrane composition, which suggests a key role for lipids in regulating GPCR coupling with subcellular precision.
To interrogate OR-proximal protein engagement in a location-resolved manner, we focused our analyses on mG probes, GRK2/3, and β-arrestins, which are cytosolic proteins and thus readily available to bind ORs in both locations. We characterize and use Gαi-based probes to study GPCR coupling and detect that mGi1, mGi2, and mGi3 rapidly bind active MOR and DOR in PM and Golgi apparatus. The mGi probes and mGo share the ability to couple with active ORs in both locations, yet it is possible that subtle Gi/o isoform-specific coupling differences of cell surface and intracellular receptors exist that our experiments did not address. While mGi/o interaction relies on agonist-driven GPCR conformational changes, β-arrestin binding additionally requires ORs to be phosphorylated (49). We here discover that Golgi-localized MOR and DOR are phosphorylated by GRK2/3 in an agonist-dependent manner; however, they do not drive β-arrestin recruitment, contrasting with PM ORs in the same cell. It conveys that additional, location-specific determinants regulate β-arrestin–OR binding (32). β-arrestins can directly interact with membrane lipids via loops in the C-domain, which contributes to the stability of select β-arrestin–GPCR complexes (50, 51). In particular, the PM phosphoinositide PIP2 is critical for β-arrestin binding to a subset of GPCRs, including MOR and DOR (52). Our data on OR–mGsi coupling suggest that lipid-based selection mechanisms may be widely relevant for GPCR–transducer coupling and provide a rationale for the absence of mGsi and β-arrestin2 engagement with MOR and DOR in the Golgi apparatus, which lacks PIP2.
The MD simulations of MOR–mGi1 and MOR–mGsi complexes embedded in membrane models with PM- or Golgi-like lipids reveal that mGsi, but not mGi1, binds MOR in a membrane-dependent manner. The findings show that two highly similar GPCR binding proteins can remarkably differ in their sensitivity to lipid regulation, which has broader implications for G protein coupling selectivity by GPCRs: mGsi is a chimeric probe that is almost entirely based on Gαs, which suggests that the lipid sensitivity may be a specific determinant of Gαs–GPCR engagement. Consistent with this, a recent study found reduced mGs recruitment to Golgi-localized dopamine D1 receptors when compared to the PM receptor pool (53). Furthermore, mass spectrometry-based experiments identified PIP2 as a lipid that enhances mGs coupling to adenosine A2A and β1 adrenergic receptors (54). It suggests that the rules for GPCR coupling selectivity may differ at intracellular organelles relative to the PM according to the lipid sensitivities of individual G proteins.
The present results, together with previous studies, establish that ORs in the Golgi apparatus are signaling competent as they engage transducer proteins (G proteins, GRKs) and inhibit cAMP production in response to permeant agonists (7, 32). Yet the transcriptomic and phosphoproteomic analyses delineate prominent differences in the downstream effects of Golgi-localized DOR relative to the PM receptor pool. A notable finding is that activation of DOR in the Golgi apparatus is uncoupled from transcriptional effects 1.5 and 6 hours after agonist addition, in stark contrast to PM DOR signaling. Yet transcription factors account for 20% of phosphoproteins modulated by Golgi-localized OR signaling. We note that several of the identified transcription factors, including signal transducer and activator of transcription 3 (STAT3) and forkhead box protein C2 (FOXC2), are known to require multisite phosphorylation for altered activity (55, 56). Multisite regulation was, however, not detected in our phosphoproteomics dataset. Therefore, we speculate that subtle phosphoregulation of transcription factors by Golgi-localized DOR signaling may not propagate to the transcriptional level due to noise filtering in the signal transduction cascade (57, 58). Golgi-localized DOR signaling also led to phosphoregulation of a large proportion of RNA binding proteins, which are involved in mRNA translation, decay, splicing, or export (59). It is therefore possible that OR signaling at the Golgi apparatus promotes unique perturbation in mRNA homeostasis and highlights an avenue of potential significance for future study of opioid drug-specific effects.
In addition to lipid-regulated mechanisms, location-specific GPCR signaling may also be promoted by differences in local transducers and effector availability. This applies to heterotrimeric G proteins, whose Gα and Gγ subunits are anchored to membranes by protein lipidation. Different Gα subunits are found at the Golgi apparatus, yet Gβγ subunits predominantly localize at the PM under basal conditions (60–62). A greater number of Gαi/o heterotrimers in the PM relative to the Golgi compartment likely affects the G protein-mediated signal strength. Notably, noncanonical signaling via Gα can occur at the Golgi apparatus, when Gα subunits signal alone or in complex with other proteins, such as guanine exchange modulators, upon GPCR activation (63). An example is the KDEL receptor, an atypical GPCR, that signals at the Golgi apparatus via Gαo subunits in complex with Rab1/Rab3 GTPases and the GDP dissociation inhibitor αGDI (64). While the downstream effects that we delineate in HEK293 cells are likely defined by the cell type-specific expression of transducer and effector proteins (65), the phosphoproteomic signature driven by Golgi-localized DOR activation suggests that noncanonical coupling mechanisms may contribute to the local signaling response. Another family of important OR-interacting proteins comprises the GRKs, of which four (GRK2, GRK3, GRK5, and GRK6) are ubiquitously expressed and subdivided into cytosolic (GRK2/3) and the PM-associated (GRK5/6) proteins (66). Our data on Golgi-localized MOR, DOR, and β2AR phosphorylation indicates that regulation by the cytosolic GRK2/3 can occur at both PM and Golgi apparatus. However, GPCRs that underlie regulation by PM-anchored GRK5/6 are unlikely to be regulated by the same kinases at internal organelles, raising the possibility of altered GPCR-kinase coupling that may further promote location-selective effects.
An important future direction is to determine how Golgi-localized OR signal transduction is controlled and terminated. We find that MOR and DOR in the Golgi apparatus remain active and phosphorylated for hours after agonist exposure without engaging β-arrestin. Previous studies have shown that membrane-permeant opioid ligands accelerate the transport of DOR from intracellular locations to the PM (17). It is possible that anterograde trafficking of ORs contributes to turning off Golgi-localized signals. Yet unknown desensitization mechanisms may control internal ORs on a more rapid time scale. Besides the Golgi apparatus, intracellular MOR and DOR also signal from endosomes following agonist-driven internalization of cell surface ORs and mediate a sustained response (7, 67). Additional studies are needed to probe if ORs in the different organelles underlie common regulatory mechanisms. The present work delineates Golgi-localized OR signaling in response to permeant opioid drugs in a simplified cellular system and focuses on effects that are initiated upon OR activation. OR family members couple to inhibitory Gi/o proteins, reduce cAMP production, inhibit calcium conductance, and promote inhibition of neuronal activity in vivo (68). Therefore, future studies should address whether Golgi-localized OR signaling downstream of opioid drugs, when paired with a separate priming stimulus, may lead to additional and synergistic signaling responses that were not captured here. This is important given the role of ORs and their ligands in the neuromodulation of dynamic, transient, and fluctuating neuronal signals.
Together, and viewed more broadly, the present results reinforce an emerging understanding that GPCRs in intracellular organelles contribute to specific agonist actions and promote signal events that differ from the canonical response initiated at the cell surface. We propose from the present observations that lipid-based regulatory mechanisms contribute to location-biased OR-transducer selection and directly affect functional selectivity. Although the physiological consequences of Golgi-localized drug-specific OR signaling remain to be elucidated, our results provide previously unknown molecular insights that advance the notion that different subcellular receptor pools contribute to functional selectivity in opioid drug action.
MATERIALS AND METHODS
Mammalian cell culture conditions and plasmids
HEK293 [CRL-1573, American Type Culture Collection (ATCC), female] and HeLa cells (CRM-CCL-2, ATCC, female) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco), supplemented with 10% fetal bovine serum (FBS, Gibco). HEK293 cells stably expressing N-terminally signal sequence FLAG (ssf)-tagged DOR (HEK293-DOR) (7) were cultured in the presence of geneticin (250 μg/ml, Gibco). For transient DNA expression, Lipofectamine 2000 (Invitrogen, catalog no. 11668019) was used, and transfection medium was exchanged after 6 hours. Transgenic HeLa cells stably expressing N-terminally signal sequence (ss) SEP-tagged DOR were generated by cloning a “CAG promoter, ssSEP-DOR, RGK promoter, PuroR” cassette into a piggyBac transposon plasmid (Addgene #84239), cotransfection of the plasmid with piggyBac transposase, followed by flow cytometry-assisted sorting of GFP-positive cells and puromycin selection. The following published plasmids were used for transient expression: ssfDOR (mouse) (7), ssfMOR (mouse) (7), ssfβ2AR (mouse) in pcDNA3.1 (5), ssfMOR (mouse)-EGFP (7), ssSEP-(GFP variant Superecliptic pHluorin) DOR (mouse) (69), β-arrestin2 (bos taurus)-mCherry (5), GRK2 (bos taurus)-EGFP in pCAGGS-SE (70), LgBiT-β-arrestin2 (human) in pNBe3 (28), ManII-BFP2 (64), and GalT-DsRed2 (6). N-terminally tagged mRuby2-mGo, mRuby2-mGsi, and mRuby2-mGs (33) were from Nevin Lambert. The following new plasmids were generated: DNA fragments for mGi1, mGi2, mGi3 probes were synthesized (Invitrogen GeneArt Synthesis) and cloned into the Clontech pEGFP-C1 vector, replacing EGFP by mRuby2 or LgBiT. SmBiT-tagged MOR and DOR were generated by PCR-amplifying murine ssfMOR and ssfDOR and replacing ssfβ2AR in the ssfβ2AR-SmBiT pcDNA3.1 vector (28). LgBiT-GRK2 (bos taurus) was cloned by replacing EGFP with LgBiT in GRK2 (bos taurus)-EGFP (70).
Ligands
DAMGO [D-Ala2, N-Me-Phe4, Gly5-ol]-enkephalin acetate salt (catalog no. E7384), DADLE [D-Ala2, N-Me-Phe4, Gly5-ol]-enkephalin acetate salt (catalog no. E7131) DPDPE [D-Pen2,5]-enkephalin hydrate (catalog no. E3888) and isoproterenol hydrochloride (catalog no. I6504) were purchased from Sigma-Aldrich. SNC80 (catalog no. 0764), ARM-390 (catalog no. 4335), ICI 174,864 (catalog no. 0820), CTAP (D-Phe-Cys-Tyr-D-Trp-Arg-Thr-Pen-Thr-NH2, catalog no. 1560), and Cmpd101 (catalog no. 5642) were purchased from Tocris. Morphine sulfate [Chemical Abstracts Service (CAS) no. 6211-15-0] and fentanyl (CAS no. 1443-54-5) were obtained from the University Hospital of Geneva with Département de la sécurité, de la population et de la santé (DSPS) authorization from the Canton of Geneva.
Confocal microscopy-based recruitment assay
HeLa cells were seeded on poly-l-lysine-coated 35-mm Cellvis glass-bottomed dishes (IBL, 220.110.022) and, after 24 hours, transfected with fluorescent protein-tagged receptors (MOR-GFP or SEP-DOR, 1 μg of DNA), ManII-BFP (0.25 μg) and (i) Ruby2-mG probes (0.6 μg) or (ii) β-arrestin2-mCherry (0.8 μg) together with GRK2-LgBiT (1 μg) using 3 μl of Lipofectamine 2000. Living cells were imaged 16 to 24 hours after transfection in HBS imaging solution (Hepes-buffered saline with 135 mM NaCl, 5 mM KCl, 0.4 mM MgCl2,1.8 mM CaCl2, 20 mM Hepes, 1 mM d-glucose, 1% FBS, adjusted to pH 7.4). Recruitment of mG probes and β-arrestin2 was monitored with a spinning disk confocal microscope (Nipkow, Zeiss) using an EC Plan Neofluar 100×/1.3 oil Ph3 objective in a temperature and CO2-controlled environment (37°C, 5% CO2). Images of the same cells were acquired before and 5 min (or 45 and 90 min) after agonist addition. Fluorescence intensities of mG probes and β-arrestin2 in the Golgi area were measured (see quantitative image analysis) and normalized to OR fluorescence. Protein recruitment to Golgi-localized ORs is represented as the agonist-induced FC relative to before agonist addition. Sustained Golgi-localized DOR activation in HEK293-DOR cells after 6 hours of ICI-SNC80 treatment (100 μM ICI and 100 nM SNC80) was probed in cells transfected with EGFP-Nb33 (0.3 μg) and GalT-DsRed (0.2 μg). Before imaging, cell surface DOR was labeled with mouse anti-FLAG (M1) antibody (1:1000; Sigma-Aldrich, catalog no. F-3040) conjugated with Alexa Fluor 647 (Thermo Fisher Scientific, catalog no. A20173).
Split NanoLuc-based protein interaction assay
HeLa cells were seeded into six-well plates, and individual wells were transfected with (i) 0.4 μg of DOR-SmBiT and 0.25 μg of LgBiT-mG probe or (ii) MOR-SmBiT, 1 μg of ssfMOR, and 0.25 μg of LgBiT-mG probe, or (iii) 0.4 μg of DOR-SmBiT, 0.25 μg of β-arrestin-LgBiT, and 0.8 μg of GRK2-GFP, using 3 μl of Lipofectamine 2000. Sixteen to 24 hours after transfection, cells were seeded into black, clear-bottom 384-well plates (20,000 cells per well) in DMEM without phenol red, containing 30 mM Hepes (pH 7.4) and the NanoLuc substrate Nano-Glo (Promega, N2012), and incubated for 45 min at 37°C. Luminescence was recorded using a FDSS/μCELL kinetic plate imager (Hamamatsu) with an integrated simultaneous dispensing head and simultaneous detection across the plate. After acquiring baseline luminescence for 3 min, agonists (see concentration in figure legends) were added to cells. Luminescence was recorded every 2 s for 5 min after agonist addition. Experiments were performed in the absence or presence of 10 μM CTAP (MOR antagonist) or 100 μM ICI (DOR antagonist) to block PM ORs (added 5 min before acquisition). Luminescence values were normalized to baseline signal (before agonist) and to vehicle-treated control cells. Recruitment kinetics are represented as the percentage of the maximal luminescence signal detected in the absence of antagonists.
Immunofluorescence-based GPCR phosphorylation assay
HeLa cells were seeded onto 15-mm glass coverslips in 12-well plates and, after 24 hours, transfected with ssf-tagged GPCRs (ssfMOR, ssfDOR, or ssfβ2AR; 0.4 μg of DNA) and ManII-BFP (0.15 μg of DNA) using 1.5 μl of Lipofectamine 2000. GRK2-GFP (0.5 μg of DNA) was cotransfected in DOR and β2AR experiments as indicated in figure legend. Sixteen to 24 hours after transfection, cells were treated with ligands (details in figure legends) for 5 min (or 45 and 90 min) and fixed using 4% formaldehyde (FA) in phosphate-buffered saline (PBS). To assess reversibility of MOR phosphorylation, morphine was washed out after 5 min, and 100 μM naloxone was added for 10 min. To probe the role of GRK2/3, 30 μM Cmpd101 was added 45 min before agonist treatment. When indicated, 100 μM ICI was added 5 min before agonist treatment. Cells were permeabilized and blocked with 0.1% Saponin and 1.5% BSA in PBS and incubated overnight at 4°C with primary antibodies: phospho-MOR (S375) (1:500; Cell Signaling Technology (CST), catalog no. 3451S), phospho-DOR (S363) (1:1000; 7TM Antibodies, catalog no. 7TM0317B), phospho-β2AR (T360/S364) (1:250; 7TM Antibodies, catalog no. 7TM0029B) antibodies, and anti-FLAG (M1) antibody (1:1000) in blocking solution. After three washes, cells were incubated with goat anti-rabbit immunoglobulin G (IgG) Alexa Fluor 647 (Thermo Fisher Scientific, catalog no. A32733) and goat anti-mouse IgG Alexa Fluor 568 (Thermo Fisher Scientific, catalog no. A11031) secondary antibodies for 45 min at room temperature. Samples were mounted using ProLong Glass Antifade Mountant (Thermo Fisher Scientific, catalog no. P36982) and imaged with a spinning disk confocal microscope (Nipkow, Zeiss) using an EC Plan Neofluar 100×/1.3 oil Ph3 objective. Signal of GPCR phosphorylation in the ManII-labeled Golgi area was normalized to Golgi-localized OR signal and to nontreated control cells. DOR localization in the Golgi of HEK293-DOR cells was assessed by immunofluorescence as described above. Permeabilized cells were incubated with recombinant human anti-giantin (1:100; Geneva antibody facility, ABCD_AA341) and mouse anti-FLAG (M1) primary antibodies followed by incubation with goat anti-human IgG Alexa Fluor 647 (Thermo Fisher Scientific, catalog no. A21445) and goat anti-mouse IgG Alexa Fluor 568 secondary antibodies. Cells were imaged with spinning disk confocal microscopy using a Plan Apo 63×/1.4 oil DICIII objective.
TIR-FM–based protein recruitment assay
HeLa cells were seeded on poly-l-lysine–coated 35 mm glass-bottomed dishes and transfected with ssfDOR or ssfMOR (1 μg of DNA) and mG probes (0.6 to 0.8 μg of DNA) using 3 μl of Lipofectamine 2000. Sixteen to 24 hours after transfection, surface ORs were labeled for 10 min with Alexa Fluor 647–conjugated anti-FLAG (M1) antibody (1:1000), and medium was changed to HBS imaging solution. Cells were imaged at 37°C using a Nikon Eclipse Ti microscope using a 100× 1.49 oil CFI Apochromat TIR-FM objective, temperature chamber, objective heater, perfect focus system, and an ORCA-Fusion BT Digital CMOS camera. Images were acquired before and 5 min after agonist addition. Protein relocalization (∆F) was calculated as F(t)/F0 with F(t) = mG signal after ligand addition and F0 = mG signal before ligand addition, both normalized to OR fluorescence.
Quantitative fluorescence image analysis
Unprocessed images were analyzed using ImageJ (2.1.0). Phosphorylation of GPCRs in the Golgi apparatus was quantified using a custom-written macro. The ManII signal was used as the Golgi mask, and within the mask, the mean fluorescence signal of (i) phosphorylated GPCRs (pGPCR) and (ii) total GPCRs was measured. A zone around the ManII mask was generated to measure the cytosolic background signal. We then calculated the ratio of background-subtracted pGPCR/GPCR signals for all acquired cells. Quantification of mG recruitment to ORs in living cells imaged with spinning disk confocal microscopy before and after agonist addition was performed in a similar manner: ManII was used as Golgi mask, and fluorescence of (i) mG and (ii) OR was measured in all cells. A region outside the cells was used as the background, and background-subtracted mG/OR ratios were calculated. The mG recruitment level F/F0 was determined by dividing the mG/OR level after agonist addition by the level before agonist addition. β-arrestin2 recruitment to ORs was measured as follows: Spinning disk confocal microscopy images with focus on the Golgi plane were used to quantify β-arrestin and DOR signals in the ManII-defined Golgi mask. β-arrestin/DOR and DOR signals were measured before (F0) and after (F) agonist addition, and recruitment signal was calculated as F/F0. Confocal images with a focus on the cell surface were used to quantify β-arrestin and DOR signals at the PM. A mask encompassing each cell was generated, and F/F0 was calculated similar to the Golgi recruitment. To quantify mG recruitment to the PM based on TIR-FM images, a mask encompassing each cell was generated. The fluorescence intensity ratio of background-subtracted mG/OR in this mask was calculated. A region outside the cell was used as the background, and quantification was performed before (F0) and 5 min after (F) agonist addition. Cells with OR levels close to background noise or aberrantly high OR expression and cells with aberrant cell and organelle morphology (i.e., extreme Golgi fragmentation and cell apoptosis) were detected rarely (<5%) and excluded from the analysis.
Statistics of image quantification
Quantification of data is presented as mean ± SD based on at least three biologically independent experiments with the precise number indicated in the figure legends. Statistical analysis was performed using unpaired two-tailed Student’s t test, Welch’s t test, or one-way analysis of variance (ANOVA) test as indicated, using the software Prism (GraphPad 9.1.1).
Sample preparation for RNA-seq
HEK293-DOR cells were treated for 1.5 or 6 hours with (i) vehicle control, (ii) 100 nM DPDPE, (iii) 100 nM SNC80, or (iv) 100 nM SNC80 after 5-min preincubation with 100 μM ICI (ICI present during entire experiment duration), with N = 3 for all samples. Total RNA was extracted using the RNeasy Mini Kit (Qiagen, catalog no. 74104), genomic DNA was removed using the RNase-free DNase kit (Qiagen, catalog no. 79254), and samples were analyzed using a bioanalyzer. Gene expression in 1.5-hour samples was measured using 3′ RNA-seq (UC Davis Genome Centre) and in 6-hour samples using whole transcript RNA-seq (iGE3 Genomics Platform, University of Geneva). Multiplexed libraries for the 1.5-hour samples were generated using the Lexogen QuantSeq 3′ mRNA-Seq Library Prep Kit and for the 6-hour samples using TruSeqHT Stranded Total RNA Library Prep protocol (Illumina).
RNA-seq and analyses
Libraries generated for the 1.5-hour treatment were sequenced using the Illumina NextSeq 500 or HiSeq 4000 devices with a single-end read length of 80 to 90 nucleotides yielding at least 3 million mapped reads per sample. Sequences were mapped to the human genome GRC h38.12 using GENCODE v28 annotation with STAR v. 2.6.0c. Generated raw counts were normalized, and genes with low expression were filtered out leaving 15,946 genes. Differential expression analyses and related statistics between control and treated conditions were conducted using the Limma-voom R package 3.38.2. Libraries generated for the 6-hour treatment were sequenced using the Illumina HiSeq 4000 device with a single-end read length of 50 nucleotides yielding around 22 million mapped reads per sample. Fastq files were generated using Illumina bcl2fastq v. 2.20.0 and mapped to the human genome UCSC hg38 using STAR v. 2.7.0 f. Aligned reads were quantified with the Python software htseq-count v. 0.9.1. Genes were filtered down to 14,467 after removal of low expressers and normalized using the R package EdgeR 3.28.1. Differential gene expression between control and treated conditions was computed, and statistics were performed using the General Linear Model, quasi-likelihood F-test (QL F-test) with false discovery rate (FDR) and Benjamini and Hochberg correction. Any gene with FC over 1.5 or below −1.5 and adjusted P < 0.05 was considered as significantly regulated.
Sample preparation for mass spectrometry
HEK293-DOR cells were treated for 5 or 25 min with 100 nM SNC80 in the presence or absence of 100 μM ICI (5-min preincubation, present throughout the experiment), with N = 3 for all samples. Cells were then washed with ice-cold PBS and spun down, and dried pellets were snap-frozen in N2. Thawed pellets were lysed in a buffer containing 50 mM tris (pH 7.4), 25 mM NaCl, 2% SDS, 2.5 mM EDTA, 20 mM tris(2-carboxyethyl)phosphine (TCEP), and 2% SDS, supplemented with Halt-protease and Halt-phosphatase inhibitors (Thermo Fisher Scientific). Samples were vortexed and heated at 95°C for 10 min with 400 rpm mixing on a thermomixer. DNA was sheared by sonication. Samples were centrifuged for 30 min at 17,000g, and the supernatant was collected and incubated with 0.5 M iodoacetamide for 1 hour at room temperature. Proteins were digested on the basis of the filter aided sample preparation (FASP) method using Amicon Ultra-4, 30-kDa centrifugal filter units (Millipore). Trypsin (Promega) was added at 1:75 enzyme/protein ratio, and digestion was performed overnight at room temperature. The resulting peptide samples were desalted with C18 macrospin columns (Harvard Apparatus) and then completely dried under speed-vacuum. Each sample was labeled with corresponding TMT10plex reagent dissolved in 110 μl of 36% CH3CN and 200 mM EPPS (pH 8.5). Reaction was performed during 1 hour at room temperature and quenched by adding hydroxylamine to a final concentration of 0.3% (v/v). Labeled samples were pooled, dried, and desalted with a peptide desalting spin column (Thermo Fisher Scientific). Phosphopeptides were enriched using the High-Select Fe-NTA Phosphopeptide Enrichment Kit (Thermo Fisher Scientific). Phosphopeptide and the flow-through fractions were desalted with a C18 macrospin column (Harvard Apparatus) and completely dried under speed-vacuum. The flow through fraction was then fractionated into 13 fractions using the Pierce High pH Reversed-Phase Peptide Fractionation Kit (Thermo Fisher Scientific).
Quantitative real-time PCR
HEK293-DOR cells were treated with 100 nM DADLE for 1.5 hours. Total RNA was extracted and genomic DNA was removed as described above. RNA samples were quantified by NanoDrop (Thermo Fisher Scientific) and subjected to reverse transcription using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, catalog no. 4368814). RT-qPCR was performed using the PowerUp SYBR green reagent (Applied Biosystems, catalog no. A25741), and relative RNA quantification was performed using the Delta-Delta Ct method. The list of used primers is available in the Yareta repository.
ERK1/2 activation assay
HEK293-DOR cells were treated with 100 nM SNC80 in the presence or absence of 100 μM ICI (5-min preincubation, present throughout experiment) for the indicated times. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer [10 mM tris (pH 7.4), 1× Triton, 150 mM NaCl, 0.1% SDS, and 2 mM EDTA] supplemented with protease and phosphatase inhibitors (Roche), and lysates were sonicated followed by quantification using the Pierce BCA protein assay (Thermo Fisher Scientific). Protein samples were mixed with NuPAGE LDS sample buffer and 100 mM dithiothreitol and heated for 10 min at 70°C. Lysates were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using 4 to 12% bis-tris plus gels (Thermo Fisher Scientific) and transferred to an isopropanol-activated polyvinylidene difluoride (PVDF) membrane. After blocking in tris-buffered saline (TBS) with 5% BSA, membranes were incubated overnight at 4°C with mouse anti-ERK1/2 (CST, catalog no. 4696S) and rabbit anti-pERK1/2 (T202/Y204) (CST, catalog no. 4370S) primary antibodies (1:2000) in TBS-Tween (0.1% v/v) with 5% BSA. After washing, horseradish peroxidase (HRP)-coupled anti-mouse (1:10,000; Sigma-Aldrich, catalog no. A5278) and anti-rabbit secondary antibodies (1:4000; Sigma-Aldrich, catalog no. A8275) were added for 1 hour at room temperature. Membranes were imaged in the presence of Pierce ECL SuperSignal West Pico plus (Thermo Fisher Scientific) using the iBright 1500 device (Invitrogen). ERK protein levels were quantified using ImageJ, and pERK/ERK was plotted.
mG expression levels
Relative expression of mG probes was assessed by Western blot and in-gel fluorescence. HeLa cells transfected with LgBiT-mG or mRuby2-mG probes were lysed in RIPA buffer and sonicated, and total protein amounts were quantified. Western blot assays followed the procedure described under the “ERK1/2 activation assay” section. Membranes were incubated overnight at 4°C with mouse anti-LgBiT (1:500; Promega, catalog no. N710A) or goat anti-mRuby (1:2000; St John’s Laboratory, catalog no. STJ140251) primary antibodies in TBS-Tween (0.1% v/v) with 5% BSA. After washing, HRP-coupled anti-mouse and anti-goat (1:10,000; Jackson ImmunoResearch, catalog no. 705-035-003) secondary antibodies were added for 1 hour at room temperature, and the membranes were imaged as described above. Tubulin, probed with a recombinant mouse IG2A anti-alpha tubulin antibody (1:5000; Geneva Antibody Facility, ABCD_AA345), served as loading control. For in-gel fluorescence, lysates of HeLa cells expressing mRuby2-mG probes were mixed with NuPAGE LDS sample buffer and loaded directly on a 4 to 12% bis-tris plus gel for SDS-PAGE separation. Gels were then imaged with the Odyssey M (LI-COR) reader, and mRuby2 was detected at 520 nm excitation. Proteins were transferred to an isopropanol-activated PVDF membrane and stained with Ponceau S as loading control.
Mass spectrometry and analyses
Phosphopeptides were reconstituted in loading buffer (5% CH3CN and 0.1% FA), and 1 μg was injected into the column. LC-ESI-MS/MS was performed on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) equipped with an Easy nLC1200 liquid chromatography system (Thermo Fisher Scientific) at the Proteomics Core Facility, University of Geneva. Peptides were trapped on a Acclaim pepmap100, C18, 3 μm, 75 μm by 20 mm nano trap column (Thermo Fisher Scientific) and separated on a 75 μm by 500 mm, C18 ReproSil-Pur from Dr. Maisch GmBH, 1.9 μm, 100 Å, home-made column. The analytical separation was run for 125 min using a gradient of 99.9%/0.1% H2O/FA (solvent A) and 80%/0.1% CH3CN/FA (solvent B). Data-dependent acquisition was performed with MS1 full scan at a resolution of 120,000 full width at half maximum followed by as many subsequent MS2 scans on selected precursors as possible within 3-s maximum cycle time. High pH reversed-phase peptide fractions were treated similarly. Raw data were processed using Proteome Discoverer 2.3 software (Thermo Fisher Scientific). Briefly, spectra were extracted and searched against the Human Reference Proteome database (release 11_2019, 20,660 entries) combined with an in-house database of common contaminants using Mascot (Matrix Science, London, UK; version 2.5.1). Trypsin was selected as the enzyme, with one potential missed cleavage. Precursor ion tolerance was set to 10 ppm and fragment ion tolerance to 0.02 Da. Carbamidomethylation of cysteine (+57.021) as well as TMT10plex (+229.163) on lysine residues and on peptide N termini were specified as fixed modification. Oxidation of methionine (+15.995) as well as phosphorylated (+79.966) serine, threonine, and tyrosine were set as variable modifications. Search results were validated with a Target Decoy peptide-spectrum match (PSM) validator. PSM and peptides were filtered with an FDR of 1% and then grouped to proteins with again an FDR of 1% and using only peptides with high confidence level. Both unique and razor peptides were used for quantitation, and protein and peptide abundances values were based on S/N values of reporter ions. The abundances were normalized on “Total Peptide Amount” and then scaled with “On all Average.” All the protein ratios were calculated from the medians of the summed abundances of replicate groups, and associated P values were calculated with an ANOVA test based on individual proteins or peptides. Any phosphopeptide with an FC ratio over 1.2 or below −1.2 and P < 0.05 was considered as significantly regulated.
Phosphoproteomics analyses and upstream kinase predictions
Upstream kinase predictions were performed using the KSEA app (v. 1.0) (71) fed with a pooled list of unique phosphopeptides significantly regulated at 5 and 25 min upon SNC80 or ICI-SNC80 treatment. Kinase plots were generated with a NetworKIN score cutoff set to 1, a substrate count cutoff set to 5, and a P value cutoff set to 0.05.
MD simulations
The three-dimensional structures of the MOR complexed with mGi1 and mGsi were built using “Alphafold2-multimer” (72). Subsequently, the two heterodimers were embedded into tailored phospholipid bilayers using CHARMM-GUI (73) to obtain the following four systems: (i) MOR-mGi1 in PM, (ii) MOR-mGsi in PM, (iii) MOR-mGi1 in Golgi-like membrane (Golgi), and (iv) MOR-mGsi in Golgi. To reproduce the properties of PM and Golgi (36, 37), two different phospholipidic compositions were used (Fig. 5A). All systems were solvated using a box of TIP3P water model and a salinity of 0.15 M NaCl. The N- and C-termini of MOR were capped with an acetyl and a metil-amino protecting group, respectively. The CHARMM36m force field was used for the MD simulations, which were run using the GROMACS 2021.5 engine. Each simulation box underwent a thermalization cycle with smoothly decreasing restraints on heavy atoms to gently equilibrate the structure. We used the following protocol: heating the system from 100 to 300 K by increasing the temperature by 50 K each step composed of 1 ns of canonical ensemble (i.e. NVT) simulation followed by 1 ns of isothermal-isobaric ensemble (i.e. NPT) simulation. During the thermalization, the V-rescale thermostat was used, whereas the Langevin dynamics temperature control scheme was used in the production runs. Periodic boundary conditions were applied, and the particle-mesh-Ewald method was used to treat long-range electrostatic interaction. For short-range interactions, a cutoff distance of 1.0 nm was applied. The pressure was fixed at a reference value equal to 1 bar by means of the Parrinello-Rahman barostat.
Cluster analysis
Cluster analyses on the MD trajectories were performed using GROMACS’s “gmx cluster” routine. The cluster families of MOR-mGi1 and MOR-mGsi were obtained by aligning on secondary structure’s Cα atoms of MOR and computing the RMSD for the secondary structure’s Cα of both mGi1 and mGsi. The RMSD cutoff was set at 1.5 Å.
Binding interface evaluation
The protein-protein interactions established by the residues at the MOR-mGi1 and MOR-mGsi interface were assessed by calculating their frequency of occurrence using the “PLOT NA” routine of “Drug Discovery Tool” (74) and displayed as histograms. We set a distance cutoff value of 4.0 Å to define two interacting residues.
Acknowledgments
We thank M. von Zastrow for valuable discussions and advice. We thank members of the Bioimaging Core Facility, the RE.A.D.S. Unit, the Proteomics Core Facility, and the iGE3 Genomics Platform at the University of Geneva and the Genome Centre at UC Davis for helpful discussions. We thank N. Lambert for sharing the mGo-mRuby2 construct. We thank our colleagues and group members at the University of Geneva for critical comments on the project and manuscript.
Funding: The work was supported by the Swiss National Science Foundation Eccellenza Professorial Fellowship to M.S. (PCEFP3_181282) and a grant to M.S. from the Olga Mayenfish Stiftung. We acknowledge PRACE and the Swiss National Supercomputing Centre (CSCS) for supercomputer time allocations on Piz Daint (project IDs: pr126 and s1169). F.L.G. is supported by the Swiss National Science Foundation and the BRIDGE program (project numbers 204795 and 203628). A part of this work was supported at an early stage through NIH DA10711 and DA012864 to M. von Zastrow at UCSF.
Author contributions: Conceptualization: A.R.-M., L.O., S.A., F.L.G., and M.S. Methodology: A.R.-M., L.O., S.A., and M.S. Investigation: A.R.-M., L.O., S.A., and M.S. Supervision: F.L.G. and M.S. Writing—original draft: A.R.-M., L.O., S.A., F.L.G., and M.S., Writing—review and editing: A.R.-M., L.O., S.A., F.L.G., and M.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The RNA-seq datasets are available in the Gene Expression Omnibus repository (GSE221762). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE repository (PXD038921). All other data are available in the Yareta depository (10.26037/yareta:g4kupsxgebdn7mowwd6dpfrvuq). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Requests for materials should be directed to M.S. (miriam.stoeber@unige.ch).
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Legends for movies S1 and S2
Legends for tables S1 and S2
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S2
Tables S1 and S2
REFERENCES AND NOTES
- 1.K. Sriram, P. A. Insel, G protein-coupled receptors as targets for approved drugs: How many targets and how many drugs? Mol. Pharmacol. 93, 251–258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.A. S. Hauser, M. M. Attwood, M. Rask-Andersen, H. B. Schiöth, D. E. Gloriam, Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Y.-J. I. Jong, S. K. Harmon, K. L. O’Malley, GPCR signalling from within the cell. Br. J. Pharmacol. 175, 4026–4035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D. Jullié, Z. Valbret, M. Stoeber, Optical tools to study the subcellular organization of GPCR neuromodulation. J. Neurosci. Methods 366, 109408 (2022). [DOI] [PubMed] [Google Scholar]
- 5.R. Irannejad, J. C. Tomshine, J. R. Tomshine, M. Chevalier, J. P. Mahoney, J. Steyaert, S. G. F. Rasmussen, R. K. Sunahara, H. El-Samad, B. Huang, M. von Zastrow, Conformational biosensors reveal GPCR signalling from endosomes. Nature 495, 534–538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.R. Irannejad, V. Pessino, D. Mika, B. Huang, P. B. Wedegaertner, M. Conti, M. von Zastrow, Functional selectivity of GPCR-directed drug action through location bias. Nat. Chem. Biol. 13, 799–806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M. Stoeber, D. Jullié, B. T. Lobingier, T. Laeremans, J. Steyaert, P. W. Schiller, A. Manglik, M. von Zastrow, A genetically encoded biosensor reveals location bias of opioid drug action. Neuron 98, 963–976.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Y.-J. I. Jong, I. Sergin, C. A. Purgert, K. L. O’Malley, Location-dependent signaling of the group 1 metabotropic glutamate receptor mGlu5. Mol. Pharmacol. 86, 774–785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.T. Che, J. English, B. E. Krumm, K. Kim, E. Pardon, R. H. J. Olsen, S. Wang, S. Zhang, J. F. Diberto, N. Sciaky, F. I. Carroll, J. Steyaert, D. Wacker, B. L. Roth, Nanobody-enabled monitoring of kappa opioid receptor states. Nat. Commun. 11, 1145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.J. M. Kunselman, A. Gupta, I. Gomes, L. A. Devi, M. A. Puthenveedu, Compartment-specific opioid receptor signaling is selectively modulated by different dynorphin peptides. eLife 10, e60270 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.L. Bao, S.-X. Jin, C. Zhang, L.-H. Wang, Z.-Z. Xu, F.-X. Zhang, L.-C. Wang, F.-S. Ning, H.-J. Cai, J.-S. Guan, H.-S. Xiao, Z.-Q. D. Xu, C. He, T. Hökfelt, Z. Zhou, X. Zhang, Activation of delta opioid receptors induces receptor insertion and neuropeptide secretion. Neuron 37, 121–133 (2003). [DOI] [PubMed] [Google Scholar]
- 12.H. Wang, V. M. Pickel, Preferential cytoplasmic localization of delta-opioid receptors in rat striatal patches: Comparison with plasmalemmal mu-opioid receptors. J. Neurosci. 21, 3242–3250 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.E. Erbs, L. Faget, G. Scherrer, A. Matifas, D. Filliol, J.-L. Vonesch, M. Koch, P. Kessler, D. Hentsch, M.-C. Birling, M. Koutsourakis, L. Vasseur, P. Veinante, B. L. Kieffer, D. Massotte, A mu–delta opioid receptor brain atlas reveals neuronal co-occurrence in subcortical networks. Brain Struct. Funct. 220, 677–702 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.G. Scherrer, P. Tryoen-Tóth, D. Filliol, A. Matifas, D. Laustriat, Y. Q. Cao, A. I. Basbaum, A. Dierich, J.-L. Vonesh, C. Gavériaux-Ruff, B. L. Kieffer, Knockin mice expressing fluorescent delta-opioid receptors uncover G protein-coupled receptor dynamics in vivo. Proc. Natl. Acad. Sci. U.S.A. 103, 9691–9696 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.A. Ozawa, G. Brunori, D. Mercatelli, J. Wu, A. Cippitelli, B. Zou, X. S. Xie, M. Williams, N. T. Zaveri, S. Low, G. Scherrer, B. L. Kieffer, L. Toll, Knock-In mice with NOP-eGFP receptors identify receptor cellular and regional localization. J. Neurosci. 35, 11682–11693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.K.-A. Kim, M. von Zastrow, Neurotrophin-regulated sorting of opioid receptors in the biosynthetic pathway of neurosecretory cells. J. Neurosci. 23, 2075–2085 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.U. E. Petäjä-Repo, M. Hogue, S. Bhalla, A. Laperrière, J.-P. Morello, M. Bouvier, Ligands act as pharmacological chaperones and increase the efficiency of delta opioid receptor maturation. EMBO J. 21, 1628–1637 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A. D. White, K. A. Peña, L. J. Clark, C. S. Maria, S. Liu, F. G. Jean-Alphonse, J. Y. Lee, S. Lei, Z. Cheng, C.-L. Tu, F. Fang, N. Szeto, T. J. Gardella, K. Xiao, S. H. Gellman, I. Bahar, I. Sutkeviciute, W. Chang, J.-P. Vilardaga, Spatial bias in cAMP generation determines biological responses to PTH type 1 receptor activation. Sci. Signal. 14, eabc5944 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A. R. B. Thomsen, B. Plouffe, T. J. Cahill 3rd, A. K. Shukla, J. T. Tarrasch, A. M. Dosey, A. W. Kahsai, R. T. Strachan, B. Pani, J. P. Mahoney, L. Huang, B. Breton, F. M. Heydenreich, R. K. Sunahara, G. Skiniotis, M. Bouvier, R. J. Lefkowitz, GPCR-G protein-β-arrestin super-complex mediates sustained G protein signaling. Cell 166, 907–919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A. Godbole, S. Lyga, M. J. Lohse, D. Calebiro, Internalized TSH receptors en route to the TGN induce local Gs-protein signaling and gene transcription. Nat. Commun. 8, 443 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D. Calebiro, I. Maiellaro, cAMP signaling microdomains and their observation by optical methods. Front. Cell. Neurosci. 8, 350 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C. A. Nash, W. Wei, R. Irannejad, A. V. Smrcka, Golgi localized β1-adrenergic receptors stimulate Golgi PI4P hydrolysis by PLCε to regulate cardiac hypertrophy. eLife 8, e48167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.N. G. Tsvetanova, M. Trester-Zedlitz, B. W. Newton, G. E. Peng, J. R. Johnson, D. Jimenez-Morales, A. P. Kurland, N. J. Krogan, M. von Zastrow, Endosomal cAMP production broadly impacts the cellular phosphoproteome. J. Biol. Chem. 297, 100907 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.N. G. Tsvetanova, M. von Zastrow, Spatial encoding of cyclic AMP signaling specificity by GPCR endocytosis. Nat. Chem. Biol. 10, 1061–1065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.B. Carpenter, Current applications of mini G proteins to study the structure and function of G protein-coupled receptors. AIMS Bioengineering 5, 209–225 (2018). [Google Scholar]
- 26.B. Carpenter, C. G. Tate, Engineering a minimal G protein to facilitate crystallisation of G protein-coupled receptors in their active conformation. Protein Eng. Des. Sel. 29, 583–594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R. Nehmé, B. Carpenter, A. Singhal, A. Strege, P. C. Edwards, C. F. White, H. Du, R. Grisshammer, C. G. Tate, Mini-G proteins: Novel tools for studying GPCRs in their active conformation. PLOS ONE 12, e0175642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.C. Laschet, N. Dupuis, J. Hanson, A dynamic and screening-compatible nanoluciferase-based complementation assay enables profiling of individual GPCR–G protein interactions. J. Biol. Chem. 294, 4079–4090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. T. Williams, S. L. Ingram, G. Henderson, C. Chavkin, M. von Zastrow, S. Schulz, T. Koch, C. J. Evans, M. J. Christie, Regulation of μ-opioid receptors: Desensitization, phosphorylation, internalization, and tolerance. Pharmacol. Rev. 65, 223–254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.C. Doll, F. Pöll, K. Peuker, A. Loktev, L. Glück, S. Schulz, Deciphering μ-opioid receptor phosphorylation and dephosphorylation in HEK293 cells. Br. J. Pharmacol. 167, 1259–1270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A. Mann, S. Liebetrau, M. Klima, P. Dasgupta, D. Massotte, S. Schulz, Agonist-induced phosphorylation bar code and differential post-activation signaling of the delta opioid receptor revealed by phosphosite-specific antibodies. Sci. Rep. 10, 8585 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. E. Crilly, W. Ko, Z. Y. Weinberg, M. A. Puthenveedu, Conformational specificity of opioid receptors is determined by subcellular location irrespective of agonist. eLife 10, e67478 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Q. Wan, N. Okashah, A. Inoue, R. Nehmé, B. Carpenter, C. G. Tate, N. A. Lambert, Mini G protein probes for active G protein–coupled receptors (GPCRs) in live cells. J. Biol. Chem. 293, 7466–7473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.N. Okashah, Q. Wan, S. Ghosh, M. Sandhu, A. Inoue, N. Vaidehi, N. A. Lambert, Variable G protein determinants of GPCR coupling selectivity. Proc. Natl. Acad. Sci. U.S.A. 116, 12054–12059 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.A. Koehl, H. Hu, S. Maeda, Y. Zhang, Q. Qu, J. M. Paggi, N. R. Latorraca, D. Hilger, R. Dawson, H. Matile, G. F. X. Schertler, S. Granier, W. I. Weis, R. O. Dror, A. Manglik, G. Skiniotis, B. K. Kobilka, Structure of the μ-opioid receptor-Gi protein complex. Nature 558, 547–552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.S. J. Marrink, V. Corradi, P. C. T. Souza, H. I. Ingólfsson, D. P. Tieleman, M. S. P. Sansom, Computational modeling of realistic cell membranes. Chem. Rev. 119, 6184–6226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.H. Koldsø, D. Shorthouse, J. Hélie, M. S. P. Sansom, Lipid clustering correlates with membrane curvature as revealed by molecular simulations of complex lipid bilayers. PLoS Comput. Biol. 10, e1003911 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.S. Schneider, D. Provasi, M. Filizola, How oliceridine (TRV-130) binds and stabilizes a μ-opioid receptor conformational state that selectively triggers G protein signaling pathways. Biochemistry 55, 6456–6466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. Ray, F. Gräter, L. Thukral, Probing molecular forces in multi-component physiological membranes. Phys. Chem. Chem. Phys. 20, 2155–2161 (2018). [DOI] [PubMed] [Google Scholar]
- 40.F.-X. Yu, B. Zhao, N. Panupinthu, J. L. Jewell, I. Lian, L. H. Wang, J. Zhao, H. Yuan, K. Tumaneng, H. Li, X.-D. Fu, G. B. Mills, K.-L. Guan, Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.M. Piechota, M. Korostynski, W. Solecki, A. Gieryk, M. Slezak, W. Bilecki, B. Ziolkowska, E. Kostrzewa, I. Cymerman, L. Swiech, J. Jaworski, R. Przewlocki, The dissection of transcriptional modules regulated by various drugs of abuse in the mouse striatum. Genome Biol. 11, R48 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.T. Shoda, K. Fukuda, H. Uga, H. Mima, H. Morikawa, Activation of mu-opioid receptor induces expression of c-fos and junB via mitogen-activated protein kinase cascade. Anesthesiology 95, 983–989 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Y. Lin, J. M. Wozniak, N. J. Grimsey, S. Girada, A. Patwardhan, O. Molinar-Inglis, T. H. Smith, J. D. Lapek, D. J. Gonzalez, J. Trejo, Phosphoproteomic analysis of protease-activated receptor-1 biased signaling reveals unique modulators of endothelial barrier function. Proc. Natl. Acad. Sci. U.S.A. 117, 5039–5048 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.A. Schäfer, E. Gjerga, R. W. Welford, I. Renz, F. Lehembre, P. M. Groenen, J. Saez-Rodriguez, R. Aebersold, M. Gstaiger, Elucidating essential kinases of endothelin signalling by logic modelling of phosphoproteomics data. Mol. Syst. Biol. 15, e8828 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.J. J. Liu, K. Sharma, L. Zangrandi, C. Chen, S. J. Humphrey, Y.-T. Chiu, M. Spetea, L.-Y. Liu-Chen, C. Schwarzer, M. Mann, In vivo brain GPCR signaling elucidated by phosphoproteomics. Science 360, eaao4927 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.J. J. Liu, Y.-T. Chiu, K. M. DiMattio, C. Chen, P. Huang, T. A. Gentile, J. W. Muschamp, A. Cowan, M. Mann, L.-Y. Liu-Chen, Phosphoproteomic approach for agonist-specific signaling in mouse brains: mTOR pathway is involved in κ opioid aversion. Neuropsychopharmacology 44, 939–949 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.V. Dozio, Y. Daali, J. Desmeules, J.-C. Sanchez, Deep proteomics and phosphoproteomics reveal novel biological pathways perturbed by morphine, morphine-3-glucuronide and morphine-6-glucuronide in human astrocytes. J. Neurosci. Res. 100, 220–236 (2022). [DOI] [PubMed] [Google Scholar]
- 48.J. Wu, Q. Jiang, H. Zhu, Y. Zhou, D. Lu, X. Liu, X. Chen, J. Chen, Y. Wang, J. Liu, R. Song, R. Huang, H. Zhou, Uncovering kappa-opioid receptor agonist-induced PAK1/2 phosphorylation by quantitative phosphoproteomics. Biochem. Biophys. Res. Commun. 516, 320–326 (2019). [DOI] [PubMed] [Google Scholar]
- 49.E. Miess, A. B. Gondin, A. Yousuf, R. Steinborn, N. Mösslein, Y. Yang, M. Göldner, J. G. Ruland, M. Bünemann, C. Krasel, M. J. Christie, M. L. Halls, S. Schulz, M. Canals, Multisite phosphorylation is required for sustained interaction with GRKs and arrestins during rapid μ-opioid receptor desensitization. Sci. Signal. 11, eaas9609 (2018). [DOI] [PubMed] [Google Scholar]
- 50.C. C. M. Lally, B. Bauer, J. Selent, M. E. Sommer, C-edge loops of arrestin function as a membrane anchor. Nat. Commun. 8, 14258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D. P. Staus, H. Hu, M. J. Robertson, A. L. W. Kleinhenz, L. M. Wingler, W. D. Capel, N. R. Latorraca, R. J. Lefkowitz, G. Skiniotis, Structure of the M2 muscarinic receptor–β-arrestin complex in a lipid nanodisc. Nature 579, 297–302 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.J. Janetzko, R. Kise, B. Barsi-Rhyne, D. H. Siepe, F. M. Heydenreich, K. Kawakami, M. Masureel, S. Maeda, K. C. Garcia, M. von Zastrow, A. Inoue, B. K. Kobilka, Membrane phosphoinositides regulate GPCR-β-arrestin complex assembly and dynamics. Cell 185, 4560–4573.e19 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.N. M. Puri, G. R. Romano, T.-Y. Lin, Q. N. Mai, R. Irannejad, The organic cation transporter 2 regulates dopamine D1 receptor signaling at the Golgi apparatus. eLife 11, e75468 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.H.-Y. Yen, K. K. Hoi, I. Liko, G. Hedger, M. R. Horrell, W. Song, D. Wu, P. Heine, T. Warne, Y. Lee, B. Carpenter, A. Plückthun, C. G. Tate, M. S. P. Sansom, C. V. Robinson, PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature 559, 423–427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.C. I. Holmberg, S. E. F. Tran, J. E. Eriksson, L. Sistonen, Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem. Sci. 27, 619–627 (2002). [DOI] [PubMed] [Google Scholar]
- 56.S. Aznar, P. F. Valerón, S. V. del Rincon, L. F. Pérez, R. Perona, J. C. Lacal, Simultaneous tyrosine and serine phosphorylation of STAT3 transcription factor is involved in Rho A GTPase oncogenic transformation. Mol. Biol. Cell 12, 3282–3294 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.M. Grundmann, E. Kostenis, Temporal bias: Time-encoded dynamic GPCR signaling. Trends Pharmacol. Sci. 38, 1110–1124 (2017). [DOI] [PubMed] [Google Scholar]
- 58.J. E. Ladbury, S. T. Arold, Noise in cellular signaling pathways: Causes and effects. Trends Biochem. Sci. 37, 173–178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.J. K. Nussbacher, R. Tabet, G. W. Yeo, C. Lagier-Tourenne, Disruption of RNA metabolism in neurological diseases and emerging therapeutic interventions. Neuron 102, 294–320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.I. Masuho, N. K. Skamangas, B. S. Muntean, K. A. Martemyanov, Diversity of the Gβγ complexes defines spatial and temporal bias of GPCR signaling. Cell Syst. 12, 324–337.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.S. P. Denker, J. M. McCaffery, G. E. Palade, P. A. Insel, M. G. Farquhar, Differential distribution of alpha subunits and beta gamma subunits of heterotrimeric G proteins on Golgi membranes of the exocrine pancreas. J. Cell Biol. 133, 1027–1040 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D. Michaelson, I. Ahearn, M. Bergo, S. Young, M. Philips, Membrane trafficking of heterotrimeric G proteins via the endoplasmic reticulum and Golgi. Mol. Biol. Cell 13, 3294–3302 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.P. Ghosh, P. Rangamani, I. Kufareva, The GAPs, GEFs, GDIs and…now, GEMs: New kids on the heterotrimeric G protein signaling block. Cell Cycle 16, 607–612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.G. P. Solis, O. Bilousov, A. Koval, A.-M. Lüchtenborg, C. Lin, V. L. Katanaev, Golgi-resident Gαo promotes protrusive membrane dynamics. Cell 170, 939–955.e24 (2017). [DOI] [PubMed] [Google Scholar]
- 65.B. K. Atwood, J. Lopez, J. Wager-Miller, K. Mackie, A. Straiker, Expression of G protein-coupled receptors and related proteins in HEK293, AtT20, BV2, and N18 cell lines as revealed by microarray analysis. BMC Genomics 12, 14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.E. V. Gurevich, J. J. G. Tesmer, A. Mushegian, V. V. Gurevich, G protein-coupled receptor kinases: More than just kinases and not only for GPCRs. Pharmacol. Ther. 133, 40–69 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.N. N. Jimenez-Vargas, J. Gong, M. J. Wisdom, D. D. Jensen, R. Latorre, A. Hegron, S. Teng, J. J. DiCello, P. Rajasekhar, N. A. Veldhuis, S. E. Carbone, Y. Yu, C. Lopez-Lopez, J. Jaramillo-Polanco, M. Canals, D. E. Reed, A. E. Lomax, B. L. Schmidt, K. W. Leong, S. J. Vanner, M. L. Halls, N. W. Bunnett, D. P. Poole, Endosomal signaling of delta opioid receptors is an endogenous mechanism and therapeutic target for relief from inflammatory pain. Proc. Natl. Acad. Sci. U.S.A. 117, 15281–15292 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.R. Al-Hasani, M. R. Bruchas, Molecular mechanisms of opioid receptor-dependent signaling and behavior. Anesthesiology 115, 1363–1381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.D. Jullié, C. Benitez, T. A. Knight, M. S. Simic, M. von Zastrow, Endocytic trafficking determines cellular tolerance of presynaptic opioid signaling. eLife 11, e81298 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.M. Stoeber, D. Jullié, J. Li, S. Chakraborty, S. Majumdar, N. A. Lambert, A. Manglik, M. von Zastrow, Agonist-selective recruitment of engineered protein probes and of GRK2 by opioid receptors in living cells. eLife 9, e54208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.D. D. Wiredja, M. Koyutürk, M. R. Chance, The KSEA app: A web-based tool for kinase activity inference from quantitative phosphoproteomics. Bioinformatics 33, 3489–3491 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.M. Mirdita, K. Schütze, Y. Moriwaki, L. Heo, S. Ovchinnikov, M. Steinegger, ColabFold: Making protein folding accessible to all. Nat. Methods 19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.J. Lee, X. Cheng, J. M. Swails, M. S. Yeom, P. K. Eastman, J. A. Lemkul, S. Wei, J. Buckner, J. C. Jeong, Y. Qi, S. Jo, V. S. Pande, D. A. Case, C. L. Brooks 3rd, A. D. MacKerell Jr., J. B. Klauda, W. Im, CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.S. Aureli, D. Di Marino, S. Raniolo, V. Limongelli, DDT—Drug Discovery Tool: A fast and intuitive graphics user interface for docking and molecular dynamics analysis. Bioinformatics 35, 5328–5330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Legends for movies S1 and S2
Legends for tables S1 and S2
Movies S1 to S2
Tables S1 and S2