Abstract
Pre-existing cross-reactive antibodies have been implicated in both protection and pathogenesis during subsequent infections with different dengue virus (DENV) serotypes (DENV1–4). Nonetheless, humoral immune correlates and mechanisms of protection have remained elusive. Using a systems serology approach to evaluate humoral responses, we profiled plasma collected before inapparent or symptomatic secondary DENV3 infection from our pediatric cohort in Nicaragua. Children protected from symptomatic infections had more anti-envelope (E) and anti-nonstructural protein 1 (NS1) total IgG, IgG4, and greater Fc effector functions than those with symptoms. Fc effector functions were also associated with protection from hemorrhagic manifestations in the pre-symptomatic group. Furthermore, in vitro virological assays using these plasma samples revealed that protection mediated by antibody-dependent complement deposition was associated with both lysis of virions and DENV-infected cells. These data suggest that E- and NS1-specific Fc functions may serve as correlates of protection, which can be potentially applied toward the design and evaluation of dengue vaccines.
One-Sentence Summary:
Antibody Fc characteristics and effector functions correlate with protection from symptomatic DENV3 infection.
More than 3.9 billion people worldwide are estimated to live in areas at risk for dengue virus (DENV) infection (1). Four serotypes (DENV1–4) are transmitted through the bite of Aedes mosquitoes, accounting for up to 100 million infections annually, with 50 million being symptomatic and up to 4 million requiring hospitalization (2). Symptomatic infections can present as classical dengue fever or the more severe dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS) (3). Primary DENV infections generate serotype-specific neutralizing antibodies (nAbs) that confer long-term protection against homotypic infections and cross-reactive nAbs that can protect against heterotypic infections(4). However, non-protective cross-reactive antibodies can sensitize individuals to secondary heterotypic symptomatic infection and severe disease via antibody-dependent enhancement (ADE) by mediating infection of Fcγ receptor (FcγR)-bearing myeloid cells (5–7). Further, high-affinity binding of anti-DENV afucosylated IgG1 antibodies to FcγRIIIA has been associated with severe disease outcomes (8–10). As such, anti-DENV antibodies play roles in both protection and pathogenesis.
Our limited understanding of the mechanisms and correlates of protection for dengue disease has hampered the development and assessment of vaccine candidates. Of note, the Sanofi Pasteur Dengvaxia® clinical trials showed that DENV vaccines can be protective in DENV-seropositive recipients but can increase the risk of hospitalization in DENV-seronegative recipients during a subsequent infection (11, 12). Neutralization assays performed on samples from these trials found that the correlation of nAb titers with vaccine efficacy and protection from dengue disease varied by serostatus, DENV serotype, and type-specific versus cross-reactive antibodies(13–16). Given the complex nature and technical challenges of nAb-based assays, additional immune correlates of protection need to be explored. Beyond antigen-specific properties, antibodies can contribute to protection using other mechanisms, including leveraging antiviral activities of the innate immune system through engagement of the antibody constant (Fc) region. Fc effector functions are critical antibody features for protection against disease caused by several viruses, including HIV, influenza virus, Ebola virus, and SARS-CoV-2 (17–20). Although Fc-effector functions have been implicated in enhancing DENV disease (8–10), a role for antibody Fc-mediated mechanisms of protection remains largely unexplored for DENV.
Results
Binding antibody titers are associated with protection from symptomatic DENV infection
We undertook an unbiased, comprehensive, systematic profiling of DENV-specific humoral immune responses to probe for correlates of protection against secondary DENV infection. Antibody-associated correlates of protection were determined in samples selected during resting state months or years after one or more previous DENV infection(s) but before a subsequent heterotypic secondary infection. We assessed a panel of antibody features in plasma samples from previously DENV-infected children (4–13 years old) prior to subsequent DENV3 infection from our long-standing cohort study in Managua, Nicaragua (21) (Fig. 1A and Table 1). The samples were classified into two groups: pre-inapparent infections (children with a >4-fold increase in binding antibody titers in paired annual samples with no reported dengue-related symptoms in the intervening year, n=30) and pre-symptomatic infections (children with laboratory-confirmed dengue cases, n=29). The history of infections was classified as prior primary or secondary infection and was determined based on longitudinal analysis of nAb titers, envelope domain III (EDIII) binding antibodies, epidemiological data, and real-time RT-qPCR and virus isolation of symptomatic infections. For children who enrolled in the cohort study with pre-existing anti-DENV antibody titers (n=13), we assessed their probable history of infection based on an EDIII Luminex assay, as anti-EDIII antibodies have been associated with higher levels of type-specific antibodies and can be useful for determining past infecting serotypes (Table 1, table S1, and data file S1). DENV1, DENV2 and/or DENV4 were identified as the previous infecting serotype(s). A few samples were classified as ‘indeterminate’ due to high cross-reactivity, likely associated with previous secondary infection and high binding antibody titers. Moreover, only two samples were associated with subsequent severe disease (DHF), with the remaining individuals presenting with classical dengue fever (data file S1). Hence, this sample set allowed us to investigate cross-reactive antibody responses against a subsequent heterotypic secondary DENV3 infection to analyze immune correlates associated with inapparent versus symptomatic infection.
Fig. 1. Biophysical and Fc effector functions of anti-DENV polyclonal antibodies.
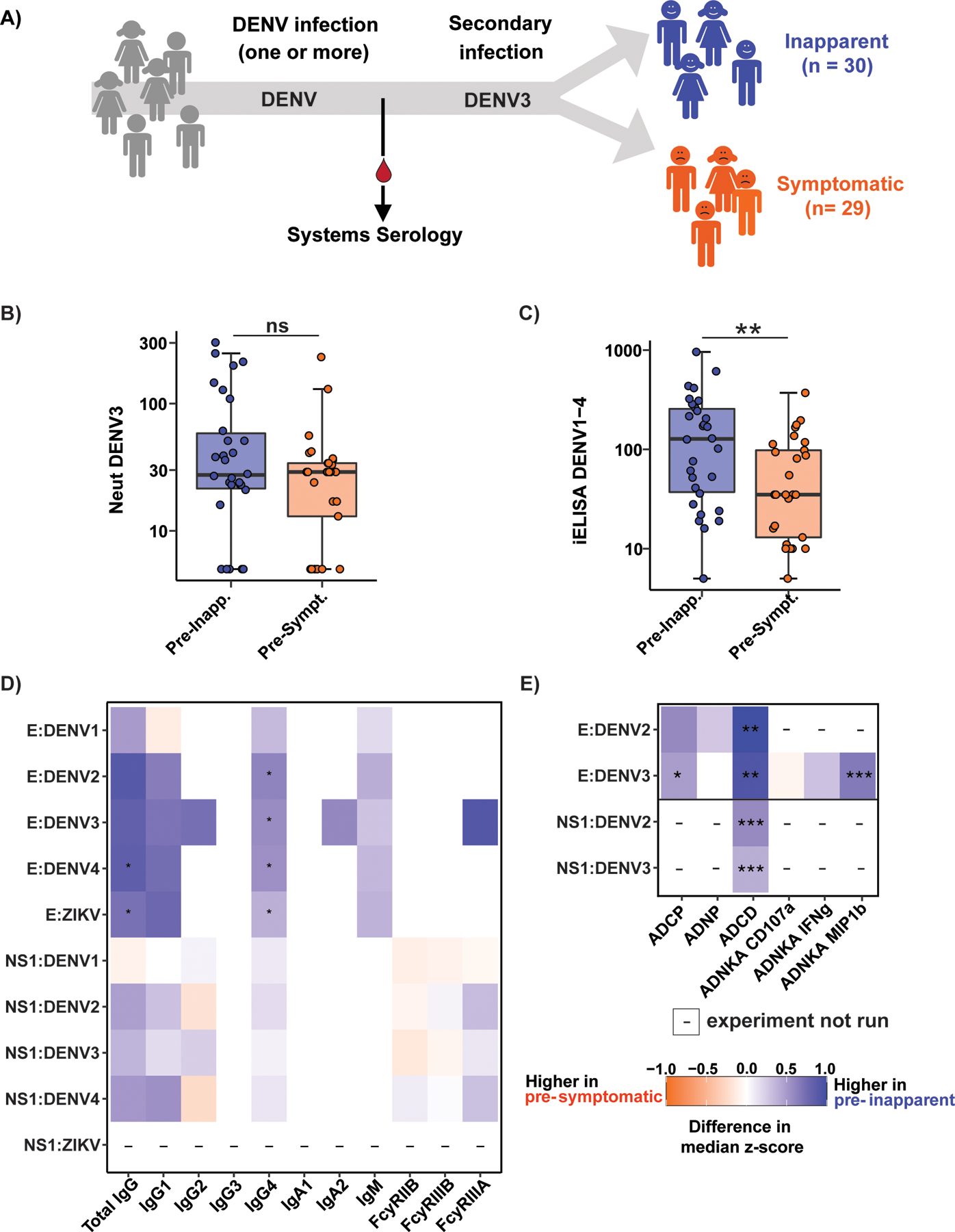
A) Pre- inapparent (blue) and pre-symptomatic (orange) secondary DENV3 infection plasma samples were collected for systems serology. Neutralization (Neut) (B) and inhibition enzyme-linked immunosorbent assay (iELISA) binding antibody titers (C) were determined. (D-E) Heatmap showing the antigen-specific antibody isotype and Fc-receptor binding (D), as well as Fc effector functions (E) of antibodies against envelope (E) and non-structural 1 (NS1) proteins of DENV1–4 and ZIKV. Antibody isotype and Fc receptor binding were measured by a Luminex-based assay and are shown as median fluorescence intensity (MFI). Heatmaps show univariate analysis for biophysical (D) and Fc effector function (E) profiling of polyclonal antibodies against envelope (E) and non-structural 1 (NS1) proteins of DENV1–4 and ZIKV. Fc effector functions investigated were: Antibody-dependent cellular phagocytosis (ADCP), neutrophil phagocytosis (ADNP), complement deposition (ADCD) in the presence of guinea pig complement, and NK cell activation (ADNKA). The values were z-scored across all samples, and the tiles indicate the difference in median z-score within the group. Asterisks indicate Benjamini-Hochberg adjusted p-values for Mann-Whitney U tests (*p<0.05, **p<0.01, ***p<0.001). “-“ indicates that this combination of antigen and antibody feature was not measured. MIP1b: macrophage inflammatory protein 1β; IFNg: Interferon-γ (gamma). B, C, and D were performed in technical duplicates. For effector functions in ‘E’, data represents an average from primary neutrophils or NK cells from two donors (ADNP and ADNKA) or average of technical duplicates (ADCP and ADCD).
Table 1.
Demographics and DENV infection history of pre-secondary heterotypic DENV3 infection cohort samples.
| Male (n) | Female (n) | Age range (y) | Average age (y) | Probable prior primary DENV infection | Probable prior secondary DENV infection | ||
|---|---|---|---|---|---|---|---|
| Pre-inapparent | 21 | 9 | 5–13 | 9.7 | 16 | 14 | |
| Pre-symptomatic | 10 | 19 | 4–13 | 9 | 21 | 8 | |
We compared nAb and inhibition enzyme-linked immunosorbent assay (iELISA) binding antibody(5) titers in samples from pre-inapparent versus pre-symptomatic infection groups (Fig. 1B–C). Anti-DENV3 nAb titers trended higher in pre-inapparent infection samples, though not statistically significant, likely due to the smaller sample size than our earlier study showing nAb titers associated with protection from symptomatic infection (4). However, iELISA binding antibody titers were significantly higher (p=0.0028) in pre-inapparent infection samples (Fig. 1C), consistent with our previous observations (5, 7). These findings suggest that anti-DENV binding antibodies may have functions other than neutralization that contribute to mechanisms of protection. We addressed this possibility by exploring Fc profiles of anti-DENV antibodies (data file S1).
Profiling of antibody Fc biophysical characteristics and effector functions reveals correlates of protection from DENV3 symptomatic infection
We first profiled Fc biophysical properties of antibodies against recombinant envelope (E) protein and nonstructural protein 1 (NS1) of DENV 1–4 and the antigenically related Zika virus (ZIKV). These proteins are highly immunogenic and induce type-specific and cross-reactive antibodies during natural infection and vaccination (21). We assessed antibody isotype titers (IgA1, IgA2, IgM, total IgG, IgG1, IgG2, IgG3, and IgG4) and binding to recombinant FcγRs (FcγRIIB, FcγRIIIA, and FcγRIIIB). Univariate analyses revealed that anti-E total IgG and IgG4 antibodies were significantly enriched in pre-inapparent infection samples, with antibody binding to FcγRIIIA trending towards protection, though not statistically significant (Fig. 1D). DENV and ZIKV E-reactive IgG1 and IgM were also enriched in the pre-inapparent group. No variables were enriched in pre-symptomatic samples. Features that were identified across different DENV serotypes and ZIKV suggest a protective role for cross-reactive antibodies. IgG4 antibodies represent more terminally class-switched isotypes and may represent a more mature humoral immune response, induced after extended antigen-exposure, late in the process of antibody affinity maturation (22–24). High-affinity IgG4 has diminshed Fc functionality, and IgM does not bind FcγR. Both IgG4 and IgM may compete with IgG1 for antigenic binding sites, thus tempering binding to FcγRs involved in potential enhancement (22, 25). These data suggest that pre-existing total IgG and IgG4 antibodies, and the ability of Abs to bind FcγRs may be key mechanisms of protection preventing symptomatic secondary DENV infection.
We next aimed to deconvolute the specific Fc functions that contribute to protection in secondary DENV3 infection. Antibody functional profiling against DENV E proteins revealed higher levels in pre-inapparent infection samples of antibody-dependent cellular phagocytosis (ADCP) and antibody-dependent NK cell activation (ADNKA) specifically able to induce chemokine (macrophage inflammatory protein 1β, MIP-1β) secretion (Fig. 1E). In support of this, another study using pre-infection samples found that NK-dependent antibody-dependent cellular cytoxicity correlated with low viremia in DENV3 secondary infections (26). In addition, antibody-dependent complement deposition (ADCD) of guinea pig complement against E and NS1 proteins was higher in pre-inapparent than pre-symptomatic infection samples (Fig. 1E). Together these data point to the presence of an expanded polyfunctional DENV-specific humoral immune response, including phagocytic, NK cell-activating, and complement-depositing antibodies, that may represent possible Fc-mediated mechanisms of protection from symptomatic DENV infections.
Multivariate analysis suggests possible antibody-mediated mechanisms of protection
We next aimed to define whether a minimal multivariate humoral signature may exist that could provide further mechanistic insights into correlates of protection. To define antibody features that were uniquely enriched and most distinct across the pre-symptomatic and pre-inapparent infection groups, we used least absolute shrinkage and selection operator (LASSO) coupled to a partial least squares discriminant analysis (PLSDA) (Fig. 2A–C). LASSO is a regression technique that was used to select the minimal set of features that best discriminate pre-inapparent from pre-symptomatic infection samples. PLSDA is a supervised dimensionality-reduction technique used to then visualize the multivariate data in two-dimensional space. We first used LASSO to select the set of features that best contributed to the model and then used PLSDA to visualize the differences between pre-inapparent and pre-symptomatic infection samples with the LASSO-selected features. The LASSO/PLSDA showed a robust separation between pre-inapparent and pre-symptomatic individuals (10-fold cross-validation accuracy = 0.69) and outperformed models built using random features or permuted labels (Fig. 2D). As few as 5 of the total 65 antibody features were sufficient to discriminate between pre-inapparent and pre-symptomatic infections. The 5 discriminatory features pointed to two potential modes of protection, one involving anti-E antibodies, characterized by high levels of IgG4, ADCD and all Fc effector functions (polyfunctionality), and the other involving anti-NS1 antibodies, marked by total IgG and ADCD (Fig 2A–E).
Fig. 2. Antibody features differentiate pre-inapparent and pre-symptomatic DENV3 infections.
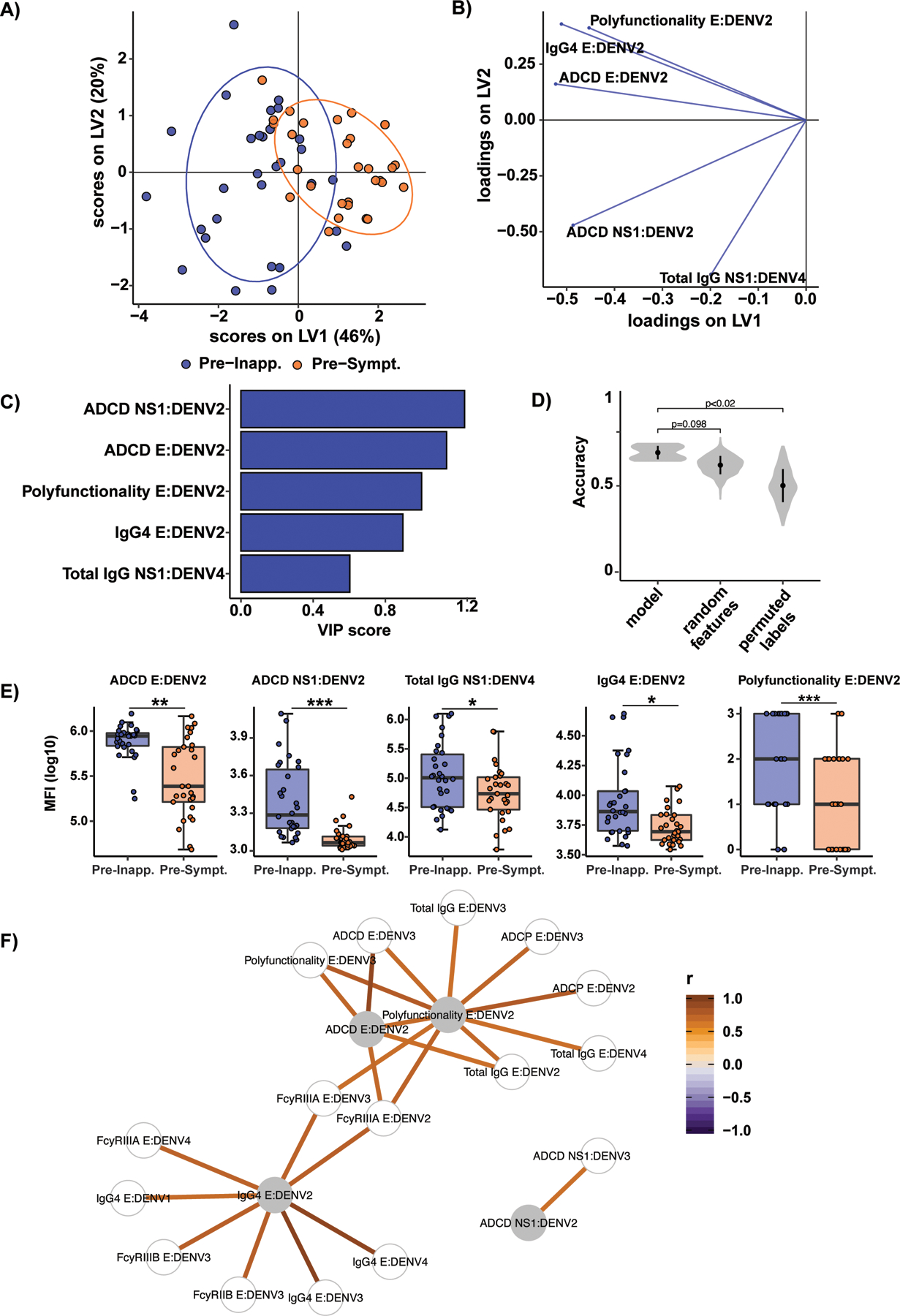
A) All 65 antibody features were analyzed for each pre-inapparent (blue dots) and pre-symptomatic (orange dots) subjects using partial least squares discriminant analysis (PLSDA). PLSDA score plot shows latent variable (LV) 1 and 2. LV2 was only calculated for visualization purposes and was not significant. Ellipses show 80% confidence regions assuming a multivariate t distribution. B) Corresponding loading plot. Each line represents one of the least absolute shrinkage and selection operator (LASSO)-selected features. The loading plots show the influence of features on latent variables. C) Bar graph showing the variable importance in projection (VIP) score of each LASSO-selected feature from the PLSDA in A. Blue bars indicate the pre-inapparent infection group in which the features are enriched, i.e. higher median values. D) Accuracies from 10 repetitions of 10-fold cross-validation for the actual model and models based on size-matched random features and shuffled labels. Random feature and permuted labels models are repeated 50 times for each cross-validation; p-values are determined based on the probability that the accuracy of the permuted and random feature model is higher than for the actual model. E) Whisker boxplots for LASSO-selected features. Asterisks indicate Benjamini-Hochberg adjusted p-values for Mann-Whitney U tests (*p<0.05, **p<0.01, ***p<0.001). F) Co-correlate network illustrating features significantly correlated with LASSO-selected features (gray nodes) with a Spearman correlation coefficient (|r|>0.65). Line color indicates the correlation coefficient. ADCP: Antibody-dependent cellular phagocytosis; ADCD: Antibody-dependent complement deposition; E: envelope protein; NS1: non-structural protein 1; MFI: Mean fluorescence intensity.
Since LASSO selects a subset of significant features to prevent overfitting, we built a correlation network to assess how other Fc features correlate with LASSO-selected features (Fig. 2F and fig. S1). We found that two main correlation networks were formed based on responses against the E or NS1 proteins (Fig. 2F). Several anti-E antibody features highly correlated with one another, whereas anti-NS1 antibodies mediating ADCD formed a separate cluster. Additional correlation matrices built using either all DENV2 or DENV3 E antigens showed that neutralization did not strongly correlate with other antibody features (fig. S1A and B). However, iELISA binding titers correlated with ADCD, among other features, suggesting possible Fc functions for binding antibodies.
To gain further mechanistic insight by which anti-E and anti-NS1 antibodies may contribute to protection, we built two additional LASSO/PLSDA models, one with only anti-E features and one with only anti-NS1 features (fig. S2). Both models showed separation between pre-inapparent and pre-symptomatic infection samples, suggesting that combinations of key functions may be critical for protective antibodies. Analysis of E-specific antibody features reinforced the important role of ADCD in pre-inapparent infections and retained IgG4, polyfunctionality, and ADNKA as key discriminants of infection outcome, while additionally including IgM among the top five variables of importance (fig. S2A and B). For the NS1-specific model, the top variables of importance did not change, with DENV3 NS1 ADCD and DENV2 NS1 IgG levels enriched in pre-inapparent individuals, pointing to a role for cross-reactive ADCD in rapid control of viral infection (Fig. 2C and D). This analysis reveals that there are two routes of protection against symptomatic DENV3 infection, by inducing polyfunctional IgG against E or ADCD against NS1.
Moreover, we analyzed possible correlations between antibody features and symptoms within the pre-symptomatic group (fig. S3 and data file S1). The development of hemorrhagic manifestations produced the most significant model (p<0.02) with a LASSO-PLSDA analysis revealing that four antibody features, namely ADCD E:DENV3, Total IgG NS1:DENV1, ADCP E:DENV3 and ADNKA IFNg E:DENV3, were able to separate symptomatic cases who experienced hemorrhagic manifestations and those who did not (fig. S3A and B). All LASSO-selected features, except for ADNKA IFNg E:DENV3, were elevated in the group that did not experience hemorrhagic manifestations (fig. S3C and D), suggesting that opsonophagocytic functions are vital for protection against hemorrhagic manifestations. Furthermore, a correlation network of additional features associated with the LASSO-selected features revealed a linked selective enrichment of ADCP E:DENV3, FcγRIIIA and IgG subclasses linked to DENV3-ADCP (fig. S3E). This analysis highlights that antibodies that induce effector functions not only protect against symptomatic dengue but also the development of more hemorrhagic manifestations.
Antibodies may mediate protection via complement deposition leading to lysis of DENV virions and infected cells
Given that E- and NS1-specific polyclonal antibodies suggested protective roles for ADCD and that the complement cascade has been associated with both protection and immunopathology of dengue disease (27–29), we next sought to characterize possible mechanisms by which complement deposition could confer protection. We confirmed that complement deposition in the ADCD assay was associated with fresh exogenous serum as a source of complement but not with exogenous heat-inactivated serum so as to rule out possible intrinsic complement activation derived from the plasma samples (fig. S4). We then performed a series of in vitro virological assays to determine whether complement can increase neutralization potency or enable lysis of virions and/or DENV-infected cells. A standard Focus Reduction Neutralization Test (FRNT) on Vero cells in a subset of samples revealed increased neutralization titers in the presence of fresh versus heat-inactivated exogenous complement, as expected (fig. S5). However, we found no significant differences in FRNT titers between pre-inapparent and pre-symptomatic infection groups when assessing the full sample set under these conditions. This may be due to specific assay conditions, and future studies can explore key variables, such as virion maturation state and cell substrate.
We assessed whether pre-inapparent and pre-symptomatic infection plasma samples could differentially contribute to virion lysis and associated protection. We incubated DENV3 virions with plasma samples, fresh guinea pig complement and RNase A, then determined the ability of RNase A to access and degrade the viral genomic RNA upon virion lysis. The majority of plasma samples from both groups triggered some degree of virion lysis, but we found pre-inapparent infection samples contained higher virion lysis activity (Fig. 3A and B), which was identified after performing a serial dilution of the plasma samples (fig. S6A–F). Furthermore, we showed that the virolysis assay functions in the presence of human complement and is dependent on C1q, C3, and C5 (fig. S6G). As NS1 and structural proteins prM and E can be detected on the surface of DENV- and other flavivirus-infected cells(30–32), we asked whether complement could accumulate on the membranes of infected cells, leading to membrane permeability due to pore formation. We incubated plasma from both sample groups plus exogenous complement with BHK-21 cells that were previously infected with DENV3 or mock-infected and measured the release of lactate dehydrogenase (LDH) in cell supernatants as a measure of antibody-dependent complement-mediated lysis (ADCML). We found that DENV3-infected BHK-21 cells, but not mock-infected cells, released higher amounts of LDH in the presence of pre-inapparent infection plasma (Fig. 3C). In agreement, previous studies have associated protective mechanisms with complement-mediated lysis of flavivirus-infected cells for Japanese encephalitis, West Nile, and yellow fever viruses (33–35). Altogether, these assays indicate that anti-E and anti-NS1 polyclonal antibodies can lead to complement deposition on the surface of virions and infected cells, and this mechanism may contribute to protection from symptomatic infections.
Fig. 3. Complement is associated with protection through lysis of virions and infected cells.
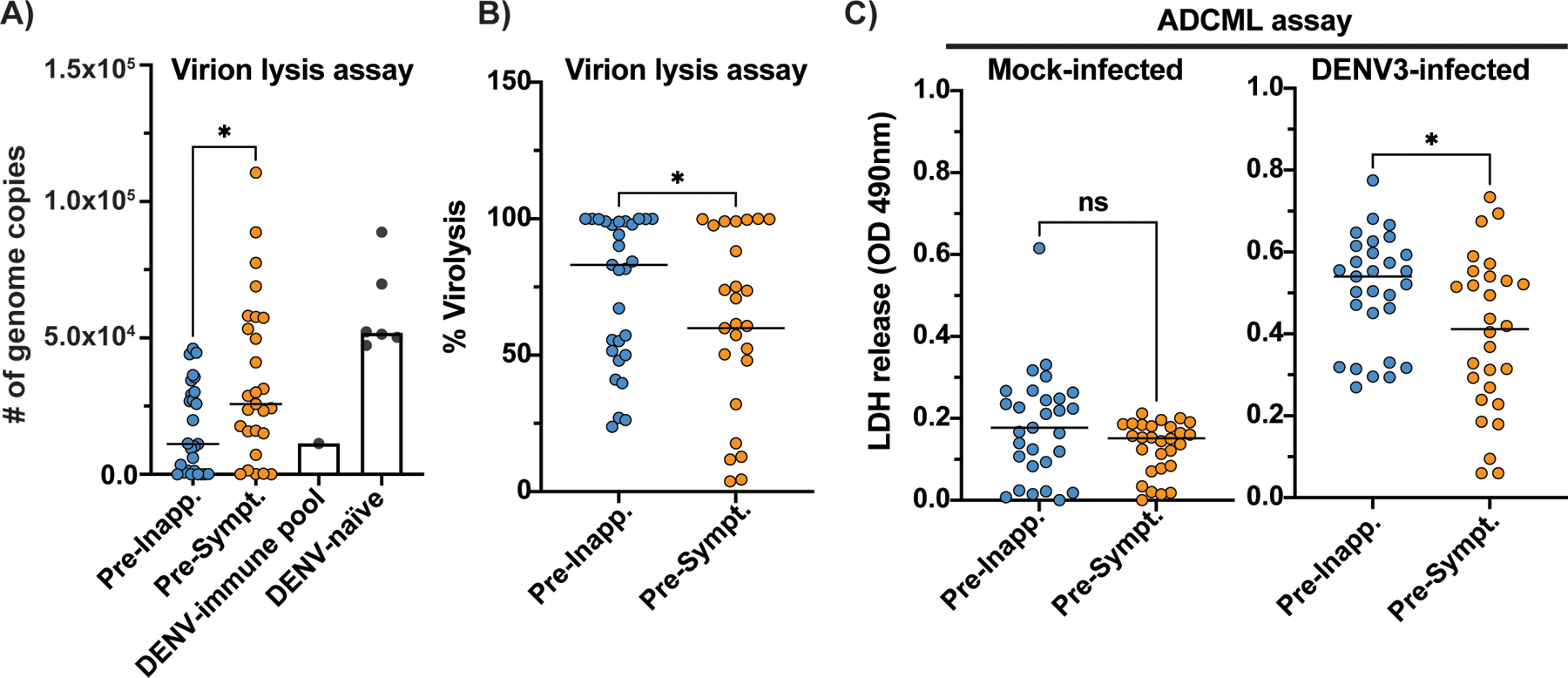
Pre-inapparent (blue dots) and pre-symptomatic (orange) secondary infection plasma samples were incubated with DENV3 virions in the presence of fresh guinea pig complement and RNase A (A-B). Virion lysis was determined by measuring viral RNA of intact virions by RT-qPCR (# of genome copies) and calculated as percentage (%) of lysis based on the average of five DENV-naïve plasma samples and one medium-only (no plasma) sample (DENV-naïve). A pool of DENV-immune plasma samples was used as positive control (DENV-immune pool). C) Antibody-dependent complement-mediated lysis (ADCML) of BHK-21 cells was detected by measuring the optical density (OD) values associated with lactacte dehydrogenase (LDH) release in the supernants of mock- or DENV3-infected cells incubated with the plasma samples and fresh exogenous human complement. Non-parametric Mann-Whitney t test, *p<0.05; ns, non-significant. Data represents average of experiments done in technical triplicates.
Considering that domains and antigenic sites within the E and NS1 proteins could differentially facilitate complement activation and deposition, we investigated which domains were associated with complement-associated protection. Regarding the E protein, our analysis focused on domain III, as recombinant proteins of this domain have been shown to fold correctly and contain antigenic sites that are accessible on the virion (36). We found that complement deposition in response to antibodies targeting EDIII was higher in pre-inapparent infection samples (Fig. 4A and B). This observation suggests a complement-mediated protective role for anti-EDIII antibodies without excluding the possibility that anti-EDI and -EDII antibodies may also be associated with protection via complement deposition or other effector functions; these domains were not available for testing here. ADCD activity against both full-length NS1 and the wing domain, but not the ß-ladder domain, was higher in plasma from children protected from symptomatic DENV3 infections (Fig. 4C and D). These findings point to particular targets of complement-activating polyclonal antibodies involved in driving protection from symptomatic infection.
Fig. 4. Domains of DENV envelope (E) protein and non-structural protein 1 (NS1) are associated with antibody-dependent complement deposition (ADCD).
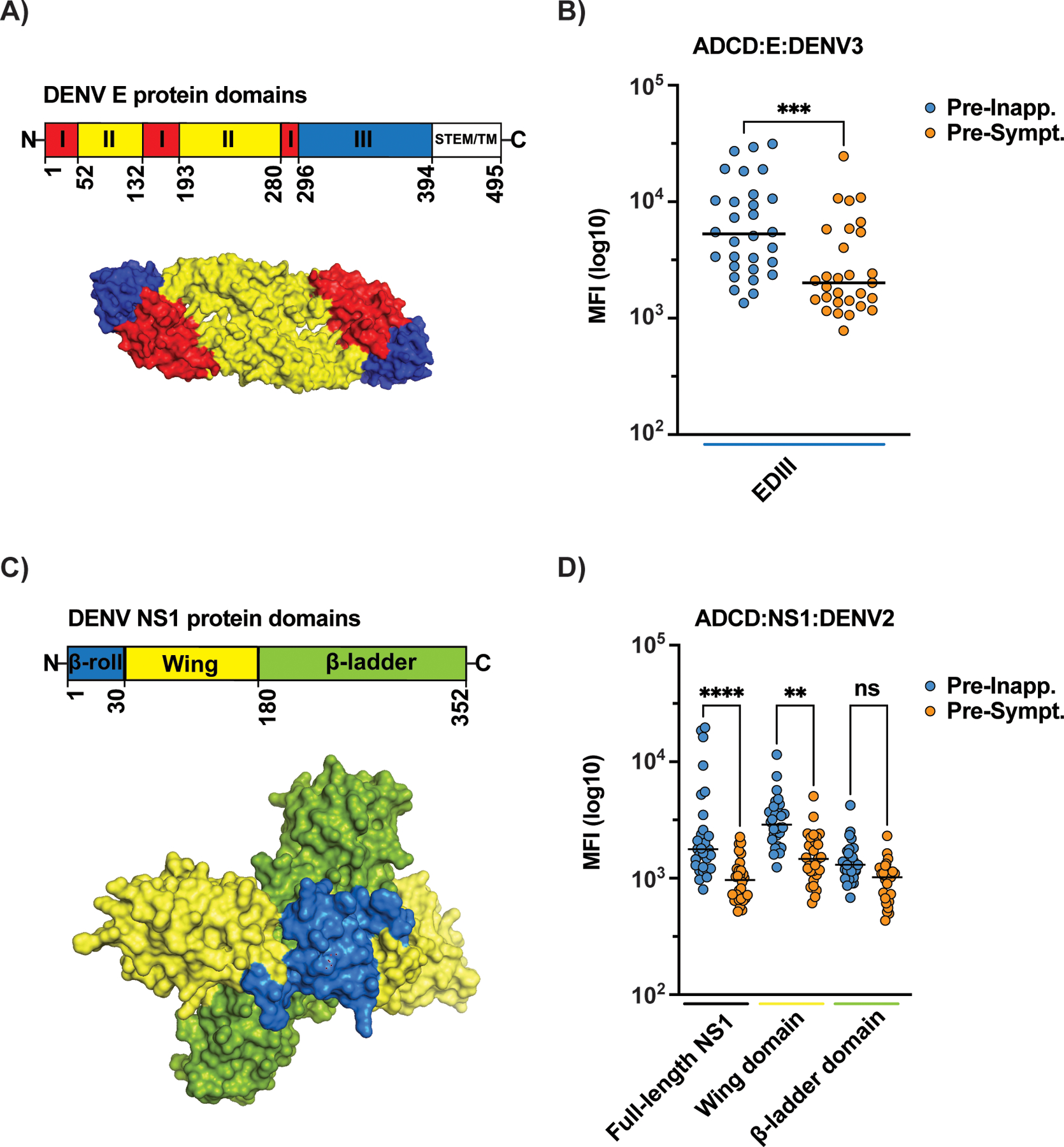
Recombinant DENV3 E domain III (EDIII) (A-B) or full-length NS1 and its wing and ß-ladder domains (C-D) were used to assess levels of ADCD in the presence of guinea pig complement with the pre-inapparent (blue dots) and pre-symptomatic (orange dots) infection plasma samples. B) Non-parametric Mann-Whitney t test was used for statistical analysis: ***p<0.001; ns, non-significant. D) One-way ANOVA was used for multiple comparisons among all groups. Relevant comparisons are shown. ****p<0.0001; **p<0.01; ns, non-significant. Structural schematics in A (E protein – PDB:1OAN) and C (NS1 protein – PBD: 4O6B) are shown as dimers. Data represents average of experiments done in technical duplicates.
Discussion
DENV vaccine development has experienced dramatic set-backs over the past two decades, with vaccine failures due to our incomplete understanding of the immunologic correlates of protection(37). Pre-infection nAb titers, as well as high levels of binding antibody titers, have been associated with protection from secondary symptomatic infections; however, nAbs are relative correlates that do not predict protection in all instances (4, 13–16). Here, we found that individuals presenting with higher iELISA binding antibody titers and specific Fc biophysical characteristics and effector functions had a striking association with protection, whereas nAb titers only trended towards protection in this sample set. Antibody functions may be associated with multiple antibody properties. For example, complement activation and deposition can be associated with levels of total IgG, IgG1, and IgM, as well as antibody Fc glycosylation status. Taking into account both binding antibody titers and functionality, our multivariate analysis (Fig. 2 and fig. S2) selected functions (ADCD, ADCP, and ADNKA) as key discriminators of children with secondary DENV3 inapparent versus symptomatic infections, highlighting the greater difference in antibody quality, over quantity, in immunity to DENV. Thus, while titers can augment antibody functions, titers were not the most significant predictors of protection, especially when analyzed alongside and compared to other antibody variables. Moreover, the correlation network on LASSO-selected features included multiple functional and Fc receptor binding humoral immune responses, as well as IgG titers, clearly linking titers and a highly functional humoral immune response as a unique antibody profile present in children who experience inapparent infection when exposed to another DENV serotype. Further studies are required to define the specific antibody subpopulations involved in driving complement activation and other Fc effector functions. Altogether, it is possible that nAbs, binding antibody titers and Fc effector functions may have distinct and/or complementary roles in limiting virus infection and replication. Given the variability of neutralization assays, key variables need to be taken into account when attempting to define correlates of protection, including virion maturation state and cell substrate (expressing different FcγRs, complement receptors, and/or DC-SIGN), as well as depletion of type-specific or cross-reactive antibodies (38). These variables can be assessed in future studies using samples derived from cohort and vaccine studies.
Our unbiased approach identified two potential routes for protection against symptomatic DENV infection involving Fc biophysical and functional features of anti-E and anti-NS1 antibodies. While previous studies showed that high affinity antibody binding to FcγRIIIA and ADE were associated with DHF/DSS during secondary infection, analysis of antibody profiles prior to secondary infection here demonstrated that a distinct set of antibody characteristics are predictors of protection against symptomatic DENV infection. This protective signature may reflect the presence of functional antibodies prior to infection capable of recognizing and rapidly eliminating the virus upon infection. A strength of our approach is the identification of combinations of antibody features that may work in cooperation. As an example, IgM antibodies are known to deposit complement and mediate viral lysis most efficiently compared to other antibody isotypes (39), and this pre-infection Fc feature was identified in combination with ADCD in the top discriminatory features. In contrast, individuals with low or less functional antibodies may instead develop a highly inflammatory response following infection that may result in pathology. Within this context, we showed that pre-infection human antibodies able to mediate complement deposition, phagocytosis and NK cell activation were associated with protection from symptomatic infections and that complement deposition leads to lysis of virions and DENV-infected cells, which may contribute to virus clearance.
Complement in dengue disease has been associated with both protection and immunopathology (27, 28, 40). Most studies suggesting a detrimental role for complement were investigated in post-infection samples in the context of DHF/DSS (28, 41, 42). These studies report the detection of complement in autopsy samples and activation of complement components during DSS. In our study, we show that anti-E and anti-NS1 antibodies promoting complement deposition are correlated with protection from symptomatic secondary DENV3 infections in a pediatric cohort. In line with this finding, complement has been associated with protection in flavivirus infections in vitro and in animal studies, including by preventing ADE, improving neutralization potential, and increasing IgG avidity (27, 43–45). Although the alternative and lectin pathways have also been implicated in protection or immunopathogenesis in dengue disease (46–49), we show here the protective role of the classical complement cascade.
The main limitations of our study were related to sample size and volume. We initially selected well-characterized pre-DENV3 infection samples for which information on infection history was available through characterization using iELISA binding antibody titers and neutralization assays. Future studies can use larger sample sets, as well as pre-secondary infection samples for other DENV serotypes to determine whether correlates of protection may vary according to the incoming serotypes. The limited sample volume from our pediatric subjects did not permit depletion assays (e.g., to determine a possible mechanistic role for IgG4 antibodies) and virological assays to validate other Fc effector functions (e.g., NK cell activation and phagocytosis), which are warranted when larger sample volumes are available.
Defining sites of virus vulnerability within particular domains of viral proteins can direct the rational design of vaccines and antibody-based therapeutic candidates. Along these lines, we find that ADCD activity is associated with specific domains of both E and NS1 proteins. Overall, identification of alternative correlates of protection is urgently needed to guide the development and evaluation of current and next-generation vaccines. This systematic approach also provides potential mechanistic insights into the nature of protective antibody responses in DENV infection.
Materials & Methods
Study Design
To investigate the correlates of protection associated with a secondary natural DENV3 infection, we selected 30 pre-inapparent and 29 pre-symptomatic infection plasma samples that were collected before a DENV3 epidemic in Managua, Nicaragua (2009–2011). The criteria for sample selection were an annual sample from a child post-primary or post-secondary DENV infection collected prior to a subsequent heterotypic DENV3 infection. Hence, individuals had been exposed in the past to other DENV serotypes, but not to DENV3, and samples were collected at a time-point that is months or years after a previous infection, as well as months or years before a subsequent infection. Samples were not randomized nor blinded but selected based on sample characteristics, taking into account infection history, epidemiological data, clinical features, diagnostic information, and neutralization and iELISA binding titers, as outlined below. All samples were analyzed at least in duplicate for all assays. All data generated have been made available in the Supplementary Materials (data file S1).
The The Pediatric Dengue Cohort Study (PDCS) was reviewed and approved by the institutional review boards of the University of California, Berkeley (protocol: 2010–09-2245), the University of Michigan (study ID: HUM00091606), and the Nicaraguan Ministry of Health (protocol NIC-MINSA/CNDR CIRE-09/03/07–008). Parents or legal guardians of all subjects provided written informed consent, and subjects ≥6 years old provide assent.
The Pediatric Dengue Cohort Study (PDCS) and Sample Characterization
The PDCS in Managua, Nicaragua, is based at the Health Center Sócrates Flores Vivas (HCSFV), the primary health care facility for District II of Managua. The cohort has followed ~3,800 children aged 2–14 years since 2004 (50). Each year, ~300 children aged 2 years old as well as additional children 3–11 years old are invited to participate to maintain the balanced age structure of the cohort. For this study, children who presented with fever were screened for signs and symptoms of dengue. Cases were classified as: A) 1997 and/or 2009 WHO criteria for dengue; B) undifferentiated fever; C) acute febrile illness diagnosed as another disease based on symptoms; or D) other non-febrile conditions. All cases meeting definitions A and B were evaluated for acute DENV infection, and at each medical appointment, clinical and laboratory information were collected (n=189 variables). Acute blood samples were collected at first contact with the patient (>90% within the first 72 hours post-illness onset), and a convalescent sample was collected 14–21 days post-illness onset for >95% of cases. All diagnostic tests were developed and performed at the National Virology Laboratory in the Centro Nacional de Diagnóstico y Referencia (CNDR). Acute samples were tested for DENV by RT-PCR, and virus isolation in C6/36 cells was performed on RT-PCR-positive samples (51). Paired acute/convalescent samples were tested for IgM seroconversion using a DENV in-house IgM capture (MAC)-enzyme-linked immunosorbent assay (ELISA) (51, 52) and for a 4-fold or greater increase in DENV-specific antibody titer between acute and convalescent sera using the DENV inhibition ELISA (iELISA) (5, 53, 54). The iELISA is a competition assay that detects heterotypic antibody titers binding to DENV1–4 antigens (5).
Each year in July, a healthy annual blood sample was collected from all participants. Samples were prepared either as serum or, for a subset of participants, as plasma/peripheral blood mononuclear cells, and paired annual samples were tested for total anti-DENV antibodies by DENV iELISA. Symptomatic infections were defined as laboratory-confirmed cases (see above), while inapparent infections were defined as children who did not report symptoms but presented with a ≥4-fold increase in iELISA binding antibody titers in paired annual samples. In a subset of samples, neutralizing antibody titers were measured using DENV1–4 reporter virus particles in a flow cytometry-based neutralization assay using human Raji-DC-SIGNR cells (4, 55). To select pre-inapparent DENV3 infection samples for this study, we also analyzed the neutralization titers against all four DENV serotypes to confirm a >4-fold increase in titers against DENV3 in paired annual samples. A primary infection represents children who enrolled in the cohort without DENV iELISA binding titers but experienced a laboratory-confirmed DENV infection or seroconverted in intervening years. Secondary infections represent children who entered the cohort DENV-naïve and experienced two or more documented infections or children who entered the cohort with previous DENV iELISA binding titers and experienced new infections after enrollment.
Viral antigens
Recombinant envelope (E) and nonstructural protein 1 (NS1) from DENV1–4 and ZIKV were commercially acquired from Meridian Life Sciences (USA) and Native Antigen (UK), respectively, and used for biophysical and Fc effector function assays. The recombinant domain III of DENV1, 2, 3, 4 and ZIKV E protein (EDIII) were site-specifically biotinylated and provided as a gift by Dr. Aravinda de Silva and Dr. Premkumar Lakshmanane (University of North Carolina, Chapel Hill, USA). The full-length recombinant NS1 and its wing and ß-ladder domains from DENV2 (Thai strain 16681) used in Fig. 4D were a kind gift from Drs. Janet Smith and W. Clay Brown (University of Michigan, USA) (56). In Fig. 4A and C, structural models were generated using the PyMol software with E (PDB: 1OAN) and NS1 (PDB: 4O6B) amino acid sequences.
EDIII Serotyping
In addition to the methods above (annual iELISA binding antibody titers and neutralization titers), the serotype of previous infections was assessed using a multiplexed Luminex assay. Site-specific biotinylated EDIII from DENV1, 2, 3, 4 and ZIKV were conjugated to avidin-coated Magplex Luminex beads. Immune complexes were formed in 384-well plates by mixing appropriately diluted plasma (six four-fold dilutions) with antigen-coupled microspheres for 90 minutes at 37°C, shaking at 1200 rpm. After incubation, plates were washed using an automatic magnetic washer (Tecan Hydrospeed) with PBS containing 0.1% BSA and 0.02% Tween-20. Antigen-specific antibody binding was detected using PE-coupled mouse anti-human detection antibodies against total IgG (Southern Biotech). Fluorescence was acquired using an iQue3 (Intellicyt) machine.
Antibody isotype and Fcγ receptor binding
Antibody isotype and Fcγ receptor (FcγR) binding was determined using a multiplexed Luminex assay, as previously described (57). Briefly, antigens were covalently coupled to carboxyl-modified Magplex Luminex beads via covalent N-hydroxysuccinimide (NHS)–ester linkages utilizing EDC [1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride] (Thermo Scientific) and sulfo-NHS (Thermo Scientific) according to the manufacturer’s instructions. Immune complexes were formed by mixing appropriately diluted plasma with antigen-coupled microspheres, and 384-well plates were incubated overnight at 4°C, shaking at 700 rpm. After incubation, plates were washed with 0.1% BSA, 0.02% Tween-20. Antigen-specific isotype binding was detected using PE-coupled mouse anti-human detection antibodies against IgA1 (Southern Bio #9130–09), IgA2 (Southern Bio #9140–09), IgM (Southern Bio #9020–09), total IgG (Southern Bio #9040–09), IgG1 (Southern Bio #9052–09), IgG2 (Southern Bio #9060–09), IgG3 (Southern Bio #9210–09), and IgG4 (Southern Bio #9200–09). For FcγR-binding, FcγRs with an AviTag (Duke Human Vaccine Institute) were biotinylated using BirA500 kit (Avidity) according to the manufacturer’s instructions. Biotinylated FcγRs were fluorescently labeled using R-streptavidin-PE (Agilent) and added to immune complexes. Fluorescence was acquired using an iQue3 (Intellicyt) machine. Antigen-specific antibody isotype and FcγR-binding is reported as Median Fluorescence Intensity (MFI). This experiment was run in duplicate, and the results reflect the average of the two replicates.
Antibody Fc functional assays
Antibody-dependent cellular phagocytosis (ADCP), antibody-dependent neutrophil phagocytosis (ADNP), antibody-dependent complement deposition (ADCD), and antibody-dependent NK cell activation (ADNKA) were performed as previously described (58–61). For ADCP, ADNP, and ADCD, antigen was biotinylated and coupled to yellow-green or red (ADCD) neutravidin beads (Invitrogen). Immune complexes were formed by mixing coupled beads and appropriately diluted plasma and incubating for 2 hours at 37°C. For ADCP, human monocytic THP-1 cells (ATCC: TIB-202) were added to immune complexes at a concentration of 1.25×105 cells/mL, and cells and immune complexes were incubated overnight at 37°C. For ADNP, white blood cells were isolated from fresh peripheral blood from healthy donors by performing ammonium-chloride-potassium lysis of red blood cells. The isolated white blood cells were then added to 96-well plates at a concentration of 2.5×105 cells/mL, and neutrophils were identified using anti-CD66b PacBlue (Biolegend 305112). For ADCD, lyophilized low-toxicity guinea pig complement (Cedarlane) diluted in gelatin veronal buffer with calcium and magnesium (GVB++, Sigma) was added to immune complexes and incubated for 20 minutes at 37°C. Fluorescein-conjugated goat IgG anti-guinea pig complement C3 detection antibody (MP Biomedicals #MP0855385) was used to detect complement deposition.
For ADNKA, ELISA plates were coated with antigen and blocked in BSA. Diluted plasma was added to the ELISA plates to form immune complexes and incubated for 2 hours at 37°C. NK cells were isolated from fresh peripheral blood (MGH Blood Bank) from two healthy donors by addition of RosetteSep (Stem Cell Technologies) to the blood, followed by Ficoll separation. NK cells were isolated on the day of the assay. Plates were washed with PBS, and NK cells were added at a concentration of 2.5×105 cells/mL in media supplemented with GolgiStop (BD), Brefeldin A (BFA, Sigma Aldrich) and anti-CD107a PE-Cy5 (BD #555802). Plates were incubated with NK cells for 5 hours at 37°C. NK cells were then fixed using Perm A (Life Tech) and stained for surface markers using anti-CD3 PacBlue (BD #558117), anti-CD16 APC-Cy7 (BD #557758), and anti-CD56 PE-Cy7 (BD #557747). Cells were then permeabilized with Perm B (Life Tech) and stained with anti-MIP-1b PE (BD #550078) and anti-IFN-γ FITC (BD #340449).
Fluorescence was acquired using an iQue3 machine. For ADCP and ADNP, a phagocytic score was calculated using the following formula: (percentage of bead-positive cells)×(geometric mean of MFI of bead-positive cells)/10,000. ADCD activity is reported as the MFI of C3 deposition. For ADNKA, NK cells were gated as CD56+CD16+CD3−, and activity was defined as the percent of NK cells positive for CD107a, IFN-γ or MIP-1b. ADNP and ADNKA were run using fresh peripheral blood from two healthy donors. ADCP and ADCD were run in duplicate. The data presented reflects the average of the biological (ADNP and ADNKA) or technical (ADCP and ADCD) replicates.
Virological assays in the presence of complement
For the virological assays, viral stocks of two strains of DENV3 were prepared in the mosquito C6/36 cell line. One strain corresponds to an infectious clone of DENV3 (UNC3001) (62) and was a kind gift from Dr. Ralph Baric (UNC, USA). The other strain, DENV3 6629.10a1SPD3, is a Nicaraguan clinical isolate and was maintained at low passage (third passage). Virus stocks were titrated on green monkey kidney Vero cells (ATCC: CCL-81) and expressed as plaque-forming units per ml (PFU/ml). In some experiments, guinea pig or human sera, as a source of complement, were used either fresh or heat-inactivated (HI) at 56°C for 30 minutes.
Focus reduction neutralization test (FRNT) was performed using Vero cells. Plasma samples were serially diluted at least seven times at 1:3 dilutions. Diluted plasmas were mixed with the Nicaraguan DENV3 clinical isolate 6629, 10% fresh or HI human serum complement (Complement sera human, Sigma) and pre-incubated at 37°C for 30 min. These mixtures were then allowed to adsorb onto Vero cells at 37°C for 1 hour. Cell monolayers were washed once with PBS and incubated at 37°C for 48 hours in the presence of overlay medium (0.8% carboxymethylcellulose [Sigma] and 5% fetal bovine serum containing DMEM [Gibco]). Cells were then fixed with 4% methanol-free formaldehyde (Thermo Fisher), permeabilized with 0.1% Triton X-100 (Sigma), incubated with the anti-DENV envelope protein mAb 4G2 (produced in-house) followed by incubation with a horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG mAb (Biolegend #9–3009). Foci were developed with KPL TrueBlue® peroxidase substrate (SeraCare) and counted with the assistance of an ImmunoSpot® Analyzer (Cellular Technology Limited, CTL). Relative infection was determined using the last plasma dilution as the maximum amount of virus for each sample and used to calculate FRNT50 titers. The plasma dilutions were transformed to log10 and plotted against relative infection values to fit a sigmoidal dose-response curve. Quality control measures were applied considering the absolute sum of squares (<0.2) and the coefficient of determination (R2) of the non-linear regression (>0.9). FRNT50 titers below 10 were considered non-neutralizing and were set to 5.
The virion lysis assay was performed as previously described (63). Briefly, plasma samples were diluted and used at final concentrations of 20, 4, or 1.3% and mixed with fresh guinea pig complement (resuspended in 1ml of GVB++, Sigma; final concentration of 50%), DENV3 strain 6629 virions, and RNase A (Invitrogen; 0.2mg/ml final concentration) in RPMI 1640 (Gibco). Mixtures were incubated at 37°C for 3 hours and frozen overnight at −20°C. Samples were then thawed, mixed with RNase A (0.77mg/ml final concentration) and incubated at 37°C for 1 hour. Reactions were terminated by incubating with Proteinase K (0.71mg/ml final concentration) at 50°C for 1 hour. RNA was extracted and purified using the RNeasy® Mini Kit (Qiagen). Absolute quantitation of viral RNA was determined by quantitative RT-PCR (RT-qPCR) using the following primer and probe sequences: Forward (GGACTGGATACACGCACCCA), Reverse (CATGTCTCTACCTTCTCGACTTGTCT), and Probe (FAM-ACCTGGATGTCGGCTGAAGGAGCTTG) (64). The Verso 1-step RT-qPCR kit (Thermo Fisher) was used following the manufacturer’s instructions. Standard curves were prepared using seven 10-fold dilutions of a DENV3 UNC3009 in vitro-transcribed RNA. Reaction mixtures were run in a QuantiStudio 3 thermocycler (Applied Biosystems), and threshold cycle (Ct) values were used for analyses. Percentages of virion lysis were calculated using the average Ct of five dengue-naïve plasma samples that were used as negative controls. Plasma samples derived from several dengue-immune individuals were mixed to form one pool or two of DENV positive plasma samples and were used as positive control for the assay. Moreover, the virion lysis assay was performed with plasma from dengue-immune individuals or normal human sera (NHS) in the presence of exogenous human serum (complement) that was depleted for C1q (Complement Technology: A300), C3 (Millipore EMD: 234403), or C5 (Millipore EMD: 234405). C1q-depleted human serum was also reconstituted with recombinant human C1q protein (Complement Technology: A099) at a final concentration of 70μg/ml.
For the antibody-dependent complement-mediated lysis (ADCML) assay, baby hamster kidney BHK-21 cells (ATTC: CCL-10) were mock-infected or infected with a multiplicity of infection (MOI) 0.1 of the DENV3 infectious clone UNC3001 for 40–48h prior to the assay. Human complement (Complement sera human, Sigma) was resuspended in 1ml of GVB++, and both complement and plasma samples were diluted in GVB++. 50ul of diluted complement (1:100) was mixed with 50ul of diluted plasma (1:200) and incubated with BHK-21 cells at 37°C for 1 hour. 50ul of cell supernatants were used to measure lactate dehydrogenase (LDH) release with CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega) following the manufacturer’s instructions. Optical density (OD) values were determined, and samples were analyzed by subtracting the OD values from wells containing GVB++ and lacking plasma and complement (negative control).
Statistical Analyses
Univariate analyses were performed in R or using Graphpad Prism software, version 8.0. The data is shown as the average of two replicates or donors. Measurements for isotypes/subclasses, FcγR-binding levels, and ADCD were log10-transformed. Outliers were removed prior to visualization and statistical analysis. Significance was determined using a Mann-Whitney U test, and p-values were adjusted using the Benjamini-Hochberg method. When indicated in the figure legends, one-way ANOVA was used to determine the statistical significance of multiple comparisons.
Multivariate analysis was performed using R, version 4.0.3. In the multivariate analyses, measurements for isotypes/subclasses, FcγR-binding levels and ADCD were log10-transformed before analysis. In addition to the antibody measurements, we calculated and included polyfunctionality scores for DENV2 and DENV3 for each subject as the number of functional readouts that were above the median across all subjects in both groups. To find a set of most discriminating features between the two groups, we used a least absolute shrinkage and selection operator (LASSO)-based selection procedure (65). First, the measurements were z-scored across all subjects. An inner cross-validation was performed for LASSO to determine the penalization parameter. This procedure was repeated 10 times, and features were selected that had a non-zero coefficient in more than a pre-defined fraction of repetitions (0.75). Using the LASSO-selected features, a partial least square discriminant analysis (PLSDA) model was built to discriminate the pre-inapparent and pre-symptomatic infection samples. For the score plots, ellipses indicate the 80% confidence regions assuming a multivariate t distribution. For the model, either the R2 for the second latent variable was < 0.01 or the Q2 < 0.05, and, thus, the component would not be included in the model and was only calculated for visualization purposes. The overall modeling approach (LASSO-PLS-DA) was validated with permutation testing. Two types of control models were generated: 1) ‘random features’, which selected fold-specific random feature sets of the same size of the features set selected by the actual modeling approach, and 2) ‘permuted labels’, for which the whole modeling approach was applied to data with shuffled group labels. This procedure was done for 50 permutations for each of the 10 cross-validation replicates. The p-values for the modeling approach were obtained from the tail probability of the generated null distribution, i.e., the distribution of classification accuracies of the control models. LASSO was performed using the R package ‘glmnet’, and the PLS-DA was generated with the R package ‘ropls’. For the correlation networks, only significant (Benjamini-Hochberg adjusted p-value <cc0.05) Spearman correlations to a selected feature with |r|>0.65 are shown. Correlation matrices were generated by calculating the Spearman correlation between features and were visualized using the corrplot package.
Data and materials availability:
Data to understand and assess the conclusions of this research are available in the main text and Supplementary Materials. Materials may be shared with outside investigators subject to availability and following UC Berkeley IRB approval. Please contact the UC Berkeley Center for the Protection of Human Subjects (ophs@berkeley.edu) and Eva Harris (eharris@berkeley.edu) to arrange for access. The materials used in this study are covered by standard material transfer agreements. There was no unique code generated for this manuscript. Code from this manuscript was adapted form https://github.com/LoosC/systemsseRology.
Supplementary Material
Acknowledgments:
We thank past and present members of the study team based at the Centro de Salud Sócrates Flores Vivas, the National Virology Laboratory in the Centro Nacional de Diagnóstico y Referencia, and the Sustainable Sciences Institute in Nicaragua for their dedication and high-quality work. We are very grateful to the Pediatric Dengue Cohort Study participants and their families. We thank Drs. W. Clay Brown and Janet Smith (University of Michigan, USA), Drs. Premkumar Lakshmanane and Aravinda de Silva (University of North Carolina Chapel Hill, USA), and Dr. Ralph Baric (University of North Carolina Chapel Hill, USA) for providing us with DENV3 E domain III, full-length recombinant DENV2 NS1 and NS1 wing and β-ladder domains, and a DENV3 infectious clone, respectively. We are also thankful to Dr. Scott Biering for providing helpful feedback on this manuscript; Dr. Fausto Bustos for discussions on statistical analyses and data presentation; and Jose Victor Zambrana for providing demographic and infection history data about the samples.
Funding:
This research was funded by National Institutes of Health (NIH) grants P01AI106695 (to E.H.) and NIH U19-AI135995 (to D.A.L.). Dr. Alter’s work was supported by Terry and Susan Ragon, and the SAMANA Kay MGH Research Scholars award, the Ragon Institute of MGH, MIT and Harvard, the NIH (3R37AI080289–11S1), the Gates Foundation Global Health Vaccine Accelerator Platform funding (OPP1146996 and INV-001650), and the Musk Foundation. The Pediatric Dengue Cohort Study was supported by the Pediatric Dengue Vaccine Initiative grant VE-1 (to E.H.) from the Bill and Melinda Gates Foundation and NIH subcontract HHSN2722001000026C (to E.H., A.B.).
Footnotes
Competing interests: G.A. is a founder of Systems Seromyx and an employee of Leyden Labs. E.H.’s laboratory received research funds from Takeda Vaccines Inc. to analyze samples from vaccine recipients. E.H. served on one-time advisory boards for Merck and Takeda.The other authors declare that they have no competing interests.
References and notes
- 1.Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, Hoen AG, Moyes CL, Farlow AW, Scott TW, Hay SI, Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl. Trop. Dis 6, e1760 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cattarino L, Rodriguez-Barraquer I, Imai N, Cummings DAT, Ferguson NM, Mapping global variation in dengue transmission intensity. Sci. Transl. Med 12, eaax4144 (2020). [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization, Ed., Dengue haemorrhagic fever: diagnosis, treatment, prevention and control (Geneva, 2. ed., 1997). [Google Scholar]
- 4.Katzelnick LC, Montoya M, Gresh L, Balmaseda A, Harris E, Neutralizing antibody titers against dengue virus correlate with protection from symptomatic infection in a longitudinal cohort. Proc. Natl. Acad. Sci 113, 728–733 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katzelnick LC, Gresh L, Halloran ME, Mercado JC, Kuan G, Gordon A, Balmaseda A, Harris E, Antibody-dependent enhancement of severe dengue disease in humans. Science 358, 929–932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halstead S, O’Rourke E, Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J. Exp. Med 146, 201–217 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katzelnick LC, Narvaez C, Arguello S, Lopez Mercado B, Collado D, Ampie O, Elizondo D, Miranda T, Bustos Carillo F, Mercado JC, Latta K, Schiller A, Segovia-Chumbez B, Ojeda S, Sanchez N, Plazaola M, Coloma J, Halloran ME, Premkumar L, Gordon A, Narvaez F, de Silva AM, Kuan G, Balmaseda A, Harris E, Zika virus infection enhances future risk of severe dengue disease. Science 369, 1123–1128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bournazos S, Vo HTM, Duong V, Auerswald H, Ly S, Sakuntabhai A, Dussart P, Cantaert T, Ravetch JV, Antibody fucosylation predicts disease severity in secondary dengue infection. Science 372, 1102–1105 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang TT, Sewatanon J, Memoli MJ, Wrammert J, Bournazos S, Bhaumik SK, Pinsky BA, Chokephaibulkit K, Onlamoon N, Pattanapanyasat K, Taubenberger JK, Ahmed R, Ravetch JV, IgG antibodies to dengue enhanced for FcγRIIIA binding determine disease severity. Science 355, 395–398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thulin NK, Brewer RC, Sherwood R, Bournazos S, Edwards KG, Ramadoss NS, Taubenberger JK, Memoli M, Gentles AJ, Jagannathan P, Zhang S, Libraty DH, Wang TT, Maternal Anti-Dengue IgG Fucosylation Predicts Susceptibility to Dengue Disease in Infants. Cell Rep 31, 107642 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capeding MR, Tran NH, Hadinegoro SRS, Ismail HIHM, Chotpitayasunondh T, Chua MN, Luong CQ, Rusmil K, Wirawan DN, Nallusamy R, Pitisuttithum P, Thisyakorn U, Yoon I-K, van der Vliet D, Langevin E, Laot T, Hutagalung Y, Frago C, Boaz M, Wartel TA, Tornieporth NG, Saville M, Bouckenooghe A, Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. The Lancet 384, 1358–1365 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Villar L, Dayan GH, Arredondo-García JL, Rivera DM, Cunha R, Deseda C, Reynales H, Costa MS, Morales-Ramírez JO, Carrasquilla G, Rey LC, Dietze R, Luz K, Rivas E, Miranda Montoya MC, Cortés Supelano M, Zambrano B, Langevin E, Boaz M, Tornieporth N, Saville M, Noriega F, Efficacy of a Tetravalent Dengue Vaccine in Children in Latin America. N. Engl. J. Med 372, 113–123 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Sridhar S, Luedtke A, Langevin E, Zhu M, Bonaparte M, Machabert T, Savarino S, Zambrano B, Moureau A, Khromava A, Moodie Z, Westling T, Mascareñas C, Frago C, Cortés M, Chansinghakul D, Noriega F, Bouckenooghe A, Chen J, Ng S-P, Gilbert PB, Gurunathan S, DiazGranados CA, Effect of Dengue Serostatus on Dengue Vaccine Safety and Efficacy. N. Engl. J. Med 379, 327–340 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Henein S, Adams C, Bonaparte M, Moser JM, Munteanu A, Baric R, de Silva AM, Dengue vaccine breakthrough infections reveal properties of neutralizing antibodies linked to protection. J. Clin. Invest 131, e147066 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moodie Z, Juraska M, Huang Y, Zhuang Y, Fong Y, Carpp LN, Self SG, Chambonneau L, Small R, Jackson N, Noriega F, Gilbert PB, Neutralizing Antibody Correlates Analysis of Tetravalent Dengue Vaccine Efficacy Trials in Asia and Latin America. J. Infect. Dis 217, 742–753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J, Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. The Lancet 380, 1559–1567 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Selva KJ, van de Sandt CE, Lemke MM, Lee CY, Shoffner SK, Chua BY, Davis SK, Nguyen THO, Rowntree LC, Hensen L, Koutsakos M, Wong CY, Mordant F, Jackson DC, Flanagan KL, Crowe J, Tosif S, Neeland MR, Sutton P, Licciardi PV, Crawford NW, Cheng AC, Doolan DL, Amanat F, Krammer F, Chappell K, Modhiran N, Watterson D, Young P, Lee WS, Wines BD, Mark Hogarth P, Esterbauer R, Kelly HG, Tan H-X, Juno JA, Wheatley AK, Kent SJ, Arnold KB, Kedzierska K, Chung AW, Systems serology detects functionally distinct coronavirus antibody features in children and elderly. Nat. Commun 12, 2037 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiLillo DJ, Tan GS, Palese P, Ravetch JV, Broadly neutralizing hemagglutinin stalk–specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med 20, 143–151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saphire EO, Schendel SL, Fusco ML, Gangavarapu K, Gunn BM, Wec AZ, Halfmann PJ, Brannan JM, Herbert AS, Qiu X, Wagh K, He S, Giorgi EE, Theiler J, Pommert KBJ, Krause TB, Turner HL, Murin CD, Pallesen J, Davidson E, Ahmed R, Aman MJ, Bukreyev A, Burton DR, Crowe JE, Davis CW, Georgiou G, Krammer F, Kyratsous CA, Lai JR, Nykiforuk C, Pauly MH, Rijal P, Takada A, Townsend AR, Volchkov V, Walker LM, Wang C-I, Zeitlin L, Doranz BJ, Ward AB, Korber B, Kobinger GP, Andersen KG, Kawaoka Y, Alter G, Chandran K, Dye JM, Systematic Analysis of Monoclonal Antibodies against Ebola Virus GP Defines Features that Contribute to Protection. Cell 174, 938–952.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung AW, Kumar MP, Arnold KB, Yu WH, Schoen MK, Dunphy LJ, Suscovich TJ, Frahm N, Linde C, Mahan AE, Hoffner M, Streeck H, Ackerman ME, McElrath MJ, Schuitemaker H, Pau MG, Baden LR, Kim JH, Michael NL, Barouch DH, Lauffenburger DA, Alter G, Dissecting Polyclonal Vaccine-Induced Humoral Immunity against HIV Using Systems Serology. Cell 163, 988–998 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallichotte EN, Baric RS, de Silva AM, in Dengue and Zika: Control and Antiviral Treatment Strategies, Advances in Experimental Medicine and Biology Hilgenfeld R, Vasudevan SG, Eds. (Springer Singapore, Singapore, 2018), vol. 1062, pp. 63–76. [DOI] [PubMed] [Google Scholar]
- 22.Collins AM, Jackson KJL, A Temporal Model of Human IgE and IgG Antibody Function. Front. Immunol 4 (2013), doi: 10.3389/fimmu.2013.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson KJL, Wang Y, Collins AM, Human immunoglobulin classes and subclasses show variability in VDJ gene mutation levels. Immunol. Cell Biol 92, 729–733 (2014). [DOI] [PubMed] [Google Scholar]
- 24.King HW, Orban N, Riches JC, Clear AJ, Warnes G, Teichmann SA, James LK, Single-cell analysis of human B cell maturation predicts how antibody class switching shapes selection dynamics. Sci. Immunol 6, eabe6291 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Malafa S, Medits I, Aberle JH, Aberle SW, Haslwanter D, Tsouchnikas G, Wölfel S, Huber KL, Percivalle E, Cherpillod P, Thaler M, Roßbacher L, Kundi M, Heinz FX, Stiasny K, Impact of flavivirus vaccine-induced immunity on primary Zika virus antibody response in humans. PLoS Negl. Trop. Dis 14, e0008034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laoprasopwattana K, Libraty DH, Endy TP, Nisalak A, Chunsuttiwat S, Ennis FA, Rothman AL, Green S, Antibody‐Dependent Cellular Cytotoxicity Mediated by Plasma Obtained before Secondary Dengue Virus Infections: Potential Involvement in Early Control of Viral Replication. J. Infect. Dis 195, 1108–1116 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Shresta S, Role of Complement in Dengue Virus Infection: Protection or Pathogenesis? mBio 3, e00003–12, mBio.00003–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halstead SB, Antibody, Macrophages, Dengue Virus Infection, Shock, and Hemorrhage: A Pathogenetic Cascade. Clin. Infect. Dis 11, S830–S839 (1989). [DOI] [PubMed] [Google Scholar]
- 29.Avirutnan P, Punyadee N, Noisakran S, Komoltri C, Thiemmeca S, Auethavornanan K, Jairungsri A, Kanlaya R, Tangthawornchaikul N, Puttikhunt C, Pattanakitsakul S, Yenchitsomanus P, Mongkolsapaya J, Kasinrerk W, Sittisombut N, Husmann M, Blettner M, Vasanawathana S, Bhakdi S, Malasit P, Vascular Leakage in Severe Dengue Virus Infections: A Potential Role for the Nonstructural Viral Protein NS1 and Complement. J. Infect. Dis 193, 1078–1088 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Sun P, Morrison BJ, Beckett CG, Liang Z, Nagabhushana N, Li A, Porter KR, Williams M, NK cell degranulation as a marker for measuring antibody-dependent cytotoxicity in neutralizing and non-neutralizing human sera from dengue patients. J. Immunol. Methods 441, 24–30 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Michlmayr D, Andrade P, Gonzalez K, Balmaseda A, Harris E, CD14+CD16+ monocytes are the main target of Zika virus infection in peripheral blood mononuclear cells in a paediatric study in Nicaragua. Nat. Microbiol 2, 1462–1470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhakdi S, Kazatchkine MD, Pathogenesis of dengue: an alternative hypothesis. Southeast Asian J. Trop. Med. Public Health 21, 652–657 (1990). [PubMed] [Google Scholar]
- 33.Mehlhop E, Whitby K, Oliphant T, Marri A, Engle M, Diamond MS, Complement Activation Is Required for Induction of a Protective Antibody Response against West Nile Virus Infection. J. Virol 79, 7466–7477 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlesinger JJ, Brandriss MW, Walsh EE, Protection against 17D yellow fever encephalitis in mice by passive transfer of monoclonal antibodies to the nonstructural glycoprotein gp48 and by active immunization with gp48. J. Immunol. Baltim. Md 1950 135, 2805–2809 (1985). [PubMed] [Google Scholar]
- 35.Krishna VD, Rangappa M, Satchidanandam V, Virus-Specific Cytolytic Antibodies to Nonstructural Protein 1 of Japanese Encephalitis Virus Effect Reduction of Virus Output from Infected Cells. J. Virol 83, 4766–4777 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wahala WMPB, Kraus AA, Haymore LB, Accavitti-Loper MA, de Silva AM, Dengue virus neutralization by human immune sera: Role of envelope protein domain III-reactive antibody. Virology 392, 103–113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katzelnick LC, Harris E, Baric R, Coller B-A, Coloma J, Crowe JE, Cummings DAT, Dean H, de Silva A, Diamond MS, Durbin A, Ferguson N, Gilbert PB, Gordon A, Gubler DJ, Guy B, Halloran ME, Halstead S, Jackson N, Jarman R, Lok S, Michael NL, Ooi EE, Papadopoulos A, Plotkin S, Precioso AR, Reiner R, Rey FA, Rodríguez-Barraquer I, Rothman A, Schmidt AC, Screaton G, Sette A, Simmons C, St AL. John, W. Sun, S. Thomas, J. Torresi, J. S. Tsang, K. Vannice, S. Whitehead, A. Wilder-Smith, I. Kyu Yoon, Immune correlates of protection for dengue: State of the art and research agenda. Vaccine 35, 4659–4669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katzelnick LC, Bos S, Harris E, Protective and enhancing interactions among dengue viruses 1–4 and Zika virus. Curr. Opin. Virol 43, 59–70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schiela B, Bernklau S, Malekshahi Z, Deutschmann D, Koske I, Banki Z, Thielens NM, Würzner R, Speth C, Weiss G, Stiasny K, Steinmann E, Stoiber H, Active Human Complement Reduces the Zika Virus Load via Formation of the Membrane-Attack Complex. Front. Immunol 9, 2177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Avirutnan P, Mehlhop E, Diamond MS, Complement and its role in protection and pathogenesis of flavivirus infections. Vaccine 26, I100–I107 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bokisch VA, Top FH, Russell PK, Dixon FJ, Müller-Eberhard HJ, The Potential Pathogenic Role of Complement in Dengue Hemorrhagic Shock Syndrome. N. Engl. J. Med 289, 996–1000 (1973). [DOI] [PubMed] [Google Scholar]
- 42.Aye KS, Charngkaew K, Win N, Wai KZ, Moe K, Punyadee N, Thiemmeca S, Suttitheptumrong A, Sukpanichnant S, Prida M, Halstead SB, Pathologic highlights of dengue hemorrhagic fever in 13 autopsy cases from Myanmar. Hum. Pathol 45, 1221–1233 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Mehlhop E, Whitby K, Oliphant T, Marri A, Engle M, Diamond MS, Complement activation is required for induction of a protective antibody response against West Nile virus infection. J. Virol 79, 7466–7477 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mehlhop E, Ansarah-Sobrinho C, Johnson S, Engle M, Fremont DH, Pierson TC, Diamond MS, Complement Protein C1q Inhibits Antibody-Dependent Enhancement of Flavivirus Infection in an IgG Subclass-Specific Manner. Cell Host Microbe 2, 417–426 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mehlhop E, Nelson S, Jost CA, Gorlatov S, Johnson S, Fremont DH, Diamond MS, Pierson TC, Complement Protein C1q Reduces the Stoichiometric Threshold for Antibody-Mediated Neutralization of West Nile Virus. Cell Host Microbe 6, 381–391 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiemmeca S, Tamdet C, Punyadee N, Prommool T, Songjaeng A, Noisakran S, Puttikhunt C, Atkinson JP, Diamond MS, Ponlawat A, Avirutnan P, Secreted NS1 Protects Dengue Virus from Mannose-Binding Lectin–Mediated Neutralization. J. Immunol 197, 4053–4065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nascimento EJM, Silva AM, Cordeiro MT, Brito CA, Gil LHVG, Braga-Neto U, Marques ETA, Rénia L, Ed. Alternative Complement Pathway Deregulation Is Correlated with Dengue Severity. PLoS ONE 4, e6782 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cabezas S, Bracho G, Aloia AL, Adamson PJ, Bonder CS, Smith JR, Gordon DL, Carr JM, Diamond MS, Ed. Dengue Virus Induces Increased Activity of the Complement Alternative Pathway in Infected Cells. J. Virol 92, e00633–18, /jvi/92/14/e00633–18.atom (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuchs A, Lin T-Y, Beasley DW, Stover CM, Schwaeble WJ, Pierson TC, Diamond MS, Direct Complement Restriction of Flavivirus Infection Requires Glycan Recognition by Mannose-Binding Lectin. Cell Host Microbe 8, 186–195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuan G, Gordon A, Aviles W, Ortega O, Hammond SN, Elizondo D, Nunez A, Coloma J, Balmaseda A, Harris E, The Nicaraguan Pediatric Dengue Cohort Study: study design, methods, use of information technology, and extension to other infectious diseases. Am. J. Epidemiol 170, 120–129 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balmaseda A, Sandoval E, Pérez L, Gutiérrez CM, Harris E, Application of molecular typing techniques in the 1998 dengue epidemic in Nicaragua. Am J Trop Med Hyg 61, 893–897 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Balmaseda A, Zambrana JV, Collado D, García S, Saborío N, Elizondo D, Mercado JC, Gonzalez K, Cerpas C, Nuñez A, Waggoner D, Corti JJ, Kuan G, Burger-Calderon R, Harris E, Vázquez Y-W, Tang, Ed. Comparison of Four Serological Methods and Two Reverse Transcription-PCR Assays for Diagnosis and Surveillance of Zika Virus Infection. J. Clin. Microbiol 56, e01785–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernández RJ, Vázquez S, Serological diagnosis of dengue by an ELISA inhibition method(EIM) . Mem. Inst. Oswaldo Cruz 85, 347–351 (1990). [DOI] [PubMed] [Google Scholar]
- 54.Balmaseda A, Hammond SN, Tellez Y, Imhoff L, Rodriguez Y, Saborío SI, Mercado JC, Perez L, Videa E, Almanza E, Kuan G, Reyes M, Saenz L, Amador JJ, Harris E, High seroprevalence of antibodies against dengue virus in a prospective study of schoolchildren in Managua, Nicaragua. Trop. Med. Int. Health TM IH 11, 935–42 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Montoya M, Gresh L, Mercado JC, Williams KL, Jose M, Gutierrez G, Kuan G, Gordon A, Balmaseda A, Harris E, Vargas MJ, Gutierrez G, Kuan G, Gordon A, Balmaseda A, Harris E, Symptomatic versus inapparent outcome in repeat dengue virus infections is influenced by the time interval between infections and study year. PLoS Negl. Trop. Dis 7, e2357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biering SB, Akey DL, Wong MP, Brown WC, Lo NTN, Puerta-Guardo H, Tramontini F Gomes de Sousa, C. Wang, J. R. Konwerski, D. A. Espinosa, N. J. Bockhaus, D. R. Glasner, J. Li, S. F. Blanc, E. Y. Juan, S. J. Elledge, M. J. Mina, P. R. Beatty, J. L. Smith, E. Harris, Structural basis for antibody inhibition of flavivirus NS1–triggered endothelial dysfunction. Science 371, 194–200 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown EP, Dowell KG, Boesch AW, Normandin E, Mahan AE, Chu T, Barouch DH, Bailey-Kellogg C, Alter G, Ackerman ME, Multiplexed Fc array for evaluation of antigen-specific antibody effector profiles. J. Immunol. Methods 443, 33–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boudreau CM, Yu W-H, Suscovich TJ, Talbot HK, Edwards KM, Alter G, Selective induction of antibody effector functional responses using MF59-adjuvanted vaccination. J. Clin. Invest 130, 662–672 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ackerman ME, Crispin M, Yu X, Baruah K, Boesch AW, Harvey DJ, Dugast A-S, Heizen EL, Ercan A, Choi I, Streeck H, Nigrovic PA, Bailey-Kellogg C, Scanlan C, Alter G, Natural variation in Fc glycosylation of HIV-specific antibodies impacts antiviral activity. J. Clin. Invest 123, 2183–2192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fischinger S, Fallon JK, Michell AR, Broge T, Suscovich TJ, Streeck H, Alter G, A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J. Immunol. Methods 473, 112630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karsten CB, Mehta N, Shin SA, Diefenbach TJ, Slein MD, Karpinski W, Irvine EB, Broge T, Suscovich TJ, Alter G, A versatile high-throughput assay to characterize antibody-mediated neutrophil phagocytosis. J. Immunol. Methods 471, 46–56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Messer WB, Yount B, Hacker KE, Donaldson EF, Huynh JP, de Silva AM, Baric RS, Rothman AL, Ed. Development and Characterization of a Reverse Genetic System for Studying Dengue Virus Serotype 3 Strain Variation and Neutralization. PLoS Negl. Trop. Dis 6, e1486 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huber M, Fischer M, Misselwitz B, Manrique A, Kuster H, Niederöst B, Weber R, von Wyl V, Günthard HF, Trkola A, Emerman M, Ed. Complement Lysis Activity in Autologous Plasma Is Associated with Lower Viral Loads during the Acute Phase of HIV-1 Infection. PLoS Med 3, e441 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson BW, Russell BJ, Lanciotti RS, Serotype-Specific Detection of Dengue Viruses in a Fourplex Real-Time Reverse Transcriptase PCR Assay. J. Clin. Microbiol 43, 4977–4983 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hastie T, Tibshirani R, Wainwright M, Statistical Learning with Sparsity: The Lasso and Generalizations (Chapman and Hall/CRC, ed. 0, 2015; https://www.taylorfrancis.com/books/9781498712170). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


