Abstract
Chromatin insulators have been proposed to play an important role in chromosome organization and local regulatory interactions. In Drosophila , one of these insulators is known as Wari. It is located immediately downstream of the 3’ end of the white transcription unit. Wari has been proposed to interact with the white promoter region, thereby facilitating recycling of the RNA polymerase machinery. We have tested this model by deleting the Wari insulator at the endogenous white locus and could not detect a significant effect on eye pigmentation.
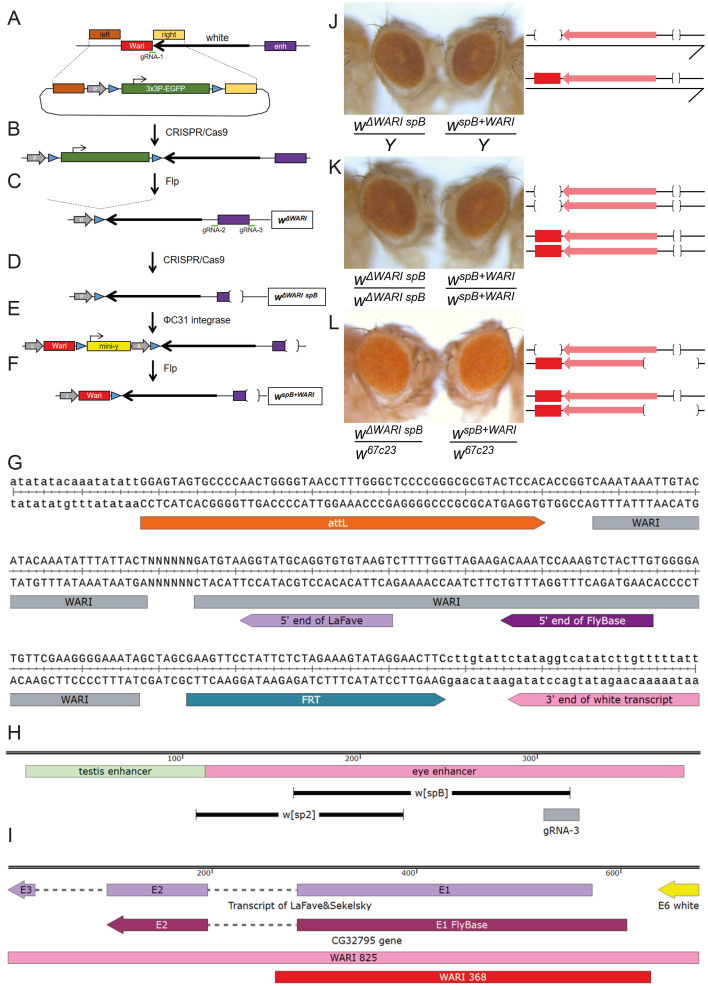
Figure 1. Genetic engineering of new white alleles and their effect on eye pigmentation .
(A) Generation of w ΔWari-GFP : The white gene (black arrow), Wari (red box) and the white enhancer (purple box) are shown. The position of gRNA-1 is indicated by a green line to the right of Wari. In addition, the 500 bp feet required for homologous recombination are shown on either side of Wari (brown and yellow boxes). Below, the relevant genetic elements present on donor plasmid pMM085 are depicted: grey arrow represents an attP site, blue arrowheads are FRT sites, green box corresponds to 3x3P-EGFP marker. Note that genetic elements in 1A-F are not drawn to scale. (B) Generation of w ΔWari-GFP by knock-in of 3x3P-EGFP marker in place of Wari. (C) Generation of w ΔWari by Flp-induced deletion of 3x3P-EGFP marker. (D) Generation of w ΔWari-spB by CRISPR/Cas9-mediated deletion of eye enhancer. The gRNAs used for this purpose are indicated in Figure 1C (green lines below purple enhancer element labeled with gRNA-2 and gRNA-3). (E) Generation of w spB+Wari-yellow by integrase-mediated insertion of reentry plasmid pDB154. Grey arrows indicate attL and attR sites. Yellow box depicts the mini-yellow marker. (F) Generation of w spB+Wari by Flp-mediated deletion of the mini-yellow marker. The names of the three alleles described in the text are indicated in (C) w ΔWARI , (D) w ΔWari spB and (F) w spB+Wari . (G) In w spB+Wari , the 364 bp Wari element (in grey) is flanked by an attL (in orange) and an FRT site (in blue). The latter is located 8 bp downstream of the 3’ end of the single white transcript w-RA (indicated in pink; sequence information on the white transcript taken from FlyBase genome release R6.47). Note that 249 bp of Wari are missing at the NNNNNN position. The small arrows in light and dark magenta indicate the 5’ end of CG32795 transcripts of LaFave and Sekelsky (2011) and FlyBase, respectively. (H) The location of gRNA target site gRNA-3 (in gray), the white testis and eye enhancers (in green and pink, respectively; indicated according to Qian et al. 1992) and of the two deletions w sp2 and w spB (black lines indicate the region missing in these deletions) are shown. w spB initiates in the gRNA-3 target site and extends distally where it overlaps by 62 bp with w sp2 . gRNA-2 target site not indicated because it is located distally to testis enhancer. Flanking DNA of w sp2 : AACGTCAATG and TGTGTGTTTG . Flanking DNA of w spB : CTTATGGGGC and AAAAATGGCA. (I) The 5’ end of gene CG32795 is located immediately downstream to the 3’ end of the white gene. At the top is a scale bar for the 676 bp genomic fragment shown in this panel. Below, the 3’ end of white (in yellow; E(xon)6 of white transcript) is shown. To its left, the 5’ end of a transcript is depicted in light purple. It was detected in several independently isolated hypomorphic P-element alleles (LaFave and Sekelsky 2011). Below, the first two exons of gene CG32795 (in dark purple) are shown. This transcript corresponds to isoform CG32795-RD (sequence information from genome release R6.47). At the bottom, the distal part of Wari 825 (in pink) and Wari 368 (in red) are shown. Wari 825 was the starting point for the dissection of the insulator activity detected downstream of mini-white , which resulted in the definition of Wari 368 (Chetverina et al. 2008). The most distal 46 bp of Wari 825 are part of pCaSpeR-2’s adjacent P-element end and thus not indicated. Consequently, the distal end of Wari 825 corresponds to that of the LaFave transcript. Note that the transcription start sites of the FlyBase and the LaFave transcripts are contained in Wari 368. Transcription start sites of other CG32795 isoforms (not indicated) are further downstream and not included in Wari 825 nor in Wari 368. ( J) Males with genotypes w ΔWari spB /Y and w spB+Wari /Y are shown. (K) Females with genotypes w ΔWARI spB /w ΔWari spB and w spB+Wari /w spB+Wari are shown. (L) Females with genotypes w ΔWari spB /w 67c23 and w spB+Wari /w 67c23 are shown. To the right of each picture, diagrams of the two genotypes are depicted (upper diagram left fly, bottom diagram right fly). Note that compared to Oregon-R , eye pigmentation is considerable reduced in w ΔWari spB males and females, indicating that the w spB enhancer deletion is largely disabling the eye enhancer. However, dosage compensation is still taking place since homozygous females have clearly darker eyes than heterozygous sisters (compare eye colors in Figure 1K and 1L). In none of the three genotypes, the presence of the Wari element leads to a significant increase in eye pigmentation. Approximate location of Df(1)w 67c23 break points at 5’ end of white indicated according to Moschetti et al. (2004).
Description
The Drosophila white ( w ) gene has been at the heart of several important discoveries in genetics and molecular biology (reviewed in Hazelrigg 1987; Green 2010; Mohr 2018). It is an X-linked gene with a rather simple structure. A single transcript is expressed under the control of a few tissue-specific enhancers. For example, a 270 bp eye enhancer is located about 1200 bp upstream of the white transcription start site. This enhancer is essential for efficient expression of white in the eye where it plays an essential role in pigment deposition (Davison et al. 1985; Qian et al. 1992) . While wild-type flies have red eyes, white null alleles have white eyes. Hypomorphic white mutants can have a continuum of allele-specific intermediate eye colors ranging from pale yellow to almost wild-type red. These differences in eye pigmentation are very useful for scoring phenotypes in a semi-quantitative manner. The easy-to-score white phenotype was employed for the construction of numerous vectors used for the generation of transgenic flies. Such vectors contain the so-called mini-white gene, a derivative of the endogenous white gene from which a large part of the first intron has been deleted and which lacks all known tissue-specific enhancers (Pirrotta 1988) .
Thanks to its simple organization and its obvious phenotype, mini-white in combination with its eye enhancer became the workhorse of researchers interested in chromatin insulators (Kellum and Schedl 1991, 1992; reviewed in Geyer 1997) . Insulators were proposed to subdivide chromatin into separate looped domains by pairwise association of two insulators. Furthermore, it was proposed that regulatory elements located in separate chromatin loops are unable to interact with each other. Therefore, putative insulators were tested by insertion in between the mini-white gene and the white eye enhancer. Transgenic flies were generated and the eye color phenotype was scored. The less pigmented the eye appeared, the stronger the effect of the insulator was ranked.
When more sophisticated arrangements of insulators and reporter genes were analyzed, Chetverina et al (2008) found that the mini-white reporter itself was associated with an insulator-like element. It is confined to a 368 bp fragment that is located only 8 bp distal to the 3’ end of the white gene (see Figure 1I ) and is called Wari (acronym for w hite a butting r esident i nsulator). Later studies reported that in S2 cells, Wari was bound by insulator proteins such as CP190. In addition, in transgene inserts, Wari was found to functionally interact with the white promoter (Erokhin et al. 2010, 2011) . These authors proposed that insulators like Wari that are located downstream of a gene could support a loop that brings together a promoter and a transcriptional terminator. They further suggested that such loop formation might be a common feature of gene activation that serves to promote efficient transcriptional elongation and reinitiation by facilitating RNAP II recycling from the terminator to the promoter.
A simple prediction from this model is a noticeable effect on eye pigmentation caused by deletion of Wari at the endogenous white locus. To test such a scenario, we deleted Wari by CRISPR/Cas9-mediated homologous recombination in Oregon-R flies (for details on the generation of all alleles presented in this study, see Figure 1A -F and Materials). The eye color of w ΔWari flies was indistinguishable from that observed in Oregon-R controls. It is known that the eye color of homozygous w + and heterozygous w + /w - females is essentially identical. Thus, we reasoned that in w ΔWari flies, a phenotypic effect could be masked by the recessive nature of the white locus. Therefore, we sensitized the white locus by introducing a 156 bp deletion in the white eye enhancer in the context of the w ΔWari allele. The new allele is called w ΔWari-spB ( spB stands for “ spotted-Basel ”; see Figure 1H ). As expected, compared to w ΔWari , the eye color of these flies was significantly reduced (see Figure 1J -L). This observation is of utmost importance for the interpretation of our experiment. With the introduction of w spB , the eye color of w ΔWari-spB has become dosage-dependent; eye pigmentation of homozygous and heterozygous w ΔWari-spB females were easily distinguishable (compare eye colors in Figures 1K and 1L). This strongly suggests that upon introduction of Wari into w ΔWari-spB , even a moderate increase in eye pigmentation should become detectable if Wari indeed contributes to white transcriptional activity. Towards that end, the attP landing site on w ΔWari-spB was used to re-introduce Wari at the 3’ end of the white gene. This allele is referred to as w spB+Wari . Importantly, we observed no significant difference between the eye color of w ΔWari-spB and w spB+Wari flies, suggesting that Wari does not play a significant role in white gene transcription (see Figure 1J -L).
A vast body of literature on insulator function has accumulated over the last two decades (reviewed in Kyrchanova and Georgiev 2013). Apart from detailed studies on insulator elements in the bithorax complex (reviewed in Maeda and Karch 2015), essentially all these studies relied on the analysis of transgenic constructs. The data suggests that insulators are not necessarily organizers of higher order chromatin structure but rather facilitators of local enhancer-promoter interactions. We have now tested this model for the Wari insulator at its endogenous genomic location. Our observations suggest that the proposed loop-formation between Wari and the white promoter does not play a significant role in white gene transcription in the eye. This raises the question why in transgenic eye color assays, Wari can act as an enhancer blocker or interact with other insulators. Based on the most recent fly genome release R6.47, the 5’ end of gene CG32795 is located immediately distal to the 3’ end of white (see Figure 1I ). In the context of our assay system (see Figure 1G ), FlyBase-annotated transcript CG32795-RD starts 23 bp distal of the FRT, the transcript of LaFave 57 bp distal to it (LaFave and Sekelsky 2011) . Drosophila promoters are in general less than 50 bp long (reviewed in Vo Ngoc et al. 2019). Hence, there should be space enough to accommodate the RNAP II machinery as well as insulator binding proteins that are frequently also bound to promoters. It is therefore conceivable that the presence of such proteins on Wari are responsible for its insulator-like behavior on transgenes. However, when analyzed at the endogenous locus, it appears as Wari has no obvious role in white gene regulation.
Methods
Generation of new white alleles
w ΔWari-GFP : For CRISPR/Cas9, the following three plasmids were injected into Oregon-R embryos: 1. Donor plasmid pMM085 (see Figure 1A ; synthesized by Genewiz). It consists of (1) 500 bp left foot, (2) 55 bp attP, (3) 34 bp FRT, (4) 1262 bp 3x3P-EGFP as selectable marker, (5) 34 bp FRT and (6) 500bp right foot. 2. gRNA-1 plasmid pDB149 (“gRNA-1” in Figure 1A ; gRNA sequence is: 5’-GGGAAATACTTGTATTCTAT-3’). 3. Cas9 helper plasmid pBS-Hsp70-Cas9 (Gratz et al. 2014) . In a successful CRISPR/Cas9 event, homologous recombination leads to the deletion of a 364 bp Wari fragment and the integration of an attP-FRT-3x3P-EGFP-FRT cassette (see Figure 1B ). Following injection in Oregon-R embryos, injectees were grown to adulthood. 30 G0 crosses were set up, of which 18 were fertile. One G0 male produced many GFP + larvae, indicating a high probability for the success of the planed homologous recombination. GFP + larvae were transferred into a fresh vial and grown to adulthood. As expected for an X-linked genome editing event in the male germline, all adult flies emerging from this vial were females. They were used to establish an FM7c balanced stock. PCR and sequencing confirmed successful genome editing according to the design on donor plasmid pMM085. The new white allele is referred to as w ΔWari-GFP . Like all alleles described in this work, it is homozygous viable and maintained as a homozygous stock.
w ΔWari : the 3x3P-FGFP marker was deleted by Flp treatment.
w ΔWari-spB : a deletion of the white enhancer was attempted with two gRNAs flanking the eye enhancer of the white gene (gRNA-2 and gRNA-3 in Figure 1C ). They are: gRNA-2 (on plasmid pDB151; gRNA sequence is: 5’-ATTTGCTGACGACGATTAAG-3’) and gRNA-3 (on plasmid pDB152; gRNA sequence is: 5’- GCAGCGAAAGAGCTGAAAAA-3’). These two plasmids were injected together with the Cas9 helper plasmid pBS-Hsp70-Cas9 into w ΔWari embryos. From the surviving injectees, 14 fertile crosses were obtained. The progeny of these were screened for flies with the typical w sp eye color phenotype (Davison et al. 1985) . Two such candidates could be isolated from 1/14 crosses and FM7c balanced stocks were established. In addition to the deletion at the 3’ end of white , PCR and sequencing proved the existence of an identical 156 bp deletion in both candidates. This deletion removes about 60% of white eye enhancer. It overlaps by 62 bp with the 117 bp w sp2 deletion (see Figure 1H ). The new allele is referred to as w ΔWari-spB . In preparation for the next step, a y 1 w ΔWari-spB recombinant was generated by meiotic recombination.
w spB+Wari-yellow : the attP landing site present on the y 1 w ΔWari-spB chromosome was used to reintroduce the 364 bp Wari element normally present at the 3’ end of the white gene. Compared to the Wari 368 of Chetverina et al. (2008), the most distal four bases are missing (GGAG). The reentry plasmid pDB154 was injected together with the ΦC31 integrase helper plasmid pBS130 into y 1 w ΔWari-spB embryos. The reentry plasmid pDB154 was constructed in the following way. First, a cassette consisting of attP-Wari-FRT followed by unique KpnI and SalI sites was synthesized by Genewiz. This plasmid is called pDB153 and lacks a selectable marker. Therefore, pDB153 was cut with KpnI and SalI and ligated with a mini-yellow KpnI/SalI-fragment isolated from piBLLFY. The resulting plasmid is pDB154 and was used to generate a Wari insert in landing site w ΔWari-spB . Unfortunately, the mini-yellow marker on pDB154 was not detected after surviving G0 flies were crossed with y w partners. Whether this marker remains silent because of a position effect or whether it contains an unknown lesion in the mini-yellow gene has not been further analyzed. Rather, 43 F1 flies were selected at random and crossed back with y w partners. After a few days, the 43 candidates were sacrificed for PCR. One of the 43 candidates was identified as the desired transgenic. It is called w spB+Wari-yellow .
w spB+Wari : in a last step, the mini-yellow marker present in w spB+Wari-yellow flies was removed by Flp treatment. This allele is called w spB+Wari . The sequence details in the Wari and white enhancer regions of this stock are depicted in Figure 1G and 1H.
Scoring of eye pigmentation
Necessary crosses were reared on standard cornmeal food and kept at 25 degrees Celsius. In the next generation, flies were collected in a four-hour window and aged for 7 days before eye pigmentation was scored. Pictures were taken with a Leica M125 binocular equipped with a Leica flexacam C3 camera.
Deficiency w 67c23 corresponds to the one contained in Bloomington stock 6599 ( y 1 w 67c23 ).
Acknowledgments
Acknowledgments
We thank Cindy Reinger for help with figure 1. We are indebted to Karin Mauro, Bernadette Bruno and Gina Evora for constant and reliable production of world’s best fly food.
Funding Statement
This study was supported by grants from the Kantons Basel-Stadt and Basel-Land, and the Swiss National Science Foundation to MA. M.Metzler was supported by a Fellowships of Excellence from the Biozentrum of the University of Basel.
References
- Chetverina D, Savitskaya E, Maksimenko O, Melnikova L, Zaytseva O, Parshikov A, Galkin AV, Georgiev P. Red flag on the white reporter: a versatile insulator abuts the white gene in Drosophila and is omnipresent in mini-white constructs. Nucleic Acids Res. 2007 Dec 17;36(3):929–937. doi: 10.1093/nar/gkm992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison D, Chapman CH, Wedeen C, Bingham PM. Genetic and physical studies of a portion of the white locus participating in transcriptional regulation and in synapsis-dependent interactions in Drosophila adult tissues. Genetics. 1985 Jul 1;110(3):479–494. doi: 10.1093/genetics/110.3.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erokhin M, Parshikov A, Georgiev P, Chetverina D. E(y)2/Sus1 is required for blocking PRE silencing by the Wari insulator in Drosophila melanogaster. Chromosoma. 2010 Jan 15;119(3):243–253. doi: 10.1007/s00412-009-0253-1. [DOI] [PubMed] [Google Scholar]
- Erokhin M, Davydova A, Kyrchanova O, Parshikov A, Georgiev P, Chetverina D. Insulators form gene loops by interacting with promoters in Drosophila. Development. 2011 Sep 1;138(18):4097–4106. doi: 10.1242/dev.062836. [DOI] [PubMed] [Google Scholar]
- Geyer PK. The role of insulator elements in defining domains of gene expression. Curr Opin Genet Dev. 1997 Apr 1;7(2):242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O'Connor-Giles KM. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014 Jan 29;196(4):961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MM. 2010: A century of Drosophila genetics through the prism of the white gene. Genetics. 2010 Jan 1;184(1):3–7. doi: 10.1534/genetics.109.110015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelrigg T. 1987. The Drosophila white gene: a molecular update. Trends in Genetics 3(2): 43-47.
- Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991 Mar 8;64(5):941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol. 1992 May 1;12(5):2424–2431. doi: 10.1128/mcb.12.5.2424-2431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O, Georgiev P. Chromatin insulators and long-distance interactions in Drosophila. FEBS Lett. 2013 Nov 5;588(1):8–14. doi: 10.1016/j.febslet.2013.10.039. [DOI] [PubMed] [Google Scholar]
- LaFave MC, Sekelsky J. Transcription initiation from within P elements generates hypomorphic mutations in Drosophila melanogaster. Genetics. 2011 Apr 28;188(3):749–752. doi: 10.1534/genetics.111.129825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda RK, Karch F. The open for business model of the bithorax complex in Drosophila. Chromosoma. 2015 Jun 12;124(3):293–307. doi: 10.1007/s00412-015-0522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr SE. 2018. First in Flies. Harvard University Press.
- Moschetti R, Marsano RM, Barsanti P, Caggese C, Caizzi R. FB elements can promote exon shuffling: a promoter-less white allele can be reactivated by FB mediated transposition in Drosophila melanogaster. Mol Genet Genomics. 2004 Apr 2;271(4):394–401. doi: 10.1007/s00438-004-1007-7. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Vectors for P-mediated transformation in Drosophila. Biotechnology. 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- Qian S, Varjavand B, Pirrotta V. Molecular analysis of the zeste-white interaction reveals a promoter-proximal element essential for distant enhancer-promoter communication. Genetics. 1992 May 1;131(1):79–90. doi: 10.1093/genetics/131.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo Ngoc L, Kassavetis GA, Kadonaga JT. The RNA Polymerase II Core Promoter in Drosophila. Genetics. 2019 May 1;212(1):13–24. doi: 10.1534/genetics.119.302021. [DOI] [PMC free article] [PubMed] [Google Scholar]