Abstract
Among the HPV-mediated cervical cancers, cellular factor BRN3A has gained considerable attention due to its role in promoting an anti-apoptotic cellular environment and in facilitating epitheliotropic transformations of the host. The majority of previous studies looked at BRN3A's molecular characteristics; however, the possibility of genetic variations in BRN3A's auto-regulatory region in relation to cervical cancer risk has been underestimated until now. In a retrospective study in the Eastern UP population, India, we detected genetic variations in the cis-regulatory proximal enhancer region located around 5.6 kb upstream of transcription start site of BRN3A. Our analysis of PCR and DNA sequencing confirmed this novel SNP (BRN3A g.60163379A>G) within the auto-regulatory region of BRN3A. As compared to control subjects, cancer cases exhibited a 1.32-fold higher allele frequency (χ2 = 6.315, p = 0.012). In homozygous (GG) but not in heterozygous conditions, odds ratio (OR) analysis suggests a significant association of cancer risk with the SNP (OR = 2.60, p ≤ 0.004). We further confirmed using the functional analysis that this SNP increased the luciferase gene activity in HPV-positive cervical cancer SiHa cells that were exposed to progesterone. As a result of the association of polymorphisms in a non-coding region of an oncogene with increased cancer risks, we are suggesting that this genetic variation in non-coding region can be used in prediction, diagnosis, or predicting the progression of the disease.
Key Words: Cervical cancer, HPV, BRN3A, BRN3A proximal enhancer region, India, g. 6016 3379A >G
Introduction
Worldwide, uterine cervix cancer is the fourth most common female cancer and the leading cause of death among women. It accounts for approximately 6.6% of the total incidence of gynecological cancer cases globally, and 85% of which, occurs in developing countries (1). In India, 96922 cases of invasive cervix cancer were identified during the period of one year - 2018 (2). Nevertheless, an intimate association of cervical cancer with various risk factors, including smoking, age, parity, compromised immune system, and infection by Chlamydia trachomatis, Trichomonas and human papillomaviruses (HPVs), have also been established (3-5). HPVs are a large group of epitheliotropic viruses (with >200 genotypes known), and more than 95% of cervical cancer cases are connected to HPVs infection (6, 7). A typical HPV genome measures 8 kb, containing early genes (E1, E2, E4, E5, E6 and E7), late genes (L1 and L2), and an upstream regulatory region (URR). In the viral genome, E6 and E7 are the two critical genes that interact with host cellular machinery. Stable expression of E6 oncoproteins causes the degradation of p53 product, likewise E7 interacted with and inactivated pRb protein in tumorigenesis. In fact, the HPV-URR region is largely responsible for regulating expression levels of these genes (8, 9).
Studies on epidemiology and virology have provided important insights into the complexity of cancer development, however, many other cellular factors also play a critical role in infection as well as epitheliotropic transformation (10). BRN3A is one of the most extensively studied transcription factors among members of the POU protein family, which binds to the URR of HPV-16 and HPV-18 and induces E6 and E7 expression (11-13). In the cervical cancer, BRN3A is highly expressed (300-fold), but the underlying molecular mechanism is largely unknown. BRN3A expression is mainly regulated by its enhancer, which is located between five and nine kilobases upstream of the transcription start site. There are two highly conserved sequences, known as proximal and distal enhancer regions, located respectively, five to six kb and nine kb, upstream of the transcription start site. Despite, both the regulatory sequences are important and involved in BRN3A auto-regulation, the proximal enhancer region shows a comparatively higher affinity for BRN3A binding (14-18) and plays a major role in auto-regulation.
Genetic polymorphisms contribute to genome diversity, but the underlying mutation is driven by complex gene-environment interactions over generations (19). Nevertheless, some variations may further complicate the situation. Epidemiological studies show that genetic variation contributes significantly to cancer susceptibility. Polymorphisms in tumor suppressor genes are associated with the persistence of HPVs and their progression to cancer (20). For example, inactivating the oncogenic potential of HPV proteins is one of the functions of the tumor suppressor gene TP53 (21). Previous studies linked genetic polymorphisms in TP53 gene coding and non-coding regions with cancer (22, 23). Furthermore, rs2981578 polymorphism in the enhancer region of fibroblast growth factor receptor 2 (FGFR2) gene was identified as one of the major risk alleles associated with breast cancer. As well, the rs12343867 T>C variant found in intron 14 of Janus kinase 2 is linked to myeloproliferative neoplasms reviewed in Deng et al. (24). Briefly, there is ample evidence on the association of polymorphism in the regulatory region with cancer susceptibility. Yet, despite a very thorough understanding of the molecular mechanisms involved in BRN3A auto-regulatory enhancer region -mediated regulation, no previous study has investigated polymorphisms in the auto-regulatory region of BRN3A and their association with cervical cancer.
In contrast to searching mutations in the coding region, exploring the causal variants in regulatory regions and confirming how they influence gene expression remains a significant challenge. Since the advent of second-generation sequencing technology and the parallel development of bioinformatics tools, noncoding variations have been recognized as a major factor in both Mendelian and common genetic diseases (25). To identify the likely signature of polymorphism(s) in BRN3A's auto-regulatory proximal enhancer region in control and cervical cancer samples, we attempted to analyze this region of BRN3A. We identified a novel variation near the proximal enhancer of BRN3A. Based on our association study (controls and cases), we show that this mutation represents a risk allele and based on our functional analysis using luciferase reporter gene assay, we verify that this mutation could potentially increase cancer risk.
Materials and Methods
Sample collection and DNA isolation
This study was approved by the university's institutional ethics committee. Informed consent was obtained from each participant in their native language. The guidelines outlined in the ICMR ethical guidelines 2017 were strictly followed. A total of 770 (143 case and 627 control) women aged 17 to 55 years were recruited from an eastern Indian population (Purvanchal region of Uttar Pradesh) for the study from 2010 to 2017.
Biopsy and hysterectomy samples were collected from women whose cervical cancer was clinically and pathologically confirmed. Pap smear samples were taken from normal control women who agreed to get these tests done and were interested in participating in this study. The sample was collected in 5 ml ice-chilled 1X PBS (phosphate buffer saline). DNA isolation was performed as described elsewhere (26). The samples were vortexed briefly and centrifuged at 3000 rpm and the pellet was re-suspended in 600 μL of lysis buffer (0.3% SDS, 1xTE) and incubated with 80 μg of proteinase K at 55 °C for 16 h. Thereafter, a mixture of phenol:chloroform:isoamyl alcohol (25:24:1) followed by chloroform:isoamyl alcohol were added to promote the partitioning of lipids and cellular debris into the organic phase, leaving isolated DNA in the aqueous phase. DNA was precipitated with 1/10 volume of 3 M sodium acetate, and 0.7 volume of isopropanol, washed two times with 1 ml of 70% ethanol, then dried and dissolved in TE buffer (10 mM Tris, 1 mM EDTA, pH 8). To determine the concentration of DNA, samples were electrophoresed on 0.8 % agarose gels, followed by concentration measurement using a spectrophotometer (NanoDrop 2000®, Wilmington, DE 19810, USA).
Cell culture
SiHa cell line was obtained from NCCS, Pune, India, and maintained in Dulbecco's Modified Eagle's Medium containing 10 % fetal bovine serum (Gibco™, Waltham, USA) and 1X antibiotic solution containing penicillin/streptomycin cocktail. Cells were grown in CO2 incubators (5 % CO2) at 37 °C, which were sub-cultured three times every three days. As part of the experiment, cells were washed in 1X PBS (GibcoTM, Waltham, USA) then trypsinized at 37 °C in 0.25 % trypsin-EDTA solution to facilitate surface detachment. Media was added after 2 min to neutralize trypsin activity, followed by transfer of cells into 1.5 mL microfuge tubes and pelleting at 1000 rpm. By aspirating and resuspending cells in fresh media, single cells were prepared which were stained with Trypan blue and counted using a hemocytometer prior to seeding into cultures plates to meet experimental requirements.
Genetic screening of BRN3A proximal enhancer region for variant analysis
A 302 bp DNA fragment of BRN3A's proximal enhancer region was amplified by PCR with BRN3A-Seq-Enahncer primers (Table 1). PCR conditions were as follows: initial denaturation at 95 °C for 5 min, followed by 30 cycles of 30 sec at 95 °C, 30 sec at 58 °C, and 30 sec at 72 °C, and then a final extension at 72 °C for 10 min. The PCR products were eluted from agarose gel using a gel-extraction kit (Fermentas Life Sciences, Waltham, USA) according to the manufacturer’s protocol and then performed the sequencing of both forward and reverse DNA strands using the same set of primers employed initially for the PCR. Fluorescent dye-termination Sanger sequencing was performed using ABI BigDye Terminator kit ver-1.1 (Applied Biosystems, USA) on a 4-capillary automated DNA-sequencer (3130 Genetic AnalyzerÒ; ABI, USA). Cycle sequencing was performed according to the manufacturer's protocol using a ready reaction mix and 10 X PCR buffer (Applied Biosystems, USA). Analysis of the sequences was conducted using the Chromas Lite (Version-2.01) software. Putative variations were analyzed in silico using 'nucleotide-BLAST' (https://ncbi.nlm.nih.gov). To ensure the accuracy of the variant identification and its pathogenicity, we manually reviewed the sequence assemblies and polymorphisms and analyzed them in silico with online tools. Several databases were searched to determine the novelty and clinical relevance of this variant, including GnomAD (https://gnomad.broadinstitute.org/), IndiGenomes (http://clingen.igib.res.in/indigen/), INDEX-db (http://indexdb.ncbs.res.in/) and ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/).
Table 1.
Primers list used in the SNP genotyping and cloning
| Primer | Sequence (5’- 3’) | Amplicon Size (bp) | Annealing Temp (In ° C) |
|---|---|---|---|
| β-globin | F: GAA GAG CCA AGG ACA GGT AC | 268 | 55 |
| R: CAA CTT CAT CCA CGT TAC ACC | |||
| BRN3A- Seq- Enahncer | F: GCCCTGCGGTTTCCTTTCATT | 302 | 58 |
| R: CACAAATCAAAGCCCCTTGCGAG | |||
| BRN3A- En-pGL3-Luc | F(W): TATAGGTACCTTAACCTTGCACACAATCCT | 221 | 55 |
| F(M): TATAGGTACCTTAACCTTGCACACAGTCCT | |||
| R: ATATGGTACCACAAATCAAAGCCCCTTGCG | |||
| pGL3 | RV3: CTAGCAAAATAGGCTGTCCCCA | - | - |
| BRN3A (A>G) 60163379 ARMS | F(W): CTTTCCACCTTAACCTTGCACACAA | 227 | 68 |
| F(M): CTTTCCACCTTAACCTTGCACACAG | |||
| R: CTAGCAAAATAGGCTGTCCCCA |
Genotyping study
ARMS (Amplification refractory mutation detection system)-PCR was designed to screen the novel variant BRN3A g.60163379 A>G. Primer3 (https://primer3.ut.ee/) was used to design allele-specific forward primers (one for each allele). The allele-specific forward primers were designed so that their last nucleotide at the 3'-end was an allele of the novel variant position (i.e., 'A' in one forward primer and 'G' in another). Hence, we used these two forward primers (see primer list Table 1) in two separate reaction mixtures, though the reverse primer was the same in both. We amplified the same template in the both reaction mixtures. PCR conditions were: 95 °C for 2 min, then 30 cycles on 95 °C for 30 sec, 68 °C for 30 sec, 72 °C for 30 sec, and final extension on 72° C for 10 min. We ran the PCR products on a 2 % agarose gel. Amplification of genotype AA was carried out with an A-specific primer, but that of genotype GG only with a G-specific primer. The AG genotypes were, however, amplified in both sets of reactions. The results of ARMS-PCR were validated by Sanger sequencing on some of the random DNA samples.
RFLP-PCR
BRN3A enhancer region variant g.60163379 A>G was PCR amplified using BRN3A enhancer region primers that produced 221 bp long PCR product in 25 µl reactions volume containing PCR buffer and 1U of Taq polymerase (Geneaid, Taiwan), dNTP (Sigma, USA), 2.5 mM of each primer and 1µL of template genomic DNA. PCR conditions for the amplification was standardized as follows: 95°C for 2 min, then 30 cycles with 95°C for 30 sec, 68°C for 30 sec, 72°C for 30 sec and final extension at 72°C for 10 min. From the PCR products 5 µl volume was used to confirm by gel electrophoresis. Then 10 µl aliquot of the resulting PCR product was digested in 20 µL volume with 0.5 µL restriction enzyme HpyCH4III (TaaI) and 1X Tango buffer and incubated using manufacturer recommendation (Thermo Fisher, USA). Digested products were assessed on 2.5 % agarose gels for homozygous AA, heterozygous AG and homozygous GG.
Cell viability assays
A cell viability test was performed to optimize progesterone concentrations in luciferase assays. Cell viability assay was performed by using MTT (3-(4, 5-Dimethylthiazol-2-yl)-2, 5- diphenyltetrazolium bromide) assay kit (HiMedia®, India), following the manufacturer's instructions. In the MTT assay, 5×103 SiHa cells were seeded into each well of 96-well plates. After 24 h of incubation, progesterone was added at various concentrations (1, 2.5, 5, 10, 20, 25, 30, 50 µM) in the 96 well plates separately, for 6, 12, and 24 h. To measure the cell death after progesterone treatment in the cells, 100 μL of MTT solution (0.5 mg/ml in 1 X PBS) was added to each well, followed by 2 h incubation in a CO2 incubator at 37 °C. Thereafter, MTT solution was replaced with 100 μL of DMSO (Dimethyl sulfoxide) and plates were incubated for another 15 min. To quantify the death of cells, the color reaction at 570 nm was measured using a multiplate reader (Bio-RAD 680, USA).
Cloning strategies
A reporter gene assay was used to study the functional significance of the variation (g.60163379 A>G) in the BRN3A auto-regulatory enhancer region. The wild-type and mutant sequences were cloned in the promoter vector pGL3 (Promega, Madison, USA). The restriction sequence ‘GGTAC’, along with additional four nucleotides (TATA) adaptors, was inserted at the 5' end of the primer set BRN 3A-En-pGL3-Luc (Table 1). For the mutant insert, PCR-based site-directed mutagenesis was used to incorporate desired nucleotide change using mutant-specific forward primer (Table 1). pGL 3 luciferase promoter vector and the inserts were digested with KpnI and ligated with T4 DNA ligase at 16 °C overnight incubation followed by transformation and then selection of E. Coli DH5α cells for the positive clones. The clones were screened with restriction digestion using BglII and XbaI, PCR, and restriction digestion with HpyCH4 III for wild-type and mutant confirmation. The wild-type and mutant clones were validated using Sanger sequencing with the pGL3 RV3 primer (Table 1).
Transfection and luciferase assay
First, we standardized the efficiency of transfection with FuGENE® 6 (Promega, Madison, USA) Transfection Reagent in wild-type and mutant cells. For luciferase assay, SiHa cells were seeded in 12 well plates, and transfection was done for 48h by using transfection reagent and then artificial progesterone (SUSTEN) was added 6h before harvesting the cells. pRL (Renilla luciferase) was used as transfection control in 1/10th ratio. Luciferase assay was done through the Dual-Luciferase® (Promega, Madison, USA) reporter assay system following the manufacturer’s instructions. The activity of the reporter gene was measured using a multiplate luminometer (Synergy HT, Biotek, Vermont, U.S.A.).
In silico and statistical analysis
The screening of clones and confirmation of variations was performed using Chromas Lite (Version-2.01) and NCBI-BLAST (27, 28). We used the New England Biolabs (NEBcutter 2.0) online tool for checking in silico restriction sites, and the UCSC in silico PCR tool for confirming primers and amplicons. For each genotype, allele frequencies were estimated, and the difference in allele frequencies between control and case samples was determined by a Chi-square test using SigmaPlot (Version11.0). To verify that the studied samples follow Hardy-Weinberg equilibrium, a goodness of fit test was performed (see, supplementary data S1). The odds ratios (OR) with confidence intervals (95% CI) and risk assessment were computed with R software using the epitool CRAN package (R) and online tool (http://www.hutchon.net/confidor.htm). One-way ANOVA with Bonferroni correction-posthoc test was performed in STATISTICA software (version 12.0) to differentiate in the mean activity of luciferase reporter gene in wild-type as well as in mutant clones at different progesterone concentrations. p-values corrected for FDR are used to estimate significance levels.
Results
Identification of a novel variation ( BRN3A g.60163379 A>G) in the BRN3A auto-regulatory proximal enhancer region
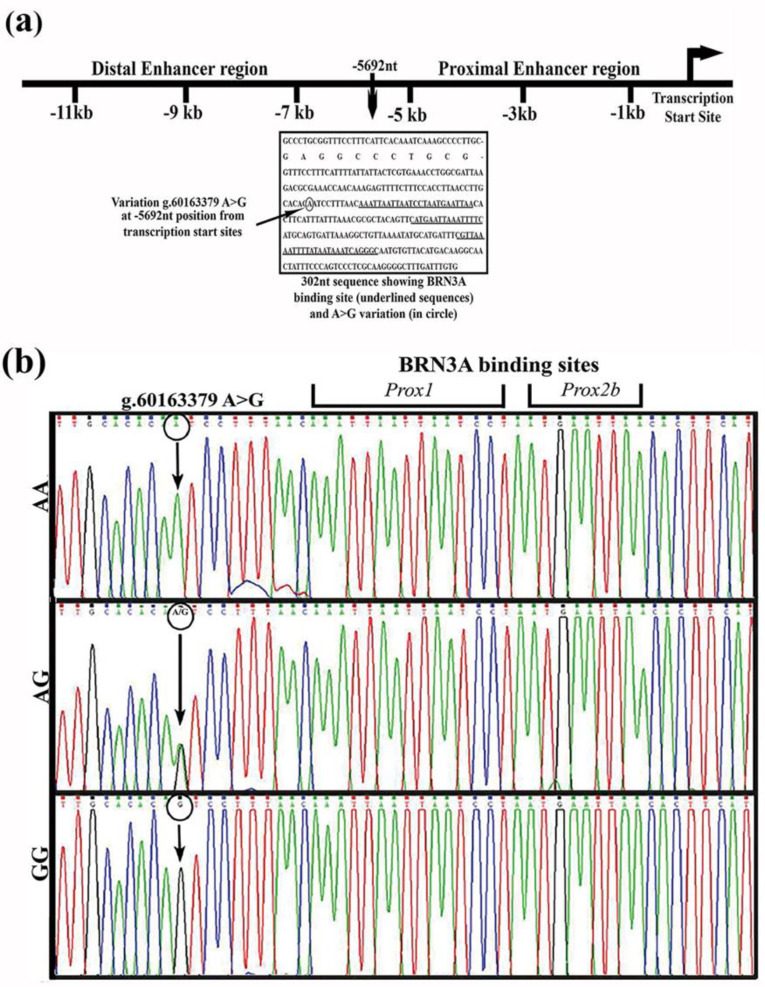
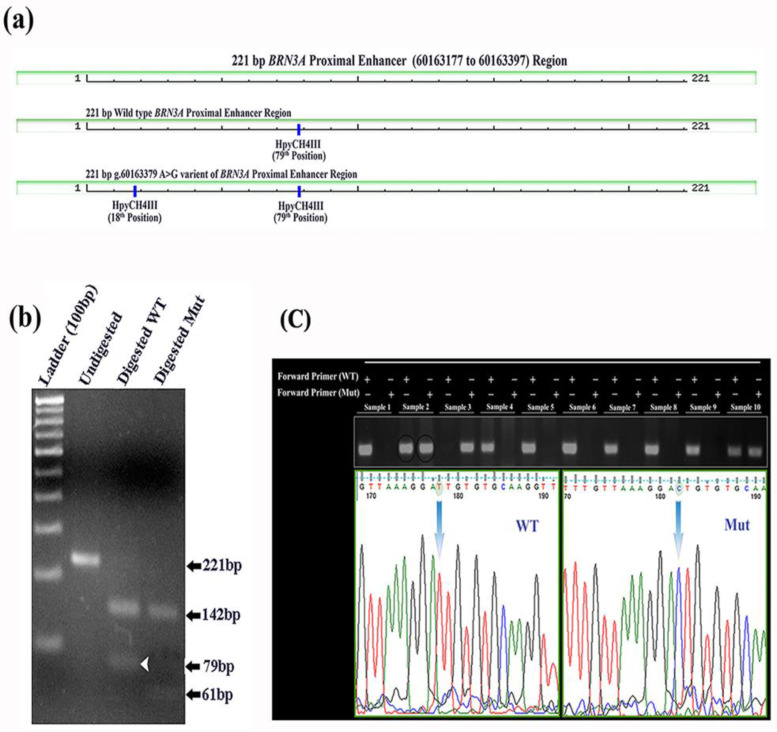
We examined a 302 bp segment of the BRN3A auto-regulatory proximal enhancer region in DNA from cervical cancer biopsy samples of 33 patients, as well as, DNA from pap smear samples of 93 matched controls (women without cervical cancer; Fig. 1a). In sequencing, only one variant BRN3A g.60163379A>G was detected. This was a novel variant located 5692 bases upstream of the transcription start site (TSS), and is the 10 bases upstream of the first binding site of the BRN3A protein (Figure 1a and 1b) in the BRN3A auto-regulatory loop. This A>G transition creates a new restriction site (ACAGT) for HpyCH4 III endonuclease (Figure 2). This mutation has been deposited in the SNP database (ss289117722 and rs1555813).
Fig.1.
Proximal enhancer region of BRN3A shows novel A>G variation in cervical tissue samples. Schematic representation of BRN3A regulatory enhancer regions and the novel variation within 302 bp proximal enhancer region through DNA sequencing, (a) Black rectangular box represents a sequence of 302 bp proximal enhancer region exhibiting multiple autoregulatory BRN3A protein binding sites (underlined sequences) and novel g.60163379A>G variation (mutation site shown in black circle) located 5692 bp from the transcription start site (b) Representative electropherogram of the automated sequencing highlights a single nucleotide substitution, g.60163379A>G (represented through black arrow) at 10 bp from 1st BRN3A binding site (Prox1) in the proximal enhancer of BRN3A
Fig.2.
Genotyping study through RFLP, ARMS-PCR and Sangar sequencing. Digestion pattern of HpyCH4III restriction endonuclease within 221 bp segment of BRN3A proximal enhancer region. (a) Schematic presentation of HpyCH4III restriction endonuclease map in the reverse direction along with fragment size in wild-type and mutant variant. Mutation created an extra HpyCH4III restriction endonuclease digestion site. (b) Agarose gel image showing the 221 bp PCR product digested with HpyCH4III. Wild-type segment with A allele yields two DNA fragments of 142 bp and 79 bp (shown with white arrow in wild-type and absent in mutant) whereas, a mutant sequence containing G allele produces three DNA fragments of 142 bp, 61 bp, and 18 bp. (c) A 227bp ARMS-PCR amplified DNA product resolved on 2% agarose gel in different lanes with wild-type and mutant samples. In odd lane PCR products showing band with wild type specific forward primer whereas in even lanes amplified PCR product displaying bands with mutant specific forward. Interpretation of three genotype AA, GG and AG is based on the bands appeared in odd and even lane. Heterozygous (AG) is also confirmed through sanger DNA sequencing with reverse primer, here sample 2 is used for validation. DNA sequencing was done with reverse primer and thus chromatogram displaying reverse complementary sequences where T is indicating for A nucleotide and C is indicating for G nucleotide
The novel variation ( BRN3A g.60163379A>G) is associated with cervical cancer
An ARMS-PCR, as well as a PCR followed by restriction digestion (PCR-RFLP) method (fig 2), were designed in addition to the DNA sequencing method, to screen the novel A>G variant identified within g.60163379. The variant was screened in DNA from cervical cancer biopsy samples from 143 patients and in DNA from pap smear samples from 627 healthy matched controls. Normal matched controls were women without a family history of cancer for at least three generations and recruited from the same geographic area as the study subjects. In cases, the minor allele frequency (MAF) of the G allele was 30.07 %, and the frequency of the A allele was 69.93 %, whereas in normal individuals (control samples), the MAF was 22.81 % and 77.19 % respectively. It was therefore found that the MAF of G allele was significantly associated with the disease phenotype and was identified as a risk allele (χ2 = 6.315, p ≤ 0.012; OR = 1.46, 95% CI = 1.094 to 1.936, p = 0.0099) (Table 2) for cervical cancer. The genotype frequencies of cases in AA, AG, and GG were 51.65 %, 36.36 %, and 11.89 %, respectively, while in controls it was 59.65 %, 35.09 %, and 5.26 %, respectively. The GG genotype showed a significantly higher risk for cervical cancer compared to the AA (χ2 = 9.17, p ≤ 0.027; OR = 2.60, 95% CI =1.378 to 4.918), or the GG + AG collectively (OR = 1.4, 95% CI = 0.957- 1.985). The risk associated in the case of the recessive model (GG vs AG + AA) was identified OR= 2.42, 95% CI =1.3118-4.4962 however, in the dominant model (AG vs AA) and co-dominant model (AG + GG vs AA) the risk contribution was not found significant (Table 2). The result suggests that G alleles and GG genotypes are significantly associated with disease phenotype and may serve as a risk factor for the development of cervical cancer in HPV-infected females.
Table 2.
Distribution and association study of SNP g.60163379 A>G in normal and cervical cancer sample
| BRN3A g.60163379 A>G | Cancer (%) | Control (%) | ꭓ 2 | p |
|---|---|---|---|---|
| Allele frequency | ||||
| A | 200 (69.93) | 968 (77.19) | 6.315 | 0.012 |
| G | 86 (30.07) | 286 (22.81) | ||
| Genotype frequency | ||||
| AA | 74 (51.75) | 374 (59.65) | ||
| AG | 52 (36.36) | 220 (35.09) | ||
| GG | 17 (11.89) | 33 (5.26) | 9.173 | 0.027 |
| Risk assessment | OR score | 95% CI | ||
| G vs A | 1.46 | 1.094 to 1.936 | ||
| AG vs AA | 1.19 | 0.807 -1.768 | ||
| GG vs AA | 2.60 | 1.378 - 4.918 | ||
| (AG+GG) vs AA | 1.38 | 0.957 to 1.985 | ||
| GG vs (AG+ AA) | 2.42 | 1.3118-4.4962 | ||
Table showing distribution of SNP g.6063379 A>G. Allele A distribution in normal population is 77.19 % and 69.93 % in cervical cancer whereas distribution of G allele in normal individual is 22.81% and 30.07 % in cervical cancer case. Allele frequency showing statistically significant differences (ꭓ2 = 6.315, p = 0.012) in the distribution of G allele in control and case. Genotypic frequency of AA, AG, GG genotypes in control sample was observed 59.65 %, 35.09 %, 5.26 % and 51.75 %, 36.36 %, 11.89 % distribution observed in normal population respectively. Comparing genotypic frequency in case and control showing statistically significant differences (ꭓ2 = 9.17, p = 0.027) in the distribution. Odd ratio (OR) assessment identified GG genotype as risk allele for developing cervical cancer (OR = 2.6 p = 0.004, 95 % CI = 1.378 - 4.918). OR in recessive model (GG vs AG + AA) was identified as a risk allele with OR =2.42 however not as a significant risk contributor in dominant (AG vs AA) and co-dominant (AG+GG vs AA) model.
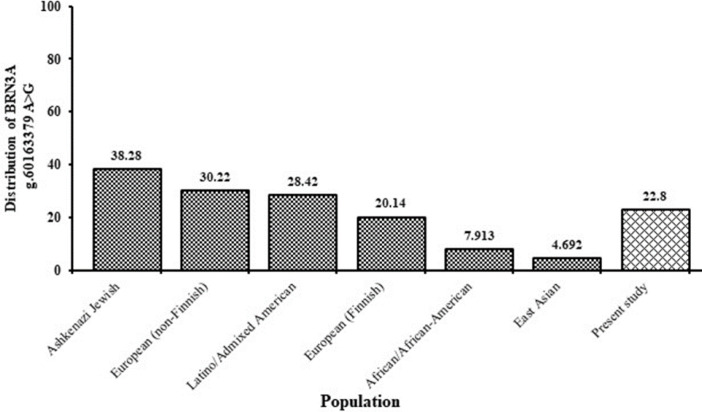
The MAF of the G allele in cases was further compared with the controls of the IndiGenomes database and was found to be significantly linked with the phenotype (OR= 1.63, 95%CI= 1.235 to 2.138) (Table 3). The significant risk was found to be associated with genotypes of cases of the present study when compared with the controls of IndiGenomes database in an association study in GG vs AA (OR= 3.02, 95% CI= 1.7434 to 5.8597) and recessive model, GG vs AG + AA (OR= 2.86, 95% CI= 1.5892 to 5.1354) (Table 3). Although, the risk association with the dominant (AG vs AA) and the co-dominant model (AG + GG vs AA) with OR=1.25, 95 % CI= 0.9222 to 1.9661 and OR= 1.57, 95 % CI= 1.1047 to 2.2323 respectively, are not found significant. Further, the MAF of the 'G' allele was retrieved from the GnomAD database for a normal population across the world and compared with the normal samples of the present study (Figure 3). MAF of the ‘G’ allele in the normal samples of our study was close to the European Finnish population (MAF = 20.14%). However, its distribution was considerably low when compared to the Ashkenazi Jewish population (MAF = 38.28 %), European non-Finnish population (MAF = 30.22%), and Latino/Admixed American population (MAF=28.42%). Interestingly, the distribution of ‘G’ in the East Asian population was markedly low (MAF = 4.6%) in comparison to our report (MAF = 22.8%). However, it was not reported for the South Asian population. This variant was not reported in INDEX-db and ClinVar databases.
Table 3.
Comparison of cases of present study vs. IndiGenomes database control samples for SNP BRN3A g.60163379 (A>G)
| BRN3A g.60163379 A>G | Cancer (%) | Indigenomes (distribution in %) | ꭓ 2 | p |
|---|---|---|---|---|
| Allele frequency | ||||
| A | 200 (69.93) | 1610 (78.92) | 11.68 | 0.001 |
| G | 86 (30.07) | 426 (20.88) | ||
| Genotype frequency | ||||
| AA | 74 (51.75) | 640 (62.75) | ||
| AG | 52 (36.36) | 334 (32.75) | ||
| GG | 17 (11.89) | 46 (4.51) | 17.38 | 0.001 |
| Risk assessment | OR score | 95% CI | ||
| G vs A | 1.63 | 1.235 to 2.138 | ||
| AG vs AA | 1.35 | 0.9222 to 1.9661 | ||
| GG vs AA | 3.20 | 1.7434 to 5.8597 | ||
| (AG+G/G) vs AA | 1.57 | 1.1047 to 2.2323 | ||
| GG vs (AG+ AA) | 2.86 | 1.5892 to 5.1354 | ||
Distribution of SNP g.60163379 A>G in cervical cancer sample and I ndigenomes database normal sample. Comparing allelic and genotypic frequency in case and control showing statistically significant differences in the distribution. Risk association study through Odd ratio identified GG genotype as risk allele for developing cervical cancer (OR = 3, 95 % CI = 1.7434 to 5.8597). OR in recessive model (GG vs AG + AA) was identified as a risk allele with OR =2.86, 95 % CI = 1.5892 to 5.1354, however, not as a significant risk contributor in dominant (AG vs AA) and co-dominant (AG+GG vs A/A) model.
Fig. 3.
Distribution of SNP g.60163379 A>G variant in different ethnic group. Graph showing the distribution of SNP g.60163379 A>G variant in different ethnic group in the world retrieved through GnomAD database and compared with the control sample of the present study
The novel variation ( BRN3A g.60163379, A>G) shows increased progesterone dependent luciferase activity
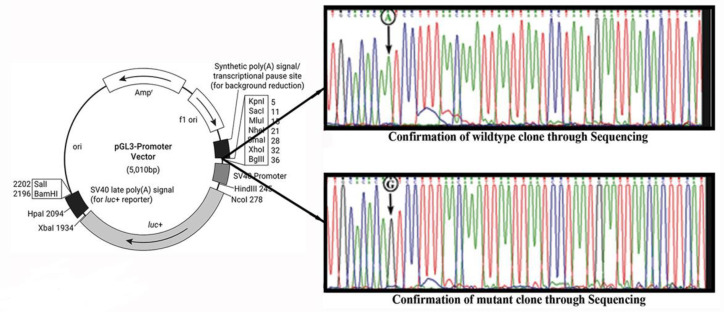
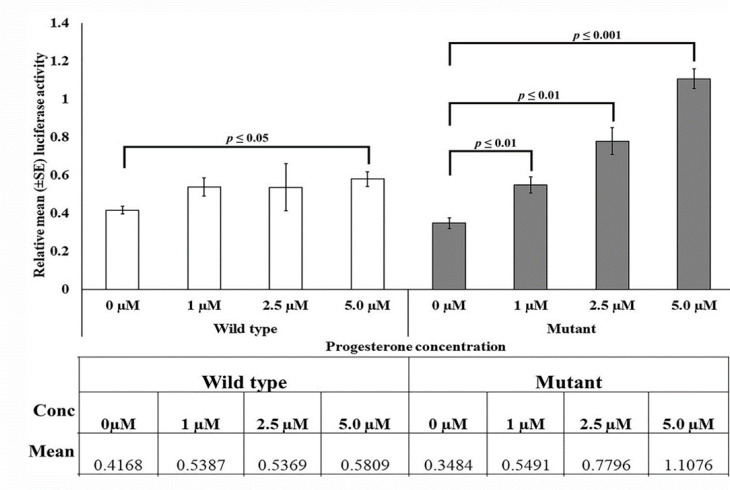
The referred sequence series were also obtained that had the progesterone receptor-β (PR-β) binding site in the AGTCCTTT sequence in mice uteroglobin gene (Bailly et al, 1986). In the present study, two clones were constructed - one containing wild-type allele (g.60163379A), and the other containing mutant allele (g.60163379 G), after inserting a 221 bp fragment from BRN3A auto-regulatory proximal enhancer region in pGL3 promoter vector upstream of SV40 promoter and luciferase gene was used as a reporter gene in pGL3 promoter vector (Figure 4). In SiHa cell-line cultures, different concentrations of progesterone were added to induce the expression of PR-β (PR-β expression is progesterone-ligand dependent) (29). By determining the optimal concentration range of progesterone using a MTT assay, luciferase gene activity was evaluated in the presence of various concentration levels of progesterone in SiHa cells transfected with wild-type and mutant clones. To transfect clones in SiHa cells 2µg plasmids were used for 48 - 72 h, and progesterone at different concentrations (1 µM, 2.5 µM, and 5 µM) was added 6 h before harvesting the cells. pRL was used as transfection control in the 1/10 ratio to the main cloned vector. Luciferase assay was performed with the Dual-Luciferase Reporter Assay System (Promega, Madison, USA). The mean luciferase activity was plotted on Y-axis against the normalized value of pRL (Figure 5). In cloned cultures containing the major allele 'A,' no change in mean luciferase activity was observed with the increase (due to addition) of progesterone concentration. In contrast, the mean luciferase activity gradually increased in the cultures containing minor allele 'G'. Our results on one-way analysis of variance indicated a non-significant change in luciferase activity at 1 µM and 2.5 µM progesterone levels in cloned cultures containing major allele 'A' (1 µM: F = 6.147, p = 0.048; 2.5 µM: F = 0.95, p = 0.367), while a narrow significant change at 5 µM (F = 14.26, p = 0.009) in the wild-type. Whilst, there was a dramatic increase in luciferase activity (1.5 to 3.5-fold for 1 to 5 µM) in the cultures containing minor allele 'G'; as a consequence of increase in progesterone concentration (1 µM: F = 16.582, p = 0.0067; 5µM: F = 165.225, p = 0.000014). The results suggest that mutant proximal enhancer region can exhibit variable responses to changes in progesterone concentration and infer that mutant proximal enhancer regions have a higher affinity for progesterone.
Fig. 4.
Cloning strategy for the wild-type (A) and mutant (G) variants in pGL3 promoter vector for the functional characterization of SNP g.60163379 A>G. Wild-type (g.60163379A) and mutant (g.60163379G) variant clones were generated and confirmed through Sanger sequencing shown in the electropherogram. The wild-type and mutant segments were cloned upstream of SV40 promoter and luciferase gene. Due to A>G point mutation, creation of any functional binding site was measured by differential luciferase activity in comparison to wild-type
Fig. 5.
Functional characterization of SNP g.60163379 A>G variant through luciferase assay in cervical cancer cells. Dual-luciferase assay was performed after the transfection of wild-type and mutant clones in SiHa cells. Progesterone was given externally 6 h before harvesting the cells in different concentrations (1 µM, 2.5 µM, and 5 µM). The bar diagram represents the relative luciferase activity of the wild-type and mutant variant clones transfected in SiHa cells under different concentrations of progesterone. Each bar is showing the mean (±) SE of merged data of at least three independent experiments with three replicates. Groups compared are connected by a line displaying significance value above the bar. The table below the graph showing the relative mean luciferase activity in wild-type and mutant under given progesterone concentrations
Discussion
BRN3A is a TATA-less transcription factor, therefore its regulation is mainly determined by its enhancer region, located five to nine kilobases upstream of its transcription start site. Proximal enhancer region of BRN3A has higher binding affinity to the BRN3A transcription factor and acts in its positive autoregulation (15, 16, 18). In a previous study from our laboratory, we demonstrated that the proximal enhancer region was involved in maintaining BRN3A expression at a constant level under genotoxic stress in cervical cancer (30). This stress was imposed by cisplatin (5µg/ml) that activates HIPK2, a known co-repressor of BRN3A and its target genes (31, 32). Despite elevated HIPK2 levels, BRN3A expression in cervical cancer did not change, suggesting either HIPK2 is unable to interact physically with BRN3A, or another key player plays a role in maintaining BRN3A expression in cervical cancer. As we have shown earlier, chromatin immunoprecipitation assays performed with an anti-HIPK2 antibody allowed us to pull the 302 bp proximal enhancer region considered in this study. Also, the anti-BRN3A antibody pulled the same sequences of proximal enhancer regions. These findings altogether showed that the auto-regulatory region is a key target of BRN3A and its co-repressor, HIPK2 (30).
In the present study, a novel variation in BRN3A was identified; g.60163379A>G (or c.-5692A>G), near the auto-regulatory loop of BRN3A (10 bp from the multiple BRN3A binding site). This variation is due to the A>G transition at genomic position g.60163379. This variation is submitted in dbSNP (ss289117722 further assigned rs1555813). We examined the distribution of variant MAF of G in normal and cervical cancer samples collected from the East Uttar Pradesh population, and MAF of G showed a much higher frequency in the cancer samples than in the normal samples, as well as indicating a potential cancer risk allele. The genotype study revealed that 'GG' was a risk factor for developing cervical cancer (OR = 2.60; p = 0.004). The comparison of the frequency of this variation with the cases from the present study and the frequency of normal population derived using IndiGenomes data for Indian population confirms that the G allele is associated with substantial risk for the development of disease. An analysis of the distribution of this variation in a population across the world revealed highly variable occurrences across different ethnic groups. According to the data obtained from GnomAD database for different ethnic groups, the frequency of this variation is highest among Ashkenazi Jewish populations (MAF= 38.28%), followed by European (non-Finnish) populations (MAF= 30.22%), but lowest among East Asian populations. However, it was not reported for South Asians. The present study gives insight into the distribution of this variation in the South Asian population. It is arguable that the location of the observed SNP (i.e., on the auto-regulatory enhancer loop of BRN3A) and its association with cancer provide new opportunities to examine how this SNP regulates expression of BRN3A.
Bailly et al. (33) reported the high binding affinity of Progesterone Receptor (PR-β) to AGTCCTTT sequences in the uteroglobin gene of mice. PR genes contain estrogen-responsive promoter, thus their expression is largely controlled by estrogen. Importantly, estrogen and progesterone function as ligands in a synergistic manner for the PR (34) Noteworthy that, expression of PR-β is predominant in gynecological cancers including cervical and ovarian cancers (35) To assess whether the observed variation has a functional significance, a cell-based luciferase reporter assay was performed under varying levels of progesterone. The results showed that this variation impacts regulatory function. In transfected SiHa cell lines with clones carrying a mutant allele, there was a progesterone-dependent increase in luciferase gene expression compared to transfected cells with a wild-type allele. The results indicate that the SNP in proximal enhancer region might affect BRN3A expression through a progesterone-dependent pathway.
Conclusions
The present study reports a novel polymorphism BRN3A g.60163379 A>G in the auto-regulatory region of the BRN3A oncogene and this variant significantly associated with an increased risk of cervical cancer. Reporter gene assays indicated that this variant resulted in higher luciferase expression in cervical cancer cells in the presence of progesterone. This study highlights that the genetic variations in non-coding cis-regions of oncogenes are critical and future studies can be extended to explore these variations' relationships with disease susceptibility and potential use as biomarkers for disease diagnosis, prediction, and prognosis.
Conflict of Interest
The authors declare no conflict of interest.
Data availability
The SNP data can be found in the NCBI repository - dbSNP: SS 289117722, rs ID: rs1555813. The genotype data that support the findings of this study are available from the corresponding author upon request.
Acknowledgements
The authors thank the individuals who participated in the study and their family members. We also thank DBT-funded Centre for Genetic Disorders for providing the automated sequencing facility, and Dr. A. C. Banerjea, NII- New Delhi for providing luminometer facility. We thank Dr. Yashvant Patel and Dr. Mukulika Ray, Research Associates from Department of Zoology, BHU, Varanasi for technical support. The equipment facilities in the department and institute supported by UGC-CAS, DST-FIST and the financial support from CSIR, DBT, IoE to JKR; fellowships from ICMR, UGC to AP and Lady Tata Memorial Trust, Mumbai, India to BPDP are greatly appreciated.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Bruni L, Albero G, Serrano B, et al. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre), Human Papillomavirus and Related Diseases in India. Summary Report . 2019 [Google Scholar]
- 3.Xi LF, Koutsky LA, Castle PE, et al. Relationship between cigarette smoking and human papilloma virus types 16 and 18 DNA load. Cancer Epidemiol Biomarkers Prev. 2009;18:3490–3496. doi: 10.1158/1055-9965.EPI-09-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srivastava S, Gupta S, Roy JK. High prevalence of oncogenic HPV-16 in cervical smears of asymptomatic women of eastern Uttar Pradesh, India: a population-based study. J Biosci. 2012;37:63–72. doi: 10.1007/s12038-012-9181-y. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh I, Mandal R, Kundu P, et al. Association of Genital Infections Other Than Human Papillomavirus with Pre-Invasive and Invasive Cervical Neoplasia. J Clin Diagn Res. 2016;10:XE01–XE06. doi: 10.7860/JCDR/2016/15305.7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng ZM, Baker CC. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci. 2006;11:2286–2302. doi: 10.2741/1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gul S, Murad S, Javed A. Prevalence of High risk Human Papillomavirus in cervical dysplasia and cancer samples from twin cities in Pakistan. Int J Infect Dis. 2015;34:14–19. doi: 10.1016/j.ijid.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 8.Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeo-Teh NSL, Ito Y, Jha S. High-Risk Human Papillomaviral Oncogenes E6 and E7 Target Key Cellular Pathways to Achieve Oncogenesis. Int J Mol Sci. 2018;19:1–27. doi: 10.3390/ijms19061706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mammas IN, Sourvinos G, Giannoudis A, et al. Human papilloma virus (HPV) and host cellular interactions. Pathol Oncol Res. 2008;14:345–354. doi: 10.1007/s12253-008-9056-6. [DOI] [PubMed] [Google Scholar]
- 11.Ndisang D, Budhram-Mahadeo V, Latchman DS. The Brn-3a transcription factor plays a critical role in regulating human papilloma virus gene expression and determining the growth characteristics of cervical cancer cells. J Biol Chem. 1999;274:28521–28527. doi: 10.1074/jbc.274.40.28521. [DOI] [PubMed] [Google Scholar]
- 12.Ndisdang D, Morris PJ, Chapman C, et al. The HPV-activating cellular transcription factor Brn-3a is overexpressed in CIN3 cervical lesions. J Clin Invest. 1998;101:1687–1692. doi: 10.1172/JCI1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ndisang D, Budhram-Mahadeo V, Singer A, Latchman DS. Widespread elevated expression of the human papilloma virus (HPV)-activating cellular transcription factor Brn-3a in the cervix of women with CIN3 (cervical intraepithelial neoplasia stage 3) Clin Sci (Lond) 2000;98:601–602. [PubMed] [Google Scholar]
- 14.Ndisang D, Budhram-Mahadeo V, Pedley B, et al. The Brn-3a transcription factor plays a key role in regulating the growth of cervical cancer cells in vivo. Oncogene. 2001;20:4899–4903. doi: 10.1038/sj.onc.1204634. [DOI] [PubMed] [Google Scholar]
- 15.Trieu M, Rhee JM, Fedtsova N, et al. Autoregulatory sequences are revealed by complex stability screening of the mouse brn-3 0 locus. J Neurosci. 1999;19:6549–6558. doi: 10.1523/JNEUROSCI.19-15-06549.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trieu M, Ma A, Eng SR, et al. Direct autoregulation and gene dosage compensation by POU-domain transcription factor Brn3a. Development. 2003;130:111–121. doi: 10.1242/dev.00194. [DOI] [PubMed] [Google Scholar]
- 17.Gruber CA, Rhee JM, Gleiberman A, et al. POU domain factors of the Brn-3 class recognize functional DNA elements which are distinctive, symmetrical, and highly conserved in evolution. Mol Cell Biol. 1997;17:2391–2400. doi: 10.1128/mcb.17.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gascoyne DM, Dunne J, Behjati S, et al. EWS/ETS proteins promote expression and regulate function of the homeodomain transcription factor BRN3A. Oncogene. 2010;29:3134–3145. doi: 10.1038/onc.2010.72. [DOI] [PubMed] [Google Scholar]
- 19.Wright AF. Genetic Variation: Polymorphisms and Mutations. eLS . 2005:1–9. [Google Scholar]
- 20.Erichsen HC, Chanock SJ. SNPs in cancer research and treatment. Br J Cancer. 2004;90:747–751. doi: 10.1038/sj.bjc.6601574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Travé G, Zanier K. HPV-mediated inactivation of tumor suppressor p53. Cell Cycle. 2016;15:2231–2232. doi: 10.1080/15384101.2016.1191257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hrstka R, Coates PJ, Vojtesek B. Polymorphisms in p53 and the p53 pathway: roles in cancer susceptibility and response to treatment. J Cell Mol Med. 2009;13:440–453. doi: 10.1111/j.1582-4934.2008.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Dumont P, Della Pietra A, et al. The codon 47 polymorphism in p53 is functionally significant. J Biol Chem. 2005;280:24245–24251. doi: 10.1074/jbc.M414637200. [DOI] [PubMed] [Google Scholar]
- 24.Deng N, Zhou H, Fan H, et al. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget. 2017;8:110635–110649. doi: 10.18632/oncotarget.22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F, Lupski JR. Non-coding genetic variants in human disease. Hum Mol Genet. 2015;24:R102–110. doi: 10.1093/hmg/ddv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gravitt PE, Peyton CL, Apple RJ, et al. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J Clin Microbiol. 1998;36:3020–3027. doi: 10.1128/jcm.36.10.3020-3027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson M, Zaretskaya I, Raytselis Y, et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel B, Elguero S, Thakore S, et al. Role of nuclear progesterone receptor isoforms in uterine pathophysiology. Hum Reprod Update. 2015;21:155–73. doi: 10.1093/humupd/dmu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye J, Coulouris G, Zaretskaya I, et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das Purkayastha BP, Roy JK. Molecular analysis of oncogenicity of the transcription factor, BRN3A, in cervical cancer cells. J Cancer Res Clin Oncol. 2011;137:1859–1867. doi: 10.1007/s00432-011-1059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Stefano V, Rinaldo C, Sacchi A, et al. Homeodomain-interacting protein kinase-2 activity and p53 phosphorylation are critical events for cisplatin-mediated apoptosis. Exp Cell Res. 2004;293:311–320. doi: 10.1016/j.yexcr.2003.09.032. [DOI] [PubMed] [Google Scholar]
- 32.Wiggins AK, Wei G, Doxakis E, et al. Interaction of Brn3a and HIPK2 mediates transcriptional repression of sensory neuron survival. J Cell Biol. 2004;167:257–267. doi: 10.1083/jcb.200406131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailly A, Page CL, Rauch M, et al. Sequence-specific DNA binding of the progesterone receptor to the uteroglobin gene: effects of hormone, antihormone and receptor phosphorylation. The EMBO Journal. 1986;5:3235–3241. doi: 10.1002/j.1460-2075.1986.tb04634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Remoue F, Jacobs N, Miot V, et al. High intraepithelial expression of estrogen and progesterone receptors in the transformation zone of the uterine cervix. Am J Obstet Gynecol. 2003;189:1660–1665. doi: 10.1016/s0002-9378(03)00852-4. [DOI] [PubMed] [Google Scholar]
- 35.Fujimoto J, Ichigo S, Hori M, et al. Expression of progesterone receptor form A and B mRNAs in gynecologic malignant tumors. Tumour Biol. 1995;16:254–260. doi: 10.1159/000217942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The SNP data can be found in the NCBI repository - dbSNP: SS 289117722, rs ID: rs1555813. The genotype data that support the findings of this study are available from the corresponding author upon request.