Abstract
Congenital Heart Disease (CHD) is the most common birth defect and leading cause of infant mortality, yet molecular mechanisms explaining CHD remain mostly unknown. Sequencing studies are identifying CHD candidate genes at a brisk rate including MINK1, a serine/threonine kinase. However, a plausible molecular mechanism connecting CHD and MINK1 is unknown. Here, we reveal that mink1 is required for proper heart development due to its role in left-right patterning. Mink1 regulates canonical Wnt signaling to define the cell fates of the Spemann Organizer and the Left-Right Organizer, a ciliated structure that breaks bilateral symmetry in the vertebrate embryo. To identify Mink1 targets, we applied an unbiased proteomics approach and identified the high mobility group architectural transcription factor, Hmga2. We report that Hmga2 is necessary and sufficient for regulating Spemann’s Organizer. Indeed, we demonstrate that Hmga2 can induce Spemann Organizer cell fates even when β-catenin, a critical effector of the Wnt signaling pathway, is depleted. In summary, we discover a transcription factor, Hmga2, downstream of Mink1 that is critical for the regulation of Spemann’s Organizer, as well as the LRO, defining a plausible mechanism for CHD.
Keywords: MINK1, HMGA2, Wnt signaling, congenital heart disease, Xenopus, left-right patterning, β-catenin, Spemann Organizer, gastrulation
Graphical Abstract:
INTRODUCTION
Congenital Heart Disease (CHD) is the most common birth defect globally and the leading cause of infant mortality and morbidity (Pierpont et al., 2018). Despite this impact, the molecular basis for most CHD remains undefined. In CHD cohorts, whole exome sequencing studies are identifying de novo and inherited alleles that become CHD candidate genes (Homsy et al., 2015; Jin et al., 2017; Zaidi et al., 2013). However, while gene identification is a critical step, it does not establish plausible disease mechanisms, which requires testing in animal models. Recent sequencing efforts of multiple parent-offspring trios identified de novo single nucleotide variations in MINK1 considered to be likely damaging (Jin et al., 2017; Zaidi et al., 2013). MINK1 was one of 12 genes where the number of de novo mutations reached genome wide significance (Jin et al., 2017).
MINK1, a serine-threonine kinase, has not previously been implicated in heart development (Hu et al., 2004). In Xenopus, a role for MINK1 has been described in convergent extension cell movements during gastrulation via the Wnt/ planar cell polarity (PCP) pathway (Daulat et al., 2012; Mikryukov & Moss, 2012; Paricio et al., 1999; Su et al., 1998). PCP signaling is dictated by the asymmetric accumulation of core PCP proteins (Wallingford, 2012). mink1 knockdown in embryos led to a loss of phosphorylation on the core PCP protein Prickle disrupting protein localization (Daulat et al., 2012). Unfortunately, despite this knowledge of MINK1 function, we do not have a plausible mechanism that could explain a role for MINK1 in heart development and CHD.
Here, we show that depletion of mink1 in Xenopus tropicalis leads to heart malformations. Specifically, mink1 crispant hearts have reversal of the outflow tract along the left-right (LR) axis. Indeed, depletion of mink1 leads to global disruption of LR patterning.
In most vertebrates, the LR axis is established at a conserved structure called the Left-Right Organizer (LRO) (Blum et al., 2009; Hamada & Tam, 2014). The LRO is a transient structure formed from the posterior mesoderm towards the end of gastrulation. Motile cilia in the LRO drive fluid flow leftward which breaks initial bilateral symmetry. Earlier in embryonic development, before gastrulation, the LRO precursors form in the superficial mesoderm of the Spemann Organizer (Blum et al., 2009). Therefore, mispatterning of the mesoderm along the dorsal-ventral axis can affect both gastrulation and LR patterning. In fact, defects in canonical Wnt signaling can alter the dorsal-ventral axis and have been implicated also in LR patterning (Griffin et al., 2018; Walentek et al., 2012). In our analysis of mink1 crispants, we find a loss of dorsal mesoderm patterning and disruption in canonical Wnt signaling, establishing an additional function for mink1 beyond the Wnt/PCP signaling previously implicated.
To improve our understanding of the molecular mechanism that connects mink1 to dorsal-ventral patterning of the mesoderm, we sought to identify downstream targets of Mink1 using unbiased proteomic approaches. By testing a subset of proteins identified as less phosphorylated when mink1 was depleted, Hmga2 emerged as an attractive potential downstream effector of Mink1. HMGA2 belongs to the high-mobility group family of chromatin-associated transcription factors. In Xenopus, hmga2 is required for neural crest cell specification (Vignali & Marracci, 2020).
During our analysis of Hmga2, we discovered that Hmga2 is necessary and sufficient for the cell fate specification of Spemann’s Organizer. In fact, Hmga2 appears essential downstream of the canonical Wnt signaling molecule, β-catenin, to specify Spemann’s Organizer. Therefore, our results identify a transcription factor downstream of canonical Wnt signaling that is essential for specification of Spemann’s Organizer. Starting with the analysis of a disease candidate gene from CHD patients, we connect cardiac development, LR patterning, and specification/maintenance of Spemann’s Organizer to mink1. Additionally, via proteomics, we identify Hmga2 as a downstream effector of Mink1 and discover an unexpected role of this transcription factor in regulating Spemann Organizer cell fates.
METHODS
Xenopus Husbandry
Xenopus tropicalis were raised, housed, and cared for in according to established protocols that were approved by Yale IACUC.
Xenopus Embryonic Manipulations
Using standard protocols, we carried out embryonic injections at the one-cell stage (Khokha et al., 2002). For mink1, CRISPR sgRNAs with the following target sequences (CR1: 5’-AGGGCCGCCATGTCAAGACTGGG-3’, CR2: 5’-GCTGGCCGCTATCAAGGTCATGG-3’, CR3: 5’-AGCAGATGGACGTCCTTGAGGGG-3’) were designed from the v9.0 model of the Xenopus tropicalis genome using CRISPRSCAN (Moreno-Mateos et al., 2015). sgRNAs (500 pg) and Cas9 protein (1.2 ng) were mixed, incubated at 37°C, and then were injected into one cell embryos along with a fluorescent tracer (Alexa-488). For hmga2, CRISPR sgRNAs with the following targeting sequences (CR1: 5’-CGGAGCAACCTGCATCCCCC-3’, CR2: 5’-GACCTCGGGGTAGACCAAAA -3’) were designed and injected as described above. We obtained a full length human MINK1 cDNA (IMAGE clone: 4384442) and human HMGA2 ORFeome clone (ID:100064147) from Horizon Discovery. We generated in vitro capped mRNA using mMessage mMachine Sp6 Transcription Kit (Invitrogen) according to the manufacturer’s instructions. Variants (MINK1 K54R, HMGA2 S18D, HMGA2 S43D) were generated using Q5 site directed mutagenesis (NEB) according to manufacturer’s instructions. Overexpression studies were performed by injecting MINK1 mRNA (500pg) or HMGA2 mRNA (600pg) into one-cell embryos with an RFP mRNA fluorescent tracer. Rescue studies were performed by injecting MINK1 mRNA (75pg), β catenin-GFP (50pg, Addgene #16839), or HMGA2 mRNA (3pg) in a subsequent injection to CRISPR-Cas9.
Cardiac Looping
Stage 45 Xenopus embryos were paralyzed with tricaine and scored for cardiac looping by inspection with light stereomicroscopy. Looping was determined by the position of the outflow tract compared to the anterior-posterior axis of the embryo. Normal outflow tract looping was defined as looping to the right of the embryo, abnormal looping was defined as looping to the left of the embryo or remaining unlooped along the midline.
Genotyping
We amplified the CRISPR cut site using PCR with Phusion High-Fidelity DNA Polymerase (NEB) or Platinum SuperFi II Taq Polymerase (ThermoFisher). Amplification conditions for Phusion High-Fidelity DNA Polymerase were 98°C for 2 minutes, followed by 30 cycles at 98°C for 15 seconds, 65°C for 30 seconds and 72°C for 30 seconds. Amplification conditions for SuperFi II Taq Polymerase were 98°C for 30 seconds, followed by 30 cycles at 98°C for 10 seconds, 60°C for 15 seconds and 72°C for 15 seconds. Final extension was at 72°C for 10 minutes. PCR products were purified using PCR & DNA Cleanup Kit (Monarch) and sequenced with one of the amplification primers. Primers for amplification were:
| sgRNA | Forward Primer | Reverse Primer |
|---|---|---|
| mink1 CR1 | gcaaccaaactgcactcctg | cccagagaacagctttttggg |
| mink1 CR2 | gcaaccaaactgcactcctg | cccagagaacagctttttggg |
| mink1 CR3 | agctgaaatacgaagcccgt | tgatggtgtctgcactccaa |
| hmga2 CR1 | ccatcccacagtgaggcatt | gaggagcagctcagcctatc |
Whole Mount in situ Hybridization
Whole mount in situ hybridization was carried out as previously described (Khokha et al., 2002). Briefly, whole Xenopus embryos were collected and fixed in MEMFA overnight at 4° and dehydrated stepwise into ethanol. For LROs, stage 14 or 19 embryos were dissected to reveal the internal posterior mesoderm tissue. Mink1 crispants with open blastopores were included in this assay. After rehydration, embryos were then hybridized with digoxigenin-labeled antisense RNA probes. Antisense probes for hmga2: XGC7558101 (Dharmacon), and the remaining from NCBI goosecoid: TGas129E16, nodal3: TGas011k18, foxj1: Tneu058M03, pitx2c: TNeu083k20, dand5: TEgg007d24, nodal1: TGas124h10 were in vitro transcribed with T7 High Yield RNA Synthesis Kit (NEB). Embryos were then washed and blocked prior to incubation with anti-DIG-Fab fragments (Roche) for 4 hours at room temperature. BM purple (Sigma) was used to visualize mRNA expression prior to post-fixation in 4% paraformaldehyde with 0.1% glutaraldehyde. To eliminate pigments, embryos were bleached and then imaged in brightfield using a Canon EOS camera DS126201.
Phosphopeptide Mass Spectrometry
Protein Isolation
Embryos were microinjected with mink1 sgRNA (CR 3)/Cas9 protein and fluorescent dye 488 as described above. Wildtype and mink1 Crispant embryos were collected at stages 10, 11 and 12, and subjected to lysis in 1x RIPA buffer (Sigma R0278) with 1x Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific). Lysates were subjected to centrifugation at 14,000 rpm for 30 minutes at 4°C. Supernatants were taken as the soluble fraction and the pellet discarded. Protein concentrations were determined using the Bradford method (Bradford, 1976).
Phosphopeptide Enrichment, MS Data Analysis and Protein Identification
The samples were subjected to TiO2-based phosphopeptide enrichment using TopTip MicroSpin Columns (Glygen Corp., USA) as described previously (Goel et al., 2018; Krishnan et al., 2012; Rich et al., 2016). All samples were analyzed on the LTQ-Orbitrap XL mass spectrometer (Thermo Scientific). The mass spectrometer was operated in the Data-Dependent mode with the Orbitrap operating at 60,000 FWHM (Full Width at Half Maximum) and 17,500 FWHM for MS and MS/MS, respectively. 5μL of each sample (0.05 μg/μL) was randomly injected, interspersed with control samples, allowing for correction due to potential batch effects. All MS/MS spectra were analyzed using Mascot. Mascot was set up to search Xenopus tropicalis v9.0. Mascot was searched with fragment ion mass tolerance of 0.020 Da and parent ion tolerance of 10.0 PPM. Phosphorylation of serine, threonine and tyrosine were specified as variable modifications. Scaffold v4.8.9 was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 90% probability by the 0.2% false discovery rate. Protein identifications were accepted if they could be established at greater than 95% probability and contained a minimum of 2 identified peptides. Protein probabilities were assigned by the Portein Prophet algorithim (Nesvizhskii et al., 2003). Peptide data deposited to the ProteomeXchange consortium via the PRIDE partner repository.
Post Translational Modifications Site Localization
Scaffold PTM (Proteome Software, Portland, Oregon, USA) was used to annotate PTM sites derived from MS/MS sequencing results obtained using Scaffold (version Scaffold_4.8.9). Using the site localization algorithm developed by Sean A Beausoleil, Judit Villén, Scott A Gerber, John Rush & Steven P Gygi, Nature Biotechnology 24, 1285 – 1292 (2006), Scaffold PTM re-analyzes Scaffold PTM re-analyzes MS/MS spectra identified as modified peptides and calculates Ascore values and site localization probabilities to assess the level of confidence in each PTM localization (Beausoleil et al., 2006). Scaffold PTM then combines localization probabilities for all peptides containing each identified PTM site to obtain the best estimated probability that a PTM is present at that particular site.
Western Blot
Total protein lysates were extracted from uninjected and injected embryos as explained above. Western blotting was carried out with 20 μg of protein loaded in each lane of 4–12% NuPAGE Bis-Tris Bolt gels. Anti β-catenin (Santa Cruz-7963, 1:1,000 in 5% milk) and anti-GAPDH (Invitrogen AM4300, 1:10,000 in PBST containing 5% dry milk) primary antibodies were used. Anti-mouse or anti-rabbit HRP conjugated secondary antibodies were used (Jackson Immuno Research Laboratories, 715-035-150 or 211-032-171 1:10,000 dilution). Western blot quantifications were performed in ImageJ by a protocol developed by Hossein Davarinejad.
RESULTS
Mink1 in Heart Development
To study Mink1’s role in heart development, we depleted mink1 via G0 CRISPR in Xenopus tropicalis. Xenopus is an ideal model to study heart development for numerous reasons. Xenopus is a high-throughput system with year-round breeding (Blum et al., 2009). Compared to mammalian models, Xenopus has rapid heart development and can be readily manipulated with gain- and loss-of-function approaches (Blum et al., 2009; Garfinkel & Khokha, 2017). Additionally, the large clutch sizes are advantageous in order to generate sufficient numbers for analysis; this is particularly relevant for LR development since mouse mutants of most LR genes are only 50% penetrant. Finally, compared to other aquatic systems such as zebrafish, Xenopus has greater evolutionary conservation to human as evidenced both by cardiac and genome structure (Hellsten et al., 2010; Warkman & Krieg, 2007).
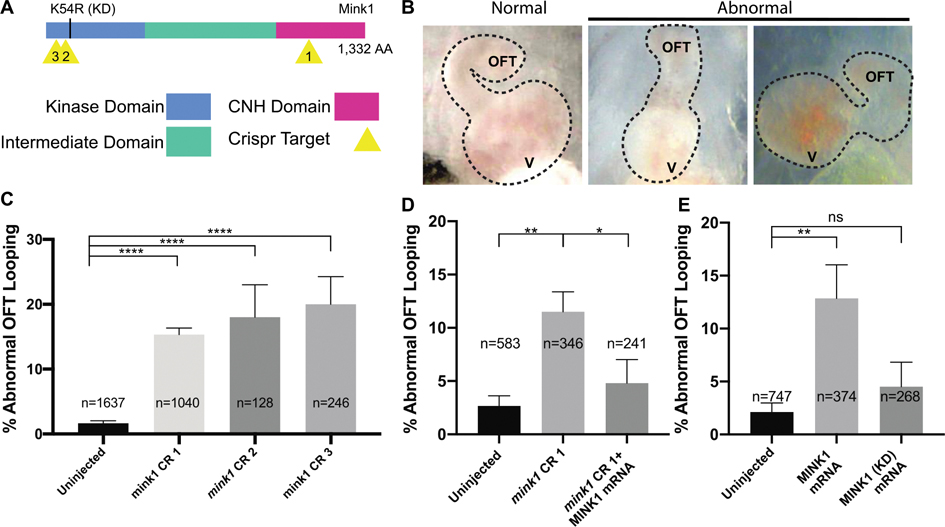
In early cardiac development, the heart tube initially forms in the midline before undergoing morphogenesis to loop to the right. In Xenopus, cardiac looping can be easily scored by comparing the orientation of the outflow tract (OFT) in relation to the ventricle, which is easily visualized through the transparent ventral skin of the stage 45 tadpole. To deplete mink1, we tested three, non-overlapping sgRNAs targeting different sites (Figure 1A) (Bhattacharya et al., 2015). In uninjected control embryos, only a small portion (1%) of tadpoles had abnormal OFT looping; most tadpoles had proper rightward looped outflow tracts. G0 CRISPR based depletion of mink1 led to a statistically significant fraction of tadpoles having abnormal OFT looping (CR1: 20%, CR2: 18% and CR3: 15%), either looping to the left or remaining unlooped (Figure 1B,C).
Figure 1: Loss of mink1 Induces Abnormal Heart Development.
A) Schematic of Xenopus tropicalis Mink1 protein domains, site of kinase-dead (KD) mutation, and Crispr target sites (yellow). Protein homology between human and X. tropicalis Mink1 is 76%. B) Representative images of Xenopus tropicalis normal and abnormal cardiac outflow tracts (ventral views with anterior at top) at stage 45. OFT: outflow tract, V: ventricle. C) Quantification of abnormal cardiac outflow tract looping in mink1 crispants. 5, 3 and 12 biological replicates included for CRISPRs 1, 2 and 3, respectively. D) Rescue of mink1 depletion cardiac looping phenotype with human MINK1 mRNA. 5 biological replicates. E) Quantification of abnormal cardiac OFT looping in embryos with overexpression of wild-type and kinase-dead human MINK1 RNA. Lower dose of MINK1 RNA used for rescue than for overexpression (see Methods). 7 and 4 biological replicates included. P < 0.0001= ****, p < 0.005= **, p < 0.05= * by two-tailed T-test.
To evaluate the specificity and efficacy of our depletion phenotype, we employed several tests. First, we show that three different non-overlapping sgRNAs targeting mink1 all gave similar cardiac phenotypes (Fig 1C). Second, we rescued the mink1 loss of function phenotype with co-injection of human MINK1 mRNA (Figure 1D). Finally, we confirmed targeting of the appropriate locus by our sgRNAs using PCR amplification of the cut sites and ICE (Inference of Crispr Edits) analysis (Supplementary Fig 1) (Conant et al., 2022). Using ICE, we detected genome editing (>73% indels) for each Crispr sgRNA. Crispr sgRNA 1 (CR1) resulted in an early stop codon which may lead to complete protein degradation or a deletion of the majority of the Citron Homology (CNH) domain. Very little is known about the function of the CNH domain of Mink1, but overexpression of the Mink1 CNH has been shown to accumulate in the nucleus and suppress secondary axis formation induced by ectopic β-catenin expression (Mikryukov & Moss, 2012). CR2 resulted in multiple frame shift mutations likely resulting in protein truncation, and CR3 resulted in deletion of two highly conserved amino acids within the N-terminal kinase domain.
We also tested the impact of mink1 overexpression on cardiac looping. We microinjected human MINK1 mRNA into one-cell embryos and examined cardiac looping at stage 45. Compared to uninjected control embryos, overexpression of 500pg of MINK1 led to statistically significant abnormalities in OFT looping suggesting that MINK1 must be carefully titrated for proper patterning of the heart (Figure 1B,E). In order to test if this effect depends on MINK1 kinase activity, we overexpressed 500pg of MINK1 K54R mRNA, which codes for a kinase-dead variant of MINK1 (Hyodo et al., 2012). Overexpression of MINK1 K54R mRNA led to lower levels of OFT looping defects compared to overexpression of the wildtype MINK1 mRNA (13% vs 4%) (Figure 1E). Additionally, overexpression of MINK1 K54R mRNA was unable to rescue abnormal OFT looping defects induced by mink1 depletion (Supplementary Figure 2). In summary, from these tests, we conclude that Mink1 regulates the direction of cardiac outflow tract looping which appears to be dependent on its kinase activity.
MINK1 is Required for Left-Right (LR) Axis Patterning
Cardiac looping is dependent on morphogenesis of the heart tube and global patterning of the LR axis. In order to distinguish between these two possibilities in mink1 depleted embryos, we examined molecular markers of the LR axis. In most vertebrates, the LR axis is established in the Left-Right Organizer (LRO), which forms in the posterior mesoderm near the end of gastrulation (Blum et al., 2014; Shook et al., 2004). In the early gastrula, cells from the superficial Spemann Organizer will give rise to the LRO (Pohl & Knochel, 2004; Stubbs et al., 2008). The cells of the LRO are ciliated, with a subset of cilia beating in a manner that creates leftward extracellular fluid flow (Hamada, 2016; McGrath et al., 2003; Tabin & Vogan, 2003). This leftward flow is sensed by immotile cilia leading to the suppression of dand5 mRNA expression (Schweickert et al., 2010; Vonica & Brivanlou, 2007). Prior to cilia-mediated flow, at stage 14, dand5 and nodal1 are symmetrically expressed at the margins of the LRO. Dand5 is an extracellular antagonist of Nodal1. Suppression of dand5 expression on the left derepresses Nodal signaling on this side of the LRO. This nodal signal is then transmitted to the left lateral plate mesoderm, activating pitx2c expression only on the left side (Kawasumi et al., 2011; Logan et al., 1998; Vonica & Brivanlou, 2007). Therefore, both dand5 and pitx2c are useful markers of global LR patterning.
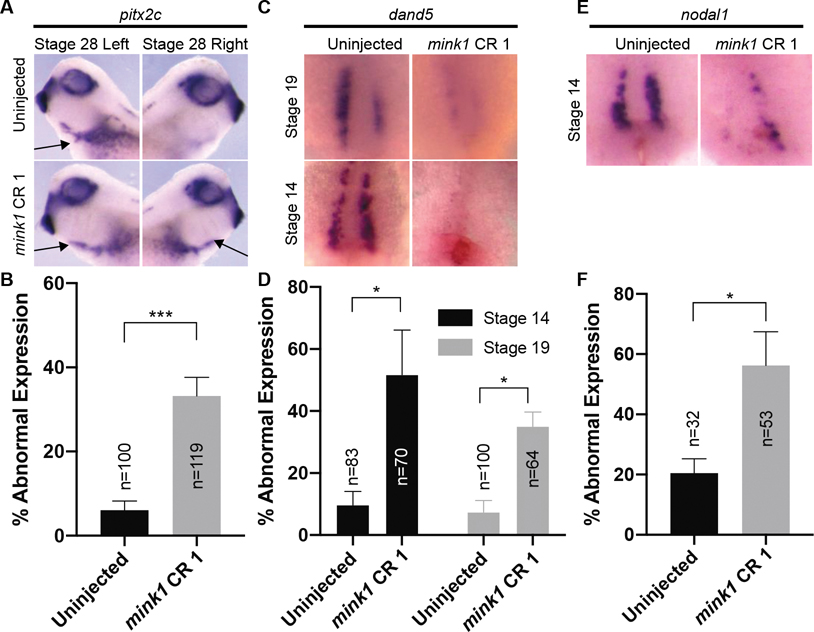
We performed in situ hybridization to assess the expression of dand5 mRNA and pitx2c mRNA in mink1 depleted embryos (Figure 2 A–D). Comparing mink1 depleted tadpoles to uninjected controls, 33% of mink1 crispants had abnormal expression of pitx2c compared to just 6% in controls. In mink1 crispants, the most common pitx2c expression pattern abnormality was bilateral pitx2c (52% of abnormal pitx2c expression) compared to left-sided only expression seen in controls (Figure 2A). Other patterns of abnormal expression seen in mink1 crispants were bilaterally absent pitx2c and pitx2c expression only on the right side (approximately split between the two phenotypes). Upstream of pitx2c, we also detected dand5 expression abnormalities in 35% of mink1 depleted embryos compared to just 7% in controls. Of note, in mink1 depleted embryos, dand5 expression was predominantly reduced bilaterally (75% of abnormal dand5 expression) (Figure 2C). Other patterns of abnormal dand5 expression seen in mink1 crispants were no reduction in dand5 within the LRO or a reduction of dand5 on the right side only. Based on these studies, we conclude that global LR patterning is affected in mink1 depleted embryos.
Figure 2: Mink1 is Required for LRO Patterning.
A) Representative images of pitx2c expression at stage 19. pitx2c is normally expressed on the left lateral plate mesoderm (arrow, top row). pitx2c expression is impaired in mink1 crispants (typically bilateral, arrows lower row). B) Quantification of abnormal pitx2c in mink1 crispants. 6 biological replicates included. C) Representative images of dand5 expression at stage 19 and stage 14. Anterior is to the top and posterior is to the bottom; ventral view. dand5 expression is absent in the left-right Organizer of mink1 crispants after initiation of flow, stage 19 (top row), and before the initiation of flow, stage 14 (bottom row). D) Quantification of abnormal dand5 expression in mink1 crispants at stages 14 and 19. 4 and 3 biological replicates included for stage 19 and 14, respectively. E) Representative images of nodal1 expression at stage 14. Anterior is to the top and posterior is to the bottom; ventral view. Nodal1 expression is also absent in the left-right Organizer of mink1 crispants before the initiation of flow, stage 14. F) Quantification of abnormal nodal1 expression in mink1 crispants. 4 biological replicates included. P < 0.0005=***, p < 0.05= * by two-tailed T-test.
Two mechanisms have been proposed to explain loss of dand5 bilaterally at stage 19. First, cilia driven flow in the LRO may be chaotic and go both left and right, repressing dand5 on both sides of the LRO (van Veenendaal et al., 2013). Alternatively, the LRO may fail to form properly (Beyer et al., 2012; Griffin et al., 2018; Walentek et al., 2012). To distinguish between these two mechanisms, we examined dand5 and nodal1 expression at stage 14 (Figure 2 C–F). At stage 14, both dand5 and nodal1 are normally expressed bilaterally at the lateral borders of the LRO (Blum et al., 2009). However, in 52% and 56% of mink1 depleted embryos, dand5 and nodal1 expression, respectively, were reduced compared to 10% and 20% in respective controls. This indicates that expression of these markers was compromised prior to flow, suggesting broader disruption of the LRO. From these results, we hypothesized that depletion of mink1 perturbs LR patterning due to a required role during patterning of the LRO.
Gastrulation Defects Precede LR Patterning Defects in mink1 Crispants
The LRO is derived from the superficial mesoderm that involutes during gastrulation (Shook et al., 2004). Thus, failure to properly gastrulate can result in patterning defects of the LRO. Interestingly, 22% of mink1 crispants failed to gastrulate normally (compared to 1% in controls) (Supplementary Figure 3 A,B). Indeed, defects in gastrulation were positively correlated with cardiac looping and LR patterning defects (Supplementary Figure 3 C,D). 24% of mink1 crispants with gastrulation defects had abnormal outflow tract looping compared to only 6% of mink1 crispants without gastrulation defects. 70% of mink1 crispants with gastrulation defects also had abnormal pitx2c expression compared to only 18% of mink1 crispants without gastrulation defects (2% abnormal pitx2c expression in uninjected controls). This finding, combined with our previous finding of absence of dand5 and nodal1 in the LRO, led us to hypothesize that gastrulation defects in mink1 crispants disrupt patterning of the LRO leading to downstream LR and outflow tract looping defects.
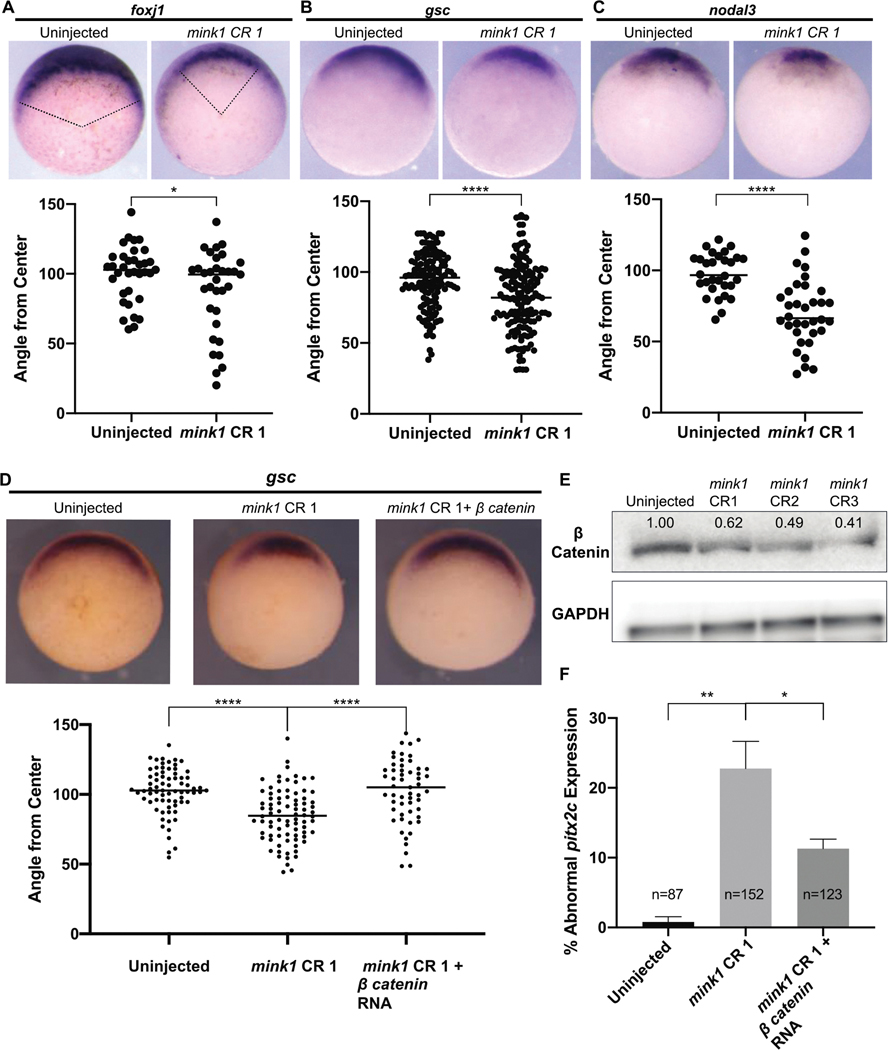
For gastrulation to proceed normally, cells must be specified properly and then engage in morphogenetic movement (Huang & Winklbauer, 2018). Gastrulation defects in mink1 crispants led us to wonder whether cell fate specification or morphogenetic movements were affected due to loss of mink1. First, we assayed cell fate specification since it is required for and precedes morphogenesis in gastrulation. We began by examining the patterning of the Spemann Organizer by assaying Organizer cell fates that are marked by foxj1, goosecoid and nodal3 in stage 10.5 embryos. The expression of each of these markers was reduced in mink1 crispants compared to controls (Figure 3 A–C). To quantitate the reduction in expression, we measured the angle corresponding to the arc of expression of each gene. For example, foxj1 expression swept an arc on average of 100° in wild-type embryos compared to an average of 87° in mink1 crispants (Figure 3A). Foxj1 specifically marks the superficial dorsal mesoderm that will form the LRO (Shook et al., 2004; Stubbs et al., 2008). Similarly, the expression of goosecoid (82° in mink1 crispants compared to 94° in controls) and nodal3 (70° in mink1 crispants compared to 98° in controls) were reduced (Figure 3 B–C). Based on our findings at stage 10.5, we conclude that Mink1 regulates Spemann Organizer cell fate specification and/or maintenance during gastrulation.
Figure 3: Mink1 Regulates Canonical Wnt Signaling.
A) Representative images of foxj1 expression in stage 10.5 uninjected embryos and mink1 crispants along with quantification. Pink lines represent angle from center measurements made in ImageJ. 3 biological replicates B) Representative images of goosecoid expression in stage 10.5 uninjected embryos and mink1 crispants along with quantification. 9 biological replicates. C) Representative images of nodal3 expression in stage 10.5 uninjected embryos and mink1 crispants along with quantification. 3 biological replicates. D) Injection of β-catenin RNA rescues loss of Spemann Organizer gene expression in mink1 crispants. Representative images of goosecoid expression along with quantification. 4 biological replicates. E) Levels of total β-catenin protein are reduced in mink1 crispants at stage 10 as assayed by western blot. F) Injection of β-catenin RNA also rescues abnormal pitx2c expression in mink1 crispants. 3 biological replicates. P < 0.0001= ****, p < 0.005= **, p < 0.05= * by two-tailed T-test.
mink1: Regulator of β-catenin-Dependent Wnt Signaling
Specification of Spemann’s Organizer cell fates depends on canonical Wnt signaling (Fagotto et al., 1997; Harland & Gerhart, 1997; Heasman et al., 1994; Khokha et al., 2005). In the Wnt signaling pathway, ligand activation leads to the sequestration of proteins that degrade β-catenin leading to its stabilization and nuclear entry. In the nucleus, β-catenin complexes with transcription factors to activate a program of Wnt-dependent genes (Liu et al., 2005; Molenaar et al., 1996). In the context of dorsal-ventral development, depletion of β-catenin eliminates the specification of Spemann’s Organizer, while overexpression of β-catenin can induce a secondary axis (Funayama et al., 1995; Heasman et al., 1994; Khokha et al., 2005).
We speculated that reduction in Spemann’s Organizer genes in mink1 depleted embryos may be due to loss of Wnt signaling. First, to determine if mink1 is functioning in canonical Wnt signaling, we performed a western blot to measure total β-catenin protein levels in stage 10.5 uninjected and mink1 crispant embryos (Figure 3E). We found that mink1 depletion by all three CRISPR sgRNAs led to β-catenin protein loss.
While a reduction in β-catenin protein in mink1 depleted embryos at st 10.5 suggests a role in Wnt signaling, in the specification of the Organizer, β-catenin plays a critical role at an earlier stage. Therefore, we sought to test our hypothesis directly with a rescue experiment. We injected a dose of β-catenin mRNA just below the threshold (50pg) to induce secondary axes into mink1 crispants and assayed stage 10.5 embryos for goosecoid gene expression. Embryos injected with β-catenin mRNA displayed a full rescue of goosecoid expression to 103°, similar to uninjected embryos, compared to 85° in mink1 crispants (Figure 3D). The loss of β-catenin in mink1 crispants and the rescue of dorsal cell fates of the Spemann Organizer with β-catenin mRNA indicated that mink1 functions upstream of β-catenin in canonical Wnt signaling.
We previously demonstrated an association between gastrulation defects and LR patterning defects possibly due to failure to properly specify the LRO (Figure 2; Supplementary Figure 3). We now sought to test if rescue of Spemann Organizer gene expression with β-catenin in mink1 depleted embryos would also rescue LR patterning defects. We depleted mink1 by G0 CRISPR, injected 50pg of β-catenin mRNA into a subset of these embryos, and then compared the expression of pitx2c between these two subsets. We observed a reduction of abnormal pitx2c expression from 23% to 11% in mink1 crispants versus mink1 crispants subsequently injected with β-catenin mRNA (1% abnormal pitx2c in uninjected controls), respectively (Figure 3F). This supports our hypothesis that LR patterning defects in mink1 crispants result from Spemann Organizer cell fate specification and/or maintenance defects due to misregulation of Wnt signaling.
Global Phosphoproteomic Analysis Reveals HMGA2 As a Potential Downstream MINK1 Effector
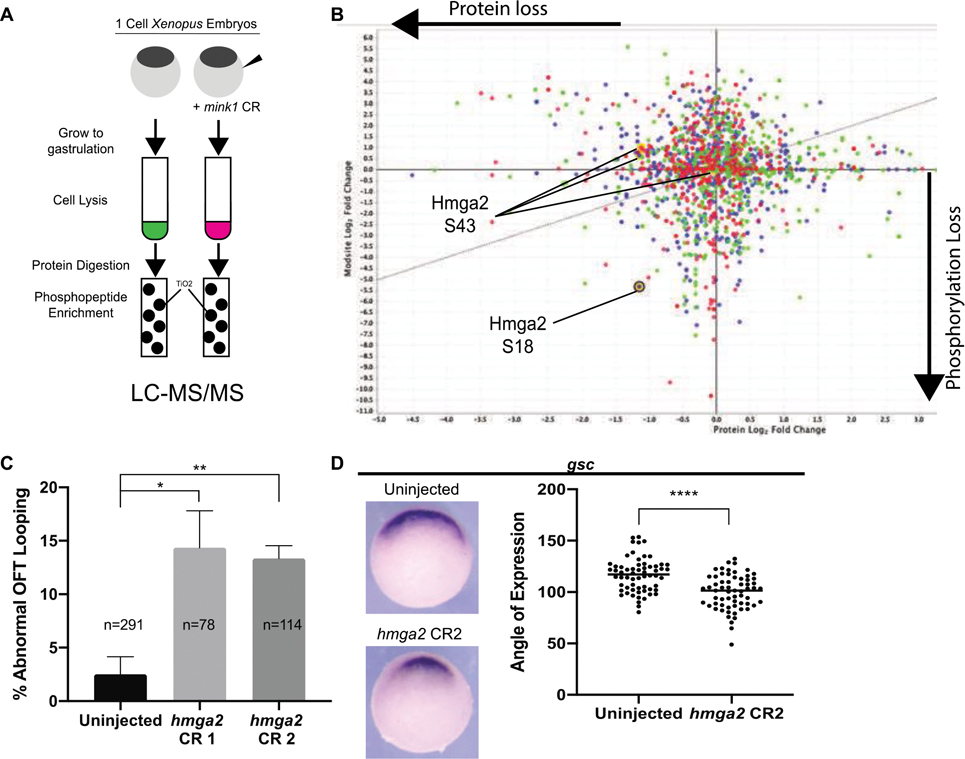
Our work identifies a role for mink1 during patterning of the early embryonic mesoderm at the onset of gastrulation that then goes on to affect LR patterning. This role for mink1 in development appears to be kinase dependent (Figure 1D–E); therefore, to identify targets of Mink1, we performed an unbiased quantitative phosphoproteomic analysis. We collected uninjected and mink1 crispant embryos at stages 10, 11 and 12, which span gastrulation. We extracted protein lysates and digested with trypsin, and then the phosphopeptides were enriched using TiO2 affinity. Both flow-through and TiO2 enriched samples were analyzed by LTQ-Orbitrap XL mass spectrometer and raw MS data were searched and quantified with Mascot. Using a 95% minimum protein confidence interval and minimum of two peptides per protein, our analysis revealed 2,248 differentially phosphorylated and expressed proteins (Figure 4 A,B). This experiment was performed once as a target discovery tool. Next, we planned to experimentally validate proteins of interest.
Figure 4: Global Phosphoproteomic Analysis Reveals HMGA2 As a Potential Downstream MINK1 Effector.
A) Schematic of experimental pathway for phosphoenriched mass spectrometry experiment. B) Hmga2 exhibited loss of protein and phosphorylation at S18 residue due to depletion of mink1. Graphic representation of global phospoproteome due to mink1 depletion, X-axis represents protein log2 fold change, Y-axis represents phosphorylation log2 fold change. Red represents stage 10, green represents stage 11, blue represents stage 12. Yellow circles represent Hmga2 protein level changes. C) Quantification of abnormal outflow tract looping in hmga2 crispants. 3 biological replicates. D) Hmga2 is required for Spemann Organizer gene expression. Representative images and quantification of reduced goosecoid expression in hmga2 crispants. 3 biological replicates. P < 0.0001= ****, p < 0.005= **, p < 0.05= * by two-tailed T-test.
Of note, β-catenin protein was mildly depleted during all stages of gastrulation in mink1 crispants coinciding with increased phosphorylation at serine-552 (Supplementary Figure 4). This served as further confirmation of our finding that β-catenin protein was reduced due to mink1 depletion, but that β-catenin is not a target of mink1 kinase activity. GSK3, an important component of the β-catenin degradation complex, also had 6.4x protein enrichment.
Since we depleted mink1, we searched the data for depleted phosphosites. Several candidates for downstream effectors of Mink1 emerged, and we tested a subset of these by CRISPR based depletion for mink1 LOF phenocopy (Supplementary Figure 4,5). For each candidate, we quantified defects of Spemann Organizer cell fates induced by gene knockdown. HMGA2 emerged as an attractive candidate due to a substantial, 40x reduction in phosphorylation at residue S18 with a corresponding 2.2x loss of protein. Hmga2 depletion by G0 CRISPR had statistically significant defects in OFT looping and Spemann Organizer cell fates (Figure 4 C,D). 14% of hmga2 crispants displayed abnormal OFT looping while only 2% of uninjected controls had similar defects. Goosecoid expression was also reduced to 99° on average in hmga2 crispants from 116° in uninjected controls. These results suggest that Hmga2 is necessary for specification and/or maintenance of the Spemann Organizer, and we next sought to test if Hmga2 acts downstream of Mink1.
Hmga2 Rescues mink1 LOF
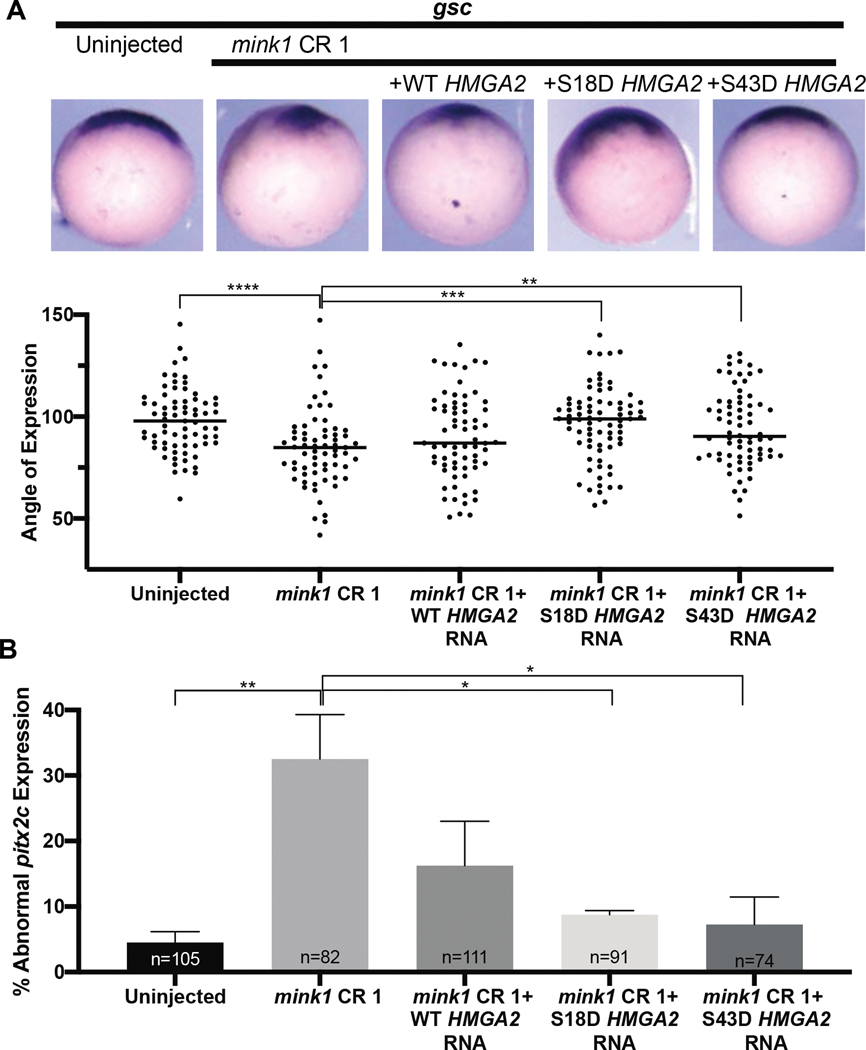
To test our hypothesis that hmga2 functions downstream of mink1 during specification and/or maintenance of the Spemann Organizer, we attempted to rescue mink1 depletion phenotypes with HMGA2 mRNA. We identified two phosphosites from our data: S18 and S43. S43 has been previously described (Sgarra et al., 2009; Xiao et al., 2000; Zou & Wang, 2007) validating our assay. Of note, phosphorylation at this site was not reduced in our mink1 depletion study. On the other hand, phosphorylation at S18 is reduced with mink1 depletion. While the S43 phosphosite does not appear affected with mink1 depletion, this phosphosite is well-characterized and alters the ability of Hmga2 to bind DNA; therefore we constructed phosphomimetic constructs of both S18D and S43D for comparison along with the wild-type (WT) HMGA2 mRNA. We injected mRNA coding for each of the HMGA2 constructs independently into mink1 crispants and observed goosecoid expression at stage 10.5. Both phosphomimetic HMGA2 constructs were able to rescue loss of goosecoid expression from 85° in mink1 crispants to 97° (S18D) and 94° (S43D) (compared to 98% in controls) (Figure 5A). Interestingly, injection of wildtype HMGA2 mRNA in mink1 crispants did not result in statistically significant rescue of goosecoid expression (89°). This suggests that Hmga2 regulates Spemann Organizer cell fate downstream of Mink1 and requires phosphorylation for its activity. Additionally, the hmga2 S18D phosphomimetic acts similarly to the well-characterized S43D suggesting that they may have a similar function.
Figure 5: Phosphomimetic HMGA2 rescues mink1 LOF phenotypes.
A) Representative images goosecoid gene expression in mink1 crispants injected with WT, S18D, or S43D HMGA2 mRNA. 4 biological replicates B) Quantification of pitx2c expression in mink1 crispants injected with WT, S18D, or S43D HMGA2 mRNA. 4 biological replicates P < 0.0001= ****, p < 0.0005= ***, p < 0.005= **, p < 0.05= * by two-tailed T-test.
Next, we tested rescue of LR patterning defects in mink1 depleted embryos. Injection of wildtype HMGA2 mRNA in mink1 crispants led to 16% abnormal pitx2c compared to 33% abnormal pitx2c in mink1 crispants alone which was not significantly different statistically. However, mRNAs coding for either the S18D or S43D HMGA2 phophomimetics were able to rescue abnormal pitx2c expression to 8% and 7%, respectively (Figure 5B). Uninjected embryos had only 5% abnormal pitx2c expression. Based on these experiments, we believe Hmga2 functions downstream of Mink1 during specification and/or maintenance of the Spemann Organizer and left-right patterning in a phosphorylation-dependent manner.
HMGA2 is Downstream of β-catenin in Specifying Spemann Organizer Cell Fates
Hmga2 transcripts are highly expressed in the developing embryo but largely absent in adult tissue (Hock et al., 2006; Zhou et al., 1996). Based on our results, we sought to determine where hmga2 transcripts were expressed during gastrulation. Using an antisense probe against hmga2, we found it to be expressed throughout the animal pole of a stage 10.5 embryo (Supplementary Figure 6). We speculated that hmga2 transcript expression may be dependent on Wnt signaling; however, hmga2 transcript localization remained unchanged in β-catenin depleted embryos (Supplementary Figure 6).
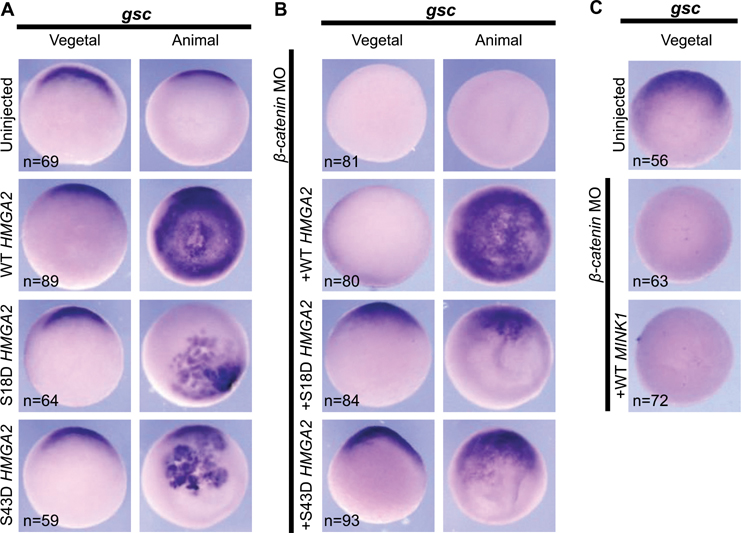
Our results demonstrate that Hmga2 is necessary for Spemann Organizer cell fates (Fig 4D), we next sought to determine if Hmga2 is also sufficient. When overexpressed, wild-type HMGA2 mRNA induced ectopic goosecoid expression throughout the animal pole of stage 10.5 embryos (Figure 6A). Interestingly, the HMGA2 phosphomimetics S18D and S43D also induced ectopic goosecoid expression, albeit in more localized areas of the animal pole.
Figure 6: HMGA2 Rescues Spemann Organizer Cell Fates in β-catenin Depleted Embryos.
A) Overexpression of WT HMGA2 is sufficient to induce broad ectopic goosecoid expression in the animal pole of stage 10.5 embryos. Overexpression of S18D and S43D HMGA2 induces localized expression of goosecoid. 3 biological replicates. B) In β-catenin morphants, HMGA2 overexpression can rescue Spemann Organizer gene expression. Wild-type HMGA2 rescues goosecoid expression in a broad pattern on the animal pole, while S18D and S43D rescue in a more localized pattern 3 biological replicates. C) MINK1 overexpression cannot rescue Spemann Organizer gene expression in β-catenin morphants. 3 biological replicates.
Evaluating our results thus far, we have demonstrated that mink1 is necessary for proper patterning of the Spemann Organizer and that hmga2 plays a role as well. Since Hmga2 is a transcription factor, we wondered what its role might be compared to β-catenin. Therefore, we depleted β-catenin by injecting a translation-blocking morpholino oligos (MO) and then injected HMGA2 mRNA and examined Spemann Organizer gene expression (Khokha et al., 2002). Remarkably, HMGA2 mRNA was able to rescue goosecoid expression in the context of β-catenin-depletion (Figure 6B). In those β-catenin depleted embryos where we expressed wildtype HMGA2 mRNA, the embryos expressed goosecoid throughout the animal pole. In contrast in the β-catenin-depleted embryo, S18D and S43D HMGA2 mRNAs led to goosecoid expression in localized areas reminiscent of goosecoid expression seen in uninjected controls. These results suggest that Hmga2 can regulate Spemann Organizer gene expression even when β-catenin is depleted.
DISCUSSION
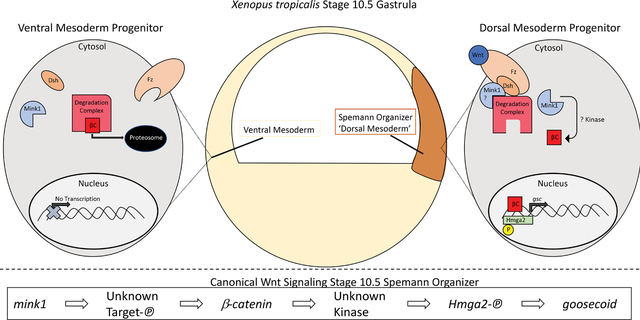
Integrating the sum of our data, we develop a model whereby Mink1 regulates β-catenin-dependent Wnt signaling during specification and/or maintenance of the Spemann Organizer leading to proper LR pattering and cardiac development. We created a Xenopus tropicalis G0 CRISPR model for the CHD candidate gene mink1, which uncovered heart malformations, abnormalities in left-right patterning and gastrulation defects. The gastrulation defects we saw were consistent with those previously reported in mink1 depleted Xenopus laevis (Daulat et al., 2012; Mikryukov & Moss, 2012). Based on the significant correlation between gastrulation defects, LR patterning and heart malformations in mink1 crispants (Supplementary Figure 3), we hypothesized that the disruption of gastrulation could explain the downstream tissue patterning abnormalities. In support of this hypothesis, we found mispatterning of the LRO and Spemann Organizer linking the gastrulation defects to the LR defects.
MINK1 has been shown to regulate cell movements during gastrulation via non-canonical Wnt/PCP signaling (Daulat et al., 2012). Based on our evidence, we postulated it may also regulate cell fate specification and/or maintenance via canonical Wnt signaling. There are multiple lines of evidence that support this hypothesis: 1) depletion of mink1 leads to reduction in the expression of nodal3 (a direct target of Wnt signaling) as well as goosecoid. 2) β-catenin rescues the mink1 loss of function phenotypes including the Spemann Organizer marker goosecoid as well LR patterning 3) depletion of mink1 leads to reduction in β-catenin levels (assayed at st 10.5). These multiple lines of evidence support a hypothesis that mink1 functions upstream of β-catenin in Wnt signaling to regulate Spemann Organizer cell fates and LR patterning. To establish a role for Mink1 in the initial specification of the Spemann Organizer, we would need to analyze a maternal-zygotic mink1 mutant, an ongoing study in the lab. Future studies are also necessary to determine the mechanism by which Mink1 regulates β-catenin protein levels. One possibility is that Mink1 leads to the degradation of GSK3β, which we found to be elevated in mink1 crispants (Supplementary Figure 4). Elevation of GSK3β could then degrade β-catenin in mink1 crispants. Interestingly, we noted a slight increase in phosphorylation at S-552 in β-catenin, which could possibly be activating (Chowdhury et al., 2015), so there are multiple findings that need to be addressed in further studies.
Mechanistically, we took advantage of proteomic tools for enriching for phophopeptides, to identify downstream targets for Mink1 and identified numerous candidates including Nucks1, Rpl30 and Chaf1a (Supplementary Figure 4). Each candidate exhibited significant phosphorylation loss. Hmga2 emerged as a strong candidate for our studies as a result of heart malformation and cell fate specification defects in hmga2 crispants that phenocopied mink1 depletion.
Our proteomics data identified two phosphosites on Hmga2, S18 and S43. As validation, S43 has been previously described as a substrate for PKC. Phosphorylation at this site reduces HMGA2 DNA binding affinity (Sgarra et al., 2009; Xiao et al., 2000; Zou & Wang, 2007). Our results also identify phosphorylation at S18, which showed a 6-fold reduction in phosphorylation in mink1 depleted embryos compared to controls. Both phosphosites are adjacent to DNA binding AT hooks suggesting that, like S43, S18 phosphorylation would reduce DNA binding activity, although this will need to be tested rigorously in future studies. We tested and confirmed that hmga2 is downstream of mink1 in the context of both Spemann Organizer fate specification and LR patterning. However, whether Mink1 directly phosphorylates Hmga2 is not clear, and our results would suggest that it does not do so directly.
There is plentiful research documenting HMGA2’s interaction with the canonical Wnt signaling pathway, especially during tumor cell proliferation and cell cycle progression (Jiang et al., 2019; Shi et al., 2016; Tan & Chen, 2021; Wang et al., 2021; Wend et al., 2013; Yang et al., 2019; Zha et al., 2013). However, whether HMGA2 acts agonistically or antagonistically to the Wnt pathway appears to be context dependent. In the developing lung, Hmga2−/− mice exhibited enhanced canonical Wnt signaling (Singh et al., 2014). However other studies have shown that HMGA2 has an agonistic role in numerous cancers (Jia et al., 2020; Ou et al., 2017; Zha et al., 2013). In these studies, overexpression of HMGA2 activates canonical Wnt signaling to enhance oncogenic properties, while depletion had the opposite effect. Our work indicates that, in the context of the late blastula embryo, Hmga2 appears to act agonistically.
Our results indicate that Hmga2 is both necessary and sufficient for Wnt-dependent Spemann Organizer cell fates. Importantly, overexpression of HMGA2 can rescue Spemann Organizer gene expression in β catenin-depleted embryos (Figure 6). This, in combination with the fact that hmga2 crispants do not display loss of β-catenin protein (Supplementary Figure 5), leads us to hypothesize that Hmga2 may function either downstream of β-catenin in Wnt signaling or an in independent, parallel pathway to induce goosecoid expession. Since Mink1 appears upstream of β-catenin, this would suggest that Hmga2 is not a direct target of Mink1. An interesting avenue of future research venture would be to identify the kinase directly responsible for phosphorylation of HMGA2 at S18 and/or S43.
Interestingly, the wildtype and phosphomimetic variants of HMGA2 appear to behave differently. Wildtype HMGA2 induces goosecoid expression broadly, while S18D and S43D HMGA2 phosphomimetics create a more localized expression. S18D and S43D phosphomimetics also rescue laterality defects in mink1 depleted embryos, but the wildtype construct does not. In the late blastula embryo, hmga2 transcripts are broadly expressed across the ectoderm/mesoderm of the embryo which is unaffected by β-catenin depletion. However, goosecoid expression is confined to the dorsal mesoderm. A possible explanation is that Hmga2 protein, including the phosphorylated form, is more narrowly localized than Hmga2 mRNA. Our hypothesis is that Hmga2 is phosphorylated at S18 and/or S43 in the dorsal region of the gastrulating embryo leading to induction of Spemann Organizer cell fates. The fact that phosphomimetic S43D, which is known to reduce DNA binding, behaves similarly to S18D indicates that the phosphorylation site S18 discovered in our study may also play a role in this process. Further study will be required to test this hypothesis and to understand the mechanisms by which the domain of Hmga2 protein is restricted.
We began our studies investigating a disease candidate gene identified in patients with CHD. By using our high-throughput animal model, Xenopus tropicalis, we identified LR patterning and Spemann Organizer specification and/or maintenance defects in mink1 crispant embryos. Given our desire to understand a molecular mechanism for mink1 we identified a downstream effector, Hmga2, by phosphor-enriched proteomics. We elucidated a new role for hmga2 in regulating Wnt-dependent gene expression downstream of β-catenin in the late blastula embryo.
Supplementary Material
MINK1 is a novel regulator of left-right patterning and heart development.
MINK1 regulates canonical Wnt signaling during induction of the Spemann Organizer.
HMGA2 is an indirect target of MINK1 phosphorylation.
HMGA2 functions downstream of b-catenin during induction of Spemann Organizer.
Acknowledgement
We would like to thank Maura Lane and Michael Slocum for animal husbandry. We would like to thank Emily Mis for her guidance and support of this project. We would like to thank Dr. TuKiet T. Lam (Department of Molecular Biophysics and Biochemistry and MS & Proteomics Resource, WM Keck Foundation Biotechnology Resource Laboratory) for his support.
Funding
This work was supported in part by the NIH grants T32GM007499-43, F31HL140791 and 1R01HD102186.
Footnotes
Availability of data and materials
The mass spectrometry proteomics data have been deposited to the ProteomeXchange consortium via the PRIDE partner repository with: Project accession: PXD028827 Project DOI: 10.6019/PXD028827.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP, 2006. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24 (10), 1285–1292. 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Beyer T, Danilchik M, Thumberger T, Vick P, Tisler M, Schneider I, Bogusch S, Andre P, Ulmer B, Walentek P, Niesler B, Blum M, Schweickert A, 2012. Serotonin signaling is required for Wnt-dependent GRP specification and leftward flow in Xenopus. Curr. Biol. 22 (1), 33–39. 10.1016/j.cub.2011.11.027. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Marfo CA, Li D, Lane M, Khokha MK, 2015. CRISPR/Cas9: an inexpensive, efficient loss of function tool to screen human disease genes in Xenopus. Dev. Biol. 408 (2), 196–204. 10.1016/j.ydbio.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M, Beyer T, Weber T, Vick P, Andre P, Bitzer E, Schweickert A, 2009. Xenopus, an ideal model system to study vertebrate left-right asymmetry. Dev. Dynam. 238 (6), 1215–1225. 10.1002/dvdy.21855. [DOI] [PubMed] [Google Scholar]
- Blum M, Feistel K, Thumberger T, Schweickert A, 2014. The evolution and conservation of left-right patterning mechanisms. Development 141 (8), 1603–1613. 10.1242/dev.100560. [DOI] [PubMed] [Google Scholar]
- Bradford MM, 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chowdhury MK, Montgomery MK, Morris MJ, Cognard E, Shepherd PR, Smith GC, 2015. Glucagon phosphorylates serine 552 of beta-catenin leading to increased expression of cyclin D1 and c-Myc in the isolated rat liver. Arch. Physiol. Biochem. 121 (3), 88–96. 10.3109/13813455.2015.1048693. [DOI] [PubMed] [Google Scholar]
- Conant D, Hsiau T, Rossi N, Oki J, Maures T, Waite K, Yang J, Joshi S, Kelso R, Holden K, Enzmann BL, Stoner R, 2022. Inference of CRISPR Edits from sanger trace data. CRISPR J 5 (1), 123–130. 10.1089/crispr.2021.0113. [DOI] [PubMed] [Google Scholar]
- Daulat AM, Luu O, Sing A, Zhang L, Wrana JL, McNeill H, Winklbauer R, Angers S, 2012. Mink1 regulates beta-catenin-independent Wnt signaling via Prickle phosphorylation. Mol. Cell Biol. 32 (1), 173–185. 10.1128/MCB.06320-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Guger K, Gumbiner BM, 1997. Induction of the primary dorsalizing center in Xenopus by the Wnt/GSK/beta-catenin signaling pathway, but not by Vg1, Activin or Noggin. Development 124 (2), 453–460. https://www.ncbi.nlm.nih.gov/pubmed/9053321. [DOI] [PubMed] [Google Scholar]
- Funayama N, Fagotto F, McCrea P, Gumbiner BM, 1995. Embryonic axis induction by the armadillo repeat domain of beta-catenin: evidence for intracellular signaling. J. Cell Biol. 128 (5), 959–968. 10.1083/jcb.128.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel AM, Khokha MK, 2017. An interspecies heart-to-heart: using Xenopus to uncover the genetic basis of congenital heart disease. Curr Pathobiol Rep 5 (2), 187–196. 10.1007/s40139-017-0142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel RK, Meyer M, Paczkowska M, Reimand J, Vizeacoumar F, Vizeacoumar F, Lam TT, Lukong KE, 2018. Global phosphoproteomic analysis identifies SRMS-regulated secondary signaling intermediates. Proteome Sci. 16, 16. 10.1186/s12953-018-0143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JN, Del Viso F, Duncan AR, Robson A, Hwang W, Kulkarni S, Liu KJ, Khokha MK, 2018. RAPGEF5 regulates nuclear translocation of beta-catenin. Dev. Cell 44 (2), 248–260. 10.1016/j.devcel.2017.12.001 e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H, 2016. Roles of motile and immotile cilia in left-right symmetry breaking. In: Nakanishi T, Markwald RR, Baldwin HS, Keller BB, Srivastava D, Yamagishi H (Eds.), Etiology and Morphogenesis of Congenital Heart Disease: from Gene Function and Cellular Interaction to Morphology, pp. 57–65. 10.1007/978-4-431-54628-3_7. [DOI] [Google Scholar]
- Hamada H, Tam PP, 2014. Mechanisms of left-right asymmetry and patterning: driver, mediator and responder. F1000Prime Rep 6, 110. 10.12703/P6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland R, Gerhart J, 1997. Formation and function of spemann’s Organizer. Annu. Rev. Cell Dev. Biol. 13, 611–667. 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, Kintner C, Noro CY, Wylie C, 1994. Overexpression of cadherins and underexpression of beta-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell 79 (5), 791–803. 10.1016/0092-8674(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, Blitz IL, Blumberg B, Dichmann DS, Dubchak I, Amaya E, Detter JC, Fletcher R, Gerhard DS, Goodstein D, Graves T, Grigoriev IV, Grimwood J, Kawashima T, Lindquist E, Lucas SM, Mead PE, Mitros T, Ogino H, Ohta Y, Poliakov AV, Pollet N, Robert J, Salamov A, Sater AK, Schmutz J, Terry A, Vize PD, Warren WC, Wells D, Wills A, Wilson RK, Zimmerman LB, Zorn AM, Grainger R, Grammer T, Khokha MK, Richardson PM, Rokhsar DS, 2010. The genome of the Western clawed frog Xenopus tropicalis. Science 328 (5978), 633–636. 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock R, Witte F, Brocher J, Schutz M, Scheer U, 2006. Expression of HMGA2 variants during oogenesis and early embryogenesis of Xenopus laevis. Eur. J. Cell Biol. 85 (6), 519–528. 10.1016/j.ejcb.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, Jin SC, Deanfield J, Giardini A, Porter GA Jr., Kim R, Bilguvar K, Lopez-Giraldez F, Tikhonova I, Mane S, Romano-Adesman A, Qi H, Vardarajan B, Ma L, Daly M, Roberts AE, Russell MW, Mital S, Newburger JW, Gaynor JW, Breitbart RE, Iossifov I, Ronemus M, Sanders SJ, Kaltman JR, Seidman JG, Brueckner M, Gelb BD, Goldmuntz E, Lifton RP, Seidman CE, Chung WK, 2015. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350 (6265), 1262–1266. 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiau T, Conant D, Rossi N, Maures T, Waite K, Yang J, Joshi S, Kelso R, Holden K, Enzmann BL, Stoner R, 2019. Inference of CRISPR Edits from sanger trace data. bioRxiv, 251082. 10.1101/251082. [DOI] [PubMed] [Google Scholar]
- Hu Y, Leo C, Yu S, Huang BC, Wang H, Shen M, Luo Y, Daniel-Issakani S, Payan DG, Xu X, 2004. Identification and functional characterization of a novel human misshapen/Nck interacting kinase-related kinase, hMINK beta. J. Biol. Chem. 279 (52), 54387–54397. 10.1074/jbc.M404497200. [DOI] [PubMed] [Google Scholar]
- Huang Y, Winklbauer R, 2018. Cell migration in the Xenopus gastrula. Wiley Interdiscip Rev Dev Biol 7 (6), e325. 10.1002/wdev.325. [DOI] [PubMed] [Google Scholar]
- Hyodo T, Ito S, Hasegawa H, Asano E, Maeda M, Urano T, Takahashi M, Hamaguchi M, Senga T, 2012. Misshapen-like kinase 1 (MINK1) is a novel component of striatin-interacting phosphatase and kinase (STRIPAK) and is required for the completion of cytokinesis. J. Biol. Chem. 287 (30), 25019–25029. 10.1074/jbc.M112.372342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia WQ, Zhu JW, Yang CY, Ma J, Pu TY, Han GQ, Zou MM, Xu RX, 2020. Verbascoside inhibits progression of glioblastoma cells by promoting Let-7g-5p and down-regulating HMGA2 via Wnt/beta-catenin signalling blockade. J. Cell Mol. Med. 24 (5), 2901–2916. 10.1111/jcmm.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Li Y, Li J, Zhang X, Niu G, Chen S, Yao S, 2019. Long noncoding RNA LSINCT5 promotes endometrial carcinoma cell proliferation, cycle, and invasion by promoting the Wnt/beta-catenin signaling pathway via HMGA2. Ther Adv Med Oncol 11, 1758835919874649. 10.1177/1758835919874649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, Zeng X, Qi H, Chang W, Sierant MC, Hung WC, Haider S, Zhang J, Knight J, Bjornson RD, Castaldi C, Tikhonoa IR, Bilguvar K, Mane SM, Sanders SJ, Mital S, Russell MW, Gaynor JW, Deanfield J, Giardini A, Porter GA Jr., Srivastava D, Lo CW, Shen Y, Watkins WS, Yandell M, Yost HJ, Tristani-Firouzi M, Newburger JW, Roberts AE, Kim R, Zhao H, Kaltman JR, Goldmuntz E, Chung WK, Seidman JG, Gelb BD, Seidman CE, Lifton RP, Brueckner M, 2017. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat. Genet. 49 (11), 1593–1601. 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasumi A, Nakamura T, Iwai N, Yashiro K, Saijoh Y, Belo JA, Shiratori H, Hamada H, 2011. Left-right asymmetry in the level of active Nodal protein produced in the node is translated into left-right asymmetry in the lateral plate of mouse embryos. Dev. Biol. 353 (2), 321–330. 10.1016/j.ydbio.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, Papalopulu N, Smith JC, Zorn AM, Harland RM, Grammer TC, 2002. Techniques and probes for the study of Xenopus tropicalis development. Dev. Dynam. 225 (4), 499–510. 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Yeh J, Grammer TC, Harland RM, 2005. Depletion of three BMP antagonists from Spemann’s Organizer leads to a catastrophic loss of dorsal structures. Dev. Cell 8 (3), 401–411. 10.1016/j.devcel.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Krishnan N, Lam TT, Fritz A, Rempinski D, O’Loughlin K, Minderman H, Berezney R, Marzluff WF, Thapar R, 2012. The prolyl isomerase Pin1 targets stem-loop binding protein (SLBP) to dissociate the SLBP-histone mRNA complex linking histone mRNA decay with SLBP ubiquitination. Mol. Cell Biol. 32 (21), 4306–4322. 10.1128/MCB.00382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, van den Broek O, Destree O, Hoppler S, 2005. Distinct roles for Xenopus Tcf/Lef genes in mediating specific responses to Wnt/beta-catenin signalling in mesoderm development. Development 132 (24), 5375–5385. 10.1242/dev.02152. [DOI] [PubMed] [Google Scholar]
- Logan M, Pagan-Westphal SM, Smith DM, Paganessi L, Tabin CJ, 1998. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell 94 (3), 307–317. 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- McGrath J, Somlo S, Makova S, Tian X, Brueckner M, 2003. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell 114 (1), 61–73. 10.1016/s0092-8674(03)00511-7. [DOI] [PubMed] [Google Scholar]
- Mikryukov A, Moss T, 2012. Agonistic and antagonistic roles for TNIK and MINK in non-canonical and canonical Wnt signalling. PLoS One 7 (9), e43330. 10.1371/journal.pone.0043330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H, 1996. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 86 (3), 391–399. 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ, 2015. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12 (10), 982–988. 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R, 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75 (17), 4646–4658. 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Ou W, Lv J, Zou X, Yao Y, Wu J, Yang J, Wang Z, Ma Y, 2017. Propofol inhibits hepatocellular carcinoma growth and invasion through the HMGA2-mediated Wnt/beta-catenin pathway. Exp. Ther. Med. 13 (5), 2501–2506. 10.3892/etm.2017.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paricio N, Feiguin F, Boutros M, Eaton S, Mlodzik M, 1999. The Drosophila STE20-like kinase misshapen is required downstream of the Frizzled receptor in planar polarity signaling. EMBO J. 18 (17), 4669–4678. 10.1093/emboj/18.17.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpont ME, Brueckner M, Chung WK, Garg V, Lacro RV, McGuire AL, Mital S, Priest JR, Pu WT, Roberts A, Ware SM, Gelb BD, Russell MW, American Heart Association Council on Cardiovascular Disease in the Y, Council on C, Stroke N, Council on G, Precision M, 2018. Genetic basis for congenital heart disease: revisited: A scientific statement from the American heart association. Circulation 138 (21), e653–e711. 10.1161/CIR.0000000000000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl BS, Knochel W, 2004. Isolation and developmental expression of Xenopus FoxJ1 and FoxK1. Dev. Gene. Evol. 214 (4), 200–205. 10.1007/s00427-004-0391-7. [DOI] [PubMed] [Google Scholar]
- Rich MT, Abbott TB, Chung L, Gulcicek EE, Stone KL, Colangelo CM, Lam TT, Nairn AC, Taylor JR, Torregrossa MM, 2016. Phosphoproteomic analysis reveals a novel mechanism of CaMKIIalpha regulation inversely induced by cocaine memory extinction versus reconsolidation. J. Neurosci. 36 (29), 7613–7627. 10.1523/JNEUROSCI.1108-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweickert A, Vick P, Getwan M, Weber T, Schneider I, Eberhardt M, Beyer T, Pachur A, Blum M, 2010. The nodal inhibitor Coco is a critical target of leftward flow in Xenopus. Curr. Biol. 20 (8), 738–743. 10.1016/j.cub.2010.02.061. [DOI] [PubMed] [Google Scholar]
- Sgarra R, Maurizio E, Zammitti S, Lo Sardo A, Giancotti V, Manfioletti G, 2009. Macroscopic differences in HMGA oncoproteins post-translational modifications: C-terminal phosphorylation of HMGA2 affects its DNA binding properties. J. Proteome Res. 8 (6), 2978–2989. 10.1021/pr900087r. [DOI] [PubMed] [Google Scholar]
- Shi Z, Li X, Wu D, Tang R, Chen R, Xue S, Sun X, 2016. Silencing of HMGA2 suppresses cellular proliferation, migration, invasion, and epithelial-mesenchymal transition in bladder cancer. Tumour Biol 37 (6), 7515–7523. 10.1007/s13277-015-4625-2. [DOI] [PubMed] [Google Scholar]
- Shook DR, Majer C, Keller R, 2004. Pattern and morphogenesis of presumptive superficial mesoderm in two closely related species, Xenopus laevis and Xenopus tropicalis. Dev. Biol. 270 (1), 163–185. 10.1016/j.ydbio.2004.02.021. [DOI] [PubMed] [Google Scholar]
- Singh I, Mehta A, Contreras A, Boettger T, Carraro G, Wheeler M, Cabrera-Fuentes HA, Bellusci S, Seeger W, Braun T, Barreto G, 2014. Hmga2 is required for canonical WNT signaling during lung development. BMC Biol. 12, 21. 10.1186/1741-7007-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisua Belmonte JC, Kintner C, 2008. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat. Genet. 40 (12), 1454–1460. 10.1038/ng.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YC, Treisman JE, Skolnik EY, 1998. The Drosophila Ste20-related kinase misshapen is required for embryonic dorsal closure and acts through a JNK MAPK module on an evolutionarily conserved signaling pathway. Genes Dev. 12 (15), 2371–2380. 10.1101/gad.12.15.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabin CJ, Vogan KJ, 2003. A two-cilia model for vertebrate left-right axis specification. Genes Dev. 17 (1), 1–6. 10.1101/gad.1053803. [DOI] [PubMed] [Google Scholar]
- Tan S, Chen J, 2021. Small interfering-high mobility group A2 attenuates epithelial-mesenchymal transition in thymic cancer cells via the Wnt/beta-catenin pathway. Oncol. Lett. 22 (2), 586. 10.3892/ol.2021.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veenendaal NR, Ulmer B, Boskovski MT, Fang X, Khokha MK, Wendler CC, Blum M, Rivkees SA, 2013. Embryonic exposure to propylthiouracil disrupts left-right patterning in Xenopus embryos. Faseb. J. 27 (2), 684–691. 10.1096/fj.12-218073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali R, Marracci S, 2020. HMGA genes and proteins in development and evolution. Int. J. Mol. Sci. 21 (2). 10.3390/ijms21020654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonica A, Brivanlou AH, 2007. The left-right axis is regulated by the interplay of Coco, Xnr1 and derriere in Xenopus embryos. Dev. Biol. 303 (1), 281–294. 10.1016/j.ydbio.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Walentek P, Beyer T, Thumberger T, Schweickert A, Blum M, 2012. ATP4a is required for Wnt-dependent Foxj1 expression and leftward flow in Xenopus left-right development. Cell Rep. 1 (5), 516–527. 10.1016/j.celrep.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Wallingford JB, 2012. Planar cell polarity and the developmental control of cell behavior in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 28, 627–653. 10.1146/annurev-cellbio-092910-154208. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang J, Wu J, 2021. Emerging roles for HMGA2 in colorectal cancer. Transl Oncol 14 (1), 100894. 10.1016/j.tranon.2020.100894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkman AS, Krieg PA, 2007. Xenopus as a model system for vertebrate heart development. Semin. Cell Dev. Biol. 18 (1), 46–53. 10.1016/j.semcdb.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wend P, Runke S, Wend K, Anchondo B, Yesayan M, Jardon M, Hardie N, Loddenkemper C, Ulasov I, Lesniak MS, Wolsky R, Bentolila LA, Grant SG, Elashoff D, Lehr S, Latimer JJ, Bose S, Sattar H, Krum SA, Miranda-Carboni GA, 2013. WNT10B/beta-catenin signalling induces HMGA2 and proliferation in metastatic triple-negative breast cancer. EMBO Mol. Med. 5 (2), 264–279. 10.1002/emmm.201201320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao DM, Pak JH, Wang X, Sato T, Huang FL, Chen HC, Huang KP, 2000. Phosphorylation of HMG-I by protein kinase C attenuates its binding affinity to the promoter regions of protein kinase C gamma and neurogranin/RC3 genes. J. Neurochem. 74 (1), 392–399. 10.1046/j.1471-4159.2000.0740392.x. [DOI] [PubMed] [Google Scholar]
- Yang S, Gu Y, Wang G, Hu Q, Chen S, Wang Y, Zhao M, 2019. HMGA2 regulates acute myeloid leukemia progression and sensitivity to daunorubicin via Wnt/beta-catenin signaling. Int. J. Mol. Med. 44 (2), 427–436. 10.3892/ijmm.2019.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, Carriero NJ, Cheung YH, Deanfield J, DePalma S, Fakhro KA, Glessner J, Hakonarson H, Italia MJ, Kaltman JR, Kaski J, Kim R, Kline JK, Lee T, Leipzig J, Lopez A, Mane SM, Mitchell LE, Newburger JW, Parfenov M, Pe’er I, Porter G, Roberts AE, Sachidanandam R, Sanders SJ, Seiden HS, State MW, Subramanian S, Tikhonova IR, Wang W, Warburton D, White PS, Williams IA, Zhao H Seidman JG, Brueckner M, Chung WK, Gelb BD, Goldmuntz E, Seidman CE, Lifton RP, 2013. De novo mutations in histone-modifying genes in congenital heart disease. Nature 498 (7453), 220–223. 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha L, Zhang J, Tang W, Zhang N, He M, Guo Y, Wang Z, 2013. HMGA2 elicits EMT by activating the Wnt/beta-catenin pathway in gastric cancer. Dig. Dis. Sci. 58 (3), 724–733. 10.1007/s10620-012-2399-6. [DOI] [PubMed] [Google Scholar]
- Zhou X, Benson KF, Przybysz K, Liu J, Hou Y, Cherath L, Chada K, 1996. Genomic structure and expression of the murine Hmgi-c gene. Nucleic Acids Res. 24 (20), 4071–4077. 10.1093/nar/24.20.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Wang Y, 2007. Mass spectrometric analysis of high-mobility group proteins and their post-translational modifications in normal and cancerous human breast tissues. J. Proteome Res. 6 (6), 2304–2314. 10.1021/pr070072q [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.