Abstract
Some viruses restructure host chromatin, influencing gene expression, with implications for disease outcome. Whether this occurs for SARS-CoV-2, the virus causing COVID-19, is largely unknown. Here, we characterized the 3D genome and epigenome of human cells after SARS-CoV-2 infection, finding widespread host chromatin restructuring that features widespread A compartmental weakening, A-B mixing, reduced intra-TAD contacts and decreased H3K27ac euchromatin modification levels. Such changes were not found following common-cold virus HCoV-OC43 infection. Intriguingly, the cohesin complex was significantly depleted from intra-TAD regions, indicating that SARS-CoV-2 disrupts cohesin loop extrusion. These altered 3D genome/epigenome structures correlated with transcriptional suppression of interferon response genes by the virus, while increased H3K4me3 was found in the promoters of pro-inflammatory genes highly induced during severe COVID-19. These findings show that SARS-CoV-2 acutely rewires host chromatin, facilitating future studies of the long-term epigenomic impacts of its infection.
INTRODUCTION
The 3D folding of mammalian chromatin influences transcription, DNA replication, recombination, or DNA damage repair1-3, with obvious effects upon how cells behave and function. Regulation of host chromatin architecture has been used by viruses to antagonize host defense or to exert long-term influences, such as viral latency4-6. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes COVID-19 and has made > 700 million infections worldwide. Several viral proteins encoded by SARS-CoV-2 have been shown to associate with chromatin or chromatin factors7,8. However, whether and how SARS-CoV-2 infection affects host chromatin architecture is under-explored.
Chromatin architecture is structured through several layers such as A/B compartments, Topological Associating Domains (TADs), and chromatin loops2,9 (Extended Data Fig.1a,b). A/B compartments largely overlap with transcriptionally active/inactive chromatin, respectively9,10. They are suggested to be formed, at least in part, via homotypic attractions between chromatin regions of similar epigenetic features9,10,11, which may function in transcription control through concentrating or sequestering specific regulatory factors11. For TADs and loops, CTCF (CCCTC-binding factor, a highly-conserved zinc-finger protein) and the cohesin complex are the two main regulators3. Growing evidence indicates that cohesin acts via a “loop extrusion” process3. These two mechanisms can crosstalk - the formation of TADs by loop extrusion appears to antagonize compartmentalization3,9,12,13. How chromatin architectures are rewired in pathological conditions including infectious diseases remains incompletely understood. Here, we sought to understand the impacts of the pandemic-causing SARS-CoV-2 on host chromatin by comprehensively mapping the chromatin architectures of human cells after infection using Hi-C 3.0 and ChIP-Seq methods. We uncovered extensive chromatin restructuring upon SARS-CoV-2 infection, with implications for understanding COVID-19 disease gene alteration and epigenetic perturbation in the host.
RESULTS
SARS-CoV-2 infection restructures the host cell genome
To study chromatin organization during SARS-CoV-2 infection, we first determined the optimal infection conditions. Our approaches use cell populations and thus require a high ratio of infection of the host cells. In human A549 cells expressing ACE2 (A549-ACE2) (Fig.1a) that were infected with SARS-CoV-2 (MOI: 0.1), approximately 90% of RNA-Seq reads aligned to the viral genome at 24 hr post-infection (24 hpi), indicating a high level of infection (Extended Data Fig.1c). Immunofluorescence of the viral spike protein verified a high infection ratio (Extended Data Fig.1d,e). We did not observe cell apoptosis (Extended Data Fig.2a-c). We therefore focused on cells 24hpi to study host chromatin changes.
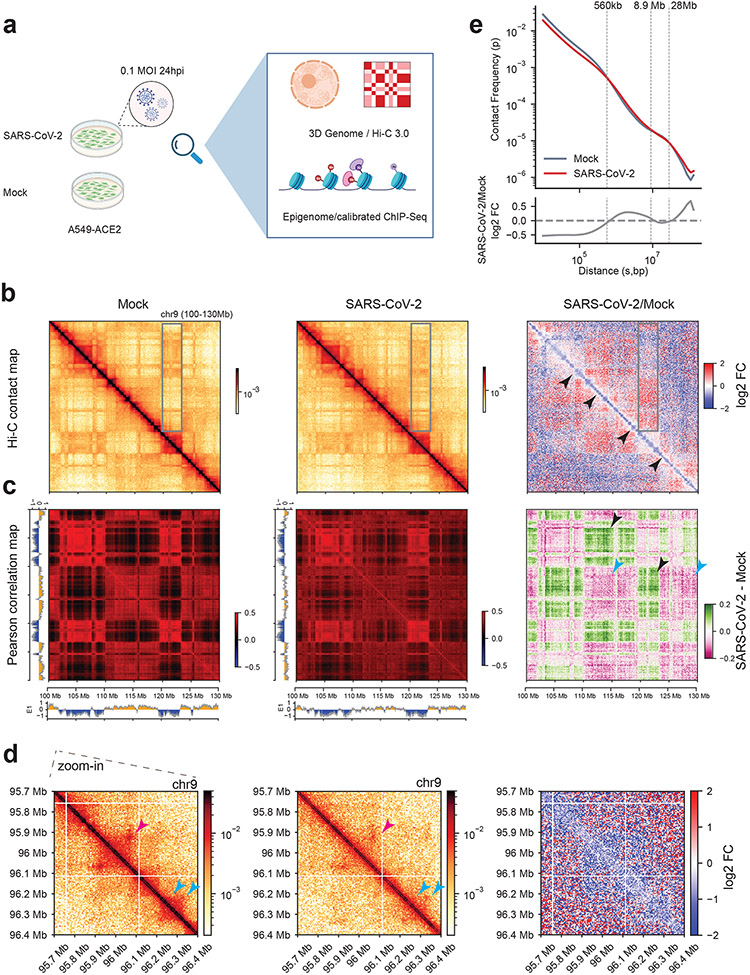
Fig. 1. SARS-CoV-2 significantly restructures the host 3D genome.
a. The experimental design of this work. Created with BioRender.com.
b. Hi-C 3.0 contact matrices of an example region (chr9:100-130Mb, hg19) in Mock or infected conditions. Black arrowheads denote reduced short-distance interactions along the diagonal. Grey boxes show regions with altered compartmentalization. Bin size = 80kb.
c. Pearson correlation matrices of Hi-C 3.0 in the same region as in panel b. Arrowheads point to regions with virus-altered A-B (black) or A-A (cyan) compartmental interactions. Bin size = 80kb. Color scales indicate Pearson correlation coefficient (left and middle), and changes of the correlation matrices after infection (SARS-CoV-2 - Mock Pearson) are shown to the right.
d. Zoom-in Hi-C snapshots of a 700kb region in panels b,c (chr9:95.7-96.4Mb, hg19). Pink and cyan arrowheads show changed dot-shaped loops or domains. Bin size = 5kb.
e. (Top) P(s) curve showing the relationship between the Hi-C contract frequency (P) of intra-chromosomal interactions ranked by genomic distance (s) in both mock (grey) and SARS-CoV-2 (red) conditions. (Bottom) Log2 FC of Hi-C contact frequency ranked by distances (infection/Mock), with dotted lines marking the crossing points of the two curves.
In panels b,d, Color scales indicate Hi-C contact frequencies (left and middle), and log2 FC of SARS-CoV-2/Mock contact frequencies (right).
We then used Hi-C 3.014, a recently improved version of in situ Hi-C (High throughput chromosome conformation capture), to analyse SARS-CoV-2-infected A549-ACE2 cells 24hpi or with mock-infected cells (Mock). We sequenced the libraries to ~2.6 billion read-pairs (Supplementary Table 1), generating ~630-770 million unique contact pairs for each condition. There is high concordance between replicates (Extended Data Fig.2d-f). We combined the replicates for the following analysis, and still refer to them as Hi-C.
Hi-C revealed a widespread alteration of the 3D genome by SARS-COV-2. The near-diagonal short-range contacts were weakened globally, as exemplified in Fig.1b (black arrowheads). In contrast, long-range chromatin contacts far away from the diagonal were often deregulated (increased or decreased for different regions, grey box) (Fig.1b). Pearson correlation map of Hi-C interactions consistently revealed these changes (Fig.1c), which also suggests altered chromatin compartmentalization. Zoom-in view of an example region of ~0.7Mb showed that rectangle-shaped chromatin domains (cyan arrowheads) are often weakened, whereas chromatin loops (“dot” off the diagonal, pink arrowheads) are deregulated (Fig.1d). A P(s) curve demonstrates that SARS-CoV-2 elicited a global reduction of short distance chromatin contact (< 560kb), a moderate increase of mid-to-long distance interactions (~560k-8.9Mb), and enhanced interactions for far-separated regions (>28Mb) (Fig.1e). Intriguingly, inter-chromosomal contacts were generally increased by viral infection, as shown by the fold changes of pairwise interactions between any two chromosomes, or by trans-v.s.-cis contact ratios (Extended Data Fig.2g,h). The enhancement of both inter-chromosomal interactions and extremely long-distance (>28Mb) intra-chromosomal interactions (Fig.1e) suggest changes in chromatin compartmentalization12,15.
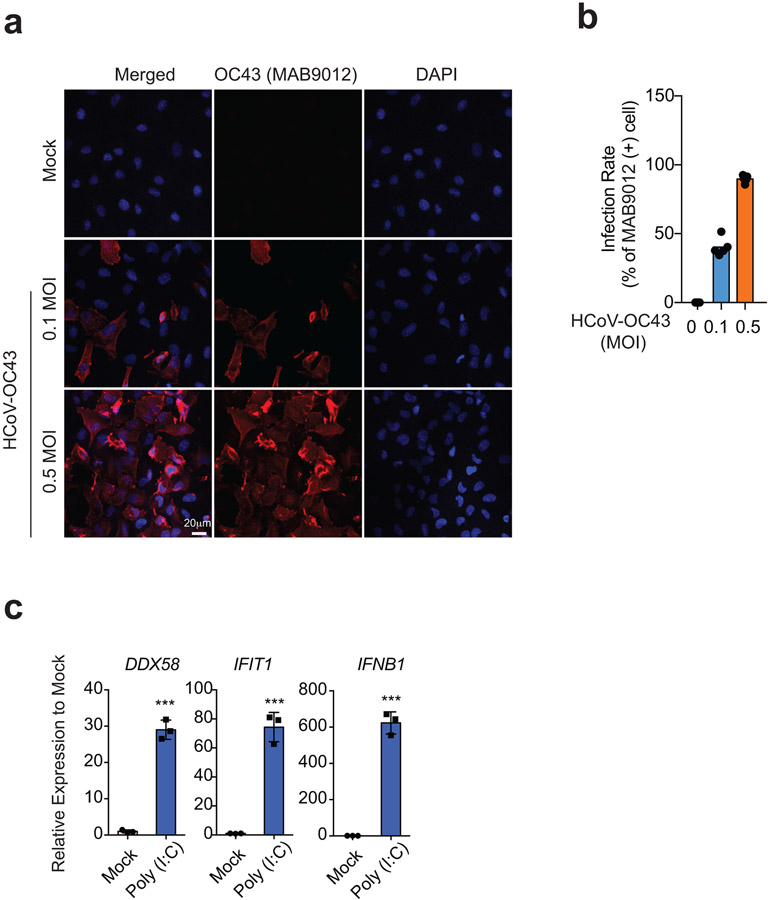
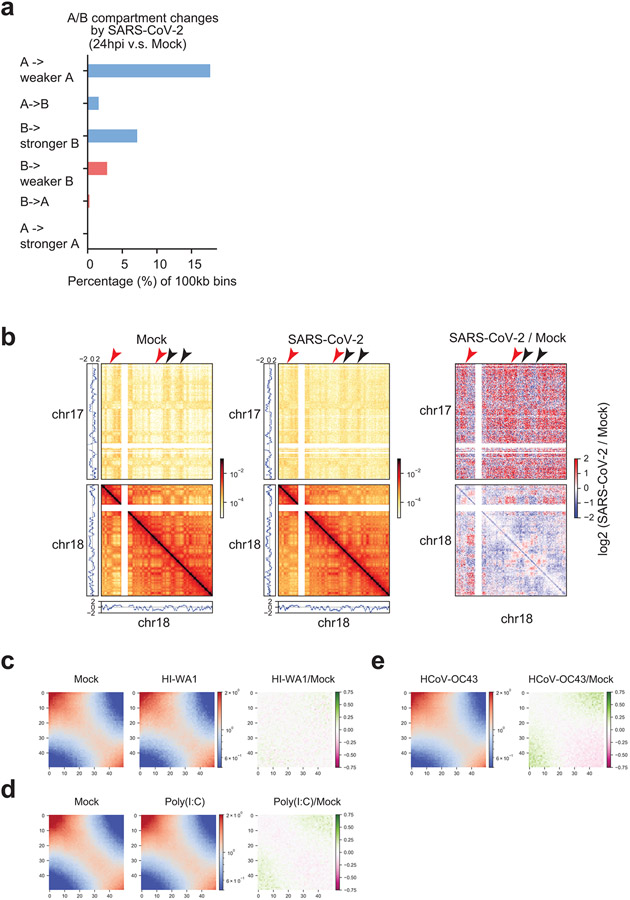
We investigated whether the observed changes are uniquely induced by SARS-CoV-2 infection, or are general phenomena upon coronavirus infection or innate immune signaling. We conducted Hi-C 3.0 after three additional treatments: 1) heat-inactivated SARS-CoV-2 virus (HI-WA1); 2) infection by a human common cold coronavirus HCoV-OC4316, which can efficiently infect A54917,18; 3) a synthetic dsRNA viral mimic, polyinosinic:polycytidylic acid or poly (I:C). We used 0.5 MOI of HCoV-OC43 to achieve >90% infection rate at 24hpi; and for poly (I:C), RT-qPCR validated the induction of target genes after treatment (Extended Data Fig. 3a-c). An initial P(s) curve analysis revealed that none of these additional treatments elicited changes similar to those observed upon SARS-CoV-2 infection (Fig 2a,b). Particularly, HI-WA1 elicited almost no change of 3D genome architecture, whereas HCoV-OC43 and Poly (I:C) infection had mild effects (Fig 2a,b).
Fig. 2. Compartmental A weakening by SARS-CoV-2.
a. P(s) curve showing the relationship between contact frequency (P) and distance (s) of mock, SARS-CoV-2, HI-WA1 (heat-inactivated SARS-CoV-2), HCoV-OC43 and poly (I:C) Hi-C datasets.
b. Log2 fold changes (FC) of P(s) curves showing the differences of Hi-C contacts between SARS-CoV-2/Mock, HI-WA1/Mock, HCoV-OC43/Mock, or poly(I:C)/Mock.
c. A scatter plot showing the compartmental E1 scores of genome-wide 100kb bins in mock or SARS-CoV-2 infected cells. Six categories of bins based on E1 changes were colored-coded.
d. A snapshot of PCA E1 tracks at an example region (chr9: 108Mb-118Mb). Rep1/Rep2 are replicates. Orange or blue indicates A- (E1 score >0) or B-compartment (E1 score < 0), respectively. Arrows and arrowheads exemplify regions with compartmental changes (with colors match panel a).
e. A diagram shows the categories of bins with changes of A/B compartmental scores, which are referred to as “A-ing” (E1 scores increase) or “B-ing” (E1 scores reduce) after virus infection.
f. Heatmaps showing enrichment of histone marks or RNA Pol2 (RPB1) on the six categories of genomic bins defined in panel e, which signifies their epigenomic features over genome background. Scale indicates log2-transformed fold enrichment.
SARS-CoV-2 infection alters chromatin compartmentalization
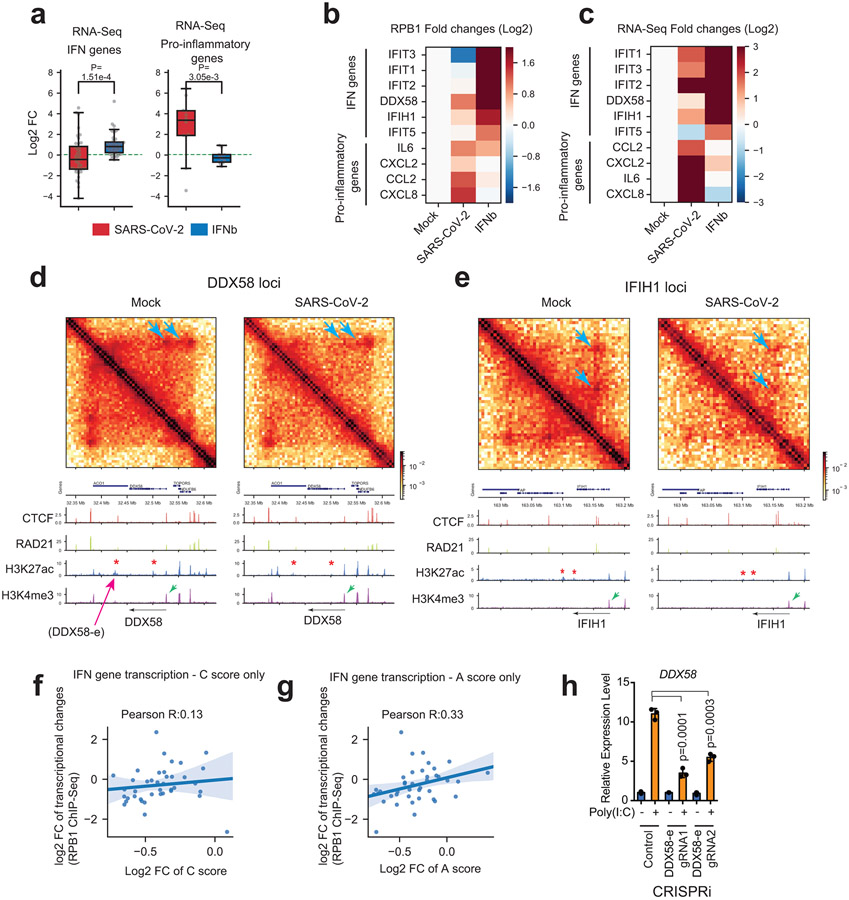
Principal component analysis (PCA) of Hi-C data can divide genomes into A/B compartments9,19, which largely overlap euchromatin/heterochromatin11. Analyzing a 100kb-binned Hi-C matrix, we found significant defects of chromatin compartmentalization in SARS-CoV-2 infected cells (Fig.2c). Overall, PCA E1 scores exhibited a general reduction in infected cells (i.e., moving below the diagonal, Fig.2c), suggesting a pervasive weakening of A compartment and/or A-to-B switching. By measuring E1 changes20, we found that ~30% of genomic regions exhibited compartmental weakening or switching after SARS-CoV-2 infection (Extended Data Fig.4a). The changes commonly displayed features of weakened A (e.g. A-to-weaker-A or A-to-B) or strengthening of B compartment (B-to-stronger-B). Among these, A-to-weaker-A is the most common (~18% of genome, Extended Data Fig.4a). Examples are shown in Fig.2d. These changes indicate that SARS-CoV-2 infection pervasively weakened the euchromatin of the host cells.
We evaluated the epigenetic features of the altered regions to understand the susceptibility to compartmental changes due to infection. By ranking E1-score changes for each genomic bin, we sorted them into six categories (Fig.2e). For those showing E1-score increase, we dubbed them “A-ing” bins and those showing decrease “B-ing”. We generated ChIP-seq data of representative histone marks (H3K27ac, H3K4me3, H3K9me3, and H3K27me3) and polymerase II (Pol2) in A549-ACE2 cells, and examined the epigenetic features of these six categories of bins. This analysis revealed that the “B-ing” genomic regions are those originally enriched in active chromatin marks (such as H3K27ac); whereas the “A-ing” genomic regions are those originally enriched in repressive histone marks (particularly H3K27me3)(Fig.2f). These results suggest that the originally well-segregated A or B compartments were losing their identity, indicating defective chromatin compartmentalization. Indeed, as shown in Fig.1c, defective compartmentalization manifests as increases of inter-compartmental interactions formed between regions of A and B and decreases of those formed within A or B (black and cyan arrowheads in Fig.1c, respectively). A saddle plot that depicts genome-wide inter-compartment interactions shows that such changes are global (Fig.3a)10,13. There is a strong reduction of A-A homotypic interactions accompanied by gain of A-B mixing, while B-B interactions were not changed (Fig.3a). Weakened compartmentalization can also be seen between chromosomes, e.g. chromosomes 17 and 18, where originally well-separated A-A/B-B homotypic interactions were significantly compromised but A-B interactions enhanced (Extended Data Fig.4b).
Fig. 3. Compartmental A-B mixing and epigenome reprogramming.
a. Saddle plots showing chromatin compartmentalization between genomic regions ranked by their E1 scores (genome was divided into 50 total bins). A-A homotypic interactions are shown in the right lower part; A-B interactions are in the right upper and left lower parts. Differential compartmental interactions are shown to the right as log2 fold changes (FC, SARS-CoV-2/Mock).
b. Western blots showing the abundances of total histone H3 or several modifications in Mock and SARS-CoV-2 infected (24hpi) cells. Quantification was done by ImageJ and shown below. Rep #1 and #2 are two replicates.
c,d. Boxplots showing the log2 FC of calibrated ChIP-Seq signals of H3K27ac (from left to right, n = 896, 1116, 16877, 25535, 3739, 3990 peaks) and H3K9me3 (from left to right, n = 1333, 1332, 10660, 10660, 1332, 1333 bins) for the six categories of genomic bins with varying compartmental changes (as in Fig.2e). For boxplots, the centre lines represent medians; box limits indicate the 25th and 75th percentiles; and whiskers extend 1.5 times the interquartile range (IQR) from the 25th and 75th percentiles.
In comparison to SARS-CoV-2, the other treatments caused little or mild changes to compartments. Saddle plot analyses showed that HI-WA1 and poly (I:C) elicited negligible changes, whereas HCoV-OC43 caused mild weakening of A-A interaction (Extended Data Fig.4c-e). We further calculated compartmental shifting in these samples at 100kb bin resolution. HI-WA1 and poly(I:C) again caused negligible changes, whereas HCoV-OC43 infection resulted in ~13% of genome bins showing compartmental alteration (Extended Data Fig.5a-c). We compared the changes due to HCoV-OC43 versus those due to SARS-CoV-2, finding limited overlaps (Extended Data Figs.5d-j). These results together support that SARS-CoV-2 infection caused unique and dramatic chromatin restructuring at A/B compartmental levels. Hereafter, we focus on SARS-CoV-2 infection and only occasionally compare the differences between SARS-CoV-2 effects and the other virus or stimulus.
SARS-CoV-2 changes the host epigenome and reduces H3K27ac
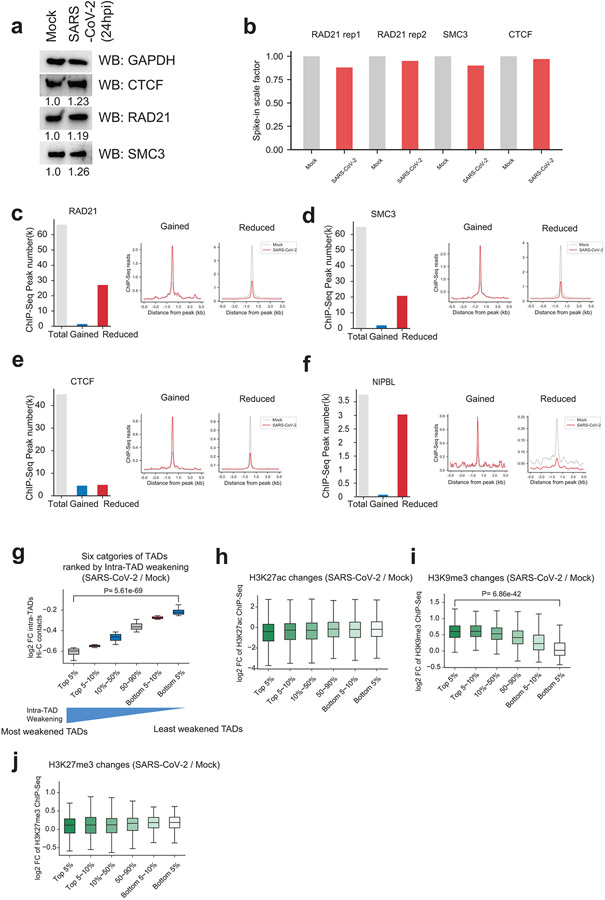
Chromatin in the A-compartment enriches for active histone modifications, while chromatin in the B-compartment enriches for repressive histone marks11,19. To mechanistically understand the observed changes, we examined active and repressive histone modifications (active: H3K4me3, H3K27ac: repressive: H3K9me3, and H3K27me3) before and after infection (Fig.1a). Western blots showed that most of these modifications remain unaltered, but surprisingly, active histone mark H3K27ac displayed a consistent and significant reduction (Fig.3b). We conducted spike-in calibrated ChIP-Seq21 to precisely quantify epigenomic changes (Extended Data Fig.6a). By calculating the ratios of ChIP-Seq reads aligned to the human versus to the mouse genome (i.e., spike-in), we consistently observed a reduction of H3K27ac on the host chromatin by ~40-45% (Extended Data Fig.6b,f), which agrees with western blots (Fig.3b). A second active mark, H3K4me3, did not exhibit overall changes, but there are moderate gains of repressive histone marks, such as the H3K9me3, after infection (Extended Data Fig.6c-i).
The epigenome reprogramming after SARS-CoV-2 infection resonates with the pervasive decrease of A-A compartmental interactions and weakening of A compartment (Figs.2c, 3a). We overlaid histone mark alterations on the six categories of genomic regions that bear compartmental changes (Fig.2e), finding that the most “B-ing” regions exhibit stronger reduction of H3K27ac (Fig.3c), whereas H3K9me3 displays the opposite trend (Fig.3d). An example is shown in Extended Data Fig.6j, where H3K27ac reduces and H3K9me3 increases, correlating with compartmental changes: weakened A-A contacts and increased A-B mixing. Because attractions between homotypic chromatin regions were suggested to be important for compartmentalization9,11,12,13, our results support the notion that SARS-CoV-2 disrupted host chromatin compartmentalization, at least in part, via reprogramming chromatin modifications.
Weakening of intra-TAD interactions on SARS-CoV-2 infection
We then examined chromatin architectures at finer scales (10kb~1Mb), namely TADs and chromatin loops2,3,9. A pronounced phenomenon (e.g., Figs.1d, 4a) is that cis-interactions within TADs (intra-TAD) were reduced by infection, whereas the contacts beyond TADs (out of the rectangles) were unchanged or increased. Examining this genome-wide, we calculated the insulation scores (IS)22 and identified 4,094 TADs. Aggregation Domain Analyses (ADA) verified the significant weakening of intra-TAD cis-interactions, which accompanies unchanged or increased cis-interactions outside of TADs (Fig.4b). Quantification of all intra-TAD contacts showed dramatic reduction (Fig.4c). Interestingly, weakened intra-TAD contacts were not accompanied by severe loss of TAD identities, i.e. the boundaries of TADs were largely unchanged (Fig.4a,b middle panels). Indeed, insulation scores of all TAD boundaries were only mildly affected by SARS-CoV-2 (Fig.4d). TAD changes due to the other viruses/stimulus are negligible; for HCoV-OC43, a trend of intra-TAD reduction was observed but the magnitude is mild (Fig.4c, Extended Data Fig.5k-m). These results indicate that strong inhibition of intra-TAD chromatin contacts is a particular feature of SARS-CoV-2.
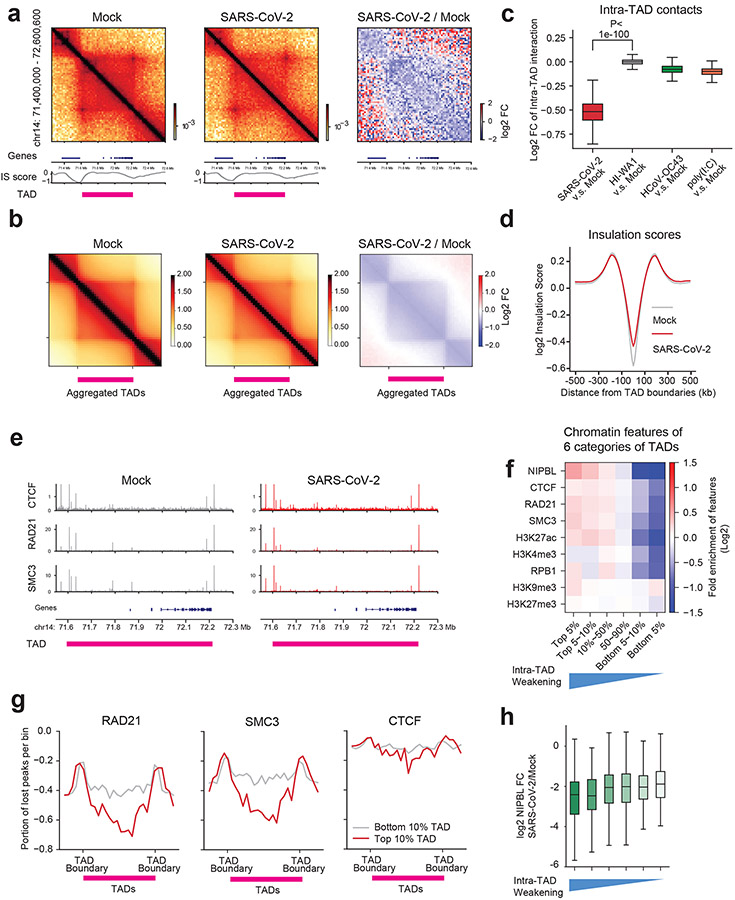
Fig. 4. SARS-CoV-2 weakens chromatin contacts and cohesin loading in intra-TAD regions.
a. Hi-C matrices (bin size: 5kb) at the indicated region (hg19), its log2 fold changes (FC) after infection (right panel), RefSeq gene tracks (bottom), the Insulation score (IS, bottom), and TAD location (pink bars). Genomic coordinate is indicated on the left.
b. Aggregated domain analyses (ADA) showing genome-wide reduction of intra-TAD interactions after infection. Right: log2 FC. TAD location is denoted by a pink bar. The plots include additional 0.5xTAD on each side for quantification.
In a and b, color scales indicate aggregated Hi-C contact frequencies (left and middle), and log2 FC of SARS-CoV-2/Mock aggregated contact frequencies (right).
c. A boxplot showing log2 fold changes of intra-TAD Hi-C interactions after several treatments in comparison to the Mock (from left to right, n = 4094, 4094, 4094, 4094). P = 0, calculated with a two-sided Mann-Whitney U test.
d. A profile plot of insulation scores calculated from all TAD boundaries in two conditions.
e. Snapshots of CTCF, RAD21, SMC3 ChIP-Seq tracks in two conditions at an example TAD region.
f. A heatmap showing the enrichment of cohesin/CTCF or epigenetic marks in six categories of TADs ranked by reduction of intra-TAD interactions (see Extended Data Fig.7g). The blue triangle below denotes the severity of intra-TAD weakening.
g. Meta-profiles showing the portions of ChIP-Seq peaks reduced by SARS-CoV-2, and the positioning of the changes in TADs. Line plots show the degree of weakened peaks relative to hosting TADs (0.25xTAD on each side out of TAD). Red lines indicate peak reduction for the top 10% weakened TADs, grey lines indicate peak reductions in the 10% least weakened TADs. The y-axis indicates the portion of lost peaks (e.g. −0.6 indicates 60% ChIP-Seq peaks in that bin were reduced).
h. Boxplots showing the log2 FC of NIPBL ChIP-Seq signals (SARS-CoV-2/Mock) in the six categories of TADs ranked by the severity of intra-TAD weakening (from left to right, n = 133, 155, 1262, 1324, 163, 108 peaks).
For boxplots in c and h, the centre lines represent medians; box limits indicate the 25th and 75th percentiles; and whiskers extend 1.5 times the interquartile range (IQR) from the 25th and 75th percentiles.
Cohesin depletion from intra-TAD regions
To further understand the reduction in intra-TAD interactions during SARS-CoV-2 infection, we examined the protein levels or chromatin binding of CTCF and cohesin, the two main regulators of TADs2,3,9. Western blots for CTCF, RAD21 showed no reduction of protein abundances (Extended Data Fig.7a). Peak calling from calibrated ChIP-seq for each factor results in ~40,000~60,000 peaks (Extended Data Fig.7b-e). Importantly, for two cohesin subunits, 40.4% (27,152/67,140) of RAD21 sites and 31.8% (20,837/65,379) of SMC3 sites were significantly reduced after SARS-CoV-2 infection (Extended Data Fig.7c,d). In contrast, only a small percentage of their binding sites were gained, i.e., 2.2% (1,510/67,140) for RAD21 sites and 3.1% (2,034/65,379) for SMC3 (Extended Data Fig.7c,d). Moderate changes of CTCF binding were observed, with 10.6% (4,853/45,530) sites lost and 10.0% (4,555/45,530) sites gained (Extended Data Fig.7e). These changes can be seen in an example TAD (Fig.4e, same as the region in Fig.4a).
We divided all TADs into six categories based on their sensitivity to SARS-CoV-2 infection (i.e., quantitative reduction of intra-TAD contacts, Extended Data Fig.7g), and examined the chromatin features of these categories. Notably, the more sensitive the TADs are to SARS-CoV-2 infection (i.e., more dramatic weakening of intra-TAD contacts), the higher enrichment of cohesin and NIPBL they bear in Mock condition (Fig. 4f), suggesting that TADs with high loads of cohesin/NIPBL make them more susceptible to cohesin depletion and intra-TAD weakening. Indeed, the top 10% most virus-weakened TADs are associated with a more dramatic loss of cohesin from the intra-TADs regions (Fig.4g). In addition, as compared to the TAD boundaries, cohesin loss was more dramatic at the intra-TAD regions (Fig.4e,g). CTCF was minimally impacted at both TAD boundaries and intra-TAD regions (Fig.4e,g). These data together indicate that SARS-CoV-2 preferentially disrupts cohesin inside TADs, but largely leaves intact the TAD structures, which is consistent with the Hi-C analysis shown in Fig.4b,d. Because NIPBL, a key factor loads cohesin to chromatin3, was found to be enriched in the most virus-weakened TADs (Fig.4f), we examined its binding by ChIP-Seq, finding dramatic inhibition by infection (Extended Data Fig.7f). NIPBL reduction correlated with the extent of intra-TAD weakening (Fig.4h). These results support that inhibited NIPBL binding to chromatin upon SARS-CoV-2 infection underlines cohesin depletion from intra-TAD regions.
Besides cohesin, H3K27ac was also more enriched in SARS-CoV-2-sensitive TADs, but heterochromatin marks did not show obvious correlation (Fig.4f). We examined the changes of epigenetic marks in these six different categories of TADs, finding that, for H3K27ac, while its level was globally reduced (Fig.3b), the reduction was consistent among all TADs (Extended Data Fig.7h). In contrast, H3K9me3, but not another heterochromatin mark H3K27me3, was gained significantly more in the most weakened TADs (Extended Data Fig.7i,j). This correlation suggests that increased H3K9me3 may play a role in cohesin depletion and intra-TAD weakening caused by SARS-CoV-2.
SARS-CoV-2 infection affects dot-shaped chromatin loops
A dot-shaped loop is a prominent feature in Hi-C that often forms between convergent CTCF sites9,19. The definition and functions of chromatin loops are debatable9. Some work has defined dot-shaped loops in Hi-C as structural loops23. In this study, we refer to dot-shaped structures as loops, and define enhancer-promoter contacts using Hi-C interaction strength (see Methods). By call-dots24, we identified 11,926 loops at 5~10 kb resolution, with examples shown below (e.g., Fig.5b). Globally, aggregation peak analyses (APA) showed that chromatin loops were not overtly affected by infection (Extended Data Fig.8a). Quantitatively (FDR<0.1), we found that 2.96% (353/11,926) of loops were weakened, whereas 4.70% (560/11,926) gained strength (Extended Data Fig.8b,c). The weakened loops are mostly short-range loops (median size 150kb), while interestingly the gained ones are much longer (median size 417.5kb, Extended Data Fig.8d). This is reminiscent of the fact that long-distance chromatin interactions were enhanced by infection (Fig.1e). As compared to loops strengthened after SARS-CoV-2 infection, the anchors of weakened loops showed more dramatic cohesin depletion after infection, and are farther away from TAD boundaries (Extended Data Fig.8e,f), suggesting that virus-weakened loops are likely a consequence of defective cohesin loop extrusion inside TADs.
Fig. 5. Chromatin restructuring and transcriptional inhibition of interferon response genes.
a. Boxplots showing fold changes RNA Pol2 (the RPB1 subunit) ChIP-Seq for IFN (n = 40) and PIF genes (n =12) due to infection (24hpi, 0.1 MOI) or IFN-ß treatment (1000u/ml, 6-hr). P-values are calculated with two-sided Mann-Whitney U test. For boxplots, the centre lines represent medians; box limits indicate the 25th and 75th percentiles; and whiskers extend 1.5 times the interquartile range (IQR) from the 25th and 75th percentiles.
b. Hi-C matrices (bin size: 5kb) and ChIP-Seq for indicated factors at the IFIT gene cluster. Top right: Mock; lower left: SARS-CoV-2. Cyan arrows denote reduced dot-shaped loops. White lines mark the TAD, with intra-TAD interactions weakened throughout. Red asterisks show virus-reduced H3K27ac peaks. Black arrows show unchanged H3K4me3 peaks on IFIT1/3 promoters. Color scales indicate Hi-C contact frequencies.
c. A diagram shows the Activity-by-contact (ABC) model of gene transcriptional outputs based on its enhancer activity and enhancer-promoter (E-P) contacts; ABC-P or ABC-P2 are revised ABC algorithms considering promoter strength.
d. A scatter plot showing the correlation between the ABC scores (x-axis) and true transcriptional changes (y-axis, SARS-CoV-2/Mock RPB1 ChIP-Seq). Error bands (light cyan) indicate the 95% confidence interval of the regression estimate. Pearson’s correlation coefficient is indicated.
e. (Upper) CRISPRi/gRNAs that target the weakened enhancers in IFIT locus (the red asterisks). The three enhancers were numerically named for simplicity. (Lower) RT-qPCR showing how CRISPRi inhibition of enhancers affects IFIT gene induction in response to poly(I:C). gRNA1 and gRNA2 are two gRNAs targeting the same enhancer. IFIT1 gene: from left to right, P = 0.0012,0.0005, 0.0051, 0.0006, 0.0004; IFIT3 gene: from left to right, P = 0.0019, 0.0042, 0.0116, 0.0018, 0.0016. Data are normalized to the -poly(I:C) in the control group, and are presented as mean values +/− SD. n = 3, representative of 2 independent experiments. P-values are calculated with a two-sided independent t-test.
f. (Upper left) Experimental design and western blots show acute RAD21 depletion and IFN-ß treatment in RAD21-mAID HCT116 cells27. Other barplots are RT-qPCR results of several IFN genes in indicated conditions. Veh: vehicle (without IAA). IAA: 5-ph-IAA (an auxin to induce RAD21 degradation). P = 2.97e-6 (IFIH1), P = 0.0125 (IFIT1), P = 3.73e-6 (IFIT2), P = 8.32e-4 (IFIT3). n = 3, representative of 2 independent experiments. Data are presented as mean values +/− SD. P-values are calculated with a two-sided independent t-test.
g. In the same region as panel b, Hi-C (bin size: 5kb) and ChIP-Seq tracks for indicated factors at the IFIT gene cluster after HCoV-OC43 infection. Top right: Mock; lower left: HCoV-OC43. The arrows and asterisks point to the same positions as in panel b. Color scales indicate Hi-C contact frequencies.
Genome/epigenome restructuring and gene deregulation
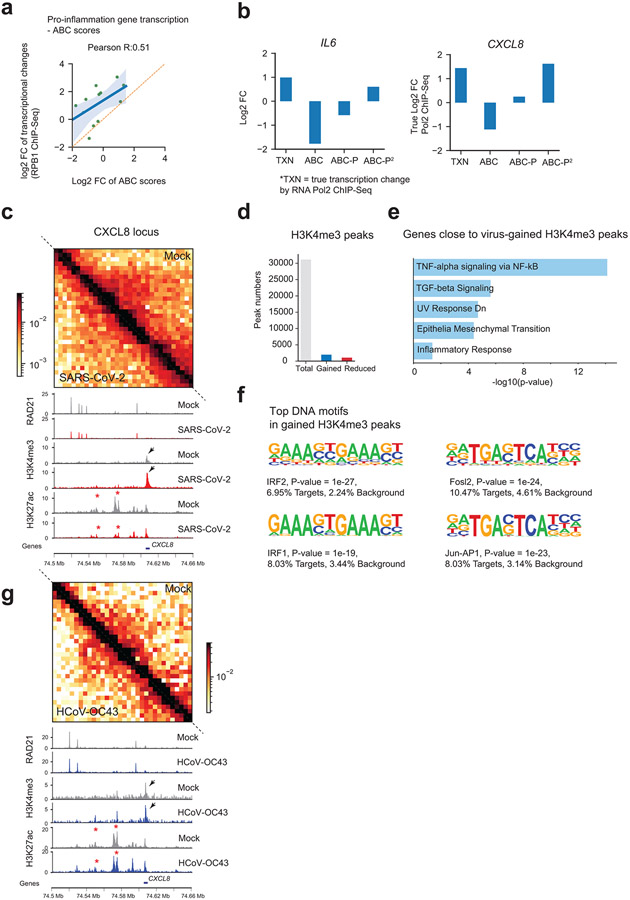
COVID-19 patients with severe symptoms often show two immuno-pathological features: a delayed or weakened innate immune response (i.e., interferon gene expression) and an exacerbated production of pro-inflammatory cytokines (e.g., IL-6)25. Our 3D genome/epigenome maps provided an opportunity to understand the deregulation of these genes. Our Pol2 ChIP-Seq recapitulated these two features (Fig.5a), indicating that their deregulation occurs transcriptionally. Their changes can also be seen by RNA-Seq (Extended Data Fig.9a). As compared to interferon-beta (IFN-ß) stimulus, SARS-CoV-2 infection elicited limited activation of interferon response genes (hereafter IFN genes) but strong increases of proinflammatory genes (hereafter PIF genes)(Fig.5a). Key examples include IFIT1/2/3/5, DDX58 (a.k.a. RIG-I) (for IFN), and IL6 or CXCL8 (for PIF)(Extended Data Fig.9b,c). Inspection of IFN gene loci revealed remarkable changes of chromatin architecture by SARS-CoV-2: 1) the dot-shaped loops were often diminished, and the chromatin contacts (which include most enhancer-promoter contacts) throughout the hosting TAD reduced; 2) cohesin occupancy within the hosting TADs was decreased; 3) H3K27ac significant reduced at many nearby putative enhancers (Fig.5b, Extended Data Fig.9d,e). Both enhancer-promoter contacts and enhancer activities may contribute to gene transcriptional outputs26, we thus modified and applied the activity-by-contact (ABC) algorithm to model how 3D genome/epigenome changes underlie virus rewiring of host transcription (Fig.5c). The ABC scores well modeled the weakened transcriptional outputs of IFN genes (Pearson’s correlation coefficient R=0.61, Fig.5d). In contrast, poor prediction was achieved if only one of the two features was considered (Extended Data Fig.9f,g). We functionally tested virus-weakened enhancers by CRISPRi in two loci, finding that IFN genes were indeed transcriptionally inhibited from responding to viral mimicry (Fig.5e, Extended Data Fig.9d,h). We validated a role of cohesin in this process by using an acute RAD21 degron system27, finding that stimulus-induced IFN gene activation was dampened (Fig.5f). This is consistent with the notion that cohesin depletion has a mild impact on basal gene transcription12,13, but can significantly affect stimuli-induced transcription28-30. These results supported that weakened 3D chromatin contacts and enhancer activity together shaped the transcriptional inhibition of IFN genes by SARS-CoV-2. Interestingly, HCoV-OC43 infection did not elicit weakening of enhancer features (H3K27ac) and chromatin contacts at IFN genes (Fig. 5g).
For PIF genes, while ABC scores showed good correlation, their true transcriptional levels after SARS-CoV-2 infection were often several-fold higher than ABC scores (Extended Data Fig.10a,b). As shown for IL6 and CXCL8, encoding key pathological cytokines, they display similar changes as IFN genes - both enhancer activities (H3K27ac) and intra-TAD contacts were reduced (Fig.6a, Extended Data Fig.10c), albeit they were strongly up-regulated (Extended Data Figs.9a-c). We therefore re-examined our epigenome data for the PIF loci, finding a unique and dramatic gain of H3K4me3 at their promoters (Fig.6a, Extended Data Fig.10c), which did not take place on IFN genes (Fig. 5b, Extended Data Fig.9d,e). H3K4me3 at promoters may play a causal role for transcription31,32. Globally, H3K4me3 exhibited relatively limited changes by SARS-CoV-2: 5.8% (1,843/31,761) of sites showed increases and 3.5% (1,104/31,761) decreases (Extended Data Fig.10d). Interestingly, genes close to gained H3K4me3 sites enrich for key pathways in COVID-19 pathology33 (e.g., TNF-alpha or TGF-beta signaling, Extended Data Fig. 10e). We revised the ABC algorithm by including H3K4me3 changes at promoters (Fig.5c), and a revised ABC-P2 score better modeled PIF genes’ transcriptional induction (Fig.6b, Extended Data Fig.10b).
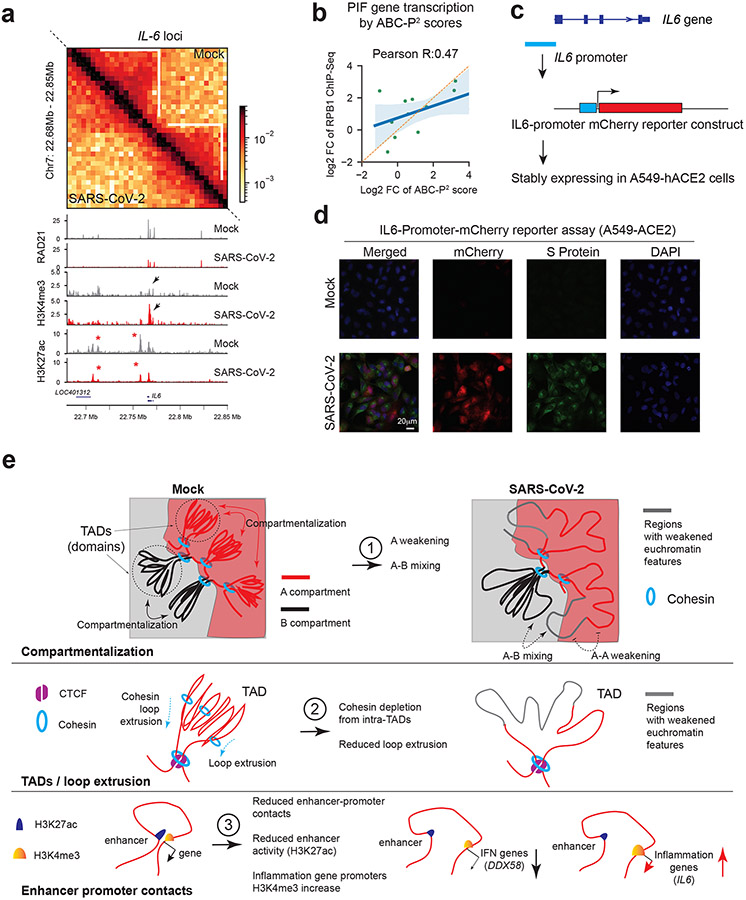
Fig. 6. Augmented promoter H3K4me3 and pro-inflammatory gene activation.
a. Hi-C matrices (bin size: 5kb) and ChIP-Seq tracks at the IL6 locus in Mock and SARS-CoV-2 conditions. White lines mark TADs, with intra-TAD interactions weakened throughout. Red asterisks show reduced H3K27ac peaks. Black arrows show enhanced H3K4me3 on IL6 promoter. Color scales indicate Hi-C contact frequencies.
b. A scatter plot showing the correlation between the ABC-P2 scores and true transcriptional changes of PIF genes (RPB1 ChIP-Seq). Fold changes denote SARS-CoV-2/Mock. Error bands indicate the 95% confidence interval of the regression estimate. Pearson’s correlation coefficient is shown.
c. A diagram of an IL6-promoter driven mCherry reporter.
d. Representative images of mCherry (red) and SARS-CoV-2 Spike protein (green) in cells carrying IL6-Promoter-mCherry reporter under Mock or SARS-CoV-2 infection (0.1 MOI, 24hpi). DAPI (blue). Scale bar is shown.
e. A model that summarizes current observations of chromatin restructuring by SARS-CoV-2 infection at the 3D genome and epigenome scale. They are categorized from top to bottom in terms of A/B compartments, TADs/loop extrusion and enhancer-promoter contacts (see Extended Data Fig.1a,b for additional information). These chromatin changes correlate with and may explain transcriptional deregulation of immuno-pathological gene deregulation in COVID-19 patients.
We tested the role of PIF promoter by using a mCherry reporter, finding that the IL6 promoter alone can elicit gene activation upon infection (Fig.6c,d). Intriguingly, promoters with virus-augmented H3K4me3 display motif enrichment of specific transcriptional factors, such as IRF1/2 or Jun/AP1 (Extended Data Fig.10f), suggesting their potential roles in PIF promoter activation. In notable contrast, HCoV-OC43 infected cells showed little or no changes of H3K4me3 at PIF promoters (Extended Data Fig.10g). These results together suggested that pro-inflammatory genes are induced by SARS-CoV-2 through a unique process involving H3K4me3 to augment promoter activity.
DISCUSSION
Here we found that SARS-CoV-2 infection significantly restructures 3D host chromatin, featuring wide-spread compartment A weakening and A-B mixing, and global reduction of intra-TAD chromatin contacts (Fig.6e). The epigenome is also altered, including a global reduction of active chromatin mark H3K27ac and a specific increase of H3K4me3 at pro-inflammatory gene promoters. Interestingly, these changes were quite unique to SARS-CoV-2 as compared to infection by common cold coronavirus or immune stimuli. What we characterized here represents a direct, cell-autonomous effect elicited by SARS-CoV-2 on the host chromatin. Consistent with our results, Ho et al. observed some compartmental changes after infection34. In parallel, SARS-CoV-2 also imposes non-cell autonomous effects on the 3D genome35. An important unanswered question is how exactly SARS-COV-2 infection restructures host chromatin and whether viral regulators are involved. In this light, ORF8 was recently found to disrupt the host epigenome7, suggesting specific viral factors could confer the chromatin restructuring we discovered in this work.
Mechanistically, SARS-CoV-2 infection depleted the cohesin complex, in a pervasive but selective manner, from intra-TAD regions. Such changes not only provide a molecular explanation to the weakening of intra-TAD interactions, but also support a notion that defective cohesin loop extrusion inside TADs releases this chromatin to engage in long-distance associations (Fig.6e). Indeed, chromatin in infected cells displayed a higher frequency of extremely long-distance intra-chromosomal and inter-chromosomal interactions (Fig.1e, Extended Data Fig.2h). Consistent with this idea, genetic depletion of cohesin induced gains of long-distance chromatin interactions, supporting the idea that active loop extrusion counteracts chromatin mixing3,12,13.
Importantly, host chromatin alterations resonate with the dichotomic deregulation of immune gene expression in COVID-19: weakened IFN responses accompany increased pro-inflammatory (PIF) gene expression25. Weakening of enhancer-promoter contacts and reduced enhancer activity correlate with inhibited IFN gene transcription, and CRISPRi data supported the regulatory roles of virus-weakened enhancers. These changes take place at critical loci, including DDX58 (coding for virus sensor RIG-I), whose inhibition is required for successful infection36. Unexpectedly, SARS-CoV-2 infection selectively modifies the H3K4me3 of PIF gene promoters, suggesting unappreciated mechanisms at these promoters that confer aberrant inflammation in COVID-1925. Additional discussion on the roles of cohesin, chromatin interactions and gene transcription, and limitations of this study can be found in Supplementary Note.
Epigenetic alteration is known to exert long-term effects on gene expression and phenotypes37,38. Given the increasingly realized high incidence of post-acute SARS-CoV-2 sequelae (long-COVID39), understanding the viral impacts on host chromatin and epigenome will not only provide new strategies to fight SARS-CoV-2 in the acute phase, but also paves the way for unraveling the molecular basis of long COVID for its intervention.
Methods:
Cell culture
Human lung adenocarcinoma cells A549 expressing human ACE2 (A549-ACE2, #NR53821) was acquired from BEI Resources (Manassas, VA). They were maintained in DMEM/F-12 (1:1, Corning) medium supplemented with 10% FBS (GeneDepot) and blasticidin (100uM). Normal A549 cells were purchased from ATCC (CCL-185) and were cultured in DMEM/F-12 (1:1, Corning) supplemented with 10% FBS. 293T cells were from ATCC and were cultured in DMEM with 10% FBS. HCT-8 cells were purchased from ATCC (CCL-224) and were cultured in RPMI1640 with 10% FBS. Vero-E6 cells were acquired from ATCC (CRL-1586). Mouse ESCs (F121-9) are a gift from David Gilbert lab (San Diego Biomedical Research Institute, CA) and were cultured following standard procedure of the 4D nucleome consortium (https://data.4dnucleome.org/biosources/4DNSRMG5APUM/). RAD21-mAID2-mClover HCT116 cells are a gift from Masato Kanemakei lab (NIG, Japan), and were cultured in McCoy's 5A Medium with 10% FBS. All these cells were cultured at 37°C with 5% CO2. Transfection of plasmids or siRNAs was performed using Lipofectamine 3000 or RNAiMAX (Life technologies) following manufacturer's instructions. For CRISPRi experiments, in order to examine enhancer functions during cell responses to RNA virus, we introduced Polyinosinic:polycytidylic acid (poly (I:C), 333 ng/ml, Sigma, P9582) into A549 cells using lipofectamine 2000 and harvested the total cellular RNAs for gene expression experiments after 4 hours. For some experiments, fluorescein-labeled poly (I:C) was acquired from Invivogen (cat# tlrl-picf). For the Hi-C 3.0 experiment, we introduced 66 ng/ml of poly (I:C) for 6hr to mimic RNA virus infection in A549-ACE2 cell line. For interferon beta (IFN-beta) treatment, we treated A549-hACE2 cells with 1000U recombinant human interferon beta (8499-IF-010/CF, R&D systems) for 6 hours. For RAD21-mAID2-mClover cells, we first pre-treated 1μM 5-ph-IAA for 2 hours and then treated 1000 U/ml interferon beta together with 1μM 5-ph-IAA for 4 hours.
SARS-CoV-2 and HCoV-OC43 infections in A549-ACE2 cells
SARS-CoV-2 isolate USA-WA1/2020 (NR-52281; BEI Resources, Manassas, VA) was employed to infect human A549-ACE2 cells (NR53821; BEI Resources). For viral infections, serum/antibiotics-free Eagle’s MEM medium supplemented with 1mM HEPES was used. Briefly, cells grown in 10-cm culture dishes at about 70-80% confluency were washed with the serum free medium, viral inoculum was added at 0.1 MOI for 1 hr. After that, non-adsorbed viral particles were gently aspirated out and the monolayers were replenished with 10% FBS containing MEM supplemented with 1mM HEPES. Infected cells were incubated at 37°C with 5% CO2 for 6hr or 24hr post-infection for experiments. For heat-inactivated SARS-CoV-2 treatment, we treated the equivalent number of the heat-inactivated virus (BEI resources, NR-52286) to 0.1 MOI based on genomic equivalence (GE). Heat-inactivated SARS-CoV-2 (HI-WA1) was obtained from the BEI Resources (NR-52286; Manassas, VA) and had 5.36x10^8 genome equivalents per ml. Heat-inactivated SARS-CoV-2 was given to cells using the same protocol as described above for SARS-CoV-2 isolate USA-WA1/2020 (NR-52281; BEI Resources, Manassas, VA), with the equivalent amount of 0.1 MOI based on genomic equivalence (GE).
HCoV-OC43 (VR-1558) was purchased from ATCC. For viral infections, serum/antibiotics-free RPMI 1640 medium was used. Briefly, A549-ACE2 cells grown in 10-cm culture dishes at about 80-90% confluency were washed two times with PBS, viruses were inoculated onto A549-ACE2 cells and allowed to adsorb for 2hr at 33 °C and 0.5 MOI. After that, non-adsorbed viral particles were gently aspirated out and the monolayers were replenished with 2% horse serum (HS) containing RPMI 1640. Infected cells were incubated at 33°C with 5% CO2 for 24hr post-infection for experiments.
Preparation of SARS-CoV-2 and HCoV-OC43 stock
The stock SARS-CoV-2 was propagated in Vero-E6 cells. Briefly, Vero-E6 cells were grown to 80% confluence in 10% FBS containing MEM medium supplemented with 1mM HEPES and 1X antibiotics and antimycotics. Prior to infection, Vero-E6 cells were washed once with PBS and the viral inoculum was added to the flask in the presence of 3 ml of serum-free and antibiotics-free MEM medium supplemented with 1mM HEPES, and incubated for 1 hour at 37°C with 5% CO2. At the end of incubation, non-adsorbed virus was aspirated out and cells were replenished with 25ml of MEM supplemented with 10% FBS and 1mM HEPES. Infected cells were incubated for 48 hours at 37°C with 5% CO2. At 80% of cell lysis, SARS-CoV-2 was harvested by detaching all the cells with a cell scraper and centrifuging at 300g for 3 minutes. Viral aliquots were stored in screw-cap vials at −80°C.
The stock of HCoV-OC43 was propagated in HCT-8 cells. Briefly, HCT-8 cells were grown to 80% confluence in RPMI 1640 medium containing 10% FBS. Prior to infection, HCT-8 cells were washed two times with PBS and the viral inoculum was added to the flask in the presence of 3 ml of serum-free and antibiotics-free RPMI 1640 and incubated for 2-hour at 33°C with 5% CO2. At the end of incubation, non-adsorbed virus was aspirated out and cells were replenished with 25ml of RPMI 1640 supplemented with 2% HS. Infected cells were incubated for 5 days at 33°C with 5% CO2. At 80% of cell lysis, HCoV-OC43 was harvested by detaching all the cells with a cell scraper and centrifuging at 300g for 3 minutes. Viral aliquots were stored in screw-cap vials at −80°C.
Determination of plaque forming units (PFU/ ml stock)
For the determination of infectious viral titers, plaque assays were performed using Vero-E6 cells. Briefly, Vero-E6 grown in 6-well plates were infected with 12 serial dilutions (1:10) of the SARS-CoV-2 stock in serum/BSA/antibiotic-free MEM medium with 1mM HEPES for 1 hour at 37°C with 5% CO2. At the end of incubation, non-adsorbed viral particles were aspirated and the infected cells were layered upon with MEM medium containing 0.5% agarose, 2% BSA and 1mM HEPES, and incubated for 48 hours at 37°C with 5% CO2. Fixation was carried out using 3.75% buffered formaldehyde (in PBS) for 10 minutes. After aspirating formaldehyde, the agarose layers were gently removed. Infected cells were stained with 0.3% crystal violet for 5 minutes, followed by washing once with PBS. Plates were air-dried and visible infectious plaques were counted in each dilution to determine the plaque forming units/ml of the stock.
To determine infectious viral particles for HCoV-OC43, the median Tissue Culture Infectious Dose (TCID50) was performed using HCT-8 cells as the following. HCT-8 cells were seeded into 96-well plates at a concentration of 5 × 10^4 cells/well and incubated for 24 hr to reach a confluence of 90%. Viral stock was serially diluted in RPMI 1640 containing 10% FBS in the range 10^−1 to 10^−8.
Diluted viral samples were incubated at 33°C for 5 days. Next, the media was removed and replaced with 0.3% crystal violet for 5 minutes, followed by washing once with PBS. Wells were scored for the presence or absence of visual cytopathic effect (CPE) in each dilution. The TCID50/mL was calculated by the Karber method43. For means of comparison to plaque assay results, TCID50/mL values were converted to PFU/mL by multiplying by 0.7.
Lenti-viral transduction and CRISPRi
We in-house generated a lentiviral construct expressing dCas9-KRAB-MeCP2 by PCR amplification of the dCas9-KRAB-MeCP2 (contains a domain of MeCP2) from pB-CAGGS-dCas9-KRAB-MeCP2 (Addgene 110824), and then insert it to the pLenti-EF1a-dCas9-VP64-2A-Blast backbone (Addgene 61425) to replace the dCas9-VP64. The gRNAs used in CRISPRi were cloned into the Addgene 61427 backbone using BsmBI enzyme. To generate lentivirus, 293T cells were transfected with the lentiviral transfer vector DNA, psPAX2 packaging and pMD2.G envelope plasmid DNA at a ratio of 4:3:1, respectively, by lipofectamine 2000. After 16 h, the culturing media was changed to fresh one, and the supernatants were collected twice at 48h to 72h post-transfection. The harvested lentiviral supernatants were filtered using 0.45 μm syringe filter (Fisher) and used to infect target A549 cells (polybrene was added at a final concentration of 8 μg/ml, Sigma). To infect A549 cells for CRISPRi, cells were first infected by a lentivirus expressing dCas9-KRAB-MeCP2 for 24 hours and selected with appropriate antibiotics (10 μg/ml blasticidin) for 7 days to generate a stable cell line. The stable cell line was then subjected to viral infection by individual gRNAs targeting each enhancer, and they were further selected by 100 μg/ml Zeocin for 4-7 days. These stable cells were then used for experiments. The gRNA cloning oligos are shown in Supplementary Table 2.
Promoter Reporter Assay
An IL6 promoter-driven mCherry reporter construct was acquired from GeneCopoeia (Cat# HPRM30562-LvPM02). This lentiviral plasmid was introduced into the target cell line using lentiviral transduction, which is similar to the steps described above in the lentivirus section. After 24hr of infection, the infected A549-ACE2 cells were selected under 2 μg/mL puromycin. The established stable cell line was maintained in DMEM/F-12 (1:1, Corning) medium supplemented with 10% FBS (GeneDepot), 100 μg/mL blasticidin, and 2 μg/mL puromycin. It was then used for Mock or SARS-CoV-2 infection (0.1 MOI, 24 hpi), and mCherry expression was examined by confocal imaging.
RNA Extraction and RT-qPCR
RNA extraction of SARS-CoV-2 infected A549-ACE2 cells was performed by TRIzol (Thermo Fisher Scientific, 15596-026) following manufacturer's instructions. In some other cases, RNA extraction from cells expressing CRISPRi or other transfection were performed using Quick-RNA Miniprep Kit (Zymo Research, 11-328). Reverse transcription was conducted by using Superscript™ IV first strand synthesis kit (Thermo Fisher, 18091050), and often the random hexamer primer was used to test the expression levels of target genes. Real-time qPCR (RT-qPCR) was performed using SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad, 172-5274). Primer sequences used in this study can be found in Supplementary Table 2. Relative gene expression was normalized to internal control (18S RNA).
Western blots
Cells were lysed in RIPA (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate) with cOmplete™ Mini Protease Inhibitor Cocktail (Roche, 11836153001) on ice for 30 min. Lysates were sonicated in Qsonica 800R (25% amplitude, 3 min, 10 sec on 20 sec off interval) and centrifuged at 18,407 g. The supernatants were mixed with 2x Laemmli Sample Buffer (Bio-Rad) and boiled at 95°C for 10min. The boiled proteins were separated on 4% to 15% SDS-PAGE gradient gels and transferred to LF PVDF membrane (Bio-Rad, Cat #1620260). The membranes were blocked in 5% skim milk in TBST (20 mM-Tris, 150 mM NaCl, and 0.2% Tween-20 (w/v)) for an hour and then briefly washed in TBST twice. Then, the membranes were incubated in TBST with primary antibodies (GAPDH (Proteintech, 60004-1, 1:2000 dilution), alpha-Tubulin (Sigma, T5168, 1:1000 dilution), RAD21 (Abcam, Ab992, Lot: GR214359-10, 1:1000 dilution), CTCF (Millipore, 07-729, 1:1000 dilution), SMC3 (Abcam, Ab9263, Lot:GR466-7, 1:1000 dilution), Total Histone H3 (Abcam, Ab1791, Lot:GR206754-1,1:2000 dilution), H3K4me3 (Abcam, Ab8580, Lot: GR3264490-1, 1:2000 dilution), H3K9me3 (Abcam, Ab8898, Lot: GR164977-4, 1:2000 dilution), H3K27ac (Abcam, Ab4729, Lot: GR3357415-1, 1:2000 dilution), and H3K27me3 (Cell Signaling Technology, #9733S, Lot19, 1:2000 dilution) at 4°C overnight. After washing 3 times in TBST, the blots were incubated in TBST with secondary antibody (Horseradish peroxidase (HRP)-conjugated antibody) for an hour. After 6 times of washing in TBST, the blots were developed in a Bio-Rad ChemiDoc™ Gel Imaging System. The intensity of the bands were quantified using ImageJ. In order to quantify the relative expression, protein expression normalized by the intensity of GAPDH was compared between infected and the Mock treated samples.
Immunofluorescence microscopy and TUNEL Assay
Expression of spike protein of SARS-CoV-2 was measured by immunofluorescence microscopy. A549-ACE2 cells seeded on glass slides were infected with SARS-CoV-2 at a MOI of 0.1. 24hr post infection (24hpi), cells were fixed by 4% paraformaldehyde in phosphate buffered saline (PBS) for 1hr at room temperature. The coverslips were washed with 0.1% BSA in 1xPBS (Wash buffer) and blocked with 1% BSA with 0.3% Triton-X100 in PBST (Blocking buffer) for 45min at room temperature. Cells were incubated with an antibody targeting the SARS-CoV-2 Spike glycoprotein (1:500, Abcam, Cat#ab272504) or a monoclonal antibody targeting the HCoV-OC43 (1:200, EMD Millipore, Cat# MAB9012) diluted in the blocking buffer overnight at 4 degree. Subsequently, after washes, cells were incubated with secondary antibody diluted in blocking buffer for 1hr at room temperature, followed by incubation with 4,6-diamidino-2-phenylindole (Invitrogen, Catalog#D1306) for 5 minutes at room temperature. Coverslips were mounted in antifade mounting medium (Thermo scientific, Cat#TA-030-FM) and the fluorescence images were recorded using a Leica confocal microscope or Nikon A1R. In order to assess the apoptotic dead cells in the virus infected condition, we used Click-iT TUNEL Alexa Fluor Imaging assay (Invitrogen, Cat# C10245) following manufacturer’s instructions. As a positive control of the TUNEL assay, one sample was incubated in DNase I solution for 30 min at room temperature, following the instructions by the manufacturer.
Hi-C 3.0
Hi-C 3.0 was performed based on a recent protocol14, which is largely modified based on in situ Hi-C19. Briefly, ~5 million SARS-CoV-2 infected A549-ACE2 cells were washed once with PBS to remove debris and dead cells, trypsinized off the culture plates, and were cross-linked using 1% formaldehyde for 10mins at room temperature, quenched with 0.75M Tris-HCl pH 7.5 for 5mins. These cells were further cross-linked with 3mM disuccinimidyl glutarate (DSG) for 50mins, and again quenched with 0.75M Tris-HCl pH 7.5 for 5mins. Cross-linked cell pellets were washed with cold PBS, and then resuspended in 0.5 ml ice-cold Hi-C lysis buffer (10 mM Tris-HCl, pH 8.0; 10mM NaCl, 0.2% NP-40 and protease inhibitor cocktail), and rotated at 4°C for 30 mins. Nuclei were washed once with 0.5 ml ice-cold Hi-C lysis buffer. After pelleting down the nuclei, 100 μl of 0.5% SDS was used to resuspend and permeabilize the nuclei at 62 °C for 10 mins. Then 260 μl H20 and 50 μl 10% Triton-X100 were added to quench the SDS at 37 °C for 15 mins. Subsequently, enzyme digestion of chromatin was performed at 37 °C overnight by adding an additional 50 μl of 10X NEB buffer 2, MboI (NEB, R0147M, 100U) and DdeI (NEB, R0175L, 100U). After overnight incubation, the restriction enzyme was inactivated at 62 °C for 20 mins. To fill-in the DNA overhangs and add biotin, 35U DNA polymerase I (Klenow, NEB, M0210) together with 10 μl of 1mM biotin-dATP (Jeana Bioscience), 1μl 10mM dCTP/dGTP/dTTP were added and incubated at 37°C for 1 hour with rotation. Blunt end Hi-C DNA ligation was performed using 5000U NEB T4 DNA ligase with 10X NEB T4 Ligase buffer with 10mM ATP, 90 μl 10% Triton X-100, 2.2 μl 50mg/ml BSA at room temperature for four hours with rotation. After ligation, nuclei were pelleted down and resuspended with 440 μl Hi-C nuclear lysis buffer (50mM Tris-HCl pH7.5, 10mM EDTA, 1% SDS and protease inhibitor cocktail), and further sheared using the parameter of 10/20 secs ON/OFF cycle, 25% Amp, 4 mins by a QSonica 800R sonicator. Around 10% of the sonicated chromatin was subjected to overnight decrosslinking at 65°C, proteinase K treatment, and DNA extraction. After DNA extraction, biotin labeled Hi-C 3.0 DNAs were purified by 20 μl Dynabeads® MyOne™ Streptavidin C1 beads (Thermo Fisher 65002). The biotinylated-DNA on C1 beads was used to perform on-beads library making with NEBNext® Ultra™ II DNA Library Prep Kit for Illumina (NEB, E7645L) following manufacturers instructions. The sequencing was done on a NextSeq 550 platform with PE40 mode, and demultiplexed with bcl2fastq v2.2.
Chromatin immunoprecipitation (ChIP-Seq) and spike-in calibrated ChIP-Seq
ChIP-Seq was performed as previously described with minor modifications44. For most ChIP-Seqs in A549-ACE2 cells with Mock treatment or 24-hr SARS-CoV-2 infection, ~5 to 10% of mouse ESCs (F121-9, a gift from David Gilbert) were added as spike-in controls before sonication with equal proportions to the two human cell samples (see Extended Data Fig.6a). For cell cross-linking for ChIP-Seq, briefly, the cells were trypsinized in Trypsin-EDTA (or Accutase for mESCs). After centrifugation, the cells were crosslinked by 1% formaldehyde (FA) in PBS for 10 min. The fixation steps were stopped in a quenching solution (0.75M Tris-HCl pH 7.5) for 10min. After centrifugation of the cells, we extracted the nuclei first by buffer LB1 [50 mM HEPES-KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA (pH 8.0), 10% (v/v) glycerol, 0.5% NP-40, 0.25% Triton X-100 and 1×cocktail protease inhibitor], and then by LB2 [10 mM Tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA (pH 8.0), 0.5 mM EGTA (pH 8.0) and 1×cocktail protease inhibitor]. After centrifugation, the nuclei were suspended in buffer LB3 [10 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA (pH 8.0), 0.5 mM EGTA (pH 8.0), 0.1% Na-deoxycholate, 0.5% N-lauroyl sarcosine and 1×cocktail protease inhibitor], and the chromatin was fragmented by using Q800R3 sonicator (QSonica) using conditions of 10 seconds ON, 20 seconds OFF for 7-9 mins (at 20% amplitude). Sheared chromatins were collected by centrifugation, and were incubated with appropriate antibodies (often 2-3μg) at 4°C overnight. The next morning, the antibody-protein-chromatin complex was retrieved by adding 25μl pre-washed Protein G Dynabeads (Thermo Fisher Scientific, 10004D). Immunoprecipitated chromatin DNA was de-crosslinked by 65°C heating overnight using elution buffer (1% SDS, 0.1M NaHCO3), and then treated by RNase A and proteinase K, which was finally purified by phenol chloroform. The DNAs were subjected to sequencing library construction using NEBNext® Ultra™ II DNA Library Prep Kit for Illumina (NEB, E7645L), and were deep sequenced on a NextSeq 550 platform using 40nt/40nt pair-ended mode. The antibodies used for ChIP-Seq include RNA Polymerase II (RPB1 N terminus, Cell Signaling Technology, #14958S, Lot4), RAD21 (Abcam, Ab992, Lot: GR214359-10), SMC3 (Abcam, Ab9263, Lot:GR466-7), CTCF (Millipore, 07-729), NIPBL (Bethyl, A301-779A, lot#4), H3K4me3 (Abcam, Ab8580, Lot: GR3264490-1), H3K9me3 (Abcam, Ab8898, Lot: GR164977-4), H3K27ac (Abcam, Ab4729, Lot: GR3357415-1), H3K27me3 (Cell Signaling Technology, #9733S, Lot19) and HA (Abcam, Ab9110, Lot:GR3231414-3).
Ribo-depleted total RNA sequencing (RNA-Seq)
Total RNAs from mock or virus infected A549-ACE2 cells were extracted by TRIzol, and 100-200ng of total RNAs were used for making strand-specific ribosome-RNA-depleted sequencing library by the NEB Ultra II Directional RNA library kit (E7760L) following manufacturer's instructions. Libraries were sequenced on a NextSeq 550 using 40nt/40nt pair-ended mode.
Bioinformatic analyses:
Calibrated ChIP-Seq analyses
Sequencing reads were aligned to a concatenated genome of hg19 human genome assembly and mm9 mouse genome assembly with STAR(v2.7.0)45. Duplicated reads were removed, and only unique aligned reads will be considered for later visualization and quantification. The scaling factor was calculated as the ratio of the number of reads uniquely aligned to human chromosomes versus the number of reads aligned to mouse chromosomes (Extended Data Fig.6a). Uniquely aligned human reads were extracted with samtools(v1.9)46, and normalized by the corresponding scaling factor with deeptools(v3.1.3)47. For RPB1 ChIP-Seq gene transcription quantification, hg19 RefSeq gene annotation coordinates were used. The peak-calling of most ChIP-Seq were performed with the parameters of -f BAM -q 0.01 in MACS2(v2.1.4)48. Peaks with Log 2 fold change of normalized ChIP-Seq reads ratio smaller than −1 or greater than 1, are considered as reduced or gained peaks. ChIP-Seq reads are summarized in Supplementary Table 1. A public NIPBL ChIP-Seq dataset was obtained from SRR3102878.
RNA-Seq analysis
RNA-Seq reads were aligned to the hg19 reference human genome or SARS-CoV-2 viral genome (NC_045512.2) with STAR(v2.7.0)45. The percentage of reads uniquely aligned to SARS-CoV-2 genome versus total reads was calculated to verify a high viral infection rate. For human gene quantification, only uniquely aligned reads mapped to the hg19 genome were kept for further analysis.
Hi-C 3.0 data processing
Hi-C 3.0 raw data was primarily processed with HiC-Pro (v2.11.4)49. The pair of reads were mapped to the human reference genome assembly hg19, and multi-mapped pairs, duplicated pairs, and other unvalid 3C pairs were filtered out following the standard procedure of HiC-Pro. All valid Hi-C pairs were merged between replicates (unless specified noted), and were further converted to Juicebox format50 or cooler format51 for visualization and further analyses. Hi-C contact matrices were normalized with cooler (v0.8.11) balance function. Reads numbers of Hi-C are listed in Supplementary Table 1. The SCC correlation coefficients between two replicates were calculated to assess the reproducibility of Hi-C experiments41. The P(s) curve was calculated as a function between contact frequency (P) and genomic distances(s) (Fig. 1e). Only intra-chromosomal pairs (cis) were used to calculate P(s) curve.
A/B Compartment analyses
A/B nuclear compartments were identified based on decomposed eigenvectors (E1) from 20kb or 100kb Hi-C contact matrices using cooltools (v0.4.1). A/B compartmental scores (E1) were corrected by GC densities in each bin. Saddle plot analyses were performed to measure the compartmentalization strength in a genome-wide scale using cooltools compute-saddle (similar to previous work10,13). Briefly, we first sorted the rows and the columns in the order of increasing compartmental scores within observed/expected (O/E) contact maps based on the data in Mock cells. Then we aggregated the rows and the columns of the resulting matrix into 50 equally sized aggregate bins, and plotted the aggregated observed/expected Hi-C matrices as the “saddle” plots (Fig.3a). In Fig.1c and other few places, Pearson correlation Hi-C matrices were used to emphasize the compartmental checkerboard pattern. We first calculated the observed/expected Hi-C maps as OE matrices (bin size = 80,000 bp). Each value (i,j) in Pearson matrices indicates the Pearson correlation coefficient between the i-th column of OE matrices and the j-th column of OE matrices (bin size = 80,000bp). For changes of compartmental strength (Fig.2c and Extended Data Figs.4a,5a-c), the changes for each genomic region between Mock and SARS-CoV-2 samples were identified based on 100kb-binned compartmental scores (E1) of two Hi-C 3.0 replicates, largely following a previous study20. For each 100kb, a student’s t-test was first performed on Mock and SARS-CoV-2 compartmental scores (E1). Only the 100kb bins that have ∣delta E1∣ > 0.2 and P-value < 0.05 were considered as bins with changed compartmental strength. Different categories of compartment changes (in Fig.2c) were defined as below (similar to 20): A to stronger A: (Mock-E1 - SARS-CoV-2-E1) < −0.2, Mock-E1>0.2; B to A: (Mock E1 - SARS-CoV-2 E1) <−0.2, Mock-E1 < −0.2, SARS-CoV-2-E1>0; B to weaker B: (Mock- E1 - SARS-CoV-2 E1) < −0.2, Mock-E1 <−0.2, SARS-CoV-2-E1<0; B to stronger B: (Mock-E1 - SARS-CoV-2-E1)>0.2, Mock-E1 < −0.2; A to B: (Mock-E1 - SARS-CoV-2-E1)<−0.2, Mock-E1 > 0.2, SARS-CoV-2-E1<0; A to weaker A: (Mock-E1 - SARS-CoV-2-E1)<−0.2, Mock-E1 > 0.2, SARS-CoV-2-E1>0. For enrichment of epigenetic features on compartments affected (e.g. Fig. 2f), we ranked all 100kb bins into six categories (Top 5%, top 5~10%, 10~50%, 50~90%, bottom 5~10%, bottom 5%), and then calculated epigenomic features at each 100k bin. For histone modifications or chromatin regulatory factors that have sharp peaks in ChIP-Seq (like H3K27ac, H3K4me3), we quantified the numbers of peaks in each 100kb bin. For modifications or factors that have broad ChIP-Seq patterns (like H3K9me3 and H3K27me3), we quantified the calibrated ChIP-Seq reads throughout the entire 100kb bin. The enrichment of these ChIP-seq signals were calculated by dividing the median quantification inside these six categories by the genome-wide median quantification.
TADs and insulation scores
Hi-C 3.0 data were used to identify topologically associating domains (TADs) in A549-ACE2 cells following standard 4D Nucleome consortium protocol (github.com/4dn-dcic/docker-4dn-insulation-scores-and-boundaries-caller). First, insulation scores22 and boundary strengths of each 10kb bin with a 200kb window size were measured to quantify the TAD insulation using cooltools (https://github.com/open2c/cooltools/blob/master/cooltools/cli/diamond_insulation.py). Then, we identified TAD boundaries respectively in Mock and SARS-CoV-2 infected samples by using a boundary score cutoff of 0.5. We further merged TAD boundaries identified in these two conditions, and compared insulation scores at merged TAD boundaries (Fig. 4d). Merged TAD coordinates were used to perform downstream analyses. For each TAD, we quantified its mean Hi-C contacts throughout the domain (excluding very short distant interactions <15kb), which is considered intra-TAD interaction used in the paper. Based on the log2 fold changes of intra-TAD mean Hi-C contacts (SARS-CoV-2 / Mock), we ranked all TADs into six categories (Top 5%, top 5~10%, 10~50%, 50~90%, bottom 5~10%, bottom 5%), and calculated different epigenomic features of these six categories. For histone modifications or chromatin regulatory factors that have sharp peaks in ChIP-Seq (like H3K27ac, H3K4me3, CTCF or cohesin subunits), we quantified the numbers of peaks as well as the numbers of gained or lost peaks in different TADs. For modifications or factors that have broad ChIP-Seq patterns (like H3K9me3 and H3K27me3), we quantified the calibrated ChIP-Seq reads throughout the TADs. The enrichment of these ChIP-seq signals were calculated by dividing the median quantification inside these six categories by the genome-wide median quantification.
Chromatin loop calling and enhancer-promoter contacts
For loop calling, we largely followed a recent 4DN benchmarking paper14. In brief, we used a reimplement of HICCUPS loop-calling tool, call-dots function inside cooltools (https://github.com/open2c/cooltools/blob/master/cooltools/cli/call_dots.py) to identify structural chromatin loops in different samples. We first called loops at 5kb and 10 kb resolution separately, then used the following strategy to merge 5kb and 10kb loops. 5kb loops called at both 10kb and 5kb resolution were first kept, all unique 10kb resolution loops were kept, and only unique 5kb loops that are smaller than 100kb were kept. Differential loops were identified by first quantifying the Hi-C raw contacts at 40kb resolution of each called loop, and then by performing DESeq2(v1.26.0)52 differential analyses on these raw counts. We considered loops with a DESeq2 FDR <0.1 and a log2FC >0 or <0 as virus-strengthened or weakened chromatin loops. The APA (Aggregation Peak Analysis) was performed by superimposing observed/expected Hi-C matrices on merged loops with the coolpuppy tool (v0.9.2)53.
Activity by Contact (ABC) score
ABC score calculation largely follows a previous study26 with modifications. For A score (enhancer activity) of a gene, we first identified all putative enhancers of this gene by selecting H3K27ac ChIP-Seq peaks located within 1Mb of the promoter. Then we quantified the calibrated H3K27ac ChIP-Seq signals on these putative enhancers (extended 150bp from MACS2 peaks) as A scores. The A-only quantification of enhancer activity for this gene will be the sum of the A scores for all putative enhancers. For C score (enhancer-promoter contact) between a gene and putative enhancers, we quantified the normalized Hi-C contacts formed in between the 5kb bins harboring the gene promoter and the putative enhancer. For the ABC score, we multiplied the A score of each enhancer by the C score, and generated the summation of these if multiple putative enhancers exist for a gene. P score of any gene was calculated as the calibrated H3K4me3 ChIP-Seq signal at its promoter region (+/− 2.5kb from TSS) of a gene. For ABC-P or ABC-P2 scores, we multiplied the summed ABC score of a gene by its P score (promoter H3K4me3 signal) or by the square of its P score. The transcriptional changes of any gene were calculated based on the log2 fold change of RPB1 ChIP-Seq reads over the whole gene body (average of three ChIP-Seq replicates). Pearson correlation coefficient was used to measure the correlation between ABC score change and transcriptional change. The list of interferon response (IFN) genes were obtained from GSEA molecular signature databases (Interfero_Alpha_Response), and the list of pro-inflammatory (PIF) genes was manually curated based on recent literature40 studying immuno-pathology of SARS-CoV-2 infection (see Supplementary Table 3).
Statistics
For all boxplots, the centre lines represent medians; box limits indicate the 25th and 75th percentiles; and whiskers extend 1.5 times the interquartile range (IQR) from the 25th and 75th percentiles. For qPCR data, we analyzed them by Prism (v7.00) and presented them as mean±SD, which are indicated in figure legends. At least two biological replicates were conducted for RNA-Seq, ChIP-Seq or Hi-C sequencing. Student’s t-test (two-tailed) was commonly used to compare means between two qPCR groups; p < 0.05 was considered significant, and we labeled the p values with asterisks in each figure panel (*, p < 0.05; **, p < 0.01; ***: p < 0.001). Statistical analyses for sequencing data were performed with Python (v3.6, pandas v1.1.5, numpy v1.17.3, matplotlib v3.1.2, scipy v1.5.4 ) or R (v3.6.0) scripts.
Extended Data
Extended Data Figure 1. Overview of 3D chromatin architectures and quality control of SARS-CoV-2 infection in our study.
a. A diagram showing the typical contact maps in Hi-C (and Hi-C 3.0 or other modified Hi-C approaches) that define A/B compartments, Topologically Associating Domains (TADs), chromatin loops or intra-TAD interactions (which perhaps include most enhancer-promoter contacts). This is an overall summary of these structures, but the exact definition of some structures may be subject to variable interpretation, and the terminology may not always be used consistently2,3,9,19.
Often, A/B compartmentalization is illustrated by a checkerboard pattern of Hi-C contact matrices over large genomic sizes, indicating preferential interactions between genomic regions belonging to the same type of compartments (A: euchromatin and transcriptionally active; B: heterochromatin, transcriptionally inactive). TADs or chromatin domains are often characterized as a square or triangle-like structure on contact maps, reflecting a higher contact frequency between any regions inside the same TAD than with regions outside of the TAD. Intra-TAD enhancer-promoter contacts are considered to be facilitated by TADs, while TAD boundaries prevent aberrant interaction with regions outside of TADs.
In Hi-C maps, the dot-shaped structures on the tip of domains suggests local enrichment of spatial interaction between a pair of two loci over nearby regions, and is regarded as a chromatin loop in this work. But loops may be subjected to other definitions in other studies. For example, enhancer-promoter contacts often do not appear as dot-shaped structures in Hi-C, but may be defined as loops by other work or other methods. For additional discussion, see2,9.
b. Cartoon diagrams describe A-A and B-B association preference within regions of similar epigenetic features, which compartmentalizes chromosomes into A and B (the left part of the diagram). The diagram in the middle depicts a current model of cohesin loop extrusion inside TADs that generates such structures. The right side shows a zoom-in view of a part of a TAD that harbors enhancer-promoter contact that may play roles in gene transcriptional regulation.
c. A barplot showing the percentage of RNA-Seq reads mapped to SARS-CoV-2 genome in Mock, 6-hr post infection (6 hpi, 0.1 MOI), and 24 hpi (0.1 MOI) conditions (n = 2). Mean and standard deviation were calculated based on two biological replicates of RNA-Seq. This data is consistent with previous observations40. Shorter term infection for 6-hr resulted in ~20% RNA-Seq reads attributed to the virus genome, suggesting insufficient viral infection/replication in cell populations.
d. Confocal images showing immunofluorescence staining of DAPI (DNA, blue) and the Spike protein of SARS-CoV-2 (red) in Mock and 24hpi (0.1 MOI) infected A549-ACE2 cells. Scale bars are shown.
e. Quantification of infection rates in panel d. SARS-CoV-2 Spike protein was used as a marker for SARS-CoV-2 infection. From left to right, n= 3 and 4 quantifications.
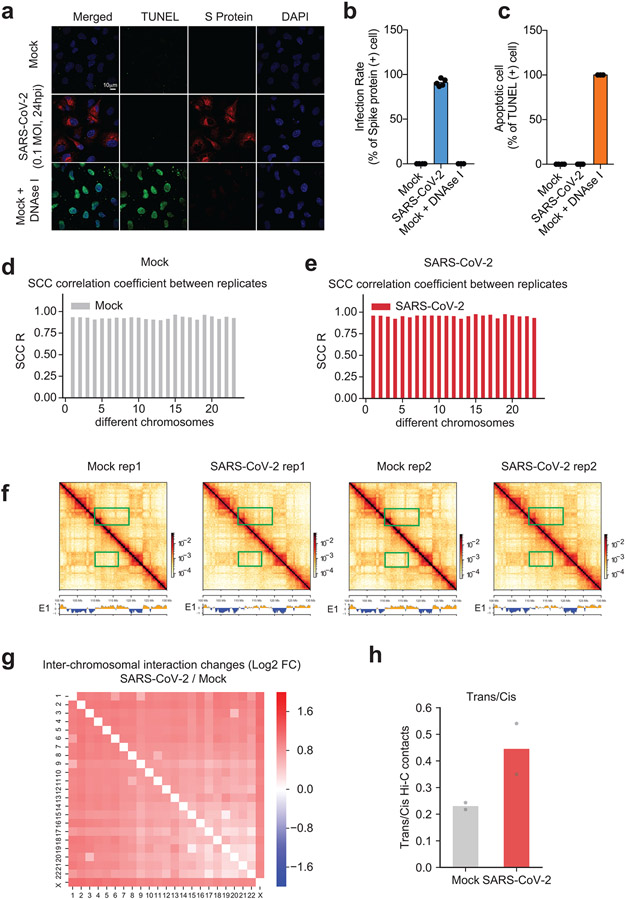
Extended Data Figure 2. Cell quality check after infection, replicate consistency of Hi-C 3.0, and the increase of trans-chromosomal interactions after infection.
a. Representative confocal images of Mock and SARS-CoV-2 infected cells (24hpi, 0.1 MOI) stained with an antibody against the Spike protein of SARS-CoV-2 (S protein, red), and by the TUNEL assay (green).
b,c. Quantification of rates of apoptosis and infection from panel a. From left to right, quantifications were conducted n = 4, 5, 3 (for panel b) and n = 4, 5, 3 (for panel c).
d,e. Barplots showing the SCC correlation coefficients41 between two Hi-C replicates of Mock or SARS-CoV-2 conditions for different chromosomes.
f. Snapshots showing two replicates of Hi-C contact matrices and compartmental score (E1) tracks in the same genomic region shown in Fig.1b. The left two matrices show data of replicate 1 (rep1), and the right two matrices show replicate 2 (rep2). Green boxes show two regions with increased A-B compartmental mixing or weakened A compartment after virus infection. Bin size = 80kb. Color scales indicate Hi-C contact frequencies.
g. A heatmap shows the log2 fold change of inter-chromosomal interactions between any two chromosomes (SARS-CoV-2/Mock).
h. A barplot showing the trans/cis Hi-C contacts ratio in two biological replicates of Hi-C in Mock or SARS-CoV-2 infected cells (n = 2). Trans contacts indicate chromatin interactions formed in between two different chromosomes.
Extended Data Figure 3. Quality check of HCoV-OC43 infection and Poly (I:C) treatment in A549-ACE2 cells.
a. To examine how many cells were infected by HCoV-OC43 at 24hpi, immunostaining using a monoclonal antibody targeting HCoV-OC43 was conducted (MAB9012). Here we show representative confocal images of Mock and HCoV-OC43 infected A549-ACE2 cells stained with MAB9012 (red) and DAPI (blue). Scale bar is shown.
b. Barplots showing quantification of infection rates of HCoV-OC43 at 0.1 and 0.5 MOIs (24hpi). Data shows n = 5 for each quantification.
c. Barplots of DDX58, IFTI1, IFNB1 gene expression (RT-qPCR) showing known antiviral responses in poly(I:C) transfected cells. n = 3, representative of two independent experiments, P = 5.59e-5 (DDX58), 2.31e-4 (IFIT1), 5.94e-5 (IFNB1), calculated with a two-sided independent t-test. Data show Mean +/− SD from replicates (***, p<0.001).
Extended Data Figure 4. Additional data of A/B compartmental changes.
a. A barplot showing the percentage of genomic bins (100kb bin size) that can be categorized into six groups based on their compartmental score changes after SARS-CoV-2 infection (E1 value change > 0.2, see Methods). These six categories are: A to weaker A, A to B, B to stronger B, B to weaker B, B to A, and A to stronger A.
b. Snapshots of inter-chromosomal Hi-C contact matrices between chr17 and chr18 (upper), and intra-chromosomal Hi-C contact matrices within chr18 (lower) in Mock and SARS-CoV-2 infected samples. On the right, differential contacts are shown as log2 fold changes of SARS-CoV-2/Mock. PCA E1 scores were put at sides to show the A or B compartments. Red and black arrowheads respectively point to increased A-B and reduced A-A interactions after SARS-CoV-2 infection. Bin size = 320kb. Color scales indicate Hi-C contact frequencies (left and middle), and log2 fold change of SARS-CoV-2/Mock contact frequencies (right).
c,d,e. Saddle plots from Hi-C 3.0 after treatments by HI-WA1 (c), poly(I:C) (d), and HCoV-OC43 infection (e), respectively, showing negligible impacts on compartmental interactions. HCoV-OC43 and HI-WA1 shared the same Mock sample. In each figure, color scales indicate observed/expected (O/E) Hi-C contact frequencies (left and middle), and log2 fold change of O/E contact frequencies (right).
Extended Data Figure 5. Limited 3D genome changes in cells infected by HCoV-OC43, treated by HI-WA1 or poly(I:C).
a,b,c. Barplots showing the percentage of genomic bins (100kb bin size) that can be categorized into six groups based on their compartmental score changes (E1 values). These six categories are: A to weaker A, A to B, B to stronger B, B to weaker B, B to A, and A to stronger A. The a,b,c panels show the changes after treatments by poly (I:C), HI-WA1 and HCoV-OC43, respectively.
d-j. Venn diagrams showing the overlaps of genomic bins displaying E1 score alteration (i.e., compartmental changes) incurred by SARS-CoV-2 (pink) versus those by HCoV-OC43 (Cyan). Numbers indicate bins with changes belong to different categories: total changed bins (d), A to weaker A(e), A to B (f), B to stronger B (g), B to weaker B(h), B to A(i), A to stronger A(j).
k-l. Aggregated domain analyses (ADA) in cells treated by HI-WA1, poly(I:C) and HCoV-OC43, in comparison to their corresponding mock samples. HI-WA1 and HCoV-OC43 samples share the same Mock. In each figure, color scales indicate aggregated Hi-C contact frequencies (left and middle), and log2 fold change of SARS-CoV-2/Mock aggregated contact frequencies (right).
Extended Data Figure 6. Calibrated ChIP-Seqs demonstrate epigenome reprogramming during SARS-CoV-2 infection.
a. A diagram illustrating the design of calibrated ChIP-Seq using mouse ESCs as spike-in for human A549 cells (with or without infection). Created with BioRender.com
b,c,d,e. Barplots showing the scale factors calculated based on human/mouse reads in both Mock and SARS-CoV-2 conditions, which permit calibrated ChIP-Seq analyses of H3K27ac, H3K4me3, H3K27me3, and H3K9me3. R1 and R2 indicate biological replicates 1 and 2.
f,g,h,i. Scatter plots show virus-caused genome-wide changes of histone mark ChIP-Seq signals at 100kb bins for H3K27ac, H3K4me3, H3K9me3 and H3K27me3. The x,y-axis are natural logarithmically scaled reads densities from calibrated ChIP-Seq data. Dotted lines denote changes by two folds.
j. Snapshots of Pearson’s correlation matrices, E1 compartmental scores, as well as ChIP-Seq tracks of H3K27ac and H3K9me3 in Mock or SARS-CoV-2 infected cells. The right side shows the difference of Pearson’s correlation matrices between SARS-CoV-2 and Mock (pink color shows decrease; green shows increase). At the bottom, red arrowheads on top of the H3K27ac peaks show strong reduction after infection, which was accompanied by quantitative increase of H3K9me3 signals in the same region (black arrowheads). Accordingly, this entire A compartment domain displays reduced compartmental strength (i.e., lower PCA E1 scores, see yellow signals in the E1 track), showing less A-A compartmental interactions within the same A compartment (black box in the rightmost panel) and increased A-B mixing with the nearby B compartment (red box in the rightmost panel). Bin size = 80kb. Color scales indicate Pearson correlation coefficient (left and middle), and the differences of Pearson correlation coefficient (right).
Extended Data Figure 7. Cohesin was specifically depleted from intra-TAD regions and epigenetic features of different groups of TADs.
a. Western blots showing the protein abundances of cohesin (RAD21, SMC3) and CTCF in Mock and SARS-CoV-2 infected cells. GAPDH was used as a loading control. Relative expression of each protein to the Mock control group was labeled under each image.
b. Barplots showing the scale factors calculated based on human/mouse reads ratio in both Mock and SARS-CoV-2 conditions that permit calibrated ChIP-Seqs of CTCF, RAD21 and SMC3. These factors were not globally affected by virus infection (the ratios are comparable in mock and infected conditions).
c. Left barplot shows the numbers of total, the gained or reduced RAD21 ChIP-Seq peaks after SARS-CoV-2 infection at 24hpi; the profile plots in the middle and right show the signals of RAD21 ChIP-Seq at the gained or reduced ChIP-Seq peaks in Mock and SARS-CoV-2 conditions.
d,e,f. Similar to panel c, these plots show analysis of ChIP-seqs of SMC3, CTCF and NIPBL in Mock and SARS-CoV-2 conditions.
g. A boxplot showing the log2 fold changes (FC) of intra-TAD Hi-C contacts for six categories of TADs. All TADs are ranked based on the quantitative changes of intra-TAD interactions and the six categories include the Top 5%, top 5~10%, 10~50%, 50~90%, bottom 5~10%, and the bottom 5%, respectively. “Top 5%” represents TADs with the most severely-weakened intra-TAD interactions after SARS-CoV-2 infection. From left to right, n = 205, 205, 1637, 1637, 205, 205 TADs. P = 5.61e-69, calculated with a two-sided Mann-Whitney U test.
h,i,j. Boxplots showing the log2 fold changes (FC) of ChIP-Seq signals of H3K27ac (from left to right, n = 1808, 1841, 18640, 22245, 3074, 3205 peaks), H3K9me3 (from left to right, n = 205, 205, 1637, 1637, 205, 205 regions), and H3K27me3 (from left to right, n = 205, 205, 1637, 1637, 205, 205 regions) in the six categories of TADs as shown in panel g. P = 6.86e-42 in H3K9me3 panel, calculated with a two-sided Mann-Whitney U test.
For all boxplots, the centre lines represent medians; box limits indicate the 25th and 75th percentiles; and whiskers extend 1.5 times the interquartile range (IQR) from the 25th and 75th percentiles.
Extended Data Figure 8. Dot-shaped chromatin loops are largely unaltered by SARS-CoV-2 infection, but a subset of them is changed.
a. Aggregated peak analysis (APA) shows the strength of all chromatin loops (observed/expected) in Mock (left) and SARS-CoV-2 (right) infected cells for 11,926 dot-shaped loops called by call-dots algorithm. Color scales indicate aggregated Hi-C observed / expected (O/E) contacts.
b. Volcano plot generated by DEseq2 using the two replicates of Hi-C that defines quantitatively changed loops after SARS-CoV-2 infection (FDR < 0.1, see Methods42).
c. APA plots for the subsets of virus-weakened and strengthened loops. The numbers of such loops are shown. For APA plots in panels a and c, the bin size for plotting the heatmap is 5kb, and the heatmaps show genomic regions +/− 100kb surrounding the loop anchors. The numbers on the heatmaps indicate the central pixel values. Color scales indicate aggregated Hi-C O/E contacts.
d. A boxplot showing the sizes of loops belonging to strengthened and weakened groups. From left to right, n = 560 and 353. P = 1.33e-66, calculated with a two-sided Mann-Whitney U test.
e. Boxplots showing the virus-induced fold changes of cohesin binding (measured by calibrated cohesin ChIP-Seq) on the loop anchors of the two groups of loops: those quantitatively strengthened or weakened. From left to right, n = 983 and 535 (RAD21); from left to right, n = 1023 and 543 (SMC3). P = 3.17e-66 (RAD21). P = 1.12e-74 (SMC3). P-values are calculated with a two-sided Mann-Whitney U test.
f. Boxplots showing the distances of each loop anchor to its closest TAD boundary for two groups of virus-affected loops: those quantitatively strengthened or weakened. From left to right, n = 1038 and 687. P = 2.35e-21, calculated with a two-sided Mann-Whitney U test.
For all boxplots, the centre lines represent medians; box limits indicate the 25th and 75th percentiles; and whiskers extend 1.5 times the interquartile range (IQR) from the 25th and 75th percentiles.
Extended Data Figure 9. SARS-CoV-2 disruption of chromatin architecture correlates with the transcriptional inhibition of interferon response genes.
a. Boxplots showing the expression deregulation of key interferon response (IFN) (n = 40) and pro-inflammatory (PIF) genes (n = 12) after SARS-CoV-2 infection or IFN-beta treatment (1000u/ml, 6-hr), as shown by RNA-Seq changes. P = 1.51e-4 and 3.05e-3, respectively, calculated with a two-sided Mann-Whitney U test. For boxplots, the centre lines represent medians; box limits indicate the 25th and 75th percentiles; and whiskers extend 1.5 times the interquartile range (IQR) from the 25th and 75th percentiles.
b,c. Heatmaps of select IFN or PIF genes showing their fold changes in Pol2 ChIP-Seq or RNA-seq after SARS-CoV-2 infection or IFN-beta treatment (1000u/ml, 6-hr).
d,e. Hi-C matrices (bin size: 5kb) and calibrated ChIP-Seq tracks for indicated factors at two key loci encoding viral RNA sensors: DDX58 (encoding RIG-I) and IFIH1 (encoding for MDA5). Left: Mock; right: SARS-CoV-2. Blue arrows point to reduced dot-shaped loops. The intra-TAD interactions were weakened throughout these two TADs. Red asterisks show reduced H3K27ac peaks by virus infection. Green arrows show H3K4me3 peaks at promoters that are not changed by SARS-CoV-2. A pink arrow in panel d points to the enhancer targeted by CRISPRi in panel h (see below). Color scales indicate Hi-C contact frequencies.
f. A scatter plot showing a poor correlation between the C score only (from Hi-C contact, x-axis) and the true transcriptional changes of IFN genes by SARS-CoV-2 (y-axis, RPB1 ChIP-Seq). Error bands indicate the 95% confidence interval of the regression estimate. Pearson’s correlation coefficient for contact only: R=0.13. X and y-axis fold changes denote SARS-CoV-2/Mock.
g. Similar to panel f, this is a scatter plot showing a poor correlation between the A score only (from enhancer H3K27ac activity, x-axis) and the true transcriptional changes of IFN genes by SARS-CoV-2 (y-axis, RPB1 ChIP-Seq). Error bands indicate the 95% confidence interval of the regression estimate. Pearson’s correlation coefficient for enhancer activity only: 0.33.
h. RT-qPCR results showing that CRISPRi inhibition of the enhancer in DDX58 locus reduced its response to poly (I:C). The DDX58 enhancer for CRISPRi is shown in panel d. Data was normalized to the -poly(I:C) in control group, and denote mean +/− SD (n=3, representative of two independent experiments); P values were calculated with a two-sided independent T-test.
Extended Data Figure 10. Pro-inflammatory genes are induced by SARS-CoV-2 infection concurrent with increased promoter activity and H3K4me3 levels.
a. A scatter plot showing good correlation between the ABC score (x-axis) and the true transcriptional changes of pro-inflammatory (PIF) genes by SARS-CoV-2 (y-axis, RPB1 ChIP-Seq). But the quantity of true changes (y axis) is much higher than the modeled changes based on ABC scores (x-axis), as all the data points are above the diagonal (also see Fig.6c for revised ABC-P2 scores). Error bands indicate the 95% confidence interval of the regression estimate.
b. Bar graphs showing the fold changes of two key PIF genes, IL6 and CXCL8, in several conditions: true RPB1 ChIP-Seq fold changes after SARS-CoV-2 infection (TXN); fold changes modeled by ABC score; modeled by ABC algorithm with inclusion of promoter H3K4me3 strength (ABC-P); modeled by ABC algorithm with inclusion of a square of promoter H3K4me3 strength (ABC-P2). Promoter strength is required to revise the ABC algorithm to better model transcriptional increases of PIF genes caused by SARS-CoV-2 infection.
c. Snapshots of Hi-C matrices (bin size: 5kb) and calibrated ChIP-Seq tracks for indicated factors at another key gene loci coding for pathologically critical PIF cytokines in COVID-19 patients: CXCL8 (encoding cytokine IL-8). Red asterisks show reduced H3K27ac peaks. Black arrows show increased H3K4me3 peaks at its promoter by SARS-CoV-2 infection. The intra-TAD interactions were weakened throughout the TAD. Color scales indicate Hi-C contact frequencies.
d. Barplot showing the numbers of total, gained or reduced H3K4me3 ChIP-Seq peaks after SARS-CoV-2 infection for 24hpi (0.1 MOI).
e. Hallmark signature analysis of genes close to H3K4me3 peaks gained in virus-infected condition show signatures associated with TNF-alpha, TGF-beta signaling or inflammatory responses, which are associated with pathological symptoms in COVID-19 patients.
f. Motif analysis of H3K4me3 peaks increased in SARS-CoV-2 infected cells shows that the top ranked motifs are those of IRF1/2 and Jun/AP1, suggesting the potential roles of these transcription factors in transcriptional activation of inflammation genes. Motif analysis was done by HOMER, and the P values and percentages of sites with motifs are shown.
g. Snapshots of Mock and HCoV-OC43 Hi-C contact matrices (bin size: 5kb) and ChIP-Seq tracks for indicated factors at the CXCL8 (encoding IL-8) loci. This is from the same region as in panel c. In comparison to SARS-CoV-2 effect in panel c, HCoV-OC43 caused no reduction of H3K27ac peaks (Red asterisks), and no obvious H3K4me3 increase at its promoter (black arrows). These asterisks and arrows point to the same peaks as those in panel c. Color scales indicate Hi-C contact frequencies.
Supplementary Material
Supplementary Table 1. A summary of the epigenomic and transcriptomic datasets generated in this study.
Supplementary Table 3. Gene lists of IFN (interferon response genes) and PIF (pro-inflammatory) genes used in this study.
Supplementary Table 2. Oligonucleotides used in this study.
Acknowledgements:
W.L. is a Cancer Prevention and Research Institute of Texas (CPRIT) Scholar. This work is supported by funding from the University of Texas McGovern Medical School, NIH “4D Nucleome” program (HL156059), NIGMS (GM132778, GM136922), CPRIT (RR160083, RP180734), Welch foundation (AU-2000-20220331) and the John S. Dunn foundation to W.L, by an NIH/NHLBI HL155950 to X.Y., by grants from NIH (HL154720, DK122796, DK109574, HL133900) and Department of Defense (W81XWH2110032) to H.K.E., and by GM083337 to D.M.G. J.-H.L. and R.W. receive support from UTHealth Innovation for Cancer Prevention Research Training Program Post- and Pre-doctoral Fellowships (RP210042). R.W. is a recipient of the Dr. John J. Kopchick Fellowship, and the John and Rebekah Harper Fellowship in Biomedical Sciences. Most of our next generation sequencing work was conducted with UTHealth Cancer Genomics Core, which received funding from CPRIT (RP180734). We thank Dr. Masato Kanemaki for generating and kindly sharing the RAD21-mAID2-mClover HCT116 cell line.
Footnotes
Disclosure
The content is solely the responsibility of the authors and does not necessarily represent the official views of the Cancer Prevention and Research Institute of Texas.
Conflict of Interests: The authors declare no competing interests.
Data availability
Data generated in this study have been deposited to NCBI GEO (GSE179184, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE179184). Raw data from fluorescence microscopy imaging are available at Mendeley (http://dx.doi.org/10.17632/czzb4jb8ss.1). Gene sets are available from GSEA molecular signature database (https://www.gsea-msigdb.org/gsea/msigdb). Source data are provided with this paper.
Code availability
Key software or algorithms used in our analysis of sequencing data are listed in methods, Hi-C data analyses tools can be found at https://github.com/open2c. This work did not report new software or mathematical algorithms.
References
- 1.Dekker J, Marti-Renom MA & Mirny LA Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet 14, 390–403 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kempfer R & Pombo A Methods for mapping 3D chromosome architecture. Nat Rev Genet 21, 207–226 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Davidson IF & Peters J-M Genome folding through loop extrusion by SMC complexes. Nat Rev Mol Cell Bio 1–20 (2021) doi: 10.1038/s41580-021-00349-7. [DOI] [PubMed] [Google Scholar]
- 4.Liu X et al. Human Virus Transcriptional Regulators. Cell 182, 24–37 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heinz S et al. Transcription Elongation Can Affect Genome 3D Structure. Cell 174, 1522–1536.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai K & Cullen BR Epigenetic and epitranscriptomic regulation of viral replication. Nat Rev Microbiol 18, 559–570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kee J et al. SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry. Nature 610, 381–388 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon DE et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mirny LA, Imakaev M & Abdennur N Two major mechanisms of chromosome organization. Curr Opin Cell Biol 58, 142–152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belaghzal H et al. Liquid chromatin Hi-C characterizes compartment-dependent chromatin interaction dynamics. Nat Genet 1–12 (2021) doi: 10.1038/s41588-021-00784-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hildebrand EM & Dekker J Mechanisms and Functions of Chromosome Compartmentalization. Trends Biochem Sci (2020) doi: 10.1016/j.tibs.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao SSP et al. Cohesin Loss Eliminates All Loop Domains. Cell 171, 305–320.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarzer W et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oksuz BA et al. Systematic evaluation of chromosome conformation capture assays. Nat Methods 18, 1046–1055 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su J-H, Zheng P, Kinrot SS, Bintu B & Zhuang X Genome-Scale Imaging of the 3D Organization and Transcriptional Activity of Chromatin. Cell (2020) doi: 10.1016/j.cell.2020.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorbalenya AE et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5, 536–544 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X et al. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc National Acad Sci 111, 6756–6761 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphries F et al. A diamidobenzimidazole STING agonist protects against SARS-CoV-2 infection. Sci Immunol 6, eabi9002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao SSP et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yusufova N et al. Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature 1–7 (2020) doi: 10.1038/s41586-020-3017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu NQ et al. WAPL maintains a cohesin loading cycle to preserve cell-type-specific distal gene regulation. Nat Genet 1–10 (2020) doi: 10.1038/s41588-020-00744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crane E et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H et al. Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature 576, 158–162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krietenstein N et al. Ultrastructural Details of Mammalian Chromosome Architecture. Mol Cell (2020) doi: 10.1016/j.molcel.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvalho T, Krammer F & Iwasaki A The first 12 months of COVID-19: a timeline of immunological insights. Nat Rev Immunol 21, 245–256 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fulco CP et al. Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat Genet 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yesbolatova A et al. The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nat Commun 11, 5701 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuartero S et al. Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat Immunol 19, 932–941 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calderon L et al. Cohesin-dependence of neuronal gene expression relates to chromatin loop length. Elife 11, e76539 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rinaldi L et al. The glucocorticoid receptor associates with the cohesin loader NIPBL to promote long-range gene regulation. Sci Adv 8, eabj8360 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cano-Rodriguez D et al. Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nat Commun 7, 12284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Policarpi C, Munafo M, Tsagkris S, Carlini V & Hackett JA Systematic Epigenome Editing Captures the Context-dependent Instructive Function of Chromatin Modifications. (2022) doi: 10.1101/2022.09.04.506519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferreira-Gomes M et al. SARS-CoV-2 in severe COVID-19 induces a TGF-β-dominated chronic immune response that does not target itself. Nat Commun 12, 1961 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho JSY et al. TOP1 inhibition therapy protects against SARS-CoV-2-induced lethal inflammation. Cell 184, 2618–2632.e17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zazhytska M et al. Non-cell-autonomous disruption of nuclear architecture as a potential cause of COVID-19-induced anosmia. Cell (2022) doi: 10.1016/j.cell.2022.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamada T et al. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat Immunol 1–9 (2021) doi: 10.1038/s41590-021-00942-0. [DOI] [PubMed] [Google Scholar]
- 37.Cavalli G & Heard E Advances in epigenetics link genetics to the environment and disease. Nature 571, 489 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Mantovani A & Netea MG Trained Innate Immunity, Epigenetics, and Covid-19. New Engl J Med 383, 1078–1080 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Nalbandian A et al. Post-acute COVID-19 syndrome. Nat Med 27, 601–615 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blanco-Melo D et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 181, 1036–1045.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang T et al. HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res 27, 1939–1949 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn JH et al. Phase separation drives aberrant chromatin looping and cancer development. Nature 1–5 (2021) doi: 10.1038/s41586-021-03662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kärber G Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn-schmiedebergs Archiv Für Exp Pathologie Und Pharmakologie 162, 480–483 (1931). [Google Scholar]
- 44.Lee J-H et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol Cell 81, 3368–3385.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramírez F, Dündar F, Diehl S, Grüning BA & Manke T deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Servant N et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durand NC et al. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst 3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abdennur N & Mirny LA Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics (2019) doi: 10.1093/bioinformatics/btz540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flyamer IM, Illingworth RS & Bickmore WA Coolpup.py: versatile pile-up analysis of Hi-C data. Bioinformatics 36, 2980–2985 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. A summary of the epigenomic and transcriptomic datasets generated in this study.
Supplementary Table 3. Gene lists of IFN (interferon response genes) and PIF (pro-inflammatory) genes used in this study.
Supplementary Table 2. Oligonucleotides used in this study.
Data Availability Statement
Data generated in this study have been deposited to NCBI GEO (GSE179184, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE179184). Raw data from fluorescence microscopy imaging are available at Mendeley (http://dx.doi.org/10.17632/czzb4jb8ss.1). Gene sets are available from GSEA molecular signature database (https://www.gsea-msigdb.org/gsea/msigdb). Source data are provided with this paper.
Key software or algorithms used in our analysis of sequencing data are listed in methods, Hi-C data analyses tools can be found at https://github.com/open2c. This work did not report new software or mathematical algorithms.