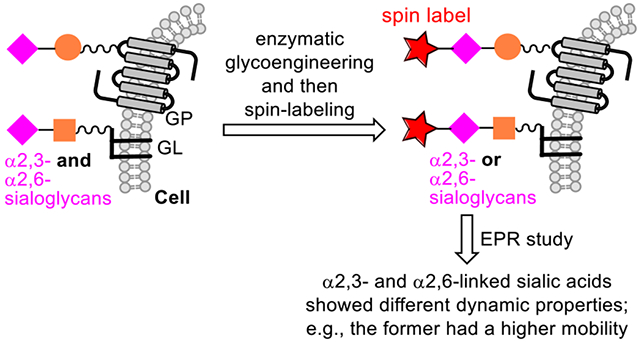
Abstract
Sialoglycans on HeLa cells were labeled with a nitroxide spin radical through enzymatic glycoengineering (EGE)-mediated installation of an azide-modified sialic acid (Neu5Ac9N3) and then click reaction-based attachment of a nitroxide spin radical. α2,6-Sialyltransferase (ST) Pd2,6ST and an α2,3-ST CSTII were used for EGE to install α2,6- and α2,3-linked Neu5Ac9N3, respectively. The spin-labeled cells were analyzed by X-band continuous wave (CW) electron paramagnetic resonance (EPR) spectroscopy to gain insights into the dynamics and organizations of cell surface α2,6- and α2,3-sialoglycans. Simulations of the EPR spectra revealed average fast- and intermediate-motion components for the spin radicals in both sialoglycans. However, α2,6- and α2,3-sialoglycans in HeLa cells possess different distributions of the two components, e.g., a higher average population of the intermediate-motion component for α2,6-sialoglycans (78%) than that for α2,3-sialoglycans (53%). Thus, the average mobility of spin radicals in α2,3-sialoglycans was higher than that in α2,6-sialoglycans. Given the fact that a spin-labeled sialic acid residue attached to the 6-O-position of galactose/N-acetyl-galactosamine would experience less steric hindrance and show more flexibility than that attached to the 3-O-position, these results may reflect the differences in local crowding/packing that restrict spin-label and sialic acid motion for the α2,6-linked sialoglycans. The studies further suggest that Pd2,6ST and CSTII may have different preferences for glycan substrates in the complex environment of extracellular matrix. The discoveries of this work are biologically important as they are useful for interpreting the different functions of α2,6- and α2,3-sialoglycans and indicate the possibility of using Pd2,6ST and CSTII to target different glycoconjugates on cells.
Keywords: Sialic acid; α2,3-linkage; α2,6-linkage; glycan; organization; electron paramagnetic resonance spectroscopy
Graphical Abstract
Introduction
Sialic acids are a group of nine-carbon acidic amino sugars derived from neuraminic acid. N-Acetylneuraminic acid (Neu5Ac) is its most common member, thus Neu5Ac is often referred to as the “sialic acid”. Neu5Ac is ubiquitous in eukaryotes and typically at the non-reducing end of glycans as the terminal sugar units. Consequently, Neu5Ac is usually exposed on the cell surface for being directly involved in cell recognition, adhesion, signaling, and other biological events.1–4 For example, the interactions of sialic acid-binding immunoglobulin-type lectins (Siglecs) with their sialylated ligands on human immune cells help the immune system distinguish self from non-self,5–6 and Siglecs are also important immune checkpoints.7
Neu5Ac possesses only three forms of linkages in humans. One has Neu5Ac C-2 α-linked to the 3-O-position of a galactose (Gal) or an >N-acetyl-galactosamine (GalNAc) residue, and the other has Neu5Ac C-2 α-linked to the Gal or GalNAc 6-O-position. The third form has Neu5Ac α-linked to the 8-O-position of another Neu5Ac. These sialyl linkages are generated by α2,3-, α2,6-, and α2,8-sialyltransferases (STs), respectively, with cytidine-5′-monophospho-N-acetylneuraminic acid (CMP-Neu5Ac) as the sialyl donor (Scheme 1). In human adults, sialyl α2,3- and α2,6-linkages are abundant whereas the α2,8-linkage is downregulated during the process of developments. However, α2,8-linkage reappears as an important molecular signature during some oncodevelopments.8–10
Scheme 1.
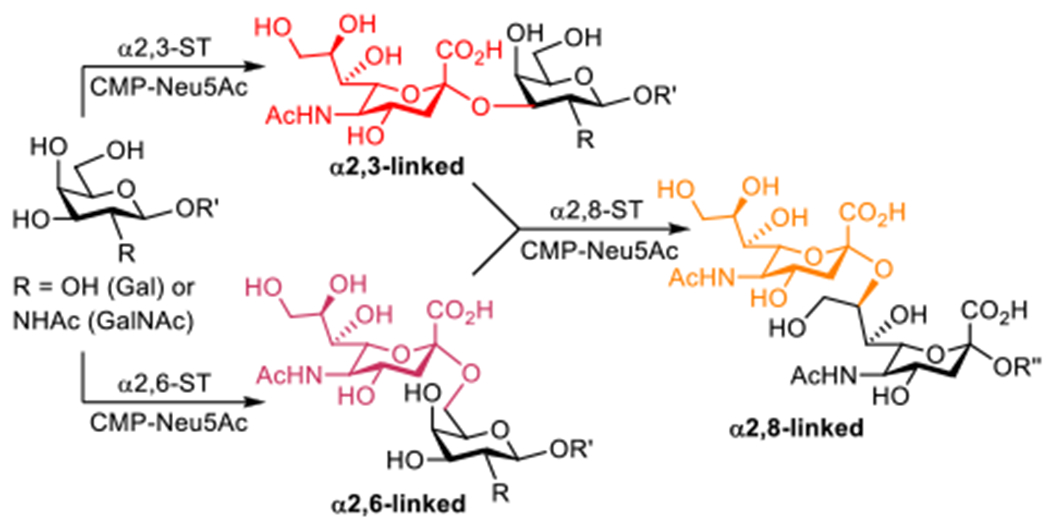
Biosynthesis of α2,3-, α2,6-, and α2,8-linked sialoglycans
Different linkage forms and expression levels of Neu5Ac in various tissues and species are closely related to biological functions.1, 11 For instance, influenza virus infection starts from the binding of the viral hemagglutinin (HA) to Neu5Ac on the hosts’ epithelial cell surface.12 Avian influenza virus binds preferentially with Neu5Acα2,3Gal, which is the principal epitope on avian epithelial cells. In contrast, human influenza virus prefers Neu5Acα2,6Gal—the main epitope on the epithelial cells in human respiratory tract.13–14 This has pathological implications. For example, for avian influenza virus to effectively infect and transmit among humans, its HA has to be mutated to recognize and bind α2,6-linked Neu5Ac.15
The presentational or organizational patterns of glycans on cells can also have a great impact on their bioactivity.16–17 For example, most tumor-associated carbohydrate antigens (TACAs) are just regular glycans overexpressed by tumors but are still closely related to cancer development and progression18–20 and useful biomarkers for cancer diagnosis and therapy.21 It is reported that although TACAs are also expressed by normal cells, the human immune system can distinguish these glycans on normal and cancer cells and selectively respond to cancer and develop TACA-specific antibodies.22 This is attributed to the different densities of these glycans on cancer and normal cells, which may affect their structure, organization, and accessibility.22,23 Hypersialylation is a hallmark for most cancers, and many TACAs are sialoglycans.19 Clearly, knowing the organization and dynamics of sialoglycans on the cell surface is important.
It is currently feasible to study the glycan structures and compositions of cell glycocalyx by a combination of advanced analytical techniques, such as nuclear magnetic resonance spectroscopy, mass spectrometry and fluorescence microscopy, with modern tools in carbohydrate chemistry and glycobiology,24–25 such as cell glycoengineering.26–27 However, deciphering the functional roles of glycan organization, including the impact of environments on their mobility, is difficult because such studies are hindered by the high diversity, complexity, and flexibility of glycans on cells. To address the issue, we developed new methods to label cell surface glycans with a nitroxide spin to enable electron paramagnetic resonance (EPR) spectroscopy studies.28–29 It is well-established that line shape analyses and simulations of continuous wave (CW)-EPR spectra of radical spins can provide metrics useful for quantifying the mobility of spin labels (SLs)30–37 and uncover the spatial alignments, flexibility, and dynamics of the labelled molecule and its adjacent environment.
Previously, we demonstrated the possibility to incorporate nitroxide SLs into cell surface glycans through metabolic glycan engineering (MGE) and then bio-orthogonal click reaction to install SLs onto the engineered glycans on various cells (Figure 1A).28 More recently, we extended this strategy to cell surface sialoglycan labeling with nitroxide radicals by enzymatic glycan engineering (EGE) and then a click reaction (Figure 1B).29 EGE of the cell glycocalyx to incorporate an azido-modified Neu5Ac into glycans was achieved with an α2,3-sialyltransferase (ST) and an azido-modified sialyl donor. Reported here, we applied the latter approach to the spin-labelling of cell surface α-2,3- or α-2,6-sialoglycans specifically and subjected the labeled cells to CW-EPR spectroscopy to study the mobility and local environments of these isomeric glycans. This work can help advance our understanding of Neu5Ac-related glycobiology, as well as the rational design of spin-probes for EPR studies of glycan mobility and packing.
Figure 1:
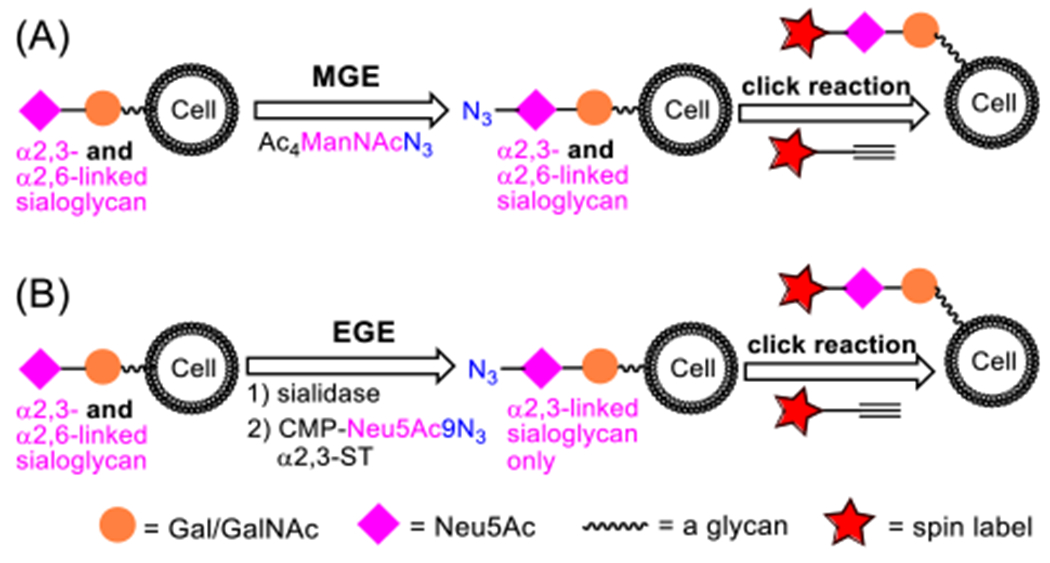
Spin labelling of cell surface sialoglycans via (A) MGE- or (B) EGE-mediated incorporation of an azido-Neu5Ac residue and then a click reaction to install the SL.
Experimental Section
Materials.
HeLa cell line was purchased from American Type Culture Collection (ATCC), USA. Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), penicillin-streptomycin (10000 U/mL), phosphate buffered saline (PBS), and other reagents were purchased from Thermo Fisher Scientific, USA. CMP-Neu5Ac9Az and DBCO-SL were synthesized according to reported methods,28, 38 and their NMR and MS data matched with that in the literature. Pd2,6ST39 and CSTII40 enzymes were prepared according to reported methods using the pET22b vector and genes custom-synthesized by GeneArt® from Thermo Fisher Scientific, USA.
EGE-based spin-labelling of cells:
HeLa cells were cultured in DMEM medium containing 10% FBS and 100 U mL−1 Penicillin–Streptomycin until 80~90% confluency. The cells were treated with trypsin and harvested as pellets via centrifugation at 600 g for 7 min. The cell pellets were washed twice with PBS buffer (pH = 7.4) containing 2% BSA and the cell number was counted by aliquoting a small amount (20 uL) of the cell suspension, treating it with Trypan blue, and then counting on a Hemocytometer. Five million cells were suspended in DMEM without FBS (50 μL), which was followed by the addition of sialidase A (100 U) in 10x Glycobuffer 1® buffer (NEB, 20 μL) per manufacturer’s instructions and then DMEM to reach 200 μL. The resulting mixture was incubated at 37 °C for 1 h with continuous shaking at 90 rpm. The enzymatic reaction was quenched with chilled DMEM, and the cells were collected through centrifugation, washed as described, and resuspended in DMEM. In the meantime, Neu5Ac9Az-CMP (0.1 mmol) in 20 mM Tris-HCl buffer containing 20 mM MgCl2 (50 μL, pH = 8) was mixed with Pd2,6ST (1 U) or CSTII (150 μg), which was adjusted with Tris-HCl buffer to 250 μL. For the controls, only Tris-HCl buffer (no ST) was added. This solution was briefly vortexed before being transferred to the cell suspension and then addition of DMEM to reach a final volume of 300 μL. The mixture was incubated at 37 °C for 30 min with continuous shaking at 90 rpm. The cells were collected and washed three times with PBS buffer containing 2% BSA (pH = 7.4). The cells were fixed using 4% (w/v) of paraformaldehyde in PBS buffer at rt for 10 min and washed with PBS and centrifuged to obtain the cell pellet. The washed cells were resuspended in PBS buffer (500 μL) containing 50 μM DBCO-SL and incubated at rt in the dark for 1 h. Finally, the cells were washed with PBS buffer containing 2% BSA three times, pelleted, and subjected to EPR analysis within 1-2 hrs after spin-labelling. The experiment was repeated 3 times under each condition.
CW-EPR analysis of spin-labelled cells.
Cell pellets containing 1 x 106 cells were resuspended in PBS buffer (40 μL) containing 0.8% agarose. The cell suspension was immediately loaded in a 50 μL-microcapillary quartz pipette tube, and the sample was allowed to solidify before CW-EPR data collection. X-Band (9.5 GHz) CW-EPR absorption spectra were collected at 30°C by a protocol reported in the literature,28 on a Magnettech MiniScope MS-5000 benchtop spectrometer equipped with a dielectric resonator. The spectra were reported as an average of 16 scans with 120 mT sweep width, 0.2 mT modulation amplitude, 100 kHz modulation frequency, and 1 mW incident microwave power. All EPR spectra were area normalized to the cell number and baseline-corrected and were processed using the LabVIEW software provided by C. Altenbach and W. Hubbell. (https://sites.google.com/site/altenbach/labview-programs).
Line shape analysis and simulation of EPR spectra.
EPR spectra were simulated using the chili and esfit functions of EasySpin. The A- and g-tensors were previously determined: gxx = 2.0070, gyy = 2.0062, gzz = 2.0033, Axx = 6.7 G, Ayy = 6.7 G, and Azz = 35 G.28 The other parameters used in EPR line shape simulations are linewidth, correlation time of motion (τc), and the ordering potential C20. Each EPR spectrum was subjected to 1-, 2-, and 3-component simulations.
Simulation of DBCO-SL mobility.
The DBCO structure was drawn in Avogadro41 and parameterized with CHARMM General Forefield (CGenFF 2.5) analog parameters employing the Paramchem web server, which includes the force fields for click-specific moieties and the nitroxide radical moiety.42 The molecule was solvated with the cTIP3P43 water model in a box with the dimensions of 41.123 Å x 47.657 Å x 40.136 Å. The CUDA accelerated AMBER2044 software implemented in the HiPerGator 3.0 high-performance computing facility at the University of Florida was used to perform the atomistic molecular dynamics simulations. The structure was energy-minimized in two stages. In the first stage, 10,000 steps of the steepest descent algorithm followed by 10,000 steps of conjugate gradient was implemented, with harmonic restraints of 50 kcal/mol on the structure and allowing the water molecules to move freely. In the second stage, all restraints were removed, and the system was subjected to 10,000 steps of the steepest descent algorithm followed by the same number of steps in the conjugate gradient algorithm. All simulations beyond this point were done with a 500 kcal/mol harmonic restraint on the carbon atom of the DBCO to mimic the covalent bond to the sialic acid residue. Particle Mesh Ewald summation was used for long-range electrostatic interactions and the non-bonded interactions cutoff was set at 12 Å. The system was heated to 300 K over 1 ns and then subjected to an equilibration step for 10 ns using an NVT ensemble. The production run was done for 500 ns in the NPT ensemble. The Langevin thermostat with a collision frequency of 1.0 ps−1 and the Berendsen barostat were used to regulate temperature and pressure, respectively. A time step of 2 fs was used along with the SHAKE45 algorithm to constrain bonds involving hydrogen atoms. The trajectory was analyzed using the cpptraj module found in the AMBERTools19 software package.46
Results and Discussion
Incorporation of nitroxide spin labels in cell surface sialoglycans.
EGE was used to selectively modify cell surface glycans with α2,3- or α2,6-linked 9-azido-9-deoxy-Neu5Ac (Neu5Ac9N3) according to the protocol depicted in Figure 2. First, cells are treated with a sialidase to delete cell surface Neu5Ac, thereby exposing Gal and GalNAc to enable more effective sialyation. Next, the treated cells are incubated with an azide-modified sialyl donor, such as CMP-Neu5Ac9N3, in the presence of either α2,3-ST or α2,6-ST to enzymatically attach Neu5Ac9N3 to the Gal/GalNAc 3-O- or 6-O-position, respectively. Subsequently, the cells were fixed and SLs are introduced to the azido-sialoglycans on the cell surface via strain-promoted alkyne-azide cycloaddition (SPAAC) using DBCO-modified nitroxide (DBCO-SL) developed for our earlier MGE and EGE studies. Finally, the spin-labelled cells are subjected to EPR studies.
Figure 2:
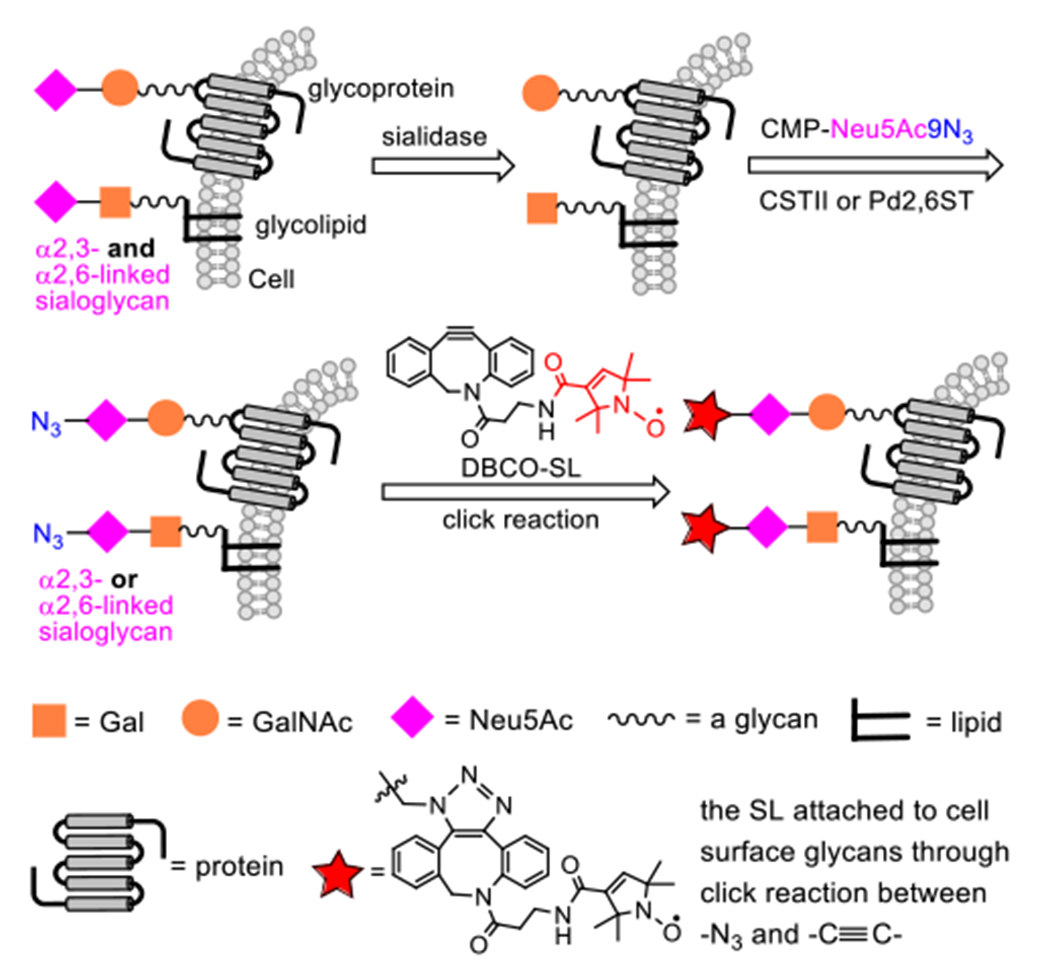
The protocol for regiospecific spin labelling of sialoglycans on cells via EGE and a click reaction. Cells were treated with a sialidase to delete both α2,3- and α2,6-linked Neu5Ac residues in glycans on the cell surface. Next, Neu5Ac9N3 was attached to the 3-O- or 6-O-position of Gal/GalNAc residues on the cell surface under the catalysis of α2,3-ST (CSTII) or α2,6-ST (Pd2,6ST), respectively. Finally, SLs were attached to the azide-marked sialoglycans via SPAAC.
Specifically, the HeLa cell line was chosen as the cell model, and Pd2,6ST and CSTII—two bacterial STs—were employed for EGE and subsequent spin labelling of cell surface α2,6- and α2,3-sialoglycans, respectively. Pd2,6ST is an α2,6-ST from Photobacterium damselae;39 CSTII is an α2,3-ST derived from Campylobacter jejuni.40, 47 Both STs are promiscuous and accept C9-modified CMP-Neu5Ac, such as CMP-Neu5Ac9N3, as sialyl donors48–50 to allow for EGE.29, 51–52 HeLa cells treated by the same protocol without ST were utilized as controls for the subtraction of non-specific spin labeling. The successful EGE of cells to bear unnatural Neu5Ac9N3 within surface sialoglycans and attachment of molecular labels to the azide group via SPAAC were verified for both CSTII29 and Pd2,6ST (Figure S-1, Supporting Information) by treating the engineered cells with DBCO-modified fluorescent labels and then analysis of the cells by flow cytometry (FACS). To further validate the labelling of cell surface glycans, we incubated the labelled cells with peptide N-glycosidase F (PNGase F), which removes N-glycans from glycoproteins, and observed a 50% decrease in the FACS signal.29 This result suggests that at least 50% of the fluorescent labels are attached to N-glycans on the cell surface. The remaining fluorescent labels might be attached to other glycans, such as the O-glycans of glycoproteins and the glycans of glycolipids.
EPR studies of spin-labelled cells.
The spin-labelled cells having SLs linked to the sialic acid C9-position of α2,6- or α2,3-sialoglycans were investigated using a benchtop 100 G X-band CW-EPR spectrometer, with the samples prepared and data collected by previously described methods.29 Briefly, counted HeLa cells (~ 1 x 106) were suspended in 0.8% agarose and loaded into an EPR tube. Agar was utilized to help suspend cells for more reproducible measurements for control subtraction. Effects of agar on the spin-label EPR spectra were minimal and within the range of variations observed for spectra obtained from multiple batches of spin-labeling cells (Figures S-2 and S-3, Supporting information). X-Band absorption EPR spectra were collected from the treated and control groups of cells with baselines corrected by the LabView software and peak areas normalized to cell numbers. A control spectrum, used to subtract out the EPR signal due to non-specific labeling, was obtained for each given batch of cells to account for any variation in cell diversity. Therefore, each control EPR spectrum was applied to only a specific set of samples. Typically, three replicate experiments were performed and analyzed.
Figure 3 shows an example of the cell count-normalized EPR spectra of Pd2,6ST-treated cells (top) and the control (middle). The EPR signal for the control likely arises from nonspecific partitioning of DBCO-SL into the cell membrane, as the signal does not decrease upon additional washing, and should not arise from any endocytosed label because the nitroxide label is expected to be reduced rapidly if it permeates into the cell.28 The intensity of the EPR signal of treated cells is remarkably higher than that of the control group (~ 35% total area), and the control-subtracted resultant spectrum (Figure 3, bottom) gives an excellent signal to noise ratio, suggesting a potent EPR signal from the Pd2,6ST-mediated spin-labelled cells, with ca. 109 to 1010 EPR active SL-sialic acid moieties per cell. These results also validated the robust glycoengineering and spin-labelling of HeLa cells using the nitroxide radical, which agreed with the FACS results using an analogous clickable fluorescent label. The line shape of the control-subtracted EPR spectrum, which is in contrast to the sharp, narrow EPR peaks of free DBCO-SL in solution,28 are diagnostic for large biomolecule-tethered nitroxide spins that have limited mobility, indicating SL attachment to cell surface glycans.
Figure 3.
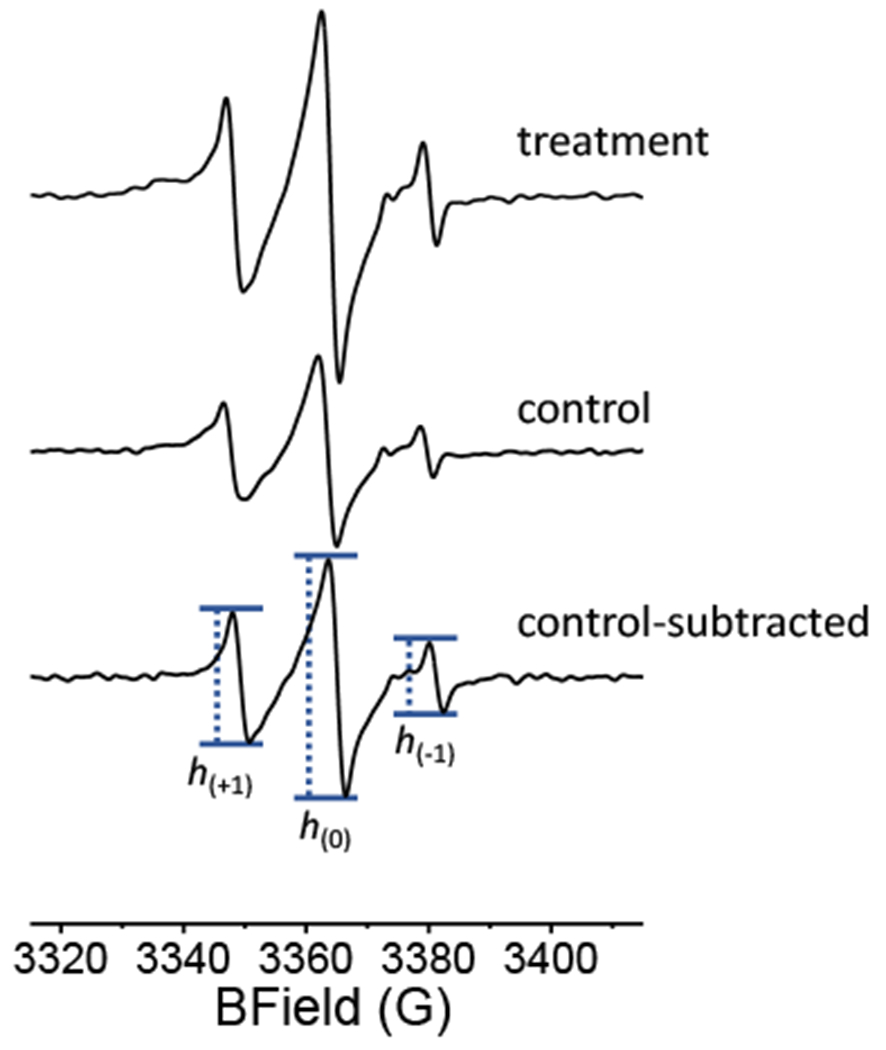
Cell count-normalized 100 G X-band CW-EPR spectra of the HeLa cells treated with (top) and without (middle, the control) Pd2,6ST, as well as the control-subtracted difference spectrum (bottom) with peak heights annotated for mobility parameter calculations.
Next, we analyzed the line shapes of the EPR spectra from the spin-labelled cells. X-Band EPR spectra from nitroxide SLs on biomolecules reflect the mobility of the SL determined by both the flexibility (rotational motion of bonds) from the SL structure and the motion (rotation of bonds, libration, etc.) of the biomolecular backbone, which in this case is the glycan. The local environment, described as tertiary packing and crowding, can also limit the mobility of the SL. For example, nitroxide SLs in fast isotropic motion produce EPR spectra with three sharp peaks of nearly equal intensity to give h(+1)/h(0) and h(−1)/h(0) values close to 1.0. As the SL motion slows down or becomes restricted, the EPR peaks broaden and show diminished intensities for h(+1) and h(−1) transitions, as compared to the central h(0) transition, leading to lower h(+1)/h(0) and h(−1)/h(0) values.53 An average h(+1)/h(0) value of 0.59 ± 0.06 for the EPR spectra of Pd2,6ST-mediated spin-labelled cells from three separate experiments implies a moderate average mobility for the SLs attached to α2,6-sialoglycans. However, the average h(+1)/h(0) value for the EPR spectra of α2,3-ST CSTII-mediated labelled cells is 0.75 ± 0.06,29 reflecting a higher mobility. This was unanticipated given the fact that spin-labeled Neu5Ac residues linked to the Gal/GalNAc C6–the carbon beyond the sugar ring–should be more flexible than spin-labeled Neu5Ac residues linked to the Gal/GalNAc C3 within the sugar ring (Figure 4). The interpretation of this finding is discussed further below.
Figure 4.
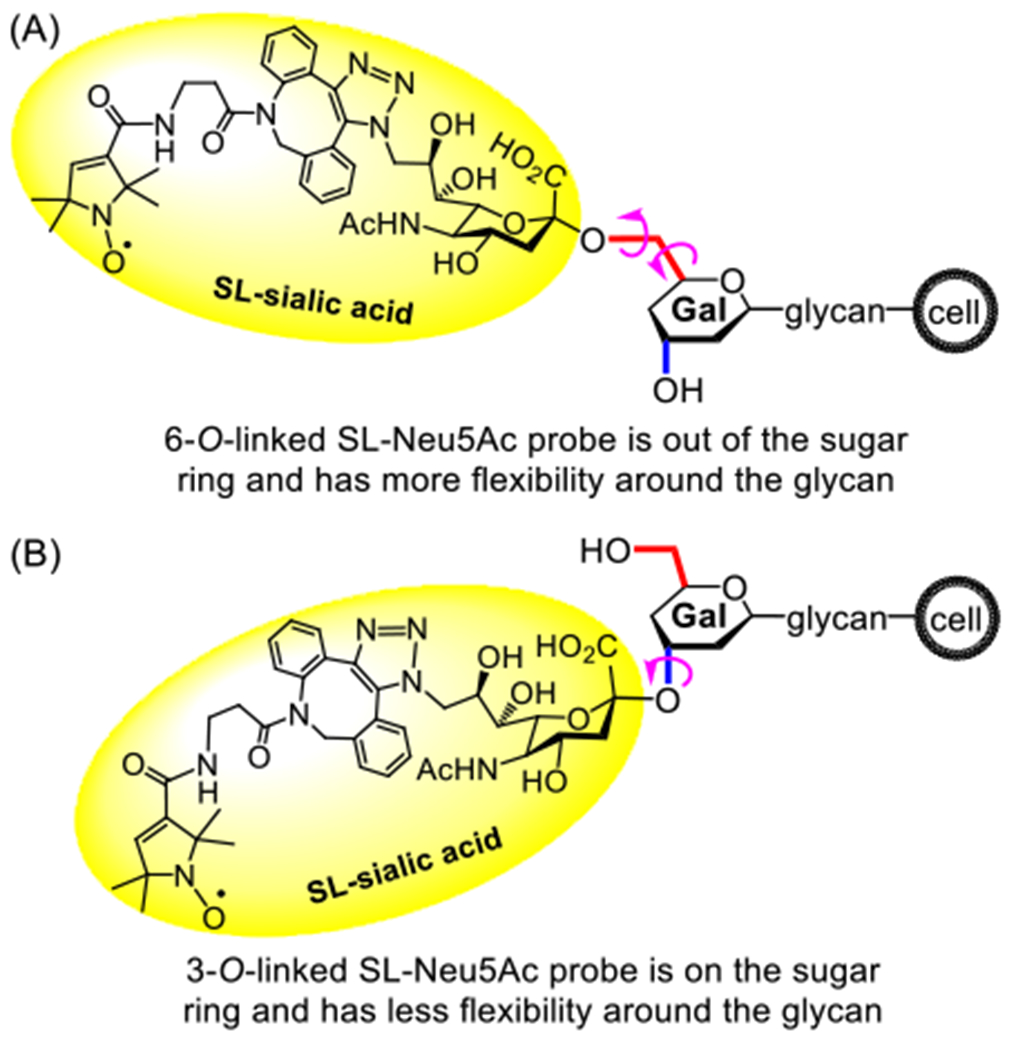
The structures of cell surface sialoglycans having spin-labeled Neu5Ac residues linked to (A) C6 and (B) C3 of Gal/GalNAc, respectively. C6 is out of the sugar ring, whereas C3 is in the sugar ring. As a result, overall, the 6-O-linked SL-Neu5Ac moiety should have a higher degree of freedom and face less steric hindrance than the 3-O-linked SL-Neu5Ac moiety.
Theoretical EPR spectral fitting.
Theoretical fitting of the EPR spectra using EasySpin software can provide insights into the environments around the SL.54 Therefore, the control-subtracted EPR spectra for Pd2,6ST- and CSTII-mediated spin-labelled cells were simulated with one-, two- and three-component theoretical spectral fits, where the lowest residuals with unique and reproducible solutions were obtained for the two-component fits in both cases, as depicted in Figure 5 (Figures S4–S5 and Tables S1–S6 in Supporting Information show one- and three-component simulations). The results revealed in average two distinct motional components for the SL α2,6-linked to the Gal/GalNAc residues on the surface of HeLa cells (Figure 5A). One is a fast-motion component (τc ~0.2-0.5 ns), and the other is an intermediate-motion component (τc ~4-8 ns) (Table 1). Interestingly, these two components are similar to those obtained from our prior simulations of the same SL α2,3-linked to cell surface glycans29 (Figure 5B, Table 1). However, the populations of these components differ for the two types of spin-labelled glycans (Table 1). The fast-motion component comprises ~22 ± 5% of the signal population for spin-labelled α2,6-sialoglycans, but this highly mobile component comprises ~47 ± 10% for spin-labelled ±2,3-sialoglycans. The changes in the relative populations of these two spectral components give rise to the difference in the average mobility derived from the h(+1)/h(0) line shape analyses.
Figure 5.
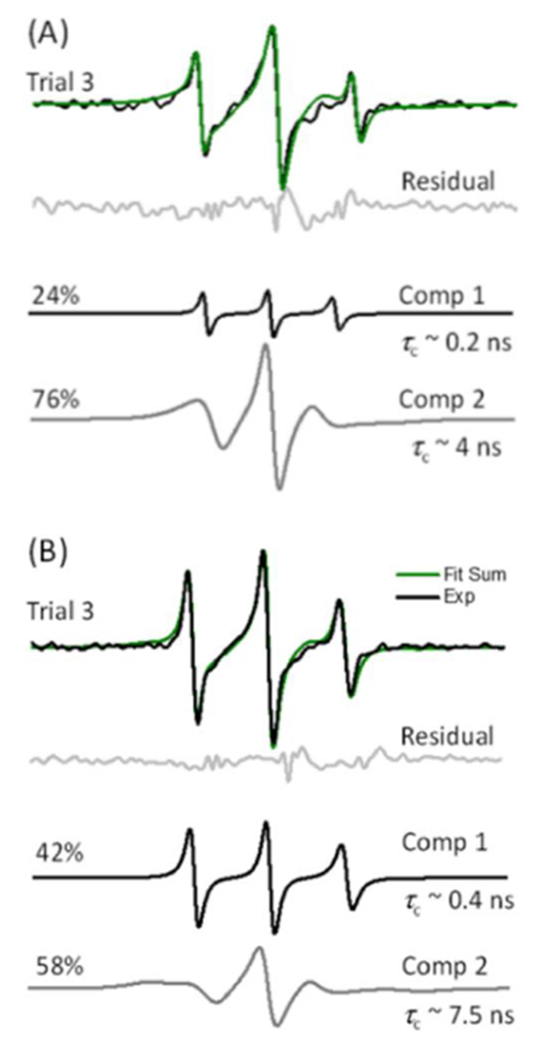
Example simulation results for X-band EPR spectra of spin-labelled HeLa cells using sialidase, CMP-Neu5Ac9N3, and (A) Pd2,6ST or (B) CSTII. The simulation spectra in green are the sums of the two component spectra at the given percentages. Residuals between the simulation and experimental spectra are shown in light grey.
Table 1.
Two-component simulation results for control-subtracted EPR spectra of spin-labelled HeLa cells and the relative populations of the two components
| Parameters | Pd2,6ST | CSTII | ||
|---|---|---|---|---|
|
| ||||
| Comp 1 | Comp 2 | Comp 1 | Comp 2 | |
| τc (ns) | ||||
| Trial 1 | 0.2 | 3.7 | 0.4 | 5.5 |
| Trial 2 | 0.5 | 7.8 | 0.3 | 9.2 |
| Trial 3 | 0.2 | 3.8 | 0.4 | 7.5 |
| average | 0.3 ± 0.1 | 5.0 ± 2.0 | 0.4 ± 0.04 | 7.0 ± 2.0 |
| Population (%) | ||||
| Trial 1 | 16 | 84 | 58 | 42 |
| Trial 2 | 25 | 75 | 41 | 59 |
| Trial 3 | 24 | 76 | 42 | 58 |
| average | 22 ± 5 | 78 ± 5 | 47 ± 10 | 53 ± 10 |
Simulation of DBCO-SL mobility.
The line shapes of X-band EPR spectra reflect the mobility of SLs due to the motional averaging of the anisotropic hyperfine interaction. Therefore, simulation of the SL mobility (rotamers) allows for the characterization of contributions that the SL makes to the obtained EPR spectra.55–57 It is common to discuss the mobility of a SL in terms of the number of rotatable bonds via dihedral angle fluctuation. For instance, Hubbell and co-workers showed that the MTSSL had limited rotation around the S-S bond due to interaction with the protein backbone. This ‘interaction” gives MTSSL its utility in reporting biomolecular motions and is referred to as the χ4/χ5 model of motion because the disulfide bond-peptide backbone interaction limits rotation around χ1/χ2/χ3.58–59 Paramagnetic 2,2,6,6-tetramethyl-N-oxyl-4-amino-4-carboxylic acid (TOAC) provides more direct information about the molecular backbone, as its molecular fluctuations are limited to ring puckering.60 Other rigid labels (for proteins and nucleic acids) are also utilized to make inferences about biomolecular motions61–63 and measure distances where narrow distribution profiles are desired.64–65
To understand the molecular origin of the two components of our EPR spectra, we performed molecular dynamic simulations of the Neu5Ac-DBCO-SL moiety to investigate its flexibility. As expected, several of the dihedral bonds (χ4, χ8 and χ9, Figure 6) show almost no rotation because of conjugation and partially conjugation (amides). The DBCO-SL segment has the highest motional flexibility around χ7 > χ6 >> χ5. In this manner, the rotational flexibility of the pyrroline ring in the SL is modulated mostly by the χ7 and χ6 dihedral angles, similar to MTSSL that also has two rotatable bonds. However, because a high flexibility is also observed in the side chain of Neu5Ac to which DBCO-SL is attached (adding rotation around χ1 > χ3 > χ2), the DBCO-SL moiety linked to Neu5Ac is expected to be flexible, thus limiting our ability to delineate the motional contribution from the sugar moiety and spin label to the motion-averaged EPR spectrum.
Figure 6.
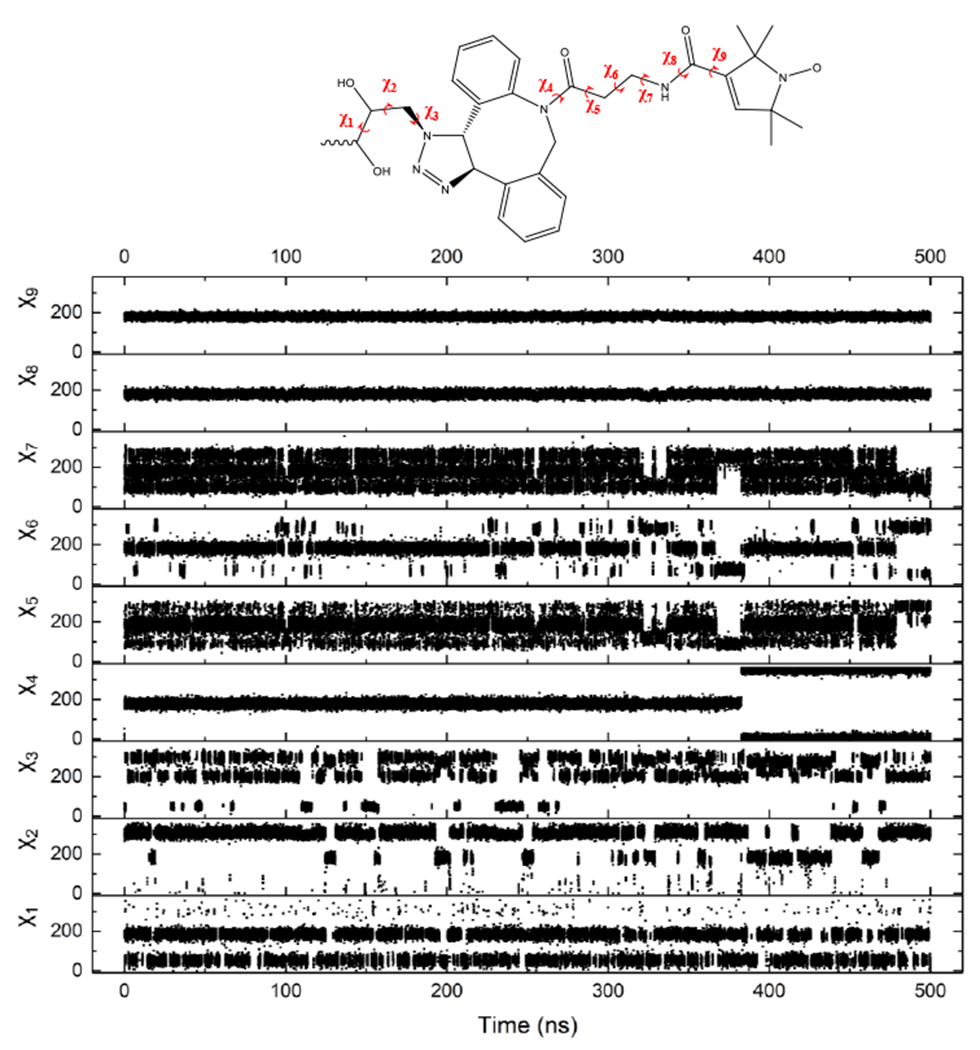
Scatter plot of the dihedral angles of the Neu5Ac-DBCO-SL moiety.
Given the substantial motion observed in our simulations around six dihedral angles we interpret the line shapes of our EPR spectra in the following way. If one assumes Neu5Ac-DBCO-SL to generate an isotropic-limit spectrum reflective of a high flexibility of the SL itself, then the broad component observed in our spectra would arise from SL interactions with the local environment (such as crowding and tertiary interactions or binding). These interactions would restrict rotation around various dihedral angles, thereby giving rise to an EPR spectrum reflective of intermediate or possibly slow motions. A highly isotropic-like signal (τc ≤1 ns) of our EPR spectra reflects high motion of the SL itself or the SLs attached to highly flexible glycans that have no strong interactions with other biomolecules, i.e., an uncrowded environment. Spectra with intermediate motion (τc ~4-8 ns) represent sites that are in contact with other moieties, i.e., a crowded environment/tertiary contact. Ideally, to probe changes in the mobility of glycans, we would desire a spin probe that can be attached to glycans by a tether with limited flexibility, e.g., a shorter tether, to essentially remove the motion from χ5/χ6/χ7.
Discussion of the EPR spectra of spin-labelled HeLa cells.
Theoretical fitting of the EPR spectra of Pd2,6ST- and CSTII-mediated spin-labelled cells showed that both spectra contained fast- and intermediate-motion components. This result is anticipated from the viewpoint of glycobiology. On the cell surface, the peripheral glycans of glycoproteins or peptidoglycans are in relatively non-crowded environments, whereas their glycans close to the cell membrane or glycans of glycolipids are deeper inside the cell glycocalyx and surrounded by the packed cell surface matrix to result in decreased molecular flexibility and mobility. Our molecular dynamic simulation results revealed that the Neu5Ac-DBCO-SL moiety is flexible around five to six dihedral angles, which further suggests that the decreased mobility of SLs seen here is likely reflective of the packing interactions of SLs with the surrounding matrix.
Pd2,6ST catalyzes the attachment of Neu5Ac9N3 and then SL to the Gal/GalNAc 6-O-position, which extends beyond the sugar ring (Figure 4). CSTII is a bifunctional enzyme with potent α2,3-ST and weak α2,8-ST activities. In this study, CSTII was employed to perform cell EGE for only a short period (30 min) to limit α2,8-sialylation.29, 47 Therefore, CSTII should have only catalyzed the attachment of Neu5Ac9N3 and SL to the Gal/GalNAc 3-O-position, which is on the sugar ring and is more restricted than the 6-O-position. Neu5Ac-DBCO-SL attached to the Gal/GalNAc 3-O-position (in plane of the sugar ring, with limited rotation) should be less flexible or mobile than that linked to the Gal/GalNAc 6-O-position (out of plane, with a higher flexibility) under the same conditions (Figure 4). On the contrary, our theoretical line shape fits of the EPR spectra suggest that the percentage of α2,6-sialoglycans with restricted mobility (75%) is greater than that of α2,3-sialoglycans in the same motion regime (58%). This indicates an overall lower mobility of α2,6-sialoglycan-linked SLs (h(+1)/h(0) value of EPR spectra: ~0.59) than that of α2,3-sialoglycan-linked SLs (h(+1)/h(0) value of EPR spectra: ~0.75). These apparently contradictory results can only be explained by the differently local environments that some of the α2,3- and α2,6-sialoglycans may have experienced on the cell surface, as all other conditions for the two experiments were identical. This observation is consistent with the above hypothesis that the difference in mobility of SLs is due to the packing interactions of SLs with the surrounding matrix.
Additionally, the EPR results also disclosed a higher population of slow-mobility SLs attached to α2,6-sialoglycans than that attached to α2,3-sialoglycans, suggesting the residence of a relatively large number of α2,6-sialoglycans in more crowded surroundings, e.g., close to the glycoprotein backbone and/or to the cell membrane. There are two plausible explanations for this finding. One explanation is that the numbers and distribution patterns of α2,6- and α2,3-sialylation sites differ on the cell surface. For example, although both Pd2,6ST and CSTII can catalyse the sialyation of terminal Gal and GalNAc residues in relatively relaxed environments, Pd2,6ST may be less stereo-sensitive and thus more effective than CSTII to catalyse Neu5Ac9N3 attachment to the relatively restricted internal N-acetyl-lactosamine (LacNAc) epitopes in the glycoprotein glycans. Literature results supporting this proposition include Pd2,6ST-catalyzed sialylation of internal LacNAc.66–68 However, CSTII is relatively selective for terminal sialic acids. It is also possible that glycolipids have more sites for α2,6-sialylation than for α2,3-sialylation; therefore, more glycolipid glycans, which are close to the cell membrane and located in crowded environments in the cell matrix, are sialylated and spin-labelled by Pd2,6ST. The other explanation is that Pd2,6ST and CSTII may have different properties. For example, Pd2,6ST may be more effective than CSTII to navigate through crowded environments to catalyse sialylation reactions. In either case, it seems that CSTII and Pd2,6ST can target different glycans and/or glycans at different locations on cells to perform α2,6- and α2,3-sialylations, which has not been observed previously. This interesting finding can be of biological importance and applications. For example, changes in the biological activity of glycans resulted from Pd2,6ST and CSTII-mediated EGE may be because of not only differences in the linkage forms of their Neu5Ac but also different molecular targets on the cells. Therefore, cautions should be used when interpreting related results.
Conclusion
In brief, sialoglycans on live HeLa cells were successfully labelled with a nitroxide radical spin through EGE-based installation of Neu5Ac9N3 to cell surface glycans and then click reaction-based attachment of the SL. In this process, Pd2,6ST and CSTII were utilized to install α2,6- and α2,3-linked Neu5Ac9N3, respectively, hence α2,6- and α2,3-sialoglycans could be separately and specifically labelled and probed on live cells. Studies on the EPR spectra of the spin-labelled cells demonstrated that both types of sialoglycans contained a slow-motion component and a fast-motion component, which are roughly defined and represent the averages of various glycan components in the diverse environments of the cell glycocalyx or extracellular matrix. Different distributions of the two motion components for α2,6- and α2,3-sialoglycans indicated the different environments of these two sialoglycans on cells, i.e., a higher population of α2,6-sialoglycan in the more restricted/crowded environments than that of α2,3-sialoglycan, as a result of Pd2,6ST- and CSTII-mediated EGE. Consequently, it seems that Pd2,6ST and CSTII can target different glycans or glycans at different locations on the cell surface. These results are not only biologically significant but also have potential biological applications. For example, Pd2,6ST and CSTII may be used to target different glycoconjugates on cells and help gain more insights into the functions of α2,6- and α2,3-linked sialoglycans. Overall, this work has validated the great potential of EGE-based spin labelling of live cells to allow for the study of different glycans on the cell surface by EPR spectroscopy. In addition, it is conceivable that this method can be useful for EPR studies of other glycans based on cell EGE using corresponding glycosyltransferases.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by NIGMS/NIH (R35GM131686; ZG), NSF (MCB-1715384; GEF), an NIH instrumentation grant (S10 RR031603) for the Bruker E500 spectrometer, and Department of Chemistry, University of Florida for the Magnettech MS-5000 benchtop spectrometer. ZG thanks Steven and Rebecca Scott endowment to support our research.
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge at: website Flow cytometry results of Pd2,6ST-mediated engineering and fluorescence labelling of cell surface sialoglycans, EPR spectra and theoretical fit results, and atomistic MD simulation results.
REFERENCES
- 1.Ghosh S, Sialic Acids and Sialoglycoconjugates in the Biology of Life, Health and Disease. Academic Press: London, UK, 2020. [Google Scholar]
- 2.Schauer R, Sialic acids as antigenic determinants of complex carbohydrates. Adv. Exp. Med. Biol 1988, 228, 47–72. [DOI] [PubMed] [Google Scholar]
- 3.Rosenburg AE, Biology of the Sialic Acid. Plenum Press: New York, USA, 1995. [Google Scholar]
- 4.Reutter W; Stasche R; Stehling P; Baum O, The biology of sialic acids: Insights into their structure, metabolism and function in particular during viral infection. In Glycosciences, Gabius H-J; Gabius S, Eds. Chapman & Hall: Weinheim, Germany, 1997; pp 245–259. [Google Scholar]
- 5.Crocker PR; Varki A, Siglecs in the immune system. Immunology 2001, 103, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crocker PR, Siglecs: Sialic-acid-binding immunoglobulin-like lectins in cell-cell interactions and signaling. Curr. Opin. Struct. Biol 2002, 12, 609–615. [DOI] [PubMed] [Google Scholar]
- 7.Duan S; Paulson JC, Siglecs as immune cell checkpoints in disease. Annu. Rev. Immunol 2020, 38, 365–395. [DOI] [PubMed] [Google Scholar]
- 8.Troy FA, Polysialylation: From bacteria to brain. Glycobiology 1992, 2, 5–23. [DOI] [PubMed] [Google Scholar]
- 9.Martersteck CM; Kedersha NL; Drapp DA; Tsi TG; Kolly KJ, Unique α2,8-polysialylated glycoproteins in breast cancer and leukemia. Glycobiology 1996, 6, 289–301. [DOI] [PubMed] [Google Scholar]
- 10.Michalides R; Kwa B; Springall D; van Zandwijk N; Koopman J; Hilkens J; Mooi W, NCAM and lung cancer. Int. J. Cancer: Suppl 1994, 8, 34–37. [DOI] [PubMed] [Google Scholar]
- 11.Schauer R; Kamerling JP, Exploration of the sialic acid world. Adv. Carbohydr. Chem. Biochem 2018, 75, 1–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabriele N; Kawaoka Y, Transmission of influenza A viruses. Virology 2015, 479–480, 234-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers GN; Paulson JC, Receptor determinants of human and animal influenza virus isolates: Differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 1983, 127, 361–373. [DOI] [PubMed] [Google Scholar]
- 14.Connor RJ; Kawaoka Y; Webster RG; J.C. P, Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 1994, 205, 17–23. [DOI] [PubMed] [Google Scholar]
- 15.Makoto O; Kawaoka Y, Cross talk between animal and human influenza viruses. Annu. Rev. Anim. Biosci 2013, 1, 21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varki A; Cummings RD; Esko JD; Freeze HH; Stanley P; Bertozzi CR; Hart GW; Etzler ME, Essentials of Glycobiology (2nd ed). Cold Spring Harbor Laboratory Press: Cold Spring Harbor, USA, 2009. [PubMed] [Google Scholar]
- 17.Bhavanandan VP; Gowda DC, Introduction to the complexity of cell surface and tissue matrix glycoconjugates. In Glycobiology of the Nervous System, Yu RK; Schengrund C-L, Eds. Springer: New York, USA, 2014; pp 1–31. [DOI] [PubMed] [Google Scholar]
- 18.Schultz MJ; Swindall AF; Bellis SL, Regulation of the metastatic cell phenotype by sialylated glycans. Cancer Metastasis Rev. 2012, 31, 501–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodrigues E; Macauley MS, Hypersialylation in cancer: Modulation of inflammation and therapeutic opportunities. Cancers 2018, 10, e207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X; Yang G; Guan F, Biological functions and analytical strategies of sialic acids in tumor. Cells 2020, 9, e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dube DH; Bertozzi CR, Glycans in cancer and inflammation - potential for therapeutics and diagnostics. Nature Rev. Drug Disc 2005, 4, 477–488. [DOI] [PubMed] [Google Scholar]
- 22.Nores GA; Dohi T; Taniguchi M; Hakomori S, Density dependent recognition of cell surface GM3 by a certain anti-melanoma antibody, and GM3 lactone as a possible immunogen: Requirements for tumor-associated antigen and immunogen. J. Immunol 1987, 139, 3171–3176. [PubMed] [Google Scholar]
- 23.Maverakis E; Kim K; Shimoda M; Gershwin ME; Patel F; Wilken R; Raychaudhuri S; Ruhaak LR; Lebrilla CB, Glycans in the immune system and the altered glycan theory of autoimmunity: A critical review. J. Autoimmun 2015, 57, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muthana S; Cao H; Chen X, Recent progress in chemical and chemoenzymatic synthesis of carbohydrates. Curr. Opin. Chem. Biol 2009, 13, 573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krasnova L; Wong C-H, Oligosaccharide synthesis and translational innovation. J. Am. Chem. Soc 2019, 141, 3735–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dube DH; Bertozzi CR, Metabolic oligosaccharide engineering as a tool for glycobiology. Curr. Opin. Chem. Biol 2003, 7, 616–625. [DOI] [PubMed] [Google Scholar]
- 27.Aguilar AL; Briard JG; Yang L; Ovryn B; Macauley MS; Wu P, Tools for studying glycans: Recent advances in chemoenzymatic glycan labeling. ACS Chem. Biol 2017, 12, 611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaiswal M; Tran T; Li Q; Yan X; Zhou M; Kundu K; Fanucci G; Guo Z, A metabolically engineered spin-labeling approach for studying glycans on cells. Chem. Sci 2020, 11, 12522–12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaiswal M; Tran T; Guo J; Zhou M; Garcia Diaz J; Fanucci G; Guo Z, Enzymatic glycoengineering-based spin labeling of cell surface sialoglycans to enable their analysis by electron paramagnetic resonance (EPR) spectroscopy. Analyst 2022, 147, 784–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubbell WL; Cafiso DS; Altenbach C, Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol 2000, 7, 735–739. [DOI] [PubMed] [Google Scholar]
- 31.Hubbell WL; McHaourab HS; Altenbach C; Lietzow MA, Watching proteins move using site-directed spin labeling. Structure 1996, 4, 779–783. [DOI] [PubMed] [Google Scholar]
- 32.McHaourab HS; Lietzow MA; Hideg K; Hubbell WL, Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry 1996, 35, 7692–7704. [DOI] [PubMed] [Google Scholar]
- 33.Steinhoff HJ; Mollaaghababa R; Altenbach C; Hideg K; Krebs M; Khorana HG; Hubbell WL, Time-resolved detection of structural changes during the photocycle of spin-labeled bacteriorhodopsin. Science 1994, 266, 105–107. [DOI] [PubMed] [Google Scholar]
- 34.Columbus L; Hubbell WL, A new spin on protein dynamics. Trends Biochem. Sci 2002, 27, 288–295. [DOI] [PubMed] [Google Scholar]
- 35.Fanucci GE; Cafiso DS, Recent advances and applications of site-directed spin labeling. Curr. Opin. Struct. Biol 2006, 16, 644–653. [DOI] [PubMed] [Google Scholar]
- 36.Roser P; Schmidt MJ; Drescher M; Summerer D, Site-directed spin labeling of proteins for distance measurements in vitro and in cells. Org. Biomol. Chem 2016, 14, 5468–5476. [DOI] [PubMed] [Google Scholar]
- 37.Gross A; Columbus L; Hideg K; Altenbach C; Hubbell WL, Structure of the KcsA potassium channel from Streptomyces lividans: A site-directed spin labeling study of the second transmembrane segment. Biochemistry 1999, 38, 10324–10335. [DOI] [PubMed] [Google Scholar]
- 38.Cheng B; Dong L; Zhu Y; Huang R; Sun Y; You Q; Song Q; Paton JC; Paton AW; Chen X, 9-Azido analogues of three sialic acid forms for metabolic remodeling of cell-surface sialoglycans. ACS Chem. Biol 2019, 14, 2141–2147. [DOI] [PubMed] [Google Scholar]
- 39.Sun M; Li Y; Chokhawala HA; Henning R; Chen X, N-Terminal 112 amino acid residues are not required for the sialyltransferase activity of Photobacterium damsela alpha2,6-sialyltransferase. Biotechnol. Lett 2008, 30, 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng J; Yu H; Lau K; Huang S; Chokhawala HA; Li Y; Tiwari VK; Chen X, Multifunctionality of Campylobacter jejuni sialyltransferase CstII: Characterization of GD3/GT3 oligosaccharide synthase, GD3 oligosaccharide sialidase, and trans-sialidase activities. Glycobiology 2008, 18, 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanwell MD; Curtis DE; Lonie DC; Vandermeersch T; Zurek E; Hutchison GR, Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J Cheminform 2012, 4, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanommeslaeghe K; Hatcher E; Acharya C; Kundu S; Zhong S; Shim J; Darian E; Guvench O; Lopes P; Vorobyov I; Mackerell AD Jr., CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem 2010, 31, 671–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price DJ; Brooks CL 3rd, A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys 2004, 121, 10096–10103. [DOI] [PubMed] [Google Scholar]
- 44.Case DA; Belfon K; Ben-Shalom IY; Brozell SR; Cerutti DS; Cheatham TE; Cruzeiro VWD; Darden TA; Duke RE; Giambasu G; et al. , AMBER 2020. University of California, San Francisco: 2020. [Google Scholar]
- 45.Ryckaert J-P; Ciccotti G; Berendsen HJC, Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys 1977, 23, 327–341. [Google Scholar]
- 46.Roe DR; Cheatham TE 3rd, PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput 2013, 9, 3084–3095. [DOI] [PubMed] [Google Scholar]
- 47.Blixt O; Vasiliu D; Allin K; Jacobsen N; Warnock D; Razi N; Paulson JC; Bernatchez S; Gilbert M; Wakarchuk W, Chemoenzymatic synthesis of 2-azidoethyl-ganglio-oligosaccharides GD3, GT3, GM2, GD2, GT2, GM1, and GD1a. Carbohydr. Res 2005, 340, 1963–1972. [DOI] [PubMed] [Google Scholar]
- 48.Yu H; Huang S; Chokhawala H; Sun M; Zheng H; Chen X, Highly efficient chemoenzymatic synthesis of naturally occurring and non-natural alpha-2,6-linked sialosides: A P. damsela alpha-2,6-sialyltransferase with extremely flexible donor-substrate specificity. Angew. Chem. Int. Ed 2006, 45, 3938–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wen L; Liu D; Zheng Y; Huang K; Cao X; Song J; Wang PG, A One-step chemoenzymatic labeling strategy for probing sialylated Thomsen-Friedenreich antigen. ACS Cent. Sci 2018, 4, 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kajihara Y; Kamitani T; Sato R; Kamei N; Miyazaki T; Okamoto R; Sakakibara T; Tsuji T; Yamamoto T, Synthesis of CMP-9’’-modified-sialic acids as donor substrate analogues for mammalian and bacterial sialyltransferases. Carbohydr. Res 2007, 342, 1680–1688. [DOI] [PubMed] [Google Scholar]
- 51.Hong S; Shi Y; Wu NC; Grande G; Douthit L; Wang H; Zhou W; Sharpless KB; Wilson IA; Xie J; Wu P, Bacterial glycosyltransferase-mediated cell-surface chemoenzymatic glycan modification. Nat. Commun 2019, 10, e1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mbua NE; Li X; Flanagan-Steet HR; Meng L; Aoki K; Moremen KW; Wolfert MA; Steet R; Boons GJ, Selective exo-enzymatic labeling of N-glycans on the surface of living cells by recombinant ST6Gal I. Angew. Chem. Int. Ed 2013, 52, 13012–13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berliner LJ, Biological Magnetic Resonance, Spin Labeling: Theory and Applications. Plenum Press: New York, USA, 1978. [Google Scholar]
- 54.Stoll S; Schweiger A, EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson 2006, 178, 42–55. [DOI] [PubMed] [Google Scholar]
- 55.Sezer D; Freed JH; Roux B, Simulating electron spin resonance spectra of nitroxide spin labels from molecular dynamics and stochastic trajectories. J. Chem. Phys 2008, 128, e165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warshaviak DT; Serbulea L; Houk KN; Hubbell WL, Conformational analysis of a nitroxide side chain in an α-helix with density functional theory. J. Phys. Chem. B 2011, 115, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin PD; Svensson B; Thomas DD; Stoll S, Trajectory-based simulation of EPR spectra: Models of rotational motion for spin labels on proteins. J. Phys. Chem. B 2019, 123, 10131–10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langen R; Oh KJ; Cascio D; Hubbell WL, Crystal structures of spin labeled T4 lysozyme mutants: Implications for the interpretation of EPR spectra in terms of structure. Biochemistry 2000, 39, 8396–8405. [DOI] [PubMed] [Google Scholar]
- 59.Guo Z; Cascio D; Hideg K; Hubbell WL, Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme. Protein Sci. 2008, 17, 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schreier S; Bozelli JC Jr.; Marín N; Vieira RF; Nakaie CR, The spin label amino acid TO AC and its uses in studies of peptides: Chemical, physicochemical, spectroscopic, and conformational aspects. Biophys. Rev 2012, 4, 45–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barhate N; Cekan P; Massey AP; Sigurdsson ST, A nucleoside that contains a rigid nitroxide spin label: A fluorophore in disguise. Angew. Chem. Int. Ed. Engl 2007, 46, 2655–2658. [DOI] [PubMed] [Google Scholar]
- 62.Fleissner MR; Bridges MD; Brooks EK; Cascio D; Kálai T; Hideg K; Hubbell WL, Structure and dynamics of a conformationally constrained nitroxide side chain and applications in EPR spectroscopy. Proc. Natl. Acad. Sci. USA 2011, 108, 16241–16246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gophane DB; Endeward B; Prisner TF; Sigurdsson ST, A semi-rigid isoindoline-derived nitroxide spin label for RNA. Org. Biomol. Chem 2018, 16, 816–824. [DOI] [PubMed] [Google Scholar]
- 64.Schnorr KA; Gophane DB; Helmling C; Cetiner E; Pasemann K; Fürtig B; Wacker A; Qureshi NS; Gränz M; Barthelmes D; Jonker HRA; Stirnal E; Sigurdsson ST; Schwalbe H, Impact of spin label rigidity on extent and accuracy of distance information from PRE data. J Biomol NMR 2017, 68, 53–63. [DOI] [PubMed] [Google Scholar]
- 65.Cunningham TF; Putterman MR; Desai A; Horne WS; Saxena S, The double-histidine Cu2+-binding motif: A highly rigid, site-specific spin probe for electron spin resonance distance measurements. Angew. Chem. Int. Ed. Engl 2015, 54, 6330–6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nycholat CM; Peng W; McBride R; Antonopoulos A; de Vries RP; Polonskaya Z; Finn MG; Dell A; Haslam SM; Paulson JC, Synthesis of biologically active N- and O-linked glycans with multisialylated poly-N-acetyllactosamine extensions using P. damsela α2-6 sialyltransferase. J. Am. Chem. Soc 2013, 135, 18280–18283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meng X; Yao W; Cheng J; Zhang X; Jin L; Yu H; Chen X; Wang F; Cao H, Regioselective chemoenzymatic synthesis of ganglioside disialyl tetrasaccharide epitopes. J. Am. Chem. Soc 2014, 136, 5205–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu Y; Fan Y; Ye J; Wang F; Nie Q; Wang L; Wang PG; Cao H; Cheng J, Successfully engineering a bacterial sialyltransferase for regioselective α2,6-sialylation. ACS Catal. 2018, 8, 7222–7227. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


