ABSTRACT
The first case of coronavirus disease 2019 (COVID-19) within the White Mountain Apache Tribe (WMAT) in Arizona was diagnosed almost 1 month after community transmission was recognized in the state. Aggressive contact tracing allowed for robust genomic epidemiology of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and subsequent phylogenetic analyses implicated only two virus introductions, which resulted in the spread of two unique viral lineages on the reservation. The phylogenies of these lineages reflect the nature of the introductions, the remoteness of the community, and the extraordinarily high attack rates. The timing and space-limited nature of the outbreaks validate the public health tracing efforts involved, which were illustrated by multiple short transmission chains over a period of several weeks, eventually resulting in extinction of the lineages. Comprehensive sampling and successful infection control efforts are illustrated in both the effective population size analyses and the limited mortality outcomes. The rapid spread and high attack rates of the two lineages may be due to a combination of sociological determinants of the WMAT and a seemingly enhanced transmissibility. The SARS-CoV-2 genomic epidemiology of the WMAT demonstrates a unique local history of the pandemic and highlights the extraordinary and successful efforts of their public health response.
IMPORTANCE This article discusses the introduction and spread of two unique viral lineages of SARS-CoV-2 within the White Mountain Apache Tribe in Arizona. Both genomic sequencing and traditional epidemiological strategies (e.g., contract tracing) were used to understand the nature of the spread of both lineages. Beyond providing a robust genomic analysis of the epidemiology of the outbreaks, this work also highlights the successful efforts of the local public health response.
KEYWORDS: COVID-19, genomic epidemiology, contact tracing, tribes, Native American
INTRODUCTION
Various attitudes and responses have emerged regarding public health efforts employed during the coronavirus disease 2019 (COVID-19) pandemic (1–6), especially regarding diverse population demographics, and some of the most impacted regions were those with the lowest population density, such as Native American (NA) reservations (3, 7–9). Excess morbidity and mortality secondary to COVID-19 were observed among tribal communities throughout the United States (3, 10–15), including those in Arizona. Arizona contains the most federally recognized NA tribes in the country (n = 22); some of these tribes (e.g., Navajo Nation) experienced case and mortality rates higher than anywhere else in the county (7, 10, 11). While COVID-19 mortality rates in some NA peoples have been disproportionately high (16, 17), the remarkably low case fatality rate in other Arizona tribal communities, specifically the White Mountain Apache Tribe (WMAT) within the Fort Apache reservation, is noteworthy (3, 18).
Although the first case of SARS-CoV-2 in Arizona was detected in late January 2020 and community transmission was confirmed in early March (19), the WMAT did not report its first case of COVID-19 until a month later (18). The Fort Apache reservation encompasses 1.67 million acres in Arizona and has over 18,000 members living in nine main communities. Multigenerational households with ≥5 members are typical, and the population is significantly impacted by high rates of obesity, heart disease, diabetes, and bacterial infections (18, 20, 21). The WMAT’s first COVID-19 case was diagnosed on 1 April 2020, and the tribe soon experienced one of the highest infection rates documented during the early days of the pandemic. By August, approximately 15% of the residents had tested positive for COVID-19 (>2,200 reported cases). Transmission of COVID-19 in the community was rapid, and secondary rates of infections among households ranged from 80 to 100% (3). However, during the peak of their 2020 COVID-19 wave, the community had a case fatality rate of just ~1.2% (22), one of the lowest recorded at the time. They sustained that low rate well into 2021 (18) despite the high rates of both comorbidities and social determinants known to increase the risk of severe disease (3, 7, 11, 12, 18, 20, 21). The low fatality rates were driven in large part by successful on-ground, rapid test-and-treat interventions by local health workers (3, 18, 22).
With the multitude of opportunities for SARS-CoV-2 to mutate due to millions of people being infected and reinfected over time, the emergence of more transmissible variants and their diverging population structure was not a surprising occurrence (23). While most mutations are not phenotypically impactful, they may serve as genotypic lineage-defining markers that allow for phylogenetic analysis at global and local levels (4, 19). That being said, some SARS-CoV-2 mutations have indeed changed the phenotype of the lineage they define; for example, the D614G spike mutation was the first to be associated with higher viral loads and faster transmission (24) and to sweep the global population (25). Subsequent mutations continually drive the epidemiology of this virus and have resulted in variants of concern (VOCs) with higher fitness (referred to here by their Pango lineage names [26, 27] and WHO labels), including B.1.1.7 (Alpha), B.1.617.2 and AY lineages (Delta), and BA.1, BA.2, BA.4, and BA.5 (Omicron) (28, 29).
With a phylogenetic analysis of the SARS-CoV-2 derived from several epidemiologically linked clusters within the WMAT, we report a surprisingly limited number of viral introductions onto the reservation during the first several months of the pandemic and describe their lineage-defining mutations. These analyses illustrate the viral and host community dynamics that first drove and then halted the outbreak.
RESULTS
The initial SARS-CoV-2 wave across the WMAT began 1 April and died out by 13 August 2020, infecting at least 15% of the community population. Although epidemiologic and clinical analyses of the outbreak pointed to five initial clusters before widespread transmission was noted, genomic analysis identified only two main introduction events of the virus onto reservation lands, which together encompass samples from the epidemiological clusters. The majority (n = 732 [88%]) of the 831 virus genomes from this wave fell into two monophyletic clades, which comprised samples only from Arizona and almost exclusively from this tribal community. Many viral genomes (n = 99 [12%]) collected from the WMAT over the same time frame did not fall into these two virus populations, but none of these outliers appeared to be transmitted to other tribal members; therefore, these were omitted to maintain focus on the outbreak lineages.
The first monophyletic clade (n = 276) (Fig. 1) represents lineage B.1.289 and is defined by one unique point mutation in the spike gene, conferring an H245Y amino acid change. The second clade (n = 456) (Fig. 2) includes lineage B.1.516, characterized by five point mutations, two of which confer the amino acid changes nucleocapsid R209I and Orf10 R24L. Both clades fall within the B.1 lineage (26) and contain the spike G614 and Orf1a I265 alleles, both of which were characteristic of the large B.1 lineage that spread globally throughout summer 2020.
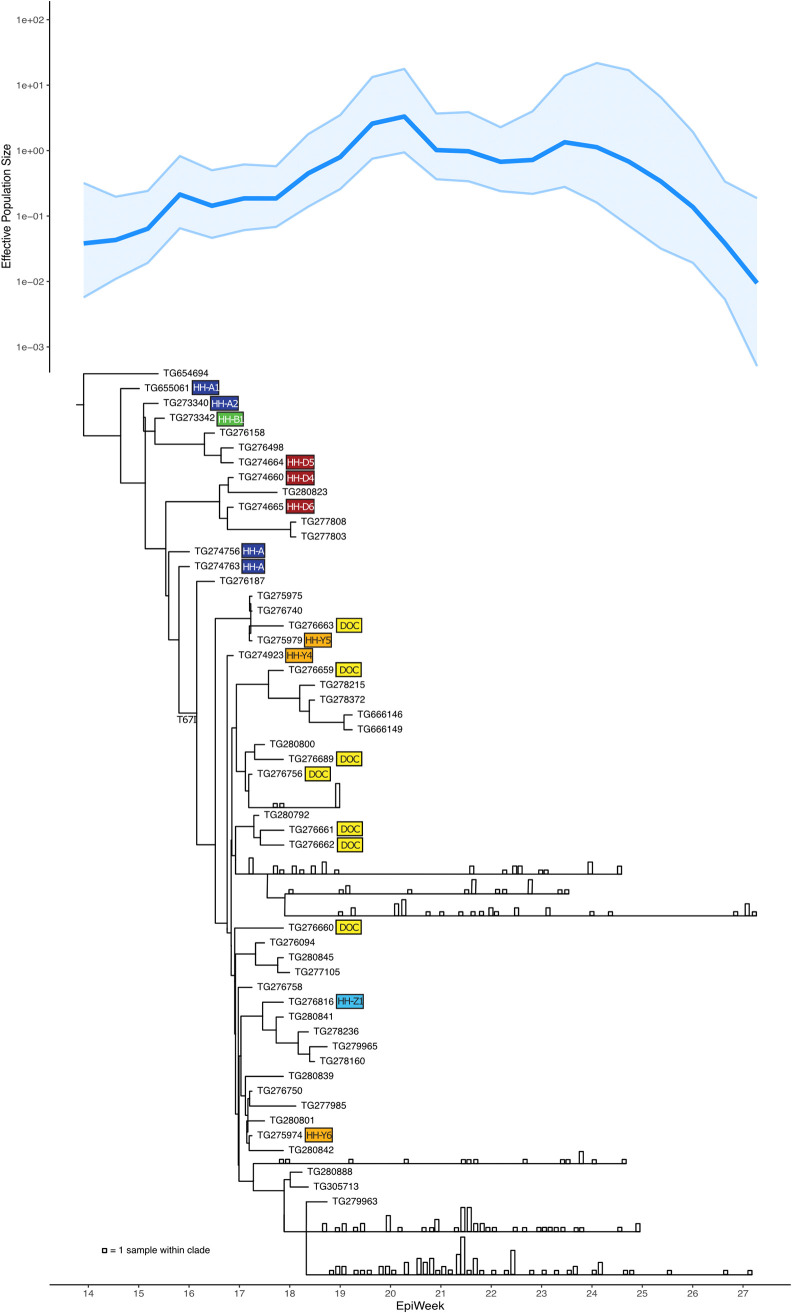
FIG 1.
Effective population size (Ne) analysis plot and phylogenetic tree of viral lineage B.1.289, characterized by spike: H245Y and Orf1b: T67I, representing the initial SARS-CoV-2 outbreak on an Arizona tribal reservation. This analysis includes 276 virus genomes, including the first known tribal case virus. Some large clades are collapsed, with vertical bars representing the number of samples in the clade collected on that date (see Fig. S1 for the full phylogenetic tree). The largely linear transmission pathway illustrated by the tree and the robust sampling and sequencing illustrated by the Ne plot reflect the unique circumstances of this lineage’s introduction and aggressive response by tribal public health. B.1.289 first appeared in April 2020, and its elimination was successful by July. Colored boxes denote SARS-CoV-2 cases associated with cluster 1 and 2, matching those depicted in the epidemiological linkage maps in Fig. 3.
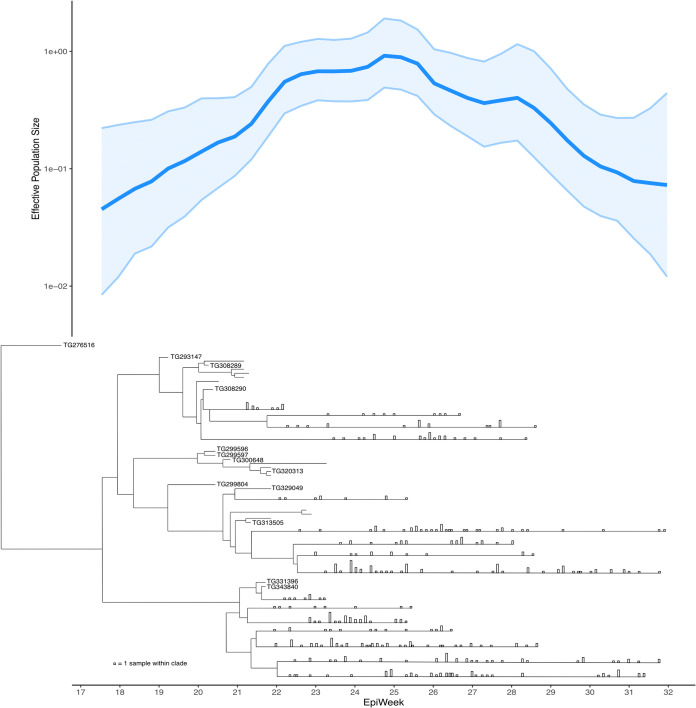
FIG 2.
Effective population size (Ne) analysis plot and phylogenetic tree of viral lineage B.1.516, characterized by nucleocapsid: R209I, representing the second introduction and outbreak of SARS-CoV-2 on an Arizona tribal reservation. This analysis includes 456 genomes. Some large clades are collapsed, with vertical bars representing the number of samples in the clade collected on that date (see Fig. S2 for the full phylogenetic tree). B.1.516 first appeared in May 2020 and quickly spread into multiple transmission pathways, which were successfully eliminated in August. As in the case of B.1.289, the tree and the robust sampling and sequencing illustrated by the Ne plot reflect the unique circumstances of this lineage’s introduction and aggressive response by tribal public health.
Full Bayesian phylogenetic tree of 276 virus genomes of SARS-CoV-2 lineage B.1.289 (as in Fig. 1 but with no clades collapsed), characterized by spike: H245Y and Orf1b: T67I. Download FIG S1, PDF file, 0.2 MB (184.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Full Bayesian phylogenetic tree of 456 virus genomes of SARS-CoV-2 lineage B.1.516 (as in Fig. 2 but with no clades collapsed), characterized by nucleocapsid: R209I. Download FIG S2, PDF file, 0.7 MB (678.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effective population size (Ne) analyses of B.1.289 (Fig. 1) and B.1.516 (Fig. 2) illustrate the rapid rise and fall of each population and the robustness of the COVID-19 outbreak case identification. Both plots show a steady increase in genomic diversity as each lineage spread and diverged, ultimately reaching a peak in viral diversity within weeks. This peak was followed by an equal decrease in diversity as each lineage was eliminated. Both plots also show tight confidence intervals, indicating near completeness in capturing the viral diversity in these localized outbreaks. By 13 August 2020, 2,335 cumulative COVID-19 cases had occurred in this community; thus, roughly 36% of known case viruses were sequenced. Successful SARS-CoV-2 genome recovery from specimens reflects the high viral loads observed for the WMAT cases, as indicated by low cycle threshold (CT) values obtained via real-time reverse transcription-PCR (rRT-PCR) (30). Of the 814 positive-case samples collected from this community with CT values available, 593 (73%) yielded CT values of ≤33.0 in the TG-N2 assay. Figures S1 and S2 in the supplemental material show complete phylogenetic trees for both H245Y and R209I mutations (lineages B.1.289 and B.1.516, respectively). Table S1 lists GISAID accession IDs for all sample genomes used in BEAST analyses.
Compilation of GISAID IDs linking to full sequence records and sample metadata used for Bayesian phylogenetic analyses. Download Table S1, PDF file, 0.09 MB (92.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
B.1.289 (S: H245Y) was seeded on 29 March 2020 with the tribe’s first COVID-19 case (TG655061, confirmed 1 April) (Fig. 1). This lineage expanded through April and May, then diminished in June, and presumably went extinct in July (Fig. 1; Fig. S1). The tree structure shows that B.1.289 followed a largely singular path for the month of April, where one branch from each node of the tree dead-ends at one or a minimal number of case genomes, while the other branch leads to a majority of all subsequent case genomes. These cases include four of the original five clusters that appeared to be separate unrelated outbreaks based on field epidemiology alone. In late April, the B.1.289 phylogeny split into four main sublineages comprising the May cases, and these sublineages remained in the population through June and early July.
The phylogeny of the second lineage, B.1.516 (N: R209I), shows that after introduction in early May, the virus diverged into several well-populated sublineages that continued into the late summer months (Fig. 2; Fig. S2). However, after these early bifurcations, the sublineages take on a branching structure somewhat similar to that of the B.1.289 phylogeny, where each sublineage follows a largely singular pathway. The B.1.516 lineage’s latest samples, collected in August, fall into only four small subclades which subsequently died out.
Three of the five epidemiologically linked clusters detected in April and May included several cases that provided viral genomes along with substantial epidemiological information. This population’s index case (A1) (Fig. 3) presented to the Emergency Department (ED) in late March 2020, with fever and low back pain, and was discharged on antibiotics. Despite recent travel to Phoenix, a location experiencing a high number of cases, the patient continued to work and reportedly refused to wear a mask. They re-presented at the ED 1 day later with additional symptoms, at a time when there were no known cases of COVID-19 in this community or surrounding localities. Thus, this patient was the first in the region to meet the Arizona state testing criteria then in use. A nasopharyngeal (NP) swab sample collected the same day tested positive for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by rRT-PCR at the Arizona State Public Health Laboratory (ASPHL) on 1 April (with a TG_N2 assay CT value of 30.7 on the remnant specimen). The patient was hospitalized for 6 days and fully recovered.
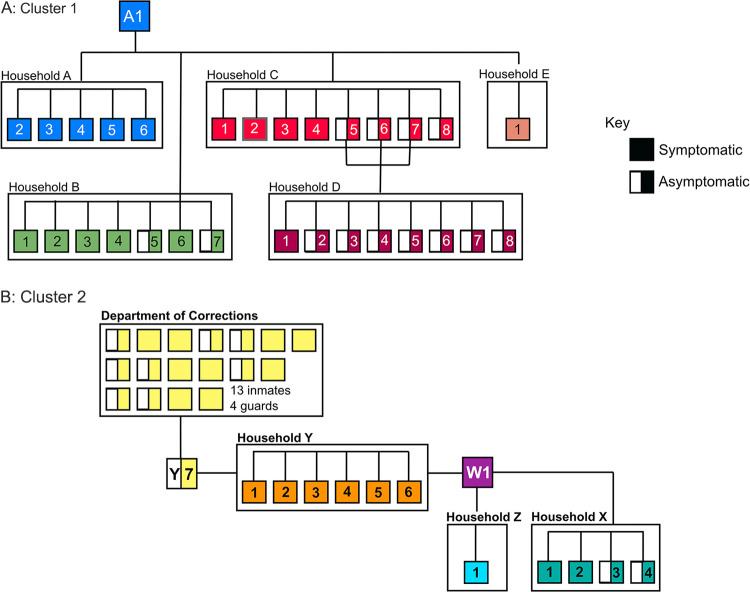
FIG 3.
Epidemiological linkages of SARS-CoV-2 cases and contacts associated with (A) cluster 1 and (B) cluster 2, identified through robust contact tracing by the White Mountain Apache Tribe public health response team. A1 (TG655061) is the index case of the B.1.289 (S:H245Y) lineage.
Patient A1 had five household (HH-A) contacts, all of whom became symptomatic and tested positive (Fig. 3). Given the high attack rate, extensive contact tracing was conducted (3, 18). Patient A1 identified family contacts living in a second household (HH-B), where seven of nine individuals tested positive with a range of reported symptoms. Two of A1’s coworkers, C1 and E1, developed mild symptoms, and C1 tested positive. In an attempt to prevent spread to the four children in HH-C (C5, C6, C7, and F1), the children were sent to stay in households HH-D and HH-F. However, the children and all but one member of HH-C subsequently tested positive. All eight members of HH-D and one of 10 members of HH-F also tested positive. A total of 49 individuals living in seven different households were evaluated and tested as part of contact tracing efforts related to cluster 1, with 32 (65%) testing positive, resulting in household attack rates of 20 to 100%.
Genomic analysis confirms that patient A1 was the index patient for the B.1.289 outbreak in the WMAT, with this virus genome (TG655061) in a basal position (Fig. 1). One virus genome (TG654694) with the S:H245Y mutation, collected in Maricopa County (outside the reservation) 2 days after the initial diagnosis of patient A1, shared an ancestor with the virus infecting patient A1. However, this genome had four additional autologous point mutations and was basal to the ingroup in a larger phylogenetic reconstruction, so it was used as the phylogenetic outgroup. No other independent instances of S:H245Y were identified in Arizona during this time period. The virus genomes from almost all cases in April and most in May from this rural community stem from A1, illustrating rapid spread of a single clone. The most recent common ancestor to this lineage (not including TG654694) was estimated to exist on 26 March, approximating the day patient A1 first transmitted the virus. SARS-CoV-2 genomes from HH-A, -B, and -D from cluster 1 were included in the phylogenetic analysis. Three of the four genomes are identical to the index case A1 genome, while one sample, TG274664 (D5), had one derived point mutation. All genomes from cluster 1 are in basal positions in the tree. This correlates with the series of events uncovered in the epidemiological investigation, including transmission by the asymptomatic children, and suggests successful isolation efforts with very limited spread except for a single line of key transmissions linking the rest of this large outbreak.
Cluster 2 was discovered when a >60-year-old tribal member (W1) presented with cough and rhinorrhea on April 13 and tested positive for SARS-CoV-2 on April 15. Contact tracing revealed numerous possible virus transmission events, including between individuals from households HH-X, -Y, and -Z, whom W1 had provided transportation to or had visited (Fig. 3). The high attack rate appeared to continue for this cluster, as three of four members of HH-X and all six members of HH-Y tested positive; the fourth member of HH-X was symptomatic but refused testing. Tracing efforts led to individual Y7, the spouse of Y1, who likely introduced the virus into HH-Y before being incarcerated April 7. Testing at the tribal department of corrections (DOC) started 16 April and revealed another 13 inmates and four guards with COVID-19. Transmission in the DOC continued the outbreak within the facility and apparently back to the community. Notably, none of the close contacts of HH-X members tested positive.
Phylogenetic evidence supports the contact tracing inferences of the viral spread and provides additional key epidemiological information. In addition to the genomic links among the individuals in cluster 2, the phylogeny also shows that cluster 2 was born out of cluster 1 and that an additional mutation, T67I in the ORF1b nonstructural replicative enzyme, occurred on or around 10 April between clusters 1 and 2 and quickly went to fixation. The first sample with this genotype (TG276187) was collected April 10, while the last sample without it was collected April 23. Our analysis also revealed that SARS-CoV-2 genomes from cluster 2 sit basal to the rest of the H245Y outbreak and that genomes from the DOC cases are interspersed with those from other cases, indicating several lines of transmission between the DOC and community. These links appear to be responsible for the change from a singular transmission path to expansion of B.1.289 into several sublineages. At least 275 cases of COVID-19 were caused by B.1.289, and 261 (95%) of those were caused by B.1.289 with Orf1b: T67I, after introduction onto the reservation.
In contrast to clusters 1 and 2, where field epidemiology initially linked cases within each cluster, rapid SARS-CoV-2 genomics first alerted the tribal public health officials to cluster 3 and to an independent introduction of a new lineage, B.1.516. This combination of patient tracking and genomic epidemiology revealed that the provenance of B.1.516 with N: R209I was around 21 April, with the congregation of the first infected individuals in a home associated with substance abuse. These cases were diagnosed 2 to 5 May, and the phylogenetic tree shows that the dispersal of these individuals and their contacts launched at least two main sublineages of B.1.516 that each spread to several members of the tribal community (Fig. 2). It is possible that the origin of B.1.516 was a correctional facility, as statewide genomic surveillance identified a sample (TG276516) collected April 14 from a correctional facility in a noncontiguous county that shared a recent ancestor with the initial cluster 3 samples. This sample was used as the phylogenetic tree’s outgroup. Additionally, one of the first case samples in the second major sublineage of B.1.516 (TG343840) (Fig. 2) was collected from a separate correctional facility. The lack of B.1.516 in other Arizona communities may counter this hypothesis of a correctional facility origin, instead suggesting introductions from members of the B.1.516 outbreak to correctional facilities (and rapid containment); in either case, the genomic linkage suggests an early connection between the individuals suffering substance abuse and incarceration. From 5 May to 13 August, at least 455 cases of this lineage were identified.
The lineages B.1.289 and B.1.516 and their defining mutations were largely specific to the WMAT, and their diagnostic real-time PCR CT values are worth noting. In >4.2 million SARS-CoV-2 global genomes in GISAID as of 31 August 2021 (~1 year after the last cases with these lineages were detected), only two instances of the B.1.289 lineage (of 400), and ten instances of the B.1.516 lineage (of 534) were found outside Arizona. We found the spike H245Y mutation in >4,400 non-Arizona genomes (including >1,300 B.1.1.7 genomes of largely British origin) and Orf1b T67I in >2,200 non-Arizona genomes (including >70 B.1.17 genomes), but these mutations did not occur together. Nucleocapsid R209I was found globally in >9,000 non-Arizona genomes of various Pango lineages, including B.1.1.222 (n = 541), B.1.560 (n = 431), and B.1.258 (n = 335). The average CT values from the WMAT (n = 1,142) throughout the study period were significantly lower than the average CT value from cases collected from other parts of Arizona (n = 1,653) during the same time period [two-sample t(df) = 3.41; P < 0.001].
DISCUSSION
COVID-19 has had significant and unprecedented impacts on human populations across the globe. In North America, NA communities have been particularly hard hit, not only in terms of morbidity and mortality (12, 16) but also in the social, cultural, and economic impacts of lockdown measures and lack of access to and delays in health care, coverage, and funding (10, 13, 31). Reservations in Arizona have been surrounded for much of the pandemic by some of the highest case rates and hospital occupancy rates in the world, which undoubtedly impacted rates in tribal communities. Early and rapid proactive measures by tribal leadership delayed the local COVID-19 epidemics and likely saved lives (3, 8, 12, 32); however, in spite of these efforts, infection rates in tribal communities described here and elsewhere eventually surged to unparalleled levels (3, 12, 13).
Through a combination of shoe-leather approaches and genomic epidemiology, we showed that only two SARS-CoV-2 viral lineages, introduced under very different circumstances approximately 1 month apart, were the cause of the multiple clusters inclusive of and exclusive to a single tribal community in Arizona. The phylogenetic patterns of both B.1.289 and B.1.516 reflect the nature of the virus introduction as well as the community’s social structure and the response by public health officials. The rapid testing by the tribe’s health care staff and the community’s unique response to a positive case (e.g., sending healthy but infected household contacts to stay at a relative’s house) may account for the singular transmission paths illustrated by the phylogenies. Initially, public health efforts were just behind the virus spread, likely preventing its expansion into multiple transmission lines. However, these efforts quickly caught up to the pace of transmission and subsequently extinguished both variants.
The introductions of B.1.289 and B.1.516 were estimated by molecular clock analysis to have occurred on 26 March and 20 April 2020, respectively, only 3 and 12 days before the first cases were reported. The tribe’s public health team used their “luxury of time” in March, when Arizona’s pandemic began but well before the tribe’s first cases, to prepare for their response (3, 22). The path of B.1.289 was largely linear from the first identified case. This case was easily visible, occurring in a health care worker and within a setting of well-informed individuals. This diagnosis came before individuals infected by this index case could spread it into a larger transmission network. In contrast, the initial cases of B.1.516 were believed to have been contracted simultaneously by congregates in a community “trap house” that then dispersed to other parts of the community, seeding several initial sublineages of this variant. It has been well documented that substance abuse and its association with a disregard for social distancing have been drivers of COVID-19 transmission (33). This hypothesis has strong merit because of the remarkable field epidemiology conducted by the tribal members and the effective population size analysis showing nearly complete viral sampling and allowing for thorough genomic epidemiology. This does not rule out possible cryptic sublineages that went undetected due to lack of sample collection (e.g., the virus spread into an unsampled host population); indeed, virus sequencing efforts significantly slowed all over the world in late summer and fall of 2020. However, subsequent comprehensive statewide SARS-CoV-2 genomic surveillance did not reveal S: H245Y and N: R209I mutations in Arizona or in the global genome database, evidencing extinction of B.1.289 and B.1.516 and refuting the possibility of a lack of sampling.
The defining mutations of these two variants, spike H245Y and Orf1b T67I in B.1.289 and nucleocapsid R209I in B.1.516, though phenotypically uncharacterized, are in regions of the SARS-CoV-2 genome considered hot spots of enhanced growth rate-associated mutations (34). Spike H245 lies in the N-terminal domain (NTD) of the protein, amid polymorphic loci associated with widespread lineages of variants of concern (e.g., A222V in Delta and D253G in Iota). Orf1b T67I falls within nsp12, the RNA-dependent RNA polymerase (RdRp) protein involved in viral replication. The mutations S202R and R203M in the linker region of the nucleocapsid protein potentially increase efficiency of SARS-CoV-2 RNA packaging and enhanced replication in lung epithelial cells (35). R209I of B.1.516 is also in this linker region. The low real-time PCR CT values and low fatality rates observed here are at least partly due to early health care interventions and aggressive contact tracing by local health care workers (3, 18), which allowed for sample collection during peak viral loads (i.e., early in the infection, which also allowed for complete genome sequencing), and the high household attack rates may be due to the nature of NA society. Whether the virus lineages’ mutation profiles might also contribute to this epidemiology or its exclusivity to a NA population remains to be established.
Transmission of COVID-19 in the community was rapid, and secondary rates of infections among households ranged from 80 to 100% (3). The low fatality rates were driven in large part by the successful, on-ground rapid test-and-treat interventions from local health workers. Despite prevention efforts, COVID-19 has permeated the landscape and successfully spread to some of the most remote communities in the United States. Taken together, the two introductions of SARS-CoV-2 into the WMAT, the confinement of each lineage to this population, and the low case fatality rate given the extreme attack rate in this community are highly noteworthy, especially given the common comorbidities and other factors predisposing NA communities to severe outcomes (3, 11, 13, 18, 21, 32, 36, 37). The relatively late introduction of COVID-19 into this region provided time for response preparations, while the involvement of tribal members in the response allowed for a more personalized approach, both of which likely limited the spread and severity of disease (1, 3). However, social circumstances unique to tribal populations in Arizona negatively impacted the public health response (3, 32); these included (i) frequent interactions among extended family members, friends, and neighbors living in close proximity, as illustrated by clusters 1 and 2, and (ii) a number of home-insecure individuals who congregate to engage in shared substance abuse activities, as illustrated by cluster 3. The rapid and aggressive contact tracing efforts and immediate clinical support activities in this community are an exemplary paradigm for public health response efforts everywhere (1, 3) and demonstrate the resilience of NA communities (13), especially given the inherent challenges in health care and response on NA reservations (10, 14, 15, 32). Ensuring highly effective field epidemiology in conjunction with rapid genomic sequencing and analysis can better prepare public health and tribal health agencies to respond effectively to future emerging pathogens.
MATERIALS AND METHODS
Ethics approval.
This work was conducted in collaboration with the tribal health department as a public health surveillance activity, involving genomic sequencing and analysis of deidentified remnant biospecimens, and therefore is exempt from needing human subject research board approval. The White Mountain Apache Tribe health board and council approved publication of this work.
Specimen collection, processing, and testing.
NP swabs were collected at health care facilities serving the WMAT or in the field during contract tracing investigations. Specimens were submitted in phosphate-buffered saline or viral transport media and tested at the TGen North Clinical Laboratory (TNCL) or ASPHL. RNA was extracted from all NP swab medium samples using the Quick Viral kit (Zymo Research). TNCL samples were run on three rRT-PCR assays to diagnose COVID-19 using Reliance one-step multiplex supermix with 450 nM primer and 150 nM probe on the CFX96 real-time PCR detection system (Bio-Rad). The three rRT-PCR assays target the SARS-CoV-2 nucleocapsid gene (TG-N2_F, TTCAGCGTTCTTCGGAATGTC; TG-N2_R, TGGCACCTGTGTAGGTCAAC; TG-N2_FAMBHQ, 6-carboxyfluorescein [FAM]-CGCATTGGCATGGAAGTCACACC-black hole quencher [BHQ]), spike gene (TG-S4_F, CCAGTTGCTGTAGTTGTCTCAAG; TG-S4_R, CTGGCTCAGAGTCGTCTTCA; TG-S4_FAMBHQ, FAM-TGTTGTTCTTGTGGATCCTGCTGC-BHQ), and human RNase P, as published by the Centers for Disease Control and Prevention (38).
SARS-CoV-2 genome sequencing and analysis.
Methods used for sample preparation and genome sequencing were described previously (19, 30). RNA was treated with DNase I (Zymo Research) and prepared for either total RNA sequencing or targeted amplicon sequencing. Total RNA was prepared with either the SmartSeq Stranded kit (TaKaRa) or the Ovation Solo kit (Nugen) without a fragmentation step and sequenced on a v2 2 × 151 high-output kit on a NextSeq system (Illumina). For targeted virus genome sequencing, cDNA was synthesized according to the nCoV-2019 sequencing protocol v2 (https://doi.org/10.17504/protocols.io.bdp7i5rn). Tiled PCR was performed with the nCov-2019/V3 primers (39) using Q5 hot start master mix (New England Biolabs [NEB]), and the amplicons were purified with Ampure XP beads (Beckman Coulter). Amplicon libraries were prepared with either Nextera Flex (Illumina) or plexWell 384 (seqWell) and sequenced with a v3 2 × 300 kit on a MiSeq system (Illumina).
Virus genome consensus sequences were built using the Amplicon Sequencing Analysis Pipeline (ASAP) as described previously (19, 30). First, adapters were trimmed from reads using bbduk (http://jgi.doe.gov/data-and-tools/bb-tools/) and mapped to the Wuhan-Hu-1 genome (GenBank accession no. MN908947) with bwa mem (40) using local alignment with soft clipping. BAM files were then processed to generate a consensus sequence and metrics on the quality of the assembly according to the following: (i) individual base calls with a quality score below 20 were discarded; (ii) remaining base calls at each position were tallied; (iii-a) if coverage was ≥10× and ≥80% of the reads agreed at a locus, a consensus call was made; (iii-b) if either of these parameters was not met, an “N” call was made; and (iv) gaps in coverage (usually the result of a missing amplicon) were filled in with a lowercase “n” to preserve alignment with the reference genome. Only consensus genomes covering at least 90% of the breadth of the reference genome with an average depth of ≥30× were used in subsequent analyses. Genomes were uploaded into GISAID (41); accession numbers are listed in Table S1.
Viruses were typed with Pangolin (Pango v.4.0.4 PLEARN-v1.3) (27). Phylogenetic analyses were conducted in Nextstrain (42), supplemented with genomes from GISAID (41) selected by the genome sampler (43) for context, and with BEAST 1.10.5 (44) to estimate the timing of relevant events. Substitution model selection was carried out with IQ-Tree (45), where GTR+F+I best fitted the data set. To determine the best-fitting clock and demographic model combinations for the data, the generalized stepping stone marginal likelihood estimator (46) was employed to compare the strict or uncorrelated lognormal (UCLN) (47) clock models combined with the exponential or Bayesian Skygrid demographic (48) models. The four model combinations were each run for 100,000,000 generations, whereas Markov chains were sampled every 10,000 generations. For both the B.1.289 and B.1.516 analyses, the combination of UCLN clock and Bayesian Skygrid outperformed the other three (Fig. S1). Using the UCLN Bayesian Skygrid model, three additional chains for 100,000,000 generations were run, with sampling every 10,000. Tracer v1.7.2 (49) was used to find convergence within and among chains. LogCombiner 1.10.4 was used to merge the four different chains, discarding the first 30% as burn in (30,000,000 generations per chain) and resampling every 30,000 generations. The resulting file from each data set was input into TreeAnnotator to produce a maximum clade credibility tree, which was visualized using FigTree v1.4.4 (50).
Data availability.
The genomic data presented are available through GISAID, a public repository (https://www.gisaid.org/). Accession IDs linking to full sequence records and sample metadata are included in Table S1, and additional data are available upon request.
ACKNOWLEDGMENTS
We acknowledge the incredible public health efforts of the White Mountain Apache Tribe, including the community health representatives and staff from the Whiteriver Indian Hospital, during the pandemic to ensure the safety and well-being of the tribal community. We also thank the Arizona State Public Health Laboratory and Translational Genomics Research Institute teams responsible for sequencing SARS-CoV-2-positive specimens and generating viral genetic sequence data. Last, we are extremely grateful to the GISAID Initiative and all its data contributors.
Funding for SARS-CoV-2 diagnostic testing, sequencing, and analysis was provided in part by the NARBHA Institute, Blue Cross and Blue Shield of Arizona, the Arizona Department of Health Services, and a grant from CORE Response to the Johns Hopkins Center for Indigenous Health.
Contributor Information
Hayley D. Yaglom, Email: hyaglom@tgen.org.
Shirit Einav, Stanford University School of Medicine.
REFERENCES
- 1.Baral SD, Mishra S, Diouf D, Phanuphak N, Dowdy D. 2020. The public health response to COVID-19: balancing precaution and unintended consequences. Ann Epidemiol 46:12–13. doi: 10.1016/j.annepidem.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuchat A, CDC COVID-19 Response Team . 2020. Public health response to the initiation and spread of pandemic COVID-19 in the United States, February 24-April 21, 2020. MMWR Morb Mortal Wkly Rep 69:551–556. doi: 10.15585/mmwr.mm6918e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Close RM, Stone MJ. 2020. Contact tracing for Native Americans in rural Arizona. N Engl J Med 383:e15. doi: 10.1056/NEJMc2023540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Worobey M, Pekar J, Larsen BB, Nelson MI, Hill V, Joy JB, Rambaut A, Suchard MA, Wertheim JO, Lemey P. 2020. The emergence of SARS-CoV-2 in Europe and North America. Science 370:564–570. doi: 10.1126/science.abc8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Messner W, Payson SE. 2021. Contextual factors and the COVID-19 outbreak rate across U.S. counties in its initial phase. Health Sci Rep 4:e242. doi: 10.1002/hsr2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallaway MS, Rigler J, Robinson S, Herrick K, Livar E, Komatsu KK, Brady S, Cunico J, Christ CM. 2020. Trends in COVID-19 incidence after implementation of mitigation measures - Arizona, January 22-August 7, 2020. MMWR Morb Mortal Wkly Rep 69:1460–1463. doi: 10.15585/mmwr.mm6940e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H. 2021. Why the Navajo Nation was hit so hard by coronavirus: understanding the disproportionate impact of the COVID-19 pandemic. Appl Geogr 134:102526. doi: 10.1016/j.apgeog.2021.102526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Humeyestewa D, Burke RM, Kaur H, Vicenti D, Jenkins R, Yatabe G, Hirschman J, Hamilton J, Fazekas K, Leslie G, Sehongva G, Honanie K, Tu’tsi E, Mayer O, Rose MA, Diallo Y, Damon S, Zilversmit Pao L, McCraw HM, Talawyma B, Herne M, Nuvangyaoma TL, Welch S, Balajee SA. 2021. COVID-19 response by the Hopi Tribe: impact of systems improvement during the first wave on the second wave of the pandemic. BMJ Glob Health 6:e005150. doi: 10.1136/bmjgh-2021-005150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins R, Burke RM, Hamilton J, Fazekas K, Humeyestewa D, Kaur H, Hirschman J, Honanie K, Herne M, Mayer O, Yatabe G, Balajee SA. 2020. Notes from the field: Development of an enhanced community-focused COVID-19 surveillance program - Hopi Tribe, June–July 2020. MMWR Morb Mortal Wkly Rep 69:1660–1661. doi: 10.15585/mmwr.mm6944a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovich H. 2020. Rural matters - Coronavirus and the Navajo Nation. N Engl J Med 383:105–107. doi: 10.1056/NEJMp2012114. [DOI] [PubMed] [Google Scholar]
- 11.Yellow HA, Yang TC, Huyser KR. 2022. Structural inequalities established the architecture for COVID-19 pandemic among Native Americans in Arizona: a geographically weighted regression perspective. J Racial Ethn Health Disparities 9:165–175. doi: 10.1007/s40615-020-00940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakol M, Upson D, Sood A. 2021. Susceptibility of southwestern American Indian tribes to coronavirus disease 2019 (COVID-19). J Rural Health 37:197–199. doi: 10.1111/jrh.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manson SM, Buchwald D. 2021. Bringing light to the darkness: COVID-19 and survivance of American Indians and Alaska Natives. Health Equity 5:59–63. doi: 10.1089/heq.2020.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirschman J, Kaur H, Honanie K, Jenkins R, Humeyestewa DA, Burke RM, Billy TM, Mayer O, Herne M, Anderson M, Bhairavabhotla R, Yatabe G, Balajee SA. 2020. A SARS-CoV-2 outbreak illustrating the challenges in limiting the spread of the virus - Hopi Tribe, May–June 2020. MMWR Morb Mortal Wkly Rep 69:1654–1659. doi: 10.15585/mmwr.mm6944a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burki T. 2021. COVID-19 among American Indians and Alaska Natives. Lancet Infect Dis 21:325–326. doi: 10.1016/S1473-3099(21)00083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arrazola J, Masiello MM, Joshi S, Dominguez AE, Poel A, Wilkie CM, Bressler JM, McLaughlin J, Kraszewski J, Komatsu KK, Peterson PX, Jespersen M, Richardson G, Lehnertz N, LeMaster P, Rust B, Keyser Metobo A, Doman B, Casey D, Kumar J, Rowell AL, Miller TK, Mannell M, Naqvi O, Wendelboe AM, Leman R, Clayton JL, Barbeau B, Rice SK, Rolland SJ, Warren-Mears V, Echo-Hawk A, Apostolou A, Landen M. 2020. COVID-19 mortality among American Indian and Alaska Native persons - 14 states, January–June 2020. MMWR Morb Mortal Wkly Rep 69:1853–1856. doi: 10.15585/mmwr.mm6949a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatcher SM, Agnew-Brune C, Anderson M, Zambrano LD, Rose CE, Jim MA, Baugher A, Liu GS, Patel SV, Evans ME, Pindyck T, Dubray CL, Rainey JJ, Chen J, Sadowski C, Winglee K, Penman-Aguilar A, Dixit A, Claw E, Parshall C, Provost E, Ayala A, Gonzalez G, Ritchey J, Davis J, Warren-Mears V, Joshi S, Weiser T, Echo-Hawk A, Dominguez A, Poel A, Duke C, Ransby I, Apostolou A, McCollum J. 2020. COVID-19 among American Indian and Alaska Native persons - 23 states, January 31–July 3, 2020. MMWR Morb Mortal Wkly Rep 69:1166–1169. doi: 10.15585/mmwr.mm6934e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stone MJ, Close RM, Jentoft CK, Pocock K, Lee-Gatewood G, Grow BI, Parker KH, Twarkins A, Nashio JT, McAuley JB. 2021. High-risk outreach for COVID-19 mortality rin an indigenous community. Am J Public Health 111:1939–1941. doi: 10.2105/AJPH.2021.306472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ladner JT, Larsen BB, Bowers JR, Hepp CM, Bolyen E, Folkerts M, Sheridan K, Pfeiffer A, Yaglom H, Lemmer D, Sahl JW, Kaelin EA, Maqsood R, Bokulich NA, Quirk G, Watts TD, Komatsu KK, Waddell V, Lim ES, Caporaso JG, Engelthaler DM, Worobey M, Keim P. 2020. An early pandemic analysis of SARS-CoV-2 population structure and dynamics in Arizona. mBio 11:e02107-20. doi: 10.1128/mBio.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sewell JL, Malasky BR, Gedney CL, Gerber TM, Brody EA, Pacheco EA, Yost D, Masden BR, Galloway JM. 2002. The increasing incidence of coronary artery disease and cardiovascular risk factors among a Southwest Native American tribe: the White Mountain Apache Heart Study. Arch Intern Med 162:1368–1372. doi: 10.1001/archinte.162.12.1368. [DOI] [PubMed] [Google Scholar]
- 21.Sutcliffe CG, Grant LR, Reid A, Douglass G, Brown LB, Kellywood K, Weatherholtz RC, Hubler R, Quintana A, Close R, McAuley JB, Santosham M, O'Brien KL, Hammitt LL. 2020. High burden of Staphylococcus aureus among Native American individuals on the White Mountain Apache tribal lands. Open Forum Infect Dis 7:ofaa061. doi: 10.1093/ofid/ofaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris A. 2021. How a local response to COVID-19 helped slow deaths on the White Mountain Apache nation. Arizona Republic, Phoenix, AZ. Retrieved 1 October 2021. https://www.azcentral.com/story/news/local/arizona-health/2021/04/04/local-response-helps-white-mountain-apache-tribe-slow-covid-19-deaths/6970608002/. [Google Scholar]
- 23.Geoghegan JL, Holmes EC. 2018. The phylogenomics of evolving virus virulence. Nat Rev Genet 19:756–769. doi: 10.1038/s41576-018-0055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Angyal A, Brown RL, Carrilero L, Green LR, Groves DC, Johnson KJ, Keeley AJ, Lindsey BB, Parsons PJ, Raza M, Rowland-Jones S, Smith N, Tucker RM, Wang D, Wyles MD. 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.E19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biswas NK, Majumder PP. 2020. Analysis of RNA sequences of 3636 SARS-CoV-2 collected from 55 countries reveals selective sweep of one virus type. Indian J Med Res 151:450–458. doi: 10.4103/ijmr.IJMR_1125_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rambaut A, Holmes EC, O’Toole Á, Hill V, McCrone JT, Ruis C, Du Plessis L, Pybus OG. 2020. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Toole A, Scher E, Underwood A, Jackson B, Hill V, McCrone JT, Colquhoun R, Ruis C, Abu-Dahab K, Taylor B, Yeats C, Du Plessis L, Maloney D, Medd N, Attwood SW, Aanensen DM, Holmes EC, Pybus OG, Rambaut A. 2021. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol 7:veab064. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao K, Tzou PL, Nouhin J, Gupta RK, de Oliveira T, Kosakovsky PS, Fera D, Shafer RW. 2021. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet 22:757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi JY, Smith DM. 2021. SARS-CoV-2 variants of concern. Yonsei Med J 62:961–968. doi: 10.3349/ymj.2021.62.11.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Folkerts ML, Lemmer D, Pfeiffer A, Vasquez D, French C, Jones A, Nguyen M, Larsen B, Porter WT, Sheridan K, Bowers JR, Engelthaler DM. 2021. Methods for sequencing the pandemic: benefits of rapid or high-throughput processing. F1000Res 10:48. doi: 10.12688/f1000research.28352.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owen MJ, Sundberg MA, Dionne J, Kosobuski AW. 2021. The impact of COVID-19 on American Indian and Alaska Native communities: a call for better relational models. Am J Public Health 111:801–803. doi: 10.2105/AJPH.2021.306219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hakim ST, Soto J, Joe G, Dotson B. 2021. Fighting the monster: how Diné College led the Navajo Nation’s response to COVID-19. Tribal College J Am Ind Higher Ed 32:4. [Google Scholar]
- 33.Taylor S, Paluszek MM, Rachor GS, McKay D, Asmundson GJG. 2021. Substance use and abuse, COVID-19-related distress, and disregard for social distancing: a network analysis. Addict Behav 114:106754. doi: 10.1016/j.addbeh.2020.106754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obermeyer F, Jankowiak M, Barkas N, Schaffner SF, Pyle JD, Yurkovetskiy L, Bosso M, Park DJ, Babadi M, MacInnis BL, Luban J, Sabeti PC, Lemieux JE. 2022. Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness. Science 376:1327–1332. doi: 10.1126/science.abm1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Syed AM, Taha TY, Tabata T, Chen IP, Ciling A, Khalid MM, Sreekumar B, Chen PY, Hayashi JM, Soczek KM, Ott M, Doudna JA. 2021. Rapid assessment of SARS-CoV-2-evolved variants using virus-like particles. Science 374:1626–1632. doi: 10.1126/science.abl6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Close RM, McAuley JB. 2020. Disparate effects of invasive group A Streptococcus on Native Americans. Emerg Infect Dis 26:1971–1977. doi: 10.3201/eid2609.181169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raifman MA, Raifman JR. 2020. Disparities in the population at risk of severe illness from COVID-19 by race/ethnicity and income. Am J Prev Med 59:137–139. doi: 10.1016/j.amepre.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu X, Wang L, Sakthivel SK, Whitaker B, Murray J, Kamili S, Lynch B, Malapati L, Burke SA, Harcourt J, Tamin A, Thornburg NJ, Villanueva JM, Lindstrom S. 2020. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tyson JR, James P, Stoddart D, Sparks N, Wickenhagen A, Hall G, Choi JH, Lapointe H, Kamelian K, Smith AD, Prystajecky N, Goodfellow I, Wilson SJ, Harrigan R, Snutch TP, Loman NJ, Quick J. 2020. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv. doi: 10.1101/2020.09.04.283077. [DOI]
- 40.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khare S, Gurry C, Freitas L, B Schultz M, Bach G, Diallo A, Akite N, Ho J, Tc Lee R, Yeo W, GISAID Core Curation Team, Maurer-Stroh S. 2021. GISAID's role in pandemic response. China CDC Wkly 3:1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolyen E, Dillon MR, Bokulich NA, Ladner JT, Larsen BB, Hepp CM, Lemmer D, Sahl JW, Sanchez A, Holdgraf C, Sewell C, Choudhury AG, Stachurski J, McKay M, Simard A, Engelthaler DM, Worobey M, Keim P, Caporaso JG. 2020. Reproducibly sampling SARS-CoV-2 genomes across time, geography, and viral diversity. F1000Res 9:657. doi: 10.12688/f1000research.24751.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol 29:2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol 4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, Suchard MA. 2013. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol Biol Evol 30:713–724. doi: 10.1093/molbev/mss265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rambaut A. 2016. FigTree v1.4.3. http://tree.bio.ed.ac.uk/software/figtree/. Retrieved 1 October 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full Bayesian phylogenetic tree of 276 virus genomes of SARS-CoV-2 lineage B.1.289 (as in Fig. 1 but with no clades collapsed), characterized by spike: H245Y and Orf1b: T67I. Download FIG S1, PDF file, 0.2 MB (184.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Full Bayesian phylogenetic tree of 456 virus genomes of SARS-CoV-2 lineage B.1.516 (as in Fig. 2 but with no clades collapsed), characterized by nucleocapsid: R209I. Download FIG S2, PDF file, 0.7 MB (678.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Compilation of GISAID IDs linking to full sequence records and sample metadata used for Bayesian phylogenetic analyses. Download Table S1, PDF file, 0.09 MB (92.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
The genomic data presented are available through GISAID, a public repository (https://www.gisaid.org/). Accession IDs linking to full sequence records and sample metadata are included in Table S1, and additional data are available upon request.