ABSTRACT
The peroxins encoded by PEX genes involved in peroxisome biogenesis play a crucial role in cellular metabolism and pathogenicity in fungi. Herein, we characterized a filamentous fungus-specific peroxin Pex14/17 in the Arthrobotrys oligospora, a representative species of nematode-trapping fungi. The deletion of AoPEX14/17 resulted in a remarkable reduction in mycelial growth, conidia yield, trap formation, and pathogenicity. Compared with the wild-type strain, the ΔAopex14/17 mutant exhibited more lipid droplet and reactive oxygen species accumulation accompanied with a significant decrease in fatty acid utilization and tolerance to oxidative stress. Transcriptomic analysis indicated that AoPEX14/17 was involved in the regulation of metabolism, genetic information processing, environmental information processing, and cellular processes. In subcellular morphology, the deletion of AoPEX14/17 resulted in a decrease in the number of cell nuclei, autophagosomes, and Woronin bodies. Metabolic profile analysis showed that AoPex14/17 affects the biosynthesis of secondary metabolites. Yeast two-hybrid assay revealed that AoPex14/17 interacted with AoPex14 but not with AoPex13. Taken together, our results suggest that Pex14/17 is the main factor for modulating growth, development, and pathogenicity in A. oligospora.
IMPORTANCE Peroxisome biogenesis genes (PEX) play an important role in growth, development, and pathogenicity in pathogenic fungi. However, the roles of PEX genes remain largely unknown in nematode-trapping (NT) fungi. Here, we provide direct evidence that AoPex14/17 regulates mycelial growth, conidiation, trap formation, autophagy, endocytosis, catalase activity, stress response to oxidants, lipid metabolism, and reactive oxygen species production. Transcriptome analysis and metabolic profile suggested that AoPex14/17 is involved in multiple cellular processes and the regulation of secondary metabolism. Therefore, our study extends the functions of PEX genes, which helps to elucidate the mechanism of organelle development and trap formation in NT fungi and lays the foundation for the development of efficient nematode biocontrol agents.
KEYWORDS: Arthrobotrys oligospora, peroxin, conidiation, trap formation, lipid droplet, reactive oxygen species, mycelial growth and development, peroxisome biogenesis
INTRODUCTION
Peroxisomes are microbodies, which are organelles enclosed by a single membrane (1). Despite the morphological structure of peroxisomes being simple, their functional diversity is unprecedented. Peroxisomes contain at least 50 kinds of enzymes, such as catalase, peroxidase, and oxidase. As a result, peroxisomes play a role in a number of metabolic processes, such as fatty acid β-oxidation, redox homeostasis, and synthesis of ether glycerolipid plasmalogens (2). Interestingly, peroxisomes have different names in different species due to the differences in their function. For example, in plants, peroxisomes are named glyoxysomes because they contain glyoxylate cycle enzymes (3). In trypanosomes, peroxisomes are called glycosomes because they are involved in the process of glycolysis (4). Furthermore, Woronin bodies (WBs), a specific kind of peroxisome found in filamentous fungi, maintain cellular integrity by shutting the septal pores in response to wounding (5). Notably, various mechanisms (ribosome read-through, differential splicing, and low-efficiency peroxisomal targeting signals) have been demonstrated to achieve partial peroxisomal targeting of glycolytic enzymes in fungi, which contributes to further understanding of the peroxisomal metabolic processes (6). Moreover, proteomic analysis has previously substantially increased our understanding of peroxisomes in some model species, such as plants, yeasts, and mammals (7). However, the distribution and functions of peroxisomes in most filamentous fungi are still poorly understood.
Peroxins are proteins involved in peroxisome biogenesis, and the genes that encode them are known as PEX genes (8). Recent advances have identified more than 30 PEX genes as crucial factors in peroxisome biogenesis in a variety of species, from yeast to humans (2). Peroxisomal matrix proteins (PMPs) are synthesized on free polyribosomes in the cytosol and posttranslationally imported into organelles (9). PMPs are delivered to peroxisomes via the following two peroxisomal targeting signals (PTSs): type1 (PTS1) and type2 (PTS2). Specific PTS receptors, Pex5 or Pex7, recognize matrix proteins containing PTS1 or PTS2 signals in the cytosol (10). The N terminus of Pex5 can mediate the cargo of the PTS1 sequence through interaction with the docking complex (Pex13, Pex14, and Pex17) (11–13). In addition, Pex18 and Pex21 interact with Pex7 and are key components for targeting the peroxisome in Saccharomyces cerevisiae (14), Pex20 in filamentous fungi (15), and the splice variant Pex5L in plants and mammals (16). Interestingly, the docking complex can also be composed of Pex13, Pex14, and Pex14/17, instead of Pex17 in filamentous fungi (17). Pex14/17, also known as Pex33, distributes in the peroxisomal membrane and interacts with Pex5 and Pex14 (18). The amino acid sequence of the N terminus of Pex14/17 is similar to that of Pex14, whereas its C terminus is similar to Pex17 (19). After docking, the receptor-cargo complex is transferred to the peroxisome matrix, and finally the target protein is released and recycled by the receptor for reuse (20). Recent studies have identified Pex14/17 as peroxin found only in filamentous fungi (21). In Penicillium chrysogenum, the loss of PEX14/17 affects conidia formation, the penicillin biosynthesis process, and the matrix protein import (19). In Podospora anserina, docked peroxins Pex14 and Pex14/17 are involved in peroxisomal import in different forms (22). In the rice blast fungus Magnaporthe oryzae, the distribution of MoPex14/17 on the peroxisomal membrane is critical for the peroxisomal targeting of PTS1-containing proteins (21). At present, the roles of Pex14/17 in filamentous fungi remain largely unknown.
Nematode-trapping (NT) fungi are a group of filamentous species that use specialized trapping devices (traps), such as constricting rings, adhesive networks, and adhesive knobs to capture, kill, and consume nematodes (23, 24). Therefore, NT fungi can act as natural antagonists of nematodes (21). Arthrobotrys oligospora is the first NT fungus whose genome has been sequenced; it can capture nematodes by producing adhesive networks (25). Previous studies revealed that the cells of the mature trap are generally crowded with “special” microbodies, called electron-dense (ED) bodies, that were not found in normal vegetative hyphae (26). Subsequently, cytochemical staining showed that EDs contained catalase and d-amino acid oxidase, thus EDs were considered as peroxisomal in nature (26). Recently, increasing numbers of studies have suggested that heterotrimeric G-proteins (G-protein) and related signals play an indispensable role in asexual development and lifestyle transition (25), and several autophagy-related genes are also involved in the trap formation of A. oligospora, such as ATG5 (27), and ATG11 (28). Recently, two PEX genes, PEX1 and PEX6, encoding an AAA-type ATPase, were identified in A. oligospora, and the absence of traps, conidia, and peroxisomes was caused by the loss of PEX1 and PEX6 (29). Thus, peroxisomes are functionally related to mycelium development and trap formation, but the functions of most PEX genes in NT fungi remain unclear.
In the current study, we identified a filamentous fungus-specific peroxin Pex14/17 (AoPex14/17) in A. oligospora via gene disruption and phenotypic comparison, and the influence of AoPex14/17 on global changes in gene transcription levels was analyzed by RNA sequencing. Meanwhile, the regulation of AoPex14/17 in the secondary metabolism was determined by the metabolome. Our analysis shows that AoPex14/17 is required for trap formation and regulates a variety of phenotypic features, including mycelial development, conidiation, fatty acid utilization, and stress response.
RESULTS
Identification of AoPEX14/17 in A. oligospora.
The compute pI/MW tool of the ExPASy server (http://web.expasy.org) predicted that the AoPEX14/17 encodes a polypeptide of 456 amino acid residues, with a predicted molecular mass of 49.27 kDa and an isoelectric point of 4.78, and AoPex14/17 contained one conserved domain of Pex14_N (IPR006785). AoPex14/17 showed a high similarity (71.83 to 95.14%) to the orthologs of four other NT fungal species, including Arthrobotrys flagrans, Dactylellina haptotyla, Drechslerella stenobrocha, and Drechslerella brochopaga, and it has a moderate similarity to the orthologs of Aspergillus nidulans (44.44%) and other filamentous fungi (29.52 to 27.70%), such as Metarhizium robertsii, Fusarium graminearum, and Aspergillus fumigatus. As expected, Pex14/17 orthologs from NT fungi clustered into an independent branch (see Fig. S1A in the supplemental material).
Neighbor-joining phylogenetic tree based on the amino acid sequence of Pex14/17 homologous proteins from different fungi and AoPEX14/17 knockout and verification in A. oligospora. (A) Phylogenetic tree. GenBank accession numbers are provided in brackets. Numbers below nodes indicate the bootstrap value. The bar marker indicates the genetic distance, which is proportional to the number. (B) Diagrammatic sketch of homologous recombination. WT, wild-type strain; MT, mutant. (C) The homologous recombinant AoPEX14/17 deletion plasmid was transformed into yeast. (D) AoPEX14/17 deletion transformants (ΔAopex14/17) were confirmed by PCR method. 2K, DNA marker. (E) Southern blotting analysis of wild-type and transformants. Download FIG S1, TIF file, 2.1 MB (2.1MB, tif) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Disruption of AoPEX14/17.
The targeted gene deletion mutants were generated by the homologous recombination method (Fig. S1B). Approximately 60 putative deletion mutants of AoPEX14/17 were selected on potato-dextrose-agar-sucrose (PDAS) regeneration medium containing hygromycin and identified by genomic PCR and Southern blotting analyses (Fig. S1C to E). Three of the confirmed mutants, ΔAopex14/17-2, ΔAopex14/17-3, and ΔAopex14/17-7, were selected for phenotypic analysis.
Transcriptomic insights into the regulatory role of AoPEX14/17.
The wild-type (WT) and ΔAopex14/17 strains were inoculated in PD broth for 5 and 7 days, and then we collected them and performed comparative RNA-seq analysis. A total of about 42.58 to 58.27 million reads were obtained per sample (see Table S1 in the supplemental material). The quality control data show that the base error rates were both less than 0.03%, while the Q20 was greater than 97.72%, the Q30 was greater than 93.62%, and the GC content was greater than 47.57% (Table S1). Principal-component analysis (PCA) suggested the gene expression patterns of WT and mutant strains were distributed in various quadrants at different time points with high similarity, indicating consistency between biological replicates (see Fig. S2A in the supplemental material). The accuracy of the transcriptome data was verified by reverse transcription-quantitative PCR (RT-qPCR). Twenty genes associated with the mitogen-activated protein kinase (MAPK) signaling pathway, glycolysis/gluconeogenesis, and ubiquitin-mediated proteolysis were selected for validation, and the results show that the RT-qPCR results were consistent with transcriptome clustering results (Fig. S2B to D).
Principal component analysis (PCA) and reverse transcription-qualitative PCR (RT-qPCR) was used to verify the transcriptome data between the WT and ΔAopex14/17 mutant strains at 5 days and 7 days. (A) Principal component analysis. Different shapes and colors represent the WT and ΔAopex14/17 mutant strains at 5 days and 7 days. (B to D) Comparison of transcription analysis of genes involved in MAPK signaling pathway (B), glycolysis/gluconeogenesis (C), and ubiquitin-mediated proteolysis (D). Download FIG S2, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Statistics of reads and mapping rate for the wild-type (WT) strain (A) and ΔAopex14/17 (H) mutant strains at different time points and statistics of reads, phred-like quality scores, and GC content for the wild-type (WT) strain (A) and ΔAopex14/17 (H) mutant at different time points. Download Table S1, DOCX file, 0.02 MB (16.7KB, docx) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
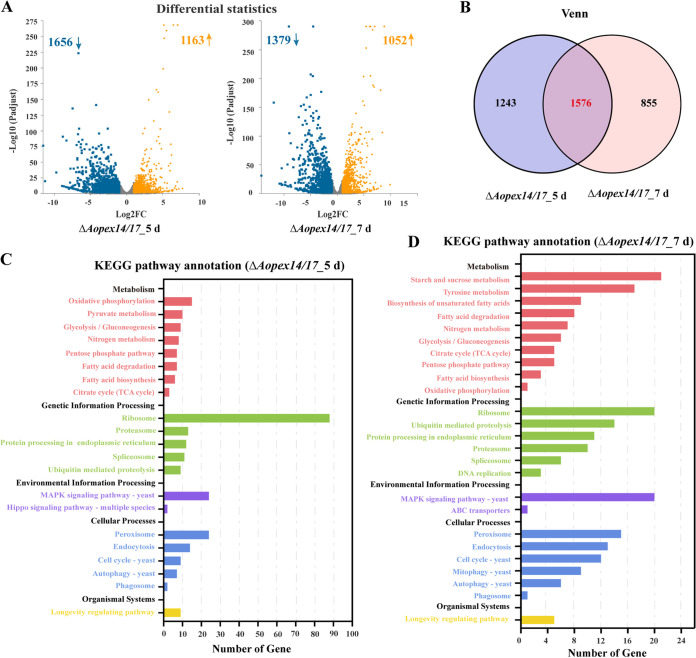
Compared with the WT strain, 2,819 and 2,431 differentially expressed genes (DEGs) were enriched in the ΔAopex14/17 strain at 5 and 7 days, respectively. The numbers of upregulated DEGs were 1,163 and 1,052, and those downregulated DEGs were 1,656 and 1,379, respectively (Fig. 1A); Venn analysis displayed that 1,576 DEGs were shared at 5 and 7 days in the ΔAopex14/17 strains (Fig. 1B). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that the DEGs involved in the metabolic pathways were divided into the following five categories: metabolism, genetic information processing, environmental information processing, cellular processes, and organismal systems. Among them, the highest proportion of pathways belonged to the metabolism category. The enrichment results were highly similar between day 5 and day 7 (Fig. 1C and D), with regard to oxidative phosphorylation, glycolysis/gluconeogenesis, nitrogen metabolism, pentose phosphate pathway, fatty acid degradation, fatty acid biosynthesis, citrate cycle (tricarboxylic acid [TCA] cycle), ribosome, proteasome, protein processing in the endoplasmic reticulum, spliceosome, ubiquitin-mediated proteolysis, the MAPK signaling pathway, peroxisome, endocytosis, cell cycle, autophagy, phagosome, and the longevity-regulating pathway. The difference was that pyruvate metabolism in the metabolism category and hippo signaling pathway—multiple species in the environmental information processing category were enriched at 5 days, while starch and sucrose metabolism, tyrosine metabolism and biosynthesis of unsaturated fatty acids, DNA replication in the genetic information processing category, and ABC transporters and mitophagy in the cellular processes category were enriched at 7 days (Fig. 1C and D).
FIG 1.
Transcription analysis of wild-type (WT) and ΔAopex14/17 mutant strains. (A) Differentially expressed genes (DEGs) on days 5 and 7. Yellow, upregulated DEGs; blue, downregulated DEGs. (B) Venn diagram analysis of DEGs in ΔAopex14/17 mutant versus WT strain at 5 and 7 days. (C) KEGG analysis of ΔAopex14/17 mutant versus WT strain at 5 days. (D) KEGG analysis of ΔAopex14/17 mutant versus WT strain at 7 days. The x axis represents the number of genes, and the y axis represents the KEGG pathway.
GO enrichment analysis of the DEGs was performed. On the 5th day, the most enriched GO term was biological process (BP), and the top three items of BP were cellular metabolic process (294 unigenes), biosynthetic process (175 unigenes), and oxidation-reduction process (99 unigenes); in the cellular component (CC) category, the top three items were intrinsic component of membrane (384 unigenes), non-membrane-bounded organelle (84 unigenes), and ribosome (72 unigenes); in the molecular function (MF) category, the top items were oxidoreductase activity (101 unigenes), transporter activity (61 unigenes), and RNA binding (41 unigenes) (see Fig. S3A and B in the supplemental material). On the 7th day, in the BP category, the most intrinsic items were metabolic process (303 unigenes), oxidation-reduction process (85 unigenes), and transmembrane transport (65 unigenes); the DEGs enriched in the CC category on day 7 were similar to the membrane intrinsic component (293 unigenes) on day 5; in the MF category, the top three items were catalytic activity (299 unigenes), oxidoreductase activity (87 unigenes), and transporter activity (51 unigenes) (Fig. S3C and D).
Gene Ontology (GO) enrichment of differentially expressed genes (DEGs) between the WT and ΔAopex14/17 mutant strains at 5 days and 7 days. (A, B) GO enrichment analysis of upregulation DEGs (A) and downregulation DEGs (B) in the ΔAopex14/17 mutant versus WT strain at 5 days. (C, D) GO enrichment analysis of upregulation DEGs (C) and downregulation DEGs (D) in the ΔAopex14/17 mutant versus WT strain at 7 days. Download FIG S3, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
AoPEX14/17 regulates vegetative growth and hyphal morphogenesis.
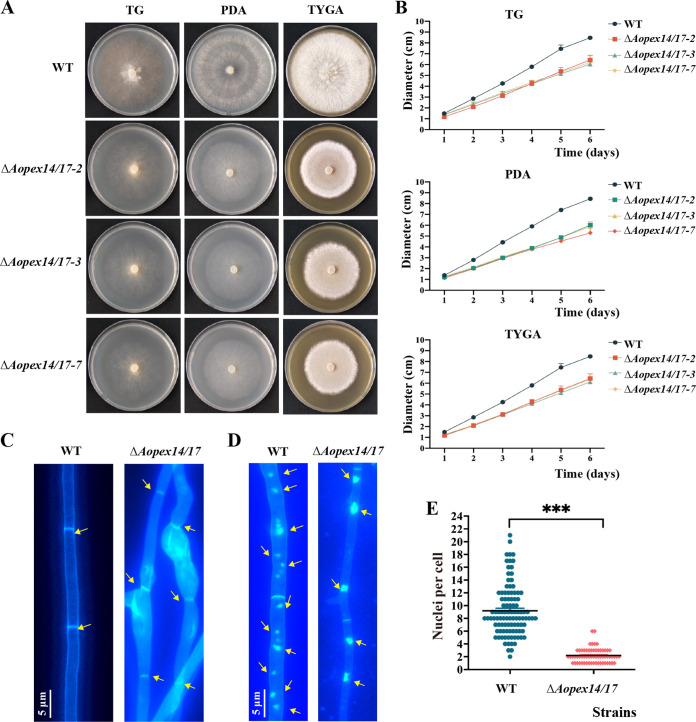
To characterize the effect of Aopex14/17 on the mycelium growth and differentiation in A. oligospora, we inoculated WT and ΔAopex14/17 mutant strains on tryptone-glucose (TG), potato dextrose agar (PDA), and tryptone yeast-extract glucose agar (TYGA) media, respectively, and compared their hyphal growth and colony morphology at 6 days postinoculation. Compared with that of the WT strain, the ΔAopex14/17 mutant showed a significant decreased radial growth on all three media (Fig. 2A and B). With regard to the defects in radial growth, we visualized the mycelia septa and morphology using calcofluor white (CFW) staining. The results indicate that the mycelia septum in the ΔAopex14/17 mutant was not significantly different from that observed in the WT strain, and partial hyphal cells of the mutant became enlarged (Fig. 2C). Furthermore, the hyphal cells of the WT strain contained 8 to 10 nuclei, whereas the mutants contained 2 to 3 nuclei (Fig. 2D and E).
FIG 2.
The role of ΔAopex14/17 in hyphal growth and morphogenesis. (A) Colony morphology of the WT and ΔAopex14/17 strains grown on TG, PDA, and TYGA, respectively. (B) Comparison of colony diameter of the WT and ΔAopex14/17 strains. (C) Hyphal septa of the WT and ΔAopex14/17 strains were stained with 20 μg/mL calcofluor white (CFW), and the fungal strains were grown for 7 days on CMY medium. Yellow arrows, hyphal septa. Scale bar, 5 μm. (D) The hyphae of the WT and ΔAopex14/17 strains were stained with CFW and DAPI, and the fungal strains were grown for 7 days on CMY medium. Yellow arrows, cell nucleus. Scale bar, 5 μm. (E) Comparison of the number of mycelial nuclei between the WT and ΔAopex14/17 strains. One hundred hyphal cells were randomly selected for counting cell nuclei. The asterisk indicates a significant difference between the mutant and the WT strains (***, P < 0.001).
AoPEX14/17 is required for conidiation, trap formation, and pathogenicity.
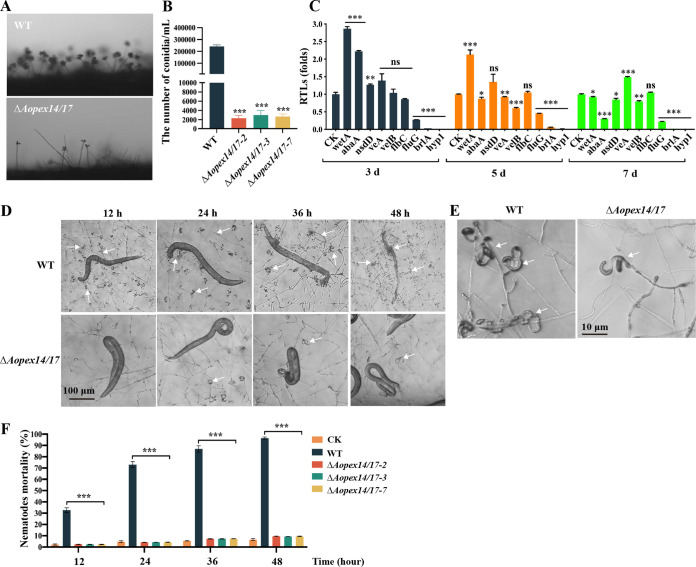
The ΔAopex14/17 mutant produced approximately 2.0 × 103 to 4.0 × 103 conidia/mL, which is only 1% of that produced by the WT strain (Fig. 3A and B). The RT-qPCR results demonstrate that the expression levels of several genes involved in conidiation from filamentous fungi, including wetA, abaA, velB, fluG, brlA, and hyp1, were significantly downregulated in the ΔAopex14/17 mutant (Fig. 3C). Moreover, we assessed the ability of WT and ΔAopex14/17 strains to produce traps by inducing with nematodes (Caenorhabditis elegans) to water agar (WA) plates. Our results demonstrate that the ΔAopex14/17 mutant was unable to produce mature traps (Fig. 3D). In the ΔAopex14/17 mutant, half rings or one ring appeared at 12 h, one and one-half ring appeared at 24 h, two rings appeared at 36 h, and three rings appeared at 48 h, but no mature traps were formed until 5 days (Fig. 3D and E). Additionally, the nematode mortality of the WT strain was as high as 96% at 48 h, whereas the nematode mortality of the ΔAopex14/17 strain was less than 10% at each time point (Fig. 3F). Thus, deletion of the AoPEX14/17 gene significantly reduced the ability of the strain to infect nematodes.
FIG 3.
Comparison of conidiation and trap formation between the WT and ΔAopex14/17 mutant strains. (A) Observation of conidiophores in the WT and mutant strains. (B) Comparison of conidia yields in the WT and mutant strains. (C) The relative transcription levels (RTLs) of the sporulation-related genes in the WT and ΔAopex14/17 mutant strains. (D) Trap formation and nematode predation at different time points. White arrows, traps. Scale bar, 100 μm. (E) Traps produced by the WT and ΔAopex14/17 mutant strains at 48 h postinduction. Scale bar, 10 μm. (F) The percentage of nematode mortality in the WT and mutant lines at different time points. CK is the negative control using the WA plate to assess the viability of the C. elegans in the absence of fungal strains (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
AoPEX14/17 contributes to stress tolerance.
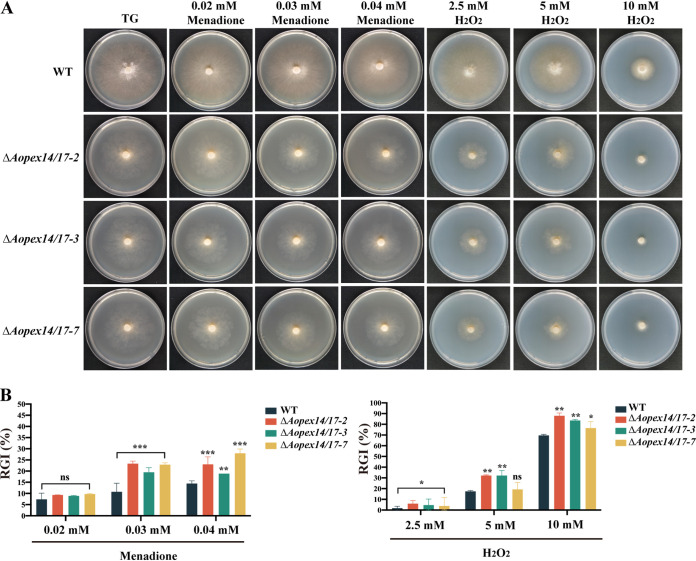
To determine the function of Aopex14/17 in stress response, we treated strains to compounds that stimulate oxidative (menadione and H2O2), cell wall (Congo red [CR] and sodium dodecyl sulfate [SDS]), and osmotic stress (NaCl and sorbitol). The ΔAopex14/17 strain was sensitive to high concentrations of menadione and H2O2, and the relative growth inhibition (RGI) values were significantly higher than those of the WT strain (Fig. 4A and B). In the transcriptome analysis, eight genes involved in oxidative stress response were significantly decreased on day 5 and day 7, including cat (AOL_s00173g374 and AOL_s00188g243), cat-2 (AOL_s00006g411 and AOL_s00076g488), nox-1, nox-2, sod, sodB, and sod-2 (Table 1).
FIG 4.
Comparison of stress tolerance to oxidative agents between the WT and ΔAopex14/17 mutant strains. (A) Colony morphologies of the WT and mutant strains incubated on TG medium supplemented with 0.02 to 0.04 mM menadione and 2.5 to 10 mM H2O2 for 6 days. (B) Relative growth inhibition (RGI) values of the WT and mutant strains incubated on TG medium supplemented with 0.02 to 0.04 mM menadione and 2.5 to 10 mM H2O2 for 6 days. The asterisk indicates a significant difference between the mutant and the WT strains (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
TABLE 1.
Transcriptional response to Aopex14/17 deletion by the genes involved in oxidation stress response and cell wall biosynthesis in comparative transcriptome analysis
| Function/locus | Function annotation | Expressional levela |
|||
|---|---|---|---|---|---|
| TPM (5 days) |
TPM (7 days) |
||||
| WT | ΔAopex14/17 strain | WT | ΔAopex14/17 strain | ||
| Genes involved in oxidation stress response | |||||
| AOL_s00173g374 | cat, catalase | 508.06 | 144.38 | 922.10 | 374.65 |
| AOL_s00188g243 | cat, catalase | 0 | 0.08 | 0 | 0.013 |
| AOL_s00006g411 | cat2, catalase | 383.91 | 60.26 | 14.17 | 15.47 |
| AOL_s00076g488 | cat2, catalase | 14.81 | 7.13 | 105.93 | 61.40 |
| AOL_s00193g69 | nox-1, NADPH oxidase | 166.42 | 52.987 | 485.51 | 43.85 |
| AOL_s00007g557 | nox-2, NADPH oxidase | 70.64 | 37.45 | 51.46 | 22.41 |
| AOL_s00054g538 | noxR, NADPH oxidase regulator | 2.68 | 6.56 | 3.81 | 11.30 |
| AOL_s00007g292 | sod, superoxide dismutase | 88.75 | 46.85 | 165.99 | 121.24 |
| AOL_s00054g687 | sodB, superoxide dismutase | 608.14 | 180.58 | 794.69 | 281.04 |
| AOL_s00170g93 | sod-2, superoxide dismutase | 8.62 | 5.65 | 5.18 | 2.27 |
| Genes involved in cell wall biosynthesis | |||||
| AOL_s00078g76 | chs-3, chitin synthases | 93.29 | 40.57 | 126.43 | 61.53 |
| AOL_s00112g89 | hex, hexokinase | 119.19 | 244.53 | 178.62 | 255.77 |
| AOL_s00075g119 | chi, chitin synthase | 66.42 | 4.86 | 28.14 | 8.70 |
| AOL_s00097g268 | trs, trehalose synthase | 246.41 | 191.29 | 156.55 | 65.34 |
| AOL_s00083g375 | glu, beta-glucosidase | 8.20 | 27.44 | 9.65 | 37.66 |
| AOL_s00054g491 | gls, 1,3-beta-glucan synthase | 184.01 | 66.87 | 245.77 | 71.90 |
TPM, transcripts per kilobase million; WT, wild-type strain; ΔAopex14/17, AoPEX14/17 deletion mutant; 5 days and 7 days, vegetative growth stage. Locus numbers and function were annotated according to the A. oligospora genome assembly (https://www.ncbi.nlm.nih.gov/).
Moreover, the RGI value of the ΔAopex14/17 mutant was higher than that of the WT strain under stress of SDS (0.01-0.03%), and the ΔAopex14/17 mutant showed a decreased RGI value compared to the WT strain at 0.1 mg/L CR (see Fig. S4A and B in the supplemental material). Similarly, the ΔAopex14/17 mutant showed a higher RGI value than the WT strain at 0.3 M NaCl, whereas no obvious defects in stress responses to sorbitol were observed (Fig. S4C and D). We also examined the expressions of genes involved in cell wall biosynthesis in the transcriptome; except for hex and glu, the expression levels of other genes were significantly decreased on day 5 and day 7, including chs-3, chs, trs, and gls (Table 1).
Comparison of stress tolerance to cell wall-perturbing agents and osmotic agents between the WT and ΔAopex14/17 mutant strains. (A) Colony morphologies of the WT and mutant strains incubated on TG medium supplemented with 0.05 to 0.1 mg/mL Congo red and 0.01 to 0.03% SDS for 6 days. (B) Relative growth inhibition (RGI) values of the WT and mutant strains incubated on TG medium supplemented with 0.05 to 0.1 mg/mL Congo red and 0.01 to 0.03% SDS for 6 days. (C) Colony morphologies of the WT and mutant strains incubated on TG medium supplemented with 0.10 to 0.30 M NaCl and 0.25 to 0.75 M sorbitol for 6 days. (D) Relative growth inhibition (RGI) values of the WT and mutant strains incubated on TG medium supplemented with 0.10 to 0.30 M NaCl and 0.25 to 0.75 M sorbitol for 6 days. The asterisk indicates a significant difference between the mutant and the WT strains (*, P < 0.05; **, P < 0.01). Download FIG S4, TIF file, 7.6 MB (7.6MB, tif) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
AoPEX14/17 regulates intracellular autophagy, ROS accumulation, and endocytosis.
Monodansylcadaverine (MDC) staining was used to monitor the autophagy process, and autophagosomes in hyphae displayed blue fluorescence after MDC staining (30). Compared with that of WT, the ΔAopex14/17 strain showed an increased accumulation of autophagosomes (Fig. 5A). Accordingly, 10 and 17 genes related to autophagy were differentially expressed at days 5 and 7, respectively, and three genes encoding cell division control protein Cdc48 (AOL_s00004g527) and serine protease (AOL_s00170g103 and AOL_s00176g50) were upregulated in the ΔAopex14/17 strain at both day 5 and day 7 by transcriptome analysis (see Fig. S5A in the supplemental material).
FIG 5.
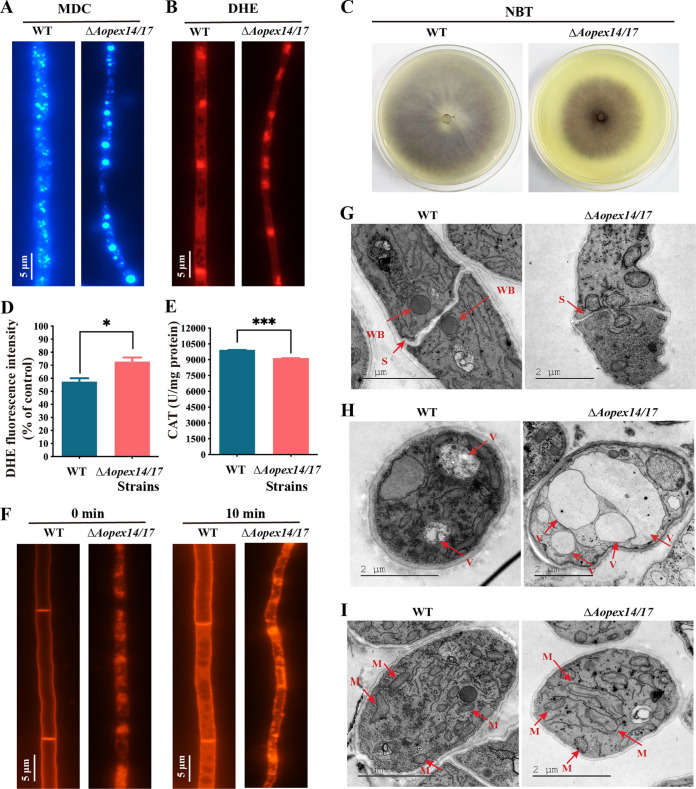
Observation of intracellular autophagosomes, ROS accumulation, endocytosis, Woronin bodies, and vacuoles between the WT and ΔAopex14/17 mutant strains. (A) The autophagosomes of the WT and ΔAopex14/17 mutant strains were stained with monodansylcadaverine (MDC). Scale bar, 5 μm. (B) Representative images of dihydroethidium (DHE) staining to detect superoxide in the WT and ΔAopex14/17 mutant strains. Scale bar, 5 μm. (C) Nitroblue tetrazolium (NBT) staining for ROS production in mycelia of the WT and ΔAopex14/17 mutant strains. The dark color of the colony indicates increased ROS production. (D) Analysis of the DHE fluorescence intensity of the same weight of mycelium in the WT and ΔAopex14/17 mutant strains via multimode microplate reader. (E) Catalase (CAT) activity assay of the same weight of mycelium in the WT and ΔAopex14/17 mutant strains via multimode microplate reader. (F) FM4-64 staining to observe endocytosis in the WT and ΔAopex14/17 mutant strains at different times. Scale bar, 5 μm. (G to I) The WT and ΔAopex14/17 mutant strains were observed by transmission electron microscopy. WB, Woronin body; S, septum; V, vacuoles; M, mitochondrion. Scale bar, 2 μm.
Differentially expressed genes (DEGs) associated with autophagy and peroxisome. Download FIG S5, TIF file, 1.2 MB (1.2MB, tif) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
After staining with dihydroethidium (DHE) and nitrotetrazolium blue chloride (NBT), the hyphae of the ΔAopex14/17 strain were more intensely stained by NBT than the WT strain, indicating greater reactive oxygen species (ROS) accumulation in the ΔAopex14/17 strain (Fig. 5B and C). Moreover, the fluorescence intensity levels of DHE in the WT and ΔAopex14/17 strains were 57.5 and 69.5, respectively (Fig. 5D). Similarly, we determined catalase (CAT) activity in the WT and ΔAopex14/17 strains. The results show that the activity of CAT was reduced in the ΔAopex14/17 strain (Fig. 5E). This is consistent with our transcriptome analysis of the significantly reduced expression levels of CAT activity-related genes cat and cat-2 (Table 1).
To clarify the role of AoPEX14/17 in endocytosis, we stained the hyphal cells of WT and ΔAopex14/17 strains with FM4-64 and followed the uptake of the dye over time. In the WT strain, the dye first appeared on the plasma membrane and was taken up by the cells after 10 min. In contrast, in the ΔAopex14/17 strain, the dye uptake was accelerated and localized to the cytoplasm, which may be mature endosomes or vacuoles (Fig. 5F).
AoPEX14/17 is required for the formation of WBs and impairs biogenesis of other organelles.
Transmission electron microscopy (TEM) revealed that WBs were observed near the septa in the hyphae of the WT, whereas they were not observed in ΔAopex14/17 hyphae (Fig. 5G), and the hyphal septum of the ΔAopex14/17 strain was broken up, consistent with the role of WBs that prevent the multiple organelles and intracellular substance from effluxing (Fig. 5G). We also found that the volume and number of vacuoles were significantly increased in the ΔAopex14/17 strain compared with those of the WT strain (Fig. 5H). However, the number of mitochondria and mitochondrial cristae decreased significantly, and the mitochondrial shape became longer, wider, and vacuolated in the ΔAopex14/17 hyphae (Fig. 5I).
Moreover, we performed cluster analysis of peroxisome-related genes. The expression levels of five genes associated with peroxisome biosynthesis were upregulated both at day 5 and day 7, including peroxisome membrane proteins (Pex4, AOL_s00078g269), Pmp47 (AOL_s00215g205), MPV17 (AOL_s00215g316), peroxin-2 (Pex2, AOL_s00080g243), peroxisomal targeting receptor (Pex5, AOL_s00043g671), and four genes relate to fatty acid degradation, including enoyl-coenzyme A (enoyl-CoA) hydratase (AOL_s00215g56 and AOL_s00043g730), acetyl-CoA acyltransferase (AOL_s00210g122), and carnitine O-acetyltransferase (AOL_s00109g5). In addition, four genes were downregulated at 5 and 7 days, namely, integral membrane protein (AOL_s00006g182), superoxide dismutase (SodB, AOL_s00054g687), and sarcosine oxidase (AOL_s00117g57), and we also clustered a marked decrease in the expression level of AoPEX14/17, consistent with which Aopex14/17 is knocked out (Fig. S5B).
AoPEX14/17 deletion led to defects in lipid metabolism.
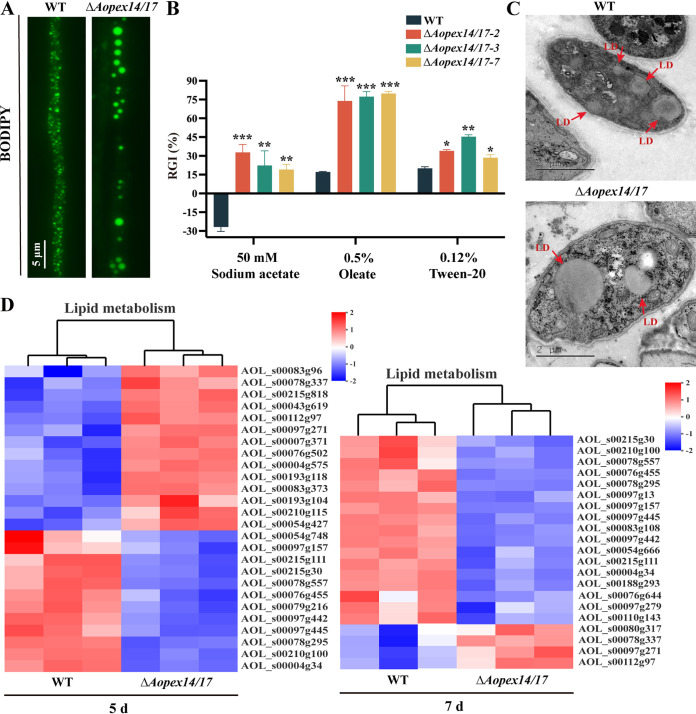
Peroxisome plays a significant role in fatty acid β-oxidation (31). We investigated whether AoPEX14/17 affects fatty acid level by staining with the neutral lipid-specific fluorescent dye boron dipyrromethene (BODIPY). The number of lipid droplets (LDs) in the ΔAopex14/17 strain was reduced, but the volume was increased compared to that of the WT strain (Fig. 6A). To investigate the role of AoPEX14/17 in fatty acid utilization, the WT and ΔAopex14/17 mutant strains were inoculated on MM plates containing short-chain (sodium acetate), medium-chain (Tween 20), or long-chain (oleate) fatty acids as the sole carbon source, respectively (Fig. 6B). After 6 days, the growth rate of the ΔAopex14/17 strain was remarkably reduced on all three plates compared to the WT strain. The RGI values of the ΔAopex14/17 strain on plates supplemented with all three fatty acids were markedly increased (Fig. 6B). Additionally, we also observed larger LDs in the hyphae of the ΔAopex14/17 strain by TEM (Fig. 6C).
FIG 6.
Comparison of lipid metabolism between the WT and ΔAopex14/17 mutant strains. (A) The LDs of the WT and ΔAopex14/17 mutant strains were stained with BODIPY. Scale bar, 5 μm. (B) Relative growth inhibition (RGI) values of fungal strains under different fatty acids as the only carbon source. (C) The lipid droplets (LDs) of the WT and ΔAopex14/17 mutant strains were observed by transmission electron microscopy. Scale bar, 2 μm. (D) Differentially expressed genes (DEGs) associated with lipid metabolism (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Moreover, we conducted a transcriptome clustering analysis of the genes involved in lipid metabolism. The results show that nine downregulated genes were shared at day 5 and day 7, including glycerophosphocholine phosphodiesterase (AOL_s00097g157), ceramide glucosyltransferase (AOL_s00215g111), phospholipase D (PldA) (AOL_s00215g30), 1-alkyl-2-acetylglycerophosphocholine esterase (AOL_s00078g557), alkaline ceramidase (AOL_s00076g455), ethanolamine phosphotransferase (AOL_s00079g216), choline kinase (AOL_s00097g442), auxin efflux carrier superfamily protein (AOL_s00097g445), ethanolamine kinase (AOL_s00078g295), phospholipase A2 (PlaA) (AOL_s00210g100), and hypothetical protein (AOL_s00004g34) (Fig. 6D).
AoPEX14/17 involvement in the regulation of the biosynthesis of secondary metabolites.
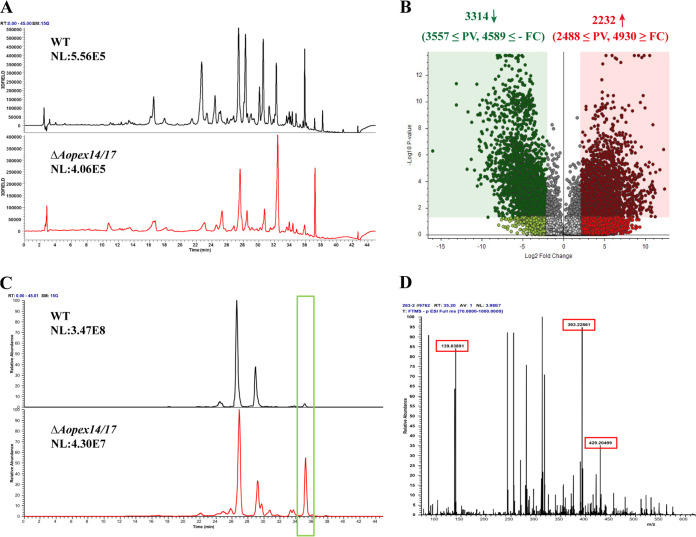
High-performance liquid chromatography (HPLC) results indicate that the metabolic abundance produced by the ΔAopex14/17 strain was significantly less than that of the WT strain, and the differential peaks were mainly concentrated at retention times (Rt) of 20 to 38 min (Fig. 7A). There were 2,232 upregulated and 3,314 downregulated compounds in the ΔAopex14/17 mutant compared with those of the WT (Fig. 7B; see also Fig. S6A and B in the supplemental material), and the top 20 differential compounds are listed in Table S2 in the supplemental material. KEGG analysis of the differential metabolic pathways between WT and ΔAopex14/17 strains showed that the upregulated pathways were involved in lipoxygenase, trichothecene biosynthesis, scopolin and esculin biosynthesis, aromatic compounds, etc. (Fig. S6C and Table S2), and the downregulated pathways mainly included aerobic toluene degradation, steroid hormone biosynthesis, rosmarinic acid degradation biosynthesis, and scopolin and esculin biosynthesis (Fig. S6D and Table S2). Ultraperformance liquid chromatography with diode array detection mass spectrometry (UPLC-DAD/MS) spectral comparison showed that the ion peak at Rt 35 min was much higher in ΔAopex14/17 strains than in WT (Fig. 7C). MS analysis of this peak revealed a diagnosis of arthrobotrisins, a specific metabolite of A. oligospora, at m/z 139, 393, and 429 (Fig. 7D).
FIG 7.
Comparison of metabolic profiling between the WT and ΔAopex14/17 mutant strains. (A) Comparison of high-performance liquid chromatography profiles of the WT and ΔAopex14/17 mutant strains. (B) Volcano plot of differential metabolites between the WT and ΔAopex14/17 mutant strains. (C) LC-MS analysis showing ion chromatogram in the WT and ΔAopex14/17 mutant strains. Green frame, the peak of arthrobotrisin. (D) Mass spectrogram of arthrobotrisins in the ΔAopex14/17 mutant strain (diagnostic fragments ion at m/z 139, 393, and 429). RT = 35.2 min.
Comparison of secondary metabolites between the WT and ΔAopex14/17 mutant strains. (A, B) Volcanograms (A) and heatmap (B) of upregulated and downregulated compounds between the WT and ΔAopex14/17 mutant strains. (C, D) The metabolic pathways of significantly upregulated (C) and downregulated (D) between the WT and ΔAopex14/17 mutant strains determined via KEGG enrichment. Download FIG S6, TIF file, 2.0 MB (2MB, tif) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The top 20 compounds with significant changes and the top 10 metabolic pathways that were significantly upregulated and downregulated. Download Table S2, DOCX file, 0.02 MB (18.4KB, docx) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
AoPex14/17 can interact with AoPex14 but not with AoPex13.
We attempted to probe the interaction between AoPex14/17 and two other docking complex members, AoPex13 and AoPex14, by yeast two-hybrid (Y2H) analysis. Firstly, we chose positive controls including plasmids pGBKT7-53 and pGADT7-T, while negative controls included pGBKT7-Lam and pGADT7-T. Furthermore, the growth of transformants was measured by serial dilution of yeast cells on SD/Trp/Leu, SD/Trp/Leu/His/Ade, and SD/Trp/Leu/His/Ade/-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) media, respectively. Ultimately, the results of Y2H experiments reflect that AoPex14/17 does not interact with AoPex13 but is able to interact with AoPex14 (see Fig. S7A and B in the supplemental material).
Yeast two-hybrid (Y2H) assays detect the interaction of AoPex14/17 with the other two members of the docking complex, AoPex13 and AoPex14, in A. oligospora. (A) Y2H analysis of AoPex14/17 and AoPex13. (B) Y2H analysis of AoPex14/17 and AoPex14. Download FIG S7, TIF file, 6.4 MB (6.4MB, tif) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
The maintenance of the peroxisome depends on the formation of the peroxisomal membrane and the import of both membrane and matrix proteins (18). The matrix proteins that are translocated to the peroxisomal membrane must interact with the docking machinery components, involving multiple peroxins, such as Pex1, Pex2, Pex5, Pex6, Pex7, Pex10, Pex12, Pex13, Pex14, and Pex26 (32, 33). Pex14/17, a filamentous fungus-specific peroxin of the docking machinery of peroxisomes, is required for the import of PMPs via binding Pex5 and Pex14 and may act as ScPex14 and ScPex17 in the docking of the peroxisomal matrix by expressing PEX14- and PEX17-similar portions of the proteins (19, 22). In this study, we first characterized the presence of homologs of PEX14/17 in the NT fungus A. oligospora and found that it is essential for the regulation of functions of peroxisomes and has a significant impact in a variety of phenotypic features through the disruption of AoPEX14/17.
Peroxisome has been linked to the growth, development, and pathogenicity of fungi (29). In Penicillium chrysogenum, the loss of PEX14/17 leads to a defect in sporulation (19). In M. oryzae, the loss of PEX14/17 delayed conidial generation and appressorial formation, reduced appressorial turgor accumulation and penetration ability in host plants, and significant decreased fungal virulence; however, PEX14/17 deletion showed no significant difference with regard to colony morphology and aerial hyphal development and only a slight reduction in radial growth (21). In M. robertsii, the conidial germination, conidiation, and fungal virulence were all greatly reduced upon PEX14/17 depletion, but there were no appreciable changes between colony phenotypes and the colony diameter of the PEX14/17 mutant strains and control strains on various conditions in the vegetative growth assay (34). In this study, we provide direct evidence that the knockout of PEX14/17 represses conidial production and germination and impairs trap formation and pathogenicity in A. oligospora, which are consistent with previous reports. Comparatively, the loss of AoPEX14/17 significantly reduces hyphae growth on the three different media. The reduced nuclei involved in the regulation of meiotic recombination (35) and the enrichment of the cell cycle indicated by transcriptome analysis indicate that AoPex14/17 may play a positive role in vegetative growth in A. oligospora. In addition, the morphology of the hyphae was changed in the ΔAopex14/17 mutants, including inflated mycelial cells. These results indicate that AoPex14/17 is necessary for mycelial growth, conidial production and germination, and the pathogenicity of A. oligospora and other filamentous fungi.
In fungi, besides common functions including the β-oxidation of fatty acids and the detoxification of hydrogen peroxide, peroxisomes are crucial for the metabolism of various unique carbon and nitrogen sources, including oleic acid, methanol, primary amines, and uric acid, and play an important role in response to stress (36, 37). Previous evidence has highlighted a decreasing ability for the control of fatty acid utilization, ROS degradation, stress tolerance, and cell wall integrity in fungi without Pex14/17 (21, 34, 38). Our results indicate that the disruption of AoPEX14/17 also resulted in a significant decrease in fatty acid utilization and tolerance to oxidative stress. In transcriptome analysis, there are nine genes involved in lipid metabolism that were downregulated in the ΔAopex14/17 strain versus WT strain, such as glycerophosphocholine phosphodiesterase, phospholipase D, alkaline ceramidase, and ethanolamine kinase; these genes are mainly involved in fatty acid catabolic processes, such as glycerophospholipid metabolic pathways and hydrolases and transferases of various lipids (39, 40). Obviously, the hyphae of ΔAopex14/17 mutants exhibited bigger lipid droplets and a much greater accumulation of ROS associated with the lower enzyme activity of CAT. Moreover, Pex14/17 plays a crucial role in peroxisome biogenesis (21). These results reveal that the downregulation of peroxisomal functions caused by a lack of AoPEX14/17 was triggered by a defect of peroxisome biogenesis in A. oligospora. However, no obvious inhibition was observed in ΔAopex14/17 mutants under chemical stressors of CR, NaCl, and sorbitol, indicating that AoPex14/17 may play a dispensable role in cell wall integrity and hyperosmotic stresses in A. oligospora.
Intriguingly, we observed that the terms of autophagy, mitophagy, and ubiquitin-mediated proteolysis were significantly enriched by transcriptome analysis, and subsequently, the remarkable increase in the level of autophagosome was confirmed by fluorescent staining in the ΔAopex14/17 mutants. Autophagy plays a vital role in the maintenance of peroxisome mass, and autophagy further regulates peroxisomes and contributes to the production of ROS (41). Peroxisome abundance can be regulated by PEX genes. The upregulation of autophagy level may be caused by the shortage of integrated peroxisome in ΔAopex14/17 mutants. Whether the regulation of PEX14/17 in autophagy and endocytosis inhibits the peroxisome function or alters matrix protein transport remains unknown. In addition, ΔAopex14/17 mutants generally increase the endocytosis process but lack WBs, which are peroxisome-related organelles necessary for sealing the septal pore in the event of hyphal damage (42, 43). Consistently, our transcriptome data suggest that Pex14/17 affects multiple metabolic pathways involved in carbohydrate metabolism, the MAPK signaling pathway, and mitochondrial function. TEM analysis revealed that the number of mitochondria and mitochondrial cristae decreased significantly in the ΔAopex14/17 mutants. Our findings support a model in which cellular energy metabolism relies heavily on the peroxisome, and the structure and function of peroxisomes and mitochondria are closely related in A. oligospora. The metabolome data also show that the abundance of secondary metabolites was significantly reduced upon PEX14/17 deletion in A. oligospora. Fungal secondary metabolites are synthesized by complex biosynthetic pathways catalyzed by enzymes located in different subcellular compartments, including the peroxisome (44). In Penicillium chrysogenum, Pex14/17 is involved in the penicillin biosynthesis process (19). Most importantly, the loss of AoPEX14/17 contributed to the accumulation of arthrobotrisins of A. oligospora. Arthrobotrisins, polyketide synthase-terpenoid synthase hybrid metabolites found in A. oligospora and other NT fungi, have been shown to inhibit hyphal growth and trap formation (35, 45). Fungal hyphal growth and pathogenicity were inhibited in ΔAopex14/17 mutants associated with the upregulated arthrobotrisin content. The mechanism underlying the ability of Pex14/17 to regulate these processes in A. oligospora awaits further analysis.
AoPex14, AoPex13, and AoPex14/17 are the components of peroxisomal docking complex, and we demonstrated by Y2H that AoPex14/17 interacted with AoPex14 but not with AoPex13. A previous study found that MoPex14 interacted with MoPex13 and MoPex14/17, but MoPex13 did not bind to MoPex14/17 in M. oryzae (46), which is consistent with our results. The Y2H data support the conclusion that AoPex14 binds primarily to AoPex14/17 and is essential for the maintenance of AoPex14/17 on the peroxisomal membrane.
In our recent work, AoPex1 and AoPex6 are required for mycelial growth, trap formation, and conidiation of A. oligospora (29), and the ΔAopex1 and ΔAopex6 mutants showed a more significant phenotypic difference than the ΔAopex14/17 mutant, such as mycelial growth and trap formation. Pex1 and Pex6 are both cytosolic and membrane-associated AAA ATPases of the peroxisomal protein import machinery, which have been demonstrated to mediate the ATP-dependent dislocation of the PTS1 receptor from the peroxisomal membrane to the cytosol (47); while Pex14/17 is a docking complex component specific to filamentous fungi and is mainly involved in the docking of receptors and cargoes during peroxisome synthesis (21). Thus, Pex1 and Pex6 are functionally different from Pex14/17, which would result in their phenotypic differences in A. oligospora.
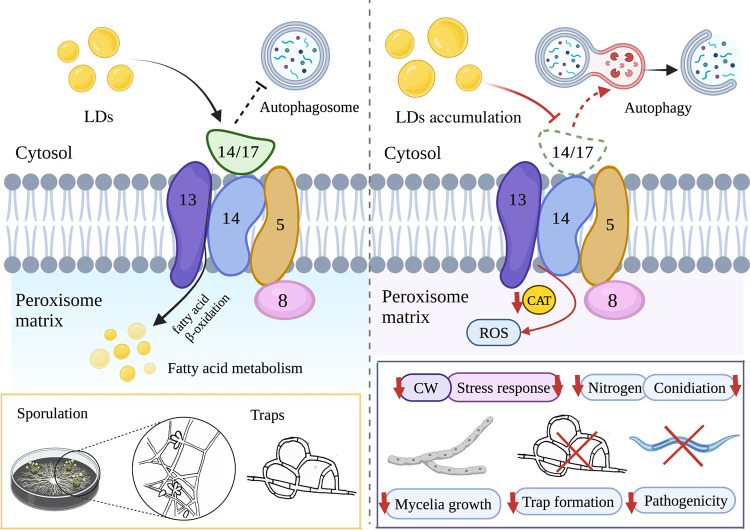
In conclusion, we characterized the function of Pex14/17 in A. oligospora involved in mycelial growth and development, conidial production, pathogenicity, lipid utilization, lipid metabolism, response to oxidative stress, ROS accumulation, autophagy, endocytosis, the maintenance of structure and function of organelles, and the regulation of secondary metabolites (Fig. 8). These findings provide a basis for exploring the features of peroxins that underlie the survival and environmental adaptations of NT fungi and contribute to developing an effective biocontrol strategy for pathogenic nematodes.
FIG 8.
Schematic illustration of the regulation of AoPEX14/17 in A. oligospora. In A. oligospora, the deletion of AoPEX14/17 inhibited the fatty acid metabolism and resulted in reduced CAT activity and increased LDs accumulation and ROS level; meanwhile, the absence of AoPEX14/17 impaired the autophagy, stress response, and endocytosis. These combined effects resulted in a reduction in mycelial growth, sporulation, trap formation, and pathogenicity. ROS, reactive oxygen species. CAT, catalase. LDs, lipid droplets. CW, cell wall.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Arthrobotrys oligospora ATCC 24927 and its derived mutants were grown on potato dextrose agar (PDA) medium in a constant-temperature incubator (28°C). The plasmid pCSN44 containing the selective marker hygromycin resistance gene (hph) was stored in the Escherichia coli strain DH5α (TaKaRa, Shiga, Japan), and the knockout vector was created using the plasmid PRS426 (48). As previously mentioned, S. cerevisiae (FY834) was cultivated in yeast extract peptone dextrose (YPD) medium for the recombinational cloning technique (49). To investigate mycelial development and associated phenotypic characteristics, the following media were used: TYGA (10 g tryptone, 5 g yeast extract, 10 g glucose, and 5 g molasses per 1 L), TG (10 g tryptone and 10 g glucose per 1 L), and CMY (20 g maizena [corn starch] and 5 g yeast extract per 1 L); additionally, 20 g agar should be added per 1 L of the above media (50). In addition, protoplast regeneration was conducted with PDAS (200 g potato, 0.3 g yeast extract, 0.6 M sucrose, and 10 g molasses per 1 L) medium. The nematode Caenorhabditis elegans was cultured in WA (20 g agar per 1 L) medium. Besides this, the utilization of carbon sources was also tested in MM (20 g glucose, 2 g NaNO3, 0.01 g FeSO4·7H2O, and 20 g agar per 1 L) medium.
Sequence and phylogenetic analyses of AoPex14/17.
The amino acid sequences for Pex14/17 homologs were obtained from the genomic databases of the model fungi S. cerevisiae, N. crassa, and A. nidulans, and the orthologous AoPex14/17 (AOL_s00081g263) was identified in A. oligospora. The NCBI online website (http://www.ncbi.nlm.nih.gov/) was used to download all protein sequences. The sequence similarity of homologous Pex14/17 from diverse fungi was analyzed by DNAman software (Lynnon Biosoft, San Ramon, USA). ClustalW was used to perform multiple sequence alignments, Mega X was used to construct a neighbor-joining tree, and the analysis of bootstrap values was conducted using 1,000 replicates (51).
Targeted gene deletion and Southern blotting.
Using the homologous recombination technique, the AoPEX14/17 gene was deleted in the wild-type (WT) strain (52). The upstream and downstream fragments of AoPEX14/17 were PCR amplified from A. oligospora using a modified yeast cloning procedure, and a hygromycin cassette (hph) was amplified using the vector pCSN44 as a template (53). The fragment, hph cassette, and gapped yeast shuttle vector pRS426 (digested with EcoRI and XhoI) were transformed into yeast strain FY834. Next, the recombinant cloned strains were screened by SC-Ura medium as described previously (53, 54). The circular construct was created by homologous recombination, and the disruption vector (pRS426-AoPEX14/17-hph) was eventually recovered through transformation into E. coli DH5α. The gene disruption target fragment was amplified using paired primers (see Table S3 in the supplemental material) and transformed into A. oligospora protoplasts. As previously mentioned, hygromycin-resistant transformants were selected on PDAS containing 200 μg/mL hygromycin (55). The candidates for the gene deletion mutant were screened from hygromycin-resistant transformants via genomic PCR and confirmed by Southern blotting.
List of primers used in this study. Download Table S3, DOCX file, 0.02 MB (20.2KB, docx) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genomic DNA was digested with NcoI enzyme (New England Biolabs, Inc., Ipswich, MA, USA) for Southern blotting. The analysis was carried out in accordance with the directions included in the North2South chemiluminescent hybridization and detection kit (Pierce, Rockford, IL, USA).
RNA sequencing analysis.
WT and mutant strains were incubated in PD (200 g potatoes and 20 g glucose per 1 L) broth medium at 28°C for 5 and 7 days, respectively. Three replicates were used for each sample. Mycelial samples were collected and frozen in liquid nitrogen, followed by sequencing by Majorbio Bio-pharm Technology Co. Ltd (Shanghai, China). Then, the Majorbio Cloud platform was used to evaluate the data (www.majorbio.com). RNA was extracted using the AxyPrep Multisource RNA miniprep kit (Axygen, Jiangsu, China), transcriptome results were verified by RT-qPCR, and the selected genes and relative primers are listed in Table S3 (23, 24). Finally, differentially expressed genes (DEGs) were screened out using P ≤ 0.05 and Log2 (fold change) ≥ 2 as thresholds. The GO and KEGG databases were used to functionally assess the selected DEGs (56).
Comparison of mycelial growth and conidia yield.
To determine mycelial growth, WT and mutant strains were inoculated onto TG, PDA, and TYGA media at 28°C for 5 days, respectively, with growth rates and colony diameters measured at 24-h intervals (57). To analyze sporulation, conidia were collected from 14-day CMY agar plates with 5 mL sterile distilled water. Conidia produced by a given colony were counted and reported as the total number of conidia present per unit area of the colony using a hemocytometer (58).
Trap formation and bioassay.
To observe trap formation, 50-μL conidia suspensions (4 × 106 conidia mL−1) of WT and mutant strains were evenly distributed on WA plates at 28°C for 3 to 4 days. Then, to induce trap formation, approximately 300 to 400 nematodes (C. elegans) were added to each WA plate (59). The number of traps produced and nematodes captured by each strain were counted using a microscope (Olympus, Japan) at 12, 24, 36, and 48 h.
Analysis of stress tolerance.
To test the sensitivity of each strain to various environmental stresses, mycelial plugs taken from the periphery of each strain were inoculated on TG medium amended with the following compounds: menadione (0.01, 0.02, and 0.03 mM) and H2O2 (2.5, 5, and 10 mM) as oxidative stressors; Congo red (CR) (0.05, 0.07, and 0.1 mg/mL) and sodium dodecyl sulfate (SDS) (0.01, 0.02, and 0.03%) as cell wall stress agents; and NaCl (0.1, 0.2, and 0.3 M) and sorbitol (0.25, 0.5, and 0.75 M) as osmotic stressors. After incubation at 28°C for 6 days, the colony diameter in each plate was measured, and the relative growth inhibition (RGI) of each colony was evaluated as described previously (60). Each experiment was repeated three times.
Analysis of fatty acid utilization.
For the lipid utilization assay, WT and mutant strains were inoculated on MM medium supplemented with sodium acetate (50 mM), oleic acid (0.12%), or Tween 20 (0.5%) as the sole carbon source. The size of the fungal colonies was measured and photographed postincubation at 28°C for 6 days, after which RGI values were determined (29).
Analyses of reactive oxygen species and endocytosis.
By staining with nitrotetrazolium blue chloride (NBT) (Solarbio, Beijing, China), the ROS levels in the hyphae were determined. WT and mutant strains were grown on PDA at 28°C for 3 days, after which plates were stained with 20 mL of a 0.2% NBT solution and incubated in the dark at 28°C for 30 min. After draining the supernatant, ethanol was used to wash the plates. The plates were reincubated in the dark at 28°C for 30 min before imaging (61).
Cellular ROS accumulation in the fungal hyphae was determined by dihydroethidium (DHE) (Beyotime, Shanghai, China) staining. Briefly, the collected mycelia were mixed with 10 μg/mL DHE solution, incubated at 37°C for 30 min, and then washed with phosphate-buffered saline (PBS) twice (62). Subsequently, ROS levels were estimated from the intensity of DHE fluorescence with the same weight of mycelium via multimode microplate reader (Molecular Devices, Shanghai, China).
Fluorescence microscopy and transmission electron microscope.
The fungal cell wall and hyphal septum were visualized by staining with 20 μg/mL calcofluor white (CFW) (Sigma-Aldrich, St. Louis, MO, USA), and mycelial cell nuclei were visualized by staining with 20 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) and 20 μg/mL CFW (63). As previously described, lipid droplets (LDs) were stained with 10 μg/mL boron dipyrromethene (BODIPY) (Thermo Fisher Scientific, Waltham, MA, USA) (29). In addition, autophagy of the hyphae was detected by 100 μg/mL monodansylcadaverine (MDC) staining.
For TEM analysis, WT and mutant strains were harvested from colonies cultured on PD broth for 3 days. Then, the mycelia were collected and fixed with 2.5% glutaraldehyde for TEM observation (30).
Reverse transcription-quantitative PCR.
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) from frozen fungal tissues. Then, the treated RNA samples were reversely transcribed into cDNA using the PrimeScript RT reagent kit (TaKaRa, Shiga, Japan). The above experiments were performed according to the manufacturer's instructions. RT-qPCR was accomplished by LightCycler 480 with SYBR green I master mix (Roche, Basel, Switzerland) and a specific primer set (Table S3). Besides this, β-tubulin (AOL_s00076g640) was used as an internal control to normalize the target gene’s expression level (64). Finally, the relative expression level of duplicated samples was analyzed by the 2−ΔΔCT method, and each reaction was independently repeated three times with three biological replicates per sample.
Untargeted metabolomics analysis.
WT and mutant strains were inoculated in 250 mL of PD broth at 28°C for 5 days. Then, the mycelium was dissolved in 250 mL ethyl acetate, and the fermentation broth was ultrasonically extracted and allowed to stand overnight. The ethyl acetate layer was dried with a rotary evaporator before being dissolved in 1 mL of chromatographic methanol. For liquid chromatography-mass spectrometry (LC-MS) analysis, the methanol solution was filtered through a 0.22-μm membrane filter. Analysis was performed using the Thermo Fisher Scientific Dionex Ultimate 3000 UHPLC system and a Thermo Fisher high-resolution Q precision focusing mass spectrometer (Thermo Fisher Scientific) (30, 65, 66). Subsequently, the Compound Discoverer 3.3 software package was used to perform metabolite deconvolutions and identifications (66, 67).
Y2H assay.
The cDNAs of Aopex14/17 were cloned into bait plasmid pGBKT7 (BD-Pex14/17), and cDNAs of Aopex13 and Aopex14 were cloned into prey plasmid pGADT7 (AD-Pex13 and AD-Pex14). This protocol was experimented and analyzed according to previously described studies (60).
Statistical analysis.
GraphPad Prism (version 9.00) was used for statistical analysis, and Tukey's multiple comparison test was applied to assess the level of difference between different strains. Data are presented as means ± standard deviation, and a P value of <0.05 was used to determine significant differences.
Data availability.
All data generated or analyzed during this study are included in the published paper and associated supplemental files. The sequencing data were deposited in the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE216978.
ACKNOWLEDGMENTS
We are grateful to the Microbial Library of the Germplasm Bank of Wild Species from Southwest China for preserving and providing experimental strains and to Guo Ying-qi (Kunming Institute of Zoology, Chinese Academy of Sciences) for her help with taking and analyzing TEM images.
This study was funded by the National Natural Science Foundation of China (31960556) and the Applied Basic Research Foundation of Yunnan Province (202001BB050004) and Yunnan University’s Research Innovation Fund for Graduate Students (KC-22222591).
Q.L. contributed Methodology, Investigation, Software, Data Curation, Writing – Original Draft. D.L. contributed Methodology, Investigation, Data Curation. N.B. and Y.Z. contributed Data Curation, Formal Analysis. J.Y. contributed Conceptualization, Funding Acquisition, Writing – Review & Editing. All authors have read and agreed to the published version of the manuscript.
We declare that no competing interests exist.
Contributor Information
Jinkui Yang, Email: jinkui960@ynu.edu.cn.
Aaron P. Mitchell, University of Georgia
REFERENCES
- 1.Veenhuis M, Salomons FA, Van Der Klei IJ. 2000. Peroxisome biogenesis and degradation in yeast: a structure/function analysis. Microsc Res Tech 51:584–600. doi:. [DOI] [PubMed] [Google Scholar]
- 2.Okumoto K, Tamura S, Honsho M, Fujiki Y. 2020. Peroxisome: metabolic functions and biogenesis. Adv Exp Med Biol 1299:3–17. doi: 10.1007/978-3-030-60204-8_1. [DOI] [PubMed] [Google Scholar]
- 3.Hayashi M, Toriyama K, Kondo M, Kato A, Mano S, De Bellis L, Hayashi-Ishimaru Y, Yamaguchi K, Hayashi H, Nishimura M. 2000. Functional transformation of plant peroxisomes. Cell Biochem Biophys 32:295–304. doi: 10.1385/cbb:32:1-3:295. [DOI] [PubMed] [Google Scholar]
- 4.Michels PA, Bringaud F, Herman M, Hannaert V. 2006. Metabolic functions of glycosomes in trypanosomatids. Biochim Biophys Acta 1763:1463–1477. doi: 10.1016/j.bbamcr.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 5.Liang L, Gao H, Li J, Liu L, Liu Z, Zhang KQ. 2017. The Woronin body in the nematophagous fungus Arthrobotrys oligospora is essential for trap formation and efficient pathogenesis. Fungal Biol 121:11–20. doi: 10.1016/j.funbio.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Freitag J, Ast J, Bolker M. 2012. Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 485:522–525. doi: 10.1038/nature11051. [DOI] [PubMed] [Google Scholar]
- 7.Gabaldon T. 2010. Peroxisome diversity and evolution. Philos Trans R Soc B 365:765–773. doi: 10.1098/rstb.2009.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farre JC, Mahalingam SS, Proietto M, Subramani S. 2019. Peroxisome biogenesis, membrane contact sites, and quality control. EMBO Rep 20:e46864. doi: 10.15252/embr.201846864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujiki Y, Lazarow PB. 1985. Post-translational import of fatty acyl-CoA oxidase and catalase into peroxisomes of rat liver in vitro. J Biol Chem 260:5603–5609. doi: 10.1016/S0021-9258(18)89065-2. [DOI] [PubMed] [Google Scholar]
- 10.Titorenko VI, Rachubinski RA. 2001. The life cycle of the peroxisome. Nat Rev Mol Cell Biol 2:357–368. doi: 10.1038/35073063. [DOI] [PubMed] [Google Scholar]
- 11.Gunkel K, van Dijk R, Veenhuis M, van der Klei IJ. 2004. Routing of Hansenula polymorpha alcohol oxidase: an alternative peroxisomal protein-sorting machinery. Mol Biol Cell 15:1347–1355. doi: 10.1091/mbc.e03-04-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otera H, Harano T, Honsho M, Ghaedi K, Mukai S, Tanaka A, Kawai A, Shimizu N, Fujiki Y. 2000. The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p.PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J Biol Chem 275:21703–21714. doi: 10.1074/jbc.M000720200. [DOI] [PubMed] [Google Scholar]
- 13.Otera H, Setoguchi K, Hamasaki M, Kumashiro T, Shimizu N, Fujiki Y. 2002. Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol Cell Biol 22:1639–1655. doi: 10.1128/MCB.22.6.1639-1655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purdue PE, Yang X, Lazarow PB. 1998. Pex18p and Pex21p, a novel pair of related peroxins essential for peroxisomal targeting by the PTS2 pathway. J Cell Biol 143:1859–1869. doi: 10.1083/jcb.143.7.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sichting M, Schell-Steven A, Prokisch H, Erdmann R, Rottensteiner H. 2003. Pex7p and Pex20p of Neurospora crassa function together in PTS2-dependent protein import into peroxisomes. Mol Biol Cell 14:810–821. doi: 10.1091/mbc.e02-08-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JR, Jang HH, Park JH, Jung JH, Lee SS, Park SK, Chi YH, Moon JC, Lee YM, Kim SY, Kim JY, Yun DJ, Cho MJ, Lee KO, Lee SY. 2006. Cloning of two splice variants of the rice PTS1 receptor, OsPex5pL and OsPex5pS, and their functional characterization using pex5-deficient yeast and Arabidopsis. Plant J 47:457–466. doi: 10.1111/j.1365-313X.2006.02797.x. [DOI] [PubMed] [Google Scholar]
- 17.Kiel JA, Veenhuis M, van der Klei IJ. 2006. PEX genes in fungal genomes: common, rare or redundant. Traffic 7:1291–1303. doi: 10.1111/j.1600-0854.2006.00479.x. [DOI] [PubMed] [Google Scholar]
- 18.Managadze D, Wurtz C, Wiese S, Schneider M, Girzalsky W, Meyer HE, Erdmann R, Warscheid B, Rottensteiner H. 2010. Identification of PEX33, a novel component of the peroxisomal docking complex in the filamentous fungus Neurospora crassa. Eur J Cell Biol 89:955–964. doi: 10.1016/j.ejcb.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Opaliński L, Kiel J, Homan TG, Veenhuis M, Klei I. 2010. Penicillium chrysogenum Pex14/17p – a novel component of the peroxisomal membrane that is important for penicillin production. FEBS J 277:3203–3218. doi: 10.1111/j.1742-4658.2010.07726.x. [DOI] [PubMed] [Google Scholar]
- 20.Pieuchot L, Jedd G. 2012. Peroxisome assembly and functional diversity in eukaryotic microorganisms. Annu Rev Microbiol 66:237–263. doi: 10.1146/annurev-micro-092611-150126. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Wang J, Chen H, Chai R, Zhang Z, Mao X, Qiu H, Jiang H, Wang Y, Sun G. 2017. Pex14/17, a filamentous fungus-specific peroxin, is required for the import of peroxisomal matrix proteins and full virulence of Magnaporthe oryzae. Mol Plant Pathol 18:1238–1252. doi: 10.1111/mpp.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peraza-Reyes L, Arnaise S, Zickler D, Coppin E, Debuchy R, Berteaux-Lecellier V. 2011. The importomer peroxins are differentially required for peroxisome assembly and meiotic development in Podospora anserina: insights into a new peroxisome import pathway. Mol Microbiol 82:365–377. doi: 10.1111/j.1365-2958.2011.07816.x. [DOI] [PubMed] [Google Scholar]
- 23.Jiang X, Xiang M, Liu X. 2017. Nematode-trapping fungi. Microbiol Spectr 5:5.1.10. doi: 10.1128/microbiolspec.FUNK-0022-2016. [DOI] [PubMed] [Google Scholar]
- 24.Zhu MC, Li XM, Zhao N, Yang L, Zhang KQ, Yang JK. 2022. Regulatory mechanism of trap formation in the nematode-trapping fungi. J Fungi 8:406. doi: 10.3390/jof8040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Wang L, Ji X, Feng Y, Li X, Zou C, Xu J, Ren Y, Mi Q, Wu J, Liu S, Liu Y, Huang X, Wang H, Niu X, Li J, Liang L, Luo Y, Ji K, Zhou W, Yu Z, Li G, Liu Y, Li L, Qiao M, Feng L, Zhang KQ. 2011. Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog 7:e1002179. doi: 10.1371/journal.ppat.1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nordbring-Hertz B, Veenhuis M, Harder W. 1984. Dialysis membrane technique for ultrastructural studies of microbial interactions. Appl Environ Microbiol 47:195–197. doi: 10.1128/aem.47.1.195-197.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou D, Zhu Y, Bai N, Yang L, Xie M, Yang J, Zhu M, Zhang KQ, Yang J. 2022. AoATG5 plays pleiotropic roles in vegetative growth, cell nucleus development, conidiation, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Sci China Life Sci 65:412–425. doi: 10.1007/s11427-020-1913-9. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Zhu M, Liu Y, Yang L, J Y. 2023. Aoatg11 and Aoatg33 are indispensable for mitophagy, and contribute to conidiation, the stress response, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Microbiol Res 266:127252. doi: 10.1016/j.micres.2022.127252. [DOI] [PubMed] [Google Scholar]
- 29.Liu Q, Li D, Jiang K, Zhang KQ, Yang J. 2022. AoPEX1 and AoPEX6 are required for mycelial growth, conidiation, stress response, fatty acid utilization, and trap formation in Arthrobotrys oligospora. Microbiol Spectr 10:e0027522. doi: 10.1128/spectrum.00275-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang KX, Liu QQ, Bai N, Zhu MC, Zhang KQ, Yang JK. 2022. AoSsk1, a response regulator required for mycelial growth and development, stress responses, trap formation, and the secondary metabolism in Arthrobotrys oligospora. J Fungi 8:260. doi: 10.3390/jof8030260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honsho M, Okumoto K, Tamura S, Fujiki Y. 2020. Peroxisome biogenesis disorders. Adv Exp Med Biol 1299:45–54. doi: 10.1007/978-3-030-60204-8_4. [DOI] [PubMed] [Google Scholar]
- 32.Ruttermann M, Gatsogiannis C. 2022. Good things come to those who bait: the peroxisomal docking complex. Biol Chem. doi: 10.1515/hsz-2022-0161. [DOI] [PubMed] [Google Scholar]
- 33.Yamashita K, Tamura S, Honsho M, Yada H, Yagita Y, Kosako H, Fujiki Y. 2020. Mitotic phosphorylation of Pex14p regulates peroxisomal import machinery. J Cell Biol 219:e202001003. doi: 10.1083/jcb.202001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Feng J, Jiang Y, Xu X, Xu L, Zhou Q, Huang B. 2021. MrPEX33 is involved in infection-related morphogenesis and pathogenicity of Metarhizium robertsii. Appl Microbiol Biotechnol 105:1079–1090. doi: 10.1007/s00253-020-11071-3. [DOI] [PubMed] [Google Scholar]
- 35.Xie M, Ma N, Bai N, Yang L, Yang X, Zhang KQ, Yang J. 2022. PKC-SWI6 signaling regulates asexual development, cell wall integrity, stress response, and lifestyle transition in the nematode-trapping fungus Arthrobotrys oligospora. Sci China Life Sci 65:2455–2471. doi: 10.1007/s11427-022-2118-0. [DOI] [PubMed] [Google Scholar]
- 36.Veenhuis M, Harder W. 1988. Microbodies in yeasts: structure, function and biogenesis. Microbiol Sci 5:347–351. [PubMed] [Google Scholar]
- 37.Kumar S, Singh R, Williams CP, van der Klei IJ. 2016. Stress exposure results in increased peroxisomal levels of yeast Pnc1 and Gpd1, which are imported via a piggy-backing mechanism. Biochim Biophys Acta 1863:148–156. doi: 10.1016/j.bbamcr.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 38.Yu W, Lin M, Peng M, Yan H, Wang J, Zhou J, Lu G, Wang Z, Shim WB. 2021. Fusarium verticillioides FvPex8 is a key component of the peroxisomal docking/translocation module that serves important roles in fumonisin biosynthesis but not in virulence. MPMI 34:803–814. doi: 10.1094/MPMI-10-20-0273-R. [DOI] [PubMed] [Google Scholar]
- 39.Chopra A, Khuller GK. 1984. Lipid metabolism in fungi. Crit Rev Microbiol 11:209–271. doi: 10.3109/10408418409105904. [DOI] [PubMed] [Google Scholar]
- 40.Inaba M, Murota K, Nikawadori M, Kishino E, Matusda R, Takagi M, Ohkubo T, Tanaka T, Terao J, Tokumura A. 2014. Extracellular metabolism-dependent uptake of lysolipids through cultured monolayer of differentiated Caco-2 cells. Biochim Biophys Acta 1841:121–131. doi: 10.1016/j.bbalip.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Cheng S, Wang Q, Manghwar H, Liu F. 2022. Autophagy-mediated regulation of different meristems in plants. Int J Mol Sci 23:6236. doi: 10.3390/ijms23116236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Managadze D, Wurtz C, Sichting M, Niehaus G, Veenhuis M, Rottensteiner H. 2007. The peroxin PEX14 of Neurospora crassa is essential for the biogenesis of both glyoxysomes and Woronin bodies. Traffic 8:687–701. doi: 10.1111/j.1600-0854.2007.00560.x. [DOI] [PubMed] [Google Scholar]
- 43.Liu F, Ng SK, Lu Y, Low W, Lai J, Jedd G. 2008. Making two organelles from one: Woronin body biogenesis by peroxisomal protein sorting. J Cell Biol 180:325–339. doi: 10.1083/jcb.200705049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin JF. 2020. Transport systems, intracellular traffic of intermediates and secretion of beta-lactam antibiotics in fungi. Fungal Biol Biotechnol 7:6. doi: 10.1186/s40694-020-00096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu X, Hu X, Pop M, Wernet N, Kirschhöfer F, Brenner-Weiß G, Keller J, Bunzel M, Fischer R. 2021. Fatal attraction of Caenorhabditis elegans to predatory fungi through 6-methyl-salicylic acid. Nat Commun 12:5462. doi: 10.1038/s41467-021-25535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang JY, Li L, Chai RY, Qiu HP, Zhang Z, Wang YL, Liu XH, Lin FC, Sun GC. 2019. Pex13 and Pex14, the key components of the peroxisomal docking complex, are required for peroxisome formation, host infection and pathogenicity-related morphogenesis in Magnaporthe oryzae. Virulence 10:292–314. doi: 10.1080/21505594.2019.1598172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Platta HW, Grunau S, Rosenkranz K, Girzalsky W, Erdmann R. 2005. Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat Cell Biol 7:817–822. doi: 10.1038/ncb1281. [DOI] [PubMed] [Google Scholar]
- 48.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 49.Jiang D, Zhou J, Bai G, Xing X, Tang L, Yang X, Li J, Zhang KQ, Yang J. 2017. Random mutagenesis analysis and identification of a novel C2H2-type transcription factor from the nematode-trapping fungus Arthrobotrys oligospora. Sci Rep 7:5640. doi: 10.1038/s41598-017-06075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie M, Bai N, Yang J, Jiang K, Zhou D, Zhao Y, Li D, Niu X, Zhang KQ, Yang J. 2020. Protein kinase Ime2 is required for mycelial growth, conidiation, osmoregulation, and pathogenicity in nematode-trapping fungus Arthrobotrys oligospora. Front Microbiol 10:3065. doi: 10.3389/fmicb.2019.03065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balogh J, Tunlid A, Rosén S. 2003. Deletion of a lectin gene does not affect the phenotype of the nematode-trapping fungus Arthrobotrys oligospora. Fungal Genet Biol 39:128–135. doi: 10.1016/s1087-1845(03)00023-9. [DOI] [PubMed] [Google Scholar]
- 53.Park G, Colot HV, Collopy PD, Krystofova S, Crew C, Ringelberg C, Litvinkova L, Altamirano L, Li L, Curilla S, Wang W, Gorrochotegui-Escalante N, Dunlap JC, Borkovich KA. 2011. High-throughput production of gene replacement mutants in Neurospora crassa. Methods Mol Biol 722:179–189. doi: 10.1007/978-1-61779-040-9_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernhards Y, Poggeler S. 2011. The phocein homologue SmMOB3 is essential for vegetative cell fusion and sexual development in the filamentous ascomycete Sordaria macrospora. Curr Genet 57:133–149. doi: 10.1007/s00294-010-0333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X, Ma N, Yang L, Zheng Y, Zhen Z, Li Q, Xie M, Li J, Zhang KQ, Yang J. 2018. Two Rab GTPases play different roles in conidiation, trap formation, stress resistance, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Appl Microbiol Biotechnol 102:4601–4613. doi: 10.1007/s00253-018-8929-1. [DOI] [PubMed] [Google Scholar]
- 56.Liu J, Yi J, Wu H, Zheng L, Zhang G. 2020. Prepupae and pupae transcriptomic characterization of Trichogramma chilonis. Genomics 112:1651–1659. doi: 10.1016/j.ygeno.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 57.Ma N, Jiang KX, Bai N, Li DN, Zhang KQ, Yang JK. 2022. Functional analysis of two affinity cAMP phosphodiesterases in the nematode-trapping fungus Arthrobotrys oligospora. Pathogens 11:405. doi: 10.3390/pathogens11040405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J, Wang ZK, Sun HH, Ying SH, Feng MG. 2017. Characterization of the Hog1 MAPK pathway in the entomopathogenic fungus Beauveria bassiana. Environ Microbiol 19:1808–1821. doi: 10.1111/1462-2920.13671. [DOI] [PubMed] [Google Scholar]
- 59.Zhang G, Zheng Y, Ma Y, Yang L, Xie M, Zhou D, Niu X, Zhang KQ, Yang J. 2019. The velvet proteins VosA and VelB play different roles in conidiation, trap formation, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front Microbiol 10:1917. doi: 10.3389/fmicb.2019.01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma Y, Yang X, Xie M, Zhang G, Yang L, Bai N, Zhao Y, Li D, Zhang KQ, Yang J. 2020. The Arf-GAP AoGlo3 regulates conidiation, endocytosis, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Fungal Genet Biol 138:103352. doi: 10.1016/j.fgb.2020.103352. [DOI] [PubMed] [Google Scholar]
- 61.Kong X, Zhang H, Wang X, van der Lee T, Waalwijk C, van Diepeningen A, Brankovics B, Xu J, Xu J, Chen W, Feng J. 2019. FgPex3, a peroxisome biogenesis factor, is involved in regulating vegetative growth, conidiation, sexual development, and virulence in Fusarium graminearum. Front Microbiol 10:2088. doi: 10.3389/fmicb.2019.02088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang L, Li X, Ma Y, Zhang K, Yang J. 2022. The Arf-GAP proteins AoGcs1 and AoGts1 regulate mycelial development, endocytosis, and pathogenicity in Arthrobotrys oligospora. J Fungi 8:463. doi: 10.3390/jof8050463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie M, Yang J, Jiang K, Bai N, Zhu M, Zhu Y, Zhang KQ, Yang J. 2021. AoBck1 and AoMkk1 are necessary to maintain cell wall integrity, vegetative growth, conidiation, stress resistance, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front Microbiol 12:649582. doi: 10.3389/fmicb.2021.649582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang L, Li X, Bai N, Yang X, Zhang KQ, Yang J. 2022. Transcriptomic analysis reveals that Rho GTPases regulate trap development and lifestyle transition of the nematode-trapping fungus Arthrobotrys oligospora. Microbiol Spectr 10:e01759-21. doi: 10.1128/spectrum.01759-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bai N, Zhang G, Wang W, Feng H, Yang X, Zheng Y, Yang L, Xie M, Zhang KQ, Yang J. 2022. Ric8 acts as a regulator of G-protein signalling required for nematode-trapping lifecycle of Arthrobotrys oligospora. Environ Microbiol 24:1714–1730. doi: 10.1111/1462-2920.15735. [DOI] [PubMed] [Google Scholar]
- 66.Xie M, Ma N, Bai N, Zhu M, Zhang KQ, Yang J. 2022. Phospholipase C (AoPLC2) regulates mycelial development, trap morphogenesis, and pathogenicity of the nematode-trapping fungus Arthrobotrys oligospora. J Appl Microbiol 132:2144–2156. doi: 10.1111/jam.15370. [DOI] [PubMed] [Google Scholar]
- 67.Chen YH, Zhang LL, Wang LJ, Yue XT, Wu QF, Jiang Y, Zhang KQ, Niu XM. 2022. Acetylation of sesquiterpenyl epoxy-cyclohexenoids regulates fungal growth, stress resistance, endocytosis, and pathogenicity of nematode-trapping fungus Arthrobotrys oligospora via metabolism and transcription. J Agric Food Chem 70:6145–6155. doi: 10.1021/acs.jafc.2c01914. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Neighbor-joining phylogenetic tree based on the amino acid sequence of Pex14/17 homologous proteins from different fungi and AoPEX14/17 knockout and verification in A. oligospora. (A) Phylogenetic tree. GenBank accession numbers are provided in brackets. Numbers below nodes indicate the bootstrap value. The bar marker indicates the genetic distance, which is proportional to the number. (B) Diagrammatic sketch of homologous recombination. WT, wild-type strain; MT, mutant. (C) The homologous recombinant AoPEX14/17 deletion plasmid was transformed into yeast. (D) AoPEX14/17 deletion transformants (ΔAopex14/17) were confirmed by PCR method. 2K, DNA marker. (E) Southern blotting analysis of wild-type and transformants. Download FIG S1, TIF file, 2.1 MB (2.1MB, tif) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Principal component analysis (PCA) and reverse transcription-qualitative PCR (RT-qPCR) was used to verify the transcriptome data between the WT and ΔAopex14/17 mutant strains at 5 days and 7 days. (A) Principal component analysis. Different shapes and colors represent the WT and ΔAopex14/17 mutant strains at 5 days and 7 days. (B to D) Comparison of transcription analysis of genes involved in MAPK signaling pathway (B), glycolysis/gluconeogenesis (C), and ubiquitin-mediated proteolysis (D). Download FIG S2, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Statistics of reads and mapping rate for the wild-type (WT) strain (A) and ΔAopex14/17 (H) mutant strains at different time points and statistics of reads, phred-like quality scores, and GC content for the wild-type (WT) strain (A) and ΔAopex14/17 (H) mutant at different time points. Download Table S1, DOCX file, 0.02 MB (16.7KB, docx) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene Ontology (GO) enrichment of differentially expressed genes (DEGs) between the WT and ΔAopex14/17 mutant strains at 5 days and 7 days. (A, B) GO enrichment analysis of upregulation DEGs (A) and downregulation DEGs (B) in the ΔAopex14/17 mutant versus WT strain at 5 days. (C, D) GO enrichment analysis of upregulation DEGs (C) and downregulation DEGs (D) in the ΔAopex14/17 mutant versus WT strain at 7 days. Download FIG S3, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of stress tolerance to cell wall-perturbing agents and osmotic agents between the WT and ΔAopex14/17 mutant strains. (A) Colony morphologies of the WT and mutant strains incubated on TG medium supplemented with 0.05 to 0.1 mg/mL Congo red and 0.01 to 0.03% SDS for 6 days. (B) Relative growth inhibition (RGI) values of the WT and mutant strains incubated on TG medium supplemented with 0.05 to 0.1 mg/mL Congo red and 0.01 to 0.03% SDS for 6 days. (C) Colony morphologies of the WT and mutant strains incubated on TG medium supplemented with 0.10 to 0.30 M NaCl and 0.25 to 0.75 M sorbitol for 6 days. (D) Relative growth inhibition (RGI) values of the WT and mutant strains incubated on TG medium supplemented with 0.10 to 0.30 M NaCl and 0.25 to 0.75 M sorbitol for 6 days. The asterisk indicates a significant difference between the mutant and the WT strains (*, P < 0.05; **, P < 0.01). Download FIG S4, TIF file, 7.6 MB (7.6MB, tif) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differentially expressed genes (DEGs) associated with autophagy and peroxisome. Download FIG S5, TIF file, 1.2 MB (1.2MB, tif) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of secondary metabolites between the WT and ΔAopex14/17 mutant strains. (A, B) Volcanograms (A) and heatmap (B) of upregulated and downregulated compounds between the WT and ΔAopex14/17 mutant strains. (C, D) The metabolic pathways of significantly upregulated (C) and downregulated (D) between the WT and ΔAopex14/17 mutant strains determined via KEGG enrichment. Download FIG S6, TIF file, 2.0 MB (2MB, tif) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The top 20 compounds with significant changes and the top 10 metabolic pathways that were significantly upregulated and downregulated. Download Table S2, DOCX file, 0.02 MB (18.4KB, docx) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Yeast two-hybrid (Y2H) assays detect the interaction of AoPex14/17 with the other two members of the docking complex, AoPex13 and AoPex14, in A. oligospora. (A) Y2H analysis of AoPex14/17 and AoPex13. (B) Y2H analysis of AoPex14/17 and AoPex14. Download FIG S7, TIF file, 6.4 MB (6.4MB, tif) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of primers used in this study. Download Table S3, DOCX file, 0.02 MB (20.2KB, docx) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All data generated or analyzed during this study are included in the published paper and associated supplemental files. The sequencing data were deposited in the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE216978.