Abstract
Small cell lung cancer (SCLC) elicits the generation of autoantibodies that result in unique paraneoplastic neurological syndromes. The mechanistic basis for the formation of such autoantibodies is largely unknown but is key to understanding their etiology. We developed a high-dimensional technique that enables detection of autoantibodies in complex with native antigens directly from patient plasma. Here we used our platform to screen 1,009 human plasma samples for 3,600 autoantibody-antigen complexes, finding that plasma from patients with SCLC harbors, on average, 4-fold higher disease-specific autoantibody signals compared to plasma from patients with other cancers. Across 3 independent SCLC cohorts, we identified a set of common but previously unknown autoantibodies that are produced in response to both intracellular and extracellular tumor antigens. We further characterized several disease-specific post translational modifications within extracellular proteins targeted by these autoantibodies including citrullination, isoaspartylation and cancer-specific glycosylation. Since most patients with SCLC have metastatic disease at diagnosis, we queried if these autoantibodies could be utilized for SCLC early detection. We created a risk-prediction model using 5 autoantibodies with an average area under the curve of 0.84 for the 3 cohorts that improved to 0.96 by incorporating cigarette smoke consumption in pack years. Taken together, our findings provide an innovative approach to identify circulating autoantibodies in SCLC with mechanistic insight into disease-specific immunogenicity and clinical utility.
One sentence summary:
Small cell lung cancer generates tumor associated antigens targeted by autoantibodies that can be exploited for tumor early detection.
Introduction
Small cell lung cancer (SCLC) is the 6th leading cause of cancer-related deaths in the United States with fewer than 6% of patients surviving 5 years after diagnosis (1-3). A critical barrier to effective, life-saving treatment is a lack of early detection as 70% of patients with SCLC are diagnosed with metastatic disease (4). More than any other cancer type, SCLC has a strong association with a group of autoimmune diseases called paraneoplastic neurological syndromes (PNS) (5, 6). Many PNS are considered clinical manifestations of an immune response against neuroendocrine antigens on tumor cells that also cross-react with proteins present in the nervous system (7). This immune activation is thought to include cytotoxic T cell responses against intracellular antigens and autoantibody (AAb) production to both intracellular and extracellular antigens (8), but the mechanistic basis and scope of AAb production in SCLC is not well understood. Strikingly, PNS symptoms precede cancer diagnosis in 80% of affected patients and result in increased detection of early-stage SCLC and improved overall survival (9, 10). Although PNS diagnoses are relatively rare, AAbs to PNS antigens can be found in patients with SCLC that do not display autoimmune symptoms (11, 12), and several PNS AAbs are associated with reduced mortality hazard ratios (13). The unique relationship of AAb production during SCLC tumorigenesis suggests AAbs to non-PNS antigens could exist and may more broadly serve as biomarkers of SCLC.
SCLC-specific AAbs that don’t cause overt autoimmune diseases like PNS are likely to be initiated by tumor alterations that increase the immunogenicity of self-antigens. Unfortunately, sequence or structure-based methods for predicting immunogenic epitopes in silico have limited power (14) and current experimental methods for AAb discovery have several limitations. Many approaches for AAb detection utilize capture reagents that have been fixed or linearized and thus exclude AAbs targeting conformational protein epitopes. Recombinant proteins produced in non-human species like bacteria or yeast have been used as conformational capture antigens but fail to detect AAbs targeting mammalian- or human-specific post-translational modifications (PTMs) or disease-specific neoepitopes (15). Methods that aim to comprehensively map potential epitopes like serum epitope repertoire analysis (16) or tumor proteome fractionation (17) can be successful in identifying disease-specific AAbs, but require labor intensive deciphering of the cognate antigen on the back end. Alternative high-dimensional approaches to capture disease-relevant AAbs are needed.
To overcome these limitations, our group has developed a large format antibody array that captures circulating AAb-antigen (AAb-Ag) complexes from human plasma. Isolating AAb-Ag complexes directly from patients uniquely enables us to detect AAbs bound to their native tumor-derived epitopes ex vivo (18-20). Here, in multiple independent cohorts of SCLC, we identified and validated a set of Ags with their corresponding AAb that are upregulated before diagnosis and in both limited and extensive stage disease. These AAbs identified tumor-associated Ags, some of which were expressed on the cell surface in most SCLC tumors examined. Detailed analysis showed some of these proteins contained immunogenic PTMs including citrullination, isoaspartylation, and cancer-specific glycosylation that, when mimicked with synthetic PTM-containing peptides, confirmed binding to AAbs from patients with SCLC. Our findings support a unique, high-throughput approach to identify tumor-specific AAbs that could easily be applied to AAb discovery in other diseases such as autoimmunity or infection. A combination of the validated AAbs yielded remarkable utility in a risk prediction model for SCLC that was further improved by incorporating smoking pack years. If applied during lung cancer screening, this model has the potential to meaningfully impact the overall survival of this deadly disease through early detection of SCLC.
Results
Circulating autoantibodies in SCLC have greater sensitivity compared to other cancers.
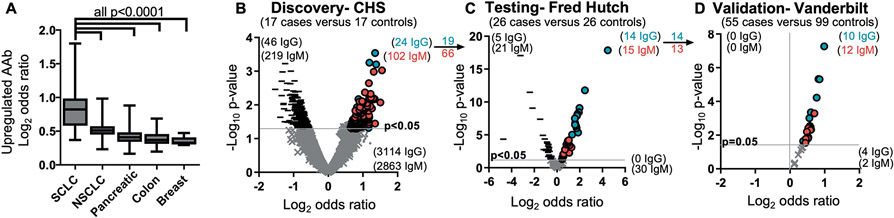
To demonstrate that cancer-specific AAbs were abundantly produced in SCLC, we measured upregulated AAb-Ag content (as a ratio between cases and controls) in a total of 1,009 plasma samples from cohorts of SCLC, non-small cell lung cancer (NSCLC), breast, colon, and pancreatic cancer. We found that both cancer-specific IgG and IgM concentrations were significantly higher (p<0.0001) in SCLC, and on average 4-fold higher, than in other cancers (Fig. 1A). This indicates a greater production of disease-specific AAb in SCLC and underscores the potential of AAbs as SCLC-specific biomarkers. We set out to identify specific AAbs consistently present across 3 independent cohorts of SCLC plasma. First, 17 case-control pairs of pre-diagnostic plasma samples from the Cardiovascular Health Study (CHS, Table 1) were analyzed for AAb-Ag complexes through an in-house discovery antibody array containing 3,600 antibody spots printed in triplicate (total 10,800 spots). We observed 46 IgG and 219 IgM downregulated and 24 IgG and 102 IgM upregulated AAb-Ag complexes (p<0.05, Fig. 1B). To further test for these specific AAb, we designed an antibody array that contained 14 independent mini-arrays on a single slide, each containing 19 of the 24 IgG and 66 of the 102 IgM capture antibodies by selecting upregulated biomarkers with the largest consistent differences between cases and controls and excluding 5 IgG and 18 IgM capture antibodies that had been discontinued without immunogen equivalent substitutions available. After testing 26 case-control pairs from the Fred Hutch diagnostic cohort (Table 1), we confirmed upregulation of 14 of the 19 IgG (73.6% confirmation) and 15 of the 66 IgM (22.7% confirmation) AAb-Ag complexes in cases compared to controls (p<0.05, Fig. 1C). To attempt to validate these results, 154 (55 SCLC cases and 99 matched controls) diagnostic plasma samples from Vanderbilt University (Table 1) were tested on mini-arrays. All AAb-Ag complexes remained upregulated, and 10 of 14 IgG (71.4%) and 12 of 13 IgM (92.3%) were validated (p<0.05, Fig. 1D). The complete list of 22 upregulated and validated AAb-Ag complexes can be found in table S1.
Figure 1. Antigen-complexed autoantibodies are highly upregulated in SCLC.
(A) The mean coefficient signal of upregulated (p<0.05) IgG and IgM autoantibodies in SCLC are higher (one-way ANOVA) than any other cancer type analyzed on antibody-antigen complex arrays. n=98 SCLC and 125 controls; n=47 NSCLC and 47 controls; n=86 pancreatic cancer and 86 controls; n=139 colon cancer and 139 controls; n=121 breast cancers and 121 controls. Box and whisker plots indicate the range of coefficients min to max for each sample. (B to D) IgG and IgM autoantibodies were sequentially discovered in the Cardiovascular Health Study (CHS) pre-diagnostic cohort (B), tested in the Fred Hutch diagnostic cohort (C), and validated in a third cohort from Vanderbilt (D). Blue circles indicate increased IgG against the target Ag; red circles indicate increased IgM against the target Ag; - indicates decreased antibodies against the target Ag; and × indicates no significant difference.
Table 1. Clinical characteristics of SCLC cohorts.
Age and pack year are averages in each group. Gender and smoking status are counts.
| Patient Characteristics |
CHS | Fred Hutch | Vanderbilt | |||
|---|---|---|---|---|---|---|
| SCLC | Control | SCLC | Control | SCLC | Control | |
| Age (year) | 70.6 | 70.8 | 63.9 | 63.9 | 70.8 | 69.2 |
| Gender | ||||||
| Male | 7 | 7 | 18 | 18 | 32 | 65 |
| Female | 10 | 10 | 8 | 8 | 23 | 35 |
| Smoking Status | ||||||
| Current | 6 | 6 | 15 | 10 | 23 | 41 |
| Former | 11 | 11 | 11 | 16 | 32 | 59 |
| Never | 0 | 0 | 0 | 0 | 0 | 0 |
| Pack years | 42.3 | 30.8 | 53.3 | 42.8 | 54.9 | 60.4 |
AAbs target tumor-associated antigens in SCLC.
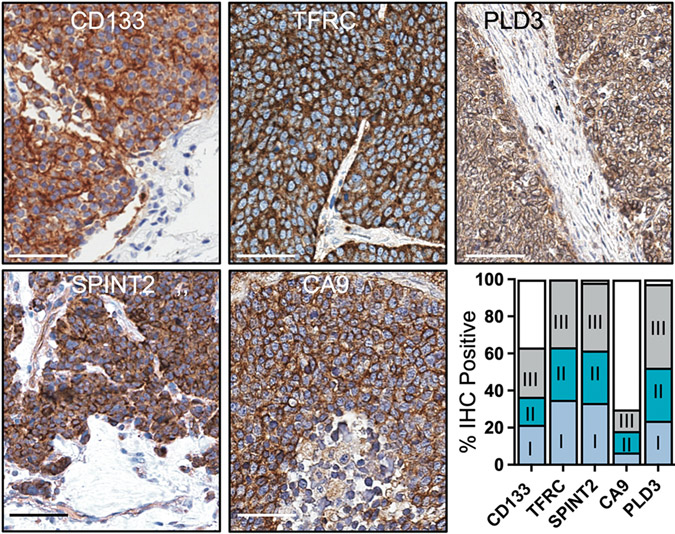
We hypothesized that the 22 validated AAbs were targeting tumor-associated antigens. To test if this was indeed the case, we prioritized 5 AAb antigens with transmembrane domains that would likely be expressed on the cell surface of tumor cells and have potential translational relevance in future applications (such as imaging or antibody drug conjugates). Using commercial antibodies for CD133, PLD3, TFRC, SPINT2 and CA9, we performed immunohistochemistry on SCLC tumor tissue microarrays to examine each protein’s expression. CD133, phospholipase D3 (PLD3), transferrin receptor (TFRC) and serine peptidase inhibitor, Kunitz type 2 (SPINT2) showed high, homogenous expression in tumor cells, but not in adjacent stroma (Fig. 2, staining controls in fig. S1). In tumors that expressed carbonic anhydrase 9 (CA9), most showed a focal staining pattern whereas approximately 20% displayed homogenous expression in tumor cells. We scored each core as positive (>5% of tumor cells stained) or negative (<5% of tumor cells stained) and found that >90% of tumor cores stained positive for TFRC, SPINT2, and PLD3, 60% for CD133, and 30% for CA9. Positive cores were further categorized into stage at diagnosis, which demonstrated that tumor associated antigens were expressed with similar frequencies in stage I, II, and III tumors. Cumulatively, this data validates our approach utilizing AAbs to discover tumor associated antigens that are expressed early and throughout tumorigenesis.
Figure 2. Antigens targeted by autoantibodies are upregulated in SCLC.
Representative immunohistochemical (IHC) images are shown of antigens targeted by peripheral autoantibodies on SCLC tissue microarrays (n=45 to 62 cases). Scale bar, 50μM. Bar graph contains positive or negative scoring of tissue microarray cores. Positive scores are further categorized by stage at diagnosis.
To determine if the identified tumor associated antigens were expressed in rare pulmonary neuroendocrine cells (PNE), a cell of origin for SCLC (21), we looked at expression using publicly available data from healthy human lung tissue (22). Only very low expression of mRNA for CD133 and CA9 was found in a low percentage of cells, moderate expression of TFRC mRNA was found in a low percentage of cells and moderate to higher expression of PLD3 and SPINT2 mRNA was found in about 30% of cells (fig. S2). In a dataset comparing PNE cells to non-NE cells isolated from mouse lungs (23), Pld3 and Spint2 mRNA had greater mRNA expression in PNE compared to non-NE in the lung, Car9 (the murine homologue for CA9), Grap2, Spink3 (the murine homologue for SPINK1), and Timp2 had no difference, and Cd133 and Tfrc had less expression in PNE (fig. S3). Thus, at the mRNA level in non-transformed lung, only PLD3 and SPINT2 are expressed in PNE cells in both mouse and human.
Despite SCLC having one of the highest tumor mutational burdens, very few SCLC cell lines or patient tumors had non-silent mutations in our 5 tumor-associated antigens. Out of 51 SCLC cell lines with publicly available data (24, 25), CD133 and CA9 were each mutated in 3, TFRC in 1 and SPINT2 and PLD3 had no SCLC cell lines with mutations (table S2). In the publicly available dataset from George et al. (26), where 110 SCLC tumor genomes were sequenced, mutations for TFRC were found in 3 samples, CD133 and CA9 in 2 samples, and no mutations were found in PLD3 and SPINT2. This low rate of mutation (0 to 5.8%) suggested that the neoantigens targeted by AAbs were likely occurring at the protein level. Since each of these proteins are exposed to the humoral immune system in normal tissues (fig. S1 to S3), we reasoned that the immune system was likely tolerant to their homeostatic expression and transformation events induced immunogenicity. This led us to investigate tumor-associated PTMs that could explain the observed production of AAbs.
Autoantibodies to CD133 target cancer-specific glycosylation motifs.
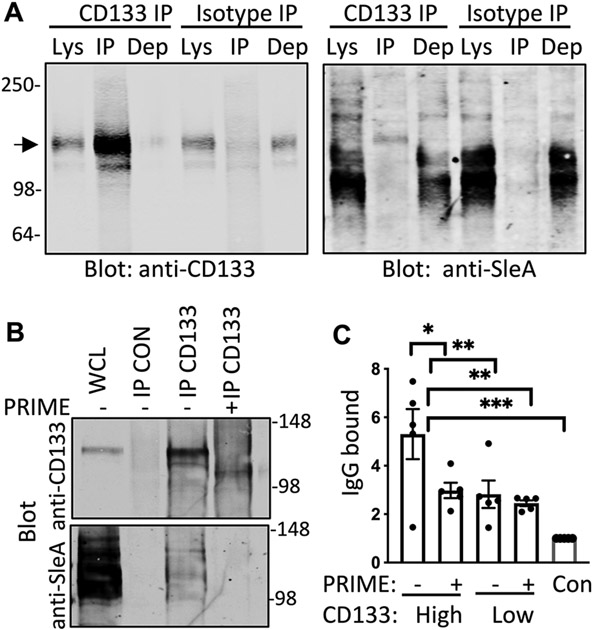
Tumor-associated glycans can induce autoantibody formation (27) and most tumor biomarkers are glycoproteins (such as prostate-specific antigen, CA125 (ovarian), CA19-9 (pancreas), α-fetoprotein (liver) and Carcinoembryonic antigen (colon)). To determine if AAb-identified antigens had tumor-specific glycan modifications, we screened for the cancer-enriched carbohydrate antigen sialyl-Lewis A (sLeA, also known as CA19-9) after immunoprecipitating CD133, PLD3, CA9 or SPINT2 from SCLC cell lines. In H82 cell lysates, sLeA appeared on multiple proteins, but CD133 immunoprecipitation yielded a lone sLeA-positive band at the correct size for CD133 (fig. S4A). This sLeA-positive, CD133-positive band was present after immunoprecipitation with either an anti-sLeA antibody or anti-CD133 antibody, but not when an anti-mouse antibody (isotype control) was used for precipitation (Fig. 3A). Immunoprecipitation of CD133 from H69, another SCLC cell line, or from LX110, FH14, and FH38 SCLC patient derived xenograft (PDX) models also showed a band positive for both SleA and CD133 (fig. S4B and C). To confirm glycosylation of CD133, we immunoprecipitated CD133 from H82 cell lines and treated with PRIME deglycosylase to remove N-linked glycans and complex sialylated structures like sLeA. PRIME treatment collapsed the migration of CD133, confirming the removal of glycan residues and eliminated sLeA detection on CD133 protein (Fig. 3B). We assessed the specificity of autoantibodies present in SCLC patient plasma for glycan modified CD133 by creating a modified sandwich enzyme-linked immunosorbent assay (ELISA) that replicated our array platform. A commercial CD133 antibody was plated to capture CD133 protein from H82 SCLC cell line lysates and, after incubation with plasma from patients with SCLC with known high or low concentration of CD133 AAbs (determined by array platform in Fig. 1C), the amount of IgG bound was verified to be different by ELISA (Fig. 3C). Treatment of H82 lysates with PRIME deglycosylase diminished autoantibody signal to the same degree as CD133-low plasma, arguing that glycan modifications were required for the increased autoantibody binding.
Figure 3. SCLC AAbs to CD133 target glycosylation motifs.
(A) Shown are immunoblots from H82 cell lysates (Lys) immunoprecipitated (IP) with CD133 antibody or isotype control and probed with anti-CD133 and anti-sLeA antibodies (N=3). Dep is depleted cell lysate after immunoprecipitation. The arrow indicates the expected protein size of CD133 at 133kDa (B) Shown are immunoblots from H82 lysates that were immunoprecipitated with isotype control antibody (CON) or CD133 antibody (N=3). Whole cell lysate (WCL) prior to immunoprecipitation is included to indicate the starting material. Lysates immunoprecipitated with CD133 antibody were then treated according to the manufacturer recommended deglycosylation protocol either with PRIME deglycosylase (+) or without enzyme (−). Immunoblots were probed with anti-CD133 and anti-sLeA antibodies. (C) ELISA quantification of autoantibody concentrations is shown for SCLC patient plasma that bound to CD133 from H82 cell lysates that had or had not been pre-treated with PRIME deglycosylase (n=5 per group). Con, negative control. Data are presented as individual values, the bar represents the mean ± SEM. *p<0.05, **p<0.005, ***p<0.0005 by student’s t test.
Autoantibodies bind to isoaspartate post translational modifications in SPINK1.
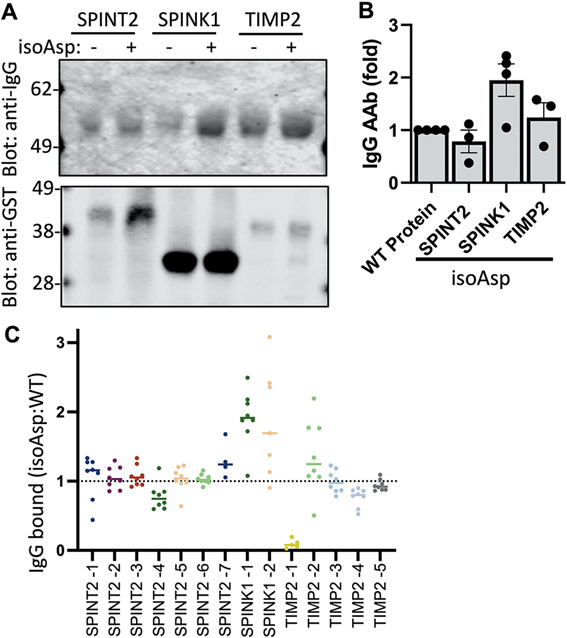
There has been a prior report that isoaspartate PTMs (isoAsp) present in ELAVL4, a PNS antigen, can be immunogenic in SCLC-associated autoimmunity (28). IsoAsp is a disease-enriched, naturally occurring PTM that is formed by deamidation of asparagine or isomerization of aspartate. To look for isoAsp-targeting AAbs, we used published in vitro methods to convert asparagine to isoAsp residues in GST fusions of the extracellular regions of SPINK1, SPINT2, and TIMP2 (29, 30). We then compared the ability of the isoAsp-modified versus the unmodified protein to pull down AAb from a pool of SCLC plasma and qualitatively showed more IgG AAb bound to isoAsp SPINK1, but not isoAsp SPINT2 or TIMP2, by immunoblotting (Fig. 4A and B). To determine specific sites of substitution, we had peptides synthesized for all the asparagine residues in the extracellular regions of SPINT2 (6 sites), SPINK1 (2 sites), and TIMP2 (4 sites) (table S3). We covalently bound both wild type (WT) asparagine or isoAsp peptides on assay plates, incubated with SCLC plasma samples, and detected AAb binding using anti-human IgG (Fig. 4C). In agreement with the immunoblotting data, we found isoAsp at both positions in SPINK1 bound at least 2-fold more AAbs than the corresponding WT peptide, whereas the other peptides bound less IgG and did not show PTM discrimination. Thus, we demonstrated AAbs in SCLC patient plasma recognize isoAsp neoepitope PTM sites on SPINK1.
Figure 4. SCLC AAb can target isoaspartylated post-translational modifications.
(A) Shown are immunoblots from GST-fusion proteins bound to glutathione beads, treated with or without isoaspartylation-inducing buffer (IsoAsp), incubated with pooled plasma from patients with SCLC (n=5 cases) and probed with anti-human IgG and anti-GST antibodies. (B) Multiple immunoblot experiments were quantified (N=3 to 4). Data are presented as individual data points and the bar represents the mean ± SEM (C) The ratio of IgG from SCLC patient plasma bound to IsoAsp compared to wild type (WT) peptides was measured by ELISA (n=4 to 8 cases). Horizontal bars indicate the mean.
SCLC AAbs recognize citrullinated TFRC not present in recombinant protein.
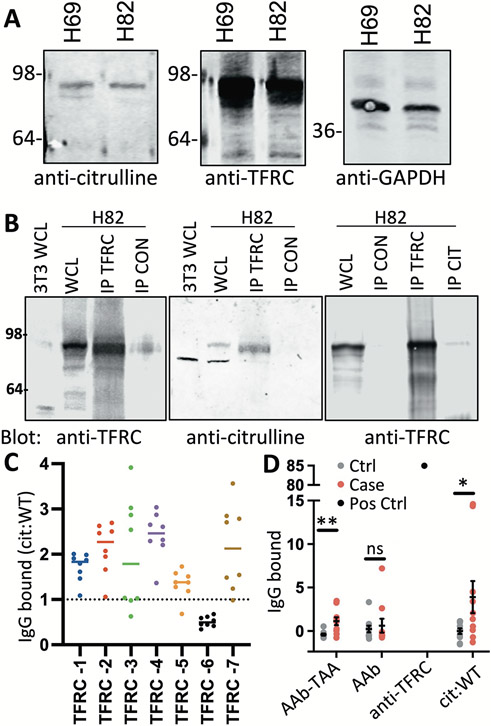
Citrulline, a unique amino acid generated by spontaneous arginine deamination, has also been shown to be immunogenic in cancer, but has yet to be described in SCLC (31-34). Therefore, we screened lysates from human SCLC H82 and H69 cells with antibodies to citrulline, TFRC, PLD3, CD133, SPINT2, and CA9 to look for any similarly sized banding patterns overlapping between citrulline and target proteins. TFRC had potential overlapping bands with citrulline that were apparent at the expected protein size (Fig. 5A and fig. S5A). To increase the sensitivity of citrulline detection, we immunoprecipitated with an anti-citrulline antibody and blotted back for target antibody expression, finding that TFRC was the only target protein detected in H82 cells (fig. S5B). We expanded these observations to show whole cell lysates from an immortalized fibroblast cell line, 3T3, had TFRC expression, but a corresponding band was not detected after probing with a citrulline antibody (Fig. 5B). Furthermore, immunoprecipitation with either anti-TFRC antibody and blotting back for citrulline or the reverse showed a matching band in H82 cells, but none in the controls, indicating TFRC has arginine residues converted to citrulline specifically in H82 cells. Immunoprecipitation of TFRC in FH14, FH38, and LX110 PDXs showed matching citrulline and TFRC bands, indicating TFRC has arginine residues converted to citrulline commonly in SCLC (fig. S5C). We considered the 32 arginines in the extracellular domain of TFRC and used published consensus motifs (35) to choose 7 likely to undergo deamidation to citrulline (table S3). Peptides representing portions of TFRC with either the wild-type arginine or citrullinated amino acid within the appropriate sequence were covalently bound to assay plates, and 5 of the 7 peptides (TFRC-1, 2, 3, 4, 7) with citrullinated residues pulled down more AAb from SCLC patient plasma than the corresponding WT peptide (Fig. 5C).
Figure 5. SCLC AAbs target citrullinated TFRC.
(A) Shown are immunoblots from H82 or H69 whole cell lysates (WCL) probed with TFRC or citruilline antibodies (N=3). GAPDH was used for a loading control. (B) Shown are immunoblots from 3T3 or H82 WCL and H82 lysates immunoprecipitated with TFRC, citrulline (CIT), or isotype control antibody (CON) and probed with anti-TFRC or anti-citrulline antibodies (N=3). (C) The ratio of IgG from SCLC patient plasma bound to citrullinated (Cit) compared to wild type (WT) peptides was measured by ELISA (n=8). (D) IgG from 10 patients with SCLC (case) and 10 controls (ctrl) captured in complex with antigen (AAb-TAA) by anti-TFRC antibody, free from antigen (AAb) by TFRC recombinant protein, or by cit/WT peptides were quantified by ELISA. An anti-TFRC antibody was used a positive control (pos ctrl) for TFRC recombinant protein. Data are presented as individual data points, the horizonal bar as mean ± SEM. *p<0.05 and **p<0.005 by paired sample t tests.
To further understand the extent that AAb could target PTM compared to unmodified protein, we looked at TFRC AAbs, either complexed to antigen or free from antigen, in a subset of plasma samples from the Fred Hutch cohort. We confirmed by ELISA that we could detect significantly (p<0.03) higher concentrations of AAb-TFRC complexes in SCLC plasma compared to controls. Conversely, no difference was detected in AAbs using TFRC recombinant protein as a capture antigen, although a commercial anti-TFRC antibody readily detected recombinant TFRC (Fig. 5D). Finally, inclusion of 2 citrullinated TFRC peptides as capture antigens bound more AAbs from SCLC plasma compared to controls, suggesting TFRC-AAbs were recognizing a citrullinated-neoantigen of TFRC not found on recombinant protein or arginine-containing WT peptides. Taken together, this data supports our AAb-Ag complex platform as a method to identify tumor-specific AAbs targeting post-translational neoantigens that would have been difficult to detect using other approaches.
Construction of a SCLC risk-prediction model.
Annual low dose computed tomography (CT) screening protocols currently in practice for early detection of NSCLC do not work for SCLC (36). However, the clinical benefit of SCLC early detection is clear: when detected at limited stage, conventional chemotherapy is curative in nearly 20% of patients, and surgical resection, generally not considered for SCLC, can be curative when combined with chemotherapy for very early stage patients (37, 38). Having demonstrated that SCLC patient AAbs target specific PTMs in SCLC antigens, we queried whether these unique AAbs could be utilized to generate a SCLC early detection risk prediction model. Therefore, we interrogated all 22 validated AAb-Ag complexes for their ability to reliably detect the presence of SCLC in plasma specimens. Since the Vanderbilt cohort had the largest sample size, we used this set as our training data and calculated the best 4 or 5 marker combinations by maximizing the area under the curve (AUC) based on logistic regression. To account for class switching of IgM to IgG over the course of an immune response, we included both AAb isotypes for each marker. This identified a 4-marker panel with the highest overall AUC of 0.872 comprised of AAb against PLD3, TIMP2, GRAP2, and SPINK1 and, with the addition of TFRC, a 5-marker panel had an AUC=0.874 (Table 2). After fixing the coefficients using the data from the Vanderbilt set, the 5-marker risk prediction model was tested in the CHS cohort yielding an AUC=0.700 and the Fred Hutch cohort with an AUC=0.950.
Table 2. Maximizing AUC Analysis to Develop a SCLC Risk Prediction Model in Vanderbilt Training Cohort.
| AAb Targets | Vanderbilt Training Cohort AUC |
|
|---|---|---|
| 4 Markers | PLD3; TIMP2; GRAP2; SPINK1 | 0.872 |
| 5 Markers | PLD3; TIMP2; GRAP2; TFRC; SPINK1 | 0.874 |
We fit a multiple logistic regression model on Vanderbilt cohort data by including in the model the 5 AAb markers yielding the highest AUC. The prediction model was applied on CHS and Fred Hutch testing datasets by partitioning cases and controls into high or low risk categories based on a positive or negative risk prediction score, respectively (Table 3). We found that the risk prediction model performed well with positive predictive values of 76.9% in the Vanderbilt cohort, 64.7% in the CHS cohort and 79.3% in the Fred Hutch cohort and negative predictive values of 85.3% in the Vanderbilt cohort, 64.7% in the CHS cohort and 86.9% in the Fred Hutch cohort. When the pre-diagnostic samples from CHS were grouped by their time to diagnosis, we had higher accuracy in flagging high risk samples within a year prior to diagnosis (7 of 11, 63.6%) than 1 to 2 years prior to diagnosis (3 of 6, 50%). Moreover, in both the Fred Hutch and Vanderbilt cohorts, high-risk designation correctly identified 23 of 26 (88.5%) limited stage SCLC and 18 of 24 (75%) extensive stage SCLC.
Table 3. The 5-AAb panel risk prediction model can accurately sort patient samples into high risk and low risk categories.
Samples were scored in each cohort as high or low risk based on a positive or negative risk prediction score, respectively.
| Vanderbilt | CHS | Fred Hutch | ||||
|---|---|---|---|---|---|---|
| control | case | control | case | control | case | |
| High Risk Score | 12 | 40 | 6 | 11 | 6 | 23 |
| Low Risk Score | 87 | 15 | 11 | 6 | 20 | 3 |
| Overall AUC | 0.874 | 0.700 | 0.950 | |||
| Overall Sensitivity | 0.727 | 0.647 | 0.885 | |||
| Overall Specificity | 0.879 | 0.647 | 0.679 | |||
| Sensitivity at 80% Specificity | 0.796 | 0.5 | 0.895 | |||
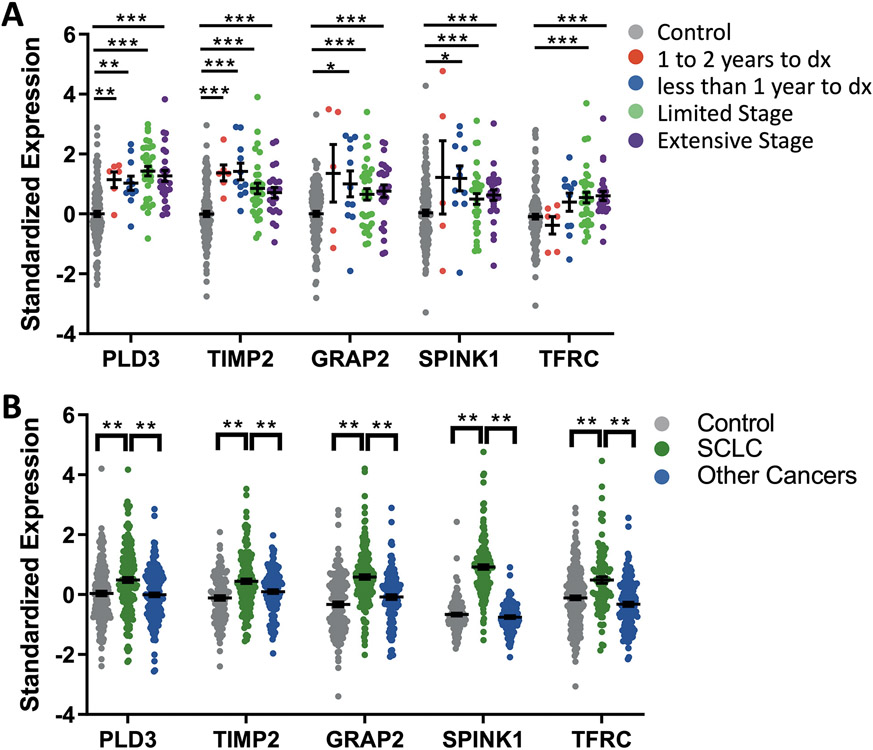
We looked at these 5 AAb individually within each cohort in greater detail. When blood was drawn within the year prior to diagnosis, AAbs targeting PLD3, TIMP2, GRAP2, and SPINK1 were significantly upregulated (p<0.05) (Fig. 6A). PLD3 and TIMP2 remained significantly upregulated (p<0.005) even up to two years prior to diagnosis. Since the cancer stage could potentially influence AAb concentrations, we separated the cases into limited and extensive stage to look at each of the markers. All 5 panel members were significantly upregulated (p<0.0005) at both limited and extensive stage disease. None of the panel markers were differentially expressed in current versus former smoker cases (fig. S6A). PLD3 was not associated with smoking pack years (fig. S6B) and TIMP2, GRAP2, SPINK1 and TFRC were only weakly associated (r<0.25) with smoking pack years (fig. S6C to F). Panel AAbs were not differentially expressed with chronic obstructive pulmonary disease (fig. S7A) or autoimmunity (fig. S7B) diagnoses. We examined at the individual performance of these markers in plasma from other cancers including non-SCLC, pancreatic cancer, and colon cancer, finding that none were upregulated in the other diseases compared to controls or SCLC (Fig. 6B), suggesting SCLC specificity. Cumulatively, these data suggest that these AAbs are specifically upregulated early in SCLC tumorigenesis and remain elevated throughout the disease process.
Figure 6. Panel autoantibodies can accurately detect SCLC.
(A) Panel autoantibodies were analyzed as a function of the time of blood draw in the CHS cohort (pre-diagnostic, 1 to 2 years prior to diagnosis (dx) or less than 1 year to dx) and Fred Hutch or Vanderbilt cohorts (diagnostic, limited or extensive stage); *p<0.05; **p<0.005, ***p<0.0005 unpaired t-test. (B) Panel autoantibodies are significantly higher in plasma from SCLC cases (n=98) compared to cases from other cancers (n=75 NSCLC, n=86 pancreatic, n=87 colon) or controls (n=175) from all cohorts **p<0.001 unpaired t-test. Data are presented as individual data points, the horizonal bar as mean ± SEM.
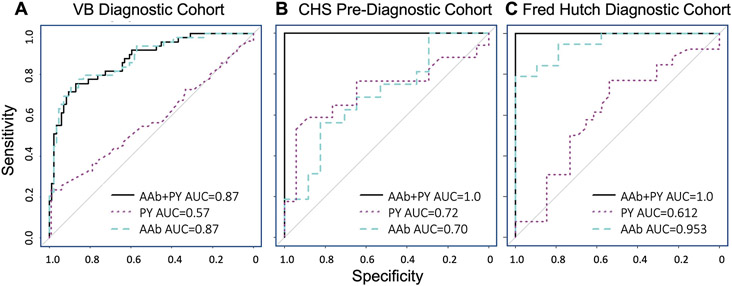
The single greatest risk factor for SCLC is current, heavy smoking, which can confer a 60 to 100-fold increase in the likelihood of developing SCLC for women and men, respectively (2). Since all three cohorts were matched on smoking history (current, former, never smokers), we incorporated pack year smoking history into the 5-panel model. The continuous variable of smoking pack years alone had an AUC equal to 0.57 in the Vanderbilt cohort, 0.72 in the CHS cohort and 0.612 in the Fred Hutch cohort (Fig. 7A to C). Combining smoking pack years with the AAb risk prediction model had minimal impact in the Vanderbilt cohort (AUC remained at 0.87) whereas the AUC increased to 1.0 in both the CHS and Fred Hutch cohorts.
Figure 7. A risk prediction model can accurately detect SCLC.
(A to C) Receiver operating characteristic (ROC) curves are shown for the 5-autoantibody (AAb, blue dashed line) complex panel identified by maximizing AUC in the Vanderbilt (VB) dataset (A), then fixing the coefficients and testing in the Cardiovascular Health Study (CHS) (B) and Fred Hutch (C) cohorts. ROC curves are also shown for pack years (PY, purple dotted line) and the combination of pack years and 5-AAb panel (black solid line).
Discussion
SCLC is one of the few malignancies with such poor outcomes that, like pancreatic cancer, it meets the definition of a “recalcitrant” cancer (39). This underscores the need for a greater understanding of the disease biology, more effective treatment strategies, and earlier intervention. In this study we show that our AAb-Ag complex capture platform takes advantage of endogenous humoral immunity to capture disease-relevant epitopes like PTMs in tumor specific antigens. By capturing AAbs bound to their immunogenic antigen, we were able to identify and validate a set of AAbs that had clinical utility in an early detection risk prediction model. Furthermore, a subset of AAbs identified tumor cell surface antigens that have the potential to be targeted in the therapeutic setting.
Although most of the 22 antigens had not been previously described as relevant to SCLC, there were a couple of interesting exceptions. CD133 overexpression has been reported in SCLC stem cells (40) and TFRC expression was higher in SCLC plasma derived microvesicles compared to controls (41). Notably, expression of antigen and production of AAb, regardless of disease stage, suggested that the AAb-identified tumor associated antigens are not lost to immunoediting. In addition to expression, our finding that AAbs target PTM epitopes adds to the growing body of evidence that tumor-specific AAbs predominantly recognize non-mutated self-proteins (42-44). We were able to identify immunogenic PTMs to 3 of our tumor-specific AAbs, but more likely exist as we only screened for a few probable PTM modifications and only at predicted sites. In autoimmunity, AAbs have been identified against many types of PTMs including phosphorylation, acetylation, hydroxylation, nitration, and carbamylation (45) and these PTM-AAbs can make highly specific biomarkers. For example, anti-citrullinated protein antibodies in the serum of patients with rheumatoid arthritis are highly diagnostic (46), prognostic (47), and can be detected years before clinical symptoms (48). Extending this observation to cancer suggests a common underlying mechanism responsible for breaching B cell tolerance in multiple disease contexts.
AAbs are uniquely well suited for early detection biomarkers as they are obtained through a simple blood draw, produced in detectable quantities (even with low antigen concentration), upregulated years prior to clinical symptoms, and highly antigen-specific (49). One of the major reasons that SCLC early detection could prove successful is that there exists a clearly defined high-risk population: heavy cigarette smokers (50). Since low-dose CT screening for heavy smokers shows a proven reduction in mortality, this uniform at-risk population is already eligible for ongoing lung cancer screening in many countries and is paid for by most insurance and Medicare in the US (51). Adding a blood draw to screening visits is feasible and a procedure accepted by patients (52, 53). However, all useful early detection approaches must demonstrate a low false positive rate to reduce the potential harms of screening. Given that smoking confers such a large relative risk for SCLC, to reduce false positives we envision only screening current, heavy smokers (those with greater than 50 pack years) through an automated AAb immunoassay to sort into high or low risk of malignancy categories. An AAb high-risk test result would flag a review or acquisition of a diagnostic CT, whereas an AAb low-risk result would indicate continued annual lung cancer screening using low-dose CT. There is also the potential of adding in additional factors to further define the patients at highest risk for development of SCLC. Germline genotyping of cancer-predisposing genes recently demonstrated that some patients with SCLC may have an inherited predisposition (54). Liquid biopsy assays that quantify a panel of somatic variants in cell free DNA (cfDNA) have shown utility in monitoring disease burden (55) and relapse (56) in patients with limited stage disease, although the panel has not been tested in a screening setting. TP53 mutations in cfDNA were present in approximately 35% of early stage SCLC but also in 11% of non-cancer controls (57) limiting its stand-alone utility. Additionally, AAbs to PNS antigens could pair well with the non-PNS AAbs discovered here. Thus, combinations with additional assays could potentially better define a risk group to be screened for specific antibodies or improve the sensitivity or specificity of the prediction rule.
Several limitations of our studies should be noted. First, each of our plasma cohorts are relatively small, although this concern is somewhat mitigated by the consistent validation of the 22 markers over 3 independent cohorts. Second, the 4- and 5-parameter risk prediction models were generated through the same samples used to select the 22 markers, so further validation in a larger, prospectively collected, screening cohort will be necessary (although such cohorts effectively do not exist today). To facilitate this, the antibody-based nature of our platforms can be easily converted to more high-throughput and lower cost platforms such as multiple smaller arrays on a single slide or ELISAs. Third, although our discovery antibody array contains over 3,600 antibodies to approximately 2,500 proteins that were selected for cancer relevance, there are almost certainly other SCLC- and PNS- specific antigens that might also perform well. Fourth, although we did not find upregulation of the panel AAbs in other types of cancer we examined, these cohorts did not include neuroendocrine subtypes. It is possible that other neuroendocrine tumors might also produce AAbs to these markers. Finally, we probed for only 3 types of PTMs that are cancer-enriched but others almost certainly exist. Additional studies with PTM-specific antibodies from both cancerous and healthy tissues will be needed to fully understand the breadth of cancer-specific neoantigens present in SCLC and specificity of the AAb response.
In summary, we developed a high-dimensional method to identify tumor-specific autoantigen-AAb complexes ex vivo. We found that isolating tumor targeted AAbs to their disease-specific antigens can contribute to accurate detection of early-stage SCLC. We also describe AAb production to non-PNS antigens and that SCLC-specific PTMs can cause AAb production to neoepitopes in proteins that one would otherwise suspect to be immune tolerant. Our findings are relevant not only for our understanding of SCLC tumorigenesis, but also to the broader field of immune related biomarkers and could lead to implementation of early detection strategies and antibody-based therapeutics.
Materials and Methods
Study Design
This study was designed to evaluate the AAb response in SCLC. We discovered AAbs in a pre-diagnostic cohort of plasma samples from SCLC and healthy controls and confirmed them in two independent diagnostic cohorts. We evaluated the expression of AAb-targeted antigens by immunohistochemistry on SCLC tissue microarrays and non-tumor tissue and designed experiments to understand the mechanisms underlying AAb production against tumor antigens with transmembrane domains. We probed for SCLC-specific AAb reaction with three types of post-translational modifications in human SCLC cell lines, PDXs or peptides. The number of replicates and statistical analyses are reported below and in the figure legends. No data points were removed as outliers.
Study Cohorts
All human plasma samples were obtained following institutional review board approvals and informed consent. This study used three independent plasma cohorts: 1) The Cardiovascular Health Study (CHS) is a population-based, longitudinal study of coronary heart disease and stroke (58). Clinical examinations and plasma samples were completed annually for up to 10 years. All 17 cases were newly diagnosed with SCLC within 24 months after a blood draw were included. These 17 cases were individually matched to controls based on age, gender, body mass index and smoking history. 2) The Fred Hutchinson Early Detection and Prevention Clinic (Fred Hutch) for pulmonary nodule evaluation provided n=26 patients with SCLC and n=26 patients with benign pulmonary nodules (active IRB protocol no. 6663). Corresponding clinicopathological data was maintained in a highly-annotated database. The controls were matched to cases based on age, gender and smoking history. 3) 55 diagnostic SCLC plasma samples and 99 unmatched control plasma samples were matched on age, gender, and smoking history and generously provided by Vanderbilt University Nashville, TN. All laboratory steps were performed blind to sample information.
AAb-Ag Complex Detection
The detection of AAb-Ag complexes using antibody arrays has been previously described (19). Briefly, antibody microarrays were printed in-house, incubated with patient plasma diluted 1:80, and probed with anti-human IgG-SeTau647 or IgM-DyLight550 antibody. Microarrays were imaged on a GenePix 4000B scanner and spot intensities measured in GenePix Pro 6.0 (Molecular Devices).
Immunohistochemistry
Tissue microarrays (TMAs) containing 62 individual SCLC cases and 6 individual lung tissue controls (RLN681A, US Biolab) were stained for CD133 (#64326, 1:1000 Cell Signaling Technologies) or the same concentration of anti-rabbit IgG isotype antibody using the automated Leica Bond RX system and detected using rabbit-horseradish peroxidase (HRP) and 3,3'-Diaminobenzidine (DAB) from Leica. The same TMAs were also used to stain for TFRC (HPA028598, 1:500 Sigma), SPINT2 (HPA011101, 1:200 Sigma), and CA9 (HPA055207, 1:1000 Sigma) after heat induced epitope retrieval in citrate pH 6 buffer. Sections were probed with biotinylated pan-specific universal antibody for 10 minutes, streptavidin-HRP for 5 minutes (PK-8800, Vector Laboratories), and counterstained with hematoxylin with washes in between each step. After RLN681A was discontinued, a different TMA was purchased from US Biomax (BS04116a) containing 45 duplicate SCLC cores and 10 lung tissue controls and stained for PLD3 (HPA012800, 1:200 Sigma) using the Vector protocol described above. Scoring of TMA staining was performed in a blinded manner where each biopsy was determined to be either positive (greater than 5% tumor cells stained) or negative (less than 5% tumor cells stained). TMAs containing 33 normal tissues (3 individuals per organ, FDA999-1, US Biolab) were also stained for each of the antibodies as positive or negative controls. Stained slides were digitized using Leica Aperio whole slide scanner with 20x magnification.
Transcriptomic Data
Single-cell RNA sequencing data for lung neuroendocrine and lung epithelial cells was analyzed using the CELLxGENE Gene Expression tool from the Chan Zuckerberg Initiative (22). RNA Sequencing (RNAseq) data from cohort GSE136580 was procured from the GEO Database (https://www.ncbi.nlm.nih.gov/geo/) (23). Whole genome sequencing was used to assess expression data and somatic mutations from the Cancer Cell Line Encyclopedia (CCLE) using the DepMap Public 21Q3 dataset (24-25), or SCLC patient sample as described by George et al. in their supplementary table 3 (26).
ELISAs
Plate-based assays were developed to quantify autoantibody concentrations to different capture approaches. WT and PTM peptides were purchased from CHI Scientific (sequences are listed in table S3) and 10μg of peptide was plated per well in maleimide 96 well plates (#15152, Pierce). White 96 well plates (#15042, Pierce) were coated with 1μg/well of anti-CD133 antibody (HB#7, Developmental Studies Hybridoma Bank) to capture CD133 from H82 lysates treated with or without PRIME glycosylase (#50-999-475, Fisher Scientific) at 1U/50 μg cell lysate. Anti-TFRC antibody (HPA028598, Sigma-Aldrich) or TFRC recombinant protein (89-760aa, #11020-H07H, Sinobiological) was plated at 0.5μg/well. After binding capture approach, all plates were washed and used as bait for autoantibodies from human plasma (1:500). Autoantibody signals were quantified using anti-human IgG-HRP secondary (I:5000, #709-065-149, Jackson ImmunoResearch) and SuperSignal ELISA Femto (#37074, Thermo Fisher Scientific) and a SpectroMax L Microplate reader (Molecular Devices).
Immunoblotting
To induce isoaspartylation, we combined GST-fusion proteins on magnetic glutathione beads (#78601, Thermo Fisher Scientific) at 1:20 with NH4HCO3 (100mM, pH 8.0) or phosphate-buffered saline (PBS) with 0.05% sodium azide and incubated at 37°C for 48 hours. After washing, treated GST-fusion proteins were incubated with human plasma at 1:10,000 4°C overnight. Samples were directly lysed in Laemmli Sample Buffer containing 1x complete protease inhibitor cocktail, 1x phosphatase inhibitor cocktail, and 2mM phenylmethylsulfonyl fluoride (all from Millipore Sigma) and sonicated. Samples were separated by SDS-PAGE, blotted to nitrocellulose, checked for load with Ponceau S, blocked in 1% non-fat dry milk and probed with 1:5000 anti-human IgG-AF680 (Jackson ImmunoResearch Labs Cat# 709-625-149, RRID:AB_2340582) and 1:500 mouse anti-GST (FHCC Antibody Technology Core) detected by anti-mouse IgG-Alexa Fluor 680 (1:10000, Thermo Fisher Scientific Cat# A32729, RRID:AB_2633278). Immunoblots were imaged on an Odyssey CLx (LiCor) with ImageStudio v5. For all immunoprecipitation experiments, whole cell lysates (WCL) were prepared from 1/30th of total lysate. CD133 protein was immunoprecipitated from H82 cells and treated with or without PRIME deglycosylase (#50-999-475, Fisher Scientific) at 1U/50 μg cell lysate at 37°C for 30 minutes and immunoblotted as described above with any differences noted below. Immunoblots were probed with antibodies to sLeA (1:2000, Fitzgerald Industries International Cat# 10-C04E, RRID:AB_1282844) and CD133 (1:1000, #6436, Cell Signaling Technologies) and detected with anti-mouse IgM-Alexa Fluor 680 and anti-rabbit IgG-Alexa Fluor 790 (1:10000, #A10038, #A10043, Thermo Fisher Scientific), respectively. Lysates from SCLC cell lines or PDXs were immunoprecipitated with 1μg of anti-TFRC (#13-6800 Invitrogen), anti-citrulline (ab100932, AbCam) or isotype control antibody (FHCC Antibody Technology Core) and protein G magnetic beads (#88847, Thermo Fisher Scientific). Immunoblots were probed with antibodies to TFRC (1:1000, #13113, Cell Signaling) or citrulline (1:500, MA5-27573, Invitrogen) and detected with anti-mouse IgG-Alexa Fluor 680 and anti-rabbit IgG-Alexa Fluor 790 (1:10000, A32729, A10043, Thermo Fisher Scientific), respectively. Uncropped western blots are presented in fig. S8 to S10.
Human SCLC Cells
The NCI-H82 and NCI-H69 human cell lines were acquired from the American Type Culture Collection (ATCC) and grown in Dulbecco’s Modified Eagle Medium supplemented with 4mM L-glutamine, 10% Fetal Bovine Serum and 1% Penicillin Streptomycin in an incubator at 37°C and 5% CO2. Cells were routinely tested negative for mycoplasma by polymerase chain reaction. The patient-derived xenografts from SCLC circulating tumor cells (FH14, FH38) or tumor biopsies (LX110) have been previously described (59).
Statistical Analyses
Raw, individual-level data are presented in data file S1. Array data contain a format identical to two-channel gene expression arrays and analysis proceeds analogously as described previously(19). The p values indicated in Fig. 1 were not corrected for multiple comparisons since we used a discovery, confirmation, validation approach (60). In other figures, two groups were compared by t-test (type of t-test noted in figure legends) and three or more groups were compared using one-way analysis of variance with Tukey’s test using GraphPad Prism. Logistic regression was used to identify the combination of multiple autoantibodies that best distinguished cases from controls. Receiver-Operator Characteristic curve was created and the Area Under the Curve was computed to evaluate the prediction capability. All related statistical analyses to the risk prediction model were performed using R statistical software (61).
Supplementary Material
Acknowledgments:
Our co-author, Dr. Pierre Massion, contributed precious samples and thoughtful advice prior to his death last year.
Funding:
This research was supported by National Institutes of Health (P50CA228944, R01CA243328) to AMH and PLD, (KL2TR002317) to KJL, (U01CA152662) to E.L.G and P.P.M., and (P30CA015704) to Fred Hutch Cancer Center. CHS samples were used with support from HHSN268201200036C, HHSN268200800007C, HHSN268201800001C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, 75N92021D00006, and U01HL080295 and U01HL130114 from the National Heart, Lung, and Blood Institute, with additional contribution from the National Institute of Neurological Disorders and Stroke, and by R01AG023629 from the National Institute on Aging. A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org.
Footnotes
Competing interests: KJL, AMH, and PDL are co-inventors on 3 patent applications: “Small cell lung cancer tumor antigens and uses thereof” (63/415,142), “Autoantibodies and autoantibody-autoantigen complexes as biomarkers of small cell lung cancer” (17/040,008), and “Antigen binding molecules for small cell lung cancer” (17/025,991). All authors declare they have no consulting, paid or unpaid, related to this work. The following authors have consulting declarations unrelated to this work: PDL is on the Scientific Advisory Board of Xequel Bio and consults on NSF grant (EECS #1930649) to PI Dr. Donglei Fan; ALF consults on the NIH R21 grant ““Genetic risk for trajectories of cognitive decline and progression to dementia among Mexican Americans and non-Hispanic whites” to PI Dr. Xueqiu Jian.
Data and materials availability: All data associated with this study are in the paper or supplementary materials. All resources generated in this study are available upon request from the lead author upon providing a completed material transfer agreement.
References and Notes
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A, Cancer statistics, 2022. CA Cancer J Clin 72, 7–33 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Wang S, Tang J, Sun T, Zheng X, Li J, Sun H, Zhou X, Zhou C, Zhang H, Cheng Z, Ma H, Sun H, Survival changes in patients with small cell lung cancer and disparities between different sexes, socioeconomic statuses and ages. Sci Rep 7, 1339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindwasser OW, Ujhazy P, Antman MA, Prindiville SA, Small Cell Lung Cancer and the Recalcitrant Cancer Research Act. J Thorac Oncol 12, S1546 (2017). [Google Scholar]
- 4.Alvarado-Luna G, Morales-Espinosa D, Treatment for small cell lung cancer, where are we now?-a review. Transl Lung Cancer Res 5, 26–38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Höftberger R, Rosenfeld MR, Dalmau J, Update on neurological paraneoplastic syndromes. Curr Opin Oncol 27, 489–495 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gozzard P, Woodhall M, Chapman C, Nibber A, Waters P, Vincent A, Lang B, Maddison P, Paraneoplastic neurologic disorders in small cell lung carcinoma. Neurology 85, 235–239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anwar A, Jafri F, Ashraf S, Jafri MAS, Fanucchi M, Paraneoplastic syndromes in lung cancer and their management. Ann Transl Med 7, 359 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darnell RB, Posner JB, Paraneoplastic Syndromes Involving the Nervous System. N Engl J Med 349, 1543–1554 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Sebastian M, Koschade S, Stratmann JA, SCLC, Paraneoplastic Syndromes, and the Immune System. J Thorac Oncol 14, 1878–1880 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Honnorat J, Antoine J-C, Paraneoplastic neurological syndromes. Orphanet J Rare Dis 2, 22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazarian M, Laird-Offringa IA, Small-cell lung cancer-associated autoantibodies: potential applications to cancer diagnosis, early detection, and therapy. Mol Cancer 10, 33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zekeridou A, Majed M, Heliopoulos I, Lennon VA, Paraneoplastic autoimmunity and small-cell lung cancer: Neurological and serological accompaniments. Thorac Cancer 10, 1001–1004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gozzard P, Chapman C, Vincent A, Lang B, Maddison P, Zhang L, Ed. Novel Humoral Prognostic Markers in Small-Cell Lung Carcinoma: A Prospective Study. PLoS ONE 10, e0143558 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jespersen MC, Mahajan S, Peters B, Nielsen M, Marcatili P, Antibody Specific B-Cell Epitope Predictions: Leveraging Information From Antibody-Antigen Protein Complexes. Front Immunol 10, 298 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyle HA, Mamula MJ, Post-translational protein modifications in antigen recognition and autoimmunity. Trends Immunol 22, 443–449 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Kamath K, Reifert J, Johnston T, Gable C, Pantazes RJ, Rivera HN, McAuliffe I, Handali S, Daugherty PS, Antibody epitope repertoire analysis enables rapid antigen discovery and multiplex serology. Sci Rep 10, 5294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu J, Choi G, Li L, Wang H, Pitteri SJ, Pereira-Faca SR, Krasnoselsky AL, Randolph TW, Omenn GS, Edelstein C, Barnett MJ, Thornquist MD, Goodman GE, Brenner DE, Feng Z, Hanash SM, Occurrence of Autoantibodies to Annexin I, 14-3-3 Theta and LAMR1 in Prediagnostic Lung Cancer Sera. J Clin Oncol 26, 5060–5066 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirus JE, Zhang Y, Li CI, Lokshin AE, Prentice RL, Hingorani SR, Lampe PD, Cross-species antibody microarray interrogation identifies a 3-protein panel of plasma biomarkers for early diagnosis of pancreas cancer. Clin Cancer Res 21, 1764–1771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rho J, Lampe PD, High-throughput screening for native autoantigen-autoantibody complexes using antibody microarrays. J Proteome Res 12, 2311–2320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lastwika KJ, Kargl J, Zhang Y, Zhu X, Lo E, Shelley D, Ladd JJ, Wu W, Kinahan P, Pipavath SNJ, Randolph TW, Shipley M, Lampe PD, Houghton AM, Tumor-derived Autoantibodies Identify Malignant Pulmonary Nodules. Am J Respir Crit Care Med 199, 1257–1266 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song J-Y, Berns A, Cell of Origin of Small Cell Lung Cancer: Inactivation of Trp53 and Rb1 in Distinct Cell Types of Adult Mouse Lung. Cancer Cell 19, 754–764 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Chan Zuckerberg CELLxGENE Discover Cellxgene Data Portal (available at https://cellxgene.cziscience.com/).
- 23.Ouadah Y, Rojas ER, Riordan DP, Capostagno S, Kuo CS, Krasnow MA, Rare Pulmonary Neuroendocrine Cells Are Stem Cells Regulated by Rb, p53, and Notch. Cell 179, 403–416.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Cell Line Encyclopedia Consortium, Genomics of Drug Sensitivity in Cancer Consortium, Pharmacogenomic agreement between two cancer cell line data sets. Nature 528, 84–87 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jané-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA, The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, Leenders F, Lu X, Fernández-Cuesta L, Bosco G, Müller C, Dahmen I, Jahchan NS, Park K-S, Yang D, Karnezis AN, Vaka D, Torres A, Wang MS, Korbel JO, Menon R, Chun S-M, Kim D, Wilkerson M, Hayes N, Engelmann D, Pützer B, Bos M, Michels S, Vlasic I, Seidel D, Pinther B, Schaub P, Becker C, Altmüller J, Yokota J, Kohno T, Iwakawa R, Tsuta K, Noguchi M, Muley T, Hoffmann H, Schnabel PA, Petersen I, Chen Y, Soltermann A, Tischler V, Choi C, Kim Y-H, Massion PP, Zou Y, Jovanovic D, Kontic M, Wright GM, Russell PA, Solomon B, Koch I, Lindner M, Muscarella LA, la Torre A, Field JK, Jakopovic M, Knezevic J, Castaños-Vélez E, Roz L, Pastorino U, Brustugun O-T, Lund-Iversen M, Thunnissen E, Köhler J, Schuler M, Botling J, Sandelin M, Sanchez-Cespedes M, Salvesen HB, Achter V, Lang U, Bogus M, Schneider PM, Zander T, Ansén S, Hallek M, Wolf J, Vingron M, Yatabe Y, Travis WD, Nürnberg P, Reinhardt C, Perner S, Heukamp L, Büttner R, Haas SA, Brambilla E, Peifer M, Sage J, Thomas RK, Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tikhonov A, Smoldovskaya O, Feyzkhanova G, Kushlinskii N, Rubina A, Glycan-specific antibodies as potential cancer biomarkers: a focus on microarray applications. Clin Chem Lab Med 58, 1611–1622 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Pulido MA, DerHartunian MK, Qin Z, Chung EM, Kang DS, Woodham AW, Tsou JA, Klooster R, Akbari O, Wang L, Kast WM, Liu SV, Verschuuren JJGM, Aswad DW, Laird-Offringa IA, Isoaspartylation appears to trigger small cell lung cancer-associated autoimmunity against neuronal protein ELAVL4. J Neuroimmunol 299, 70–78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geiger T, Clarke S, Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J Biol Chem 262, 785–794 (1987). [PubMed] [Google Scholar]
- 30.Curnis F, Longhi R, Crippa L, Cattaneo A, Dondossola E, Bachi A, Corti A, Spontaneous Formation of L-Isoaspartate and Gain of Function in Fibronectin. J Biol Chem 281, 36466–36476 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Brentville VA, Metheringham RL, Gunn B, Symonds P, Daniels I, Gijon M, Cook K, Xue W, Durrant LG, Citrullinated Vimentin Presented on MHC-II in Tumor Cells Is a Target for CD4+ T-Cell-Mediated Antitumor Immunity. Cancer Res 76, 548–560 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Cook K, Daniels I, Symonds P, Pitt T, Gijon M, Xue W, Metheringham R, Durrant L, Brentville V, Citrullinated α-enolase is an effective target for anti-cancer immunity. Oncoimmunology 7, 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulido MA, DerHartunian MK, Qin Z, Chung EM, Kang DS, Woodham AW, Tsou JA, Klooster R, Akbari O, Wang L, Kast WM, Liu SV, Verschuuren JJGM, Aswad DW, Laird-Offringa IA, Isoaspartylation appears to trigger small cell lung cancer-associated autoimmunity against neuronal protein ELAVL4. J Neuroimmunol 299, 70–78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doyle HA, Zhou J, Wolff MJ, Harvey BP, Roman RM, Gee RJ, Koski RA, Mamula MJ, Isoaspartyl post-translational modification triggers anti-tumor T and B lymphocyte immunity. J Biol Chem 281, 32676–32683 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Ju Z, Wang S-Y, Prediction of citrullination sites by incorporating k-spaced amino acid pairs into Chou’s general pseudo amino acid composition. Gene 664, 78–83 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Thomas A, Pattanayak P, Szabo E, Pinsky P, Characteristics and Outcomes of Small Cell Lung Cancer Detected by CT Screening. Chest 154, 1284–1290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lassen U, Osterlind K, Hansen M, Dombernowsky P, Bergman B, Hansen HH, Long-term survival in small-cell lung cancer: posttreatment characteristics in patients surviving 5 to 18+ years--an analysis of 1,714 consecutive patients. J Clin Oncol 13, 1215–1220 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Koletsis EN, Prokakis C, Karanikolas M, Apostolakis E, Dougenis D, Current role of surgery in small cell lung carcinoma. J Cardiothorac Surg 4, 30 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eshoo AG, Text - H.R.733 - 112th Congress (2011-2012): Recalcitrant Cancer Research Act of 2012 (2012) (available at https://www.congress.gov/bill/112th-congress/house-bill/733/text). [Google Scholar]
- 40.Sarvi S, Mackinnon AC, Avlonitis N, Bradley M, Rintoul RC, Rassl DM, Wang W, Forbes SJ, Gregory CD, Sethi T, CD133+ Cancer Stem-like Cells in Small Cell Lung Cancer Are Highly Tumorigenic and Chemoresistant but Sensitive to a Novel Neuropeptide Antagonist. Cancer Res 74, 1554–1565 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Pedersen S, Jensen KP, Honoré B, Kristensen SR, Pedersen CH, Szejniuk WM, Maltesen RG, Falkmer U, Circulating microvesicles and exosomes in small cell lung cancer by quantitative proteomics. Clin Proteomics 19, 2 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banville AC, Nelson BH, Breaching B cell tolerance in the tumor microenvironment. Cancer Cell 40, 356–358 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Zaenker P, Gray ES, Ziman MR, Autoantibody Production in Cancer—The Humoral Immune Response toward Autologous Antigens in Cancer Patients. Autoimmun Rev 15, 477–483 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Mazor RD, Nathan N, Gilboa A, Stoler-Barak L, Moss L, Solomonov I, Hanuna A, Divinsky Y, Shmueli MD, Hezroni H, Zaretsky I, Mor M, Golani O, Sabah G, Jakobson-Setton A, Yanichkin N, Feinmesser M, Tsoref D, Salman L, Yeoshoua E, Peretz E, Erlich I, Cohen NM, Gershoni JM, Freund N, Merbl Y, Yaari G, Eitan R, Sagi I, Shulman Z, Tumor-reactive antibodies evolve from non-binding and autoreactive precursors. Cell 185, 1208–1222 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Doyle HA, Mamula MJ, Autoantigenesis: the evolution of protein modifications in autoimmune disease. Curr Opin Immunol 24, 112–118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO, Birnbaum NS, Burmester GR, Bykerk VP, Cohen MD, Combe B, Costenbader KH, Dougados M, Emery P, Ferraccioli G, Hazes JMW, Hobbs K, Huizinga TWJ, Kavanaugh A, Kay J, Kvien TK, Laing T, Mease P, Ménard HA, Moreland LW, Naden RL, Pincus T, Smolen JS, Stanislawska-Biernat E, Symmons D, Tak PP, Upchurch KS, Vencovský J, Wolfe F, Hawker G, 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum 62, 2569–2581 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Meyer O, Labarre C, Dougados M, Goupille P, Cantagrel A, Dubois A, Nicaise-Roland P, Sibilia J, Combe B, Anticitrullinated protein/peptide antibody assays in early rheumatoid arthritis for predicting five year radiographic damage. Ann Rheum Dis 62, 120–126 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rantapää-Dahlqvist S, de Jong BAW, Berglin E, Hallmans G, Wadell G, Stenlund H, Sundin U, van Venrooij WJ, Antibodies against cyclic citrullinated peptide and IgA rheumatoid factor predict the development of rheumatoid arthritis. Arthritis Rheum 48, 2741–2749 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Kobayashi M, Katayama H, Fahrmann JF, Hanash SM, Development of autoantibody signatures for common cancers. Semin Immunol 47, 101388 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Kenfield SA, Wei EK, Stampfer MJ, Rosner BA, Colditz GA, Comparison of aspects of smoking among the four histological types of lung cancer. Tob Control 17, 198 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramaswamy A, Lung Cancer Screening: Review and 2021 Update. Curr Pulmonol Rep 11, 15–28 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taber JM, Aspinwall LG, Heichman KA, Kinney AY, Preferences for blood-based colon cancer screening differ by race/ethnicity. Am J Health Behav 38, 351–361 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Relton A, Collins A, Guttery DS, Gorsia DN, McDermott HJ, Moss EL, Patient acceptability of circulating tumour DNA testing in endometrial cancer follow-up. Eur J Cancer Care (Engl) 30, e13429 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tlemsani C, Takahashi N, Pongor L, Rajapakse VN, Tyagi M, Wen X, Fasaye G-A, Schmidt KT, Desai P, Kim C, Rajan A, Swift S, Sciuto L, Vilimas R, Webb S, Nichols S, Figg WD, Pommier Y, Calzone K, Steinberg SM, Wei JS, Guha U, Turner CE, Khan J, Thomas A, Whole-exome sequencing reveals germline-mutated small cell lung cancer subtype with favorable response to DNA repair–targeted therapies. Sci Transl Med 13, 578 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Almodovar K, Iams WT, Meador CB, Zhao Z, York S, Horn L, Yan Y, Hernandez J, Chen H, Shyr Y, Lim LP, Raymond CK, Lovly CM, Longitudinal Cell-Free DNA Analysis in Patients with Small Cell Lung Cancer Reveals Dynamic Insights into Treatment Efficacy and Disease Relapse. J Thorac Oncol 13, 112–123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iams WT, Kopparapu PR, Yan Y, Muterspaugh A, Zhao Z, Chen H, Cann C, York S, Horn L, Ancell K, Wyman K, Bertucci C, Shaffer T, Hodsdon LA, Garg K, Hosseini SA, Lim LP, Lovly CM, Blood-Based Surveillance Monitoring of Circulating Tumor DNA From Patients With SCLC Detects Disease Relapse and Predicts Death in Patients With Limited-Stage Disease. JTO Clinic Res Rep 1, 100024 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernandez-Cuesta L, Perdomo S, Avogbe PH, Leblay N, Delhomme TM, Gaborieau V, Abedi-Ardekani B, Chanudet E, Olivier M, Zaridze D, Mukeria A, Vilensky M, Holcatova I, Polesel J, Simonato L, Canova C, Lagiou P, Brambilla C, Brambilla E, Byrnes G, Scelo G, Le Calvez-Kelm F, Foll M, McKay JD, Brennan P, Identification of Circulating Tumor DNA for the Early Detection of Small-cell Lung Cancer. EBioMedicine 10, 117–123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A, The Cardiovascular Health Study: design and rationale. Ann Epidemiol 1, 263–276 (1991). [DOI] [PubMed] [Google Scholar]
- 59.Augert A, Eastwood E, Ibrahim AH, Wu N, Grunblatt E, Basom R, Liggitt D, Eaton KD, Martins R, Poirier JT, Rudin CM, Milletti F, Cheng W-Y, Mack F, MacPherson D, Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition. Sci Signal 12, 2922 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghosh D, Poisson LM. "Omics" data and levels of evidence for biomarker discovery. Genomics 93, 13–16 (2009). [DOI] [PubMed] [Google Scholar]
- 61.R Core Team (2021). R: A language and environment for statistical computing. (Vienna, Austria: ) (available at https://www.R-project.org/). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.