Abstract
In untargeted metabolomics, multiple ions are often measured for each original metabolite, including isotopic forms and in-source modifications, such as adducts and fragments. Without prior knowledge of the chemical identity or formula, computational organization and interpretation of these ions is challenging, which is the deficit of previous software tools that perform the task using network algorithms. We propose here a generalized tree structure to annotate ions in relationships to the original compound and infer neutral mass. An algorithm is presented to convert mass distance networks to this tree structure with high fidelity. This method is useful for both regular untargeted metabolomics and stable isotope tracing experiments. It is implemented as a Python package (khipu) and provides a JSON format for easy data exchange and software interoperability. By generalized preannotation, khipu makes it feasible to connect metabolomics data with common data science tools and supports flexible experimental designs.
Metabolomics is becoming an increasingly important tool to biomedicine. Untargeted LC-MS (liquid chromatography–mass spectrometry) metabolomics is key to performing high-coverage chemical analyses and discoveries. The term “annotation” in metabolomics often includes (i) the assignment of measured ions to their original compounds and (ii) the establishment of the identity of the compounds.1,2 For clarity, we refer to the first step as “preannotation” in this paper, which is the assignment of isotopes, adducts, and fragments to the unique compounds. Correct preannotation will greatly facilitate the later step of identification by reducing errors of analyzing and searching the redundant ions. Multiple software tools have been developed for this purpose of preannotation, including CAMERA,3 Mz.unity,4 xMSannotator,5 MS-FLO,6 MetNet,7 CliqueMS,8 Binner,9 MolNotator,10 IIMN,11 and NetID.12
In high-resolution mass spectrometry, the m/z (mass to charge ratio) difference between isotopes is usually resolved unambiguously. In the case of LC-MS experiments, ion species originating from the same compound in the ion source (adducts, isotopic ions, fragments, and conjugates) should have the same retention time. These ions are often referred to as redundant or degenerate peaks in the literature. All preannotation tools utilize the m/z differences between experimental peaks, which correspond to the theoretical mass differential between isotopes, atoms, or chemical groups. Compared to direct infusion, LC-MS brings the retention time dimension, which is critical to grouping these degenerate peaks. Some tools also use similarities in the shapes of elution peaks11 and sometimes statistical correlation between peak intensity across samples.5,9 Such correlations can be supporting evidence but are not a prerequisite.4
Most preannotation tools use a network representation of degenerate peaks. Because the pairwise relationships between peaks are established first, it is natural to connect the pairs into networks by using pairwise relationships as edges and shared peaks as nodes. Such networks still contain redundant and often erroneous edges. The main challenge remains to resolve how all peaks are generated from the same original compound, which requires a) inferring the neutral mass of the original compound and b) establishing the relationship of all peaks to the original compound. Given the difficulty of organizing this information in untargeted metabolomics, the coverage of untargeted analyses is often called to question.
A couple of notable studies tried to address the question of coverage using isotope tracing in untargeted metabolomics and suggested that a small number of metabolites are actually measured, and the majority of peaks are “junks”, either from contaminants, isotopes, or LC-MS artifacts.13,14 A new challenge also arose in that analyzing these isotope tracing data by global metabolomic is not trivial. So far, isotope tracing experiments usually require targeted metabolites and specialized software.15−18 In untargeted analysis, without prior knowledge of the chemical formulas, special experimental designs are required, and the software tools are tied to the designs, as is the case for X13CMS19,20 and PAVE.14 It is highly desirable to have a generic and flexible tool to process untargeted isotope tracing metabolomics, and to enable more flexible data analysis and modeling.
In this study, we propose a generalized tree structure to assign the relationship of each ion to the original compound and infer its neutral mass. The preannotation software tool, khipu, is freely available as a Python package. It is applicable to both regular untargeted metabolomics and stable isotope tracing data, and helps plug metabolomics data easily into common data science tools.
Experimental Section
The dry extracts of unlabeled and 13C-labeled E. coli (Cambridge Isotope Laboratories, Inc.; Catalog number: MSK-CRED-DD-KIT) were reconstituted in 100 μL of acetonitrile:H2O (1:1, v/v) then sonicated (10 min) and centrifuged (10 min at 13,000 rpm and 4 °C) before overnight incubation at 4 °C. The supernatant for each 12C/13C E. coli extract was collected and then prepared for LC-MS metabolomics analysis. Metabolite extraction was carried out using acetonitrile:methanol (8:1, v/v) containing 0.1% formic acid. All samples were vortexed and incubated with shaking at 1000 rpm for 10 min at 4 °C, followed by centrifugation at 4 °C for 15 min at 15,000 rpm. The supernatant was transferred into mass spec vials, and 2 μL were injected into the UHPLC-MS. All samples were maintained at 4 °C in the autosampler and analyzed using a Thermo Scientific Orbitrap ID-X Tribid Mass Spectrometer coupled to a Thermo Scientific Transcen LX-2 Duo UHPLC system with a HESI ionization source, using positive ionization. A Hypersil GOLDTM RP column (3 μm, 2.1 mm × 50 mm) maintained at 45 °C was used. 0.1% formic acid in water and 0.1% formic acid in acetonitrile (ACN) were used as mobile phase A and B, respectively. The following gradient was applied at a flow rate of 0.4 mL/min: 0–0.1 min: 0% B, 0.10–1.9 min: 60% B, 1.9–5.0 min: 98% B, 5.00–5.10 min: 0% B, and 4.9 min cleaning and column equilibration. The chromatographic run time was 5 min followed by 5 min washing step after each sample. The MS settings are spray voltage, 3500 V; sheath gas, 45 Arb; auxiliary gas, 20 Arb; sweep gas, 1 Arb; ion transfer tube temperature, 325 °C; vaporizer temperature, 325 °C; mass range, 80–1000 Da; maximum injection time, 100 ms; resolution 60,000. The yeast data from Chen et al.12 were retrieved from the MassIVE repository (https://massive.ucsd.edu, ID no. MSV000087434). The yeast ESI+ data contain both unlabeled and 13C isotope-labeled samples, while the ESI– data did not involve isotope tracing. The data from Mahieu and Patti13 and Wang et al.14 were not found publicly. All data sets were processed using asari21 version 1.9.2. The yeast ESI– data set was quality-filtered for a signal:noise ratio >100 to serve as a cleaner demo.
Results and Discussion
The Combination of Isotopes and Adducts Is a 2-Tier Tree
The redundant or degenerate ions in mass spectrometry can be from in-source modifications (adducts, fragments, and conjugates) on any of the isotopic forms. For simplicity, we only consider adducts in the initial steps. The combination of isotopes and adducts leads to a grid of mass values, relative to the neutral mass of M0, exemplified in Table 1. We use M0 to denote the molecules with only 12C atoms. The isotopes are denoted as 13C/12C, 13C/12C*2, etc., where the last digit is the number of 13C atoms present in each molecule.
Table 1. Combinations of Isotopes and Adducts Generate Mass Differences as a Grida.
| [M + H]+ | [M + NH4]+ | [M + Na]+ | [M + HCl + H]+ | [M + K]+ | [M + ACN + H]+ | |
|---|---|---|---|---|---|---|
| M0 | 1.007276 | 18.033826 | 22.989276 | 36.983976 | 38.963158 | 42.033825 |
| 13C/12C | 2.010631 | 19.037181 | 23.992631 | 37.987331 | 39.966513 | 43.037180 |
| 13C/12C*2 | 3.013986 | 20.040536 | 24.995986 | 38.990686 | 40.969868 | 44.040535 |
| 13C/12C*3 | 4.017341 | 21.043891 | 25.999341 | 39.994041 | 41.973223 | 45.043890 |
| 13C/12C*4 | 5.020696 | 22.047246 | 27.002696 | 40.997396 | 42.976578 | 46.047245 |
| 13C/12C*5 | 6.024051 | 23.050601 | 28.006051 | 42.000751 | 43.979933 | 47.050600 |
| 13C/12C*6 | 7.027406 | 24.053956 | 29.009406 | 43.004106 | 44.983288 | 48.053955 |
The mass values are relative to the 12C-only neutral mass. Examples are using a limited number of isotopes and in-source modifications in positive ionization.
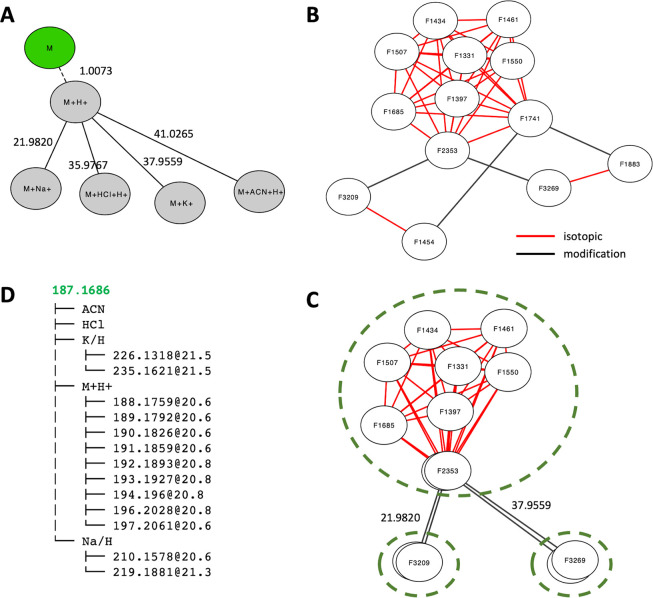
The adducts can be represented as a tree (Figure 1A) using the neutral form as the root, which is usually not measured in mass spectrometry. Each edge in the tree corresponds to a specific mass difference from the reaction forming the adduct. In fact, the full grid in Table 1 can be accommodated into the tree, using isotopes as leaves to the adducts. Two arguments favor the tree as a preferred data structure over a generic network: 1) each ion measured in mass spectrometry is formed from a specific “predecessor” and 2) the whole group of ions are from a unique compound, which is the “root”. In computational terms, a network becomes a tree once it fulfills the two requirements: each node must have 1) no more than one predecessor and 2) a unique root. The benefit of this tree representation is important, allowing automated interpretation of all ions via defined semantics.
Figure 1.
The khipu algorithm converts a mass distance network to a tree structure. A) An adduct tree based on Table 1. Mass differences on the edges are relative to the predecessor nodes. The green node is usually not observed. B) An example mass distance network from our credentialed E. coli data set, which contains both unlabeled and 13C-labeled samples. Edges in red are from isotopic patterns and edges in black from adduct patterns. C) The isotopic subnetworks can be treated as individual nodes, then the abstracted network has only adduct edges, which facilitates the alignment to the theoretical adduct tree in A. D) Resulting 2-tier tree. The root is colored in green as inferred neutral mass. No ion is assigned to ACN or HCl adducts.
Because the isotopes are present independently from each other at the time of measurement, we treat them equally as one tier of the tree here. It is noted that the generation of the isotopes may have biochemical significances in isotope tracing experiments, but that problem is outside data processing and annotation. Therefore, the combination of isotopes and adducts, as exemplified in Table 1, can be represented as a 2-tier tree. The tree can use either adducts or isotopes as tier 1. Adducts are chosen for tier 1 because a) adduct mass patterns are more distinct, and b) isotopes are often limited by abundance, resulting in only M0 ions in many compounds.
An Algorithm to Convert a Mass Distance Network to a 2-Tier Tree
Annotation methods in MS metabolomics commonly start by searching mass difference patterns, e.g. 1.0034 for 13C/12C in isotopes and 22.9893 for Na+ in adducts. Each match leads to a pair of ions (also called features), and many pairs are connected via shared ions into a network of ions (Figure 1B). During the mass difference search, additional redundancy is introduced; e.g., the mass difference between 13C and 12C is the same as between 13C/12C*2 and 13C/12C*3, and so forth. This network redundancy is apparent in the top part of the network in Figure 1B. The objective of annotation is to infer the true root (original compound) from the network, which has been challenging in previous works.
As biological reactions are not part of data annotation here, the edges in our mass distance networks belong to one of the two categories: isotopic differences or in-source modifications (Figure 1B). A key observation is that all ions connected by isotopic edges belong to the same adduct. Therefore, subnetworks per adduct can be defined from a mass distance network (Figure 1C). Once these isotopic subnetworks are abstracted into individual network nodes, we can find the best alignment between this abstracted network (Figure 1C) and the adduct tree (Figure 1A). The algorithm is designed as a two-step optimization: to obtain a tree with an optimal number of ions explained in the alignment of adduct trees, then in the alignment of isotopes (details in Note S1). The result of this algorithm on our example network is shown in Figure 1D. To match our 2-tier tree structure, the networks have to become directed acyclic graph (DAG). During this process, erroneous edges are weeded out because they do not satisfy DAG and a rooted tree. This method yields a structured and unique annotation of each ion in the tree. Based on the matched m/z values, the neutral mass of the M0 compound is obtained by a regression model. Once the core structure of a tree is established, additional adducts and fragments can be searched in the data. The algorithm is implemented into a freely available Python package khipu.
Khipu Plots Allow Intuitive Interpretation of Isotope Tracing Data
After ions are grouped into a tree for each original compound, they are recorded into transparent JSON format, as defined for empirical compounds (see examples in Note S2). An “empirical compound” refers to a tentatively defined compound in metabolomics data used in our previous projects,22,23 as the technology may not deliver definitive identification or resolve a mixture (e.g., isomers not successfully separated).
We continue using the compound in Figure 1 B–D to illustrate the khipu plotting functions. Each ion is measured with an intensity value in one or more biological samples. While the tree visualization in Figure 1D is useful, khipu includes multiple functions to visualize the features, m/z values, and intensity values as data frame tables (Note S2) to facilitate the intuitive interpretation of each compound. An enhanced visualization of the tree is demonstrated in Figure 2A, where the adducts are organized as a “trunk” and isotopes as “branches”. It is clear that several isotopes are present as the protonated ion; Na and K adducts are present for the more abundant isotopes. Because this visualization style resembles the khipu knot records used by Andean South Americans, we named our software “khipu”.
Figure 2.
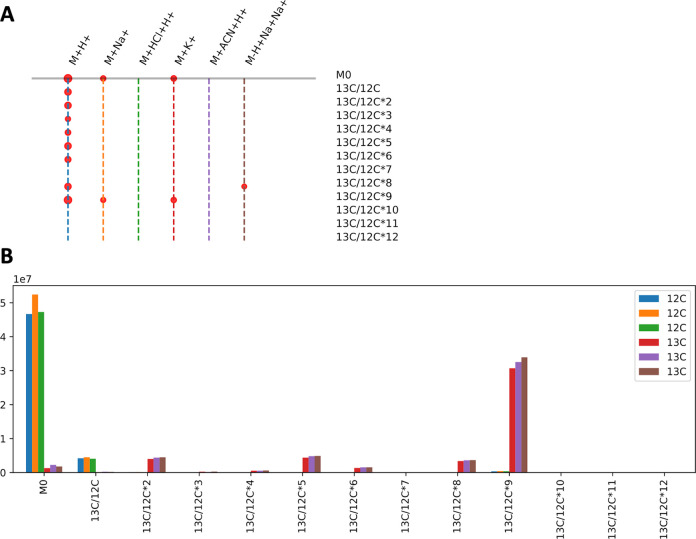
Visualization using khipu facilitates interpretation of isotope tracing data. A) An example khipugram plot for the compound in Figure 1, with its 13 ions aligned to the tree in Figure 1D. Each dot represents an ion measured in the data, with the dot size proportional to average intensity. The vertical dashed lines are colored for easy navigation, and the colors are of no particular meaning. B) Bar plot for intensity values of the [M + H]+ ion in different isotopes (x-axis) for three 12C samples and three 13C samples (in color legend) from the first branch in A.
This experimental data set was from cultured E. coli, containing three unlabeled samples and three samples grown on U-13C-glucose. Figure 2B visualizes the intensity values across samples for the [M + H]+ ion. The three unlabeled samples have high M0 peaks and smaller 13C/12C (M1) peaks due to the naturally occurring isotopes. The U-13C-labeled samples have the highest peaks at 13C/12C*9 (M9), with smaller peaks for other isotopes. This indicates that the latter samples are almost fully labeled by 13C, and the compound should contain 9 carbon atoms. The neutral mass inferred by khipu is 187.1686: it matches acetylspermidine, which has a chemical formula C9H21N3O, perfectly consistent with the isotopic pattern. Further LC-MS/MS experiment using authentic standards confirmed that it is a mixture of N1- and N8-acetylspermidine, where the isomers were not separated by chromatography.
Software Implementation and Connection to Data Science Tools
Khipu is an open-source Python 3 package, easy to incorporate into preprocessing21 and other software tools (Note S1). The common data formats in khipu are tab delimited tables and JSON (JavaScript Object Notation), which is a common format for data exchange between software programs and web applications. This enables an effective way for sharing metabolite annotation, which is human-friendly, computable, and neutral to software platforms. A snippet of a khipu export in JSON is as follows:
As a preannotation tool, khipu is positioned to feed organized data for downstream data analysis. Users can choose to model the isotopes and compute flux using other tools.24,25 Multiple Jupyter notebooks are provided as part of the software package to demonstrate how khipu is plugged into common data science tools. This gives great flexibility to people in using both regular and isotope tracing metabolomics data because the computational methods, as well as experimental designs, are no longer limited by rigid software designs. Traditional software development is often too costly, and its maintenance is even more challenging.26 Fundamentally, no software developer can meet every demand via a point-and-click interface, so scientific data analysis has to depend greatly on scripting. The combination of modular software components, transparent data structures, and Jupyter notebooks opens up many opportunities for collaborations and scientific progress.27
How Many Metabolites Do We Measure?
Proper pre-annotation is key to answering the question of how many metabolites/compounds are measured in an experiment, which is a matter that has been debated for over a decade. Many studies overestimated the coverage because the database search was inflated by redundant/degenerate features/ions. Studies from the Patti and Rabinowitz laboratories used isotope tracing techniques and suggested the numbers are around 1,000–2,000 in E. coli and yeast.13,14 Our khipu software now provides systematic and fast preannotation on metabolomic data sets.
In our E. coli data (reverse phase ESI+), 3,602 LC-MS features were measured, and khipu annotated 548 empirical compounds (trees) from 1,745 features. Among the 548 empirical compounds, 445 have multiple isotopes (Figure 3A). The remaining 1,857 features are singletons, i.e., not grouped with any other features. In two yeast data sets from the Rabinowitz lab, khipu annotation resulted in 1,775 and 908 empirical compounds in ESI+ and ESI– modes, respectively (Figure 3B–C). In the yeast data sets, we included additional adducts from Chen et al.,12 which by design did not increase the number of empirical compounds, but increased the explained ESI+ features from 6,310 to 8,049, and ESI– features from 2,601 to 2,912. These results suggest that less than 2,000 compounds were reliably measured in these experiments. Of note, closer examination of each data set should also remove contaminants, which is not part of khipu.
Figure 3.
Number of measured compounds in three metabolomic data sets. In each panel, the first bar is total number of LC-MS features, and the orange portion is referred to as “singletons”. The second bar is empirical compounds (empCpds), i.e., the number of khipu trees. The third bar is the number of khipu trees with multiple isotopes. A) Credentialed E. coli data generated in this study. Reanalysis of previously published yeast B) ESI+ and C) ESI– data sets from the Rabinowitz lab.12 Khipu annotation on these data sets took 2–6 s on a laptop computer of Intel i7 CPU.
Conclusions
Annotation of untargeted metabolomics data, including isotopic tracing data, is still not fully solved. Many current tools take a network approach but depend on assumed base ions or formulas to assign relationships between ions. We present a new algorithm here to resolve the mass distance networks into a tree structure, unambiguously defining ion relationships and inferring neutral mass. This approach shall reduce false annotations and facilitate new compound identifications and discoveries. We consider the preannotation with khipu a key step forward also because it ships with the generalized annotation format, which will greatly facilitate data exchange and software interoperability. With this foundation, future benchmarking and improvements are expected. Khipu can be easily reused by other software tools and incorporated into metabolomics workflows, where complete annotation can take into consideration contaminants, authentic libraries, and tandem mass spectrometry data.
Acknowledgments
This work was in parted funded by NIH grants (to SL) U01 CA235493 (NCI) and R01 AI149746 (NIAID).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.2c05810.
User manual and Jupyter Notebook (PDF)
Author Contributions
S.L. designed the study and wrote the khipu software and manuscript. S.Z. performed the LC-MS and LC-MS/MS experiment on credentialed E. coli samples.
The authors declare no competing financial interest.
Notes
The asari source code is available at GitHub, https://github.com/shuzhao-li/khipu, and as a Python package via https://pypi.org/project/khipu-metabolomics/. The demonstration data sets are provided as part of the source code.
Supplementary Material
References
- Blazenovic I.; Kind T.; Sa M. R.; Ji J.; Vaniya A.; Wancewicz B.; Roberts B. S.; Torbasinovic H.; Lee T.; Mehta S. S.; Showalter M. R.; Song H.; Kwok J.; Jahn D.; Kim J.; Fiehn O. Structure Annotation of All Mass Spectra in Untargeted Metabolomics. Anal. Chem. 2019, 91 (3), 2155–2162. 10.1021/acs.analchem.8b04698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Almenara X.; Montenegro-Burke J. R.; Benton H. P.; Siuzdak G. Annotation: A Computational Solution for Streamlining Metabolomics Analysis. Anal. Chem. 2018, 90 (1), 480–489. 10.1021/acs.analchem.7b03929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl C.; Tautenhahn R.; Bottcher C.; Larson T. R.; Neumann S. CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 2012, 84 (1), 283–9. 10.1021/ac202450g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahieu N. G.; Spalding J. L.; Gelman S. J.; Patti G. J. Defining and Detecting Complex Peak Relationships in Mass Spectral Data: The Mz.unity Algorithm. Anal. Chem. 2016, 88 (18), 9037–46. 10.1021/acs.analchem.6b01702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K.; Walker D. I.; Jones D. P. xMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal. Chem. 2017, 89 (2), 1063–1067. 10.1021/acs.analchem.6b01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFelice B. C.; Mehta S. S.; Samra S.; Cajka T.; Wancewicz B.; Fahrmann J. F.; Fiehn O. Mass Spectral Feature List Optimizer (MS-FLO): A Tool To Minimize False Positive Peak Reports in Untargeted Liquid Chromatography-Mass Spectroscopy (LC-MS) Data Processing. Anal. Chem. 2017, 89 (6), 3250–3255. 10.1021/acs.analchem.6b04372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naake T.; Fernie A. R. MetNet: Metabolite Network Prediction from High-Resolution Mass Spectrometry Data in R Aiding Metabolite Annotation. Anal. Chem. 2019, 91 (3), 1768–1772. 10.1021/acs.analchem.8b04096. [DOI] [PubMed] [Google Scholar]
- Senan O.; Aguilar-Mogas A.; Navarro M.; Capellades J.; Noon L.; Burks D.; Yanes O.; Guimera R.; Sales-Pardo M. CliqueMS: a computational tool for annotating in-source metabolite ions from LC-MS untargeted metabolomics data based on a coelution similarity network. Bioinformatics 2019, 35 (20), 4089–4097. 10.1093/bioinformatics/btz207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachman M.; Habra H.; Duren W.; Wigginton J.; Sajjakulnukit P.; Michailidis G.; Burant C.; Karnovsky A. Deep annotation of untargeted LC-MS metabolomics data with Binner. Bioinformatics 2020, 36 (6), 1801–1806. 10.1093/bioinformatics/btz798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier-Jimenez D.; Bouchouireb Z.; Olivier S.; Mocquard J.; Allard P.; Bernadat G.; Chollet-Krugler M.; Rondeau D.; Boustie J.; van der Hooft J.; Wolfender J.. From mass spectral features to molecules in molecular networks: a novel workflow for untargeted metabolomics. bioRxiv 2021. 10.1101/2021.12.21.473622 [DOI]
- Schmid R.; Petras D.; Nothias L. F.; Wang M.; Aron A. T.; Jagels A.; Tsugawa H.; Rainer J.; Garcia-Aloy M.; Duhrkop K.; Korf A.; Pluskal T.; Kamenik Z.; Jarmusch A. K.; Caraballo-Rodriguez A. M.; Weldon K. C.; Nothias-Esposito M.; Aksenov A. A.; Bauermeister A.; Albarracin Orio A.; Grundmann C. O.; Vargas F.; Koester I.; Gauglitz J. M.; Gentry E. C.; Hovelmann Y.; Kalinina S. A.; Pendergraft M. A.; Panitchpakdi M.; Tehan R.; Le Gouellec A.; Aleti G.; Mannochio Russo H.; Arndt B.; Hubner F.; Hayen H.; Zhi H.; Raffatellu M.; Prather K. A.; Aluwihare L. I.; Bocker S.; McPhail K. L.; Humpf H. U.; Karst U.; Dorrestein P. C. Ion identity molecular networking for mass spectrometry-based metabolomics in the GNPS environment. Nat. Commun. 2021, 12 (1), 3832. 10.1038/s41467-021-23953-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.; Lu W.; Wang L.; Xing X.; Chen Z.; Teng X.; Zeng X.; Muscarella A. D.; Shen Y.; Cowan A.; McReynolds M. R.; Kennedy B. J.; Lato A. M.; Campagna S. R.; Singh M.; Rabinowitz J. D. Metabolite discovery through global annotation of untargeted metabolomics data. Nat. Methods 2021, 18 (11), 1377–1385. 10.1038/s41592-021-01303-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahieu N. G.; Patti G. J. Systems-Level Annotation of a Metabolomics Data Set Reduces 25 000 Features to Fewer than 1000 Unique Metabolites. Anal. Chem. 2017, 89 (19), 10397–10406. 10.1021/acs.analchem.7b02380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Xing X.; Chen L.; Yang L.; Su X.; Rabitz H.; Lu W.; Rabinowitz J. D. Peak Annotation and Verification Engine for Untargeted LC-MS Metabolomics. Anal. Chem. 2019, 91 (3), 1838–1846. 10.1021/acs.analchem.8b03132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueschl C.; Kluger B.; Neumann N. K. N.; Doppler M.; Maschietto V.; Thallinger G. G.; Meng-Reiterer J.; Krska R.; Schuhmacher R. MetExtract II: A Software Suite for Stable Isotope-Assisted Untargeted Metabolomics. Anal. Chem. 2017, 89 (17), 9518–9526. 10.1021/acs.analchem.7b02518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokkathukalam A.; Jankevics A.; Creek D. J.; Achcar F.; Barrett M. P.; Breitling R. mzMatch-ISO: an R tool for the annotation and relative quantification of isotope-labelled mass spectrometry data. Bioinformatics 2013, 29 (2), 281–3. 10.1093/bioinformatics/bts674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Previs S. F.; Downes D. P. Key Concepts Surrounding Studies of Stable Isotope-Resolved Metabolomics. Methods Mol. Biol. 2020, 2104, 99–120. 10.1007/978-1-0716-0239-3_6. [DOI] [PubMed] [Google Scholar]
- Rahim M.; Ragavan M.; Deja S.; Merritt M. E.; Burgess S. C.; Young J. D. INCA 2.0: A tool for integrated, dynamic modeling of NMR- and MS-based isotopomer measurements and rigorous metabolic flux analysis. Metab Eng. 2022, 69, 275–285. 10.1016/j.ymben.2021.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.; Chen Y. J.; Cho K.; Nikolskiy I.; Crawford P. A.; Patti G. J. X13CMS: global tracking of isotopic labels in untargeted metabolomics. Anal. Chem. 2014, 86 (3), 1632–9. 10.1021/ac403384n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llufrio E. M.; Cho K.; Patti G. J. Systems-level analysis of isotopic labeling in untargeted metabolomic data by X(13)CMS. Nat. Protoc 2019, 14 (7), 1970–1990. 10.1038/s41596-019-0167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Siddiqa A.; Thapa M.; Zheng S.. Trackable and scalable LC-MS metabolomics data processing using asari. bioRxiv 2023. 10.1101/2022.06.10.495665 [DOI] [PMC free article] [PubMed]
- Li S.; Park Y.; Duraisingham S.; Strobel F. H.; Khan N.; Soltow Q. A.; Jones D. P.; Pulendran B. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013, 9 (7), e1003123 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z.; Chong J.; Li S.; Xia J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10 (5), 186. 10.3390/metabo10050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard P.; Delepine B.; Guionnet M.; Heuillet M.; Bellvert F.; Letisse F. IsoCor: isotope correction for high-resolution MS labeling experiments. Bioinformatics 2019, 35 (21), 4484–4487. 10.1093/bioinformatics/btz209. [DOI] [PubMed] [Google Scholar]
- Moseley H. N. Correcting for the effects of natural abundance in stable isotope resolved metabolomics experiments involving ultra-high resolution mass spectrometry. BMC Bioinformatics 2010, 11, 139. 10.1186/1471-2105-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. Y.; Colby S. M.; Du X.; Gomez J. D.; Helf M. J.; Kechris K.; Kirkpatrick C. R.; Li S.; Patti G. J.; Renslow R. S.; Subramaniam S.; Verma M.; Xia J.; Young J. D. A Practical Guide to Metabolomics Software Development. Anal. Chem. 2021, 93 (4), 1912–1923. 10.1021/acs.analchem.0c03581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittard W. S.; Villaveces C. K.; Li S. A Bioinformatics Primer to Data Science, with Examples for Metabolomics. Methods Mol. Biol. 2020, 2104, 245–263. 10.1007/978-1-0716-0239-3_14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.