The authors note an error in Figure 3 of the article “Evolution of factors shaping the endoplasmic reticulum”(1), which is corrected in Figure 3 below. The branches for M. brevicollis and T. adhaerens should be above the branching point from fungi, not below.
FIGURE 3.
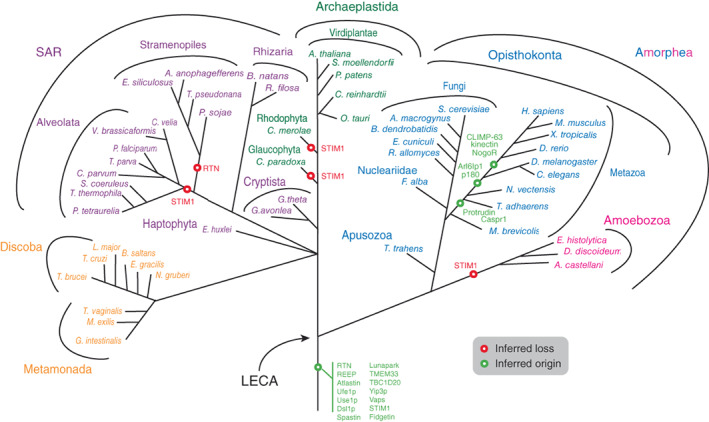
Distribution of ER morphology proteins mapped onto eukaryotic phylogeny. The most likely point of origin of each protein is indicated, based on the results of the present study. The tree omits much detail regarding losses of paralogs from specific taxa and ignores any potential lateral gene transfer. STIM1: most likely origin, but with multiple secondary losses (also see Figure 2). Sey1p, synthetic enhancer of Yop1p; LECA, last eukaryotic common ancestor; STIM1, stromal interaction molecule 1, other abbreviations as in Figure 1.
REFERENCE
- 1. Kontou A, Herman EK, Field MC, Dacks JB, Koumandou VL. Evolution of factors shaping the endoplasmic reticulum. Traffic. 2022;23(9):462‐473. [DOI] [PMC free article] [PubMed] [Google Scholar]


