Abstract
In contrast to extensively studied prokaryotic ‘small’ transcriptomes (encompassing all small noncoding RNAs), small proteomes (here defined as including proteins ≤70 aa) are only now entering the limelight. The absence of a complete small protein catalogue in most prokaryotes precludes our understanding of how these molecules affect physiology. So far, archaeal genomes have not yet been analyzed broadly with a dedicated focus on small proteins. Here, we present a combinatorial approach, integrating experimental data from small protein-optimized mass spectrometry (MS) and ribosome profiling (Ribo-seq), to generate a high confidence inventory of small proteins in the model archaeon Haloferax volcanii. We demonstrate by MS and Ribo-seq that 67% of the 317 annotated small open reading frames (sORFs) are translated under standard growth conditions. Furthermore, annotation-independent analysis of Ribo-seq data showed ribosomal engagement for 47 novel sORFs in intergenic regions. A total of seven of these were also detected by proteomics, in addition to an eighth novel small protein solely identified by MS. We also provide independent experimental evidence in vivo for the translation of 12 sORFs (annotated and novel) using epitope tagging and western blotting, underlining the validity of our identification scheme. Several novel sORFs are conserved in Haloferax species and might have important functions. Based on our findings, we conclude that the small proteome of H. volcanii is larger than previously appreciated, and that combining MS with Ribo-seq is a powerful approach for the discovery of novel small protein coding genes in archaea.
Keywords: proteomics, ribosome profiling, small protein, sORF, mass spectrometry, sprotein, Haloferax volcanii, archaea, small proteome, Ribo-seq
Introduction
Small open reading frames (sORFs) of 100 codons or fewer have long been omitted from genome annotations and proteomic analyses (Dinger et al. 2008, Storz et al. 2014). Nonetheless, the ‘small proteome’ has recently become of interest as the functional importance of a number of small proteins [here defined as those ≤70 amino acids (aa)] has been demonstrated in eukarya, bacteria, and even viruses [reviewed in Storz et al. (2014), Pueyo et al. (2016), Duval and Cossart (2017), Plaza et al. (2017), Orr et al. (2020), Steinberg and Koch (2021), Gray et al. (2022)]. Emerging evidence in bacteria suggests that small proteins can function as regulators or components of larger proteins or protein complexes, are often localized at membranes, and commonly act by protein–protein interactions (Storz et al. 2014, Garai and Blanc‐Potard 2020, Orr et al. 2020).
Their short length and absence of canonical protein domains makes small ORFs (sORFs) difficult to predict based on sequence alone (Pueyo et al. 2016). Therefore, most sORFs are omitted from annotations as false positives. Experimental identification of small proteins using biochemical and mass spectrometry (MS) approaches has traditionally been hampered by technical constraints [recently reviewed in Cassidy et al. (2021), Ahrens et al. (2022)]. Classical methods for protein analysis are biased against small proteins, which typically combine short length with limited charge and strong hydrophobicity (Weaver et al. 2019). Moreover, small proteins usually represent only a small fraction of the total protein mass within a cell (Klein et al. 2007, Miravet‐Verde et al. 2019). Recently, several improvements of classical MS sample preparation workflows, driven by the interest in small proteins, have been made (Slavoff et al. 2013, Petruschke et al. 2020). These include the generation of peptides by proteases other than trypsin to increase sequence coverage of small proteins, as more than one peptide is required for confident detection and the short length of small proteins caters poorly to this requirement (Pueyo et al. 2016, Kaulich et al. 2020). Besides experimental workflow adaptations, specialized algorithms for MS data analysis tailored to small proteins have also been developed [recently reviewed in Cassidy et al. (2021)].
Ribosome profiling (Ribo-seq) is based on sequencing of transcripts associated with translating ribosomes (‘polysomes’), and has provided widespread evidence for translation of unannotated ORFs in diverse organisms (Menschaert et al. 2013, Mumtaz and Couso 2015, Ingolia 2016). In Ribo-seq, translating ribosomes are captured along with mRNAs, which are trimmed by nonspecific RNases to generate so-called ribosome footprints, and the resulting RNA fragments are deep sequenced. Mapped reads reveal ribosome occupancy genome wide, and comparison to coverage in a transcriptome library prepared in parallel allows for the quantification of translation efficiency (ratio Ribo-seq/RNA-seq coverage) as well as definition of ORF boundaries and untranslated regions (UTRs) (Ingolia et al. 2009, Ingolia 2016). Ribo-seq addresses protein biosynthesis from an angle complementary to MS, as it relies on the high sensitivity and resolution of RNA-seq while being independent of protein biochemistry. Ribo-seq has identified sORFs in, e.g. the human cytomegalovirus (Stern-Ginossar et al. 2012), SARS-CoV-2 (Finkel et al. 2021), Escherichiacoli (Neuhaus et al. 2017, Weaver et al. 2019, Hemm et al. 2020), Salmonella Typhimurium (Baek et al. 2017, Venturini et al. 2020), Staphylococcus aureus (Fuchs et al. 2021), and diverse vertebrates (Bazzini et al. 2014, Ji et al. 2015). Ribo-seq is by now an invaluable part of integrated omics approaches aimed at the detection of novel sORFs (Menschaert et al. 2013, Miravet‐Verde et al. 2019, Venturini et al. 2020, Fuchs et al. 2021, Vazquez-Laslop et al. 2022).
Archaea represent the third domain of life and exhibit an exceptional mosaic of bacterial and eukaryotic traits. A handful of studies have identified certain small proteins in a few archaea, mostly identified coincidentally based on their role in specific physiological processes [reviewed in Weidenbach et al. (2021)]. Nonetheless, small protein biology is emerging as a growing field of study in archaea (Kubatova et al. 2020a,b, Prasse et al. 2015, Cassidy et al. 2016, 2019, Nagel et al. 2019, Kaulich et al. 2020, Gutt et al. 2021, Liao et al. 2021, Zahn et al. 2021). Apart from an early attempt for Halobacterium salinarum and recent efforts in Methanosarcina mazei, inventories of small proteomes are still missing (Klein et al. 2007, Cassidy et al. 2016, 2019, Kaulich et al. 2020, Gutt et al. 2021). Another recent study used ribosome profiling to determine characteristics of translation for Haloferax volcanii strain H98 (Gelsinger et al. 2020). Haloferax volcanii is a halophilic archaeon and a model organism for Haloarchaea. Halophilic archaea are easy to grow, because they are aerobic, grow at moderate temperatures and they only require high salt concentrations. Due to its genetic accessibility, H. volcanii is one of the few archaea where small protein functions have been experimentally addressed in. A total of three ubiquitin-like small archaeal modifier proteins (SAMP1–3 with 87, 66, and 92 aa) have been investigated for their role in protein degradation, oxidative stress responses, and sulphur metabolism (Humbard et al. 2010, Dantuluri et al. 2016). Evidence for translation, alongside structural insights, for a small iron metabolism-related protein has been reported, as well as the involvement of several small one-domain zinc-finger proteins (up to 70 aa) in diverse cellular functions, including stress-tolerance (Kubatova et al. 2020a, Nagel et al. 2019, Zahn et al. 2021). The CdrS small protein (61 aa) was shown to be a transcriptional regulator governing H. volcanii cell shape and division (Liao et al. 2021). While this limited body of information already underlines the metabolic and regulatory potential of small proteins in (halo-)archaeal physiology, an inventory of its complete small protein complement is lacking.
Here, we explored the small protein landscape of H. volcanii strain H119 by combining MS-based proteomics, tailored to small proteins, with Ribo-seq to detect translated ORFs. In this way, we provide evidence for translation of 212 annotated sORFs under standard growth conditions. We also discovered a set of 48 novel sORFs with high confidence. We also provide our Ribo-seq data in an easily accessible interactive web-based genome-browser: http://www.bioinf.uni-freiburg.de/ribobase. A subset of H. volcanii small proteins was selected for validation, and expression was confirmed in vivo by western blotting, thereby supporting the predictive power of our approach. This inventory of H. volcanii small proteins is a crucial prerequisite for functional and systems biology analysis of this model archaeon, and provides a framework for characterizing the small proteome in other archaea.
Methods
Growth of H. volcanii for MS analysis
All experiments were carried out using the H. volcanii wild-type strain H119 (Allers et al. 2004). H119 was cultivated aerobically at 45°C in YPC (for MS and Ribo-seq sample preparation) or selective media Hv-Ca with addition of tryptophan (for in vivo validation) (Allers et al. 2004).
Enrichment of small proteins for MS analysis
A method based on the interaction of small proteins with a here" was applied to enrich small proteins present in archaeal protein extracts. For this, H. volcanii cells were grown to exponential (OD650nm 0.5) or stationary phase (OD650nm 1.2–1.3) and harvested in triplicate by centrifugation. Flash-frozen pellets were thawed and suspended in TrisHCl buffer (50 mM, pH 7.4) before disruption by ultrasonication with an MS72 sonotrode (Bandelin) operated at 40 W output (4 × 60 s). A mild centrifugation step at 8000 × g for 5 min ensured minimal loss of membrane-related small proteins, while undisrupted cells and cell debris were sufficiently removed. Small proteins were enriched by solid phase enrichment (SPE) as previously described in Bartel et al. (2020). Briefly, disposable SPE columns (Phenomenex, 8B-S100-AAK) packed with an 8.5-nm pore size, modified styrene–divinylbenzene resin were equilibrated and 500 µg proteins were loaded, and unbound, potentially larger proteins were removed by washing. Finally, the enriched small protein fraction was eluted and evaporated to dryness in a vacuum centrifuge.
Proteolytic digest for MS analysis
One small protein enriched aliquot of each sample (∼20 µg) was digested with Lys-C (Bartel et al. 2020). In brief, the samples were diluted with triethylammonium bicarbonate buffer (TEAB), containing the acidlabile detergent RapiGest (Waters). All samples were subsequently reduced with Tris(2carboxyethyl) phosphine (TCEP). The reaction was quenched by adding iodoacetamide and the mixture was allowed to alkylate before Lys-C was added in a 1:100 enzyme to protein ratio. The samples were digested for 12 h and the digestion was interrupted by acidifying the mixture with hydrochloric acid. Prior to analysis with MS, peptides were purified by Pierce C18 Tips (Thermo Fisher Scientific) according to the manufacturer’s protocol and retention time calibration peptides (iRT, Biognosys) were spiked to each sample in order to monitor reproducibility of subsequent LCMS runs. In a complementary approach, an additional aliquot from each sample was prepared without digestion before MS analysis.
Liquid chromatography and electrospray MS
All samples were analyzed with a Q-Exactive coupled to an EASYnLC 1000 (both Thermo Fisher Scientific) which was equipped with an inhouse built 20 cm reversed phase column packed with 3 µm diameter C18 particles (Dr. Maisch) with integrated emitter tip. About 1 µg peptides were loaded onto the column with solvent A [0.1% (v/v) acetic acid in water], eluted by a nonlinear gradient of solvent B [0.1% (v/v) acetic acid in acetonitrile] and online infused in the MS. For analysis of the Lys-C digested samples the Q-Exactive was operated with the following parameters: Survey scan: 300–1650 thomson (Th) mass range; 70 000 resolution at m/z 200; 3 × 1e6 predictive automatic gain control target; max. 120 ms injection time; activated lock mass correction. Fragment scans: data-dependent higher energy collisional dissociation at normalized energy of 27.5 for the top 10 ions with an assigned charge state between +2 and +6; fixed first mass: 100 Th; mass range dependent on precursor m/z; 17 500 resolution at m/z 200; 1 × 1e5 predictive automatic gain control target; max. 60 ms injection time. For undigested samples the resolution of the survey and fragment scans was increased to 140 000 and 35 000, respectively, and only the top eight ions with any assigned charge state above +1 were fragmented with a stepped normalized collision energy of 27.5/32.5 omitting a fixed first mass.
MS data analysis
The mass spectrometric data were analyzed by two different strategies: First, proteins annotated in the H. volcanii genome were identified using a conventional database containing 4107 already annotated proteins. This database was complemented with 116 sequences of commonly observed laboratory contaminants and a reversed version of each protein. The final database contained 8442 entries. Second, unannotated proteins were identified based on a six-frame translation of the complete H. volcanii genome sequence. After stop codon-to-stop codon translation and in silico performed nonspecific enzymatic cleavage, 547 803 unique peptide sequences were supplemented with a shuffled decoy variant of each peptide to obtain the final search database. Unspecific enzymatic cleavage, i.e. possible cleavage between any amino acids independent of the protease's specific motif on both peptide termini, was assumed for two reasons: First, we observed a high amount of unspecific or semispecific (only one nonspecific terminus per peptide) cleavage upon SPE in our earlier work (Bartel et al. 2020) and second, this assumption ensures that all mapped peptides used to identify novel sORFs are sequence-unique and not only unique by different adjacent protease cleavage-sites.
Obtained raw files were converted to mzXML using the msconvert script of ProteoWizard (v3.0.9974) and its vendor-supplied algorithms for centroiding mass peaks. Afterwards, spectra were searched against either database using MSFragger (v2.4) (Kong et al. 2017). During the search for already annotated proteins, semispecific enzymatic cleavage (Lys-C) or unspecific enzymatic cleavage (undigested samples), up to two missed cleavage sites and optional methionine oxidation (+15.994915 Da), conversion of N-terminal glutamic acid or glutamine to pyroglutamic acid (17.026549 or 18.010565 Da, respectively), and acetylation at protein N-termini (+42.01060 Da) as variable modifications were considered. Contrary, during the search against the six-frame translation derived database, nonspecific enzymatic cleavage for all samples and no variable modifications were assumed. For the Lys-C-digested samples static carbamidomethylation (+57.021464 Da) was taken into account for both search strategies. A deviation up to 10 ppm was allowed for the precursor mass whereas fragment mass tolerance was 20 ppm. MSFragger was set up to automatically recalibrate the masses for each file separately and the option to detect optimal parameters was enabled. Identification probabilities were unified experimentwise by PeptideProphet (Keller et al. 2002) using a minimal peptide length of six aa, semiparametric modelling of a ppm-error based high mass accuracy model, reporting of decoy hits and calculated MW of proteins. Results from Lys-C digested and undigested samples were merged by InterProphet applying the peptide-length model. The ipro.pep.xml file resulting from the search against the conventional database was subsequently processed with ProteinProphet (Nesvizhskii et al. 2003) and analyzed with the filter and report options of Philosopher (v3.2.3) (da Veiga Leprevost et al. 2020). Multi-level false-discovery rates (FDR) were calculated for spectrum, peptide, and protein level using the picked FDR algorithm (Savitski et al. 2015) and a cut-off of 1.0% was applied for reporting. In accordance with (Omasits et al. 2017), annotated proteins were considered to be identified only if the number of independent spectra was at least two.
For the detection of novel proteins, spectral hits with an interprophet adjusted peptide-probability of at least 0.998 were analyzed with the Pepper proteogenomics software suite (v1.5.1) (Fuchs et al. 2021). Briefly, proteinase-K digestion (which is identical to unspecific digestion in Pepper) was assumed, peptide sequences were translated into all possible DNA sequences with NCBIs translation table 11 and mapped onto the genome sequence of H. volcanii using Peppers ‘uniq_only_smart’ option. For mapping, individual replicons were concatenated with a 100xN spacer. Potential ORFs and sORFs were then concluded following a set of rules, ensuring that already annotated ORFs are preferred. Unannotated ORFs were ranked by Pepper based on the putative start codon, the presence and location of potential ribosome-binding sites, and product length, which ensured a minimal set of potential novelties. The results were manually curated for plausibility and sORFMS8 was rejected as false-positive as it was identified with only one spectral hit, mapping to a highly hydrophobic peptide with a basic pI.
Physicochemical properties of novel and annotated small proteins were calculated by the aminoAcidProperties function of the alakazam (Gupta et al. 2015) package in R 3.6 (R Core Team 2019). The distribution of these values was further visualized by density plots, assuming a Gaussian kernel and an adjusted bandwidth (x 2 for annotated proteins, x 1.4 for novel proteins) chosen by the Sheather & Jones method. The reference data set (complete small proteome) contained 316 sequences of annotated small proteins and 47 predicted, novel small proteins that were identified by Ribo-seq.
Additionally, the MS-detectability of sORFs was analyzed based on unique peptides. For this purpose, the generate-peptides option of the crux toolkit (Park et al. 2008) was applied, assuming semispecific cleavage by Lys-C (up to two missed cleavage sites) or unspecific cleavage for the semi-top-down approach. Protein N-termini were considered with and without methionine cleavage and peptides between 6 and 30 residues length and a mass between 700 and 8000 Da were stored for analysis with in-house python scripts. Using ProteoMapper (Mendoza et al. 2018), peptide sequences were mapped to a six-frame translation of the H. volcanii genome sequence. Unique peptides were defined as those belonging to ORF variants sharing the same stop codon but could have multiple starting sites. Detectability scores of unique peptides were calculated by Deep-MS-Peptide (Serrano et al. 2020) and a score greater than 0.5 indicated that the peptide might be detectable.
Validation of MS-detected novel small proteins by synthetic peptides
To validate the identification of peptides assigned to proteins not annotated in the reference database, 26 peptides for 12 proteins were synthesized by JPT Peptide Technologies (Berlin) and analyzed by MS. These peptides were selected based on a search against a preliminary database with incomplete annotation and seven peptides originating from six proteins of these were not identified in the main search. Thus 19 peptides for six identified sORFs (sORF7, 8, 10, 11, 12, and 45) were available for validation (Table S4D, Supporting Information). Notably, the reference peptides were not limited to peptides with enzyme-specific cleavage sites but were allowed to be unspecifically cleaved. Using the same pipeline as described above, the samples were searched with MSFragger against a database containing the full length sequences of the 12 proteins to validate, 44 sequences of commonly observed laboratory contaminants, and the retention time calibration peptides, as well as a reversed version of each protein. This database finally contained 112 entries. Spectra identified with a PeptideProphet assigned probability of at least 0.99 that identified one of the synthesized peptides or a truncated version of these, were included into a spectral library built with SpectraST (version 5.0) (Lam et al. 2007), which we called synthetic peptide library. Truncated versions of synthetic peptides could appear due to the process of peptide synthesis at JPT. The synthetic peptide library was supplemented with spectra obtained at a 0.1% peptide FDR from an MSFragger database search of 48 H. volcanii samples, which were analyzed as userservice samples on the same instrument, against the reference database. Decoy spectra were created within SpectraST on the level of the merged consensus spectra. The resulting spectral library was called ‘combined spectral library’. The SEP-enriched samples were finally searched against the combined spectral library and not yet annotated proteins, which obtained at least two spectral counts at 1% FDR on spectral, peptide, and protein level, were considered validated if the identifying spectra passed a subsequent manual inspection.
Sample preparation for ribosome profiling
For Ribo-seq sample preparation, H. volcanii cells were grown aerobically at 45°C in YPC to exponential phase (OD600nm 0.4) and 40 OD600 equivalent units were harvested rapidly by fast-chilling in an ice bath to halt cell growth and translation. Briefly, cultures were rapidly placed in a prechilled flask in an ice-water bath and incubated with gentle shaking for 3 min. Cells were then immediately harvested by centrifugation at 6000 × g for 10 min before snap-freezing in liquid N2. Before centrifugation, a sample of culture for total RNA analysis was harvested, mixed with 0.2 vol stop mix [5% buffer-saturated phenol (Roth) in 95% ethanol], and snap frozen in liquid N2.
Preparation of ribosome footprints
Ribosome profiling was performed as previously described (Oh et al. 2011) with some modifications. Briefly, cell pellets were resuspended with cold lysis buffer [100 mM NH4Cl, 25 mM MgCl2, 20 mM Tris-HCl, pH 8.0, 0.1% NP-40, 0.4% Triton X-100, 150 U DNase I (Fermentas), 500 U RNase Inhibitor (MoloX, Berlin), and lysed by sonication (constant power 50%, duty cycle 50%, 3 × 30 s cycles with 30 s cooling on a water-ice bath between each sonication cycle to avoid heating of the sample)]. The lysate was clarified by centrifugation at 10 000 × g for 12 min at 4°C. Next, 15 A260 units of lysate were digested with either 20 000 U of MNase (NEB) or 200 U of RNase I (Thermo Fisher Scientific) in lysis buffer supplemented with 10 mM CaCl2 (only for MNase) and 500 U RNase Inhibitor. Polysome digestion was performed at 25°C with shaking at 1450 rpm for 90 min (MNase) or at 450 rpm for 60 min (RNase I). A mock-digested control (no enzyme added) was also included for each lysate to confirm the presence of polysomes. MNase digestion was stopped with ethylene glycol-bis(β-aminoethyl ether)-N, N, N′, N′-tetraacetic acid (EGTA, final concentration 6 mM) for the MNase treated sample. To analyze polysome profiles and recover digested monosomes, 15 A260 units were layered onto a linear 10%–55% sucrose gradient prepared in the following buffer: [25 mM MgCl2, 20 mM Tris-HCl, pH 8, 100 mM NH4Cl, 5 mM CaCl2, 2 mM dithiothreitol (DTT)], in an ultracentrifuge tube (13.2 ml Beckman Coulter SW-41). Gradients were centrifuged in a SW40-Ti rotor at 35 000 rpm for 2 h 30 min at 4°C in a Beckman Coulter Optima XPN-80 ultracentrifuge. Gradients were processed using a Gradient Station (IP, Biocomp Instruments) fractionation system with continuous absorbance monitoring at 254 nm to resolve ribosomal subunit peaks. The 70S monosome fractions were collected and subjected to RNA extraction to purify the RNA footprints. RNA was extracted from fractions or cell pellets for total RNA using hot phenol–chloroform–isoamyl alcohol (25:24:1, Roth) or hot phenol (Roth), respectively, as described previously (Sharma et al. 2007, Vasquez et al. 2014). Ribosomal RNA was depleted from 5 µg of DNase I-digested total RNA by subtractive hybridization (Pan-Bacteria riboPOOLs, siTOOLs, Germany) according to the manufacturer’s protocol with Dynabeads MyOne Streptavidin T1 beads (Invitrogen). Total RNA was fragmented with RNA Fragmentation Reagent (Ambion). Monosome RNA and fragmented total RNA were size-selected (26–34 nt) on a 15% polyacrylamide/7 M urea gel as described previously (Ingolia et al. 2012) using RNA oligonucleotides NI-19 and NI-20 as guides. RNA was cleaned up and concentrated by isopropanol precipitation with 15 μg GlycoBlue (Ambion) and dissolved in H2O. Libraries were prepared by Vertis Biotechnologie AG (Freising, Germany) using a small RNA protocol without fragmentation and sequenced on a NextSeq500 instrument (high-output, 75 cycles) at the Core Unit SysMed at the University of Würzburg.
Ribosome profiling data analysis
Ribo-seq data were analyzed using the workflow HRIBO (version 1.4.3) (Gelhausen et al. 2020), which has previously been used for analysis of bacterial Ribo-seq data (Venturini et al. 2020, Gelhausen et al. 2022). In brief, read files were processed with a snakemake (Koster and Rahmann 2012) workflow that downloads all required tools from bioconda (Grüning et al. 2018) and automatically determines the necessary processing steps. Adapters were trimmed from the reads with cutadapt (version 2.1) (Martin 2011) and then mapped against the H. volcanii genome with segemehl (version 0.3.4) (Otto et al. 2014). Reads corresponding to ribosomal RNA (rRNA), multimappers, and tRNAs were removed with SAMtools (version 1.9) (Li et al. 2009) using the rRNA and tRNA annotations. Quality control was performed by creating read count statistics for each processing step and RNA-class with Subread featureCounts (1.6.3) (Liao et al. 2014). All processing steps were analyzed with FastQC (version 0.11.8) (Wingett and Andrews 2018) and results were aggregated with MultiQC (version 1.7) (Ewels et al. 2016). ORFs were called with an adapted variant of REPARATION (Ndah et al. 2017) using blast instead of usearch (REPARATION_blast https://github.com/RickGelhausen/REPARATION_blast) and with DeepRibo (Clauwaert et al. 2019). Summary statistics for all available annotated and merged novel ORFs detected by REPARATION and DeepRibo (Ndah et al. 2017, Clauwaert et al. 2019) were computed in a tabularized form including TE, RPKM normalized read-counts, codon counts, nucleotide, and amino acid sequences, and so on. Additionally, GFF track files with the same information were created for in-depth genome browser manual inspection.
To benchmark the performance of the ribosome profiling based open reading frame prediction tools we used the RiboReport and manual inspection approaches described previously (Gelhausen et al. 2022). Asserting about translation/no translation based on the inspection of paired Ribo-seq and RNA-seq libraries (normalized to the lowest number of reads between the two) was conducted using an Integrated Genome Browser (IGB) and similar scale. Novel or annotated ORFs were called as ‘translated’ using the following three criteria. (1) The shape of the Ribo-seq coverage over the ORF, its evenness and its restriction within ORF boundaries (and ribosome footprints excluded from 5′/3′ UTRs) were called as translated, (2) the Ribo-seq signal was generally required to be comparable or higher to the transcriptome signal from the RNA-seq library (often TE >0.5–1), and (3) RNA-seq and Ribo-seq coverage was required to be, generally, at least ten reads per nucleotide normalized (RPKM) by sample size.
We manually curated all 317 annotated sORFs to obtain a set of 205 translated sORFs. Using the evaluation scripts contained in the RiboReport GitHub repository (https://github.com/RickGelhausen/RiboReport), we then created the set of positive sORF candidates that were predicted by DeepRibo, REPARATION_blast and IRSOM, respectively. The candidates were predicted with the same input genome, annotation and sequencing files already used for the HRIBO analysis. The hand curated set was automatically intersected by the RiboReport pipeline with the tool predictions resulting in performance evaluation for the tools using our current labelling and dataset. To generate a reasonable set of novel sORFs, we applied the following expression cutoffs: mean TE cut-off ≥0.5 and RNA-seq RPKM ≥10 (in both replicates). In addition, novel translated sORF candidates were required to be predicted by REPARATION or DeepRibo [DeepRibo score >0 that allows for ORF candidate ranking (Clauwaert et al. 2019)]. In the context of Ribo-seq, TE is used in order to normalize Ribo-seq coverage to available total RNA transcript levels. From this, one can infer the relative rate of translation of an ORF. TE is calculated by taking the ratio of the ORF expression in the Ribo-seq library compared to its expression in the total RNA library. For example, a highly translated ORF will have a high TE ratio (often >0.5–1), because it has coverage in both the total RNA and Ribo-seq libraries and most of the time the coverage is high in the Ribo-seq library compared to the RNA-seq library. In contrast, noncoding RNAs should have a very low TE ratio, as the coverage in the Ribo-seq library is very low compared to the RNA-seq library. Many top-hit DeepRibo predicted sORFs were not labelled as translated after manual inspection because they did not correlate with the Ribo-seq/RNA-seq coverage (e.g. ORFs harbouring uneven/strange Ribo-seq coverage possibly introduced by technical biases or the translatome structure of H. volcanii). To our knowledge, this the first time that DeepRibo has been applied to archaeal Ribo-seq datasets. We hypothesize DeepRibo falls short as this prediction tool is trained on model bacterial species with mostly leadered mRNAs and strong reliance on Shine–Dalgarno sequences (RBS). Therefore, we recommend using the DeepRibo ORF predictions combined with translation efficiency values as well as manual curation of the predictions in a genome browser [see Gelhausen et al. (2022)], before asserting translation for a given ORF or sORF.
In vivo protein validation by epitope-tagging and western blot analysis
Genes for candidate sORFs were cloned, including up to 150 nucleotides of the upstream promoter region or without promoter region if combined with the p.tna promoter, with a 3xFLAG-epitope tag sequence fused to its penultimate codon (CFLAG constructs) or a 3xFLAG-epitope-tag sequence preceding the coding region (NFLAG constructs). Construction of plasmid pTA-pnat-xxx-CFLAG was carried out using classical PCR amplification with a H. volcanii gDNA template and the oligonucleotides listed in Table S4 (Supporting Information) followed by standard enzymatic cloning using ApaI/SnaBI restriction sites and plasmid pTA927-CFLAG carrying the 3xFLAG ORF including a stop codon (oligonucleotides and plasmids are listed in Table S4, Supporting Information). Construction of plasmid pTA-ptna-NFLAG was carried out analogously but followed by standard enzymatic cloning using HindIII/BamHI and with plasmid pTA927-NFLAG carrying the 3xFLAG ORF excluding the stop codon (oligonucleotides and plasmids are listed in Table S4, Supporting Information). Plasmids were transformed into H119 as described previously (Cline et al. 1989). Strains were grown to OD650nm 0.4–0.7 (exponential phase) in selective media [Hv-Ca+Trp; (Allers et al. 2004)] and harvested by centrifugation. S70 protein extracts were prepared by 1 h ultracentrifugation at 70 000 × g and 4°C after lysis by ultrasonication in 100 mM Tris-HCl, pH 75, 10 mM EDTA.
S70 protein extracts were directly analyzed by Tricine SDS-PAGE gel electrophoresis (Schagger 2006) or subjected to immunopurification of FLAG-tagged small proteins using ANTI-FLAG® M2 agarose affinity gel according to the manufacturer’s recommendations (Sigma Aldrich). Where required, protein concentration was increased by acetone precipitation. After separation in a 16% Tricine-SDS-PAGE gel, proteins were transferred to nitrocellulose membranes with the semidry Trans-Blot Turbo Transfer System (BioRad). FLAG-tagged proteins were detected using monoclonal ANTI-FLAG® M2-HRP antibody (Sigma-Aldrich) at a dilution of 1:1000 and the signal was captured via the ChemiDoc System (BioRad).
Conservation and domain search analyses
The identification of novel small protein homologues was performed by using Blastp and tBlastn searches of the genomes of Haloferax species using the National Center for Biotechnology Information (NCBI) database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The protein sequences for the novel protein candidates identified by Ribo-seq and MS in H. volcanii H119 were used as the query sequence. For tBlastn, the following parameters were used: an E-value (Expect value) ≤100, a seed length that initiates an alignment (word size) of 6, and the filter for low complexity regions off. Novel small proteins were also analyzed for secondary structure and conserved domains using the Phyre2 server [http://www.sbg.bio.ic.ac.uk/∼phyre2/(Kelley et al. 2015)] potential cellular localization with PSORTb v3.0.2 [https://www.psort.org/psortb/, (Yu et al. 2010)].
Results
Small protein-adapted MS increases identification of H. volcanii small proteins
The H. volcanii genome is currently annotated to encode 4107 proteins (Schulze et al. 2020). Of these, 317 possess 70 aa or less (Fig. 1C; Table S1, Supporting Information). To explore the H. volcanii small proteome, we performed small protein-adapted MS (Fig. 1A) and combined this with analysis of the translatome using Ribo-seq (Fig. 2A). To provide a comprehensive insight into protein translation in the low molecular weight (MW) range, we first generated a new proteomic dataset with an adapted MS approach that aimed at optimal detection of small proteins (Fig. 1A).
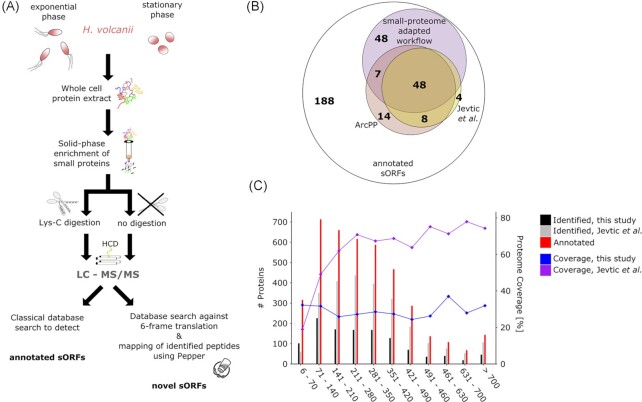
Figure 1.
MS-based proteomic detection of small proteins in H. volcanii. (A). We adjusted a standard proteomics workflow to optimize the detection of small proteins. Small proteins were enriched on a solid-phase column and either digested with Lys-C instead of trypsin or were directly measured by LC-MS/MS (liquid chromatography-tandem mass spectrometry). Small proteins were detected using both a classical database search aimed at detection of annotated small proteins and a proteogenomics search strategy to reveal novel candidates. For MS analysis, Haloferax cells grown to exponential and stationary phase were analyzed. Higher-energy collisional dissociation (HCD) was used to fragment the peptides. (B). Venn-diagram showing the overlap of detected, published and annotated sORFs. The number of annotated sORFs (outer circle) and MS-identified small proteins detected by our adapted small protein MS (purple) and previous datasets [ArcPP (rose) and Jevtić et al. (mustard) (Jevtić et al. 2019, Schulze et al. 2020)] is shown. (C). The number of proteins (left axis) for different protein length bins is compared between annotated proteins (red), small protein adapted MS (dark grey; data from this study), and standard MS [light grey; exemplified by Jevtić et al. dataset (Jevtić et al. 2019)]. Proteome coverage (right axis) achieved across length bins by this small protein adapted MS (blue) opposed to nonadjusted MS approaches [lilac; exemplified by Jevtić et al. dataset; (Jevtić et al. 2019)] is shown.
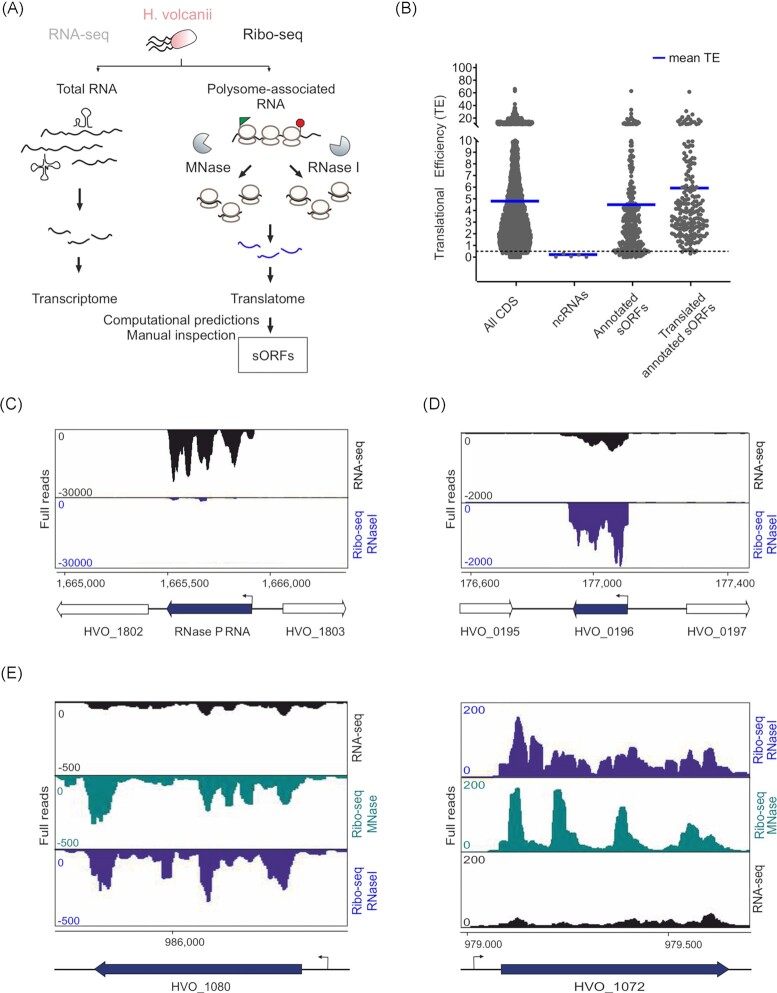
Figure 2.
Ribosome profiling distinguishes between H. volcanii coding and noncoding transcripts. (A). The setup of Ribo-seq to map the translatome of H. volcanii. Translating ribosomes (polysomes) were first captured on mRNAs by fast chilling and subsequently digested to monosomes by either MNase or RNase I treatment. Approximately, 30 nt footprints protected from digestion and copurifying with ribosomes were then subjected to cDNA library preparation and deep sequencing. A second library was generated from total RNA for standard RNA-seq. (B). Scatter plot showing global translation efficiencies computed from all H. volcanii Ribo-seq datasets for all annotated coding sequences (CDS; 4107), five selected abundant noncoding RNAs (ncRNAs; RNase P RNA, SRP RNA, and three CRISPR RNAs), annotated sORFs and the annotated sORFs that were detected as translated (after filtering and visual inspection) by Ribo-seq (205 sORFs). The blue lines indicate the mean TE for each gene class for all replicates of MNase and RNase I libraries. (C). Coverage for the RNase P RNA gene (HVO_1802R) is mostly restricted to the RNA-seq library (black track), confirming that the RNase P RNA is not translated. Ribo-seq coverage is shown in blue (obtained with RNase I). (D). A leaderless sORF (HVO_0196, uncharacterized protein, 55 aa) detected by MS was also identified as translated based on Ribo-seq data (coverage shown for Ribo-seq library obtained with RNase I digest). (E). Comparison of RNA trimming by MNase and RNase I. Read coverage for two leadered genes (HVO_1080 and HVO_1072) in the RNA-seq library (black track) and Ribo-seq libraries obtained with MNase (blue track) and RNase I (green track). The genomic position is indicated for the genes shown in panels (C), (D), and (E) at the bottom alongside a schematic representation of the genomic region (relevant genes in black). Arrows indicate the transcription start sites [TSS based on Babski et al. (2016)].
To increase the number of spectra generated from small proteins, we applied solid-phase extraction (SPE) with small pore size column material to enrich small proteins. This was combined with peptidase Lys-C treatment, or in a semi-top-down approach, without protease treatment. During the enrichment step, only small proteins can enter the pores of the column and interact with the column material while larger proteins pass the SPE column directly. This has been demonstrated to increase the absolute numbers of identified small proteins by factor two (Bartel et al. 2020). Spectra were first matched to a classical proteomics search database generated from the H. volcanii ORF annotation (Schulze et al. 2020). This modified MS workflow identified 1174 proteins, represented by 12 949 peptides, from 74 519 spectra. The majority of identified small proteins had normalized spectral abundance values (Paoletti et al. 2006) above the median NSAF (normalized spectral abundance factor) of the whole dataset, demonstrating enrichment of shorter proteins by our approach (Figure S1, Supporting Information). We detected 103 out of 317 (32.5%) annotated small proteins in the H. volcanii reference genome under the examined (standard) growth conditions (exponential and stationary phase) (Fig. 1B). The enrichment of small proteins achieved by our adapted MS approach is illustrated by the distribution of the spectra collected, as 23% of all spectra belong to proteins shorter than 70 aa. Moreover, while small proteins only represent 7.7% of the coding potential of the H. volcanii genome, they represent 8.7% of all proteins identified in our experiment. The overall proteome coverage achieved by our adapted MS analysis was stable across all size ranges (around 30%, Fig. 1C) in contrast to the coverage achieved by the previous analysis (Jevtić et al. 2019) resulting in a higher identification rate in the ≤70 aa size range in this study (Fig. 1C).
To obtain the most comprehensive set of small proteins, we compared those identified in our dataset to two additional previously published MS-based H. volcanii proteomics datasets (Jevtić et al. 2019, Schulze et al. 2020). The study of Jevtić et al. (2019) is an in-depth analysis of the H. volcanii protein complement under standard and various stress conditions using shot-gun liquid chromatography-MS/MS (LC-MS/MS) (Jevtić et al. 2019). Although the experimental and analysis workflow of this study was not specifically designed for the identification of small proteins, we re-evaluated these data for proteins ≤70 aa. This identified 60 out of 317 annotated small proteins (range 42–70 aa) with a recovery rate of only 19% even though several different stress conditions were analyzed (Fig. 1B and C, Table S1A, Supporting Information).
We next compared the small proteins identified by our adapted workflow to the Archaeal Protein Project (ArcPP) database (Schulze et al. 2020). This database stems from a joint overview project summarizing and reanalyzing MS data available for various H. volcanii strains under a variety of growth conditions, encompassing the dataset of Jevtić et al. (2019). The ArcPP database includes 77 small proteins detected in at least one of the 12 datasets (Fig. 1B). Despite covering a much larger variety of genomic backgrounds and growth conditions, only 55 out of 103 small proteins captured by our adapted MS analysis were identified in the ArcPP database, highlighting the advantage of the small protein adapted workflow for archaeal low MW proteome identification. Taken together, previously published MS data together with MS data from this study support the translation of 129 of the 317 annotated H. volcanii small proteins under at least one condition.
Establishment of ribosome profiling (Ribo-seq) in H. volcanii
Ribo-seq is a different approach for (small) protein discovery identifying the so-called ‘translatome’. Haloferaxvolcanii strain H119 was grown to exponential phase and samples were harvested for both Ribo-seq and RNA-seq (Fig. 2A). Several steps of the Ribo-seq workflow must be adapted to the physiology of the organism being studied including cell harvest, lysis, and footprint generation (Glaub et al. 2020, Vazquez-Laslop et al. 2022). To maintain the stability of ribosome–mRNA complexes, H. volcanii, cells were harvested by a ‘fast-chilling’ method (see ‘Methods’). We were able to recover polysomes using this approach even without treatment with translation inhibitors (Figure S2A, Supporting Information), which might introduce bias into Ribo-seq coverage (Gerashchenko and Gladyshev 2014, Mohammad et al. 2019, Vazquez-Laslop et al. 2022). However, even under our optimized conditions, many ribosomes split into subunits during the harvesting or lysis steps, such that only a fraction of the potential ribosome footprints were recovered (Figure S2A, Supporting Information).
To produce ribosome-protected ‘footprints’, complete trimming of mRNA not protected by ribosomes is necessary, while ribosome stability must be maintained and over-digestion limited. The broad-range nuclease RNase I, used in eukaryotic Ribo-seq (Ingolia et al. 2012), was reported to be inactive in bacteria such as E. coli (Bartholomäus et al. 2016). Consequently, prokaryotic Ribo-seq protocols, including the one used recently for archaea (H. volcanii) (Gelsinger et al. 2020), use micrococcal nuclease (MNase) instead. However, MNase can show some preference for cleavage before A or T nucleotides, as well as imprecise trimming of the mRNA on the 5´ side of the ribosome in bacteria (Bartholomäus et al. 2016, Hwang and Buskirk 2017, Mohammad et al. 2019). Therefore, we generated Ribo-seq libraries from a lysate that has been split to be digested either with MNase or RNase I (Fig. 2A).
In general, by comparing Ribo-seq coverage to a paired RNA-seq library for a given gene, ORF boundaries and 5´/3´ UTRs, if they exist, can be defined and TE (translational efficiency; Ribo-seq/total RNA ratio) can be calculated. Global inspection of TE across our dataset for different annotated gene classes (CDS: all annotated coding sequences, abundant noncoding RNAs, sORFs) showed that protein coding features had a higher mean TE when compared to noncoding RNA genes such as RNase P RNA, SRP RNA, and CRISPR RNAs (Fig. 2B). This demonstrates generally that the Ribo-seq dataset obtained can differentiate between H. volcanii protein coding and RNA coding genes. Manual inspection of coverage at single loci also showed differentiation between these two gene classes. For example, read coverage for the noncoding transcript of the RNase P RNA (HVO_1802R) was restricted to the RNA-seq library (Fig. 2C). In contrast, HVO_0196, coding for a 55-aa small protein, showed significant read coverage in the Ribo-seq library when compared to the associated RNA-seq library (Fig. 2D).
Overall, no significant difference between MNase and RNase I trimming was observed via visual inspection for trimming of known 5´ UTRs like those found for HVO_1080 and HVO_1072 (Fig. 2E). This shows that RNase I, which is not suitable for bacteria, does work well for archaea.
The observed ribosome density in the Ribo-seq libraries at the UTRs of these genes (−15 nucleotides before the start codon) is a characteristic of leadered translation and is probably generated from footprints of initiating ribosomes (Fig. 2E).
The footprints obtained from our Ribo-seq libraries showed a broad distribution in length from 12 to 40 nucleotides with the 27–30 nt footprints being predominant in both the MNase and RNase I libraries (Figure S2B, Supporting Information). In addition, metagene analysis of ribosome occupancy near all annotated start codons (ATG, GTG, and TTG) for leadered and leaderless transcripts using the 5´ end of the 27 and 30 nt footprints, respectively, showed an enriched ribosome density directly at the translation start site (Figure S2C, Supporting Information) for the leaderless transcripts and at −12 nucleotides upstream of the start codon for the leadered transcripts. This is in line with what was observed previously for H. volcanii ribosome footprints (Gelsinger et al. 2020).
Ribosome profiling reveals the small translatome of H. volcanii
Altogether, the MS datasets suggested that 129 of the 317 annotated sORFs are in fact translated into proteins under at least one condition (Fig. 3A; Table S1B, Supporting Information), providing us with a set of true positive translated sORFs to evaluate our Ribo-seq data. To benchmark our Ribo-seq data for its utility in small protein analysis, we manually inspected the read coverage of the 317 annotated sORFs in a genome browser. This suggested that 205 out of 317 annotated small proteins were translated under the analyzed growth condition (Table S1, Supporting Information). The overlap between sORFs detected as translated by Ribo-seq and all MS datasets obtained for Haloferax hitherto was 122, including all nine annotated ribosomal proteins ≤70 aa (Fig. 3A; Table S1, Supporting Information). This overlap suggests our Ribo-seq approach is a sensitive method for detecting translated small proteins. In addition, Ribo-seq detected translation of 83 annotated small proteins not detected in any proteomics dataset, suggesting that it might be more sensitive (Fig. 3A and B).
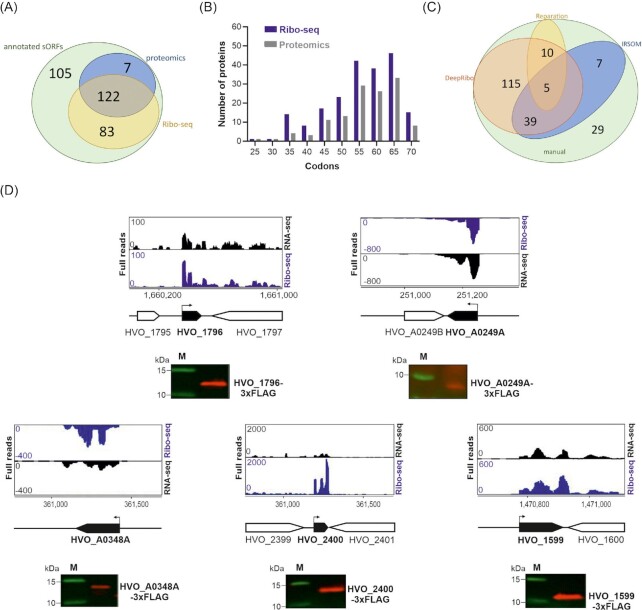
Figure 3.
Translation of the H. volcanii annotated small proteome revealed by Ribo-seq. (A). Overlap between annotated sORFs (‘annotated sORFs’, green) and translated small proteins detected by MS (all MS datasets, labelled ‘proteomics’; blue) and Ribo-seq (yellow). (B). Length distribution (in codons) of annotated small proteins identified by proteomics (all datasets; grey) and Ribo-seq (blue). (C). Comparison of sORFs detected in Ribo-seq data by manual labelling (‘manual’; green) or different automated ORF prediction tools for Ribo-seq data: REPARATION (yellow) and DeepRibo (orange); IRSOM (only RNA-seq data) (blue). (D). In vivo validation of translation for five annotated small proteins identified either by both Ribo-seq and MS (HVO_1796, 46 aa; HVO_A0348A, 63 aa; HVO_2400, 58 aa, and HVO_1599, 49 aa) or only by Ribo-seq (HVO_A0249A, 34 aa). ORFs were tagged at their N- or C-terminus with a 3xFLAG epitope and expressed under a p.tna promoter (Allers et al. 2010) or natural promoter (HVO_A0348A) from a plasmid in H. volcanii. Strains were grown to exponential phase in selective media and protein extracts were analyzed by western blotting with an anti-FLAG antibody. Analysis of a nontranslated sORF served as negative control (Figure S5D, Supporting Information). M: molecular weight marker, sizes are shown in kDa. Top: Ribo-seq (blue) and RNA-seq (black) coverage, genomic position is indicated below with a schematic representation of the genomic region (black: sORF investigated). Bent arrows indicate the transcription start sites [TSS based on Babski, Haas et al. (2016)].
However, seven small proteins were detected by MS alone (Fig. 3A; Table S1A, Supporting Information). Further inspection of Ribo-seq coverage for their corresponding genes showed that some were lowly expressed and lost upon application of TE or RNA-seq RPKM cut-offs (e.g. HVO_A0542, HVO_A0399; Figure S4A, Supporting Information) or might be wrongly annotated as sORFs and their Ribo-seq and RNA-seq read coverage would rather fit longer ORFs (e.g. HVO_A0015, HVO_1204; Figure S4B, Supporting Information).
The HRIBO workflow used to process our Ribo-seq datasets (Gelhausen et al. 2020) generates ORF predictions based on the Ribo-seq coverage using two tools [REPARATION and DeepRibo (Ndah et al. 2017, Clauwaert et al. 2019)]. These tools as well as IRSOM (Platon et al. 2018), a transcriptomic-based ORF prediction tool established in eukaryotes, were recently evaluated for their performance on Ribo-seq data from diverse bacterial species (Gelhausen et al. 2022), but have not yet been investigated for their performance with archaeal data.
We took advantage of our curated H. volcanii sORF set, manually labelled based on Ribo-seq data (Table S1B, Supporting Information), to evaluate their performance. DeepRibo detected 169 of the 205 positively labelled benchmark sORFs and none called as not translated (negatively labelled) by manual curation (Fig. 3C; Table S1B, Supporting Information). In contrast, IRSOM and REPARATION detected only 51 and 15 positively labelled sORFs, respectively, and even missed highly translated ribosomal sORFs. These results indicate that out of the tested ORF prediction tools, DeepRibo is best suited for H. volcanii Ribo-seq data.
To validate the accuracy of our MS/Ribo-seq datasets in detecting translated sORFs, we selected translated annotated sORFs for validation by an independent method in vivo. A total of five annotated sORFs with strong Ribo-seq and MS signals were tagged with a 3xFLAG epitope at their C-terminus (HVO_1796, a 46-aa uncharacterized protein; HVO_A0249A, a 34-aa uncharacterized protein, and HVO_A0348A, a 63-aa uncharacterized protein) or N-terminus (HVO_1599, a 49-aa uncharacterized protein; and HVO_2400, a 58-aa CPxCG-related zinc finger protein). Western blot analysis of cell lysates showed specific anti-FLAG signals at the expected sizes for all five protein fractions, confirming that their encoding sORFs are indeed translated in vivo (Fig. 3D). We also validated translation in a similar fashion for two additional sORFs (Figure S5, Supporting Information). Taken together, these results show that our complementary approach using MS and Ribo-seq is a powerful tool to detect the translated small proteome of H. volcanii.
Ribo-seq reveals hidden sORFs
Visual inspection of Ribo-seq data strongly suggested that genomic regions outside the annotation might encode small proteins not detected by MS. To identify strong candidates for novel sORFs, we first inspected DeepRibo predictions. To reduce the large number of predictions (8000) to a more manageable list, we applied cut-off for the TE (≥0.5) and RNA-seq expression (≥10 RPKM) based on the 205 sORFs, that were positively labelled as translated (Fig. 3A). DeepRibo also provides a score to rank candidates, and we also applied a relatively stringent score cut-off (score >0), based on values for the annotated translated small proteome (Table S2B, Supporting Information; see ‘Methods’ for details). Manual inspection of coverage for the resulting 161 predictions indicated that only 13 might be translated. The high rate of potentially false positives generated by DeepRibo was already reported in a previous study (Gelhausen et al. 2022) and can be due to the translatome structure of the organisms being studied or technical biases from cDNA library preparation. This prediction tool is trained on model bacterial species with mostly leadered mRNAs and strong reliance on Shine–Dalgarno sequences. In addition to the analysis of DeepRibo top-hits, 26 novel sORF candidates were identified by a genome-wide visual inspection of Ribo- and RNA-seq coverage performed as in a previous study (Gelhausen et al. 2022) (for details see ‘Methods’). While the TE of these candidates was generally high (>1), they exhibited a negative DeepRibo score (prediction score below 0; Table S5D, Supporting Information). However, negative scores do not per se argue against translation, as seen for some of the annotated sORFs from H. volcanii that were identified as translated in our study (Tables S5D and S6, Supporting Information) and for lowly expressed validated small proteins in E. coli (Gelhausen et al. 2022).
In line with our data, 16 out of these additional 26 predicted novel sORFs were also predicted as translated in the recently published H. volcanii Ribo-seq data (Gelsinger et al. 2020). The large number of strong candidates identified manually but missed by DeepRibo stringent cut-offs indicates that the ranking score of this tool is imperfect for archaeal Ribo-seq data.
In addition to computational predictions from Ribo-seq data, we also manually curated previously predicted sORFs and sRNAs from the literature (for details see ‘Methods’) (Babski et al. 2011, Laass et al. 2019). Most previously reported sRNAs had significant coverage only in RNA-seq libraries, suggesting they are bona fide noncoding transcripts (data not shown). However, two genes predicted to encode sRNAs had significant coverage in Ribo-seq libraries, suggesting that they might harbour translating sORFs (Table S5D, Supporting Information). We termed these genes HVO_1425A and HVO_2293A [previously termed HVO_1425n3 and HVO_2293n3 in Babski et al. (2011)]. Inspection of 16 long 5´ UTRs identified by differential RNA-seq and proposed to encode sORFs (Babski et al. 2016) added six additional candidates (Table S5D, Supporting Information).
Overall, Ribo-seq identified 47 strong candidates for novel sORFs (Fig. 4A; Tables S2B and S5D, Supporting Information) and showed that genes annotated as noncoding RNA genes might be protein coding genes.
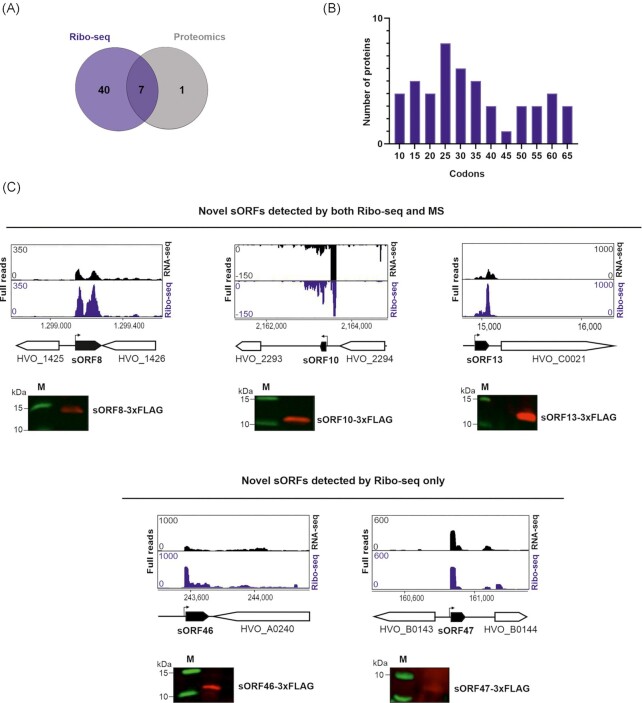
Figure 4.
Ribo-seq combined with MS expand the H. volcanii small proteome. (A). Overlap between the novel small proteins detected by Ribo-seq (blue) and MS (grey). (B). Length distribution (in codons) of the novel small proteins identified by Ribo-seq. (C). and (D). In vivo validation of translation for five novel small proteins identified either by both Ribo-seq and MS (sORF8, 56 aa; sORF10, 40 aa, and sORF13, 45 aa; top panel) (C) or only by Ribo-seq (sORF46, 42 aa and sORF47, 23 aa; bottom panel) (D). Small ORFs were C-terminally fused to a 3xFLAG tag and expressed from a plasmid in H. volcanii. Strains were grown to exponential phase in selective media and protein extracts were analyzed by western blotting. Proteins were detected with an anti-FLAG antibody. A nontranslated sORF served as negative control (Figure S5D, Supporting Information). M: molecular weight marker. Top: genome browser screenshots of read coverage from Ribo-seq(blue track)/RNA-seq(black track) libraries. Genomic positions are indicated below with a schematic representation of the genomic region (novel sORFs in black). Bent arrows indicate the transcription start sites (TSS) based on Babski et al. (2016).
De novo protein identification by MS identifies additional novel sORFs
To approach novel sORF identification from a complementary angle, the spectral data gathered by small protein adapted MS was analyzed by an additional database search following a proteogenomics approach (Fuchs et al. 2021). While traditional database searches are restricted to a search space defined by the organisms genome annotation, proteogenomics aims to overcome this limitation by extending the search space to an annotation-independent, complete translation of the genome in all reading frames, thereby enabling the identification of nonannotated proteins (Pueyo et al. 2016, Fuchs et al. 2021). This reanalysis identified eight candidate small proteins, four of which were validated by measuring synthetic peptides (sORFMS1, 2, 3, and 7) (Table S2A, Supporting Information). Inspection of Ribo-seq coverage for these eight MS candidates showed evidence of translation for seven (sORFMS1, 2, 4, 5, 6, 7, and 9). The Pepper proteogenomics software outputs the best potential ORF, but also reports second-order ORFs with start codons further downstream of the best spectral match. Comparison to the Ribo-seq data allowed us to clarify the ORF engaged by the ribosome in vivo (Table S5, Supporting Information).
To compare Ribo-seq- and MS-based detection for novel sORFs, we inspected the sets of novel sORFs predicted by each method in detail. Ribo-seq generally detected smaller ORFs, as well as basic proteins with a predicted isoelectric point up to 12 (Fig. 4B; Figure S3A and B, Supporting Information). The fraction of basic residues (His, Lys, and Arg) was 4% higher in candidates detected only by Ribo-seq compared to MS (predicted sORFs MS-identified: 12.3%; predicted sORFs Ribo-seq-identified: 16.5%; and annotated small proteins: 12.5%).
Short and basic proteins are often underrepresented in MS-based studies, as they generate only a few peptides in the MS-detectable range (Omasits et al. 2013). Although we reduced this bias by application of Lys-C, which cuts, unlike trypsin, only after lysine and not arginine, as well as by the measurement of the same samples without any protease added, the median number of unique peptides potentially generated per small protein in our data was substantially smaller for Ribo-seq only candidates (both novel and annotated) (Table S3, Supporting Information). Due to overlapping sequences, not all theoretically generated peptides are present in a sample and we therefore considered the Deep-MS-Peptide MS detectability score (Serrano et al. 2020). Peptides with a detectability score greater than 0.5 are generally considered detectable. Again, of the novel candidates detected by both, Ribo-seq and MS, six candidates passed this threshold with at least two peptides, whereas the majority of Ribo-seq identified candidates (27 of 47) did not generate at least two detectable peptides (Table S3, Supporting Information). Notably, there was no bias against membrane-associated small proteins by MS, as the predicted grand average of hydrophobicity index (GRAVY) for all three subsets (MS, Ribo-seq, and annotated) did not differ (Figure S3C, Supporting Information).
In summary, Ribo-seq and MS both detected an overlapping set of seven novel sORFs (Fig. 4A). These sORFs, in addition to those detected by epitope tagging, provide a high confidence list of novel H. volcanii small proteins. Further, MS-based identification suggests the presence of eight and Ribo-seq of 47 sORF candidates demonstrating the power of Ribo-seq in revealing novel sORFs. We confirmed the translation for five of these novel small proteins by in vivo expression and western blot analysis (Fig. 4C and D).
Characteristics of Haloferax small proteins
The combined power of small protein-adapted MS and Ribo-seq confirmed the translation of 212 of the 317 small proteins annotated in the H. volcanii genome and revealed translation for 48 novel small proteins, thereby raising the total to 260.
Next, we sought to broadly characterize the translated small proteins (annotated and novel, 260 sORFs) in terms of their potential function and the location of their genes. Small proteins detected by our analysis were distributed across the chromosome as well as all the minichromosomes (Fig. 5). Less than 20% of the 212 translated sORFs were encoded by leadered transcripts and most of them (74%) were found in intergenic regions or in operons (Fig. 6A and B). The novel sORFs (48 sORFs), however, showed a different distribution, with around half being preceded by a UTR (Fig. 6A and B). A large number of novel sORFs were also encoded in 5´/3´ UTRs, with a considerable number being encoded antisense to coding sequences (Fig. 6B).
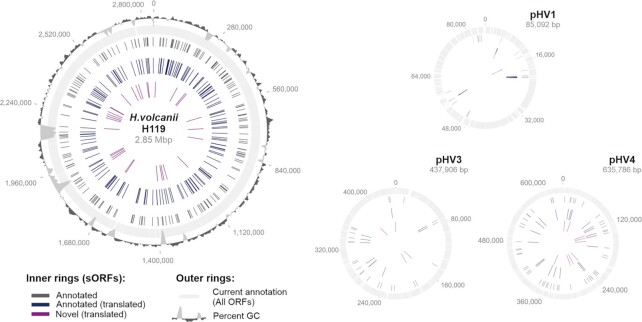
Figure 5.
Genomic distribution of H. volcanii translated small proteins. Data from MS and Ribo-seq were used to display the expanded proteome. The outer rings indicate all currently annotated ORFs (light grey) and % GC (dark grey). Black: all annotated sORFs; dark blue: all annotated translated sORFs; and purple: novel translated sORFs. The main chromosome as well as the three minichromosomes pHV1, pHV3, and pHV4 are shown.
Figure 6.
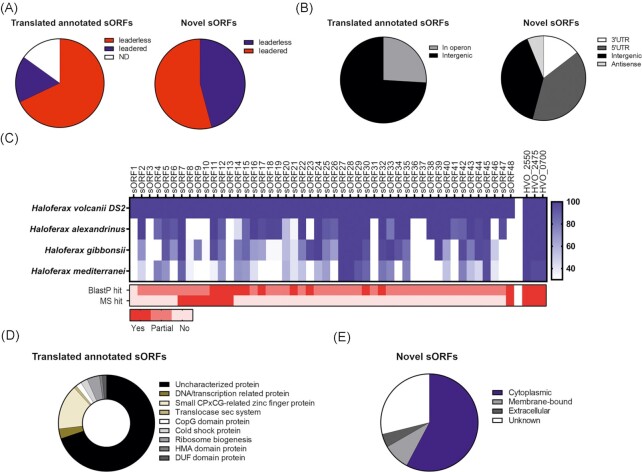
Features identified for the H. volcanii translated small proteome. (A). Pie chart indicating the proportion of leaderless (red) and leadered (blue) sORFs in the 212 translated annotated (left) and 48 novel (right) sORFs. ND: not determined. (B). Genomic location of translated annotated (left) and novel (right) sORFs relative to currently annotated genes. (C). Conservation of the 48 translated novel sORFs was determined for the genus Haloferax using tblastn (blue; depth denotes % conservation). The gradient on the right side indicates the % identity at the amino acid level. For comparison, three ribosomal proteins (HVO_2550, HVO_2475, and HVO_0700; far right) are included. Detection by blastp and MS are indicated at the bottom (red; depth indicates recovered hits). Legend at the bottom: ‘yes’: a 100% match was found to an annotated protein; ‘partial’: parts of the protein sequence match to an annotated protein, ‘no’: no matches were found for the sORF. (D). Annotated as well as predicted function (using Phyre2) for the 212 translated annotated sORFs. (E). Cellular localization predicted using PSORTb for the 48 translated novel sORFs in H. volcanii.
To gain additional confidence in our 48 novel sORF1 predictions, as well as first evidence that they might encode a functional small protein, we investigated their conservation in Haloferax species using tblastn searches (Altschul et al. 1997) (Fig. 6C). Our analysis revealed that all but 10 novel sORF candidates are conserved in at least one other Haloferax species with more than 60% amino acid identity. BlastP analysis showed perfect matches for 11 novel sORFs indicating that they are already annotated in at least one of the analyzed Haloferax species. For most of the other novel sORFs, only partial matches with annotated proteins were recovered. These partial matches correspond to annotated proteins that are either slightly shorter or longer. Most of these partial hits were to hypothetical proteins of unknown function.
Undefined function was also a hallmark we observed for almost all annotated small proteins found as translated in our study (Fig. 6D). For a handful, functions are already assigned, such as examples of ribosomal proteins, cold shock proteins, and components of the Sec transport system (Fig. 6D). We analyzed the amino acid sequences of the translated annotated small proteins of at least 30 residues length using the Phyre2 suite (Kelley et al. 2015), which informs on potential protein homology, secondary structure, and tertiary structure. This analysis identified conserved domains such as the CPxCG-related zinc finger domain, the CopG domain, or transmembrane helices (Fig. 6D; Table S5B, Supporting Information). We next performed Phyre2 analyses for the newly predicted sORFs. However, domain assignment was only possible for approximately half of them, as most were shorter than the 30 aa cut-off of Phyre2. For several of the sORFs analyzed, domains such as RuvA-domain like, transmembrane domain, zinc finger, Rmlc-like-cupin domain, ATPase domain, docking domain as well as several DUF (domain of unknown function) domains (Table S5B, Supporting Information) were found. To gain information on potential subcellular location of the newly identified small proteins, we investigated their sequences with PSORTb (Yu et al. 2010). Out of 48 novel candidates, four were predicted to be membrane-bound (sORF7, 23, 26, and 33), 26 were predicted to be cytosolic, and three examples had a predicted extracellular localization (sORF1, 27, and 47) (Fig. 6E).
Discussion
The small proteome is a vastly unexplored part of the archaeal cellular machinery. Here, we report the first use of a combination of ribosome profiling and MS-based proteomics to provide a comprehensive description of the small protein complement of the archaeon H. volcanii.
Inventory of the small proteome of H. volcanii
We approached the identification of small proteins in H. volcanii from two complementary angles: small protein adapted MS to detect their physical presence and Ribo-seq to detect and map translational events across the transcriptome. Importantly, both approaches featured open-end analysis and were not limited to a priori annotated ORFs, thereby allowing the detection of previously unannotated small proteins.
Our modified experimental and analysis MS workflow, tailored for small protein detection, captured 103 (32.5%) of 317 annotated sORFs under standard conditions (exponential and stationary phase). Other small protein adapted MS analyses detected 25% of proteins up to 10 kDa in H. salinarum (Klein et al. 2007) and 68 of approximately 1400 predicted small proteins in M. mazei (4.9%) (Kaulich et al. 2020, Gutt et al. 2021, Weidenbach et al. 2021). Our results are more comparable to the 31% recovered for S. aureus Newman by a similar MS-based approach for small proteins (here a cut-off of 100 aa was used) (Fuchs et al. 2021), suggesting that the applied modifications for small protein detection are invaluable in the investigation of archaeal small proteomes. We also re-evaluated publicly available proteomic datasets, generated without adaptations for small protein detection, for different H. volcanii strains from diverse growth conditions (Jevtić et al. 2019, Schulze et al. 2020). This added evidence for 26 additional annotated small proteins. All MS data together confirmed the expression of 129 (41%) out of 317 annotated H. volcanii small proteins under various conditions.
Small proteome characterization greatly benefits from multidisciplinary approaches (Fijalkowski et al. 2022). Thus, we added Ribo-seq analysis to map ribosome engagement for sORFs across the H. volcanii transcriptome. We made our Ribo-seq data available to the scientific community at our interactive web-based genome-browser: http://www.bioinf.uni-freiburg.de/ribobase. Ribo-seq detected translation of 205 out of 317 (65%) annotated small proteins at exponential growth phase. This coverage is similar to that observed for Salmonella (76%, Venturini et al. 2020), and clearly demonstrates the sensitivity of translatomics. No single approach can catalogue the small proteome to completeness due to immanent methodological biases. Therefore, the presence of annotated small proteins should be classified as high confidence if accounted for by both methods and as highly likely if returned by only one technique as suggested earlier (Venturini et al. 2020). Together, our data support the translation of 122 annotated H. volcanii small proteins (38%) with high confidence (detected by both methods) and 212 (67%) small proteins have strong evidence for translation under the examined growth condition.
Annotation algorithms are often biased against sORF detection (Storz et al. 2014). Thus, the small proteome of H. volcanii known to us via annotation is likely far from complete. Experimental data analysis can expand and refine proteome annotations in the low MW spectrum (VanOrsdel et al. 2018, Miravet‐Verde et al. 2019, Hemm et al. 2020, Venturini et al. 2020, Fuchs et al. 2021). The predictive power of the bioinformatic workflows may be limited on the archaeal datasets, as the underlying algorithms were optimized for bacterial genome features and need training on archaeal data for which we provide a valuable first dataset—albeit more of them will be needed. Therefore, the H. volcanii small proteome could be larger than the relatively conservative, high-confidence list presented here.
A recent study reporting Ribo-seq data for another H. volcanii strain (strain H98: ∆pyrE2 ∆hdrB) predicted 68 novel putative sORFs (with <50 codons) (Gelsinger et al. 2020). The H. volcanii strain H119 used in this study (Table S4A, Supporting Information) and strain H98 used by Gelsinger et al. (2020) are both derived from H. volcanii H26 (∆pyrE2) and only differ in the deletion of additional marker genes (H119 has additionally trpA and leuB genes deleted and H98 has an additional deletion in the hdrB gene) (Allers et al. 2004), thus data from both studies are comparable. We investigated these 68 putative sORFs and found only 16 to be translated after manual inspection of Ribo-seq coverage generated in our study. Moreover, our Ribo-seq data revealed translation of 31 additional sORFs that were not detected in strain H98. Differences in sORF translation observed between the two datasets might arise from the strain background, the growth conditions and protocols used for Ribo-seq, as well as prediction of sORFs based on start codon Ribo-seq coverage for strain H98. Despite these biological differences, we could provide independent validation (MS or tagging/western blotting) for five novel sORFs in H. volcanii, underscoring the robustness of our dataset. Altogether, application of Ribo-seq to H. volcanii suggests its small proteome is much larger than what is currently annotated.
The number of candidate novel H. volcanii sORFs detected by our analyses lies with 48 within the range of those described in other prokaryotic species using Ribo-seq, MS or combined approaches [Salmonella: 42 by Ribo-seq (Venturini et al. 2020, Fijalkowski et al. 2022), S. aureus: 24 by MS (Fuchs et al. 2021), and E. coli: 68 by Ribo-seq (Weaver et al. 2019)]. However, as study designs are so individual only a rough comparison can be drawn.
The total number of sORFs (up to 100 aa in length) has been estimated to be 16 +/− 9% of the total coding capacity in bacterial genomes (Miravet‐Verde et al. 2019). Our analysis suggests a total of 8.8% sORFs for H. volcanii, falling within the limits of the prediction, despite only considering proteins up to 70 aa (excluding sORF overlapping larger ORFs). The comparatively small number of novel sORFs we uncovered might reflect an already relatively complete coverage of sORFs in the current annotation. As small proteins may mediate adaptation to specific metabolic or stress-related states, detection of novel sORFs might also be limited in the standard conditions we applied for this proof-of concept study.
The difference between the number of translated ORFs (annotated and novel) revealed by proteomics and Ribo-seq shows that MS analysis is less sensitive than Ribo-seq in detecting translation of small proteins. Absence of detection by proteomics may be accounted for by short half-life, or specific features that interfere with MS detection (small number of peptides, low charge, and high hydrophobicity). In a study of the small proteome of S. Typhimurium with a similar experimental setup, it was recently shown that lower abundance was not a major factor contributing to the different sensitivities of Ribo-seq and MS-based detection of sORFs (Fijalkowski et al. 2022). In line with this, there was no difference in TE between small proteins detected by both methods and those only detected by Ribo-seq in our data. Novel sORFs identified by Ribo-seq alone tended to be shorter and exhibit a larger pI range, with many having a more basic isoelectric point than those identified by MS. The higher number of basic residues results in an overabundance of protease cleavage sites quenching the median number of unique peptides available. Additionally, analysis of Deep-MS-Peptide scores attests a lower MS-detectability of the generated peptides. Ribo-seq coverage accounts for ribosome occupancy, but has also been demonstrated for noncoding sequences that associate in a translation-independent fashion with the ribosome as well as a consequence of ribosome scanning, protection by RNA binding proteins, and/or RNase resistance (Wilson and Masel 2011, Pueyo et al. 2016, Fremin and Bhatt 2020), thereby increasing the number of false positives. Moreover, protein levels do not necessarily correlate with ribosome density (Hemm et al. 2020). This illustrates that concordant identification in both datasets ascertains presence in vivo but missing representation does not preclude it. However, candidates detected by only a single method should not be dismissed, but they will need further experimental support.
In the light of this, we supported the validity of our small protein identification scheme by successful in vivo detection of epitope-tagged versions of seven of the annotated proteins found at the proteomic or translational level. A similarly high rate of in vivo validation was achieved for the novel small proteins demonstrating the analytical power of our combinatorial approach for the detection of small proteins. Here, in vivo support for five novel sORFs predicted only by a Ribo-seq event strengthens confidence in ribosomal profiling-only predictions. Validation of all candidates is beyond the scope of this article but their description as potential novel sORFs is a valuable and essential first step to their future analysis. Bacterial studies, together with our analysis of H. volcanii illustrate that genome annotations are likely missing substantial portions of the small proteome in all prokaryotes. This gap must be filled to allow for genetic screens, comparative genomic studies, including small proteins (Hemm et al. 2020). To do so, systematic approaches that gather empirical evidence for missing gene annotations are called for and our analysis demonstrates for the first-time the feasibility and power of multiomic approaches to fill this gap for the archaeal domain.
Characteristics of the haloarchaeal small proteome
The functions of small proteins can be difficult to identify because they are too small to harbour known domains and encode less sequence information than longer proteins (Law et al. 2001, Storz et al. 2014). Small proteins in general tend to be less conserved and seem to be a species-specific adaptation tailored to individual needs (Wang et al. 2008, Storz et al. 2014, Venturini et al. 2020). We detected close homologs for almost all of the newly described sORF candidates within other Haloferax species, which might reflect either functional conservation or the propensity of Haloarchaea for extensive horizontal gene transfer and recombination (Papke et al. 2015) or a combination of both owed to shared evolutionary pressures. This conservation across species barriers strengthens the robustness of our predictions despite a large portion of candidates being recovered from Ribo-seq data only. Unfortunately, homologs of these are mostly annotated as hypothetical proteins, and like most small proteins, most do not harbour known domains (Wang et al. 2008, Storz et al. 2014). A large proportion of the novel sORFs we identified in 5´ UTRs are located within a few nucleotides of, or extend into, the coding region of the neighbouring gene. This is reminiscent of the upstream ORFs (uORFs) or leader peptide genes that regulate transcription or translation of the downstream gene in bacteria and eukaryotes [reviewed in Cabrera-Quio et al. (2016), Orr et al. (2020)].
Conclusion
In summary, our analysis shows that translated small proteins are found across the Haloferax genome and with evidence for, considering all datasets, 260 (212 annotated, 48 new) being translated. The lack of functional predictions is unsatisfying but leaves room for ample exploration that the comprehensive catalogue of small proteins, venturing beyond annotated coding regions, presented herein will spark. We also hope to inspire future use of this multiomics scheme in other archaeal species as the wealth of archaeal small proteins is still largely untapped.
Supplementary Material
Acknowledgements
This research was funded by the German Research Council (Deutsche Forschungsgemeinschaft, DFG) in the framework of the priority programme SPP 2002, ‘Small Proteins in Prokaryotes, an Unexplored World’ to RB (BA 2168/21–1), DB (BE 3869/5–1, BE 3869/5–2), CMS (SH 580/7–1 and 7–2 as well as SH 580/8–1 and 8–2), and AM (MA 1538/24–1). We would like to thank Stephanie Färber for technical assistance with Ribo-seq sample preparation, Susanne Schmidt for expert technical assistance, Britta Stoll for help with preparing mass spectrometry samples, as well as Jutta Brendel for help with western blots. We are also grateful to Thomas Sura who operated the QExactive mass spectrometer.
Footnotes
A total of 47 novel ORFs are predicted by Ribo-seq and an additional novel one by MS only.
Contributor Information
Lydia Hadjeras, Department of Molecular Infection Biology II, Institute of Molecular Infection Biology (IMIB), University of Würzburg, Josef-Schneider-Straße 2 / D15, 97080 Würzburg, Germany.
Jürgen Bartel, Department of Microbial Proteomics, Institute of Microbiology, University of Greifswald, Felix-Hausdorff-Str. 8, 17489 Greifswald, Germany.
Lisa-Katharina Maier, Biology II, Ulm University, Albert-Einstein-Allee 11, 89081 Ulm, Germany.
Sandra Maaß, Department of Microbial Proteomics, Institute of Microbiology, University of Greifswald, Felix-Hausdorff-Str. 8, 17489 Greifswald, Germany.
Verena Vogel, Biology II, Ulm University, Albert-Einstein-Allee 11, 89081 Ulm, Germany.
Sarah L Svensson, Department of Molecular Infection Biology II, Institute of Molecular Infection Biology (IMIB), University of Würzburg, Josef-Schneider-Straße 2 / D15, 97080 Würzburg, Germany.
Florian Eggenhofer, Bioinformatics Group, Department of Computer Science, University of Freiburg, Georges-Koehler-Allee 106, 79110 Freiburg, Germany.
Rick Gelhausen, Bioinformatics Group, Department of Computer Science, University of Freiburg, Georges-Koehler-Allee 106, 79110 Freiburg, Germany.
Teresa Müller, Bioinformatics Group, Department of Computer Science, University of Freiburg, Georges-Koehler-Allee 106, 79110 Freiburg, Germany.
Omer S Alkhnbashi, Information and Computer Science Department, King Fahd University of Petroleum and Minerals, Dhahran 31261, Saudi Arabia.
Rolf Backofen, Bioinformatics Group, Department of Computer Science, University of Freiburg, Georges-Koehler-Allee 106, 79110 Freiburg, Germany; Signalling Research Centres BIOSS and CIBSS, University of Freiburg, Schaenzlestr. 18, 79104 Freiburg, Germany.
Dörte Becher, Department of Microbial Proteomics, Institute of Microbiology, University of Greifswald, Felix-Hausdorff-Str. 8, 17489 Greifswald, Germany.
Cynthia M Sharma, Department of Molecular Infection Biology II, Institute of Molecular Infection Biology (IMIB), University of Würzburg, Josef-Schneider-Straße 2 / D15, 97080 Würzburg, Germany.
Anita Marchfelder, Biology II, Ulm University, Albert-Einstein-Allee 11, 89081 Ulm, Germany.
Conflict of interest statement
None declared.
Authors’ contributions
L.H., J.B., S.M., and V.V. performed the experiments. L.H., S.L.S., L.K.M., J.B., S.M., O.S.A., F.E., T.M., and R.G. analyzed the data. A.M., D.B., and C.M.S. conceptualized the project. L.H., S.L.S., L.K.M., and A.M. wrote the original draft. S.M., J.B., D.B., R.B., L.H., S.L.S., L.K.M., C.M.S. A.M. reviewed and edited the draft, D.B., R.B., C.M.S., and A.M. supervised the research and provided resources and funding. All authors approved the submitted version.
Data availability
MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaíno et al. 2014) and are available through the identifiers PXD031423 (searches for annotated sORFs), PXD031691 (searches for novel sORFs), PXD031460 (spectral-library based validation). Ribo-seq and RNA-seq data have been deposited in GEO under the accession number GSE208086. In addition, the Ribo-seq data can be viewed with an interactive online JBrowse instance (http://www.bioinf.uni-freiburg.de/ribobase).
References
- Ahrens CH, Wade JT, Champion MMet al. A practical guide to small protein discovery and characterization using mass spectrometry. J Bacteriol. 2022;204:e0035321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T, Barak S, Liddell Set al. Improved strains and plasmid vectors for conditional overexpression of his-tagged proteins in Haloferax volcanii. Appl Environ Microbiol. 2010;76:1759–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T, Ngo HP, Mevarech Met al. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol. 2004;70:943–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AAet al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babski J, Haas KA, Näther-Schindler Det al. Genome-wide identification of transcriptional start sites in the haloarchaeon Haloferax volcanii based on differential RNA-seq (dRNA-Seq). Bmc Genomics. 2016;17:629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babski J, Tjaden B, Voss Bet al. Bioinformatic prediction and experimental verification of sRNAs in the haloarchaeon Haloferax volcanii. RNA Biol. 2011;8:806–16. [DOI] [PubMed] [Google Scholar]
- Baek J, Lee J, Yoon Ket al. Identification of unannotated small genes in Salmonella. G3 Genes|Genomes|Genetics. 2017;7:983–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel J, Varadarajan AR, Sura Tet al. Optimized proteomics workflow for the detection of small proteins. J Proteome Res. 2020;19:4004–18. [DOI] [PubMed] [Google Scholar]
- Bartholomäus A, Del Campo C, Ignatova Z. Mapping the non-standardized biases of ribosome profiling. Biol Chem. 2016;397:23–35. [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Johnstone TG, Christiano Ret al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Quio LE, Herberg S, Pauli A. Decoding sORF translation – from small proteins to gene regulation. RNA Biol. 2016;13:1051–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy L, Kaulich PT, Maaß Set al. Bottom-up and top-down proteomic approaches for the identification, characterization, and quantification of the low molecular weight proteome with focus on short open reading frame-encoded peptides. Proteomics:2021;21:2100008. [DOI] [PubMed] [Google Scholar]
- Cassidy L, Kaulich PT, Tholey A. Depletion of high-molecular-mass proteins for the identification of small proteins and short open reading frame encoded peptides in cellular proteomes. J Proteome Res. 2019;18:1725–34. [DOI] [PubMed] [Google Scholar]
- Cassidy L, Prasse D, Linke Det al. Combination of bottom-up 2D-LC-MS and semi-top-down GelFree-LC-MS enhances coverage of proteome and low molecular weight short open reading frame encoded peptides of the Archaeon Methanosarcina mazei. J Proteome Res. 2016;15:3773–83. [DOI] [PubMed] [Google Scholar]
- Clauwaert J, Menschaert G, Waegeman W. DeepRibo: a neural network for precise gene annotation of prokaryotes by combining ribosome profiling signal and binding site patterns. Nucleic Acids Res. 2019;47:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline SW, Lam WL, Charlebois RLet al. Transformation methods for halophilic archaebacteria. Can J Microbiol. 1989;35:148–52. [DOI] [PubMed] [Google Scholar]
- da Veiga Leprevost F, Haynes SE, Avtonomov DMet al. Philosopher: a versatile toolkit for shotgun proteomics data analysis. Nat Methods. 2020;17:869–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuluri S, Wu Y, Hepowit NLet al. Proteome targets of ubiquitin-like samp1ylation are associated with sulfur metabolism and oxidative stress in Haloferax volcanii. Proteomics. 2016;16:1100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TRet al. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol. 2008;4:e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M, Cossart P. Small bacterial and phagic proteins: an updated view on a rapidly moving field. Curr Opin Microbiol. 2017;39:81–8. [DOI] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin Set al. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowski I, Willems P, Jonckheere Vet al. Hidden in plain sight: challenges in proteomics detection of small ORF-encoded polypeptides. Microlife. 2022;3:uqac005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel Y, Mizrahi O, Nachshon Aet al. The coding capacity of SARS-CoV-2. Nature. 2021;589:125–30. [DOI] [PubMed] [Google Scholar]
- Fremin BJ, Bhatt AS. Structured RNA contaminants in bacterial ribo-seq. Msphere. 2020;5:e00855–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S, Kucklick M, Lehmann Eet al. Towards the characterization of the hidden world of small proteins in Staphylococcus aureus, a proteogenomics approach. PLos Genet. 2021;17:e1009585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garai P, Blanc-Potard A. Uncovering small membrane proteins in pathogenic bacteria: regulatory functions and therapeutic potential. Mol Microbiol. 2020;114:710–20. [DOI] [PubMed] [Google Scholar]
- Gelhausen R, Müller T, Svensson SLet al. RiboReport - benchmarking tools for ribosome profiling-based identification of open reading frames in bacteria. Briefings Bioinf. 2022;23:bbab549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhausen R, Svensson SL, Froschauer Ket al. HRIBO: high-throughput analysis of bacterial ribosome profiling data. Bioinformatics. 2020;37:btaa959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelsinger DR, Dallon E, Reddy Ret al. Ribosome profiling in archaea reveals leaderless translation, novel translational initiation sites, and ribosome pausing at single codon resolution. Nucleic Acids Res. 2020;48:5201–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Gladyshev VN. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014;42:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaub A, Huptas C, Neuhaus Ket al. Recommendations for bacterial ribosome profiling experiments based on bioinformatic evaluation of published data. J Biol Chem. 2020;295:8999–9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray T, Storz G, Papenfort K. Small proteins; big questions. J Bacteriol. 2022;204:e0034121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüning B, Dale R, Sjödin Aet al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat Methods. 2018;15:475–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta NT, Vander Heiden JA, Uduman Met al. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutt M, Jordan B, Weidenbach Ket al. High complexity of glutamine synthetase regulation in Methanosarcina mazei : small protein 26 interacts and enhances glutamine synthetase activity. FEBS J. 2021;288:febs.15799. [DOI] [PubMed] [Google Scholar]
- Hemm MR, Weaver J, Storz G. Escherichia coli small proteome. EcoSal Plus. 2020;9:32385980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbard MA, Miranda HV, Lim J-Met al. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010;463:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J-Y, Buskirk AR. A ribosome profiling study of mRNA cleavage by the endonuclease RelE. Nucleic Acids Res. 2017;45:327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin Set al. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRSet al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT. Ribosome footprint profiling of translation throughout the genome. Cell. 2016;165:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtić Ž, Stoll B, Pfeiffer Fet al. The response of Haloferax volcanii to salt and temperature stress: a proteome study by label-free mass spectrometry. Proteomics. 2019;19:1800491. [DOI] [PubMed] [Google Scholar]
- Ji Z, Song R, Regev Aet al. Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife. 2015;4:e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaulich PT, Cassidy L, Weidenbach Ket al. Complementarity of different SDS-PAGE gel staining methods for the identification of short open reading frame-encoded peptides. Proteomics. 2020;20:2000084. [DOI] [PubMed] [Google Scholar]
- Keller A, Purvine S, Nesvizhskii AIet al. Experimental protein mixture for validating tandem mass spectral analysis. OMICS J Integr Biol. 2002;6:207–12. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CMet al. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C, Aivaliotis M, Olsen JVet al. The low molecular weight proteome of Halobacterium salinarum. J Proteome Res. 2007;6:1510–8. [DOI] [PubMed] [Google Scholar]
- Kong AT, Leprevost FV, Avtonomov DMet al. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat Methods. 2017;14:513–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster J, Rahmann S. Snakemake–a scalable bioinformatics workflow engine. Bioinformatics. 2012;28:2520–2. [DOI] [PubMed] [Google Scholar]
- Kubatova N, Jonker HR, Saxena Ket al. Solution structure and dynamics of the small protein HVO_2922 from Haloferax volcanii. ChemBioChem. 2020a;21:149–56. [DOI] [PubMed] [Google Scholar]
- Kubatova N, Pyper DJ, Jonker HRAet al. Rapid biophysical characterization and NMR spectroscopy structural analysis of small proteins from bacteria and archaea. ChemBioChem. 2020b;21:1178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laass S, Monzon VA, Kliemt Jet al. Characterization of the transcriptome of Haloferax volcanii, grown under four different conditions, with mixed RNA-seq. PLoS ONE. 2019;14:e0215986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Deutsch EW, Eddes JSet al. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics. 2007;7:655–67. [DOI] [PubMed] [Google Scholar]
- Law GL, Raney A, Heusner Cet al. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J Biol Chem. 2001;276:38036–43. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker Aet al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- Liao Y, Vogel V, Hauber Set al. CdrS is a global transcriptional regulator influencing cell division in Haloferax volcanii. Mbio. 2021;0:e01416–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10. [Google Scholar]
- Mendoza L, Deutsch EW, Sun Zet al. Flexible and fast mapping of peptides to a proteome with ProteoMapper. J Proteome Res. 2018;17:4337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menschaert G, Van Criekinge W, Notelaers Tet al. Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events*. Mol Cell Proteomics. 2013;12:1780–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miravet-Verde S, Ferrar T, Espadas-García Get al. Unraveling the hidden universe of small proteins in bacterial genomes. Mol Syst Biol. 2019;15:e8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Green R, Buskirk AR. A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. eLife. 2019;8:e42591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumtaz MAli S, Couso Juan P. Ribosomal profiling adds new coding sequences to the proteome. Biochem Soc Trans. 2015;43:1271–6. [DOI] [PubMed] [Google Scholar]
- Nagel C, Machulla A, Zahn Set al. Several one-domain zinc finger µ-proteins of Haloferax volcanii are important for stress adaptation, biofilm formation, and swarming. Genes. 2019;10:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndah E, Jonckheere V, Giess Aet al. REPARATION: ribosome profiling assisted (re-)annotation of bacterial genomes. Nucleic Acids Res. 2017;45:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker Eet al. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–58. [DOI] [PubMed] [Google Scholar]
- Neuhaus K, Landstorfer R, Simon Set al. Differentiation of ncRNAs from small mRNAs in Escherichia coli O157:H7 EDL933 (EHEC) by combined rnaseq and riboseq – ryhB encodes the regulatory RNA RyhB and a peptide, RyhP. Bmc Genomics. 2017;18:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Becker Annemarie H, Sandikci Aet al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U, Quebatte M, Stekhoven DJet al. Directed shotgun proteomics guided by saturated RNA-seq identifies a complete expressed prokaryotic proteome. Genome Res. 2013;23:1916–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U, Varadarajan AR, Schmid Met al. An integrative strategy to identify the entire protein coding potential of prokaryotic genomes by proteogenomics. Genome Res. 2017;27:2083–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MW, Mao Y, Storz Get al. Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Res. 2020;48:1029–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto C, Stadler PF, Hoffmann S. Lacking alignments? The next-generation sequencing mapper segemehl revisited. Bioinformatics. 2014;30:1837–43. [DOI] [PubMed] [Google Scholar]
- Paoletti AC, Parmely TJ, Tomomori-Sato Cet al. Quantitative proteomic analysis of distinct mammalian mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci. 2006;103:18928–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papke R, Corral P, Ram-Mohan Net al. Horizontal gene transfer, dispersal and haloarchaeal speciation. Life. 2015;5:1405–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Klammer AA, Kall Let al. Rapid and accurate peptide identification from tandem mass spectra. J Proteome Res. 2008;7:3022–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruschke H, Anders J, Stadler PFet al. Enrichment and identification of small proteins in a simplified human gut microbiome. J Proteomics. 2020;213:103604. [DOI] [PubMed] [Google Scholar]
- Platon L, Zehraoui F, Bendahmane Aet al. IRSOM, a reliable identifier of ncRNAs based on supervised self-organizing maps with rejection. Bioinformatics. 2018;34:i620–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza S, Menschaert G, Payre F. In search of lost small peptides. Annu Rev Cell Dev Biol. 2017;33:391–416. [DOI] [PubMed] [Google Scholar]
- Prasse D, Thomsen J, De Santis Ret al. First description of small proteins encoded by spRNAs in Methanosarcina mazei strain Gö1. Biochimie. 2015;117:138–48. [DOI] [PubMed] [Google Scholar]
- Pueyo JI, Magny EG, Couso JP. New peptides under the s(ORF)ace of the genome. Trends Biochem Sci. 2016;41:665–78. [DOI] [PubMed] [Google Scholar]
- R Core Team . R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2019. https://www.R-project.org/(19 July 2019, date last accessed). [Google Scholar]
- Savitski MM, Wilhelm M, Hahne Het al. A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol Cell Proteomics. 2015;14:2394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1:16–22. [DOI] [PubMed] [Google Scholar]
- Schulze S, Adams Z, Cerletti Met al. The Archaeal Proteome Project advances knowledge about archaeal cell biology through comprehensive proteomics. Nat Commun. 2020;11:3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano G, Guruceaga E, Segura V. DeepMSPeptide: peptide detectability prediction using deep learning. Bioinformatics. 2020;36:1279–80. [DOI] [PubMed] [Google Scholar]
- Sharma CM, Darfeuille F, Plantinga THet al. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff SA, Mitchell AJ, Schwaid AGet al. Peptidomic discovery of short open reading frame–encoded peptides in human cells. Nat Chem Biol. 2013;9:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg R, Koch HG. The largely unexplored biology of small proteins in pro- and eukaryotes. FEBS J. 2021;288:7002–24. [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N, Weisburd B, Michalski Aet al. Decoding human cytomegalovirus. Science. 2012;338:1088–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Wolf YI, Ramamurthi KS. Small proteins can no longer be ignored. Annu Rev Biochem. 2014;83:753–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanOrsdel CE, Kelly JP, Burke BNet al. Identifying new small proteins in Escherichia coli. Proteomics. 2018;18:1700064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez J-J, Hon C-C, Vanselow JTet al. Comparative ribosome profiling reveals extensive translational complexity in different Trypanosoma brucei life cycle stages. Nucleic Acids Res. 2014;42:3623–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Sharma CM, Mankin Aet al. Identifying small open reading frames in prokaryotes with ribosome profiling. J Bacteriol. 2022;204:e0029421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini E, Svensson SL, Maaß Set al. A global data-driven census of Salmonella small proteins and their potential functions in bacterial virulence. Microlife. 2020;1:uqaa002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Deutsch EW, Wang Ret al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Xiao J, Pan Let al. A systematic survey of mini-proteins in bacteria and archaea. PLoS ONE. 2008;3:e4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver J, Mohammad F, Buskirk ARet al. Identifying small proteins by ribosome profiling with stalled initiation complexes. Mbio. 2019;10:e02819–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenbach K, Gutt M, Cassidy Let al. Small proteins in Archaea, a mainly unexplored world. J Bacteriol. 2021;204:00313–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BA, Masel J. Putatively noncoding transcripts show extensive association with ribosomes. Genome Biol Evol. 2011;3:1245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingett SW, Andrews S. FastQ screen: a tool for multi-genome mapping and quality control. F1000Research. 2018;7:1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MRet al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn S, Kubatova N, Pyper DJet al. Biological functions, genetic and biochemical characterization, and NMR structure determination of the small zinc finger protein HVO_2753 from Haloferax volcanii. FEBS J. 2021;288:2042–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaíno et al. 2014) and are available through the identifiers PXD031423 (searches for annotated sORFs), PXD031691 (searches for novel sORFs), PXD031460 (spectral-library based validation). Ribo-seq and RNA-seq data have been deposited in GEO under the accession number GSE208086. In addition, the Ribo-seq data can be viewed with an interactive online JBrowse instance (http://www.bioinf.uni-freiburg.de/ribobase).