Abstract
Background
Infections with respiratory viruses [e.g. influenza and respiratory syncytial virus (RSV)] can increase the risk of severe pneumococcal infections. Likewise, pneumococcal coinfection is associated with poorer outcomes in viral respiratory infection. However, there are limited data describing the frequency of pneumococcus and SARS-CoV-2 coinfection and the role of coinfection in influencing COVID-19 severity. We, therefore, investigated the detection of pneumococcus in COVID-19 inpatients during the early pandemic period.
Methods
The study included patients aged 18 years and older, admitted to the Yale-New Haven Hospital who were symptomatic for respiratory infection and tested positive for SARS-CoV-2 during March–August 2020. Patients were tested for pneumococcus through culture-enrichment of saliva followed by RT-qPCR (to identify carriage) and serotype-specific urine antigen detection (UAD) assays (to identify presumed lower respiratory tract pneumococcal disease).
Results
Among 148 subjects, the median age was 65 years; 54.7% were male; 50.7% had an ICU stay; 64.9% received antibiotics; and 14.9% died while admitted. Pneumococcal carriage was detected in 3/96 (3.1%) individuals tested by saliva RT-qPCR. Additionally, pneumococcus was detected in 14/127 (11.0%) individuals tested by UAD, and more commonly in severe than moderate COVID-19 [OR: 2.20; 95% CI: (0.72, 7.48)]; however, the numbers were small with a high degree of uncertainty. None of the UAD-positive individuals died.
Conclusions
Pneumococcal lower respiratory tract infection (LRTI), as detected by positive UAD, occurred in patients hospitalized with COVID-19. Moreover, pneumococcal LRTI was more common in those with more serious COVID-19 outcomes. Future studies should assess how pneumococcus and SARS-CoV-2 interact to influence COVID-19 severity in hospitalized patients.
Keywords: Streptococcus pneumoniae, COVID-19, saliva, urinary antigen detection
Introduction
Most infections with SARS-CoV-2 are mild-to-moderate in severity while some progress to severe manifestations of COVID-19 including pneumonia, acute respiratory distress syndrome (ARDS), and death (Gupta et al. 2020). The factors that determine disease severity remain incompletely understood. For other viral pathogens, coinfections with bacteria influence the severity of infection. This is evident during seasonal epidemics of influenza and respiratory syncytial virus (RSV), as well as during historical pandemics (Morens et al. 2008, McCullers 2014). Streptococcus pneumoniae (pneumococcus) is one of the most common coinfecting pathogens with influenza and RSV (Klein et al. 2016). There has, therefore, been interest in the possible role of bacterial coinfections in influencing the severity of COVID-19. Clinical studies have generally reported a small percentage of COVID-19 cases with secondary bacterial pneumonia (Langford et al. 2020, Zhu et al. 2020, Dirkx et al. 2021, Lehmann et al. 2021). In contrast, the rates of COVID-19 disease, hospitalization, and death were lower among those who received pneumococcal conjugate vaccines (PCVs) compared with those who did not (Noale et al. 2020, Nunes et al. 2021, Lewnard et al. 2022). This is consistent with previous work from a randomized trial that demonstrated lower rates of hospitalization for a number of viruses among children receiving PCVs, including endemic coronaviruses (Madhi et al. 2004).
We sought to quantify the prevalence of pneumococcal infection among patients hospitalized with COVID-19 and to explore whether pneumococcal carriage or infection was associated with COVID-19 disease severity. Pneumococcus is commonly found in the upper respiratory tract of healthy children (Auranen et al. 2010) and adults (Wyllie et al. 2016a), and can be detected by bacterial culture or qPCR in samples from the upper respiratory tract. When individuals go on to develop lower respiratory tract infections (LRTIs), samples are not typically obtained from the lower respiratory tract, rather, evidence of infection is indirect. The detection of pneumococcal capsular polysaccharides in the urine has been demonstrated to be a reliable marker of LRTIs caused by pneumococcus and can distinguish between carriage and disease.
We hypothesized that pneumococcus would be detected frequently in this population, in both saliva and urine, and that individuals coinfected with pneumococcus and SARS-CoV-2 would have more severe COVID outcomes compared with individuals infected with SARS-CoV-2 alone. We used data and samples from a retrospective cohort of individuals hospitalized with COVID-19 to evaluate the prevalence of pneumococcus and to explore the association with severity. Understanding these relationships is important when designing public health response measures that could mitigate both pneumococcal disease and COVID-19.
Materials and methods
Study design
During March–August of 2020, paired saliva and urine samples were collected from hospitalized patients at the Yale-New Haven Hospital who were admitted to the hospital with symptomatic SARS-CoV-2 infection, diagnosed from a nasopharyngeal swab-based PCR test, and enrolled in the Yale IMPACT study (Wyllie et al. 2020). Signed informed consent was obtained from all study participants following the Yale University HIC-approved protocol #2000027690. Demographic and case information was collected through systematic and retrospective review of patient electronic medical records, including vaccination history and a peak measure of disease severity as described elsewhere (Lucas et al. 2020). SARS-CoV-2 positive individuals with non-COVID reasons for admission (i.e. patients admitted for injury, to give birth, and so on) and those under 18 years of age were excluded from this study.
Sample collection
Saliva self-collection was attempted every 3 days over the course of the hospital stay, using previously described methods (Ott et al.2020). Samples were transported to the Yale School of Public Health at room temperature within 5 hours of sample collection and tested for SARS-CoV-2 within 12 hours of sample collection. Urine collection was also attempted every 3 days from enrolled participants and stored at −80°C until further processing.
Detection of pneumococcal carriage using the molecular method
For all COVID-19 inpatients with at least one SARS-CoV-2 positive test, remaining saliva samples were tested for pneumococcal carriage. Thawed saliva samples were first culture-enriched with 100 µl plated on gentamicin (10%)-supplemented blood agar plates (Krone et al. 2015). Cultures were incubated overnight after which all growth was harvested and stored at −80°C (Trzciński et al. 2013). Pneumococcal detection was performed by DNA extraction of 200 µl of each sample using the MagMAX Ultra viral/pathogen nucleic acid isolation kit (ThermoFisher Scientific) on the KingFisher DNA extraction robot (ThermoFisher Scientific) following manufacturer’s protocol. Samples were classified as positive when both piaB (Wyllie et al. 2016b, Trzciński et al. 2013) and lytA (Carvalho et al. 2007) targets reported a cycle threshold (Ct) value < 40 Ct by RT-qPCR.
Detection of pneumococcal LRTI using urine antigen detection
Urine samples were thawed at room temperature and aliquoted into PIPES buffer. Aliquots were refrozen at −80°C in preparation for batch shipping on dry ice to the reference laboratory of Pfizer Vaccine Research (Pearl River, NY) for testing (Huijts et al. 2013). Upon receipt, samples were stored at −80°C until batched urine antigen testing according to the manufacturer’s protocol could be performed. All samples were tested using the serotype-specific UAD (SSUAD) test, which targets 24 of the 100 known pneumococcal serotypes and the BinaxNOW test, which targets a pan-pneumococcal antigen to determine the presence of any pneumococcus of serotype not covered by the SSUAD.
Statistical analysis
Differences in demographic data between pneumococcal status groups were tested using the Kruskal–Wallis rank sum test (continuous variable) or Fisher’s exact test (categorical variable). Estimates were considered statistically significant at P< .05. All statistical analyses were performed in RStudio v1.2.133535, using R v3.6.1.36.
Results
Study characteristics
Of the COVID-19 inpatients enrolled into the study, 156 saliva samples from 96 inpatients and 219 urine samples from 127 inpatients were available for testing (summarized in Table 1; detailed in Figure S1, Supporting Information); both saliva and urine samples were available from 75 inpatients. Overall, data from 148 inpatients were included in the analysis in this study. Patient characteristics are summarized in Table 2. The overall cohort of SARS-CoV-2 positive participants included in this study was 54.7% male, with average age 64.6 years; 69 (46.6%) individuals were categorized as having severe COVID-19 disease, based on supplemental oxygen level requirements and admission to ICU (Lucas et al. 2020). Antibiotic use at any point during hospitalization was high amongst all groups and was reported for 96/148 (64.9%; range: 64.1%–71.4%) total inpatients. A total of 10 patients had no bacterial growth following culture-enrichment of saliva; 8/10 (80.0%) of these patients had received antibiotics.
Table 1.
Distribution of patient samples tested either by urine antigen detection (UAD) and/or RT-qPCR of culture-enriched saliva.
| Urine | Saliva | |
|---|---|---|
| Total inpatients | 127 | 96 |
| Total samples | 219 | 156 |
| Positive inpatients | 14 (11%) | 3 (3.1%) |
A total of 75 inpatients had both their saliva and urine tested; none of these 75 individuals tested positive for pneumococcus on both sample types.
Table 2.
Characteristics of patients categorized by pneumococcal status.
| Characteristic | No pneumococcal detection (n = 131) | UAD positive (n = 14) | Culture positive (n = 3) | P † |
|---|---|---|---|---|
| Age (years), mean ± SD | 64.6 ± 15.6 | 64.9 ± 15.2 | 64.0 ± 21.5 | .896 |
| BMI, mean ± SD | 31.7 ± 7.4 | 28.5 ± 7.6 | 33.0 ± 19.8 | .256 |
| Family size, mean ± SD | 1.2 ± 1.6 | 0.6 ± 1.0 | 5.0 ± 4.6 | .287 |
| Days on Abx, mean ± SD | 5.1 ± 7.8 | 7.8 ± 12.5 | 3.0 ± 4.4 | .574 |
| Antibiotic use | ||||
| Yes | 84 (64.1) | 10 (71.4) | 2 (66.7) | .771 |
| No | 47 (35.9) | 4 (28.6) | 1 (33.3) | |
| Sex | ||||
| Female | 58 (44.3) | 8 (57.1) | 1 (33.3) | .407 |
| Male | 73 (55.7) | 6 (42.9) | 2 (66.7) | |
| Admitted to ICU, n (%) | .400 | |||
| No | 67 (51.1) | 5 (35.7) | 1 (33.3) | |
| Yes | 64 (48.9) | 9 (64.3) | 2 (66.7) | |
| Patient disposition, n (%) | .129 | |||
| Living | 109 (83.2) | 14 (100.0) | 3 (100.0) | |
| Deceased | 22 (16.8) | 0 (0.0) | 0 (0.0) | |
| Disease severity, n (%) | .260 | |||
| Moderate | 72 (55.0) | 5 (35.7) | 2 (66.7) | |
| Severe | 59 (45.0) | 9 (64.3) | 1 (33.3) | |
| Race/ethnicity, n (%) | .010 | |||
| Non-Hispanic White | 62 (50.0) | 3 (21.4) | 0 (0.0) | |
| Non-Hispanic Black | 42 (33.9) | 11 (78.6) | 2 (66.7) | |
| Hispanic | 20 (16.1) | 0 (0.0) | 1 (33.3) |
P-value for Kruskal–Wallis rank sum test (continuous variable) or Fisher’s exact test (categorical variable) between urine antigen detection (UAD) positive and no pneumococcal detection.
bNumbers may not sum to totals due to missing data, and column percentages may not sum to 100% due to rounding.
Detection of pneumococcal carriage or pneumococcal LRTI in participants hospitalized with COVID-19
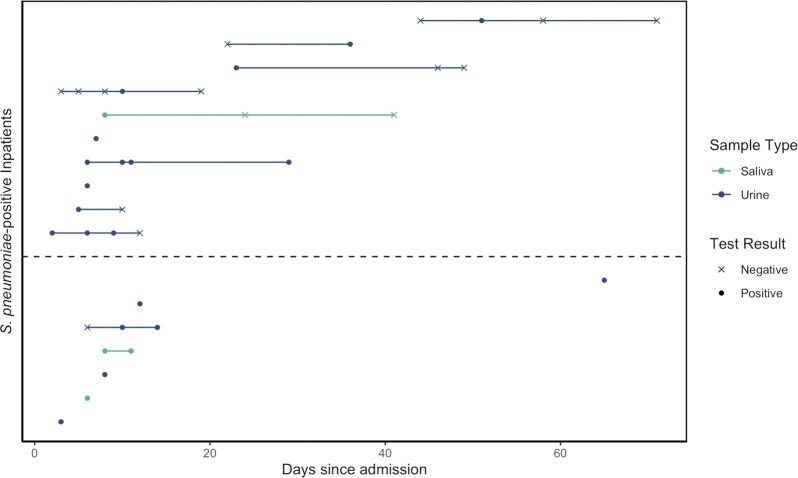
In this study population, 14/127 (11%; SSUAD, n = 8; BinaxNOW, n = 6) participants tested by urine antigen detection (UAD) had a positive result for at least one sample, indicating pneumococcal LRTI, and 3/96 (3.1%) had a positive saliva-RT-qPCR for at least one sample, indicating pneumococcal carriage (Table 1). Individuals testing positive at least once in either sample type are detailed in Fig. 1. None tested positive by both RT-qPCR and urine for pneumococcus. A total of three individuals returned indeterminate UAD results, which were treated as negative in the remaining analyses. For the eight urine samples which tested positive by SSUAD, serotypes detected included 4, 5, 17F, 22F, and 19A in one patient each and 33F in three separate patients. None of the individuals had received a pneumococcal vaccination in the 30 days prior to testing UAD-positive.
Figure 1.
Pneumococcal detection in COVID-19 inpatients. Of the COVID-19 inpatients who tested positive for pneumococcus, detection was exclusively via PCR testing of culture-enriched saliva (indicating carriage; green) or urine antigen detection (UAD, indicating pneumococcal etiology; blue). For some individuals, multiple samples were tested, as shown by points connected by the solid line. Note: a positive UAD result that follows a negative UAD does not necessarily indicate a new acquisition, but can occur due to either imperfect UAD test sensitivity or low level infection or prior carriage that was insufficient to trigger a positive UAD result. Individuals are sorted by time from admission to first positive test for pneumococcus. The dashed line indicates separation of individuals with COVID-19 cases classified as severe (top) and moderate (bottom). All COVID-19 inpatients who tested positive for pneumococcus survived.
Characteristics associated with detection of pneumococcus
With just 14 positive UAD samples, definitive associations cannot be inferred. However, there were some demographic differences between patients with and without detection of pneumococcus, including larger reported family sizes amongst those with pneumococcal carriage (Table 2). Detection of pneumococcus by both UAD and qPCR was more common in non-Hispanic Black patients.
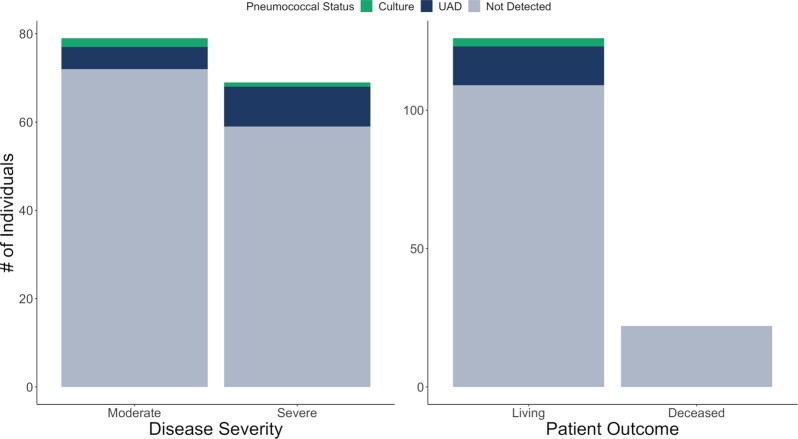
Pneumococcus was detected by UAD more commonly in severe than moderate cases of COVID-19 [Fig. 2; OR: 2.20; 95% CI: (0.72, 7.48)]. Severe disease was more frequent in non-Hispanic Black patients as compared to non-Hispanic White patients [OR: 1.92, 95% CI: (0.93, 4.02)].
Figure 2.
Outcome measures of COVID-19 infection categorized by pneumococcal status. UAD-positive individuals are disproportionately categorized as severe disease, particularly in comparison to the other pneumococcal statuses (positive by culture of saliva or not detected). Despite this, no culture—nor UAD-positive individuals died while hospitalized for COVID-19.
Death was less common in UAD-positive patients; if the case-fatality rate of 16.8% amongst those patients who did not test positive by UAD were applied to those who were UAD-positive, we would have expected 2–3 (2.7) deaths in this latter group of 14 patients. However, the numbers were small for both severe disease and death with a high degree of uncertainty.
Discussion
Based on previously identified interactions between pneumococcus and viral respiratory pathogens, there was concern that pneumococcus and SARS-CoV-2 could synergistically interact and contribute to the burden from the pandemic. With other viral–bacterial coinfections, the primary viral infection induces changes in the host (immune response, integrity of the airway epithelium), and carried bacteria—including pneumococci—which then increases the likelihood that bacteria can both proliferate and cause severe infection (Bosch et al. 2013). Moreover, increasing evidence exists suggesting that bacteria can influence host immune responses to viruses (Carniel et al. 2021, Mitsi et al. 2022). Therefore, we hypothesized that mixed infections between bacteria and viruses could influence the severity of the initial viral infection. To evaluate this hypothesis, we tested saliva and urine samples collected from COVID-19 inpatients in 2020 for markers indicative of the presence of pneumococcal carriage and infection. While detection of pneumococcus in saliva by culture was low relative to previously reported prevalence for nonelderly (Wyllie et al. 2016a) and elderly adults in the community (Krone et al. 2015), detection of pneumococcal antigens by UAD (11.0%), was similar to levels previously reported throughout the USA amongst older adults with community-acquired pneumonia (CAP; Sherwin et al. 2013). This, together with similar findings from Italy (13.0%; Desai et al. 2020) and Spain (9%; Anton-Vazquez and Clivillé 2021), suggests that secondary infections in COVID-19 patients may be on par with prepandemic rates of detection in CAP.
Despite decreased rates of invasive pneumococcal disease (IPD) reported from around the world during the study period, with respiratory viruses other than SARS-CoV-2 rarely reported both locally and internationally, LRTI with pneumococcus was still detected. While the small number of patients made it difficult to draw firm conclusions, pneumococcus was more commonly detected by UAD in individuals with severe COVID-19. Larger datasets that would allow for multivariate analysis would provide additional insights on this question. Further questions can be raised as to whether a UAD-positive CAP (vs. pneumococcal CAP with a false negative UAD) is predictive of disease severity or whether more severe cases of COVID-19 are more likely to develop a secondary pneumococcal pneumonia due to existing disease pathology.
The interaction of severe COVID-19 and pneumococcal detection may be confounded by the role of race-driven inequities, such as residential segregation and inadequate delivery of high-quality care, which can lead to increased exposure to pneumococcus and SARS-CoV-2 as well as increased severity of COVID-19 outcomes. More data are needed to adequately address the role of racial inequity, in understanding pneumococcal pneumonia in COVID-19 patients throughout the pandemic.
Amongst individuals testing positive for pneumococcus by either culture or UAD, none tested positive by both. PCR testing of culture-enriched saliva is intended for detection of carriage and requires the ability to isolate viable bacteria (Huijts et al. 2013). Culture-enrichment of saliva limits detection to living bacteria (Wyllie et al. 2014), excluding the detection of bacteria killed by antibiotic use. Prior studies suggest that antibiotic use in inpatients may be upwards of 70% (Langford et al. 2020). Our study population had a similar level of antibiotic use (64.9%) at any point during hospitalization for COVID-19. With such a high level of antibiotic usage, our estimates of the proportion of pneumonia due to pneumococcus, as well as pneumococcal carriage detected, may be underestimated and may have led to misclassification of pneumococcal status.
A total of three of eight serotypes detected by SSUAD were serotypes covered by PCV13, with seven of eight contained in PCV15, PCV20, and 23-valent pneumococcal polysaccharide vaccine (PPV23). While PPV23 is not considered to provide substantial protection against nonbacteremic pneumonia by the US CDC (Smith et al. 2013), conjugate vaccines do. If serotype distribution is confirmed by larger studies, new higher-valency PCV15 and PCV20 could provide substantial public health benefit to the older adult population.
One of the major limitations of this study is small sample size. Future studies could be expanded to include more individuals of varying disease severities and demographic and socio-ecologic backgrounds with more complete sample and data collection. This could be done both retrospectively through chart reviews and analysis of previously stored samples or prospectively with new cases and data collection. Increasing longitudinal saliva and urine collection during both inpatient and outpatient stay to monitor both existing and newly acquired carriage as well as secondary infection throughout the duration of disease could help inform important questions on the roles of these different types of pneumococcal states. Furthermore, longitudinal surveillance on both the general community as well as inpatients would best inform viral–bacterial interactions in disease pathogenesis, including the interaction between SARS-CoV-2 and pneumococcus.
In conclusion, these data confirm that patients hospitalized with COVID-19 continued to experience LRTI with S. pneumoniae despite dramatic declines in IPD and are suggestive of an association between detection of pneumococcus and severity of COVID-19. Future studies should aim to more deeply and longitudinally explore these dynamics to better assess how pneumococcal carriage may interact with SARS-CoV-2 to influence COVID-19 severity, as well as the degree to which the combination of COVID-19 and pneumococcal infection influence disease outcomes.
Role of the funder
This study was performed as a collaborative research project between researchers (D.M.W. and A.L.W.) at the Yale School of Public Health and Pfizer. The study protocol was designed by the Yale researchers in consultation with Pfizer. The decision to publish was made by the Yale researchers in consultation with Pfizer; all authors agree with the decision to publish and with the results of the study.
Supplementary Material
Acknowledgments
We thank the study participants for their contribution to our study and the Yale IMPACT biorepository team for their assistance with sample collection.
Notes
Co-senior authors.
Contributor Information
Anne Stahlfeld, Department of Epidemiology of Microbial Diseases, Yale School of Public Health, LEPH823, 60 College St, New Haven, CT 06510, United States.
Laura R Glick, Yale School of Medicine, 333 Cedar St, New Haven, CT 06511, United States.
Isabel M Ott, Department of Epidemiology of Microbial Diseases, Yale School of Public Health, LEPH823, 60 College St, New Haven, CT 06510, United States.
Samuel B Craft, Yale School of Medicine, 333 Cedar St, New Haven, CT 06511, United States.
Devyn Yolda-Carr, Department of Epidemiology of Microbial Diseases, Yale School of Public Health, LEPH823, 60 College St, New Haven, CT 06510, United States.
Christina A Harden, Department of Epidemiology of Microbial Diseases, Yale School of Public Health, LEPH823, 60 College St, New Haven, CT 06510, United States.
Maura Nakahata, Department of Epidemiology of Microbial Diseases, Yale School of Public Health, LEPH823, 60 College St, New Haven, CT 06510, United States.
Shelli F Farhadian, Yale School of Medicine, 333 Cedar St, New Haven, CT 06511, United States.
Lindsay R Grant, Medical and Scientific Affairs, Pfizer Inc, 500 Arcola Rd, Collegeville, PA 19426, United States.
Ronika Alexander-Parrish, Medical and Scientific Affairs, Pfizer Inc, 500 Arcola Rd, Collegeville, PA 19426, United States.
Adriano Arguedas, Medical and Scientific Affairs, Pfizer Inc, 500 Arcola Rd, Collegeville, PA 19426, United States.
Bradford D Gessner, Medical and Scientific Affairs, Pfizer Inc, 500 Arcola Rd, Collegeville, PA 19426, United States.
Daniel M Weinberger, Department of Epidemiology of Microbial Diseases, Yale School of Public Health, LEPH823, 60 College St, New Haven, CT 06510, United States.
Anne L Wyllie, Department of Epidemiology of Microbial Diseases, Yale School of Public Health, LEPH823, 60 College St, New Haven, CT 06510, United States.
Authors’ contributions
A.L.W. conceived the study. L.R.G., R.A.-P., A.A., B.D.G., D.M.W., and A.L.W. designed the study protocol. A.E.W., S.F.F., R.A.-P., and A.L.W. managed the study. A.E.W., I.M.O., D.Y.-C., C.A.H., and A.L.W. were responsible for sample receipt, processing, and testing. A.E.W., L.R.G., I.M.O., S.B.C., D.Y.-C., C.A.H., M.N., S.F.F., and A.L.W. collected the data. A.E.W., D.M.W., and A.L.W. performed the analyses and interpreted the data. A.E.W., D.M.W., and A.L.W. drafted the manuscript. All authors amended and commented on the final manuscript.
References
- Anton-Vazquez V, Clivillé R. Streptococcus pneumoniae coinfection in hospitalised patients with COVID-19. Eur J Clin Microbiol Infect Dis. 2021;40:1353–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auranen K, Mehtälä J, Tanskanen Aet al. Between-strain competition in acquisition and clearance of pneumococcal carriage–epidemiologic evidence from a longitudinal study of day-care children. Am J Epidemiol. 2010;171:169–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch AATM, Biesbroek G, Trzcinski Ket al. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog. 2013;9:e1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carniel BF, Marcon F, Rylance Jet al. Pneumococcal colonization impairs mucosal immune responses to live attenuated influenza vaccine. JCI Insight. 2021;6:e141088. DOI: 10.1172/jci.insight.141088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho M da GS, Tondella ML, McCaustland Ket al. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol. 2007;45:2460–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Santonocito OG, Caltagirone Get al. Effectiveness of Streptococcus pneumoniae urinary antigen testing in decreasing mortality of COVID-19 co-infected patients: a clinical investigation. Medicina. 2020;56:572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirkx KKT, Mulder B, Post ASet al. The drop in reported invasive pneumococcal disease among adults during the first COVID-19 wave in the Netherlands explained. Int J Infect Dis. 2021;111:196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Hayek SS, Wang Wet al. Factors associated with death in critically ill patients with coronavirus disease 2019 in the US. JAMA Intern Med. 2020;180:1436–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijts SM, Pride MW, Vos JMIet al. Diagnostic accuracy of a serotype-specific antigen test in community-acquired pneumonia. Eur Respir J. 2013;42:1283–90. [DOI] [PubMed] [Google Scholar]
- Klein EY, Monteforte B, Gupta Aet al. The frequency of influenza and bacterial coinfection: a systematic review and meta-analysis. Influenza Other Respir Virus. 2016;10:394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krone CL, Wyllie AL, van Beek Jet al. Carriage of Streptococcus pneumoniae in aged adults with influenza-like-illness. PLoS ONE. 2015;10:e0119875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford BJ, So M, Raybardhan Set al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020;26:1622–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann CJ, Pho MT, Pitrak Det al. Community-acquired coinfection in coronavirus disease 2019: a retrospective observational experience. Clin Infect Dis. 2021;72:1450–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewnard JA, Bruxvoort KJ, Hong VXet al. Effectiveness of pneumococcal conjugate vaccination against virus-associated lower respiratory tract infection among adults: a case-control study. J Infect Dis. 2022:jiac098. https://academic.oup.com/jid/advance-article-abstract/doi/10.1093/infdis/jiac098/6552253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C, Wong P, Klein Jet al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi SA, Klugman KP, Vaccine Trialist Group . A role for Streptococcus pneumoniae in virus-associated pneumonia. Nat Med. 2004;10:811–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol. 2014;12:252–62. [DOI] [PubMed] [Google Scholar]
- Mitsi E, Reiné J, Urban BCet al. Streptococcus pneumoniae colonization associates with impaired adaptive immune responses against SARS-CoV-2. J Clin Invest. 2022;132:e15712. DOI: 10.1172/JCI157124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198:962–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noale M, Trevisan C, Maggi Set al. The association between influenza and pneumococcal vaccinations and SARS-Cov-2 infection: data from the EPICOVID19 web-based survey. Vaccines. 2020;8:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes MC, Cutland CL, Klugman KPet al. Pneumococcal conjugate vaccine protection against coronavirus-associated pneumonia hospitalization in children living with and without HIV. MBio. 2021;12:1710–20. DOI: 10.1128/mBio.02347-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott I, Vogels C, Grubaugh Net al. Saliva collection and RNA extraction for SARS-CoV-2 detection. Berkeley: protocols.io, 2020. [Google Scholar]
- Sherwin RL, Gray S, Alexander Ret al. Distribution of 13-valent pneumococcal conjugate vaccine Streptococcus pneumoniae serotypes in US adults aged ≥50 years with community-acquired pneumonia. J Infect Dis. 2013;208:1813–20. [DOI] [PubMed] [Google Scholar]
- Smith KJ, Wateska AR, Nowalk MPet al. Modeling of cost effectiveness of pneumococcal conjugate vaccination strategies in U.S. older adults. Am J Prev Med. 2013;44:373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzciński K, Bogaert D, Wyllie Aet al. Superiority of trans-oral over trans-nasal sampling in detecting Streptococcus pneumoniae colonization in adults. PLoS ONE. 2013;8:e60520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie AL, Chu MLJN, Schellens MHBet al. Streptococcus pneumoniae in saliva of Dutch primary school children. PLoS ONE. 2014;9:e102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie AL, Fournier J, Casanovas-Massana Aet al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383:1283–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie AL, Rümke LW, Arp Ket al. Molecular surveillance on Streptococcus pneumoniae carriage in non-elderly adults; little evidence for pneumococcal circulation independent from the reservoir in children. Sci Rep. 2016a;6:34888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie AL, Wijmenga-Monsuur AJ, van Houten MAet al. Molecular surveillance of nasopharyngeal carriage of Streptococcus pneumoniae in children vaccinated with conjugated polysaccharide pneumococcal vaccines. Sci Rep. 2016b;6:23809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Ge Y, Wu Tet al. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285:198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.