Abstract
Neisseria gonorrhoeae causes the sexually transmitted disease gonorrhoea. The treatment of gonorrhoea is becoming increasingly challenging, as N. gonorrhoeae has developed resistance to antimicrobial agents routinely used in the clinic. Resistance to penicillin is wide-spread partly due to the acquisition of β-lactamase genes. How N. gonorrhoeae survives an initial exposure to β-lactams before acquiring resistance genes remains to be understood. Here, using a panel of clinical isolates of N. gonorrhoeae we show that the β-lactamase enzyme is packaged into outer membrane vesicles (OMVs) by strains expressing blaTEM-1B or blaTEM-106, which protects otherwise susceptible clinical isolates from the β-lactam drug amoxycillin. We characterized the phenotypes of these clinical isolates of N. gonorrhoeae and the time courses over which the cross-protection of the strains is effective. Imaging and biochemical assays suggest that OMVs promote the transfer of proteins and lipids between bacteria. Thus, N. gonorrhoeae strains secret antibiotic degrading enzymes via OMVs enabling survival of otherwise susceptible bacteria.
Keywords: OMV, lactamase, gonorrhoea, antimicrobial resistance, TEM-1, secretion system
The secretion of β-lactamase protects otherwise susceptible Neisseria gonorrhoeae pathogens from antibiotics.
Introduction
Neisseria gonorrhoeae is classified as a priority antimicrobial-resistant (AMR) pathogen by the World Health Organization (WHO) and the Centre for Disease Control and Prevention (CDC). This is in response to the increasing infection rates around the world, including Australia, and the rise of drug resistance to common antimicrobial agents such as penicillin, but also last-line antibiotics including third-generation cephalosporins (Lahra et al. 2021). Treating N. gonorrhoeae, and thus the second most common genital infection has become challenging (Unemo and Jensen 2017). Extremely drug-resistant N. gonorrhoeae as defined by resistance to the standard azithromycin and ceftriaxone dual therapy are rapidly emerging around the globe (Unemo and Shafer 2014). AMR in N. gonorrhoeae involves well-established mechanisms such as mutations and acquisition of drug resistance genes (Blair et al. 2015). For instance, resistance to β-lactam antibiotics such as penicillin and amoxycillin (AMX) can occur through chromosomal mutations in ponA and penA, which encode of penicillin binding proteins (Rice et al. 2017). Alternatively, the acquisition of plasmids encoding β-lactamases results in penicillinase producing N. gonorrhoeae (PPNG), which are thereby capable of inactivating the antibiotic by enzymatic hydrolysis. PPNG strains were first identified in 1976, harbouring the TEM-1 β-lactamase allele (blaTEM-1; Ashford et al. 1976, Phillips 1976). The archetypal TEM-1 β-lactamase is an ∼ 30 kDa protein synthesized with an N-terminal signal sequence that engages with the Sec translocon to deliver TEM-1 into the periplasm, allowing it to fold into its enzymatically active form (Pradel et al. 2009). Point mutations in blaTEM-1 result in alleles that have broader spectrum of activity against β-lactam antibiotics, and it is thought that the exposure to β-lactam antibiotics provides selection pressure to drive resistance mechanisms that include the emergence of β-lactamase alleles such as blaTEM-135 that can hydrolyse ceftriaxone (Datta and Kontomichalou 1965, Cole et al. 2015, Micaëlo et al. 2017). While the prevalence of PPNG strains can be as high as 100% in some regions, in other parts of the world where there is relatively low presence of β-lactamase containing strains additional mechanisms might contribute to the survival of N. gonorrhoea to β-lactam antibiotics (Bala et al. 2013, Cehovin et al. 2020).
Antibiotic treatment of susceptible bacteria triggers stress responses, which includes the release of outer membrane vesicles (OMVs; Orench-Rivera and Kuehn 2016). This is a fundamental process, with all Gram-negative bacteria studied so far found capable of releasing OMVs. OMVs are spherical structures ranging from 20 to 200 nm, bound by an asymmetric bilayer of with an outer leaflet of lipopolysaccharide (LPS) or lipooligosaccharide (LOS) and an inner leaflet of phospholipid. OMVs can carry proteins, peptidoglycan, and genetic material (DNA/RNA), which are packaged in the OMV lumen or embedded in the membrane (Schwechheimer and Kuehn 2015). It is thought that OMVs neutralize the effects of antibiotics and other antimicrobial compounds via adsorption, thus acting as decoys to prevent bacterial killing (Manning and Kuehn 2011, Kulkarni et al. 2014). This is particularly relevant for antibiotics that act on the lipid components of the outer membrane, such as daptomycin and polymyxin/colistin (Sabnis et al. 2018). In addition, OMVs can contain antibiotic resistant genes and plasmids including blaTEMas observed in N. gonorrhoeae, suggesting the possibility to transfer resistance to susceptible bacteria as observed in other pathogens (Dorward et al.1989, Chatterjee et al. 2017). The mechanism by which nucleic acids are packaged into OMVs remains controversial, as the majority of OMV-associated DNA remains susceptible to nuclease treatments, suggesting external association (Bitto et al. 2017). OMVs secreted by resistant bacteria may also contain antibiotic hydrolyzing enzymes like β-lactamase, which potentially degrade antibiotics before they reach parent and unrelated susceptible bacteria (Schaar et al. 2011, Kim et al. 2018). β-Lactamase containing OMVs protect not only the parent bacteria but unrelated species during antibiotic exposure, including commensal and pathogenic microbes (Schaar et al. 2011, Stentz et al. 2015, Bielaszewska et al. 2021). Effective sorting of β-lactamases into OMVs depends on membrane anchoring, although additional mechanisms are at play as not all membrane-bound β-lactamases are associated with OMVs (Gonzalez et al. 2016). Whether clinical isolates of N. gonorrhoeae can transmit β-lactamases via OMVs and whether this contributes to AMR is not known, and these questions are the focus of this study.
Here, we investigated a panel of recent clinical isolates of N. gonorrhoeae to determine antimicrobial resistance. Our data suggest that N. gonorrhoeae package β-lactamases inside OMVs, thereby secreting enzymatically active cargo into the surrounding milieu. The β-lactamase cargo in the OMVs were shown to confer protection against AMX treatment on what were otherwise susceptible N. gonorrhoeae isolates. Likewise, the OMVs provided cross-species protection against AMX treatment on susceptible Escherichiacoli isolates. We propose that OMVs enable the transfer of proteins, which is associated with increased survival of otherwise susceptible bacteria after antibiotic treatment.
Materials and methods
Bacterial strains and growth conditions
Clinical isolates of N. gonorrhoeae were collected from different body sites with different antimicrobial susceptibility profile as shown in Table 1. Neisseriagonorrhoeae was grown routinely in Gonococcal (GC) agar supplemented with 1% (v/v) Deakin Modified Isovitalex (DMIV) at 37°C in 5% CO2 enriched atmosphere or GC broth supplemented with 1% DMIV and 0.01 M sodium bicarbonate (NaHCO3). Escherichia coli BL21 was grown in Luria–Bertani (LB) agar or LB broth at 37°C shaking at 200 rpm.
Table 1.
Antimicrobial susceptibility profile.
| Clinical isolate | Antibiotic susceptibility | MIC (μg/ml) | |||||
|---|---|---|---|---|---|---|---|
| AMX | CIP | AZT | PEN* | AMX | CTX | CRO | |
| R72.04/N. gon-1 | R | R | ND | > 35 | 512 | 0.06 | 0.016 |
| R72.14/N. gon-2 | R | R | R | > 35 | 512 | 0.06 | 0.016 |
| R72.17/N. gon-3 | S | R | R | 0.5 | 0.25 | < 0.0325 | < 0.008 |
| R72.20/N. gon-4 | R | S | R | > 35 | 32 | < 0.0325 | < 0.008 |
*MIC was determined using E-test strip.
The MIC breakpoints of > 1 μg/ml were considered resistance against penicillin, AMX, and azithromycin or < 0.06 μg/ml susceptible against ciprofloxacin and < 0.125 μg/ml for ceftriaxone and cefotaxime as recommended by EUCAST. R—Resistant; S—Susceptible; and ND—Not defined. AMX—Amoxycillin; AZT—Azithromycin; CIP—Ciprofloxacin; CRO—Ceftriaxone; CTX—Cefotaxime, and PEN—Penicillin. Red and green colours indicate drug resistance and susceptibility, respectively.
Purification of OMVs
Neisseria gonorrhoeae or E. coli were used to inoculate 500 ml broth and cultured until an optical density at 600 nm (OD600) of ∼ 0.8 at 37°C. Bacteria were then pelleted via centrifugation at 12 000 × g for 12 min at 4°C. The supernatant was filtered through a 0.45 µm membrane (Millipore) to remove any bacterial cells and debris. The filtered supernatant was then centrifuged for 3 h at 186 000 × g at 4°C using a 45 Ti rotor. The OMV pellet was washed with phosphate-buffered saline (PBS), suspended in 900 μl PBS, filtered with a 0.2 µm sterile low protein binding membrane and stored at −80°C until further analysis.
Minimum inhibitory concentration
The minimum inhibitory concentration (MIC) of antibiotics was determined using an agar dilution assay as described previously (Wiegand et al. 2008). Briefly, serial dilutions of antibiotics were added to 20 ml autoclaved GC agar media cooled to 50°C. Bacteria were suspended in sterile PBS at an OD600 = 0.013 and 1 μl added to the antibiotic containing GC agar. The agar plates were incubated at 37°C with 5% CO2 for 18 h. Minimal inhibitory concentration (MIC) was determined as the lowest concentration of antibiotics that prevented any bacterial growth. To measure the MIC of OMV-treated bacteria, the clinical isolate N. gon-3 was grown in 5 ml GC broth in two different glass tubes to an OD600 of 0.3. Then, OMVs isolated from N. gon-2 and N. gon-3 were added at a final concentration of 50 μg/ml, and bacteria were grown at 37°C, rotating at 200 rpm for up to 4 h. Bacteria were harvested every hour, diluted to an OD600 of 0.13 to determine MIC values as described above.
Subcellular fractionation
To extract the periplasmic fraction, bacteria were harvested from GC agar plates, suspended in 1.5 ml of ice-cold PBS at an OD600 ∼ 0.6 and washed three times with PBS. The PBS was gently removed from the final bacterial pellet before 20 µl chloroform was added, vortexed and incubated for 15 min at 22°C. Next, 100 µl of ice cold 0.01M Tris HCl, pH 8.0, was added, vortexed and bacteria were pelleted by centrifugation at 12 000 × g for 5 min. The Tris HCl, containing released periplasmic material was carefully aspirated and placed in a second microfuge tube and stored at −20°C for further use.
To prepare a total membrane fraction, bacteria were suspended in spheroplast buffer (10mM Tris 0.75M sucrose, pH 7.5) and incubated with lysozyme (100 µg/ml) on ice for 2 min. Then, one tablet protease inhibitor cocktail (Sigma), two volume of lysis buffer (1.5mM EDTA, pH 7.5), and DNase (25 µg/ml) were added. The bacteria were eventually lysed with the use of Avestin Emulsiflex. The lysate was cleared by centrifugation at 10 000 × g for 10 min at 4°C. The supernatant was then subjected to ultracentrifugation in a Ti45 rotor at 186 000 × g for 90 min to pellet the membrane fraction. The membrane pellet was resuspended in PBS and stored at −20°C for further use.
Immunoblot analysis
Bacteria were grown in a 5 ml GC broth in glass tubes to an OD600 of 0.8. Then, 1 ml of the bacterial culture was pelleted at 4000 × g for 4 min, washed with PBS, and suspended in 200 µl of 1x reducing SDS-PAGE loading buffer. In addition, bacteria were washed with PBS, resuspended in 200 µl HBSS buffer containing calcium and magnesium, and treated with OMVs isolated from N. gon-2 (80 µg/ml). OMV protein concentrations were determined using the BCA assay, according to the manufacturer's instructions (Thermo scientific). After 1 h at 37°C rotating at 200 rpm, bacteria were pelleted at 4000 × g for 3 min at 4°C and washed three times with PBS, discarding the supernatants. The bacterial pellet was suspended in 1x reducing SDS-PAGE loading buffer. Analysis of purified OMVs was performed based on 150 µg of protein/ml. OMV samples were treated with 6x reducing SDS-PAGE loading buffer. Equal volumes of prepared OMVs and bacterial lysates were run at 125 V for 2 h using 15% SDS-PAGE. The separated proteins were then transferred onto nitrocellulose filter (BioRad) using 0.5x transfer buffer (380 mM glycine, 202 mM Tris, 0.02% (w/v) SDS, and 20% (v/v) methanol) at 100 V for 60 min. The filter was then blocked with 5% skimmed milk in TBST (0.2% Tween-20, 137 mM NaCl, 2.7 mM KCl, and 25 mM Tris with final pH 7.4) for 1 h at room temperature (RT). The filter was probed with 10 µg/ml of anti-β-lactamase antibody (Abcam), anti-LOS (diluted 1:5000; LOS 4C4; registry ID: AB 2617192; Developmental Studies Hybridoma Bank, University of Iowa), and anti-PorB antibody (diluted 1:5000; Deo et al. 2018) prepared in 5% skimmed milk in TBST overnight at 4°C. The filter was then washed three times with TBST and probed with secondary antibody (goat antimouse, Life Technologies) conjugated to HRP for 1 h at RT. Nitrocellulose filters were then developed with luminol-based enhanced chemiluminescence (Bio-Rad) ECL using manufacturers protocol and exposed to film (KODAK).
Measurement of β-lactamase activity
The activity of β-lactamase present in OMVs was determined based on hydrolysis of chromogenic substrate nitrocefin, a colorimetric assay (Abcam 197008). Briefly, a standard was prepared by adding the nitrocefin, hydrolysis buffer and DMSO in the ratio of 1:2:7 in an Eppendorf tube and then incubating it at 60°C for 10 min. Dilutions of hydrolyzed nitrocefin (0, 4, 8, 12, 16, and 20 nmol) were used to generate a standard curve. A volume of 100 µl of standards, 5 µl positive control (provided by manufacturer), and 10 µl OMVs, normalized to 150 μg/ml protein concentration, were distributed in a 96-well plate in duplicate. Then, 50 µl nitrocefin prepared in assay buffer (2 µl nitrocefin plus 48 µl assay buffer) was added to the samples as well as the control. The absorbance was measured immediately in Tecan microplate reader at 490 nm for 36 min at intervals of 3 min. The standard curve was generated using Microsoft Excel and activity of β-lactamase was calculated for the OMVs.
Screening for extended spectrum β-lactamase
The presence or absence of extended spectrum β-lactamase (ESBL), which could hydrolyze third-generation cephalosporins (cefotaxime and ceftriaxone) in the isolates was tested using double disk synergy test as recommended by CLSI (Clinical and Laboratory Standards Institute).
Protection assay for β-lactamase in OMVs
OMVs normalized to protein concentrations (500 μg/ml) were treated with 0.5 μg/ml and 1.0 μg/ml of proteinase K, which was reconstituted in 50 mM Tris-HCl (pH8.0), at 37°C for 1 h. In addition, some OMV samples were treated with 0.1% Triton X-100 (TX-100) in PBS at 37°C for 30 min. After TX-100 treatment, OMV samples were treated with proteinase K (1 μg/ml) or control (PBS). The equal volume was loaded on 15% SDS-PAGE and run at 125 V for 2 h. The immunoblot analysis of β-lactamase and PorB was performed as described above.
Growth of N. gonorrhoeae in presence of antibiotics and OMVs
The antibiotic susceptible clinical isolate N. gon-3 was grown overnight in 5 ml GC broth as described above. The overnight culture was then diluted into a new 5 ml preculture in GC broth until OD600 of ∼ 0.5. The culture was again diluted to OD600 = 0.005–0.01 and 200 μl of bacterial suspension was transferred into a 96-well plate in triplicate. Simultaneously, AMX (0.5 or 1.0 μg/ml) or cefotaxime (0.06 μg/ml) as well as OMVs isolated from resistant strains (N. gon-1, -2, and -4) were added to the wells at a concentration of 10, 25, or 50 μg/ml based on protein. Bacterial growth was measured by determining OD600 using Tecan microplate reader for 18 h at intervals of 60 min.
Enumeration of colony forming units
The MS11A and N. gon-3 were grown in 5 ml GC broth as described above to OD600 nm = 0.5. Bacterial cells were then challenged with different AMX concentrations (0.05, 0.1, 0.2, 0.5, 1, and 2 µg/ml). After 1 h of AMX challenge, bacteria were washed with PBS and resuspended in GC liquid media supplemented with NaHCO3 and DMIV and plated on GC plates to determine the MIC. Cultures were supplemented with either 0.5 μg/ml AMX and/or 50 μg/ml protein of OMVs isolated from N. gon-2 (NOMV-2). Colony forming units (CFUs)/ml were determined every hour for 5 h. For this, five 20-fold serial dilutions of the bacteria were prepared in PBS. A 20 μl of each dilution were plated on GC agar, tilted to generate smear, and incubated in 5% CO2 at 37°C. Then, number of colonies were counted manually and multiplied by the dilution factor to determine total numbers of bacteria. Alternatively, bacteria were incubated with NOMV2 or vehicle control for an hour and then washed three times with PBS to remove OMVs. Bacteria were subsequently challenged with 0.1 µg/ml of AMX for 1 h and surviving cells identified on antibiotic-free GC agar plates by CFUs.
Labelling bacteria utilizing fluorescent dye labelled OMVs
The protein concentration of OMVs isolated from N. gon-2 was normalized to 200 μg/ml and 500 μl of OMVs was aliquoted in an Eppendorf vial and labelled with the fluorescent dye FM 4–64 (N-(3 triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide (Thermo Fisher) at a final concentration of 5 μg/ml of the dye. After 1 h at RT, OMVs were washed three times with PBS and pelleted by ultracentrifugation at 186 000 × g for 15 min. The labelled OMVs were suspended in HBSS buffer containing calcium and magnesium and incubated with unlabelled bacteria grown to OD600nm = 0.3 as described above. Afterward, the bacteria were pelleted at 4000 × g for 3 min at 4 °C, washed three times with PBS and suspended in PBS for flow cytometry analysis. Likewise, bacteria were labelled directly with FM 4–64 at a final concentration of 5 μg/ml of the dye, washed two times, and suspended in PBS. The labelled bacteria and OMVs along with unlabelled OMVs were analyzed on a BD biosciences LSRFortessa X20 cell analyzer.
OMV fusion assay
OMVs were labelled with Octadecyl-rhodamine B chloride-R18 (R18, Thermo scientific # O246) at a concentration of 1 mg/ml for 1 h at 25°C, followed by ultracentrifugation at 128 000 × g for 30 min at 4°C, as described above and previously (Bomberger et al. 2009). R18 fluorescence is quenched at high concentrations within membranes, but not after dilution due to membrane fusion or Triton X-100 treatment. Labelled OMVs were incubated in 50 mM Na2CO3, 100 mM NaCl, pH 9 with or without N. gonorrhoeae bacteria and fluorescence intensity was determined using a Tecan plate reader for 30 min at every 2 min, at 37°C and 5% CO2 supplied.
Polymerase chain reaction assay
The PorB and β-lactamase genes were amplified with polymerase chain reaction (PCR) using primers binding to the GC PorB PQE6-PorB5’- (5’-GGA CCA CCA TGG ATG AAA AAA TCC CTG ATT GCC C-3’) and PQE6-PorB3’- (5’-CGG ACC GGA TCC GAA TTT GTT TTC GCC TTT GCC-3’), or beta-lactamase blaTEM-F (5'-TTA CGG ATG GCA TGA CAG TAA G-3') and blaTEM-R (5'-GAC TCC CCG TCG TGT AGA TAA C-3'), respectively. Neisseriagonorrhoeae bacteria, purified OMVs and culture supernatants were used in PCR assays.
Immunofluorescence analysis
Neisseria gonorrhoeae strains were assessed for β-lactamase expression using indirect immunofluorescent analysis in a similar manner as described previously (Gunasinghe et al. 2018). Briefly, bacteria were grown to mid log phase in a GC broth. A volume of 1 ml of bacterial culture was pelleted at 4000 × g at 4°C and washed with PBS twice. The bacteria were then fixed using 2% paraformaldehyde (PFA) for 10 min at RT and then washed with and suspended in 500 μl PBS. A volume of 5 μl of the bacteria containing solution were added on poly-L-lysine coated coverslips and allowed to dry. Bacteria were permeabilized with 0.1% Triton-X 100 in PBS at RT for 10 min and blocked with 5% BSA. Then, bacteria were incubated with anti-β-lactamase (Biorbyt 500404), diluted 1:100 in 5% BSA for 1 h or with anti-PorB as described previously (Deo et al. 2018). Cells were washed twice with PBS followed by addition of antirabbit or mouse immunoglobulin G (IgG)-Alexa Flour 488 (Thermo Fisher Scientific)-conjugated antibodies diluted to 1:300 in 5% BSA in PBS for 1 h at RT. Bacteria were washed twice with PBS and coverslips were mounted on glass slides of dimension 76 × 26 mm (Thermo Scientific) using fluorescence mounting medium (mowiol) that contained 4′,6-diamidino-2-phenylindole (DAPI). Cells were imaged using a Nikon Upright Confocal laser scanning microscope with 100x objective. Alternatively, E. coli were treated with NOMV-2 in LB media for 1 h. Bacteria were then washed three times with PBS and added to an 8-well chambered glass slide (Sarstedt) coated with 0.01% Poly-L-Lysine. After centrifugation, bacteria were fixed with 4% EM grade PFA. After fixation, cells were washed three times with ice-cold PBS and permeabilized with 0.2% Triton X-100 and finally blocked with 3% BSA. Antibodies recognizing LPS (Biorbyt 500404) and PorB (in house) were conjugated with the Alexa Fluor Antibody labelling Kit (Thermo Fisher) before applying to bacteria for 1 h. The images were collected using EVOS microscope using 100x oil immersion objective lens and resultant images were analyzed using Fiji.
Results
AMR phenotypes in clinical isolates of N. gonorrhoeae
We focused on four recent clinical isolates to determine the susceptibility and MICs to commonly used antibiotics including benzyl penicillin, AMX, cefotaxime, ceftriaxone, ciprofloxacin, and azithromycin. A total of three strains, containing the reference number R72.04, R72.14, and R72.20 or referred to as N. gon-1 N. gon-2, and N. gon-4, respectively, showed resistance to AMX (Table 1). In particular, the growth of N. gon-1 and N. gon-2 was only inhibited at relatively high AMX concentrations (512 µg/ml), whereas 32 µg/ml of AMX was sufficient to prevent N. gon-4 growth (Table 1). N. gon-1, N. gon-2, and N. gon-4 were also resistant to penicillin (Table 1). Only N.gon-4 remained susceptible to ciprofloxacin, whereas all four isolates were susceptible to third-generation cephalosporins (cefotaxime and ceftriaxone; Table 1). To determine the mechanism for AMX resistance, we performed whole genome sequencing, which identified two alleles of the TEM-1 encoding gene, namely blaTEM-106 in N. gon-1 and N. gon-2 and blaTEM-1Bin N. gon-4 (Williamson et al. 2019). Sequence analysis showed that these alleles encode proteins that differ by five residues in the 286 residues of the β-lactamase protein, and that both proteins have a 23 residue N-terminal signal sequence that would engage the SEC translocon for transport of the protein into the periplasm (Payne et al. 2012). Phenotypic assessment of the four strains to determine the MIC against benzyl penicillin, AMX, ceftriaxone, and cefotaxime, revealed a sensitivity in N. gon-3 suggesting that the β-lactamase expressed in N. gon-1, N. gon-2, and N. gon-4 hydrolyses penicillins, such as benzyl penicillin or AMX, but not the third-generation cephalosporins, including ceftriaxone and cefotaxime (Table 1).
Neisseria gonorrhoeae isolates secrete β-lactamases
Having identified that some clinical isolates carry alleles of blaTEM-1, we sought to investigate their expression. Immunofluorescence assays were consistent with β-lactamase production in in N.gon-2 (Fig. 1A), but not in N.gon-3 (Fig. 1C). The presence of the outer membrane protein PorB served as a control for staining in both isolates (Fig. 1B and D). While the resolution of this mode of microscopy was insufficient to determine the subcellular location of the proteins, the staining for β-lactamase was clearly confined to the profile of each bacterial cell (Fig. 1A, inset). As an independent confirmation of expression, immunoblot analysis of whole bacterial lysates indicated that the β-lactamase was more highly expressed in N. gon-1 and N.gon-2 (TEM-106), than in N.gon-4 (TEM-1B) and undetectable in N. gon-3 (Fig. 1E). The steady-state levels of these protein isoforms reflected the resistance profiles of the N. gonorrhoeae isolates as determined by MIC assessment (Table 1).
Figure 1.
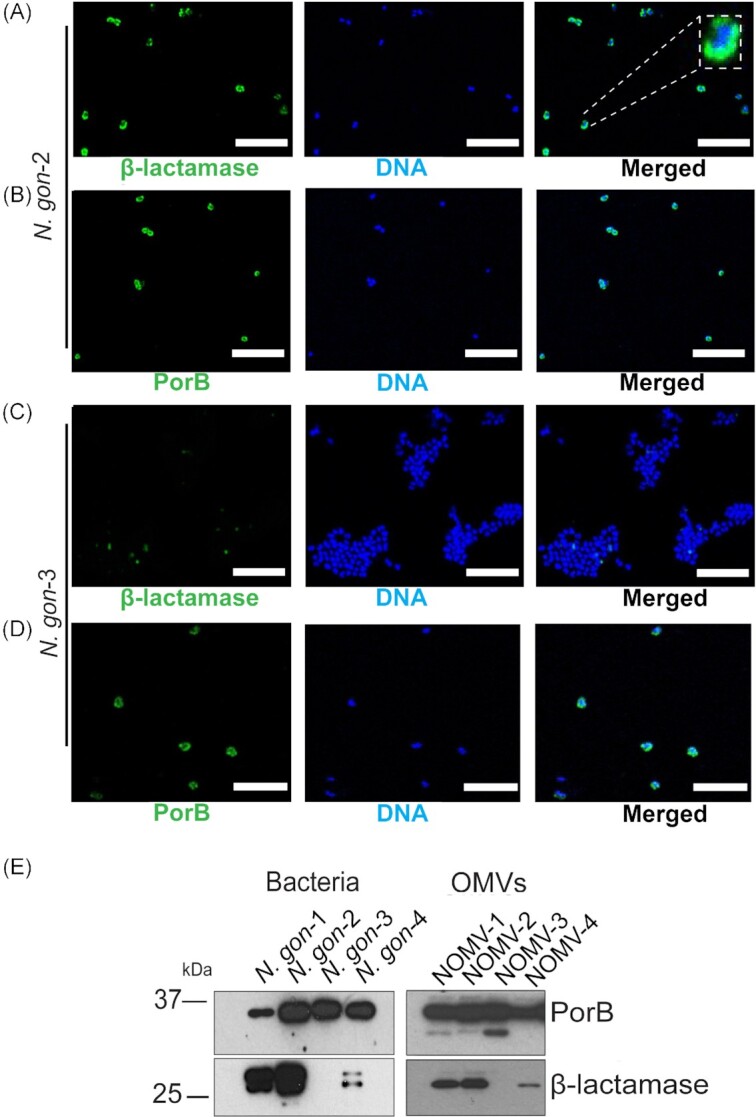
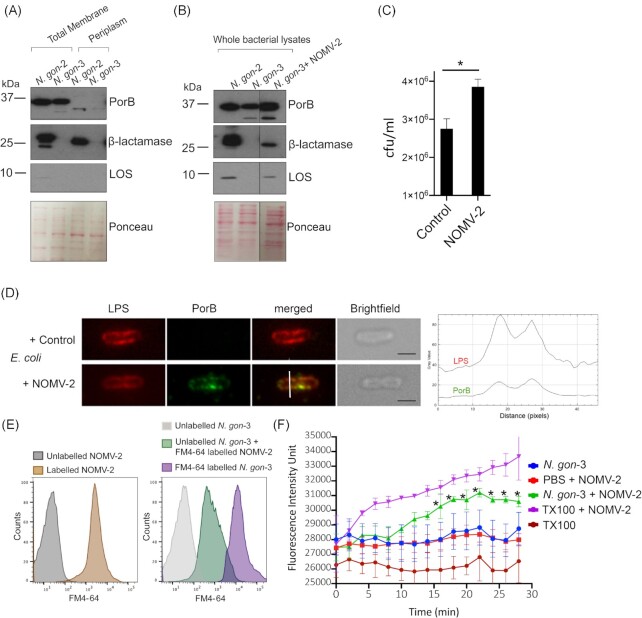
(A)–(D) Confocal laser scanning microscopy of β-lactamase positive isolate N. gon-2 (A) and (B) or β-lactamase negative isolate N. gon-3 (C) and (D) labelled with anti-β-lactamase (A) and (C) or anti-PorB (B) and (D). DAPI (blue) was used to stain DNA. Scale bar 5 μm. Representative image of more than 500 bacterial cells in two biological samples. (E) Bacterial whole cell lysates (N. gon-1, N. gon-2, N. gon-3, and N. gon-4) and corresponding OMVs (NOMV-1, -2, -3, and -4) were analyzed by SDS-PAGE, and immunoblots for PorB and β-lactamase. Loading of the lysates was normalized based on optical density at 600 nm (OD600) and OMV fractions to protein concentrations. Molecular weight markers are indicated on the left.
To determine whether β-lactamase was also secreted by N. gonorrhoeae, we next focused on OMVs released by N. gonorrhoeae. OMVs were purified from culture supernatants of the four clinical N. gonorrhoeae isolates and probed for β-lactamase. Immunoblots revealed β-lactamase to be present in OMVs derived from N-gon-1, -2, and -4, referred to as NOMV-1, -2, and -4 (Fig. 1E). Consistent with the overall expression levels in whole cells, NOMV-4 showed lower levels of β-lactamase compared to NOMV-1 and -2 (Fig. 1E). The OMVs from all four strains contained similar levels of the outer membrane protein PorB, a signature protein in N. gonorrhoeae OMVs (Fig. 1E). It has been previously noted in E. coli and in Klebsiella pneumoniae clinical isolates that evolve under drug selection express β-lactamase at such high levels resulting in the detection of both processed (periplasmic) and unprocessed (cytoplasmic and enzymatically inactive) forms (Bharathwaj et al. 2021). Here, the β-lactamase packaged into the OMVs was detected as a single band (lower) consistent in size with the processed form of the proteins, in contrast to the whole bacterial lysates, which also contained the unprocessed precursor form (Fig. 1E). To assess whether the β-lactamase present in OMVs is enzymatically active, we used the chromogenic β-lactam compound nitrocefin. These substrate hydrolysis assays revealed that the β-lactamase activity was substantial in NOMV-2 (0.015402 mmol/min/ml), ~ 5-fold and 50-fold more than in the same quantity of NOMV-1 and -4, respectively (Table 2). There was negligible β-lactamase activity detected in NOMV-3 (Table 2). Taken together, this demonstrates that the β-lactamase synthesized in N. gonorrhoeae clinical isolates is secreted via OMVs, and that this secreted protein is enzymatically active.
Table 2.
Amount of nitrocefin hydrolysed and calculated activity.
| OMVs 150 µg/ml | Nitrocefin hydrolyzed nmol (initial) | Nitrocefin hydrolyzed nmol (36 min) | Activity (nmol/min/ml) |
|---|---|---|---|
| NOMV-1 | 4.09 | 8.04 | 0.003291 |
| NOMV-2 | 23.38 | 41.8 | 0.015402 |
| NOMV-3 | 3.40 | 3.57 | 1.143E-04 |
| NOMV-4 | 3.34 | 5.16 | 0.000303 |
β-lactamases are packaged in the lumen of OMVs
To assess whether the β-lactamase is packaged as cargo within the lumen of the OMVs, Proteinase K digestion was used. Previous studies have shown that the broad cleavage specificity of Proteinase K can be used to assess exposed or membrane protected proteins, including β-lactamases (Jiang et al. 2011, Deo et al. 2018, Bharathwaj et al. 2021). As expected, Proteinase K cleaved the extramembrane loops of the β-barrel outer membrane protein PorB, resulting in multiple fragments of the protein resolved by the SDS-PAGE analysis (Fig. 2). As a control and to enable Proteinase K access to the luminal cargo, OMVs were treated with the detergent Triton X-100, resulting in further degradation of the now fully exposed PorB (Fig. 2). Through these treatments, the β-lactamases remained resistant to Proteinase K, until Triton X-100 was included (Fig. 2). Triton X-100 by itself did not affect stability of PorB or the β-lactamase (Fig. 2). This biochemical assessment suggests that β-lactamases are packaged in the lumen of OMVs.
Figure 2.
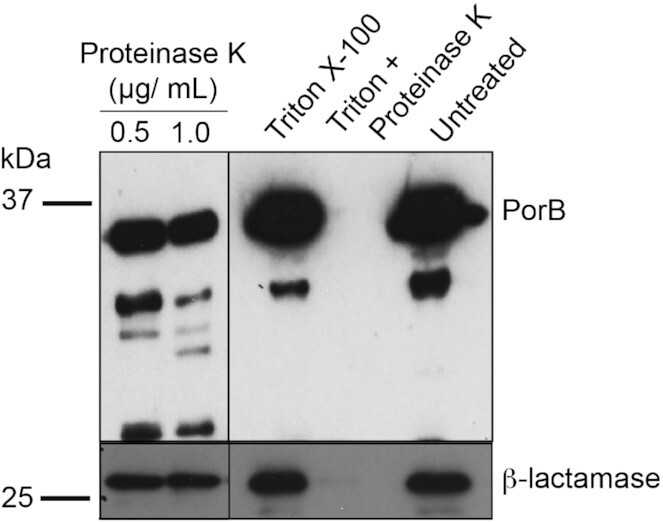
Immunoblot analysis of β-lactamase in proteinase K and Triton X-100-treated OMVs. PorB, β-barrel outer membrane protein was used as the control. OMVs were treated with 0.5 μg/ml and 1.0 μg/ml of proteinase K. Some OMVs samples were treated with 0.1% Triton X-100 and some OMVs samples were further, in addition treated with proteinase K (1 μg/ml). Molecular weight markers are indicated on the left.
OMVs can protect susceptible bacteria from antibiotics
To understand whether OMVs can protect susceptible bacteria from antibiotics, we assessed bacterial growth in antibiotic containing media, in the presence or absence of OMVs (Fig. 3A). Given that N. gon-3 was susceptible to AMX, it served as the model strain in these assays. A starting culture of N. gon-3 replicated for up to 10 h in antibiotic free media and, as expected, was completely suppressed by the addition of 0.5 µg/ml AMX (Fig. 3B). Under these established conditions, we assessed the impact of NOMVs on the growth of N. gon-3. As shown in Fig. 3(B), NOMV-1, -2, and -4 enabled N. gon-3 replication in the presence of AMX. Titration experiments also showed that growth was restored even with decreased amounts of NOMVs (Figure S1, Supporting Information). By way of control, NOMV-3 failed to restore growth of the parent bacteria in the presence of AMX (Fig. 3B). Speaking to the specificity of the β-lactamase cargo, NOMV-2 did not protect N. gon-3 from the presence of the β-lactamase resistant antibiotic cefotaxime (Fig. 3C).
Figure 3.
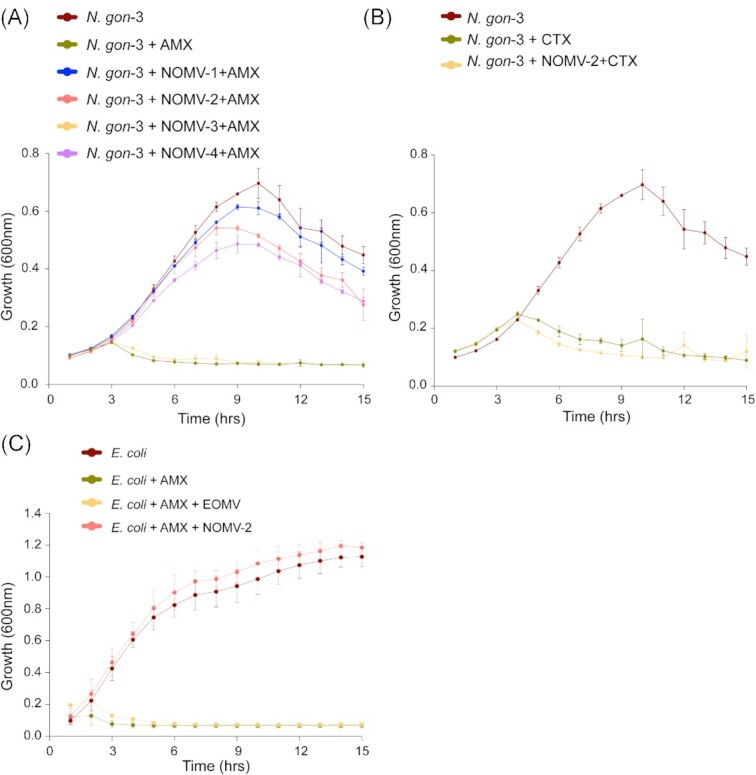
β-lactamase containing OMVs enable replication antibiotics susceptible bacteria. (A) N. gon-3 growth was determined by optical density at 600 nm, OD600, with or without 0.5 μg/ml AMX (0.5) in the presence or absence of NOMVs isolated from the clinical isolates (NOMV-1, -2, -3, and -4) in concentration of 50 μg/ml (50). (B) Growth of N. gon-3 with or without 0.006 μg/ml cefotaxime and NOMV-2 (50 μg/ml). The untreated N. gon-3 growth curve is the same as in (A), as experiments were performed simultaneously. Mean and standard deviation from three independent experiments are shown. (C) Growth (OD600) of E. coli alone or in presence of AMX (2.0 μg/ml) or in combination of AMX (2.0 μg/ml) and OMVs isolated from E. coli lacking β-lactamase (EOMVs) or NOMV-2 (50.0 μg/ml). Mean and standard deviation from three independent experiments are shown.
To address whether this OMV-mediated cross-protection is species specific, NOMV-2 was added to E. coli BL21 in the presence of AMX. Under the conditions of this assay, growth of E. coli was completely inhibited by AMX at 2 µg/ml (Fig. 3D), and addition of OMVs derived from parent E. coli strain (EOMVs) failed to protect E. coli growth in the presence of AMX (Fig. 3D). By contrast, NOMV-2 protected E. coli from AMX (Fig. 3D; Figure S2, Supporting Information). The corollary to this result is that the presence of antibiotic resistant N. gonorrhoeae could potentially protect AMX susceptible bacteria, including but not limited to drug-sensitive N. gonorrhoeae.
OMVs confer immediate protection from antibiotic exposure
To investigate whether β-lactamase containing N. gonorrhoeae OMVs protect susceptible bacteria from initial antibiotic exposure, we quantified bacterial survival immediately following AMX treatment (Fig. 4A). Over 4 h, AMX treatment increasingly killed N. gon-3, as determined by the decreased recovery of bacteria over-time using colony-forming unit assays (CFU; Fig. 4B). Under these conditions, susceptible N. gon-3 bacteria were completely protected from AMX treatment when NOMV-2 was present (Fig. 4B). Similarly, we determined the MIC of AMX in the susceptible N. gon-3 (Fig. 4A), which was 0.25 µg/ml at every time point tested (Table 3). The presence of NOMV-2 increased the MIC to 4 µg/ml when N. gon-3 were exposed to AMX for 1 or 2 h (Table 3). Subsequently, the MIC decreased with increasing exposure times (Table 3). Taken together, the data demonstrate that β-lactamase containing OMVs confer protection from antibiotics enabling survival of susceptible N. gonorrhoeae.
Figure 4.
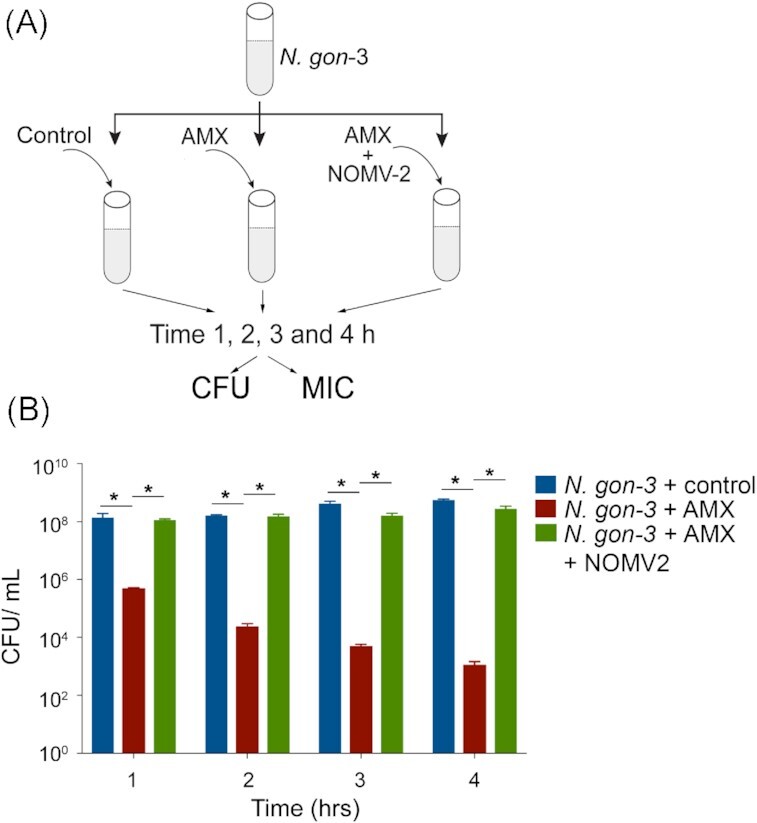
(A) Schematic of the experiments to determine CFUs and MIC of AMX. (B) Neisseria gonorrhoeae isolate N. gon-3 were incubated in control media, with AMX (0.5 μg/ml) or OMVs derived from resistant N. gon-2 (NOMV-2, 10 μg/ml). CFUs/ml were determent between 1 and 4 h. Mean and SEM from three independent experiments. Statistical analysis was determined by ANOVA (two way) and P < .01 are indicated with *.
Table 3.
MIC of AMX.
| Treatment | MICs (μg/ml) hours post treatment | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| N.gon-3 + control | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| N.gon-3 + NOMV-2 | 4 | 4 | 2 | 2 | 1 |
OMVs transfer β-lactamase to antibiotic susceptible N. gonorrhoeae
To assess the possibility that OMVs enable trafficking of β-lactamase between bacteria, we tested for the presence of β-lactamase in N. gon-3 after exposure to NOMV-2. Subcellular fractionation demonstrated that β-lactamase was present within the periplasm of N. gon-2 (Fig. 5A). The total membrane also contained a single band, which was larger in size, representing β-lactamases with secretion signal when attached to inner membrane (Fig. 5A). As expected, the total bacterial lysates of N. gon-2 contained an equal abundant longer and shorter form, which were absent in N. gon-3 (Fig. 5B). However, after exposure to NOMV-2 and subsequent washing of the bacteria to remove any associated vesicles, the total lysates of N. gon-3 contained detectable β-lactamase (Fig. 5B). In contrast to the N. gon-2 lysates, N. gon-3 exposed to NOMV-2 contained a single β-lactamase band that corresponded to the processed form (Fig. 5B). Given that only a fraction of the OMV-associated β-lactamase was transferred to bacteria, analysis of its subcellular localization remained challenging due to the low abundance. The absence of the unprocessed form in N. gon-3 exposed to NOMV-2 suggested a lack of de novo β-lactamase synthesis. Despite this, OMVs can carry nucleic acids including antimicrobial resistance genes (Uddin et al. 2020). Consistent with this notion, the PorB encoding gene was detectable in the bacterial strains MS11, N. gon-2, and N. gon-3 but also in their purified OMVs, to varying levels (Figure S3A, Supporting Information). Similarly, the β-lactamase gene present in N. gon-2 was also in NOMV-2 (Figure S3A, Supporting Information). We also noticed the β-lactamase gene in NOMV-3 and MS11-derived OMVs, even though the parent bacteria lacked the genomic information (Figure S3A, Supporting Information). This likely reflects contamination during the OMV preparation, as the β-lactamase gene was not present or markedly reduced in the MS11 and N. gon-3, but readily detectable in N. gon-2 culture supernatants (Figure S3B, Supporting Information). This would raise the possibility that NOMV-2 may transfer the β-lactamase gene to susceptible strains. However, the β-lactamase gene was not readily detectable in MS11a and N. gon-3 bacteria treated with NOMV-2 (Figure S3A, Supporting Information). Despite this, susceptible N. gonorrhoeae strains that were exposed to NOMV-2, but washed to remove OMVs, showed increased resistance to AMX (Fig. 5C). We were not able to image the presence of β-lactamase under these conditions, likely reflecting low abundance. In contrast, the most abundant OMV protein, PorB, was detectable in E. coli treated with NOMV-2, but not in control cells (Fig. 5D). We next reasoned that delivery of OMV-packaged β-lactamase would likely involve membrane fusion of vesicles with bacteria, which should also transfer vesicle-derived lipids. LOS is the major outer membrane lipid in N. gonorrhoeae, which is also enriched in OMVs. LOS was only detected in N. gon-2 but not N. gon-3 with this particular antibody (Fig. 5A). LOS was, however, detected in N. gon-3 after incubation with NOMV-2 (Fig. 5B). In addition to LOS, we further probed for membrane fusion using OMVs labelled with the lipophilic fluorescent dye FM4-64. Both OMVs and bacteria were readily labelled with FM4-64 (Fig. 5E). Incubation of unlabelled N. gon-3 with FM4-64 labelled NOMV-2 markedly increased fluorescence of the washed bacteria (Fig. 5E). Finally, we used Octadecyl-rhodamine B chloride-R18 (R18). R18 is a lipophilic dye that incorporates into lipid bilayers, but fails to emit light due to self-quenching at high concentrations. The fluorescence is dequenched when R18 is diluted such as the treatment of labelled NOMV-2 with Triton X-100 detergent (Fig. 5F). Similarly, R18 fluorescence was dequenched when NOMV-2 was incubated with N. gon-3 bacteria, but not in their absence (Fig. 5F). Taken together, the data suggests that N. gonorrhoeae OMVs fuse with bacteria to transfer protein cargo such as β-lactamase and membrane lipids.
Figure 5.
(A) The total membrane and periplasm (5 µg protein each) of N. gonorrhoeae strains N. gon-2 and -3 were probed for PorB, β-lactamase, and LOS. Ponceau staining was used a loading control. Molecular weight markers are indicated on the left. (B) Immunoblot analysis of whole bacterial lysate of N. gon-2, N. gon-3, and N. gon-2 treated with NOMV-2 adjusted to equal cell numbers (OD600). Ponceau staining was used a loading control. (C) CFUs of N. gonorrhoeae MS11 exposed to NOMV-2 or control vehicle, washed and then treated with AMX (0.1 μg/ml) for 1 h. Mean and SEM from six independent experiments. *P < .05 (student’s t-test) (D). Escherichia coli treated with NOMV-2 for 1 h were imaged for LPS and PorB. Scale bar = 1 µm. Fluorescence intensity for LPS and PorB along the white line are shown on the right. (E) Flow cytometric analysis of unlabelled and FM4-64-labelled NOMV-2 (OMVs), and N. gon-3 treated with control or FM4-64-labelled NOMV-2 or labelled directly with FM4-64 showing count of events and fluorescence intensity. (F) NOMV-2 were labelled with R18 and then incubated with N. gon-3, Triton X-100 (TX100) or PBS. Unlabelled N. gono-2 and TX100 are included as control. Fluorescence intensity was measured over time (arbitrary units). Mean and SD from triplicate samples. *P < .01. Representative of two independent experiments.
Discussion
Neisseria gonorrhoeae readily acquires plasmids from the environment, including those that encode β-lactamases, and this is a common mechanism for acquiring AMR phenotypes (Rice et al. 2017). After translation in the cytoplasm, the acquired β-lactamase is translocated via the Sec-system into the periplasm, where it is folded into an enzymatically active form that can hydrolyse β-lactam antibiotics (Pradel et al. 2009). Periplasmic β-lactamases are critical to protect the enzymes that drive the synthesis of peptidoglycan, since these enzymes are the target of β-lactam drugs. The current study suggests an alternative mechanism, which was sufficient to protect susceptible N. gonorrhoeae isolates from the β-lactam AMX. We showed that β-lactamases are packaged into N. gonorrhoeae derived OMVs, which protect otherwise susceptible bacteria from AMX. OMVs containing β-lactamase increased the MIC for AMX in susceptible N. gonorrhoeae isolates. At least part of the protection may thus be mediated via direct transfer of the β-lactamase enzyme from OMVs to susceptible bacteria, as our data indicate a membrane fusion event.
There are numerous protein secretion systems in bacteria, with as many as 10 distinct types of proteinaceous molecular machines delivering proteins from bacterial pathogens into host cells (Guerrero-Mandujano et al. 2017). Recently, OMVs have emerged as an important alternative secretion system in several bacterial lineages, with N. gonorrhoeae being an exemplar bacterium that uses OMVs during infection to communicate with host cells (Devoe and Gilchrist 1973, Deo et al. 2018, 2020). In the absence of recognizable proteinaceous secretion machineries, N. gonorrhoeae generates comparatively large numbers of OMVs in culture and in human infections, with recent reports identifying that OMVs deliver outer membrane proteins such as PorB into host cells to modulate immune responses (Deo et al. 2020). In this study, we identified a distinct role for OMVs in that clinical isolates of N. gonorrhoeae produce OMVs such that these β-lactamase-containing OMVs protect otherwise susceptible bacteria from antibiotic treatment. The isolates encoding the β-lactamase variant TEM-1106 expressed particularly high levels of the enzyme in the periplasm, this processed form was found in the OMVs, and these OMVs afforded greater protection to β-lactam drug that OMVs containing less enzyme. How periplasmic proteins are sorted into OMVs remains yet to be determined in any bacteria, but high concentrations of β-lactamase may facilitate packaging during vesicle formation. OMVs are typically enriched for outer membrane proteins, given the vesicles originate from the outer membrane, although additional sorting mechanism are at play as well.
Consistent with this notion, a previous report showed that when the β-lactamase NDM-1 was expressed in E. coli it was sorted into OMVs by a mechanism that depended on its anchorage in the outer membrane via lipidation, with a periplasmic form of the enzyme being largely excluded from OMVs (Gonzalez et al. 2016). Membrane-dependent mechanism of OMV sorting are not restricted to E. coli, since NDM-1 was also detected in OMVs in Providencia rettgeri and Enterobacter cloacae, but not inSerratia marcescens (Gonzalez et al. 2016). Other reports indicated the secretion of soluble periplasmic β-lactamase via OMVs as seen in Pseudomonas aeruginosa (Ciofu et al. 2000). Likewise, both periplasmic L1-metallo and L2-serine β-lactamase were secreted from Stenotrophomonas maltophilia packaged into OMVs, when bacteria are challenged with antibiotics (Devos et al. 2015). Intriguingly, L1 and L2 β-lactamases depend on the Sec and the Tat export system, respectively, excluding the possibility that OMV sorting is linked to a particular inner membrane translocation machinery (Devos et al. 2015). Besides lipid anchors, electrostatic interactions with the outer membrane can promote packaging of β-lactamases into OMVs (Lopez et al. 2021). Based on these and other studies, secreted β-lactamases can protect neighbouring bacteria from antibiotic exposure, by hydrolyzing β-lactam drugs extracellularly, as has been demonstrated with Moraxella catarrhalis OMVs that contain β-lactamases (Schaar et al. 2011). The degradation of extracellular antibiotics by OMVs is perhaps best understood by the cephalosporinase secreted via OMVs by gut microbiota. In this case, the β-lactamases remain exposed on the surface of OMVs readily degrading antibiotics in the environment, thus protecting commensals as well as enteric pathogens in gut (Stentz et al. 2015).
Packing of β-lactamases within OMVs enables not only the secretion of active enzymes but enables protection from extracellular proteases as shown here and previously (Gonzalez et al. 2016), which would likely lead to increased drug resistance. In addition, OMV packaging may enable transfer to other bacteria, thus spreading the resistance to neighbouring cells. In this scenario, the delivery of packaged β-lactamases to susceptible bacteria would have to invoke fusion of outer membranes with extracellular vesicles to enable release of content into the periplasm. Consistent with this notion, only the processed β-lactamase form was detected in susceptible bacteria treated with the OMVs by immunoblotting. Future work is aimed at imaging the presence of β-lactamase within the periplasm of bacteria to confirm delivery of OMV packaged proteins to bacteria. Based on our R18 dequenching experiment, we suggest that the delivery would invoke membrane fusion between OMVs and bacteria. This would also enable the transfer of lipids such as LOS and lipophilic fluorescent probes, as indicated here. The mechanism appears to also enable communication of N. gonorrhoeae-derived OMVs with non-Neisseria species, as the β-lactamase containing vesicles protected susceptible E. coli, which acquired PorB. While previous reports suggested that OMVs only interact with their parent bacteria (Tashiro et al. 2017), OMVs from P. aeruginosa and Shigella flexneri are thought to fuse with Salmonella Typhi and Typhimurium based on the transfer of species-specific LPS without compromising bacterial survival or growth (Kadurugamuwa and Beveridge 1999). The transfer of a diverse set of molecules, including hydrolases, via OMVs to other cells may promote survival of bacteria that prey on others (Evans et al. 2012). Rather than compromising E. coli survival, N. gonorrhoeae-derived OMVs appear to benefit other strains and bacterial species. Whether N. gonorrhoeae-derived OMVs promote nutrient uptake as observed in other bacteria awaits to be seen (Prados-Rosales et al. 2014, Rakoff-Nahoum et al. 2014). It has become evident, however, that OMVs can contribute to the spread of antimicrobial resistance. Besides acting as decoy for some antibiotics, OMVs may also enable horizontal transfer of genetic material including antibiotic resistance genes (Dorward et al. 1989, Chatterjee et al. 2017), although the molecular mechanisms remain incompletely understood and may not be highly efficient. The presence of β-lactamase enzymes in OMVs would provide immediate protection to otherwise susceptible bacteria. It is, thus possible that a single strain of β-lactamase producing N. gonorrhoeae can protect other strains and species during antibiotic treatment of infected individuals. Recent N. gonorrhoeae genomic analysis have indicated clusters of transmission including recurrent infections within risk groups (Williamson et al. 2019). It will be interesting to test experimentally how β-lactamase-containing OMVs affect N. gonorrhoeae infections and commensal communities within the reproductive tract during antimicrobial treatments.
Conclusion
In conclusion, clinical isolates of N. gonorrhoeae that are resistant to AMX and other β-lactam antibiotics release active β-lactamases via OMVs. β-lactamases containing OMVs protect susceptible clinic isolates from antibiotics. Current efforts to detect the presence of β-lactamases are focused on DNA-based and cellular methods, which suggest that resistance in N. gonorrhoeae isolates to be relatively common, depending on the region (Dong and Klausner 2019, Cehovin et al. 2020). β-Lactamase containing OMVs that are likely not detected by current methods may also contribute to the increased failure of β-lactam and perhaps other antibiotics to clear infections in the clinic.
Authors’ contributions
S.D. and T.N. conceived the project; S.D., P.D., M.B., K.H., J.N., M.A., I.S., S.H.C., S.D.G., and R.B. performed the experiments; J.L, T.L, and B.P.H. analyzed and interpreted the data. S.D. and T.N. drafted the manuscript. All authors revised the manuscript and provided intellectual input.
Supplementary Material
ACKNOWLEDGEMENTS
The work was supported by funds from the National Health and Medical Research Council (Ideas grant number 1183848 to T.N.) and the Australian Research Council (Future Fellowship number FT170100313 to T.N.). We thank Kerri Stevens (University of Melbourne) and Grishma Vadlamani (Monash University) for providing N. gonorrhoeae strains and β-lactamase antibodies, respectively, and Dr Alex Fulcher (Monash Micro Imaging) for expert assistance with imaging.
Contributor Information
Subhash Dhital, Infection Program, Monash Biomedicine Discovery Institute and Department of Biochemistry and Molecular Biology, Monash University, Clayton, Australia; Centre to Impact AMR, Monash University, Clayton, Australia.
Pankaj Deo, Infection Program, Monash Biomedicine Discovery Institute and Department of Biochemistry and Molecular Biology, Monash University, Clayton, Australia; Centre to Impact AMR, Monash University, Clayton, Australia.
Manasa Bharathwaj, Centre to Impact AMR, Monash University, Clayton, Australia; Infection Program, Monash Biomedicine Discovery Institute and Department of Microbiology, Monash University, Melbourne, Victoria, Australia.
Kristy Horan, Microbiological Diagnostic Unit Public Health Laboratory, Department of Microbiology and Immunology, The University of Melbourne at The Peter Doherty Institute for Infection and Immunity, Melbourne, Victoria, Australia.
Joshua Nickson, Infection Program, Monash Biomedicine Discovery Institute and Department of Biochemistry and Molecular Biology, Monash University, Clayton, Australia; Centre to Impact AMR, Monash University, Clayton, Australia.
Mohammad Azad, Centre to Impact AMR, Monash University, Clayton, Australia; Infection Program, Monash Biomedicine Discovery Institute and Department of Microbiology, Monash University, Melbourne, Victoria, Australia.
Isabella Stuart, Infection Program, Monash Biomedicine Discovery Institute and Department of Biochemistry and Molecular Biology, Monash University, Clayton, Australia; Centre to Impact AMR, Monash University, Clayton, Australia.
Seong H Chow, Infection Program, Monash Biomedicine Discovery Institute and Department of Biochemistry and Molecular Biology, Monash University, Clayton, Australia; Centre to Impact AMR, Monash University, Clayton, Australia.
Sachith D Gunasinghe, Infection Program, Monash Biomedicine Discovery Institute and Department of Microbiology, Monash University, Melbourne, Victoria, Australia; European Molecular Biology Laboratory (EMBL) Australia Node in Single Molecule Science, School of Medical Sciences, University of New South Wales, Sydney, New South Wales, Australia.
Rebecca Bamert, Centre to Impact AMR, Monash University, Clayton, Australia; Infection Program, Monash Biomedicine Discovery Institute and Department of Microbiology, Monash University, Melbourne, Victoria, Australia.
Jian Li, Centre to Impact AMR, Monash University, Clayton, Australia; Infection Program, Monash Biomedicine Discovery Institute and Department of Microbiology, Monash University, Melbourne, Victoria, Australia.
Trevor Lithgow, Centre to Impact AMR, Monash University, Clayton, Australia; Infection Program, Monash Biomedicine Discovery Institute and Department of Microbiology, Monash University, Melbourne, Victoria, Australia.
Benjamin P Howden, Microbiological Diagnostic Unit Public Health Laboratory, Department of Microbiology and Immunology, The University of Melbourne at The Peter Doherty Institute for Infection and Immunity, Melbourne, Victoria, Australia.
Thomas Naderer, Infection Program, Monash Biomedicine Discovery Institute and Department of Biochemistry and Molecular Biology, Monash University, Clayton, Australia; Centre to Impact AMR, Monash University, Clayton, Australia.
Conflict of interest
None declared.
Data availability
All data are available upon reasonable requests.
References
- Ashford WA, Golash RG, Hemming VG.. Penicillinase-producing Neisseria gonorrhoeae. Lancet North Am Ed. 1976;308:657–8. [DOI] [PubMed] [Google Scholar]
- Bala M, Kakran M, Singh Vet al. . Monitoring antimicrobial resistance in Neisseria gonorrhoeae in selected countries of the WHO South-East Asia Region between 2009 and 2012: a retrospective analysis. Sex Transm Infect. 2013;89:iv28–iv35. [DOI] [PubMed] [Google Scholar]
- Bharathwaj M, Webb CT, Vadlamani Get al. . The Carbapenemase BKC-1 from Klebsiella pneumoniae is adapted for translocation by both the tat and sec translocons. mBio. 2021;12:e0130221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielaszewska M, Daniel O, Nyc Oet al. . In vivo secretion of beta-lactamase-carrying outer membrane vesicles as a mechanism of beta-lactam therapy failure. Membranes. 2021;11:806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitto NJ, Chapman R, Pidot Set al. . Bacterial membrane vesicles transport their DNA cargo into host cells. Sci Rep. 2017;7:7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JMA, Webber MA, Baylay AJet al. . Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42–51. [DOI] [PubMed] [Google Scholar]
- Bomberger JM, Maceachran DP, Coutermarsh BAet al. . Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009;5:e1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cehovin A, Jolley KA, Maiden MCJet al. . Association of Neisseria gonorrhoeae plasmids with distinct lineages and the economic status of their country of origin. J Infect Dis. 2020;222:1826–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Mondal A, Mitra Set al. . Acinetobacter baumannii transfers the blaNDM-1 gene via outer membrane vesicles. J Antimicrob Chemother. 2017;72:2201–7. [DOI] [PubMed] [Google Scholar]
- Ciofu O, Beveridge TJ, Kadurugamuwa Jet al. . Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J Antimicrob Chemother. 2000;45:9–13. [DOI] [PubMed] [Google Scholar]
- Cole MJ, Unemo M, Grigorjev Vet al. . Genetic diversity of blaTEM alleles, antimicrobial susceptibility and molecular epidemiological characteristics of penicillinase-producing Neisseria gonorrhoeae from England and Wales. J Antimicrob Chemother. 2015;70:3238–43. [DOI] [PubMed] [Google Scholar]
- Datta N, Kontomichalou P.. Penicillinase synthesis controlled by infectious R factors in enterobacteriaceae. Nature. 1965;208:239–41. [DOI] [PubMed] [Google Scholar]
- Deo P, Chow SH, Han MLet al. . Mitochondrial dysfunction caused by outer membrane vesicles from Gram-negative bacteria activates intrinsic apoptosis and inflammation. Nat Microbiol. 2020;5:1418–27. [DOI] [PubMed] [Google Scholar]
- Deo P, Chow SH, Hay IDet al. . Outer membrane vesicles from Neisseria gonorrhoeae target PorB to mitochondria and induce apoptosis. PLoS Pathog. 2018;14:e1006945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoe IW, Gilchrist JE.. Release of endotoxin in the form of cell wall blebs during in vitro growth of Neisseria meningitidis. J Exp Med. 1973;138:1156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos S, Van Oudenhove L, Stremersch Set al. . The effect of imipenem and diffusible signaling factors on the secretion of outer membrane vesicles and associated Ax21 proteins in. Stenotrophomonas maltophilia. Front Microbiol. 2015;6:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HV, Klausner JD.. Neisseria gonorrhoeae resistance driven by antibiotic use. Nat Rev Urol. 2019;16:509–10. [DOI] [PubMed] [Google Scholar]
- Dorward DW, Garon CF, Judd RC. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J Bacteriol. 1989;171:2499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AGL, Davey HM, Cookson Aet al. . Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology. 2012;158:2742–52. [DOI] [PubMed] [Google Scholar]
- Gonzalez LJ, Bahr G, Nakashige TGet al. . Membrane anchoring stabilizes and favors secretion of New Delhi metallo-beta-lactamase. Nat Chem Biol. 2016;12:516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Mandujano A, Hernandez-Cortez C, Ibarra JAet al. . The outer membrane vesicles: secretion system type zero. Traffic. 2017;18:425–32. [DOI] [PubMed] [Google Scholar]
- Gunasinghe SD, Shiota T, Stubenrauch CJet al. . The WD40 protein BamB mediates coupling of BAM complexes into assembly precincts in the bacterial outer membrane. Cell Rep. 2018;23:2782–94. [DOI] [PubMed] [Google Scholar]
- Jiang JH, Davies JK, Lithgow Tet al. . Targeting of Neisserial PorB to the mitochondrial outer membrane: an insight on the evolution of beta-barrel protein assembly machines. Mol Microbiol. 2011;82:976–87. [DOI] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ.. Membrane vesicles derived from Pseudomonas aeruginosa and Shigella flexneri can be integrated into the surfaces of other Gram-negative bacteria. Microbiology. 1999;145:2051–60. [DOI] [PubMed] [Google Scholar]
- Kim SW, Park SB, Im SPet al. . Outer membrane vesicles from β-lactam-resistant Escherichia coli enable the survival of β-lactam-susceptible E. coli in the presence of β-lactam antibiotics. Sci Rep. 2018;8:5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni HM, Swamy Ch V, Jagannadham MV. Molecular characterization and functional analysis of outer membrane vesicles from the antarctic bacterium Pseudomonas syringae suggest a possible response to environmental conditions. J Proteome Res. 2014;13:1345–58. [DOI] [PubMed] [Google Scholar]
- Lahra MM, Hogan TR, Shoushtari Met al. . Australian gonococcal surveillance programme annual report, 2020. Commun Dis Intell. 2021;45:33934693. [DOI] [PubMed] [Google Scholar]
- Lopez C, Prunotto A, Bahr Get al. . Specific protein-membrane interactions promote packaging of metallo-beta-lactamases into outer membrane vesicles. Antimicrob Agents Chemother. 2021;65:e0050721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AJ, Kuehn MJ.. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 2011;11:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micaëlo M, Goubard A, La Ruche Get al. . Molecular epidemiology of penicillinase-producing Neisseria gonorrhoeae isolates in France. Clin Microbiol Infect. 2017;23:968–73. [DOI] [PubMed] [Google Scholar]
- Orench-Rivera N, Kuehn MJ.. Environmentally controlled bacterial vesicle-mediated export. Cell Microbiol. 2016;18:1525–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne SH, Bonissone S, Wu Set al. . Unexpected diversity of signal peptides in prokaryotes. mBio. 2012;3:e00339–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips I. Beta-lactamase-producing, penicillin-resistant gonococcus. Lancet North Am Ed. 1976;308:656–7. [DOI] [PubMed] [Google Scholar]
- Pradel N, Delmas J, Wu LFet al. . Sec- and tat-dependent translocation of β-Lactamases across the Escherichia coli inner membrane. Antimicrob Agents Chemother. 2009;3:242–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prados-Rosales R, Weinrick BC, Pique DGet al. . Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J Bacteriol. 2014;196:1250–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Coyne MJ, Comstock LE.. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol. 2014;24:40–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice PA, Shafer WM, Ram Set al. . Neisseria gonorrhoeae: drug resistance, mouse models, and vaccine development. Annu Rev Microbiol. 2017;71:665–86. [DOI] [PubMed] [Google Scholar]
- Sabnis A, Ledger EVK, Pader Vet al. . Antibiotic interceptors: creating safe spaces for bacteria. PLoS Pathog. 2018;14:e1006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar V, Nordstrom T, Morgelin Met al. . Moraxella catarrhalis outer membrane vesicles carry beta-lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrob Agents Chemother. 2011;55:3845–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Kuehn MJ.. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 2015;13:605–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stentz R, Horn N, Cross Ket al. . Cephalosporinases associated with outer membrane vesicles released by Bacteroides spp. protect gut pathogens and commensals against beta-lactam antibiotics. J Antimicrob Chemother. 2015;70:701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y, Hasegawa Y, Shintani Met al. . Interaction of bacterial membrane vesicles with specific species and their potential for delivery to target. Cells Front Microbiol. 2017;8:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin MJ, Dawan J, Jeon Get al. . The role of bacterial membrane vesicles in the dissemination of antibiotic resistance and as promising carriers for therapeutic agent delivery. Microorganisms. 2020;8:670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M, Jensen JS.. Antimicrobial-resistant sexually transmitted infections: gonorrhoea and Mycoplasma genitalium. Nat Rev Urol. 2017;14:139–52. [DOI] [PubMed] [Google Scholar]
- Unemo M, Shafer WM.. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev. 2014;27:587–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand I, Hilpert K, Hancock REW.. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163–75. [DOI] [PubMed] [Google Scholar]
- Williamson DA, Chow EPF, Gorrie CLet al. . Bridging of Neisseria gonorrhoeae lineages across sexual networks in the HIV pre-exposure prophylaxis era. Nat Commun. 2019;10:3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available upon reasonable requests.