ABSTRACT
Membrane vesicles (MVs) contribute to various biological processes in bacteria, including virulence factor delivery, antimicrobial resistance, host immune evasion and cross-species communication. MVs are frequently released from the surface of both Gram-negative and Gram-positive bacteria during growth. In some Gram-positive bacteria, genes affecting MV biogenesis have been identified, but the mechanism of MV formation is unknown. In Enterococcus faecalis, a causative agent of life-threatening bacteraemia and endocarditis, neither mechanisms of MV formation nor their role in virulence has been examined. Since MVs of many bacterial species are implicated in host–pathogen interactions, biofilm formation, horizontal gene transfer, and virulence factor secretion in other species, we sought to identify, describe and functionally characterize MVs from E. faecalis. Here, we show that E. faecalis releases MVs that possess unique lipid and protein profiles, distinct from the intact cell membrane and are enriched in lipoproteins. MVs of E. faecalis are specifically enriched in unsaturated lipids that might provide membrane flexibility to enable MV formation, providing the first insights into the mechanism of MV formation in this Gram-positive organism.
Keywords: membrane vesicles, Enterococcus faecalis, NF-kB signaling, proteomics, lipidomics, horizontal gene transfer
Enterococcal MVs possess unique proteome and lipidome and activate NF-kB signaling in macrophages.
INTRODUCTION
Membrane vesicles (MVs) are widely produced by Gram-negative and Gram-positive bacteria. While MVs confer many similar functions to Gram-positive and Gram-negative bacteria, including virulence factor delivery and host immune modulation, the mechanism of MV formation differs due to intrinsic differences in the structure of the cell wall (Ellis and Kuehn 2010; Rivera et al. 2010; Gurung et al. 2011; Yonezawa et al. 2011; Grull, Mulligan and Lang 2018). MVs are well-studied in Gram-negative bacteria where they are derived from the outer membrane and are produced under the normal growth and stress conditions as well as a result of phage-mediated lysis (Sabra, Lunsdorf and Zeng 2003; McBroom and Kuehn 2007; Turnbull et al. 2016; Zlatkov et al. 2020). Since Gram-positive bacteria lack an outer membrane and have a thicker peptidoglycan which could impact MV formation or release, MVs in Gram-positive bacteria were largely neglected until the first report in 2009 characterizing MVs from Staphylococcus aureus (Lee et al. 2009). Since then, MVs have been reported in numerous Gram-positive bacteria including Bacillus subtilis, Streptococcus pyogenes, Streptococcus mutans, Listeria monocytogenes, Enterococcus faecium, but not in Enterococcus faecalis (Lee et al. 2009; Rivera et al. 2010; Lee et al. 2013; Liao et al. 2014; Biagini et al. 2015; Wagner et al. 2018). Proposed mechanisms of MV formation in Gram-positive fall into two categories—cell blebbing or cell-death mediated (Kuehn and Kesty 2005; Toyofuku, Nomura and Eberl 2019). The proposed blebbing mechanism is independent of cell death or cell-wall digesting enzymes and can be regulated genetically. In L. monocytogenes, RNA polymerase sigma factor σB, a general stress transcription factor that activates stress response genes and ftsZ contributes to MV formation with significantly reduced vesiculation in a ΔsigB mutant (Lee et al. 2013). Similarly, MV biogenesis in M. tuberculosis is affected by VirA (vesiculogenesis and immune response regulator) and vesiculation increases in a VirA deficient mutant (Rath et al. 2013). In S. pyogenes, the two-component system CovRS negatively impacts vesiculation (Resch et al. 2016). In the cell-death mediated mechanism of MV formation, also known as ‘bubbling cell death’, phage endolysins contribute to cell-wall lysis and MV release as observed in B. subtilis (Toyofuku et al. 2017). Released endolysins and autolysins can in turn digest the cell-wall of neighbouring cells contributing to MV formation triggered from outside (Toyofuku, Nomura and Eberl 2019).
Enterococcus faecalis is an opportunistic pathogen that can cause urinary tract infections (UTI), catheter associated urinary tract infections (CAUTI), wound infection, and life-threatening bacteraemia and endocarditis (Gilmore and Clewell 2002; Vu and Carvalho 2011; Ch'ng et al. 2019). Infections by this opportunistic pathogen can be difficult to treat due to their propensity to form biofilms, as well as their frequent and multiple antibiotic resistances (Miller et al. 2016). Enterococcusfaecalis and E. faecium both cause disease in humans, but E. faecalis is more frequently isolated from clinical specimens (Gilmore and Clewell 2002).
We used mass spectrometry-based proteomics to identify the unique proteome of MVs with a high abundance of septal proteins. Using previously established multiple reaction monitoring methods, we detected an enrichment of unsaturated lipids in purified MVs (Rashid et al. 2017). Based on proteome and lipidome studies, coupled with in situ imaging of live bacteria, we propose a model for septal MV formation in E. faecalis. We show that MVs do not contribute to plasmid transfer within- or cross-species, but are capable of activating NF-kB signalling in macrophages. Understanding the mechanism of vesiculation in E. faecalis and the role MVs play in virulence may improve strategies to treat enterococcal infections
METHODS
Bacterial strains and growth conditions
Enterococcus faecalis OG1RF and Escherichia coli DH5α with or without the plasmid pGCP123 (Nielsen et al. 2012) were used in this study. Enterococcus faecalis strains were grown statically in brain heart infusion media (BHI; BD Difco, USA) or on BHI agar (BHI supplemented with 1.5% agarose (1st BASE, Singapore)) at 37°C. Enterococcus coli was grown in Luria-Bertani Broth (Miller) (LB; BD, Difco, USA) at 37°C, 200 rpm shaking. For biofilm assays, tryptone soy broth (Oxoid, UK) supplemented with 10 mM glucose (TSBG) was used. Kanamycin (kan) was used in the following concentrations where appropriate: 50 µg/mL (E. coli), 500 µg/mL (E. faecalis), unless otherwise noted.
Genetic manipulations
To construct the in-frame deletion of pp2 (OG1RF 11 046–11063), regions approximately 1Kb upstream and downstream of the genes were amplified from OG1RF using primer pairs pp2.infu.1F/R and pp2.infu.2F/R for upstream and downstream regions, respectively (Table S1, Supporting Information). These products were sewn together and amplified using pp2.infu.1F/pp2.infu.2R. The temperature sensitive plasmid pGCP213 was amplified using the pGCPinphuF/R primer pair. The pp2.infuF/R PCR product of approximately 2Kb was then cloned into the pGCP213 fragment using the In-Fusion HD Cloning Plus System (Takara Bio Inc, Shimogyō-ku, Kyoto) to generate the temperature sensitive deletion plasmid pGCPpp2.
Deletion constructs were then transformed into OG1RF by electroporation and the transformants were selected at 30°C on agar plates with kan. Chromosomal integrants were selected by growth at 42°C on the agar plate in the presence of kan. Selection for excision of the integrated plasmid by homologous recombination was accomplished by growing the bacteria at 30°C in the absence of kan in the broth. Loss of the pp2 locus in kanamycin-sensitive bacteria was demonstrated by PCR using primer pair infu_check_pp2F/R.
Membrane vesicle isolation
A single E. faecalis colony was inoculated into 100 mL of BHI broth and grown overnight. The resulting overnight culture was diluted 1:10 with warm fresh BHI and grown for 2 hours to reach late exponential phase of growth with OD600nm∼0.8. Cells were harvested using a Beckman centrifuge and JA 12.650 rotor at 4000 × g for 15 min at 4°C. Supernatants were filtered through a 0.45 µm vacuum filter and concentrated to 70 mL using a VIVAFLOW 100 000 MWCO Hydrosart (Sartorius, Germany). Concentrated supernatants were then subjected to ultracentrifugation at 160 000 × g for 2 hours at 4°C using a Beckman Ti 45 rotor (Beckman, Germany). Supernatants were completely removed and the pellet containing the crude MV fraction was resuspended in 400 µL of chilled PBS. MVs were further purified using density gradient centrifugation as previously described (Sjöström et al. 2015). Briefly, 400 μL of PBS containing crude MVs was layered over an OptiPrep density gradient in a 4.2 mL tube in the following order from bottom to top: 400 µL (45% OptiPrep), 500 µL (35% OptiPrep), 600 µL (30% OptiPrep), 600 µL (25% OptiPrep), 600 µL (20% OptiPrep), 500 µL (15% OptiPrep) and 600 µL (10% OptiPrep). The 4.2 mL tubes were centrifuged in a SW60 Ti rotor (Beckman Coulter, USA) for 3 hours at 160 000 × g at 4°C. After centrifugation, 200 µL OptiPrep gradient aliquots were removed from the top and transferred into 21 Eppendorf tubes (21 fractions in total). Collected fractions were subjected to SDS-PAGE and silver staining. Fractions 13–16 containing MVs were combined and used within two days or stored at –80°C until further analysis. For Nanosight, proteomics, and lipidomic analyses, samples were further purified from OptiPrep by ultracentrifugation at 160 000 × g for 2 hours and the pellet was resuspended in 50 µL of PBS.
Transmission electron microscopy (TEM) analysis
For negative staining, 4 µL of sample was deposited onto glow-discharged carbon-coated copper grids (NTU, Singapore). After 1 min, the sample was blotted with a filter paper and 4 µL of 1% uranyl acetate was added onto the grid. After 1 min, excess uranyl acetate was removed. Samples were visualized using a T12 120kV TEM (Fei, USA).
Live imaging
To visualize MV formation in situ, we imaged live late-log phase E. faecalis mounted on agar pads and stained with the hydrophobic membrane dye Nile Red (Strahl, Bürmann and Hamoen 2014; Spahn et al. 2018). Cells were washed in PBS, stained with Nile Red (10 µg/mL) for 10 mins at (Lew et al. 2011) room temperature, washed again in PBS, spotted on 1 mm thick BHI agar (1.5%) pad prepared on a glass slide and mounted with coverslip (glass-agar-glass). Sample was then imaged using Zeiss Axio Observer 7 widefield epifluorescence microscope, equipped with 100x/1.4NA PlanApochromat oil-immersion objective and Hamamatsu Orca Flash 4.0 detector. Images were acquired with 2 s interval with minimal excitation light intensity but sufficient for imaging, to minimize photobleaching and phototoxicity effects. Images were then processed using ImageJ/FIJI.
Nanosight analysis
Purified MV samples were analysed with a NanoSight NS300 (Malvern Instruments, UK) according to the manufacturer's protocol. Briefly, the system was first washed 3 times with 1 mL sterile water. Next, the sample was diluted 1:10 in 1X PBS and loaded into the chamber with a sterile syringe to ensure no bubbles were trapped in the installed tubing. All samples were captured under the same settings and concentration and size were calculated automatically by NanoSight NS300 Control Software (Malvern Instruments, UK).
MV plasmid transfer assay
250 μL of purified MV sample was added to 104 CFU/mL of recipient OG1RF or DH5α in 250 μL of 2X BHI (for E. faecalis OG1RF) or 2X LB (for E. coli DH5α) broth. Two hours after incubation, 100 μL of recipient E. faecalis or E. coli DH5α was plated on BHI agar (kan 1000 μg/mL) for E. faecalis or LB agar E. coli (kan 50 μg/mL) in duplicates. The remaining volume of recipient E. faecalis OG1RF or E. coli DH5α was serially diluted plated on plain BHI/LB agar, incubated overnight for CFU enumeration.
DNA quantification
DNA concentration in MV samples before and after heating at 95°C for 10 mins was measured by Qubit (Qubit® 2.0 Fluorometer, Invitrogen™, Thermo Fisher Scientific) as per user manufacturer's instructions.
DNAse treatment
Five μL of crude or purified MV sample was treated with 1 μL DNAse in a 50 μL reaction volume for 30 min at 37°C. 10 ng of purified pGCP123 was added to control samples. Where indicated, DNase was inactivated at 98°C for 10 min prior to addition of the sample and PCR reaction using primers to amplify pGCP123 plasmid: M13 Forward (5′-GTAAAACGACGGCCAGTG-3′)/Reverse (5′-CAGGAAACAGCTATGAC-3′).
Cell culture and NF-κB reporter assay
RAW-Blue cells derived from RAW 264.7 macrophages (Invivogen), containing a plasmid encoding a secreted embryonic alkaline phosphatase (SEAP) reporter under transcriptional control of an NF-κB-inducible promoter, were cultivated in Dulbecco Modified Eagle medium containing 4500 mg/L high glucose (1X) with 4.0 nM L-glutamine without sodium pyruvate (Gibco), and supplemented with 10% fetal bovine serum (FBS) (Gibco) supplemented with 200 µg/mL Zeocin at 37°C in 5% CO2. RAW-Blue cells were seeded in a 96 well plate at 100 000 cells/well in 200 µL of antibiotic-free cell culture media. Following overnight incubation, the cells were washed once with PBS and fresh media was added. Cells were stimulated using LPS purified from E. coli O111: B4 (Sigma Aldrich) (100 ng/mL) as a positive control, or cell culture media alone or OptiPrep alone as negative controls. MVs purified in OptiPrep (1000 particles/macrophage) were added to RAW-Blue cells and incubated for 6 hours with or without simultaneous LPS stimulation. With an average rate of 2–5 MVs per bacteria Post-infection, 20 µL of supernatant was added to 180 μL of QUANTI-Blue reagent (Invivogen) and incubated overnight at 37°C. SEAP levels were determined at 640 nm using a TECAN M200 microplate reader.
Protein content analysis
Purified MV samples or mid-log phase intact bacterial cells, washed in PBS and incubated in lysis buffer with 1 mg/mL lysozyme for 1 hour at 37°C, were boiled in 1X NuPAGE® LDS sample buffer (Invitrogen, USA) and 0.1 M dithiothreitol (DTT) at 95°C for 10 min. Samples were run on 10% NuPAGE® Bis-Tris mini gel (ThermoFisher Scientific, USA) in 1x MOPS SDS running buffer in XCellSureLock® Mini-Cell for 10 min at 150 V. Gel lanes containing silver stained proteins were cut out and sent to the Harvard Medical School Taplin Mass Spectrometry facility for mass spectrometry (MS) analysis (Harvard, USA). From the list of identified proteins within each sample, proteins containing fewer than 3 unique peptides were excluded from the analysis (Table S2 and S3, Supporting Information), the full list of all of the identified proteins is found under supplementary data (Table S4, Supporting Information). All the other proteins were assigned an abundance score (A*) based on the number of unique peptides per protein divided by the total number of unique peptides. We averaged A* within triplicates and sorted proteins in descending order based on the A*. The unique abundant proteins are the most abundant proteins within MV fraction that were not identified in WCL.
Lipid content analysis
Lipids were extracted from lyophilised membrane vesicles or whole cell pellets, using a modified Bligh & Dyer method as previously described (Bligh and Dyer 1959; Rashid et al. 2017). Prior to extraction, samples were spiked with known amounts of internal standards for phosphatidylglycerol (PG) and lysyl-phosphatidylglycerol (Lys-PG) or external calibration standard, monoglucosyl-diacylglycerol (MGDAG 34:1) (Avanti polar lipids, Alabaster, AL, USA) and were run alongside the samples (Table S5, Supporting Information).
PG and L-PG in WCL and MVs were quantified by LC-MS/MS using multiple reaction monitoring (MRM) that we previously established (Rashid et al. 2017). An Agilent 6490 or 6495A QqQ mass spectrometer connected to a 1290 series chromatographic system was used together with electrospray ionization (ESI) for lipid ionisation. Each lipid molecular species was analyzed using a targeted MRM approach containing transitions for known precursor/product mass-to-charge ratio (m1/m3). Signal intensities were normalized to the spiked internal standards (PG 14:0 and L-PG 16:0) to obtain relative measurements, as described previously (13). Due to an absence of suitable internal standards, semi-quantitative analysis of diglucosyl-diacylglycerol (DGDAG) was carried out instead. Lipid extraction was performed without spiking of internal standards and DGDAG lipid species were analysed by LC-MS/MS using MRMs using monoglucosyl-diacylglycerol (MGDAG) 34:1 as a surrogate standard (Table S5, Supporting Information) for external calibration curves. Measurements of MGDAG 34:1 dilution, from 0.2 ng/mL to 1000 ng/mL were used to construct external calibration curves to estimate the levels of DGDAG. The MRM transitions for DGDAG molecular species and MGDAG 34:1 are listed in Table S6 (Supporting Information). All lipid species abundances were expressed as a percentage of their respective lipid class.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software (Version 6.05 for Windows, California, United States). All experiments were performed at least in three biological replicates and the mean value was calculated. All graphs indicate standard deviation from independent experiments. Statistical analysis was performed by unpaired t-test using GraphPad (* P < 0.05, ** P < 0.01, *** P < 0.001; ****P < 0.0001, ns: P > 0.05). SEAP assays were analyzed using one-way ANOVA with Tukey's multiple comparison. P-values less than 0.05 were deemed significant.
RESULTS
Enterococcus faecalis produces MVs with a size range of 50–400 nm
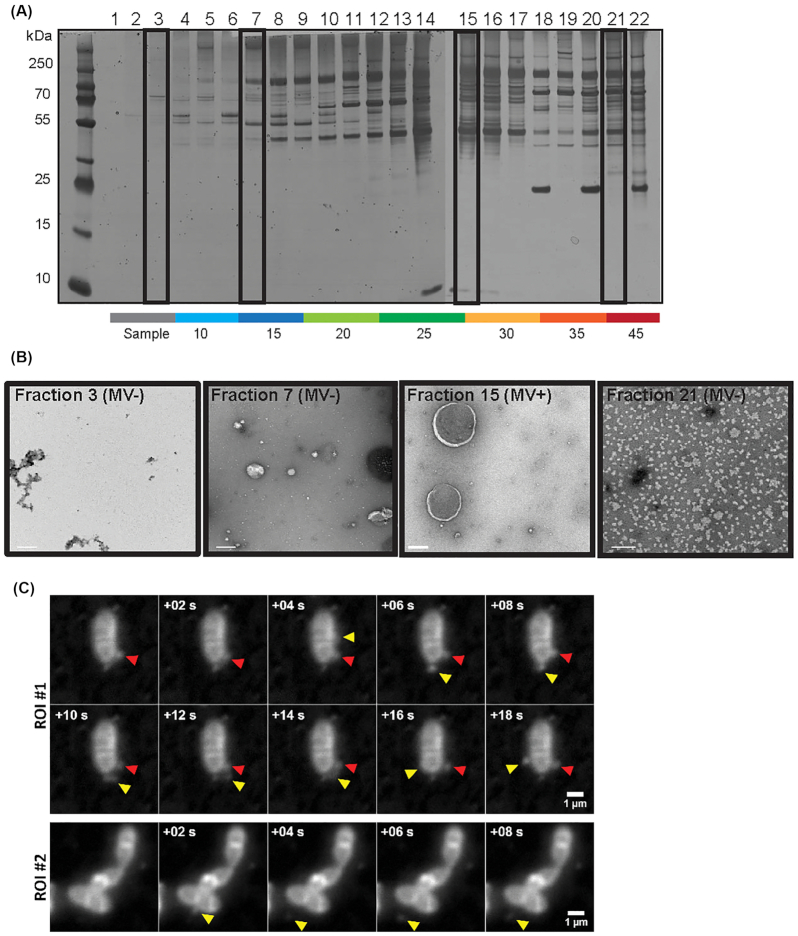
To determine whether E. faecalis produces MVs, we used methods previously described for the isolation of MVs from Gram-positive bacteria (Lee et al. 2009). We collected cell-free supernatants of late-log phase cultures and performed ultracentrifugation to obtain a crude pellet. We then separated the crude pellet by OptiPrep gradient centrifugation, from which we collected 22 fractions followed by SDS PAGE and silver stain (Fig. 1A). We observed a denser staining pattern with altered protein abundances in fractions 13–16, at approximately 25% OptiPrep, a similar density at which MVs were found for S. aureus and E. coli (Beveridge 1999; Lee et al. 2008; Lee et al. 2009).
Figure 1.
Enterococcus faecalis produces MVs ranging from 50–400 nm in size. (A) 22 fractions consisting of 200 μL each were collected from the top of a 4.4 mL OptiPrep gradient, separated by SDS-PAGE, and silver stained. (B) Selected fractions (# 3, 7, 15, 21 indicated in the boxes in panel A) were negatively stained and viewed by TEM. Scale bar is 200 nm. (C) In-situ imaged live E. faecalis on the agar pad. Panels of consecutive frames (+0.2 s) from two distinct areas on the pad. The existing vesicle is indicated with a red arrow, the new vesicle—with a yellow arrow. Scale bar 1 μL.
To determine whether fractions 13–16 contained MVs, we performed transmission electron microscopy (TEM) on fraction 15 (MV fraction) as well as 3 other fractions chosen to represent the crude sample after the centrifugation (fraction 3), and fractions before (fraction 7) and after (fraction 21) those predicted to contain MVs in the gradient. We observed spherical, double membrane particles resembling MVs described in other Gram-positive species in fraction 15, but not in the other three fractions (Fig. 1B) (Gurung et al. 2011; Biagini et al. 2015). To further characterize MVs observed in fraction 15, we combined fractions 13–16 in order to increase our sample mass, and measured the size and concentration of MVs by dynamic light scattering using a Nanosight NS300 instrument. MVs varied in size with a diameter ranging from 50 to 400 nm (Fig. S1, Supporting Information), similar to that reported for other Gram-positive bacteria (Rivera et al. 2010; Jiang et al. 2014; Wagner et al. 2018). We also observed three smaller peaks with a larger diameter of 450–760 nm: however, since a 450 nm cut-off membrane filter was used in MV isolation, the larger peaks might be fused MVs or air bubbles. To visualize MV formation in situ, we imaged live E. faecalis mounted on agar pads and stained with Nile Red in late-log phase. Acquired time series revealed increasing number of small (resolution limited) vesicles that appear to detach from the cell body, and continue to diffuse in close bacterial proximity, most likely trapped in a confined volume around the cell between agar and glass slide (Fig. 1C, Supplementary video 1A, 1B).
Enterococcus faecalis MVs are enriched in unsaturated lipids
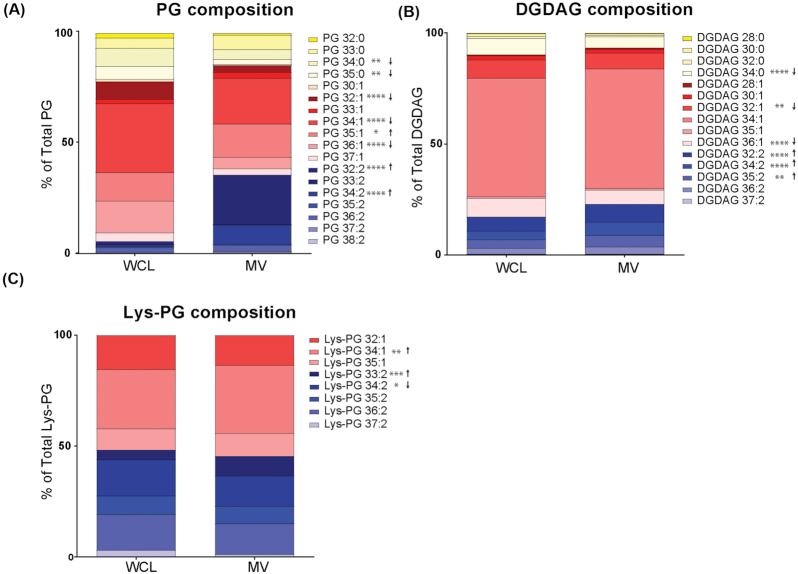
The bacterial membrane is non-homogenous, containing distinct microdomains associated with functions including bacterial secretion and virulence, and which may serve as targets for antimicrobials (Rashid, Veleba and Kline 2016; Lopez and Koch 2017). Since the E. faecalis membrane also contains lipid microdomains where secretion and virulence factor assembly functions are enriched (Kandaswamy et al. 2013; Tran et al. 2013; Rashid, Veleba and Kline 2016) and since MVs are associated with distinct lipid repertoires in some bacteria (Resch et al. 2016), we considered the possibility that E. faecalis MVs are also released from specific lipid microdomains on the bacterial membrane. To address whether E. faecalis MVs are composed of distinct lipid subsets, we compared the lipidomes of purified MVs to lipidomes from the entire membrane of late log phase bacterial cells. We analysed the lipid composition from five independent biological samples by liquid chromatography tandem mass spectrometry (LC-MS) using multiple reaction monitoring (MRM) methods previously established for (PG) and lysyl-phosphatidylglycerol (Lys-PG), as well as newly developed MRMs for diglucosyl-diacylglycerol (DGDAG) (Rashid et al. 2017).
Within the PG species, which are the predominant lipid in the E. faecalis lipid membrane (Rashid et al. 2017), we observed a significant increase in the overall abundance of polyunsaturated lipids (PG 32:2; PG 34:2), and reduction in monounsaturated and saturated lipids (PG 34:0, PG 35:0, PG 32:1, PG 34:1, PG 36:1) in MV samples as compared to whole cell lysates (WCL) (Fig. 2A). For the less abundant DGDAG lipid species, the trend is similar in that we observe an increase in overall polyunsaturated species (Fig. 2B). While we observed differences in individual species of Lys-PG (increased 34:1 and 33:2 in MVs, decreased 34:2 in MVs), the overall levels of saturation for Lys-PG were similar between MV and WCL (Fig. 2C).
Figure 2.
MVs are enriched in polyunsaturated PG species. Constituent distribution of individual lipid species from whole cell lysate (WCL) or MVs within the analysed lipid classes (A), phosphatidylglycerol (PG), (B), diglucosyl-diacylglycerol (DGDAG), and (C), lysyl-phosphatidylglycerol (Lys-PG). Each stack represents the mean from 6 biological replicates. *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001; Fisher's LSD test for one-way ANOVA.
Enterococcus faecalis MVs possess a unique proteome
To further characterize the properties of E. faecalis MVs, we performed proteomic analysis on OptiPrep-purified MVs and from late-log phase whole cells using LC/MS/MS (Elias and Gygi 2007). Proteins for which three or more unique peptides were identified were considered in the analysis. In total, we identified 225 proteins from MV samples and 397 proteins from whole cell lysates, and sorted the proteins based on their calculated average abundance score (Tables S2 and S3, Supporting Information). Abundance was calculated as the number of unique peptides for a given protein divided by the total number of peptides within each sample, and the average abundance for each protein in the three biological replicates is reported. Next, we sorted the 10 most abundant proteins that were also found in the WCL fraction, and the 10 most abundant unique MV proteins that were not found in WCL fraction (Table 1). Within those are IreK (OG1RF_12 384), a eukaryote-like Ser/Thr kinase and phosphatase; penicillin binding proteins Pbp1A, Pbp1B and PenA; a S1 family extracellular protease OG1RF_12 235; lipoprotein (OG1RF_12 508); CnaB and 2 ABC superfamily ATP binding cassette transporters (OG1RF 10 124 and OG1RF_12 508).
Table 1.
Function and rank position of the most abundant and 10 unique proteins identified in MVs by MS. Where n/p—not present in WCL within 397 identified proteins.
| Protein | Position in MVs (of 225) | Position in WCL (of 397) | Function |
|---|---|---|---|
| Aad | 1 | 8 | Aldehyde-alcohol dehydrogenase |
| GroL | 2 | 15 | Chaperonin |
| OG1RF_12 384 IreK | 3 | 168 | Non-specific serine/threonine protein kinase |
| OG1RF_10 124 | 6 | 151 | ABC superfamily ATP binding cassette transporter, binding protein |
| OG1RF_12 203 | 8 | 155 | ABC superfamily ATP binding cassette transporter, substrate-binding protein |
| OG1RF_10 125 | 9 | 67 | ABC superfamily ATP binding cassette transporter, binding protein |
| OppA | 11 | 33 | Oligopeptide ABC superfamily ATP binding cassette transporter, binding protein |
| TraC2 | 12 | 93 | Peptide ABC superfamily ATP binding cassette transporter, binding protein |
| PrsA | 15 | 118 | Peptidyl-prolyl cis-trans isomerase |
| RplB | 16 | 44 | 50S ribosomal protein L2 |
| Pbp1A | 4 | n/p | Penicillin binding proteins 1A |
| CnaB | 5 | n/p | Cna protein B-type domain protein |
| OG1RF_12 508 | 7 | n/p | Thiamine biosynthesis lipoprotein |
| PenA | 10 | n/p | Penicillin-binding protein 2 gene |
| OG1RF_12 235 | 13 | n/p | S1 family extracellular protease |
| Pbp1B | 14 | n/p | Penicillin-binding protein 1B |
| MreC | 19 | n/p | Rod shape-determining protein MreC |
| MetQ | 20 | n/p | ABC superfamily ATP binding cassette transporter, binding protein |
| OG1RF_11 061 | 26 | n/p | Phage tail protein |
| LytR | 32 | n/p | Response regulator |
In addition to a phage tail protein OG1RF_11061 that was found among the 10 most abundant proteins not found in WCL, we detected additional 4 phage proteins encoded at a previously described phage02 locus that are only present in MV fraction and are not found in a whole cell lysate (Table S2, Supporting Information). The phage02-encoding operon (OG1RF 11046–11063) is a part of the E. faecalis core genome and considered cryptic because it lacks crucial genes for DNA packaging and excision (McBride et al. 2007). However, we observed phage tail-like structures in the purified MV fraction by TEM (Fig. 3A). In Pseudomonas aeruginosa MVs can be formed as a result of explosive cell lysis mediated by cell-wall modifying phage endolysins that degrade the bacterial membrane to burst the bacterial cell, resulting in MV formation and spontaneous membrane reannealing (Turnbull et al. 2016). We hypothesized that phage tails could be assembled inside E. faecalis cells and released through explosive cell lysis, concurrently contributing to MV formation. We performed TEM on mid-log phase bacteria and observed MVs closely associated with the bacterial surface (Fig. 3B), similar to bacteria-associated MVs observed in P. aeruginosa upon phage-mediated cell lysis (Turnbull et al. 2016). To test whether phage-mediated explosive cell lysis occurred in E. faecalis, we first generated a strain in which the entire phage02 operon was deleted (Δpp2), including the predicted phage structural proteins, holin, and endolysin genes. We performed TEM on MVs isolated from WT and Δpp2 and confirmed absence of the phage tails in the Δpp2 phage deletion mutant (data not shown).
Figure 3.
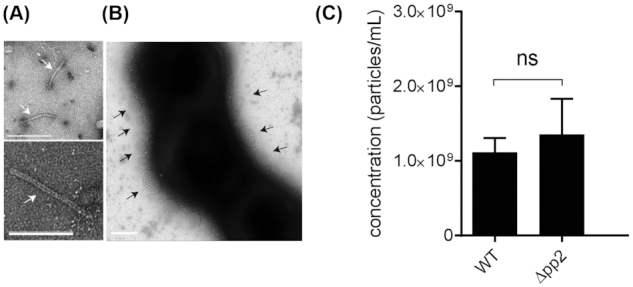
Phage tails co-purify with MVs, but phage tail production does not contribute to MV abundance. (A) TEM on assembled phage tails that are present within purified MVs. Scale bar is 100 nm. (B) TEM image of negatively stained E. faecalis, where MVs are associated with the cells surface. Scale bar is 100 nm. (C) The concentration of MVs in WT and the Δpp2 mutant as determined by Nanosight from three independent experiments. Statistical analysis was performed by the unpaired t-test. ns: P > 0.05.
To determine whether the phage02-encoded factors, such as an endolysin, contributed to cell lysis and MV formation, we quantified MVs from WT and Δpp2, but observed no difference in MV numbers between the two strains (Fig. 3C). Since phage02 is the only predicted phage element in the OG1RF genome, these findings suggested that a phage-mediated mechanism, such explosive cell lysis, does not significantly contribute to E. faecalis MV formation.
Enterococcus faecalis MVs do not mediate intra-species or cross-genera horizontal gene transfer in vitro
OMVs from Gram-negative bacteria may contain DNA from chromosomal, plasmid, or viral origin (Domingues and Nielsen 2017). Plasmid and chromosomal DNA carrying antibiotic resistance genes can be transferred via OMVs to cells of the same or different genera to confer antibiotic resistance in the recipient (Dorward, Garon and Judd 1989; Rumbo et al. 2011). Among Gram-positive bacteria, Clostridium perfringens and S. mutans pack chromosomal DNA in MVs (Liao et al. 2014; Jiang et al. 2014). A single report on MV-mediated horizontal gene transfer (HGT) in Gram-positive bacteria demonstrated the ability of a MV-containing fraction from WT Ruminococcus sp. strain YE71 to transform and permanently restore cellulose-degrading activity to a Cel− mutant that was otherwise unable to degrade cellulose (Klieve et al. 2005). We tested if E. faecalis can pack plasmids inside MVs for HGT to another E. faecalis strain or to another species. We first isolated MVs from plasmid-carrying OG1RF pGCP123 (Nielsen et al. 2012), a 3045 bp, non-conjugative plasmid that can stably replicate in both Gram-positive and Gram-negative bacteria and which encodes kanamycin-resistance.
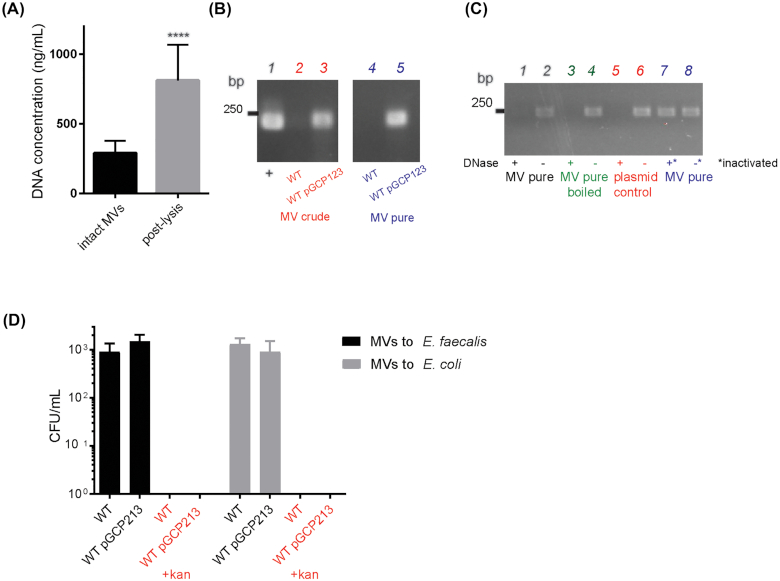
To determine if E. faecalis MVs contain DNA, we first quantified the DNA concentration in intact MVs, and subsequently in the lysed MV fraction. We observed a ∼170% increase in DNA concentration post-lysis, indicating that bacterial DNA was present within MV lumen and was released upon MV lysis (Fig. 4A). To determine if plasmid DNA is present within the MV prep, we performed PCR with plasmid-specific primers on the crude and purified MV fractions, using purified pGCP123 plasmid as a control. We observed a plasmid-specific PCR product of 229 bp in the plasmid control and MV sample from plasmid-carrying strain (Fig. 4B), indicating that plasmid DNA is present in both crude and purified MV fractions. In principle, plasmid DNA can be either packed inside MVs or associated with the exterior of MVs (and therefore co-purified with MVs). To address localization of pGCP123 with respect to E. faecalis MVs, we exposed MVs to DNase in order to degrade any extracellular plasmid, followed by DNase inactivation, MV lysis and PCR. DNAse treatment resulted in a loss of plasmid-specific PCR product. To ensure that DNase was fully inactivated after incubation with MVs prior PCR reaction, we mixed MVs with DNase followed by immediate DNase inactivation by boiling at 98°C for 10 min. We observed a plasmid-specific PCR product in the inactivated DNase sample suggesting that DNase was fully inactivated before the PCR reaction (Fig. 4C). To control for efficiency of DNase treatment, we used purified plasmid and lysed MVs as controls. We observed a plasmid-specific PCR product only in DNase untreated samples, indicating that DNase efficiently digests purified and MV-associated plasmid. Our results suggest, that plasmid DNA is not present within the MV lumen, but rather co-purifies with MVs. However, chromosomal DNA is present within MVs, since we observed an increase in DNA concentration upon MV lysis.
Figure 4.
Plasmid DNA co-purifies with MVs but is not transferred to E. faecalis or E. colicells. (A) DNA concentration was measured by Qubit in intact MV samples and in MVs lysed by boiling. Data shown from three independent experiments. Statistical analysis was performed by the unpaired t-test using GraphPad. ****, P < 0.0001. (B) Agarose gel showing PCR product amplified with plasmid-specific primers on crude and purified MV fractions of WT and WT pGCP123. The expected plasmid PCR product is 240 bp. (C) Agarose gel showing PCR product amplified with plasmid-specific primers on intact and lysed MVs from WT pGCP123, subjected to DNAse treatment or treatment with inactivated DNAse prior to PCR. DNAse treated pGCP123 serves as a control. (D) The number of transformants following E. faecalis and E. coli incubation with MVs extracted from WT or WT pGCP123 determined by CFU enumeration on non-selective BHI or selective media with kanamycin. Statistical analysis was performed by the one-way ANOVA using one-way ANOVA test with Tukey's multiple comparison test. ****, P < 0.0001.
To determine if plasmids co-purified with MVs could mediate HGT, we exposed purified MVs to plasmid free strains of E. faecalis and E. coli for 2 hours to allow for plasmid transfer prior plating bacteria on selective plates containing kanamycin. However, we detected no resistant colonies on the selective plates suggesting that plasmid was not transferred to E. coli or E. faecalis by MVs under these experimental conditions (Fig. 4D).
MVs modulate the NF-κB response in macrophages
Proteomic analysis identified 5 lipoproteins (OG1RF_11390, OG1RF_11130, OG1RF_11506, OG1RF_12508, OG1RF_12509) enriched in MVs, similar to reports of lipoprotein-rich MVs from S. pyogenes and M. tuberculosis (Rath et al. 2013; Biagini et al. 2015) (Tables S3 and S4). Bacteria and pathogen-associated molecular patterns (PAMPs) stimulate the NF-κB response and inflammation in macrophages through various cellular immune receptors, and lipoproteins activate NF-kB through TLR1, TLR2 and TLR6 (Shimizu, Kida and Kuwano 2008; Schenk, Belisle and Modlin 2009; Rahman and McFadden 2011). Mycobacteria-derived MVs include two lipoproteins which are demonstrated agonists of the TLR2 receptor and that activate an inflammatory response in mice (Rath et al. 2013). By contrast, MVs from S. pyogenes do not activate TLR2, indicating that the immunogenic potential of MVs varies between species (Prados-Rosales et al. 2011; Rath et al. 2013; Biagini et al. 2015). We have previously shown that E. faecalis activates macrophages at low multiplicities of infection (MOI) and is immunosuppressive to macrophages at high MOI (Tien et al. 2017). Since MVs carry bacterial DNA, lipoproteins and other PAMPs, we asked whether E. faecalis MVs may contribute to host-pathogen interactions, by testing the NF-κB response of macrophages upon exposure to E. faecalis MVs. We purified MVs using OptiPrep and added them to RAW-blue macrophages (1000 MVs/macrophage) with or without LPS to assess their ability to activate NF-κB signalling on their own, or suppress LPS-mediated activation. Purified lipopolysaccharide (LPS) served as a positive control for maximal NF-κB activation and 25% OptiPrep media as a negative control. We also combined the first 10 fractions from the OptiPrep gradient after centrifugation to serve as a MV-free concentrated supernatant for a secondary negative control. In this assay, we observed that E. faecalis MVs activate NF-κB signalling in macrophages (Fig. 5). Based on Nanosight MV concentration calculations and the number of bacterial cells used to isolate MVs, we calculated an average rate of 0.3 MV/CFU, which does not account for MVs that are still attached to the cells (as observed by TEM and in situ imaging) or lost during the purification process. We therefore roughly estimate 1–2 MVs/CFU are produced under the tested conditions in vitro, while the in vivo relevance and what might be vesiculation rate during the infection in the hostile host environment remains unclear.
Figure 5.
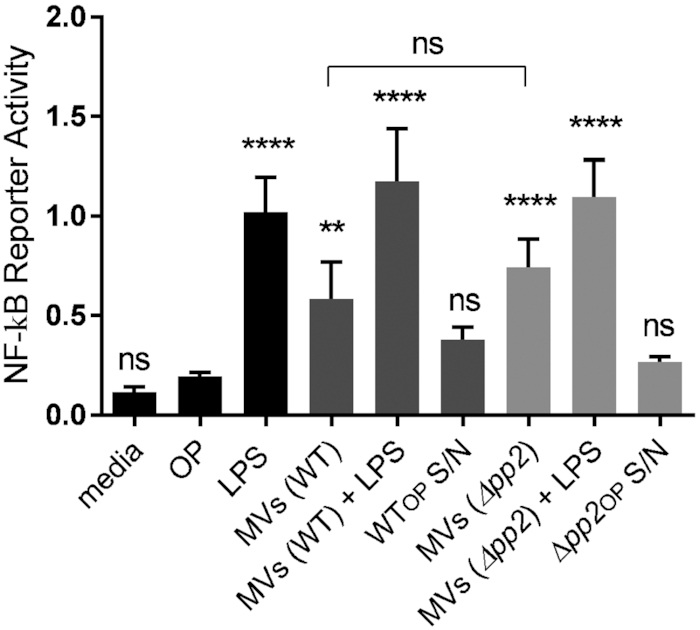
MVs but not phage tails activate NF-kB pathway in macrophages. RAW-blue cells derived from RAW267.4 macrophages were stimulated with lipopolysaccharide (LPS) at 100 ng/mL (positive control), OptiPrep (OP) (negative control), MV-free concentrated supernatant in OptiPrep from WT and Δpp2 (WTOP S/N and Δpp2OP S/N) (secondary controls), MVs derived from WT and Δpp2 at 1000 particles/macrophage, and LPS + MVs. Six hours after stimulation, the NF-kB response was measured by secreted embryonic alkaline phosphatase reporter activity, transcribed from the plasmid under NF-kB inducible promoter. Statistical analysis was performed by the one-way ANOVA using one-way ANOVA test with Tukey's multiple comparison test. ****, P < 0.0001, ** P < 0.001, ns: P > 0.05 among all of the conditions as compared to OptiPrep negative control.
Since we observed that the MV fraction from WT E. faecalis co-purified with bacteriophage tails (Fig. 3), and since fully assembled bacteriophages can be immunomodulatory and suppress phagocytosis and LPS-induced phosphorylation of NF-κBp65 (Van Belleghem et al. 2017; Zhang et al. 2018; Sweere et al. 2019), we next dissected whether phage tails contribute to MV-mediated activation of NF-κB in macrophages (Fig. 5). We isolated MVs from Δpp2 lacking the whole prophage operon and compared the NF-κB response to the response elicited by MVs from WT. We observed no statistical difference in NF-κB reporter activity of Δpp2 mutant (MVs only) compared to WT (MVs + phage tails), suggesting that MVs alone are immunostimulatory and can induce NF-κB signalling in macrophages, regardless of the presence of co-purified phage tails (Fig. 5).
DISCUSSION
Vesicle shedding from bacteria is a common process in Gram-negative and Gram-positive bacteria. Yet the mechanisms of MV formation and function, especially in Gram-positive species are largely unknown. MVs of Gram-positive bacteria possess unique proteomes, are immunogenic, and in some cases contribute to host cells death by carrying toxins (Rivera et al. 2010; Gurung et al. 2011; Prados-Rosales et al. 2011; Pathirana and Kaparakis-Liaskos 2016). In this study, we report that the opportunistic pathogen E. faecalis strain OG1RF produces MVs ranging in size from 50 to 400 nm. E. faecalis MVs are enriched for a unique subset of proteins, unsaturated lipids, and are capable of activating NF-kB signalling in macrophages.
Several studies have compared the lipid profile of MVs to that of the complete bacterial membrane. In Group A Streptococcus, MVs are enriched in PG and significantly reduced in CL while the saturation levels of fatty acids remained unchanged (Resch et al. 2016). In Propionibacterium acnes, MVs also possess a distinct lipid profile with levels of triacylglycerol significantly lower compared to the cell membrane (Jeon et al. 2018). Both reports suggest that lipid composition and distribution in MVs can be very different from that of the bacterial cell membrane, supporting the hypothesis that MV formation is not a random process. In E. faecalis, we observed a significant increase in the levels of unsaturated PG in MVs as compared to the whole cell membrane. Unsaturated lipids enhance membrane fluidity and partition to ordered domains in more fluid regions of the bilayer (Mukherjee, Soe and Maxfield 1999; Jeon et al. 2018). In model membranes, more flexible unsaturated lipids become concentrated in tubular regions pulled from a vesicle and unsaturated lipids are sorted into pathways involving highly curved tubular intermediates (Cooke and Deserno 2006). Therefore, E. faecalis microdomains with polyunsaturated PGs might provide additional flexibility for vesicle formation.
In addition to a distinct lipid profile, E. faecalis MVs possess a unique proteome. Among top abundant and unique proteins are penicillin binding proteins, Pbp1A, Pbp1B, MreC and PenA, which are enriched in the septal region during the cell division (Land and Winkler 2011; Lee et al. 2016; Boes et al. 2019). These observations are consistent with a model in which E. faecalis MVs might be formed from the Pbp-enriched septal region during cell division, where peptidoglycan is thinnest (Li et al. 2018). Consistent with this hypothesis, within the most abundant MV proteins, we detected the serine-threonine kinase IreK that monitors cell wall integrity and mediates adaptive responses to cell wall-active antibiotics (Kristich, Wells and Dunny 2007; Labbe and Kristich 2017). An IreK homologue in S. pneumoniae—StkP—is localized at the division septum via penicillin-binding protein and serine/threonine kinase associated (PASTA) domains linked to un-crosslinked PG, the same unique domains that are present in enterococcal IreK (Beilharz et al. 2012; Zucchini et al. 2018). The abundance of likely septum-localized proteins within E. faecalis MVs suggests that MV formation is a spatiotemporally organized process, where IreK kinase might be involved in MV biogenesis similar to CovS histidine kinase in GAS (Resch et al. 2016). Moreover, by in situ live imaging MVs appeared to be derived from the septal region, further supporting septal vesiculation model.
Supplementary to MV release from septal polyunsaturated PG microdomains, it was formally possible that E. faecalis MVs might be formed through explosive cell lysis mediated by phage tail release, similar to MV formation during bacteriophage release in P. aeruginosa (Turnbull et al. 2016). Indeed, we observed that MVs co-purified with fully-assembled phage tails encoded by a cryptic phage. While we didn't observe significantly changed MV numbers between the WT and ∆pp2 phage mutant, the question of what function these phage tails might confer to E. faecalis now stands. Moreover, in P. aeruginosa, a population of MVs were observed to be attached to cells adjacent to those which underwent lysis, which would not be reflected in MV quantification experiments. Therefore, we cannot yet rule out partial contribution of explosive cell lysis to MV formation in E. faecalis. Stimulating the phage lytic cycle through DNA damage-inducible SOS response and comparing MV concentrations between induced and uninduced bacteria may further address if endolysins contribute to MV formation in E. faecalis.
Finally, we observed that lipoproteins are enriched in MVs, similar to MVs from S. pyogenes and M. tuberculosis (Rath et al. 2013; Biagini et al. 2015). Lipoproteins are potent inducers of the host inflammatory responses through TLR2 receptors, and lipoprotein-rich MVs from M. tuberculosis and C. perfringens activate macrophages, leading to the release of inflammatory cytokines (Prados-Rosales et al. 2011; Jiang et al. 2014; Nguyen et al. 2020). In L. monocytogenes, pheromone cAD1, the homologue of pheromone cAD1 lipoprotein that is enriched in enterococcal MVs, enhances bacterial escape from host cell vacuoles and bacterial virulence (Xayarath, Alonzo and Freitag 2015). We hypothesize that E. faecalis lipoprotein-enriched MVs may contribute to their ability to promote NF-kB activation in macrophages. In addition, it is possible that other MV-associated PAMPs, such as unmethylated prokaryotic DNA which is recognized by TLR9 receptors on host immune system cells, may also lead to NF-kB activation (Du et al. 2000; Dalpke et al. 2006). The precise roles for enterococcal MVs during infection, whether they confer benefits via immune modulation or promoting bacterial growth and survival, are the subject of ongoing investigation.
In summary, we identified that E. faecalis MVs are composed of distinct protein and lipid profiles. The presence of septal proteins and abundance of more flexible polyunsaturated lipids suggest that MVs arise via a regulated process from the septum, where the cell-wall is thinnest and the membrane is enriched in more flexible polyunsaturated microdomains (Doughty et al. 2014; Li et al. 2018). To support the septal biogenesis hypothesis, we observed MVs to appear from the septal region by live imaging. We propose a model for septal MV biogenesis in E. faecalis (Fig. 6). Even though plasmid DNA co-purifies with MVs, plasmids are not packaged within MVs and do not contribute to HGT within- or cross-species. Functionally, MVs activate NF-kB signalling in macrophages, possibly due to the abundance of immunogenic lipoproteins within MVs. Future work on mechanisms of MV formation and comparing vesiculation rate and function of MVs from both commensal and pathogenic E. faecalis strains will enhance our understanding in enterococcal pathogenesis and host–pathogen interactions.
Figure 6.
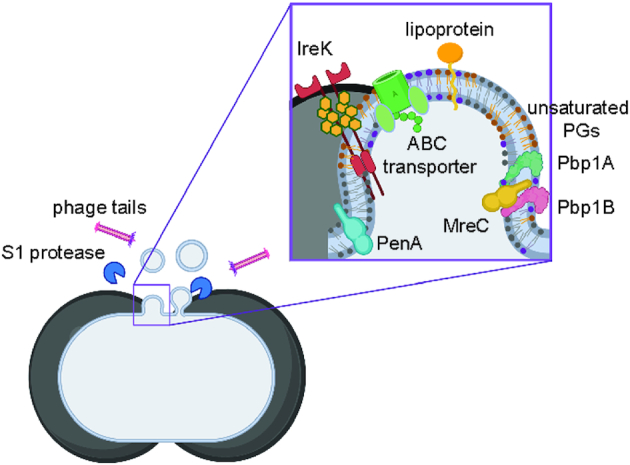
Septal model for MV formation in E. faecalis. Cartoon depiction for MV formation from the septal region, where peptidoglycan is thinnest, during the cell divison. Enterococcus faecalisMVs are enriched in more flexible unsaturated PGs, and have a high abundance of septal proteins—Pbp1A, Pbp1B, MreC, PenA, that are involved in cell division, and IreK kinase, that is bound to un-crosslinked peptidoglycan through PASTA domains. Phage tails and S1 extracellular protease co-purify with MV fraction.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation and Ministry of Education Singapore under its Research Centre of Excellence Programme, as well as the National Research Foundation under its Singapore NRF Fellowship programme (NRF- NRFF2011–11). This work was also supported by a Tier 1 grant sponsored by the Singapore Ministry of Education (MOE2017-T1–001–269).
We are grateful to Jenny Dale and Gary Dunny for supplying us with E. faecalis OG1RF transposon mutants used in this study. We also thankful to the Taplin Biological Mass Spectrometry Facility and Ross Tomaino for the MS analysis. The TEM work was undertaken at the NTU Institute of Structural Biology Cryo-EM lab in Nanyang Technological University, Singapore. We thank Andrew Wong for the assistance in sample prep and imaging. We also thank Annika Sjöström and Monica Persson from Umea University, Sweden for advice on MV isolation and atomic force microscopy. The cartoon model in Fig. 6 was created with BioRender.com.
Contributor Information
Irina Afonina, Singapore Centre for Environmental Life Science Engineering, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore; School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore; Singapore–MIT Alliance for Research and Technology, Antimicrobial Resistance Interdisciplinary Research Group, 1 Create Way, Singapore 138602.
Brenda Tien, Singapore Centre for Environmental Life Science Engineering, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore; School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore.
Zeus Nair, Singapore Centre for Environmental Life Science Engineering, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore; Interdisciplinary Graduate School, Nanyang Technological University, 61 Nanyang Drive, Singapore 637335.
Artur Matysik, Singapore Centre for Environmental Life Science Engineering, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore; School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore.
Ling Ning Lam, Singapore Centre for Environmental Life Science Engineering, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore; School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore.
Mark Veleba, Singapore Centre for Environmental Life Science Engineering, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore; School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore.
Augustine Koh Jing Jie, Singapore Centre for Environmental Life Science Engineering, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore; School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore.
Rafi Rashid, Singapore Centre for Environmental Life Science Engineering, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore; Graduate School for Integrative Sciences & Engineering, National University of Singapore, 21 Lower Kent Ridge Rd, Singapore 119077.
Amaury Cazenave-Gassiot, Singapore Lipidomics Incubator, National University of Singapore, 28 Medical Dr, Singapore 117456; Department of Biochemistry, National University of Singapore, 8 Medical Drive, Block MD7, 117597, Singapore.
Marcus Wenk, Singapore Lipidomics Incubator, National University of Singapore, 28 Medical Dr, Singapore 117456; Department of Biochemistry, National University of Singapore, 8 Medical Drive, Block MD7, 117597, Singapore; Department of Biological Sciences, Faculty of Science, National University of Singapore, 16 Science Drive 4, Singapore 117558.
Sun Nyunt Wai, Umeå Centre for Microbial Research (UCMR), Department of Molecular Biology, Umeå University, 90187 Umeå, Sweden.
Kimberly A Kline, Singapore Centre for Environmental Life Science Engineering, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore; School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore.
Conflict of interest
None declared.
REFERENCES
- Beilharz K, Nováková L, Fadda Det al. Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proc Natl Acad Sci. 2012;109:E905– 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol. 1999;181:4725– 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagini M, Garibaldi M, Aprea Set al. The human pathogen Streptococcus pyogenes releases lipoproteins as Lipoprotein-rich Membrane Vesicles. Mol Cell Proteomics. 2015;2138–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911– 7. [DOI] [PubMed] [Google Scholar]
- Boes A, Olatunji S, Breukink Eet al. Regulation of the peptidoglycan polymerase activity of PBP1b by antagonist actions of the core divisome proteins FtsBLQ and FtsN. mBio. 2019;10:e01912–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch'ng J-H, Chong KKL, Lam LNet al. Biofilm-associated infection by enterococci. Nat Rev Microbiol. 2019;17:82–94. [DOI] [PubMed] [Google Scholar]
- Cooke IR, Deserno M. Coupling between lipid shape and membrane curvature. Biophys J. 2006;91:487–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalpke A, Frank J, Peter Met al. Activation of toll-like receptor 9 by DNA from different bacterial species. Infect Immun. 2006;74:940–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues S, Nielsen KM. Membrane vesicles and horizontal gene transfer in prokaryotes. Curr Opin Microbiol. 2017;38:16–21. [DOI] [PubMed] [Google Scholar]
- Dorward DW, Garon CF, Judd RC. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J Bacteriol. 1989;171:2499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doughty DM, Dieterle M, Sessions ALet al. Probing the subcellular localization of hopanoid lipids in bacteria using NanoSIMS. PLoS One. 2014;9:e84455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Poltorak A, Wei Yet al. Three novel mammalian toll-like receptors: gene structure, expression, and evolution. Eur Cytokine Netw. 2000;11:362–71. [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–14. [DOI] [PubMed] [Google Scholar]
- Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. 2010;74:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore MS, Clewell DB. The Enterococci: pathogenesis, molecular biology, and antibiotic resistance, Vol. 10, Washington, DC: ASM press, 2002, 439. [Google Scholar]
- Grull MP, Mulligan ME, Lang AS. Small extracellular particles with big potential for horizontal gene transfer: membrane vesicles and gene transfer agents. FEMS Microbiol Lett. 2018;365. [DOI] [PubMed] [Google Scholar]
- Gurung M, Moon DC, Choi CWet al. Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. PLoS One. 2011;6:e27958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J, Park SC, Her Jet al. Comparative lipidomic profiling of the human commensal bacterium Propionibacterium acnes and its extracellular vesicles. RSC Adv. 2018;8:15241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Kong Q, Roland KLet al. Membrane vesicles of Clostridium perfringens type A strains induce innate and adaptive immunity. Int J Med Microbiol. 2014;304:431–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandaswamy K, Liew TH, Wang CYet al. Focal targeting by human beta-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc Natl Acad Sci. 2013;110:20230–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klieve AV, Yokoyama MT, Forster RJet al. Naturally occurring DNA transfer system associated with membrane vesicles in cellulolytic ruminococcus spp. of ruminal origin. Appl Environ Microbiol. 2005;71:4248–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristich CJ, Wells CL, Dunny GM. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc Natl Acad Sci. 2007;104:3508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005;19:2645–55. [DOI] [PubMed] [Google Scholar]
- Labbe BD, Kristich CJ. Growth- and Stress-Induced PASTA Kinase Phosphorylation in Enterococcus faecalis. J Bacteriol. 2017;199:e00363–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land AD, Winkler ME. The requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. J Bacteriol. 2011;193:4166–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EY, Choi DS, Kim KPet al. Proteomics in gram-negative bacterial outer membrane vesicles. Mass Spectrom Rev. 2008;27:535–55. [DOI] [PubMed] [Google Scholar]
- Lee EY, Choi DY, Kim DKet al. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics. 2009;9:5425–36. [DOI] [PubMed] [Google Scholar]
- Lee JH, Choi CW, Lee Tet al. Transcription factor σ B plays an important role in the production of extracellular membrane-derived vesicles in Listeria monocytogenes. PLoS One. 2013;8:e73196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TK, Meng K, Shi Het al. Single-molecule imaging reveals modulation of cell wall synthesis dynamics in live bacterial cells. Nat Commun. 2016;7:13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew MD, Lee SF, Ptacin JLet al. Three-dimensional superresolution colocalization of intracellular protein superstructures and the cell surface in live Caulobacter crescentus. Proc Natl Acad Sci. 2011;108:E1102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Klein MI, Heim KPet al. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol. 2014;196:2355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Yuan X-X, Sun H-Met al. Atomic force microscopy of side wall and septa peptidoglycan from bacillus subtilis reveals an architectural remodeling during growth. Front Microbiol. 2018;9:620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Koch G. Exploring functional membrane microdomains in bacteria: an overview. Curr Opin Microbiol. 2017;36:76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride SM, Fischetti VA, LeBlanc DJet al. Genetic diversity among Enterococcus faecalis. PLoS One. 2007;2:e582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63:545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WR, Murray BE, Rice LBet al. Vancomycin-Resistant Enterococci: therapeutic Challenges in the 21st Century. Infect Dis Clin North Am. 2016;30:415–39. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Soe TT, Maxfield FR. Endocytic sorting of lipid analogues differing solely in the chemistry of their hydrophobic tails. J Cell Biol. 1999;144:1271–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M-T, Matsuo M, Niemann Set al. Lipoproteins in Gram-Positive bacteria: abundance, function, fitness. Front Microbiol. 2020;11:2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HV, Guiton PS, Kline KAet al. The metal ion-dependent adhesion site motif of the Enterococcus faecalis EbpA pilin mediates pilus function in catheter-associated urinary tract infection. MBio. 2012;3:e00177–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathirana RD, Kaparakis-Liaskos M. Bacterial membrane vesicles: biogenesis, immune regulation and pathogenesis. Cell Microbiol. 2016;18:1518–24. [DOI] [PubMed] [Google Scholar]
- Prados-Rosales R, Baena A, Martinez LRet al. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J Clin Invest. 2011;121:1471–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MM, McFadden G. Modulation of NF-κB signalling by microbial pathogens. Nat Rev Microbiol. 2011;9:291–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid R, Cazenave-Gassiot A, Gao IHet al. Comprehensive analysis of phospholipids and glycolipids in the opportunistic pathogen Enterococcus faecalis. PLoS One. 2017;12:e0175886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid R, Veleba M, Kline KA. Focal targeting of the bacterial envelope by antimicrobial peptides. Front Cell Dev Biol. 2016;4:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath P, Huang C, Wang Tet al. Genetic regulation of vesiculogenesis and immunomodulation in Mycobacterium tuberculosis. Proc Natl Acad Sci. 2013;110:E4790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch U, Tsatsaronis JA, Rhun ALet al. A Two-Component regulatory system impacts extracellular Membrane-Derived vesicle production in group a streptococcus. mBio. 2016;7:e00207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera J, Cordero RJB, Nakouzi ASet al. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc Natl Acad Sci. 2010;107:19002–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbo C, Fernández-Moreira E, Merino Met al. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55:3084–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabra W, Lunsdorf H, Zeng AP. Alterations in the formation of lipopolysaccharide and membrane vesicles on the surface of Pseudomonas aeruginosa PAO1 under oxygen stress conditions. Microbiology. 2003;149:2789–95. [DOI] [PubMed] [Google Scholar]
- Schenk M, Belisle JT, Modlin RL. TLR2 looks at lipoproteins. Immunity. 2009;31:847–9. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Kida Y, Kuwano K. A triacylated lipoprotein from Mycoplasma genitalium activates NF-kappaB through Toll-like receptor 1 (TLR1) and TLR2. Infect Immun. 2008;76:3672–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöström AE, Sandblad L, Uhlin BEet al. Membrane vesicle-mediated release of bacterial RNA. Sci Rep. 2015;5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CK, Glaesmann M, Grimm JBet al. A toolbox for multiplexed super-resolution imaging of the E. coli nucleoid and membrane using novel PAINT labels. Sci Rep. 2018;8:14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl H, Bürmann F, Hamoen LW. The actin homologue MreB organizes the bacterial cell membrane. Nat Commun. 2014;5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweere JM, Van Belleghem JD, Ishak Het al. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science. 2019;363:eaat9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien BYQ, Goh HMS, Chong KKLet al. Enterococcus faecalis Promotes Innate Immune Suppression and Polymicrobial Catheter-Associated Urinary Tract Infection. Infect Immun. 2017;85:e00378–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku M, Cárcamo-Oyarce G, Yamamoto Tet al. Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat Commun. 2017;8:481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku M, Nomura N, Eberl L. Types and origins of bacterial membrane vesicles. Nat Rev Microbiol. 2019;17:13–24. [DOI] [PubMed] [Google Scholar]
- Tran TT, Panesso D, Mishra NNet al. Daptomycin-Resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. mBio. 2013;4:e00281–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull L, Toyofuku M, Hynen ALet al. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun. 2016;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belleghem JD, Dąbrowska K, Vaneechoutte Met al. Pro- and anti-inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Sci Rep. 2017;7:8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu J, Carvalho J. Enterococcus: review of its physiology, pathogenesis, diseases and the challenges it poses for clinical microbiology. Front Biol. 2011;6:357–66. [Google Scholar]
- Wagner T, Joshi B, Janice Jet al. Enterococcus faecium produces membrane vesicles containing virulence factors and antimicrobial resistance related proteins. J Proteomics. 2018;187:28–38. [DOI] [PubMed] [Google Scholar]
- Xayarath B, Alonzo F III, Freitag NE. Identification of a Peptide-Pheromone that Enhances Listeria monocytogenes Escape from Host Cell Vacuoles. PLoS Pathog. 2015;11:e1004707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa H, Osaki T, Woo Tet al. Analysis of outer membrane vesicle protein involved in biofilm formation of Helicobacter pylori. Anaerobe. 2011;17:388–90. [DOI] [PubMed] [Google Scholar]
- Zhang L, Hou X, Sun Let al. Staphylococcus aureus bacteriophage suppresses LPS-induced inflammation in MAC-T bovine mammary epithelial cells. Front Microbiol. 2018;9:1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatkov N, Nadeem A, Uhlin BEet al. Eco-evolutionary feedbacks mediated by bacterial membrane vesicles. FEMS Microbiol Rev. 2020;1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchini L, Mercy C, Garcia PSet al. PASTA repeats of the protein kinase StkP interconnect cell constriction and separation of Streptococcus pneumoniae. Nat Microbiol. 2018;3:197–209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.