ABSTRACT
Extracellular vesicles (EVs) are lipidic nanosized particles that deliver a highly complex molecular cargo between cells and organisms and may serve numerous functions in intercellular communication, thereby influencing the evolution of microbial communities. Their roles in infectious diseases have been studied for a long time, comprising viral, bacterial, parasitic and to a less extent, fungal infections. Over the last few years, fungal EVs have become an increasingly active research field. Nevertheless, the understanding of EV functions during fungal infections poses challenging points, comprising the genetics regulating EV release, the EV structural and compositional complexity, the heterogeneity of the EV populations and their impact on host-pathogen interactions. This review explores the state-of-the-art investigations on fungal EVs and how this fast-evolving field can impact the development of new tools to fight fungal infections.
Keywords: fungi, extracellular vesicles, pathogens, intercellular communication, virulence, interkingdom communication
The study of fungal extracellular vesicles is a fast evolving field which will impact the development of new tools to fight fungal infections in the future.
INTRODUCTION
Extracellular vesicles (EVs) are membranous compartments that enclose diverse biomolecules and are released by virtually all living cells in the three domains of life (Gill, Catchpole and Forterre 2019). EVs isolated in different studies vary in size, cargo, density and morphology and originate either from the plasma membrane or the endocytic trafficking pathway (Mathieu et al. 2019). Extracellular vesicles have been suggested as universal communication players in the microbial world (Tsatsaronis et al. 2018). They were reported as circulating particles during the development of different human pathologies and infectious diseases (Schorey and Harding 2016; Szempruch et al. 2016; Hosseini-Beheshti and Grau 2018; Shah, Patel and Freedman 2018), as well as in different environmental niches, such as marine ecosystems (Biller et al. 2014). Three major EV subtypes have been studied and discussed by several groups: exosomes, microvesicles and apoptotic bodies. Exosomes (30–150 nm) are formed in the endosomal pathway through the inward budding of endosomal membranes during their maturation into multivesicular bodies (MVB) that bind to the plasma membrane and release the intraluminal vesicles (ILVs) through exocytosis (Gandham et al. 2020). Microvesicles (100–1000 nm) are formed by the outward budding of the plasma membrane, and apoptotic bodies are vesicles formed during apoptosis which are usually larger than 1000 nm (Gandham et al. 2020).
EVs were first described as ‘platelet dust’ by Peter Wolf in 1967, and for a long time, they were considered debris of damaged cells (Wolf 1967). However, over the past few decades, EVs were shown to be not simply inert cellular products but biologically active particles instead, regulating cell physiology, delivering virulence-associated molecules, mediating the long-distance transfer of genetic information, among other complex functions (Mathieu et al. 2019; Pegtel and Gould 2019). Extracellular vesicle release has been reported in various microorganisms, such as bacteria, archaea, protozoa, virus-infected cells and fungi and the impact of EV production during host–pathogen interactions is an active research area and has been extensively reviewed (Szempruch et al. 2016; Raab-Traub and Dittmer 2017; Freitas et al. 2019; Gill, Catchpole and Forterre 2019; de Souza and Barrias 2020; Torrecilhas et al. 2020; Rizzo, Rodrigues and Janbon 2020a; Piffer et al. 2021; Sabatke et al. 2021).
In fungi, ultrastructural analysis of Aspergillus nidulans protoplasts carried out early in the 1970s showed EV-like particles pinching off from fungal plasma membrane, then referred to as ‘subprotoplasts’ (Gibson and Peberdy 1972). The presence of secreted vesicles was then indicated in 1973 in the fungal pathogen Cryptococcus neoformans (Takeo et al. 1973). After these initial microscopical evidences, EV structure and composition were first described more than 30 years later in C. neoformans (Rodrigues et al. 2007, 2008) and the number of studies characterizing EVs in fungi have been incredibly increasing since then. Fungal EVs have been described in more than 50 independent studies comprising pathogenic and nonpathogenic species, and the timeline concerning most of these reports was recently reviewed (de Oliveira et al. 2020; Rizzo, Rodrigues and Janbon 2020a).
Extracellular vesicle production is a relevant mechanism of virulence-associated secretion of molecules. It has been described in most of the primary, opportunistic and emerging fungal pathogens, comprising species from the genera Candida, Cryptococcus, Histoplasma, Aspergillus, Sporothrix, Paracoccidiodes, Malassezia, Rhizopus, Exophiala, Pichia, Trichophyton and Talaromyces (Albuquerque et al. 2008; Gehrmann et al. 2011; da Silva et al. 2015; Gil-Bona et al. 2015; Bitencourt et al. 2018; Leone et al. 2018; Liu et al. 2018; Peres da Silva et al. 2019; Souza et al. 2019; Brauer et al. 2020; Karkowska-Kuleta et al. 2020; Lavrin et al. 2020; Rizzo et al. 2020b; Yang et al. 2021). EVs were also characterized in many phytopathogens such as Alternaria infectoria (Silva et al. 2014), Trichoderma reesei (de Paula et al. 2019), Fusarium oxysporum f. sp. vasinfectum (Bleackley, Dawson and Anderson 2019), Zymoseptoria tritici (Hill and Solomon 2020) and Penicillium digitatum (Costa et al. 2021). They have also been described in numerous nonpathogenic species such as Saccharomyces cerevisiae (Albuquerque et al. 2008), the industrial fungus T. reesei (de Paula et al. 2019) and different wine yeast species, including Torulaspora delbrueckii, Lachancea thermotolerans, Hanseniaspora uvarum, Candida sake and Metschnikowia pulcherrima (Mencher et al. 2020).
The increasing number of publications highlights the interest of the scientific community in fungal EVs and the relevance of these particles in many contexts, including fungal pathophysiology and host interactions (Rodrigues and Casadevall 2018; Zamith-Miranda et al. 2018; de Oliveira et al. 2020; Rizzo, Rodrigues and Janbon 2020a; Piffer et al. 2021), as well as food biotechnology (Morales et al. 2021). However, studies on fungal EV functions are hindered by the limited understanding of their biogenesis, composition and structural diversity. This review explores the recent advances on fungal EVs, focusing on EVs derived from human fungal pathogens.
EVs IN FUNGI: BIOGENESIS, COMPOSITION, STRUCTURE AND CARGO LOADING REGULATION
EV biogenesis
Based on the analogy to EV biogenesis in mammalian cells and the analysis of their structure and cargo, it has been hypothesized that fungal EVs could have different cellular sites of origin as reported earlier (Bielska and May 2019; de Oliveira et al. 2020; Rizzo, Rodrigues and Janbon 2020a). Electron microscopic approaches revealed that these sites could include the plasma membrane, with events of plasma membrane reshaping and cytoplasmic subtraction (Rodrigues et al. 2013) or the direct vesicle shedding (Rizzo, Rodrigues and Janbon 2020a). It was also suggested that EVs could be originated from vesicles-containing vacuoles or via endosome-derived MVB that fuse with the plasma membrane and release EVs to the extracellular space (Rodrigues et al. 2008). It was also recently suggested that other organelles, such as the intracellular vesicles clusters (IVCs), are important sites for selecting the EV-associated cargo proteins in S. cerevisiae (Winters, Hong-Brown and Chiang 2020).
Despite many reports suggesting potential mechanisms of EV production in fungi, mutant strains that cannot produce EVs are yet to be described, suggesting either functional redundancy of genes and pathways implicated in their formation or essentiality of EV production for membrane biosynthesis and consequently for fungal life (Coelho and Casadevall 2019). Nevertheless, many studies suggest that intracellular trafficking might regulate EV formation. In this regard, some members of the Endosomal Sorting Complex Required for Transport (ESCRT) machinery (Hurley 2015) might be implicated in the control of EV production in fungi, as shown in mammalian cells (Mathieu et al. 2019). For instance, Vps2, Vps23, Vps27, Vps36 and Snf7, among others, were shown to regulate the formation, selection of cargo or proper release of EVs in different fungal models such as S. cerevisiae (Oliveira et al. 2010b, Zhao et al. 2019), Candida albicans (Zarnowski et al. 2018) and C. neoformans (Park et al. 2020). Accordingly, some of the corresponding mutant strains produced less EVs than the wild type (WT) without being completely deficient in EV production. Similarly, the autophagy regulator Atg7 and the Golgi reassembly stacking (GRASP) were also suggested to affect EV size. GRASP was also reported to be involved in the transport of vesicular mRNA (Peres da Silva et al. 2018). Recently, the endocytic adaptor protein Cin1 involved in intracellular vesicular traffic in Cryptococcus deneoformans has been shown to regulate EV-mediated RNA release (Liu et al. 2020). Also, the anti-fungal drug turbinmicin has been shown to inhibit EV production in C. albicans and biofilm formation (Zhao et al. 2021). The mechanism of action of turbinmicin is not completely identified. Still, it seems to target a Sec14-dependent intracellular vesicle trafficking pathway, suggesting that the post-Golgi secretory pathways could also be involved in EV production in this yeast (Zhang et al. 2020).
Other proteins implicated in lipid metabolism and transport, such as the lipid flippase APT1 (Rizzo et al. 2014) and the lipid scramblase AIM25 (Reis et al. 2019) in Cryptococcus, as well as the lipid biosynthetic phosphatidylserine decarboxylase encoding genes, PSD1 and PSD2 (Wolf et al. 2015) in C. albicans were also reported to regulate EV diversity and composition. Similarly, the sterol C-24 methyltransferase encoding gene ERG6, involved in ergosterol biogenesis, regulates EV size and cargo in C. neoformans (de Oliveira et al. 2020). Overall, the above studies suggest that several EV production pathways as well as complex cargo sorting mechanisms exist in fungi.
EV composition
Fungal EVs enclose a highly diverse cargo, comprising lipids, proteins, carbohydrates, pigments, nucleic acids, prions, toxins and other small molecules (Rodrigues et al. 2007, 2008; Albuquerque et al. 2008; Vallejo et al. 2012b; da Silva et al. 2015; Gil-Bona et al. 2015; Kabani and Melki 2015; Vargas et al. 2015; Costa et al. 2021; Reis et al. 2021). However, despite significant advances in new isolation strategies and characterization methods, such as powerful electron microscopy techniques and high-sensitivity nanoparticle analyses (Gandham et al. 2020), the characterization of fungal EV composition is still in its infancy. The major components identified in EVs from the most relevant fungal pathogens are described below and summarized in Fig. 1. It is important to highlight that most compositional studies on fungal EVs have not used any enrichment analysis, which can hinder downstream studies. Moreover, although different gradient-based approaches have been suggested to increase the EV purity in mammals (Roux et al. 2020), only a few reports using such strategies have been published in fungi (Rodrigues et al. 2007; Yang et al. 2021). Thus, most published EV components should be considered better as EV-associated until their genuine EV nature could be more carefully addressed. In this sense, a standardization of fungal EV purification protocols is necessary in order to identify fungal EVs markers as well as for compositional and functional analyses.
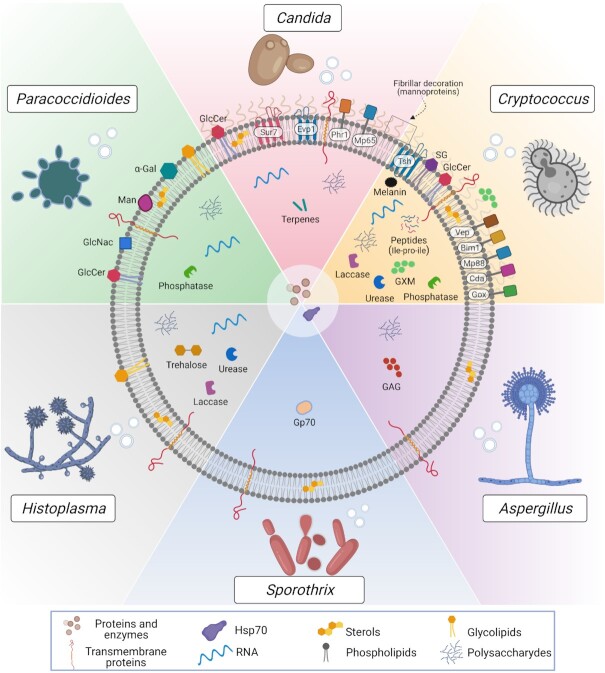
Figure 1.
Compositional aspects of EVs released by the main fungal pathogens. EVs are nanoscale lipid bags enclosing a highly complex molecular cargo. EVs have been studied in many pathogenic fungi, including yeast and filamentous species, such as Cryptococcus, Candida, Paracoccidioides, Histoplasma, Sporothrix and Aspergillus. The structure and compositional characterization of fungal EVs are still far from being complete. However, in this scheme, we aimed to summarize the major surface and intraluminal EV components, comprising proteins, polysaccharides, lipids, nucleic acids, small molecules and pigments. Some of the proteins are predicted to be GPI-anchored, such as Phr1 (in C. albicans), and Mp88, Vep (Vesicles enriched protein-4 and 5) and other members of the chitin deacetylase family (Cda), the putative glyoxal oxidase family (Gox) and the BCS-inducible membrane protein 1 (Bim1) in Cryptococcus. Interestingly, many of these proteins have already been described as highly immunogenic. Other transmembrane proteins, such as Sur7 (C. albicans) and Tsh (Cryptococcus), are tetraspanins, considered biomarkers of mammalian EVs. Many of those membrane/GPI-anchored proteins are mannosylated and suggested as the molecular composition of the fibrillar decoration on the surface of C. albicans and C. neoformans EVs revealed by electron microscopy. This mannoprotein-based decoration is not exclusive of pathogenic species since it was also detected in the model yeast S. cerevisiae. The presence of this feature needs to be further explored for other pathogens. Carbohydrates involved with stress response were found in Histoplasma capsulatum EVs, such as trehalose, and immunogenic polysaccharides were described in fungal EVs, such as the major capsular polysaccharide Glucuronoxylomannan (GXM) in Cryptococcus and the exopolysaccharide Galactosaminogalactan (GAG) in Aspergillus fumigatus. Glucosyl residues were found on the surface of Paracoccidiodes EVs, such as mannose (Man) and N-acetylglucosamine (GlcNAc), together with the immunogenic α-linked galactopyranosyl (α-Gal). Some enzymes were biochemically described as active in fungal EVs, including urease, laccase and phosphatase. Besides the presence of laccase, it has been suggested that its final product, the pigment melanin, was also enclosed in C. neoformans EVs. Actually, a model of EV-based melanization has been proposed in C. neoformans (Eisenman et al. 2009). However, additional experiments are needed to confirm it. Similarly, we cannot exclude the presence of other enzymes detected by proteomics, such as chitinases and cell wall remodeling enzymes, but their activities remain to be confirmed. Some other possible EV-luminal components, such as the 70-kDa glycoprotein (Gp70) in Sporotrix brasiliensis were shown to be antigenic, and the peptide leucine-proline-isoleucine (Ile-Pro-Ile) was reported to be biologically active during C. neoformans in vivo infection. Among the lipids, many phospholipids, sterols and glycolipids were described, comprising the glycosphingolipid Glucosylceramide (GlcCer) and the sterylglucoside (SG); the latter was detected only in C. neoformans. Moreover, terpenes were suggested as the major EVs components inhibiting yeast-to-hyphae transition in C. albicans. However, it is unknown if this class of lipids would be associated with membranes or in the lumen of the vesicles. Moreover, although more abundantly found in C. albicans, some derivatives were also present in H. capsulatum and S. cerevisiae EVs. Few other EVs-associated structural lipids were not explored as their biological functions are not deciphered so far. By analyzing the EV-associated proteins among different pathogenic species, it is suggested that the ortholog group of the Heat-shock protein (Hsp70) was the only one found in all these species. For clarity, all the RNA sub-classes identified in fungal EVs were not explored in this figure. The vesicular RNA diversity is detailed in plain text.
Proteins
Different studies exploring fungal EV proteomics were recently discussed (Bleackley, Dawson and Anderson 2019; de Toledo Martins et al. 2019). Overall, proteins related to cell wall biogenesis, plasma membrane, stress responses, pathogenesis, transport and signaling are the most commonly identified in EVs from multiple species (Nimrichter et al. 2016; Bleackley, Dawson and Anderson 2019). Notably, the activity of many virulence-associated enzymes was identified in fungal EVs. In C. neoformans, laccase, as well as urease and acid phosphatase activities, were described to be associated with EVs (Rodrigues et al. 2008). Interestingly, it has been recently shown that Vps27, a key protein of the ESCRT-0 complex essential for virulence in C. neoformans, is required for the laccase transport to the cell wall (Park et al. 2020). A vps27Δ mutant also accumulated MVB and showed a partial defect in EV production, releasing EVs with larger size distribution than wild type, emphasizing the hypothesis that EV-mediated transport of enzymes is an important mechanism for fungal pathogenesis. The presence of active enzymes associated with EVs was also observed in other species, such as a laccase in Paracoccidoides brasiliensis (Vallejo et al. 2012a), as well as a laccase and an urease in H. capsulatum (Baltazar et al. 2018). A β-glucosidase was also reported to be associated with EVs in T. reesei(de Paulaet al.2019).
A total of two recent EV-protein enrichment analyses suggest the existence of EV-marker proteins in fungi. In C. albicans, a list of 22 proteins as EV markers, including the claudin-like Sur7 family proteins (including Sur7 and Evp1) and the GPI-anchored glycosidase Phr1, have been suggested (Dawson et al. 2020). In Cryptococcus, 17 EV-associated proteins in all three species, C. neoformans, C. deneoformans and Cryptococcus deuterogattii represent potential cryptococcal EV markers (Rizzo et al. 2021). Additionally, six membrane-associated protein families were described as typically associated with these EVs, including the chitin deacetylase (Cda) family, the Ricin-type beta-trefoil lectin domain-containing protein (Ril) family, the putative glyoxal oxidase (Gox) family, the tetraspanin membrane proteins containing a SUR7/PalI family motif (Tsh), the pr4/barwin4 domain protein (Blp) family and the multicopper oxidase (Cfo) (Rizzo et al. 2021). Interestingly, some of these proteins were described as immunogenic, such as Mp65 in C. albicans (Gomez et al. 2000) and Mp88 in C. neoformans (Levitz et al. 2001; Huang et al. 2002). Antigenic proteins were also identified for other fungal EVs, such as the 70-kDa glycoprotein (Gp70) in S. brasiliensis (Ikeda et al. 2018), which was reported as an important virulence factor in Sporothrix (de Almeida et al. 2015).
Using an orthology approach, a comparative analysis of EV protein content from multiple fungal species has been recently conducted (Parreira et al. 2021). The data were compiled in a novel free web repository created for fungal EV datasets, named ExVe (http://exve.icc.fiocruz.br). Interestingly, the OG6_100083 (Hsp70 Pfam domain) was the only ortholog group found in all considered species, suggesting that heat-shock related proteins might play a universal role in fungal EVs (Parreira et al. 2021).
Lipids
The lipid composition of fungal EVs is still poorly described. The published data are also difficult to compare and integrate due to the diversity of strategies employed in these analyses. Nevertheless, lipid analysis by mass spectrometry revealed that virulence-associated glycosphingolipids, such as glucosylceramide (GlcCer) and sterols are enriched in cryptococcal EVs (Rodrigues et al. 2007). Similarly, in C. albicans EVs, the major lipids detected by thin-layer chromatography were sterols (ergosterol and lanosterol) and GlcCer (Vargas et al. 2015)., Mass spectroscopy identified many phospholipids in H. capsulatum EVs (Albuquerque et al. 2008). More recently, lipidomic analysis detected on hundred different lipids, comprising phospholipids, triacylglycerides (TG), diacylglycerides (DG), ceramides, sphingomyelins and fatty acids (FA), suggesting a very complex organization of the lipidic membrane of EVs (Cleare et al. 2020). Interestingly, the EV lipid profile obtained from cells grown in distinct media was remarkably different (Cleare et al. 2020). Similarly, a detailed EV lipidomic analysis performed in P. brasiliensis revealed two species of monohexosylceramide and 33 phospholipids, together with glycolipids such as GlcCer, and sterols like brassicasterol, ergosterol and lanosterol (Vallejo et al. 2012a).
Thus, the knowledge of the lipid content in fungal EVs appears today as a very long list of lipid classes without any associated function. A better understanding of fungal lipid components is fundamental since many EV-associated lipids can be closely linked to virulence during infection, as for the immunogenic glycolipid sterylglucoside (SG), which seems to be enriched in C. neoformans EVs and can induce protection of the invertebrate host Galleria mellonella against a lethal challenge with C. neoformans (Colombo et al. 2019). Similarly, C. albicans EVs can inhibit yeast-to-hyphae differentiation, this effect being mediated by EV associated lipids (Honorato et al. 2021).
Carbohydrates
Approaches to characterize the fungal EV glycol-profile have also been undertaken. In C. neoformans immunogold labeling using a specific monoclonal antibody suggested that the presence of the capsular polysaccharide glucuronoxylomannan (GXM) associated with EVs (Rodrigues et al. 2007). Similarly, in 2011, enzyme-linked immunosorbent assays (ELISA) revealed the presence of immunogenic α-linked galactopyranosyl (α-Gal) epitopes associated with P. brasiliensis EVs (Vallejo et al. 2011). Subsequently, Paracoccidioides EV fractions were exhibited to contain residues of glucose (Glc), mannose (Man) and galactose (Gal), comprising a high molecular mass α-glucan and a galactofuranosylmannan, which were resolved through high-resolution analyses and determined to dominate the population of EV polysaccharides (da Silva et al. 2015). The presence of small amounts of cell wall-associated polysaccharides such as (1-3)-glucan and (1-6)-glucan was also confirmed. Moreover, plant lectin microarray profiling revealed terminal Man and N-acetylglucosamine (GlcNAc) residues exposed at the EV surface (da Silva et al. 2015).
It was also suggested that the vesicular polysaccharide content (specifically mannan and glucan) could be a major source of the extracellular polymeric matrix material in C. albicans biofilms (Zarnowski et al. 2018). In this study, exogenous wild type EVs were able to rescue the matrix production in ESCRT mutant biofilms. In A. fumigatus, it was also shown that protoplasts submitted to cell-wall regeneration change their carbohydrate-associated EV profile compared to non-regeneration conditions (Rizzo et al. 2020b). Thus, the immunogenic polysaccharide galactosaminogalactan (GAG) was detected in EVs during cell wall synthesis but not in control, revealing that the EV carbohydrate composition changes during fungal cell wall biosynthesis. The amount and diversity of EV-associated carbohydrates is also dependent on the conditions of fungal growth. For instance, EVs obtained from H. capsulatum cells grown on Ham's medium presented the highest amount of carbohydrates associated with energy production and storage, whereas trehalose, a stress tolerance related carbohydrate, was more abundant in EVs obtained from cells grown on BHI medium (Cleare et al. 2020).
Prions, small molecules and mycotoxins
In fungi, the EV metabolome is generally poorly described. Nervertheless, the presence of EV-associated toxins was suggested in few fungal models, such as the cotton pathogen F. oxysporum f. sp. vasinfectum (Bleackley, Dawson and Anderson 2019). In the phytopathogen P. digitatum, untargeted metabolomics have been identified as EV-metabolite, including tryptoquialanines and the mycotoxin fungisporin, which are important small molecules for plant pathogenesis (Costa et al. 2021). By using chromatographic and spectrometric approaches and metabolomic techniques, small molecules were identified in EVs obtained from the pathogen C. deuterogattii. One of these vesicular peptides, chemically characterized as leucine-proline-isoleucine (Ile-Pro-Ile), was reported as biologically active during fungal infection in the G. mellonella model (Reis et al. 2021). These recent studies suggest that EV-associated small molecules could play major roles in host–pathogen interactions and pave the way for further discoveries in this field.
Finally, it has been demonstrated that EVs can also export proteinaceous infectious particles (prions), such as the prion Sup35p (Kabani and Melki 2015). Sup35p is exported in S. cerevisiae EVs in its soluble and aggregated infectious states, indicating that prions can also be transmitted between cells through fungal EVs.
RNA
For more than a decade, the composition, function and diversity of RNA molecules in EVs has been the focus of a large number of studies in mammals and in some microorganisms. Accordingly, in 2015, a solid sequencing approach was used to identify several EV-associated RNA sub-classes by C. neoformans, P. brasiliensis, C. albicans and S. cerevisiae (da Silva et al. 2015). They identified mRNAs, microRNA-like (miRNA-like) and several sub-classes of non-coding RNAs (ncRNAs), including transfer RNA (tRNA), ribosomal RNA (rRNA), small nucleolar RNA (snoRNAs) and small nuclear RNA (snRNA)s. Since then, few studies reported additional RNA sub-classes in fungal EVs depending on the employed sequencing strategy. For instance, Malassezia sympodialis produces exosome-like vesicles called MalaEx, which carry very small RNAs of unknown function in a size range of 16–20 nucleotides (Rayner et al. 2017). In H. capsulatum, RNA molecules larger than 200 nucleotides such as mRNAs, rRNAs, tRNAs and snRNAs, and snoRNAs were identified (Alves et al. 2019). A group of small RNAs (25–40 nucleotides) was also identified in the same study. These RNAs antisense of several mRNAs encoding proteins of unknown function has been suggested to belong to the miRNA sub-class (Alves et al. 2019). More recently, the transcriptome of C. deneoformans EVs was characterized, and mRNAs and miRNAs-like associated with these particles were confirmed (Liu et al. 2020). This study also identified siRNAs and lncRNAs as the major components of the C. deneoformans extracellular transcriptome (Liu et al. 2020). Accordingly, several RNA binding proteins (RBPs) including the main components of the RISC complex were identified in the EV proteome from H. capsulatum (Alves et al. 2019).
Although data sets describing the fungal EV transcriptome are still small, several reports suggest its plasticity in response to environmental cues and genetic mutations, as well as a large diversity between fungal species and even strains. Thus, several differences in the composition and abundances of ncRNAs and mRNAs have been shown between the EV transcriptome of two P. brasiliensis strains and the one of Paracoccidoides lutzii (Peres da Silva et al. 2019). Similarly, among the 124 mRNAs identified in H. capsulatum EVs, 93 were enriched in the highly virulent G217B strain and 31 in the less virulent G186AR strain (Alves et al. 2019). Very recently, the analysis of Candida auris EV transcriptomes in the presence or absence of caspofungin revealed a dynamic composition of EV specific transcripts (Zamith-Miranda et al. 2020). Moreover, mutations in GRASP, ATG7 or CIN1 have been shown to affect the EV-transcriptome in Cryptococcus, suggesting a tight control of its composition (Peres da Silva et al. 2018; Liu et al. 2020).
As for other mammalian cell-derived EVs, identifying diverse RNA sub-classes associated with EVs raises many questions regarding the life cycle of exRNAs in fungi, including the molecular mechanisms underlying their specific loading in EVs and their biological functions (Gruner and McManus 2021). These recent reports also highlight the need for more studies on the characterization of exRNAs associated with different EV subpopulations in fungi.
EV structure
Despite initial reports describing differences in the morphology of fungal EV populations (Albuquerque et al. 2008; Rodrigues et al. 2008), the EV structural diversity remained largely unknown until recently. Thus, recent Cryo-electron microscopy (Crypo-EM) analyses revealed that most Cryptococcus, C. albicans and S. cerevisiae EV lipid bilayer are superficially covered by a mixture of molecules assuming a fibrillar decoration, mostly composed of membrane-associated mannoproteins (Rizzo et al. 2021). Yet, about 10% of the EVs are not decorated. On average, these EVs are smaller, suggesting different biosynthetic pathways (Rizzo et al. 2021). In A. fumigatus, electron microscopy (EM) analysis suggested the presence of fibril-like structure of unknown composition covering EVs extracted from cells that grow under conditions that stimulate cell wall synthesis, suggesting that EV structural changes might also be associated with the cellular physiological status (Rizzo et al. 2020b).
The presence of mannoproteins in EV preparations was also reported in other fungal pathogens (Dawson et al. 2020; Karkowska-Kuleta et al. 2020). However, in M. sympodialis, which is largely devoid of GPI-anchor proteins and many other fungal typical mannoproteins (Gioti et al. 2013), EVs exhibit no evidence of decoration on their surface (Johansson et al. 2018). This highlights significant structural diversity in fungal EVs. Many of those fungal-EV associated mannoproteins were previously described as immunogenic proteins (Gomez et al. 2000; Specht et al. 2017), suggesting that these surface decorations might impact host–pathogen interactions and disease development. Glycans are also a common feature for many types of other non-fungal cell-derived EVs (Williams et al. 2018) and different structures covering the vesicular surfaces were also revealed by Cryo-EM; for instance, in myxobacteria (Goes et al. 2020), virus-infected cells (Yang et al. 2020), human cell lines (Noble et al. 2020) and EVs obtained from human fluids such as urine (Wang et al. 2019). However, the molecular composition and the biological relevance of these structures remain to be explored.
The fungal EVs size ranges from 10 to 1000 nm, but the observed distribution depends on the technique employed for size measurements (Bielska and May 2019; Piffer et al. 2021). It is important to note that larger vesicles (>400 nm) and very small EVs (<50 nm) are not well quantified in most Nanoparticle Tracking Analyses (NTA), and small EVs are not easy to detect by most common flow cytometers. Therefore, a combination of complementary techniques providing single EV imaging and single-particle analyzers is recommended to explore the diversity of EV structure (Théry et al. 2018). The most reliable technique to estimate EV size is Cryo-EM, with the caveat that it can only be performed on a limited number of samples. In C. neoformans, for instance, the combination of high-resolution flow cytometry and Cryo-EM techniques allowed the identification of a prominent population of EVs smaller than 100 nm, contrasting to the previous NTA and DLS analyses (Rizzo et al. 2021).
EV cargo loading regulation
Although the underlying mechanisms of EV composition regulation remain unknown, the differential loading of specific or enriched EV molecules seems to be influenced by numerous conditions, such as the environment and nutrition status, including conditions posed by the host cells. For instance, in the dimorphic fungus H. capsulatum, distinct media conditions alters EV characteristics and payloads, such as the sorting of organic molecules (Cleare et al. 2020). Similarly, in C. deneoformans, nutrient availability interferes with EV size and cargo (Marina et al. 2020). It has been shown that EVs obtained from cells grown on nutrient-poor medium have a higher hydrodynamic diameter, increased activity of virulence-associated enzymes and increased amount of capsular polysaccharide GXM, as compared to EVs obtained from cells grown on rich-medium (Marina et al. 2020). H. capsulatum EV protein load and release are modulated by specific antibody binding to fungal cell surface proteins (Matos Baltazar et al. 2016). The binding of these monoclonal antibodies eventually alter EV-associated laccase activity, thereby influencing the pathophysiological roles of these EVs during the host-pathogen interaction (Baltazar et al. 2018).
Some other host factors can also affect EV stability. For instance, serum albumin could lead to the disruption of C. neoformans EVs (Wolf, Rivera and Casadevall 2012). Moreover, the mammalian β-galactoside-binding protein Galectin-3 (Gal-3), which is increased during cryptococcal infection, was described to inhibit C. neoformans growth and exerts a direct lytic effect on fungal EVs, thereby reducing EV stability and possibly changes the course of infection (Almeida et al. 2017). Similarly, increasing concentrations of Gal-3 disrupted P. brasiliensis EVs in a dose-dependent manner (Hatanaka et al. 2019).
EVs MEDIATING INTERCELLULAR AND INTER-KINGDOM COMMUNICATION PROCESSES
By analogy with their mammalian counterparts, fungal EVs are thought to be a mean for communication in vivo and in vitro between cells of the same species and with the host during infection. A few examples of EV-based communication in fungi have been reported. In 2018, transwell assays were used to demonstrate that EVs released by C. deuterogattii virulent strains can be taken up by infected macrophages where they promote intracellular yeast replication of a less virulent strain. This effect appears to be dependent on some proteins and RNAs, as suggested through proteinase K and RNAse EV treatments (Bielska et al. 2018). These results suggest EV-based long-distance pathogen-to-pathogen communication regulating virulence in this fungal pathogen. This hypothesis is enforced by in vivo studies, where pre-incubation of poorly virulent C. neoformans cells with EVs produced by a hypervirulent strain increases the virulence of the recipient cells when tested in a G. mellonella larvae model of infection (Hai et al. 2020).
In C. albicans, EVs produced by yeast cells can inhibit filamentation and biofilm formation, suggesting that growth conditions and the status of the cells impact the biological properties of EVs in fungi (Honorato et al. 2021). This study also reported that yeast cells previously incubated with C. albicans EVs, loose their ability to invade the agar and became avirulent in G. mellonella larvae (Honorato et al. 2021). In contrast, EVs produced from C. albicans cells grown under biofilm conditions were shown to complement the extracellular matrix accumulation defect and fluconazole hypersensitivity of ESCRT mutants (Zarnowski et al. 2018). These studies suggest different properties and composition of EVs isolated from yeast cells and filaments in C. abicans. Interestingly, adding exogenous EVs reduced the efficacy of combined therapy (turbinmicin–fluzonazole) on C. albicans biofilms (Zhao et al. 2021).
Because EV-associated RNA species play central roles in intercellular communication in other models (Tsatsaronis et al. 2018), and because of the wide range of RNA molecules identified in fungal EVs, it is predictable that EV RNAs are among these mediators in fungi. Accordingly, in P. brasiliensis EVs, several exonic sRNA-like sequences were identified as mapping to the α-amylase coding gene, which is involved in the synthesis of α- and β-glucans, suggesting a potential role of EVs in the regulation of the expression of these virulence factors (Peres da Silva et al. 2019). Moreover, two spectacular examples of fungal extracellular RNAs interacting with the host in a cross-kingdom communication have been reported. The first one describes an extracellular miRNA produced by the entomopathogenic fungus Beauveria bassiana, which binds to an argonaute protein of the host (Anopheles stephensi) and eventually suppresses the host immunity by silencing the expression of a toll receptor ligand (Cui et al. 2019). Yet, it is important to note that the loading of this miRNA in fungal EVs remains to be demonstrated. Similarly, an extracellular miRNA produced by the phytopathogenic fungus Botrytis cinerae was shown to hijack the Arabidopsis RISC complex, suppressing the host immunity (Weiberg et al. 2013). Although the delivery pathway was not identified, it is reminiscent of an EV-based host-pathogen interaction. Interestingly, in that model, EVs produced by the host were shown to contain siRNAs together with several Arabidopsis RNA-binding proteins that were able to penetrate fungal cells and inhibit the expression of fungal virulence factors (Cai et al. 2018; He et al. 2021). Other examples of fungal-plant interaction are from the studies on the cotton pathogen F. oxysporum f. sp. vasinfectum, which was shown to release EVs regulating phytotoxic response in the leaves of Nicotiana benthamiana (Bleackley, Dawson and Anderson 2019). Recently it has been shown that EVs from P. digitatum regulate the germination of Citrus sinensis seeds by releasing EV-associated small molecules produced during plant infection, such as the alkaloid tryptoquialanine A (Costa et al. 2021).
MECHANISMS OF EV-BASED TRANSFER OF INFORMATION
The mechanisms by which EVs can transfer information from a donor fungal cell to recipient cells (either fungal or host cells) remain poorly understood. For instance, the model of EV based long-distance communication regulating intracellular replication of C. deuterogattii (Bielska et al. 2018) implies that EVs cross the cell wall from the donor cells, the membrane of the macrophage, reach the phagolysosome in which these yeasts can replicate, and finally the cell wall and plasma membrane of the recipient yeast cells.
Several hypotheses raise the concern on the mechanisms by which fungal EVs can bidirectionally cross the fungal cell wall (Rodrigues and Casadevall 2018). Nevertheless, liposomes containing the antifungal drug amphotericin B were shown to cross the fungal cell wall, suggesting that EVs could also do the same (Walker et al. 2018). Moreover, proteomic analysis of S. cerevisiae EVs revealed enrichment in enzymes implicated in cell wall biosynthesis, and EV-based complementation assays suggested potential roles for yeast EVs in the cell wall remodeling. This enrichment of cell wall-related proteins in EV proteome was also observed in other fungi suggesting common properties. Nevertheless, the mechanisms through which EVs can cross the fungal cell wall and plasma membrane remain to be described.
Some experiments using actin polymerization inhibitors or cholesterol depleting agents suggested that fungi EV uptake by the host cell is an active rather than a passive process (Bielska et al. 2018). However, the impact of clathrin-mediated or clathrin-independent endocytosis (macropinocytosis and phagocytosis) or the endocytosis via caveolae and lipid rafts for the EV internalization, previously described as the main mechanisms of EV uptake by host cells in other models, remain to be elucidated in fungi. In the last few years, few studies started to explore the mechanisms regulating these processes. Thus, C. albicans EVs were shown to co-localize with the plasma membrane ganglioside GM1 of bone marrow-derived macrophages and dendritic cells. Since GM1 is a raft marker, it was suggested that EV internalization might be mediated by lipid rafts. M. sympodialis EVs were internalized by both keratinocytes and monocytes at 37°C but not at 4°C, suggesting that EVs could actively be engulfed by endocytosis. Co-localization assays using C. neoformans EVs and FITC-labeled cholera toxin subunit B suggested that fungal EVs are not fusing with the plasma membrane of the macrophages (Oliveira et al. 2010a). However, it has been demonstrated that EVs from the same fungal pathogen fuse with human brain microvascular endothelial cells (HBMECs) membranes and potentiate fungal brain invasion (Huang et al. 2012). Although mammalian lectin microarray profiling assays suggested that DC-SIGN receptors could recognize surface carbohydrates in P. brasiliensis and P. lutzii EVs, very little information is available concerning the surface recognition of fungal EVs by the host cells.
IN VITRO AND IN VIVO EV-MEDIATED FUNGAL–HOST INTERACTIONS
With the notable exceptions of Malassezia spp and C. albicans, human fungal infections are mostly due to pathogens living in the environment (Janbon et al. 2019). In these cases, the main route of contamination is the respiratory tract, leading to lung infection. Therefore, the first lines of defense against the pulmonary fungal infection are the alveolar macrophages. In the later stages of infection, phagocytes, including monocytes, macrophages, dendritic cells and neutrophils, are involved in nearly all the steps of the host–pathogen interactions. In the past few years, many studies and reviews focused on these immune cells in which fungal pathogens, once internalized, can be either eliminated or survived and multiplied during the infection (Gilbert, Wheeler and May 2014; Heung 2020; Weerasinghe and Traven 2020).
Because of their complex composition, fungal EVs are studied for their impact on the host immune cells. Here again, due to the lack of standardization for fungal EVs purification protocols and to the absence of identified EV specific markers, the following discussed studies need to be regarded as using EV extracts more than purified EVs. Therefore, the associated conclusions should be taken cautiously and might be challenged by experiments using purified EVs in the future. Nevertheless, RAW 264.7 murine macrophage cell line was used to suggest that C. neoformans EVs are biologically active and can stimulate immune cells (Oliveira et al. 2010a). In this study, C. neoformans EVs were shown to be uptaken by macrophages modulating nitric oxide (NO) concentration and pro-inflammatory cytokines TNF-α production. This overall M1 polarization of the EV-treated macrophages was, as expected, associated with an increase in the phagocyte fungicidal activity (Oliveira et al. 2010a). Surprisingly, EVs treatment also induces anti-inflammatory cytokine response (IL-10 and TGF-β). This apparent conflicting effect was only partly dependent on the presence of capsular polysaccharide, known to be associated with C. neoformans EVs (Rodrigues et al. 2007).
C. albicans EVs were shown to be internalized by both bone marrow‐derived murine macrophages (BMDM) and dendritic cells, in which they modulate both pro-inflammatory and anti-inflammatory cytokines synthesis as well as NO production, in a dose-dependent manner (Vargas et al. 2015). Similar data were recently obtained using Talaromyces marneffei EVs (Yang et al. 2021). In M. sympodialis, EVs were shown to induce both TNF-α and IL-4 cytokines production from peripheral blood mononuclear cells (PBMC), albeit the production of only IL-4 was significantly higher when PBMC from patients suffering from atopic eczema were used as compared to healthy controls (Gehrmann et al. 2011).
All these observations suggest that the very complex composition of fungal EVs can result in multiple effects on immune cells, depending on the organism from which EVs are isolated and on the patient's status. For instance, EVs extracted from H. capsulatum decrease phagocytosis of this yeast by 35% and no significant increase of NO production was observed upon treating BMDM with EVs (Baltazar et al. 2018). In contrast, for some organisms, the effect of EVs appears to be only directed towards a Th1 type response. Thus, in recent years, M1 polarization of macrophages upon incubation with fungal EVs has been reported in P. brasiliensis (da Silva et al. 2016), Trichophyton interdigitale (Bitencourt et al. 2018) and Aspergillus flavus (Brauer et al. 2020). Similarly, in A. fumigatus, EVs increase macrophage phagocytosis by more than 10% and strongly induced macrophage pro-inflammatory mediators TNF-α and CCL2 production (Souza et al. 2019). A similar effect was reported using bone marrow-derived neutrophils, which are primed by A. fumigatus EVs to increase their phagocytic capacity (Souza et al. 2019) and on dendritic cells using S. brasiliensis EVs (Ikeda et al. 2018). As produced by pathogens responsible for skin infection, M. sympodialis and T. interdigitale EVs interactions with keratinocytes have also been studied (Bitencourt et al. 2018; Johansson et al. 2018; Vallhov et al. 2020). M. sympodialis EVs are actively internalized by human keratinocytes, inducing up-regulation of ICAM-1 expression (Johansson et al. 2018; Vallhov et al. 2020). T. interdigitale EVs were shown to induce NO, TNF-α, IL-6, IL-1β and IL-8 from human keratinocyte cell line HaCaT cells in a dose-dependent manner.
The above results suggest that fungal EVs could modulate the innate immune response in vivo. This hypothesis was tested using the G. mellonella larvae model of fungal infection. In agreement with the variable effect of fungal EVs on phagocytes, the outcomes of these assays are diverse. A pre-treatment of G. mellonella larvae with C. albicans EVs previously stored at –80, –20 or 4°C reduces the fungal burden in this model without statistically significant increase of the larvae survival (Vargas et al. 2015, 2020). Strikingly, the same experiment performed with freshly prepared EVs prolonged larvae survival (Vargas et al. 2020). In C. neoformans, EVs isolated from an acapsular mutant strain does not protect the infected larvae, whereas wild type EVs accelerated their death (Colombo et al. 2019). In contrast, prior stimulation of G. mellonella larvae with A. flavus EVs promoted decreased fungal burden and increased survival of the larvae (Brauer et al. 2020). These results further illustrate the complexity of the message carried by fungal EVs.
PERSPECTIVES—EV-BASED VACCINE FOR FUNGAL INFECTIONS
In bacteria and parasites, EVs have been shown to stimulate adaptive immunity, and several vaccine candidates have been tested in the murine model of infection and clinical assays (Choi et al. 2015; Shears et al. 2018; Launay et al. 2019). In bacteria, EVs have been used for vaccine development since the end of the 1980s, and several EV-based vaccines are already licensed against meningococcal B disease (Acevedo et al. 2014). Several observations suggest that fungal EVs could also modulate adaptive immune response (Silverman et al. 2010; Pathirana and Kaparakis-Liaskos 2016). For instance, DC exposed to C. albicans EVs produce a higher level of CD86 and MHC-II compared to the untreated control (Vargas et al. 2015). Similar data recently obtained using macrophages incubated with T. marneffei EVs, suggest that EVs could be involved in T cell activation (Yang et al. 2021).
The analysis of EV proteome isolated from different species also revealed major known antigens associated with these vesicles. For instance, MP65 is one of the most prominent C. albicans EV protein and a major antigen in this pathogenic yeast (Gomez et al. 2000; Vargas et al. 2015). Similarly, M. sympodialis EVs are enriched in the two allergens Mala s 1 and s 7 (Zargari et al. 1994; Johansson et al. 2018). Also, in C. neoformans, EV proteome analysis revealed a spectacular enrichment of several major antigens such as the mannoproteins MP88 and MP98 (Levitz et al. 2001; Huang et al. 2002; Rizzo et al. 2021). Similar results were obtained in A. fumigatus and S. brasiliensis, suggesting that EV proteins can be antigenic and sensitize the host immune system (Ikeda et al. 2018; Souza et al. 2019).
Mice pre-treated using C. albicans EVs were demonstrated to fully protect against C. albicans, whereas non-vaccinated mice died after two weeks (Vargas et al. 2020). Immunization was shown to induce specific profiles of cytokines and is associated with a reduced fungal burden in different organs (Vargas et al. 2020). Similar data were obtained in C. neoformans, although pre-treatment with EVs only prolonged the survival of the mice (Rizzo et al. 2021). Interestingly, mice vaccinated with EVs extracted from an acapsular mutant survived longer than the ones vaccinated using wild type EVs, suggesting that the structure and composition of the EVs can also impact their vaccination properties (Rizzo et al. 2021).
In contrast, for some species under some conditions, pre-treatment with EVs appeared to be harmful for the disease outcome in mouse model of infection. Thus, in a mouse model of sporotrichosis, skin lesions and fungal burden are enhanced by a pre-subcutaneous injection of EVs (Ikeda et al. 2018). More surprisingly, some data suggested that EVs could favor the passage of C. neoformans through the blood-brain barrier and thus the virulence of this yeast (Huang et al. 2012). This paradoxical impact of fungal EVs during the infection remains poorly understood (Piffer et al. 2021) even though it is predictable that they are produced and can interact with the host cells in vivo.
CONCLUDING REMARKS AND PERSPECTIVES ON FUNGAL EV RESEARCH
The field of fungal EVs is only in the early stage, and the knowledge basis regarding their structure, composition and function needs to be improved. Although a general picture of the fungal EV starts to be revealed, it remains unclear due to several challenging points. First, the diversity of fungal species studied for EVs, and the expected diversity of fungal EVs render a general comparison complicated. In this context, the creation of other public databases, similar to the ExVe (Parreira et al. 2021), designed for fungal EV-proteins that could list and compare fungal EV components among different species, would be instrumental. Second, as it has been done for mammalian EVs, the efforts for standardization of purification and characterization protocols of fungal EVs need to be undertaken aiming to identify specific markers to validate the purity of the EV extracts. Large sets of data related to fungal EVs will undoubtedly be produced soon, but standardized methods would be fundamental for a better knowledge of fungal EV structure and composition. This will also greatly impact the study of the fungal EV functionality. Indeed, the biological function of fungal EVs and their involvement in fungal infections remain obscure. Fungal EVs are expected to participate in many aspects of cell-to-cell communication and host-pathogen interactions. Still, only a few examples of EV functional roles have been published, as discussed in this review and summarized in Fig. 2. Yet, the perspectives are huge, although optimized tools and functional assays remain to be set up. Similar to other EV fields, the diversity of fungal EVs is still poorly studied. For instance, it is known that fungal EVs have different sizes and decorations, but the analysis of the heterogeneity of their cargo has not been fully addressed.
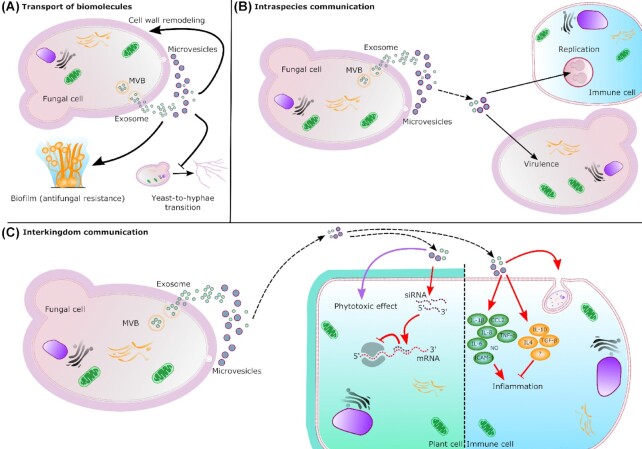
Figure 2.
Biosynthesis and roles of fungal EVs. EVs are transported across the cell wall to the extracellular space. Exosomes result from the fusion of multi-vesicular bodies (MVB) with the plasma membrane and microvesicles bud out from the plasma membrane. (A) Transport of biomolecules. EVs can carry and release several biomolecules. In S. cerevisiae, the enrichment of several cell wall related enzymes in EVs suggests an impact on cell wall remodeling. In C. albicans, EVs are able to promote biofilm formation depending on their origin. In other hand, yeast-derived EVs inhibit yeast-to-hyphae differentiation. (B) Intraspecies communication. EVs released by Cryptococcus donor cells (dashed arrow) can also regulate intracellular replication and virulence of receptor cells. (C) Interkingdom communication. Fungal EVs are also implicated in interkingdom interactions influencing immune response (red arrow) during the infection. Fungal siRNAs can enter the plant cells though EVs where their association with the host Argonaute protein inhibits the immune response. Fungal EVs can also have phytotoxic effect (purple arrow) carrying small molecules. In immune cell, fungal EVs can promote the phagocytosis and regulate anti-inflammatory and pro-inflammatory (NO: nitric oxide) responses.
Single EV analyses have started to be implemented for mammalian EVs (Chiang and Chen 2019). This type of analysis demands specific markers and adapted protocols. It is expected to see the fungal EV field following this path as well soon. Similarly, the biosynthetic pathways regulating fungal EVs are also mostly unknown, although few analyses suggest a link between intracellular trafficking and the release of EVs. The existence of microvesicles and exosomes has been hypothesized in fungi by analogy with the mammalian EVs. Indeed, several microscopic and flow cytometry observations tend to support this model. However, the genetic and the mechanisms regulating their production are still underexplored.
Fungi are very well suited to genetic screens. Together with adapted biochemistry and cell biology assays, they might assist in deciphering the mechanisms by which EVs are produced and how they transmit a message to another cell and how this message is read. Additionally, the strength of fungal genetics applied to EV research should help, in some instances, to propose models for metazoan EVs.
Several EV-based vaccines have already been licensed in bacteria. Accordingly, fungal EVs have been considered as vaccine candidates in several studies. It is promising, as no antifungal vaccine yet to be licensed. Fungi are usually very easy to grow and, most of the times, easy to amenable to genetic approaches. Adapted protocols are now accessible to produce fungal EVs on large scales and at a low cost. Specific modification of EVs to improve their vaccine capacities should thus be possible soon and pave the way for developing antifungal vaccines. Overall, the study of EVs in the biology of fungal pathogens and the pathophysiology of fungal infections are evolving fast, which might deeply change the scientific approaches in this field soon.
ACKNOWLEDGEMENTS
This work was supported by the CAPES-COFECUB Franco-Brazilian Research Exchange Program (88887.357947/2019–00) (GJ). JR is supported by Pasteur-Roux-Cantarini Fellowship (Institut Pasteur, Paris, France). AT is funded by a PhD Fellowship from Ecole Doctorale BioSPC, Université de Paris, 75006 Paris, France.
Contributor Information
Juliana Rizzo, Unité Biologie des ARN des Pathogènes Fongiques, Département de Mycologie, Institut Pasteur, F-75015 Paris, France.
Adam Taheraly, Unité Biologie des ARN des Pathogènes Fongiques, Département de Mycologie, Institut Pasteur, F-75015 Paris, France; Ecole Doctorale BioSPC, Université de Paris, 75006 Paris, France.
Guilhem Janbon, Unité Biologie des ARN des Pathogènes Fongiques, Département de Mycologie, Institut Pasteur, F-75015 Paris, France.
AUTHORS CONTRIBUTIONS
JR contributed to the conception and writing of this review and the design of Fig. 1, which was created using BioRender. AT contributed to the writing of this review and the design of Fig. 2. GJ contributed to the conception and writing of this review.
Conflict of interest
None declared.
REFERENCES
- Acevedo R, Fernandez S, Zayas Cet al. . Bacterial outer membrane vesicles and vaccine applications. Front Immunol. 2014;5:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque PC, Nakayasu ES, Rodrigues MLet al. . Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell Microbiol. 2008;10:1695–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida F, Wolf JM, da Silva TAet al. . Galectin-3 impacts Cryptococcus neoformans infection through direct antifungal effects. Nat Commun. 2017;8:1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves LR, Peres da Silva R, Sanchez DAet al. . Extracellular vesicle-mediated RNA release in Histoplasma capsulatum. mSphere. 2019;4:e00176–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltazar LM, Zamith-Miranda D, Burnet MCet al. . Concentration-dependent protein loading of extracellular vesicles released by Histoplasma capsulatum after antibody treatment and its modulatory action upon macrophages. Sci Rep. 2018;8:8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielska E, May RC. Extracellular vesicles of human pathogenic fungi. Curr Opin Microbiol. 2019;52:90–9. [DOI] [PubMed] [Google Scholar]
- Bielska E, Sisquella MA, Aldeieg Met al. . Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Nat Commun. 2018;9:1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Schubotz F, Roggensack SEet al. . Bacterial vesicles in marine ecosystems. Science. 2014;343:183–6. [DOI] [PubMed] [Google Scholar]
- Bitencourt TA, Rezende CP, Quaresemin NRet al. . Extracellular vesicles from the dermatophyte Trichophyton interdigitale modulate macrophage and keratinocyte functions. Front Immunol. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleackley MR, Dawson CS, Anderson MA. Fungal extracellular vesicles with a focus on proteomic analysis. Proteomics. 2019;19:1800232. [DOI] [PubMed] [Google Scholar]
- Brauer VS, Pessoni AM, Bitencourt TAet al. . Extracellular vesicles from Aspergillus flavus induce M1 polarization in vitro. mSphere. 2020;5:e00190–00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Qiao L, Wang Met al. . Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science. 2018;360:1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C-Y, Chen C. Toward characterizing extracellular vesicles at a single-particle level. J Biomed Sci. 2019;26:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SJ, Kim M-H, Jeon Jet al. . Active immunization with extracellular vesicles derived from Staphylococcus aureus effectively protects against staphylococcal lung infections, mainly via Th1 cell-mediated immunity. PLoS ONE. 2015;10:e0136021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleare LG, Zamith D, Heyman HMet al. . Media matters! Alterations in the loading and release of Histoplasma capsulatum extracellular vesicles in response to different nutritional milieus. Cell Microbiol. 2020;22:e13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho C, Casadevall A. Answers to naysayers regarding microbial extracellular vesicles. Biochem Soc Trans. 2019;47:1005–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo AC, Rella A, Normile Tet al. . Cryptococcus neoformans glucuronoxylomannan and sterylglucoside are required for host protection in an animal vaccination model. mBio. 2019;10:e02909–02918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa JH, Bazioli JM, Barbosa LDet al. . Phytotoxic tryptoquialanines produced in vivo by Penicillium digitatum are exported in extracellular vesicles. mBio. 2021;12:e03393–03320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Wang Y, Liu Jet al. . A fungal pathogen deploys a small silencing RNA that attenuates mosquito immunity and facilitates infection. Nat Commun. 2019;10:4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva RP, Heiss C, Black Iet al. . Extracellular vesicles from Paracoccidioides pathogenic species transport polysaccharide and expose ligands for DC-SIGN receptors. Sci Rep. 2015;5:14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva TA, Roque-Barreira MC, Casadevall Aet al. . Extracellular vesicles from Paracoccidioides brasiliensis induced M1 polarization in vitro. Sci Rep. 2016;6:35867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson CS, Garcia-Ceron D, Rajapaksha Het al. . Protein markers for Candida albicans EVs include claudin-like Sur7 family proteins. J Extracell Ves. 2020;9:1750810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida JR, Kaihami GH, Jannuzzi GPet al. . Therapeutic vaccine using a monoclonal antibody against a 70-kDa glycoprotein in mice infected with highly virulent Sporothrix schenckii and Sporothrix brasiliensis. Med Mycol. 2015;53:42–50. [DOI] [PubMed] [Google Scholar]
- de Oliveira HC, Castelli RF, Reis FCGet al. . Pathogenic delivery: the biological roles of cryptococcal extracellular vesicles. Pathogens. 2020;9:754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paula RG, Antoniêto ACC, Nogueira KMVet al. . Extracellular vesicles carry cellulases in the industrial fungus Trichoderma reesei. Biotechnol Biofuels. 2019;12:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza W, Barrias ES.. Membrane-bound extracellular vesicles secreted by parasitic protozoa: cellular structures involved in the communication between cells. Parasitol Res. 2020;119:2005–23. [DOI] [PubMed] [Google Scholar]
- de Toledo Martins S, Szwarc P, Goldenberg Set al. . Extracellular vesicles in fungi: composition and functions. Curr Top Microbiol Immunol. 2019;422:45–59. [DOI] [PubMed] [Google Scholar]
- Eisenman HC, Frases S, Nicola AMet al. . Vesicle-associated melanization in Cryptococcus neoformans. Microbiology. 2009;155:3860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas MS, Bonato VLD, Pessoni AMet al. . Fungal extracellular vesicles as potential targets for immune interventions. mSphere. 2019;4:e00747–00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandham S, Su X, Wood Jet al. . Technologies and standardization in research on extracellular vesicles. Trends Biotechnol. 2020;38:1066–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrmann U, Qazi KR, Johansson Cet al. . Nanovesicles from Malassezia sympodialis and host exosomes induce cytokine responses–novel mechanisms for host-microbe interactions in atopic eczema. PLoS One. 2011;6:e21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson RK, Peberdy JF. Fine structure of protoplasts of Aspergillus nidulans. Microbiology. 1972;72:529–38. [DOI] [PubMed] [Google Scholar]
- Gil-Bona A, Llama-Palacios A, Parra CMet al. . Proteomics unravels extracellular vesicles as carriers of classical cytoplasmic proteins in Candida albicans. J Proteome Res. 2015;14:142–53. [DOI] [PubMed] [Google Scholar]
- Gilbert AS, Wheeler RT, May RC. Fungal pathogens: survival and replication within macrophages. Cold Spring Harb Perspect Med. 2015;5:a019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S, Catchpole R, Forterre P.. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol Rev. 2019;43:273–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioti A, Nystedt B, Li Wet al. . Genomic insights into the atopic eczema-associated skin commensal yeast Malassezia sympodialis. mBio. 2013;4:e00572–00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goes A, Lapuhs P, Kuhn Tet al. . Myxobacteria-derived outer membrane vesicles: potential applicability against intracellular infections. Cells. 2020;9:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez MJ, Maras B, Barca Aet al. . Biochemical and immunological characterization of MP65, a major mannoprotein antigen of the opportunistic human pathogen Candida albicans. Infect Immun. 2000;68:694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner HN, McManus MT. Examining the evidence for extracellular RNA function in mammals. Nat Rev Genet. 2021;22:448–58. [DOI] [PubMed] [Google Scholar]
- Hai TP, Tuan TL, Van Anh Det al. . The virulence of the Cryptococcus neoformans VNIa-5 lineage is highly plastic and associated with isolate background. bioRxiv. 2020. DOI: 2020.2002.2024.962134. [Google Scholar]
- Hatanaka O, Rezende CP, Moreno Pet al. . Galectin-3 inhibits Paracoccidioides brasiliensis growth and impacts paracoccidioidomycosis through multiple mechanisms. mSphere. 2019;4:e00209–00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Cai Q, Qiao Let al. . RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat Plant. 2021;7:342–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heung LJ. Monocytes and the host response to fungal pathogens. Front Cell Infect Microbiol. 2020;10. DOI: 10.3389/fcimb.2020.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill E, Solomon P. Extracellular vesicles from the apoplastic fungal wheat pathogen Zymoseptoria Tritici. Fungal Biol Biotechnol. 2020; 18. Research Square. DOI:10.1186/s40694-020-00103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honorato L, Demetrio JF, Ellis CCet al. . Extracellular vesicles regulate yeast growth, biofilm formation, and yeast-to-hypha differentiation in Candida albicans. bioRxiv. 2021. DOI: 2021.2001.2021.427696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini-Beheshti E, Grau GER. Extracellular vesicles as mediators of immunopathology in infectious diseases. Immunol Cell Biol. 2018;96:694–703. [DOI] [PubMed] [Google Scholar]
- Huang C, Nong SH, Mansour MKet al. . Purification and characterization of a second immunoreactive mannoprotein from Cryptococcus neoformans that stimulates T-cell responses. Infect Immun. 2002;70:5485–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S-H, Wu C-H, Chang YCet al. . Cryptococcus neoformans-derived microvesicles enhance the pathogenesis of fungal brain infection. PLoS One. 2012;7:e48570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34:2398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda MAK, de Almeida JRF, Jannuzzi GPet al. . Extracellular vesicles from Sporothrix brasiliensis are an important virulence factor that induce an increase in fungal burden in experimental sporotrichosis. Front Microbiol. 2018;9. DOI: 10.3389/fmicb.2018.02286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon G, Quintin J, Lanternier Fet al. . Studying fungal pathogens of humans and fungal infections: fungal diversity and diversity of approaches. Genes Immun. 2019;20:403–14. [DOI] [PubMed] [Google Scholar]
- Johansson HJ, Vallhov H, Holm Tet al. . Extracellular nanovesicles released from the commensal yeast Malassezia sympodialis are enriched in allergens and interact with cells in human skin. Sci Rep. 2018;8:9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M, Melki R. Sup35p in its soluble and prion states is packaged inside extracellular vesicles. mBio. 2015;6. DOI: 10.1128/mbio.01017-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkowska-Kuleta J, Kulig K, Karnas Eet al. . Characteristics of extracellular vesicles released by the pathogenic yeast-like fungi Candida glabrata, Candida parapsilosis and Candida tropicalis. Cells. 2020;9:1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launay O, Ndiaye AGW, Conti Vet al. . Booster vaccination with GVGH Shigella sonnei 1790GAHB GMMA vaccine compared to single vaccination in unvaccinated healthy European adults: results from a phase 1 clinical trial. Front Immunol. 2019;10:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrin T, Konte T, Kostanjšek Ret al. . The neurotropic black yeast Exophiala dermatitidis induces neurocytotoxicity in neuroblastoma cells and progressive cell death. Cells. 2020;9:963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone F, Bellani L, Muccifora Set al. . Analysis of extracellular vesicles produced in the biofilm by the dimorphic yeast Pichia fermentans. J Cell Physiol. 2018;233:2759–67. [DOI] [PubMed] [Google Scholar]
- Levitz SM, Nong S, Mansour MKet al. . Molecular characterization of a mannoprotein with homology to chitin deacetylases that stimulates T cell responses to Cryptococcus neoformans. Proc Natl Acad Sci. 2001;98:10422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Bruni GO, Taylor CMet al. . Comparative genome-wide analysis of extracellular small RNAs from the mucormycosis pathogen Rhizopus delemar. Sci Rep. 2018;8:5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang Z, Ding Cet al. . Transcriptomic analysis of extracellular RNA governed by the endocytic adaptor protein Cin1 of Cryptococcus deneoformans. Front Cell Infect Microbiol. 2020;10:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marina CL, Bürgel PH, Agostinho DPet al. . Nutritional conditions modulate C. neoformans extracellular vesicles' capacity to elicit host immune response. Microorganisms. 2020;8:1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M, Martin-Jaular L, Lavieu Get al. . Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9–17. [DOI] [PubMed] [Google Scholar]
- Matos Baltazar L, Nakayasu ES, Sobreira TJet al. . Antibody binding alters the characteristics and contents of extracellular vesicles released by Histoplasma capsulatum. mSphere. 2016;1. DOI: 10.1128/mSphere.00085-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencher A, Morales P, Valero Eet al. . Proteomic characterization of extracellular vesicles produced by several wine yeast species. Microb Biotechnol. 2020;13:1581–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales P, Mencher A, Tronchoni Jet al. . Extracellular vesicles in food biotechnology. Microb Biotechnol. 2021;14:8–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimrichter L, de Souza MM, Del Poeta Met al. . Extracellular vesicle-associated transitory cell wall components and their impact on the interaction of fungi with host cells. Front Microbiol. 2016;7:1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble JM, Roberts LM, Vidavsky Net al. . Direct comparison of optical and electron microscopy methods for structural characterization of extracellular vesicles. J Struct Biol. 2020;210:107474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DL, Freire-de-Lima CG, Nosanchuk JDet al. . Extracellular vesicles from Cryptococcus neoformans modulate macrophage functions. Infect Immun. 2010;78:1601–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DL, Nakayasu ES, Joffe LSet al. . Characterization of yeast extracellular vesicles: evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS ONE. 2010b;5:e11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y-D, Chen SH, Camacho Eet al. . Role of the ESCRT pathway in laccase trafficking and virulence of Cryptococcus neoformans. Infect Immun. 2020;88:e00954–00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parreira VdSC, Santos LGC, Rodrigues MLet al. . ExVe: the knowledge base of orthologous proteins identified in fungal extracellular vesicles. Comput Struct Biotechnol J. 2021;19:2286–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathirana RD, Kaparakis-Liaskos M. Bacterial membrane vesicles: biogenesis, immune regulation and pathogenesis. Cell Microbiol. 2016;18:1518–24. [DOI] [PubMed] [Google Scholar]
- Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487–514. [DOI] [PubMed] [Google Scholar]
- Peres da Silva R, Longo LGV, Cunha Jet al. . Comparison of the RNA content of extracellular vesicles derived from Paracoccidioides brasiliensis and Paracoccidioides lutzii. Cells. 2019;8:765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres da Silva R, Martins SdT, Rizzo Jet al. . Golgi Reassembly and Stacking Protein (GRASP) participates in vesicle-mediated RNA export in Cryptococcus Neoformans. Genes. 2018;9:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffer AC, Kuczera D, Rodrigues MLet al. . The paradoxical and still obscure properties of fungal extracellular vesicles. Mol Immunol. 2021;135:137–46. [DOI] [PubMed] [Google Scholar]
- Raab-Traub N, Dittmer DP. Viral effects on the content and function of extracellular vesicles. Nat Rev Microbiol. 2017;15:559–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis FCG, Borges BS, Jozefowicz LJet al. . A novel protocol for the isolation of fungal extracellular vesicles reveals the participation of a putative scramblase in polysaccharide export and capsule construction in Cryptococcus gattii. mSphere. 2019;4:e00080–00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis FCG, Costa JH, Honorato Let al. . Small molecule analysis of extracellular vesicles produced by Cryptococcus gattii: identification of a tripeptide controlling cryptococcal infection in an invertebrate host model. Front Immunol. 2021;12. DOI: 10.3389/fimmu.2021.654574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo J, Chaze T, Miranda Ket al. . Characterization of extracellular vesicles produced by Aspergillus fumigatus protoplasts. mSphere. 2020;5:e00476–00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo J, Oliveira DL, Joffe LSet al. . Role of the Apt1 protein in polysaccharide secretion by Cryptococcus neoformans. Eukaryotic Cell. 2014;13:715–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo J, Rodrigues ML, Janbon G. Extracellular vesicles in fungi: past, present, and future perspectives. Front Cell Infect Microbiol. 2020a;10. DOI: 10.3389/fcimb.2020.00346.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo j, Wong SSW, Gazi ADet al. . Revisiting cryptococcus extracellular vesicles properties and their use as vaccine platforms. bioRxiv. 2021. DOI: 2020.2008.2017.253716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Casadevall A. A two-way road: novel roles for fungal extracellular vesicles. Mol Microbiol. 2018;110:11–5. [DOI] [PubMed] [Google Scholar]
- Rodrigues ML, Franzen AJ, Nimrichter Let al. . Vesicular mechanisms of traffic of fungal molecules to the extracellular space. Curr Opin Microbiol. 2013;16:414–20. [DOI] [PubMed] [Google Scholar]
- Rodrigues ML, Nakayasu ES, Oliveira DLet al. . Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryotic Cell. 2008;7:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nimrichter L, Oliveira DLet al. . Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryotic Cell. 2007;6:48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux Q, Van Deun J, Dedeyne Set al. . The EV-TRACK summary add-on: integration of experimental information in databases to ensure comprehensive interpretation of biological knowledge on extracellular vesicles. J Extracell Ves. 2020;9:1699367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatke B, Gavinho B, Coceres Vet al. . Unveiling the role of EVs in anaerobic parasitic protozoa. Mol Immunol. 2021;133:34–43. [DOI] [PubMed] [Google Scholar]
- Schorey JS, Harding CV. Extracellular vesicles and infectious diseases: new complexity to an old story. J Clin Invest. 2016;126:1181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R, Patel T, Freedman JE. Circulating extracellular vesicles in human disease. N Engl J Med. 2018;379:958–66. [DOI] [PubMed] [Google Scholar]
- Shears RK, Bancroft AJ, Hughes GWet al. . Extracellular vesicles induce protective immunity against Trichuris muris. Parasite Immunol. 2018;40:e12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva BMA, Prados-Rosales R, Espadas-Moreno Jet al. . Characterization of Alternaria infectoria extracellular vesicles. Med Mycol. 2014;52:202–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Clos J, Horakova Eet al. . Leishmania exosomes modulate innate and adaptive immune responses through effects on monocytes and dendritic cells. J Immunol. 2010;185:5011–22. [DOI] [PubMed] [Google Scholar]
- Souza JAM, Baltazar LdM, Carregal VMet al. . Characterization of Aspergillus fumigatus extracellular vesicles and their effects on macrophages and neutrophils functions. Front Microbiol. 2019;10:2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht CA, Lee CK, Huang Het al. . Vaccination with recombinant cryptococcus proteins in glucan particles protects mice against cryptococcosis in a manner dependent upon mouse strain and cryptococcal species. mBio. 2017;8:e01872–01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szempruch AJ, Sykes SE, Kieft Ret al. . Extracellular vesicles from Trypanosoma brucei mediate virulence factor transfer and cause host anemia. Cell. 2016;164:246–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo K, Uesaka I, Uehira Ket al. . Fine structure of Cryptococcus neoformans grown in vitro as observed by freeze-etching. J Bacteriol. 1973;113:1442–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C, Witwer KW, Aikawa Eet al. . Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Ves. 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrecilhas AC, Soares RP, Schenkman Set al. . Extracellular vesicles in trypanosomatids: host cell communication. Front Cell Infect Microbiol. 2020;10:602502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsatsaronis JA, Franch-Arroyo S, Resch Uet al. . Extracellular vesicle RNA: a universal mediator of microbial communication?. Trends Microbiol. 2018;26:401–10. [DOI] [PubMed] [Google Scholar]
- Vallejo MC, Matsuo AL, Ganiko Let al. . The pathogenic fungus Paracoccidioides brasiliensis exports extracellular vesicles containing highly immunogenic α-Galactosyl epitopes. Eukaryotic Cell. 2011;10:343–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo MC, Nakayasu ES, Longo LVGet al. . Lipidomic analysis of extracellular vesicles from the pathogenic phase of Paracoccidioides brasiliensis. PLoS ONE. 2012a;7:e39463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo MC, Nakayasu ES, Matsuo ALet al. . Vesicle and vesicle-free extracellular proteome of Paracoccidioides brasiliensis: comparative analysis with other pathogenic fungi. J Proteome Res. 2012;11:1676–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallhov H, Johansson C, Veerman REet al. . Extracellular vesicles released from the skin commensal yeast Malassezia sympodialis activate human primary keratinocytes. Front Cell Infect Microbiol. 2020;10. DOI: 10.3389/fcimb.2020.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas G, Honorato L, Guimarães AJet al. . Protective effect of fungal extracellular vesicles against murine candidiasis. Cell Microbiol. 2020;10:e13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas G, Rocha JDB, Oliveira DLet al. . Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cell Microbiol. 2015;17:389–407. [DOI] [PubMed] [Google Scholar]
- Walker L, Sood P, Lenardon MDet al. . The viscoelastic properties of the fungal cell wall allow traffic of AmBisome as intact liposome vesicles. mBio. 2018;9:e02383–02317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kojima K, Mobley JAet al. . Proteomic analysis of urinary extracellular vesicles reveal biomarkers for neurologic disease. EBioMedicine. 2019;45:351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasinghe H, Traven A.. Immunometabolism in fungal infections: the need to eat to compete. Curr Opin Microbiol. 2020;58:32–40. [DOI] [PubMed] [Google Scholar]
- Weiberg A, Wang M, Lin FMet al. . Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Royo F, Aizpurua-Olaizola Oet al. . Glycosylation of extracellular vesicles: current knowledge, tools and clinical perspectives. J Extracell Ves. 2018;7:1442985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters CM, Hong-Brown LQ, Chiang HL.. Intracellular vesicle clusters are organelles that synthesize extracellular vesicle-associated cargo proteins in yeast. J Biol Chem. 2020;295:2650–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JM, Espadas J, Luque-Garcia Jet al. . Lipid biosynthetic genes affect Candida albicans extracellular vesicle morphology, cargo, and immunostimulatory properties. Eukaryotic Cell. 2015;14:745–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JM, Rivera J, Casadevall A.. Serum albumin disrupts Cryptococcus neoformans and Bacillus anthracis extracellular vesicles. Cell Microbiol. 2012;14:762–73. [DOI] [PubMed] [Google Scholar]
- Wolf P. The nature and significance of platelet products in human plasma. Br J Haematol. 1967;13:269–88. [DOI] [PubMed] [Google Scholar]
- Yang B, Wang J, Jiang Het al. . Extracellular vesicles derived from Talaromyces marneffei yeasts mediate inflammatory response in macrophage cells by bioactive protein components. Front Microbiol. 2021;11. DOI: 10.3389/fmicb.2020.603183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JE, Rossignol ED, Chang Det al. . Complexity and ultrastructure of infectious extracellular vesicles from cells infected by non-enveloped virus. Sci Rep. 2020;10:7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamith-Miranda D, Nimrichter L, Rodrigues MLet al. . Fungal extracellular vesicles: modulating host-pathogen interactions by both the fungus and the host. Microbes Infect. 2018;20:501–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zargari A, Härfast B, Johansson Set al. . Identification of allergen components of the opportunistic yeast Pityrosporum orbiculare by monoclonal antibodies. Allergy. 1994;49:50–6. [DOI] [PubMed] [Google Scholar]
- Zarnowski R, Sanchez H, Covelli ASet al. . Candida albicans biofilm–induced vesicles confer drug resistance through matrix biogenesis. PLoS Biol. 2018;16:e2006872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Zhao M, Braun DRet al. . A marine microbiome antifungal targets urgent-threat drug-resistant fungi. Science. 2020;370:974–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Bleackley M, Chisanga Det al. . Extracellular vesicles secreted by Saccharomyces cerevisiae are involved in cell wall remodelling. Commun Biol. 2019;2:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Zhang F, Zarnowski Ret al. . Turbinmicin inhibits Candida biofilm growth by disrupting fungal vesicle–mediated trafficking. J Clin Invest. 2021;131. DOI: 10.1172/JCI145123.. [DOI] [PMC free article] [PubMed] [Google Scholar]