Abstract
The recent discovery that giant viruses encode proteins related to sugar synthesis and processing paved the way for the study of their glycosylation machinery. We focused on the proposed Megavirinae subfamily, for which glycan-related genes were proposed to code for proteins involved in glycosylation of the layer of fibrils surrounding their icosahedral capsids. We compared sugar compositions and corresponding biosynthetic pathways among clade members using a combination of chemical and bioinformatics approaches. We first demonstrated that Megavirinae glycosylation differs in many aspects from what was previously reported for viruses, as they have complex glycosylation gene clusters made of six and up to 33 genes to synthetize their fibril glycans (biosynthetic pathways for nucleotide-sugars and glycosyltransferases). Second, they synthesize rare amino-sugars, usually restricted to bacteria and absent from their eukaryotic host. Finally, we showed that Megavirinae glycosylation is clade-specific and that Moumouvirus australiensis, a B-clade outsider, shares key features with Cotonvirus japonicus (clade E) and Tupanviruses (clade D). The existence of a glycosylation toolbox in this family could represent an advantageous strategy to survive in an environment where members of the same family are competing for the same amoeba host. This study expands the field of viral glycobiology and raises questions on how Megavirinae evolved such versatile glycosylation machinery.
Keywords: giant virus, proposed Megavirinae subfamily, viral glycosylation, chemical analysis, bioinformatics
The fibrils decorating the viral capsids of members of the Megavirinae are glycosylated by clade-specific machineries.
Introduction
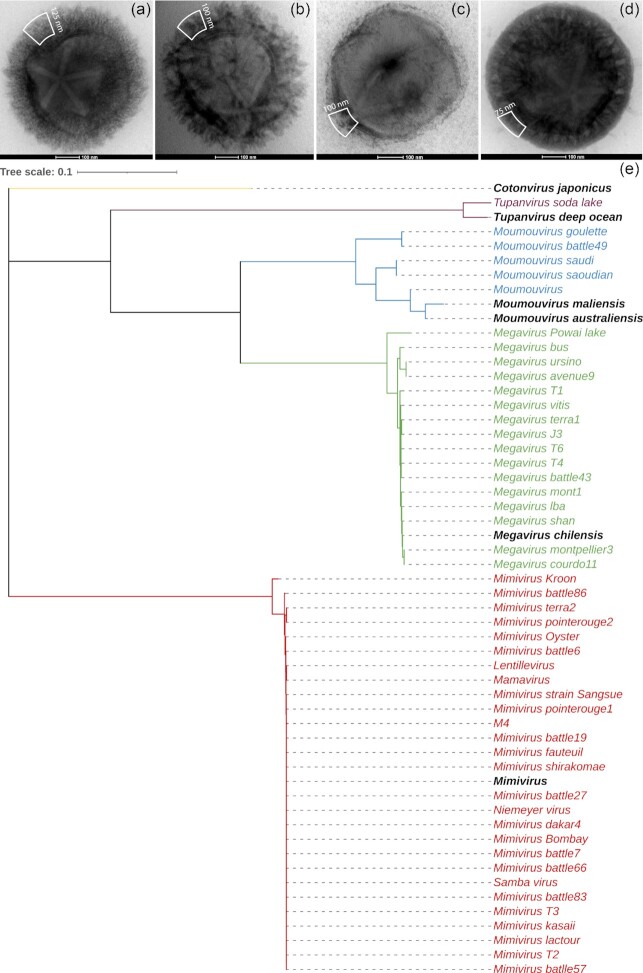
The general perception of viruses as small and simple entities has been challenged with the discovery of giant viruses (B. La Scola et al. 2003, Abergel et al. 2015). Giant viruses are endowed with dsDNA genomes up to 2.5 Mb that can encode up to 1500 proteins while more conventional viruses have much smaller genomes and sometimes just a handful of genes (Lu et al. 2020). Giant virus capsids are so large (up to 2 µm) that they can easily be seen by light microscopy. These viruses are larger than the smallest bacteria (Mycoplasma genitalium <0.3 μm) and archaea (Nanoarchaeum equitans, 0.4 μm) and contain more genes than the smallest parasitic eukaryote encephalitozoon (Philippe et al. 2013). They all infect unicellular eukaryotes and are major players in the environment (Suttle 2005, Brussaard et al. 2008) where they regulate protist populations. Given their genomic complexity, they contain genes never encountered before in viruses (B. La Scola et al. 2003, Renesto et al. 2006), such as those related to protein translation (Abergel et al. 2007, Raoult 2004, Jeudy et al. 2012) and glycan synthesis (Parakkottil Chothi et al. 2010, Piacente et al. 2012, 2014a,b,2017a). Currently, several families of giant viruses have been discovered, such as the Mimiviridae and the proposed Pandoraviridae, Molliviridae and Pithoviridae (Abergel et al. 2015). The present study is focused on the Mimiviridae family and in particular on the proposed Megavirinae subfamily (Gallot-Lavallée et al. 2017), which encompasses five clades, all infecting Acanthamoebasp. (Fig. 1): Mimiviruses (A-clade), Moumouviruses (B-clade), Megaviruses (C-clade), Tupanviruses (D-clade) (Abrahão et al. 2018) and the recently isolated Cotonvirus japonicus (E-clade) (Takahashi et al. 2021). As of today, all members of the proposed Megavirinae are characterized by a fibril layer that differs in thickness and length among the clades, as shown by negative staining transmission electron microscopy (NS-TEM) (Fig. 1). Interestingly, NS-TEM images of Moumouvirus australiensis (Fig. 1c) and Moumouvirus maliensis (Fig. 1b) also presented marked differences in their fibril layer thickness while supposedly belonging to the same clade.
Figure 1.
Top: Transmission electron microscopy (TEM) images of negative-stained (NS) virions of the proposed Megavirinaesubfamily. (a)Mimivirus (A-clade); (b)Moumouvirus maliensis (B-clade); (c)Moumouvirus australiensis (B-clade); (d)Megavirus chilensis (C-clade). White boxes delineate the fibril layers. Thickness of the layer was measured on TEM images of resin-embedded infected cells when mature particles were visible, as NS-TEM on dehydrated samples can induce fibril collapse or shrinking; (e)phylogenetic tree of the proposed Megavirinae subfamily. This family encompasses five clades: Mimiviruses (A-clade, in red), Moumouviruses (B-clade, in blue), Megaviruses (C-clade, in green), Tupanviruses (D-clade, in violet) and Cotonvirus (E-clade, in yellow). For each clade, the prototypes are in black and in bold. The tree is based on the concatenation and alignment of seven protein markers (see Methods).
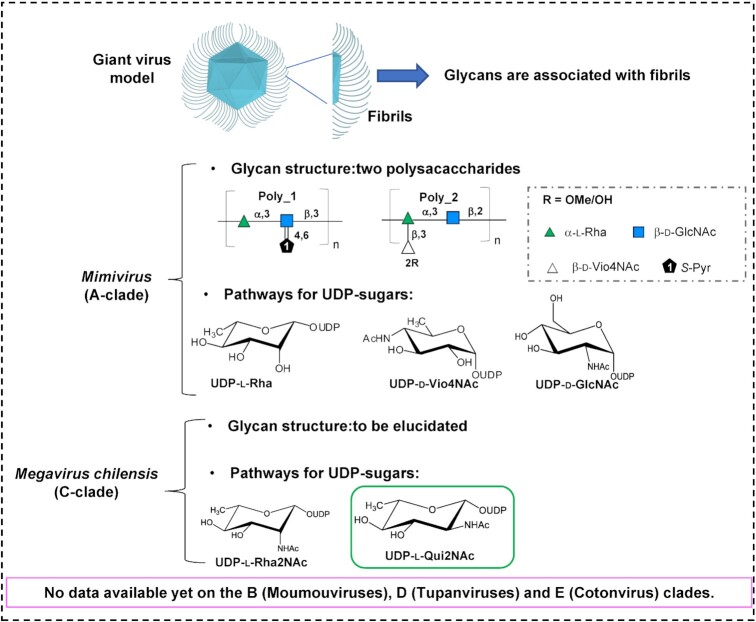
Recent studies on Mimivirus (A-clade) revealed that complex glycans with unique structures made of two large polysaccharides were branched to the fibrils (Fig. 2). These carbohydrate polymers are made of up to 20 units of sugars that are not synthetized by the amoeba host (Notaro et al. 2021). This result challenged the common belief that eukaryotic viruses divert the host machinery to decorate their envelope proteins with small oligosaccharides containing two to 10 sugar units (Bagdonaite and Wandall 2018). For Megavirus chilensis (C-clade) virions, preliminary analysis revealed that its fibrils were also composed of rare amino-sugars not synthesized by the amoeba host (Piacente et al. 2014b).
Figure 2.
Glycosylation in the proposed Megavirinae subfamily. A hallmark of the members of the family is the presence of a fibril layer surrounding the icosahedral capsids with evidence for some of the associated sugars. The glycan structure of Mimivirus (A-clade) has been elucidated: poly_1 has a linear repeating unit made of 3)-α-l-Rha-(1→3)-β-d-GlcNAc-(1→, with GlcNAc modified by pyruvic acid at position 4,6; poly_2 has a branched repeating unit made of 2)-α-l-Rha-(1→3)-β-d-GlcNAc-(1→ in the linear backbone; Rha is further branched with a terminal β-d-2OMeVio4NAc, which is 75% methylated. The biosynthetic pathways for UDP-l-rhamnose (UDP-l-Rha), UDP-d-N-acetyl-glucosamine (UDP-d-GlcNAc) and UDP-d-4N-acetyl-viosamine (UDP-d-Vio4NAc) have been predicted and experimentally validated. For Megavirus chilensis (C-clade) amino-sugars are found on the fibrils, but the glycan structure is still unknown. The biosynthetic pathways for UDP-l-rhamnosamine (UDP-l-Rha2NAc) and UDP-l-quinovosamine were predicted (UDP-l-Qui2NAc, green box), but experimental validation was only performed for the UDP-l-Rha2NAc pathway. No data are available for the other clades.
Similarly, in PBCV-1, a large dsDNA virus of the Phycodnaviridae family, the Major Capsid Protein was shown to be decorated with an oligosaccharide that was different from those found in the three domains of life (De Castro et al. 2013). The unique nature of giant viruses glycans compared to their hosts is made possible by the presence of genes involved in glycosylation in their complex genomes. In PBCV-1, two biosynthetic pathways for sugars activated as nucleotides (donor-sugars) have been found (Piacente et al. 2015), along with a great number of predicted and experimentally validated glycosyltransferases (Noel et al. 2021, Speciale et al. 2020). Similarly, for the proposed Megavirinae subfamily, biosynthetic pathways for nucleotide-sugars along with glycosyltransferases (Fig. 2) have been discovered for Mimivirus and Megavirus chilensis (Parakkottil Chothi et al. 2010, Piacente et al. 2012, 2014a,b, 2017a). Functional pathways for UDP-l-rhamnose (UDP-L-Rha), UDP-d-glucosamine (UDP-d-GlcNAc) and UDP-d-4N-acetyl-viosamine (UDP-d-Vio4NAc) in Mimivirus (Parakkottil Chothi et al. 2010; Piacente et al. 2012, 2014a,b,2017a) and a synthesis pathway for UDP-l-rhamnosamine (UDP-l-Rha2NAc) in Megavirus chilensis (Piacente et al. 2014b) have been experimentally validated. However, none of the predicted glycosyltransferases (GT) have been experimentally characterized, and there are no data available on glycans and glycogenes for the other clades of the family (Fig. 2).
A global view of glycosylation in the proposed Megavirinae subfamily is a prerequisite to make progress in the nascent field of viral glycosylation and increase our knowledge on carbohydrates and their biosynthesis. Key questions include (i) What is the viral machinery (glycogenes) used to synthesize these complex sugars in the different clades of the proposed Megavirinae subfamily? (ii) What is the nature of the glycans synthesized by other members of this subfamily? (iii) Do these glycans share a common architecture, as seen in Chloroviruses (De Castro et al. 2016), or is their nature and architecture clade specific? To answer these questions, the present study aimed at comparing glycosylation of the giant viruses of the proposed Megavirinae subfamily by exploring the composition of their glycans and searching their genomes to identify possible genes involved in their biosynthesis.
To do this, we combined carbohydrate chemistry with bioinformatic analyses for members of each clade. Mimivirus (Scola 2003) and Megavirus chilensis (Arslan et al. 2011) were used as prototypes for the A- and C-clades, respectively, while we used both Moumouvirus australiensis and maliensis for the B-clade. For each, we chemically characterized their glycan composition, and performed an in silico comparative analysis to evaluate similarities/differences within each clade and within the entire family. As dense fibrils also decorate Tupanviruses (D-clade) and Cotonvirus japonicus (E-clade), we searched their genomes for putative glycogenes and included these data to provide a comprehensive comparative analysis covering all members of the family.
Methods
Phylogeny of the proposed Megavirinae subfamily
The phylogenetic tree of the proposed Megavirinae subfamily is based on concatenation and alignment of the following seven protein markers: Asp-Synthase, Helicase, mRNA Capping Enzyme, MutS, Packaging ATPase, PolyA polymerase and VLTF3. Fifty-five genomes of members of the different clades were used to generate the phylogenetic tree using the command line: mafft (v7.307) for the alignment, trimal (v1.4. rev22) for the gap filtering and iqtree (v1.6.9), with default settings at each step.
Production and purification of the virions
All viruses described here as prototypes of the different clades have been isolated by our laboratory. The protocol adopted to propagate and purify the viral particles is the same for all members of the different clades. Briefly, viral particles were propagated using A. castellanii (Douglas) Neff (American Type Culture Collection 30010TM) cells, which were infected with viruses at a multiplicity of infection of 0.25. After 2 days of incubation at 32°C, the infection was complete and led to cell lysis and release of viral particles into the culture medium. Viral particles were then purified by removing cell debris by centrifugation at 500 g for 10 min at 20°C. Subsequently, the supernatant containing viral particles was spun at 6,800 g, 45 min at 20°C. The pellet containing the virions was washed with water, resuspended in CsCl 1.2 density, deposited on a discontinuous CsCl gradient made of successive layers of 1.3/1.4/1.5 densities (g/ml) and finally spun for 20 h at 100 000 g. The white band corresponding to viral particles was recovered with a syringe and washed three times with water. Purified virions were imaged by light microscopy (ZEISS) and the concentration of the virion was estimated on a spectrophotometer (Eppendorf) at OD 600 nm.
NS-TEM of the virions
Viral particles were visualized by NS-TEM, as reported previously (Notaro et al. 2021). Briefly, viral particles were fixed in glutaraldehyde (2.5% v/v in water) for 1 h at room temperature. Samples were centrifuged at 5000 g for 10 min and pellets were washed twice with water. The structure of the fibrils was visualized by NS-TEM using methyl cellulose (M6385 Sigma) and uranyl acetate (2% v/v in water) as reported (Notaro et al. 2021). Viral particles were observed by TEM on a TECNAI G2°200 KV.
Sugar composition
Monosaccharide composition analysis (as acetylated methyl glycoside) and determination of their absolute configuration (as octyl-glycosides derivatives) was performed on 1×1010 viral particles, as previously reported (De Castro et al. 2010). Gas chromatography-mass spectrometry (GC-MS) analyses were performed on an Agilent instrument (GC instrument Agilent 6850 coupled to MS Agilent 5973) equipped with a SPB-5 capillary column (Supelco, 30 m × 0.25 i.d., flow rate, 0.8 mL min–1) and He as the carrier gas. Electron impact mass spectra were recorded with an ionization energy of 70 eV and an ionizing current of 0.2 mA. The temperature program used for analyses was as follows: 150°C for 5 min, 150 to 280°C at 3°C/min, 300°C for 5 min. Interpretation of these data is based on the following concept: each sugar that is derivatized as acetylated methyl-glycoside or octyl-glycoside is eluted in a specific range of the chromatogram and each peak corresponds to a specific fragmentation pattern, which enables identification of the monosaccharide (Lönngren and Svensson 1974). Elution time is compared with those of standard monosaccharides and allows discrimination between sugars of the same class (i.e. glucose and mannose) and between sugars with different absolute configuration (D or L).
Identification of new genes encoding proteins involved in fibril glycosylation for each clade
We used an in silico approach to search for genes coding for potential proteins involved in glycosylation, hereafter referred to as glycogenes. Because giant virus genetics was not yet available, we could not validate these predictions by mutagenesis. Instead, we considered that identification of predicted sugars by chemical analysis of the viral particles validated the presence of a functional pathway encoded by the virus, because the amoeba host does not synthesize these rare bacteria-like sugars. The general approach used to attribute a specific function to a gene consisted in the comparison of the translated sequences with reference annotated protein sequences by a multiple alignment based on structural information, using the Expresso Server (Armougom et al. 2006) (http://tcoffee.crg.cat/apps/tcoffee/do:expresso). Multiple alignments were submitted to the ESPript server (http://espript.ibcp.fr/ESPript/ESPript/) to illustrate sequence similarities and display secondary structure elements (Gouet et al. 2003).
Multiple alignments allowed to verify if catalytic residues were conserved in candidate proteins compared with the reference protein for which the function and structure were known. Conservation of the catalytic site was a prerequisite to attribute a specific function. In addition, we used the HHpred server (Hildebrand et al. 2009) for remote homology detection to annotate hypothetical proteins. We only kept those for which the confidence level was above 99%.
To understand if identified glycogenes were recently acquired from other microorganisms, such as bacteria on which the amoeba feeds, we compared the GC content of these genes with those of the seven marker genes (Asp Synthase, Helicase, mRNA Capping Enzyme, MutS, Packaging ATPase, PolyA polymerase and VLTF3) as representatives of whole genome GC content. GC content was computed using geecee (Emboss package v6.6.0, https://www.bioinformatics.nl/cgi-bin/emboss/geecee).
Conservation of the proteins involved in fibril glycosylation in the proposed Megavirinae
Tblastn was used to assess the presence and conservation level of all proteins possibly involved in fibril glycosylation in all members of the three clades. We restricted the study to those for which complete genome sequences were available in the NCBI database (Table S1). Results are presented as conservation heatmaps, which were built using an in-house developed software. Construction of the heatmap was based on the following steps:
tblastn of N proteins (protein file) against M genomes (genome file), so that each protein sequence (query) is compared with the six-frame translations of nucleotide genome sequences;
for each protein, recovery of the best scoring ORF in each genome, with normalization by the size of the protein (score/autoscore), which defines the ‘conservation score’ in heatmaps;
generation of the matrix (N best scores) x (M genomes) and plot of the heatmap (library pheatmap in R, with default parameters).
NCBI accession numbers of complete genome sequences and of protein sequences (query) are reported in Tables S1 and S2, respectively.
Results
Sugar composition varies in fibrils from viruses in different clades
Mimivirus (A-clade), Moumouvirus australiensis and maliensis (B-clade) and Megavirus chilensis (C-clade) were used as prototypes to investigate the glycan composition of their fibrils and identify conserved and clade-specific features. The nature of two complex polysaccharides (Fig. 2) composing Mimivirus fibrils has recently been characterized (Notaro et al. 2021), and they involve three different sugar moieties: rhamnose (Rha), N-acetyl-glucosamine (GlcNAc) and N-acetyl-viosamine (Vio4NAc) (Fig. 3a). Here, we characterized all sugar moieties constituting the fibrils of representative members of B- and C-clades.
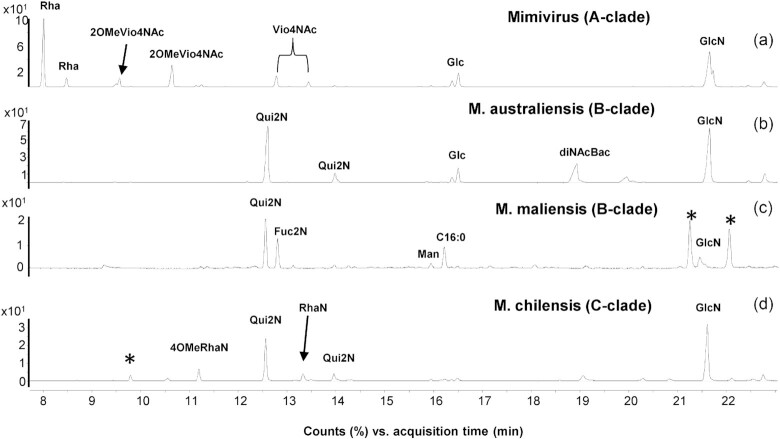
Figure 3.
Sugar composition of the fibrils for members of the proposed Megavirinae subfamily by GC-MS. Chromatogram profiles of the sugars composing the fibrils of (a) Mimivirus (Notaro et al. 2021), (b) Moumouvirus australiensis, (c) Moumouvirus maliensis and (d) Megavirus chilensis. Known standards have been used to assign peaks to the corresponding monosaccharides. The * indicates impurities.
First, chemical characterization of B-clade Moumouvirus australiensis and maliensis fibrils revealed different sugar compositions. Fibrils from Moumouvirus australiensis (Jeudy et al. 2020) contained glucosamine (GlcN), quinovosamine (Qui2N) and bacillosamine (diNAcBac) as major components and glucose (Glc) as a minor component (Fig. 3b). The peak at 19 min was identified as diNAcBac by applying the fragmentation rules of the acetylated methylglycosides derivatives (Lönngren and Svensson 1974); indeed, the EI-MS spectrum contained a fragment at m/z 271 consistent with the oxonium ion of a six-deoxy-sugar with two amino functions (Fig. S1).
By contrast, fibrils from Moumouvirus maliensis contained fucosamine (Fuc2N) and Qui2N as major components, while mannose (Man) and GlcN were present in small amounts (Fig. 3c).
Previous analysis performed on intact virions from Megavirus chilensis, the prototype of C-clade, had revealed the presence of GlcN, rhamnosamine (RhaN) and RhaN methylated at position 4 (4-OMe-RhaN) as the main components of the fibrils together with an unidentified rhamnosamine epimer (Piacente et al. 2014b). Here, we completed this analysis and identified the nature of this component as Qui2N, in agreement with previous results revealing the biosynthetic pathways encoded by the virus (Piacente et al. 2014b). In addition, our current analysis confirmed the presence of GlcN, RhaN and 4-OMe-RhaN (Fig. 3d).
We determined absolute configurations for most sugars for members of all clades. The absolute configuration of GlcN was experimentally confirmed to be d for Mimivirus (Fig. S2), in agreement with the presence of a biosynthetic pathway for UDP-d-GlcNAc (Piacente et al. 2014a). Similarly, we showed that glycans in Moumouvirus australiensis and Megavirus chilensis contained d-GlcNAc (Fig. S2), and that Qui2N was in the l configuration. By contrast, we determined that Qui2N and Fuc2N were both in the d configuration for Moumouvirus maliensis (Fig. S2). We could not determine the absolute configuration of Rha2N and diNAcBac due to the lack of appropriate standards. However, it is likely that the absolute configuration of Rha2N is l, because the UDP-l-Rha2NAc biosynthetic pathway was validated in vitro for Megavirus chilensis (Piacente et al. 2014b). In bacteria, diNAcBac is in the d configuration (Morrison and Imperiali 2014), suggesting that this sugar could adopt the same configuration in Moumouvirus australiensis glycans.
Filling the gap between experimental and genomic data for Mimivirus (A-clade)
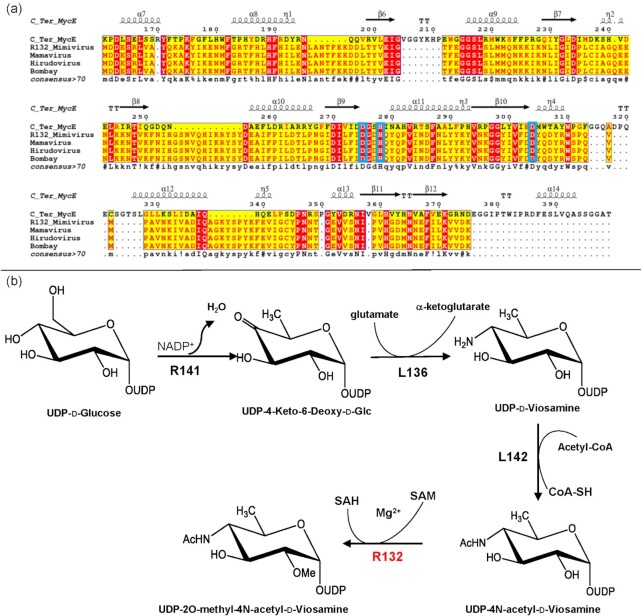
The elucidation of the glycan structures of Mimivirus fibrils (Fig. 2) (Notaro et al. 2021) prompted the search for genes coding for proteins that could be involved in the biosynthesis of these polysaccharides. A previous study had proposed a nine-gene cluster (Piacente et al. 2012) that includes genes encoding proteins necessary for the biosynthesis of Vio4NAc (R141, L136, L142), a gene annotated as pyruvyltransferase (L143), several genes encoding glycosyltransferases (L137, L138, R139, L140 and the C-ter of L142) and a gene encoding a GMC-type oxidoreductase (R135). However, a gene coding for a Vio4NAc methyltransferase was missing, raising the question on the origin of this sugar modification. Viosamine is not produced by the amoeba host and is only encountered in bacteria. For example, Pseudomonas syringe possesses a Vio-island including all the genes involved in viosamine production, methylation and transfer (Yamamoto et al. 2011). Thus, we hypothesized that a similar organization could exist in the Mimivirus genome. We narrowed the search within 2 kbp of the nine-gene cluster (Piacente et al. 2012, 2017b) and identified R132 as a promising candidate. The R132 gene is predicted to encode a 221-amino-acid protein belonging to class I SAM-dependent methyltransferases. Its closest homologs are bacterial methyltransferases from Rizhobiales (WP_112 557 168.1), Lelliottia (WP_107 702 876.1) and Pantoea dispersa (WP_021 509 887.1), which share 35 to 39% sequence identity with R132 on the entire length of the protein sequence. The R132 protein was modeled using Phyre2 with 100% confidence based on several sugar-methyltransferases structures and confirmed by AlphaFold (Jumper et al. 2021). The best ranked model was obtained using the C-terminal catalytic domain of MycE as template (residues 161–399, 20% sequence identity). This SAM and metal-dependent methyltransferase domain is responsible for methylation of the 6-deoxyallose sugar moiety of mycinamicins (Akey et al. 2011). A multiple-alignment of R132 and its orthologs in A-clade members with the C-terminal domain of MycE (Fig. 4a) revealed that all catalytic residues are conserved, suggesting that R132 could encode a functional methyltransferase (Fig. 4a). Further support of this prediction comes from two additional observations. First, previous studies showed that timing and expression level of R132 were comparable with those of the nine genes belonging to the glycan formation cluster (Legendre et al. 2010). Second, the R132 gene is absent from the genome of Mimivirus M4. This virus is the result of a 150-times subculture of Mimivirus in a germ-free amoeba host that led to the dramatic reduction of its genome from 1.20 to 0.993 Mbp due to two large deletions, mainly at the two extremities of the genome. One of these deletions includes genes encoding for proteins known to be involved in sugar biosynthesis (Boyer et al. 2011a, Piacente et al. 2012) as well as structural proteins composing the fibrils such as GMC-oxidoreductase R135 (Klose et al. 2015). Taken together, our results suggest a possible role for R132 in O-2 methylation of Vio4NAc. We thus propose that R132 is part of the gene cluster encoding a biosynthetic pathway for the sugars of structural proteins making Mimivirus fibrils.
Figure 4.
R132 methyltransferase, a candidate for viosamine methylation. (a) Multiple alignment of the C-Ter of MycE (PDB 3SSN), R132 of Mimivirus (YP_003 986 624), its homolog in Mamavirus (AEQ60310.1), Hirudovirus strain Sangsue (AHA45742.1) and Mimivirus Bombay (AMZ02580.1). The blue rectangles indicate the residues involved in catalytic activity: the two aspartates coordinating the magnesium D275 and D304 (Akey et al. 2011) are conserved in R132 and A-clade (D127 and D156); the histidine residue (H278) involved in deprotonation of the 2-OH is also conserved (H138). (b) Proposed biosynthetic pathway for the UDP-d-2O-methyl-4N-acetyl-viosamine. The functions of the first three enzymes have been experimentally validated and R132, based on in silico analysis, would be the missing methyltransferase.
As proposed in a previous study, the methylation reaction should occur after the acetylation of the amino function at position 4 (Fig. 3b), due to the steric hindrance caused by the 2OMe on the viosamine moiety that would impair acetylation by the L142 enzyme (Piacente et al. 2017b). We still do not know if methylation occurs on the UDP-sugar, as reported in Fig. 4 or on viosamine inside the polysaccharide chain, this point being only addressable experimentally.
Biosynthetic pathways for nucleotide-sugars in Moumouvirus australiensis(B-clade)
Here, we identified the monosaccharides composing Moumouvirus australiensis fibrils (Fig. 3b) as GlcN, Qui2N and diNAcBac. To determine if its genome encodes proteins necessary for biosynthesis of these sugars as activated nucleotides, we searched for genes and proteins similar to those already reported for the Mimivirus and Megavirus chilensis biosynthetic pathways for GlcN (Piacente et al. 2014a), Qui2N (Piacente et al. 2014b) and to those of Campylobacter jejuni for the biosynthesis of diNAcBac (Olivier and Imperiali 2008, Morrison and Imperiali 2014, Riegert et al. 2015, 2017). Sequence identity values with reference sequences are reported in Table 1. Structural multiple alignments were performed to assess conservation of the catalytic sites for each enzyme of the pathway and to infer whether Moumouvirus australiensis candidate enzymes could be functional.
Table 1.
Identification of proteins responsible for nucleotide-sugars synthesis in Moumouvirus australiensis (B-clade). Based on known proteins involved in biosynthetis pathways for quinovosamine (UDP-l-Qui2N), bacillosamine (UDP-d-diNAcBac) and glucosamine (UDP-d-GlcNAc), we identified corresponding proteins in Moumouvirus australiensis and its orthologs in other viral genomes, such as Tupanviruses and Cotonvirus japonicus. For each pathway we reported proteins used as references, homologs in Moumouvirus australiensis and its closest orthologs in other viral genomes. For each protein, we show Accession number (NCBI database), % of identity (% Id.) and % of query coverage (% Cover).
| Reference protein | Ortholog in Moumouvirus australiensis | Orthologs in the proposed Megavirinae subfamily | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pathway | Source | Name | Accession | Name | Accession | % Id. | %Cover | Source | Accession | % Id. | % Cover |
| UDP-l-Qui2N | Megavirus chilensis | Mg534 | PDB: 4TQG | Ma458 | AVL94844 | 79.50 | 99 | Cotonvirus | BCS83197 | 78.26 | 99 |
| Hyperionvirus | AYV83503 | 61.84 | 93 | ||||||||
| KNV1 | ARF11838 | 57.01 | 99 | ||||||||
| CTV1 | ARF08716 | 57.85 | 99 | ||||||||
| Mg535 | YP_004894586 | Ma459 | AVL94845 | 59.48 | 99 | Cotonvirus | BCS83198 | 65.19 | 99 | ||
| KNV1 | ARF11657 | 48.91 | 98 | ||||||||
| CTV1 | ARF08710 | 45.59 | 99 | ||||||||
| Mg536 | YP_004894587 | Ma460 | AVL94846 | 70.00 | 99 | Cotonvirus | BCS83199 | 72.78 | 100 | ||
| KNV1 | ARF11840 | 60.38 | 98 | ||||||||
| UDP-d-diNAcBac | Campylobacter jejuni | PglF | PDB:5BJU | Ma467 | AVL94853 | 27.95 | 79 | Moumouvirus | YP_007354434 | 69.18 | 100 |
| Moumouvirus | YP_007354431 | 52.54 | 98 | ||||||||
| M. goulette | AGF85279 | 67.38 | 100 | ||||||||
| M. monve | AEX62730 | 52.54 | 98 | ||||||||
| Cotonvirus | BCS83207 | 79.57 | 100 | ||||||||
| T. deep ocean | QKU33921 | 78.49 | 100 | ||||||||
| T. soda lake | QKU35168 | 78.49 | 100 | ||||||||
| PglE | PDB:1O61 | Ma465 | AVL94851 | 33.19 | 60 | Cotonvirus | BCS83209 | 75.32 | 99 | ||
| T. deep ocean | QKU33915 | 66.93 | 98 | ||||||||
| T. soda lake | QKU35164 | 67.19 | 99 | ||||||||
| PglD | PDB:3BSS | Ma466 | AVL94852 | 30.69 | 95 | Cotonvirus | BCS83208 | 68.78 | 98 | ||
| T. deep ocean | QKU33920 | 63.94 | 99 | ||||||||
| T. soda lake | QKU35167 | 64.08 | 98 | ||||||||
| Mimivirus | YP_003986634 | 45.19 | 99 | ||||||||
| UDP-d-GlcNAc | Mimivirus | L619 | YP_003987136 | Ma652 | AVL95038 | 61.58 | 100 | Cotonvirus | BCS83440 | 63.01 | 100 |
| M. chilensis | YP_004894796 | 58.62 | 100 | ||||||||
| T. deep ocean | QKU33475 | 58.12 | 100 | ||||||||
| T. soda lake | QKU34707 | 58.28 | 100 | ||||||||
| Hyperionvirus | AYV83493 | 41.28 | 100 | ||||||||
| L316 | YP_003986819 | Ma514 | AVL94900 | 58.22 | 98 | Cotonvirus | BCS83270 | 63.27 | 99 | ||
| M. chilensis | YP_004894641 | 56.16 | 98 | ||||||||
| T. deep ocean | QKU33820 | 61.64 | 98 | ||||||||
| T. soda lake | QKU35067 | 66.21 | 97 | ||||||||
| R689 | YP_003987216 | Ma192 | AVL94578 | 66.94 | 96 | Cotonvirus | BCS82870 | 61 | 96 | ||
| M. chilensis | YP_004894291 | 65.59 | 96 | ||||||||
| T. deep ocean | QKU34490 | 68.31 | 95 | ||||||||
| T. soda lake | QKU35828 | 65.74 | 98 | ||||||||
UDP-d-N, N’-diacetyl-bacillosamine pathway (UDP-d-diNAcBac)
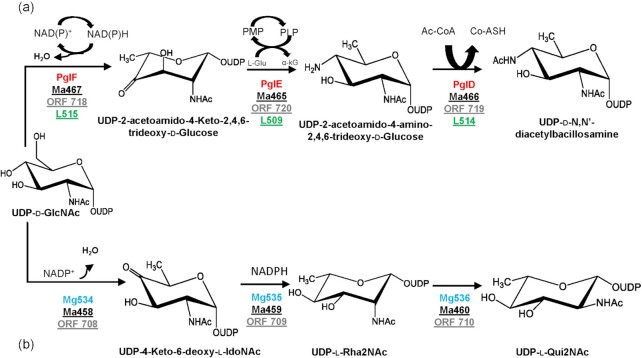
The UDP-d-diNAcBac pathway has been characterized in Campylobacter jejuni by determining catalytic activities and 3D structures of all its enzymes (Olivier and Imperiali 2008, Morrison and Imperiali 2014, Riegert et al. 2015, 2017). Synthesis of diNAcBac begins with GlcNAc and involves a three-step reaction: (1) dehydration (PglF), (2) transamination (PglE) and (3) acetylation (PglD). A Tblastn search using C. jejuni enzymes as queries (Table 2) enabled identification of Ma467, Ma465 and Ma466 as the first, second and third enzymes of the Moumouvirus australiensis pathway (Fig. 5a, Table 1).
Table 2.
Identification of proteins responsible for nucleotide-sugars synthesis in Moumouvirus maliensis (B-clade). Based on known proteins involved in biosynthesis pathways for quinovosamine (UDP-d-Qui2N) and fucosamine (UDP-d-Fuc2N), we identified corresponding proteins in Moumouvirus maliensis and its closest orthologs in other giant virus genomes. For each pathway, we report proteins used as references, homologs in Moumouvirus maliensis and its orthologs in the proposed Megavirinae subfamily and in Pseudomonas aeruginosa. For each protein, we show Accession number (NCBI database), % of identity (% Id.) and % of query coverage (% Cover).
| Reference protein | Ortholog in Moumouvirus maliensis | Orthologs in the proposed Megavirinae subfamily | Orthologs in Pseudomonas aeruginosa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pathway | Source | Name | Accession | Name | Accession | % Id. | % Cover | Source | Accession | % Id. | % Cover | Accession | % Id. | % Cover |
| UDP-d-Qui2Nor UDP-d_Fuc2N | C. jejuni | PglF | PDB:5BJU | Mm422 | QGR53991 | 30.05 | 59 | M. australiensis | AVL94853 | 67,74 | 100 | AAF72960 | 24,66 | 73 |
| T. deep ocean | QKU33921 | 68,46 | 100 | |||||||||||
| T. soda lake | QKU35168 | 68,46 | 100 | |||||||||||
| A. thermoaerophilus | Rmd | PDB:2PK3 | Mm421 | QGR53990 | 26,32 | 99 | T. deep ocean | AUL79276 | 63,92 | 98 | AAC45865 | 21,48 | 82 | |
| T. soda lake | QKU35170 | 62,46 | 98 | AF23991 | 22,85 | 98 | ||||||||
| Mm419 | QGR53988 | 31 | 29 | T. deep ocean | QKU33921 | 53,99 | 98 | AAC45865 | 19,5 | 52 | ||||
| T. soda lake | QKU35168 | 53,99 | 98 | AF23991 | 21,18 | 52 | ||||||||
| UDP-d-GlcNAc | Mimivirus | L619 | YP_003987136 | Mm595 | QGR54170 | 61,25 | 100 | M. chilensis | YP_004894796 | 58.62 | 100 | — | — | — |
| T. deep ocean | QKU33475 | 58.12 | 100 | |||||||||||
| T. soda lake | QKU34707 | 58.28 | 100 | |||||||||||
| Hyperionvirus | AYV83493 | 41.28 | 100 | |||||||||||
| L316 | YP_003986819 | Mm467 | QGR54037 | 57,53 | 98 | M. chiliensis | YP_004894641 | 56.16 | 98 | — | — | — | ||
| T. deep ocean | QKU33820 | 61.64 | 98 | |||||||||||
| T. soda lake | QKU35168 | 66.21 | 97 | |||||||||||
| R689 | YP_003987216 | Mm152 | QGR53719 | 67,35 | 96 | M. chiliensis | YP_004894291 | 65.59 | 96 | — | — | — | ||
| T. deep ocean | QKU34490 | 68.31 | 95 | |||||||||||
| T. soda lake | QKU35828 | 65.74 | 98 | |||||||||||
Figure 5.
Proposed Moumouvirus australiensis(a) UDP-d-diNAcBac pathway shared with Tupanviruses and (b) UDP-l-QuiNAc synthetic pathway shared with Cotonvirus japonicus. Campylobacter jejuni proteins (Pgl, red) are used as reference for the bacillosamine pathway and Megavirus chilensis proteins (Mg, blue) for the quinovosamine pathway. The corresponding Moumouvirus australiensis (Ma, black), Cotonvirus japonicus (ORF, dark gray) and Tupanvirus deep ocean (L, light green) proteins are underlined. The NCBI accession number of protein sequences, their lengths and functions are reported in Table 1.
Ma467 is conserved in all B-clade members (Table 1), while Ma465 and Ma466 have no homolog inside the B-clade. Surprisingly, their closest homologs are found in the Tupanvirus strains (D-clade) and Cotonvirus japonicus (E-clade) (Table 1). By contrast, lower sequence identities were found with the corresponding enzymes of C. jejuni (Table 1) (Riegert et al. 2017). Our in silico analyses of Ma467, Ma465 and Ma466 revealed that all catalytic residues are conserved (Figs. S3-S5), suggesting that these enzymes are functional and responsible for the biosynthesis of UDP-d-diNAcBac in Moumouvirus australiensis as suggested by the presence of diNAcBac in the fibrils of Moumouvirus australiensis (Fig. 3b). In addition, the identified biosynthetic pathway supports a d configured bacillosamine. Finally, in Moumouvirus gp464 and Moumouvirus Monve mvR525 proteins, which are homologues of Ma467, one of the catalytic aspartates is replaced by an asparagine (Fig. S3). This change has been associated with a loss of activity in C. jejuni D396N-PglF mutant (Riegert et al. 2017), suggesting that these strains (and the B-clade) could be in the process of losing the pathway.
UDP-l-N-acetyl-quinovosamine pathway (UDP-l-Qui2NAc)
Previous studies had shown that two Megavirus chilensis proteins (Mg534, Mg535) were involved in UDP-l-Rha2NAc biosynthesis (Fig. 5b) and had predicted that a third protein (Mg536) could convert Rha2NAc into Qui2NAc (Piacente et al. 2014b). Although the function of Mg536 was not experimentally validated, the bioinformatic prediction is based on the identification of Qui2NAc as a component of the glycans decorating Megavirus chilensis fibrils (Fig. 3d).
Synthesis of Qui2NAc starts from GlcNAc and involves a five-step reaction and three enzymes (Fig 5b): (1 and 2) 4,6-dehydration and 5-epimerization (Mg534); (3 and 4) 4-reduction and 3-epimerization (Mg535); and (5) 2-epimerization (Mg536). Using these three proteins as queries, we identified corresponding orthologs in Moumouvirus australiensis (Ma458, Ma459 and Ma460) as the ones possibly involved in Qui2NAc production (Figs 5b and S6, Table 1). Ma458 corresponds to a 324-amino acid protein homologous to the Megavirus chilensis inverting UDP-GlcNAc 4,6-dehydratase, for which structure (PDB 4TQG) and activity were determined (Piacente et al. 2014b). Ma459 and Ma460 correspond respectively to 270- and 376-amino acid proteins with their closest homologues found in Megavirus chilensis (Mg535 and Mg536, respectively) (Table 1). Taken together, these results support the presence of a functional pathway for Qui2NAc in Moumouvirus australiensis (Fig. 5).
Ma458 is also present in Cotonvirus japonicus and in giant DNA viruses identified from metagenomics data (Klosneuvirus KNV1, Catovirus CTV1 and Hyperionvirus) (Schulz et al. 2017), while Ma459 is present in Cotonvirus, KNV1 and CTV1, and Ma460 is only present in Cotonvirus and in KNV1 (Table 1). Therefore, these results also support a functional pathway for Qui2NAc in Cotonvirus and KNV1 (Table 1). By contrast, CTV1 seems to only have the two first enzymes of the pathway, consequently restrained to Rha2N production.
UDP-d-N-acetyl-glucosamine pathway (UDP-d-GlcNAc)
Although the amoeba host produces GlcNAc using typical eukaryotic enzymes, Mimivirus and Megavirus chilensis also possess necessary enzymes to synthesize this sugar (Piacente et al. 2014a) with intermediate properties between the eukaryotic and prokaryotic pathways. Using Mimivirus proteins (L619, L316 and R689) as queries, we identified corresponding enzymes in Moumouvirus australiensis (Ma652, Ma514 and Ma192, respectively; Table 1). Our in silico analyses suggested that the pathway for UDP-d-GlcNAc is conserved in members of all clades (Table 1) and that corresponding genes are scattered along the genome.
Biosynthetic pathways for nucleotide-sugars in Moumouvirus maliensis (B-clade)
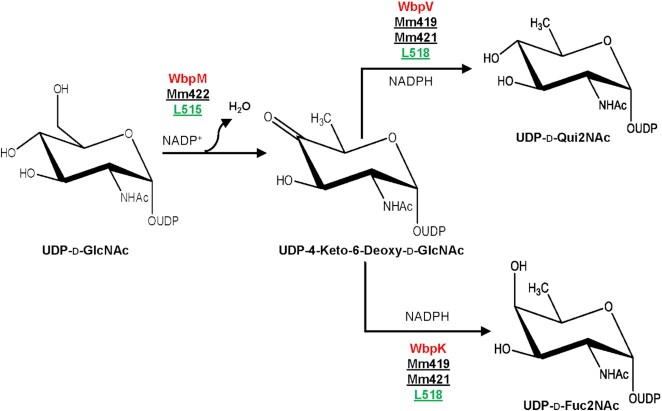
The chemical analysis of Moumouvirus maliensis virions revealed the presence of d-Fuc2N and d-Qui2N (Fig. 3c). Based on this finding, we searched for genes encoding proteins responsible for UDP-d-Qui2N and UDP-d-Fuc2N biosynthesis. Previous studies in bacteria showed that the precursor for both Qui2N and Fuc2N was UDP-d-GlcNAc (Burrows et al. 2000, Li et al. 2014) and that their synthesis involved a two-step reaction: (1) dehydration (UDP-GlcNAc 4,6 dehydratase), resulting in the intermediate UDP-4-keto-2-N-Acetyl-d-GlcNAc; and (2) reduction (4-reductase). Depending on the stereospecificity of the reductase, UDP-d-Qui2NAc or its C4-epimer UDP-d-Fuc2NAc are obtained (Fig. 6). Using bacterial enzymes as queries (Burrows et al. 2000, Li et al. 2014), we were able to identify corresponding enzymes in Moumouvirus maliensis (Fig. 6 and Table 2).
Figure 6.
Proposed biosynthetic pathways for UDP-d-Qui2NAc and UDP-d-Fuc2NAc in B-clade (except Moumouvirus australiensis) and Tupanviruses. Pseudomonas aeruginosa reference proteins are in red and the corresponding Moumouvirus maliensis (black) and Tupanvirus deep ocean (light green) proteins are underlined. The NCBI accession number of the protein sequences, lengths and functions are reported in Table 2.
We found two proteins (Mm419, Mm422) homologous to C. jejuni 4,6-dehydratase (Table 2) (Riegert et al. 2017), but only Mm422 appeared to be functional, while Mm419 presented the mutation of the catalytic aspartate with asparagine (Fig. S7), which was reported to induce loss of activity (Riegert et al. 2017).
By using the A. thermoaerophilus 4-reductase (King et al. 2009) as a reference gene, we identified two putative 4-reductase enzymes (Mm419 and Mm421), which both have the typical consensus sequence for NADP binding and the catalytic triad common to all 4-reductase enzymes (Fig. S8, Table 2). At this stage, it is not possible to discriminate between the two 4-reductases to reach a complete understanding of the pathways for d-Fuc2N and d-Qui2N, as the stereospecificity of the two enzymes can only be assessed experimentally. In addition to the Fuc2N/Qui2N pathway, all the genes responsible for the GlcN production are also present and conserved compared with experimentally validated enzymes (Table 2). The low amount of GlcN revealed by our chemical analysis of Moumouvirus maliensis could possibly be due to conversion of most GlcN into Fuc2N and Qui2N.
Biosynthetic pathways for nucleotide-sugars in Cotonvirus japonicus and Tupanviruses
In contrast to other clades (Fig. 3), the presence of sugars associated with the fibrils of Cotonvirus japonicus and Tupanviruses has not been experimentally characterized. Here, we searched their genomes for genes encoding for possible biosynthetic pathways for nucleotide-sugars. We used protein sequences of enzymes involved in the pathways identified for the A- (Piacente et al. 2012, 2014a, 2017b), B- (Tables 1 and 2) and C-clades (Piacente et al. 2014b) as queries. We found that Cotonvirus japonicus encodes a pathway for UDP-l-Qui2NAc shared with Megavirus chilensis and Moumouvirus australiensis (Figs. 5b and S6, Table 1). Both Cotonvirus japonicus and Tupanvirus strains share the biosynthetic pathway for UDP-d-diNAcBac with Moumouvirus australiensis (Table 1, Fig. 5a), with conservation of all catalytic residues suggesting all enzymes can be functional (Figs S3, S4 and S5). In addition, Tupanviruses possess the biosynthetic pathway for UDP-d-Qui2N/Fuc2N (Figs 6 and S7, Table 2) conserved in members of the B-clade, except in Moumouvirus australiensis. The first enzyme of the pathway (L515) is also the first enzyme in the diNAcBac pathway and is conserved in Moumouvirus australiensis (Fig. S7). However, in contrast to Moumouvirus australiensis, Tupanviruses possess the second enzyme (L518) corresponding to the 4-reductase in Moumouvirus maliensis (Mm421). The catalytic triad of this enzyme is conserved (Table 2, Fig. S8), suggesting that the entire pathway should be functional in Tupanviruses.
In the vicinity of the genes encoding the diNAcBac pathway in Tupanviruses, we identified the R520 gene as encoding an UDP-glucose-6-dehydrogenase enzyme. This result suggests that Tupanvirus could convert UDP-d-glucose (UDP-d-Glc) into UDP-d-glucuronic acid (UDP-d-GlcA) (Fig. S9a). The closest homologs of R520 are found in KNV1 and Cafeteria roenbergensis virus (CroV), which also belongs to the Mimiviridae family (Fischer et al. 2010). Using HHpred for remote homology and structure prediction (Hildebrand et al. 2009), we found that Burkholderia cepacia UDP-glucose-6-dehydrogenase (PDB:2Y0C) was homologous to R520 (30% of identity and 100% confidence). Comparison of their sequences revealed that all catalytic residues were conserved in R520, suggesting that it is a functional enzyme (Fig. S9). In contrast to the other clades that use UDP-d-Glc from their host, Tupanviruses possess the L502 gene encoding a 634-amino acid protein with a predicted glucose-1P-uridyltransferase N-terminal domain (confidence 99%). This enzyme uses glucose-1P to synthesize UDP-d-Glc, allowing Tupanviruses to be completely independent from the host for glycosylation. The biosynthetic pathway for UDP-d-Glc is present in the amoeba host and is expressed during the late stage of the infection, as is seen for the viral pathway. Therefore, we cannot exclude that viruses could also recruit the host UDP-d-Glc to build their own glycans.
Genomic organization of glyco-related genes in the different clades
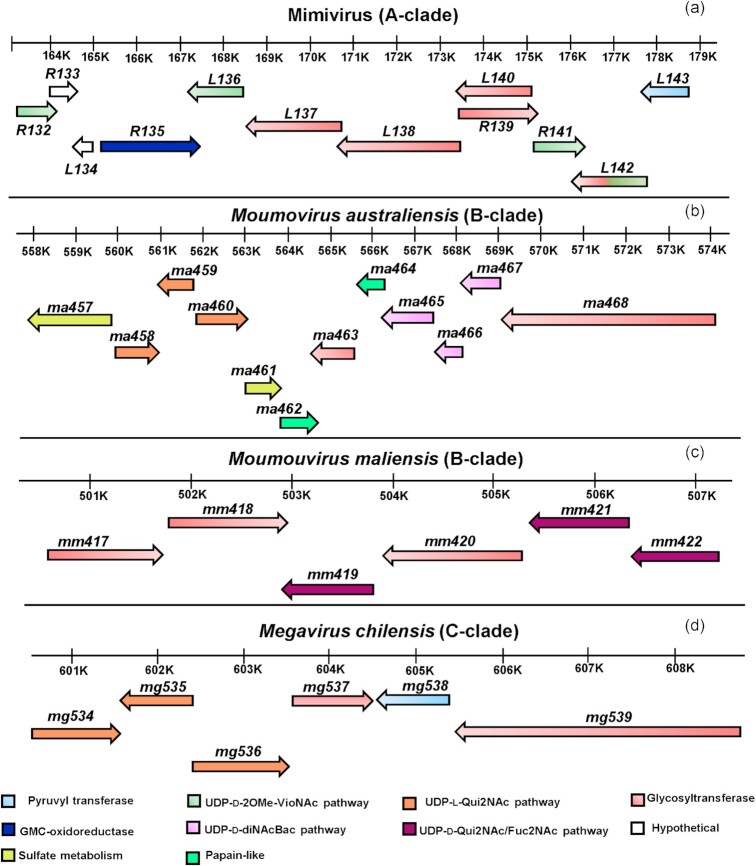
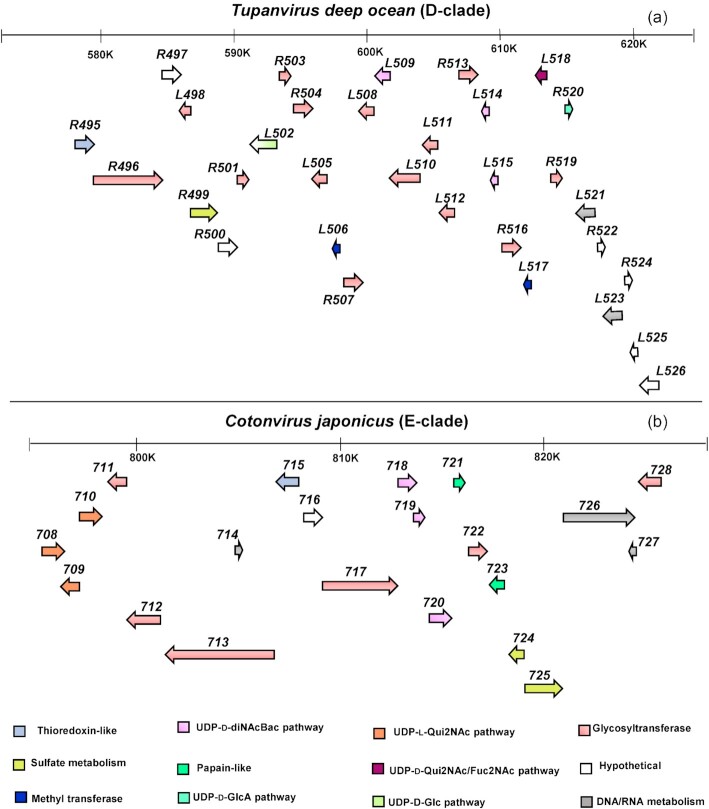
Our analysis of the genomes of prototypical viruses from different clades revealed that most genes responsible for the synthesis of nucleotide-sugars, along with others likely involved in the glycosylation process, are located within the same region of the genome, namely in gene clusters (Figs. 7 and 8, Tables 3, 4 and 5).
Figure 7.
Organization of the glycosylation gene clusters of (a)Mimivirus, involved in the biosynthesis of two polysaccharides (Notaro et al. 2021) and used as prototype for A-clade; (b) for B-clade outsider Moumouvirus australiensis and (c) for the B-clade prototype Moumouvirus maliensis; (d) for C-clade prototype Megavirus chilensis. The arrows' direction indicates the coding strand. The function of each gene is color-coded (explained in the legend) and reported in Table 3.
Figure 8.
Organization of the glycosylation gene clusters of (a)Tupanvirus deep ocean as prototype of the two Tupanviruses strains. (b)Cotonvirus japonicus as prototype of a new clade isolated in 2021. The arrows' direction indicates the coding strand. The function of each gene is color-coded (explained in the legend) and it is reported in Tables 4 and 5.
Table 3.
Glycosylation gene clusters for Mimivirus, Moumouvirus australiensis, Moumouvirus maliensis and Megavirus chilensis. For each virus, the NCBI Accession number of the complete genome and the genes names in each cluster are reported as well as the Accession number of the encoded protein, its length and expected function.
| Source | Genome accession # | Gene | Protein accession # | Length (aa) | Predicted function |
|---|---|---|---|---|---|
| Mimivirus (A-clade) | NC_014649 | R132 | YP_003 986 624 | 221 | Methyltransferase |
| R133 | YP_003 986 625 | 127 | Hypothetical | ||
| L134 | YP_003 986 626 | 163 | Hypothetical | ||
| R135 | YP_003 986 627 | 702 | GMC-type oxidoreductase | ||
| L136 | YP_003 986 628 | 352 | Pyridoxal phosphate-dependent transferase | ||
| L137 | YP_003 986 629 | 732 | Glycosyltransferase | ||
| L138 | YP_003 986 630 | 883 | Glycosyltransferase | ||
| R139 | YP_003 986 631 | 248 | Glycosyltransferase | ||
| L140 | YP_003 986 632 | 304 | Glycosyltransferase | ||
| R141 | YP_003 986 633 | 323 | 4,6-dehydratase | ||
| L142 | YP_003 986 634 | 490 | N-ter:acetyltransferase, C-ter: glycosyltransferase | ||
| L143 | YP_003 986 635 | 287 | Piruvyltransferase | ||
| Moumouvirus australiensis (B-clade) | MG807320 | ma457 | AVL94843 | 642 | Adenylyl-sulfate kinase |
| ma458 | AVL94844 | 324 | 4,6-dehydratase, 5-epimerase | ||
| ma459 | AVL94845 | 270 | 4-reductase | ||
| ma460 | AVL94846 | 376 | 2-epimerase | ||
| ma461 | AVL94847 | 263 | Sulfotransferase* | ||
| ma462 | AVL94848 | 188 | Papain-like* | ||
| ma463 | AVL94849 | 325 | Glycosyltransferase* | ||
| ma464 | AVL94850 | 204 | Papain-like* | ||
| ma465 | AVL94851 | 384 | Aminotransferase | ||
| ma466 | AVL94852 | 209 | Acetyltransferase | ||
| ma467 | AVL94853 | 278 | 4,6-dehydratase | ||
| ma468 | AVL94854 | 1666 | Glycosyltransferase | ||
| Moumouvirus maliensis (B-clade) | MK978772 | mm417 | QGR53986 | 389 | Glycosyltransferase (GT-B) |
| mm418 | QGR53987 | 386 | Glycosyltransferase (GT-B) | ||
| mm419 | QGR53988 | 278 | 4-reductase | ||
| mm420 | QGR53989 | 467 | Glycosyltransferase | ||
| mm421 | QGR53990 | 321 | 4-reductase | ||
| mm422 | QGR53991 | 279 | 4,6-dehydratase | ||
| Megavirus chilensis (C-clade) | NC_016 072 | mg534 | PDB: 4TQG | 323 | 4,6-dehydratase, 5-epimerase |
| mg535 | YP_004 894 586 | 270 | 4-reductase | ||
| mg536 | YP_004 894 587 | 370 | 2-epimerase | ||
| mg537 | YP_004 894 588 | 312 | Putative GT* | ||
| mg538 | YP_004 894 589 | 277 | Piruvyltransferase | ||
| mg539 | YP_004 894 590 | 1097 | Glycosyltransferase |
indicates that the protein function was predicted with >99% confidence by HHpred.
Table 4.
Glycosylation gene clusters for Tupanviruses. The NCBI Accession number of the complete genomes and the names of the genes in each cluster are reported as well as the Accession number of the encoded protein, its length and expected function.
| Tupanvirus deep ocean | Tupanvirus soda lake | ||||||
|---|---|---|---|---|---|---|---|
| Genome accession # | Gene | Protein accession # | Length (aa) | Predicted function | Protein accession | %ID | %Cover |
| MF405918 | R496 | QKU33902 | 1707 | Glycosyltransferase | QKU35148 | 83.63 | 100 |
| R497 | QKU33903 | 475 | Hypothetical | QKU35149. | 85.09 | 96 | |
| L498 | QKU33904 | 266 | Glycosyltransferase* | QKU35150 | 82.17 | 86 | |
| R499 | QKU33905 | 655 | Adenylyl-sulfate kinase | QKU35151 | 88.85 | 100 | |
| R500 | QKU33906 | 453 | Hypothetical | QKU35152 | 80.49 | 99 | |
| R501 | QKU33907 | 291 | Glycosyltransferase* | QKU35153 | 80.80 | 94 | |
| L502 | QKU33908 | 639 | Glycosyltransferase | QKU35154 | 90.27 | 99 | |
| R503 | QKU33909 | 334 | Glycosyltransferase* | QKU35155. | 84.73 | 100 | |
| R504 | QKU33910 | 510 | Glycosyltransferase* | QKU35156 | 83.53 | 100 | |
| L505 | QKU33911 | 365 | Glycosyltransferase* | QKU35157 | 74.52 | 100 | |
| L506 | QKU33912 | 219 | Methyltransferase* | QKU35158 | 84.72 | 98 | |
| R507 | QKU33913 | 425 | Glycosyltransferase* | QKU35159 | 80.19 | 99 | |
| L508 | QKU33914 | 373 | Glycosyltransferase* | QKU35161 | 61.20 | 97 | |
| L509 | QKU33915 | 388 | Aminotransferase | QKU35164. | 87.99 | 98 | |
| L510 | QKU33916 | 761 | Multi GT domains* | QKU35165 | 57.49 | 93 | |
| L511 | QKU33917 | 367 | Multi GT domains | QKU35162 | 73.00 | 100 | |
| L512 | QKU33918 | 448 | Glycosyltransferase* | QKU35165 | 55.19 | 81 | |
| R513 | QKU33919 | 509 | Glycosyltransferase* | QKU35166 | 59.49 | 100 | |
| L514 | QKU33920 | 209 | Acetyltransferase | QKU35167 | 79.13 | 98 | |
| L515 | QKU33921 | 279 | 4-epimerase | QKU35168 | 87.81 | 100 | |
| R516 | QKU33922 | 546 | Glycosyltransferase* | QKU35169 | 63.08 | 83 | |
| L517 | QKU33923 | 194 | Methyltransferase* | — | — | — | |
| L518 | AUL79276 | 321 | 4-reductase | QKU35170 | 90.88 | 99 | |
| R519 | QKU33924 | 360 | Glycosyltransferase | QKU35171 | 87.53 | 100 | |
| R520 | QKU33925 | 268 | UDP-glucose 6-dehydrogenase | QKU35172 | 91.76 | 95 | |
| L521 | QKU33926 | 506 | Bifunctional glutamate/proline tRNA-synthetase | QKU35173 | 86.17 | 100 | |
| R522 | QKU33927 | 172 | Hypothetical | AUL77993 | 43.20 | 94 | |
| L523 | QKU33928 | 445 | Histidine tRNA synthetase | QKU35174 | 85.20 | 100 | |
| R524 | QKU33929 | 188 | Hypothetical | QKU35176 | 85.11 | 100 | |
| L525 | AUL79282 | 200 | Hypothetical | QKU35177 | 81.59 | 100 | |
| L526 | QKU33930 | 511 | Hypothetical | QKU35178 | 62.44 | 66 | |
indicates that the protein function was predicted with >99% confidence by HHpred.
Table 5.
Glycosylation gene cluster for Cotonvirus japonicus. The NCBI Accession number of the complete genome and the name of the genes in the cluster are reported as well as the Accession number of the encoded protein, its length and expected function.
| Genome accession # | Gene | Protein accession # | Length (aa) | Predicted function |
|---|---|---|---|---|
| AP024483 | ORF_708 | BCS83197 | 324 | 4,6-dehydratase, 5-epimerase |
| ORF_709 | BCS83198 | 271 | 4-reductase | |
| ORF_710 | BCS83199 | 372 | 2-epimerase | |
| ORF_711 | BCS83200 | 285 | Glycosyltransferase | |
| ORF_712 | BCS83201 | 564 | Glycosyltransferase* | |
| ORF_713 | BCS83202 | 1802 | Glycosyltransferase | |
| ORF_714 | BCS83203 | 138 | ABC transporter permease protein YphD | |
| ORF_715 | BCS83204 | 361 | Thioredoxin-like | |
| ORF_716 | BCS83205 | 286 | Hypothetical | |
| ORF_717 | BCS83206 | 1196 | Glycosyltransferase | |
| ORF_718 | BCS83207 | 279 | 4-epimerase | |
| ORF_719 | BCS83208 | 210 | Acetyltransferase | |
| ORF_720 | BCS83209 | 386 | Aminotransferase | |
| ORF_721 | BCS83210 | 208 | Papain-like | |
| ORF_722 | BCS83211 | 336 | Glycosyltransferase* | |
| ORF_723 | BCS83212 | 265 | Papain-like | |
| ORF_724 | BCS83213 | 264 | Sulfotransferase | |
| ORF_725 | BCS83214 | 624 | Adenylyl-sulfate kinase | |
| ORF_726 | BCS83215 | 1092 | Muts-like protein | |
| ORF_727 | BCS83216 | 73 | DNA-directed RNA polymerase | |
| ORF_728 | BCS83217 | 371 | Glycosyltransferase* |
indicates that the protein function was predicted with >99% confidence by HHpred.
For Mimivirus (A-clade), we identified R132, which encodes a Vio4NAc methyltransferase (Fig. 4), and expanded the original cluster from nine (Piacente et al. 2012) to 12 genes (Fig. 7a, Table 3). This larger cluster also includes R133 and L134, which encode two ORFans proteins whose function in glycosylation remains to be determined (Fig. 7a, Table 3).
For Moumouvirus australiensis (B-clade), we identified gene clusters responsible for l-Qui2NAc (ma458, ma459, ma460) and d-diNAcBac (ma465, ma466, ma467) synthesis. Next to these genes, we found others that are also related to glycosylation, allowing us to define a larger cluster containing 12 genes (Fig. 7b, Table 3). Two of these genes encode proteins involved in sulfate metabolism (ma457, ma461), with Ma457 predicted as a bifunctional Sulfate adenylyltransferase/Adenosine-5'-phosphosulfate kinase and Ma461 as a sulfotransferase. This result suggests that sugars could be further modified by a sulfate group, but experimental validation is needed. In the same region, we found two predicted papain-like proteins (Ma462 and Ma464) that are not strictly related to glycosylation and a predicted glycosyltransferase (Ma463), which is related to glycosyltransferases (GTs) from Streptococcus sanguinis (5V4A) and S. parasanguinis (4PHR). In addition, ma468 encodes a 1666-amino acids protein with four GT domains. Moumouvirus australiensis thus contains the genes necessary for its nucleotide-sugars production as well as the GTs responsible for their assembly into oligosaccharides or polysaccharides.
For Moumouvirus maliensis (B-clade), we identified a smaller six-gene cluster (Fig. 7c, Table 3) that includes genes for UDP-d-Qui2NAc and UDP-d-Fuc2NAc biosynthesis, and three genes (mm417, mm418 and mm420) encoding putative GT enzymes.
For Megavirus chilensis (C-clade), we had also identified a six-gene cluster (Piacente et al. 2014b), which encodes proteins involved in Rha2NAc (Mg534, Mg535) and l-Qui2NAc (Mg534, Mg535, Mg536) production, as well as a protein with three GT domains (Mg539), a hypothetical protein (Mg537) and a pyruvyltransferase (Mg538) which could play the same role as Mimivirus L143 (Fig. 7d, Table 3). We found that the closest homologs of Mg537 are Streptococcus GTs (PDB codes: 4PHR and 5V4A, 99% confidence), also identified above for Ma463. These results suggest that Mg537 and Mg539 could correspond to GTs involved in the C-clade glycosylation pathway.
In accordance with the complexity of their decorated capsids and tails, Tupanviruses (D-clade) also have a bigger and more complex glycosylation gene cluster covering a region of 49 Kb with up to 33 genes (Fig. 8a, Table 4). This cluster includes several biosynthetic pathways for nucleotide-sugars such as UDP-d-diNAcBac (L515, L509 and L514), UDP-d-Qui2N/Fuc2N (L515 and L518) and UDP-d-glucuronic acid (R520), suggesting that fibrils decorating the capsids and tails may also be glycosylated. Their sugars could be further modified by sugar methyltransferases (L506 and L517) present in the cluster (Fig. 8a). We also found a SAT/APS kinase homolog of Ma457, but the homolog of the sulfotransferase (Ma461) is missing. As a result, experimental validation of these modifications is needed. Compared with other clades (Fig. 7), Tupanviruses present an increased number of GTs (Fig. 8, Table 4), with 11 genes encoding a single GT domain and two genes (R496, L510) encoding four and two GT domains, respectively. Final glycan structures could be very complex and heterogeneous reflecting the presence of fibrils decorating both capsids and tails. Two additional enzymes (L521 and L523) clearly related to RNA metabolism (see Table 4 for details) were identified in the cluster as well as ORFans proteins (R497, R500, R522, R524, L525 and L526). Their function remains to be determined.
For Cotonvirus japonicus (E-clade), biosynthetic pathways for UDP-l-Qui2NAc (ORFs 708, 709 and 710) and UDP-d-diNAcBac (ORFs 718, 719 and 720) are conserved and next to other glycogenes. This allowed us to define a 21-gene cluster in Cotonvirus (Fig. 8b, Table 5). This cluster includes seven GTs with some homology to Moumouvirus australiensis GTs. For instance, GT 713 and 717 share 58 and 40% identities with Ma468 with a coverage of 70 and 95%, respectively, while GT 722 and 712 share 72 and 29% identity with Ma463 on the entire protein sequences. In addition, two enzymes involved in the sulfate metabolism were identified as ORFs 724 and 725 that correspond to Ma461 and Ma457 enzymes, with identities up to 60% along the entire protein sequences (Table 5). Inside the cluster, there are genes not related to glycosylation such as two predicted papain-like proteins (ORFs 721 and 723), a permease, MutS and an RNA polymerase subunit (ORFs 714, 726 and 727) (Table 5).
Except for Mimivirus, gene clusters involved in proposed Megavirinae glycosylation are located between a conserved helicase and a thioredoxin-like gene (Fig. S10). The number of genes in this region ranges from six genes for B- and C-clades to 12 genes for Moumouvirus australiensis and up to 33 genes for Tupanviruses. In Mimivirus, this region is only 3.5 kb long and only includes one GT (R363; Fig. S10), while the 12-gene cluster (Fig. 7a) is located in a different genomic region between a putative transcription factor gene (R131) and an orfan gene (L144). In Cotonvirus japonicus, only seven glycogenes of the glycosylation gene cluster (Fig. 8b) lie between the helicase and thioredoxin-like genes (Fig. S10). This part of the cluster presents strong homology with the Megavirus chilensis cluster and includes the l-Qui2N pathway and one GT. After the thioredoxin-like gene (715), there are the remaining 14 glycogenes that are part of the 21-gene cluster (Figs 8b and S10).
Expanding the glycosylation feature of each prototype to the clade
To gain further insights into the evolution of the glycosylation machinery in the proposed Megavirinae subfamily, we examined conservation of the gene clusters (Figs 7 and 8) within and among clades. We only used clade members for which the genome sequence was complete (Figs 9, 10, 11 and S11;,Table S1).
Figure 9.
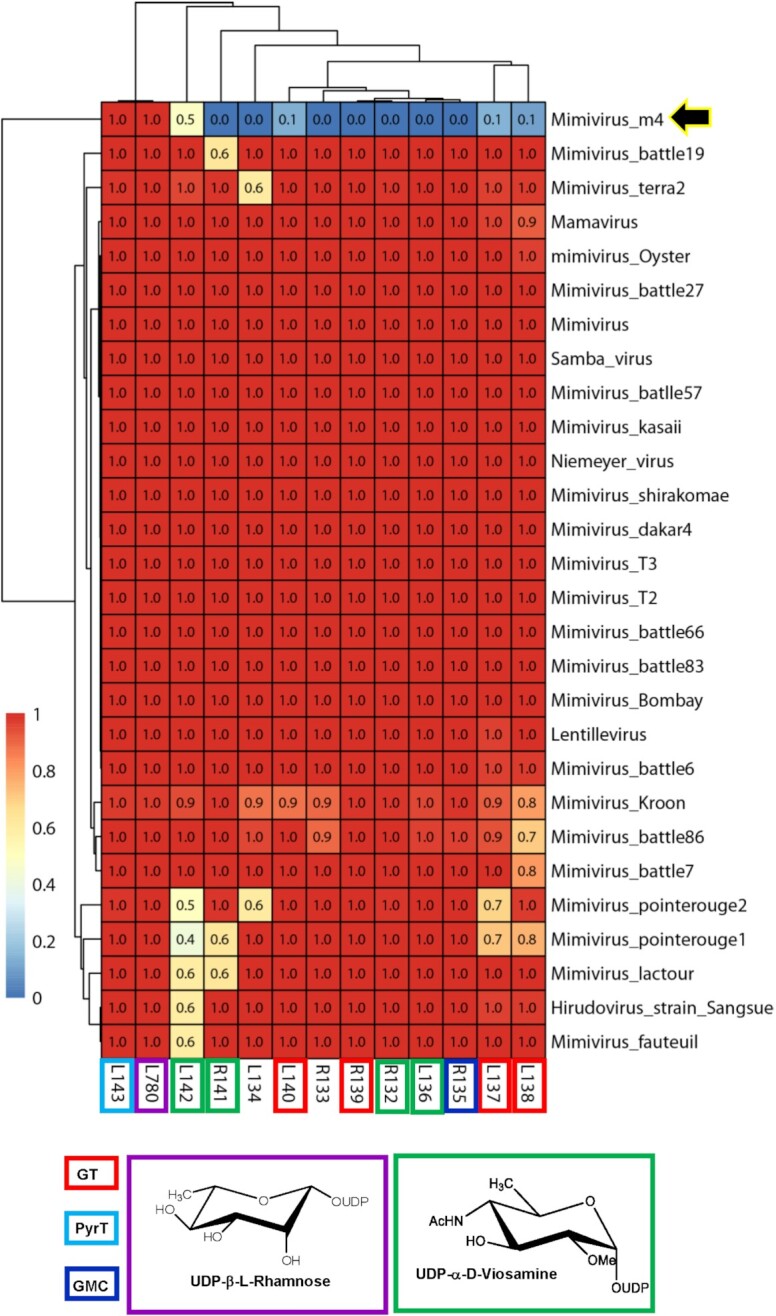
Heatmap based on the level of conservation of the Mimivirus 12-glycogenes cluster in the A-clade. The reference Mimivirus L780 (in violet) and R141 responsible for Rha production have also been included. Proteins involved in UDP-d-2OMeVio4NAc biosynthesis are marked in green; the pyruvyltransferase (L143) involved in modification of GlcNAc in poly_1 of Mimivirus (Fig. 2) is shown in light blue; all glycosyltransferases (GTs) are labeled in red. R135 (dark blue) is the GMC-oxidoreductase composing the fibrils. R133 and L134 are ORFans. The black arrow indicates M4, which lacks the gene cluster. The conservation score for each protein in the different genomes ranges from 0 to 1 (low to high: blue to red).
Figure 10.
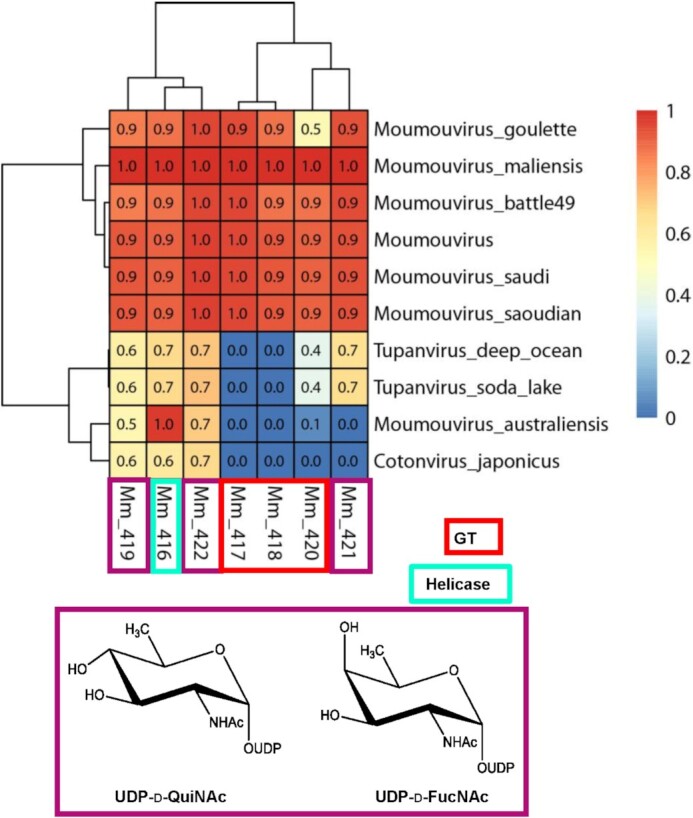
Heatmap of the conservation level of the Moumouvirus maliensis 6-glycogene cluster in the B-clade, D-clade (Tupanviruses) and E-clade (Cotonvirus japonicus). Proteins involved in UDP-d-Qui2NAc/FucNAc biosynthesis are marked in violet, while glycosyltransferases (GTs) are in red. The helicase Mm416 (light green) is used for reference of the conservation level between clades. The conservation score for each protein in the different genomes ranges from 0 to 1 (low to high: blue to red).
Figure 11.
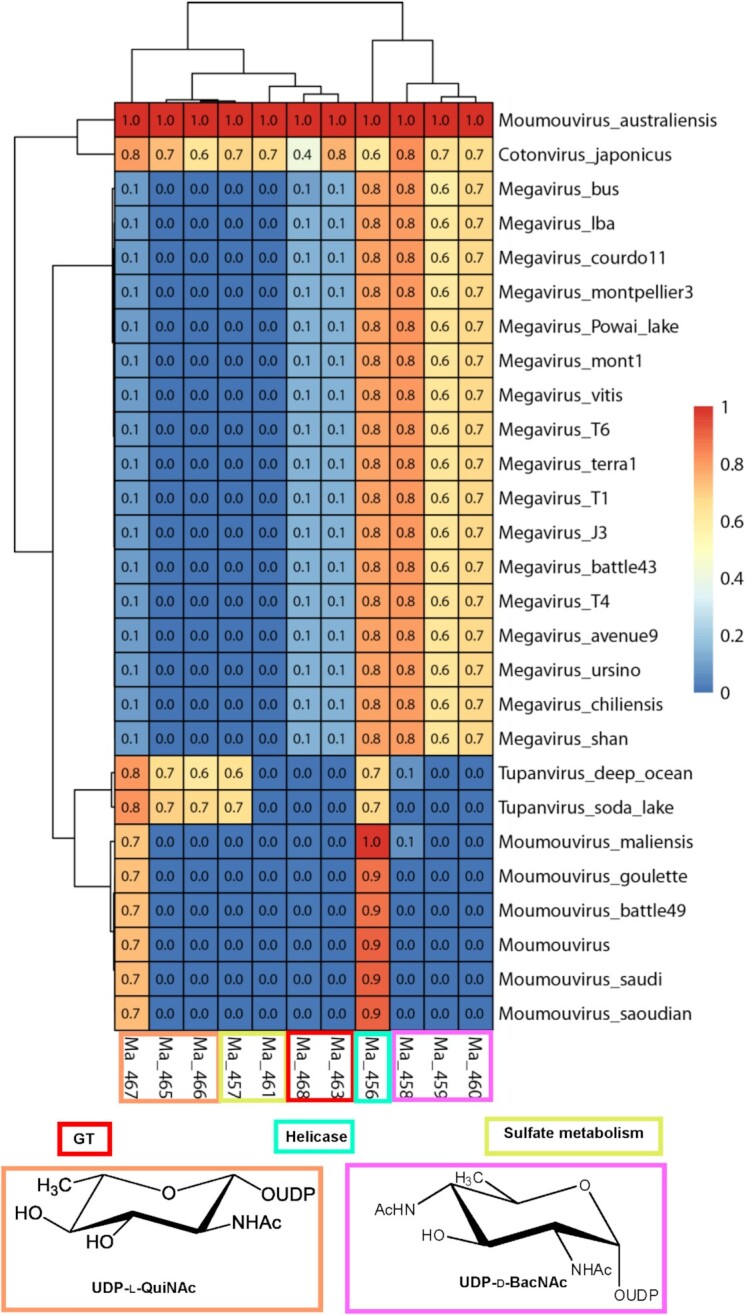
Heatmap of the conservation level of the Moumouvirus australiensis 12-glycogene cluster in the B-clade, C-clade, D-clade (Tupanviruses) and E-clade (Cotonvirus japonicus). Proteins involved in the UDP-l-Qui2NAc and UDP-d-BacNAc biosynthetic pathways are in orange and pink, respectively. The GTs are in red, while proteins responsible for sulfate metabolism are in dark yellow. The helicase Ma456 (in cyan) is used for reference of the conservation level between clades. The two papain-like proteins (Ma462, Ma464) are not included because they are not directly related to fibrils glycosylation. The conservation score for each protein in the different genomes ranges from 0 to 1 (low to high: blue to red).
For the A-clade, our analysis shows that enzymes for UDP-l-Rha (R141, L780) and UDP-d-2OMe-Vio4NAc (R132, L136, R141 and L142) biosynthesis are conserved (Fig. 9). While L780 (YP_003 987 312; 289 aa) does not belong to the gene cluster (Fig. 7a), it was included because it is the second enzyme in the Rha biosynthesis pathway (Parakkottil Chothi et al. 2010). The highest divergence was obtained for L142 (Table 3), which is a bifunctional enzyme with an N-terminal N-acetyltransferase domain responsible for Viosamine acetylation and a C-terminal putative GT domain, which could transfer Vio4NAc onto its acceptor (Piacente et al. 2017a). This divergence can be explained by the split (or fusion) of the L142 gene leading to two genes in Mimivirus M4 (L142a and L142b) and in Hirudovirus strain sangsue (HIUR_S825 and HIUR_S826), where the first ones correspond to the N-acetyltransferase domain and the second ones to the GT domain. Moreover, all GTs in the Mimivirus gene cluster (L137, L138, L139, R140 and the C-ter L142), predicted as type 2 GTs in the Carbohydrate-Active enZYmes database (CAZY) (Cantarel et al. 2009), are specific to the A-clade and are likely involved in building the two polysaccharides constituting Mimivirus fibrils (Fig. 2) (Notaro et al. 2021). They share a high level of sequence identity (Table S3), as expected because the repeated units of the two glycans include Rha and GlcN linked in a different way (Fig. 2) (Notaro et al. 2021). R133 and L134 are hypothetical proteins that seem to be specific of the A-clade and at this stage it is not possible to conclude on their possible contribution to fibril glycosylation or maturation. Inside the A-clade, the laboratory strain Mimivirus M4, which lacks fibrils (Boyer et al. 2011a), is the only one without a glycosylation cluster (Fig. 9, yellow arrow) and without the proteins constituting Mimivirus fibrils.
In line with the A-clade, the Moumouvirus maliensis six-gene cluster is conserved in B-clade except for Moumouvirus australiensis (Fig. 10).
Moumouvirus australiensis, Cotonvirus japonicus and the two Tupanvirus strains share with the B-clade the 4,6-dehydratase enzyme (Mm422) involved both in diNAcBac and Qui2N/Fuc2N production (Fig. 11) as well as the non-functional Mm419 protein.
Tupanviruses share the UDP-d-Qui2NAc/Fuc2NAc biosynthetic pathway with the B-clade, as evidenced by the presence of a 4-reductase enzyme homolog to Moumouvirus maliensis Mm421, as well as a glycosyltransferase homolog to Mm420. Despite clearly belonging to the B-clade phylogenetic tree when using conserved core genes (Fig. 1), Moumouvirus australiensis appears as an outsider of the B-clade for its glycosylation genes (Figures 10 and 11). Enzymes involved in UDP-l-Qui2NAc biosynthesis (Ma458, Ma459 and Ma460) and glycosyltransferases (Ma463 and Ma468) are completely absent from the B-clade.
Even more surprisingly in terms of evolution, Moumouvirus australiensis shares the UDP-l-Qui2N biosynthesis pathway with the entire C-clade and Cotonvirus japonicus (Fig. 11), while it shares the UDP-d-diNAcBac biosynthesis pathway with Cotonvirus japonicus and the more distant Tupanvirus strains (Fig. 11). Enzymes involved in sulfate modification of sugars (Ma457 and Ma461) are both conserved in Cotonvirus japonicus. By contrast, only one enzyme remains in Tupanviruses (Ma457) (Fig. 11). The Moumouvirus australiensis gene cluster is completely conserved in Cotonvirus japonicus (Fig. 11), but the gene cluster of Cotonvirus counts 21 genes instead of 12 and includes several additional glycosyltransferases and hypothetical proteins (Fig. 8, Table 4). These data allowed us to address the evolutionary position of Moumouvirus australiensis between B-clade, Cotonvirus japonicus and Tupanviruses with whom it appears closely related in terms of glycosylation (Fig. 11).
For the C-clade, all enzymes involved in UDP-l-RhaNAc (Mg534, Mg 535) and Qui2NAc synthesis (Mg534, Mg535 and Mg536 homologues of Ma458, Ma459 and Ma460) are conserved inside the clade. In addition, the two GTs (Mg537, Mg539) are also conserved and distant from the Moumouvirus australiensis ones (Ma463, Ma468) (Fig. 11).
This analysis confirmed our initial hypothesis that the glycosylation machinery is clade-specific and suggests that Moumouvirus australiensis likely represents an intermediate prototype for the evolution of a new glycosylation cassette.
Finally, the UDP-d-GlcNAc biosynthesis pathway is conserved in the five clades and the corresponding genes are not arranged into clusters (Fig. S11). In addition, the A-clade, C-clade and Cotonvirus japonicus share a putative pyruvyltransferase (Fig. S11) that could be involved in GlcNAc modification with pyruvic acid, as is observed in Mimivirus polysaccharides (Fig. 2) (Notaro et al. 2021). Even if GlcNAc is present in the amoeba host, all giant DNA viruses encode their own proteins to synthesize this sugar, which is also the precursor of other 6-deoxy-amino-sugars (Rha2NAc, Qui2NAc, diNacBac and Fuc2NAc) that constitute the capsid fibrils of the different members of the three clades.
Discussion
Here, we show that complex gene clusters are involved in glycosylation of the fibril layer surrounding the viral particles in the proposed Megavirinae subfamily (Figs 7 and 8). However, fibril glycosylation occurs in a clade-specific manner, suggesting a high degree of variability in glycan structures for the different members of the family. Our analyses raise two important questions: (i) What are the possible evolutionary implications of organizing glycosylation genes into clusters? and (ii) What is the role played by sugars constituting the fibrils for the different clades? Answering these questions is essential for a clear understanding of glycosylation in giant viruses.
Implications of clustering glycogenes
Organization of glycogenes into clusters is reminiscent of what occurs in bacteria. For example, genes involved in biosynthesis of lipopolysaccharide (LPS), the main component of the Gram-negative bacteria outer membrane, are organized in a cluster controlled by a unique promoter defining the waa operon in Escherichia coli K12 (Gronow and Brade 2001). Similarly, genes responsible for the biosynthesis of the N-/O-glycans that decorate flagellin or pili are organized into operons (all controlled by the same promoter). Campylobacter jejuni presents a cluster of genes responsible for flagellin glycosylation, called pgl, which is further organized in two operons: operon I includes pglB, pglA, pglC and pglD, while operon II contains pglE, pglF and pglG (Szymanski et al. 1999). Interestingly, pglE and pglF are co-transcribed, while pglD transcription is regulated by another operon system (Szymanski et al. 1999), despite all being involved in UDP-d-diNAcBAc biosynthesis. Because operons are often controlled by a single promoter, levels and timing of expression are comparable for all the genes.
It is currently unknown how transcription and expression of glycogenes occur in giant viruses. The transcriptomes of Mimivirus (Legendre et al. 2010) and Megavirus chilensis (Arslan et al. 2011) being the only ones available, they are used as reference for the entire family. We had previously showed that there are three main expression classes: early (from T = 0 to T = 3 h), intermediate (from T = 3 h to T = 6 h) and late (from T = 6 h to T = 12 h). Only early and late promoters have been identified, with a highly stringent early promoter (AAAATTGA) (Suhre et al. 2005) and a less stringent late promoter (Legendre et al. 2010). All glycogenes are expressed in the late stage of the infection cycle whether they are organized into a cluster or not. Mimivirus glycosylation genes are clustered (Fig. 7a) and all have their own late promoter, except R133, which encodes a hypothetical protein. All are expressed in the late phase (6–12 hours post infection ) of the infectious cycle, supporting a role for this cluster in fibril glycosylation. On the contrary, for genes in the Megavirus chilensis glycosylation cluster (Fig. 7d), only mg535, mg536 and mg539 have a promoter. Given the low conservation of the late promoter, it could have been missed by the annotation or these genes are expressed as polycistronic mRNAs, as shown previously for some Mimivirus genes (Byrne et al. 2009).
Previous studies (Piacente et al. 2012, 2014a, 2015) and this one show that it is highly challenging to establish the origin of these glycosylation gene clusters. For the viosamine synthesis pathway, the patchwork of bacterial-like and eukaryotic-like genes composing the cluster precludes the hypothesis that they were all acquired at once from a single organism (Piacente et al. 2012). To detect putative horizontal gene transfer, we compared the GC content of glycogenes (Figs 7 and 8) with Mimiviridae core genes (Fig. S12). We showed that glycogenes are as AT-rich as the overall genome, suggesting that they could have been acquired early in evolution so that their GC content had time to evolve. Giant viruses possess up to five genes encoding transposases that might catalyze integrations of foreign DNA into their genomes, but these genes are far (more than 30 Kb) from the genomic region containing the glycogenes. In addition, the fact that glycogenes have the typical viral promoter (Legendre et al. 2010) could also suggest that giant viruses have evolved their own glycosylation machinery prior to the radiation of eukaryotes.
We showed that glycosylation gene-clusters of the B-clade (Moumouviruses), C-clade (Megaviruses), D-clade (Tupanviruses) and E-clade (Cotonvirus japonicus) are all located between conserved helicase and thioredoxin-like genes (Fig. S10). This result suggests that giant viruses could have exchanged these glycogenes through homologous recombination events or horizontal gene transfer inside the amoeba host, by analogy with bacteria (Aydanian et al. 2011). However, further studies are required to experimentally prove that these genes can be exchanged between giant viruses belonging to different clades and eventually with bacteria.
Bacteria-like sugars as markers of proposed Megavirinae clades
From a structural point of view, the type of sugars found in giant viruses is drastically different from what was reported for eukaryotic viruses. For example, SARS-Cov-2 virus exhibits oligosaccharides of discrete size made by sugar units typical of the eukaryotic world, such as glucosamine, galactosamine, fucose and sialic acid (Zhao et al. 2021). By contrast, members of the proposed Megavirinae subfamily possess their own glycosylation machinery to synthesize and decorate their fibrils with rare amino sugars that are only encountered in bacteria and are absent from their amoeba host. The organization of these glycogenes into clusters also increases the evolvability of the glycosylation machinery resulting in different sugars between clades and even inside the same clade, as was observed for Moumouvirus australiensis.
By comparing the conservation level of glyco-enzymes, we identified one or two sugars as markers of a specific clade. A-clade is characterized by the rare sugar d-viosamine, which is a component of Pseudomonas syringe flagellin (Yamamoto et al. 2011) and has also been identified in the O-chain of several E. coli strains. In addition to viosamine, the A-clade is the only clade to have rhamnose, a deoxy-sugar found as a component of the O-antigen of several bacteria, such as S. enterica (Samuel and Reeves 2003), in the N-glycans of archaea (Kaminski and Eichler 2014) and plant cell-wall polysaccharides. Interestingly, the A-clade follows the plant way instead of the microbial pathway to synthetize rhamnose (Parakkottil Chothi et al. 2010). The precursor for both these sugars is UDP-d-Glc, which is the only UDP-sugar for which the A-clade does not appear to have a dedicated biosynthetic machinery, thus relying on its host.
Except for Moumouvirus australiensis, B-clade members have two rare amino sugars, d-Qui2NAc and d-Fuc2NAc, whose biosynthetic pathways are closely interconnected. d-Fuc2NAc was found in the LPS of Pseudomonas aeruginosa O5 (Burrows et al. 1996) and in the O-Chain of the marine bacterium Pseudoalteromonas agrivorans (Perepelov et al. 2000). d-Qui2NAc was also found in the O-chain of Gram-negative bacteria, such as Rizobium etli CE3 (D'Haeze et al. 2007) and Pseudomonas aeruginosa O10 (Knirel et al. 1986).
C-clade viruses, unlike B-clade, decorate their fibrils with L-RhaNAc and l-Qui2NAc that is also a component of Moumouvirus australiensis fibrils and is shared with Cotonvirus japonicus. l-Qui2NAc is a component of the O-antigen of several pathogenic bacteria, such as Yersinia enterocolitica serotype O11, 23 and O11, 24 (Marsden et al. 1994), Vibrio cholerae O37 (Kneidinger et al. 2003) and Proteous penneri 26 (Shashkov et al. 1998) and has also been found in the capsular polysaccharide (CPS) of Bacteroides fragilis NCT 9343 and ATCC 23 745 (Kasper et al. 1983). Rhamnosamine is a rare sugar found only in Proteous vulgaris TG 155 O-antigen (Kondakova et al. 2003), E. coli O3 LPS (Jaan and Jann 1968) and Vibrio vulnificus CPS (Reddy et al. 1993).
Finally, Moumouvirus australiensis (outsider of the B-clade) along with Cotonvirus japonicus and Tupanviruses are characterized by another rare amino sugar, d-diNacBac, which has also been found in pathogenic bacteria (Morrison and Imperiali 2014) as the reducing sugar of N-linked and O-linked glycoproteins of C. jejuni and Neisseria gonorrhoeae, respectively. It is also a component of the O-Chain of Pseudomonas reactans and Vibrio cholerae, and of the CPS of Alteromonas(Perepelov et al.2000, Aydanian et al.2011).
The precursor of the rare amino-sugars characteristic of B- and C-clades, Cotonvirus japonicus and Tupanviruses is UDP-d-GlcNAc, for which all clades possess the biosynthetic pathway. Consequently, in contrast to the A-clade, they could be completely independent from their host for synthesis of their glycans, which could possibly extend the range of hosts that they can replicate in.
Chemical analyses of the fibrils in Tupanvirus and Cotonvirus japonicus were not performed and gene expression data are also lacking. However, our results showing conservation of the biosynthetic pathways for Qui2N and diNAcBac in Cotonvirus suggest that its fibrils could be decorated with the corresponding glycans, while those in Tupanviruses could be composed of diNAcBac, Fuc2N/Qui2N and GlcA. The UDP-GlcNAc biosynthetic pathway identified in the A-, B- and C-clades is also conserved in Cotonvirus japonicus and in Tupanviruses (Table 1). Thus, further studies are needed to determine whether their fibrils could be glycosylated using virus-encoded enzymes.
Why are giant viruses covered by rare amino sugars and why are these different depending on the clade? The presence of a highly glycosylated fibril layer for all members of proposed Megavirinae isolated so far suggests an essential role played by these glycans in the environment. Regarding the first question, we think that the presence of bacterial-like sugars on the surface of giant viruses can be considered as a strategy to mimic the bacteria that the amoeba feeds on, thus facilitating competition with other parasites for the same host in the natural environment. We could also speculate that they play an important role in the physiology of these viruses. In fact, it has been established that glycans play a crucial role in the adhesion process to the host cell, while they do not affect dramatically the viral replication even if they appear to have a fitness cost (Boyer et al. 2011b). In this context, amino sugars are essential because their physiochemical properties enable them to create a highly viscous or sticky surface (Salton 1965), which is suitable for adhesion on cell surfaces. In addition, glycans constitute a protective barrier both against the natural environment in which they must propagate and the enzymes of the host cell. Finally, these viruses are targets of viral infection by virophages, which stick to the giant virus fibrils (Duponchel and Fischer 2019; La Scola et al. 2008), allowing them to be transported along into the host cell. Some virophages, like Sputnik (La Scola et al. 2008), are deleterious to the giant virus, while others, like Zamilon (Jeudy et al. 2020), appear to be commensal. Evolving different sugar compositions for fibril glycosylation could thus impair virophage interaction with the fibrils and be advantageous against pathogenic virophages.
The answer to the second question comes from the organization of the glycogenes in hot-spot mutation areas that could be essential to introduce variability in glycan composition. Because fibril glycans play a pivotal role in the interaction with the host cell, an arms race is taking place between the host and the giant viruses, but also among giant viruses that compete for the same host in the natural environment. Having a flexible toolbox for glycan synthesis could reflect the need to continuously adjust the set of glycans decorating the capsids to trigger increased phagocytosis and outcompete other parasites. It is tempting to compare giant virus glycosylation clusters with antibiotic resistance cassettes in bacteria. The fact that this complex glycosylation machinery is lost in laboratory conditions, as was shown for M4 (Boyer et al. 2011a), also suggests that there must be a fitness cost for such complex glycan synthesis, again echoing what is observed for antibiotic resistance genes in bacteria.
Conclusions
This in-depth study of glycosylation in the proposed Megavirinae subfamily revealed that giant DNA viruses are different from other eukaryotic viruses in several aspects. First, they possess a complex glycosylation machinery consisting of clusters of six to 33 genes responsible for the formation of glycans constituting their fibrils. This results in glycosylated fibrils with sugars different from those found in the host and from those of other eukaryotic viruses. Giant viruses produce rare amino sugars, yet there is a clade-specific glycosylation trend, although some exceptions could occur, as is shown for Moumouvirus australiensis that could be in the process of evolving a new glycosylation cassette (losing and/or gaining new glycosylation genes).
This variegated glycosylation between the different clades could be the result of adaptation to the host they infect and/or competition with other viruses and bacteria and could also affect their ability to be infected by virophages.
Studying the glycobiology of these giant DNA viruses is important as it may reveal new enzymes encoded by genes of unknown function located within these clusters and could provide clues to the origin of the various elements of their glycosylation machinery, the glycosylation machinery of their ancestor that may predate the radiation of eukaryotes, and ultimately lead to advances for the glycobiology discipline in general. Finally, this work could be considered as a pilot study, which can be extended to other giant and large DNA viruses, such as the atypical Pandoraviruses, Pithoviruses and Molliviruses.
Supplementary Material
ACKNOWLEDGEMENTS
The electron microscopy analyses were performed on the PiCSL-FBI core facility (Nicolas Brouilly, Fabrice Richard, Aïcha Aouane, IBDM, AMU-Marseille), member of the France-BioImaging national research infrastructure (ANR-10-INBS-04) and on the IMM imaging platform (Artemis Kosta). We also acknowledge the use of the PACA Bioinfo platform. We thank Jean-Marie Alempic whom isolated the Moumouvirus australiensis and Moumouvirus maliensis viruses for his help and advices in producing the virions.
Contributor Information
Anna Notaro, University of Naples Federico II, Department of Agricultural Sciences, Via Università100, 80055, Portici, Naples, Italy; Aix-Marseille University and Centre National de la Recherche Scientifique and Institut de Microbiology de la Méditerranée; IGS Unité Mixte de Recherche 7256, FR3479, IM2B, 13288 Marseille Cedex 9, France.
Olivier Poirot, Aix-Marseille University and Centre National de la Recherche Scientifique and Institut de Microbiology de la Méditerranée; IGS Unité Mixte de Recherche 7256, FR3479, IM2B, 13288 Marseille Cedex 9, France.
Elsa D Garcin, Aix-Marseille University and Centre National de la Recherche Scientifique and Institut de Microbiology de la Méditerranée; IGS Unité Mixte de Recherche 7256, FR3479, IM2B, 13288 Marseille Cedex 9, France.
Sebastien Nin, Aix-Marseille University and Centre National de la Recherche Scientifique and Institut de Microbiology de la Méditerranée; IGS Unité Mixte de Recherche 7256, FR3479, IM2B, 13288 Marseille Cedex 9, France.
Antonio Molinaro, Department of Chemical Sciences, University of Naples Federico II, Via Cinthia 26, 80126, Naples, Italy.
Michela Tonetti, Department of Experimental Medicine and Center of Excellence for Biomedical Research, University of Genova, Viale Benedetto XV, 1, 16132, Genova, Italy.
Cristina De Castro, University of Naples Federico II, Department of Agricultural Sciences, Via Università100, 80055, Portici, Naples, Italy.
Chantal Abergel, Aix-Marseille University and Centre National de la Recherche Scientifique and Institut de Microbiology de la Méditerranée; IGS Unité Mixte de Recherche 7256, FR3479, IM2B, 13288 Marseille Cedex 9, France.
Funding
AN acknowledges UIF Vinci program 2015 (contract C3_90) and 2019 (contract C4-180), and the STAR2-2017 project for financial support. EG has received funding from the Excellence Initiative of Aix-Marseille Université—A*Midex, a French ‘Investissements d'Avenir’ program, AMX-18-ACE-004. SN's salary was partially supported by the Bettencourt-Schueller foundation (AGDI_122 861_01_0447_2016). CA received funding from the European Research Council (ERC) under the European Union′s Horizon 2020 research and innovation program (grant agreement No 832 601). This work was also partially supported by the French National Research Agency ANR-16-CE11-0033-01.
Conflict of interest statement. None declared.
References
- Abergel C, Joëlle R-T, Richard Get al. Virus-encoded aminoacyl-tRNA synthetases: structural and functional characterization of Mimivirus TyrRS and MetRS. J Virol. 2007;81:12406–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abergel C, Legendre M, Claverie J-M. The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol Rev. 2015;39:779–96. [DOI] [PubMed] [Google Scholar]
- Abrahão J, Lorena S, Ludmila SSet al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat Commun. 2018;9:749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey DL, Li S, Konwerski JRet al. A new structural form in the SAM/Metal-Dependent O–Methyltransferase family: Myce from mycinamicin biosynthetic pathway. J Mol Biol. 2011;413:438–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armougom F, Sébastien M, Olivier Pet al. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34:W604–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydanian A, Tang Li, Morris JGet al. Genetic diversity of O-antigen biosynthesis regions in Vibrio cholerae. Appl Environ Microbiol. 2011;77:2247–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdonaite I, Wandall HH. Global aspects of viral glycosylation. Glycobiology. 2018;28:443–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M, Azza S, Barrassi Let al. Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc Natl Acad Sci. 2011a;108:10296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M, Saïd A, Lina Bet al. Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc Natl Acad Sci. 2011b;108:10296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussaard CP, Steven WW, Frede Tet al. Global-scale processes with a nanoscale drive: the role of marine viruses. ISME J. 2008;2:575–8. [DOI] [PubMed] [Google Scholar]
- Burrows LL, Charter DF, Lam JS. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO1) B-band lipopolysaccharide gene cluster. 1996;22:481–95. [DOI] [PubMed] [Google Scholar]
- Burrows LL, Urbanic RV, Lam JS. Functional conservation of the polysaccharide biosynthetic protein WbpM and its homologues in Pseudomonas aeruginosa and other medically significant bacteria. Infect Immun. 2000;68:931–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne D, Grzela R, Lartigue Aet al. The polyadenylation site of Mimivirus transcripts obeys a stringent ‘hairpin rule’. Genome Res. 2009;19:1233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Haeze W, Leoff C, Freshour Get al. Rhizobium etli CE3 bacteroid lipopolysaccharides are structurally similar but not identical to those produced by cultured CE3 bacteria. J Biol Chem. 2007;282:17101–13. [DOI] [PubMed] [Google Scholar]
- De Castro C, Molinaro A, Piacente Fet al. Structure of N-linked oligosaccharides attached to chlorovirus PBCV-1 major capsid protein reveals unusual class of complex N-glycans. Proc Natl Acad Sci. 2013;110:13956–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro C, Parrilli M, Holst Oet al. Microbe-associated molecular patterns in innate immunity. Methods Enzymol. 2010;480:89–115. [DOI] [PubMed] [Google Scholar]
- De Castro C, Speciale I, Duncan Get al. N-linked glycans of chloroviruses sharing a core architecture without precedent. Angew Chem Int Ed. 2016;55:654–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defne A, Legendre M, Seltzer Vet al. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc Natl Acad Sci. 2011;108:17486–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duponchel S, Fischer MG. Viva lavidaviruses! Five features of virophages that parasitize giant DNA viruses. PLoS Pathog. 2019;15:e1007592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer MG, Allen MJ, Wilson WHet al. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci. 2010;107:19508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallot-Lavallée L, Blanc G, Claverie J-M. Comparative genomics of chrysochromulina ericina virus and other microalga-infecting large DNA viruses highlights their intricate evolutionary relationship with the established mimiviridae family. J Virol. 2017;91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Robert X, Courcelle E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronow S, Brade H. Invited review: Lipopolysaccharide biosynthesis: which steps do bacteria need to survive?. J Endotoxin Res. 2001;7:3–23. [PubMed] [Google Scholar]
- Hildebrand A, Remmert M, Biegert Aet al. Fast and accurate automatic structure prediction with HHpred. Proteins Struct Funct Bioinf. 2009;77:128–32. [DOI] [PubMed] [Google Scholar]
- Jaan B, Jann K. 2-Amino-2, 6-Dideoxy-L-Mannose (L-Rhamnosamine) isolated from the lipopolysaccharide of escherichia coli 03: k2ab(L):H2. 1968;5:173–7. [DOI] [PubMed] [Google Scholar]
- Jeudy S, Abergel C, Claverie J-Met al. Translation in giant viruses: a unique mixture of bacterial and eukaryotic termination schemes. PLos Genet. 2012;8:e1003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeudy S, Bertaux L, Alempic J-Met al. Exploration of the propagation of transpovirons within mimiviridae reveals a unique example of commensalism in the viral world. ISME J. 2020;14:727–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel Aet al. Highly accurate protein structure prediction with alphafold. Nature. 2021;596:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski L, Eichler J. Haloferax volcanii N-glycosylation: delineating the pathway of dTDP-rhamnose biosynthesis. PLoS One. 2014;9:e97441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper DL, Weintraub A, Lindberg AAet al. Capsular polysaccharides and lipopolysaccharides from two bacteroides fragilis reference strains: chemical and immunochemical characterization. J Bacteriol. 1983;153:991–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JD, Poon KKH, Webb NAet al. The structural basis for catalytic function of GMD and RMD, two closely related enzymes from the GDP-d-rhamnose biosynthesis pathway. FEBS J. 2009;276:2686–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose T, Herbst DA, Zhu Het al. A Mimivirus enzyme that participates in viral entry. Structure. 2015;23:1058–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneidinger B, Larocque S, Brisson J-Ret al. Biosynthesis of 2-acetamido-2,6-dideoxy-L-hexoses in bacteria follows a pattern distinct from those of the pathways of 6-deoxy-L-hexoses. Biochem J. 2003;371:989–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirel YA, Vinogradov EV, Shashkov ASet al. Somatic antigens of Pseudomonas aeruginosa. The structure of O-specific polysaccharide chains of P. aeruginosa O10 (Lányi) lipopolysaccharides. Eur J Biochem. 1986;157:129–38. [DOI] [PubMed] [Google Scholar]
- Kondakova AN, Kolodziejska K, Zych Ket al. Structure of the N-acetyl-L-rhamnosamine-containing O-polysaccharide of Proteus vulgaris TG 155 from a new proteus serogroup, O55. Carbohydr Res. 2003;338:1999–2004. [DOI] [PubMed] [Google Scholar]
- La Scola B, Audic S, Robert Cet al. A giant virus in amoebae. Science. 2003;299:2033. [DOI] [PubMed] [Google Scholar]
- La Scola B, Desnues C, Pagnier Iet al. The virophage as a unique parasite of the giant Mimivirus. Nature. 2008;455:100–4. [DOI] [PubMed] [Google Scholar]
- Legendre M, Audic S, Poirot Oet al. mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. Genome Res. 2010;20:664–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, et al. In vitro biosynthesis and chemical identification of UDP- N -acetyl-d-quinovosamine (UDP-d-QuiNAc). J Biol Chem. 2014;289:18110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lönngren J, Svensson S. Mass spectrometry in structural analysis of natural carbohydrates. Adv Carbohydr Chem Biochem. 1974;29:41–106. [Google Scholar]
- Lu R, Zhao X, Li Jet al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet North Am Ed. 2020;395:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden BJ, Bundle DR, Perry MB. Serological and structural relationships between Escherichia coli O:98 and Yersinia enterocolitica O:11,23 and O:11,24 lipopolysaccharide O-antigens. Biochem Cell Biol. 1994;72:163–8. [DOI] [PubMed] [Google Scholar]
- Morrison MJ, Imperiali B. The renaissance of bacillosamine and its derivatives: pathway characterization and implications in pathogenicity. Biochemistry. 2014;53:624–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel E, Notaro A, Speciale Iet al. Chlorovirus PBCV-1 multidomain protein A111/114R has three glycosyltransferase functions involved in the synthesis of atypical N-Glycans. Viruses. 2021;13:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaro A, Couté Y, Belmudes Let al. Expanding the occurrence of polysaccharides to the viral world: the case of Mimivirus. Angew Chem Int Ed Engl. 2021;60:19897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier NB, Imperiali B. Crystal structure and catalytic mechanism of PglD from Campylobacterjejuni. J Biol Chem. 2008;283:27937–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parakkottil Chothi M, Duncan GA, Armirotti Aet al. Identification of an L-rhamnose synthetic pathway in two nucleocytoplasmic large DNA viruses. J Virol. 2010;84:8829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perepelov AV, Senchenkova S'N, Shashkov ASet al. First application of triflic acid for selective cleavage of glycosidic linkages in structural studies of a bacterial polysaccharide from Pseudoalteromonas sp. KMM 634 †. J Chem Soc, Perkin Trans 1. 2000;1:363–6. [Google Scholar]
- Philippe N, Legendre M, Doutre Get al. Pandoraviruses: Amoeba viruses with genomes up to 2.5 MB reaching that of Parasitic Eukaryotes. Science. 2013;341:281–6. [DOI] [PubMed] [Google Scholar]
- Piacente F, Bernardi C, Marin Met al. Characterization of a UDP-N-acetylglucosamine biosynthetic pathway encoded by the giant DNA virus Mimivirus. Glycobiology. 2014a;24:51–61. [DOI] [PubMed] [Google Scholar]
- Piacente F, De Castro C, Jeudy Set al. Giant virus Megavirus chilensis encodes the biosynthetic pathway for uncommon acetamido sugars. J Biol Chem. 2014b;289:24428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacente F, de Castro C, Jeudy Set al. The rare sugar N-acetylated viosamine is a major component of Mimivirus fibers. J Biol Chem. 2017a;292:7385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacente F, Gaglianone M, Elena Laugieri Met al. The autonomous glycosylation of large DNA viruses. Int J Mol Sci. 2015;16:29315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacente F, Marin M, Molinaro Aet al. Giant DNA virus Mimivirus encodes pathway for biosynthesis of unusual sugar 4-amino-4,6-dideoxy-D-glucose (Viosamine). J Biol Chem. 2012;287:3009–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D. The 1.2-Megabase genome sequence of Mimivirus. Science. 2004;306:1344–50. [DOI] [PubMed] [Google Scholar]
- Reddy GP, Hayat U, Bush CAet al. Capsular polysaccharide structure of a clinical isolate of Vibrio vulnificus strain BO62316 determined by heteronuclear NMR spectroscopy and high-performance anion-exchange chromatography. Anal Biochem. 1993;214:106–15. [DOI] [PubMed] [Google Scholar]
- Renesto P, Abergel C, Decloquement Pet al. Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J Virol. 2006;80:11678–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegert AS, Thoden JB, Schoenhofen ICet al. Structural and biochemical investigation of PglF from Campylobacterjejuni reveals a new mechanism for a member of the short chain dehydrogenase/reductase superfamily. Biochemistry. 2017;56:6030–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegert AS, Young NM, Watson DCet al. Structure of the external aldimine form of PglE, an aminotransferase required for n, n ’-diacetylbacillosamine biosynthesis: structure of the Aminotransferase PglE'. Protein Sci. 2015;24:1609–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton MRJ. Chemistry and function of amino sugars and derivatives. Annu Rev Biochem. 1965;34:143–74. [DOI] [PubMed] [Google Scholar]
- Samuel G, Reeves P. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr Res. 2003;338:2503–19. [DOI] [PubMed] [Google Scholar]
- Schulz F, Yutin N, Ivanova NNet al. Giant viruses with an expanded complement of translation system components. Science. 2017;356:82–5. [DOI] [PubMed] [Google Scholar]
- Shashkov AS, Paramonov NA, Veremeychenko SPet al. Somatic antigens of Pseudomonads: structure of the O-specific polysaccharide of Pseudomonas fluorescens biovar B, strain IMV 247. Carbohydr Res. 1998;306:297–303. [DOI] [PubMed] [Google Scholar]
- Speciale I, Laugieri ME, Noel Eet al. Chlorovirus PBCV-1 protein A064R has three of the transferase activities necessary to synthesize its capsid protein N-linked glycans. Proc Natl Acad Sci. 2020;117:28735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Audic S, Claverie JM. Mimivirus gene promoters exhibit an unprecedented conservation among all eukaryotes. Proc Natl Acad Sci. 2005;102:14689–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA. Viruses in the sea. Nature. 2005;437:356–61. [DOI] [PubMed] [Google Scholar]
- Szymanski CM, Yao R, Ewing CPet al. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol Microbiol. 1999;32:1022–30. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Fukaya S, Song Cet al. Morphological and taxonomic properties of the newly isolated Cotonvirus japonicus, a new lineage of the subfamily Megavirinae. J Virol. 2021;95:Jvi0091921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Ohnishi-Kameyama M, Nguyen CLet al. Identification of genes involved in the glycosylation of modified viosamine of flagellins in Pseudomonas syringae by mass spectrometry. Genes. 2011;2:788–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Chen H, Wang H. Glycans of SARS-CoV-2 spike protein in virus infection and antibody production. 2021;8:629873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.