Abstract
Tracking SARS-CoV-2 variants in wastewater is primarily performed by detecting characteristic mutations of the variants. Unlike the Delta variant, the emergence of the Omicron variant and its sublineages as variants of concern has posed a challenge in using characteristic mutations for wastewater surveillance. In this study, we monitored the temporal and spatial variation of SARS-CoV-2 variants by including all the detected mutations and compared whether limiting the analyses to characteristic mutations for variants like Omicron impact the outcomes. We collected 24-hour composite samples from 15 wastewater treatment plants (WWTP) in Hesse and sequenced 164 wastewater samples with a targeted sequencing approach from September 2021 to March 2022. Our results show that comparing the number of all the mutations against the number of the characteristic mutations reveals a different outcome. A different temporal variation was observed for the ORF1a and S gene. As Omicron became dominant, we observed an increase in the overall number of mutations. Based on the characteristic mutations of the SARS-CoV-2 variants, a decreasing trend for the number of ORF1a and S gene mutations was noticed, though the number of known characteristic mutations in both genes is higher in Omicron than Delta.
Keywords: COVID-19, Germany, mutations, Omicron, wastewater-based epidemiology, genome sequencing
Introduction
Genomic sequencing has played a central role during the COVID-19 pandemic to support public health agencies to determine the occurrence pattern of the mutations in the SARS-CoV-2 genome. The sequencing of clinical samples has enabled to monitor emerging variants in time and spatial scale (Priesemann et al. 2021). The clinical data in Germany showed that the Delta variant was dominant until the first positive case of Omicron in Germany on 26 November 2021 was reported. An exponential increase in the relative share of the Omicron variant was visible in the following weeks, until the point where the Omicron variant completely replaced Delta as dominant variant of concern (VOC) in week 6 of 2022 (Robert Kock Institute 2022). The rapid surge in COVID-19 cases due to the emergence of Omicron stressed the public health department (ÖGD) in Germany to further monitor COVID-19 cases. This included infection prevention, infection source identification, and contact tracing, and quarantine management of infected and suspected cases. An impaired detection rate of Omicron variants due to the limited availability of test kits and financial resources has resulted in an incomplete picture about the real Omicron case scenario (Osterman et al. 2022).
Wastewater-based epidemiology (WBE) is a suitable tool to monitor the prevalence, genetic diversity, and geographical distribution of the virus (Kitajima et al. 2020) to supplement the clinical data. Since the beginning of the pandemic, surveillance of SARS-CoV-2 in wastewater has become popular and a complementary tool to the clinical testing either using qPCR/dPCR or sequencing approaches. Studies using qPCR- or dPCR-based approaches for monitoring SARS-CoV-2 in wastewater used either universal assays to capture the total SARS-CoV-2 concentration (Ahmed et al. 2020, Sherchan et al. 2020, Calderón-Franco et al. 2022) or characteristic mutation assays to identify different SARS-CoV-2 variants, respectively (Heijnen et al. 2021, Herold et al. 2021, Boogaerts et al. 2022, La Rosa et al. 2022). Targeted sequencing approaches, in which overlapping amplicons covering the SARS-CoV-2 genome are sequenced (Gohl et al. 2020), are used for the surveillance of the individual SARS-CoV-2 lineages. However, monitoring of an individual SARS-CoV-2 variant in wastewater samples is challenging, as it contains a mixture of lineages circulating in the sewershed. To overcome this challenge, the currently used approach is to monitor lineages by tracing the characteristic mutations of each lineage (Crits-Christoph et al. 2021, Herold et al. 2021, Wurtz et al. 2021, Agrawal et al. 2022a, Ahmed et al. 2022a, Pérez-Cataluña et al. 2022), which can provide information about the circulating VOCs (McClary-Gutierrez et al. 2021, Amman et al. 2022). A recent study suggests that the sequencing surveillance of SARS-CoV-2 could provide more information, not just limited to monitoring VOCs, variant of interest (VOI), and variant under monitoring (VUMs) based on the characteristic mutations (Smyth et al. 2022). In this study, researchers were able to detect mutations that had been rarely detected in clinical samples, suggesting the presence of novel cryptic SARS-CoV-2 lineages not available in GISAID's EpiCoV database (Smyth et al. 2022).
In this study, we monitored the occurrence patterns of SARS-CoV-2 variants and their respective mutations in 15 WWTPs in the state of Hesse in Germany, through high-throughput sequencing. The aim of the study was to determine the temporal and spatial variation of the emerging SARS-CoV-2 variants. Also, we focused on evaluating the impact of using the characteristic mutations in comparison to including all the detected mutations on the temporal and spatial variation of the SARS-CoV-2 variants circulating in the sewersheds.
Material and methods
Sampling sites
From the 15 wastewater treatment plants (WWTP) in this study (Table S1, Supporting Information), 24-hour time proportional composite samples were taken from the influent of the WWTPs or the influent to the primary treatment (after the grid chamber) either monthly or in a biweekly sampling cycle. The collected wastewater samples were stored at 4°C and further processed in the laboratory within 24–48 hours. Samples were collected between September 2021 and March 2022.
RNA extraction, sequencing, and variant calling
A volume of 200 ml of the untreated wastewater was concentrated by ultrafiltration in 100 kDa Centricon® Plus-70 centrifugal ultrafilters (Merck) and RNA was extracted using the Ultra Microbiome kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. cDNA synthesis was performed using SuperScript™ VILO™ Master Mix (Thermo Fisher Scientific), followed by library preparation using the Ion AmpliSeq SARS-CoV-2 Research Panel (Thermo Fisher Scientific) according to manufacturers’ instructions. This panel consists of 237 primer pairs, resulting in an amplicon length range of 125–275 bp, which cover the near-full genome of SARS-CoV-2. Libraries were multiplexed and sequenced using an Ion Torrent 530 chip on an Ion S5 sequencer (Thermo Fisher Scientific) according to manufacturers’ instructions. We performed multiple sequencing runs on an Ion S5 sequencer (Thermo Fisher Scientific) to achieve a high number of reads per sample. Then, the generated sequences (reads) were compared with the reference genome (Wuhan-Hu-1-NC_045512/MN908947.3) for mutation detection.
We used the SARS-CoV-2 Research Plug-in Package, which we installed in our Ion Torrent Suite software (v5.12.2) of Ion S5 sequence. We also used the SARS_CoV_2_coverageAnalysis (v5.16) plugin, which maps the generated reads to a SARS-CoV-2 reference genome (Wuhan-Hu-1-NC_045512/MN908947.3), using TMAP software included in the Torrent Suite. For mutation calls, additional Ion Torrent plugins were used, similar to our previous study (Agrawal et al. 2022b). First, all single nucleotide variants (SNVs) were called using Variant Caller (v5.12.0.4) with ‘Generic—S5/S5XL (510/520/530)—Somatic—Low Stringency’ default parameters. The main variant caller parameter settings were: (1) minimum allele frequency > 1% for indel as well as snp; (2) minimum coverage on both strands to be > 20 reads for snp and > 40 reads for indels; and (3) minimum variant score for snp was 15 and 20 for the indels. A detailed parameter setting file for the variant caller is provided as additional supplementary file. Then variant caller removes alleles resulting from sequencing error using a sequence variant baseline (SVB) BED file (additional supplementary file). The SVB BED file has been created to avoid biased positions while using the Ion AmpliSeq SARS-CoV-2 Research Panel (Thermo Fisher Scientific) and the Ion Torrent sequencing platform. Only variants whose frequencies were above the threshold frequencies provided in the SVB BED file are called. After this quality filtering step, for annotation and determination of the base substitution effect, COVID19AnnotateSnpEff (v1.3.0.2) (Cingolani et al. 2012), a plugin developed explicitly for SARS-CoV-2, was used.
For characterization of the SARS-CoV-2 variants, i.e. Alpha (20I or B.1.1.7), Beta (20H or B.1.351), Delta (21A, 21I, and 21 J), Epsilon (21C), Gamma (20 J or P.1), and Omicron (21 K or BA.1 and 21 L or BA.2), based on the characteristic mutations we used our in-house database and the defining list of mutations from the Covariants.org (Hodcroft 2021). For the in-house database, we regularly downloaded the variant surveillance package from GISAID (https://www.gisaid.org). This package contains information about each sequence uploaded in GISAID, including information about the identified lineage and their associated list of mutations. Before downstream database creation, incomplete and nonhuman sequences were discarded. Then, we determined the frequency of each mutation reported for each variant. And finally, in the database, we retained the top 50 mutations reported in association with each variant. Furthermore, these mutations were compared with the list of defining mutations present at Covariants.org, to verify the association to the respective variant, which were then used as ‘characteristic mutations’ for each variant in this study. The database and the script used for generating the database are provided on Zenodo under https://doi.org/10.5281/zenodo.7319647 and https://doi.org/10.5281/zenodo.7319702, and the list of defining mutations is provided in Table S2 (Supporting Information). We used a two-indicator approach to report the presence of a SARS-CoV-2 variant in wastewater samples, as shown in Agrawal et al. (2022a). In this two-indicator approach, first, we compare the mutations of the variant caller output of each wastewater sample with the in-house characteristic mutation database. At least 10 mutations associated with each target variant should be present in the wastewater sample for the following second step. In the second step, we look at the presence of at least one unique mutation for each target variant. A variant in the sample is reported only if wastewater samples pass through these two steps.
The abundance analysis of a variant (RA) is based on the ratio of the sum of the occurrence of mutations (Ms) of a variant divided by the sum of the occurrence of all mutations (Ma) found in a sample.
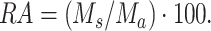 |
Ms: sum of the occurrence (number of reads X allele frequency) of the characteristic mutations of a variant.
Ma: sum of the occurrence (number of reads X allele frequency) of all mutations found in a sample.
Data analysis
Data analysis was performed in R (v.3.6.2) using the ggplot package (v.3.3.3.) and vegan (v.2.5.7) for data visualization and pheatmap (v.1.0.12) for hierarchy clustering and heatmap construction. Nonmetric multidimensional scaling (NMDS) was performed on the mutations detected in the wastewater samples.
All data are available in the main text or the Supplementary materials. We sequenced 164 wastewater samples in total during this project. Based on our sequencing approach, we were able to generate very high genomic coverage of SARS-CoV-2 genomes (97 ± 2% of target base coverage at 500X depth). The sequencing data for the samples are summarized in Table S3 (Supporting Information). Raw metagenomic sequence data are available from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject number PRJNA856091.
Results and discussion
Sampling
In the framework of the wastewater-based surveillance of SARS-CoV-2 in Hesse, composite samples were taken from 15 WWTPs from different cities all over Hesse, resulting in capturing information from 2.475.663 inhabitants (Table S1, Supporting Information). With 6.293.154 inhabitants living in Hesse in 2020, the 15 sampled WWTPs covered approximately 39% of Hesse’s inhabitants (Statista 2021). Samples were taken over a period of 6 months from September 2021 to March 2022. During this sampling period, initially the Delta variant dominated as VOC (Figure S1, Supporting Information). At the end of the summer 2021, a change from Delta to the emerging Omicron became apparent in the winter in Germany.
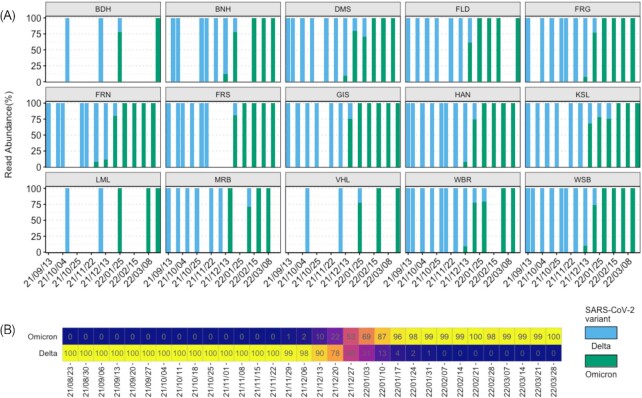
Similar to clinical data, Fig. 1 presents the genome sequencing results of wastewater samples focusing on Delta (21A, 21I, and 21 J) and Omicron (21 K or BA.1 and 21 L or BA.2) variants. At the beginning of the sampling period, until the 22nd of November 2021, Delta was dominant in the wastewater (Fig. 1). On 29 November 2021, Omicron was detected for the first time in the influent of the FRN WWTP (Agrawal et al. 2022b). According to the clinical data of the Robert Koch Institute (RKI) (compare Figure S1, Supporting Information), a strong increase of ∼50% of the Omicron content can be seen in the period from 13 December 2021 (week 50/2021 with ∼23% Omicron) to 10 January 2021 (week 2/2022 with ∼73% Omicron). On the 14th of February 2022, Omicron was 100% predominant and the Delta variant was fully replaced. A similar scenario was captured from the wastewater data: the fraction of Omicron increased from 13 December 2022 onwards at all WWTPs. Looking at the WWTPs in northern Hesse, Omicron could only be detected in the VHL WWTP already on 06 December 2021. In the samples from FRN, FRG, FRS, and WSB, the Omicron variant was already the predominant by 31 January 2022. Among the northern located WWTP, GIS was the only WWTP where the Delta variant was completely replaced by 08 February 2022. At the WWTPs plants MRB, KSL, and FLD a small fraction of Delta could still be detected on 08 Februar 2022. This variation in the emergence of Omicron among different WWTPs shows, that a spatial variation within Hesse existed.
Figure 1.
(A) Abundance of Sars-CoV-2 variants across the samples during the sampling period. WWTP city: (1) Bad Hersfeld (BDH), (2) Bensheim (BNH), (3) Darmstadt (DMS), (4) Fulda/Gläserzell (FLD), (5) Frankfurt Griesheim (FRG), (6) Frankfurt Niederrad (FRN), (7) Frankfurt Sindlingen (FRS), (8) Gießen (GIS), (9) Hanau (HAN), (10) Kassel (KSL), (11) Limburg a.d. Lahn (LML), (12) Marburg (MRB), (13) Vöhl/Thalitter (VHL), (14) Wiesbaden Biebrich (WBR), and (15) Wiesbaden Hauptklärwerk (WSB). (B) Fraction of Omicron and Delta reported by RKI, based on the sequencing analysis of the clinical samples during the period of this study. Color gradient from blue to yellow defines 0%–100% fraction.
When comparing the sequencing data of the clinical samples reported by RKI in GISAID and the wastewater sequencing data from this study, very similar courses can be observed with regard to the distributions of the virus variants over time (Figure S1, Supporting Information). Other studies show that trends and developments can be detected by the WBE 10–14 days before the clinically collected data, because viral RNA can be detected in the wastewater even before the onset of symptoms of those infected with SARS-CoV-2 (Ahmed et al. 2020, Medema et al. 2020). In this work, no such trends could be identified prior to clinical studies. However, the main limitation here was the low sampling frequency; samples were taken only bi-weekly at the most.
Occurrence pattern of the characteristic mutations
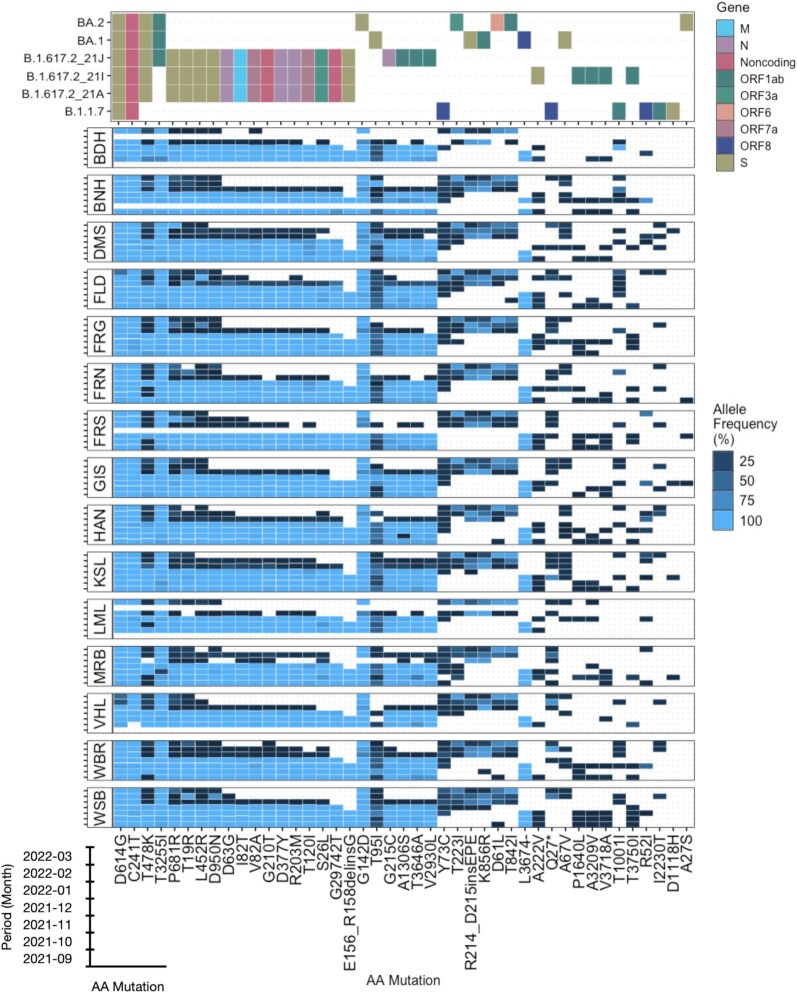
In the beginning of September 2021, the mutations that are associated with Alpha and Delta were prevalent in the samples with allele frequencies of 100% (Fig. 2 and Figure S2, Supporting Information, provide information about the detected mutations and their association with the variants). For Delta, A1306S, D63G, D950N, G210T, G215C, I82T, T120I, T19R, V2930L, D377Y, and V82A were detected from September 2021 until January 2022 in all WWTPs. Between December 2021 and January 2022, there was a marked change with respect to the occurrence of certain mutations (Fig. 2). D377Y, A1306S, G29742T, I82T, and S26 L were detected with a high allele frequency of 100% at every sampling point until December 2021. Afterwards, these mutations were either not detected or had lower allele frequency (<25%). All these mutations are typical mutations for the Delta variant and have also been used as characteristic reference mutations for Delta (Hodcroft 2021). The mutations D61L, A67V, T842I, and K856R, occurred consistently since the first sampling in 2022 on 10 January 2022. All of these mutations are characteristic mutations for Omicron (Hodcroft 2021). Overall, the decrease in prevalence of characteristic mutations of Delta was observed from beginning of 2022 and the subsequent increase in the prevalence of characteristic mutations of Omicron followed.
Figure 2.
Allele frequency of characteristic AA mutations associated to Alpha (20I or B.1.1.7), Delta (21A, 21I, and 21 J), and Omicron (21 K or BA.1 and 21 L or BA.2) over the sampling period for each WWTP. WWTP city: (1) Bad Hersfeld (BDH), (2) Bensheim (BNH), (3) Darmstadt (DMS), (4) Fulda/Gläserzell (FLD), (5) Frankfurt Griesheim (FRG), (6) Frankfurt Niederrad (FRN), (7) Frankfurt Sindlingen (FRS), (8) Gießen (GIS), (9) Hanau (HAN), (10) Kassel (KSL), (11) Limburg a.d. Lahn (LML), (12) Marburg (MRB), (13) Vöhl/Thalitter (VHL), (14) Wiesbaden Biebrich (WBR), and (15) Wiesbaden Hauptklärwerk (WSB)
C241T, T3255I, G142D, and D614G were detected in all samples at all sampling points with an allele frequency of 100%. This might be due to the fact that high-frequency mutations in the SARS-CoV-2 genome include C241T, C1059T, C3037T, C14408T, A23403G, G25563T, and G28883C, out of which the C241T has been found in ∼50% of clinical sequences by October 2020 (Hadfield et al. 2018). Additionally D614G, with glycine substituted for aspartic acid at position 614 in the S protein, was one of the most frequent mutations present in a majority of the circulating SARS-CoV-2 strains (Jiang et al. 2020, Korber et al. 2020). T3255I and D614G are known to be associated with the Delta as well as the Omicron variant, which explains the high occurrence and prevalence across all the samples (Shanmugaraj et al. 2022).
Characteristic mutations vs. all mutations in wastewater
Most of the attention is given to known characteristic mutations to evaluate the sequencing results and these mutations also serve as principal targets for qPCR or dPCR analysis. Studies stated the hypothesis that even to monitor the shift from BA.1 (21 K) to BA.2 (21 L), assays based on characteristic mutations for variant sublineages would be enough for WBE (Boehm et al. 2022, Wolfe et al. 2022). However, we believe that limiting the focus mainly to characteristic mutations could limit the information about the circulating lineages and may even lead to false-negative outcomes. This is especially critical for Omicron sublineages, which have very few unique mutations (Hodcroft 2021) and due to plausible uneven coverage of the SARS-CoV-2 genome resulting from possible fragmentation of SARS-CoV-2 RNA in wastewater (Fontenele et al. 2021, Smyth et al. 2022).
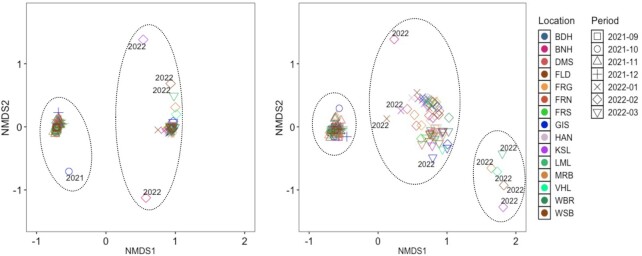
A NMDS ordination analysis based on all mutations shows the formation of two clusters (Fig. 3). The smaller cluster combines all samples from 2021, whereas the larger cluster includes all samples taken in 2022, but with two outliers representing samples taken in February 2022 in KSL and FNR. These results clearly show the switch from Delta as the dominant variant to Omicron in December 2021 to early January 2022. However, within each sampling month the samples’ cluster varied (Figure S3, Supporting Information), emphasizing the spatial variation. Also, the analysis based on the characteristic mutations shows a smaller cluster with all samples from 2021 and a bigger one with samples from 2022. But additionally, we see a second smaller cluster with samples from February 2022 including VHL, MRB, KSL, and WSB as well as WBR from March 2022.
Figure 3.
(Left) NMDS plot considering all the mutations. (Right) NMDS plot based on the characteristic mutations. WWTP city: (1) Bad Hersfeld (BDH), (2) Bensheim (BNH), (3) Darmstadt (DMS), (4) Fulda/Gläserzell (FLD), (5) Frankfurt Griesheim (FRG), (6) Frankfurt Niederrad (FRN), (7) Frankfurt Sindlingen (FRS), (8) Gießen (GIS), (9) Hanau (HAN), (10) Kassel (KSL), (11) Limburg a.d. Lahn (LML), (12) Marburg (MRB), (13) Vöhl/Thalitter (VHL), (14) Wiesbaden Biebrich (WBR), and (15) Wiesbaden Hauptklärwerk (WSB)
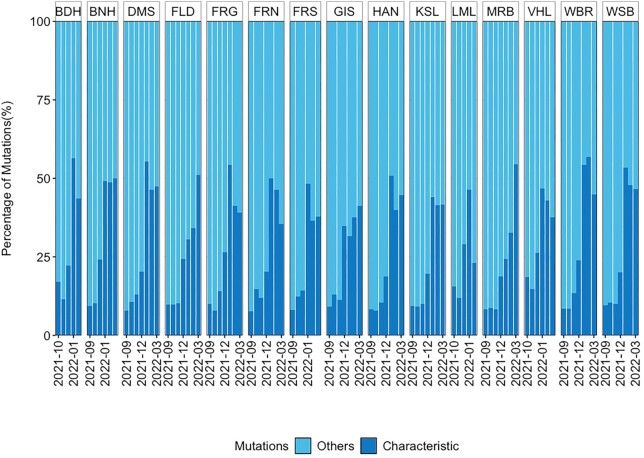
Characteristic mutations, key mutations or signature mutations play an important role in the analysis of wastewater samples, as they are either the basis for the development of qPCR/dPCR assays (Boehm et al. 2022) or for tracking specific variants in NGS datasets for WBE. The S gene reveals the highest number of characteristic mutations, especially for the pango lineages BA.1, BA.2, BA.2.12.1, BA.4, and BA.5, as well as the noncoding genomic region with around 30–40 characteristic mutations (Figure S2, Supporting Information). Variants before Omicron have around 10 characteristic mutations in the S gene and around 5–7 in the noncoding region. The origin of immune-compromised individuals or animals have been suggested to be the reason for the increase of mutations in the Omicron variant (40 residue changes vs. 10 on average in all the previous dominant VOCs) (Fig. 4; Corey et al. 2021, Wei et al. 2021, Zhang et al. 2022) shows the fraction of characteristic mutations among the total mutations found in each sample. During the sampling period from September 2021 to March 2022, there was an increasing percentage of characteristic mutations in all WWTPs. In September the percentage was around 10%–15%, and it increased after the emergence of Omicron between December 2021 and January 2022 to 25% and even to 50% until March 2022 for all locations. This is due to the fact, that Omicron in general is bearing an unusually high number of mutations compared to other variants (Zhang et al. 2022). Thus, analysis based on the characteristic mutations may vary with the change in number of known characteristic mutations of a variant.
Figure 4.
Fraction of characteristic mutations among the total mutations found in each sample during the sampling period. WWTP city: (1) Bad Hersfeld (BDH), (2) Bensheim (BNH), (3) Darmstadt (DMS), (4) Fulda/Gläserzell (FLD), (5) Frankfurt Griesheim (FRG), (6) Frankfurt Niederrad (FRN), (7) Frankfurt Sindlingen (FRS), (8) Gießen (GIS), (9) Hanau (HAN), (10) Kassel (KSL), (11) Limburg a.d. Lahn (LML), (12) Marburg (MRB), (13) Vöhl/Thalitter (VHL), (14) Wiesbaden Biebrich (WBR), and (15) Wiesbaden Hauptklärwerk (WSB)
Impact of selecting characteristic mutations on the different genomic region of the SARS-CoV-2
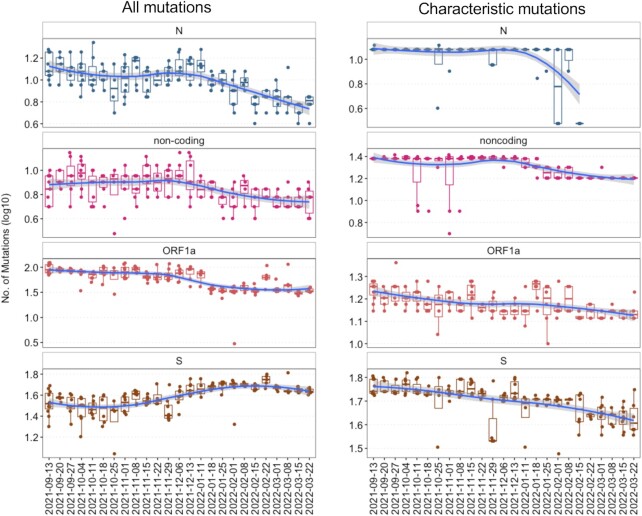
Here, we focused only on the S, noncoding, ORF1a, and N gene because the number of mutations in these genes is highest among the lineages (Figure S2, Supporting Information). We compared temporal shifts in the number of all the mutations against the number of the characteristic mutations. The results underline that focusing on the characteristic mutations reveals a different outcome than considering all mutations (Fig. 5). For the N gene, there is a slow decrease in the number of mutations over time, whereas for the known characteristic mutations after mid of February 2022, there were no characteristic mutations detectable. For the noncoding mutations, the results showed an inverted trend. For the ORF1a and S gene, considering all mutations there was an increase in the number of mutations at the end of March 2022, whereas when considering only the number of the characteristic mutations a decrease at the end of March (Fig. 5) was observable.
Figure 5.
(Left) Number of all the mutations, which are associated with S, noncoding, ORF1a, and N gene of SARS-CoV-2, detected in wastewater samples during the sampling period. (Right) Number of all the known characteristic mutations which are associated with S, noncoding, ORF1a, and N gene of SARS-CoV-2, detected in wastewater samples during the sampling period.
This study demonstrates the consistency between the evolution of the Delta and Omicron variant in Hesse based on clinical data and wastewater surveillance (Fig. 1 and Figure S1, Supporting Information). The data suggest that surveillance of SARS-CoV-2 variants based on the characteristic mutation does provide valuable information about the temporal and spatial variation. However, monitoring SARS-CoV-2 variants in wastewater requires existing knowledge about the variants to be classified. Moreover, while limiting the focus to the characteristic mutations of known variants could be challenging. For example, in the early phase of the emergence of Omicron, the del(H69-V70) mutation was considered to be one of the characteristic mutations for Omicron. But some initial data reported the absence of the del(H69-V70) mutation, leading to unclarity about the detection of the Omicron variant until the designation of BA.1 (21 K) and BA.2 (21 L). Also, during early emergence of Omicron, researchers reported lower coverage of the ORF1ab and spike protein region (Ahmed et al. 2022b), thus limited detection of the characteristic mutations. One approach to overcome this challenge is to include all the mutations detected in the wastewater samples. The results in this study clearly show that the temporal and spatial variation were better captured including all the mutations. In this study, we achieved at least >97% SARS-CoV-2 genome coverage, however, we observed an effect on the temporal and spatial variations when based on characteristic mutations. The difference in capturing trends based on characteristic mutations compared to including all the mutations could be more critical in case of lower coverage of specific genomic region. Including all mutations also allows to include cryptic mutations, which are undetected in clinical samples, which might have animal reservoirs.
Supplementary Material
Acknowledgements
This work was funded by the Hessian Ministry of Higher Education, Research, Science, and the Arts, grant numbers 408/05.001 and 451/06.006-(0003). We would also like to thank all WWTP operators for providing wastewater samples and members of our research group for their support with sample collection. We gratefully acknowledge the contribution from the originating laboratories responsible for obtaining the specimens and the submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative (https://www.gisaid.org)
Contributor Information
Shelesh Agrawal, Technical University of Darmstadt, Institute IWAR, Chair of Water and Environmental Biotechnology, Franziska-Braun-Straße 7, 64287 Darmstadt, Germany.
Laura Orschler, Technical University of Darmstadt, Institute IWAR, Chair of Water and Environmental Biotechnology, Franziska-Braun-Straße 7, 64287 Darmstadt, Germany.
Kira Zachmann, Technical University of Darmstadt, Institute IWAR, Chair of Water and Environmental Biotechnology, Franziska-Braun-Straße 7, 64287 Darmstadt, Germany.
Susanne Lackner, Technical University of Darmstadt, Institute IWAR, Chair of Water and Environmental Biotechnology, Franziska-Braun-Straße 7, 64287 Darmstadt, Germany.
Conflicts of interest statement
None declared.
References
- Agrawal S, Orschler L, Schubert Set al. Prevalence and circulation patterns of SARS-CoV-2 variants in European sewage mirror clinical data of 54 European cities. Water Res. 2022a;214:118162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S, Orschler L, Tavazzi Set al. Genome sequencing of wastewater confirms the arrival of the SARS-CoV-2 Omicron variant at Frankfurt airport but limited spread in the city of Frankfurt, Germany, in November 2021. Microbiol Resour Announc. 2022b;11:e01229–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Angel N, Edson Jet al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020;728:138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Bivins A, Metcalfe Set al. RT-qPCR and ATOPlex sequencing for the sensitive detection of SARS-CoV-2 RNA for wastewater surveillance. Water Res. 2022a;220:118621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Bivins A, Smith WJMet al. Detection of the Omicron (B.1.1.529) variant of SARS-CoV-2 in aircraft wastewater. Sci Total Environ. 2022b;820:153171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman F, Markt R, Endler Let al. Viral variant-resolved wastewater surveillance of SARS-CoV-2 at national scale. Nat Biotechnol. 2022;40:1814–22., [DOI] [PubMed] [Google Scholar]
- Boehm AB, Hughes B, Wolfe MKet al. Regional replacement of SARS-CoV-2 variant Omicron BA.1 with BA.2 as observed through wastewater surveillance. Environ Sci Technol Lett. 2022;9:575–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boogaerts T, Van den Bogaert S, Van Poelvoorde LAEet al. Optimization and application of a multiplex digital PCR assay for the detection of SARS-CoV-2 variants of concern in Belgian influent wastewater. Viruses. 2022;14:610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón-Franco D, Orschler L, Lackner Set al. Monitoring SARS-CoV-2 in sewage: toward sentinels with analytical accuracy. Sci Total Environ. 2022;804:150244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LLet al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey L, Beyrer C, Cohen MSet al. SARS-CoV-2 variants in patients with immunosuppression. N Engl J Med. 2021;385:562–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crits-Christoph A, Kantor RS, Olm MRet al. Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. MBio. 2021;12:e02703–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenele RS, Kraberger S, Hadfield Jet al. High-throughput sequencing of SARS-CoV-2 in wastewater provides insights into circulating variants. Water Res. 2021;205:117710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl DM, Garbe J, Grady Pet al. A rapid, cost-effective tailed amplicon method for sequencing SARS-CoV-2. BMC Genomics. 2020;21:863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J, Megill C, Bell SMet al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijnen L, Elsinga G, de Graaf Met al. Droplet digital RT-PCR to detect SARS-CoV-2 signature mutations of variants of concern in wastewater. Sci Total Environ. 2021;799:149456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold M, d'Hérouël AF, May Pet al. Genome sequencing of SARS-CoV-2 allows monitoring of variants of concern through wastewater. Water. 2021;13:3018. [Google Scholar]
- Hodcroft EB. CoVariants: SARS-CoV-2 mutations and variants of interest. GISAID, 2021. [Google Scholar]
- Jiang X, Zhang Z, Wang Cet al. Bimodular effects of D614G mutation on the spike glycoprotein of SARS-CoV-2 enhance protein processing, membrane fusion, and viral infectivity. Sig Transduct Target Ther. 2020;5:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M, Ahmed W, Bibby Ket al. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci Total Environ. 2020;739:139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Fischer WM, Gnanakaran Set al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G, Iaconelli M, Veneri Cet al. The rapid spread of SARS-COV-2 Omicron variant in Italy reflected early through wastewater surveillance. Sci Total Environ. 2022;837:155767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClary-Gutierrez JS, Mattioli MC, Marcenac Pet al. SARS-CoV-2 wastewater surveillance for public health action. Emerg Infect Dis. 2021;27:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G, Heijnen L, Elsinga Get al. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ Sci Technol Lett. 2020;7:511–6. [DOI] [PubMed] [Google Scholar]
- Osterman A, Badell I, Basara Eet al. Impaired detection of omicron by SARS-CoV-2 rapid antigen tests. Med Microbiol Immunol. 2022;211:105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cataluña A, Chiner-Oms Á, Cuevas-Ferrando Eet al. Spatial and temporal distribution of SARS-CoV-2 diversity circulating in wastewater. Water Res. 2022;211:118007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priesemann V, Balling R, Brinkmann MMet al. An action plan for pan-European defence against new SARS-CoV-2 variants. The Lancet. 2021;397:469–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert Kock Institute . Wöchentlicher Lagebericht des RKI zur Coronavirus-Krankheit-2019 (COVID-19). 2022.
- Shanmugaraj B, Malla A, Khorattanakulchai Net al. SARS-CoV-2 omicron variant: could it be another threat?. J Med Virol. 2022;94:1284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan SP, Shahin S, Ward LMet al. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci Total Environ. 2020;743:140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DS, Trujillo M, Gregory DAet al. Tracking cryptic SARS-CoV-2 lineages detected in NYC wastewater. Nat Commun. 2022;13:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statista . Einwohnerzahl in Hessen bis 2021. 2021.
- Wei C, Shan K-J, Wang Wet al. Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. J Genet Genomics. 2021;48:1111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M, Hughes B, Duong Det al. Detection of SARS-CoV-2 variants Mu, Beta, Gamma, Lambda, Delta, Alpha, and Omicron in wastewater settled solids using mutation-specific assays is associated with regional detection of variants in clinical samples. Appl Environ Microbiol. 2022;88:e00045–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz N, Revol O, Jardot Pet al. Monitoring the circulation of SARS-CoV-2 variants by genomic analysis of wastewater in Marseille, South-East France. Pathogens. 2021;10:1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cai Y, Lavine CLet al. Structural and functional impact by SARS-CoV-2 Omicron spike mutations. Cell Rep. 2022;39:110729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.