Abstract
In the last decade, investigating white matter microstructure and connectivity via diffusion MRI (dmri) has become a crucial cornerstone in neuroimaging studies. However, even modern dmri sequences have inherently a low signal-to-noise ratio and long acquisition times, depending on the spatial resolution. Furthermore, many types of artifacts complicate the appropriate analysis of dmri, necessitating appropriate quality control (QC) procedures, including exclusion and/or correction of inappropriate/erroneous dmri data. Our group has been developing and promoting QC procedures and tools to the community to enable appropriate dmri analyses. Since its development in 2011, our DTIPrep QC tool has become a major tool due its ease of use and dmri QC performance. Over the years, novel developments in acquisition and artifact correction methods have led to a need to modernize DTIPrep. Here, we present a novel diffusion MRI analysis environment called dtiplayground with a fully redesigned and significantly enhanced QC module dmriprep, and its graphical user interface dmriprep-ui, building on in-house developed code, FSL and dipy. The user interface is designed to be a unified, user friendly tool for thorough QC of dMRI data.Artifacts addressed by dmriprep include eddy-currents, head motion, bed vibration and pulsation, venetian blind artifacts, slice-wise and gradient-wise intensity inconsistencies, and susceptibility artifacts. It further provides an user interface for visual QC of gradients and automated tractography. In summary, our work presents a novel open-source framework for modern comprehensive dmri QC.
Keywords: diffusion MRI, open source, quality control, preprocessing
1. INTRODUCTION
The collection and analysis of diffusion MRI (dmri) datasets are a crucial cornerstone of neuroimaging studies, illuminating white matter pathology and effects as potential biomarkers in studies of autism, schizophrenia, Huntington’s disease, Alzheimer’s disease, Parkinson’s disease, and many other conditions. The tool of choice for studying WM is dmri, which extends the capabilities of conventional MRI methods by measuring the diffusion properties of the tissue via fitting microstructure models in a voxel-wise fashion. The most common model employed is the diffusion tensor with property maps of fractional anisotropy (FA), axonal (AD), radial (RD) and mean diffudivity (MD),1 but models such as neurite orientation dispersion and density imaging (NODDI)2 and other multi-compartment models are increasingly in standard use.3
dmri has enormous potential, but it suffers from a unique and complex set of image quality problems, limiting the sensitivity of dmri studies and reducing the accuracy of findings. Its data is obtained by acquiring a series of images using non-colinear diffusion-sensitizing gradients (called DWI,Diffusion Weighted Images) with longer overall acquisition time than conventional MRI due to the need for multiple DWI acquisitions, with higher order diffusion models necessitating often over 100 DWI volumes at various diffusion shells. Figure 1 visualizes examples of some observed dmri intensity artifacts. In addition, motion, eddy-current induced and susceptibility induced artifacts (see Figure 2) are commonly present. Therefore, unlike conventional structural MRI data, dmri data needs to be carefully checked and corrected to remove these artifacts, making a quality control (QC) procedure absolutely necessary for dmri studies.
Figure 1.
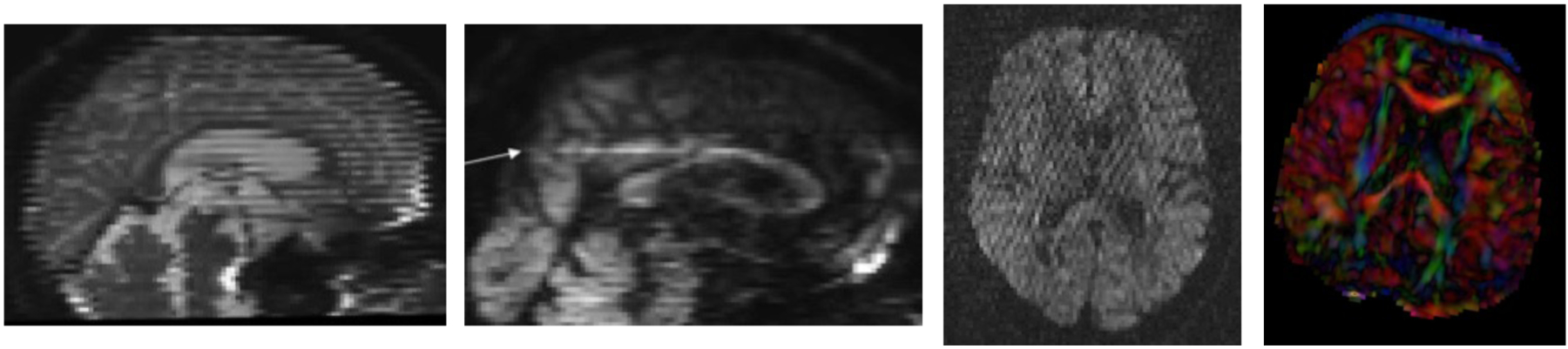
Examples of intensity artifacts detected with DTIPrep. Left: Venetian blind artifacts. Mid left: inter-slice intensity artifact. Mid right: checkerboard artifact. Right: Directional over-saturation artifact (commonly red shift on colorized FA maps), which are commonly not detectable on individual DWI volumes.
Figure 2.
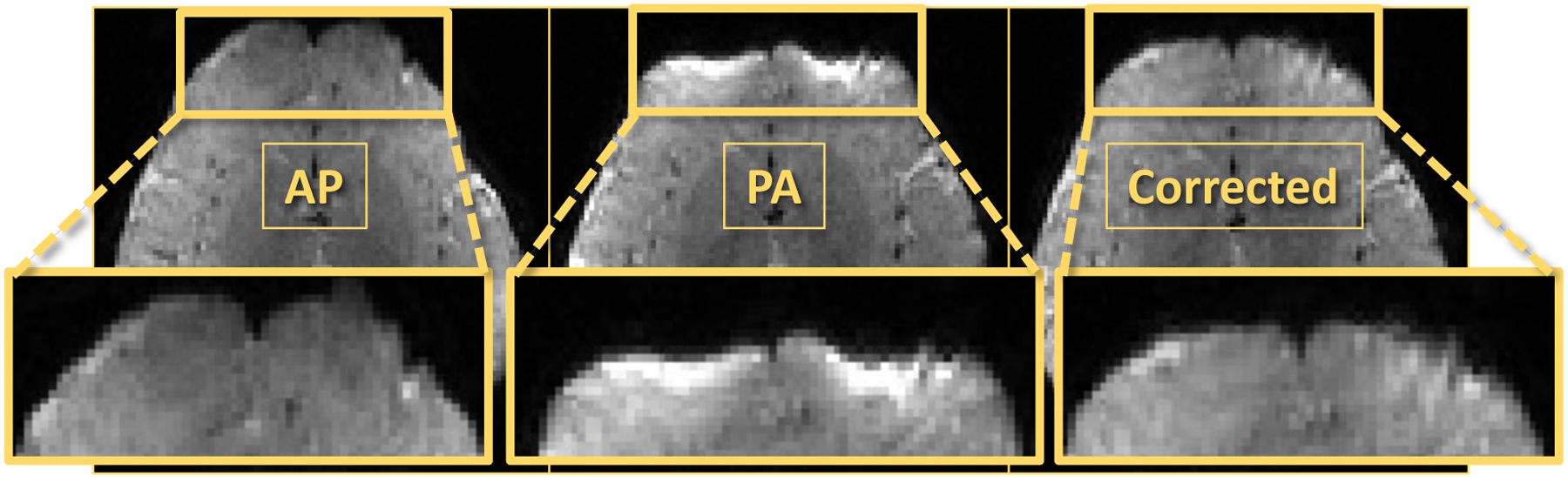
Susceptibility artifact in AP (anterior-posterior) and PA (posterior-anterior) polarity encoding, as well as the combined average image after correction with topup.
Our group has a history of developing and promoting dmri QC procedures and tools to the neuroimaging community to enable appropriate dmri analyses. Since its development in 2011, our DTIPrep QC tool4 has become a major tool due its ease of use and dmri QC performance,5,6 addressing most of the above mentioned dmri artifacts. Yet, over the years, novel developments in acquisition and artifact correction methods have led to a need to modernize DTIPrep. As the original DTIPrep source code has a non-modular design that is difficult to expand, we opted for a fully redesigned open-source implementation, which incorporates not just dmri QC but also fiber-based dmri analysis7 in a modular, easy to expand and customize fashion. This python-based dmri framework is called dtiplayground and its first major module is a fully redesigned and significantly enhanced QC module called dmriprep, with its graphical user interface dmriprep-ui, built based on in-house developed code, FSL,8 dipy,3 and PyQt. The goal of dmriprep is to provide an easy-to-use diffusion MRI QC tool for both non-technical and technical users.
The contribution of this work is not novel methodology, but rather a novel open-source software framework for comprehensive dmri QC.
2. METHODS
2.1. The dtiplayground framework
The proposed dmriprep QC module is built on the open-source dtiplayground *, a novel, python based dmri workflow pipeline software currently including dmriprep, dmriatlasbuilder (for building diffusion tensor atlases), and dmrifiberanalyzer (for generating micro-structure property profiles along WM fiber bundles). dtiplayground is under development since 2020 and currently available in a pre-release, beta version. dtiplayground and dmriprep are currently distributed as a Python package (via pip installation) and a Docker container is in preparation. The python framework is based on dipy,3 simpleITK, and nibabel. Currently two separate UIs are supported in separate versiouns: a) one based on PyQt, and supports deep learning based modules (currently via ANTsPyNet), and b) a web-based flask UI using a server based approach to access the UI. In addition, FSL version 6.X is employed for several of the processing steps, and can be installed separately, or as part of the dtiplayground installation.
Note that the feature rich dmri preprocessing and analysis framework qsiprep9 has been independently developed at the same time as we started development of dtiplayground. As both frameworks are python based and built on similar underlying libraries, we expect that future development of dtiplayground will include cross-framework compatibility and interaction with qsiprep.
2.2. dmriprep and dmriprep-ui
Similar to DTIPrep, dmriprep is protocol based. Its graphical user interface (dmriprep-ui) largely serves as an easy-to-use way to both assemble the preprocessing/QC protocol and then run it on individual datasets. The protocol is stored in YML format on disk and a copy is stored in the processing output folder to ensure proper reporting. Within a single neuroimaging study, the user is advised to use the same QC protocol. The protocol is modular in design and implementation, and it is easy to extend dmriprep with new modules as is evident by the number of already implemented modules (see also Figure 3 Left):
Slice wise checking: allows the detection and rejection of slice-wise artifacts, such as signal loss due to motion, or hyperintense slices due to flow artifacts.
Interlace checking: allows the detection and rejection of significant within-volume motion
Susceptibility correction: allows the correction of susceptibility artifacts via FSL 6.X
Eddy and motion correction: allows for the correction of eddy current and motion artifacts via FSL eddy openmp tool. If susceptibility information is present, then that information is incorporated into this correction module. The eddy current correction module via FSL allows also for the replacement of outlier measurements10 and the correction of within volume movement.11
Brain mask: computes a brain mask (excluding extra-axial cerebrospinal fluid), by applying FSL bet to either a) the isotropically weighted diffusion image (the geometric average over all non-b=0 images), b) the FA image, or c) the average baseline b=0 image.
Full brain tractography: automatic full brain tractography for the purpose of QC, available tractography methods include deterministic principal tensor axis tractography, deterministic maximum-peak constant solid angle (CSA) tractography, and deterministic maximum-peak QBall tractography. The whole brain tractography module provides a quick visual assessment whether directional artifacts are present, as such artifacts are expected to lead to unusually distorted fiber tracts (see Fig 4 right).
Tensor model estimation: allows the computation of tensor images, multiple estimation options are available (we suggest the traditional weighted least square estimate or using RESTORE?)
B-shell filtering: allows the removal or selection of individual diffusion b-shells. This is particularly useful if high b-value shells are removed prior to tensor estimation
Tensor image (DTI) registration: allows the registration of a prior DTI atlas for the purpose of QC
QC report: generates PDF and CSV based reports of the quality of the data for use in study analyses.
Removal of DWI volumes: this interactive gradient exclusion module is commonly performed following the automated QC process and allows users to visually identify DWI volumes with remaining, significant artifacts so that they can be removed prior to further analysis (see Fig 4 right).
Figure 3.
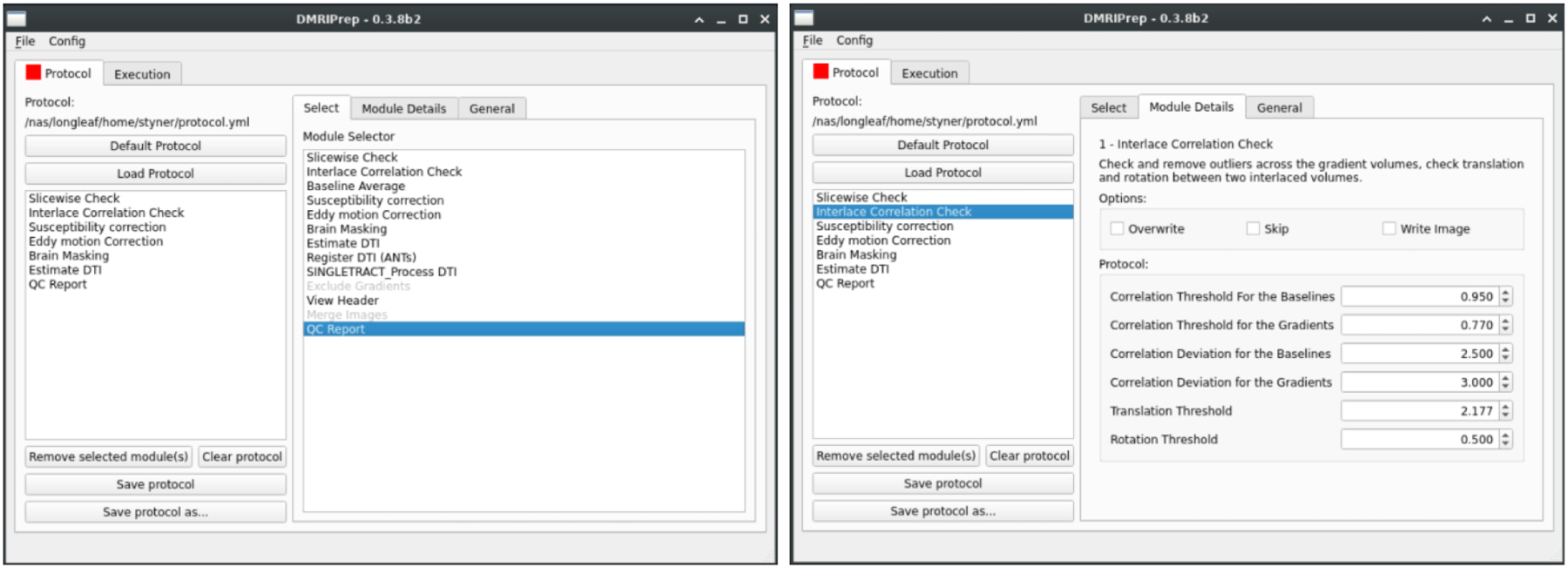
Graphical user interface for dmriprep (dmriprep-ui). Left: current list of modules (Module selector) and an example list of modules comprising a protocol (protocol area). Right: example of parameter settings (here: interlace correction module).
Figure 4.
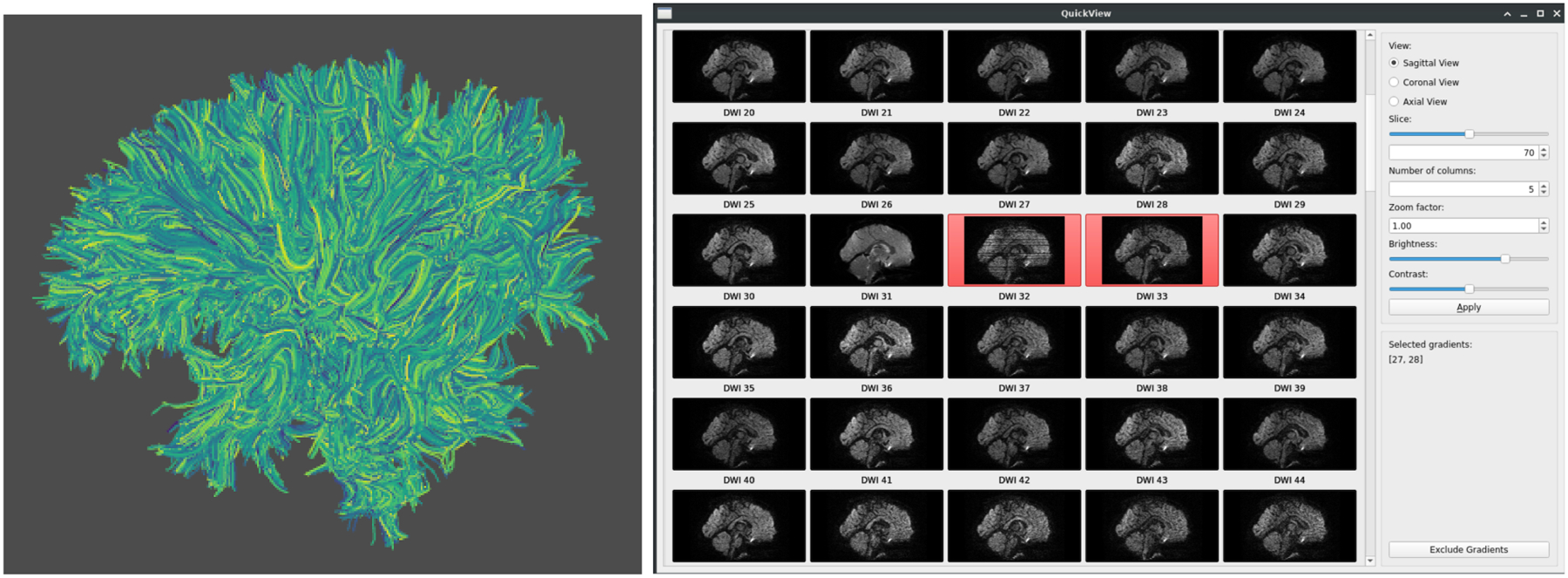
Visualization of automated full brain tractography for visual QC inspection (Left), and DWI QC inspection in dmriprep with interactive exclusion of bad DWI volumes (Right). Two artifact rich DWI volumes are selected for exclusion.
In order to have broad applicability, dtiplayground and dmriprep aim at both the FSL/NIFTI community as well as the 3D Slicer community and thus supports both NIFTI and NRRD formats and coordinate systems. DICOM conversion is currently not included in dmriprep and the user is expected to have converted the DICOM data to either NIFTI or NRRD beforehand.
2.3. Discussion
While dmriprep is an open-source framework under active development, with updated releases on a monthly basis, it is robust and stable enough to be applied to neuroimaging studies. In the authors’ lab, all 500+ dMRI dataset that are publicly available as part of the Baby Connectome Project12 on the National Institute of Mental Health Data Archive (NDA). With a few mouse clicks in dmriprep-UI, the QC processing protocol was assembled, with default values chosen by dmriprep, and then applied to all datasets via the command line interface for easy batch processing. Final visual QC of the whole brain tractography and all the corrected DWI volumes is currently ongoing.
dmriprep is the first major tool in the dtiplayground framework, with several additional tools under construction to provide an end-to-end solution for atlas based fiber profile analysis similar to the UNC-Utah NAMIC framework.7
ACKNOWLEDGMENTS
This research has been supported by the following NIH grants: R01HDO55741, U54HDO79124, R01EB021391, P50HD103573.
Footnotes
Source code and installation information: https://github.com/NIRALUser/DTIPlayground
REFERENCES
- [1].Pierpaoli C, Jezzard P, Basser PJ, Barnett A, and Chiro GD, “Diffusion tensor MR imaging of the human brain.,” Radiology 201, 637–648 (12 1996). [DOI] [PubMed] [Google Scholar]
- [2].Zhang H, Schneider T, Wheeler-Kingshott CA, and Alexander DC, “NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain.,” NeuroImage 61, 1000–1016 (07 2012). [DOI] [PubMed] [Google Scholar]
- [3].Fick RHJ, Wassermann D, and Deriche R, “The Dmipy Toolbox: Diffusion MRI Multi-Compartment Modeling and Microstructure Recovery Made Easy,” Frontiers in Neuroinformatics 13, 64 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Oguz I, Farzinfar M, Matsui J, Budin F, Liu Z, Gerig G, Johnson HJ, and Styner M, “DTIPrep: quality control of diffusion-weighted images,” Frontiers in Neuroinformatics 8, 4 (00 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kreilkamp BAK, Zacà D, Papinutto N, and Jovicich J, “Retrospective head motion correction approaches for diffusion tensor imaging: Effects of preprocessing choices on biases and reproducibility of scalar diffusion metrics.,” Journal of magnetic resonance imaging : JMRI (06 2015). [DOI] [PubMed] [Google Scholar]
- [6].Liu B, Zhu T, and Zhong J, “Comparison of quality control software tools for diffusion tensor imaging.,” Magnetic resonance imaging 33, 276–285 (04 2015). [DOI] [PubMed] [Google Scholar]
- [7].Verde AR, Budin F, Berger J-B, Gupta A, Farzinfar M, Kaiser A, Ahn M, Johnson H, Matsui J, Hazlett HC, Sharma A, Goodlett C, Shi Y, Gouttard S, Vachet C, Piven J, Zhu H, Gerig G, and Styner M, “UNC-Utah NA-MIC framework for DTI fiber tract analysis,” Frontiers in Neuroinformatics 7, 51 (00 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, and Smith SM, “FSL.,” NeuroImage 62, 782–790 (08 2012). [DOI] [PubMed] [Google Scholar]
- [9].Cieslak M, Cook PA, He X, Yeh F-C, Dhollander T, Adebimpe A, Aguirre GK, Bassett DS, Betzel RF, Bourque J, Cabral LM, Davatzikos C, Detre JA, Earl E, Elliott MA, Fadnavis S, Fair DA, Foran W, Fotiadis P, Garyfallidis E, Giesbrecht B, Gur RC, Gur RE, Kelz MB, Keshavan A, Larsen BS, Luna B, Mackey AP, Milham MP, Oathes DJ, Perrone A, Pines AR, Roalf DR, Richie-Halford A, Rokem A, Sydnor VJ, Tapera TM, Tooley UA, Vettel JM, Yeatman JD, Grafton ST, and Satterthwaite TD, “QSIPrep: an integrative platform for preprocessing and reconstructing diffusion MRI data,” Nature Methods 18(7), 775–778 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Andersson JLR, Graham MS, Zsoldos E, and Sotiropoulos SN, “Incorporating outlier detection and replacement into a non-parametric framework for movement and distortion correction of diffusion MR images.,” NeuroImage 141, 556–572 (11 2016). [DOI] [PubMed] [Google Scholar]
- [11].Andersson JLR, Graham MS, Drobnjak I, Zhang H, Filippini N, and Bastiani M, “Towards a comprehensive framework for movement and distortion correction of diffusion MR images: Within volume movement.,” NeuroImage 152, 450–466 (05 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Howell BR, Styner MA, Gao W, Yap P-T, Wang L, Baluyot K, Yacoub E, Chen G, Potts T, Salzwedel A, Li G, Gilmore JH, Piven J, Smith JK, Shen D, Ugurbil K, Zhu H, Lin W, and Elison JT, “The UNC/UMN Baby Connectome Project (BCP): An overview of the study design and protocol development,” NeuroImage 185, 891–905 (01 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


