Abstract
Ongoing emergence of SARS-CoV-2 Omicron subvariants and their rapid worldwide spread pose a threat to public health. From November 2022 to February 2023, newly emerged Omicron subvariants, including BQ.1.1, BF.7, BA.5.2, XBB.1, XBB.1.5, and BN.1.9, became prevalent global strains (>5% global prevalence). These Omicron subvariants are resistant to several therapeutic antibodies. Thus, the antiviral activity of current drugs such as remdesivir, molnupiravir, and nirmatrelvir, which target highly conserved regions of SARS-CoV-2, against newly emerged Omicron subvariants need to be evaluated. We assessed the antiviral efficacy of the drugs using the half-maximal inhibitory concentration (IC50) against human isolates of 23 Omicron subvariants and four former SARS-CoV-2 variants of concern (VOCs) and compared it with the antiviral efficacy of these drugs against the SARS-CoV-2 reference strain (hCoV/Korea/KCDC03/2020). Maximal IC50-fold changes of remdesivir, molnupiravir, and nirmatrelvir were 1.9 (BA.2.75.2), 1.2 (B.1.627.2), and 1.4 (BA.2.3), respectively, compared to median IC50 values of the reference strain. Moreover, median IC50-fold changes of remdesivir, molnupiravir, and nirmatrelvir against the Omicron variants were 0.96, 0.4, and 0.62, respectively, similar to the 1.02, 0.88, and 0.67, respectively, median IC50-fold changes for previous VOCs. Although K90R and P132H in Nsp 5, and P323L, A529V, G671S, V405F, and ins823D in Nsp 12 mutations were identified, these amino acid substitutions did not affect drug antiviral activity. These results indicate that current antivirals retain antiviral efficacy against newly emerged Omicron subvariants. It is important to continue active surveillance and testing of new variants for drug resistance to enable early identification of drug-resistant strains.
Keywords: SARS-CoV-2, Omicron subvariant, Remdesivir, Molnupiravir, Nirmatrelvir, Antiviral activity
The ongoing emergence of SARS-CoV-2 Omicron subvariants is accompanied by rapid spread worldwide. Since its appearance in November 2021, the Omicron variant, B.1.1.529, has outcompeted other variants of concern (VOCs) with 26–32 amino acid changes in the spike protein, and Omicron subvariants have become the dominant strains worldwide (Guo et al., 2022). The globally prevalent Omicron sublineages (>5% global preference) from November 2022 to February 2023 were the BQ.1.1, BF.7, BA.5.2, XBB.1, XBB.1.5, and BN.1.9 variants (Khare et al., 2021). The emergence of these Omicron subvariants is concerning because of the possibility of resistance to current antiviral drugs.
Two main classes of targeted antiviral drugs have been developed for treating COVID-19. The first are monoclonal antibodies, which bind directly to the SARS-CoV-2 spike protein and inhibit viral infection (Brady et al., 2022). Although several monoclonal antibodies showed effective virus neutralisation early in the pandemic, they were less or not effective against the emerging Omicron subvariants (Arora et al., 2023).
The second are small-molecule drugs that target viral RNA-dependent RNA polymerase (RdRp) or main protease (Mpro) (Brady et al., 2022). Remdesivir (Veklury) and molnupiravir (Lagevrio) are nucleoside analogue prodrugs that target the RdRp of SARS-CoV-2, and nirmatrelvir/ritonavir (Paxlovid) is a peptidomimetic inhibitor of the SARS-CoV-2 Mpro. Remdesivir became the first US Food and Drug Administration (FDA)-approved drug in October 2020, and molnupiravir and nirmatrelvir/ritonavir received emergency use authorisation by the FDA in December 2021 (Brady et al., 2022). Despite these drugs targeting highly conserved regions of the virus genome, genetic mutations could occur in the spike protein and elsewhere in the viral genome (Kim et al., 2020b). Thus, the antiviral efficacy of the drugs for treating COVID-19 needs to be assessed against other SARS-CoV-2 variants.
In this study, we assessed the antiviral efficacy of the drugs by comparing the fold-change ratio of the in vitro half-maximal inhibitory concentration (IC50) and median IC50 value against human isolates of 23 new Omicron subvariants and four former SARS-CoV-2 VOCs to that of the Korean reference strain (hCoV/Korea/KCDC03/2020), which shows high homology of >99.5% with first isolated SARS-CoV-2 sequence in Wuhan, China (Kim et al., 2020a).
To measure the IC50 of the drugs against authentic virus infection, 0.1 multiplicity of infection of each virus was inoculated onto Vero E6 cells (ATCC, Manassas, VA, USA). Subsequently, seven concentrations of the drugs were treated with two-fold serial dilution (remdesivir and nirmatrelvir [antiviral component of Paxlovid]: 20 to 0.31 μM, molnupiravir: 40 to 0.62 μM) for 48 h. The cell infectivity ratio between the drug treatment and virus only-treated groups was assessed through high-content imaging (HCI) analysis to determine the number of infected cells (N protein-expressing cells) using immunofluorescence images with viral N-specific antibody and total cells (number of nuclei) using DAPI staining (Supplementary Fig. S1A; methods described in the Supplementary Information.) The IC50 and 95% confidence interval (CI) values were determined from dose-response curves based on treatment with seven concentrations of each drug, using Prism 7 (GraphPad Software, San Diego, CA, USA), and the quality of the assay was assessed using a Z′ factor >0.5 (Table 1 and Supplementary Fig. S1BC).
Table 1.
IC50 values of remdesivir, molnupiravir, and nirmatrelvir against SARS-CoV-2 variants.
| Drug |
Remdesivir |
Molnupiravirc |
Nirmatrelvir |
|||
|---|---|---|---|---|---|---|
| Viral strain | IC50; μΜa (95% CI) | Fold changeb | IC50; μΜ (95% CI) | Fold change | IC50; μΜ (95% CI) | Fold change |
| Reference (S) | 2.24 (1.83–2.64) | 11.08 (9.51–12.66) | 1.51 (1.34–1.79) | |||
| Alpha (B.1.1.7) | 1.75 (1.53–2.01) | 0.78 | 6.19 (5.23–7.33) | 0.56 | 1.02 (0.94–1.10) | 0.68 |
| Beta (B.1.351) | 1.84 (1.53–2.194) | 0.82 | 7.97 (6.26–10.03) | 0.72 | 0.98 (0.94–1.07) | 0.65 |
| Gamma (P.1) | 2.73 (2.47–3.03) | 1.22 | 11.62 (10.8–12.49) | 1.05 | 1.371 (1.31–1.42) | 0.91 |
| Delta (B.1.627.2) | 3.70 (3.44–3.98) | 1.65 | 12.96 (10.67–14.51) | 1.17 | 1.01 (0.97–1.06) | 0.67 |
| BA.1 | 1.42 (1.22–1.63) | 0.63 | 4.66 (4.10–5.22) | 0.42 | 1.11 (1.02–1.22) | 0.74 |
| BA.2.2 | 1.95 (1.81–2.1) | 0.87 | 4.63 (4.23–5.07) | 0.42 | 0.84 (0.76–0.93) | 0.56 |
| BA.2 | 2.77 (2.39–3.21) | 1.24 | 8.09 (7.19–9.1) | 0.42 | 2.07 (1.89–2.26) | 0.56 |
| BA.2.12.1 | 1.28 (1.15–1.42) | 0.57 | 9.67 (8.09–11.55) | 0.87 | 1.25 (1.05–1.54) | 0.83 |
| BA.2.3 | 1.07 (0.94–1.22) | 0.48 | 7.53 (6.57–8.60) | 0.68 | 2.11 (1.90–2.35) | 1.40 |
| BA.4.1.1 | 3.34 (3.23–3.47) | 1.49 | 9.19 (8.74–9.66) | 0.83 | 1.83 (1.72–1.95) | 1.21 |
| BA.5.2.1 | 1.91 (1.51–2.37) | 0.85 | 1.94 (1.64–2.23) | 0.18 | 0.51 (0.49–0.53) | 0.30 |
| BA.2.75 | 1.59 (1.38–1.84) | 0.71 | 7.31 (6.18–8.64) | 1.16 | 0.54 (0.52–0.56) | 0.34 |
| BA.4.6 | 1.82 (1.52–2.17) | 0.81 | 4.74 (2.91–7.26) | 0.43 | 0.92 (0.76–1.11) | 0.61 |
| BA.2.75.2 | 4.34 (4.04–4.65) | 1.94 | 2.92 (2.66–3.20) | 0.26 | 1.27 (1.2–1.36) | 0.84 |
| BF.7 | 2.45 (2.35–2.54) | 1.09 | 9.4 (9.01–9.81) | 0.85 | 1.32 (1.26–1.39) | 0.87 |
| BQ.1.5 | 3.04 (2.59–3.57) | 1.36 | 4.43 (4.11–4.76) | 0.40 | 1.32 (1.17–1.49) | 0.87 |
| BQ.1.1 | 2.00 (1.90–2.11) | 0.89 | 6.62 (6.31–6.95) | 0.60 | 1.30 (1.19–1.42) | 0.86 |
| BJ.1 | 3.22 (2.52–4.11) | 1.44 | 1.56 (1.35–1.80) | 0.14 | 1.18 (1.03–1.35) | 0.78 |
| XBB.1 | 2.76 (2.32–3.25) | 1.23 | 3.87 (3.08–4.85) | 0.35 | 0.94 (0.80–1.11) | 0.62 |
| BA.2.3.20 | 2.94 (2.82–3.06) | 1.31 | 4.27 (3.97–4.60) | 0.39 | 1.15 (1.11–1.19) | 0.76 |
| BA.2.75.5 | 2.15 (1.91–2.40) | 0.96 | 1.92 (1.66–2.21) | 0.17 | 0.83 (0.79–0.87) | 0.55 |
| XBB | 1.90 (1.76–2.05) | 0.85 | 2.70 (2.38–3.05) | 0.24 | 0.69 (0.63–0.76) | 0.46 |
| XAY.1 | 3.44 (3.18–3.72) | 1.54 | 4.48 (4.13–4.85) | 0.40 | 0.75 (0.71–0.79) | 0.50 |
| XBC | 0.92 (0.85–0.99) | 0.41 | 1.89 (1.74–2.33) | 0.17 | 0.73 (0.68–0.78) | 0.48 |
| BN.1.9 | 2.19 (1.84–2.58) | 0.98 | 2.36 (2.13–2.62) | 0.21 | 0.59 (0.56–0.62) | 0.39 |
| BN.1.5 | 1.67 (1.46–1.89) | 0.75 | 4.21 (3.63–4.86) | 0.38 | 0.69 (0.64–0.74) | 0.46 |
| XBB.1.5 | 2.19 (2.04–2.36) | 0.98 | 5.18 (4.61–5.80) | 0.47 | 1.20 (1.13–1.28) | 0.79 |
The in vitro half inhibitory concentration (IC50) values and 95% confidence interval (CI) were determined using high-content imaging analysis, which calculated infectivity ratio (infected cells/total cells) based on the seven concentrations of each drug treatment, and fold changes were calculated compared with the reference strain. The median IC50 and 95% CI values for remdesivir (n = 21), molnupiravir (n = 12), and nirmatrelvir (n = 13) against the reference strain were determined, and the normality test of the data was performed using the D'Agostino-Pearson normality test. All the experiments were performed in triplicate, and the quality of the high-content imaging assay was assessed by Z′ factor (Z > 0.5) using Harmony 4.9 (PerkinElmer Software).
The IC50-fold change value was represented as a relative value compared with the reference strain.
EIDD-2801 (an orally bioavailable prodrug of EIDD-1931).
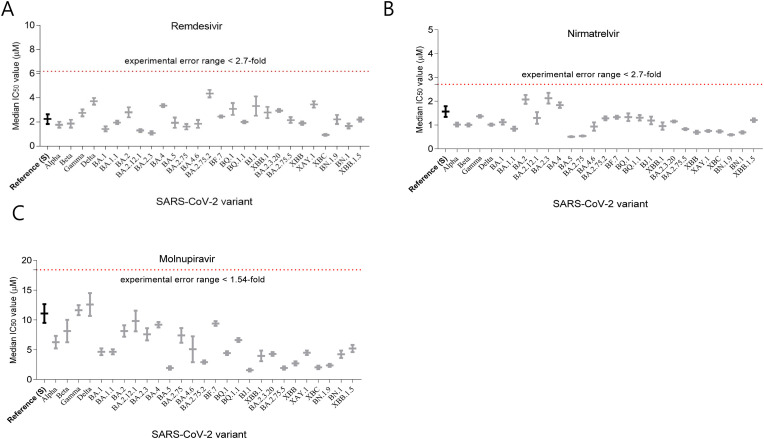
The maximal IC50-fold-change values of remdesivir, molnupiravir, and nirmatrelvir against all the SARS-CoV-2 variants were 1.9 (BA.2.75.2), 1.2 (B.1.627.2), and 1.4 (BA.2.3), respectively, compared to the median IC50 values of the reference strain for remdesivir, molnupiravir, and nirmatrelvir (Fig. 1 and Table 1). Moreover, the median IC50-fold change values in remdesivir, molnupiravir, and nirmatrelvir for the Omicron variants were 0.96, 0.4, and 0.62, respectively, which were similar to the median IC50-fold change values of 1.02, 0.88, and 0.67, respectively, for the former VOCs (Table 1).
Fig. 1.
Median IC50 value of the drugs in VeroE6 cells
Median IC50 (left axis) of (A) remdesivir, (B) nirmatrelvir, and (C) molnupiravir. Vero E6 cells were infected with indicated SARS-CoV-2 variants at 0.1 multiplicity of infection for 1 h. Subsequently, seven concentrations of the drugs were treated with two-fold serial dilution (remdesivir and nirmatrelvir [antiviral component of Paxlovid]: 20 to 0.31 μM, molnupiravir: 40 to 0.62 μM) for 48 h. The cell infectivity ratio between the drug treatment and virus only-treated groups was assessed through high-content imaging analysis to determine the number of infected cells (N protein-expressing cells) using immunofluorescence images with viral N-specific antibody and total cells (number of nuclei) using DAPI staining. The IC50 values were determined from dose-response curves and median IC50 value was calculated. The experiments were performed in triplicate (the reference strain for remdesivir [n = 21], nirmatrelvir [n = 13], and molnupiravir [n = 12]).
Using GISAID sequence analysis, BA.2.75.2 and Delta (B.1.627.2) have P323L and G671S amino acid substitution in nonstructural protein (Nsp) 12, and BA.2.3 has P132H amino acid substation in Nsp 5 (Supplementary Table S1). However, it has been reported that the high-prevalence changes, P323L (>99% in Omicron variant) and G671S (>99% in Delta variant) substitution in Nsp 12, and P132H substitution in Nsp 5, do not affect the antiviral activity of nirmatrelvir due to the distance of drug binding location (Pitts et al., 2022; Ullrich et al., 2022). Moreover, the maximal-fold changes of the drugs were within experimental error range in our experiment condition in which the 95% and 5% percentile ratio of the IC50 for remdesivir (n = 21), molnupiravir (n = 13), and nirmatrelvir (n = 12) of the reference strain were approximately 2.7-, 1.5-, and 1.9-fold, respectively (Supplementary Fig. S1B) (Vangeel et al., 2022).
Other amino acid substitutions such as V405F (BA.2.75), Y273H (BQ.1.1 and BQ.1.5), A529V (XAY.1 and XBC), and ins823D (XBC) were identified in Nsp 12. A529V, V405F, X823D, and Y273H substitutions which have been reported previously, with a frequency of approximately 1%, do not affect the binding of remdesivir (Pitts et al., 2022; Vangeel et al., 2022; Kumar et al., 2022; Imai et al., 2023). Moreover, they did not include the most important residues for RdRp binding (Nguyen et al., 2020). In Nsp 5, K90R substitutions were previously reported in Beta (B.1.351) and XBB VOCs; however, the substitution does not change the antiviral activity of nirmatrelvir (Imai et al., 2023; Hu et al., 2022). Thus, the drugs are likely to retain their antiviral activity against newly emerging Omicron subvariants.
In our results, IC50 value of the drugs was approximately 10-fold higher than that in similar studies (Ullrich et al., 2022; Vangeel et al., 2022). In drug treatment, EIDD-2801 (an orally bioavailable prodrug of EIDD-1931) and EIDD-1931 (an active form of molnupiravir) treatment in Vero E6 cells do not show significant IC50 value changes (Vangeel et al., 2022). Thus, the enhanced IC50 values by 10–100-fold for nirmatrelvir and remdesivir in Vero E6 cells under experimental conditions may be attributable to culture without P-glycoprotein inhibitor (drug efflux inhibitor) (Vangeel et al., 2022; Imai et al., 2023; Fiaschi et al., 2022; Zhu et al., 2022).
In this study, we assessed the antiviral activity of remdesivir, molnupiravir, and nirmatrelvir against newly emerging Omicron subvariants and confirmed that the antiviral activity was maintained. Moreover, it has previously been shown that the mutations of Nsp 5 and Nsp 12 in the current Omicron subvariants do not affect the antiviral activity of the drugs. Our results provide comprehensive information that antiviral efficacy of remdesivir, molnupiravir, and nirmatrelvir against 27 SARS-CoV-2 variants is maintained and thus these drugs can continue to be used for treatment of COVID-19. However, new SARS-CoV-2 variants will continue to emerge and there is an ongoing possibility that new SARS-CoV-2 variants will emerge that are resistant to current drugs. Thus, it is important to continue active surveillance and testing of new variants for drug resistance to enable early identification of new drug-resistant strains.
Funding
This work was supported by the Korea National Institute of Health [grant numbers 2021-NI-026-02, 6634-325-210].
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2023.105609.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Arora P., Kempf A., Nehlmeier I., Schulz S.R., Jäck H.M., Pöhlmann S., Hoffmann M. Omicron sublineage BQ.1.1 resistance to monoclonal antibodies. Lancet Infect. Dis. 2023;23:22–23. doi: 10.1016/S1473-3099(22)00733-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady D.K., Gurijala A.R., Huang L., Hussain A.A., Lingan A.L., Pembridge O.G., Ratangee B.A., Sealy T.T., Vallone K.T., Clements T.P. A guide to COVID‐19 antiviral therapeutics: a summary and perspective of the antiviral weapons against SARS‐CoV‐2 infection. FEBS J. 2022:1–31. doi: 10.1111/febs.16662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiaschi L., Dragoni F., Schiaroli E., Bergna A., Rossetti B., Giammarino F., Biba C., Gidari A., Lai A., Nencioni C., Francisci D., Zazzi M., Vicenti I. 2022. Efficacy of Licensed Monoclonal Antibodies and Antiviral Agents against the SARS-CoV-2 Omicron Sublineages BA.1 and BA. Viruses. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Han J., Zhang Y., He J., Yu W., Zhang X., Wu J., Zhang S., Kong Y., Guo Y., Lin Y., Zhang J. SARS-CoV-2 Omicron variant: epidemiological features, biological characteristics, and clinical significance. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Lewandowski E.M., Tan H., Zhang X., Morgan R.T., Zhang X., Jacobs L.M.C., Butler S.G., Gongora M.V., Choy J., Deng X., Chen Y., Wang J. 2022. Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Ito M., Kiso M., Yamayoshi S., Uraki R., Fukushi S., Watanabe S., Suzuki T., Maeda K., Sakai-Tagawa Y., Iwatsuki-Horimoto K., Halfmann P.J., Kawaoka Y. Efficacy of antiviral agents against omicron subvariants BQ.1.1 and XBB. N. Engl. J. Med. 2023;388:89–91. doi: 10.1056/NEJMc2214302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S., Gurry C., Freitas L., Schultz M.B., Bach G., Diallo A., Akite N., Ho J., Lee R.T., Yeo W. GISAID's role in pandemic response. China CDC Wkly. 2021;3:1049–1051. doi: 10.46234/ccdcw2021.255. GISAID Core Curation Team, Maurer-Stroh S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Jang J.H., Kim J.M., Chung Y.S., Yoo C.K., Han M.G. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Publ. Health Res. Perspect. 2020;11:3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Jang J.H., Kim J.M., Chung Y.S., Yoo C.K., Han M.G. Genome-wide identification and characterization of point mutations in the SARS-CoV-2 genome. Osong Publ. Health Res. Perspect. 2020;11:101–111. doi: 10.24171/j.phrp.2020.11.3.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Kumari K., Azad G.K. Emerging genetic diversity of SARS-CoV-2 RNA dependent RNA polymerase (RdRp) alters its B-cell epitopes. Biologicals. 2022;75:29–36. doi: 10.1016/j.biologicals.2021.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.L., Thai N.Q., Truong D.T., Li M.S. Remdesivir strongly binds to both RNA-dependent RNA polymerase and main protease of SARS-CoV-2: evidence from molecular simulations. J. Phys. Chem. B. 2020;124:11337–11348. doi: 10.1021/acs.jpcb.0c07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts J., Li J., Perry J.K., Du Pont V., Riola N., Rodriguez L., Lu X., Kurhade C., Xie X., Camus G., Manhas S., Martin R., Shi P.Y., Cihlar T., Porter D.P., Mo H., Maiorova E., Bilello J.P. Remdesivir and GS-441524 retain antiviral activity against Delta, Omicron, and other emergent SARS-CoV-2 variants. Antimicrob. Agents Chemother. 2022;66 doi: 10.1128/aac.00222-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich S., Ekanayake K.B., Otting G., Nitsche C. Main protease mutants of SARS-CoV-2 variants remain susceptible to nirmatrelvir. Bioorg. Med. Chem. Lett. 2022;62 doi: 10.1016/j.bmcl.2022.128629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangeel L., Chiu W., De Jonghe S., Maes P., Slechten B., Raymenants J., André E., Leyssen P., Neyts J., Jochmans D. Remdesivir, molnupiravir and nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antivir. Res. 2022;198 doi: 10.1016/j.antiviral.2022.105252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Binder J., Yurgelonis I., Rai D.K., Lazarro S., Costales C., Kobylarz K., McMonagle P., Steppan C.M., Aschenbrenner L., Anderson A.S., Cardin R.D. Generation of a VeroE6 Pgp gene knock out cell line and its use in SARS-CoV-2 antiviral study. Antivir. Res. 2022;208 doi: 10.1016/j.antiviral.2022.105429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.