Abstract
The Arabidopsis thaliana ASYMMETRIC LEAVES2 (AS2) gene is responsible for the development of flat, symmetric, and extended leaf laminae and their veins. The AS2 gene belongs to the plant-specific AS2-LIKE/LATERAL ORGAN BOUNDARIES (LOB)-domain (ASL/LBD), which consists of 42 proteins in Arabidopsis with a conserved amino-terminal domain known as the AS2/LOB domain, and a variable carboxyl-terminal region. AS2/LOB domain consists of an amino-terminal (N-terminal) that contains a cysteine repeat (the C-motif), a conserved glycine residue, and a leucine-zipper-like. AS2/LOB domain has been characterised in plants such as A. thaliana, Zea mays, and Oryza sativum. Nevertheless, it remains uncharacterised in cassava (Manihot esculenta). Characterisation and identification of cassava ASL/LBD genes using the computational algorithms, hidden Markov model profiles (PF03195), determined 55 ASL/LBD genes (MeASLBD1 to MeASLBD55). The gene structure and motif composition were conserved in MeASLBDs, while the expression profiles of these genes were highly diverse, implying that they are associated with diverse functions. Weighted gene co-expression network analysis (WGCNA) of target genes and promoter analysis suggest that these MeASLBDs may be involved in hormone and stress responses. Furthermore, the analysis of cis-regulatory elements in promoter regions suggested that MeASLBDs may be involved in the plant phytohormone signal response. The transcriptome data of cassava under biotic and abiotic stresses revealed that MeASLBD46 and MeASLBD47 greatly respond to disease and drought. The MeASLBD47 gene was selected for functional analysis. The result indicated that MeASLBD47 significantly mitigated the virulence of cassava bacterial blight (XamCHN11) through Real-Time Quantitative Reverse Transcription PCR (qRT-PCR) and Virus-induced gene silencing (VIGS). These findings provided a comprehensive analysis of ASL/LBD genes and laid the groundwork for future research to understand ASL/LBD genes.
Introduction
Organ development in multicellular systems is governed by the activation of a new genetic program and the repression of a previously active program, which is largely governed by epigenetic systems. The prime model for studying such positive and negative control programs is the Arabidopsis thaliana leaf developmental process with an adaxial-abaxial (dorsal-ventral) polarity that is epigenetically regulated by the repressor complex ASYMMETRIC LEAVES1 (AS1)-AS2 [1–4]. AS2 gene has received the most extensive investigation in genetic and molecular research [5]. The AS2 gene in Arabidopsis is a key regulator in the development of flat symmetric leaves with vascular bundles and fine networks of venation systems, the morphology of which appears to have evolved suitably for efficient photosynthesis [5]. For instance, Arabidopsis AS2 regulates leaf adaxial-abaxial partitioning by suppressing the expression of the abaxial-determining gene ETTIN/AUXIN RESPONSE FACTOR3 (ETT/ARF3), implying that AS2 is involved in epigenetic repression of ETT/ARF3 by gene body DNA methylation [6]. AS2 specifically binds the CpG-containing sequence in exon 1 of ETT/ARF3, and the binding requires the zinc-finger-like motif in AS2, which is structurally similar to the zinc-finger-CxxC domain in vertebrate DNA methyltransferase1 [6]. In addition, AS2 is involved in epigenetic repression of the abaxial ARF4 and class 1 KNOX homeobox genes by forming a complex with the MYB protein. The cooperative action of various modifier genes significantly boosts the repressed expression of these genes by AS2 [6].
Transcription factor (TF) families play critical roles in plant growth, development, and environmental stress responses [7]. The ASYMMETRIC LEAVES2-LIKE/LATERAL ORGAN BOUNDARIES DOMAIN (ASL/LBD) family is an important plant-specific TF family [8]. These members are also named the Lateral Organ Boundary (LOB) Domain (LBD). ASL/LBDs are defined by a highly conserved LOB domain of approximately 100 amino acids (aa) [9]. AS2 gene belongs to the plant-specific AS2/LOB protein family, which contains 42 members in Arabidopsis with a conserved amino-terminal domain known as the AS2/LOB domain, and a variable carboxyl-terminal region; however, little is known about their functions [9–13]. The AS2/LOB domain contains (i) a zinc-finger-like motif (CX2CX6CX3C) required for binding specific DNA sequences (5’ GCGGCG 3’), and the interaction with basic helix-loop-helix (bHLH) proteins could reduce this affinity; (ii) an invariant glycine residue, Gly-Ala-Ser (GAS), critical for the biological function of AS2/LOB proteins in Arabidopsis; and (iii) a leucine-zipper-like (LZL-region; LX6LX3LX6L) which may play a role in the protein-protein interactions to form homo and/or hetero multimers [9–11, 14]. Importantly, the crystal structure of the homodimeric LOB domain of Ramosa2 from wheat (TtRa2LD) was crystallized and determined [15]. The structure mainly consists of a zinc finger, a GAS motif consisting of two α-helices, a highly conserved five-residue motif (Asp-Pro-Val-Tyr-Gly, known as DPVYG motif), and an amphipathic α-helix with the feature of leucine zipper-like coiled-coil element. According to biochemical, molecular modeling, and small-angle X-ray scattering analysis, dimerization is important for cooperative DNA binding and palindromic DNA discrimination through a molecular calipers mechanism [15]. Based on the presence of the LZL-region, the ASL/LBD gene family can be classified at least into two classes: “Class I” contains all these conserved domain motifs, whereas “Class II” contains only structural motifs similar to the zinc-finger-like motif [9–11, 16]. “Class I” is further divided into two subclasses (Ia and Ib) [5]. A base substitution mutation in the conserved glycine codon of ASL/LBD5 causes the typical phenotypic alterations observed in numerous ASL/LBD mutants, making this residue essential for ASL/LBD function [11, 14]. In addition, the ASL/LBD5 mutant, which contains a mutation in the region encoding the carboxy-terminal (C-terminal) half of ASL/LBD, results in the typical mutant phenotype; thus, the C-terminal half also plays a role in ASL/LBD function [17].
The first ASL/LBD gene (AT5G63090) was identified in A. thaliana [9]. Since then, ASL/LBD genes have been continually reported in various plants and associated with diverse functions (S1 Table in S1 File). For instance, AS2 (AS2/LOB6) in Arabidopsis is involved in a regulatory loop that maintains the shoot meristem and controls leaf polarity and flower development by interacting with an MYB motif encoded by AS1, recently defined as the SANT domain [12, 14, 18]. In addition, it was found that the loss-of-function of AS2/LOB20 in Arabidopsis enhanced resistance to the root-infecting vascular wilt pathogen Fusarium oxysporum [19]. Furthermore, the expression profiles of A. thaliana (At) LBD members indicated that pathogen inoculation induced the expression of AS2/LOB37 and AS2/LOB38 [19]. CsLOB1, a homolog of AS2/LOB1 and AS2/LOB11, acts as a target of transcription activator-like (TAL) effectors following infection with bacterial canker disease [20]. These findings imply that the ASL/LBD genes play important roles in plant defence responses.
Cassava (2n = 36; Manihot esculenta Crantz) is a major staple crop in tropical regions and the third most consumed grain after rice and maize [21–23]. M. esculenta Crantz originated from its wild ancestor, M. esculenta ssp. Flabellifolia [24]. The cassava root crop provides staple food for over 700 million people worldwide [23, 25]. Cassava is highly drought-resistant, and its storage roots can be preserved in the soil for a few years, making it an essential carbohydrate source to alleviate global famine [26]. Besides, it is an ideal feedstock crop for bioenergy, biomaterials, and animal feeds due to its advantageous agricultural characteristics and high starch quantity and quality [27, 28]. Nonetheless, cassava production is constrained by several plant pathogens that threaten the food security of millions of people worldwide [29–31]. Cassava’s most serious bacterial disease is Cassava Bacterial Blight (CBB), caused by Xanthomonas axonopodis pv. manihotis (Xam) [32]. CBB threatens food security in tropical regions and can generate up to 100 percent losses under favourable climatic conditions (CABI, 2015; FAO, 2008) [33]. The rapid spread of CBB in some cassava-producing regions and the emergence of new disease reports in regions where cassava is a staple crop highlight the necessity of developing novel methods to control this plant disease [30, 34–36]. Cassava genome publication lays the groundwork for genome-wide analysis of new gene resources [37]. Nonetheless, studies on the molecular mechanisms of CBB resistance are scarce [38]. The characteristics underlying the biotic stress response of cassava remain largely unknown. This investigation was conducted to identify the ASL/LBD gene family in cassava and determine LOB expression profiles under pathogen stress. Identifying and characterising the ASL/LBD genes and determining their expression profiles under biotic stress could provide valuable insight for plant disease control.
This study identified 55 M. esculenta ASL/LBDs (MeASLBDs) from the cassava genome. The phylogeny, conserved motifs, gene structures, and expression profiles of MeASLBDs were thoroughly determined. Moreover, potential target genes were identified via promoter analysis, and a co-expression network was constructed along with MeASLBDs based on a transcriptome dataset derived from pathogen treatment. The MeASLBD47 gene expression was highly induced following inoculation with cassava bacterial blight pathogen (XamCHN11) and was selected for functional analysis. The results indicated that MeASLBD47 significantly mitigated the virulence of cassava bacterial blight (XamCHN11) through qRT-PCR and Virus-induced gene silencing (VIGS). These findings provide invaluable insight for further physiological and functional studies of the AS2/LOB domain in cassava and highlight the possible functions in pathogen response.
Materials and methods
Plant material and growth conditions
The cassava cultivar used in this study were SC8, Arg7, W14, and KU50. The stems were cut into three-node sections and grown in a pest-free sterile phytotron at temperatures ranging from 35°C to 25°C under a 16/8-h photoperiod and 80% relative humidity. The leaves (30-day-old) were inoculated with Xanthomonas axonopodis pv. Manihotis (XamCHN11) [32]. The leaves of five cassava plants (nine leaves per plant) were inoculated by dropping 10‐μL of XamCHN11 suspension of 1 × 108 CFU/mL [optical density at 600 nm (OD600) = 0.1] into a 2‐mm‐diameter ring. Cassava seedlings were sampled at 1 h, 3 h, 6 h, and 9 h post-inoculation. All samples were frozen in liquid nitrogen and stored at -80°C until use. To quantify the size of the lesion, typical Xam symptoms, such as necrosis and chlorosis around the inoculation point, were considered. At 3 days post‐inoculation (dpi), lesions were measured on eight leaves per treatment and then plotted. The experiments were conducted at least twice with identical results.
RNA isolation, qRT-PCR analysis, and statistics analysis
Total RNA was isolated with the RNAprep Pure Plant Kit (DP441, TIANGEN, Beijing, China). RNA concentration determination, DNase I, RNase-free (1 U/μL) treatment, cDNA synthesis, qRT-PCRs, and data analysis were performed following published protocols with minor modifications [39, 40]. Photometric UV-Vis RNA/DNA quantification was performed using NanoDrop One/OneC. The cDNA was synthesized with the FastKing RT Kit (TIANGEN, Beijing, China). The gene-specific primers were designed with the Primer 3.0 program (S2 Table in S1 File). qRT-PCR was performed in a reaction system of 20 μL containing 10 μL TB Green Premix Ex Taq II, 1 μL 10 μM forward primer, 1 μL 10 μM reverse primer, 2 μL cDNA, and 6 μL ddH2O. The PCR amplification conditions were set as follows: denaturation at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 60°C for 35 s, 95°C for 15 s, and 60°C for 1 min. The EF1α gene was selected to calculate the relative fold differences using the 2-ΔΔCT method [ΔΔCT = (Cttarget gene—CtEF1α)] [40]. Statistics analysis was conducted using SPSS software. Rstudio software was used to infer the heatmap. Three sets of biological replicates and three sets of technical replicates were analysed.
Generation of pCsCMV VIGS construct and agroinfiltration
VIGS tool (https://vigs.solgenomics.net/) was applied to analyse the CDS sequence of the gene and obtain the optimal silencing fragment for the construction of VIGS vector [41, 42]. 487 bp cassava PDS (Manes.05G193700) and 237 bp MeASLBD47 (Manes.12G110600) fragments were amplified using cassava cDNAs as a template and the corresponding primer pairs (S2 Table in S1 File). Subsequently, the amplified fragments were cloned into pCsCMV vector to generate pCsCMV-PDS and pCsCMV-AS2/LOB47 using Nimble Cloning [43]. pCsCMV-NC was utilised as a negative control (NC). The pCsCMV-NC-based constructs were transformed into Agrobacterium tumefaciens (strain GV3101). A single colony of A. tumefaciens (GV3101) for each strain was incubated for 20–36 h at 28°C in Luria–Bertani (LB) medium (10 mL) containing 25 mg/mL kanamycin and 25 mg/mL rifampicin. The bacterial cultures were then centrifuged at 4,000g for 5 min and were resuspended in agroinfiltration buffer [10 mM MgCl2, 10 mM 2-(N-Morpholino) ethanesulfonic acid, and 100 μM acetosyringone] for reaching an optical density of 1.0 at 600 nm (OD600) [44]. The agrobacterium suspension was kept at room temperature for 3 h in the dark before inoculation. 2-week old cassava plants were used for agroinfiltration using a 1-mL needleless syringe. The back sides of the healthy and fully developed leaves from the middle of each plant were selected for agroinfiltration. Inoculations were administered at 8–10 spots on both sides of the main vein per leaf to increase the infiltrated leaf area.
Identification and sequence analysis of MeASLBD genes
The cassava genome sequence was downloaded from Phytozome (http://www.phytozome.net) [45]. The AS2/LOB protein sequences of A. thaliana were retrieved from TAIR 12.0 (www.arabidopsis.org) [46]. The hidden Markov model (HMM) -based profile (Pfam: PF03195) of the putative AS2/LOB domain was built using the HMMER program (http://hmmer.org/) [47, 48]. AS2/LOB domain obtained from the Pfam database (http://pfam.xfam.org/) [49] was used as a query to identify all potential AS2/LOB protein sequences using a BLASTp search E-value threshold of 1.0 × 10−10. The putative AS2/LOB domain of the candidate sequences was further confirmed by the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/) and the SMART database (http://smart.embl-heidelberg.de/). The MW and pI of cassava AS2/LOB proteins (MeASLBDs) were calculated with the online program ExPASy (https://www.expasy.org/) [50].
Phylogenetic tree construction, conserved motifs, and structure analysis
To determine the evolutionary relationships between M. esculenta and A. thaliana ASL/LBD gene family, phylogenetic analysis was performed with MEGA 7.0 by constructing a minimum evolution (ME) phylogenetic tree using substitution models G + I with 1,000 bootstrap replicates to support statistical reliability [51]. The full-length amino acid sequences of M. esculenta and A. thaliana AS2/LOB protein sequences were aligned using the ClustalX program with the default parameters. The results were displayed with DNAMAN 9.0 (Lynnon Biosoft) software. The Multiple Em for Motif Elucidation (MEME, version 5.4.1.) tool (http://meme-suite.org/tools/meme) was used to predict the conserved motifs of the MeASLBD protein sequences [52]. The relative parameters were set to an optimum motif width of 25–50 and a maximum number of 10 motifs. The exon-intron structure of MeASLBD genes was analysed using the Gene Structure Display Server (http://gsds.gao-lab.org/) [53].
Chromosomal location, gene duplication event, gene collinearity, and synteny analysis
The chromosomal location information of 55 MeASLBD genes was extracted from the cassava genomic annotation file GFF3 (general feature format), retrieved from Ensembl Plants (http://plants.ensembl.org/index.html). The chromosomal distribution of MeASLBD genes was visualised using TBtools software [54]. Duplication detection for MeASLBD genes was performed using the Multiple Collinearity Scan Toolkit (MCScanX) with an E-value of 10−5 [55]. The homologous regions of the MeASLBD genes were identified with the MCScanX (http://chibba.pgml.uga.edu/mcscan2/) (https://github.com/wyp1125/MCScanX) program to determine the level of synteny [55]. Tandem and segmental duplication events were identified and visualised with Circos software (http://circos.ca/) version 0.69 [56]. The Ka/Ks value was calculated as the ratio of the number of nonsynonymous substitutions per nonsynonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks) over a given period.
Cis-acting element analysis and identification of MeASLBDs target gene
The promoter regions of 55 MeASLBD genes, the upstream (2000 bp) of the translation initiation codons, were extracted from the Phytozome database of the cassava genome (https://phytozome.jgi.doe.gov/). These sequences were applied in PlantCARE database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html) [57] to predict the cis-acting elements present in the promoter regions of the MeASLBDs, and visualised with TBtools. The genes containing the motif (5’ GCGGCG 3’) in the upstream promoter region are candidate target genes for MeASLBD. Blast2GO tool was used to annotate the candidate target genes [58].
Gene expression profiles of MeASLBDs in cassava
The expression profiles of 55 MeASLBD genes in root and leaf tissues of cassava cultivars Argentina 7 (Arg7), wild subspecies 14 (W14), and Kasetsart University 50 (KU50) were analysed. RNA-seq data of the Arg7, W14, and KU50 were retrieved from NCBI database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93098) (S7 Table in S1 File). HISAT2 (https://github.com/DaehwanKimLab/hisat2) software [59] was applied with default parameters to map the reads. Under default parameters, the StringTie program [60] was applied to assemble transcripts and compute reads per kilobase million (RPKM) values. The cassava AS2/LOB family heatmaps were established with log2-transformed RPKM values and visualised with Tbtools [54]. The min-max method was used to normalise the RPKM values.
Co-expression analysis of MeASLBDs and identification of MeASLBD interacting genes by WGCNA
A co-expression analysis of MeASLBD and its target genes was generated using 55 selected transcriptomes (S3 Table in S1 File). The RPKM values of MeASLBDs and their predicted target genes were screened, and genes with an RPKM value < 1 were removed. Weighted Gene Co-expression Network Analysis (WGCNA) was performed using the Rstudio environment to identify the selected co-expression module [61]. The selected co-expression module is considered a cluster of highly related genes. The module genes showing expression patterns consistent with MeASLBDs were selected for Cytoscape visual analysis [62].
Results
Identification and chromosome localisation of cassava ASL/LBD genes
55 ASL/LBD genes were identified in cassava and named MeASLBD (MeASLBD1 to MeASLBD55). The predicted protein products of MeASLBDs ranged from 116 to 429 aa, with the molecular weight (MW) varying from 13.14 (MeASLBD28) to 46.78 kDa (MeASLBD55). The isoelectric point (pI) of their proteins varied from 4.67 (MeASLBD38) to 9.02 (MeASLBD16). Through chromosomal localisation analysis, 54 MeASLBD genes were unevenly distributed on the 18 cassava chromosomes (S1 Fig in S1 File). However, MeASLBD19 was localized on an unanchored scaffold004600 (S1 Fig in S1 File). The largest number of MeASLBD genes was found on chromosome 7 (7, 12.7%), followed by chromosomes 12 (6, 10.9%), 5 (5, 9%), 10 (5, 9%), 14 (5, 9%), 6 (4, 7.2%), and 13 (4, 7.2%). The remaining chromosomes contained one or two MeASLBD genes.
Evolutionary relationship and gene structure analysis of the MeASLBD genes
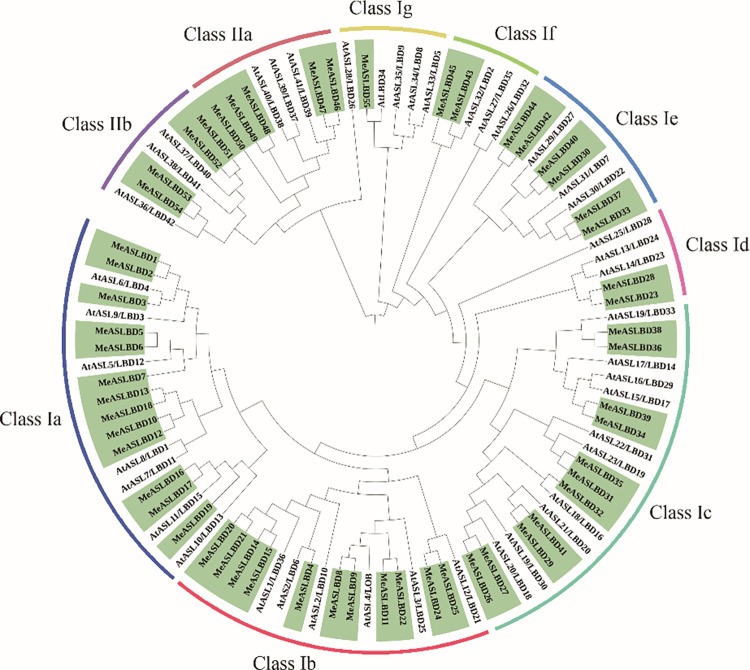
To investigate the evolutionary relationship of the MeASLBD genes, 43 AS2/LOB proteins of A. thaliana (AtASL/LBD) and 55 MeASLBD were used to construct a ME tree (Fig 1) MeASLBDs were classified into two major classes: Class I (46) and Class II (9). Class I ASL/LBD genes contain a conserved CX2CX6CX3C zinc-finger-like motif and an LX6LX3LX6L LZL-region, while Class II ASL/LBD genes contain only a conserved zinc-finger-like motif. Forty-six MeASLBDs (83%) were clustered into Class I. Class I was subdivided into seven subgroups (Class Ia to Class Ig). Class Ia contained the highest number of MeASLBD (13), followed by 11, 11, 6, 2, 2, and 1 MeASLBDs belonging to Class Ib, Ic, Ie, Id, and If, respectively. Nine MeASLBDs belonged to Class II, subdivided into two distinct subgroups: Class IIa (4) and Class IIb (5). Interestingly, this result is consistent with Arabidopsis ASL/LBD genes classification [9]. Previous studies showed that many AtASL/LBDs genes (Class I) were involved in lateral organ development [10]. Class II ASL/LBD genes are related to metabolism [10]. The phylogenetic analysis of A. thaliana and M. esculenta indicated that MeASLBD genes might have similar biological functions as AtASL/LBDs.
Fig 1. Phylogenetic analysis of AS2/LOB proteins in M. esculenta and A. thaliana.
The proteins AtASL/LBD and MeASLBD sequences were used to construct a minimum evolution (ME) tree. The tree was divided into two classes (Class I and II) and nine subclasses (Ia-g and IIa-b).
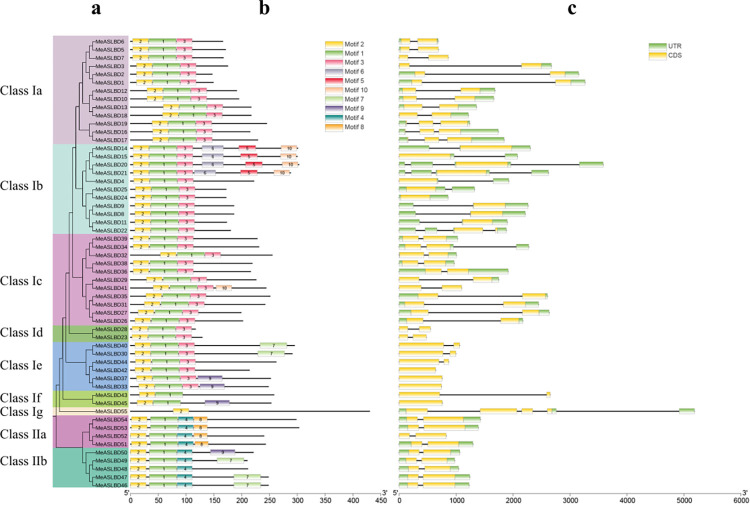
Gene structure analysis (Fig 2A) showed that MeASLBD genes have a simple structure with 2 to 4 introns and 1 to 4 exons. The majority (45) of the MeASLBD (82%) genes contains two exons, three MeASLBD genes (5%) contain one exon, three MeASLBD genes (5%) contain four exons and one MeASLBD gene (8%), clustered in Class Ig, contains five exons (Fig 2C). 40 MeASLBD coding regions contained a splicing site in the AS2/LOB domain (Fig 2C). 11 MeASLBD noncoding regions (UTR) contained an intron (Fig 2C). Ten putative motifs were identified using the MEME tool (Fig 2B). The conserved motifs of the MeASLBD protein sequences ranged from 25 to 50 aa (motifs 8 and 1, respectively) in length and contained between one (MeASLBD55) to six (MeASLBD14, MeASLBD15, MeASLBD20 and MeASLBD21) motifs. Motifs 1 and 2 were conserved in all MeASLBD protein sequences except MeASLBD55 which contained only motif 2. Cluster analysis showed that Class Ia, Id, Ig, and II have each similar gene structures. Motifs 3, 5, 6, and 10 were only conserved in MeASLD proteins of Class I, whereas motifs 8 and 4 were only conserved in MeASLDs proteins of Class II, suggesting that they are evolutionary divergent. Notably, motif 2 is conserved in all MeASLBD proteins. The MeASLBD proteins with similar motif compositions and gene lengths indicate they may share similar functions.
Fig 2. Phylogenetic analysis of ASL/LBD genes and the structures and relative motif positions of the ASL/LBD gene family members in M. esculenta.
(a) Minimum evolution (ME) tree of AS2/LOB proteins in M. esculenta. The phylogenetic tree was constructed using MEGA 7.0 software with the bootstrap method (1000 replicates). Branches of the different classes are shown in different colours. (b) Conserved motifs of MeASLBD proteins predicted by MEME. (c) Exon-intron structures of the MeASLBD genes.
Conserved motif analysis of MeASLBD genes
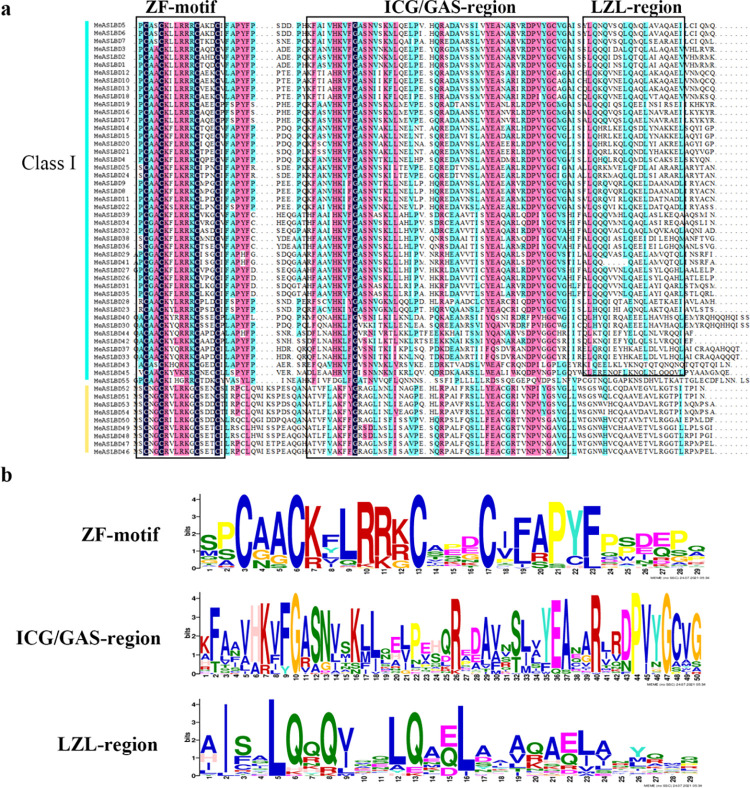
Multiple sequence alignment of the MeASLBD protein sequences using ClustalX showed that all sequences contained a zinc-finger-like motif (CX2CX6CX3C) at the N-terminus (Fig 3). The LZL-region (LX6LX3LX6L) was only found in MeASLBD proteins of Class Ⅰ, and 20 MeASLBD protein sequences (MeASLBD4, MeASLBD8, MeASLBD9, MeASLBD10, MeASLBD11, MeASLBD12, MeASLBD14, MeASLBD15, MeASLBD20, MeASLBD21, MeASLBD22, MeASLBD24, MeASLBD25, MeASLBD26, MeASLBD27, MeASLBD29, MeASLBD31, MeASLBD32, MeASLBD35, MeASLBD45) contained a complete motif, while the remaining MeASLBDs contained an uncomplete motif. Moreover, most MeASLBD genes contained a complete ICG/GAS-region, except for MeASLBD55.
Fig 3. Conserved domains of MeASLBD proteins.
(a) Multiple sequence alignment of MeASLBD conserved domains by DNAMAN. (b) Visualisation of the CX2CX6CX3C motif, ICG/GAS-region motif, and LX6LX3LX6L LZL-region.
Duplication, Collinearity, and synteny analysis
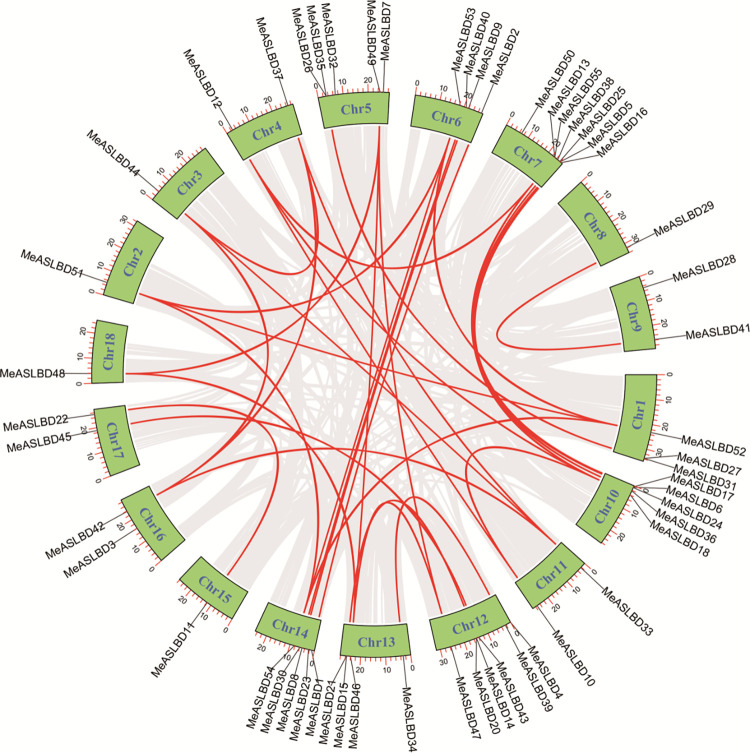
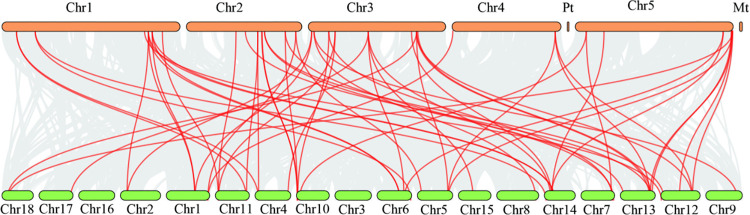
Genomic segmental and tandem duplications represent the two main driving forces of gene family expansion. MCScanX was applied to explore the gene duplication events in the MeASLBD gene family (Fig 4). Seven pairs of tandem duplicated genes were identified on chromosomes 1 (MeASLBD31/27), 5 (MeASLBD26/35), 7 (MeASLBD13/55), 10 (MeASLBD24/6), 12 (MeASLBD20/14), and 13 (MeASLBD21/15). 42 MeASLBD genes were segmentally duplicated, implying segmental duplication was a major driving force in the evolution of the MeASLBD gene family compared to tandem duplication. In addition, the Ka/Ks for 34 ASL/LBD gene pairs were determined (S4 Table in S1 File). The Ka/Ks ratio of ASL/LBD gene pairs varied from 0.0708 to 0.5342. The Ka/Ks ratios were less than 1.0, indicating that these genes might have undergone purifying selection during the evolution process. Collinearity analysis revealed chromosomal duplication, translocation, and inversion in the MeASLBD gene family (Fig 5). Genome Synteny and Collinearity Analysis show that ASL/LBD is relatively conserved in Arabidopsis and cassava. AtLBD has a high degree of height with MeASLBD.
Fig 4. Chromosomal distribution and interchromosomal relationships of MeASLBD genes.
The black lines show the position distribution of MeASLBD genes on 18 chromosomes. The red lines show the collinear gene pairs of MeASLBDs.
Fig 5. Colinearity analysis.
Orange represents the chromosomes (Chr) of Arabidopsis thaliana, Green represents the chr of cassava, Red lines represent genes containing Homologous. Pt represents the Chloroplast, Mt represents mitochondrion. The collinearity analysis was conducted using MCScanX and visualized with TBtools.
Analysis of putative cis-elements of the MeASLBD gene
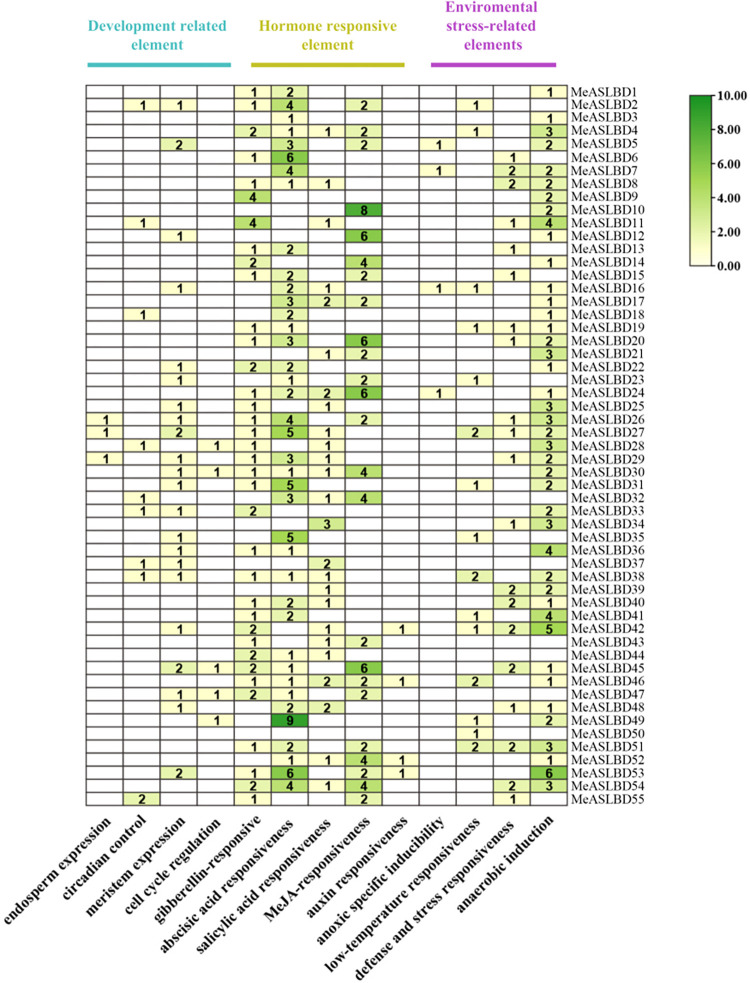
Analysis of cis-regulatory elements in promoter regions helps understand the mode of gene regulation and predicts the functions of genes. The online tool Plant CARE was used to investigate the cis-elements of the MeASLBD genes within a 2000-bp upstream region (Fig 6). Among the seven known elements, the hormone-responsive, environmental stress-related, and development-related elements were the core physiological processes represented by the regulatory elements. The hormone-responsive elements present in the MeASLBD gene family were found to be related to auxin-producing genes (TGA-element, AuxRE, and AuxRR-core), gibberellin (GARE-motif, P-box, and TATC-box), salicylic acid (TCA-element), abscisic acid (ABRE), methyl jasmonic acid (CGTCA-motif, TGACG-motif), and ethylene (ERE), the elements related to ethylene, jasmonic methyl acid, and abscisic acid were abundant, indicating that these genes may respond to phytohormone signals and/or abiotic stresses. Environmental stress-related elements, including anaerobic induction, defence and stress responsiveness (TC-rich repeats), low-temperature responsiveness, and enhancer-like elements, are involved in anoxic-specific inducibility (GC motif). In addition, meristem expression was found in development-related elements, suggesting that these genes might be involved in the development of plant meristems.
Fig 6. The cis-elements of 2000 bp sequences upstream of MeASLBD gene analysed with the online tool PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html).
Tbtools software was used to search the cassava promoter elements, count the number of promoters with the same function, and used TBtools for visualization. The color bar represents the number of the putative cis-elements of the MeASLBD gene ranging from light green to dark green.
Expression analysis of MeASLBD genes in different tissues and organs
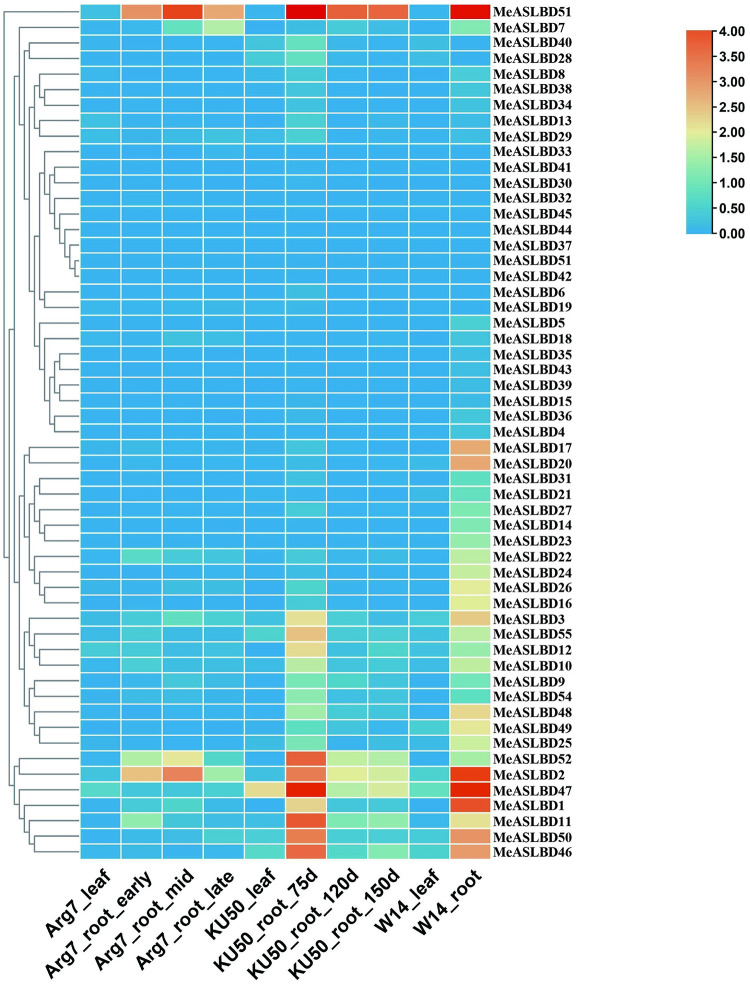
The expression profiles of MeASLBD genes in different tissues (leaves and roots) of the cassava cultivars Arg7, W14, and KU50 were analysed (Fig 7). In cultivars Arg7, KU50, and W14; 1% (5/55), 9% (16/55), and 47% (26/55), respectively, of the MeASLBD genes were expressed in leaves and roots. MeASLBD1, MeASLBD2, MeASLBD11, MeASLBD46, MeASLBD47 and MeASLBD51 were highly expressed (RPKM value > 10) in roots and leaves, while 26 MeASLBDs had low or no expression in the three cassava cultivars.
Fig 7. Expression pattern analysis of MeASLBD transcription factors in roots and leaves.
Red indicates high expression, and blue indicates low expression. The colour scale shows RPKM values normalised by log2. Different tissues (leaves and roots) of the cassava cultivars Arg7, W14, and KU50 were analysed. The Arg7_leaf, KU50_leaf, and W14_leaf stand for leaves. And Arg7_root, KU50_root, and W14_root stand for root. Early, mid and late represent the stages. d = days.
Several genes displayed differential expression profiles between tissues or different cultivars. MeASLBD1 and MeASLBD2 were highly expressed in the roots of W14 but at low levels in the leaves. MeASLBD47 and MeASLBD51 displayed higher expression in W14_root and KU50_root_75d. In addition, MeASLBD47 was also expressed in KU50_leaf. These results suggest the genotype-dependent tissue expression of these genes.
Expression profiles of the MeASLBD genes in response to drought and XamCHN11 infection
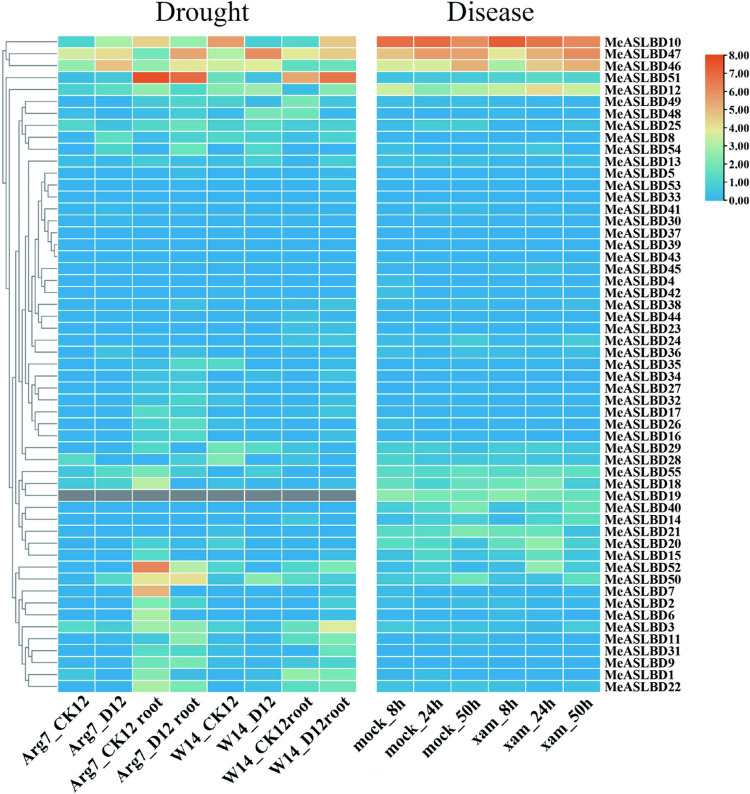
The expression profiles of the 55 MeASLBDs in root and leaf tissues of two cassava cultivars (Arg7 and W14) under drought stress were investigated (Fig 8). The MeASLBD gene was expressed higher in roots than in leaves for both cultivars. Six MeASLBDs (MeASLBD7, MeASLBD10, MeASLBD46, MeASLBD47, MeASLBD51 and MeASLBD52) had significant differential expression under drought stress. The MeASLBD51 expression was significantly induced in Arg7, but decreased in W14 for the root tissues. Meanwhile, the expressions of MeASLBD10, MeASLBD46, and MeASLBD47 were significantly altered in both roots and leaves. MeASLBD7 and MeASLBD52 were highly expressed in Arg7_CK12 root but were significantly downregulated under drought stress.
Fig 8. Expression profiles of MeASLBD genes under drought and XamCHN11 infection.
The ratio of the experimental group to the mock group was log2 transformed. W14_ck12l, W14D12, Arg7_ck12, and W14_CK12 represent leaves. And 14_ck12, W14D12 root, Arg7_ck12 root, and W14_CK12 root stands for root (S7 Table in S1 File). D stands for drought. (https://www.ncbi.nlm.nih.gov/biosample?Db=biosample&DbFrom=bioproject&Cmd=Link&LinkName=bioproject_biosample&LinkReadableName=BioSample&ordinalpos=1&IdsFromResult=246428).
Besides the drought stress, the expressions of some MeASLBDs were also influenced under XamCHN11 stress, including MeASLBD10, MeASLBD12, MeASLBD46, and MeASLBD47. Among these four genes, the MeASLBD10 gene expression was steadily downregulated over infection time, whereas the MeASLBD46 and MeASLBD47 gene expressions were upregulated. Analyses of the transcriptome data of the cassava cultivars (Arg7 and W14) under stress treatments showed that some ASL/LBD genes respond significantly to abiotic and biotic stresses.
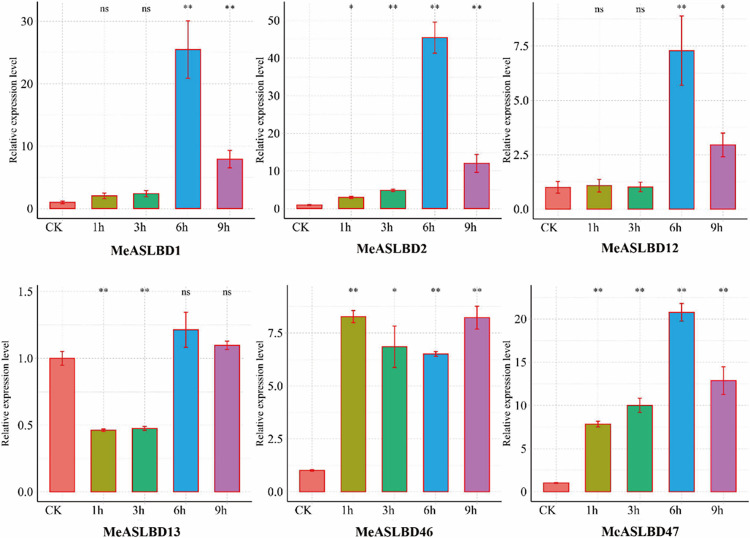
Validation of six MeASLBD genes in response to XamCHN11 via qRT-PCR
Six MeASLBD genes were randomly selected for qRT-PCR validation under a pathogenic bacterium (Xam) stress. The results of qRT-PCR showed that the MeASLBD genes (MeASLBD1, MeASLBD2, MeASLBD12, MeASLBD13, MeASLBD46, and MeASLBD47) had a different expression level at 1 h, 3 h, 6 h, and 9 h post-inoculation (Fig 9). The expression of MeASLBD13 was upregulated at 6 h and 9 h post-inoculation but was downregulated at 1 h and 3 h post-inoculation. MeASLBD46 was significantly upregulated at 1 h, 3 h, 6 h, and 9 h post-inoculation. MeASLBD47 was upregulated progressively after inoculation but decreased slightly at 9 h post-inoculation. The expression levels of MeASLBD1, MeASLBD2, and MeASLBD12 increased significantly at 6 h post-inoculation and then decreased at 9 h post-inoculation. These gene expression levels were consistent with the transcriptomic data. The results indicated that the most significant response of MeASLBD genes to pathogens occurred at 6 h post-inoculation. MeASLBD46, MeASLBD13, and MeASLBD47 expression levels were significantly expressed following treatment with XamCHN11.
Fig 9. Expression patterns of 6 ASL/LBD genes in different samples of M. esculenta (1h, 3h, 6h, 9h, and CK).
Accumulation levels of MeASLBD transcripts were determined by qRT-PCR (n = 3). EF1α was the internal control. Transcription levels were analysed using the comparative “2−ΔΔCt” method. Symbols represent the mean ± SD. Groups designated by the same letter are not significantly different, while those with different letters (“*” or “**”). “**” (p < 0.01) and “*” (0.01 < p < 0.05).
Identification of candidate genes coexpressed with MeASLBD genes
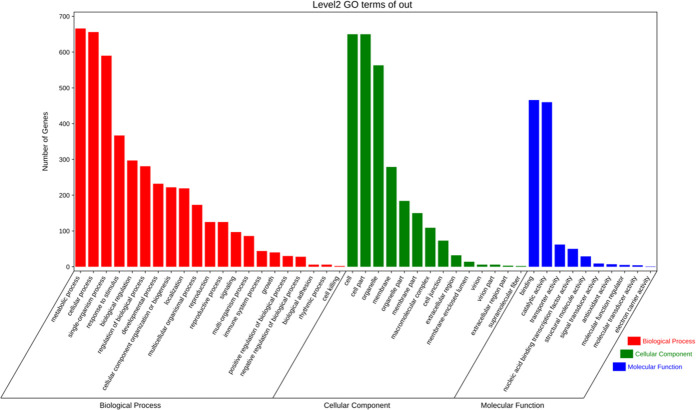
One thousand four hundred forty genes contain a 5’ GCGGCG 3’ motif in the promoter of the cassava genome, a putative motif bound by ASL/LBD genes. GO enrichment analysis revealed that these genes are mainly involved in metabolic, cellular, and biological processes. These genes are mainly allocated to cellular components for cells, cell parts, and organelles. Binding and catalytic activity are the main categories under molecular functions (Fig 10).
Fig 10. GO enrichment analysis of MeASLBDs target genes.
Red is biological processes, green is cell composition, and blue is molecular functions.
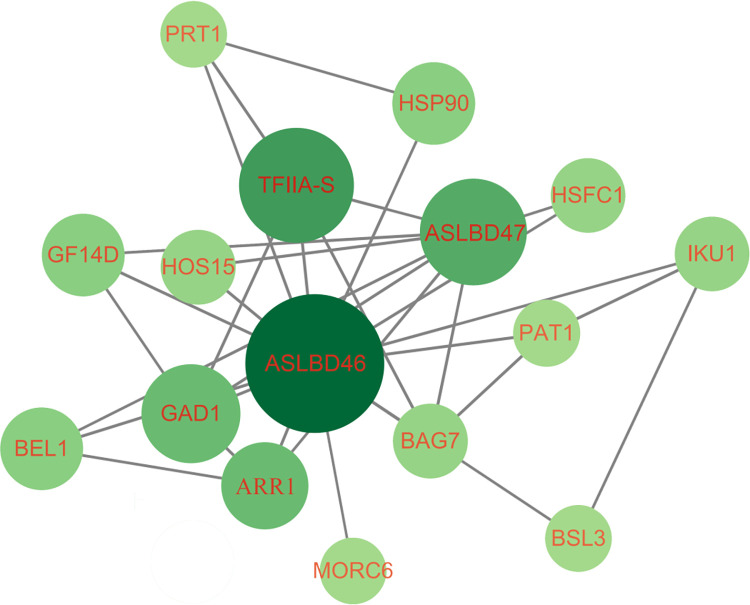
To further analyse the relationship between MeASLBD genes and the predicted downstream target genes, a set of 55 transcriptomes derived from XamCHN11 infected cassava leaves was utilised to perform WGCNA co-expression analysis. The RPKM values of 55 MeASLBD genes and 1,450 target genes were determined. 347 genes with the expression coefficient of variation (CV) >1 and RPKM >1 were retained for WGCNA (S5 Table in S1 File). Highly interconnected gene sets known as modules were obtained via WGNCA analysis. The 347 genes could be divided into four modules via WGCNA, namely blue (67), brown (37), turquoise (217), and grey (25) (S6 Table in S1 File). Grey was considered irrelevant to the sample; therefore, this module was removed. The gene co-expression modules for the MeASLBD and target genes are shown in Fig 11. The enrichment analysis showed that 16 genes were involved in plant-pathogen interaction, 6 in the MAPK signalling pathway, and 14 in defence.
Fig 11. The network of MeASLBDs and target genes in the turquoise module.
The genes in the WGCNA module showed expression patterns and consistent with those of MeASLBDs.
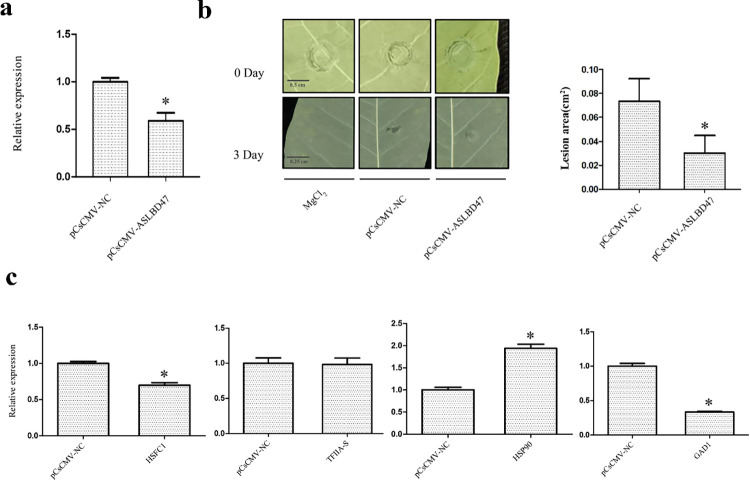
Silencing of MeASLBD47 reduced the lesion area and enabled differential expression of target genes
VIGS approach was applied to silence MeASLBD47 gene. The silencing efficiency was examined using qRT-PCR. The results showed that the abundance of MeASLBD47 in cassava species was reduced compared with NC (Negative Control), indicating that MeASLBD47 was efficiently silenced (Fig 12A). Cassava was inoculated with pCsCMV-AS2/LOB47, pCsCMV-NC and 10 mM MgCl2 (Mock) for 3 days (Fig 12B). After three days, lesions appeared on the NC leaves, whereas pCsCMV-AS2/LOB47 showed no obvious lesions. The result suggests that MeASLBD47 silencing enhances plant resistance by reducing lesion areas. Through the co-expression network constructed using WGCNA, the possible downstream genes regulated by MeASLBD genes were predicted. The genes expressed in the regulatory network were detected based on VIGS (Fig 12C). Among them, the expressions of HSFC1 and GAD1 were significantly downregulated, whereas HSP90 was significantly upregulated, indicating that they were affected by the effect of VIGS. After three days, TFIIA-S expression was slightly affected. Overall, MeASLBD47 might affect plant resistance by regulating HSFC1 and GAD1.
Fig 12. Silencing of MeASLBD47 in cassava plants using a cassava common mosaic virus (Cscmv)-based Virus-induced gene silencing (VIGS) system.
(a) Validation of the silencing efficiency of MeASLBD by qRT-PCR, pCsCMV-NC represents an empty vector. (b) The phenotypes of the cassava leaf infected with XamCHN11 following inoculation with pCsCMV-AS2/LOB47 or empty vector (pCsCMV-NC), 10 mM MgCl2 (mock for XamCHN11). The lesion area of cassava leaves was calculated using the ImageJ tool at 200 pixels/cm. (c) Differential expression of target genes in the WGCNA network upon silencing of MeASLBD47. “*”: significantly different.
Discussion
Cassava (M. esculenta. Crantz) is a starch-rich, woody tuberous root crop that is important for food and as a potential bioenergy crop. Despite its importance in food and bioenergy, cassava cultivation is fraught with difficulties (abiotic and biotic stresses), resulting in decreased crop yield. Most are susceptible to pests and diseases threatening existing cultivars [63, 64]. AS2/LOB genes are widely present in plants and are important plant-specific process regulators [13]. To date, several ASL/LBD genes have been identified in A. thaliana (43) [9], Eucalyptus grandis (46) [65], Glycine max (90) [66], and other species. However, systematic studies of MeASLBD genes have not been reported so far. The availability of whole-genome sequences of M. esculenta helps in the genome-wide characterisation of AS2/LOB genes, which may be used to improve crop yield in the field. In this study, 55 MeASLBD genes were identified, indicating that the AS2/LBD gene family has largely retained a fixed function in the genetic evolution of different species. Two types of terminology were listed side by side (ASLs and LBDs). Such terminology might provide an advantage in discussing the evolutionary developmental biology of the AS2/LOB protein family [5]. The AS2/LOB gene family in M. esculenta is similar to the estimates for other reported plant species. The MeASLBD genes were divided into two classes (I and II) and nine subclasses (Ia~Ig, IIa, and IIb) based on the structure of the LOB domain (Fig 1). Forty-six MeASLBD genes (83%) belonged to Class I, and nine MeASLBD genes (17%) belonged to Class II. Previous studies also reported that 84% and 16% of the ASL/LBD genes in A. thaliana belonged to Classes I and II, respectively [11]. 98 ASL/LBD genes from A. thaliana and M. esculenta were further classified into nine subclasses (Ia-If and IIa-IIb) based on their phylogenetic relationships (Fig 1), and were consistent with those previously reported [67]. Homologous genes in the same class or subclass might share the same function. For instance, ASL15/LBD17 was proposed to play a similar role to ASL18/LBD16, ASL16/LBD29, ASL24/LBD33, and ASL20/LBD18 involved in auxin-induced lateral root formation, all of which belong to the close narrow clades of the tree [68–71]. These results imply that our finding of homologous genes may share similar functions.
The origin and evolutionary history of the ASL/LBD proteins remain largely unexplored. ASL/LBD proteins are streptophyte-specific proteins and likely evolved from the charophycean green algae [72]. Few TF families evolved within land plants, implying the origin of a new TF family is not critical for land plant evolution. Nevertheless, the increased diversity of several TF families, such as ASL/LBD, suggests that they may have been instrumental in terrestrialization [73]. Paleobotanical studies indicate that roots evolved at least twice independently during the evolution of land plants, once in lycophytes and once in euphyllophytes. Auxin promotes the initiation of postembryonic roots in both groups but from different cell types. ASL/LBD proteins act directly downstream of auxin in several euphyllophytes, including Arabidopsis, maize, and rice, and are conserved elements required for root initiation [72]. However, a clade with no lycophyte sequence specifically associated with root development was inferred, implying that root initiation in lycophytes and euphyllophytes is mediated by distinct genetic mechanisms downstream of auxin [17]. In contrast, leaves evolved independently in lycophytes and euphyllophytes [74]. The MeALSBDs evolutionary relationships provide an understanding and could contribute to determining the origin of the ASL/LBD protein family.
Structural analysis effectively extracts valuable information about gene duplication events and phylogenetic relationships within gene families. Gene duplication events, followed by neofunctionalisation, were enough to drive molecular diversification [73, 75]. MeASLBD genes have simple gene structures (Fig 2B). Most MeASLBD members within the same subgroup showed different exon/intron structure and gene length (Fig 2C), slightly different from A. thaliana ASL/LBD genes [9]. Therefore, ASL/LBD gene structures might not be relatively conserved in different species. In addition, cassava contained 55 ASL/LBD genes, 12 more than Arabidopsis and 35 fewer than soybean [60, 66]. Therefore, ASL/LBD genes vary from one species to another, consistent with previously reported ASL/LBD genes [9]. The Ka/Ks ratio indicated that purifying selection had affected the cassava AS2/LOB domain (S4 Table in S1 File). In Arabidopsis ASL/LBD protein sequences comparison of the cloned cDNAs with those of the corresponding genes revealed that these genes were divided into five subtypes based on the positions of the introns in the coding regions (subtypes A to E). 40 MeASLBD genes contained splicing sites in their coding regions (Fig 2C). In comparison, 29 Arabidopsis ASL/LBD genes contained introns in their coding regions.
The ASL/LBD gene family is an important TF family in plant species. Therefore, the function of MeASLBD genes was investigated. Through promoter analysis, most of the MeASLBD promoter positions consisted of ABA, MeJA, and GA response cis-elements (Fig 6), which indicated that MeASLBDs might be involved in the plant phytohormone signal response. In A. thaliana, AtASL/LBD20 is thought to play a role in the JA response [76], and AtASL/LBD40 is downregulated under gibberellin treatment [77]. In cotton, GhLBD123 was significantly upregulated under MeJA treatment [78]. These findings indicate that MeASLBD genes may be involved in hormone and stress responses.
The expression profiles of MeASLBD genes in different tissues (Fig 7) revealed that MeASLBD genes are mainly expressed in the roots, such as MeASLBD2, MeASLBD47, and MeASLBD51, indicating that they might be involved in cassava root development. The transcriptome expression profile of cassava under drought and disease stress was used to study the expression profiles of MeASLBDs (Fig 8). The results revealed that MeASLBD46 and MeASLBD47 responded to drought stress and XamCHN11 infection. MeASLBD46 and MeASLBD47 showed a strong response through qRT-PCR, indicating that they are associated with Xam infection response (Fig 9). The genes MeASLBD46, MeASLBD47, AtASL/LBD37, and AtASL/LBD38 are homologous. MeASLBD46 and MeASLBD47 may thus be involved in nitrogen metabolism, anthocyanin synthesis [79], and stress response.
AS2 can form complexes with other proteins to control different aspects of plant growth and development [1, 11, 80, 81]. AS2 physically associates with AS1 to form a repressor complex that regulates leaf polarity and morphology, inflorescence architecture and fertility, and shoot apical meristem differentiation [1, 11, 80, 81]. AtASL2/LBD10 and AtLBD28 are classified in class I in Arabidopsis [82]. In this study, AtLBD28 was classified in class Id (Fig 1) alongside MeASLBD28 and MeALSBD23. AtLBD28 may influence leaf polarity and morphology, as well as shoot apical meristem differentiation [83]. Therefore, AtLBD28, MeASLBD28, and MeALSBD23 may share similar functions. Moreover, AtASL2/LBD10 participates in both microspore polarization before the first asymmetric division, and germ cell mitosis [84] AtASL2/LBD10 mutants had a 12.7% aborted pollen grain yield, indicating that AtASL2/LBD10 is important for Arabidopsis pollen development [85]. AtASL2/LBD10 was classified in Class Ib (Fig 1), indicating that members such as MeASLBD4 may share similar functions. In addition to their roles in plant development, some Arabidopsis ASL/LBD members play important roles in response to fungal pathogens (e.g., Fusarium wilt) and soil nematodes (e.g., Meloidogyne incognita) [76, 86]. For instance, AtLBD20, primarily expressed in the roots, is a negative regulator of Fusarium wilt resistance and a subset of jasmonate (JA) responses [76]. In this study, AtLBD20, MeASLBD29, and MeASLBD41 were subgrouped into Class Ic (Fig 1), indicating that they may share the same function.
WGCNA is an effective tool for investigating the relationships between different gene sets (modules) [61]. Gene expression modules based on specific species have been efficiently used in diverse varieties. One thousand four hundred fifty genes were identified through promoter analysis. GO enrichment analysis (Fig 11) showed that these genes participate in different regulatory networks. Therefore, AS2/LOB proteins may regulate lateral root growth and plant signal transduction, metabolic processes, and stress responses. MeASLBD46 and MeASLBD47 responded to XamCHN11 infection. In addition, fourteen genes were identified as target genes for MeASLBD regulation via the WGCNA co-expression network (Fig 11). In Arabidopsis, over 350 AtLBD29 target genes have been identified to participate in the regulation of cell reprogramming during callus formation [13, 87, 88]. Some MeASLBD target genes are related to the plant pathogenic bacteria, such as HSP90 [89], TFIIA [90], and GAD1 [91].
VIGS tool was used to silence MeASLBD47 gene and inoculate cassava with XamCHN11 [42]. After 3 days of disease treatment, the water spots on leaves were reduced, and lesions developed slower than NC (Fig 12B). This indicates that the silencing of MeASLBD47 enhances plant resistance against XamCHN11. Simultaneously, qRT-PCR validation was performed on the co-expression network genes (Fig 12A). The result showed that GAD1 and HSFC1 expression was inhibited and, on the contrary, enhanced the expression of HSP90 (Fig 12C). MeASLBD47 might regulate downstream target genes GAD1, HSFC1, and HSP90 to enhance plant resistance. In addition, transcriptome analysis of AS2/LOB37 mutant material in Arabidopsis [79] found that LRR (At1g66090) was significantly upregulated when AS2/LOB37 was deleted. Therefore, the LRR gene in cassava was detected, and the results were consistent with Arabidopsis (S2 Fig in S1 File). These findings provide useful clues for further investigations into gene regulatory networks involved in cassava disease resistance.
Conclusions
Fifty-five ASL/LBD genes were identified from M. esculenta and distributed unevenly among the chromosomes. The MeASLBD gene family is classified into two categories based on gene structure and phylogeny: Class I (46) and Class II (9). Collinearity analysis and the Ks/Ks values indicated that purifying selection was the main force driving the evolution of the MeASLBD gene family. According to the transcriptome data of cassava under biological and abiotic stress, MeASLBD46 and MeASLBD47 were found to have a strong disease and drought response. To better understand the regulatory function of MeASLBDs, the target genes of MeASLBDs were screened and revealed the disease-related genes of HSP90, TFIIA-S, and GAD1. The latter result indicates that MeASLBD46 and MeASLBD47 may participate in the plant response to stress by regulating these target genes. Furthermore, these findings provide valuable information for subsequent elucidation of the role of the ASL/LBD genes.
Supporting information
(DOCX)
Acknowledgments
The authors thank AJE (https://www.aje.cn) for providing linguistic assistance during the preparation of this manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was supported by National Key Research and Development Program (2018YFD1000500), the Natural Science Foundation of Hainan Province (320QN193) and China Agriculture Research System (CARS-11-hncyh). National Key Research and Development Program (2018YFD1000500) Funding had role in study design and material collection; the Natural Science Foundation of Hainan Province (320QN193) had role in the research of qPCR data collection and analysis; China Agriculture Research System (CARS-11-hncyh) founders had role in RNA-Seq data collection and analysis, manuscript preparation.
References
- 1.Guo M, Thomas J, Collins G, Timmermans MC. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. The Plant Cell. 2008; 20(1):48–58. 10.1105/tpc.107.056127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang JY, Iwasaki M, Machida C, Machida Y, Zhou X, Chua NH, et al. βC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes & Development. 2008; 22(18):2564–77. 10.1101/gad.1682208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwasaki M, Takahashi H, Iwakawa H, Nakagawa A, Ishikawa T, Tanaka H, et al. Dual regulation of ETTIN (ARF3) gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in Arabidopsis. Development. 2013; 140(9):1958–69. 10.1242/dev.085365 [DOI] [PubMed] [Google Scholar]
- 4.Lodha M, Marco CF, Timmermans MC. The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes & Development. 2013; 27(6):596–601. 10.1101/gad.211425.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Machida Y, Suzuki T, Sasabe M, Iwakawa H, Kojima S, Machida CJ. Arabidopsis ASYMMETRIC LEAVES2 (AS2): roles in plant morphogenesis, cell division, and pathogenesis. Journal of Plant Research. 2022; 135(1):3–14. 10.1007/s10265-021-01349-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vial-Pradel S, Keta S, Nomoto M, Luo L, Takahashi H, Suzuki M, et al. Arabidopsis zinc-finger-like protein ASYMMETRIC LEAVES2 (AS2) and two nucleolar proteins maintain gene body DNA methylation in the leaf polarity gene ETTIN (ARF3). Plant and Cell Physiology. 2018; 59(7):1385–97. 10.1093/pcp/pcy031 [DOI] [PubMed] [Google Scholar]
- 7.Riechmann JL, Ratcliffe OJ. A genomic perspective on plant transcription factors. Current Opinion in Plant Biology. 2000; 3(5):423–34. doi: 10.1016/s1369-5266(00)00107-2 [DOI] [PubMed] [Google Scholar]
- 8.Gong W, Shen YP, Ma LG, Pan Y, Du YL, Wang DH, et al. Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiology. 2004; 135(2):773–82. 10.1104/pp.104.042176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shuai B, Reynaga-Pena CG, Springer PS. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiology. 2002; 129(2):747–61. 10.1104/pp.010926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majer C, Hochholdinger F. Defining the boundaries: structure and function of LOB domain proteins. Trends in Plant Science. 2011; 16(1):47–52. 10.1016/j.tplants.2010.09.009 [DOI] [PubMed] [Google Scholar]
- 11.Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant and Cell Physiology. 2002; 43(5):467–78. 10.1093/pcp/pcf077 [DOI] [PubMed] [Google Scholar]
- 12.Husbands A, Bell EM, Shuai B, Smith HM, Springer PS. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Research. 2007; 35(19):6663–71. 10.1093/nar/gkm775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu C, Luo F, Hochholdinger F. LOB domain proteins: beyond lateral organ boundaries. Trends in Plant Science. 2016; 21(2):159–67. 10.1016/j.tplants.2015.10.010 [DOI] [PubMed] [Google Scholar]
- 14.Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development. 2001; 128(10):1771–83. 10.1242/dev.128.10.1771 [DOI] [PubMed] [Google Scholar]
- 15.Chen WF, Wei XB, Rety S, Huang LY, Liu NN, Dou SX, et al. Structural analysis reveals a “molecular calipers” mechanism for a LATERAL ORGAN BOUNDARIES DOMAIN transcription factor protein from wheat. Journal of Biological Chemistry. 2019; 294(1):142–56. 10.1074/jbc.RA118.003956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee HW, Kim- J, Park MY, Han KH, Kim J. The conserved proline residue in the LOB domain of LBD18 is critical for DNA-binding and biological function. Molecular Plant. 2013; 6(5):1722–25. 10.1093/mp/sst037 [DOI] [PubMed] [Google Scholar]
- 17.Matsumura Y, Iwakawa H, Machida Y, Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. The Plant Journal. 2009; 58(3):525–37. 10.1111/j.1365-313X.2009.03797.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uchida N, Townsley B, Chung KH, Sinha N. Regulation of SHOOT MERISTEMLESS genes via an upstream-conserved noncoding sequence coordinates leaf development. Proceedings of the National Academy of Sciences. 2007; 104(40):15953–58. doi: 10.1073/pnas.0707577104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, et al. Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 2003; 130(17):4097–107. 10.1242/dev.00622 [DOI] [PubMed] [Google Scholar]
- 20.Xu B, Li Z, Zhu Y, Wang H, Ma H, Dong A, et al. Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary-specifying genes to promote sepal and petal development. Plant Physiology. 2008; 146(2):566. 10.1104/pp.107.113787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okogbenin E, Setter TL, Ferguson M, Mutegi R, Ceballos H, Olasanmi B, et al. Phenotypic approaches to drought in cassava. Frontiers in Physiology. 2013; 4:93. 10.3389/fphys.2013.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliveira E, Santana F, Oliveira L, Santos V. Genetic parameters and prediction of genotypic values for root quality traits in cassava using REML/BLUP. Genetics and Molecular Research. 2014; 13(3):6683–700. doi: 10.4238/2014.August.28.13 [DOI] [PubMed] [Google Scholar]
- 23.Muiruri SK, Ntui VO, Tripathi L, Tripathi JN. Mechanisms and approaches towards enhanced drought tolerance in cassava (Manihot esculenta). Current Plant Biology. 2021; 28:100227. 10.1016/j.cpb.2021.100227 [DOI] [Google Scholar]
- 24.Allem AC. The origin of Manihot esculenta crantz (Euphorbiaceae). Genetic resources and crop Evolution. 1994; 41(3):133–50. 10.1007/BF00051630 [DOI] [Google Scholar]
- 25.De Souza AP, Massenburg LN, Jaiswal D, Cheng S, Shekar R, Long SP. Rooting for cassava: insights into photosynthesis and associated physiology as a route to improve yield potential. New Phytologist. 2017; 213(1):50–65. 10.1111/nph.14250 [DOI] [PubMed] [Google Scholar]
- 26.Raheem D, Chukwuma C. Foods from cassava and their relevance to Nigeria and other African countries. Agriculture and Human Values. 2001; 18(4):383–90. 10.1023/A:1015233606665 [DOI] [Google Scholar]
- 27.Balat M, Balat H. Recent trends in global production and utilization of bio-ethanol fuel. Applied Energy. 2009; 86(11):2273–82. 10.1016/j.apenergy.2009.03.015 [DOI] [Google Scholar]
- 28.Schmitz P, Kavallari A. Crop plants versus energy plants—On the international food crisis. Bioorganic & Medicinal Chemistry. 2009; 17(12):4020–21. 10.1016/j.bmc.2008.11.041 [DOI] [PubMed] [Google Scholar]
- 29.Legg JP, Kumar PL, Makeshkumar T, Tripathi L, Ferguson M, Kanju E, et al. Cassava virus diseases: biology, epidemiology, and management. Advances in Virus research. 2015; 91:85–142. doi: 10.1016/bs.aivir.2014.10.001 [DOI] [PubMed] [Google Scholar]
- 30.López CE, Bernal AJ. Cassava bacterial blight: using genomics for the elucidation and management of an old problem. Tropical Plant Biology. 2012; 5(1):117–26. 10.1007/s12042-011-9092-3 [DOI] [Google Scholar]
- 31.Patil BL, Legg JP, Kanju E, Fauquet CM. Cassava brown streak disease: a threat to food security in Africa. Journal of General Virology. 2015; 96(5):956–68. doi: 10.1099/vir.0.000014 [DOI] [PubMed] [Google Scholar]
- 32.Lozano JC. Cassava bacterial blight: a manageable disease. Plant Disease. 1986; 70(12):1989–93. [Google Scholar]
- 33.Lozano J, Sequiera L. Bacterial blight of cassava in Colombia: epidemiology and control. Phytopathology. Fao; 1974. [Google Scholar]
- 34.Joseph J, Elango F. The status of cassava bacterial blight caused by Xanthomonas campestris pv. manihotis in Trinidad. Journal of Phytopathology. 1991; 133(4):320–26. 10.1111/j.1439-0434.1991.tb00167.x [DOI] [Google Scholar]
- 35.Koné D, Dao S, Tekete C, Doumbia I, Koita O, Abo K, et al. Confirmation of Xanthomonas axonopodis pv. manihotis causing cassava bacterial blight in Ivory Coast. Plant Diseases. 2015; 99: 10.1094/PDIS-02-15-0172-PDN [DOI] [Google Scholar]
- 36.Wonni I, Ouedraogo L, Dao S, Tekete C, Koita O, Taghouti G, et al. First report of cassava bacterial blight caused by Xanthomonas axonopodis pv. manihotis in Burkina Faso. Plant Disease. 2015; 99(4):551. 10.1094/PDIS-03-14-0302-PDN [DOI] [Google Scholar]
- 37.Bredeson JV, Lyons JB, Prochnik SE, Wu GA, Ha CM, Edsinger-Gonzales E, et al. Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nature Biotechnology. 2016; 34(5):562–70. doi: 10.1038/nbt.3535 [DOI] [PubMed] [Google Scholar]
- 38.Huang S, Tang Z, Zhao R, Hong Y, Zhu S, Fan R, et al. Genome-wide identification of cassava MeRboh genes and functional analysis in Arabidopsis. Plant Physiology and Biochemistry. 2021; 167:296–308. 10.1016/j.plaphy.2021.07.039 [DOI] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001; 25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 40.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nature Protocols. 2008; 3(6):1101–108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 41.Fernandez-Pozo N, Rosli HG, Martin GB, Mueller LA. The SGN VIGS tool: user-friendly software to design virus-induced gene silencing (VIGS) constructs for functional genomics. Molecular Plant. 2015; 8(3):486–88. doi: 10.1016/j.molp.2014.11.024 [DOI] [PubMed] [Google Scholar]
- 42.Jupin I. A protocol for VIGS in Arabidopsis thaliana using a one-step TYMV-derived vector. In Virus-Induced Gene Silencing: Springer. 2013; 197–210. 10.1007/978-1-62703-278-0_15 [DOI] [PubMed] [Google Scholar]
- 43.Yan P, Zeng Y, Shen W, Tuo D, Li X, Zhou P. Nimble cloning: a simple, versatile, and efficient system for standardized molecular cloning. Frontiers in Bioengineering and Biotechnology. 2020; 7:460. doi: 10.3389/fbioe.2019.00460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuo D, Fu L, Shen W, Li X, Zhou P, Yan P. Generation of stable infectious clones of plant viruses by using Rhizobium radiobacter for both cloning and inoculation. Virology. 2017; 510:99–103. 10.1016/j.virol.2017.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research. 2012; 40(D1):D1178–86. doi: 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Research. 2003; 31(1):224–28. 10.1093/nar/gkg076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eddy SR. Accelerated profile HMM searches. PLoS Computational Biology. 2011; 7(10):e1002195. doi: 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sonnhammer EL, Eddy SR, Birney E, Bateman A, Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Research. 1998; 26(1):320–2. doi: 10.1093/nar/26.1.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, et al. The Pfam protein families database in 2019. Nucleic Acids Research. 2019; 47(D1):D427–32. doi: 10.1093/nar/gky995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, De Castro E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Research. 2012; 40(W1):W597–603. doi: 10.1093/nar/gks400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016; 33(7):1870–74. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic acids research. 2015; 43(W1):W39–W49. doi: 10.1093/nar/gkv416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015; 31(8):1296–97. doi: 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant. 2020; 13(8):1194–202. doi: 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research. 2012; 40(7):e49. doi: 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Research. 2009; 19(9):1639–45. doi: 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lescot M, Dthais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research. 2002; 30(1):325–27. doi: 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005; 21(18):3674–76. doi: 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- 59.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature biotechnology. 2019; 37(8):907–15. doi: 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nature protocols. 2016; 11(9):1650–67. doi: 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Statistical Applications in Genetics and Molecular Biology. 2005; 4(1). doi: 10.2202/1544-6115.1128 [DOI] [PubMed] [Google Scholar]
- 62.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003; 13(11):2498–504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fathima AA, Sanitha M, Tripathi L, Muiruri S. Cassava (Manihot esculenta) dual use for food and bioenergy: A review. Food and Energy Security. 2022; e380. 10.1002/fes3.380 [DOI] [Google Scholar]
- 64.Ogwok E, Ilyas M, Alicai T, Rey ME, Taylor NJ. Comparative analysis of virus-derived small RNAs within cassava (Manihot esculenta Crantz) infected with cassava brown streak viruses. Virus Research. 2016; 215:1–11. 10.1016/j.virusres.2016.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu Q, Shao F, Macmillan C, Wilson IW. Genome-wide analysis of the lateral organ boundaries domain (LBD) gene family in Eucalyptus grandis reveals members that differentially impact secondary growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang H, Shi G, Du H, Wang H, Zhang Z, Hu D, et al. Genome‐Wide Analysis of Soybean LATERAL ORGAN BOUNDARIES Domain‐Containing Genes: A Functional Investigation of GmLBD12. The plant genome. 2017; 10(1):plantgenome2016.07.0058. 10.3835/plantgenome2016.07.0058 [DOI] [PubMed] [Google Scholar]
- 67.Song B, Tang Z, Li X, Li J, Zhang M, Zhao K, et al. Mining and evolution analysis of lateral organ boundaries domain (LBD) genes in Chinese white pear (Pyrus bretschneideri). BMC genomics. 2020; 21(1):644. 10.1186/s12864-020-06999-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee HW, Kim NY, Lee DJ, Kim JJ. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiology. 2009; 151(3):1377–89. 10.1104/pp.109.143685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berckmans B, Vassileva V, Schmid SP, Maes S, Parizot B, Naramoto S, et al. Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. The Plant Cell. 2011; 23(10):3671–83. 10.1105/tpc.111.088377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goh T, Joi S, Mimura T, Fukaki HJ. The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development. 2012; 139(5):883–93. 10.1242/dev.071928 [DOI] [PubMed] [Google Scholar]
- 71.Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka MJ. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. The Plant Cell. 2007; 19(1):118–30. 10.1105/tpc.106.047761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coudert Y, Dievart A, Droc G, Gantet PJ. ASL/LBD phylogeny suggests that genetic mechanisms of root initiation downstream of auxin are distinct in lycophytes and euphyllophytes. Molecular Biology and Evolution. 2013; 30(3):569–72. ttps://doi.org/10.1093/molbev/mss250 [DOI] [PubMed] [Google Scholar]
- 73.Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 2017; 171(2):287–304. e15. 10.1016/j.cell.2017.09.030 [DOI] [PubMed] [Google Scholar]
- 74.Harrison CJ, Corley SB, Moylan EC, Alexander DL, Scotland RW, Langdale JA. Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature. 2005; 434(7032):509–514. doi: 10.1038/nature03410 [DOI] [PubMed] [Google Scholar]
- 75.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biology. 2004; 4:10. 10.1186/1471-2229-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thatcher LF, Powell JJ, Aitken EA, Kazan K, Manners JM. The lateral organ boundaries domain transcription factor LBD20 functions in Fusarium wilt susceptibility and jasmonate signaling in Arabidopsis. Plant Physiology. 2012. b; 160(1):407–18. 10.1104/pp.112.199067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. The Plant Cell. 2007; 19(10):3037–57. 10.1105/tpc.107.054999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu J, Xie Q, Li C, Dong Y, Zhu S, Chen J. Comprehensive characterization and gene expression patterns of LBD gene family in Gossypium. Planta. 2020; 251(4):1–16. 10.1007/s00425-020-03364-8 [DOI] [PubMed] [Google Scholar]
- 79.Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. The Plant Cell. 2009; 21(11):3567–84. 10.1105/tpc.109.067041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin W-c, Shuai B, Springer PS. The Arabidopsis LATERAL ORGAN BOUNDARIES–domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. The Plant Cell. 2003; 15(10):2241–52. doi: 10.1105/tpc.014969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen X, Wang H, Li J, Huang H, Xu LJ. Quantitative control of ASYMMETRIC LEAVES2 expression is critical for leaf axial patterning in Arabidopsis. Journal of Experimental Botany. 2013; 64(16):4895–905. 10.1093/jxb/ert278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y, Li Z, Ma B, Hou Q, Wan XJ. Phylogeny and functions of LOB domain proteins in plants. International Jounal of Molecular Sciences. 2020; 21(7):2278. 10.3390/ijms21072278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang L, Wang W, Xu D, Chang Z, Cui XJ. Overexpression of Arabidopsis ASL25/LBD28 gene affects leaf morphogenesis. Acta Botanica Boreali-Occidentalia Sinica. 2010; 30(5):888–93. [Google Scholar]
- 84.Kim MJ, Kim M, Kim JJ. Combinatorial interactions between LBD10 and LBD27 are essential for male gametophyte development in Arabidopsis. Plant signaling & Behaviour. 2015. a; 10(8):e1044193. 10.1080/15592324.2015.1044193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim MJ, Kim M, Lee MR, Park SK, Kim JJ. LATERAL ORGAN BOUNDARIES DOMAIN (LBD) 10 interacts with SIDECAR POLLEN/LBD 27 to control pollen development in Arabidopsis. The Plant Journal. 2015. b; 81(5):794–809. 10.1111/tpj.12767 [DOI] [PubMed] [Google Scholar]
- 86.Thatcher LF, Kazan K, Manners JM. Lateral organ boundaries domain transcription factors: New roles in plant defense.Plant Signaling & Behavior. 2012. a; 7(12):1702–804. 10.4161/psb.22097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fan M, Xu C, Xu K, Hu Y. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell research. 2012; 22(7):1169–80. 10.1038/cr.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sugimoto K, Jiao Y, Meyerowitz EM. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Developmental cell. 2010; 18(3):463–71. 10.1016/j.devcel.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 89.Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, et al. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. The EMBO journal. 2003; 22(21):5679–89. 10.1093/emboj/cdg547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jiang GH, Xia ZH, Zhou YL, Wan J, Li DY, Chen RS, et al. Testifying the rice bacterial blight resistance gene xa5 by genetic complementation and further analyzing xa5 (Xa5) in comparison with its homolog TFIIAγ1. Molecular Genetics and Genomics. 2006; 275(4):354–66. 10.1007/s00438-005-0091-7 [DOI] [PubMed] [Google Scholar]
- 91.Deng X, Xu X, Liu Y, Zhang Y, Yang L, Zhang S, et al. Induction of γ‐aminobutyric acid plays a positive role to Arabidopsis resistance against Pseudomonas syringae. Journal of integrative plant biology. 2020; 62(11):1797–812. 10.1111/jipb.12974 [DOI] [PMC free article] [PubMed] [Google Scholar]