Abstract
Introduction:
Hand, foot, and mouth disease (HFMD) is a common childhood infectious disease, caused by enteroviruses (EVs) which can present with typical or atypical lesions. Although the disease is self-limiting, it can also lead to serious complications. In the era of polio eradication, it is important to understand the population dynamics of enteroviruses causing HFMD as one of the circulating strains may become dominant.
Methods:
It was a collaborative study carried out in the Department of Dermatology and Microbiology of a tertiary care teaching hospital. The throat swabs were collected from 132 suspected HFMD cases. Real-time polymerase chain reaction (PCR) was performed to detect the presence of pan enteroviruses, followed by genotype-specific PCR targeting Human Enterovirus 71 (HEV-71) and Coxsackie virus A16 (CVA-16) and CVA-6 for pan Enterovirus-positive samples. Follow-up samples were collected from 14 children in the 2nd week and subjected to molecular testing to detect enteroviruses.
Results:
Among 132 children suspected to have HFMD, 44 were girls and 88 were boys, and the majority of them 76.5% (101/132) were under 2 years of age. A history of exposure to a similar clinical presentation was present in 15 children. Of 132 suspected cases, 60 samples (45.5%) were positive for pan Enterovirus. The predominantly circulating genotype was found to be CVA-6 (31.6% [19/60]). There were about 10 cases (16.6%) which had co-infection with both HEV71 and CVA-6. Rash with fever was the most common presentation (57%). In most of the cases with HEV 71, 92.3% (12/13) presented within 3 days of illness to the health-care facility. Of 60 positive cases, 25% (15/60) of children had the atypical distribution of rashes in the face, trunk, genitalia, thigh, neck, and axilla and 16.7% of children (10/60) had the atypical type of lesion either only papular lesions or erythema multiforme. Out of 14 follow-up samples, 13 were negative for EVs; one was positive for pan EV in the 2nd week, however, the patient lost to follow-up after that.
Conclusion:
HFMD outbreaks in our region were caused by various genotypes of enteroviruses. No severe complications were seen in the affected children. Nearly 30% had atypical presentation either in the form of lesion or site. Robust molecular epidemiological surveillance of HFMD is required to know the strain variations and other emerging genotypes in our setup.
Keywords: Coxsackie virus-A16, Coxsackie virus-A6, enteroviruses, hand- foot and mouth disease, Human Enterovirus-71
INTRODUCTION
Hand, foot, and mouth disease (HFMD) is an emerging disease of childhood, most commonly caused by enteroviruses (EVs) serotypes – Human Enterovirus 71 (HEV-71) and Coxsackie virus A16 (CVA-16), of which HEV 71 is associated with fatal complications.[1-4] Other serotypes of EVs such as CVA-A4-A10, A24, B1-B5, and Echovirus – 4, 7, 9, 11, 18, 19, 25, and 30 can also be frequently identified in recent years.[2,5] Out of these, CVA-6 and CVA-10 were most frequently detected and even become the predominant serotype than EV71 or CVA-16 in China.[6] After the fatal outbreak of HFMD in 2008, it has been identified as one of the notifiable diseases in China under category “C.”[7]
In India, the first outbreak of HFMD was reported in Calicut, Kerala in 2013.[8] Subsequently, outbreaks were reported from Tamil Nadu, Odisha, Karnataka, and West Bengal by multiple serotypes.[2,9-12] In India, the disease was relatively unknown until recently due to low suspicion or common diseases such as chicken pox and mosquito bite have similar clinical presentations leading to misdiagnosis or due to the fact that the disease is usually self-limiting with uneventful recovery which leads to underestimated disease burden.[9,13]
Given the fact that only a single Enterovirus multiplies in the gut at any given point of time, in the era of polio eradication, infections due to other enteroviruses are gaining importance.[9,14] Co-circulation of multiple serotypes of Enterovirus has complicated the HFMD epidemic and poses many difficulties for disease control.[15,16] Furthermore, apart from HEV-71, some serotypes have reported with severe or fatal HFMD in recent years.[17] To control HFMD, it is necessary to know the various clinical presentation and molecular epidemiology of the disease based on the geographic pattern. Furthermore, very little is known globally about the viral excretion period of enteroviruses causing HFMD.[18] Since there is no effective antiviral therapy or vaccine covering all strains currently available, the knowledge of viral shedding patterns in affected children will help in planning the refraining period of these children.
The purpose of our study was to know the regional molecular epidemiology of HFMD disease caused by the most common genotypes such as HEV-71, CVA-16, CVA-6, and the viral shedding patterns of EVs in affected children.
METHODS
This collaborative study was carried out in the Department of Microbiology and Dermatology in a tertiary care hospital. Any child of age 6 months to 18 years with mouth/tongue ulcer and maculopapular rashes and/or vesicles on palms or soles with or without fever suspected of HFMD attending the outpatient services of the Department of Dermatology and Venereology between January 2016 and November 2017 were included in the study.
Case recruitment and clinical examination
Suspected cases were recruited after getting written informed consent from parents/guardians. The structured proforma was filled with the demographic, clinical, and investigational details of the patient. Lesions in hands/feet/elbows/knees/buttocks/mouth and vesicles/vesiculobullous lesions with or without papular lesions were taken as typical presentation and lesions in other areas or only papular lesions or erythema multiforme with or without vesicles were grouped as atypical presentation.[19]
Sample collection and transport
Throat swabs were collected from both the tonsils and posterior pharyngeal wall using a commercially available sterile nylon flocked swab, placed in the viral transport medium (Hi-Media, Mumbai, India), and transported to the laboratory maintaining the cold chain.
Molecular characterization
The samples in the viral transport medium were centrifuged at 10,000 rpm for 5 min at 4°C. RNA was extracted from the supernatant fluid as per the manufacturer’s instructions (Helini biomolecules viral nucleic acid).
Pan Enterovirus real-time polymerase chain reaction
Real-time reverse transcriptase–polymerase chain reaction (PCR) for identification of pan Enterovirus was performed using the primers FP: 5’- TACTTTGGGTGTCCGTGTTT-3’, RP: 5’- TGGCCAATCCAATAGCTATATG-3’, PP: FAM-AYTGGCTGCTTATGGTGACRAT-BHQ1 targeting conserved 5’Untranslated region on Cobas Z 480 (Manufacturer Roche) using a final reaction mixture volume of 25 μl. The cycling conditions were reverse transcription at 42°C for 30 min, followed by Taq enzyme activation at 95°C for 15 min and 45 cycles of initial denaturation at 95°C for 20 s, annealing at 56°C for 20 s, and extension at 72°C for 20 s.[20]
Polymerase chain reaction for Enterovirus genotyping
The samples that tested positive for pan Enterovirus were genotyped. The genotyping was done by amplification of the VP1 region for HEV-71, CVA-16, and CVA-6 using the primers listed in Table 1.
Table 1.
Enterovirus genotyping polymerase chain reaction primers
| Genotype | Primers | Product size (bp) |
|---|---|---|
| Human Enterovirus - 71 | FP : GCAGCCCAAAAGAACTTCAC RP : ATTTCAGCAGCTTGGAGTGC |
243 |
| Coxsackievirus - A6 | FP: CAAGC TGCAG AAACG GGAG RP: GCTCC ACACT CGCCT CATT |
106 |
| Coxsackievirus - A16 | FP: ATTGGTGCTCCCACTACAGC RP : TCAGTGTTGGCAGCTGTAGG |
225 |
All the extracted RNA was converted to cDNA on thermal cycler BioRad (model no: C1000 TOUCH) with cycling conditions of priming at 25°C for 10 min, reverse transcription at 37°C for 120 min, RT inactivation at 85°C for 5 min with the final volume of 20 μl and stored at − 20°C till further processing. PCR was performed on the thermal cycler BioRad (model no: C1000 TOUCH) using a final reaction mixture volume of 20 μl.
Cycling conditions were as follows: 3 min at 950°C for the initial denature step, 33 cycles for 30 s at 950°C for denature, 30 s at 500°C (HEV-71), 600°C (CVA-6), 580°C (CVA-16) for annealing, 40 s at 720°C for extension, and a final extension at 720°C for 7 min.
The amplicons were detected by electrophoresis on agarose gel (2%) with ethidium bromide and visualized with the help of a gel documentation system (Bio-Rad Gel Doc XR system, USA). Specific band regions were looked for HEV-71-243 bp, CVA-16-225 bp, and CVA-6-106 bp.
Statistical methods
The distribution of categorical data such as age, gender, clinical profile, sociodemographic profile, and Enterovirus genotypes was expressed in frequency and percentages. The association between the categorical data mentioned above with the Enterovirus genotypes was assessed using the Chi-square test. All statistical analysis were carried out at a 5% level of significance, and P < 0.05 was considered significant.
Statistical analysis was performed using SPSS 19.0 (IBM Corp., Armonk, NY, USA) software.
RESULTS
A total of 132 children suspected to have HFMD were included in the study. Among them, 44 were girls and 88 were boys. The majority of our cases 76.5% (101/132) were under 2 years of age. Out of 132 suspected cases, 15 children had given a history of exposure to a similar presentation from a neighbor (9), relative (3), school (2), and hospital (1).
Molecular detection
Of the 132 suspected samples tested, 60 samples (45.5%) turned out to be positive for EV by pan Enterovirus real-time polymerase chain reaction (RT-PCR) [Figure 1].
Figure 1.
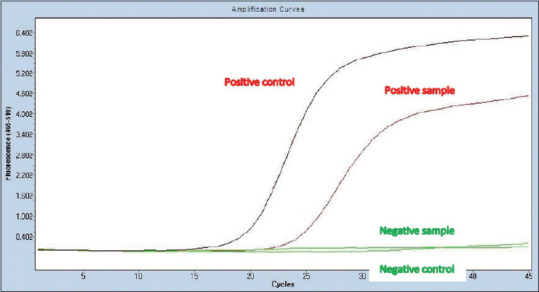
Amplification curve in real-time RT-PCR showing positive and negative samples for pan enteroviruses. RT-PCR: Real-time polymerase chain reaction
Genotyping of all pan EV-positive samples by conventional PCR revealed 19 samples (31.6%) positive for CVA-6 (9 as single genotype and 10 circulated with HEV-71); 13 detected (21.6%) as HEV-71 (3 as single genotype and 10 circulated with CVA-6); 1 identified as CVA-16, and 37 samples (61.6%) were not genotyped [Figure 2].
Figure 2.
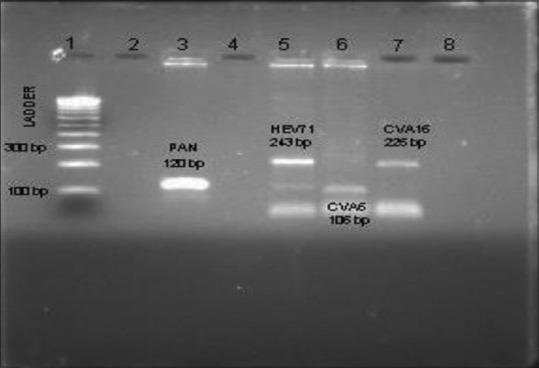
Genotyping shows positivity for pan enteroviruses (120 bp), HEV-71 (243 bp), CVA-6 (106 bp), and CVA-16 (225 bp). HEV: Human Enterovirus, CVA: Coxsackie virus
Clinical presentation and its association with genotypes
The most common presentation among the cases was rash with fever (57%). Other symptoms seen were sore throat and cold in six, pruritus in five, irritability in three, malaise in two, rhinitis, cough, headache, nausea, vomiting, anorexia, diarrhea, and constipation in one case only. The health-care facility (HCF) was sought within a week by most of the cases (85%). Among the positive cases, 60% (36/60) of cases presented within 3 days of illness. However, there was no statistical significance noted between the day of the presentation to HCF and the detection of EVs [Table 2].
Table 2.
Day of presentation to health care facility and detection of Enteroviruses
| Day of presentation | EV positive, n (%) | EV negative, n (%) | Total, n (%) | P |
|---|---|---|---|---|
| 1-3 | 36 (27.3) | 35 (26.5) | 71 (53.8) | 0.077 |
| 4-6 | 15 (11.4) | 12 (9.1) | 27 (20.5) | |
| 7-10 | 7 (5.3) | 19 (14.4) | 26 (19.7) | |
| >10 | 2 (1.5) | 6 (4.5) | 8 (6.1) | |
| Total | 60 (45.5) | 72 (54.5) | 132 (100) |
EV: Enterovirus
However, there was a statistically significant difference found between the genotype presentations to HCF. The majority 92.3% (12/13) of HEV-71 (both single as well as co-infection) came to HCF within 3 days of the onset of illness when compared to 57% of CVA-6 (11/19) (both single as well as co-infection) [Table 3].
Table 3.
Day of presentation to health care facility and Enterovirus genotypes
| Day of presentation | HEV 71 | CVA 6 | Both (HEV 71 and CVA 6) | P |
|---|---|---|---|---|
| 1-3 | 3 | 2 | 9 | 0.002 |
| 4-6 | 0 | 6 | 1 | |
| Total | 3 | 8 | 10 |
P<0.05 considered significant. HEV 71: Human Enterovirus 71, CVA 6: Coxsackie virus A6
Typical and atypical distribution and type of lesion
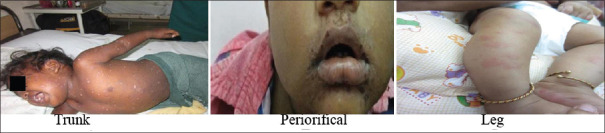
The atypical distribution of the lesion over the face, trunk, genitalia, thigh, neck, and axilla was found in 15 children (25%) while 10 children (16.7%) had the atypical type of lesion [Figure 3] either only papular lesions or erythema multiforme [Table 4].
Figure 3.
Atypical presentation of HFMD. HFMD: Hand, foot, and mouth disease
Table 4.
Lesion site and type among suspected, Pan polymerase chain reaction positive and Human Enterovirus 71; Coxsackie virus A6 cases
| Site of lesion | Total (n=132), n (%) | Positive for Pan Enterovirus (n=60), n (%) | Positive for HEV 71 (n=3), n (%) | Positive for CVA 6 (n=9), n (%) | Positive for HEV 71 and CVA 6 (n=10), n (%) |
|---|---|---|---|---|---|
| Typical site Hands/feet/elbows/knees/buttocks/mouth | 83 (62.9) | 45 (75) | 2 (66.6) | 5 (55.5) | 10 (100) |
| Atypical site Lesions in other areas - face, trunk, genitalia, thigh, neck, axilla, periorificial | 49 (37.1) | 15 (25) | 1 (33.3) | 4 (44.4) | - |
| Typical type Vesicles/vesiculobullous lesions with or without papular lesions | 56 (42.4) | 50 (83.3) | 2 (66.6) | 6 (66.6) | 10 (100) |
| Atypical type Only papular lesions or erythema multiforme | 72 (57.6) | 10 (16.7) | 1 (33.3) | 3 (33.3) | - |
HEV 71: Human Enterovirus 71, CVA 6: Coxsackie virus A6
Follow-up
Only 14 children responded with further follow-up on 2nd-week. Almost 13 children were negative for pan Enterovirus at 2nd week while 1 child turned out to be positive for the HEV-71 genotype. The child who was positive at 2nd week, however, lost to follow-up thereafter.
DISCUSSION
Although mostly HFMD is self-limited, some serotypes such as HEV-71 infections may lead to severe neurological complications and pulmonary edema, especially in young children.[2,4] Cutaneous lesions usually provide clues for the early diagnosis of infectious diseases in children. HFMD also largely relies on typical clinical manifestations for early diagnosis.[9,12] However, some serotypes such as CVA-6 infections may cause atypical manifestations of HFMD which leads to an underestimation of the actual disease burden.[13] Hence, for closely monitoring the trends of Enterovirus infections in terms of clinical presentations, genotype surveillance is essential to combat the raising EV illness.
In our study, we found that around 76.5% (101/132) of the suspected cases and 90% (54/60) of EV-confirmed cases were in the age group below 2 years. These findings are consistent with other studies carried out in Taiwan, China, Thailand, and southern Karnataka.[1,13,21-23] The increased frequency of disease in children <5 years of age is because >50% of the children in this age group lack neutralizing antibodies to the virus. Furthermore, maternal antibodies provided immunity only for the first 6 months which eventually decreases over time and also due to the lack of cross-protection by polio vaccines.
We observed that the male-to-female ratio of children affected was 2:1; similar trends were reported in a study conducted in West Bengal between 2013 and 2014, where male cases were predominant.[2] The increase in the incidence in boys can be related to their increased physical activities which promote the spread of the disease or the probable gender preference in seeking health care for children.
Only 15 (11%) cases were reported with exposure history which indirectly indicates poor awareness among the public. Since it is known that transmission happens in settings away from homes such as schools or daycare centers, there is a need to increase awareness among the caretakers and teachers regarding the disease and restricting the attendance of children during illness.
Molecular detection
Of the 132 throat swabs collected, 60 (45.5%) samples tested positive for enteroviruses. This detection rate was similar to the detection rates observed during a large outbreak involving 1844 suspected cases of HFMD in Central China, where EV was detected in 45% of cases.[21] This again emphasizes the need for robust screening of HFMD in our setup which is an essential tool to take precautionary action in this polio eradication era.
Genotype analysis revealed that CVA-6 (31.6%) was the predominant strain, followed by HEV-71 (21.6%). Only one case of CVA-16 was reported. Ten children had co-infection with HEV-71 and CVA-6 (16.6%). Our study findings were different from other studies in India, as many of them either reported HEV-71/CVA-16 as a predominant circulating strain.[8-12] Our study finding of co-infection with HEV71 and CVA-6 was also different from previously reported co-infection (CVA-16 and CVA-6) from Navi, Mumbai.[24] This emphasizes the importance of detecting the genotypes at regular intervals which helps us to know the currently circulating strains as genomic recombination is the ongoing process in EVs.
Clinical presentation
The most common presentation among the cases was rash with fever (57%) while around 43% of the cases presented with rash only without any prodromal symptoms.
The HCF was sought within a week by most of the cases (85%). Among the pan EV-positive children, 60% (36/60) of cases presented within 3 days of illness. There was a statistically significant difference found between the genotype presentation to HCF as a majority 92.3% (12/13) of children with HEV 71 (both single as well as co-infection) came to HCF within 3 days of the onset of illness when compared to only 57% of children with CVA-6 (11/19) (both single as well as co-infection). Of children positive with a single genotype, all three HEV-71 cases (100%) and only two cases of CVA-6 (25%) have presented to HCF within 3 days of illness. This finding reveals that HEV-71 was associated with moderate–severe symptoms for which medical attention was sought early compared to CVA-6 which caused milder symptoms.
In our study, 25% of children had the atypical distribution and 16.7% had the atypical type of lesion. Even though no statistical significance was noted between the genotypes causing disease in the typical and atypical presentation, nearly 30%–40% of both HEV71 and CVA-6 presented with atypical presentation either in atypical site or type of lesion which makes the clinical diagnosis more difficult. This again emphasizes the importance of epidemiological surveillance to document these atypical cases in the near future in developing countries like India for better early diagnosis and treatment.
We had only 26% of cases who presented with lesions in the oral cavity while a similarly low incidence of oral lesions was also seen in the surveillance done in France and North America.[25,26] This was in contrast to other studies, where oral lesions were observed in all the cases reported in the outbreak.[13,19,22] The reason for the low incidence of oral lesions in our population is not known.
Viral shedding
Viral excretion pattern has not been studied well in the Indian population so far; we attempted to study these parameters. Follow-up samples at 2nd week were collected from 14 children who tested positive in the first sample. Thirteen children were reported to be negative by 2 weeks and one child tested positive by 2nd week but lost to follow-up thereafter. Teng et al. documented the viral shedding in stool samples of children infected with HEV-71 and CVA-16 and found that the excretion was 10 weeks and 6 weeks, respectively.[27] The virus was known to shed for a longer period in stool than in other secretions. Ooi et al. demonstrated the viral shedding to be 2 weeks and 11 weeks in throat swabs and stool samples, respectively.[28] The difference in the viral shedding pattern may be due to the difference in the samples collected and we were able to collect only throat swabs from our follow-up cases, which would have led to the shorter viral excretion period in our study.
Limitation
The study was conducted on the samples collected from the children presented to the hospital during the outbreaks. As the disease is self-limiting in our region, the number of children requiring hospital visits was less. Furthermore, due to financial constraints, we performed genotyping using conventional PCR which may have led to lower detection of subtypes. This mandates future studies to look for other genotypes. Only 14 children responded with follow-up. A robust system to trace out these follow-up cases in the community like the field mobile unit will warrant the early diagnosis and follow-up of many other illnesses.
CONCLUSION
There is a circulation of various genotypes of enteroviruses in our region, CVA-6 is the predominant genotype, and co-infection with HEV-71 and CVA-6 is also noted. No severe complications were seen in the affected children. Majority of HEV-71 cases were reported within 3 days of illness compared to other genotypes. Atypical presentation was seen in nearly 30%–40% of all genotype cases emphasize the need for robust molecular epidemiological surveillance of HFMD in our setup.
Research quality and ethics statement
This study was approved by the Institutional Ethics Committee (JIP/IEC/SC/2015/23/832). We followed applicable EQUATOR Network guidelines during the conduct of this research project.
We also certify that the Corresponding Author is a member of the Global Advisory Board of the Journal of Global Infectious Diseases.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent forms. In the form, the legal guardian has given his consent for images and other clinical information to be reported in the journal. The guardian understands that names and initials will not be published and due efforts will be made to conceal identity, but anonymity cannot be guaranteed.
Financial support and sponsorship
The study was supported by Institute Intramural Research Fund, JIPMER.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
We are very thankful to our Professor and Head for providing support and encouragement and to all technicians in Regional VRDL, JIPMER who provided technical support.
REFERENCES
- 1.Kumar KB, Kiran AG, Kumar BU. Hand, foot and mouth disease in children: A clinico epidemiological study. Indian J Paediatr Dermatol. 2016;17:7–12. [Google Scholar]
- 2.Sarma N, Chakraborty S, Dutta A, Sadhukhan PC. Hand, foot and mouth disease in West Bengal, India: A preliminary report on clinicovirological trend over 3 successive years (2013-2015) Indian J Dermatol. 2017;62:486–90. doi: 10.4103/ijd.IJD_381_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu LJ, Xu HM, Li XJ, Wang J, Wang XJ, Ding SJ, et al. Co-detection in the pathogenesis of severe hand-foot-mouth disease. Arch Virol. 2012;157:2219–22. doi: 10.1007/s00705-012-1396-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He SJ, Han JF, Ding XX, Wang YD, Qin CF. Characterization of enterovirus 71 and Coxsackievirus A16 isolated in hand, foot, and mouth disease patients in Guangdong, 2010. Int J Infect Dis. 2013;17:e1025–30. doi: 10.1016/j.ijid.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Guan H, Wang J, Wang C, Yang M, Liu L, Yang G, et al. Etiology of multiple non-EV71 and non-CVA16 enteroviruses associated with hand, foot and mouth disease in Jinan, China, 2009-June 2013. PLoS One. 2015;10:e0142733. doi: 10.1371/journal.pone.0142733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu M, Su L, Cao L, Zhong H, Dong N, Xu J. Enterovirus genotypes causing hand foot and mouth disease in Shanghai, China: A molecular epidemiological analysis. BMC Infect Dis. 2013;13:489. doi: 10.1186/1471-2334-13-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, Sun Y, Lin C, Jia L, Wu Q, Li X, et al. Acase-control study to identify environmental risk factors for hand, foot, and mouth disease outbreaks in Beijing. Jpn J Infect Dis. 2014;67:95–9. doi: 10.7883/yoken.67.95. [DOI] [PubMed] [Google Scholar]
- 8.Sasidharan CK, Sugathan P, Agarwal R, Khare S, Lal S, Jayaram Paniker CK. Hand-foot-and-mouth disease in Calicut. Indian J Pediatr. 2005;72:17–21. doi: 10.1007/BF02760573. [DOI] [PubMed] [Google Scholar]
- 9.Vijayaraghavan PM, Chandy S, Selvaraj K, Pulimood S, Abraham AM. Virological investigation of hand, foot, and mouth disease in a tertiary care center in South India. J Glob Infect Dis. 2012;4:153–61. doi: 10.4103/0974-777X.100572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kar BR, Dwibedi B, Kar SK. An outbreak of hand, foot and mouth disease in Bhubaneswar, Odisha. Indian Pediatr. 2013;50:139–42. doi: 10.1007/s13312-013-0033-0. [DOI] [PubMed] [Google Scholar]
- 11.Rao DC, Naidu JR, Maiya PP, Babu A, Bailly JL. Large-scale HFMD epidemics caused by Coxsackievirus A16 in Bangalore, India during 2013 and 2015. Infect Genet Evol. 2017;55:228–35. doi: 10.1016/j.meegid.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 12.Gopalkrishna V, Patil PR, Patil GP, Chitambar SD. Circulation of multiple enterovirus serotypes causing hand, foot and mouth disease in India. J Med Microbiol. 2012;61:420–5. doi: 10.1099/jmm.0.036400-0. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi M, Makino T, Hanaoka N, Shimizu H, Enomoto M, Okabe N, et al. Clinical manifestations of Coxsackievirus A6 infection associated with a major outbreak of hand, foot, and mouth disease in Japan. Jpn J Infect Dis. 2013;66:260–1. doi: 10.7883/yoken.66.260. [DOI] [PubMed] [Google Scholar]
- 14.Rao PK, Veena K, Jagadishchandra H, Bhat SS, Shetty SR. Hand, foot and mouth disease: Changing Indian scenario. Int J Clin Pediatr Dent. 2012;5:220–2. doi: 10.5005/jp-journals-10005-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Sun Y, Du Y, Yan Y, Huo D, Liu Y, et al. Characterization of Coxsackievirus A6 and enterovirus 71-associated hand foot and mouth disease in Beijing, China, from 2013 to 2015. Front Microbiol. 2016;7:391. doi: 10.3389/fmicb.2016.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreuter JD, Barnes A, McCarthy JE, Schwartzman JD, Oberste MS, Rhodes CH, et al. Afatal central nervous system enterovirus 68 infection. Arch Pathol Lab Med. 2011;135:793–6. doi: 10.5858/2010-0174-CR.1. [DOI] [PubMed] [Google Scholar]
- 17.Lee CJ, Huang YC, Yang S, Tsao KC, Chen CJ, Hsieh YC, et al. Clinical features of Coxsackievirus A4, B3 and B4 infections in children. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087391. e87391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Lin C, Qu M, Li X, Gao Z, Zhang X, et al. Excretion of enterovirus 71 in persons infected with hand, foot and mouth disease. Virol J. 2013;10:31. doi: 10.1186/1743-422X-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryu WS, Kang B, Hong J, Hwang S, Kim J, Cheon DS. Clinical and etiological characteristics of enterovirus 71-related diseases during a recent 2-year period in Korea. J Clin Microbiol. 2010;48:2490–4. doi: 10.1128/JCM.02369-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, Wang J, Yan Q, He S, Zhou W, Ge S, et al. Aone-step, triplex, real-time RT-PCR assay for the simultaneous detection of enterovirus 71, coxsackie A16 and pan-enterovirus in a single tube. PLoS One. 2014;9:e102724. doi: 10.1371/journal.pone.0102724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W, Wu S, Xiong Y, Li T, Wen Z, Yan M, et al. Co-circulation and genomic recombination of Coxsackievirus A16 and enterovirus 71 during a large outbreak of hand, foot, and mouth disease in Central China. PLoS One. 2014;9:e96051. doi: 10.1371/journal.pone.0096051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang YC, Chu YH, Yen TY, Huang WC, Huang LM, Cheng AL, et al. Clinical features and phylogenetic analysis of Coxsackievirus A9 in Northern Taiwan in 2011. BMC Infect Dis. 2013;13:33. doi: 10.1186/1471-2334-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatproedprai S, Theanboonlers A, Korkong S, Thongmee C, Wananukul S, Poovorawan Y. Clinical and molecular characterization of hand-foot-and-mouth disease in Thailand, 2008-2009. Jpn J Infect Dis. 2010;63:229–33. [PubMed] [Google Scholar]
- 24.Dharmapalan D, Saxena VK, Pawar SD, Qureshi TH, Surve P. Clinical and molecular investigations of hand, foot and mouth disease outbreak in Navi Mumbai, India. Indian Pediatr. 2019;56:1052–4. [PubMed] [Google Scholar]
- 25.Mirand A, le Sage FV, Pereira B, Cohen R, Levy C, Archimbaud C, et al. Ambulatory pediatric surveillance of hand, foot and mouth disease as signal of an outbreak of Coxsackievirus A6 infections, France, 2014-2015. Emerg Infect Dis. 2016;22:1884–93. doi: 10.3201/eid2211.160590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YJ, Chang SC, Tsao KC, Shih SR, Yang SL, Lin TY, et al. Comparative genomic analysis of Coxsackievirus A6 strains of different clinical disease entities. PLoS One. 2012;7:e52432. doi: 10.1371/journal.pone.0052432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng S, Zhao SY, Wei Y, Shao QM, Jiang MY, Cui DW, et al. Observation on virus shedding periods of enterovirus-71 and Coxsackievirus A 16 monitored by nucleic acids determination in stool samples of children with hand, foot and mouth disease. Zhonghua Er Ke Za Zhi. 2013;51:787–92. [PubMed] [Google Scholar]
- 28.Ooi MH, Solomon T, Podin Y, Mohan A, Akin W, Yusuf MA, et al. Evaluation of different clinical sample types in diagnosis of human enterovirus 71-associated hand-foot-and-mouth disease. J Clin Microbiol. 2007;45:1858–66. doi: 10.1128/JCM.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]